Abstract
Outbreaks of avian influenza A virus infection, particularly the H5N1 strains that have affected birds and some humans for the past 15 years, have highlighted the need for increased surveillance and disease control. Such measures require diagnostic tests to detect and characterize the different subtypes of influenza virus. In the current study, a simple method for producing reference avian influenza virus antisera to be used in diagnostic tests was developed. Antisera of nine avian influenza A virus neuraminidases (NA) used for NA subtyping were produced using a recombinant baculovirus. The recombinant NA (rNA) proteins were expressed in Sf9 insect cells and inoculated intramuscularly into specific-pathogen-free chickens with the ISA70 adjuvant. The NA inhibition antibody titers of the rNA antiserum were in the ranges of 5 to 8 and 6 to 9 log2 units after the primary and boost immunizations, respectively. The antisera were subtype specific, showing low cross-reactivity against every other NA subtype using the conventional thiobarbituric acid NA inhibition assay. These results suggest that this simple method for producing reference NA antisera without purification may be useful for the diagnosis and surveillance of influenza virus.
INTRODUCTION
Avian influenza viruses (AIVs) are enveloped, single-stranded, segmented, negative-sense RNA viruses belonging to the subfamily Influenzavirus A. AIVs can be divided into subtypes according to the antigenic properties of the major hemagglutinin (HA) and neuraminidase (NA) glycoproteins, with 16 HA subtypes (H1 to H16) and nine NA subtypes (N1 to N9) identified to date (1). AIV diversity is caused by antigenic drift and shift (2). Antigenic drift is caused by mutations within HA and/or NA, resulting in conformational changes of antigenic sites, which may allow the mutant virus to evade the host's immune system. Antigenic shift is caused by reassortment, in which multiple influenza viruses infecting one cell can exchange genetic material to generate novel influenza viruses. These two phenomena can contribute to novel virus epidemics and pandemics (3). Therefore, rapid diagnosis and characterization of newly emerging AIVs is an important part of surveillance and disease control strategies. AIVs are generally isolated from migratory or infected animals using specific-pathogen-free (SPF) eggs. Characterization of the isolated viruses is then achieved using molecular and serological methods.
Molecular methods, such as reverse transcription-PCR (RT-PCR) (4, 5) and real-time RT-PCR (6–8), are rapid and sensitive for detection of known viruses. However, method development, including sequencing and redesign of primers, may be required for the detection of novel AIV subtypes. Serological methods, such as the hemagglutination inhibition assay and the neuraminidase inhibition (NI) assay, are useful tools; however, they require reference antisera to be developed for each HA or NA subtype. Reference NA antisera are usually produced by immunizing animals with highly purified antigens or the whole inactivated virus (WIV). Monospecific NA antisera can also be prepared by immunizing goats with purified NA proteins (9). However, this technique requires highly purified proteins and can show broad cross-reactivity. Polyclonal NA antisera are commonly produced by WIV immunizations. WIV antiserum preparation is easy and convenient but can produce antibodies for other viral proteins, such as HA, which may interfere with serological test responses. In a previous study, HA was found to be superior to NA as an antigen in mice (10). The presence of HA antibodies can produce unreliable results in the NI assay because the anti-NA response may be masked by the anti-HA response (9). Moreover, because WIV antiserum preparation requires the handling of live viruses, there are problems with safety and the availability of suitable laboratory facilities.
In the current study, homogenates containing recombinant NA (rNA) proteins were produced for each of the nine NA subtypes using a baculovirus (BV) expression system. The homogenates were used to prepare NA antisera.
MATERIALS AND METHODS
Viruses.
Nine different reference AIVs, each with a different NA subtype, were selected for producing the rNA proteins (Table 1). An additional 59 AIVs were used to validate the antisera in the NI assay. Of these 68 viruses, only A/chicken/Kr/Gimje/08 (H5N1) was highly pathogenic. Most of the viruses were isolated from wild birds and some from poultry in Korea from 2004 to 2012 (Table 1). All subtypes were determined by RT-PCR and sequencing.
Table 1.
