Abstract
Some of the most devastating plant and animal pathogens belong to the oomycete class. The cell walls of these microorganisms represent an excellent target for disease control, but their carbohydrate composition is elusive. We have undertaken a detailed cell wall analysis in 10 species from 2 major oomycete orders, the Peronosporales and the Saprolegniales, thereby unveiling the existence of 3 clearly different cell wall types: type I is devoid of N-acetylglucosamine (GlcNAc) but contains glucuronic acid and mannose; type II contains up to 5% GlcNAc and residues indicative of cross-links between cellulose and 1,3-β-glucans; type III is characterized by the highest GlcNAc content (>5%) and the occurrence of unusual carbohydrates that consist of 1,6-linked GlcNAc residues. These 3 cell wall types are also distinguishable by their cellulose content and the fine structure of their 1,3-β-glucans. We propose a cell wall paradigm for oomycetes that can serve as a basis for the establishment of cell wall architectural models and the further identification of cell wall subtypes. This paradigm is complementary to morphological and molecular criteria for taxonomic grouping and provides useful information for unraveling poorly understood cell wall carbohydrate biosynthetic pathways through the identification and characterization of the corresponding enzymes.
INTRODUCTION
The oomycete class comprises a large number of eukaryotic saprobes and parasites that are widely distributed worldwide. Some of these microbial species have important environmental and economic impacts as pathogens of plants or animals, in both natural habitats and industrial cultures. Typical examples are the plant pathogen Phytophthora infestans, responsible for the late blight disease in potato (1), and the salmonid pathogen Saprolegnia parasitica (2). Annual losses in the agriculture and aquaculture industries due to pathogenic oomycetes represent tens of billions of dollars globally (3, 4).
Although oomycetes grow in a vegetative mycelial form as true fungi, they are classified in a completely different taxonomic group, the Stramenopiles, together with diatoms and brown algae (5). Oomycetes and fungi can be biochemically distinguished by their cell wall polysaccharide composition. As opposed to fungi, which contain chitin as a cell wall scaffold, oomycetes are classically described as containing no or very small amounts (<1%) of this carbohydrate. Instead, cellulose occurs in oomycetes together with a high proportion of other β-glucans (6–9). This distinction between fungal and oomycete cell walls was established in the 1960s and 1970s on the basis of techniques of limited analytical performance, such as paper chromatography. In addition, this pioneering work was restricted to a few species only, thereby providing limited information on the diversity of the cell wall composition across different oomycete orders. Since then, and as opposed to the extracellular matrices of true fungi, oomycete cell walls have been understudied and their fine composition and structure remain elusive.
Microbial cell walls are vital for the pathogens and thus represent an interesting target for disease control. This is illustrated by the case of pathogenic fungi, for instance, Candida albicans, in which 1,3-β-glucan biosynthesis can be specifically blocked by drugs from the echinocandin family (10), thereby leading to cell death. In addition, our group has recently demonstrated the potential of targeting the biosynthesis of the minor chitin component in the oomycete Saprolegnia monoica for disease control (11). In this context, deciphering the details of carbohydrate structures within the three-dimensional oomycete cell wall network is of great relevance. Indeed, cell wall analysis can be combined with the identification of novel biosynthetic enzymes that can be blocked to specifically inhibit the growth of the pathogens. Furthermore, extending these cell wall studies across different orders of oomycetes may allow the development of species-specific disease control processes. This will be greatly facilitated by the availability of an increasing number of sequenced genomes from important pathogenic oomycetes distributed in different taxonomic groups. Typical examples of sequenced genomes are the genomes of the Peronosporales Phytophthora sojae (12), Phytophthora ramorum (12), P. infestans (13), and Pythium ultimum (14) and the Saprolegniales S. parasitica (Saprolegnia Sequencing Project, Broad Institute of Harvard and MIT; http://www.broadinstitute.org). In addition to the area of disease control, unraveling cell wall carbohydrate structures across oomycete orders may provide important information complementary to conventional morphological and molecular approaches and can be used as an additional criterion for taxonomic distinction.
The objective of our work was to gain further insight into the cell wall carbohydrate composition in oomycetes as a first step toward the elaboration of three-dimensional architectural models. With this goal in mind, we have undertaken a detailed polysaccharide analysis of the mycelial walls of 10 oomycete species from two major orders, the Peronosporales and Saprolegniales. The selected microorganisms are representative of the saprobe and the plant and animal parasite groups. An experimental work flow that can be applied to any microbial fungus-like cells was implemented. The procedure involves carbohydrate fractionation, and the analytical part integrates quantitative monosaccharide and linkage determinations. This study unveiled the existence of three different cell wall types across the Peronosporales and Saprolegniales. Criteria allowing the distinction between these three cell wall types are presented, and evidence that these can be combined with morphological and molecular profiling for taxonomic grouping is provided.
MATERIALS AND METHODS
Strains and culture methods.
All analyses were performed on the mycelial cell walls from 10 different Peronosporales and Saprolegniales strains representing saprobes and plant and animal pathogens. The strains were from (i) the Centraal Bureau voor Schimmel Culture (CBS; Baarn, The Netherlands) [P. infestans (Mont.) de Bary 1876 (CBS 120920; GenBank accession number JX418021), S. monoica Pringsheim 1858 (CBS 539.67; GenBank accession number JX418016), and S. parasitica Coker 1923 (CBS 223.65; GenBank accession number JX418013)] or (ii) the culture collection of the Real Jardín Botánico (Madrid, Spain) [Saprolegnia ferax (Gruith) Kütz 1843 (SAP0479; GenBank accession number JX418014), Saprolegnia anisospora de Bary 1888 (SAP1148; GenBank accession number JX418015), Achlya caroliniana Coker 1923 (SAP1240; GenBank accession number JX418018), Leptolegnia sp. (SAP0772; GenBank accession number JX418017), and Dictyuchus sp. (SAP1135; GenBank accession number JX418019)] or (iii) provided by A. Bottin (University of Toulouse, Toulouse, France) [Aphanomyces euteiches Drechsler 1925 (ATCC 201684; GenBank accession number JX418020) and Phytophthora parasitica Dastur 1913 (GenBank accession number JX418022)].
All strains were maintained on potato dextrose agar (Sigma-Aldrich). For cell wall analyses, the mycelia were grown in the liquid medium of Machlis (15) for 5 to 7 days at 25°C, as previously described (16).
DNA amplification and sequencing.
DNA extraction and PCR were performed as described earlier (17). The internal transcribed spacer (ITS) regions were amplified using universal primers for eukaryotes, namely, un-SSU-1766 (ITS5, 5′-GGAAGTAAAAGTCGTAACAAGG-3′) and un-LSU-0041 (ITS4, 5′-TCCTCCGCTTATTGATATGC-3′) (18). The amplified products were sequenced (3730xl automated DNA sequencer; Applied Biosystems, Macrogen, The Netherlands), and their consensus sequences were assembled and edited using the Sequencher program (version 4.2; Gene Codes Corporation).
Phylogenetic analyses.
Additional sequences of the following pathogens were included in the phylogenetic analysis: Peronospora sparsa Berk. 1862 (GenBank accession number AF266783), Pythium irregulare Buisman 1927 (GenBank accession number HQ643659), and Lagenidium caudatum G. L. Barron 1976 (GenBank accession number HQ643136). The Haliphthoraceae Halodaphnea panulirata (Kitanch. & Hatai) M. W. Dick 1998 (GenBank accession number AB285512) was used as an outgroup. Multiple alignments were performed using the default settings of the MAFFT (version 6.903b) program (19, 20) and the L-INS-i algorithm (21). The alignments were adjusted by visual examination using the software Se-Al (version 2.0a11 Carbon).
