Abstract
Intracellular trafficking is a vital component of both virulence mechanisms and drug interactions in Trypanosoma brucei, the causative agent of human African trypanosomiasis and n'agana of cattle. Both maintaining the surface proteome composition within a life stage and remodeling the composition when progressing between life stages are important features of immune evasion and development for trypanosomes. Our recent work implicates the abundant transmembrane invariant surface glycoproteins (ISGs) in the uptake of first-line therapeutic suramin, suggesting a potential therapeutic route into the cell. RME-8 is a mediator of recycling pathways in higher eukaryotes and is one of a small cohort of intracellular transport gene products upregulated in mammal-infective trypanosomes, suggesting a role in controlling the copy number of surface proteins in trypanosomes. Here we investigate RME-8 function and its contribution to intracellular trafficking and stability of ISGs. RME-8 is a highly conserved protein and is broadly distributed across multiple endocytic compartments. By knockdown we find that RME-8 is essential and mediates delivery of endocytic probes to late endosomal compartments. Further, we find ISG accumulation within endosomes, but that RME-8 knockdown also increases ISG turnover; combined with previous data, this suggests that it is most probable that ISGs are recycled, and that RME-8 is required to support recycling.
INTRODUCTION
Trypanosoma brucei is the causative agent of African sleeping sickness, also known as human African trypanosomiasis and n'agana in other mammals. Trypanosomiasis results in tens of thousands of deaths a year and severe economic losses due to affected livestock (1). T. brucei infects the mammalian bloodstream and lymphatic system and is transmitted by the bite of the tsetse fly, endemic to sub-Saharan Africa. The major mechanism of immune evasion in the mammalian host is antigenic variation, where the variant surface glycoprotein (VSG) covers the cell surface to a very high density, and shields invariant epitopes from recognition by the acquired immune system. A sophisticated VSG gene-switching mechanism ensures that immunologically distinct VSG proteins are expressed as the infection proceeds, allowing the population to escape immune detection (2, 3). Additional mechanisms include antibody clearance from the surface, and likely extend the period of immune avoidance between VSG switches (4–7). Both maintaining the VSG coat and antibody surface clearance depend on clathrin-mediated endocytosis (CME). Antibody-capped VSG molecules are rapidly delivered to the flagellar pocket, endocytosed, and trafficked through the endocytic system; antibodies are sorted for degradation in late endosomal compartments, while VSG is recycled to the plasma membrane (8, 9). Recently, the trafficking and functions of a family of invariant surface glycoproteins (ISGs) have been described. These proteins have type I transmembrane topology and intercalate with VSG on the surface, but are rapidly turned over by a ubiquitylation-dependent mechanism (10–12). Most recently, the ISG75 subfamily have been implicated in the uptake of the first-line defense compound suramin, indicating an important role in drug interactions and also that ISGs may have physiological roles as receptors, although the ligand(s) remains to be identified (13). Hence, differential trafficking of VSGs and ISGs is highly important for the parasite in vivo, both for immune evasion and for interaction with the external environment.
Endocytic activity is highly upregulated in the T. brucei mammalian bloodstream stage compared with the insect form of the parasite, suggesting direct involvement in pathogenicity and immune evasion (14). Trafficking factors, including clathrin and multiple Rab GTPases, are developmentally regulated, and likely mediate this shift in activity. We previously analyzed the T. brucei transcriptome to identify additional transcripts, and hence proteins, that may be upregulated in the bloodstream stage, and hence may mediate alterations in the activity of trafficking pathways associated with mammalian infectivity. Of several hundred transcripts analyzed, the ortholog of metazoan RME-8 was significantly upregulated in blood-stage parasites (15) [RME-8 stands for “required for receptor-mediated endocytosis 8”; the gene is also known as “DnaJ (Hsp40) homolog, subfamily C, member 13,” or DNAJC13].
RME-8 was identified by a genetic screen in Caenorhabditis elegans for endocytic factors essential for fluid-phase and receptor-mediated endocytosis, development, and viability (16, 17). Orthologs are present in many multicellular organisms, and studies in Drosophila, mammals, and plants confirm an important, conserved role in CME and endosomal trafficking (17–20). RME-8 is a >200-kDa protein with a central DnaJ domain, characteristic of Hsp70 chaperone binding partners, flanked on either side by two IWN-repeat regions of unknown function, conserved between metazoan and plant orthologs (17). RME-8 interacts as cochaperone with Hsc70 (21, 22), and is thought to control clathrin dynamics, mediating uncoating of endosomal clathrin coats via stimulating the weak ATPase activity of Hsc70. However, there is no evidence for direct RME-8 interaction with clathrin and no canonical clathrin-binding site; the precise mechanism and location of RME-8 remain unclear, with apparent differences in essentiality in distinct taxa. As many trafficking pathways intersect at the late endosome/multivesicular body (MVB) mediating sorting to the lysosome or retrograde transport to the Golgi apparatus, it has been difficult to dissect the precise roles of RME-8. Evidence for roles in both lysosomal sorting and promoting retrograde transport via interactions with retromer and Hsc70 has been previously described (23–26).
In C. elegans, RME-8 localizes to the limiting membrane of MVBs and is involved in prelysosomal trafficking (17), while Drosophila RME-8 colocalizes with multiple compartments, including clathrin-, Rab5-, and Rab7-positive structures, suggesting a function in organizing or maintaining multiple compartments (18). Mammalian RME-8 associates with early, recycling, and late endosomes (19, 27). RME-8 mutants in Drosophila reveal disrupted endocytosis and clathrin distribution, and confirm the interaction between the RME-8 DnaJ domain and Hsc70 (18). In Arabidopsis, RME-8 has a nonessential role, but mutants have defects in vacuolar membrane dynamics (20, 24). In mammals, knockdown of RME-8 causes clathrin depletion, reduced epidermal growth factor (EGF) uptake, and defects in lysosomal enzyme and mannose-6-phosphate receptor trafficking. However, the constitutive endocytosis pathway exemplified by the transferrin receptor is unaffected (19). Nevertheless, expression of truncated RME-8 led to effects consistent with a role in early endosomal trafficking, but no effects on clathrin or EGF were observed (27).
Recently, RME-8 was found to associate with retromer component SNX-1 at early endosomes in C. elegans and mammalian cells (25, 26). Retromer is required for retrograde transport from endosomes to the Golgi apparatus, and endosomal clathrin is required via formation of endosomal subdomains to sort cargo to the degradative pathway through association with ubiquitin- and clathrin-binding protein Hrs. RNA interference (RNAi) depletion of SNX-1, RME-8, or Hsc70 leads to accumulation of endosomal clathrin and mistargeting of cargo (25, 26). Retromer may thus be involved in the regulation of clathrin dynamics via RME-8 and Hsc70, possibly through SNX-1 bridging RME-8 and clathrin or relieving RME-8 autoinhibition. Here we examined the evolutionary history of RME-8, and to elucidate functional conservation, we explored RME-8 location and functions in T. brucei. As T. brucei is very distantly related to mammals and yeast and possesses a complete, yet simplified, trafficking system, these insights provide further evidence for the role of RME-8 in control of endosomal dynamics (28). Furthermore, we present evidence for a recycling pathway for transmembrane domain proteins in trypanosomes.
MATERIALS AND METHODS
Taxonomic homology survey.
To examine conservation of RME-8 across the eukaryotes, taxon sampling of 35 species, representing all eukaryotic supergroups sensu Adl, was used (29). Where possible, at least two representative taxa were included from each supergroup, to facilitate detection of secondary losses versus absence within an entire group, and to minimize detection failure because of species-specific divergence. Details of databases selected are given in Table S1 in the supplemental material. In addition, predicted proteomes for most species were downloaded by File Transfer Protocol from the respective databases for local analysis. Initial queries for RME-8 typically used the Homo sapiens predicted protein and BLAST (30) using the BLOSUM62 amino acid matrix with manual cutoff. Where initial searches failed to recover a candidate, query sequences from a taxon closely related to the target genome were used, if a clear ortholog was available. If that also failed to recover a candidate, HMMer was also used with a template composed of the whole set of recovered proteins. All recovered sequences were subjected to reverse BLAST searches using BLOSUM62, against the original genome (H. sapiens), and against the nr database, for confirmation of orthology. A candidate ortholog was considered retrieved if reverse BLAST recovered the original query, or annotated orthologs from other species, within the top five hits. Sequences were also parsed through the NCBI conserved domain database (CDDB) using default parameters and analyzed by alignment for the presence of significant sequence similarities and conservation of overall length and domain architecture. In addition, candidates were subjected to phylogeny to confirm orthology. Failure to meet all of these criteria resulted in an assignment of “not found.” Further, returned candidate sequences were aligned and subjected to phylogenetic analysis to confirm both extensive sequence homology and monophyly (see Fig. S1 and S2 in the supplemental material).
Plasmid constructs for RNA interference and chromosome tagging.
