Abstract
In the budding yeast Saccharomyces cerevisiae, Rho4 GTPase partially plays a redundant role with Rho3 in the control of polarized growth, as deletion of RHO4 and RHO3 together, but not RHO4 alone, caused lethality and a loss of cell polarity at 30°C. Here, we show that overexpression of the constitutively active rho4Q131L mutant in an rdi1Δ strain caused a severe growth defect and generated large, round, unbudded cells, suggesting that an excess of Rho4 activity could block bud emergence. We also generated four temperature-sensitive rho4-Ts alleles in a rho3Δ rho4Δ strain. These mutants showed growth and morphological defects at 37°C. Interestingly, two rho4-Ts alleles contain mutations that cause amino acid substitutions in the N-terminal region of Rho4. Rho4 possesses a long N-terminal extension that is unique among the six Rho GTPases in the budding yeast but is common in Rho4 homologs in other yeasts and filamentous fungi. We show that the N-terminal extension plays an important role in Rho4 function since rho3Δ rho4Δ61 cells expressing truncated Rho4 lacking amino acids (aa) 1 to 61 exhibited morphological defects at 24°C and a growth defect at 37°C. Furthermore, we show that Rho4 interacts with Bem2, a Rho GTPase-activating protein (RhoGAP) for Cdc42 and Rho1, by yeast two-hybrid, bimolecular fluorescence complementation (BiFC), and glutathione S-transferase (GST) pulldown assays. Bem2 specifically interacts with the GTP-bound form of Rho4, and the interaction is mediated by its RhoGAP domain. Overexpression of BEM2 aggravates the defects of rho3Δ rho4 mutants. These results suggest that Bem2 might be a novel GAP for Rho4.
INTRODUCTION
Rho family GTPases are small GTP-binding proteins of the Ras superfamily widespread in eukaryotes from yeasts to humans. They are widely implicated in cellular processes, including cytoskeletal reorganization, vesicular trafficking, and gene transcription (1, 2). Rho GTPases act as molecular switches, cycling between the GTP- and GDP-bound states. However, only in the GTP-bound state can they bind to downstream effectors to transduce signals. The activities of Rho proteins are regulated by guanine nucleotide exchange factors (GEFs), GTPase-activating proteins (GAPs), and guanine nucleotide dissociation inhibitors (GDIs). RhoGEFs promote the binding of Rho proteins to GTP, leading to their activation. Conversely, RhoGAPs inactivate Rho proteins through accelerating their intrinsic GTPase activity. RhoGDIs also negatively regulate Rho GTPases by inhibiting the dissociation of bound GDP and by extracting Rho GTPases from membranes into the cytosol (1, 2).
The budding yeast Saccharomyces cerevisiae reproduces by budding, a process that is heavily dependent on a polarized actin cytoskeleton and secretion (3). As in animal cells, actin organization in yeast cells is controlled by Rho GTPases. S. cerevisiae has six Rho GTPases: Cdc42 and Rho1 to Rho5 (4). Cdc42 plays a master role in yeast budding by regulating actin organization, septin organization, and polarized secretion. Rho1 and Rho2 are involved in actin organization and cell wall integrity. Rho3 and Rho4 are functionally related, though they do not share a high degree of amino acid sequence identity with each other. RHO3 appears to be more essential than RHO4 since deletion of RHO4 did not produce a detectable defect whereas deletion of RHO3 caused slow growth, which can be suppressed by high-copy-number RHO4 (5). Rho3 and Rho4 are thought to play a critical role in the maintenance, but not in the initiation, of bud growth based on the observation that deletion of RHO3 and RHO4 together caused lethality at 30°C, and the cells lysed at the small-budded stage with a large, round morphology and a depolarized actin cytoskeleton (6).
The essential role of Rho3 and Rho4 in bud growth appears to activate Bni1 and Bnr1, two formins responsible for the assembly of actin cables required for polarized secretion, since Rho3 and Rho4 bind to the formins (7, 8) and the lethality of rho3Δ rho4Δ cells could be rescued by the expression of Bni1 or Bnr1 mutants lacking the Rho-binding domain (9). Rho3 also plays a direct role in polarized secretion, a membrane trafficking process crucial for bud growth, as the rho3E51V mutant bearing a point mutation in the effector-binding domain had normal actin organization but displayed a post-Golgi secretory defect (10). This function of Rho3 is thought to require the interaction of Rho3 with Myo2, a type V myosin, and with Sec3 and Exo70, two subunits of the exocyst (11). Rho4 is known to interact with Sec3 and Exo70 but not with Myo2 (11). It is likely that Rho4 may also play a role in polarized secretion independently of its role in actin organization.
