Abstract
In yeast (Saccharomyces cerevisiae) and animals, the sulfhydryl oxidase Erv1 functions with Mia40 in the import and oxidative folding of numerous cysteine-rich proteins in the mitochondrial intermembrane space (IMS). Erv1 is also required for Fe-S cluster assembly in the cytosol, which uses at least one mitochondrially derived precursor. Here, we characterize an essential Erv1 orthologue from the protist Trypanosoma brucei (TbERV1), which naturally lacks a Mia40 homolog. We report kinetic parameters for physiologically relevant oxidants cytochrome c and O2, unexpectedly find O2 and cytochrome c are reduced simultaneously, and demonstrate that efficient reduction of O2 by TbERV1 is not dependent upon a simple O2 channel defined by conserved histidine and tyrosine residues. Massive mitochondrial swelling following TbERV1 RNA interference (RNAi) provides evidence that trypanosome Erv1 functions in IMS protein import despite the natural absence of the key player in the yeast and animal import pathways, Mia40. This suggests significant evolutionary divergence from a recently established paradigm in mitochondrial cell biology. Phylogenomic profiling of genes also points to a conserved role for TbERV1 in cytosolic Fe-S cluster assembly. Conversely, loss of genes implicated in precursor delivery for cytosolic Fe-S assembly in Entamoeba, Trichomonas, and Giardia suggests fundamental differences in intracellular trafficking pathways for activated iron or sulfur species in anaerobic versus aerobic eukaryotes.
INTRODUCTION
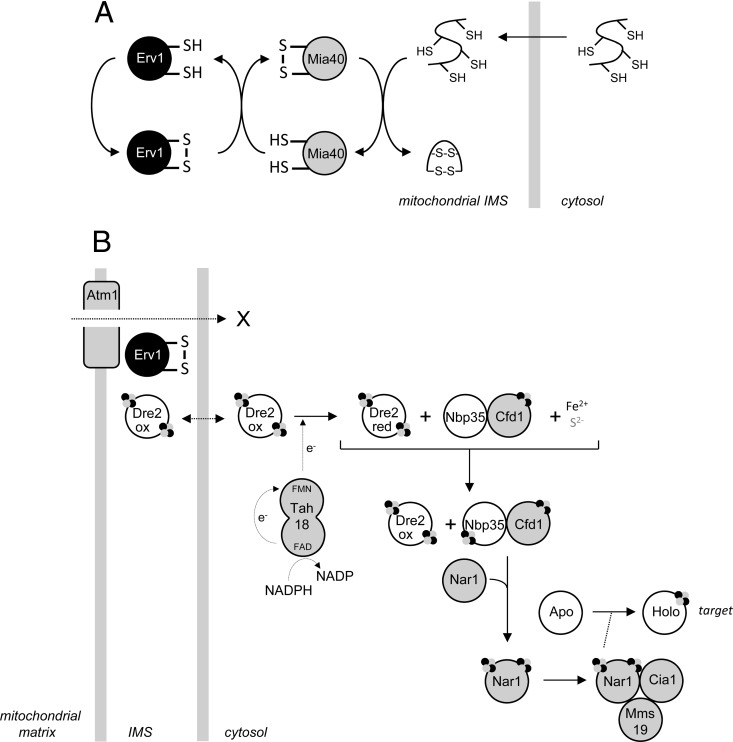
Erv1 is a highly conserved protein in eukaryotes, required for essential mitochondrial processes. In animals and yeast (Saccharomyces cerevisiae), mitochondrial import and oxidative folding within the mitochondrial intermembrane space (IMS) of the small translocase of the inner membrane chaperone family (small TIM) and other cysteine-rich IMS proteins are dependent upon a disulfide relay that requires two essential proteins—Mia40 and Erv1 (1). Mia40 acts as a receptor for and catalyst of disulfide bond formation in newly imported cysteine-rich substrates, and the sulfhydryl oxidase Erv1 is the oxidant for reduced Mia40 (Fig. 1A). Mia40-dependent mitochondrial import and folding of small TIMs and other cysteine-rich proteins is commonly referred to as the “mitochondrial disulfide relay” or the “mitochondrial intermembrane space assembly (MIA) pathway.” The second essential process in which Erv1 functions, at least in yeast, is the export from the mitochondrial matrix to the cytosol of precursor(s) for cytosolic Fe-S assembly (2, 3). This export pathway (which is incompletely defined) and the subsequent assembly of cytosolic Fe-S clusters (known as the CIA pathway) involve the proteins shown in Fig. 1B (3–5).
Fig 1.
Erv1-dependent processes. (A) The pathway of Erv1-Mia40-dependent import of cysteine-rich proteins into the mitochondrial IMS. (B) Current understanding of the pathway for precursor delivery and assembly of cytosolic Fe-S clusters based on genetic analysis of yeast and mammalian cells. In panel A, only the redox-active cysteines of Erv1 and Mia40 are highlighted. In panel B, the interactions between the proteins shown is summarized from references 4 and 5; the Fe-S clusters present in Dre2, Cfd1, and holo Nar1, assembled on Nbp35 and transferred to cytosolic/nuclear targets by holo Nar1, are also shown. X represents the unknown CIA precursor exported from mitochondria.
Previously, we reported the conservation of Erv1 in all aerobic eukaryotes and widespread conservation of Mia40 homologs in at least three eukaryotic supergroups: the opisthokonts (including animals and fungi), the Amoebozoa, and the Plantae (6). Curiously, however, there are protist groups, notably the chromalveolates (a large group that includes ciliates, dinoflagellates, diatoms, and apicomplexan parasites, such as Plasmodium falciparum and Toxoplasma gondii) and the trypanosomatids, in which Erv1 and cysteine-rich substrates of the MIA pathway are present, but there is no trace of a Mia40 homolog encoded within complete nuclear genome sequences (6–8). In trypanosomatids, a virtual absence of introns, coupled to excellent gene prediction algorithms covering multiple complete genome sequences, makes the absence of Mia40 difficult to question. The trypanosomatids are a flagellate parasite family that includes the sleeping sickness parasite Trypanosoma brucei, the etiological agent of Chagas' disease, Trypanosoma cruzi, and numerous pathogenic Leishmania species.
The absence of Mia40 from trypanosomatids raises obvious questions. For instance, (i) how are small TIMs and other cysteine-rich proteins imported into the mitochondrial IMS of trypanosomatids? (ii) What is the function of the trypanosomatid Erv1 homolog? (iii) Is it retained only for export from the mitochondrial matrix of a precursor for the cytosolic iron-sulfur cluster assembly (CIA) pathway (Fig. 1B), or does it function in protein import into the mitochondrial IMS? From an evolutionary perspective, the trypanosomatids belong to a diverse protist group, the Excavata, in which key aspects of mitochondrial function in both free-living and parasitic taxa are considered closer to the sorts of molecular characteristics one could reasonably predict for the proto-mitochondrion than is the case in other eukaryotes: e.g., a mitochondrial genome in free-living Reclinomonas americana that more closely resembles a eubacterial genome in terms of architecture and gene content than do other mitochondrial genomes (9) and a protein translocase of the mitochondrial outer membrane in trypanosomatids that appears to more closely resemble protein translocases of the bacterial Omp85 family than it does the Tom40 proteins found in other eukaryotes (9, 10). While the root of eukaryotic evolution remains open to debate (11, 12), at least one multigene phylogeny supports the divergence of Excavata relative to other eukaryotic groups (13) and that multigene phylogeny (13) and also the evolutionary distribution of various molecular traits (14) are at least consistent with the idea of divergence for the last common ancestor of extant Excavata at an early point in eukaryotic evolution. Thus, what implications do the answers to the three questions posed earlier have for our understanding of the evolution of the Erv1/Mia40 mitochondrial IMS protein import pathway that is currently considered to be a conserved and fundamental feature of mitochondrial cell biology? The significance of these questions was also highlighted by Deponte and coworkers in their recent survey of the evolutionary conservation of mitochondrial protein import pathways (8). Here we investigate the roles and biochemistry of Erv1 in T. brucei with respect to import of cysteine-rich proteins into the mitochondrial IMS and synthesis of cytosolic Fe-S clusters. Set in a wider context, our analysis of a highly conserved protein, little studied in protists (which account for the bulk of eukaryotic diversity), emphasizes variations that exist for (recently discovered) ubiquitous mitochondrial import and export pathways.
MATERIALS AND METHODS
Production of active recombinant T. brucei Erv1.
The full-length open reading frame (ORF) corresponding to Tb09.160.4440 was amplified by PCR from T. brucei genomic DNA and cloned in frame into NdeI/BamHI-digested pET14b (Novagen) to create pTbErv1, a construct consisting of T. brucei ERV1 (TbERV1) with an N-terminal hexahistidine tag. All plasmid constructs generated in this work were sequenced by ABI Prism technology before use.
Expression of TbERV1 in several Escherichia coli strains produced only a mixture of six major soluble forms, truncated from the C terminus. This mixture was purified from E. coli by nickel affinity chromatography and used to raise anti-TbERV1 antisera (Covalab). When TbERV1 was expressed in E. coli strain C43-DE3 (15), ERV1 inclusion bodies formed. When solubilized, the majority of the ERV1 was observed at the expected size on Western blots (confirmed by electrospray ionization mass spectrometry [ESI-MS)]). Protein was therefore purified from this strain following growth and protein induction in Terrific Broth supplemented with 20 mM flavin adenine dinucleotide disodium salt hydrate (FAD) (Sigma-Aldrich).
Cells were harvested and disrupted using a French press at 16,000 lb/in2. Then, 0.5% (vol/vol) Triton X-100 (Sigma-Aldrich) was added, and the suspension was incubated at room temperature, under gentle agitation for 30 min, followed by centrifugation at 25,000 × g for 10 min at 4°C. The inclusion body pellet was solubilized in 8 M guanidine·HCl–50 mM Tris·HCl (pH 7.5) by gentle agitation for 30 min at 4°C. Unsolubilized material was removed by centrifugation at 257,000 × g for 30 min at 4°C. The supernatant was applied to Fast Flow chelating Sepharose (Amersham Biosciences), charged with Ni2+, and preequilibrated with 6 M guanidine·HCl–50 mM Tris·HCl (pH 7.5). The column was washed extensively with the equilibration buffer, and the bound protein was eluted with 6 M guanidine·HCl–50 mM Tris·HCl–500 mM imidazole (pH 7.5). Dithiothreitol (DTT) (Melford) was added at a final concentration of 100 mM, and the eluted protein solution was incubated under gentle agitation for 30 min at 4°C. Subsequently, the protein solution was acidified (pH 2) using concentrated HCl and dialyzed against acidified water (pH 2) overnight. The dialysate was concentrated and filtered, and following batch adsorption of secreted protein to reverse-phase C18 particles and elution with 50% acetonitrile–H2O–0.1% trifluoroacetic acid, the protein was lyophilized.