Primers for amplification of an RNAi target fragment were designed using RNAit (31) so as to amplify a region outside the DnaJ domain and unique to the RME-8 gene compared to the rest of the T. brucei genome. RNAi target fragments were PCR amplified using Taq DNA polymerase with gene-specific primers (TbRME-8-RNAi-F [AACGCCTGAACAATTTACCG] and TbRME-8-RNAi-R [AATTCACGGTAGTTCCACGC]) and cloned into the tetracycline-inducible RNAi expression vector p2T7TAblue (32), linearized with Eam1105I. The construct was verified by standard sequencing methods (Geneservice Ltd.) prior to introduction into trypanosomes. TbRME-8 was chromosome tagged with hemagglutinin (HA) or green fluorescent protein (GFP) using the protocol and vectors described in reference 33 and the following primers: TbRME-8-CTF (CAACCATCAGCAACAGTTACTTTCGATTACACTTGCTTCAACAAGTGCCGTTGGAAATGATGACGAGCCACCACCGATAggtACCgggCCCcccCTGgag) and TbRME-8-CTR (CGTATGCTCCGGATTTCTATGCTTTTTTTTTGCAAATTCCTCCACTCCGCAATTTAACCCTTCTCACCGCATAAtggCGGccgCTCtagAACtagTGGat) (the last 21 bases of the forward primer correspond to nucleotides of the plasmid backbone immediately preceding the tag sequence, and alternating uppercase and lowercase letters specify the reading frame; the last 26 bases of the reverse primer correspond to nucleotides of the plasmid backbone immediately downstream of the antibiotic resistance gene). The HA-TbVps23 vector was a gift from Ka-Fai Leung (Cambridge, United Kingdom) (34).
Transfection of bloodstream-form T. brucei.
Culturing of bloodstream-form T. brucei was performed as described previously (34). Cells were maintained at densities between 105 and 2 × 106 cells/ml. The single marker bloodstream (SMB) line was used for expression of tetracycline-inducible constructs (35). A total of 3 × 107 cells were harvested at 800 × g for 10 min at 4°C, and the Amaxa nucleofector protocol (36) was used for transfection with 10 to 25 μg of linearized DNA (either linearized plasmid for the RNAi lines, or ethanol-precipitated PCR product for chromosome-tagged lines). Cells were transferred to a flask containing HMI-9 complete medium and allowed to recover at 37°C for 6 h. Antibiotics were added for selection, and cells were subdivided into a 24-well plate. Positive clones were selected 5 to 6 days posttransfection. Chromosome-tagged lines, and ectopic expression of plasmid constructs for RNAi, were maintained using antibiotic selection at the following concentration: G418 and hygromycin B at 2.5 μg/ml. Induction of RNAi and growth analysis were as described previously (34). For colocalization with Vps23, an established cell line with ectopic expression of a C-terminal HA-tagged version of T. brucei Vps23 (TbVps23) was transfected with p2T7-RME8 RNAi.
qRT-PCR.
A total of 1 × 108 cells were harvested at 800 × g for 10 min at 4°C and washed with ice-cold phosphate-buffered saline (PBS) and quick-frozen in dry ice for 1 min. RNA was purified using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. The RNA concentration was quantified using a ND-1000 spectrophotometer and NANODROP software (Nanodrop Technologies). qRT-PCR was performed using iQ-SYBR green Supermix on a MiniOpticon real-time PCR (RT-PCR) detection system (Bio-Rad), and the results were quantified using OPTICON3 software (Bio-Rad), as described in reference 15. The following primers were used for quantitative real-time PCR (qRT-PCR): RME8-RTF (AGAGGAATGTGGTCGCTCTG) and RME-8-RTR (GGGCATATCCAAGGGAAACT). Data were normalized against β-tubulin (Tub-RTF [CAAGATGGCTGTCACCTTCA] and Tub-RTR [GCCAGTGTACCAGTGCAAGA]) and TbRab11 (Rab11-RTF [ATCGGCGTGGAGTTTATGAC] and Rab11-RTR [GTGGTAAATCGAACGGGAGA]) as examples of high- and low-abundance mRNAs.
Immunofluorescence analysis.
Preparation of samples and acquisition and processing of images were as described previously (34). For intracellular staining, cells were permeabilized: specifically, after harvesting, fixing, and applying the cells to polylysine microscope slides (VWR International), and before blocking or applying any antibodies, cells were incubated with 0.1% Triton X-100–PBS for 10 min at room temperature and washed three times for 5 min with PBS. To detect cell-surface staining of ISG or VSG, nonpermeabilized cells were used; i.e., the Triton X-100 treatment step was omitted. Immunofluorescence images are for permeabilized cells unless indicated otherwise in the figure legend. Antibodies were used at the following dilutions: mouse and rabbit anti-HA epitope immunoglobulin G (IgG) (both from Santa Cruz Biotechnology Inc.) at 1:1,000, rabbit anti-yellow fluorescent protein (YFP) (from Michael Rout, Rockefeller University) at 1:500, rabbit anti-Rab5A at 1:200, rabbit anti-Rab11 at 1:400, mouse anti-p67 (from J. Bangs, University of Wisconsin—Madison) at 1:1,000, rabbit anti-GRASP (from Graham Warren, Vienna, Austria) at 1:500, rabbit anti-clathrin at 1:250, and rabbit anti-ISG65 and rabbit anti-ISG75 (from Mark Carrington, Cambridge, United Kingdom) at 1:1,000. Secondary antibodies were anti-mouse Oregon Green 488 (Molecular Probes) at 1:1,000 and anti-rabbit Alexa Fluor 568 (Invitrogen) at 1:1,000. Images were captured using METAMORPH software (Universal Imaging Corp.) and processed using Adobe Photoshop (Adobe Systems Inc.). Quantification of the areas of fluorescence and integration was performed, either for distinct puncta in the case of intracellular ISG65 and ISG76 (see Fig. 6C) or per cell over the whole visible surface of each cell (see Fig. 6D), using the raw data, from identical exposures, as appropriate, and the Metamorph Region Measurements function.
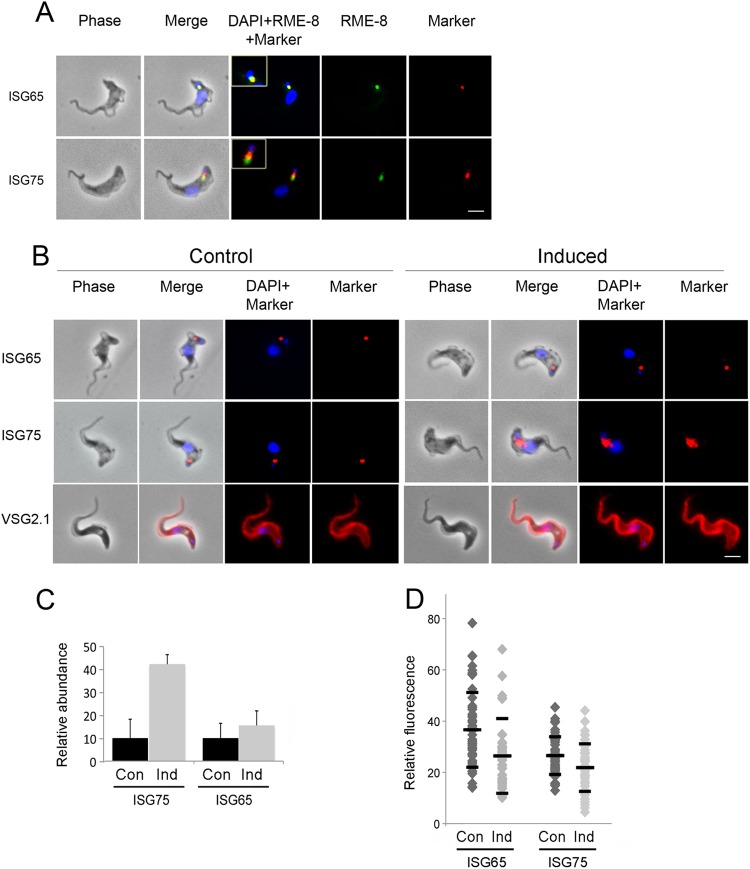
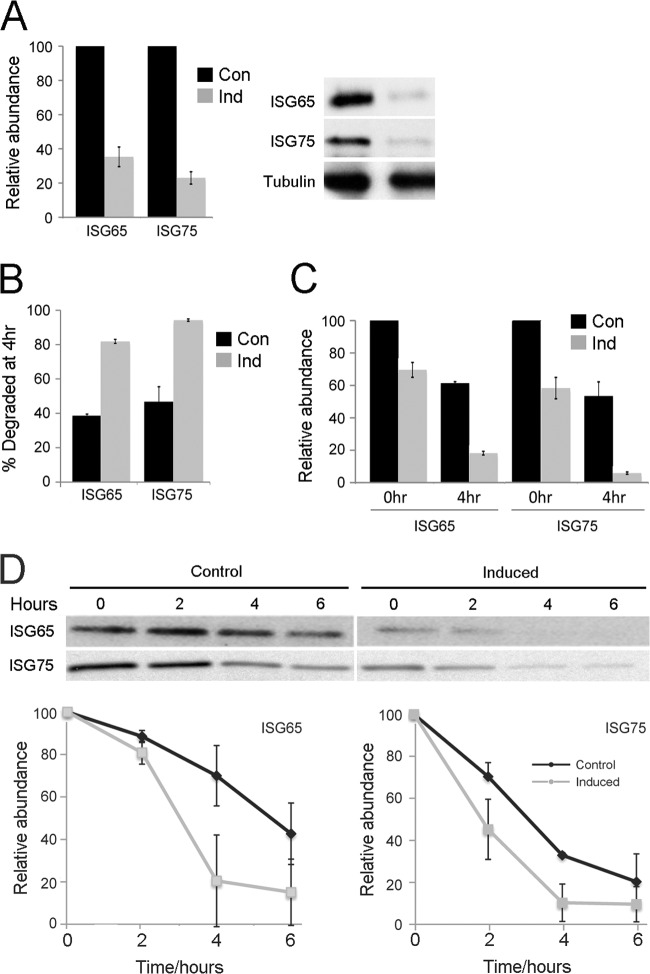
Fig 6.