Rho4's role in actin organization and polarized secretion appears to be conserved in other yeasts and in the filamentous fungi. In the fission yeast Schizosaccharomyces pombe, deletion of the SpRHO4 homolog resulted in defective actin organization at the growing cell ends and a cell separation defect at elevated temperature (12, 13). The cell separation defect was thought to result from inefficient polarized secretion of the endo-1,3-glucanases SpEng1 and SpAgn1 to the septation sites (14). Similarly, in the pathogenic yeast Candida albicans, deletion of CaRHO4 also led to a cell separation defect, which can be partially rescued by overexpression of CaENG1 (15). Remarkably, Rho4 homologs in the filamentous fungi Neurospora crassa and Aspergillus nidulans are essential for cytokinesis, as deletion of NcRHO4 or AnRHO4 resulted in a complete abolishment of actomyosin contractile ring formation and septum construction (16, 17).
Rho4 was found to be regulated by Rdi1 (18), the sole RhoGDI in the budding yeast. Rdi1 overexpression caused the dissociation of Rho4 from the membrane compartment. In addition, Rdi1 and Ygk3 (a glycogen synthase kinase 3β [GSK-3β] kinase) promoted the degradation of Rho4 through proteolysis (18). So far, the only know GAP for Rho4, and Rho3 as well, is Rgd1 (19). No GEFs for Rho4 or Rho3 have been identified. Little is known about how Rho4 activity is regulated by Rdi1-independent mechanisms.
Here, we show that overexpression of constitutively active rho4 mutants affects polarized growth. We also show that the long N-terminal extension of Rho4 plays an important role in Rho4 function. In addition, we identify an interaction between Rho4 and Bem2, a RhoGAP for Cdc42 and Rho1. Our data suggest that Bem2 might be a novel GAP regulating Rho4 activity.
MATERIALS AND METHODS
Strains, media, and genetic methods.
Yeast strains used in this study are listed in Table 1. Standard culture media and genetic techniques were used except where noted (22). Escherichia coli strains DH12S (Life Technologies, Gaithersburg, MD) and DH5α (TaKaRa, Japan) were used as hosts for plasmid manipulation. Oligonucleotide primers for PCR were purchased from Sangon Biotech (Shanghai, China). The nucleotide sequences of primers are available upon request. SC-Ura medium contained 2% glucose (Dex). SRG-Ura medium that contained 1% raffinose and 2% galactose was used to overexpress genes driven by the GAL1 promoter. 5-Fluoroorotic acid (5-FOA; Sangon Biotech, China) was added to culture medium at 1 mg/ml to select for cells that have lost URA3-carrying plasmids.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| YEF473A | MATa his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 | 20 |
| YEF3593 | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 rho4Δ::kanMX | Research Genetics |
| THY521 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 rdi1Δ::KlTRP1 | 18 |
| ABY1910 | MATa ade2-101 his3 leu2 lys2 ura3 rho3Δ::Kanrrho4Δ::Kanr [pRS316-RHO3] | 9 |
| JGY2024 | MATa ade2-101 his3 leu2 lys2 ura3 rho3Δ::Kanrrho4Δ::Kanr [rho4-1 LEU2 CEN] | This study |
| JGY2025 | MATa ade2-101 his3 leu2 lys2 ura3 rho3Δ::Kanr rho4Δ::Kanr [rho4-2 LEU2 CEN] | This study |
| JGY2093 | MATa ade2-101 his3 leu2 lys2 ura3 rho3Δ::Kanrrho4Δ::Kanr [rho4-3 LEU2 CEN] | This study |
| JGY2097 | MATa ade2-101 his3 leu2 lys2 ura3 rho3Δ::Kanrrho4Δ::Kanr [rho4-4 LEU2 CEN] | This study |
| JGY2242 | MATa ade2-101 his3 leu2 lys2 ura3 rho3Δ::Kanrrho4Δ::Kanr [rho4Δ61 LEU2 CEN] | This study |
| JGY2243 | MATa ade2-101 his3 leu2 lys2 ura3 rho3Δ::Kanrrho4Δ::Kanr [RHO4LEU2 CEN] | This study |
| JGY2617 | MATa ade2-101 his3 leu2 lys2 ura3 rho3Δ::Kanrrho4Δ::Kanr [rho4Δ42 LEU2 CEN] | This study |
| JGY2727 | MATa ade2-101 his3 leu2 lys2 ura3 rho3Δ::Kanrrho4Δ::Kanr [rho4Q131LLEU2 CEN] | This study |
| pJ69-4A | MATa his3-Δ200 leu2-3,112 trp1-901 ura3-52 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | 21 |
| pJ69-4α | MATα his3-Δ200 leu2-3,112 trp1-901 ura3-52 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | 21 |
Plasmid and strain construction.