The phylogenetic analysis was performed with MrBayes (version 3.2) software (22). The program jModeltest (version 0.1.1) (23) was used to select the optimal model of the nucleotide evolution (HKY+G) and the appropriate parameters for building the phylogenetic trees. The following settings were used: the number of substitution types (nst) was 2; the number of rate categories for the gamma distribution (ncat) was 4; rates were gamma; state frequencies (Statefreqpr) were 0.2500, 0.1752, 0.2305, and 0.3443; the gamma shape parameter (Shapepr) was 0.5740; the proportion of invariable sites (Pinvarpr) was 0; the number of generations (Ngen) was 10,000,000; 2 independent analyses (Nruns) with 4 chains (Nchains) were performed with a sampling every 1,000th generation; burn-in was 25%; and the deviation of split frequencies was <0.01. The final tree was edited with the FigTree (version 1.3.1) and Adobe Illustrator programs.
Preparation and fractionation of cell walls.
Cell wall polysaccharides were extracted and fractionated from purified cell walls as outlined in Fig. 1. The mycelia from the different species grown in the liquid medium of Machlis (15) were washed extensively with distilled water and immediately frozen in liquid nitrogen. The cells were disrupted in liquid nitrogen using a mortar and pestle. The resulting fine powders were subjected to 3 consecutive extractions of 6, 12, and 8 h in 70% (vol/vol) ethanol. The ethanol-insoluble materials recovered after each step were concentrated by centrifugation at 4,000 × g for 10 min at 4°C. The suspensions were then successively filtered through glass-fiber filters (GF/A; Whatman), washed 6 times with 70% ethanol and 6 times with acetone, and dried under vacuum (SpeedVac Plus; Savant) at room temperature. Proteins in the resulting alcohol-insoluble residues (AIRs) were removed by heating the samples at 80°C for 10 min in a 50 mM Tris-HCl buffer (pH 7.8) containing 2% (wt/vol) sodium dodecyl sulfate (SDS), 40 mM 2-mercaptoethanol, and 10 mM EDTA. This extraction step was repeated 3 times, and the remaining suspensions were filtered through glass-fiber filters, as described above. The residues recovered after final washes in 70% ethanol and acetone, as described above, corresponded to the purified cell walls. The latter were dried under vacuum and fractionated into an alkali-soluble fraction (ASF) and an alkali-insoluble fraction (AIF). For this purpose, the cell walls were resuspended in 80% (vol/vol) methanol containing 5% (wt/vol) KOH and 0.1% (wt/vol) NaBH4. After each of 3 consecutive treatments of 15 min at 100°C in this solution, the ASF and AIF samples from each strain were recovered by centrifugation at 5,000 × g for 10 min and pooled. The AIFs were washed 3 times with 0.5 M acetic acid and water. The first wash was pooled with the corresponding ASF, and the pH was adjusted to 6.0 using glacial acetic acid. The ASF and AIF samples were freeze-dried after a dialysis step (molecular mass cutoff, 3,500 Da; Spectra/Por; Spectrum Laboratories) against deionized water to remove solutes of a small molecular mass (the dialysis tubings were thoroughly washed before use to eliminate any contaminants potentially associated with the membranes). The presence of glycogen/starch-like polymers in the samples was ruled out by performing carbohydrate analyses at different steps of the fractionation procedure, before and after enzymatic hydrolysis in the presence of amylases (24). Since identical total sugar and glycosidic linkage profiles were obtained from all samples analyzed before and after enzymatic treatment, the amylase hydrolytic step was omitted in the final procedure to avoid the introduction of undesirable proteins in the cell wall extracts.
Fig 1.
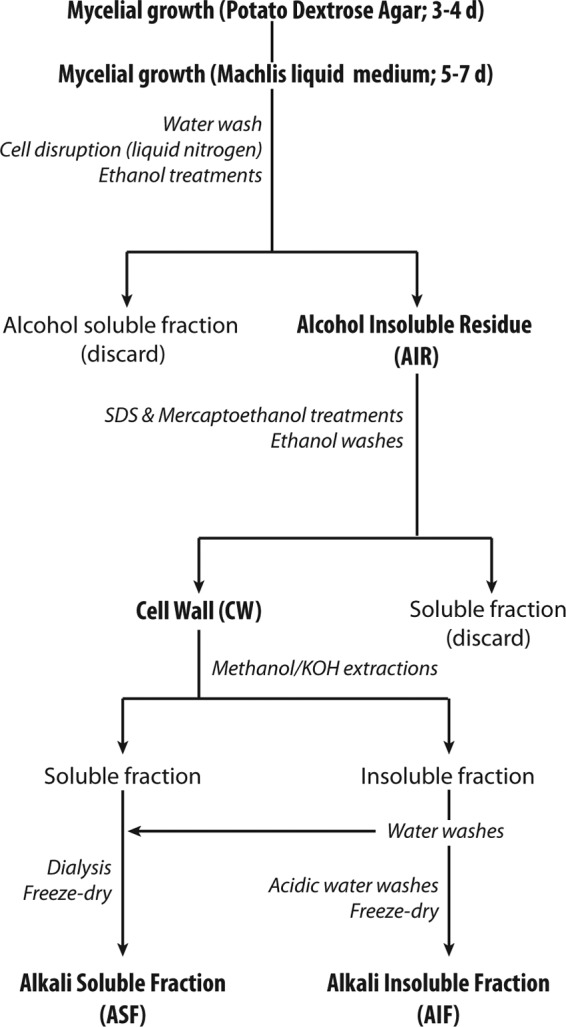
Preparation and fractionation of cell wall carbohydrates.
Monosaccharide analysis.
Total sugar analysis was performed as described by Albersheim et al. (25) with some modifications. The freeze-dried ASF and AIF samples (1 mg) were hydrolyzed in the first instance in the presence of 2 M trifluoroacetic acid (TFA) at 121°C for 3 h. The TFA-resistant materials were further hydrolyzed with 6 N HCl at 100°C for 16 h. myo-Inositol was used as an internal standard. The resulting monosaccharides were converted to alditol acetates (25), separated, and analyzed by gas chromatography (GC) on a SP-2380 capillary column (30 m by 0.25 mm [inner diameter]; Supelco) using an HP-6890 GC system and an HP-5973 electron-impact mass spectrometer (EI-MS) as a detector (Agilent Technologies). The temperature program increased from 180°C to 230°C at a rate of 1.5°C min−1.
Determination of uronic acid contents was performed on the TFA hydrolysates by high-performance anion-exchange chromatography (HPAEC) on a CarboPac PA-10 anion-exchange column (4.6 by 250 mm; Dionex) using a pulsed amperometric detector (PAD; Dionex ICS 3000 system). Monosaccharides were eluted at 1 ml min−1 using a linear saline gradient: 100 mM NaOH to 100 mM NaOH–300 mM sodium acetate over 20 min.
Glycosidic linkage analysis.
Polysaccharide networks in the dry ASF and AIF samples (0.5 mg) were first swollen in 200 μl dry dimethyl sulfoxide (DMSO). Ten microliters of DMSO containing 0.3 mg liter−1 sulfur dioxide and 5 μl of diethylamine was added, and the samples were subsequently sonicated for 20 min and stirred under argon at room temperature for 3 h. Methylation reactions were performed using the NaOH/CH3I method (26), by repeating 5 times the methylation step on each sample, thereby avoiding any risk of undermethylation. Partially methylated polysaccharides were hydrolyzed in the presence of 2 M TFA at 121°C for 2 h and further derivatized to permethylated alditol acetates (25). The latter were separated and analyzed by GC/EI-MS on a CP-Sil 5 CB capillary column (30 m by 0.25 mm [inner diameter]; Agilent Technologies) with a temperature program increasing from 160°C to 210°C at a rate of 1°C min−1. The mass spectra of the fragments obtained from the permethylated alditol acetates (EI-MS) were compared with those of reference derivatives and by comparison to available data (27).