AIV strains used in the studya
| Strain nameb | Subtype | Phenotyped |
|---|---|---|
| Chicken/Kr/Gimje/08c | H5N1 | HP |
| Wild duck/Kr/CSM 21-1/08 | H4N1 | LP |
| Wild duck/Kr/CSM 4-12/09 | H5N1 | LP |
| Wild duck/Kr/PSC 30--20/10 | H1N1 | LP |
| Wild duck/Kr/SH 38-16/10 | H10N1 | LP |
| Wild duck/Kr/PSC 2-17/10 | H9N1 | LP |
| Wild duck/Kr/CSM 4-6/10 | H3N1 | LP |
| Wild duck/Kr/SH 37-8/11 | H1N1 | LP |
| Wild duck/Kr/SH 37-27/11 | H6N1 | LP |
| Wild duck/Kr/PSC 26-16/12 | H7N1 | LP |
| Chicken/Kr/01310CE20/01c | H9N2 | LP |
| Spot billed duck/CSM 31-19/09 | H5N2 | LP |
| Wild duck/Kr/PSC 2-5/09 | H9N2 | LP |
| Wbf/Kr/MHC 5-7/09 | H4N2 | LP |
| Bean goose/Kr/CSM 8-24/09 | H9N2 | LP |
| Wild duck/Kr/PSC 10-14/09 | H6N2 | LP |
| Wild duck/Kr/SH 13-26/10 | H3N2 | LP |
| Whooper swan/Kr/CSM 20-20/10 | H11N2 | LP |
| Mandarin/Kr/PSC 24-13/10 | H5N2 | LP |
| Pink footed goose/Kr/SH 37-49/11 | H1N2 | LP |
| Duck/Kr/BC 10/07c | H7N3 | LP |
| Wbf/Kr/SH 29/06 | H1N3 | LP |
| Wbf/Kr/SYG 41/06 | H5N3 | LP |
| Wbf/Kr/NHG 185/08 | H1N3 | LP |
| Wild duck/Kr/SH 38-48/10 | H11N3 | LP |
| Wild duck/Kr/PSC 26-36/12 | H11N3 | LP |
| Wild duck/Kr/MHC 21-46/12 | H1N3 | LP |
| Wild duck/Kr/CSM 24-48/12 | H7N3 | LP |
| Wbf/Kr/PSC 13-43/08c | H8N4 | LP |
| Wbf/Kr/GG 82/04 | H8N4 | LP |
| Wbf/Kr/GS 66/05 | H10N4 | LP |
| Wbf/Kr/SH 16/06 | H10N4 | LP |
| Wild duck/Kr/SH 12-7/08c | H10N5 | LP |
| Wbf/Kr/CW 79/05 | H6N5 | LP |
| Wbf/Kr/PJ 83/05 | H6N5 | LP |
| Wild duck/Kr/SH 38-10/10 | H6N5 | LP |
| Duck/Kr/U 10-2/07c | H3N6 | LP |
| Mallard/Kr/SH 26-15/09 | H4N6 | LP |
| Mallard/Kr/CSM 27-12/09 | H7N6 | LP |
| Spot billed duck/CSM 27-15/09 | H4N6 | LP |
| Wild duck/Kr/PSC 2-11/09 | H6N6 | LP |
| Wild duck/Kr/PSC 2-3/10 | H3N6 | LP |
| Wild duck/Kr/MHC 7-7/10 | H4N6 | LP |
| Wild duck/Kr/MHC 25-1/12 | H1N6 | LP |
| Wild duck/Kr/CSM 28-22/12 | H4N6 | LP |
| Wild duck/Kr/CSM 28-37/12 | H4N6 | LP |
| Magpie/Kr/YJD 174/07c | H7N7 | LP |
| Common teal/Kr/MHC 5-8/09 | H7N7 | LP |
| Wild duck/Kr/SH 38-36/10 | H10N7 | LP |
| Wild duck/Kr/SH 41-33/11 | H7N7 | LP |
| Wild duck/Kr/CSM 42-36/11 | H10N7 | LP |
| Wild duck/Kr/CSM 42-36/11 | H10N7 | LP |
| Duck/Kr/GJ 79/07c | H3N8 | LP |
| Wild duck/Kr/PSC 2-21/09 | H3N8 | LP |
| Wild duck/Kr/MHC 5-33/09 | H3N8 | LP |
| Wild duck/Kr/SH 11-1/09 | H3N8 | LP |
| Pacific black duck/Kr/PSC 10-27/10 | H3N8 | LP |
| Wild duck/Kr/PSC 18-15/10 | H5N8 | LP |
| Wild duck/Kr/PSC 26-1/12 | H1N8 | LP |
| Wild duck/Kr/PSC 26-28/12 | H1N8 | LP |
| Wild duck/Kr/PSC 26-43/12 | H1N8 | LP |
| Wild duck/Kr/SH 27-1/12 | H3N8 | LP |
| Wild duck/Kr/SH 20-27/08c | H7N9 | LP |
| Wild duck/Kr/PSC 22-3/09 | H11N9 | LP |
| Wild duck/Kr/PSC 18-28/10 | H11N9 | LP |
| Wild duck/Kr/SH 19-27/10 | H7N9 | LP |
| Wild duck/Kr/MHC 39-13/11 | H7N9 | LP |
| Wild duck/Kr/CSM 42-1/11 | H7N9 | LP |
All viruses were isolated from wild birds or poultry in Korea.