Lyophilized protein (which was stable and could be stored conveniently) was reconstituted in 8 M guanidine·HCl–100 mM Tris·HCl–10 mM β-mercaptoethanol (pH 7.5). The pH of the resulting solution was adjusted to 7.5, FAD was added at final concentration of 2 mM, and the solution was incubated under gentle agitation for 1 h at 4°C and then dialyzed overnight against 100 mM Tris·HCl (pH 7.5). The dialysate was filtered to remove particulates, treated with β-mercaptoethanol at a final concentration of 10 mM, and applied to Fast Flow chelating Sepharose charged with Ni2+ and preequilibrated with 100 mM Tris·HCl–300 mM NaCl–10 mM β-mercaptoethanol–20 mM imidazole (pH 7.5). The resin was washed sequentially with 10 column volumes of the equilibration buffer, and then 100 mM Tris·HCl–50 mM NaCl–10 mM β-mercaptoethanol–20 mM imidazole (pH 7.5), and finally 100 mM Tris·HCl–50 mM NaCl–20 mM imidazole (pH 7.5). Then the resin was further washed with 3 column volumes of 100 mM Tris·HCl–100 mM imidazole (pH 7.5), and the protein was eluted with 100 mM Tris·HCl–300 mM imidazole (pH 7.5). Fractions containing native and active TbERV1 (yellow) were combined, concentrated, and stored at 4°C, where the protein was stable for several months.
Site-directed mutants of TbERV1 were prepared using pTbERV1 as the template, Pfu polymerase (Stratagene), and the Quikchange method. When expressed in E. coli C43-DE3, each mutant behaved as the wild type (WT) (i.e., they formed inclusion bodies of predominantly full-length protein with some lower-molecular-weight/degradation species also present). The mutants were therefore purified and refolded as for wild-type TbERV1 (described above).
Assays of Erv1 activity and protein biochemistry.
Refolded, recombinant, full-length TbERV1 and variants were used to assay potential electron acceptors. Reduction of O2 was measured in a Clark oxygen electrode (Rank Brothers) using DTT or tris(2-carboxyethyl)phosphine (TCEP) as the artificial substrate. The O2 electrode was set at −0.6 V, and samples were assayed in a volume of 1.5 ml in 50 mM phosphate buffer (pH 7.5) (unless stated otherwise). O2 reduction was initiated by adding TCEP or DTT via a Hamilton syringe to the reaction mixture. Assays were carried out in air-saturated solutions (approximately 250 μM O2). Cytochrome c was purified from the trypanosomatid Crithidia fasciculata (16) and assayed spectrophotometrically as an electron acceptor from TbERV1. Blank reactions were undertaken without Erv1 to ascertain the background rate of cytochrome c reduction by TCEP, which was subtracted from the observed rate of reduction of cytochrome c when Erv1 was present. Unless stated, all solutions were fully deoxygenated prior to the assays and gas-tight cuvettes were used. Erv1 and cytochrome c were mixed in the cuvette, and the reaction was initiated by addition of TCEP via a Hamilton syringe. The absorbance change at 555 nm was used to calculate a rate of reduction, using a difference extinction coefficient (between reduced and oxidized cytochrome c) of 19,500 M−1 cm−1.
Experiments were also undertaken in the presence of both cytochrome c and O2 and monitored in either the Clark electrode or the spectrophotometer. In the O2 electrode, TCEP was added to a solution of Erv1 to initiate the reaction, the rate of oxygen reduction was monitored, and then C. fasciculata cytochrome c was added. In the spectrophotometer, both O2 and cytochrome c were present in the reaction mixture, and the reaction was initiated by addition of TCEP. This reaction was carried out in the presence of superoxide dismutase (Sigma-Aldrich) (added to 20 U/ml) to ensure that any reduction of cytochrome c was by Erv1 rather than by superoxide produced from O2.
For in vitro biochemistry, immunoblot detection of TbERV1 used a SnapID protein detection system (Millipore) in conjunction with either PentaHis horseradish peroxidase-conjugated antibody (Qiagen) or TbERV1 antisera followed by anti-rabbit alkaline phosphatase conjugate (Sigma-Aldrich). Visible absorption spectra were recorded using a PerkinElmer Lambda 2 spectrophotometer. The type of flavin present on TbERV1 was identified by the method of Faeder and Siegel (17). Fluorescence spectra were recorded using fluorescence quartz cuvettes in a PerkinElmer LS 50B fluorimeter. Protein concentrations were determined using a bicinchoninic acid (BCA) assay kit (Thermo Scientific, Pierce). FAD occupancy was estimated spectrophotometrically by comparing protein spectra with the spectrum of FAD standard solutions. Circular dichroism spectroscopy was carried out using a Jasco J-720 spectropolarimeter with 1-mm path-length quartz cuvettes. Electrospray ionization mass spectrometry (ESI-MS) was performed using a Micromass Bio-Q II-ZS triple-quadrupole mass spectrometer: 10-μl protein samples in 1:1 water-acetonitrile with 1% formic acid at a concentration of 20 pmol μl−1 were injected into the electrospray source at a flow rate of 10 μl min−1.
In vitro assay of small Tim protein folding by recombinant TbERV1.
Tb927.3.1600 encodes a putative T. brucei small Tim protein with two CX3C pairs. An ORF flanked by NdeI and BamHI restriction sites was synthesized (Genscript), restriction digested, and ligated into pET14b that had also been digested with NdeI and BamHI. Protein (TbTIM9) was expressed in E. coli CD43-DE3 cells and purified from cell extracts using the N-terminal polyhistidine tag. DTT was added to purified TbTIM9 to a concentration of 10 mM, and the mixture was incubated at 4°C overnight to reduce the protein. The DTT was dialyzed out, and TbTIM9 was stored in 50 mM sodium acetate buffer (pH 5) at −20°C to minimize reoxidation.
Folding of TbTIM9 by TbERV1 was assayed in the presence of O2 ± cytochrome c. To monitor the oxidation state of TbTIM9, AMS (4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid), which selectively binds to free cysteine thiols but not disulfide bonds, was used; each AMS molecule has a mass of ∼600 Da, so binding produced a shift on SDS-PAGE gels. Control experiments showed reduced TbTIM9 could be AMS labeled in this way and shifted by the expected mass (∼2,500 Da) on SDS-PAGE. The level of reduction of cytochrome c was also monitored spectrophotometrically. Folding of TbTIM9 by TbERV1 was assayed in 50 mM phosphate buffer (pH 7.5) with Erv1 added in catalytic amounts. Where cytochrome c was used, this was present at a >4-fold-higher level than TbTIM9.
Trypanosome cell culture and RNAi.
Procyclic T. brucei (line 29-13) (18) was cultured in SDM-79 containing heat-inactivated fetal bovine serum and hemin as described previously (19); bloodstream-form T. brucei (line 90-13) (18) was cultured in HMI-9 medium (20). For RNA interference (RNAi), coding sequence from TbERV1 was amplified by PCR using the primer combinations 5′-AATAGGATCCATGTCGAAACAGGAGCC-3′ and 5′-GGCTCTAGACGCTGATGATTTCTTATCATCAGG-3′ and AAGTCTAGACTCGCTAGTTGTATCGGTCC-3′ and 5′-CTTAAGCTTATGTCGAAACAGGAGCC-3′, respectively. (Restriction sites added to 5′-primer ends are underlined.) Purified PCR amplicons were digested with either BamHI and XbaI or XbaI and HindIII, purified, and ligated into the vector p2T7-177 (21), which had been digested with BamHI and HindIII. In this way, inducible expression of hairpin intramolecular double-stranded RNA (dsRNA) was under the control of opposing tetracycline-regulated T7 RNA promoters. Stable transformation of procyclic T. brucei was achieved using a BTX electroporator, as described previously (22), and transformation of bloodstream-form T. brucei used an Amaxa Nucleofector II electroporator (23). Tetracycline (1 μg ml−1) was added to cultures to induce RNAi; cell densities were monitored using a Beckman Coulter Z2 counter. For immunoblot validation of TbERV1 depletion, cells were collected by centrifugation, washed with phosphate-buffered saline (PBS), and resuspended in hot Laemmli sample buffer, and 5 × 106 cell equivalents per lane were separated by SDS-PAGE. Polyclonal rabbit antisera raised against recombinant TbERV1 and T. brucei enolase were used at dilutions of 1:1,000 and 1:150,000, respectively.
Digitonin extraction of procyclic T. brucei.
Cells were collected by centrifugation (800 × g for 10 min), washed in PBS, and resuspended at 6.5 × 108 cells ml−1 in 250 mM sucrose–25 mM Tris (pH 7.4)–1 mM EDTA–150 mM NaCl (STE-NaCl buffer). Aliquots (150 μl) of this resuspension were then taken to a final volume of 300 μl containing increasing concentrations of digitonin (0 to 1.5 mM final concentration) and incubated at room temperature for 4 min. After immediate centrifugation at 13,000 rpm for 2 min, supernatants were added to boiling hot 2× Laemmli buffer containing 0.2 M DTT. Samples were separated by SDS-PAGE, Western blotted to nitrocellulose membranes, and labeled with antisera raised against TbERV1, mitochondrial RNA binding protein 2 (MRP2) (used at a 1:1,000 dilution), prohibitin (1:1,000 dilution), triosephosphate isomerase (1:1,000 dilution), aldolase (1:1,000 dilution), or enolase, respectively.
TEM.
For transmission electron microscopy (TEM), cells were fixed in 2.5% (vol/vol) glutaraldehyde in 0.1 M sodium cacodylate (SC) buffer, pH 7.4, for 1 h at 4°C, followed by a short wash in SC buffer and postfixation with 2% (wt/vol) osmium tetroxide in the SC buffer for 1 h at room temperature. Next, cells were dehydrated in ethanol and embedded in Epon Araldite resin. Thin sections were stained with lead citrate and uranyl acetate and observed under a JEOL JEM 1010 microscope.
Bioinformatics.
Orthology of candidate Erv1 and CIA pathway proteins was assessed using reciprocal BLAST (24) and sequence alignment by ClustalW (25), followed by manual evaluation, and analysis of domain architecture was performed using SMART (26). When searches of protein databases failed to return hits, tBLASTn analyses were made.
RESULTS
Expression and in vitro folding of recombinant TbERV1.
