Abstract
The human fungal pathogen Candida albicans can grow at temperatures of up to 45°C. Here, we show that at 42°C substantially less biomass was formed than at 37°C. The cells also became more sensitive to wall-perturbing compounds, and the wall chitin levels increased, changes that are indicative of wall stress. Quantitative mass spectrometry of the wall proteome using 15N metabolically labeled wall proteins as internal standards revealed that at 42°C the levels of the β-glucan transglycosylases Phr1 and Phr2, the predicted chitin transglycosylases Crh11 and Utr2, and the wall maintenance protein Ecm33 increased. Consistent with our previous results for fluconazole stress, this suggests that a wall-remodeling response is mounted to relieve wall stress. Thermal stress as well as different wall and membrane stressors led to an increased phosphorylation of the mitogen-activated protein (MAP) kinase Mkc1, suggesting activation of the cell wall integrity (CWI) pathway. Furthermore, all wall and membrane stresses tested resulted in diminished cell separation. This was accompanied by decreased secretion of the major chitinase Cht3 and the endoglucanase Eng1 into the medium. Consistent with this, cht3 cells showed a similar phenotype. When treated with exogenous chitinase, cell clusters both from stressed cells and mutant strains were dispersed, underlining the importance of Cht3 for cell separation. We propose that surface stresses lead to a conserved cell wall remodeling response that is mainly governed by Mkc1 and is characterized by chitin reinforcement of the wall and the expression of remedial wall remodeling enzymes.
INTRODUCTION
Candida albicans is an opportunistic fungal pathogen of humans and other warm-blooded animals. It is one of the leading causes of fungal infections among immunocompromised patients, which are often fatal if not diagnosed in time (1, 2). During infection, C. albicans encounters stresses from host defenses (e.g., fever and oxidative and nitrosative stress), environmental niches (e.g., hypoxia in the gut and antimicrobial peptides in saliva and in epithelial layers), and antifungal intervention (e.g., azoles and echinocandins). Many of these stresses directly affect the cell surface, which has distinct fungal features compared to mammalian cells. The most important difference is the presence of a cell wall.
The cell wall is the initial site of host-pathogen interaction and is composed of a skeletal layer of carbohydrates, mainly β-glucans and chitin, which is covered with an external layer of covalently anchored mannoproteins. These mannoproteins have been shown to serve a variety of functions, from immune evasion (3, 4) and nutrient acquisition (5, 6) to adhesion, biofilm formation (7, 8), and tissue degradation (9). Many cell wall proteins also directly modulate the wall composition and architecture as carbohydrate-active enzymes (10). Reinforcement of the cell wall in response to antifungal stresses is well described, especially with respect to an increase in chitin content as a result of increased chitin synthesis (11, 12). The cell wall proteome itself is highly dynamic (13) and adaptable in response to external conditions (14, 15) as well as morphological changes (16).
This dynamic surface is crucial for an opportunistic pathogen, enabling it to colonize different niches in a variety of hosts. Sites of infection differ dramatically in, for example, oxygen levels, pH, and available nutrients. Another environmental factor that has a major impact on the fungal surface and growth is temperature, which can vary considerably depending on the host species. While most fungi are not able to grow above 40°C (17), C. albicans causes infections in many animals (18), among them birds (e.g., penguins and pigeons), whose body temperatures are in this range (19, 20). As a consequence of prolonged thermal stress, both Saccharomyces cerevisiae and C. albicans cells have been shown to accumulate trehalose, which facilitates proper protein folding under stress conditions (21, 22). In S. cerevisiae thermal stress also leads to the activation of the cell wall integrity (CWI) pathway which, in turn, affects the composition of the wall and its proteins (reviewed in reference 23). In addition, Mkc1, the C. albicans ortholog of Slt2 in S. cerevisiae and a key signal transducer in the CWI pathway of C. albicans, has been suggested to be required for growth at elevated temperatures (24). Another member of the CWI pathway, Pkc1, was shown to be involved in the response to fluconazole (25), a widely used antimycotic agent that leads to the depletion of ergosterol in the fungal membrane (26), resulting in increased membrane fluidity (27, 28).
We recently established that fluconazole does not only elicit membrane stress but also leads to a stressed cell wall. Since membrane fluidity is correlated with temperature (29), we hypothesized that a similar effect as seen for fluconazole could be achieved by growth at elevated temperatures. In this study, we examined the effect of thermal stress on the wall composition, the secretome, and the cell wall proteome as well as underlying regulatory proteins. Relative quantification revealed a set of wall-remodeling proteins that was induced during thermal stress. Intriguingly, the same set of proteins was shown previously to be involved in the response to other surface stressors (14, 30), suggesting a general stress response. Therefore, we investigated the phosphorylation status of three stress-responsive mitogen-activated protein (MAP) kinases, Hog1, Cek1, and Mkc1. This analysis revealed that strongly increased Mkc1 phosphorylation is a hallmark of prolonged thermal stress as well as long-term exposure to wall and membrane stress. We also show that high-temperature stress as well as antifungal treatment leads to cell clustering, which is linked to reduced secretion of chitinases, since the clusters are dispersible by exogenous chitinase but not by sodium dodecyl sulfate (SDS) treatment or vortexing. To cope with prolonged surface stress, a highly conserved wall remodeling response, probably mainly mediated by Mkc1 signaling, is activated, and the wall is reinforced with chitin through increased chitin synthesis and reduced chitin degradation. The identification of proteins involved in wall stress adaptation opens the door to the development of strategies to improve clinical outcomes of antifungal treatment.
MATERIALS AND METHODS
Strains, growth conditions, biomass measurements, and microscopy.
All chemicals were obtained from Sigma-Aldrich unless otherwise stated. C. albicans wild-type (Wt) SC5314 or mutant strains (Table 1) were precultured overnight at 30°C in liquid YPD medium (10 g/liter yeast extract, 20 g/liter peptone, and 20 g/liter glucose) in a rotary shaker at 200 rpm. The next day, flasks containing 50 ml of YNB-S (6.7 g/liter yeast nitrogen base [YNB], 20 g/liter sucrose) either buffered at pH 7.4 using 75 mM MOPSO [3-(N-morpholino)-2-hydroxypropanesulphonic acid] or at pH 4 using 75 mM tartaric acid were inoculated from the overnight cultures to an optical density at 600 nm (OD600) of 0.05. Sucrose was used as a carbon source to avoid glucose repression. MOPSO and tartaric acid were selected as buffers because they are hydrophilic and nonmetabolizable, with pKa values close to the desired pH (36). The cultures were stirred at 200 rpm and incubated for 18 h at either 25°C, 30°C, 37°C, or 42°C. For analysis of the response to different wall or membrane stresses, the culture medium was supplemented with calcofluor white (CFW), Congo red (CR), fluconazole (FCZ), or sodium dodecyl sulfate (SDS). To increase biomass yield for wall purification of cells grown at 42°C, multiple flasks were pooled per biological replicate. To determine biomass of the cultures after 18 h of growth, cultures were spun down, and the pellets were dried at 60°C overnight and weighed the next day. For morphological analysis, cells were stained with CFW and photographed using fluorescence microscopy. For staining, 3 μl of the culture was mixed on a glass slide with 3 μl of a 1:1,000 dilution of a 10 mg/ml stock of CFW, incubated for 5 min, and visualized using an Axiovert 40 CFL microscope (Zeiss, United Kingdom). To determine the effect of exogenous chitinase on various strains, the cells were incubated for 3 h at room temperature in the presence of 0.5 U of chitinase purified from Trichoderma viride (C8241) in 50 mM phosphate buffer (pH 6.1) and then stained and visualized as described above.
Table 1.
Candida albicans strains used in this study
| Name | Parent | Relevant genotype | Abbreviation | Reference |
|---|---|---|---|---|
| SC5314 | NAa | Wild type | Wt | 31 |
| DSY1768 | CAF-2 | cht2Δ::hisG-URA3-hisG/cht2Δ::hisG | Δ/Δcht2 | 32 |
| SPY24 | CAF-2 | cht3Δ::hisG-URA3-hisG/cht3Δ::hisG | Δ/Δcht3 | 32 |
| DSY1741 | CAF-2 | cht2Δ::hisG-URA3-hisG/cht2Δ::hisG cht3Δ::hisG/cht3Δ::hisG | Δ/Δcht2/3 | 32 |
| CNC63 | CAF-2 | eng1Δ::hisG-URA3-hisG/eng1::hisG | Δ/Δeng1 | 33 |
| MK106 | SC5314 | ace2Δ::FRT/ace2Δ::FRT | Δ/Δace2 | 34 |
| CAMM-292-4 | CAI4 | ura3/ura3 cbk1Δ::hisG/cbk1Δ::hisG-URA3-hisG | Δ/Δcbk1 | 35 |
Not applicable.
Spectrometric assay of relative sedimentation times.
C. albicans cells were cultured as described above. C. albicans wild-type SC5314 or mutant strains (Table 1) were grown for 18 h at 37°C or 42°C in the presence or absence of fluconazole or Congo red. Subsequently, selected strains (indicated with a plus sign in Fig. 5) were treated with chitinase as described above. The OD600 from 800 μl of a culture with or without chitinase treatment was measured each min for 15 min. Each strain and growth condition was measured in duplicate, and the time required for the optical density to decline to 80% of the original value, designated the sedimentation time, was determined for each time course. The sedimentation time of wild-type cells grown at 37°C was set at 100%, and the sedimentation times of all other conditions and strains were expressed as a percentage of this value.