Kr, Korea; Wbf, wild bird feces.
Viruses used as reference AIVs for expressing the recombinant neuraminidase proteins. Accession numbers of NA sequences: N1, GQ412064; N2, JX094860; N3, JX679161; N4, JX679162; N5, JX679163; N6, JN087162; N7, FJ750856; N8, JN087250; N9, JX679164.
HP, highly pathogenic influenza virus; LP, low-pathogenic influenza virus.
Construction of rNA expression vectors.
Vectors containing the genes for the relevant rNAs from the reference AIVs were constructed using the pFastBac HT A vector (pFB) (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Briefly, viral RNAs were extracted from reference viruses using a Viral Gene-Spin Kit (iNtRON Biotechnology, Daejeon, South Korea). The nine full-length NA genes were amplified with restriction enzyme site-tagged primer sets by RT-PCR and cloned into the pFB. Successful cloning was confirmed by sequencing. The pFB-NA plasmids were transformed into Escherichia coli DH10Bac competent cells (Invitrogen) to generate recombinant bacmids. The insertion of 6×His-tagged NA genes was confirmed by PCR using pUC/M13 primers and sequencing of the product.
Preparation of the rNA proteins using a baculovirus expression system.
Sf9 cells were transfected with recombinant bacmids to generate the rNA baculoviruses. At 72 h posttransfection, the supernatants were harvested and centrifuged to remove cell debris. Each of the nine rNA baculoviruses was amplified in fresh Sf9 cells over a total of three passages. The Sf9 cells were seeded in three 150-cm2 flasks at 14 × 106 cells/flask with 30 ml of Sf-900 II SFM medium (Invitrogen). The cells were infected with the rNA baculoviruses at a multiplicity of infection of 1 to 3. The infected Sf9 cells were incubated for 72 h at 27°C in an incubator. After incubation, the cells were collected by centrifugation for 5 min at 1,000 × g, and the medium was removed. The pellets were resuspended in 2 ml of PBS buffer, and the cells were disrupted by sonication on ice. The sonicated cell preparations were centrifuged at 13,000 × g for 10 min at 4°C, and the supernatants were collected and stored at −70°C. These homogenates were used as antigens for NA subtype-specific antiserum production. A nonrecombinant BV was used as a negative control. The BV protein was prepared with the BV gene in E. coli DH10Bac competent cells according to the procedures described above.
Western blot analysis.
The nine rNA proteins were analyzed by Western blotting, using a Penta-His monoclonal antibody (Qiagen, Hilden, Germany). The protein concentrations of the nine rNA homogenates were determined using the SMART BCA Protein Assay Kit (iNtRON Biotechnology). Thirty micrograms of each rNA was separated on a Novex 4 to 12% Bis-Tris gel [1× 2-(N-morpholino)ethanesulfonic acid (MES) running buffer; Invitrogen] and transferred to a polyvinylidene difluoride (PVDF) membrane using the iBlot Gel Transfer Stacks PVDF mini (Invitrogen). The membrane was blocked for 1 h in Tris-buffered saline (TBS) containing 5% skim milk and 0.05% Tween 20 at room temperature. The membrane was then washed twice with 0.05% Tween 20-TBS and incubated for 1 h with Penta-His monoclonal antibody at a 1:1,000 dilution in 2.5% skim milk-TBS. The membrane was rinsed and incubated for 1 h with a 1:2,000 dilution of horseradish peroxidase-conjugated anti-mouse IgG antibody (KPL, Gaithersburg, MD). After the incubation, the membrane was washed twice for 10 min each time and developed at room temperature with Sigma Fast 3,3′-diaminobenzidine (DAB) tablets (Sigma, St. Louis, MO) according to the manufacturer's instructions.
Immunization of chickens to produce rNA antisera.
Eight-week-old SPF chickens were used to prepare the rNA antisera, with three chickens immunized with each rNA protein. The 300-μg rNA homogenates were mixed with ISA70 oil adjuvant at a 3:7 (wt/wt) ratio. Each animal was immunized intramuscularly with 0.5 ml of rNA homogenate-ISA70 mixture (150 μl rNA homogenate with 350 μl ISA70). After 3 weeks, the birds were bled to collect antisera. The animals were boosted with a 150-μg homogenate-ISA70 inoculation. Two weeks after the boost, antisera were collected again from each animal. For the negative-control antisera, chickens were immunized with the BV homogenate and antisera were prepared according to the procedures described above.
Standardization of the NA activity assay.