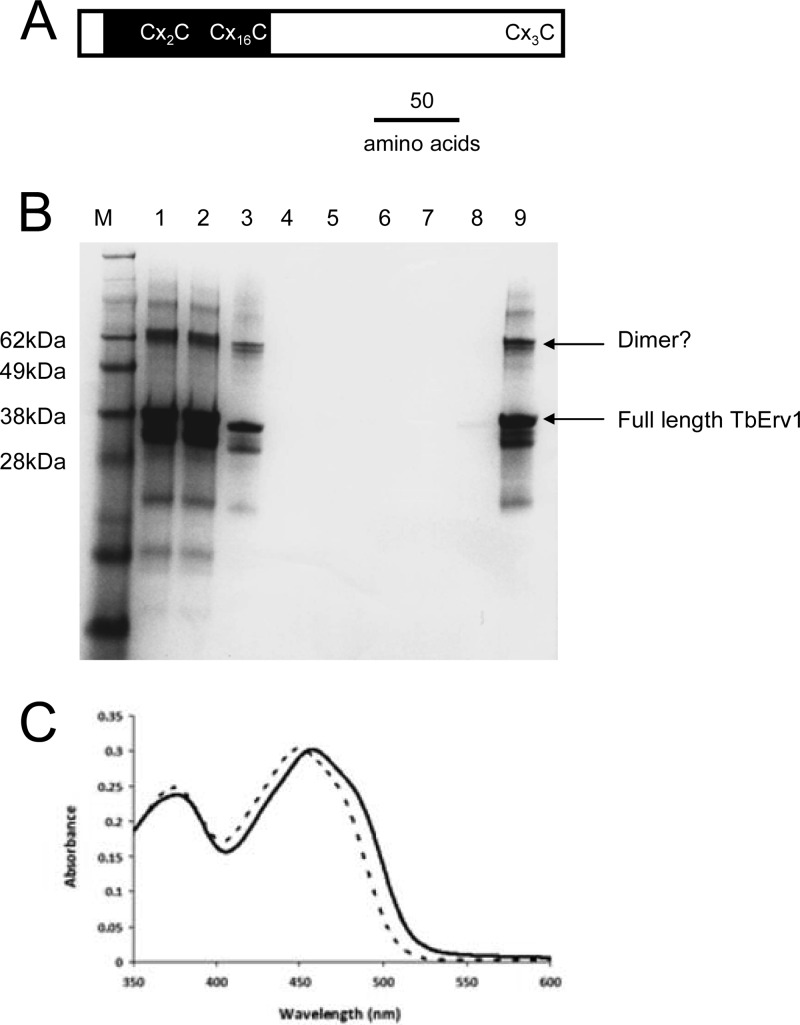
Initial attempts at expression produced only truncated forms of TbERV1, but this was used (as described in Materials and Methods) to demonstrate the presence of bound FAD, rather than flavin mononucleotide (FMN), as expected for a member of the Erv1/Ero2 enzyme family and for the generation of polyclonal TbERV1-specific antibodies. Subsequently, we produced full-length TbERV1, refolding the protein around the FAD from inclusion bodies using a lengthy and complex protocol (Fig. 2B and C). This protein was used for biochemical analyses.
Fig 2.
Isolation of soluble recombinant TbERV1. (A) Schematic of TbERV1 architecture; the relative positions of the conserved core domain (black shading) containing the flavin-binding site and conserved Cys pairs are highlighted. (B) SimplyBlue-stained SDS-PAGE gel documenting in vitro folding of recombinant TbERV1. Lanes: M, molecular mass markers; 1, resuspended high-performance liquid chromatography (HPLC)-purified freeze-dried protein; 2, guanidine–β-mercaptoethanol solubilized protein; 3, solution from lane 2 following incubation with FAD and postdialysis; 4, flowthrough from the Ni-affinity column; 5, 20 mM imidazole–300 mM NaCl–10 mM β-mercaptoethanol wash; 6, 20 mM imidazole–50 mM NaCl–10 mM β-mercaptoethanol wash; 7, 20 mM imidazole–50 mM NaCl wash; 8, 100 mM imidazole–50 mM NaCl wash; 9, 300 mM imidazole–50 mM NaCl elution. Electrospray MS revealed major protein masses at 32,522 ± 6 Da and 29,396 Da; the calculated mass of His6-tagged TbERV1 minus the initial Met is 32,519 Da. Even after the extensive purification and refolding protocol, some low-molecular-mass TbERV1 degradation products remain. (C) Visible absorption spectra of folded TbERV1 and free FAD; dashed line, free FAD; solid line, TbERV1. Absorption maxima of free flavin at ∼375 nm and ∼450 nm shifted to 458 nm for TbERV1, which, together with the shoulder of the 458-nm peak, indicates protein-bound flavin. Flavin occupancy in folded TbERV1 was estimated at 100%, and bleaching of FAD absorption by dithionite treatment confirmed the flavin was redox active.
TbERV1 readily reduces either cytochrome c or O2.
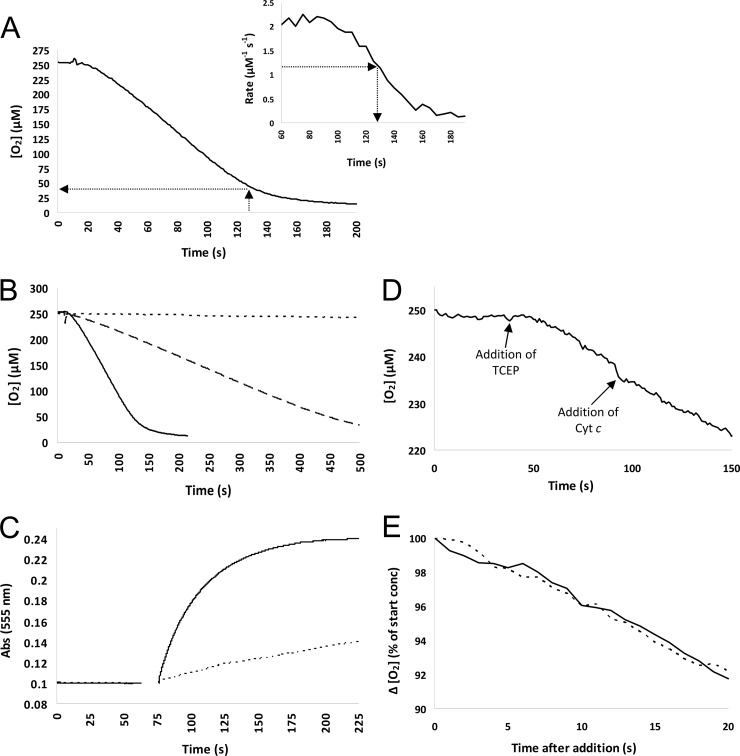
Erv1 family proteins form disulfide bonds in a substrate protein and therefore become reduced. Electrons must then be transferred from the flavin group of Erv1 to an electron acceptor to complete the catalytic cycle (Fig. 1A). The physiological oxidant for the Erv1 of S. cerevisiae and mammalian cells is cytochrome c (27, 28); in these organisms O2 is a poor electron acceptor, whereas Erv1 from Arabidopsis thaliana (29) readily reduces O2. The situation in T. brucei was not clear a priori since procyclic T. brucei trypanosomes found in the tsetse fly vector possess an active mitochondrial respiratory chain with cytochrome c present, but long slender bloodstream-form trypanosomes that infect mammals lack the capacity for cytochrome-dependent respiration. We used full-length, recombinant, folded TbERV1 for assays of potential electron acceptors. Using either DTT (5 mM) or TCEP (3.5 mM) as artificial substrates (30) in air-saturated solutions (∼ 250 μM O2), we measured sulfhydryl oxidase activities for TbERV1 with Km values for O2 of 41 μM or 124 μM and kcat values of 30.6 min−1 or 9.52 min−1, respectively (Table 1; Fig. 3A). As expected for an enzyme-catalyzed reaction, the initial rate of reaction was proportional to enzyme concentration (Fig. 3B). To quantify a rate of cytochrome c reduction by TbERV1, we performed assays using cytochrome purified from the trypanosomatid Crithidia fasciculata (16) (84% identical and 92% similar to T. brucei cytochrome c). We found that DTT reduced trypanosomatid cytochrome c quite rapidly. Thus, we used TCEP as a substrate (Fig. 3C) and, in the absence of oxygen, determined a Km for C. fasciculata cytochrome c of 13 μM and a kcat of 16.7 min−1 (Table 1).
Table 1.
Kinetic parameters for Erv1 proteins from T. brucei, S. cerevisiae, and H. sapiens using different combinations of electron donor and acceptor
| Organism | kcat (min−1) | Km (μM) | kcat/Km (M−1 s−1) | Electron donor/acceptor | Reference |
|---|---|---|---|---|---|
| T. brucei | 30.6 | 41 | 1.2 × 104 | DTT/O2 | This work |
| 9.5 | 124 | 1,300 | TCEP/O2 | This work | |
| 16.7 | 13 | 2.1 × 104 | TCEP/cytochrome c | This work | |
| S. cerevisiae | 78 | 57 | 2.3 × 104 | DTT/O2 | 30 |
| 66 | 27 | 4.1 × 104 | TCEP/O2 | 30 | |
| H. sapiens | |||||
| ALR | 60.5 | 240 | 4,200 | DTT/O2 | 55 |
| ALR long form | 61 | 3,000 | 330 | DTT/O2 | 73 |
| ALR | 270 | 10 | 4.5 × 105 | DTT/cytochrome c | 55 |
Fig 3.
TbERV1 readily reduces either O2 or cytochrome c. (A) Determination of the Km for TbERV1-catalyzed oxygen consumption using DTT as the electron donor. The main graph shows the time course of O2 consumption with 4 μM TbERV1 in the presence of 5 mM DTT; the inset shows the reaction rate during the assay, derived from the main curve. Km was estimated according to the method of Ang and Lu (30): the time corresponding to half the maximal rate was estimated from the graph inset and used to extrapolate to the O2 concentration at that time in the main figure (dashed arrows). (B) The initial rate of TbERV1-catalyzed O2 consumption is proportional to the amount of protein present. Readings were taken using a Clark-type oxygen electrode, and the reaction was started by the introduction of DTT, after ∼10 s, to a final concentration of 5 mM, using a Hamilton syringe. Short dashes, 5 mM DTT only (note that this shows a background rate of oxygen consumption as the electrode itself consumes oxygen during measurement); long dashes, 1 μM TbERV1; solid line, 4 μM TbERV1. (C) Anaerobic reduction of trypanosomatid cytochrome c by TbERV1. A time course for TbERV1-catalyzed oxidation of TCEP is shown. Reduction of C. fasciculata cytochrome c was monitored by following absorbance (Abs) at 555 nm. TCEP was added, and mixing of solutions occurred between 65 and 75 s. The final reaction mixture in deoxygenated 50 mM phosphate buffer (pH 7.5) contained 3.5 mM TCEP and 10 μM cytochrome c. Dashed line, no TbERV1 present (all reduction of cytochrome c due to TCEP, representing the background rate); solid line, 1 μM TbERV1. (D and E) The rate of TbERV1-catalyzed O2 consumption is unaffected by the addition of cytochrome c (Cyt c). (D) Assays were carried out in a Clark electrode using 50 mM phosphate buffer (pH 7.5) and 1 μM TbERV1; TCEP and C. fasciculata cytochrome c were added to final concentrations of 3.5 mM and 15 μM, respectively, at the time points indicated. Panel E shows the rate of O2 consumption in the initial seconds after the additions of TCEP (solid line) and cytochrome c (dashed line). (Data in panel E are from panel D.)
Although enzymology of cytochrome c oxidase (unlike TbERV1, a topologically fixed membrane protein) has shown that use of a heterologous cytochrome c can influence kinetic parameters determined in vitro (31), it was not feasible to carry out our kinetic analysis of T. brucei ERV1 using T. brucei cytochrome c as an oxidant. In part, this was due to the absence of a suitable expression system to generate recombinant T. brucei cytochrome c (which has an exceptional heme attachment posttranslational modification) (16, 32, 33), and in part it was due to the impracticality of growing sufficiently large volumes of procyclic T. brucei (>100 liters) to isolate the milligram quantities of cytochrome c that would have been required for our experiments. It is therefore possible that the kinetic values we report for cytochrome c are uncertain because the Crithidia cytochrome we used does not interact as effectively with TbERV1 as T. brucei cytochrome c. However, in preliminary assays using C. fasciculata cytochrome c, commercial preparations of horse or Saccharomyces cerevisiae cytochrome c (both Sigma-Aldrich), or a recombinant variant of T. brucei cytochrome c with heme attached through a conventional binding motif (32), we saw little difference in the rates of cytochrome reduction by TbERV1; we are therefore confident in the validity of the kinetic values reported in Tables 1 and 2.