RESULTS
Evolutionary relationship of the 10 selected oomycete species.
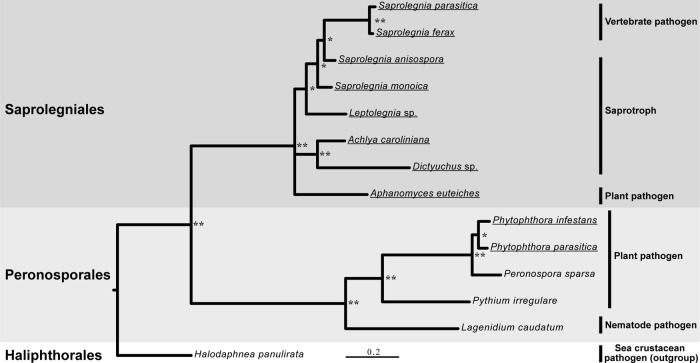
A phylogenetic analysis based on ITS regions from 10 different saprobe and pathogen species from the Peronosporales and Saprolegniales orders was performed to confirm their taxonomic grouping and evolutionary relationship. As expected, high values of posterior probabilities verified the existence of a divergent evolution between members of the 2 orders (Fig. 2). Each of the Saprolegniales and Peronosporales orders was subdivided into groups of species with different lifestyles, namely, vertebrate pathogens, phytopathogens, and saprotrophs (Fig. 2). In the next part of this work, we investigated whether the cell wall carbohydrate composition of the 10 selected species (Fig. 2) reflects or is independent from the taxonomic grouping.
Fig 2.
Phylogenetic relations between oomycete species from the Peronosporales and Saprolegniales orders. Phylogenetic relationships based on 14 ITS ribosomal DNA sequences showing the distribution of 10 selected oomycete plant and animal pathogens are presented. The Haliphthorales Halodaphnea panulirata was used as an outgroup. Posterior probabilities are provided for branches receiving 0.5 or more support. **, posterior probabilities higher than 0.9; *, posterior probabilities lower than 0.9. The names of the species selected for cell wall analyses are underlined.
Saprolegniales, but not Peronosporales, contain different types of GlcNAc-based carbohydrates.
In order to obtain a detailed insight into the extracellular carbohydrate composition of oomycetes from the orders Saprolegniales and Peronosporales, cell wall polysaccharides from the 10 selected species were fractionated on the basis of their solubility in alkali, following the procedure presented in Fig. 1. For all species analyzed, cell walls consisted, on average, of ∼70% alkali-insoluble polysaccharides and 30% alkali-soluble carbohydrates. The AIFs of all oomycetes contained a high proportion of glucans, i.e., from 87% in the Saprolegniales A. euteiches to up to 99% in the Peronosporales P. parasitica (Fig. 3A). A distinguishing feature of the two oomycete orders was the occurrence of N-acetylglucosamine (GlcNAc)-based saccharides in the Saprolegniales and the absence of such carbohydrates in the Peronosporales (Fig. 3 and 4) (see Table 1 for a list of the abbreviations used for monosaccharides and the corresponding permethylated alditol acetate derivatives). With the exception of A. euteiches, which exhibited a particularly high GlcNAc content in both the AIF (∼12%) and the ASF (∼5%), all other Saprolegniales contained 2.5 to 7% GlcNAc in the AIF and no more than 1.6% of the monosaccharide in the ASF (Fig. 3A and 4A).
Fig 3.
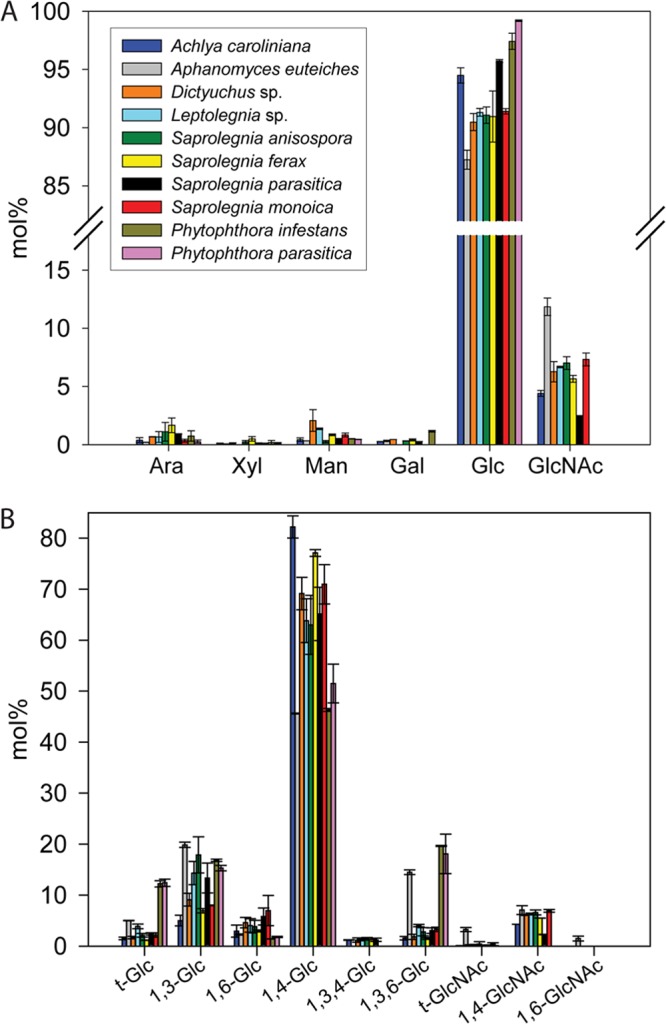
Carbohydrate analysis of the AIF samples from the 10 selected oomycete species. (A) Monosaccharide composition. Values are means ± SDs of six determinations from 2 independent experiments. Ara, arabinose; Xyl, xylose; Man, mannose; Gal, galactose; Glc, glucose; GlcNAc, N-acetylglucosamine. (B) Glycosidic linkage analysis. Values are means ± SDs of 5 determinations from 2 independent experiments. The different glycosidic linkages were identified by EI-MS.
Fig 4.
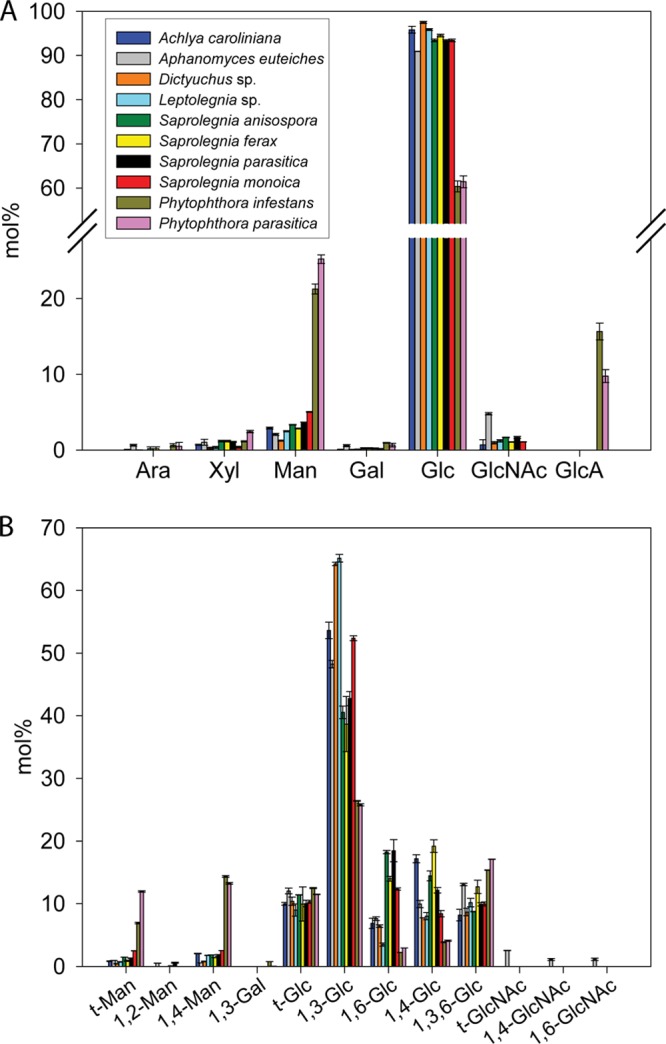
Carbohydrate analysis of ASF samples from the 10 selected oomycete species. (A) Monosaccharide composition. Values are means ± SDs of six determinations from 2 independent experiments. GlcA, glucuronic acid. The definitions of the other monosaccharides are presented in the legend to Fig. 3A. (B) Glycosidic linkage analysis. Values are means ± SDs of 5 determinations from 2 independent experiments. The different glycosidic linkages were identified by EI-MS.