Effect of TbRME-8 RNAi on ISG65 and ISG75 expression levels. (A) Immunofluorescence staining of TbRME-8–HA and ISG65/ISG75 under permeabilized conditions shows that TbRME-8 partially colocalizes with the endocellular pools of ISG65 and ISG75, two transmembrane proteins known to be marked by ubiquitylation and to traffic through the MVB. The scale bar, bottom right, is 2 μm. (B) The localization and relative expression levels of ISG65, ISG75, and VSG were assessed by immunofluorescence with specific antibodies, in control cells and 24 h postinduction of TbRME-8-RNAi. Staining for intracellular levels of ISG65 and ISG75 was performed in permeabilized cells, and an increase in intracellular levels of ISG75 was observed. Staining for VSG expression at the cell surface was performed in nonpermeabilized cells, and showed no significant alteration. The scale bar, bottom right, is 2 μm. (C) Quantification of the immunofluorescence signal of the cells shown in panel B. A modest but significant increase of 50% in the ISG65 fluorescence and a significant increase of 300% in the ISG75 fluorescence were observed 24 h postinduction of RNAi for TbRME-8 in 1N1K cells. Quantification of the areas of fluorescence and integration was performed for distinct puncta, using the raw data from identical exposures, and the Metamorph Region Measurements function. The data represent means (± standard deviations [SD]) of the results determined for at least 25 cells per sample (ISG65, P = 0.023; ISG75, P < 0.001 [as measured by Welch's unpaired t tests for the induced compared with the noninduced cells]). Con, control; Ind, induced. (D) Quantification of the ISG surface pool. Staining for ISG65 and ISG75 under nonpermeabilized conditions was quantified by immunofluorescence 24 h postinduction. A modest but significant decrease in the surface signal was observed for ISG65 and ISG75 in 1N1K cells. Quantification of the areas of fluorescence and integration was performed, per cell, for the whole visible surface of each cell, using the raw data from identical exposures, and the Metamorph Region Measurements function. The data represent means (± SD) of the results determined for at least 50 cells per sample (ISG65, P < 0.001; ISG75, P = 0.006 [as measured by Welch's unpaired t tests for the induced compared with the noninduced cells]).
ConA uptake assay.
The assay was as described previously (34). Briefly, 1.5 × 106 cells were harvested at 800 × g for 10 min at 4°C, washed with serum-free HMI-9 medium supplemented with 1% bovine serum albumin (BSA), and incubated in serum-free HMI-9 medium with 1% BSA for 20 min at 4°C, 12°C, or 37°C. Fluorescein isothiocyanate (FITC)-conjugated concanavalin A (ConA) (Molecular Probes) (50 mg/ml) was added, and the cells were incubated for a further 30 min at the above temperatures to allow uptake. Cells were washed with Voorheis' modified PBS and harvested by centrifugation at 800 × g for 10 min at 4°C, fixed, and costained for immunofluorescence as described above. To check ConA uptake dynamics, the same procedure was followed, except that cells were harvested by centrifugation, fixed, and stained, after 0, 10, 20, 30, and 60 min of ConA incubation at 37°C.
Western blot analysis.
To verify TbRME-8 chromosome tagging with HA and GFP, 1 × 107 cells were harvested by centrifugation at 800 × g for 10 min at 4°C. Cells were washed in ice-cold PBS and then resuspended in SDS sample buffer and incubated at 95°C for 10 min. Samples were subjected to electrophoresis on 12.5% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes, and proteins were detected by Western immunoblotting. Antibodies were used at the following dilutions: rabbit anti-HA (Santa Cruz Biotechnology Inc.) and rabbit anti-GFP (from Michael Rout, Rockefeller University) at 1:5,000 and anti-rabbit IgG (A0545; Sigma) (peroxidase conjugated) at 1:10,000. Densitometry quantification was achieved using IMAGEJ software (National Institutes of Health).
Protein turnover.
Protein synthesis was blocked by the addition of cycloheximide (10 μg/ml), and samples were analyzed by Western blotting, as described previously (34). Antibodies were used at the following dilutions: rabbit anti-ISG65 and rabbit ISG75 antibodies at 1:5,000, rabbit anti-BiP antibody at 1:10,000, and horseradish peroxidase-conjugated anti-(rabbit IgG) and anti-(mouse IgG) (A0545; Sigma) at 1:10,000. Densitometry quantification was achieved using ImageJ (NIH).
RIPAs.
Radioimmunoprecipitation assays (RIPAs) were performed as described previously (12). Briefly, 1 × 107 cells were washed in PBS, and resuspended in 500 μl of methionine-cysteine (Met/Cys)-free RPMI 1640 (Sigma) medium supplemented with 10% dialyzed fetal bovine serum (FBS), followed by incubation at 37°C for 1 h. Cells were pulse-labeled for 1 h with EasyTAg EXPRESS35S protein labeling mix (PerkinElmer) at a specific activity of 200 μCi/ml and chased by addition of 4.5 ml HMI-9 medium for 4 h. Cells were washed in ice-cold PBS, lysed in 100 μl RIPA buffer (25 mM Tris-Cl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) for 15 min on ice, and incubated for 5 min at 95°C in RIPA buffer containing 1% SDS. Lysates were precleared for 1 h with Pansorbin (Calbiochem) and incubated overnight with polyclonal antibodies against ISG65 and ISG75. Immunocomplexes were isolated by incubation with protein A-Sepharose (Sigma) for 1 h and subjected to SDS-PAGE. Gels were dried on Whatman paper and exposed to autoradiographic films for 24 to 48 h.
RESULTS
RME-8 is well conserved throughout the eukaryotes but with multiple secondary losses.
Given the presence of RME-8 in multicellular metazoan organisms and in the highly divergent trypanosomes, we examined whether the protein is conserved across the eukaryotes. We used comparative genomics to search for RME-8 orthologs in 35 species selected to represent all eukaryotic supergroups, and where genome data are well assembled, using several criteria to ensure detection of orthologs (see Materials and Methods). An RME-8 ortholog could be detected in all supergroups, predicting an early origin prior to the radiation of modern eukaryote lineages (Fig. 1). Further, the domain architecture and sequence identity between RME-8 orthologs indicated considerable selective pressure in those taxa where the gene is retained (E-values below e-110 were obtained in BLAST between the H. sapiens protein and orthologs of Drosophila melanogaster, C. elegans, Tetraodon nigroviridis, Nematostella vectensis, Monosiga brevicollis, Entamoeba histolytica, Dictyostelium discoideum, Arabidopsis thaliana, Oryza sativa, Populus trichocarpa, T. brucei, and Leishmania major [see Table S1 and Fig. S1 in the supplemental material]). Significantly, few paralogs suggesting gene duplication were detected (see Table S1 and Fig. S2 in the supplemental material), while multiple secondary losses were evident, based on BLAST searches and confirmed with PSI-BLAST and HMMer. While some of these losses may be due to annotation problems, since a variety of species were sampled for each lineage, most predicted losses are likely valid. The absence of RME-8 from all six fungal species and all five alveolates examined suggests secondary losses comparatively early in the evolution of these major groupings. This evolutionary distribution clearly indicates a variability in requirements for RME-8 in endosomal trafficking pathways in different taxa and asks what determines the importance of RME-8 in certain species but not in others.
Fig 1.
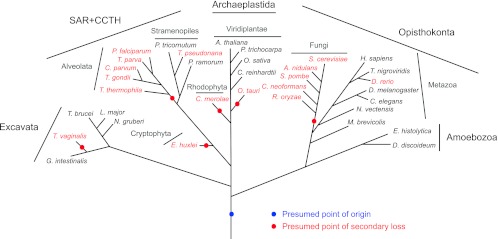
RME-8 is well conserved throughout eukaryotes but with multiple secondary losses. The presence (black taxa) and absence (red taxa) of RME-8 across 35 species representing all eukaryotic lineages are mapped on a schematic phylogenetic tree showing evolutionary relationships. Based on the presence of RME-8 in multiple lineages (Metazoa, Archaeplastida, Excavata), the origin of RME-8 is indicated by a blue dot before the diversification of eukaryotes. The multiple presumed secondary losses are indicated by red dots. Data are based on BLAST, PSI-BLAST, and HMMer results, together with alignments and phylogenetic analysis. Accession numbers are provided in Table S1 in the supplemental material. The sequence found in Danio rerio is abnormally short, suggesting possible misannotation.
TbRME-8 localizes to the endocytic system.