To generate the pGAL2 (2μ URA3 PGAL1-TCYC1) vector used for the GAL1-driven overexpression of RHO4 and rho4 mutants, the GAL1 promoter (nucleotides −541 to −5 relative to the A of the start codon) was amplified by PCR from pBM272 (23) and inserted into SacI- and BamHI-digested pRS426 (2μ URA3). Then, the SmaI-KpnI fragment of the CYC1 transcription terminator from pUG36 was subcloned into it, yielding pGAL2. RHO4 and rho4 mutants were amplified by PCR using plasmids pCM185-RHO4, pCM185-RHO4Q131L, pCM185-RHO4T86N, and pRS305-3HA-RHO4G81V kindly provided by François Doignon, Marc Crouzet, and Thomas Höfken (18, 24) as the templates and inserted into BamHI- and EcoRI-digested pGAL2, yielding pGAL2-RHO4, pGAL2-RHO4Q131L, pGAL2-RHO4T86N, and pGAL2-RHO4G81V. Plasmid pEGKT316-BEM2 used for BEM2 overexpression was generated by inserting the BEM2 open reading frame (ORF) into SmaI- and SalI-digested pEGKT316 (25). YIp211-CDC3-GFP (26) was used to examine Cdc3-green fluorescent protein (GFP) localization.
Plasmids pRS315-RHO4 and pRS315-RHO4-5′,3′-UTR were used for the generation of temperature-sensitive rho4-Ts mutants. pRS315-RHO4 was constructed by inserting the RHO4 gene (including the 647-bp promoter region and the 279-bp 3′ untranslated region [UTR]) amplified by PCR from yeast genomic DNA into SacI- and XhoI-digested pRS315 (CEN LEU2). To generate pRS315-RHO4-5′,3′-UTR, the 647-bp RHO4 promoter was amplified by PCR from genomic DNA and ligated into SacI- and BamHI-digested pRS315. Then, the 279-bp 3′ UTR of RHO4 was PCR amplified and ligated into HindIII- and XhoI-digested pRS315-RHO4-5′UTR, yielding pRS315-RHO4-5′,3′-UTR. Plasmid pRS315-RHO4-Δ61 was generated by inserting the RHO4 segment encoding amino acids (aa) 62 to 291 and the RHO4 3′ UTR into BamHI- and XhoI-digested pRS315-RHO4-5′UTR. pRS315-RHO4-Δ42, which carries the RHO4 ORF encoding aa 43 to 291, was constructed in a similar way. pRS315-RHO4-Q131L was generated by the insertion of the RHO4Q131L ORF region (carrying the CAAX box) amplified from pODB80-RHO4Q131L,ΔC into BamHI- and HindIII-digested pRS315-RHO4-5′,3′-UTR. pUG34-RHO4Q131L was generated by inserting RHO4Q131L into BamHI- and EcoRI-digested pUG34 (CEN HIS3 PMET25-yEGFP3-TCYC1).
Yeast two-hybrid bait vector pODB80 (2μ TRP GAL4-BD) and constructs pODB80-RHO1Q68L,ΔC, pODB80-RHO2Q65L,ΔC, pODB80-RHO3Q74L,ΔC, pODB80-RHO4Q131L,ΔC, pODB80-RHO5Q91L,ΔC, and pODB8-CDC42G12V,ΔC were kindly provided by François Doignon and Marc Crouzet (19). The negative-control plasmid pODB80-S was generated by introducing a stop codon after the NcoI site in frame with the Gal4-DBD ORF in pODB80 to avoid extra peptide translation. Bait plasmids pGBDU-RHO4ΔC, pGBDU-RHO4G81V,ΔC, pGBDU-RHO4Q131L,ΔC, pGBDU-RHO4Q131L,Δ61,ΔC, and pGBDU-RHO4T86N,ΔC were generated by PCR amplification of RHO4 or the respective rho4 mutant without the C-terminal CAAX box using plasmids pCM185-RHO4, pODB80-RHO4Q131L,ΔC, and pCM185-RHO4T86N (19, 24) and plasmid pRS305-3HA-RHO4G81V (18) as the templates and ligated into BamHI- and SalI-digested pGBDU-C1 (2μ URA3 GAL4-BD) (21).
For the bimolecular fluorescence complementation (BiFC) assay, plasmid pVN1 (CEN URA3 PMET25-Venus-N-TCYC1) was generated by replacing the yEGFP3 gene (an XbaI-BamHI fragment) in pUG36 with the XbaI-BamHI fragment of Venus-N encoding aa 1 to 173 of Venus, a GFP variant. Similarly, pVC1 (CEN HIS3 PMET25-Venus-C-TCYC1) was generated by replacing the yEGFP3 gene in pUG34 with the XbaI-BamHI fragment of Venus-C encoding aa 155 to 238 of Venus. A linker encoding four alanines was added after Venus-N and Venus-C. Venus-N and Venus-C genes were amplified from plasmids kindly provided by Won-Ki Huh (27). pVN1-RHO4Q131L was generated by inserting RHO4Q131L into BamHI- and EcoRI-digested pVN1. pVC1-BEM2 was generated by inserting the BEM2 ORF into SmaI- and SalI-digested pVC1.