The reference viruses of the N1 to N9 subtypes were amplified in 9- to 11-day-old SPF eggs. The NA activity assay was conducted based on a thiobarbituric acid (TBA) assay, as previously described (9, 11, 12), with some modifications. The nine reference viruses were diluted serially in 2-fold steps. The optimal dilution values were 1.33 to 3.83 log2 units (N1, 1.39; N2, 2.80; N3, 2.66; N4, 1.33; N5, 1.96; N6, 1.60; N7, 3.74; N8, 3.83; N9, 3.27 log2 units). The diluted viruses were mixed with 10 μl fetuin (25 mg/ml; Sigma) in a PCR tube and incubated at 37°C for 3 h. After cooling for 5 min at 10°C, 5 μl of periodate reagent was added and incubated for 20 min. A 25-μl aliquot of arsenite reagent was added to stop the reaction. Then, 50 μl TBA was added, and the mixtures were incubated at 99°C for 15 min. After cooling, 75 μl of Warrenoff reagent was added. The mixtures were vortexed and centrifuged for 5 min at 1,200 rpm. Finally, 50 μl of the butanol phase was transferred to a Nunc-Immuno 96-well plate (Nunc, Rochester, NY). The optical density (OD) was measured at 549 nm, and the NA activity dose of each virus was determined at an OD of 1.0 as the standard to be used in the NI assay.
NI assay.
We determined the NI antibody titer of the rNA antisera. The chicken sera that had been collected were serially diluted in 2-fold steps to a range of dilution values from 4 to 11 log2 units. A 5-μl aliquot of diluted serum was mixed with 5 μl of a defined standard NA activity dose. Following a 1-h incubation at room temperature, 10 μl fetuin (25 mg/ml; Sigma) was added. The remaining procedures were performed as described for the NA assay. The NI antibody titer in the antisera was determined as the half-maximal inhibitory concentration (IC50), i.e., the antiserum concentration required to inhibit NA activity by 50%.
RESULTS
Expression of rNA proteins in insect cells.
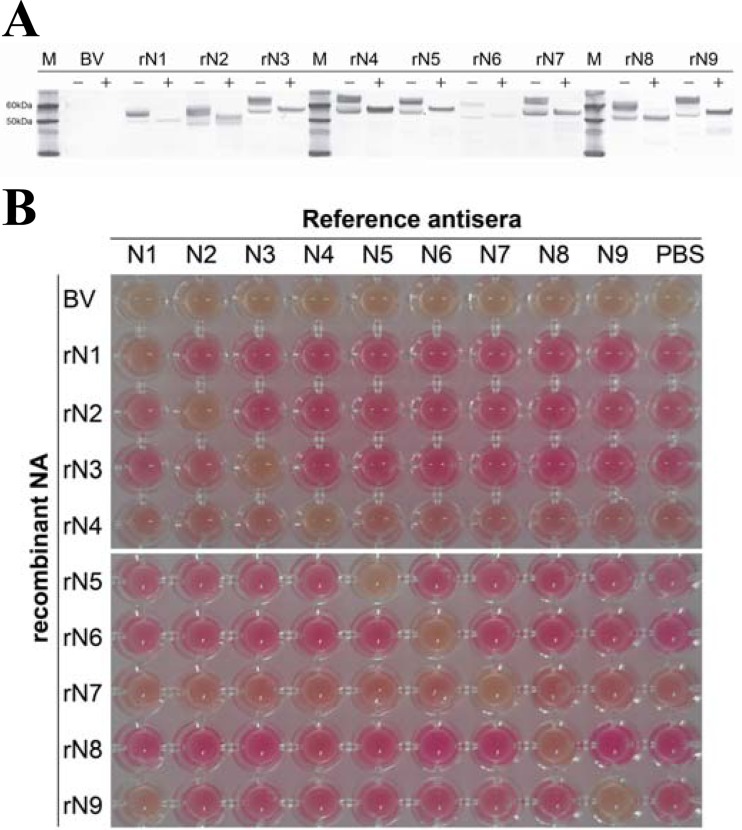
We expressed the rN1 to rN9 proteins using a baculovirus expression system to produce NA-specific antisera. The molecular masses and serological activities of the rNA proteins were determined by Western blotting and NI assay. As expected, the rNA proteins in homogenates were approximately 50 to 60 kDa (Fig. 1A). The rNA proteins were observed as two bands in the Western blot, indicating glycosylated (upper band) and nonglycosylated (lower band) rNA proteins in the Sf9 cells. We confirmed NA activity and NA-specific inhibition of rNA proteins in the NI assay using commercial NA reference antisera (Fig. 1B). All rNA homogenates had NA activity and were inhibited only by subtype-specific antisera, except for the rN9 protein, which was also inhibited by anti-N1. The NA amino acid sequences of the commercial NA reference antisera were between 89.0% and 95.1% similar to those of the rNA homogenates for each NA subtype.