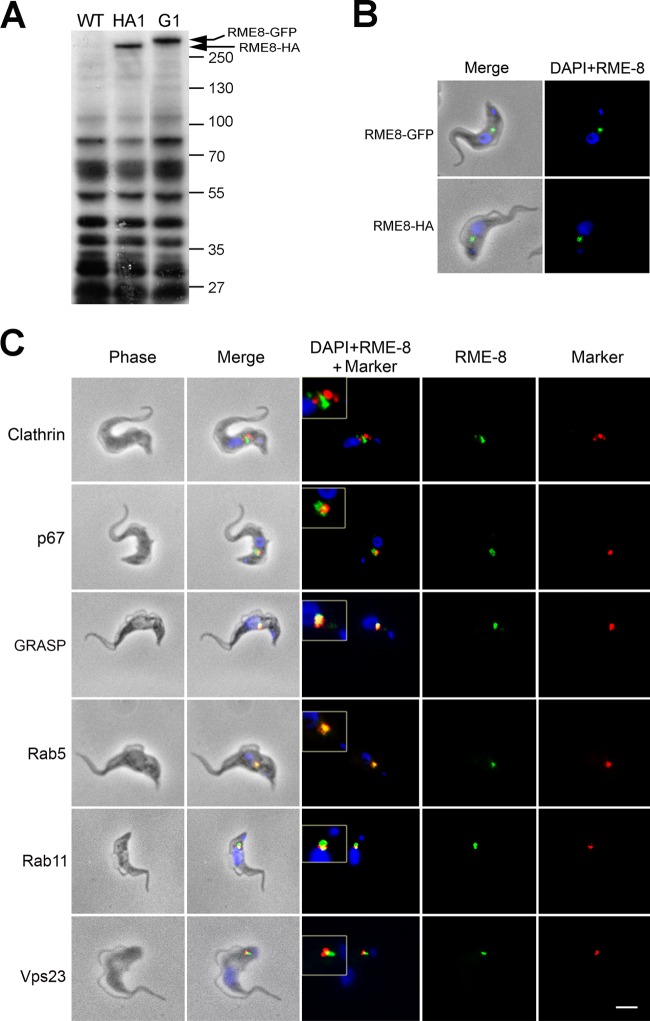
To elucidate functional conservation, we explored the location and activity of RME-8 in T. brucei, chosen for a divergent evolutionary position, a well-described endocytic system, and tractability. We generated cell lines with C-terminal chromosome-tagged versions of TbRME-8 with either a GFP or HA epitope. Expression of the tagged gene products was verified by Western blotting, using anti-HA and anti-GFP antibodies, and detected antigens with the predicted molecular weights (Fig. 2A; note that the high background is likely a result of extended transfer times required for the large TbRME-8 protein and the high titers of primary antibodies needed to detect tagged TbRME-8). Immunofluorescence of permeabilized cells harboring these tagged genes exhibited punctate staining between the nucleus and kinetoplast (Fig. 2B), the region of the cell containing the endosomal apparatus (28). Staining of untagged cells using either antibody combination, i.e., anti-GFP or anti-HA primary antibody, resulted in only background staining, indicating that the staining is specific for the tagged protein (data not shown). Costaining with several established markers for subcellular organelles of trypanosomes showed TbRME-8 juxtaposed with TbGRASP, a Golgi marker (37), and partially colocalized with Rab5A and Rab11 markers for the early and recycling endosomes, respectively (38, 39). TbRME-8 was also juxtaposed to the lysosomal marker p67 (40) and Vps23, a marker for the multivesicular body (34), as well as the clathrin heavy chain (CHC). The extensive localization of CHC does not allow formal discrimination between endosomes or post-Golgi compartments, although the endosomal membranes tend to locate at a position posterior to the Golgi complex (Fig. 2C). Overall, these data are highly indicative of an extensive endosomal location for trypanosomal RME-8, in agreement with observations from both metazoan and A. thaliana cells.
Fig 2.
TbRME-8 localizes to the endocytic system. (A) Western blot analysis using anti-HA and anti-GFP antibodies confirms chromosome tagging with HA and GFP in transfected bloodstream-form (BSF) lines after selection. Lane WT, BSF wild-type line; lane HA1, chromosome-tagged BSF clone; lane G1, chromosome-tagged BSF clone. Protein size markers are shown on the side, along with the positions of the HA-tagged and the GFP-tagged TbRME-8. (B) TbRME-8 was chromosome tagged with HA and GFP in BSF. The intracellular location was determined by immunofluorescence, and cells were counterstained with DAPI for DNA (blue). (C) Colocalization of TbRME-8–HA with a repertoire of endosomal markers to the T. brucei clathrin heavy chain, GRASP, Rab5A, Rab11, and Vps23 and of TbRME-8–GFP with p67 shows that TbRME-8 localizes to the posterior of the cell, between the nucleus and cytoplasm, which corresponds to the location of the endocytic apparatus. The scale bar, bottom right, is 2 μm. Inserts are electronic magnifications of regions of interest.
Knockdown of TbRME-8 suggests functions in membrane transport and endocytosis.
An RNAi cell line was generated using the p2T7 vector for tetracycline-inducible knockdown of RME-8. As shown by qRT-PCR, TbRME-8 mRNA levels were significantly reduced by about 50% within 24 h after induction,when normalized to β-tubulin and TbRab11 mRNA levels (Fig. 3A). This modest impact in knockdown is a reflection in part of the choice of time point, where impact on cell viability and morphology and secondary effects are minimal, but is similar to many other studies. Absence of a significant impact on β-tubulin and TbRab11 levels indicates that the RNAi is specific against RME-8 (data not shown).
Fig 3.
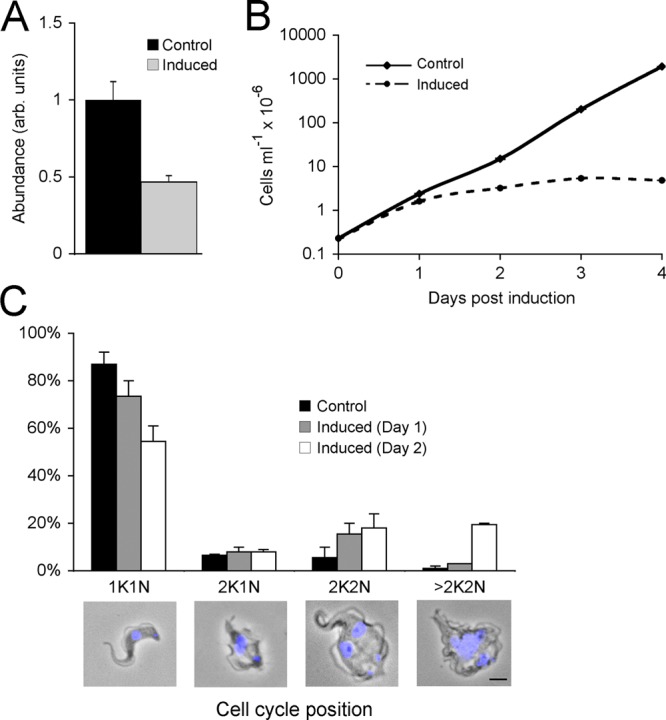
TbRME-8 RNAi causes a severe proliferative defect, associated with a block in cytokinesis. (A) qRT-PCR of control and tetracycline-induced RNAi lines for TbRME-8 shows a 50% decrease in the RME-8 mRNA within 24 h of induction. arb., arbitrary. (B) Growth curves of control and tetracycline-induced RNAi lines for TbRME-8 show a severe growth defect within 24 h postinduction. Representative results for one of the two clonal cell lines studied are shown (all subsequent experiments were performed using this cell line). Cultures were diluted daily to maintain cell densities between 105 and 2 × 106 cells/ml, but the cumulative effect is shown. (C) Quantitative analysis of copy numbers of nuclei and kinetoplasts following RNAi induction for TbRME-8. A modest disruption in cell cycle progression (increase in 2K2N postmitotis cells) was observed within 24 h after RNAi induction (Day 1), and abnormal >2K2N cells accumulated within 48 h after RNAi induction (Day 2). The data and error bars are based on analysis of 100 cells each from two different clones. Examples of cells in each division state are shown: phase images merged with DAPI staining for DNA in the nucleus and the kinetoplast. The scale bar, bottom right, is 2 μm.
To determine the importance of TbRME-8 to cell viability, cell proliferation over a period of 4 days postinduction was compared with a control cell population. A severe proliferative defect was observed within 24 h postinduction, consistent with the specificity of RNAi against TbRME-8 and closely coupled to the loss of RME-8 expression (Fig. 3B). A large proportion of the induced cells exhibited abnormal motility and cellular morphology. To quantify the frequency of cells at various points along the cell cycle, control and induced cells were further examined using epifluorescence microscopy with 4′,6′diamidino-2-phenylindole (DAPI) staining. We observed a significant increase in the proportion of cells with two kinetoplasts and two nuclei (2K2N) at day 1 and day 2 postinduction (Fig. 3C). About 20% of the induced cells on day 2 also showed prominent defects in cell morphology, with multiple kinetoplasts and nuclei (>2K2N cells, Fig. 3C), suggesting a defect in cytokinesis but, as discrete nuclei and kinetoplasts were seen, with ongoing mitosis and kinetoplast division. It is possible that loss of viability is partly due to this cytokinesis defect as a result of a general defect in cellular processes. Cytokinesis defects are a common feature of knockdowns of trafficking factors, and hence these phenotypes are consistent with a role in membrane trafficking, although other systems, for example, the cytoskeleton, also impact cytokinesis.