Isolation of temperature-sensitive rho3Δ rho4-Ts mutants.
RHO4 was randomly mutagenized by an error-prone PCR method (28). The PCR-amplified RHO4 genes with mutations were mixed with BamHI- and HindIII-digested plasmid pRS315-RHO4-5′,3′-UTR in a 10:1 ratio to cotransform yeast strain ABY1910 (a rho3Δ rho4Δ pRS316-RHO3). Transformants were grown on SC-Leu plates at 24°C to allow gap repair between the PCR products and the linearized plasmid vector. Transformants were replica plated onto SC-Leu plates twice and then onto SC-Leu plates containing 5-FOA twice at 24°C to select for the loss of pRS316-RHO3. The Leu+ Ura− transformants were replicated onto two sets of SC-Leu plates. One plate was incubated at 24°C, and the other one was incubated at 37°C, to allow the identification of temperature-sensitive mutants. pRS315-RHO4-Ts plasmids that carried the temperature-sensitive mutations were retrieved from yeast clones and reintroduced into the yeast strain ABY1910 to verify that the temperature-sensitive growth defect was due to mutations on the plasmid-borne RHO4 gene.
Yeast two-hybrid screen.
A yeast two-hybrid pOAD-cDNA library was transformed into yeast strain pJ69-4A carrying pGBDU-RHO4Q131L,ΔC, a bait construct that expresses the active GTP-bound form of Rho4. Transformants were grown on SC-Leu-Ura-His plates supplemented with 2 mM 3-amino-1,2,4-triazole (3-AT; Sigma-Aldrich) at 30°C. The plates were then replica plated onto SC-Leu-Ura-Ade plates to allow the identification of candidate clones. The pOAD-prey library plasmids were retrieved, and the cDNA inserts were sequenced. We screened ∼200,000 transformants and isolated 67 positive yeast clones. DNA sequencing and PCR showed that we have isolated a ribosomal subunit-encoding RPS0A gene 28 times. Two N-terminally truncated BEM2 clones were among the several genes that we isolated. One BEM2 clone encodes the region of aa 1846 to 2167, and the other one encodes the region of aa 1927 to 2167.
Yeast two-hybrid assay.
The pODB80-based (2μ TRP1) or pGBDU-C1-based (2μ URA3) bait plasmids were transformed into yeast strain pJ69-4α. The prey plasmid vector pOAD (CEN LEU2) (21) and the two pOAD-BEM2 clones isolated from the library were transformed into yeast strain pJ69-4A. Haploid pJ69-4A and pJ69-4α strains carrying plasmids were mated on yeast extract-peptone-dextrose (YPD) plates and then replica plated onto SC-Leu-Trp or SC-Leu-Ura plates to select for diploid cells that harbor both bait and prey plasmids. Diploid cells from these plates were replica plated onto SC-Leu-Trp-His plus 2 mM 3-AT and SC-Leu-Trp-Ade plates or SC-Leu-Ura-His plus 2 mM 3-AT and SC-Leu-Ura-Ade plates to check for growth.
GST pulldown assay.
The glutathione S-transferase (GST) assay followed a previously described protocol (25, 26).
Microscopy.
An Olympus BX51 microscope (Tokyo, Japan) and a Retiga 2000R charge-coupled device (CCD) camera (QImagine Corporation, Canada) were used to visualize cell morphology and GFP-tagged proteins by differential interference contrast (DIC) and fluorescence microscopy. The budding pattern of a yeast strain was determined by staining the bud scars with calcofluor white (Sigma-Aldrich). At least 200 cells that exhibit three or more bud scars were counted. For the examination of F-actin and DNA, yeast cells were stained with 1.0 μg/ml phalloidin-TRITC (tetramethyl rhodamine isocyanate) (Sigma-Aldrich) and 1.0 μg/ml of Hoechst 33258 (Polysciences, Inc.) at room temperature for 30 min and washed three times with 1× phosphate-buffered saline (PBS) (pH 8.0).
RESULTS
Effects of overexpressing RHO4 and constitutively active or inactive rho4 mutants on polarized growth.