Spot assays to test resistance to cell wall-perturbing agents.
C. albicans wild-type or mutant strains were grown overnight in liquid YPD medium at 200 rpm and 30°C. The overnight culture was 1/10 serially diluted, and 5 μl of undiluted culture and each consecutive 10-fold diluted culture were spotted on plates containing YNB-S medium (pH 7.4) solidified with 1.5% agar. For treatment with cell wall- or plasma membrane-perturbing agents, the plates were supplemented with CFW, CR, or SDS. After incubation at 37°C or 42°C for 2 days, the plates were photographed.
Intracellular protein extraction and immunoblot analysis.
C. albicans was cultured in YNB-S medium at 37° or 42°C as described above. After 18 h the cells were harvested by centrifugation at 4°C and washed once with cold phosphate-buffered saline (PBS). Intracellular proteins were prepared as described previously (37). Briefly, cells were resuspended in cold lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% of the detergent NP-40, 2 mg/liter leupeptin, 2 mg/liter pepstatin, 1 mM phenylmethylsulfonyl fluoride [PMSF], 2 mM Na3VO4, 50 mM NaF). Cold glass beads with a diameter of 0.25 to 0.50 mm were added, and cells were disintegrated using a Precellys 24 homogenizer (Bertin Technologies, France). After centrifugation to remove glass beads and cell debris, the protein concentration of the supernatants was determined and normalized using a Bradford assay (38). Proteins were separated on a 4 to 12% Bis-Tris gel (Invitrogen, San Diego, CA) and transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA). Membranes were blocked in a Tris-buffered saline–Tween solution (TBS-T) with 10% bovine serum albumin (BSA) and 50 mM NaF at room temperature for 30 min. They were further incubated overnight at 4°C with either anti-phospho-p38 MAP kinase (MAPK) (Thr180/Tyr182) antibody or anti-phospho-p44/42 MAPK (Thr202/Tyr204) antibody (both from New England BioLabs, United Kingdom) in TBS-T with 5% BSA and 50 mM NaF. As a loading control, antibodies against 3-phosphoglycerate kinase (Acris, Germany) were used. After the PVDF membranes were washed five times for 5 min in TBS-T, they were incubated with peroxidase-conjugated goat-anti rabbit antiserum (Thermo Fisher Scientific, Waltham, MA) in TBS-T with 5% BSA and 50 mM NaF at room temperature for 1 h. Subsequently, the membranes were washed three times for 5 min in TBS-T. Proteins were visualized using ECL Plus detection reagents (GE Healthcare, Waukesha, WI).
Secretome analysis.
Secretome analysis was performed as described before (14, 36). Cells were separated from the growth medium by centrifugation. In order to remove remaining cells, the supernatant was passed through a 0.2-μm-pore-size filter and then concentrated using 10-kDa-cutoff filters (Amicon Ultra-15 centrifugal filter units; Millipore, Billerica, MA). Secreted proteins were quantified using a bicinchoninic acid (BCA) assay and BSA as a standard and normalized to dry weight. For mass spectrometric analysis, the concentrated proteins were further reduced, alkylated, and digested with trypsin as described for wall proteins.
Cell wall purification, reduction, and alkylation.
Cell walls were prepared as described elsewhere (10) with some modifications. Briefly, the cell pellet was washed several times with PBS and then subjected to breakage in a Precellys 24 homogenizer with glass beads (0.25 to 0.50 mm) in the presence of a protease inhibitor cocktail. Full breakage was controlled by light-microscopic inspection. The pellet was washed several times with 1 M NaCl and then washed twice with MilliQ-water. The pellets were resuspended in an appropriate volume of SDS extraction buffer (150 mM NaCl, 20 g/liter SDS, 100 mM Na-EDTA, 100 mM β-mercaptoethanol, 50 mM Tris-HCl, pH 7.8) and incubated with shaking at 200 rpm at 37°C for 30 min. The pellets were spun down and resuspended in fresh SDS extraction buffer and incubated overnight under the same conditions. These steps ensure thorough and stringent washing to retain only covalently anchored wall proteins. The following day, the pellets were boiled four times for 10 min in SDS extraction buffer, washed several times with MilliQ-water, and lyophilized overnight.
The purified wall pellets were stored at −80°C until needed or directly reduced and S-alkylated as described elsewhere (39). Briefly, treatment of the wall pellets with reducing solution (10 mM dithiothreitol in 100 mM NH4HCO3) for 1 h at 55°C is followed by alkylation (65 mM iodoacetamide in 100 mM NH4HCO3) for 45 min at room temperature in the dark and subsequent quenching of the reaction by the addition of 55 mM dithiothreitol/100 mM NH4HCO3 for 5 min. To eliminate excess iodoacetamide and dithiothreitol, the pellets were washed six times with 50 mM NH4HCO3. Similarly, the secretome samples were reduced and alkylated on 10-kDa-cutoff spin filters (Amicon, Billerica, MA) (36). Reduced and alkylated samples were either directly trypsinized or stored at −80°C until used.
Determination of cell wall protein and chitin content.
The wall protein content and the chitin content of the cell walls were determined as described previously (40). Four milligrams of freeze-dried cell walls was suspended in 100 μl of 1 M NaOH and boiled for 10 min. After the suspension was cooled to room temperature, it was neutralized by the addition of 100 μl of 1 M HCl and centrifuged to get rid of insoluble material. Using a BCA assay, the protein content of the supernatant was determined and compared to bovine serum albumin as a standard.
For chitin determination, the insoluble cell wall components obtained after the NaOH extraction step were hydrolyzed in 1 ml of 6 M HCl for 17 h at 100°C. After evaporation of the liquid under a stream of nitrogen, the pellet was resuspended in 1 ml of demineralized water. Samples of 100 μl were mixed with 100 μl of 1.5 M Na2CO3 in 4% acetylacetone, and the mixtures were boiled for 20 min. After cooling to room temperature, the reaction mixtures were supplemented and mixed with 700 μl of 96% ethanol and 100 μl of a p-dimethyl-aminobenzaldehyde solution (1.6 g in 30 ml of concentrated HCl and 30 ml of 96% ethanol). After a 1-h incubation at room temperature, absorbance at 520 nm was measured and compared to a standard of glucosamine.
Metabolic 15N labeling of the reference cultures.
Two different culturing conditions were used to generate a 15N metabolically labeled reference culture. Cells grown at pH 4 and 30°C and at pH 7.4 and 37°C were labeled as described previously (16). Briefly, optimal 15N loading was ensured by preculturing twice in YNB-S medium with 15N-labeled ammonium sulfate as the sole nitrogen source (15N content of >99%; Spectra Stable Isotopes) with a starting OD600 of 0.05. The final preculture was then used to inoculate the appropriate medium for the reference culture, and cells were grown at 30°C or 37°C and 200 rpm for 18 h. The 15N-labeled cells of both cultures were harvested; their walls were isolated, freeze-dried, combined in a 1:1 ratio based on dry weight, divided into 2-mg aliquots, and stored at −80°C.
Sample preparation for MS analysis and 14N/15N mixing.
Sample preparation for mass spectrometry (MS) analysis was performed as described previously (14, 16, 36). For quadrupole time of flight liquid chromatography-tandem mass spectrometry (Q-TOF LC-MS/MS) analysis of secreted proteins, alkylated proteins were digested for 18 h using 2 μg of Trypsin Gold (Promega, Madison, WI) and subsequently desalted using a C18 tip column (Varian, Palo Alto, CA) according to the supplier's instructions. For relative quantification, 2 mg of the freeze-dried, reduced, and alkylated 14N-labeled query walls was mixed 1:1 based on protein content with 15N-labeled reference walls (with 60% reference walls at pH 7.4 and 80% walls at pH 4), resuspended in 50 mM NH4HCO3, and then digested and desalted as described above. To eliminate acetonitrile, all samples were evaporated using a Speedvac (Genevac, Ipswich, England), and the peptides were resolubilized in 20 μl of 0.1% trifluoroacetic acid (TFA). The amount of peptides in all samples was determined using a NanoDrop ND-1000 (Isogen Life Science, IJsselstein, The Netherlands) at 205 nm as described before (14, 16, 41, 42).
LC-MS/MS (Q-TOF) analysis and protein identification.