Fig 1.
Expression and serological activity of rNA proteins expressed using a baculovirus system. (A) The rNA proteins were analyzed by Western blotting using an anti-Penta-His monoclonal antibody. −, not treated with N-glycosidase F; +, treated with N-glycosidase F; M, marker; BV, nonrecombinant baculovirus-infected cell lysate; rN1 to -9, recombinant neuraminidase baculovirus-infected cell lysates. (B) Conventional thiobarbituric acid neuraminidase inhibition assay conducted using a 1:10 dilution of commercial reference antisera. N1 (A/Chicken/Vietnam/8/2004 [H5N1]; AAHL), N2 (A/Pakistan/136-2/99 [H9N2]; CVL), N3 (A/TY/Oregon/71 [H7N3]; NVSL), N4 (A/TY/ONT/6118/67 [H8N4]; NVSL), N5 (A/Duck/Alb/60/76 [H12N5]; NVSL), N6 (A/Duck/England/1/56 [H11N6]; NVSL), N7 (A/Chicken/German “N”/49 [H10N7]; NVSL), N8 (NWS-Eq2 [H10N8]; NVSL), and N9 (A/Pintail/Alberta/293/77 [H2N9]; NVSL), PBS (negative control). AAHL, Australian Animal Health Laboratory; CVL, Central Veterinary Laboratory; NVSL, National Veterinary Services Laboratory.
Immune responses of chickens to the rNA proteins.
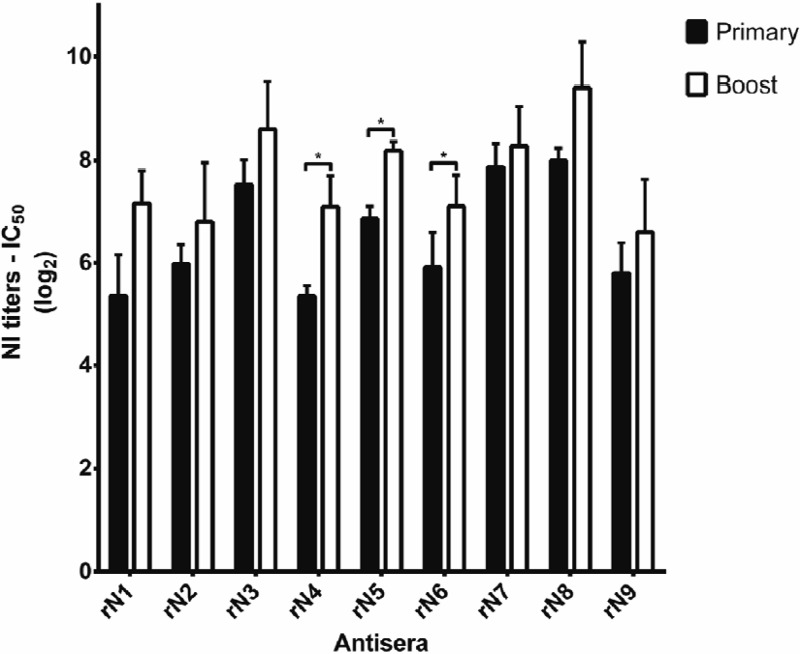
Chicken antisera were collected after both the primary and boost immunizations and evaluated by the NI assay (Fig. 2). All sera induced the NI antibody in the ranges 5 to 8 and 6 to 9 log2 units after the primary and boost immunizations, respectively. The average NI antibody titers of the boosted antisera were higher than those following the primary immunization; however, there was evidence that only rN4, rN5, and rN6 antisera were statistically significant after administering a boost immunization.
Fig 2.
NI antibody titration of chicken antisera to rNA. The NI antibody titer was measured in the sera collected from chickens that had been immunized with rNA homogenates. The antibody titers were determined using the thiobarbituric acid NI assay. The primary serum samples were collected 3 weeks after primary immunization; the boost immunization was then administered, and the boost immunization serum samples were collected 2 weeks later. Each NA subtype antiserum was tested in triplicate, with three chickens inoculated with each rNA subtype. The data are presented as means and standard deviations. Differences in antibody titers between primary and boost immunizations were tested using two-tailed paired Student's t tests, with the asterisks indicating statistical significance (P < 0.05).
Cross-reactivity of the rNA antisera.
