Abstract
Biofilm formation is associated with the ability of Candida albicans, the major human fungal pathogen, to resist antifungal therapies and grow on tissues, catheters, and medical devices. In order to better understand the relationship between C. albicans morphology and biofilm formation, we examined biofilms generated in response to expression of UME6, a key filament-specific transcriptional regulator. As UME6 levels rise, C. albicans cells are known to transition from yeast to hyphae, and we also observed a corresponding increase in the level of biofilm formation in vitro. In addition to forming a biofilm, we observed that a C. albicans strain expressing constitutive high levels of UME6 promoted tissue invasion in a reconstituted human three-dimensional model of oropharyngeal candidiasis. Confocal microscopy indicated that both the top and bottom layers of the biofilm generated upon high-level constitutive UME6 expression consist primarily of hyphal cells. UME6-driven biofilm formation was reduced upon deletion of Hgc1, a cyclin-related protein important for hyphal development, as well as Sun41, a putative cell wall glycosidase. Constitutive high-level UME6 expression was also able to completely bypass both the filamentation and biofilm defects of a strain deleted for Efg1, a key transcriptional regulator of these processes. Finally, we show that both Sun41 and Efg1 affect the ability of UME6 to induce certain filament-specific transcripts. Overall, these findings indicate a strong correlation between increased C. albicans hyphal growth and enhanced biofilm formation and also suggest functional relationships between UME6 and other regulators of biofilm development.
INTRODUCTION
Candida albicans is a major human fungal pathogen responsible for a wide variety of both systemic and mucosal infections (1). Candida species are the fourth leading cause of nosocomial infections in the United States, with a mortality rate of 30 to 50% (2–5). Immunocompromised individuals, such as AIDS patients, cancer patients on chemotherapy, and organ transplant recipients, are especially susceptible to infections (6–10). Many of these infections are associated with the formation of biofilms on tissues, catheters, and implanted medical devices. C. albicans biofilms are known to provide protection from host immune defenses and are also extremely resistant to antifungal treatments. In addition, these biofilms can serve as reservoirs for infection, since cells dispersed from the biofilm can traverse the bloodstream and eventually establish secondary sites of infection (11–14).
In C. albicans, biofilm formation involves adhesion of single cells to a surface (biotic or abiotic), proliferation, hyphal development, and generation of exopolymeric material (14–18). A variety of adhesins, including the glycosylphosphatidylinositol (GPI)-linked cell wall protein Eap1 and the agglutinin-like protein Als1, appear to play important roles in the initial attachment of C. albicans cells to surfaces (16, 19–21). Als3 and Hwp1, a mammalian transglutaminase substrate mimic, also function as complementary adhesins and are most likely involved in cell-cell and cell-surface interactions of hyphae in biofilms (16, 22–24). Several additional cell surface proteins have been implicated in C. albicans biofilm formation, the most notable of which is Sun41, a putative glycosidase. sun41Δ/Δ mutants have defects in cytokinesis, cell wall biogenesis, and adhesion to host cells and are highly attenuated for virulence in mouse models of disseminated and oropharyngeal candidiasis (25–27). Most of these phenotypes are believed to be attributed to a cell wall defect since the sun41Δ/Δ mutant is very sensitive to cell wall inhibitors, such as Congo red, and shows altered expression of several cell wall damage repair genes (25–27). Interestingly, this mutant shows defective hypha formation on solid medium, and one group reported that the sun41Δ/Δ strain forms aberrant hyphae in liquid medium as well (25, 26). Although Sun41 has been extensively characterized at the phenotypic level, very little information is available to link this cell wall protein to known biofilm development pathways.
The ability to form hyphae appears to be particularly important for biofilm formation, since a strain genetically manipulated to grow exclusively in the yeast form is highly defective in generating biofilms, and a variety of C. albicans mutants defective for hypha formation also show biofilm defects (16, 28, 29). One such mutant strain bears a homozygous deletion of Efg1, a major transcriptional regulator of C. albicans filamentous growth (30). The efg1Δ/Δ mutant grows as elongated yeast cells under most conditions and is highly defective for biofilm formation in vitro (28). Recently, this mutant was also observed to be defective for biofilm formation in both rat catheter and denture in vivo models, and Efg1 was shown to function as a component of a master transcriptional network that controls C. albicans biofilm formation (31). Although many direct targets for Efg1 were identified by this analysis, few downstream genes have specifically been shown to be important for the ability of Efg1 to promote biofilm formation.
While a number of transcription factors have been identified which, like Efg1, are required to generate C. albicans biofilms, considerably less is known about regulators whose expression enhances biofilm formation. A recent screen of a C. albicans overexpression library identified four such regulators: GAT2, TEC1, CPH1, and UME6 (32). Our studies have focused on UME6, which encodes a key filament-specific transcriptional regulator of C. albicans hyphal development and virulence (33). UME6 is a downstream target of multiple filamentous growth signaling pathways, and we have previously shown that constitutive high-level expression of UME6 is sufficient to drive complete hypha formation in the absence of filament-inducing conditions (34, 35). Interestingly, as UME6 levels rise, cells sequentially transition from yeast cells to pseudohyphae to hyphae, and there is a corresponding increase in the number of filament-specific genes expressed as well as their levels of expression. ume6Δ/Δ mutants are attenuated for virulence, are defective for hyphal extension, and also show a defect in biofilm formation (33). In addition, we have demonstrated that a strain expressing constitutive high levels of UME6 generates a very filamentous biofilm and is highly defective for biofilm dispersion (36).
We have previously shown that UME6 drives hyphal development via transcriptional induction of HGC1, which encodes a cyclin-related protein (37). Hgc1 is known to form a cyclin/Cdk complex with Cdc28 kinase, which, in turn, is important for septin phosphorylation, inhibition of cell separation, and activation of the Cdc42 master polarity regulator (involved in septin ring organization, vesicle transport to the hyphal tip, and actin polymerization) (38–44). Expression of UME6 in an hgc1Δ/Δ mutant strain results in shorter filaments with constrictions at septal junctions (37). Although Hgc1 is directly involved in a variety of mechanisms important for driving C. albicans hyphal growth, a role for Hgc1 in biofilm formation has not yet been reported.
Because biofilms generated by our UME6 expression strain contain a significantly high proportion of hyphal cells (36), this strain provides a powerful strategy to determine the specific role(s) of hyphae in biofilm formation. Here, we demonstrate that increased UME6 expression is correlated with enhanced biofilm formation. In order to gain a better understanding of the molecular mechanism(s) involved in UME6-driven increased biofilm formation, we examine the roles of a key biofilm transcriptional regulator (Efg1), a cyclin-related protein specifically important for the physical process of hyphal development (Hgc1), and a putative cell wall glycosidase (Sun41) in this process. Our results suggest that important functional relationships among these different proteins play a significant role in the ability of C. albicans hyphae to promote biofilm formation.
MATERIALS AND METHODS
Strain and plasmid constructions.