Table 2.
Activity of recombinant TbERV1 in comparison with that of site-directed mutants
| Enzyme | Substrate | Reduction (μM min−1 μM TbERV1−1) ofa: |
|
|---|---|---|---|
| O2 | Cytochrome c | ||
| WT TbERV1 | TCEP | 11.97 ± 3.3 | 19.92 ± 5.0 |
| DTT | 22.91 ± 3.7 | NDb | |
| Distal Cys pair mutant | TCEP | 30.03 ± 2.7 | 26.67 ± 1.5 |
| DTT | 30.43 ± 3.8 | NDb | |
| Proximal Cys pair mutant | TCEP | 0.38 ± 0.1 | 1.08 ± 1.9 |
| DTT | 0.14 ± 0.2 | NDb | |
| O2 channel mutant | TCEP | 7.24 | NDc |
| DTT | NDc | ||
Rates were determined using 3.5 mM electron donor under either aerobic conditions or under anaerobic conditions in the presence of 15 μM C. fasciculata cytochrome c. Errors are standard deviations from at least 3 independent measurements. The values for WT TbERV1 are similar to those quoted in Table 1; note that the Erv1 used for experiments summarized in Table 2 was purified, refolded, and assayed independently from the TbERV1 prepared for the experiments summarized in Table 1.
Not determined (ND) due to efficient reduction of cytochrome c by DTT.
Not determined due to a lack of protein (see the text).
To assess the relative preference of O2 or C. fasciculata cytochrome c as the oxidant for TbERV1, we also undertook assays in the presence of both oxygen and cytochrome c. In the Clark electrode, the addition of cytochrome c had no effect on the rate of O2 consumption when TCEP was used as the substrate for sulfhydryl oxidase activity (Fig. 3D and E). When reduction of cytochrome c by TbERV1 was assayed in the spectrophotometer under aerobic conditions, cytochrome c was reduced at 58% of the rate observed under anaerobic conditions (data not shown). The latter experiments were conducted in the presence of superoxide dismutase to ensure any cytochrome c reduction was by TbERV1 directly, rather than by superoxide derived from reduction of O2 by TbERV1. Thus, surprisingly, TbERV1 can reduce both cytochrome c and O2 simultaneously, which has not been previously observed for members of the Erv1 family.
One possible explanation for the apparent O2 preference of TbERV1 is the conservation of residues predicted to form a hydrophobic channel from the enzyme surface to the N5 nitrogen of the FAD isoalloxazine ring in other Erv1/Ero2 family members that readily reduce O2 (34). The putative conserved residues are His-66 and Tyr-70 in the trypanosome protein; in Erv1 proteins thought to lack this hydrophobic channel, Asp typically replaces His and the identity of the amino acid four residues downstream is variable (see Fig. S1 in the supplemental material). To look further at O2 utilization by TbERV1 and test experimentally the hypothesis of a hydrophobic O2 channel in Erv1, we prepared an H66D Y70R variant by site-directed mutagenesis and assayed purified folded protein for sulfhydryl oxidase activity. Even though our channel mutant was clearly unstable—only tiny amounts of protein could be prepared even from large cell growths—circular dichroism spectroscopy showed that mutation did not detectably affect the protein fold (see Fig. S2 in the supplemental material), and in a single experiment using TCEP as the substrate, the protein was clearly very active (Table 2).
We also expressed and in vitro folded other site-directed mutant TbERV1 proteins, focusing on mutation of cysteine pairs previously shown to participate in the electron shuttle from substrate to FAD cofactor in Erv1 family proteins from other organisms (30) (analogous to the proximal and distal cysteine pairs highlighted in Fig. 2A). C60S C63S (proximal) and C261S C265S (distal) TbERV1 variants expressed in E. coli C43-DE3 refolded similarly to the wild-type protein purified from inclusion bodies; this was evident from spectroscopic analyses of the in vitro folded proteins, confirming that the mutations did not detectably affect the protein fold (see Fig. S2 in the supplemental material) and that flavin occupancy was close to 100% (data not shown). The absorption spectrum of the proximal cysteine pair mutant showed small shifts in the FAD absorption maxima compared with wild-type TbERV1, consistent with the mutagenesis of residues spatially close to the bound flavin (data not shown) (30). Analysis of purified, in vitro folded proteins by SDS-PAGE revealed that the proportion of protein present as full-length versus degradation products was different in each ERV1 variant (see Fig. S3 in the supplemental material). Similar to S. cerevisiae Erv1 and the mammalian ortholog ALR (30, 35, 36), mutation of cysteines adjacent to the FAD cofactor (the proximal cysteines) rendered TbERV1 virtually inactive for the oxidation of either DTT or TCEP and the reduction of either cytochrome c or O2 (Table 2). As with yeast Erv1 and ALR, mutation of the distal cysteine pair did not abolish the activity of TbERV1 with DTT, but in contrast to Erv1 orthologs from other organisms (30, 37), as well as related sulfhydryl oxidases exhibiting a similar core FAD-binding domain Ero1 and Erv2 (38, 39), the distal cysteine variant of TbERV1 was also active with TCEP as the substrate for sulfhydryl oxidation. In yeast Erv1, the distal cysteine pair is responsible for electron transfer from the substrate to the proximal cysteine pair and the distal cysteines interact directly with reduced Mia40 (40, 41). Small substrates such as DTT, however, can access the active site directly. The observation that the C261S C265S TbERV1 mutant retained activity toward the larger TCEP substrate suggests that the active site of the trypanosome enzyme adopts a more open or accessible conformation than the Erv1 enzymes from yeast and humans.
TbERV1 is an essential gene required for normal mitochondrial morphology.
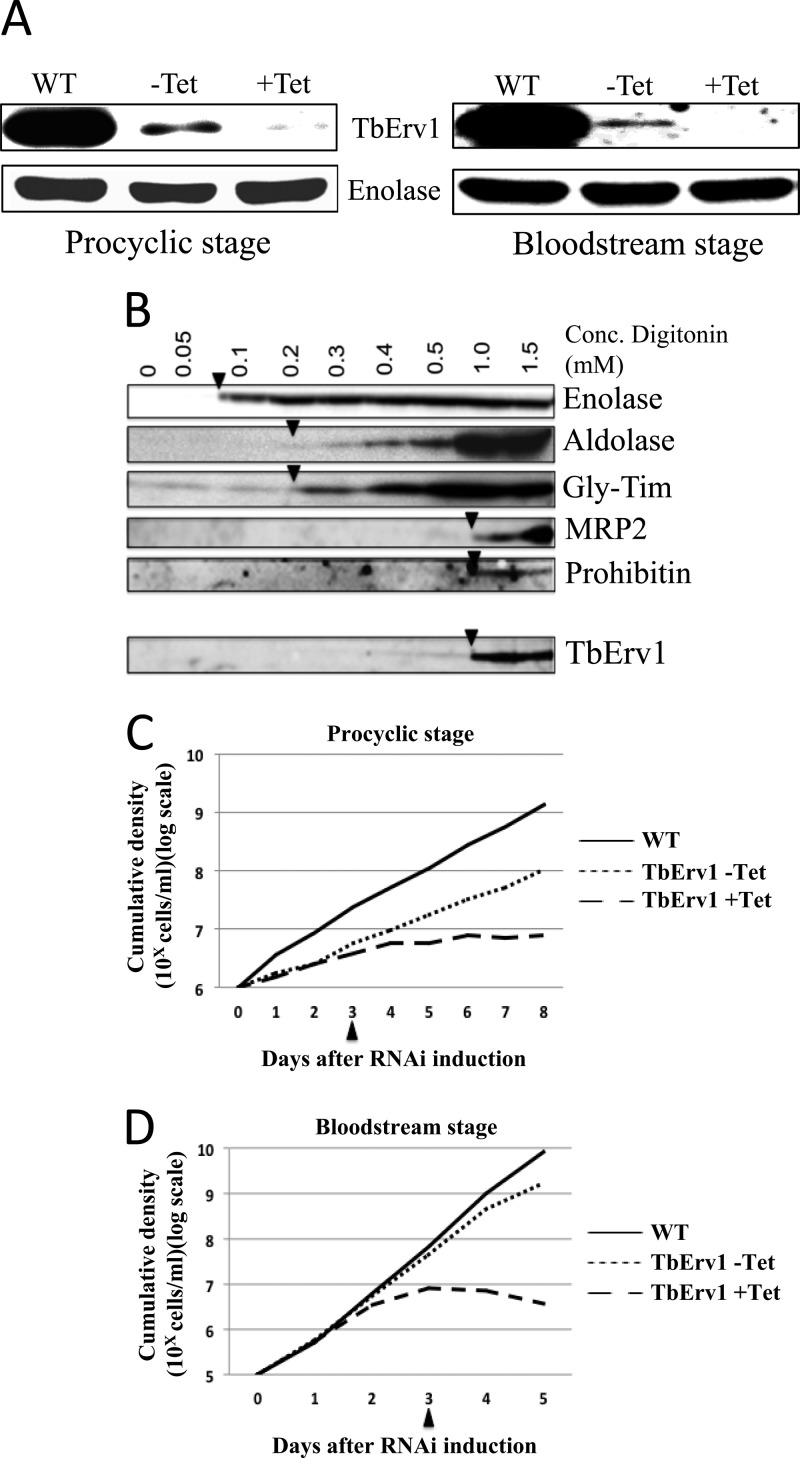
By enzymological characterization of recombinant TbERV1, we have shown that the protein is a sulfhydryl oxidase that catalyzes the transfer of electrons from a substrate to both cytochrome c and O2. We next investigated its expression and function in vivo in T. brucei. Immunoblot analysis revealed TbERV1 expression in both procyclic (i.e., tsetse fly) and bloodstream (mammalian)-form trypanosomes (Fig. 4A). Subcellular fractionation of procyclic T. brucei confirmed TbERV1 is a mitochondrial protein (Fig. 4B).
Fig 4.
TbERV1 is essential in procyclic and bloodstream-form T. brucei. (A) Immunoblot analysis reveals constitutive expression of TbERV1 in procyclic and bloodstream stages of T. brucei. WT, wild-type parental cell lines from which RNAi mutants were derived; −Tet, RNAi mutants grown in the absence of tetracycline; +Tet, RNAi mutants 72 h postinduction of RNAi. Blots were labeled with antibodies recognizing either TbERV1 or enolase, which served as a loading control. (B) Subcellular fractionation of procyclic cells by digitonin extraction reveals TbERV1 to be a mitochondrial protein. Soluble fractions from the digitonin extraction were blotted and probed with antibodies detecting the cytosolic marker enolase, the glycosomal markers aldolase and triosephosphate isomerase, the mitochondrial markers MRP2 and prohibitin, or TbERV1. Arrowheads indicate the minimal concentration of digitonin required to release the designated protein. (C and D) Growth of procyclic (C) or bloodstream-form (D) TbERV1 RNAi mutants in the absence (dotted line) or presence (dashed line) of tetracycline (1 μg ml−1); the solid line denotes the growth of the parental wild-type cell line from which the RNAi mutants were derived.