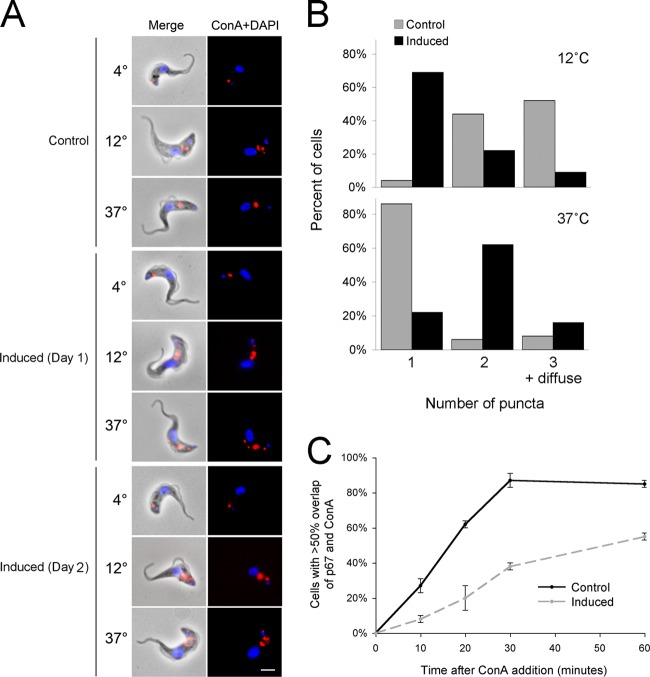
Having established that trypanosomal RME-8 is localized within the endocytic system, the effect of the RME-8 knockdown on the dynamics of endocytosis was studied by following the uptake of a fluorescently labeled lectin, concanavalin A (ConA) (Fig. 4). Cells were incubated at three different temperatures, enabling internalization of ConA into different organelles of the endocytic system. For control cells, ConA localized at the flagellar pocket at 4°C, which is visualized as one discrete spot and which lies close to the DNA-containing kinetoplast or mitochondrial genome to which it is physically connected (kinetoplast is the small blue spot in Fig. 4). At 12°C, ConA largely located to both the flagellar pocket and the early endosomes, the latter present as additional small fluorescent puncta that are closer to the nucleus than the flagellar pocket. Finally, at 37°C, ConA occupied the lysosome, which appears as a large structure that is more proximal to the nucleus than either the flagellar pocket or the endosomes (Fig. 4A) (39). Cells depleted of TbRME-8 after 1 day also showed one discrete spot at the flagellar pocket at 4°C (Fig. 4A), suggesting that the flagellar pocket remains intact. At 12°C, 69% of induced cells still showed the discrete flagellar pocket location compared to 4% of control cells (Fig. 4B), suggesting a delay in early trafficking to the endosomes. At 37°C, these induced cells showed a wide variety of phenotypes, including lysosomal or endosomal location and diffuse localization between the kinetoplast and the nucleus, whereas 86% of the control cells stained only the lysosome (Fig. 4A and B), suggesting decreased trafficking to the lysosome. An additional experiment to measure the dynamics of ConA trafficking confirmed that induced cells were substantially delayed at all stages of the endocytic route, from uptake at the flagellar pocket to lysosomal delivery (Fig. 4C; see also Fig. S3 in the supplemental material). Collectively, these results imply that the knockdown of RME-8 generates a defect in endocytic flux and delivery to the lysosome.
Fig 4.
TbRME-8 RNAi delays endocytosis in BSF. (A) Control and tetracycline-induced RNAi lines for TbRME-8 were allowed to take up FITC-conjugated ConA (red) at the indicated temperatures, fixed, and counterstained with DAPI for DNA (blue). ConA staining was assessed by immunofluorescence. In control cells, at 4°C ConA is not internalized and remains at the flagellar pocket, at 12°C ConA is preferentially blocked at the early endosome, and at 37°C ConA stains the lysosome (39). Induced RNAi lines for TbRME-8 were analyzed at 24 (Day 1) and 48 h (Day 2) postinduction. Representative images show a delay in delivery of ConA to the lysosome at 37°C in the RNAi lines. The scale bar, bottom right, is 2 μm. (B) Quantification of ConA trafficking at 12°C and 37°C, assessed by counting ConA puncta in 50 cells per sample, 1 day postinduction. Most control cells show two or three ConA puncta at 12°C, representing staining at the flagellar pocket and early endosomal compartments. In TbRME-8 RNAi cells, there is an overrepresentation of cells with only one ConA spot, possibly representing a delay of trafficking from the flagellar pocket. Furthermore, most control cells show a single ConA spot at 37°C, representing staining restricted to the lysosome. In TbRME-8 RNAi cells, there is an overrepresentation of cells with two or three ConA spots, likely representing a delay in trafficking through the endosomes. (C) Quantification of ConA lysosomal uptake dynamics at 37°C, 1 day postinduction. In control cells, ConA staining overlaps with the lysosomal marker p67 in the majority of cells within 30 min of exposure to ConA, while in the RNAi-induced cell line (day 1: 24 h postinduction), there is a marked delay in ConA uptake and overlap of p67. Data and error bars represent results for two different TbRME-8 RNAi clones; at least 30 cells were examined for each sample at each time point. Representative images are shown in Fig. S3 in the supplemental material.
TbRME-8 mediates trafficking of ISG65 and ISG75.
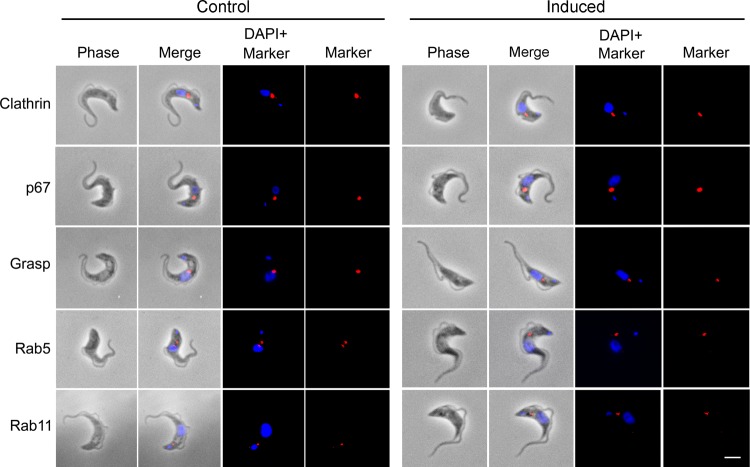
To further investigate the functions of TbRME-8, we examined the effects of knockdown on lysosomal, Golgi, and endocytic compartments by immunofluorescence, staining with specific antibodies under permeabilized conditions. There was no mislocalization of these markers, at least in 1N1K cells that retained normal morphology (Fig. 5). Importantly, this included the late endosome/lysosomal marker p67, which suggests that both lysosomal morphology and the trafficking of components to the terminal compartment are not substantially affected; therefore, it is likely that the lysosome remains functional in these knockdown cells. However, a substantial increase in the endocytic pool of ISG75 and a more moderate increase of the internal ISG65 pool were detected in the induced cell line (Fig. 6B and C), prompting us to examine the role of TbRME-8 in the trafficking of ISGs in more detail. Given the evidence for a normal lysosome, the increase in ISG levels cannot be simply ascribed to defective degradation.
Fig 5.
Effect of TbRME-8 RNAi on endocytic markers. The localization and relative expression levels of endocytic markers were assessed by immunofluorescence with specific antibodies, in control cells and 24 h postinduction of RNAi for TbRME-8. No significant change was observed for the flagellar pocket and early endosome markers (clathrin heavy chain, Rab5), the recycling endosome (Rab11), the lysosome (p67), or the Golgi complex (Grasp). The scale bar, bottom right, is 2 μm.
ISG65 and ISG75 are the major transmembrane proteins residing on the trypanosome plasma membrane; they are present throughout the surface coat, but also traffic through the endosomal system as they undergo rapid ubiquitylation-dependent degradation; both have half lives substantially shorter than the generation time of the trypanosome cell (10–12, 34). Immunofluoresence in nonpermeabilized cells detects ISGs on the cell surface, whereas staining permeabilized cells reveals the intracellular ISG localization. When permeabilized cells were stained for RME-8 and either ISG65 or ISG75, ISG65 colocalized with TbRME-8, while ISG75 partially colocalized (Fig. 6A), suggesting that the two proteins may take somewhat distinct trafficking routes, but that both localize in close proximity to RME-8-positive structures. While TbRME-8 knockdown led to a moderate increase in ISG65 and a significant increase in ISG75 intracellular pools (Fig. 6B and C), ISG65 and ISG75 steady-state levels both decreased 3-fold as assessed by Western blotting of whole-cell lysates (Fig. 7A). An increase in the internal ISG pool, accompanied by a decrease of the total steady-state level, suggests that ISG recycling to the plasma membrane is inhibited by TbRME-8 knockdown, i.e., that proportionately more internalized ISG is diverted toward the lysosome than recycled to the plasma membrane. To verify this hypothesis, we immunostained control and induced cells under nonpermeabilized conditions and quantified ISG levels on the cell surface. Induced RME-8 RNAi cells show moderately reduced ISG65 and ISG75 surface levels compared to control cells, consistent with decreased recycling (Fig. 6D).
Fig 7.
Influence of RME-8 on ISG65 and ISG75 stability. (A) Expression of ISG65 and ISG75 in RME-8 RNAi cells 24 h postinduction of TbRME-8 knockdown detected by Western blotting. Steady-state ISG expression is decreased by approximately 3-fold compared to control cells as assessed in three independent experiments and normalized to tubulin expression. (B) ISG turnover was analyzed in radioimmunoprecipitation assays (RIPAs). Cells were pulse-labeled with [35S]methionine-cysteine for 1 h and chased for 4 h. In RME-8 RNAi cells 70% to 90% of labeled ISGs were degraded after a 4-h chase compared to 40% to 50% in control cells, indicating an accelerated ISG turnover as an effect of RME-8 depletion. (C) ISG biosynthesis as analyzed by 35S incorporation for 1 h in RIPAs was decreased to 70% for ISG65 and 60% for ISG75 24 h postinduction compared to control cells (0-h time point). After a 4-h chase (4-h time point), the induced RNAi lines showed much lower abundance of ISG65 and ISG75 than control cells, indicative of faster turnover, as shown in panel B. (D) ISG65 and ISG75 are destabilized after RNAi knockdown of TbRME-8 as seen by Western blotting of whole-cell lysates after treatment with cycloheximide. The ISG half-life was reduced from ∼4.5 h in control cells to ∼2.5 h in induced cells. Data represent the means of the results of two independent experiments normalized to the values for tubulin; standard deviations are indicated.