A total of 250 ng of peptides in 10 μl of 0.1% TFA was injected onto an Ultimate 2000 nano-HPLC (high-performance liquid chromatography) system (LC Packings, Amsterdam, The Netherlands) equipped with a PepMap100 C18 reversed-phase column (75-μm inner diameter, 25-cm length; Dionex, Sunnyvale, CA). A linear gradient with increasing acetonitrile concentration (0% to 50%) over 45 min and an elution flow rate of 0.3 μl/min was used for LC separation. The eluate was online ionized by electrospray in a Q-TOF instrument (Micromass, Whyttenshawe, United Kingdom). Survey scans were acquired from m/z 350 to 1,500. For low-energy collision-induced dissociation (MS/MS), the most intense ions were selected in a data-dependent mode. Pkl (peak list) files were generated using the MaxEnt3 algorithm, included in the Masslynx Proteinlynx software. Proteins were identified by submitting the pkl files to an internally licensed version of MASCOT (Matrix Science, Great Britain), searching against a complete open reading frame (ORF) translation of the C. albicans genome. In MASCOT two miscleavages and a tolerance of 0.5 Da for peptides and MS/MS were allowed. Based on probabilistic MASCOT scoring, a P value of <0.05 was considered significant for peptide identification. Four independently obtained biological samples (the biological replicates) were analyzed for each condition. Each biological sample was subjected to two MS/MS runs (technical replicates). All identified proteins were subjected to signal peptide prediction using SignalP, version 3.0 (43), and prediction of a glycosylphosphatidylinositol (GPI) anchor sequence using a BIG-PI fungal predictor (44). For a semiquantitative analysis of our data, we calculated for each growth condition the percent peptide identifications. Per biological replicate, the number of peptide identifications per protein was divided by the total number of identified peptides. The percent spectral counts was averaged per growth condition.
LC-FT-MS/MS data acquisition.
MS/MS data were acquired using an ApexUltra Fourier transform (FT) ion cyclotron resonance mass spectrometer (Bruker Daltonic, Bremen, Germany) equipped with a 7T magnet and a nano-electrospray Apollo II DualSource coupled to an Ultimate 3000 (Dionex, Sunnyvale, CA) HPLC system. Samples containing 80 ng of the 14N- and 15N-labeled tryptic peptide mixtures were injected as a 3-μl 0.1% trifluoroacetic acid aqueous solution and loaded onto the PepMap100 C18 (5-μm particle size, 100-Å pore size, 300-μm inner diameter by 5-mm length) precolumn. Following injection, the peptides were eluted via an Acclaim PepMap 100 C18 (3-μm particle size, 100-Å pore size, 75-μm inner diameter by 250-mm length) analytical column (Thermo Scientific, Etten-Leur, The Netherlands) using a linear gradient from 0.1% formic acid/6% CH3CN/94% H2O to 0.1% formic acid/40% CH3CN/60% H2O over a period of 120 min at a flow rate of 300 nl/min.
Data-dependent Q-selected peptide ions were fragmented in a hexapole collision cell at an Argon pressure of 6 × 10−4 Pa (measured at the ion gauge), and the fragment ions were detected in the ion cyclotron resonance (ICR) cell at a resolution of up to 60,000. Mass calibration was better than 5 ppm over all MS and MS/MS spectra in the LC-MS/MS chromatogram. MS/MS rate was about 1.3 Hz. This yielded more than 9,000 MS/MS spectra over the 120-min LC-MS/MS chromatogram.
LC-FT-MS/MS data processing and relative protein quantification.
Raw FT-MS/MS data were processed with the MASCOT DISTILLER program, version 2.4.3.1 (64 bits) and MDRO, version 2.4.3.0 (MATRIX science, London, United Kingdom), including the Search toolbox and the Quantification toolbox. Peak-picking for both MS and MS/MS spectra were optimized for the mass resolution of up to 60,000. Peaks were fitted to a simulated isotope distribution with a correlation threshold of 0.7, with a minimum signal-to-noise ratio of 2. MS/MS spectra from precursor peptides within a mass tolerance of 0.03 Da and within a retention time window of maximum 60 s were added before peak picking.
The processed data were searched with the MASCOT server program 2.3.02 (Matrix Science, London, United Kingdom) against a complete Candida albicans ORF translation (6,210 entries) obtained from the Candida Genome Database (CGD) (www.candidagenome.org). Trypsin was used as the enzyme, and two missed cleavages were allowed. Carbamidomethylation of cysteine was used as a fixed modification, and oxidation of methionine was used as a variable modification. Semitryptic peptides were allowed. The peptide mass tolerance was set to 5 ppm, and the peptide fragment mass tolerance was set to 0.01 Da. The quantification method was set to the metabolic 15N-labeling method to enable MASCOT to identify both 14N- and 15N-labeled peptides.
Using the quantification toolbox, the isotopic ratios for all identified proteins were determined as the average of the isotopic ratios of the corresponding light over heavy (L/H) peptides. Selected critical settings were as follows: method was set to average protein ratio type, requiring bold red and a significance threshold of 0.05 with the precursor protocol type selected; the 15N-loading value was set to 99.4 via the components correction for elements, and the report ratio was set to L/H. For integration, Simpson's integration method with the survey integration source was used. Elution time shift was allowed with an elution time delta of 20 s and a standard error threshold of 0.15. The correlation threshold based on isotopic distribution fit was set to 0.98. All charge states were used with an extracted ion chromatogram (XIC) threshold of 0.1 and a maximal XIC width of 200 s. Only unique peptides were used for quantification, and a peptide threshold of 0.05 together with the least-homology threshold for peptides was used. Normalization mode was set to none, and outliers were automatically removed.
LC-FT-MS/MS data were acquired and analyzed of four replicas of both 37°C and 42°C cell cultures at pH 4 and of three replicas of both 37°C and 42°C cell cultures at pH 7.4. For all these 14 analyses the comprehensive MASCOT DISTILLER quantification reports are published as Excel documents in the supplemental material. Only bona fide wall proteins previously verified were used for quantification (see Fig. 2 and Tables S1 and S2 in the supplemental material). These reports show that over all analyses, cell wall proteins are quantified as L/H isotopic ratios that were obtained by averaging the L/H ratios of up to 25 corresponding tryptic peptides. From the geometric standard deviations, it can be estimated that errors in the protein mean average isotopic ratios are limited to less than 15%.
For Als1, Phr2, Sap9, and Tos1, the changes in the protein levels between the 37°C and 42°C culture were so large that for the two conditions different tryptic peptide sets were identified. To obtain more overlap between these tryptic peptide sets for the two conditions, the corresponding missing peptides were manually identified from the chromatogram MS trace, based on their accurate mass and LC retention, and their L/H isotopic ratios were determined. These peptide isotopic ratios were then included in the averaged isotopic ratios for the above proteins.
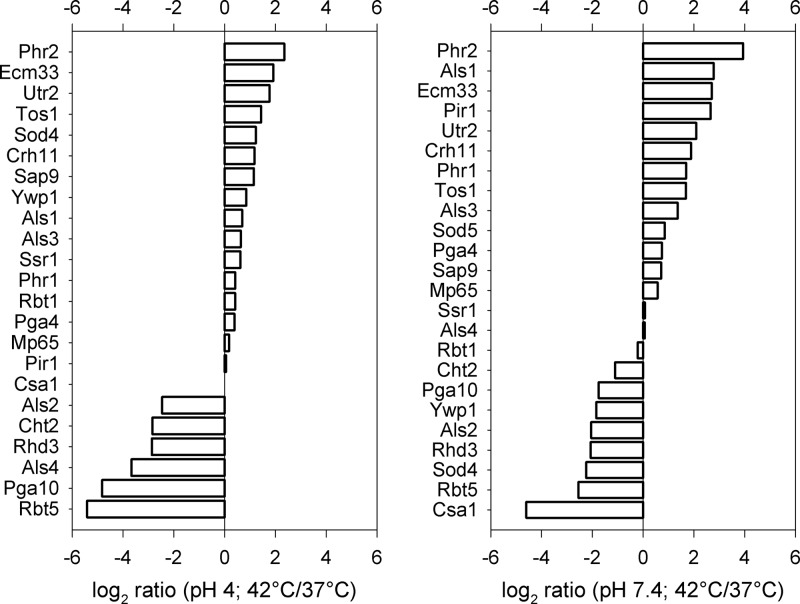
Tables S1 and S2 in the supplemental material summarize the cell wall protein L/H isotopic ratios of each replica sample. The results show that the isotopic ratios over the replicas are very consistent. This indicates that the biological variation is not dominant, that the cell culture and cell processing are reproducible, and that the 14N/15N cell culture mixing errors are limited. Therefore, the final protein 14N/15N isotopic ratios in Tables S1 and S2 are replica averages. The log2 values of the resulting 14N/15N protein isotopic ratios are listed in the histograms of Fig. 2, which show the perturbation in the cell wall proteome upon increasing the temperature from 37°C and 42°C for pH 4 and pH 7.4.
RESULTS
Prolonged high-temperature stress causes cell wall stress.