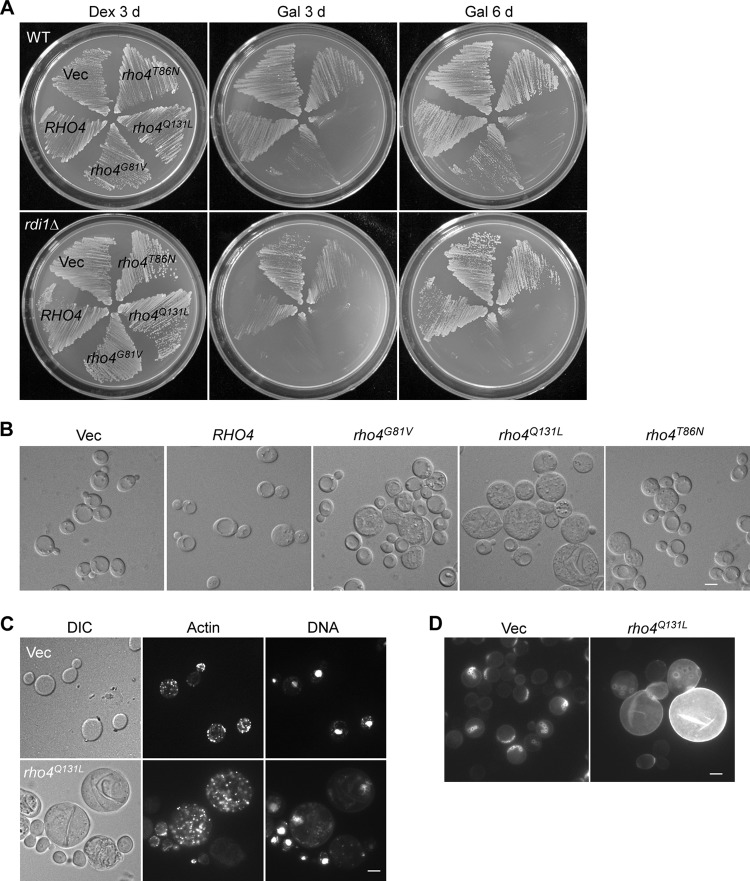
Previous studies have shown that RHO4 plays a role in polarized growth, since deletion of RHO4 in a rho3Δ strain caused lethality at 30°C and the cells often died at the small-budded stage with a depolarized actin cytoskeleton (5, 6). As a complementary approach to the RHO4 deletion study, we examined the effects of overexpressing wild-type RHO4 as well as rho4 mutants analogous to H-ras mutants defective in GTP hydrolysis or GTP binding on polarized growth. To this end, RHO4, rho4G81V, rho4Q131L, and rho4T86N mutants (18, 24, 29) were expressed under the control of the galactose-inducible GAL1 promoter on high-copy-number plasmids in wild-type cells. We observed that overexpression of RHO4 or the rho4T86N mutant did not impair growth. However, overexpression of the constitutively active rho4G81V or rho4Q131L mutant drastically reduced growth (Fig. 1A, upper panel). rho4Q131L appears to be more robust in inhibiting growth than rho4G81V as cells overexpressing rho4Q131L did not grow into colonies at 30°C in 3 days and formed tiny colonies only in 6 days whereas cells overexpressing rho4G81V formed tiny colonies in 3 days and colonies of moderate size in 6 days (Fig. 1A, upper panel). Examination of cell morphology under the microscope revealed that rho4G81V and rho4Q131L overexpression caused a slight increase of cell size in a small fraction of cells (data not shown). This finding is consistent with a previous report that overexpression of rho4Q131L caused cell enlargement and round shape (29).
Fig 1.
Phenotypes of yeast cells overexpressing RHO4 and rho4 mutants. (A) Cells of YEF473A (wild-type [WT]) and THY521 (rdi1Δ) strains carrying pGAL2 (Vec), pGAL2-RHO4, pGAL2-RHO4G81V, pGAL2-RHO4Q131L, or pGAL2-RHO4T86N overexpression plasmids were streaked and incubated on SC-Ura (Dex) and SRG-Ura (Gal) plates at 30°C. Photographs were taken after 3 or 6 days. (B) DIC images of rdi1Δ cells carrying pGAL2 (Vec) or other overexpression plasmids as in panel A were taken after growth on an SRG-Ura plate at 30°C for 3 days. (C) rdi1Δ cells carrying pGAL2 (Vec) or pGAL2-RHO4Q131L were grown in SRG-Ura liquid medium at 30°C for 15 h. Cells were fixed and stained for F-actin and DNA. (D) Cells as in panel C were stained for chitin with calcofluor. Bars, 5 μm.