Genotypes for all strains used in this study are shown in Table 1. The wild-type tetR control strain (PCY87) as well as tetO-UME6 (MBY38) and tetO-UME6 hgc1Δ/Δ (PCY50) strains were described previously (34, 37). In order to construct the tetO-UME6 efg1Δ/Δ strain (PCY21), primer pairs 1/2 and 3/4 (see Table S1 in the supplemental material for a list of all primers used in this study) were used to generate PCR products corresponding to the 5′ and 3′ flanking regions (just outside the open reading frame), respectively, of EFG1. The 5′ flank was digested with KpnI and XhoI, and the 3′ flank was digested with NotI and SacII. These fragments were then cloned stepwise into plasmid pSFS2 (46). The resulting construct was digested with KpnI and SacII to release an efg1Δ::SAT1 fragment, which was used to transform the tetO-UME6 strain (MBY38). Homozygous deletion mutants were generated by using the SAT flipper method (46), and whole-cell PCR was used to verify correct integration at the 5′ and 3′ disruption junctions as well as the absence of the open reading frame. A similar strategy was used to construct the tetO-UME6 sun41Δ/Δ strain (MBY179), using primer pairs 5/6 and 7/8 (see Table S1 in the supplemental material) to generate 5′ and 3′ flanking regions, respectively, by PCR. A different version of the tetO-UME6 strain (MBY208), along with a wild-type tetR control strain (JKC915), was used exclusively in the experiment involving the reconstituted three-dimensional model of the human oral mucosa. In order to generate MBY208, PCR fragments corresponding to positions −650 to −122 (relative to the UME6 start ATG) and positions −45 to +443 (relative to the UME6 start ATG) were obtained by using primers 29/30 and 31/32, respectively (see Table S1 in the supplemental material). The UME6 fragment at positions −650 to −122 was digested with KpnI and ApaI, and the UME6 fragment at positions −45 to +443 was digested with SacII and NcoI. These fragments were then cloned stepwise into plasmid pJK1000 (45). The resulting construct was digested with KpnI and NcoI to release an FLP-CaNAT1-tetO-UME6 fragment, which was used to transform tetR parent strain JKC915. Whole-cell PCR was used to confirm the integration of the tetO cassette at the UME6 locus.
Table 1.
Strains used in this study
| Straina | Genotype | Reference |
|---|---|---|
| PCY87 (WT) | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 rps1::URA3/RPS1 | 37 |
| MBY38 (tetO-UME6) | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 tetO-UME6::URA3/UME6 | 34 |
| PCY50 (tetO-UME6 hgc1Δ/Δ) | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 hgc1Δ::frt/hgc1Δ::SAT1 tetO-UME6::URA3/UME6 | 37 |
| PCY21 (tetO-UME6 efg1Δ/Δ) | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 efg1Δ::frt/efg1Δ::SAT1 tetO-UME6::URA3/UME6 | This study |
| MBY179 (tetO-UME6 sun41Δ/Δ) | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 sun41Δ::frt/sun41Δ::SAT1 tetO-UME6::URA3/UME6 | This study |
| JKC915 (WT) | HIS1/his1::FRT tetR | 45 |
| MBY208 (tetO-UME6) | HIS1/his1::FRT tetR UME6/FLP-CaNAT1 tetO-UME6 | This study |
WT, wild type.
Media and growth conditions.
Standard growth conditions for all strains included solid or liquid yeast extract-peptone-dextrose (YEPD) medium at 30°C (47) in the presence or absence of 20 μg/ml doxycycline (Dox) (Sigma-Aldrich, St. Louis, MO), unless otherwise indicated. Liquid cultures were grown overnight at 30°C and harvested at an optical density at 600 nm (OD600) of ∼1.0 for both differential interference contrast (DIC) microscopy and RNA preparation, as described previously (37). Cells for static biofilm formation assays were initially grown overnight in YEPD medium at 30°C in the presence of 20 μg/ml Dox. In these assays, either Lee's medium, pH 6.8, or minimal medium (yeast nitrogen base [YNB] without amino acids plus 2% dextrose) was used for biofilm formation, as indicated. Biofilms used for confocal scanning laser microscopy (CSLM) were formed by using YNB with amino acids plus 2% dextrose medium.
Biofilm development assays.
A standard 96-well assay was used to assess static biofilm formation, as described previously (48, 49). Briefly, cell suspensions grown overnight were washed twice in phosphate-buffered saline (PBS) and, based on OD600 readings, diluted to a concentration of 1 × 106 cells/ml. One hundred microliters of each diluted cell suspension was added to single wells of a 96-well polystyrene plate containing the indicated medium and incubated at 30°C for 24 h. Each well was washed twice with 200 μl of PBS, and the level of biofilm formation was determined by using a standard semiquantitative colorimetric 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) reduction assay, as reported previously (18, 48). Biofilms for CSLM were developed under static conditions at 37°C for 24 h in 6-well tissue culture-treated polystyrene plates.
RNA preparation and Northern analysis.
RNA extractions were performed by using a hot acid phenol protocol (50). Northern analysis was carried out and blot images were visualized as described previously (33). ACT1 and rRNA were used as loading controls. Primers used to generate probes for the Northern analysis are shown in Table S1 in the supplemental material.
Confocal scanning laser microscopy.
CSLM was performed by using biofilms stained for 1 h in the dark at 37°C with 25 μg/ml concanavalin A-Alexa Fluor 594 conjugate (catalog number C-11253; Molecular Probes, Eugene, OR), as described previously (29). Biofilms were visualized by CSLM using a Zeiss LSM 510 upright confocal microscope with a Zeiss Achroplan 40×, 0.8-W objective (excitation wavelength of 543 nm). Zeiss LSM Image Browser v.4.2 was used to assemble CSLM microscopy images.
Reconstituted three-dimensional model of the human oral mucosa.
The three-dimensional model of the human oral mucosa used in this study was described previously (51–54). This system is composed of gingival fibroblasts embedded in a biomatrix of collagen type I and overlaid by a multilayer of oral epithelial cells. To study C. albicans invasion in this system, the three-dimensional model of the oral mucosa was challenged with 1 × 105 C. albicans yeast cells in 100 μl of airlift medium (keratinocyte serum-free medium [KSFM] containing 5% fetal bovine serum, 1.88 mM CaCl2, and 0.025 mM dextrose) in the presence or absence of 20 μg/ml Dox (inoculum for the MBY208 tetO-UME6 strain was prepared from cells grown in the presence of 20 μg/ml Dox). Airlift medium with or without 20 μg/ml Dox was also added to the uninfected controls. At 24 h postinfection, the cultures were fixed with 10% formaldehyde–PBS and embedded in paraffin. Formalin-fixed paraffin-embedded sections (thickness, 5 μm) of three-dimensional oral mucosal cultures were stained with hematoxylin and eosin (H&E). Stained sections were visualized by using a Leica DM RB microscope connected to a digital camera.
RESULTS
Increased UME6 expression is correlated with enhanced C. albicans biofilm formation.