Table 1.
Abbreviations used for monosaccharides and the corresponding permethylated alditol acetates and deduced linkages
| Monosaccharide | Permethylated alditol acetate | Deduced linkage |
|---|---|---|
| Mannopyranose (Man) | 1,5-Di-O-acetyl-2,3,4,6-tetra-O-methyl mannitol | t-Man |
| 1,2,5-Tri-O-acetyl-3,4,6-tri-O-methyl mannitol | 1,2-Man | |
| 1,4,5-Tri-O-acetyl-2,3,6-tri-O-methyl mannitol | 1,4-Man | |
| Galactopyranose (Gal) | 1,3,5-Tri-O-acetyl-2,4,6-tri-O-methyl galactitol | 1,3-Gal |
| Glucopyranose (Glc) | 1,5-Di-O-acetyl-2,3,4,6-tetra-O-methyl glucitol | t-Glc |
| 1,3,5-Tri-O-acetyl-2,4,6-tri-O-methyl glucitol | 1,3-Glc | |
| 1,5,6-Tri-O-acetyl-2,3,4-tri-O-methyl glucitol | 1,6-Glc | |
| 1,4,5-Tri-O-acetyl-2,3,6-tri-O-methyl glucitol | 1,4-Glc | |
| 1,3,4,5-Tetra-O-acetyl-2,6-di-O-methyl glucitol | 1,3,4-Glc | |
| 1,3,5,6-Tetra-O-acetyl-2,4-di-O-methyl glucitol | 1,3,6-Glc | |
| N-Acetylglucopyranosamine (GlcNAc) | 1,5-Di-O-acetyl-(2-deoxy,2-N-methylacetamido)-3,4,6-tri-O-methyl glucitol | t-GlcNAc |
| 1,4,5-Tri-O-acetyl-(2-deoxy,2-N-methylacetamido)-3,6-di-O-methyl glucitol | 1,4-GlcNAc | |
| 1,5,6-Tri-O-acetyl-(2-deoxy,2-N-methylacetamido)-3,4-di-O-methyl glucitol | 1,6-GlcNAc | |
| Xylopyranose (Xyl) | 1,4,5-Tri-O-acetyl-2,3-di-O-methyl xylitol | 1,4-Xyl |
The types of linkages present in the Saprolegniales GlcNAc-based carbohydrates were determined by methylation analysis (GC/EI-MS) (Fig. 3B and 4B). The AIFs of all species contained 1,4-linked GlcNAc residues, pointing to the occurrence of chitin-like polysaccharides (Fig. 3B). Interestingly, as opposed to all other species analyzed, the AIF and ASF from the cell wall of A. euteiches contained 1,6-linked GlcNAc. This type of residue has never been reported in any other eukaryotic microorganism. Its occurrence was evidenced by a specific retention time on the gas chromatogram (Fig. 5A) and a characteristic MS fragment of 189 (m/z) arising from the cleavage between carbons 3 and 4 (Fig. 5D). In addition, clear differences in the relative abundance of 45, 99, 129, 142, 145, 173, 202, 205, and 233 (m/z) fragments could be used to further distinguish the 1,6-linked GlcNAc residue from the 1,4-linked GlcNAc and t-GlcNAc derivatives (Fig. 5B to D) (28).
Fig 5.

Methylation analysis of carbohydrates containing N-acetylglucosamine residues. (A) Gas chromatograph of the permethylated alditol acetates from methylation analysis of the N-acetylglucosamine-based carbohydrates; (B) mass spectrum and fragmentation pattern (inset) obtained by EI-MS analysis of 1,5-di-O-acetyl-(2-deoxy, 2-N-methylacetamido)-3,4,6-tri-O-methyl glucitol (deduced linkage, t-GlcNAc); (C) as in panel B but for 1,4,5-tri-O-acetyl- (2-deoxy, 2-N-methylacetamido)-3,6-di-O-methyl glucitol (deduced linkage, 1,4-GlcNAc); (D) as in panel B but for 1,5,6-tri-O-acetyl-(2-deoxy, 2-N-methylacetamido)-3,4-di-O-methyl glucitol (deduced linkage, 1,6-GlcNAc). The larger numbers above the MS fragments in panels B to D correspond to the m/z signals that were used to distinguish the different GlcNAc derivatives. OAc, acetate; MeOAc, methyl acetate; OMe, methoxy.
Although all Saprolegniales species contained GlcNAc residues in their ASF (Fig. 4A), no corresponding permethylated alditol acetates were detected after methylation analysis of these fractions, except in the case of A. euteiches (Fig. 4B). This is due to the originally low GlcNAc content (≤1.6%) of these fractions (Fig. 4A) and the detection limit of the method, which is of ∼1%. 1,4-Linked (7%) and 1,6-linked (1.5%) GlcNAc residues, together with t-GlcNAc (3.2%), were present in the AIF from A. euteiches, whereas 1,4-linked residues (2.2 to 6.9%) and only traces of t-GlcNAc were observed in the corresponding fractions from the other Saprolegniales. Altogether, these observations point toward a lower degree of polymerization of the alkali-insoluble GlcNAc-based carbohydrates in A. euteiches than other Saprolegniales (if we assume that all GlcNAc residues arise from homosaccharidic GlcNAc chains).
Conditions used to release monosaccharides from cell wall carbohydrates by acid hydrolysis typically involve a 2- to 4-h treatment at 120°C in the presence of 2 to 4 M TFA. Under such hydrolytic conditions, no more than 25 to 73% of the GlcNAc from the Saprolegniales AIF samples was released (Fig. 6). It is only when the residual material obtained after TFA hydrolysis was subjected to further acid hydrolysis with HCl (6 M, 16 h at 100°C) that additional GlcNAc was released from the AIF samples (Fig. 6). The cell walls of A. euteiches and S. parasitica exhibited the highest proportion of TFA-extractable GlcNAc (73 and 63%, respectively), whereas S. anisospora and S. monoica showed the opposite trend, i.e., the lowest percentage of TFA-sensitive GlcNAc-based carbohydrates in AIF (25 and 34%, respectively). These differences most likely reflect the occurrence of various structural forms of GlcNAc-based carbohydrates in different species.
Fig 6.