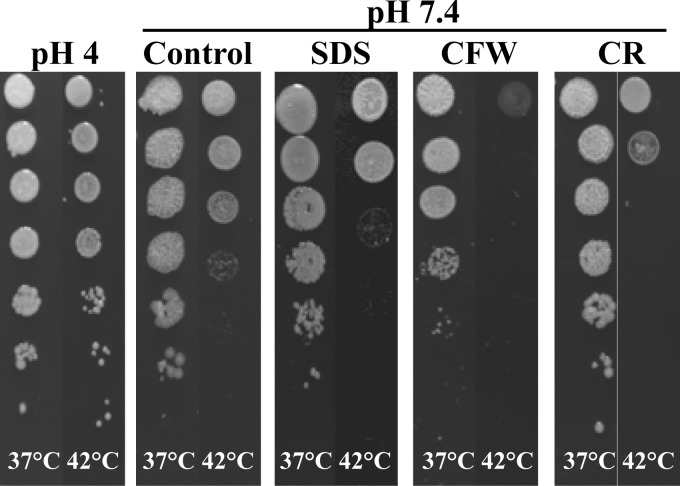
C. albicans is able to withstand and grow under conditions many other fungi cannot survive. This suggests that C. albicans has evolved strategies to minimize the impact of these stresses. We previously showed that the widely used antifungal fluconazole leads to both membrane as well as wall stress, which the cell tries to counteract by increased chitin incorporation as well as the increase of repair-associated proteins in the wall (14). Similar to fluconazole, temperature affects the membrane composition and fluidity, which could in turn affect the polysaccharide composition of the cell wall and the incorporation levels of its covalently anchored proteins. When analyzing the effects of temperature on growth at two pHs, we found that liquid cultures grew faster at pH 4 than at pH 7.4 at all temperatures tested and that at both pHs growth was peaking at 30°C (Table 2). Compared to growth at 30°C, growth at 42°C was reduced by ∼70% at pH 4 and by >80% at pH 7.4. Interestingly, when grown on solid medium at 42°C and pH 4, the cells were considerably more resistant to this temperature than at pH 7.4 (Fig. 1).
Table 2.
Biomass yields after 18 h at pH 7.4 and pH 4
| Growth temp (°C) | Biomass (mg of dry wt/ml) at:a |
|
|---|---|---|
| pH 7.4 | pH 4 | |
| 25 | 1.7 ± 0.2 | 2.2 ± 0.3 |
| 30 | 4.4 ± 0.3 | 5.0 ± 0.4 |
| 37 | 2.0 ± 0.1 | 3.7 ± 0.1 |
| 42 | 0.7 ± 0.1 | 1.4 ± 0.5 |
Mean ± standard deviation (n = 3).
Fig 1.
Spot assays of Candida albicans grown at 37°C and 42°C in the presence and absence of chemical stressors. SC5314 overnight cultures in 1:10 serial dilutions were spotted onto YNB-S plates at pH 4 or pH 7.4 and supplemented with 100 mg/liter SDS, 50 mg/liter calcofluor white, or 1 mg/liter Congo red, respectively, and grown for 2 days.
It is known that cells respond to wall stress by increasing the chitin levels in the wall, thus strengthening the weakened cell wall (45). We therefore analyzed the chitin content of walls from cells grown at 37°C and 42°C, both at pH 7.4 and pH 4. At pH 7.4 chitin levels increased about 1.7-fold, and at pH 4 the amount of chitin in the cell wall even doubled at 42°C compared to amount in the 37°C control (Table 3). Overall, growth at pH 4 led to higher wall chitin levels than at pH 7.4 (Table 3), which probably contributes to the higher heat resistance at low pH and might be related to morphotype (Fig. 1). Furthermore, when C. albicans was incubated at 42°C in the presence of the wall-perturbing compounds calcofluor white and Congo red or the membrane-perturbing SDS (Fig. 1), higher sensitivity was observed, supporting the notion that cell wall integrity is compromised at elevated temperatures.
Table 3.
Cell wall chitin content after growth for 18 h
| pH and temp (°C) | Chitin content (μg of chitin/mg of wall)a |
|---|---|
| pH 4 | |
| 37 | 47.4 ± 9.2 |
| 42 | 97.4 ± 8.0** |
| pH 7.4 | |
| 37 | 33.0 ± 4.3 |
| 42 | 54.8 ± 9.0** |
Values are means ± standard deviations (n ≥ 3). **, P < 0.01 versus growth at 37°C.
Prolonged high-temperature stress affects the wall proteome.
Subsequently, we analyzed the cell wall proteome at 42°C by applying relative quantification using a metabolically labeled 15N reference culture and FT-MS (14, 16). Combining technical and biological variance, the median coefficient of variation of our measurements was 11.2% and 13.1% for pH 4 and pH 7.4, respectively. Comparing the wall proteomes obtained at 37°C and 42°C revealed a strong impact of prolonged heat treatment at pH 7.4, while the changes were less pronounced at pH 4 (Fig. 2). At both pHs the wall levels of Sap9, involved in maintaining wall integrity (46), increased. The wall levels of the chitin transglycosylases Crh11 and Utr2 (47) and of a protein of unknown function involved in wall integrity, Ecm33 (48), also increased significantly at both pHs. The levels of other GPI-anchored wall proteins clearly decreased at both pHs upon heat stress, namely, the chitinase Cht2, the adhesin Als2, and the small temperature-regulated protein Rhd2. Interestingly, the set of proteins that showed reduced wall levels upon thermal stress also included the GPI-anchored wall proteins Csa1, Pga10, and Rbt5. They contain one or more cysteine-containing fungal extracellular membrane (CFEM) domains, consisting of eight cysteines separated by conserved spacing (49), and have been linked to iron starvation and heme/hemoglobin binding (6, 50). Fascinatingly, both of the antagonistically pH-regulated transglycosylases Phr1 and Phr2 are strongly increased in abundance at pH 7.4, suggesting a relief of the Rim101-regulated repression of PHR2 at this pH during wall stress (15, 51). In summary, our observations reveal that upon prolonged thermal stress, the wall proteome changes dramatically, showing significantly increased levels of wall proteins involved in wall maintenance and transglycosylation. This same set has been also identified during fluconazole stress (14), hinting at a more general stress response.
Fig 2.
Relative quantification of wall proteins using FT-MS and 15N labeling. The graphs represent log2 ratios of wall proteins at 42°C versus 37°C, at both pH 4 (median coefficient of variation, 13.1%) and pH 7.4 (median coefficient of variation, 11.2%). Positive values represent an increase and negative values represent a decrease of the corresponding protein at 42°C compared to the 37°C control.
Thermal stress leads to Mkc1 phosphorylation.
Next, we wanted to investigate the involvement of the major MAP kinase pathways in C. albicans in the late response to continuous thermal stress in comparison to the late responses elicited by known wall- and membrane-perturbing compounds. We therefore carried out immunoblot analysis of the phosphorylated forms of Mkc1, Cek1, and Hog1 after 18 h of growth under the tested conditions. As shown in Fig. 3 the phosphorylated form of Mkc1 became more abundant after prolonged thermal stress and even more so in the presence of fluconazole, Congo red, and calcofluor white. This is consistent with earlier observations that loss of MKC1 results in a strongly decreased resistance to Congo red and calcofluor white (52). The levels of phosphorylated Cek1 (Cek1∼P) tended to increase only slightly, whereas Hog1∼P did not seem to increase significantly at 42°C and decreased in the presence of the other stress conditions. Under our growth conditions Mkc1 seems to play an important role in the late responses to membrane and wall stress while Cek1 and Hog1 play only minor roles. Our observations do not, however, exclude a role for these MAP kinases during earlier responses to thermal stress (22).
Fig 3.
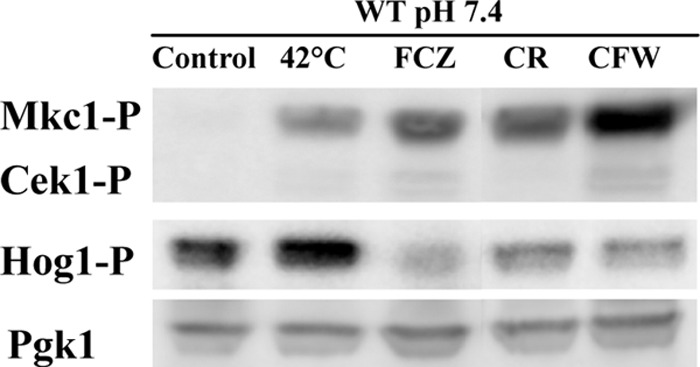
Immunoblot analysis of phosphorylated MAP kinases. The cultures were grown for 18 h at pH 7.4 and 37°C in the presence of various chemical stressors (0.5 mg/liter fluconazole, 125 mg/liter SDS, 5 mg/liter Congo red, and 40 mg/liter calcofluor white) or subjected to thermal stress at 42°C. Fifteen micrograms of protein extracts was loaded per lane. Pgk1 was used as a loading control.
Stress-induced cell clustering is linked to decreased chitinase abundance.