It has been reported that the protein level of Rho4 is modulated by Rdi1 as rdi1Δ cells contained a higher level of Rho4 than did wild-type cells. In addition, Rdi1 extracted wild-type Rho4 and the Rho4G81V mutant equally well (18). We then repeated the overexpression experiment in an rdi1Δ strain. Our result showed that overexpression of the rho4G81V or rho4Q131L mutant in rdi1Δ cells caused a severe growth defect. Overexpression of RHO4 or the rho4T86N mutant also led to a marked, albeit milder, reduction of growth (Fig. 1A, lower panel). Examination of the morphology of cells grown at 30°C for 3 days showed that most cells overexpressing RHO4 or rho4T86N had a morphology identical to that of control cells. Only a small fraction of cells became slightly enlarged and round. In contrast, cells overexpressing rho4Q131L were much larger and rounder than control cells (Fig. 1B). About 20% of the cells (n = 120) were moderately enlarged, while 30% of the cells were greatly enlarged (several times bigger than control cells). Most of the enlarged cells (80%, n = 175) were unbudded. Within the population of greatly enlarged cells, all of them (n = 66) displayed a depolarized actin cytoskeleton and most of them were unbudded (Fig. 1C), indicative of a defect in bud emergence. Interestingly, nearly all of them contained a single nucleus (Fig. 1C). Overexpression of rho4G81V also caused a significant increase of cell size, but the effect was less pronounced than that of rho4Q131L.
rho4Q131L overexpression also caused delocalized chitin deposition, particularly in the cells that became enlarged (Fig. 1D). In addition, rho4Q131L overexpression changed the budding pattern from an axial pattern to random. Haploid rdi1Δ cells harboring an empty vector exhibited axial budding in 83% of cells. In contrast, the percentage of axial budding cells was reduced to about 20% upon rho4Q131L overexpression. The majority of the cells (78%) budded randomly. The defective bud-site selection likely results from depolarized actin organization.
Together, these results showed that an excess of Rho4 activity affects polarized growth in a dosage-dependent manner. Particularly, the overexpression of the rho4Q131L mutant in rdi1Δ cells could block bud emergence. These findings support the notion that Rho4 is an important player in the control of polarized growth.
Temperature-sensitive rho3Δ rho4-Ts mutants display defects in polarized growth.
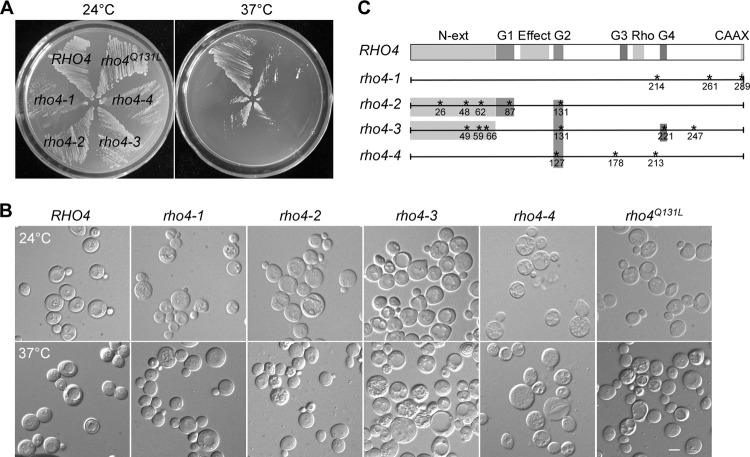
To characterize the function of Rho4 further, we generated temperature-sensitive rho4-Ts mutant alleles in a rho3Δ rho4Δ strain by error-prone PCR. Four alleles, rho4-1, rho4-2, rho4-3, and rho4-4, which could confer a growth defect to rho3Δ rho4Δ cells at 37°C, were isolated. Among these alleles, the rho4-3 and rho4-4 alleles conferred a more penetrant growth defect than did the other two alleles at 37°C. The rho4-1 and rho4-4 alleles also conferred a growth defect at 24°C (Fig. 2A). Examination of cell morphology revealed that cells carrying all four alleles showed an enlargement of cell size but in various proportions (Fig. 2B). rho3Δ rho4-3 cells had the most defective cell morphology at 37°C. The cells became large and round, but most cells still had a bud, similar to rho3Δ rho4Δ cells. DNA staining showed that most cells had one nucleus and that only a low percentage of cells contained two nuclei (data not shown). In addition to a loss of cell polarity, rho3Δ rho4-3 and rho3Δ rho4-4 cells were prone to cell lysis, suggesting that the cell wall may also have defects.
Fig 2.
Phenotypes of temperature-sensitive rho3Δ rho4-Ts mutants. (A) Cells of strains JGY2243 (rho3Δ RHO4), JGY2024 (rho3Δ rho4-1), JGY2025 (rho3Δ rho4-2), JGY2093 (rho3Δ rho4-3), JGY2097 (rho3Δ rho4-4), and JGY2727 (rho3Δ rho4Q131L) were streaked onto SC-Leu plates and grown at 24°C and 37°C for 4 days. (B) DIC images of cells as shown in panel A after 2 days. Bar, 5 μm. (C) The positions of amino acid substitutions in rho4-1, rho4-2, rho4-3, and rho4-4 alleles. The Rho4 precursor has 291 aa. Listed domains and positions are N-terminal extension (N-ext, aa 1 to 73), effector domain (Effect, aa 95 to 120), Rho insert (Rho, aa 192 to 202), G1 box (aa 74 to 90), G2 box (aa 123 to 132), G3 box (aa 181 to 188), and G4 box (aa 216 to 222).