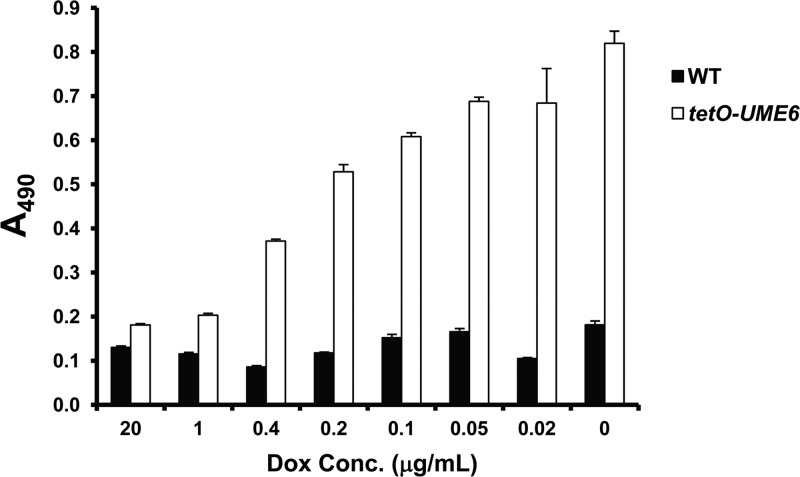
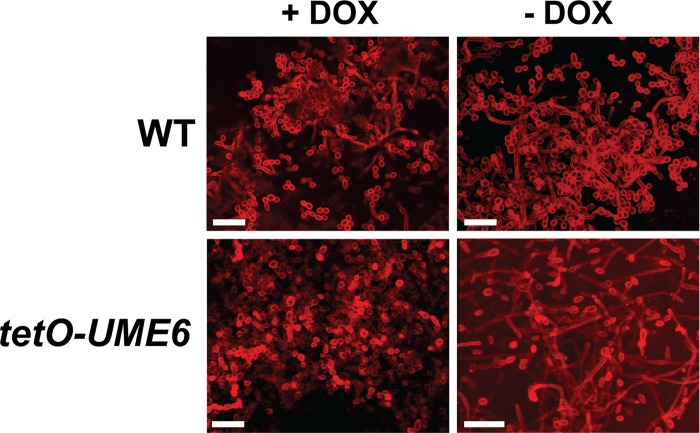
In order to determine the effect of the UME6 expression level on C. albicans biofilm formation, we used a strain in which one allele of UME6 is placed under the control of the Escherichia coli tet operator (tetO) (34). This strain also expresses constitutive levels of an E. coli tetR DNA-binding domain–Saccharomyces cerevisiae HAP4 activation domain fusion protein (55). In the absence of Dox, the tetR-HAP4 transactivator binds as a dimer to the tet operator and directs high levels of UME6 expression. In the presence of Dox, the transactivator no longer dimerizes, and the UME6 allele is not expressed. Using this system, we have previously shown that when UME6 is not expressed in the presence of 20 μg/ml Dox, cells grow as yeast. As the Dox concentration is reduced and UME6 levels rise, cells gradually transition from yeast to pseudohyphae to hyphae (34). A similar experiment was performed to monitor biofilm formation in vitro as UME6 levels increased. As demonstrated in Fig. 1, the tetO-UME6 strain showed a level of biofilm formation equivalent to that of the wild-type control strain (which expresses the tetR-HAP4 transactivator but lacks a tet operator) at 20 μg/ml Dox. However, as Dox levels decreased, there was a gradual increase in the level of biofilm formation by the tetO-UME6 strain, whereas the wild-type control strain showed nearly constant biofilm levels and was not affected by Dox. Confocal microscopy indicated that cells in the bottommost layer of the tetO-UME6 biofilm showed a significantly higher proportion of hyphae in the absence of Dox (−Dox) than in the presence of Dox (+Dox) and than cells of the wild-type control strain (+Dox or −Dox) (Fig. 2). Taken together, these results indicate a direct correlation between UME6 expression levels and C. albicans biofilm formation and suggest that increasing the number of hyphal cells in a biofilm can lead to significantly enhanced overall biofilm growth.
Fig 1.
UME6 expression level is correlated with increased biofilm formation. Suspensions of 1 × 106 cells/ml of the indicated strains were allowed to form biofilms on 96-well polystyrene plates in Lee's pH 6.8 medium at 30°C either in the absence of Dox or in the presence of the indicated Dox concentrations. Biofilm formation was assessed by using a standard colorimetric XTT reduction assay (18, 48). Error bars represent standard deviations (n = 4).
Fig 2.
UME6 expression promotes hyphal growth in C. albicans biofilms. CSLM was used to visualize cells in the bottommost layer of biofilms formed on 6-well polystyrene plates by the indicated strains in the presence or absence of 20 μg/ml Dox. C. albicans cells were stained with concanavalin A for 1 h in the dark at 37°C. Bar = 25 μm.
UME6 expression promotes tissue invasion in a reconstituted human model of oropharyngeal candidiasis.
In order to determine the effect of C. albicans UME6 expression on biofilm formation and tissue invasion of a host mucosal surface, we used a 3-dimensional model of the human oral mucosa (51–54). A total of 1 × 105 cells of both the tetO-UME6 and wild-type control strains were used to challenge the epithelial cell layer of this model in the presence or absence of Dox for a 24-h infection period. As indicated in Fig. 3, the level of biofilm formation appeared to be roughly equivalent on cell layers infected with the wild-type strain (+Dox and −Dox) and the tetO-UME6 strain in the presence of Dox. In the absence of Dox, the tetO-UME6 strain still formed a biofilm, and a significant proportion of C. albicans cells clearly appeared to invade the oral epithelium. Indeed, a large number of hyphal filaments completely penetrated the oral epithelium and reached the subepithelial layer of collagen-embedded fibroblasts. Increased invasion and penetration of the three-dimensional model of the human oral mucosa by the tetO-UME6 strain in the absence of Dox were also observed during a 36-h infection (data not shown). These results indicate that constitutive high-level UME6 expression alone is sufficient to promote tissue invasion in a reconstituted human model of oropharyngeal candidiasis and are consistent with our previous findings using a mouse systemic model (34).
Fig 3.
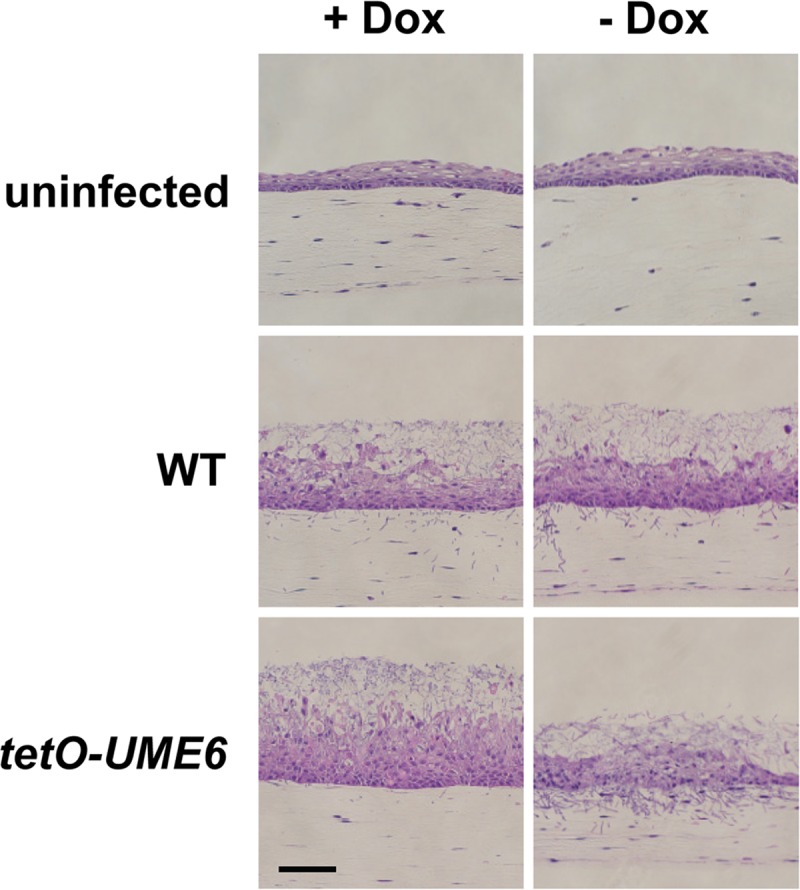
UME6 expression drives C. albicans tissue invasion in a reconstituted three-dimensional model of oropharyngeal candidiasis. A three-dimensional organotypic model of the human oral mucosa was infected with 1 × 105 cells of the indicated C. albicans strains in the presence or absence of 20 μg/ml Dox. Following a 24-h infection period, cultures were fixed in formaldehyde, embedded in paraffin, stained with H&E, and visualized by light microscopy. Bar = 100 μm.
Hgc1 and Sun41 are important for enhanced biofilm formation in response to UME6 expression.