We checked the specificity of rN1 antiserum prepared from the rN1 homogenate (Fig. 3). The figures showed that rN1 antiserum did not demonstrate any cross-reactivity with other subtypes, in contrast to the cross-reactivity shown by commercial reference antisera produced by immunization with WIV (Fig. 1).
Fig 3.
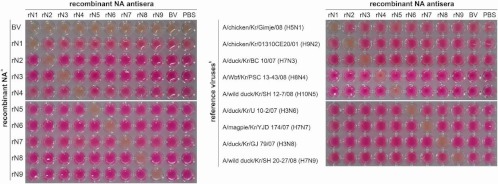
The specificity of rNA antiserum responses to rNA homogenates and reference viruses. The responses of rNA antisera were tested for specificity with rNA homogenates and reference viruses. The conventional thiobarbituric acid neuraminidase inhibition assay was conducted using a 1:10 dilution of rNA antisera. (Left) All rNA homogenates were prepared as a 1:100 dilution; (right) nine reference viruses indicated in Table 1. These viruses were tested with 16 to 32 HA units.
All antisera produced by the rNA proteins were evaluated for cross-reactivity with other virus subtypes. The test viruses were determined to be subtypes by RT-PCR and conventional NI assay with commercial reference antisera made previously by WIV. Although none of the antisera caused visible cross-reactivity, some cross-reactivity was apparent from OD values (Table 2). The rN1 serum showed cross-reactivity with N2, N4, and N5 subtype viruses. In addition, several antisera (rN1, -2, -3, -5, -6, -8, and -9) showed cross-reactivity with the A/wbf/Kr/GG 82/04 (H8N4) virus. Despite the evidence of cross-reactivity, the antigen/antiserum of the same NA subtypes always showed the highest inhibition.
Table 2.
Cross-reactivity of antisera determined by OD values from neuraminidase inhibition assays
| NA subtype | No. of virusesa | AIVs showing cross-reactivity with rNA antisera by OD valueb |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rN1 | rN2 | rN3 | rN4 | rN5 | rN6 | rN7 | rN8 | rN9 | ||
| N1 | 9 | + | − | − | − | − | − | − | − | − |
| N2 | 9 | 2 | + | − | − | − | − | − | − | − |
| N3 | 7 | − | − | + | − | − | − | − | − | − |
| N4 | 3 | 1 | 1 | 1 | + | 1 | 1 | − | 1 | 1 |
| N5 | 3 | 1 | − | − | − | + | − | − | − | − |
| N6 | 9 | − | − | − | − | − | + | − | − | − |
| N7 | 5 | − | − | − | − | − | − | + | − | − |
| N8 | 9 | − | − | − | − | − | − | − | + | − |
| N9 | 5 | − | − | − | − | − | − | − | − | + |
These AIVs are presented in Table 1, except for the reference AIVs.
Antisera were produced in this study. We defined cross-reactivity as a neuraminidase inhibition rate that was more than 30% with a 1:100 dilution of serum. +, significant inhibition; −, no cross-reactivity. The numbers indicate the numbers of AIVs among the subtypes that showed cross-reactivity with the antiserum.
DISCUSSION
The hemagglutination inhibition and NI assays have been widely used as serological methods for subtyping AIV (9, 13). These assays require monospecific or polyclonal reference antisera for each of the 16 HA and nine NA subtypes of AIV. WIV immunization is commonly used to develop a polyclonal antibody as a reference antiserum. However, this method has several problems. First, WIV must be prepared by amplification of live viruses at a suitable biosafety level facility (14). Second, WIV immunization often induces a less robust NI antibody response because of competition from dominant anti-HA antibody responses (15, 16). The HA antibody induced by WIV immunization can inhibit residual NA activity in the NI assay (9). Therefore, alternative ways of generating reference antisera that are more specific for NI assays are needed. To achieve this, we expressed rNA proteins using a recombinant baculovirus expression system (Fig. 1). The rNA homogenates induced NA-specific antibody responses in chickens and presented low visible cross-reactivity (Fig. 2 and Table 2).
Previous studies have focused on producing HA-specific reference antisera, using methods such as immunization with recombinant HA proteins or HA-encoding DNA (14, 17, 18). One study on producing NA-specific reference antisera described the preparation of N2-specific antiserum by chromatographic separation, which was tested using the agar gel precipitation test (19). Alphavirus-vectored rN2, DNA vaccines encoding rN2, and baculovirus-derived rN2 have also been tested for immunogenicity and vaccine efficacy in chickens and have been found to be N2 specific (20). We developed N1 to N9 reference antisera using a baculovirus expression system that expressed rNA proteins in Sf9 cells. When we checked the molecular weights of the rNA expressed proteins, they presented as two differently sized bands in a Western blot (Fig. 1A). Previously, several other studies had demonstrated that proteins of two sizes were generated as glycosylated mature proteins (upper band) and nonglycosylated immature proteins (lower band) (21, 22). When we treated the rNA homogenates with N-glycosidase F, which hydrolyzes N-glycan chains, the rNA proteins presented as one band (Fig. 1A). Our results, therefore, show that the baculovirus expression system properly produces both mature and immature rNA glycoproteins.