DNA sequencing showed that rho4-2 and rho4-3 alleles each carried three point mutations that caused amino acid substitutions in the N-terminal region as well as two mutations in the G1, G2, or G4 box involved in GTP binding and GTP hydrolysis (Fig. 2C). They also shared a Q131R mutation in the G2 box. The rho4-4 allele carried three point mutations, one in the G2 box and two in the C-terminal half of the protein. The rho4-1 allele carried three point mutations in the C-terminal half, but none of them were in the G boxes or the Rho insert.
Because rho4-2 and rho4-3 alleles shared a Q131R mutation that may render Rho4 constitutively active based on the analogous oncogenic N-rasQ61R mutation (30), we generated a similar constitutively active rho4Q131L mutant to investigate if the Q131 mutation alone may be responsible for the phenotypes of rho3Δ rho4-2 and rho3Δ rho4-3 cells. We observed that rho3Δ rho4Q131L cells did not show a dramatic increase of cell size at 37°C compared to rho3Δ rho4-2 and rho3Δ rho4-3 cells (Fig. 2B). However, rho3Δ rho4Q131L cells indeed had a moderate growth defect at 37°C (Fig. 2A). Moreover, rho3Δ rho4Q131L cells showed a defect in bud-site selection at 37°C, as only 26% of cells budded axially and 52% of cells budded randomly, whereas rho3Δ RHO4 control cells displayed axial budding in 92% of cells. These findings indicate that the rho4Q131L mutant was indeed not fully functional. However, the mutation at Q131 alone could not explain all the defects shown by rho4-2 and rho4-3 alleles. It is possible that other mutations such as the three point mutations in the N-terminal extension of Rho4 may also contribute to the defects in rho4-2 and rho4-3 alleles.
The N-terminal extension of Rho4 plays an important role in Rho4 function.
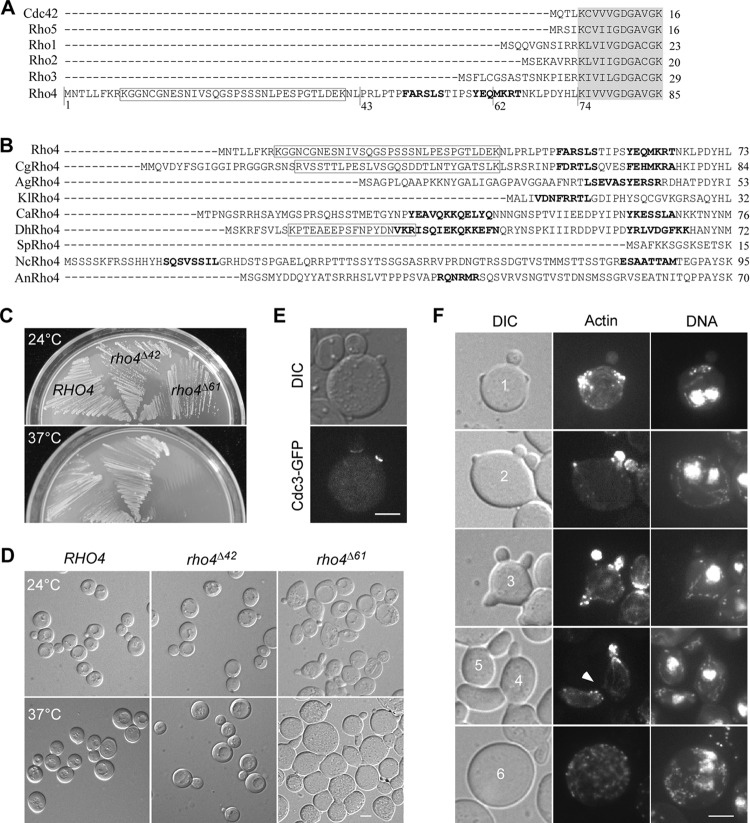
Compared to the other five Rho GTPases in the budding yeast, Rho4 has an unusually long N-terminal extension upstream of the conserved G1 box, which is 69 aa longer than that of Cdc42 and Rho5 (Fig. 3A). Interestingly, Rho4 homologs in most yeast species and filamentous fungi also have a long N-terminal extension (Fig. 3B). The N-terminal extension of Rho4 contains two short α-helices at amino acid residues 49 to 54 and 59 to 65, as predicted by SWISS-MODEL Workspace (http://swissmodel.expasy.org/workspace/) (31). In addition, near the beginning of the N-terminal extension, there is a potential PEST motif at amino acid residues 9 to 40 (score 4.82), which is a short hydrophilic stretch responsible for the rapid degradation of some proteins (32).