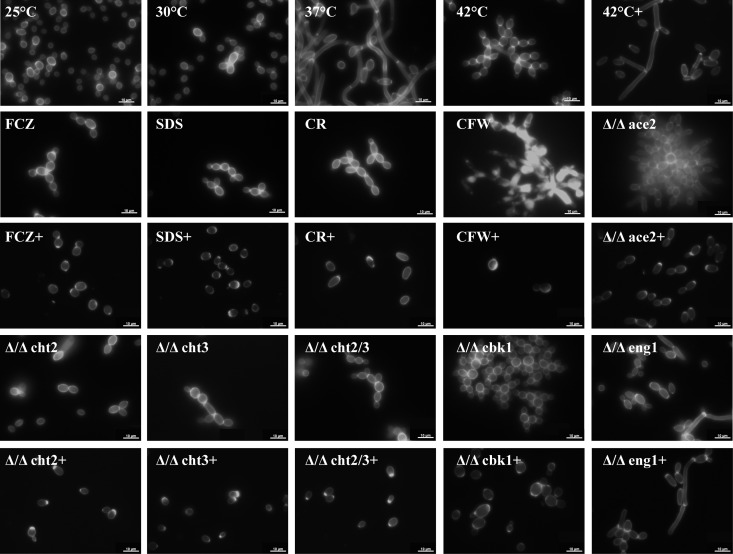
Fluorescence microscopy in combination with calcofluor white staining revealed significant formation of cell clusters at 42°C compared to growth at lower temperatures (Fig. 4). These cell clusters could not be disrupted by SDS treatment or sonication, suggesting a covalent linkage between the cells. The same holds for chitinase knockout strains and homozygous ace2 and cbk1 (Δ/Δace2 and Δ/Δcbk1, respectively) mutants, which are involved in the regulation of cell separation (data not shown). This is also consistent with the intense staining by the chitin-binding dye calcofluor white of the septal region (Fig. 4). Mass spectrometric analysis of the secretome from cells grown at pH 4 revealed a significant decrease of the cell separation-related secretory proteins Cht3 and Eng1 at 42°C compared to growth at temperatures (Table 4; for all of the >40 detected proteins see Tables S3 and S4 in the supplemental material) Due to the large number of intracellular proteins in the secretome of cells grown at 42°C in pH 7.4 medium, these data were not used for comparison (data not shown). The presence of a large number of intracellular proteins is most likely due to increased breakage or lysis of stressed cells and hyphae, hinting at a higher resilience of pH 4-grown yeast cells. Interestingly, as already mentioned above, the GPI-anchored, wall-bound chitinase Cht2 also decreased upon thermal stress, especially at pH 4 (Fig. 2).
Fig 4.
Temperature, stress conditions, and chitinase treatment influence the morphology of C. albicans wild-type and mutant strains. C. albicans wild-type and mutants were grown in YNB-S medium at pH 7.4 and were stained with calcofluor white. All cultures were grown at 37°C unless labeled otherwise. +, treatment with 0.5 U of chitinase for 3 h. FCZ, fluconazole (0.5 mg/liter); CFW, calcofluor white (50 mg/liter); CR, Congo red (2 mg/liter); SDS, sodium dodecyl sulfate (100 mg/liter). Scale bar, 10 μm.
Table 4.
Semiquantitative analysis of selected medium proteins
| Protein group and name | Relative abundance (%) by growth conditionsa |
||||||
|---|---|---|---|---|---|---|---|
| pH 7.4 |
pH 4 |
||||||
| 25°C | 30°C | 37°C | 25°C | 30°C | 37°C | 42°C | |
| Cell separation-associated proteins | |||||||
| Cht1 | 3.3 | 2.5 | ND | ND | 2.1 | 0.4 | ND |
| Cht3 | 5.7 | 8.4 | 5.2 | 10.7 | 8.6 | 7.0 | 3.3 |
| Eng1 | 3.5 | 6.9 | 4.3 | ND | 4.3 | 6.3 | ND |
| Stress response-associated protein | |||||||
| Msb2 | ND | 0.3 | 3.0 | 1.6 | 0.7 | 2.1 | 4.2 |
| Core set of abundantly secreted proteins | |||||||
| Cht3 | 5.7 | 8.4 | 5.2 | 10.7 | 8.6 | 7.0 | 3.3 |
| Mp65 | 12.9 | 11.8 | 9.1 | 7.8 | 11.5 | 8.9 | 8.7 |
| Scw11 | 9.9 | 7.3 | 8.0 | 0.3 | 8.6 | 6.5 | 4.6 |
| Sim1 | 9.0 | 8.1 | 8.7 | 9.8 | 5.0 | 4.7 | 7.1 |
| Sun41 | 7.2 | 7.9 | 5.4 | 12.5 | 10.0 | 9.1 | 8.3 |
| Tos1 | 8.3 | 9.2 | 9.3 | 11.2 | 7.6 | 5.6 | 10.3 |
| Xog1 | 7.1 | 7.3 | 11.1 | 3.5 | 4.5 | 6.1 | 5.8 |
The values represent the average relative abundances of protein identifications of selected medium proteins per growth condition as estimated by peptide counting from Q-TOF analysis. For information on all of the >40 identified proteins, see Tables S3 and S4 in the supplemental material. ND, not detected.
Cht3 and Eng1, both less abundant during heat stress, are involved in the dissolution of wall carbohydrates after the formation of a primary septum, allowing cell separation. Cht2 and Cht3 are both chitinases, close homologs to the only chitinase in Saccharomyces cerevisiae, Cts1, with Cht3 being the major chitinase in Candida albicans (53). Eng1 is an endo-1,3-beta-glucanase and is also involved in cell separation (33). Knockout mutants of cht3 and the homozygous diploid mutant strain cht2/cht3 (Δ/Δcth2/3) showed cellular nonseparation even at 37°C (Fig. 5). The protein kinase Cbk1 and the transcription factor Ace2, which participate in the RAM (regulation of Ace2 and morphogenesis) pathway (54), are major regulators of wall and secreted proteins. The expression levels of both CHT3 and ENG1 decrease about 3-fold when ACE2 is knocked out (55). The complete absence of cell separation in the ace2 deletion strain (34) suggests that Cht3 and Eng1 act synergistically during cell separation (Fig. 4). Like the knockouts or cells exposed to thermal stress, cells treated with the membrane-perturbing compounds fluconazole and SDS as well as with the cell wall-perturbing agents calcofluor white and Congo red displayed similar nonseparation phenotypes (Fig. 4), suggesting that this phenotype is associated with plasma membrane and cell wall stress. To corroborate that decreased chitin degradation is the reason for the cell separation defect as observed during cell wall stress, we treated C. albicans with exogenous chitinase for 3 h. As shown in Fig. 4, treatment with chitinase led to the dispersal of cell clusters, most notably for Δ/Δace2 and Δ/Δcbk1 cells.
Fig 5.
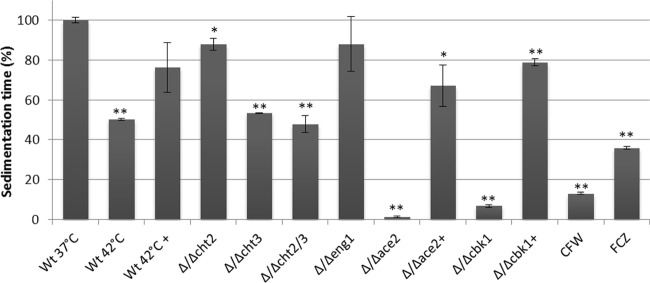
Relative sedimentation times of different C. albicans mutant strains and wild-type cells cultured under various stress conditions. The sedimentation times of wild-type cells grown at 37°C without (Wt 37°C) or with 0.5 mg/liter fluconazole (FCZ) or 40 mg/liter calcofluor white (CFW), of wild-type cells grown at 42°C, or of various mutant strains at 37°C were compared. Strains that were treated with chitinase are indicated (+). The time needed for the optical density to decline to 80% of the initial optical density was determined and is expressed as a percentage of wild-type cells grown at 37°C (relative sedimentation time). The sedimentation time of wild-type cells was 16.2 min. Bars represent averages ± standard deviations of two experiments. *, P < 0.05; **, P < 0.01 (relative to the wild-type control at 37°C).
For a comparative analysis of cell clustering, we introduced a spectrophotometric assay to measure relative sedimentation times. For each culture, the time required for the OD600 to decline to 80% of its initial value was measured, and the sedimentation time was expressed as a percentage of the sedimentation time of wild-type cells grown at 37°C. As expected, Δ/Δace2 and Δ/Δcbk1 cells, which cluster strongly, sedimented much faster than wild-type cells grown at 37°C (Fig. 5). Cultures grown in the presence of calcofluor white or fluconazole also showed shorter relative sedimentation times, as did the Δ/Δcht3 and Δ/Δcht2/3 homozygous diploid mutant strain, but not the Δ/Δcht2 strain, underlining the importance of Cht3 for cell separation and the dispersion of cell clusters. Importantly, treatment of the Δ/Δace2 and Δ/Δcbk1 strains with exogenous chitinase led to strongly increased sedimentation times. Cells grown at 42°C also sedimented significantly faster than control cells, and this phenotype could be largely restored by treating them with exogenous chitinase. This supports the notion that cell clustering during membrane and cell wall stress is linked to the decrease in chitinase levels in the medium.
DISCUSSION
In this study, we have shown that prolonged thermal stress is causing wall stress, which activates the Mkc1 signaling pathway. Chitin levels of the cell wall become higher, leading to reinforcement of the wall, whereas chitinase levels in the medium decrease. This probably reflects decreased formation and secretion of Cht3, leading to incomplete degradation of the primary chitinous septum during cytokinesis, thus leading to reduced cell separation. Additionally, a set of wall maintenance and repair proteins is strongly increased on the wall. All these responses seem to be a general consequence of membrane and wall stress.
As previously demonstrated for the antifungal fluconazole, we could validate that prolonged thermal stress is causing wall stress. Both fluconazole and high temperatures are affecting membrane fluidity (29, 56), which probably indirectly weakens the cell wall. Cells with a wall defect are less resistant to the chitin-binding dye calcofluor white, the β-1,3-glucan-binding Congo red, and SDS (57). Since C. albicans grown at 42°C showed less resistance to these compounds, this supports the idea that thermal stress causes wall stress.