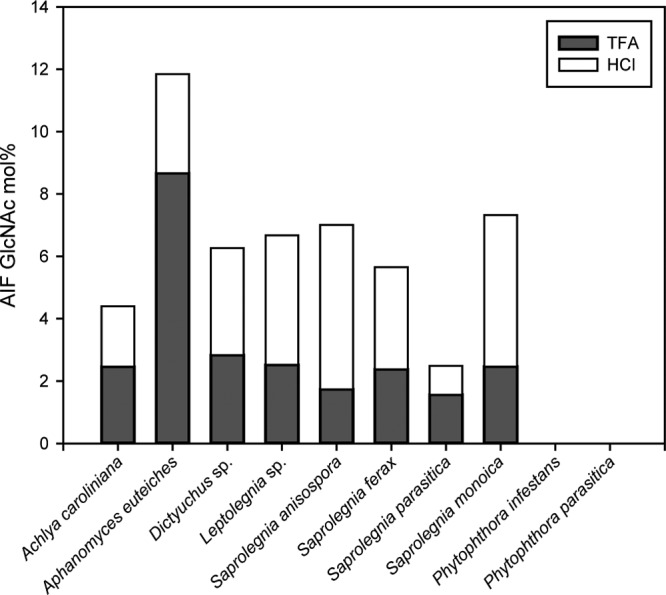
Extractability of GlcNAc residues from the AIF samples under different acid hydrolysis conditions. AIF samples were first subjected to acid hydrolysis for 3 h at 120°C in the presence of 2 M TFA. The TFA-resistant material was then further hydrolyzed with 6 M HCl for 16 h at 100°C. The amount of GlcNAc released after each step was quantified in the form of alditol acetates as described in the Materials and Methods section.
Analysis of cell wall glucans.
The lack of GlcNAc in the AIF isolated from the walls of Peronosporales species was counterbalanced by a higher glucose content (Fig. 3A). Indeed, in the case of P. parasitica and P. infestans, the AIF consisted of 99% glucose (Fig. 3A), ∼50% of which was 1,4-linked and most likely arose from cellulose (Fig. 3B). Interestingly, A. euteiches was the only Saprolegniales for which a similar proportion of cellulose (46%) was measured in AIF, whereas the samples from all other species contained more of this insoluble polymer (63 to 82% of the AIF) (Fig. 3B). The lower cellulose content in A. euteiches and the Peronosporales was counterbalanced by a higher proportion of glucans containing 1,3- and 1,3,6-linked glucosyl residues (Fig. 3B). In addition, the Peronosporales and, to a much lesser extent, A. euteiches exhibited the highest proportion of t-Glc residues. Altogether, these data show that the AIFs from A. euteiches and the Peronosporales contain a larger amount of 1,3-glucans whose degree of branching at the 6 position is higher than that in the other species. Furthermore, since cellulose is typically known to exhibit a high degree of polymerization (>100) and since the sensitivity of the GC/MS method is ∼1%, it can be assumed that most of the t-Glc detected in AIFs arises from 1,3-glucans. In this case, the higher t-Glc abundance in the Peronosporales is indicative of a lower degree of polymerization of the 1,3-glucans compared to that in the other species. The AIF samples from A. euteiches and the Peronosporales also contained traces of 1,2,3- and 1,2,3,6-linked glucosyl residues (not shown) as well as comparable amounts (1.4 to 2.3%) of 1,6-linked glucosyl residues (Fig. 3B). With the exception of A. euteiches and the Peronosporales and in addition to various amounts of the above-mentioned glucosyl linkages, all other AIF samples analyzed contained 1 to 1.5% 1,3,4-linked glucose, suggesting the occurrence of residues cross-linking 1,3-glucans and cellulose.
Analyses of the glucan content of the ASF samples revealed that 1,3-glucans are the most abundant alkali-soluble glucans in all species (Fig. 4). However, as opposed to Saprolegniales, Peronosporales were characterized by a lower proportion of 1,3-, 1,6-, and 1,4-linked glucosyl residues but a higher content of 1,3,6-linked glucose (Fig. 4B). This suggests the occurrence of a higher number of branching points at position 6 in the 1,3-glucans of the Peronosporales. Furthermore, as opposed to the AIF, the glucans in the Peronosporales ASF were devoid of 1,3,4-linked glucosyl residues. The ASFs from all species analyzed contained 1,4-linked glucose, but the proportion of these residues was 48 to 80% lower in the Peronosporales (Fig. 4B).
Interestingly, the ASFs from the 4 species that belong to the Saprolegnia genus (S. anisospora, S. ferax, S. parasitica, and S. monoica) exhibited a higher proportion of 1,6-linked glucosyl residues (2- to 9-fold increase) than the other Saprolegniales species and the Peronosporales (Fig. 4B). Apart from S. monoica, the other 3 members of the Saprolegnia genus were characterized by a lower content of 1,3-linked glucosyl residues than the other Saprolegniales species (Fig. 4B).
Peronosporales species exhibit a specific alkali-soluble carbohydrate profile.
A clear distinction between Saprolegniales and Peronosporales is the composition of their ASF. Indeed, in addition to a markedly lower glucan content, the Peronosporales ASF samples were characterized by a much higher proportion of Man (4- to 20-fold increase) than their Saprolegniales counterpart (Fig. 4A). In the ASF samples from all species, the 1,4-linked Man residues were quantitatively dominant, but only slightly more dominant than the t-Man residues (Fig. 4B). This indicates that the chain length of the oomycete mannans is no longer than a few residues. In some species (A. euteiches, S. ferax, and S. parasitica), only traces of 1,2-linked Man were detected (Fig. 4B). All species also contained minor amounts of Xyl and Gal residues (Fig. 4A). Due to the low abundance of these monosaccharides, only 1,4-linked Xyl (not shown) and 1,3-linked Gal (Fig. 4B) could be detected and unequivocally identified during methylation analysis.
Another distinguishing feature of the Peronosporales ASF samples was the occurrence of glucuronic acid (GlcA). While the Saprolegniales cell walls were devoid of such residues, the Peronosporales contained up to 15.7% GlcA (Fig. 4A). However, the type of glycosidic bond linking GlcA could not be determined because the corresponding carbohydrate chains are typically degraded by β elimination in the type of alkali-based extraction and methylation procedures that we used (29).
DISCUSSION
Pioneering work on oomycetes revealed that the cell walls of this class of microorganisms consist essentially of 1,3-β- and 1,6-β-glucans (7–9). As opposed to true fungi, whose cell walls are devoid of cellulose, oomycetes have historically been described as containing a rather low proportion of cellulose (4 to 20%) but no chitin (6–8, 30). It is only in later work that some species have been shown to contain minor amounts of chitin (<0.5%) or up to 10% of other GlcNAc-based carbohydrates (16, 31, 32). From these data, a general concept that all oomycete cell walls are similar has emerged. It should be kept in mind, however, that the information available is restricted to a limited number of species and based in many instances on data obtained from global or superficial analytical methods with rather poor resolution. For this reason, even in the case of Phytophthora species, which have been the most studied (6, 33, 34), knowledge on cell wall composition is still elusive. There is therefore a need to revisit the composition of the cell walls of the most studied species and to extend these investigations to a larger group of microorganisms to identify order- or species-specific structural features. This knowledge can be exploited for uncovering new enzymes involved in the biosynthesis of specific cell wall carbohydrates and the formation of stabilizing cross-links between structural polymers. With this information, new molecular approaches targeting cell wall stability may be devised for disease control.