TbRME-8 knockdown alters the stability of ISG65 and ISG75.
Since the drastic decrease in ISG steady-state levels cannot be explained solely by a moderate decrease in surface ISG levels, we asked if the rate of de novo ISG biosynthesis was affected by RME-8 knockdown, using a radioimmunoprecipitation assay (RIPA). We metabolically labeled cells with [35S]methionine-cysteine, chased for 4 h, and immunoprecipitated ISGs with ISG65- and ISG75-specific antibodies to monitor both steady-state synthesis and the turnover of these two proteins. Incorporation of [35S]methionine-cysteine was decreased by 30% for ISG65 and 40% for ISG75 in the TbRME-8 RNAi cells 24 h postinduction (Fig. 7C, 0 h time point). Analysis of unfractionated whole-cell lysates demonstrated that overall protein biosynthesis was not significantly affected by RME-8 knockdown (data not shown). Further, after chasing for 4 h, >80% of labeled ISGs were degraded in the induced cells, compared to ∼45% degradation in uninduced cells, indicating increased turnover of ISG65 and ISG75 in RME-8 RNAi cells (Fig. 7B and C, 4-h time point).
To confirm this observation, we also determined the half-life of ISG65 and IGS75 following blockade of ongoing protein synthesis with cycloheximide and followed protein levels in whole-cell lysates by Western blotting. In control cells, ISG65 and ISG75 had a half-life of ∼4.5 h, consistent with previous reports (12), but the half-life of ISG65 was reduced to 3 h, and the half-life of ISG75 to 2 h in TbRME-8 RNAi cells 24 h postinduction (Fig. 7D). Taken together, these analyses suggest that accelerated turnover likely accounts for much of the reduced steady-state ISG levels under conditions of TbRME-8 knockdown. Hence, these data indicate that a perturbation in trafficking of ISGs is likely associated with accumulation within the endosomal system. This is accompanied by accelerated breakdown at the lysosome, resulting from decreased recycling to the cell surface, and hence a reduction in whole-cell ISG levels. Although the precise itinerary of ISG65 and ISG75 is not yet known, these data clearly indicate that TbRME-8 participates in trafficking of surface proteins en route through the endosomal system and, in particular, is consistent with a role in recycling.
DISCUSSION
In T. brucei, developmental remodeling of the surface proteome, as well as activity within the endocytic pathway, has been well documented and is likely part of the program for life cycle adaptation (14). However, a surprisingly small cohort of genes involved in surface biosynthesis or turnover are developmentally regulated, based on transcriptome analysis (15, 41). One of the few gene products that does appear to be subject to developmental regulation is trypanosome RME-8, which was found to be significantly upregulated ∼3-fold in the bloodstream versus the insect stage (15), prompting the present investigation.
Comparative genomics demonstrated overall retention of RME-8 across the eukaryotes, indicating an ancient origin. Indeed, the sequence conservation in distant species is quite striking given that no clearly identified functional domains are present; we presume that such domain/fold architecture is present, but remains unrecognized. However, at least seven, and likely many more, secondary losses were detected, such that distinct species in different lineages are lacking an RME-8 ortholog, e.g., T. vaginalis in the Excavata, while present in neighboring taxa. The gene encoding RME-8 appears to have been lost early in the evolution of alveolates and fungi, as we were unable to identify an ortholog in any of the taxa we examined from these lineages. This pattern of partial conservation indicates both a requirement for RME-8 in many species and the presence of RME-8-independent endocytic systems, which have clearly arisen on multiple occasions. This is a phenomenon similar to that of Rab4, which we previously reported to also exhibit patchy distribution, with evidence for multiple secondary losses (42), as well as the recently described AP-5 complex (43). However, the secondary losses of Rab4 and AP-5 do not correlate with the pattern seen here for RME-8, ruling out coevolution. Eukaryotes have multiple DnaJ domain-containing proteins (22); of these, mammalian auxilin and yeast Swa2 have been shown to be involved in clathrin uncoating (44–46). Since RME-8 is also thought to be involved in clathrin uncoating at endosomes, orthologs of auxilin or Swa2 may compensate for RME-8 in those species where it is absent. It is, however, interesting that in at least some taxa where RME-8 is present, the gene is essential, as demonstrated in the present study, for example, and also in D. melanogaster (18).
Trypanosome RME-8 localizes to the endosomal system and partially colocalizes with Rab5A and Rab11 markers for the early and recycling endosomes, respectively. TbRME-8 was also juxtaposed to the clathrin heavy chain, the lysosomal marker p67, and Vps23, a marker for the multivesicular body. These data agree with observations of the localization of RME-8 in other organisms, where the protein has a presence on multiple endosomal structures, including early, recycling, and late endosomes (17–19, 27). Notably, TbRME-8 also colocalized with TbGRASP, a Golgi marker, which is a novel location, but not inconsistent with a role for RME-8 in retromer retrograde traffic from the endosomes to the Golgi apparatus (25, 26).
TbRME-8 is required for proliferation of bloodstream-form cells, as RNAi knockdown resulted in a severe proliferative defect and a cytokinesis block. It is tempting to suggest that the effect on cytokinesis is due to the role of RME-8 in clathrin uncoating: depletion of RME-8 can increase the amount of clathrin associated with membranes in Drosophila, C. elegans, and mammalian cells (18, 19, 26) and thus affect membrane dynamics required for cytokinesis. However, the cytokinesis block may also result from a general defect in transport, as it is a common phenotype after RNAi in T. brucei, especially in membrane trafficking (32). To avoid confounding direct and secondary consequences of TbRME-8 knockdown, for most functional analyses we examined cells after only 1 day of RNAi induction.
TbRME-8 clearly functions in endocytosis, as knockdown resulted in a delay in trafficking through the endosomal system and a partial block to delivery of ConA to the lysosome. However, TbRME-8 depletion does not result in a “BigEye” phenotype, as seen for T. brucei clathrin and several other major players in endocytosis, including Rab5A and Rab11 (47–49). This suggests that there is no bulk membrane defect at the flagellar pocket, the site of endocytic uptake for trypanosomes, but the knockdown is rather more similar to that obtained by knockdown of T. brucei EpsinR, a clathrin-associated protein, or CLASP (50). Further, we have recently obtained evidence for an interaction between clathrin and RME-8 based on coimmunoprecipitation (V. Adung'a, C. Gadelha, and M. C. Field, unpublished data), suggesting that RME-8, at least in trypanosomes, is a bona fide CLASP. However, staining with antibodies against antigens for the major endocytic compartments uncovered no abnormal morphology or targeting of these markers, indicating that TbRME-8 knockdown does not affect the structure, maintenance, or location of the major known endocytic compartments, in agreement with studies in other organisms (18, 19, 26). Therefore, despite the interaction with clathrin, there is no evidence for a major structural role for RME-8 in trypanosomes, but these data rather imply that RME-8 functions downstream of clathrin.
The trypanosome cell surface is unusual, and dominated by the GPI-anchored VSG. It is well established that VSG itself is recycled, but the major transmembrane domain surface antigens, ISG65 and ISG75, have not been formally demonstrated to do so; nonetheless, colocalization with Rab11 indicates that they associate with recycling endosomes (12). Recycling would be consistent with the half-life of the ISGs, which is more than 4 h, much in excess of the time required for delivery to the surface (∼40 min), recycling (∼15 min), and/or lysosomal delivery (∼15 min), suggesting that ISGs can experience multiple rounds of endocytosis and re-export prior to being turned over, similarly to both VSG and the transferrin receptor (8, 9, 11, 12, 28, 34, 51–54). The importance of these proteins has been difficult to ascertain, as both are nonessential in in vitro culture based on knockdowns, but their high abundance and rapid turnover indicates that they are major endocytic cargoes. Further, they are capable of being recognized by antisera and moreover appear to be important in mediating the uptake of the trypanocide suramin (11, 13, 55). Hence, fuller characterization of ISG trafficking is of importance to understanding immune evasion and drug interactions in these organisms.
Our data indicate a major role for RME-8 in ISG trafficking, and lead to the conclusion that these proteins are most likely recycled as the most parsimonious interpretation of the present data and earlier work. An increase in intracellular pools of ISG65 and ISG75 suggests defective prelysosomal trafficking in RME-8 knockdown. In whole-cell extracts, total levels of both ISG65 and ISG75 were decreased in TbRME-8 RNAi cells and cycloheximide treatment showed increased turnover rates for both proteins. The more rapid turnover and lower whole-cell levels indicate an overall increase in the proportion of ISG65 and ISG75 delivered to the lysosome (Fig. 8). This is in agreement with data from mammalian cells, where there is accelerated EGF receptor degradation after RME-8 knockdown (23). Therefore, we suggest that RNAi against TbRME-8 leads to a block in the RME-8-dependent recycling routes from the endosomes, such that despite an increased presence within the endosomal population, these internal pools of ISG65 and ISG75 are preferentially targeted to the degradative arm of the endocytic apparatus, accounting for decreased stability (Fig. 8). In mammalian cells, RME-8 interacts with the sorting nexin component of the retromer complex, which functions in retrograde traffic from the endosome to the Golgi apparatus, and RME-8 depletion affects retrograde traffic (25, 26).