Another hallmark of a stressed cell wall is an increase in wall chitin levels. This wall reinforcement in response to various stresses is a well-described phenomenon in fungi (12, 58–61). While chitin is only a minor constituent of the cell wall, a small increase in the percentage of chitin leads to a more robust wall. For example, in the presence of the β-1,3-glucan inhibitor caspofungin, the resulting loss of structural cell wall integrity is compensated by an increase of chitin in the wall (12). An increase of chitin can be achieved by either increased expression of the chitin synthases (CHS), a decrease in the degradation of chitin by the chitinases (CHT), or, most likely, both. A variety of pathways are involved in cell wall maintenance including the Mkc1, Hog1, Msb2-Cek1, and calcineurin pathways as well as the chaperone Hsp90, which all have been shown to be involved in the regulation of chitin synthase expression (11, 25, 62).
Less attention has been focused on the degradation of chitin and its role in stress resistance. Cells grown at 42°C for 18 h showed a severe cell separation defect. Similarly, cell aggregates were also described for several mutants that show a defective wall, like the glycosylation mutant mnn9, pmt1, and pmt4 strains, the wall protein ecm33, pga10, and pga13 mutants, and the wall-associated transglycosylase bgl2 mutants (48, 63–66). Gregori et al. assigned the cell separation defect in caspofungin-treated cells to an increased expression of adhesins, mainly Als1 (67). We also observed an increase of Als1 in stressed cells but found that the lack of cell separation is not primarily due to protein-protein interactions since the cell clusters could not be dispersed by SDS and vortexing or by mild sonication. In addition, our mass spectrometric analysis did reveal a distinct decrease of Cht3 and Eng1 in the secretome (Table 4), and cell clusters could be dispersed by adding exogenous chitinase (Fig. 4).
For full cell separation of mother and daughter cells, the β-glucan layer needs to be severed, and the primary septum needs to be dissolved. The observed reduced levels of chitinases and endoglucanase are presumably not sufficient any longer to ensure proper cell segregation, thus resulting in cell clustering. We hypothesized that, generally, wall and membrane stress leads to an increase of chitin by a concerted increase of chitin synthesis as well as reduced degradation. We therefore also tested SDS and calcofluor white and observed the same clustering phenotype (Fig. 4). In conjunction with our previous analyses of fluconazole- and Congo red-treated cells (14), this suggests that the decreased abundance of cell separation enzymes, the increase of chitin in the wall and the absence of cell separation in stressed cells are directly linked and represent a general stress response. Furthermore, we observed a similar clustering phenotype when testing knockout mutants of the chitinase Cht3 and endoglucanase Eng1 and severe clustering of their regulators, Ace2 and Cbk1 (Fig. 5). Cht3 has been described as the major chitinase in C. albicans (53), and since Ace2 regulates both ENG1 and CHT3 (55), the more severe phenotype of the Ace2 knockout mutant suggests a synergistic effect between Cht3, Eng1, and other potential Ace2 targets. Strikingly, an ace2 mutant is less sensitive to calcofluor white, SDS, and azoles (68). Possibly, within cell aggregates a protective microenvironment can develop, which might be further facilitated by the formation of extracellular matrix material (69). The growth rates of ace2 mutant cells and wild-type cells at 42°C are, however, similar (68).
In a recent study in which C. albicans was treated with micafungin, as expected an increased expression of the chitin synthases was observed, but in addition a marked downregulation of CHT2 and CHT3 was reported (70). Resistance to echinocandins is mainly mediated by the mutation of FKS1, the targeted subunit of β-1,3-glucan synthase (71). Intriguingly, a recent analysis of echinocandin-resistant strains without FKS1 mutations also identified mutations in both CHT2 and CHT3, with two point mutations in CHT3 directly located in the active site and the substrate binding cleft, conceivably severely limiting the activity of the enzyme (72). These strains also exhibited a strongly increased chitin content, which has been shown to be leading to a higher resistance to caspofungin in vivo (45). This suggests that mutations that render chitinases less active lead to increased resistance to wall stress. Taken together, we propose that different forms of wall and membrane stress result in diminished cell separation. Not just the increased expression of chitin synthases but also the concerted reduction of chitin degradation by reduced abundance of chitinases result in wall reinforcement with chitin and diminished cell separation. This is supported by the dispersal of cell clusters in the presence of chitinase and the associated slower sedimentation.
Increased chitin incorporation is only one of a number of stress responses at the disposal of C. albicans. At any given time, carbohydrate-active enzymes are remodeling the wall structure and architecture (10). Among these are the aforementioned chitinases (Cht2 and Cht3), the calcineurin-dependent chitin transglycosylases (Crh11 and Utr2) (57), β-glucan transglycosylases (Pga4, Phr1, and Phr2) and the β-1,3-glucan cross-linking Pir1 and the yapsin-like protease Sap9. Intriguingly, while the chitinases are reduced in abundance in response to stress, all of the others are strongly increased in abundance during thermal stress (Fig. 2). We also observed a marked increase of these proteins in response to fluconazole and Congo red stress (14). The yapsin-like protease Sap9 has been implicated to have a major role in cell wall integrity and host-pathogen interaction (46). Consistent with our data presented here and previously (14), SAP9 expression has been reported to be strongly increased in response to antifungal treatment (73). Pir1 has been shown to be crucial for cell wall maintenance (74), while the chitin transglycosylases Crh11 and Utr2, both belonging to the same protein family, are required for cell wall biogenesis. Although Phr1 and Phr2 have been reported to be differently expressed according to pH (75), our data show that both proteins are strongly increased at pH 7.4 during stress conditions, consistent with our previous results (14), suggesting a relief of Rim101 control of PHR2 at neutral pH. Taking these results together and including the results of our previous work (14) and transcriptional data obtained for cells treated with a variety of antifungal agents (30, 76), we hypothesize that this set of proteins participates in a highly conserved response to wall and membrane stress.
Secreted aspartyl proteases, most notably Sap8, have also been implicated in processing Msb2 (77–79), a transmembrane protein which upon cleavage activates the Cek1 MAP kinase pathway. A recent study also showed not only that proteolytic cleavage of Msb2 is important for signaling but also that its extracellular part can directly interact with human antimicrobial peptides (80), hinting at an important role in host-induced stresses. We observed strong Mkc1 phosphorylation under the stress conditions tested. Mkc1 and its homolog Slt2 in S. cerevisiae and Candida glabrata have been shown to be involved in heat resistance (24), virulence (81), wall reinforcement (58), and caspofungin resistance (82). We show here that Mkc1 is not only transiently activated by wall and membrane stress but that Mkc1 also continues to be activated in the presence of stress. This suggests that C. albicans adapts to long-term cell surface stress with a continuous activation of Mkc1, resulting in increased expression of remedial proteins, including the previously mentioned carbohydrate-active enzymes.
Concluding remarks.
In our previous work (14, 36), we identified a core set of abundantly secreted proteins (Cht3, Mp65, Scw11, Sim1, Sun41, Tos1, and Xog1) under virtually all growth conditions tested. When cells were grown at 25°C, 30°C, and 37°C both at pH 7.4 or pH 4 and at pH 4 at 42°C, most of these proteins were secreted in abundance as well (Table 3). Nevertheless, Cht3 was considerably less abundant at pH 4 and 42°C and Scw11 at pH 4 and 25°C. Since this core set of proteins seems to be consistently present and since ample peptide identifications suggest abundance, these proteins or antibodies raised against them might be suitable potential infection markers. The increase of repair-associated proteins during heat stress (Fig. 2) has also been observed during clinically induced stresses like fluconazole (14) or caspofungin treatment (30). They seem to be a general and relevant response to membrane and wall stress, which C. albicans also encounters in the host in the form of various antifungal peptides, like histatins, alpha- and beta-defensins, and cathelicidins.
We have shown that thermal stress causes wall stress in C. albicans, supported by the increased chitin levels and sensitivity to wall-perturbing agents at 42°C. Furthermore, we have shown that the wall stress response to thermal treatment seems to be similar to various surface stressors, leading to the activation of Mkc1 signaling, an increase of certain repair-associated wall proteins, reduction in cell-separation enzymes, most importantly Cht3, leading to wall reinforcement with chitin, and a correlated decrease in cell separation.
Supplementary Material
ACKNOWLEDGMENTS
We thank the reviewers for their constructive comments. A.G.S. and C.J.H. are grateful for the support by all members of the FINSysB consortium.
F.M.K. was supported by the EU Program FP7-214004-2 FINSysB.