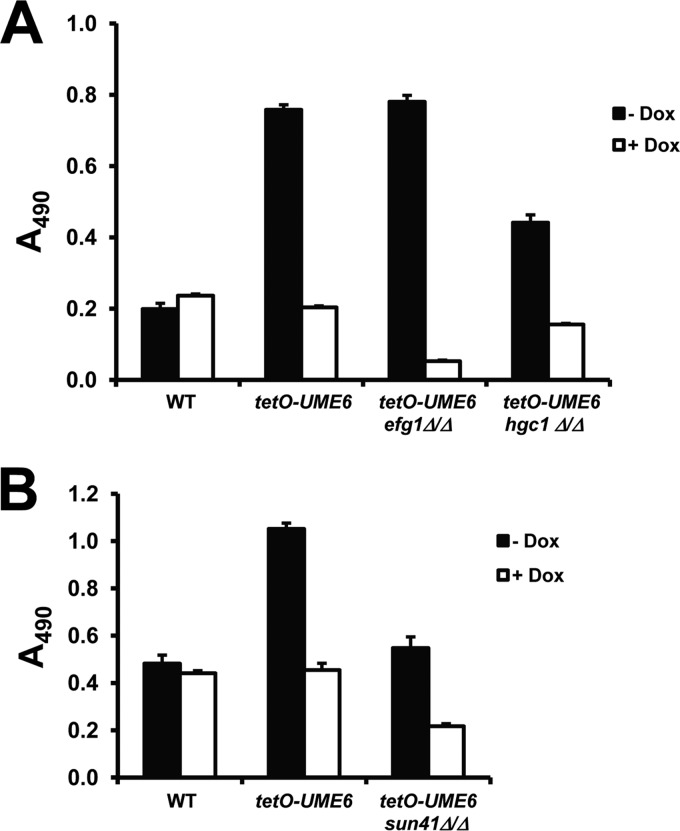
We next sought to determine the role of hypha-specific and cell wall components in mediating UME6-directed enhanced biofilm formation. Homozygous deletion mutations of Efg1 and Sun41 were generated in the tetO-UME6 strain background, and along with a previously generated tetO-hgc1Δ/Δ mutant (37), biofilm formation ability was assessed in the presence and absence of Dox. The tetO-UME6 efg1Δ/Δ strain showed a significant biofilm defect in the presence of Dox compared to the parent tetO-UME6 strain in the absence of Dox or the wild-type control strain in the presence or absence of Dox (Fig. 4A). However, the tetO-UME6 efg1Δ/Δ mutant continued to show enhanced biofilm formation, equivalent to that of the parent tetO-UME6 strain, upon high-level UME6 expression in the absence of Dox. This result indicates that expression of UME6 alone is sufficient to bypass the efg1Δ/Δ biofilm defect and suggests that UME6 functions downstream of the Efg1 regulator in C. albicans biofilm development. In the absence of Dox, the level of enhanced biofilm formation by the tetO-UME6 hgc1Δ/Δ mutant was clearly reduced, although not completely abolished, compared to that of the parent tetO-UME6 strain. In the presence of Dox, deletion of Hgc1 appeared to have very little, if any, effect on biofilm formation by the tetO-UME6 strain. Deletion of Sun41 caused an overall decrease in the level of tetO-UME6 biofilm formation (Fig. 4B). While the tetO-UME6 sun41Δ/Δ mutant still showed an increase in the level of biofilm formation in the absence versus the presence of Dox, the level of biofilm formation in the absence of Dox was equivalent to that of the wild-type control strain (+Dox or −Dox) and did not show an overall enhancement. As previously observed for the sun41Δ/Δ strain (26, 27), the tetO-UME6 sun41Δ/Δ mutant also showed a hypersensitivity to Congo red, strongly suggesting a cell wall defect (see Fig. S1 in the supplemental material). Taken together, these results indicate that both Hgc1 and Sun41 play important roles in the ability of UME6 expression to enhance overall C. albicans biofilm formation relative to that of a wild-type strain and suggest that both hypha-dependent and cell wall-dependent mechanisms are involved in this process.
Fig 4.
Hgc1 and Sun41 are important for the ability of UME6 expression to cause enhanced biofilm formation. Suspensions of 1 × 106 cells/ml of the indicated strains were allowed to form biofilms for 24 h on 96-well polystyrene plates in Lee's medium, pH 6.8, (A) or in minimal medium (B) at 30°C in the presence or absence of 20 μg/ml Dox. Biofilm formation was assessed by using a standard colorimetric XTT reduction assay (18, 48). Error bars represent standard deviations (n = 8).
Hgc1 and Sun41 are important for UME6-driven hyphal growth in biofilms.
In order to specifically examine the effect of efg1Δ/Δ, sun41Δ/Δ, and hgc1Δ/Δ mutations on UME6-driven hyphal growth within biofilms, we used confocal microscopy. Interestingly, as shown in Fig. 5, the top layer of the tetO-UME6 efg1Δ/Δ biofilm showed increased hypha formation compared to that of the parent tetO-UME6 strain upon UME6 expression in the absence of Dox. In the presence of Dox, when UME6 was not expressed, this mutant showed a mostly yeast biofilm, as expected. The tetO-UME6 sun41Δ/Δ biofilm showed reduced hypha formation in the absence of Dox and appeared to have a substrate adherence defect in the presence of Dox, as previously observed for the sun41Δ/Δ mutant (27). Also consistent with a previous report (25), we observed a reduction in the hyphal compartment length of the tetO-UME6 strain upon deletion of Sun41 when this strain was grown as a biofilm in either the presence or absence of Dox (see Fig. S2 in the supplemental material). While the tetO-UME6 hgc1Δ/Δ mutant biofilm still formed hyphae upon UME6 expression in the absence of Dox, these filaments were shorter and not as prevalent as those observed in the parent tetO-UME6 strain biofilm (Fig. 5).
Fig 5.
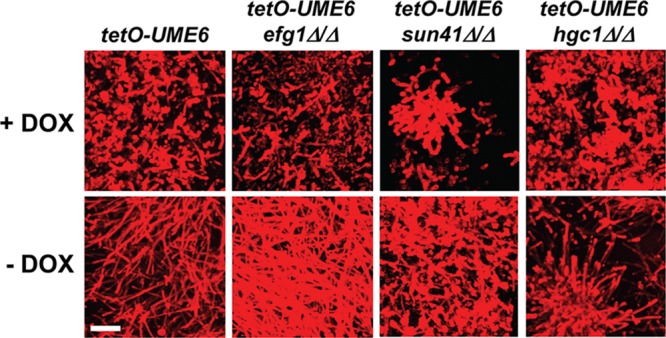
Hgc1 and Sun41 are important for UME6-driven hyphal growth in C. albicans biofilms. CSLM was used to visualize cells in the top layer of biofilms formed on 6-well polystyrene plates by the indicated strains in the presence or absence of 20 μg/ml Dox. C. albicans cells were stained with concanavalin A for 1 h in the dark at 37°C. Bar = 25 μm.