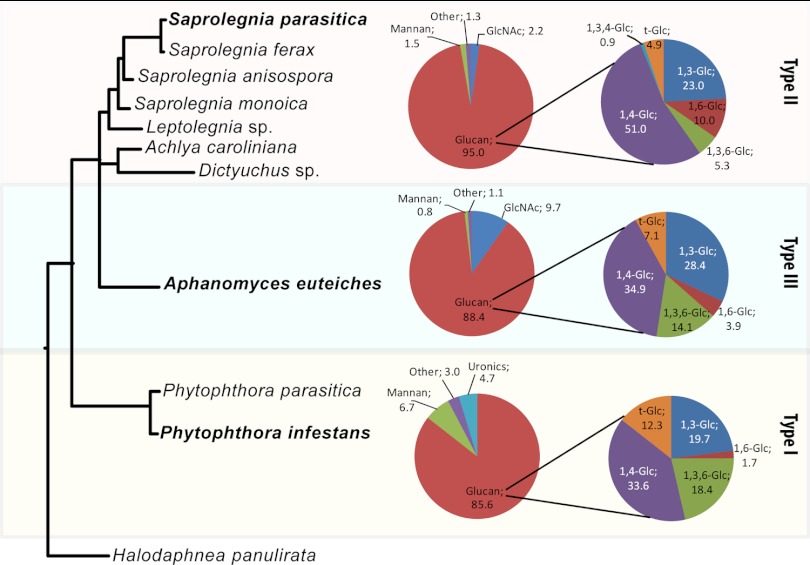
The primary objective of our work was to gain further insight into the cell wall structure of important oomycetes, including some vertebrate and plant pathogens. For this purpose, we have undertaken carbohydrate analyses on the mycelia of 10 selected representative species belonging to the Peronosporales and Saprolegniales orders. This has allowed us to (i) demonstrate the existence of so far unknown linkages and carbohydrates in the walls of some oomycete species only, (ii) reveal the occurrence of order-specific monosaccharide units in oomycete cell walls, (iii) uncover the existence of several types of GlcNAc-based carbohydrates in the walls of Saprolegniales, and (iv) determine precisely the relative abundance of the different types of β-glucans that are typically considered most abundant in oomycete cell walls but whose detailed analysis has so far remained elusive. A synthesis of these data has allowed us to unveil the existence of 3 clearly distinguishable cell wall types and propose a new paradigm for cell wall composition and structure in oomycetes. Our new cell wall classification is primarily based on GlcNAc content: type I corresponds to species devoid of GlcNAc, type II cell walls contain up to 5% GlcNAc, and type III cell walls are characterized by more than 5% GlcNAc. In addition to this primary criterion, each cell wall type can be further distinguished from the others by several additional compositional features, as summarized in Fig. 7 and Table 2. While Fig. 3 and 4 represent the carbohydrate compositions of the AIF and ASF samples, respectively, the summary presented in Fig. 7 corresponds to the carbohydrate content of the whole cell walls; i.e., the values given have been corrected by taking into account the fact that the AIF and ASF samples from each species represented, on average, 70% and 30% of the total cell walls, respectively. The following paragraphs discuss the properties of the different cell wall types on the basis of the synthetic data in Fig. 7 and Table 2.
Fig 7.
Typical carbohydrate composition of the 3 different cell wall types identified among 10 representative oomycete species. Values (mol%) from a species representative of each cell wall type (bold font) were calculated by taking into account the relative proportion of the AIF (∼70%) and ASF (∼30%) samples from each species. The precise phylogenetic distances between species are shown in Fig. 2.
Table 2.
Distinguishing criteria of the 3 major oomycete cell wall types a
| Cell wall type (representative species) | Carbohydrate content (mol%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Primary criterion, GlcNAc | Secondary criteria |
Tertiary criteria |
|||||||
| 1,6-GlcNAc | GalA | Man | 1,4-Glc | t-Glc | 1,6-Glc | 1,3,4-Glc | 1,3,6-Glc | ||
| I (P. infestans) | − | − | + | >3 | <40 | >10 | <5 | − | >10 |
| II (S. parasitica) | <5 | − | − | <3 | >40 | <6 | >5 | + | <10 |
| III (A. euteiches) | >5 | + | − | <3 | <40 | <10 | <5 | − | >10 |
The data correspond to the carbohydrate content of whole cell walls. The values have been corrected by taking into account the relative proportion of the AIF (∼70%) and ASF (∼30%) samples from each species. +, detected; −, not detected.
Species from the Peronosporales order are representative of type I cell walls, as exemplified here with P. infestans and P. parasitica. In addition to being devoid of GlcNAc, type I cell walls can be distinguished from types II and III by the occurrence of GlcA and a higher Man content (>3% of the total cell wall composition) in alkali-soluble carbohydrates. Man has previously been reported in the cell wall of P. parasitica and Phytophthora cinnamomi, but the amount reported did not exceed 0.6% of the cell walls (33). It is noteworthy that in the latter case the alkali-soluble fraction was discarded. Therefore, the mannose content reported was underestimated compared to that in our work.
Glucosamine was reported in the cell walls of P. parasitica and P. cinnamomi (33). However, since proteins were not removed prior to cell wall analysis, it was suggested that glucosamine most likely arose from proteins (33). This is further supported by our own data, in which no glucosamine was detected in our protein-depleted cell walls. Interestingly, analysis of the Phytophthora infestans genome has revealed the existence of a putative chitin synthase gene in this species (13, 35). Since the cell walls of Phytophthora species are devoid of chitin and GlcNAc-based polysaccharides, as confirmed in our work, the function of this gene remains elusive. It is possible that it is involved in processes other than chitin biosynthesis while still encoding an N-acetylglucosaminyltransferase. Possible functions consistent with this hypothesis are the biosynthesis of glycan moieties of glycoproteins or the formation of glycosylphosphatidylinositol (GPI) anchors in GPI proteins, as both processes require an N-acetylglucosaminyltransferase activity. Further experimental work is being developed in our laboratory to determine the actual function of the Phytophthora chitin synthase gene.
The total amount of cellulose in type I cell walls is about 32 to 35%, assuming that all 1,4-linked glucosyl residues quantified arose from cellulose (as specified in the Materials and Methods section, no 1,4-α-linked glucan corresponding to glycogen-like polysaccharides was detected). This cellulose content is significantly higher than the 15% typically reported in other Phytophthora species (7). This discrepancy is most likely due to the use of different approaches for cellulose quantification. Indeed, as opposed to the approaches used by others, which preferentially measure crystalline cellulose only, our analyses based on GC/MS do not distinguish between crystalline and noncrystalline forms of cellulose. Since cellulose from Saprolegniales is known to be poorly crystalline (16), the 15% cellulose content reported in the literature is underestimated compared to that from our analyses.
Type II cell walls are represented by members of the Saprolegniales orders, including the genera Achlya, Dictyuchus, Leptolegnia, and Saprolegnia. These cell walls contain less than 5% GlcNAc, which was shown to correspond to α-chitin according to previous work performed in S. monoica (16). A unique feature of type II cell walls is the presence of 1,3,4-linked glucosyl residues in the alkali-insoluble fraction, which is indicative of the occurrence of cross-links between cellulose and 1,3-β-glucans. To our knowledge, this type of cross-link has never been reported in any other organism. In addition, type II cell walls can be distinguished from type I cell walls by the occurrence of a higher proportion of glucans, namely, cellulose, 1,6-glucans, and, to a lesser extent, 1,3-glucans (∼10% altogether). Interestingly, since significantly smaller contents of t-Glc and 1,3,6-linked glucosyl residues were measured in the AIFs of type II walls, it can be inferred that some glucans in these fractions exhibit a higher degree of polymerization and a lower number of branches in the 6 position than the alkali-insoluble glucans from type I cell walls. As opposed to the reports available in the literature, which merely mention and discuss glucan contents globally (30, 36–39), we provide here the first comprehensive quantitative analysis of fractionated carbohydrates from type II cell walls, including cellulose and other glucans.