Footnotes
Published ahead of print 14 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00278-12
REFERENCES
- 1. Vincent JL, Rello J, Marshall J, Silva E, Anzueto A, Martin CD, Moreno R, Lipman J, Gomersall C, Sakr Y, Reinhart K. 2009. International study of the prevalence and outcomes of infection in intensive care units. JAMA 302:2323–2329 [DOI] [PubMed] [Google Scholar]
- 2. Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
- 3. Frohner IE, Bourgeois C, Yatsyk K, Majer O, Kuchler K. 2009. Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol. Microbiol. 71:240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Luo G, Ibrahim AS, Spellberg B, Nobile CJ, Mitchell AP, Fu Y. 2010. Candida albicans Hyr1p confers resistance to neutrophil killing and is a potential vaccine target. J. Infect. Dis. 201:1718–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, Edwards JE, Filler SG, Hube B. 2008. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 4:e1000217 doi:10.1371/journal.ppat.1000217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weissman Z, Kornitzer D. 2004. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol. Microbiol. 53:1209–1220 [DOI] [PubMed] [Google Scholar]
- 7. Hoyer LL, Payne TL, Bell M, Myers AM, Scherer S. 1998. Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr. Genet. 33:451–459 [DOI] [PubMed] [Google Scholar]
- 8. Staab JF, Bradway SD, Fidel PL, Sundstrom P. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535–1538 [DOI] [PubMed] [Google Scholar]
- 9. Schaller M, Borelli C, Korting HC, Hube B. 2005. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses 48:365–377 [DOI] [PubMed] [Google Scholar]
- 10. de Groot PW, de Boer AD, Cunningham J, Dekker HL, de Jong L, Hellingwerf KJ, de Koster C, Klis FM. 2004. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot. Cell 3:955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Munro CA, Selvaggini S, de Bruijn I, Walker L, Lenardon MD, Gerssen B, Milne S, Brown AJ, Gow NA. 2007. The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol. Microbiol. 63:1399–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Walker LA, Munro CA, de Bruijn I, Lenardon MD, McKinnon A, Gow NA. 2008. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 4:e1000040 doi:10.1371/journal.ppat.1000040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Klis FM, Brul S, De Groot PW. 2010. Covalently linked wall proteins in ascomycetous fungi. Yeast 27:489–493 [DOI] [PubMed] [Google Scholar]
- 14. Sorgo AG, Heilmann CJ, Dekker HL, Bekker M, Brul S, de Koster CG, de Koning LJ, Klis FM. 2011. Effects of fluconazole on the secretome, the wall proteome, and wall integrity of the clinical fungus Candida albicans. Eukaryot. Cell 10:1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sosinska GJ, de Koning LJ, de Groot PW, Manders EM, Dekker HL, Hellingwerf KJ, de Koster CG, Klis FM. 2011. Mass spectrometric quantification of the adaptations in the wall proteome of Candida albicans in response to ambient pH. Microbiology 157:136–146 [DOI] [PubMed] [Google Scholar]
- 16. Heilmann CJ, Sorgo AG, Siliakus AR, Dekker HL, Brul S, de Koster CG, de Koning LJ, Klis FM. 2011. Hyphal induction in the human fungal pathogen Candida albicans reveals a characteristic wall protein profile. Microbiology 157:2297–2307 [DOI] [PubMed] [Google Scholar]
- 17. Robert VA, Casadevall A. 2009. Vertebrate endothermy restricts most fungi as potential pathogens. J. Infect. Dis. 200:1623–1626 [DOI] [PubMed] [Google Scholar]
- 18. Jacobsen MD, Bougnoux ME, d'Enfert C, Odds FC. 2008. Multilocus sequence typing of Candida albicans isolates from animals. Res. Microbiol. 159:436–440 [DOI] [PubMed] [Google Scholar]
- 19. Prinzinger R. 1982. The energy costs of temperature regulation in birds: the influence of quick sinusoidal temperature fluctuations on the gaseous metabolism of the Japanese quail (Coturnix coturnix japonica). Comp. Biochem. Physiol. A Comp. Physiol. 71:469–472 [DOI] [PubMed] [Google Scholar]
- 20. Schleucher E, Withers PC. 2002. Metabolic and thermal physiology of pigeons and doves. Physiol. Biochem. Zool. 75:439–450 [DOI] [PubMed] [Google Scholar]
- 21. Arguelles JC. 1997. Thermotolerance and trehalose accumulation induced by heat shock in yeast cells of Candida albicans. FEMS Microbiol. Lett. 146:65–71 [DOI] [PubMed] [Google Scholar]
- 22. Mayer FL, Wilson D, Jacobsen ID, Miramon P, Slesiona S, Bohovych IM, Brown AJ, Hube B. 2012. Small but crucial: the novel small heat shock protein Hsp21 mediates stress adaptation and virulence in Candida albicans. PLoS One 7:e38584 doi:10.1371/journal.pone.0038584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Levin DE. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Navarro-Garcia F, Sanchez M, Pla J, Nombela C. 1995. Functional characterization of the MKC1 gene of Candida albicans, which encodes a mitogen-activated protein kinase homolog related to cell integrity. Mol. Cell. Biol. 15:2197–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. LaFayette SL, Collins C, Zaas AK, Schell WA, Betancourt-Quiroz M, Gunatilaka AA, Perfect JR, Cowen LE. 2010. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90. PLoS Pathog. 6:e1001069 doi:10.1371/journal.ppat.1001069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mysiakina IS, Funtikova NS. 2007. The role of sterols in morphogenetic processes and dimorphism in fungi. Mikrobiologiia 76:5–18. (In Russian.) [PubMed] [Google Scholar]
- 27. Abe F, Usui K, Hiraki T. 2009. Fluconazole modulates membrane rigidity, heterogeneity, and water penetration into the plasma membrane in Saccharomyces cerevisiae. Biochemistry 48:8494–8504 [DOI] [PubMed] [Google Scholar]
- 28. Cruz MC, Goldstein AL, Blankenship JR, Del Poeta M, Davis D, Cardenas ME, Perfect JR, McCusker JH, Heitman J. 2002. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 21:546–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Welker S, Rudolph B, Frenzel E, Hagn F, Liebisch G, Schmitz G, Scheuring J, Kerth A, Blume A, Weinkauf S, Haslbeck M, Kessler H, Buchner J. 2010. Hsp12 is an intrinsically unstructured stress protein that folds upon membrane association and modulates membrane function. Mol. Cell 39:507–520 [DOI] [PubMed] [Google Scholar]
- 30. Bruno VM, Kalachikov S, Subaran R, Nobile CJ, Kyratsous C, Mitchell AP. 2006. Control of the C. albicans cell wall damage response by transcriptional regulator Cas5. PLoS Pathog. 2:e21 doi:10.1371/journal.ppat.0020021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gillum AM, Tsay EY, Kirsch DR. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179–182 [DOI] [PubMed] [Google Scholar]
- 32. Selvaggini S, Munro CA, Paschoud S, Sanglard D, Gow NA. 2004. Independent regulation of chitin synthase and chitinase activity in Candida albicans and Saccharomyces cerevisiae. Microbiology 150:921–928 [DOI] [PubMed] [Google Scholar]
- 33. Esteban PF, Rios I, Garcia R, Duenas E, Pla J, Sanchez M, de Aldana CR, Del Rey F. 2005. Characterization of the CaENG1 gene encoding an endo-1,3-beta-glucanase involved in cell separation in Candida albicans. Curr. Microbiol. 51:385–392 [DOI] [PubMed] [Google Scholar]
- 34. Kelly MT, MacCallum DM, Clancy SD, Odds FC, Brown AJ, Butler G. 2004. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol. Microbiol. 53:969–983 [DOI] [PubMed] [Google Scholar]
- 35. McNemar MD, Fonzi WA. 2002. Conserved serine/threonine kinase encoded by CBK1 regulates expression of several hypha-associated transcripts and genes encoding cell wall proteins in Candida albicans. J. Bacteriol. 184:2058–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sorgo AG, Heilmann CJ, Dekker HL, Brul S, de Koster CG, Klis FM. 2010. Mass spectrometric analysis of the secretome of Candida albicans. Yeast 27:661–672 [DOI] [PubMed] [Google Scholar]
- 37. Millar JB, Buck V, Wilkinson MG. 1995. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 9:2117–2130 [DOI] [PubMed] [Google Scholar]
- 38. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 39. Yin QY, de Groot PW, Dekker HL, de Jong L, Klis FM, de Koster CG. 2005. Comprehensive proteomic analysis of Saccharomyces cerevisiae cell walls: identification of proteins covalently attached via glycosylphosphatidylinositol remnants or mild alkali-sensitive linkages. J. Biol. Chem. 280:20894–20901 [DOI] [PubMed] [Google Scholar]
- 40. Kapteyn JC, ter Riet B, Vink E, Blad S, De Nobel H, Van Den Ende H, Klis FM. 2001. Low external pH induces HOG1-dependent changes in the organization of the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 39:469–479 [DOI] [PubMed] [Google Scholar]
- 41. Buerth C, Heilmann CJ, Klis FM, de Koster CG, Ernst JF, Tielker D. 2011. Growth-dependent secretome of Candida utilis. Microbiology 157:2493–2503 [DOI] [PubMed] [Google Scholar]
- 42. Desjardins P, Hansen JB, Allen M. 2009. Microvolume spectrophotometric and fluorometric determination of protein concentration. Curr. Protoc. Protein Sci., chapter 3, unit 3.10 doi:10.1002/0471140864.ps0310s55 [DOI] [PubMed] [Google Scholar]