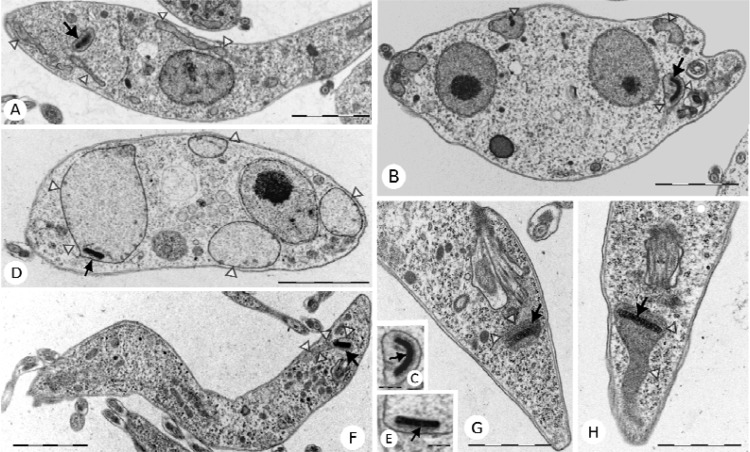
We used tetracycline-inducible RNAi to deplete TbERV1. In both procyclic and bloodstream-form mutants, TbERV1 abundance was decreased even prior to the induction of RNAi, indicating considerable leakiness to our inducible system. Depletion of protein did not result in a major growth phenotype in the bloodstream-form RNAi mutant (Fig. 4D), but in noninduced procyclic RNAi cells, leaky TbERV1 depletion correlated with a modest growth defect (Fig. 4C). Addition of tetracycline to induce RNAi resulted in the virtual disappearance of TbERV1 and a cessation of cell growth in both bloodstream (mammalian)-form and procyclic (tsetse fly midgut) cultures (Fig. 4A, C, and D). Thus, TbERV1 is essential in both bloodstream-form and procyclic T. brucei. To look further at the effect of RNAi, we fixed cells for analysis by TEM. Following RNAi induction, massively swollen mitochondria were apparent (Fig. 5D). Even in the absence of tetracycline, mitochondrial swelling was evident (Fig. 5B; presumably as a consequence of reduced TbERV1 content [Fig. 4A]), but not to the extent seen following RNAi induction.
Fig 5.
TbERV1 depletion causes mitochondrial swelling. (A) The thin-section electron micrograph of a longitudinal section through a parental T. brucei procyclic cell shows the normal morphology of the mitochondrion. (B) A thin-section micrograph reveals discernible mitochondrial swelling of a procyclic TbERV1 RNAi mutant in the noninduced state. (C) Intact kinetoplast DNA disk in a noninduced procyclic TbERV1 RNAi mutant. (D) Massive mitochondrial swelling in TbERV1 procyclic RNAi mutants 72 h after the induction of RNAi. (E) Intact kinetoplast DNA disk in a procyclic TbERV1 RNAi mutant 72 h after RNAi induction. (F) Longitudinal section revealing normal mitochondrial morphology in a bloodstream-form trypomastigote cell. (G) Thin-section micrograph showing normal mitochondrial morphology of a bloodstream-form TbERV1 RNAi mutant in the noninduced state. (H) Conspicuous mitochondrial swelling in a TbERV1 bloodstream-form RNAi mutant 72 h post-RNAi induction. Scale bars represent 1 μm (A, B, D, F, G, and H) and 200 nm (C and E). Arrows point to the kinetoplast; open triangles designate the mitochondrion.
In S. cerevisiae, mitochondrial swelling is seen in a variety of mutants defective in mitochondrial protein import, including Erv1-deficient yeast (42, 43). Yet, at least in yeast, Erv1 is also required for maturation of cytosolic and nuclear Fe-S cluster-containing proteins. Thus, we examined ultrastructure in procyclic T. brucei mutants where RNAi had been used to selectively deplete different components of mitochondrial and cytosolic Fe-S cluster assembly pathways. For essential components of the mitochondrial Fe-S cluster assembly pathway, we analyzed mutants depleted for either T. brucei frataxin (TbFRAT) (44) or both Isa1 and Isa2 (45). In the TbISA1 TbISA2 RNAi mutant, assembly of mitochondrial, but seemingly not cytosolic, Fe-S proteins is affected (45), whereas RNAi of TbFRAT strongly affects Fe-S cluster assembly in both the mitochondrion and cytosol. For a component of the mitochondrion-to-cytosol Fe-S delivery pathway, we looked at a TbATM1 RNAi mutant (P. Changmai and J. Lukeš, unpublished data); Atm1 is a transporter of the inner mitochondrial membrane, which like Erv1 is essential for the maturation of cytosolic Fe-S proteins (46). Finally, we looked at RNAi mutants of Nbp35 and Cfd1 (47), both of which act downstream of Erv1 in the cytosolic Fe-S cluster assembly pathway (Fig. 1B). The RNAi phenotypes of all of these mutants are lethal; thus, mutants were fixed for analysis by TEM shortly before the time points at which the growth of RNAi-induced cultures ceased. Mitochondrial swelling was not evident in any of the Fe-S cluster assembly-defective RNAi mutants analyzed (see Fig. S4 in the supplemental material). This suggests that the extensive degree of mitochondrial swelling seen in TbERV1 RNAi mutants is not due to a defect in the export from the mitochondrial matrix of the precursor(s) for cytosolic Fe-S cluster assembly or a pleiotrophic phenotype common to dying cells. Instead, it is likely that the mitochondrial swelling derives directly from a role for TbERV1 in mitochondrial protein import.
Failure of recombinant TbERV1 to fold a cysteine-rich small Tim protein in vitro.
T. brucei lacks a homolog of Mia40, but its Erv1 contains an unusually extended C-terminal domain that includes the distal cysteine pair (see Fig. S1 in the supplemental material). One hypothesis is that TbERV1 acts directly as the oxidant for small, cysteine-rich proteins imported into the mitochondrial IMS: i.e., TbERV1 could combine the roles of yeast Erv1 and Mia40. If TbERV1 acts directly on IMS import substrates, it should be able to oxidize the cysteine thiols of TbTIM9 (the homolog of a yeast/animal Mia40 substrate) in the presence of either O2 or cytochrome c. Based on the approach of Bien and coworkers (41), we prepared assay mixtures containing recombinant TbERV1, initially reduced TbTIM9 (Tb927.3.1600), and O2 ± cytochrome c. To monitor the oxidation state of TbTIM9, we used AMS, which selectively binds to free cysteine thiols, but not disulfide bonds, in conjunction with SDS-PAGE gel shift assays. We also monitored the reduction of cytochrome c spectrophotometrically.
The presence of TbERV1 affected neither the extent of TbTIM9 oxidation nor cytochrome c reduction. TbTIM9 was present in 10- to 40-fold stoichiometric excess over TbERV1 (such that TbERV1 was present in catalytic amounts), and cytochrome c and/or O2 was present in excess over the thiols of TbTIM9. Reactions were allowed to proceed for >2 h (cf. in vitro assay times of typically up to 15 to 45 min in the study by Bien et al. [41]). Thus, we conclude that, at least under these assay conditions, TbERV1 does not directly oxidize this mitochondrial IMS protein. Cytochrome c was slowly reduced to some extent when mixed with TbTIM9, but the level and rate of reduction were independent of TbERV1. This, presumably direct cytochrome c-TbTIM9 interaction did not lead to full oxidation of TbTIM9.
TbERV1 and cytosolic Fe-S cluster assembly.
The single mitochondrion in trypanosomatids extends throughout the cell body and is notoriously fragile under most standard subcellular fractionation conditions. Given the likely extreme fragility of swollen mitochondria following induction of TbERV1 RNAi, we felt the direct study of a possible role of TbERV1 in cytosolic Fe-S cluster assembly would yield equivocal data. Thus, we used a comparative genomics approach to ask whether proteins known to function in the delivery or assembly of cytosolic Fe-S cluster precursors (Fig. 1B) are conserved in trypanosomatid parasites. A similar phylogenomic profiling strategy of genes required for mitochondrial Fe-S cluster assembly pointed toward a direct and key role for frataxin in Fe-S cluster assembly sometime before the function of that then enigmatic protein was revealed experimentally (48).
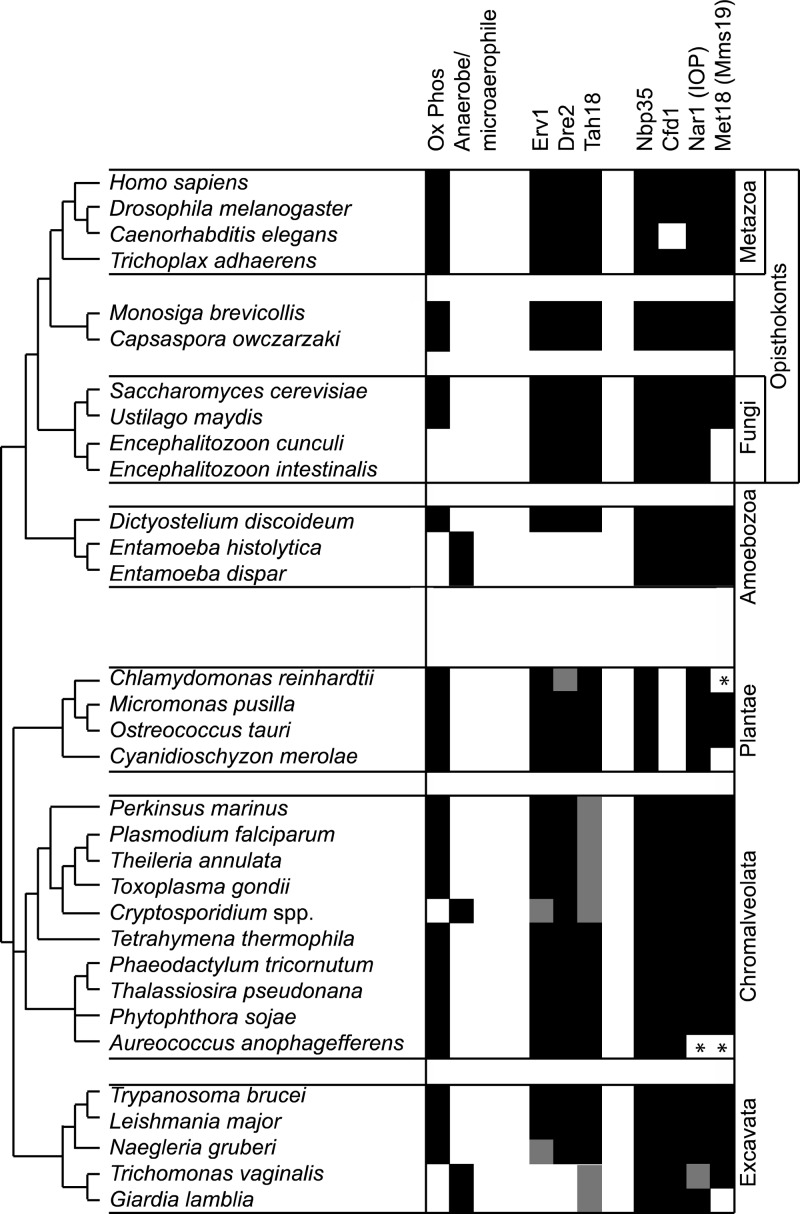
The results of our analysis (Fig. 6) revealed conservation of genes encoding proteins required for cytosolic Fe-S cluster biogenesis in aerobic eukaryotes, including trypanosomatids. Some component absences (marked with asterisks in Fig. 6) likely reflect gaps in draft sequence assembly. In the microsporidian parasites Encephalitozoon cuniculi and Encephalitozoon intestinalis, which possess the most highly reduced nuclear genomes known in terms of gene content (49, 50), the absence of Mia40 and known substrates of the MIA pathway (6) means Erv1 can obviously function in cytosolic Fe-S cluster assembly only. Known CIA proteins, except for Met18, are conserved in these highly divergent parasites. However, our analysis also produced unexpected and striking observations relating to anaerobic eukaryotes. The obligate anaerobic protists Entamoeba, Trichomonas, and Giardia not only lack Erv1 but also Dre2, and in Entamoeba, the Dre2-interacting protein Tah18 (51) is also absent. Tah18 is a flavoprotein that transfers electrons from NADPH to the Fe-S cluster(s) of Dre2 (4). Neither canonical-looking Tah18 nor Nar1 is present in Trichomonas, but in this microaerophile, we observed a unique architecture corresponding to Nar1-like Fe-hydrogenase-related proteins fused at the C terminus to flavodoxin and FAD-binding domains conserved in Tah18 homologs. Since genome coverage is available for multiple species and isolates of Entamoeba, Trichomonas, and Giardia and a paucity of introns in these organisms reduces difficulties in ORF annotation, predicted absences of Erv1, Dre2, and Tah18 are very likely to be real. Unexpected outcomes from our phylogenomic profiling of cytosolic Fe-S cluster assembly are therefore indications of moderation and variation in the intracellular trafficking pathways for activated iron or sulfur species in anaerobic protists versus eukaryotes that reside in O2-replete environments.