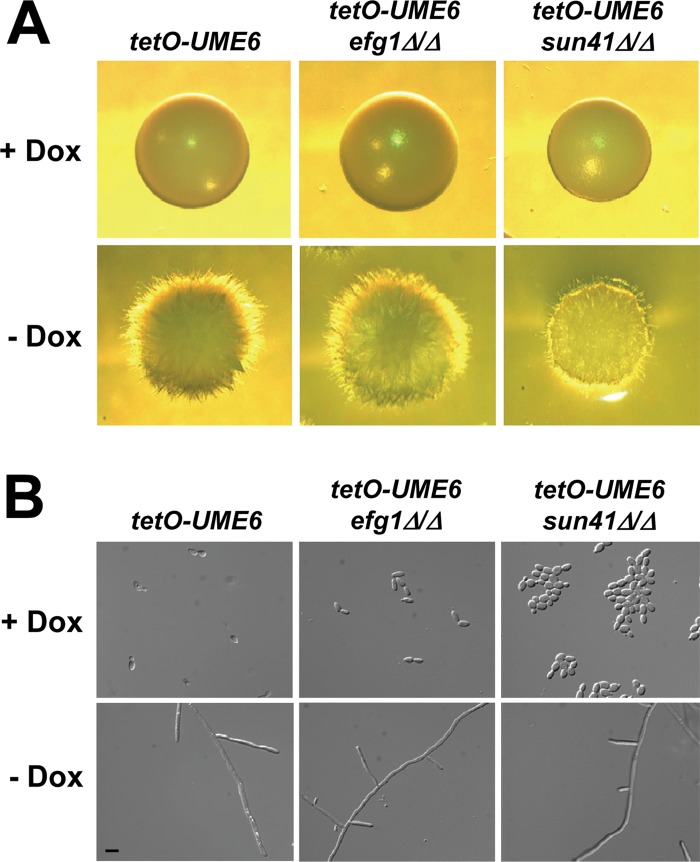
We also examined the effect of efg1Δ/Δ and sun41Δ/Δ mutations on UME6-driven filamentation under both solid and liquid non-filament-inducing conditions (we have previously shown that Hgc1 plays an important role in this process [37]). As indicated in Fig. 6A and B, constitutive high-level expression of UME6 was able to completely bypass the efg1Δ/Δ filamentation defect in either solid or liquid medium. Consistent with our biofilm results (Fig. 5), the tetO-UME6 sun41Δ/Δ mutant appeared to show a filamentation defect under solid growth conditions (Fig. 6A). Interestingly, however, under liquid growth conditions, this mutant appeared to show normal hyphal growth in response to UME6 expression in the absence of Dox (Fig. 6B), although we cannot exclude the possibility that the tetO-UME6 sun41Δ/Δ strain shows a reduction in the hyphal compartment length, as we have observed in biofilms (see Fig. S2 in the supplemental material) and as Firon et al. (25) observed previously in liquid medium. In the presence of Dox, this strain also showed a cell separation defect (Fig. 6B) characteristic of sun41Δ/Δ mutants growing in the yeast form (25, 26). Altogether, our results, combined with previous findings (37), indicate that Sun41 and Hgc1, but not Efg1, are important for UME6-driven filamentation in biofilms.
Fig 6.
Sun41 is important for UME6-driven filamentous growth under solid, non-filament-inducing conditions. (A) Colonies of the indicated strains were grown on solid YEPD medium at 30°C for 2 days in the presence or absence of 20 μg/ml Dox and visualized by light microscopy. (B) The indicated strains were grown in liquid YEPD medium at 30°C overnight to an OD600 of ∼1.0 in the presence or absence of 20 μg/ml Dox and visualized by using DIC microscopy. Bar = 10 μm.
Sun41 and Efg1 affect the ability of Ume6 to induce certain filament-specific transcripts.
Because Sun41 and Efg1 have different effects on UME6-driven hyphal growth, we next sought to determine the role that these proteins play in the ability of Ume6 to induce filament-specific transcripts (we have previously shown that an hgc1Δ/Δ mutation does not affect this process [37]). tetO-UME6 efg1Δ/Δ and tetO-UME6 sun41Δ/Δ mutants, as well as the tetO-UME6 parent strain, were grown under non-filament-inducing conditions in the presence and absence of Dox. Northern analysis was used to examine the levels of a variety of known filament-specific transcripts. As observed previously (34, 37), all of these transcripts were induced in the tetO-UME6 parent strain upon UME6 expression in the absence, but not the presence, of Dox (Fig. 7). Interestingly, five transcripts (HYR1, HWP1, RBT4, ECE1, and ALS3) showed significantly reduced expression levels in the tetO-UME6 efg1Δ/Δ mutant but significantly increased expression levels in the tetO-UME6 sun41Δ/Δ mutant. Levels of the RBT1 transcript showed a minor increase in the tetO-UME6 efg1Δ/Δ strain and a small decrease in the tetO-UME6 sun41Δ/Δ mutant. In contrast, induction of PHR1 and HGC1 by Ume6 did not appear to be significantly affected in the tetO-UME6 efg1Δ/Δ and tetO-UME6 sun41Δ/Δ mutant backgrounds. These results suggest, unexpectedly, that both Efg1 and Sun41 play important roles in the ability of Ume6 to induce certain filament-specific transcripts and that additional functional relationships may exist among these biofilm regulators.
Fig 7.
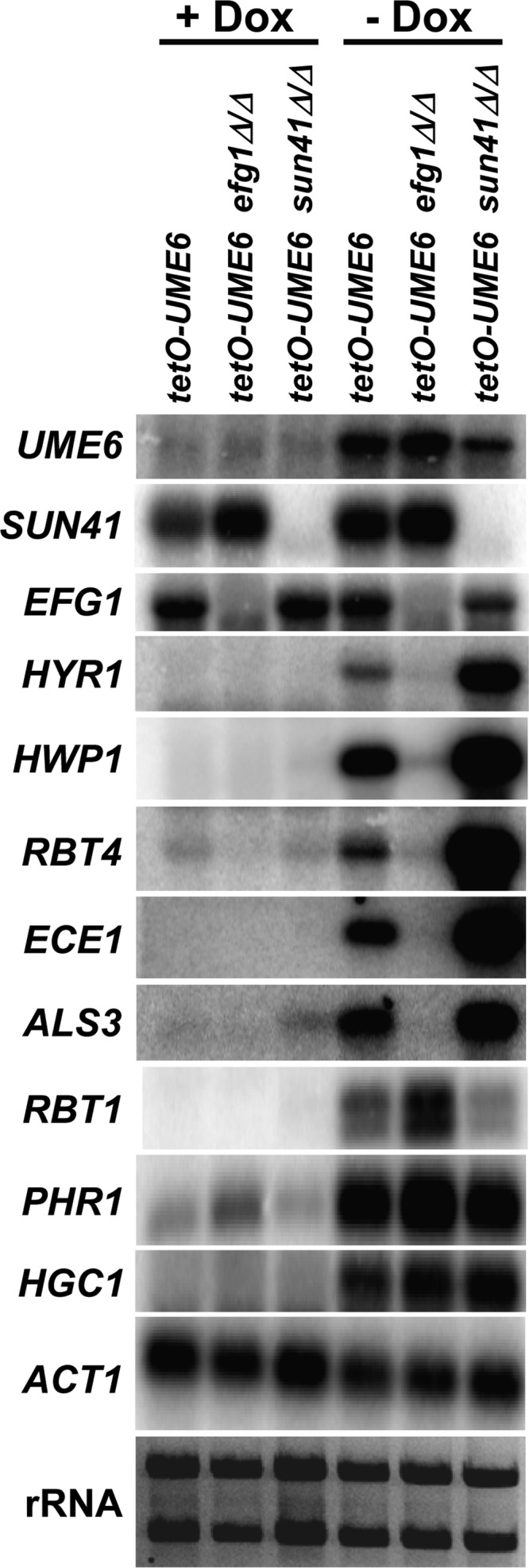
Efg1 and Sun41 differentially affect the ability of UME6 to induce certain filament-specific transcripts. The indicated strains were grown as described in the legend of Fig. 6B. Cells were harvested, and total RNA was prepared for Northern analysis. Blots were probed for the indicated transcripts. Each lane was loaded with 3 μg of total RNA. ACT1 and rRNA are shown as loading controls.
DISCUSSION
Correlation of increased hyphal growth with enhanced C. albicans biofilm formation.
While several previous studies identified regulators and pathways that are required or important for both C. albicans filamentation and biofilm formation (16, 28, 29, 33, 56–59), considerably less is known about mechanisms that promote enhanced formation of biofilms. Here, we identify one such mechanism, which is mediated by the transcriptional regulator Ume6. As UME6 levels rise and C. albicans transitions from yeast to a nearly complete hyphal population, we demonstrate that there is a corresponding increase in biofilm formation above levels typically observed for a wild-type strain. This result suggests a direct correlation between increased hyphal growth (and/or increased expression levels of genes associated with hyphal growth) and enhanced C. albicans biofilm formation. Our findings are consistent with previous observations that while the initiation stage of C. albicans biofilms involves attachment of yeast cells to a surface, subsequent biofilm growth and development involve the generation of pseudohyphal and hyphal filaments as well as the formation of a dense exopolymeric matrix (14–18). Interestingly, while most wild-type C. albicans biofilms maintain a bottom layer of yeast cells, our tetO-UME6 biofilm appears to be nearly completely hyphal, even in the bottom layer, when UME6 is expressed at high levels. Because the tetO-UME6 strain is typically grown in the yeast form (in the presence of Dox) prior to the start of biofilm assays, our results suggest that following the initiation step, the yeast form appears to be dispensable for enhanced C. albicans biofilm formation. It is important to note, however, that the ability of C. albicans cells within a biofilm to transition from filaments to yeast appears to be critical for biofilm dispersion, since we have previously demonstrated that constitutive high-level UME6 expression significantly inhibits this process (36).