- 43. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 44. Eisenhaber B, Schneider G, Wildpaner M, Eisenhaber F. 2004. A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae and Schizosaccharomyces pombe. J. Mol. Biol. 337:243–253 [DOI] [PubMed] [Google Scholar]
- 45. Lee KK, Maccallum DM, Jacobsen MD, Walker LA, Odds FC, Gow NA, Munro CA. 2012. Elevated cell wall chitin in Candida albicans confers echinocandin resistance in vivo. Antimicrob. Agents Chemother. 56:208–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Albrecht A, Felk A, Pichova I, Naglik JR, Schaller M, de Groot P, Maccallum D, Odds FC, Schafer W, Klis F, Monod M, Hube B. 2006. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J. Biol. Chem. 281:688–694 [DOI] [PubMed] [Google Scholar]
- 47. Cabib E. 2009. Two novel techniques for determination of polysaccharide cross-links show that Crh1p and Crh2p attach chitin to both beta(1-6)- and beta(1-3)glucan in the Saccharomyces cerevisiae cell wall. Eukaryot. Cell 8:1626–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Martinez-Lopez R, Monteoliva L, Diez-Orejas R, Nombela C, Gil C. 2004. The GPI-anchored protein CaEcm33p is required for cell wall integrity, morphogenesis and virulence in Candida albicans. Microbiology 150:3341–3354 [DOI] [PubMed] [Google Scholar]
- 49. Kulkarni RD, Kelkar HS, Dean RA. 2003. An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 28:118–121 [DOI] [PubMed] [Google Scholar]
- 50. Weissman Z, Shemer R, Conibear E, Kornitzer D. 2008. An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol. Microbiol. 69:201–217 [DOI] [PubMed] [Google Scholar]
- 51. De Bernardis F, Muhlschlegel FA, Cassone A, Fonzi WA. 1998. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect. Immun. 66:3317–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Navarro-Garcia F, Eisman B, Fiuza SM, Nombela C, Pla J. 2005. The MAP kinase Mkc1p is activated under different stress conditions in Candida albicans. Microbiology 151:2737–2749 [DOI] [PubMed] [Google Scholar]
- 53. Dunkler A, Walther A, Specht CA, Wendland J. 2005. Candida albicans CHT3 encodes the functional homolog of the Cts1 chitinase of Saccharomyces cerevisiae. Fungal Genet. Biol. 42:935–947 [DOI] [PubMed] [Google Scholar]
- 54. Saputo S, Chabrier-Rosello Y, Luca FC, Kumar A, Krysan DJ. 2012. The RAM network in pathogenic fungi. Eukaryot. Cell 11:708–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mulhern SM, Logue ME, Butler G. 2006. Candida albicans transcription factor Ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryot. Cell 5:2001–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Prasad T, Chandra A, Mukhopadhyay CK, Prasad R. 2006. Unexpected link between iron and drug resistance of Candida spp.: iron depletion enhances membrane fluidity and drug diffusion, leading to drug-susceptible cells. Antimicrob. Agents Chemother. 50:3597–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pardini G, De Groot PW, Coste AT, Karababa M, Klis FM, de Koster CG, Sanglard D. 2006. The CRH family coding for cell wall glycosylphosphatidylinositol proteins with a predicted transglycosidase domain affects cell wall organization and virulence of Candida albicans. J. Biol. Chem. 281:40399–40411 [DOI] [PubMed] [Google Scholar]
- 58. Cota JM, Grabinski JL, Talbert RL, Burgess DS, Rogers PD, Edlind TD, Wiederhold NP. 2008. Increases in SLT2 expression and chitin content are associated with incomplete killing of Candida glabrata by caspofungin. Antimicrob. Agents Chemother. 52:1144–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fortwendel JR, Juvvadi PR, Pinchai N, Perfect BZ, Alspaugh JA, Perfect JR, Steinbach WJ. 2009. Differential effects of inhibiting chitin and 1,3-β-d-glucan synthesis in ras and calcineurin mutants of Aspergillus fumigatus. Antimicrob. Agents Chemother. 53:476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pfaller M, Riley J. 1992. Effects of fluconazole on the sterol and carbohydrate composition of four species of Candida. Eur. J. Clin. Microbiol. Infect. Dis. 11:152–156 [DOI] [PubMed] [Google Scholar]
- 61. Popolo L, Gualtieri T, Ragni E. 2001. The yeast cell-wall salvage pathway. Med. Mycol. 39(Suppl 1): 111–121 [PubMed] [Google Scholar]
- 62. Kelly J, Rowan R, McCann M, Kavanagh K. 2009. Exposure to caspofungin activates Cap and Hog pathways in Candida albicans. Med. Mycol. 47:697–706 [DOI] [PubMed] [Google Scholar]
- 63. Gelis S, de Groot PW, Castillo L, Moragues MD, Sentandreu R, Gomez MM, Valentin E. 2012. Pga13 in Candida albicans is localized in the cell wall and influences cell surface properties, morphogenesis and virulence. Fungal Genet. Biol. 49:322–331 [DOI] [PubMed] [Google Scholar]
- 64. Perez A, Pedros B, Murgui A, Casanova M, Lopez-Ribot JL, Martinez JP. 2006. Biofilm formation by Candida albicans mutants for genes coding fungal proteins exhibiting the eight-cysteine-containing CFEM domain. FEMS Yeast Res. 6:1074–1084 [DOI] [PubMed] [Google Scholar]
- 65. Prill SK, Klinkert B, Timpel C, Gale CA, Schroppel K, Ernst JF. 2005. PMT family of Candida albicans: five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol. Microbiol. 55:546–560 [DOI] [PubMed] [Google Scholar]
- 66. Southard SB, Specht CA, Mishra C, Chen-Weiner J, Robbins PW. 1999. Molecular analysis of the Candida albicans homolog of Saccharomyces cerevisiae MNN9, required for glycosylation of cell wall mannoproteins. J. Bacteriol. 181:7439–7448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gregori C, Glaser W, Frohner IE, Reinoso-Martin C, Rupp S, Schuller C, Kuchler K. 2011. Efg1 controls caspofungin-induced cell aggregation of Candida albicans through the adhesin Als1. Eukaryot. Cell 10:1694–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Homann OR, Dea J, Noble SM, Johnson AD. 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 5:e1000783 doi:10.1371/journal.pgen.1000783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Joshi KR, Wheeler EE, Gavin JB. 1973. Scanning electron microscopy of colonies of six species of Candida. J. Bacteriol. 115:341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kaneko Y, Ohno H, Kohno S, Miyazaki Y. 2010. Micafungin alters the expression of genes related to cell wall integrity in Candida albicans biofilms. Jpn. J. Infect. Dis. 63:355–357 [PubMed] [Google Scholar]
- 71. Perlin DS. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 10:121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Drakulovski P, Dunyach C, Bertout S, Reynes J, Mallie M. 2011. A Candida albicans strain with high MIC for caspofungin and no FKS1 mutations exhibits a high chitin content and mutations in two chitinase genes. Med. Mycol. 49:467–474 [DOI] [PubMed] [Google Scholar]
- 73. Copping VM, Barelle CJ, Hube B, Gow NA, Brown AJ, Odds FC. 2005. Exposure of Candida albicans to antifungal agents affects expression of SAP2 and SAP9 secreted proteinase genes. J. Antimicrob. Chemother. 55:645–654 [DOI] [PubMed] [Google Scholar]
- 74. Martinez AI, Castillo L, Garcera A, Elorza MV, Valentin E, Sentandreu R. 2004. Role of Pir1 in the construction of the Candida albicans cell wall. Microbiology 150:3151–3161 [DOI] [PubMed] [Google Scholar]
- 75. Muhlschlegel FA, Fonzi WA. 1997. PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of pH-dependent expression. Mol. Cell. Biol. 17:5960–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Liu TT, Lee RE, Barker KS, Wei L, Homayouni R, Rogers PD. 2005. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 49:2226–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Puri S, Kumar R, Chadha S, Tati S, Conti HR, Hube B, Cullen PJ, Edgerton M. 2012. Secreted aspartic protease cleavage of Candida albicans Msb2 activates Cek1 MAPK signaling affecting biofilm formation and oropharyngeal candidiasis. PLoS One 7:e46020 doi:10.1371/journal.pone.0046020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Roman E, Cottier F, Ernst JF, Pla J. 2009. Msb2 signaling mucin controls activation of Cek1 mitogen-activated protein kinase in Candida albicans. Eukaryot. Cell 8:1235–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Vadaie N, Dionne H, Akajagbor DS, Nickerson SR, Krysan DJ, Cullen PJ. 2008. Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J. Cell Biol. 181:1073–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Szafranski-Schneider E, Swidergall M, Cottier F, Tielker D, Roman E, Pla J, Ernst JF. 2012. Msb2 shedding protects Candida albicans against antimicrobial peptides. PLoS Pathog. 8:e1002501 doi:10.1371/journal.ppat.1002501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Diez-Orejas R, Molero G, Navarro-Garcia F, Pla J, Nombela C, Sanchez-Perez M. 1997. Reduced virulence of Candida albicans MKC1 mutants: a role for mitogen-activated protein kinase in pathogenesis. Infect. Immun. 65:833–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wiederhold NP, Kontoyiannis DP, Prince RA, Lewis RE. 2005. Attenuation of the activity of caspofungin at high concentrations against Candida albicans: possible role of cell wall integrity and calcineurin pathways. Antimicrob. Agents Chemother. 49:5146–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.