Type II and type III cell walls can primarily be distinguished on the basis of their GlcNAc content, which is 2 to 4 times higher in type III cell walls. The Aphanomyces genus is representative of the latter cell wall type, as exemplified here with A. euteiches, which contains nearly 10% GlcNAc. Although a similar GlcNAc content has previously been reported in this species (32), our work is the first to reveal the unique structural features of this newly defined cell wall type. A specificity of type III cell walls is the occurrence of carbohydrates that consist of 1,6-linked GlcNAc residues in both the ASF and AIF samples. This unusual type of polysaccharide has never been reported in any eukaryotic microorganism. It is only in extracellular biofilms produced by the bacterial genera Escherichia and Staphylococcus that some 1,6-linked GlcNAc residues have been described to be part of linear homopolysaccharides (40, 41). The fine structure of the GlcNAc-based carbohydrates in the type III cell walls remains to be determined. However, our data suggest the occurrence of both 1,6-GlcNAc-based saccharides and 1,4-linked polymers of GlcNAc that most likely correspond to chitin of a low degree of polymerization. This somewhat contradicts the earlier conclusion that A. euteiches cell walls are devoid of chitin. Indeed, in the report from Badreddine et al. (32), it was proposed that the cell wall GlcNAc residues in A. euteiches arise from soluble carbohydrates that may be covalently linked to glucans rather than from free crystalline chitin.
Of all cell wall types, type III contains the highest proportion of 1,3-glucans. On the basis of the 1,3,6-linked Glc content, these polysaccharides exhibit a similar degree of 1,6 branching as the 1,3-glucans from type I cell walls but a higher number of 1,6 branches than type II cell walls. In addition, as for type I cell walls, type III cell walls contain less cellulose and 1,6-glucans than type II cell walls.
Oomycetes are traditionally described as exhibiting the same cell wall composition, structure, and organization, regardless of the order and species considered. The salient finding of our study is the existence of clearly distinguishable types of cell walls within this class of eukaryotic microorganisms. The 3 types of oomycete cell walls defined here can be used as a complement to the morphological criteria and molecular markers typically used for distinguishing major taxonomic groups. This chemical approach has allowed the discrimination between groups of species from the Saprolegniales and Peronosporales orders, complementing the data from molecular approaches, such as phylogenetic analysis (Fig. 2). It is expectable that further cell wall carbohydrate analyses on other oomycete orders would uncover variations (subtypes) of the 3 main types of cell walls described here, instead of leading to the identification of completely new cell wall types. Indeed, although the same types of major carbohydrates (glucans) are expected across different orders, specific novel structures may be identified as more detailed data become available. This is illustrated by our discovery of two new types of glycosidic linkages that (i) suggest the existence of cross-links between cellulose and 1,3-glucans in type II cell walls and (ii) show the occurrence of polymers of 1,6-GlcNAc in type III cell walls. As opposed to all reports available in the literature, our work shows that cellulose is not a minor cell wall component in oomycetes but represents at least 30% of the total cell wall carbohydrates.
At this stage, it is not possible to relate genes that encode putative biosynthetic enzymes to carbohydrate structures, as the function of most glycosyltransferases identifiable through genome analysis has not been demonstrated. We are currently addressing this question by combining genome and proteome analyses in Phytophthora and S. parasitica with the biochemical functional characterization of putative carbohydrate synthases. In conclusion, the new structural knowledge presented here, together with the identification and characterization of cell wall polysaccharide biosynthetic enzymes corresponding to newly uncovered glycosidic linkages, paves the way for the future development of disease control measures targeted to the cell walls of some of the most devastating oomycete pathogens.
ACKNOWLEDGMENTS
This work was supported by grants to V.B. from the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS) (2009-515 and 2010-1807), to V.B. and J.D.-U. from the European Union (ITN-SAPRO-238550), and to J.D.-U. from the Spanish Ministry of Science and Innovation (CGL2009-10032).
We are grateful to A. Bottin (University of Toulouse, Toulouse, France) for providing the A. euteiches and P. parasitica strains and to Francisco Vilaplana (KTH, Sweden) for technical assistance with the HPAEC-PAD analysis of uronic acids.
Footnotes
Published ahead of print 30 November 2012
REFERENCES
- 1. Kamoun S. 2001. Nonhost resistance to Phytophthora: novel prospects for a classical problem. Curr. Opin. Plant Biol. 4:295–300 [DOI] [PubMed] [Google Scholar]
- 2. Phillips AJ, Anderson VL, Robertson EJ, Secombes CJ, van West P. 2008. New insights into animal pathogenic oomycetes. Trends Microbiol. 16:13–19 [DOI] [PubMed] [Google Scholar]
- 3. Haverkort AJ, Boonekamp PM, Hutten R, Jacobsen E, Lotz LAP, Kessel GJT. 2008. Societal costs of late blight in potato and prospects of durable resistance through cisgenic modification. Potato Res. 51:47–57 [Google Scholar]
- 4. van West P. 2006. Saprolegnia parasitica, an oomycete pathogen with a fishy appetite: new challenges for an old problem. Mycologist 20:99–104 [Google Scholar]
- 5. Beakes GW, Glockling SL, Sekimoto S. 2012. The evolutionary phylogeny of the oomycete “fungi.” Protoplasma 249:3–19 [DOI] [PubMed] [Google Scholar]
- 6. Aronson JM. 1965. The Fungi, p 49–76 In Ainsworth GC, Sussman AS. (ed). Academic Press, New York, NY [Google Scholar]
- 7. Aronson JM, Barbara A, Cooper BA, Fuller MS. 1967. Glucans of oomycete cell walls. Science 155:332–335 [DOI] [PubMed] [Google Scholar]
- 8. Bartnicki-García S. 1968. Cell wall chemistry, morphogenesis, and taxonomy of fungi. Annu. Rev. Microbiol. 22:87–108 [DOI] [PubMed] [Google Scholar]
- 9. Wessels JGH, Sietsma JH. 1981. Fungal cell walls: a survey, p 352–394 In Tanner W, Loewus FA. (ed), Encyclopedia of plant physiology. Plant carbohydrates II. Springer-Verlag, Berlin, Germany [Google Scholar]
- 10. Pfaller MA, Boyken L, Hollis RJ, Kroeger J, Messer SA, Tendolkar S, Diekema DJ. 2008. In vitro susceptibility of invasive isolates of Candida spp. to anidulafungin, caspofungin, and micafungin: six years of global surveillance. J. Clin. Microbiol. 46:150–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guerriero G, Avino M, Zhou Q, Fugelstad J, Clergeot PH, Bulone V. 2010. Chitin synthases from Saprolegnia are involved in tip growth and represent a potential target for anti-oomycete drugs. PLoS Pathog. 6:e1001070 doi:10.1371/journal.ppat.1001070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL, Chapman J, Damasceno CM, Dorrance AE, Dou D, Dickerman AW, Dubchak IL, Garbelotto M, Gijzen M, Gordon SG, Govers F, Grunwald NJ, Huang W, Ivors KL, Jones RW, Kamoun S, Krampis K, Lamour KH, Lee MK, McDonald WH, Medina M, Meijer HJ, Nordberg EK, Maclean DJ, Ospina-Giraldo MD, Morris PF, Phuntumart V, Putnam NH, Rash S, Rose JK, Sakihama Y, Salamov AA, Savidor A, Scheuring CF, Smith BM, Sobral BW, Terry A, Torto-Alalibo TA, Win J, Xu Z, Zhang H, Grigoriev IV, Rokhsar DS, Boore JL. 2006. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313:1261–1266 [DOI] [PubMed] [Google Scholar]