In the clinical setting, biofilm formation can occur on both abiotic surfaces (e.g., catheters, denture materials, and implanted medical devices) as well as mucosal surfaces (11–14). While UME6 expression and increased hypha formation appear to clearly enhance biofilm formation on solid abiotic surfaces (equivalent to catheters and implanted medical devices), we have also shown that in the context of a biotic surface, the reconstituted three-dimensional model of the human oral mucosa, a C. albicans strain expressing constitutive high levels of UME6 shows significantly increased tissue invasion. These findings are consistent with our previous observation that UME6 expression promotes hypha formation, tissue invasion, and virulence in a mouse model of systemic candidiasis (34) and also suggest that shifting the morphology of cells in a C. albicans biofilm to hyphae is an important step in the pathogenesis of mucosal candidal infections.
Hypha-dependent and cell wall-dependent mechanisms important for UME6-driven enhanced biofilm formation.
There are a number of mechanisms that may account for our observation that UME6-driven hyphal growth promotes C. albicans biofilm formation. UME6 encodes a key transcriptional regulator of C. albicans hyphal development and is known to induce a variety of filament-specific transcripts (33, 35). Several of these target transcripts encode key adhesins, such as the Als3 agglutinin-like protein and Hwp1, a mammalian transglutaminase substrate mimic, which are known to play critical roles in biofilm formation, most likely by functioning as mediators of cell-cell adherence (16, 22–24, 60, 61). Overexpression of either ALS3 or HWP1 was previously shown to rescue the biofilm defect of a strain deleted for BCR1, an important transcriptional regulator of biofilm development (61, 62). Induction of ALS3 and HWP1 in response to UME6 expression could therefore possibly contribute to enhanced biofilm formation. However, based on our gene expression analysis (Fig. 7), neither adhesin may be solely required for this process (see below).
In addition to ALS3, HWP1, and other genes whose expression is associated with filamentous growth, UME6 is also known to control at least one gene, HGC1, important for the physical process of hyphal development (37). Hgc1 is known to promote hyphal development by septin phosphorylation, inhibition of cell separation, and activation of the Cdc42 master polarity regulator (38–44). We have previously demonstrated that UME6 directs hyphal growth via the Hgc1 pathway and that a tetO-UME6 strain bearing a homozygous deletion of HGC1 is defective for extended hyphal development and true septum formation (37). Our current finding that this mutant is also defective for biofilm formation is significant because it indicates that mechanisms specifically associated with the physical process of hypha formation play an important role in promoting biofilm development (Fig. 8).
Fig 8.
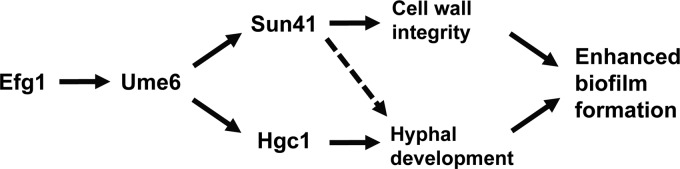
Model for roles of Efg1, Hgc1, and Sun41 in UME6-driven enhanced C. albicans biofilm formation. Ume6 functions downstream of Efg1 and upstream of both Hgc1 and Sun41 to promote biofilm development. UME6 expression is known to cause transcriptional induction of the Hgc1 cyclin-related protein, which, in turn, directs hyphal development via septin phosphorylation, inhibition of cell separation genes, and activation of the Cdc42 master polarity regulator (38–44). UME6 expression also appears to cause a slight increase in SUN41 transcript levels. SUN41, in turn, may function indirectly in a positive-feedback loop to increase UME6 expression levels (not shown); however, the relevance of these transcriptional effects for biofilm formation has not yet been established. In either case, Sun41, a putative cell wall glycosidase, is known to be primarily involved in maintaining cell wall integrity (27). Both the physical process of hyphal development and Sun41-mediated cell wall integrity therefore appear to play important roles in UME6-driven enhanced biofilm formation. In addition, we cannot exclude the possibility that Sun41 at least partly contributes to UME6-driven biofilm growth by playing a role in hyphal development (dashed line). Finally, it is important to note that an additional mechanism(s), which at this point has not yet been determined, may also contribute to UME6-driven enhanced biofilm formation.
The Sun41 cell wall putative glycosidase has been clearly shown to be important for C. albicans biofilm formation (25–27). Although we cannot exclude the possibility that the sun41Δ/Δ biofilm defect is partially due to a filamentation defect, the available evidence suggests that this mutant shows reduced biofilm formation primarily as a consequence of a severe cell wall defect (27). Our finding that enhanced biofilm formation in response to UME6 expression is significantly reduced upon deletion of SUN41 strongly suggests that general mechanisms important for cell wall integrity play an important role in this process (Fig. 8). Consistent with this hypothesis, the tetO-UME6 sun41Δ/Δ mutant, like the sun41Δ/Δ mutant, shows a clear cell wall defect, as indicated by hypersensitivity to Congo red. Interestingly, both our Northern analysis as well as a recent DNA microarray analysis (66) indicate that the SUN41 transcript is very mildly induced upon UME6 expression. In addition, we have observed that deletion of Sun41 causes a slight decrease in both UME6 and EFG1 transcript levels in the tetO-UME6 strain in the absence of Dox (although unlikely, we cannot exclude the possibility that these effects are due to a reduction in transactivator levels) (Fig. 7). These findings suggest that, at a transcriptional level, UME6 and SUN41 may function in a feedback loop as mildly positive regulators of each other; in addition, SUN41 may also function as a slight positive regulator of EFG1 expression when UME6 is expressed at high constitutive levels (since Sun41 is a cell wall component, transcriptional effects directed by this protein would most likely be indirect [see below]). However, the mild reduction in UME6 and EFG1 transcript levels in the tetO-UME6 sun41Δ/Δ mutant generally did not appear to be sufficient to cause a decrease in UME6 target gene expression levels; instead, most UME6 target genes appeared to be induced at equivalent or higher levels in this strain. In addition, while the overall level of biofilm formation by the tetO-UME6 sun41Δ/Δ mutant was not significantly enhanced compared to that of a wild-type control strain, it was still increased upon UME6 expression. This observation suggests that additional mechanisms associated with UME6 expression may still have a limited capacity to overcome the sun41Δ/Δ biofilm defect.
In summary, both hypha-dependent and cell wall-dependent mechanisms, mediated via the Hgc1 cyclin-related protein and the Sun41 putative glycosidase, respectively, appear to play important roles in promoting UME6-driven enhanced biofilm formation (Fig. 8). At this point, we cannot exclude the possibility that an additional downstream mechanism(s), which has not yet been determined, may also be involved in this process.
Relationship between EFG1 and UME6 with respect to biofilm formation, filamentation, and filament-specific gene expression.