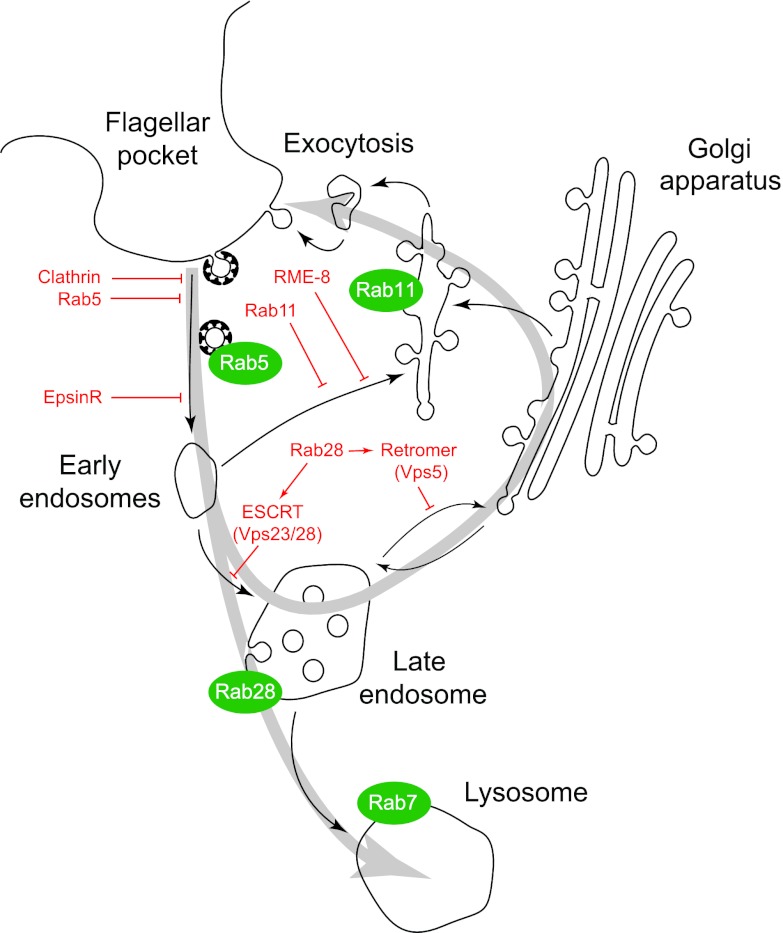
Fig 8.
A model for endocytic trafficking in trypanosomes. A schematic focused on endocytic and recycling pathways is shown, with major Rab GTPases represented as green lozenges, and routes between compartments shown with black arrows. Proposed points of inhibition of specific steps are shown for selected gene products in red. Evidence is summarized from the following previous studies: clathrin-mediated endocytosis at the flagellar pocket (47); Rab5, early endosome (38); EpsinR (50); Rab 11, recycling endosome (39, 53); Vps23/28, multivesicular body (34); Rab28 (58); Vps5, retromer (57); and Rab7, late endosome (54, 59) (also see reference 28). The present study places RME-8 as a mediator of recycling, with ISG65 and ISG75 both accumulating within the endosomal system, and as a consequence of a lack of access to the plasma membrane for internalized molecules, being turned over more rapidly under knockdown conditions. The gray arrows indicate the major routes for ISG trafficking based on the data presented here and in earlier studies (11, 12, 55).
A “counting” mechanism has been suggested to exist in trypanosomes for regulating levels of ISG and VSG on the cell surface and maintaining surface architecture (12, 56). Thus, it is possible that the cell can sense an increased ISG load within the endosomes, interpret this as meaning that there is excess extracellular ISG, and consequently downregulate synthesis and increase turnover to reduce surface ISG levels. Interestingly, accelerated turnover of ISG75 has also been observed after knockdown of retromer complex components in T. brucei (57), where a cytokinesis defect similar to that seen here was observed after TbRME-8 RNAi. The retromer complex is involved in endosome-to-Golgi retrograde traffic, but, in that case, trafficking to the lysosome was unaffected and, therefore, lower intracellular levels of ISG were observed; this argues against a general signaling mechanism linking endosomal load to lysosomal turnover. Further, we have also observed increased ISG65 and ISG75 intracellular pools in Rab28 knockdowns accompanied by decreased turnover rates (58); therefore, a trafficking model is preferred to explain the data (Fig. 8), but a counting mechanism cannot be ruled out. As Rab28 appears to interact with both the retromer and late endosomal ESCRT systems, increased ISG65 and ISG75 intracellular pools support the idea of a function of RME-8 in targeting these surface antigens to the lysosome (58). But since Rab28 is involved not only in regulation of ESCRT-mediated degradation pathways but also in retromer-dependent retrograde traffic from endosomes to the Golgi apparatus, there is evidence that ISGs are not solely destined for destruction but travel through the endocytic system back to the surface (Fig. 8). Taken together, these earlier reports and the present one provide strong evidence for active recycling pathways in trypanosomes and, in particular, for the trafficking of the ISG proteins. The postulated involvement of TbRME-8 in ISG recycling, which is present only in the bloodstream stage, could also explain the upregulated expression that was observed for this gene in the bloodstream versus the procyclic stage (15), and which prompted the current analysis.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Wellcome Trust (program grant 082813 to M.C.F.). We are also indebted to CamPOD for provision of equipment funds and the CamGrid computational resource on which some of the phylogenetic analyses were performed.
We thank James Bangs for anti-p67 antibody, Michael Rout for anti-GFP/YFP antibody, and Graham Warren for anti-TbGRASP antibody.
Footnotes
Published ahead of print 21 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00273-12.
REFERENCES
- 1. Barrett MP, Burchmore RJ, Stich A, Lazzari JO, Frasch AC, Cazzulo JJ, Krishna S. 2003. The trypanosomiases. Lancet 362:1469–1480 [DOI] [PubMed] [Google Scholar]
- 2. Horn D, Barry JD. 2005. The central roles of telomeres and subtelomeres in antigenic variation in African trypanosomes. Chromosome Res. 13:525–533 [DOI] [PubMed] [Google Scholar]
- 3. Boothroyd CE, Dreesen O, Leonova T, Ly KI, Figueiredo LM, Cross GA, Papavasiliou FN. 2009. A yeast-endonuclease-generated DNA break induces antigenic switching in Trypanosoma brucei. Nature 459:278–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barry JD. 1979. Capping of variable antigen on Trypanosoma brucei, and its immunological and biological significance. J. Cell Sci. 37:287–302 [DOI] [PubMed] [Google Scholar]
- 5. O'Beirne C, Lowry CM, Voorheis HP. 1998. Both IgM and IgG anti-VSG antibodies initiate a cycle of aggregation-disaggregation of bloodstream forms of Trypanosoma brucei without damage to the parasite. Mol. Biochem. Parasitol. 91:165–193 [DOI] [PubMed] [Google Scholar]
- 6. Sternberg JM. 2004. Human African trypanosomiasis: clinical presentation and immune response. Parasite Immunol. 26:469–476 [DOI] [PubMed] [Google Scholar]
- 7. Gadelha C, Holden JM, Allison HC, Field MC. 2011. Specializations in a successful parasite: what makes the bloodstream-form African trypanosome so deadly? Mol. Biochem. Parasitol. 179:51–58 [DOI] [PubMed] [Google Scholar]
- 8. Overath P, Engstler M. 2004. Endocytosis, membrane recycling and sorting of GPI-anchored proteins: Trypanosoma brucei as a model system. Mol. Microbiol. 53:735–744 [DOI] [PubMed] [Google Scholar]
- 9. Pal A, Hall BS, Jeffries TR, Field MC. 2003. Rab5 and Rab11 mediate transferrin and anti-variant surface glycoprotein antibody recycling in Trypanosoma brucei. Biochem. J. 374:443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ziegelbauer K, Multhaup G, Overath P. 1992. Molecular characterization of two invariant surface glycoproteins specific for the bloodstream stage of Trypanosoma brucei. J. Biol. Chem. 267:10797–10803 [PubMed] [Google Scholar]
- 11. Chung WL, Leung KF, Carrington M, Field MC. 2008. Ubiquitylation is required for degradation of transmembrane surface proteins in trypanosomes. Traffic 9:1681–1697 [DOI] [PubMed] [Google Scholar]
- 12. Leung KF, Riley FS, Carrington M, Field MC. 2011. Ubiquitylation and developmental regulation of invariant surface protein expression in trypanosomes. Eukaryot. Cell 10:916–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alsford S, Eckert S, Baker N, Glover L, Sanchez-Flores A, Leung KF, Turner DJ, Field MC, Berriman M, Horn D. 2012. High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature 482:232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Natesan SK, Peacock L, Matthews K, Gibson W, Field MC. 2007. Activation of endocytosis as an adaptation to the mammalian host by trypanosomes. Eukaryot. Cell 6:2029–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koumandou VL, Natesan SK, Sergeenko T, Field MC. 2008. The trypanosome transcriptome is remodelled during differentiation but displays limited responsiveness within life stages. BMC Genomics 9:298 doi:10.1186/1471-2164-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grant B, Hirsh D. 1999. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell 10:4311–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Y, Grant B, Hirsh D. 2001. RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol. Biol. Cell 12:2011–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang HC, Hull M, Mellman I. 2004. The J-domain protein Rme-8 interacts with Hsc70 to control clathrin-dependent endocytosis in Drosophila. J. Cell Biol. 164:1055–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Girard M, Poupon V, Blondeau F, McPherson PS. 2005. The DnaJ-domain protein RME-8 functions in endosomal trafficking. J. Biol. Chem. 280:40135–40143 [DOI] [PubMed] [Google Scholar]
- 20. Silady RA, Kato T, Lukowitz W, Sieber P, Tasaka M, Somerville CR. 2004. The gravitropism defective 2 mutants of Arabidopsis are deficient in a protein implicated in endocytosis in Caenorhabditis elegans. Plant Physiol. 136:3095–3103; discussion 3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ungewickell E, Ungewickell H, Holstein SE. 1997. Functional interaction of the auxilin J. domain with the nucleotide- and substrate-binding modules of Hsc70. J. Biol. Chem. 272:19594–19600 [DOI] [PubMed] [Google Scholar]