Fig 6.
Phylogenomic distribution in eukaryotes of Erv1 and known components of the CIA pathway. Relationships between different eukaryotic groups represent a consensus of current opinion (11). Conservation of candidate orthologs is denoted by black shading, absence of shading denotes the absence of candidates, and gray shading indicates where there is a clear deviation from a simple binary classification of presence or absence (e.g., atypical domain architecture). Asterisks mark unexpected absences that probably reflect gaps in draft genome coverage. For example, in C. reinhardtii, no Dre2 is predicted from the genome assembly, but Dre2 is present in the closely related Volvox carteri, and a Chlamydomonas expressed sequence tag (EST) with a sequence match to Dre2 is available. For Tah18, homologs lacking conservation to all three domains of the yeast protein are evident in the dinoflagellate Perkinsus marinus, apicomplexan parasites, and Giardia. Curiously, Erv1 can only be found in Cryptosporidium muris, but not C. hominis or C. parvum; a partial hit to Erv1 is evident only from tBLASTn searches in Naegleria gruberi, suggesting problems in prediction of a gene model due to introns. In Trichomonas, canonical-looking Nar1 and Tah18 are not present, but a novel architecture of Nar1 fused to Tah18-related domains is seen.
DISCUSSION
With this study, we have followed our previous in silico analysis of the phylogenomic distribution of genes encoding substrates for and components of the MIA pathway (6) with (i) experimental analyses of Erv1 function in the African sleeping sickness parasite T. brucei and (ii) a new in silico analysis aimed at clarifying the involvement of Erv1 in cytosolic Fe-S cluster assembly within evolutionarily diverse protists. From our earlier analysis (6), we proposed a two-step model for evolution of the MIA pathway, whereby in early emerging eukaryotes the import of disulfide-bonded proteins into the mitochondrial IMS was dependent upon Erv1, but not Mia40. Novel results present here are (i) evidence that trypanosome ERV1 is indeed involved in the import of cysteine-rich proteins into the mitochondrial IMS, despite the absence of Mia40, (ii) a kinetic analysis of recombinant T. brucei that (a) raises the novel possibility in the Erv1 family of simultaneous reduction of two electron acceptors and (b) challenges the essentiality of conserved amino acids within a putative “oxygen channel” present in Erv1 orthologs capable of readily using O2 as an electron acceptor, and (iii) the observation that obligate secondary transition to growth in microoxic or anaerobic environments correlates with marked variations in the pathway that leads to cytosolic Fe-S cluster assembly in model aerobic eukaryotes. These points are expanded upon in the discussion below.
In animals and yeast, Mia40 is essential for the IMS import and folding of numerous small, cysteine-rich proteins (1). In the model plant Arabidopsis thaliana, however, the Mia40 gene is nonessential, although its absence results in the loss of some cysteine-rich proteins that would be substrates for the MIA pathway in yeast (52). Interestingly, Erv1 abundance is increased in mitochondria isolated from Δmia40 plants, leading to speculation that in the absence of Mia40, Erv1 might function alone in IMS import of small Tim proteins and other essential cysteine-rich proteins (52). The dramatic mitochondrial swelling that occurs following RNAi against TbERV1 was not phenocopied by a variety of RNAi mutants defective in mitochondrial and/or cytosolic Fe-S cluster assembly. However, it is reminiscent of the globular mitochondria seen by epifluoresence microscopy following RNAi against trypanosome small Tim proteins (53). Small Tims are well known for their chaperone function in sorting mitochondrial membrane proteins. Thus, the available data suggest a direct role for TbERV1 in mitochondrial protein import, most probably in the import of cysteine-rich proteins into the IMS. Unfortunately, antipeptide antibodies generated against T. brucei homologs of two classic MIA substrates did not yield reagents that were tractable for monitoring the abundance of cysteine-rich mitochondrial IMS proteins following induction of TbERV1 RNAi. Since in our assays recombinant TbERV1 did not catalyze oxidative folding of a small Tim in vitro, it is not clear whether trypanosomes operate a streamlined pathway for oxidative protein import into the mitochondrial IMS (as hypothesized in reference 6) or if TbERV1 functions in concert with (a yet to be identified) protein nonhomologous to Mia40, but related in function.
In vitro enzyme assays of TbERV1 with TCEP and DTT as electron donors suggest possible differences in active site accessibility between TbERV1 and S. cerevisiae Erv1 (ScErv1). Does this point toward the possibility that TbERV1 acts directly (i.e., without a Mia40-like partner) in the oxidative folding of small Tim proteins or other cysteine-rich proteins in trypanosome mitochondria (discussed further in references 6 and 8) and that our failure to see direct oxidative folding of recombinant TbTIM9 in vitro by TbERV1 is a consequence of technical limitations? Conceivably, the long, predicted disordered C-terminal domain separating the FAD-binding core helix bundle and distal cysteine pairs in TbERV1 (disorder predicted using the “Regional Order Neural Network” [54]) could guide the interaction with diverse cysteine-rich substrates in much the same way the hydrophobic cleft of human Mia40 acts as a receptor for numerous substrates, including the flexible N-terminal domain of the human Erv1 ortholog ALR (40). Further work is needed to test this possibility.
Erv1 proteins from S. cerevisiae (ScErv1) and humans (ALR) utilize cytochrome c as their preferred electron acceptor, and addition of cytochrome c to an oxygen electrode containing reacting ScErv1 or ALR suppresses the use of O2 until the oxidized cytochrome is consumed (28, 55). In contrast, in our work, the rate of O2 reduction by TbERV1 was unaffected by the addition of cytochrome c, although, unexpectedly, cytochrome c could be reduced in addition to the O2 reduction that was taking place—i.e., simultaneously and independently. The ∼40% decrease in cytochrome c reduction under aerobic, compared with anaerobic, conditions suggests O2 may be the preferred electron acceptor for TbERV1. At first glance, this conclusion is supported by the observation that residues implicated in high-affinity electron transport to O2 by microsomal sulfhydryl oxidases are conserved at equivalent positions in TbERV1 (6, 34). While an H66D Y70R double mutation severely destabilized the protein (potentially because of destabilization of the nearby FAD binding site), limited kinetic analysis of the few micrograms of soluble protein that we were able to prepare showed it was clearly active with O2 as the electron acceptor (∼60% of the activity seen with wild-type protein) (Table 2). If the hypothesis of an O2 channel from the protein surface to the FAD, defined simply by HxxxΦ residues, was correct (34), one might expect to see greater, perhaps almost complete, loss of activity toward O2 in the H66D Y70R TbERV1 mutant.
The use of both O2 and cytochrome c as viable electron acceptors for TbERV1 is consistent with the requirement for TbERV1 in both bloodstream-form and procyclic T. brucei. In bloodstream-form parasites, when cytochromes are not available, O2 is the obvious suitable electron acceptor. In vivo oxygen levels in mammalian mitochondria are thought to be ∼2 to 10 μM (e.g., see references 56 and 57), which is at least 4-fold lower than the calculated Km of TbERV1 for O2 (Table 1). Yet, bloodstream-form T. brucei radically downregulates mitochondrial metabolism (58); the net activity of Erv1 in IMS protein import and Fe-S cluster export is therefore likely to be (very) low and not compromised by an apparently kinetically unfavorable Km for O2. Moreover, the leakiness of our RNAi system, in which an ∼90% depletion of protein is evident even in the absence of tetracycline but cell growth was only slightly affected (Fig. 4), suggests that Erv1 abundance is conceivably well above the minimal threshold for function. In procyclic T. brucei, cytochrome c, which is likely to be present within the mitochondrial IMS in the millimolar range (0.5 to 5 mM) (cf. data published in references 59 and 60), is presumably a physiologically significant electron acceptor for TbERV1, as it is in yeast and mammals.
Regarding the extent of leakiness observed in both our procyclic and bloodstream-form RNAi mutants, it should be noted that extreme examples of leakiness, which give rise to a partial phenotype even in the absence of RNAi induction, while unusual, are not without precedent (e.g., see reference 61). There are also reported instances where ∼5% of normal enzyme activity is still sufficient to support normal trypanosome growth, but nonetheless the alleles encoding that enzyme are refractory to complete deletion from the diploid T. brucei genome (62). In initial experiments to silence the expression of TbERV1, neither a conventional gene-specific insert placed between opposing tetracycline-regulatable promoters nor a gene-specific “stem-loop” placed downstream of a single inducible promoter yielded observable growth or morphology phenotypes following RNAi induction. It was only with the leaky presentation of a TbERV1-specific “stem-loop” between opposing inducible promoters that we were able to observe the essentiality of TbERV1 and its requirement for normal mitochondrial morphology. (We note that use of a stem-loop between opposing promoters has in other instances also proved leaky [e.g., reference 63], although not to the extent seen in our TbERV1 mutants.)