- 13. Haas BJ, Kamoun S, Zody MC, Jiang RH, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T, Bozkurt TO, Ah-Fong AM, Alvarado L, Anderson VL, Armstrong MR, Avrova A, Baxter L, Beynon J, Boevink PC, Bollmann SR, Bos JI, Bulone V, Cai G, Cakir C, Carrington JC, Chawner M, Conti L, Costanzo S, Ewan R, Fahlgren N, Fischbach MA, Fugelstad J, Gilroy EM, Gnerre S, Green PJ, Grenville-Briggs LJ, Griffith J, Grünwald NJ, Horn K, Horner NR, Hu CH, Huitema E, Jeong DH, Jones AM, Jones JD, Jones RW, Karlsson EK, Kunjeti SG, Lamour K, Liu Z, MA L, Maclean D, Chibucos MC, McDonald H, McWalters J, Meijer HJ, Morgan W, Morris PF, Munro CA, O'Neill K, Ospina-Giraldo M, Pinzón A, Pritchard L, Ramsahoye B, Ren Q, Restrepo S, Roy S, Sadanandom A, Savidor A, Schornack S, Schwartz DC, Schumann UD, Schwessinger B, Seyer L, Sharpe T, Silvar C, Song J, Studholme DJ, Sykes S, Thines M, van de Vondervoort PJ, Phuntumart V, Wawra S, Weide R, Win J, Young C, Zhou S, Fry W, Meyers BC, van West P, Ristaino J, Govers F, Birch PR, Whisson SC, Judelson HS, Nusbaum C. 2009. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461:393–398 [DOI] [PubMed] [Google Scholar]
- 14. Lévesque CA, Brouwer H, Cano L, Hamilton JP, Holt C, Huitema E, Raffaele S, Robideau GP, Thines M, Win J, Zerillo MM, Beakes GW, Boore JL, Busam D, Dumas B, Ferriera S, Fuerstenberg SI, Gachon CM, Gaulin E, Govers F, Grenville-Briggs L, Horner N, Hostetler J, Jiang RH, Johnson J, Krajaejun T, Lin H, Meijer HJ, Moore B, Morris P, Phuntmart V, Puiu D, Shetty J, Stajich JE, Tripathy S, Wawra S, van West P, Whitty BR, Coutinho PM, Henrissat B, Martin F, Thomas PD, Tyler BM, De Vries RP, Kamoun S, Yandell M, Tisserat N, Buell CR. 2010. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 11:R73 doi:10.1186/gb-2010-11-7-r73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Machlis L. 1953. Growth and nutrition of water molds in the subgenus Euallomyces. II. Optimal composition of the minimal medium. Am. J. Bot. 40:449–460 [Google Scholar]
- 16. Bulone V, Chanzy H, Gay L, Girard V, Fèvre M. 1992. Characterization of chitin and chitin synthase from the cellulosic cell wall fungus Saprolegnia monoica. Exp. Mycol. 16:8–21 [Google Scholar]
- 17. Diéguez-Uribeondo J, Fregeneda-Grandes JM, Cerenius L, Pérez-Iniesta E, Aller-Gancedo JM, Tellería MT, Söderhäll K, Martín MP. 2007. Re-evaluation of the enigmatic species complex Saprolegnia diclina-Saprolegnia parasitica based on morphological, physiological and molecular data. Fungal Genet. Biol. 44:585–601 [DOI] [PubMed] [Google Scholar]
- 18. White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322 In Innis MA, Gelfand DH, Sninsky JJ, White TJ. (ed), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA [Google Scholar]
- 19. Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT, p 39–64 In Posada D. (ed), Bioinformatics for DNA sequence analysis. Humana Press, Hatfield, United Kingdom: [DOI] [PubMed] [Google Scholar]
- 21. Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61:539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256 [DOI] [PubMed] [Google Scholar]
- 24. Mélida H, García-Angulo P, Alonso-Simón A, Encina A, Alvarez J, Acebes JL. 2009. Novel type II cell wall architecture in dichlobenil-habituated maize calluses. Planta 229:617–631 [DOI] [PubMed] [Google Scholar]
- 25. Albersheim P, Nevins PD, English PD, Karr A. 1967. A method for the analysis of sugars in plant cell wall polysaccharides by gas liquid chromatography. Carbohydr. Res. 5:340–345 [Google Scholar]
- 26. Ciucanu I, Kerek F. 1984. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131:209–217 [Google Scholar]
- 27. Carpita NC, Shea EM. 1989. Linkage structure of carbohydrates by gas chromatography-mass spectrometry (GCMS) of partially methylated alditol acetates, p 157–216 In Biermann CJ, McGinnis GD. (ed), Analysis of carbohydrates by GLC and MS. CRC Press, Boca Raton, FL [Google Scholar]
- 28. Schwarzmann GOH, Jeanloz RW. 1974. Separation by gas-liquid chromatography, and identification by mass spectrometry, of the methyl ethers of 2-deoxy-2-(N-methylacetamido)-d-glucose. Carbohydr. Res. 34:161–168 [Google Scholar]
- 29. Björndal H, Hellerqvist CG, Lindberg B, Svensson S. 1970. Gas-liquid chromatography and mass spectrometry in methylation analysis of polysaccharides Angew. Chem. Int. Ed. Engl. 9:610–619 [Google Scholar]
- 30. Novaes-Ledieu M, Jiménez-Martínez A, Villanueva JR. 1967. Chemical composition of hyphal wall of Phycomycetes. J. Gen. Microbiol. 47:237–245 [DOI] [PubMed] [Google Scholar]
- 31. Dietrich SMC. 1973. Carbohydrates from the hyphal wall of some oomycetes. Biochim. Biophys. Acta 313:95–98 [DOI] [PubMed] [Google Scholar]
- 32. Badreddine I, Lafitte C, Heux L, Skandalis N, Spanou Z, Martinez Y, Esquerré-Tugayé MT, Bulone V, Dumas B, Bottin A. 2008. Cell wall chitosaccharides are essential components and exposed patterns of the phytopathogenic oomycete Aphanomyces euteiches. Eukaryot. Cell 7:1980–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bartnicki-García S. 1966. Chemistry of hyphal walls of Phytophthora. J. Gen. Microbiol. 42:57–69 [DOI] [PubMed] [Google Scholar]
- 34. Tokunaga J, Bartnicki-García S. 1971. Structure and differentiation of the cell wall of Phytophthora palmivora: cysts, hyphae and sporangia. Arch. Mikrobiol. 79:293–310 [DOI] [PubMed] [Google Scholar]
- 35. Ospina-Giraldo MD, Griffith JG, Laird EW, Mingora C. 2010. The CAZyome of Phytophthora spp.: a comprehensive analysis of the gene complement coding for carbohydrate-active enzymes in species of the genus Phytophthora. BMC Genomics 11:525 doi:10.1186/1471-2164-11-525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Crook EM, Johnston IR. 1962. The qualitative analysis of the cell walls of selected species of fungi. Biochem. J. 83:325–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parker BC, Preston RD, Fogg GE. 1963. Studies of the structure and chemical composition of the cell walls of Vaucheriaceae and Saprolegniaceae. Proc. R. Soc. Lond. 158:435–445 [Google Scholar]
- 38. Sietsma JH, Eveleigh DE, Haskins RH. 1969. Cell wall composition and protoplast formation of some Oomycete species. Biochim. Biophys. Acta 184:306–317 [DOI] [PubMed] [Google Scholar]
- 39. Cameron DS, Taylor IE. 1976. Quantitative microanalysis of cell walls of Saprolegnia diclina Humphrey and Tremella mesenterica Fries. Biochim. Biophys. Acta 444:212–222 [DOI] [PubMed] [Google Scholar]
- 40. Götz F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367–1378 [DOI] [PubMed] [Google Scholar]
- 41. Itoh Y, Wang X, Hinnebusch BJ, Preston JF, III, Romeo T. 2005. Depolymerization of β-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 187:382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]