- 22. Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. 2004. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 5:567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Girard M, McPherson PS. 2008. RME-8 regulates trafficking of the epidermal growth factor receptor. FEBS Lett. 582:961–966 [DOI] [PubMed] [Google Scholar]
- 24. Silady RA, Ehrhardt DW, Jackson K, Faulkner C, Oparka K, Somerville CR. 2008. The GRV2/RME-8 protein of Arabidopsis functions in the late endocytic pathway and is required for vacuolar membrane flow. Plant J. 53:29–41 [DOI] [PubMed] [Google Scholar]
- 25. Popoff V, Mardones GA, Bai SK, Chambon V, Tenza D, Burgos PV, Shi A, Benaroch P, Urbe S, Lamaze C, Grant BD, Raposo G, Johannes L. 2009. Analysis of articulation between clathrin and retromer in retrograde sorting on early endosomes. Traffic 10:1868–1880 [DOI] [PubMed] [Google Scholar]
- 26. Shi A, Sun L, Banerjee R, Tobin M, Zhang Y, Grant BD. 2009. Regulation of endosomal clathrin and retromer-mediated endosome to Golgi retrograde transport by the J-domain protein RME-8. EMBO J. 28:3290–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fujibayashi A, Taguchi T, Misaki R, Ohtani M, Dohmae N, Takio K, Yamada M, Gu J, Yamakami M, Fukuda M, Waguri S, Uchiyama Y, Yoshimori T, Sekiguchi K. 2008. Human RME-8 is involved in membrane trafficking through early endosomes. Cell Struct. Funct. 33:35–50 [DOI] [PubMed] [Google Scholar]
- 28. Field MC, Carrington M. 2009. The trypanosome flagellar pocket. Nat. Rev. Microbiol. 7:775–786 [DOI] [PubMed] [Google Scholar]
- 29. Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, Karpov S, Kugrens P, Krug J, Lane CE, Lewis LA, Lodge J, Lynn DH, Mann DG, McCourt RM, Mendoza L, Moestrup O, Mozley-Standridge SE, Nerad TA, Shearer CA, Smirnov AV, Spiegel FW, Taylor MF. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52:399–451 [DOI] [PubMed] [Google Scholar]
- 30. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Redmond S, Vadivelu J, Field MC. 2003. RNAit: an automated Web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol. Biochem. Parasitol. 128:115–118 [DOI] [PubMed] [Google Scholar]
- 32. Subramaniam C, Veazey P, Redmond S, Hayes-Sinclair J, Chambers E, Carrington M, Gull K, Matthews K, Horn D, Field MC. 2006. Chromosome-wide analysis of gene function by RNA interference in the African trypanosome. Eukaryot. Cell 5:1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oberholzer M, Morand S, Kunz S, Seebeck T. 2006. A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol. Biochem. Parasitol. 145:117–120 [DOI] [PubMed] [Google Scholar]
- 34. Leung KF, Dacks JB, Field MC. 2008. Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic 9:1698–1716 [DOI] [PubMed] [Google Scholar]
- 35. Wirtz E, Leal S, Ochatt C, Cross GA. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101 [DOI] [PubMed] [Google Scholar]
- 36. Burkard G, Fragoso CM, Roditi I. 2007. Highly efficient stable transformation of bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 153:220–223 [DOI] [PubMed] [Google Scholar]
- 37. He CY, Ho HH, Malsam J, Chalouni C, West CM, Ullu E, Toomre D, Warren G. 2004. Golgi duplication in Trypanosoma brucei. J. Cell Biol. 165:313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pal A, Hall BS, Nesbeth DN, Field HI, Field MC. 2002. Differential endocytic functions of Trypanosoma brucei Rab5 isoforms reveal a glycosylphosphatidylinositol-specific endosomal pathway. J. Biol. Chem. 277:9529–9539 [DOI] [PubMed] [Google Scholar]
- 39. Jeffries TR, Morgan GW, Field MC. 2001. A developmentally regulated rab11 homologue in Trypanosoma brucei is involved in recycling processes. J. Cell Sci. 114:2617–2626 [DOI] [PubMed] [Google Scholar]
- 40. Alexander DL, Schwartz KJ, Balber AE, Bangs JD. 2002. Developmentally regulated trafficking of the lysosomal membrane protein p67 in Trypanosoma brucei. J. Cell Sci. 115:3253–3263 [DOI] [PubMed] [Google Scholar]
- 41. Jensen BC, Sivam D, Kifer CT, Myler PJ, Parsons M. 2009. Widespread variation in transcript abundance within and across developmental stages of Trypanosoma brucei. BMC Genomics 10:482 doi:10.1186/1471-2164-10-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Field MC, Gabernet-Castello C, Dacks JB. 2007. Reconstructing the evolution of the endocytic system: insights from genomics and molecular cell biology. Adv. Exp. Med. Biol. 607:84–96 [DOI] [PubMed] [Google Scholar]
- 43. Hirst J, Barlow LD, Francisco GC, Sahlender DA, Seaman MN, Dacks JB, Robinson MS. 2011. The fifth adaptor protein complex. PLoS Biol. 9:e1001170 doi:10.1371/journal.pbio.1001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gall WE, Higginbotham MA, Chen C, Ingram MF, Cyr DM, Graham TR. 2000. The auxilin-like phosphoprotein Swa2p is required for clathrin function in yeast. Curr. Biol. 10:1349–1358 [DOI] [PubMed] [Google Scholar]
- 45. Lemmon SK. 2001. Clathrin uncoating: auxilin comes to life. Curr. Biol. 11:R49–R52 [DOI] [PubMed] [Google Scholar]
- 46. Eisenberg E, Greene LE. 2007. Multiple roles of auxilin and hsc70 in clathrin-mediated endocytosis. Traffic 8:640–646 [DOI] [PubMed] [Google Scholar]
- 47. Allen CL, Goulding D, Field MC. 2003. Clathrin-mediated endocytosis is essential in Trypanosoma brucei. EMBO J. 22:4991–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hall B, Allen CL, Goulding D, Field MC. 2004. Both of the Rab5 subfamily small GTPases of Trypanosoma brucei are essential and required for endocytosis. Mol. Biochem. Parasitol. 138:67–77 [DOI] [PubMed] [Google Scholar]
- 49. Hall BS, Smith E, Langer W, Jacobs LA, Goulding D, Field MC. 2005. Developmental variation in Rab11-dependent trafficking in Trypanosoma brucei. Eukaryot. Cell 4:971–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gabernet-Castello C, Dacks JB, Field MC. 2009. The single ENTH-domain protein of trypanosomes; endocytic functions and evolutionary relationship with epsin. Traffic 10:894–911 [DOI] [PubMed] [Google Scholar]
- 51. Bangs JD, Andrews NW, Hart GW, Englund PT. 1986. Posttranslational modification and intracellular transport of a trypanosome variant surface glycoprotein. J. Cell Biol. 103:255–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kabiri M, Steverding D. 2000. Studies on the recycling of the transferrin receptor in Trypanosoma brucei using an inducible gene expression system. Eur. J. Biochem. 267:3309–3314 [DOI] [PubMed] [Google Scholar]
- 53. Grünfelder CG, Engstler M, Weise F, Schwarz H, Stierhof YD, Morgan GW, Field MC, Overath P. 2003. Endocytosis of a glycosylphosphatidylinositol-anchored protein via clathrin-coated vesicles, sorting by default in endosomes, and exocytosis via RAB11-positive carriers. Mol. Biol. Cell 14:2029–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Engstler M, Thilo L, Weise F, Grunfelder CG, Schwarz H, Boshart M, Overath P. 2004. Kinetics of endocytosis and recycling of the GPI-anchored variant surface glycoprotein in Trypanosoma brucei. J. Cell Sci. 117:1105–1115 [DOI] [PubMed] [Google Scholar]
- 55. Chung WL, Carrington M, Field MC. 2004. Cytoplasmic targeting signals in transmembrane invariant surface glycoproteins of trypanosomes. J. Biol. Chem. 279:54887–54895 [DOI] [PubMed] [Google Scholar]
- 56. Wang YN, Wang M, Field MC. 2010. Trypanosoma brucei: trypanosome-specific endoplasmic reticulum proteins involved in variant surface glycoprotein expression. Exp. Parasitol. 125:208–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Koumandou VL, Klute MJ, Herman EK, Nunez-Miguel R, Dacks JB, Field MC. 2011. Evolutionary reconstruction of the retromer complex and its function in Trypanosoma brucei. J. Cell Sci. 124:1496–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lumb JH, Leung KF, Dubois KN, Field MC. 2011. Rab28 function in trypanosomes: interactions with retromer and ESCRT pathways. J. Cell Sci. 124:3771–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Silverman JS, Schwartz KJ, Hajduk SL, Bangs JD. 2011. Late endosomal Rab7 regulates lysosomal trafficking of endocytic but not biosynthetic cargo in Trypanosoma brucei. Mol. Microbiol. 82:664–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.