Chickens needed to be immunized at least three times to produce reference antisera from recombinant HA proteins and HA-encoding DNA (14, 18). In addition, recombinant proteins usually need to be purified by complex methods, such as polypeptide tag affinity purification and chromatography (17, 23–25), extending the amount of time need to produce antisera via these methods. In the current study, we prepared rNA proteins as homogenates, which were sonicated and centrifuged. These homogenates induced NA-specific antibodies following the primary immunization. Antibody titers against all NAs were increased by the boost immunization (Fig. 2), although this increase was significant only for rN4, rN5, and rN6 antisera. It was supposed to be caused by the amounts of rNA protein contained in each homogenate that did not equilibrated. Although the NI antibody titers of antisera generated by the primary immunization were less than those of the antisera following the boost immunization, the primary-immunization antisera showed enough antibody titers for use in the NI assay. Generally, NA reference antiserum production prepared via WIV or purified NA proteins requires several boosting stages. Therefore, the production of reference antisera using a baculovirus expression system can save time. Moreover, this baculovirus system is safer than production of reference antisera by WIV immunization and can be produced in a low-level biosafety facility.
In the cross-reactivity test, antisera prepared with the rNA homogenates produced subtype-specific reactions, although several antisera also showed nonspecific reactions according to the OD measurements (Table 2). We verified that less cross-reactivity occurred in the NI assay with the rNA antisera than with the WIV antisera (Fig. 1 and 3). It is likely that this lower cross-reactivity reflects the absence of interference by HA antibodies. These results indicate that NA reference antisera generated with the rNA proteins can be used in the NI assay with subtype specificity.
In conclusion, we demonstrated that generating rNA proteins in a baculovirus expression system could be useful as a substitute preparation method for conventional antisera. The baculovirus expression system could be used to produce antisera for various emerging NA subtype viruses.
ACKNOWLEDGMENT
This work was supported by a grant from the Animal, Plant and Fisheries Quarantine Inspection Agency (C-AD15-2010-12-02), Republic of Korea.
Footnotes
Published ahead of print 5 December 2012
REFERENCES
- 1. Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 79:2814–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McHardy AC, Adams B. 2009. The role of genomics in tracking the evolution of influenza A virus. PLoS Pathog. 5:e1000566 doi:10.1371/journal.ppat.1000566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fereidouni SR, Starick E, Grund C, Globig A, Mettenleiter TC, Beer M, Harder T. 2009. Rapid molecular subtyping by reverse transcription polymerase chain reaction of the neuraminidase gene of avian influenza A viruses. Vet. Microbiol. 135:253–260 [DOI] [PubMed] [Google Scholar]
- 5. Lee MS, Chang PC, Shien JH, Cheng MC, Shieh HK. 2001. Identification and subtyping of avian influenza viruses by reverse transcription-PCR. J. Virol. Methods 97:13–22 [DOI] [PubMed] [Google Scholar]
- 6. Spackman E, Senne DA, Bulaga LL, Myers TJ, Perdue ML, Garber LP, Lohman K, Daum LT, Suarez DL. 2003. Development of real-time RT-PCR for the detection of avian influenza virus. Avian Dis. 47:1079–1082 [DOI] [PubMed] [Google Scholar]
- 7. Stone B, Burrows J, Schepetiuk S, Higgins G, Hampson A, Shaw R, Kok T. 2004. Rapid detection and simultaneous subtype differentiation of influenza A viruses by real time PCR. J. Virol. Methods 117:103–112 [DOI] [PubMed] [Google Scholar]
- 8. Suwannakarn K, Payungporn S, Chieochansin T, Samransamruajkit R, Amonsin A, Songserm T, Chaisingh A, Chamnanpood P, Chutinimitkul S, Theamboonlers A, Poovorawan Y. 2008. Typing (A/B) and subtyping (H1/H3/H5) of influenza A viruses by multiplex real-time RT-PCR assays. J. Virol. Methods 152:25–31 [DOI] [PubMed] [Google Scholar]
- 9. World Health Organization 2002. WHO manual on animal influenza diagnosis and surveillance. World Health Organization, Geneva, Switzerland: http://www.who.int/csr/resources/publications/influenza/whocdscsrncs20025rev.pdf Accessed 5 June 2012 [Google Scholar]