Efg1, an important transcription factor which controls C. albicans filamentous growth, is also known to function as a key regulator of biofilm formation (28, 30). Our observation that constitutive high-level UME6 expression is sufficient to completely bypass the severe efg1Δ/Δ mutant biofilm and filamentation defects is consistent with previous findings (35) and strongly suggests that UME6 functions downstream of EFG1 with respect to these processes (Fig. 8). These results are also consistent with previous findings that Efg1 is important for transcriptional induction of UME6 as well as downregulation of NRG1, which encodes a negative regulator of UME6, in response to serum at 37°C (33, 35, 63). Our finding that deletion of EFG1 increases the density of hypha formation in the top layers of the tetO-UME6 biofilm was unexpected but may be related to previous reports that Efg1 functions as a negative regulator of C. albicans filamentation under embedded/matrix conditions (64) (as it is conceivable that the dense hyphal mat generated by constitutive high-level UME6 expression may lead to similar microaerophilic conditions within the biofilms). Interestingly, despite the fact that the density of hyphal filaments was increased in the tetO-UME6 strain upon deletion of Efg1, there was not a corresponding further increase in overall biofilm formation. This result suggests that while increased hyphal growth is generally correlated with enhanced biofilm formation, there may be a limit above which the density of hyphal filaments in a biofilm has no additional effect.
Given that UME6 appears to function downstream of Efg1 with respect to biofilm and filament formation, our finding that Efg1 is also required for the ability of UME6 to transcriptionally induce certain hypha-specific genes was also surprising. Of the five UME6 target genes whose induction is affected by the efg1Δ/Δ mutation, three (HYR1, ALS3, and HWP1) encode cell wall proteins, one is associated with cell elongation (ECE1), and one encodes a secreted virulence factor (RBT4). Interestingly, all five genes also show increased induction by UME6 upon deletion of SUN41 (and no significant induction in the absence of UME6 expression). These results suggest that perturbation of the C. albicans hyphal cell wall could indirectly trigger a compensatory increase in the expression level of cell wall/secreted proteins and are consistent with previous findings that deletion of Sun41 causes altered expression of cell wall biogenesis genes and results in a compensatory regulation of other glycosidases (26, 27). Efg1 is a major regulator of cell wall genes (65) and may also play an indirect role in this pathway, although deletion of Efg1 does not appear to cause a cell wall defect (see Fig. S1 in the supplemental material). In either case, since the tetO-UME6 efg1Δ/Δ mutant still showed increased biofilm formation and filamentation relative to that of a wild-type control strain, our gene expression results strongly suggest that HYR1, HWP1, RBT4, ECE1, and ALS3 are largely dispensable for these processes; instead, additional target genes, which are induced by UME6 in an Efg1-independent manner, are likely to function in UME6-driven enhanced biofilm formation and filamentous growth. Recently, Nobile et al. found that Efg1 functions as a component of a large and complex regulatory network that controls C. albicans biofilm development (31). Chromatin immunoprecipitation with microarray technology (ChIP-chip) data from this study indicate that UME6 appears to be a downstream target of many transcriptional regulators in the network, including Efg1. Perturbations in this network may also at least partially explain the differential effects of efg1Δ/Δ and sun41Δ/Δ mutations on the induction of specific UME6 target genes.
While the complex pathways which control both C. albicans hyphal growth and biofilm formation have yet to be fully elucidated, these studies provide new information about the mechanistic relationship between these two processes. Future work in this area using strains (such as tetO-UME6) which can be genetically manipulated to alter morphology is likely to significantly expand our knowledge of and provide greater insight into this complex, but extremely important, relationship.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Brian Wickes for useful advice and suggestions during the course of the experiments.
P.L.C. was supported by a COSTAR fellowship (T32DE14318-07) and by a NRSA predoctoral fellowship from the National Institute of Dental and Craniofacial Research (5F31DE020214-03). P.U. was supported by a postdoctoral fellowship from the American Heart Association (10POST4280033). This work was also supported by grant R21AI080930 from the National Institute of Allergy and Infectious Diseases to J.J.L.-R., award number UL 1RR025767 from the National Center for Research Resources to C.C.V., as well as a Voelcker Young Investigator award from the Max and Minnie Tomerlin Voelcker Fund and National Institute of Allergy and Infectious Diseases grant 5RO1AI083344 to D.K.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, the National Institute of Dental and Craniofacial Research, or the National Institutes of Health.
Footnotes
Published ahead of print 7 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00163-12.
REFERENCES
- 1. Odds FC. 1988. Candida and candidosis. Baillière Tindall, London, United Kingdom [Google Scholar]
- 2. Beck-Sague C, Jarvis WR. 1993. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980-1990. National Nosocomial Infections Surveillance System. J. Infect. Dis. 167:1247–1251 [DOI] [PubMed] [Google Scholar]
- 3. Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239–244 [DOI] [PubMed] [Google Scholar]
- 4. Pfaller MA, Lockhart SR, Pujol C, Swails-Wenger JA, Messer SA, Edmond MB, Jones RN, Wenzel RP, Soll DR. 1998. Hospital specificity, region specificity, and fluconazole resistance of Candida albicans bloodstream isolates. J. Clin. Microbiol. 36:1518–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
- 6. Cannon RD, Chaffin WL. 1999. Oral colonization by Candida albicans. Crit. Rev. Oral Biol. Med. 10:359–383 [DOI] [PubMed] [Google Scholar]
- 7. Dongari-Bagtzoglou A, Wen K, Lamster IB. 1999. Candida albicans triggers interleukin-6 and interleukin-8 responses by oral fibroblasts in vitro. Oral Microbiol. Immunol. 14:364–370 [DOI] [PubMed] [Google Scholar]
- 8. Dupont PF. 1995. Candida albicans, the opportunist. A cellular and molecular perspective. J. Am. Podiatr. Med. Assoc. 85:104–115 [DOI] [PubMed] [Google Scholar]
- 9. Filler SG, Kullberg BJ. 2002. Deep-seated candidal infections, p 341–348 In Calderone R. (ed), Candida and candidiasis. ASM Press, Washington, DC [Google Scholar]
- 10. Weig M, Gross U, Muhlschlegel F. 1998. Clinical aspects and pathogenesis of Candida infection. Trends Microbiol. 6:468–470 [DOI] [PubMed] [Google Scholar]
- 11. Douglas LJ. 2002. Medical importance of biofilms in Candida infections. Rev. Iberoam. Micol. 19:139–143 [PubMed] [Google Scholar]
- 12. Douglas LJ. 2003. Candida biofilms and their role in infection. Trends Microbiol. 11:30–36 [DOI] [PubMed] [Google Scholar]
- 13. Kojic EM, Darouiche RO. 2004. Candida infections of medical devices. Clin. Microbiol. Rev. 17:255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ramage G, Saville SP, Thomas DP, Lopez-Ribot JL. 2005. Candida biofilms: an update. Eukaryot. Cell 4:633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183:5385–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Finkel JS, Mitchell AP. 2011. Genetic control of Candida albicans biofilm development. Nat. Rev. Microbiol. 9:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lopez-Ribot JL. 2005. Candida albicans biofilms: more than filamentation. Curr. Biol. 15:R453–R455 doi:10.1016/j.cub.2005.06.020 [DOI] [PubMed] [Google Scholar]
- 18. Ramage G, Vandewalle K, Wickes BL, Lopez-Ribot JL. 2001. Characteristics of biofilm formation by Candida albicans. Rev. Iberoam. Micol. 18:163–170 [PubMed] [Google Scholar]
- 19. Hoyer LL. 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9:176–180 [DOI] [PubMed] [Google Scholar]
- 20. Li F, Palecek SP. 2003. EAP1, a Candida albicans gene involved in binding human epithelial cells. Eukaryot. Cell 2:1266–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li F, Svarovsky MJ, Karlsson AJ, Wagner JP, Marchillo K, Oshel P, Andes D, Palecek SP. 2007. Eap1p, an adhesin that mediates Candida albicans biofilm formation in vitro and in vivo. Eukaryot. Cell 6:931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, Mitchell AP. 2008. Complementary adhesin function in C. albicans biofilm formation. Curr. Biol. 18:1017–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Staab JF, Bradway SD, Fidel PL, Sundstrom P. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535–1538 [DOI] [PubMed] [Google Scholar]