We showed previously that the absence of Erv1 from the genome of a microbial eukaryote cosegregated with the absence of orthologs for the cysteine-rich substrates of the MIA pathway, pointing toward the importance of Erv1 in mitochondrial protein import (6). The validity of that analysis is emphasized by the recent identification in Trichomonas, which lacks Erv1 and Mia40, of small Tim proteins without disulfide bonds (64). Here, we also took a comparative genomics approach to question whether a conserved role for TbERV1 in the assembly of cytosolic Fe-S clusters was likely. (As noted above, the large, fragile mitochondria of trypanosomes would make obtaining such data experimentally very challenging.) The presence of conserved Dre2 and Tah18 proteins, together with other known CIA components in trypanosomatid genomes allows us to make a confident prediction that TbERV1 is a multifunctional protein with roles in both mitochondrial IMS protein import and maturation of cytosolic Fe-S proteins. It is striking that the available data indicate roles for TbERV1 in both IMS protein import and cytosolic Fe-S cluster biogenesis, even though Mia40, a paradigm partner protein of Erv1 in yeast and animals, is absent from trypanosomatids. This result was not necessarily obvious; it may well have been, for example, that in the absence of Mia40, IMS import used a pathway wholly independent of TbERV1.
Our results were also unexpectedly revealing for organisms well beyond trypanosomatids and indicate independent variations of precursor delivery for the CIA pathway (for cytosolic Fe-S cluster biosynthesis) in anaerobic protists. In the protists lacking Erv1, Dre2, or Tah18, and among anaerobic protists generally, mitochondrial function is highly degenerate (65, 66). In these organisms, relic mitochondria do not generate ATP and contain only a handful of proteins in comparison with classical mitochondria. Yet, in taxa examined thus far, relic mitochondria from anaerobes do function in Fe-S cluster biogenesis (67–69). In Entamoeba, loss of Erv1, Dre2, and Tah18 coincides with the unique replacement of the mitochondrial Fe-S cluster assembly pathway by a bacterial type “Nif”-type system (70–72). However, in Giardia and Trichomonas, Erv1 and Dre2 are absent, but mitochondrial Fe-S cluster assembly uses proteins conserved in aerobic eukaryotes. Patterns of Erv1 and Dre2 retention and loss (Fig. 6) are understandable if the currently unknown, mitochondrially derived precursor for cytosolic Fe-S cluster assembly is either O2 labile (requiring Erv1 and Dre2 for its protection) or its Erv1/Dre2-dependent export to the cytosol is O2 dependent. That unknown precursor has been postulated to be a sulfur-containing molecule (3). In the examples of Giardia and Trichomonas, adaptation to obligate microoxic environments would either bypass the requirement for Erv1 and Dre2 if the precursor is O2 labile, or if precursor delivery was O2 dependent could have resulted in innovation of an Erv1/Dre2-independent export pathway. In the example of Entamoeba, adaptation to a microoxic environment may have had the same consequence, but equally, replacement of the mitochondrial Fe-S cluster assembly pathway present in other eukaryotes could have resulted in loss of an ability to produce the intermediate(s) for which trafficking is dependent upon Erv1 and Dre2 function. The likely fusion of Nar1 and Tah18 homologs in Trichomonas is particularly intriguing since these proteins are not thought to act sequentially in the CIA pathway (Fig. 1B), perhaps suggesting the organization of CIA proteins in a stable multiprotein complex. Collectively, these data highlight the value of turning toward evolutionarily and ecologically diverse protists for studying fundamental processes in eukaryotic cells and the subtle, yet informative, variations inherent in those processes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paul A. M. Michels (Universidad de los Andes, Mérida, Venezuela/University of Edinburgh, United Kingdom) for the gift of enolase, aldolase, and glycosomal triosephosphate isomerase antibodies, André Schneider (University of Bern, Bern, Switzerland) for the gift of Nbp35 and Cfd1 RNAi constructs, and Julie Kovářová (University of South Bohemia, České Budějovice [Budweis], Czech Republic) for help with digitonin fractionation.
This work was supported by grants from the Royal Society (to M.L.G., who was a Royal Society University Research Fellow) and the Biotechnology and Biological Sciences Research Council (grant BB/D019753/1 to J.W.A.A., who was a BBSRC David Phillips Fellow), and the Grant Agency of the Czech Republic (P305/11/2179), Ministry of Education of the Czech Republic (AMVIS LH12104), as well as the Praemium Academiae award to J.L., who is also a Fellow of the Canadian Institute for Advanced Research.
J.W.A.A., J.L., and M.L.G. designed and directed the study. S.B., J.C.L., N.D., D.A.I.M., K.H.T., A.D.G., and M.L.G. undertook experiments. S.B., M.L.G., J.L., and J.W.A.A. wrote the paper.
The authors declare they have no conflicts of interest.
ADDENDUM IN PROOF
Following the revision of our manuscript, a study was published describing characterization of Erv1 from another trypanosomatid, Leishmania tarentolae (E. Eckers, C. Petrungaro, D. Gross, J. Riemer, K. Hell, and M. Deponte, J. Biol. Chem., in press). Some of the approaches used and results obtained complement data shown in this work.
Footnotes
Published ahead of print 21 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00304-12.
REFERENCES
- 1. Herrmann JM, Riemer J. 2012. Mitochondrial disulfide relay: redox-regulated protein import into the intermembrane space. J. Biol. Chem. 287:4426–4433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lange H, Lisowsky T, Gerber J, Muhlenhoff U, Kispal G, Lill R. 2001. An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins. EMBO Rep. 2:715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lill R. 2009. Function and biogenesis of iron-sulphur proteins. Nature 460:831–838 [DOI] [PubMed] [Google Scholar]
- 4. Netz DJ, Stumpfig M, Dore C, Muhlenhoff U, Pierik AJ, Lill R. 2010. Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat. Chem. Biol. 6:758–765 [DOI] [PubMed] [Google Scholar]
- 5. Stehling O, Vashisht AA, Mascarenhas J, Jonsson ZO, Sharma T, Netz DJ, Pierik AJ, Wohlschlegel JA, Lill R. 2012. MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science 337:195–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Allen JW, Ferguson SJ, Ginger ML. 2008. Distinctive biochemistry in the trypanosome mitochondrial intermembrane space suggests a model for stepwise evolution of the MIA pathway for import of cysteine-rich proteins. FEBS Lett. 582:2817–2825 [DOI] [PubMed] [Google Scholar]
- 7. Carrie C, Murcha MW, Whelan J. 2010. An in silico analysis of the mitochondrial protein import apparatus of plants. BMC Plant Biol. 10:249 doi:10.1186/1471-2229-10-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eckers E, Cyrklaff M, Simpson L, Deponte M. 2012. Mitochondrial protein import pathways are functionally conserved among eukaryotes despite compositional diversity of the import machineries. Biol. Chem. 393:513–524 [DOI] [PubMed] [Google Scholar]
- 9. Harsman A, Niemann M, Pusnik M, Schmidt O, Burmann BM, Hiller S, Meisinger C, Schneider A, Wagner R. 2012. Bacterial origin of a mitochondrial outer membrane protein translocase: new perspectives from comparative single channel electrophysiology. J. Biol. Chem. 287:31437–31445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pusnik M, Schmidt O, Perry AJ, Oeljeklaus S, Niemann M, Warscheid B, Lithgow T, Meisinger C, Schneider A. 2011. Mitochondrial preprotein translocase of trypanosomatids has a bacterial origin. Curr. Biol. 21:1738–1743 [DOI] [PubMed] [Google Scholar]
- 11. Embley TM, Martin W. 2006. Eukaryotic evolution, changes and challenges. Nature 440:623–630 [DOI] [PubMed] [Google Scholar]
- 12. Katz LA. 2012. Origin and diversification of eukaryotes. Annu. Rev. Microbiol. 66:411–427 [DOI] [PubMed] [Google Scholar]
- 13. Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AG, Roger AJ. 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups.” Proc. Natl. Acad. Sci. U. S. A. 106:3859–3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cavalier-Smith T. 2010. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol. Lett. 6:342–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miroux B, Walker JE. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels, J. Mol. Biol. 260:289–298 [DOI] [PubMed] [Google Scholar]
- 16. Fulop V, Sam KA, Ferguson SJ, Ginger ML, Allen JW. 2009. Structure of a trypanosomatid mitochondrial cytochrome c with heme attached via only one thioether bond and implications for the substrate recognition requirements of heme lyase. FEBS J. 276:2822–2832 [DOI] [PubMed] [Google Scholar]
- 17. Faeder EJ, Siegel LM. 1973. A rapid micromethod for determination of FMN and FAD in mixtures. Anal. Biochem. 53:332–336 [DOI] [PubMed] [Google Scholar]
- 18. Wirtz E, Leal S, Ochatt C, Cross GA. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101 [DOI] [PubMed] [Google Scholar]
- 19. Brun R, Schonenberger M. 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36:289–292 [PubMed] [Google Scholar]
- 20. Hirumi H, Hirumi K. 1989. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 75:985–989 [PubMed] [Google Scholar]
- 21. Wickstead B, Ersfeld K, Gull K. 2002. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 125:211–216 [DOI] [PubMed] [Google Scholar]
- 22. Vondrušková E, van den Burg J, Zíková A, Ernst NL, Stuart K, Benne R, Lukeš J. 2005. RNA interference analyses suggest a transcript-specific regulatory role for mitochondrial RNA-binding proteins MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei. J. Biol. Chem. 280:2429–2438 [DOI] [PubMed] [Google Scholar]
- 23. Burkard G, Fragoso CM, Roditi I. 2007. Highly efficient stable transformation of bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 153:220–223 [DOI] [PubMed] [Google Scholar]
- 24. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 26. Letunic I, Doerks T, Bork P. 2012. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40:D302–D305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bihlmaier K, Mesecke N, Terziyska N, Bien M, Hell K, Herrmann JM. 2007. The disulfide relay system of mitochondria is connected to the respiratory chain. J. Cell Biol. 179:389–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dabir DV, Leverich EP, Kim SK, Tsai FD, Hirasawa M, Knaff DB, Koehler CM. 2007. A role for cytochrome c and cytochrome c peroxidase in electron shuttling from Erv1. EMBO J. 26:4801–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Levitan A, Danon A, Lisowsky T. 2004. Unique features of plant mitochondrial sulfhydryl oxidase. J. Biol. Chem. 279:20002–20008 [DOI] [PubMed] [Google Scholar]