- 10. Johansson BE, Moran TM, Bona CA, Popple SW, Kilbourne ED. 1987. Immunologic response to influenza virus neuraminidase is influenced by prior experience with the associated viral hemagglutinin. II. Sequential infection of mice simulates human experience. J. Immunol. 139:2010–2014 [PubMed] [Google Scholar]
- 11. Fritz R, Sabarth N, Kiermayr S, Hohenadl C, Howard MK, Ilk R, Kistner O, Ehrlich HJ, Barrett PN, Kreil TR. 2012. A Vero cell-derived whole-virus H5N1 vaccine effectively induces neuraminidase-inhibiting antibodies. J. Infect. Dis. 205:28–34 [DOI] [PubMed] [Google Scholar]
- 12. Sandbulte MR, Gao J, Straight TM, Eichelberger MC. 2009. A miniaturized assay for influenza neuraminidase-inhibiting antibodies utilizing reverse genetics-derived antigens. Influenza Other Respi. Viruses 3:233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. World Organization for Animal Health 2009. Avian influenza. Manual of diagnostic tests and vaccines for terrestrial animals 2009. OIE, Paris, France: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.03.04_AI.pdf Accessed 5 June 2012 [Google Scholar]
- 14. Lee CW, Senne DA, Suarez DL. 2006. Development and application of reference antisera against 15 hemagglutinin subtypes of influenza virus by DNA vaccination of chickens. Clin. Vaccine Immunol. 13:395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johansson BE, Pokorny BA, Tiso VA. 2002. Supplementation of conventional trivalent influenza vaccine with purified viral N1 and N2 neuraminidases induces a balanced immune response without antigenic competition. Vaccine 20:1670–1674 [DOI] [PubMed] [Google Scholar]
- 16. Kendal AP, Bozeman FM, Ennis FA. 1980. Further studies of the neuraminidase content of inactivated influenza vaccines and the neuraminidase antibody responses after vaccination of immunologically primed and unprimed populations. Infect. Immun. 29:966–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cui S, Wu C, Zhou H, Zhao R, Guo L, Wang J, Hung T. 2011. Secretory expression of all 16 subtypes of the hemagglutinin 1 protein of influenza A virus in insect cells. J. Virol. Methods 177:160–167 [DOI] [PubMed] [Google Scholar]
- 18. Shahsavandi S, Salmanian AH, Ghorashi SA, Masoudi S, Fotouhi F, Ebrahimi MM. 2011. Specific subtyping of influenza A virus using a recombinant hemagglutinin protein expressed in baculovirus. Mol. Biol. Rep. 38:3293–3298 [DOI] [PubMed] [Google Scholar]
- 19. Shirvan AN, Moradi M, Aminian M, Madani R. 2007. Preparation of neuraminidase-specific antiserum from the H9N2 subtype of avian influenza virus Turk. J. Vet. Anim. Sci. 31:219–223 [Google Scholar]
- 20. Sylte MJ, Hubby B, Suarez DL. 2007. Influenza neuraminidase antibodies provide partial protection for chickens against high pathogenic avian influenza infection. Vaccine 25:3763–3772 [DOI] [PubMed] [Google Scholar]
- 21. Barman S, Nayak DP. 2000. Analysis of the transmembrane domain of influenza virus neuraminidase, a type II transmembrane glycoprotein, for apical sorting and raft association. J. Virol. 74:6538–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yongkiettrakul S, Boonyapakron K, Jongkaewwattana A, Wanitchang A, Leartsakulpanich U, Chitnumsub P, Eurwilaichitr L, Yuthavong Y. 2009. Avian influenza A/H5N1 neuraminidase expressed in yeast with a functional head domain. J. Virol. Methods 156:44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bright RA, Carter DM, Daniluk S, Toapanta FR, Ahmad A, Gavrilov V, Massare M, Pushko P, Mytle N, Rowe T, Smith G, Ross TM. 2007. Influenza virus-like particles elicit broader immune responses than whole virion inactivated influenza virus or recombinant hemagglutinin. Vaccine 25:3871–3878 [DOI] [PubMed] [Google Scholar]
- 24. Schmidt PM, Attwood RM, Mohr PG, Barrett SA, McKimm-Breschkin JL. 2011. A generic system for the expression and purification of soluble and stable influenza neuraminidase. PLoS One 6:e16284 doi:10.1371/journal.pone.0016284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Upadhyay C, Ammayappan A, Vakharia VN. 2009. Detection of NP, N3 and N7 antibodies to avian influenza virus by indirect ELISA using yeast-expressed antigens. Virol. J. 6:158. [DOI] [PMC free article] [PubMed] [Google Scholar]