- 24. Zhao X, Oh SH, Cheng G, Green CB, Nuessen JA, Yeater K, Leng RP, Brown AJ, Hoyer LL. 2004. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 150:2415–2428 [DOI] [PubMed] [Google Scholar]
- 25. Firon A, Aubert S, Iraqui I, Guadagnini S, Goyard S, Prevost MC, Janbon G, d'Enfert C. 2007. The SUN41 and SUN42 genes are essential for cell separation in Candida albicans. Mol. Microbiol. 66:1256–1275 [DOI] [PubMed] [Google Scholar]
- 26. Hiller E, Heine S, Brunner H, Rupp S. 2007. Candida albicans Sun41p, a putative glycosidase, is involved in morphogenesis, cell wall biogenesis, and biofilm formation. Eukaryot. Cell 6:2056–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Norice CT, Smith FJ, Jr, Solis N, Filler SG, Mitchell AP. 2007. Requirement for Candida albicans Sun41 in biofilm formation and virulence. Eukaryot. Cell 6:2046–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramage G, VandeWalle K, Lopez-Ribot J, Wickes B. 2002. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol. Lett. 214:95 doi:10.1111/j.1574-6968.2002.tb11330.x [DOI] [PubMed] [Google Scholar]
- 29. Uppuluri P, Pierce CG, Thomas DP, Bubeck SS, Saville SP, Lopez-Ribot JL. 2010. The transcriptional regulator Nrg1p controls Candida albicans biofilm formation and dispersion. Eukaryot. Cell 9:1531–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stoldt VR, Sonneborn A, Leuker CE, Ernst JF. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148:126–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Du H, Guan G, Xie J, Sun Y, Tong Y, Zhang L, Huang G. 2012. Roles of Candida albicans Gat2, a GATA-type zinc finger transcription factor, in biofilm formation, filamentous growth and virulence. PLoS One 7:e29707 doi:10.1371/journal.pone.0029707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Banerjee M, Thompson DS, Lazzell A, Carlisle PL, Pierce C, Monteagudo C, Lopez-Ribot JL, Kadosh D. 2008. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol. Biol. Cell 19:1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carlisle PL, Banerjee M, Lazzell A, Monteagudo C, Lopez-Ribot JL, Kadosh D. 2009. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc. Natl. Acad. Sci. U. S. A. 106:599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zeidler U, Lettner T, Lassnig C, Muller M, Lajko R, Hintner H, Breitenbach M, Bito A. 2009. UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS Yeast Res. 9:126–142 [DOI] [PubMed] [Google Scholar]
- 36. Uppuluri P, Chaturvedi AK, Srinivasan A, Banerjee M, Ramasubramaniam AK, Kohler JR, Kadosh D, Lopez-Ribot JL. 2010. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 6:e1000828 doi:10.1371/journal.ppat.1000828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carlisle PL, Kadosh D. 2010. Candida albicans Ume6, a filament-specific transcriptional regulator, directs hyphal growth via a pathway involving Hgc1 cyclin-related protein. Eukaryot. Cell 9:1320–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gonzalez-Novo A, Correa-Bordes J, Labrador L, Sanchez M, Vazquez de Aldana CR, Jimenez J. 2008. Sep7 is essential to modify septin ring dynamics and inhibit cell separation during Candida albicans hyphal growth. Mol. Biol. Cell 19:1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Park HO, Bi E. 2007. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 71:48–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sinha I, Wang YM, Philp R, Li CR, Yap WH, Wang Y. 2007. Cyclin-dependent kinases control septin phosphorylation in Candida albicans hyphal development. Dev. Cell 13:421–432 [DOI] [PubMed] [Google Scholar]
- 41. Wang A, Raniga PP, Lane S, Lu Y, Liu H. 2009. Hyphal chain formation in Candida albicans: Cdc28-Hgc1 phosphorylation of Efg1 represses cell separation genes. Mol. Cell. Biol. 29:4406–4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Y. 2009. CDKs and the yeast-hyphal decision. Curr. Opin. Microbiol. 12:644–649 [DOI] [PubMed] [Google Scholar]
- 43. Zheng X, Wang Y, Wang Y. 2004. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 23:1845–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zheng XD, Lee RT, Wang YM, Lin QS, Wang Y. 2007. Phosphorylation of Rga2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J. 26:3760–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shen J, Cowen LE, Griffin AM, Chan L, Kohler JR. 2008. The Candida albicans pescadillo homolog is required for normal hypha-to-yeast morphogenesis and yeast proliferation. Proc. Natl. Acad. Sci. U. S. A. 105:20918–20923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reuss O, Vik A, Kolter R, Morschhauser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127 [DOI] [PubMed] [Google Scholar]
- 47. Guthrie C, Fink GR. 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, CA [Google Scholar]
- 48. Pierce CG, Uppuluri P, Tristan AR, Wormley FL, Jr, Mowat E, Ramage G, Lopez-Ribot JL. 2008. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat. Protoc. 3:1494–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ramage G, Vande Walle K, Wickes BL, Lopez-Ribot JL. 2001. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob. Agents Chemother. 45:2475–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. (ed). 1992. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, NY [Google Scholar]
- 51. Dongari-Bagtzoglou A, Kashleva H. 2006. Development of a highly reproducible three-dimensional organotypic model of the oral mucosa. Nat. Protoc. 1:2012–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dongari-Bagtzoglou A, Kashleva H. 2006. Development of a novel three-dimensional in vitro model of oral Candida infection. Microb. Pathog. 40:271–278 [DOI] [PubMed] [Google Scholar]
- 53. Villar CC, Kashleva H, Mitchell AP, Dongari-Bagtzoglou A. 2005. Invasive phenotype of Candida albicans affects the host proinflammatory response to infection. Infect. Immun. 73:4588–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Villar CC, Kashleva H, Nobile CJ, Mitchell AP, Dongari-Bagtzoglou A. 2007. Mucosal tissue invasion by Candida albicans is associated with E-cadherin degradation, mediated by transcription factor Rim101p and protease Sap5p. Infect. Immun. 75:2126–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nakayama H, Mio T, Nagahashi S, Kokado M, Arisawa M, Aoki Y. 2000. Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus Candida albicans. Infect. Immun. 68:6712–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP. 2010. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 6:e1000752 doi:10.1371/journal.ppat.1000752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Goyard S, Knechtle P, Chauvel M, Mallet A, Prevost MC, Proux C, Coppee JY, Schwartz P, Dromer F, Park H, Filler SG, Janbon G, d'Enfert C. 2008. The Yak1 kinase is involved in the initiation and maintenance of hyphal growth in Candida albicans. Mol. Biol. Cell 19:2251–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kelly MT, MacCallum DM, Clancy SD, Odds FC, Brown AJ, Butler G. 2004. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol. Microbiol. 53:969–983 [DOI] [PubMed] [Google Scholar]
- 59. Kumamoto CA. 2005. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc. Natl. Acad. Sci. U. S. A. 102:5576–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ene IV, Bennett RJ. 2009. Hwp1 and related adhesins contribute to both mating and biofilm formation in Candida albicans. Eukaryot. Cell 8:1909–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nobile CJ, Nett JE, Andes DR, Mitchell AP. 2006. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot. Cell 5:1604–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, Edwards JE, Filler SG, Mitchell AP. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2:e63 doi:10.1371/journal.ppat.0020063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Braun BR, Kadosh D, Johnson AD. 2001. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20:4753–4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Giusani AD, Vinces M, Kumamoto CA. 2002. Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics 160:1749–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sohn K, Urban C, Brunner H, Rupp S. 2003. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol. Microbiol. 47:89–102 [DOI] [PubMed] [Google Scholar]
- 66. Carlisle PL, Kadosh D. 14 December 2012, posting date. A genome-wide transcriptional analysis of morphology determination in Candida albicans. Mol. Biol. Cell [Epub ahead of print.] doi:10.1091/mbc.E12-01-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.