- 30. Ang SK, Lu H. 2009. Deciphering structural and functional roles of individual disulfide bonds of the mitochondrial sulfhydryl oxidase Erv1p. J. Biol. Chem. 284:28754–28761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rodriguez-Roldan V, Garcia-Heredia JM, Navarro JA, Hervas M, De la Cerda B, Molina-Heredia FP, De la Rosa MA. 2006. A comparative kinetic analysis of the reactivity of plant, horse, and human respiratory cytochrome c towards cytochrome c oxidase. Biochem. Biophys. Res. Commun. 346:1108–1113 [DOI] [PubMed] [Google Scholar]
- 32. Allen JW, Ginger ML, Ferguson SJ. 2004. Maturation of the unusual single-cysteine (XXXCH) mitochondrial c-type cytochromes found in trypanosomatids must occur through a novel biogenesis pathway. Biochem. J. 383:537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ginger ML, Sam KA, Allen JW. 2012. Probing why trypanosomes assemble atypical cytochrome c with an AxxCH haem-binding motif instead of CxxCH. Biochem. J. 448:253–260 [DOI] [PubMed] [Google Scholar]
- 34. Fass D. 2008. The Erv family of sulfhydryl oxidases. Biochim. Biophys. Acta 1783:557–566 [DOI] [PubMed] [Google Scholar]
- 35. Hofhaus G, Lee JE, Tews I, Rosenberg B, Lisowsky T. 2003. The N-terminal cysteine pair of yeast sulfhydryl oxidase Erv1p is essential for in vivo activity and interacts with the primary redox centre. Eur. J. Biochem. 270:1528–1535 [DOI] [PubMed] [Google Scholar]
- 36. Wu CK, Dailey TA, Dailey HA, Wang BC, Rose JP. 2003. The crystal structure of augmenter of liver regeneration: a mammalian FAD-dependent sulfhydryl oxidase. Protein Sci. 12:1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vitu E, Bentzur M, Lisowsky T, Kaiser CA, Fass D. 2006. Gain of function in an ERV/ALR sulfhydryl oxidase by molecular engineering of the shuttle disulfide. J. Mol. Biol. 362:89–101 [DOI] [PubMed] [Google Scholar]
- 38. Sevier CS, Kaiser CA. 2006. Disulfide transfer between two conserved cysteine pairs imparts selectivity to protein oxidation by Ero1. Mol. Biol. Cell 17:2256–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vala A, Sevier CS, Kaiser CA. 2005. Structural determinants of substrate access to the disulfide oxidase Erv2p. J. Mol. Biol. 354:952–966 [DOI] [PubMed] [Google Scholar]
- 40. Banci L, Bertini I, Calderone V, Cefaro C, Ciofi-Baffoni S, Gallo A, Kallergi E, Lionaki E, Pozidis C, Tokatlidis K. 2011. Molecular recognition and substrate mimicry drive the electron-transfer process between MIA40 and ALR. Proc. Natl. Acad. Sci. U. S. A. 108:4811–4816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bien M, Longen S, Wagener N, Chwalla I, Herrmann JM, Riemer J. 2010. Mitochondrial disulfide bond formation is driven by intersubunit electron transfer in Erv1 and proofread by glutathione. Mol. Cell 37:516–528 [DOI] [PubMed] [Google Scholar]
- 42. Becher D, Kricke J, Stein G, Lisowsky T. 1999. A mutant for the yeast scERV1 gene displays a new defect in mitochondrial morphology and distribution. Yeast 15:1171–1181 [DOI] [PubMed] [Google Scholar]
- 43. Stojanovski D, Rissler M, Pfanner N, Meisinger C. 2006. Mitochondrial morphology and protein import—a tight connection? Biochim. Biophys. Acta 1763:414–421 [DOI] [PubMed] [Google Scholar]
- 44. Long S, Jirků M, Mach J, Ginger ML, Šuták R, Richardson D, Tachezy J, Lukeš J. 2008. Ancestral roles of eukaryotic frataxin: mitochondrial frataxin function and heterologous expression of hydrogenosomal Trichomonas homologues in trypanosomes. Mol. Microbiol. 69:94–109 [DOI] [PubMed] [Google Scholar]
- 45. Long S, Changmai P, Tsaousis AD, Skalický T, Verner Z, Wen YZ, Roger AJ, Lukeš J. 2011. Stage-specific requirement for Isa1 and Isa2 proteins in the mitochondrion of Trypanosoma brucei and heterologous rescue by human and Blastocystis orthologues. Mol. Microbiol. 81:1403–1418 [DOI] [PubMed] [Google Scholar]
- 46. Kispal G, Csere P, Prohl C, Lill R. 1999. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18:3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bruske EI, Sendfeld F, Schneider A. 2009. Thiolated tRNAs of Trypanosoma brucei are imported into mitochondria and dethiolated after import. J. Biol. Chem. 284:36491–36499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huynen MA, Snel B, Bork P, Gibson TJ. 2001. The phylogenetic distribution of frataxin indicates a role in iron-sulfur cluster protein assembly. Hum. Mol. Genet. 10:2463–2468 [DOI] [PubMed] [Google Scholar]
- 49. Corradi N, Pombert JF, Farinelli L, Didier ES, Keeling PJ. 2010. The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis. Nat. Commun. 1:77 doi:10.1038/ncomms1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, Delbac F, El Alaoui H, Peyret P, Saurin W, Gouy M, Weissenbach J, Vivares CP. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414:450–453 [DOI] [PubMed] [Google Scholar]
- 51. Soler N, Delagoutte E, Miron S, Facca C, Baille D, D'Autreaux B, Craescu G, Frapart YM, Mansuy D, Baldacci G, Huang ME, Vernis L. 2011. Interaction between the reductase Tah18 and highly conserved Fe-S containing Dre2 C-terminus is essential for yeast viability. Mol. Microbiol. 82:54–67 [DOI] [PubMed] [Google Scholar]
- 52. Carrie C, Giraud E, Duncan O, Xu L, Wang Y, Huang S, Clifton R, Murcha M, Filipovska A, Rackham O, Vrielink A, Whelan J. 2010. Conserved and novel functions for Arabidopsis thaliana MIA40 in assembly of proteins in mitochondria and peroxisomes. J. Biol. Chem. 285:36138–36148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gentle IE, Perry AJ, Alcock FH, Likic VA, Dolezal P, Ng ET, Purcell AW, McConnville M, Naderer T, Chanez AL, Charriere F, Aschinger C, Schneider A, Tokatlidis K, Lithgow T. 2007. Conserved motifs reveal details of ancestry and structure in the small TIM chaperones of the mitochondrial intermembrane space. Mol. Biol. Evol. 24:1149–1160 [DOI] [PubMed] [Google Scholar]
- 54. Yang ZR, Thomson R, McNeil P, Esnouf RM. 2005. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics 21:3369–3376 [DOI] [PubMed] [Google Scholar]
- 55. Farrell SR, Thorpe C. 2005. Augmenter of liver regeneration: a flavin-dependent sulfhydryl oxidase with cytochrome c reductase activity. Biochemistry 44:1532–1541 [DOI] [PubMed] [Google Scholar]
- 56. Gnaiger E, Kuznetsov AV. 2002. Mitochondrial respiration at low levels of oxygen and cytochrome c. Biochem. Soc. Trans. 30:252–258 [DOI] [PubMed] [Google Scholar]
- 57. Jones DP. 1986. Intracellular diffusion gradients of O2 and ATP. Am. J. Physiol. 250:C663–C675 [DOI] [PubMed] [Google Scholar]
- 58. Hellemond JJ, Bakker BM, Tielens AG. 2005. Energy metabolism and its compartmentation in Trypanosoma brucei. Adv. Microb. Physiol. 50:199–226 [DOI] [PubMed] [Google Scholar]
- 59. Forman HJ, Azzi A. 1997. On the virtual existence of superoxide anions in mitochondria: thoughts regarding its role in pathophysiology. FASEB J. 11:374–375 [DOI] [PubMed] [Google Scholar]
- 60. Oursler MJ, Bradley EW, Elfering SL, Giulivi C. 2005. Native, not nitrated, cytochrome c and mitochondria-derived hydrogen peroxide drive osteoclast apoptosis. Am. J. Physiol. Cell Physiol. 288:C156–C168 [DOI] [PubMed] [Google Scholar]
- 61. Dean S, Marchetti R, Kirk K, Matthews KR. 2009. A surface transporter family conveys the trypanosome differentiation signal. Nature 459:213–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Spitznagel D, Ebikeme C, Biran M, Nic a' Bhaird N, Bringaud F, Henehan GT, Nolan DP. 2009. Alanine aminotransferase of Trypanosoma brucei—a key role in proline metabolism in procyclic life forms. FEBS J. 276:7187–7199 [DOI] [PubMed] [Google Scholar]
- 63. Coustou V, Biran M, Besteiro S, Riviere L, Baltz T, Franconi JM, Bringaud F. 2006. Fumarate is an essential intermediary metabolite produced by the procyclic Trypanosoma brucei. J. Biol. Chem. 281:26832–26846 [DOI] [PubMed] [Google Scholar]
- 64. Rada P, Doležal P, Jedelský PL, Bursac D, Perry AJ, Šedinová M, Smíšková K, Novotný M, Beltrán NC, Hrdý I, Lithgow T, Tachezy J. 2011. The core components of organelle biogenesis and membrane transport in the hydrogenosomes of Trichomonas vaginalis. PLoS One 6:e24428 doi:10.1371/journal.pone.0024428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Muller M, Mentel M, van Hellemond JJ, Henze K, Woehle C, Gould SB, Yu RY, van der Giezen M, Tielens AG, Martin WF. 2012. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 76:444–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shiflett AM, Johnson PJ. 2010. Mitochondrion-related organelles in eukaryotic protists. Annu. Rev. Microbiol. 64:409–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Šuták R, Doležal P, Fiumera HL, Hrdý I, Dancis A, Delgadillo-Correa M, Johnson PJ, Muller M, Tachezy J. 2004. Mitochondrial-type assembly of FeS centers in the hydrogenosomes of the amitochondriate eukaryote Trichomonas vaginalis. Proc. Natl. Acad. Sci. U. S. A. 101:10368–10373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tovar J, Leon-Avila G, Sanchez LB, Šuták R, Tachezy J, van der Giezen M, Hernandez M, Muller M, Lucocq JM. 2003. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature 426:172–176 [DOI] [PubMed] [Google Scholar]
- 69. Tsaousis AD, Ollagnier de Choudens S, Gentekaki E, Long S, Gaston D, Stechmann A, Vinella D, Py B, Fontecave M, Barras F, Lukeš J, Roger AJ. 2012. Evolution of Fe/S cluster biogenesis in the anaerobic parasite Blastocystis. Proc. Natl. Acad. Sci. U. S. A. 109:10426–10431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ali V, Shigeta Y, Tokumoto U, Takahashi Y, Nozaki T. 2004. An intestinal parasitic protist, Entamoeba histolytica, possesses a non-redundant nitrogen fixation-like system for iron-sulfur cluster assembly under anaerobic conditions. J. Biol. Chem. 279:16863–16874 [DOI] [PubMed] [Google Scholar]
- 71. Maralikova B, Ali V, Nakada-Tsukui K, Nozaki T, van der Giezen M, Henze K, Tovar J. 2010. Bacterial-type oxygen detoxification and iron-sulfur cluster assembly in amoebal relict mitochondria. Cell. Microbiol. 12:331–342 [DOI] [PubMed] [Google Scholar]
- 72. van der Giezen M, Cox S, Tovar J. 2004. The iron-sulfur cluster assembly genes iscS and iscU of Entamoeba histolytica were acquired by horizontal gene transfer. BMC Evol. Biol. 4:7 doi:10.1186/1471-2148-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Daithankar VN, Schaefer SA, Dong M, Bahnson BJ, Thorpe C. 2010. Structure of the human sulfhydryl oxidase augmenter of liver regeneration and characterization of a human mutation causing an autosomal recessive myopathy. Biochemistry 49:6737–6745 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.