Fig 3.
The N-terminal extension of Rho4 is important for its function. (A) Alignment of the amino acid sequences of the N-terminal regions of the six Rho GTPases in the budding yeast. The conserved G1 boxes (partial sequence) were highlighted in gray. The potential PEST motif and the two short α-helices in Rho4's N-terminal extension were boxed and indicated by bold letters, respectively. (B) Alignment of the N-terminal extensions (upstream of the G1 box) of Rho4 homologs in several yeasts and filamentous fungi. The short α-helices were indicated by bold letters. The potential PEST motifs in Rho4 (score 4.82), Candida glabrata CgRho4 (score 1.36), and Debaryomyces hansenii DhRho4 (score 4.77) were boxed. The other Rho4 homologs in the sequence alignment include Ashbya gossypii AgRho4, Kluyveromyces lactis KlRho4, Candida albicans CaRho4, Schizosaccharomyces pombe SpRho4, Neurospora crassa NcRho4, and Aspergillus nidulans AnRho4. (C) Cells of strains JGY2243 (rho3Δ RHO4), JGY2242 (rho3Δ rho4Δ61), and JGY2617 (rho3Δ rho4Δ42) were streaked on an SC-Leu plate and incubated at 24°C and 37°C for 3 days. (D) DIC images of cells as in panel C were taken after growth at 24°C and 37°C for 3 days. (E) The multiple buds on rho3Δ rho4Δ61 cells have septins at the bud neck. Cells of JGY2242 (rho3Δ rho4Δ61) carrying integrative YIp211-CDC3-GFP were grown at 24°C and examined for Cdc3-GFP localization. (F) Cells of JGY2242 (rho3Δ rho4Δ61) were grown in liquid SC-Leu medium at 24°C overnight and then shifted to 37°C for 4 h. Cells were fixed and stained for F-actin and DNA. The arrowhead shows a bud defective in cell separation. Bars, 5 μm.
Given that the temperature-sensitive rho4-2 and rho4-3 alleles both contained point mutations in the N-terminal extension, we wanted to investigate if the N-terminal extension is required for Rho4 function. For this purpose, we generated an N-terminally truncated rho4Δ61 mutant, whose coding region for amino acid residues 1 to 61 was removed. The rho4Δ61 mutant was introduced into a rho3Δ rho4Δ strain by plasmid shuffling to evaluate its ability to support growth. We found that rho3Δ rho4Δ61 cells grew well at 24°C (Fig. 3C). However, examination of cell morphology revealed that rho3Δ rho4Δ61 cells were larger than rho3Δ RHO4 control cells. In addition, about 20% of cells carried two or more (mostly two) buds, instead of just one, on the same mother cell (Fig. 3D). Each bud displayed a Cdc3-GFP (a septin subunit) localization at the bud neck (Fig. 3E), indicating that they were true buds, not just surface protrusions. rho3Δ rho4Δ61 cells also displayed a defect in bud-site selection. At 24°C, haploid rho3Δ RHO4 control cells showed an axial budding pattern in 92% of cells. In contrast, only 10% of rho3Δ rho4Δ61 cells showed an axial budding pattern. The majority of these cells (82%) budded randomly. Remarkably, rho3Δ rho4Δ61 cells failed to grow at 37°C (Fig. 3C). The majority of cells (∼70%) became much larger and rounder than rho3Δ RHO4 control cells, and about 40% of budded cells were multibudded (Fig. 3D, E, and F, cells 1, 2, and 3). Moreover, ∼10% of cells displayed pointed or slightly elongated buds (Fig. 3F, cells 4 and 5), which was not seen in rho3Δ RHO4 control cells. These findings indicate that the removal of the first 61 aa of the N-terminal extension renders Rho4 defective.
To get a hint of the defective cellular functions, we stained rho3Δ rho4Δ61 cells grown at 37°C for F-actin and DNA. We observed that within the population of large and round cells, 27% (n = 175) were unbudded. DNA staining showed that 68% of unbudded cells (n = 47) had one nucleus, while the rest of the population (32%) had two or more nuclei (Fig. 3F, cell 6), suggesting that some rho3Δ rho4Δ61 cells may have a defect in bud emergence. rho3Δ rho4Δ61 cells also appear to have a defect in the segregation of nucleus into the bud, as 15% of cells (n = 106) carried at least one bud but had more than one nucleus (Fig. 3F, cells 1 and 2), which was not seen in control cells.
Since a large number of large and round rho3Δ rho4Δ61 cells carried more than one bud, we further examined these cells to understand the cause of the multibudded phenotype. We observed that, in the population of multibudded cells, ∼50% of cells (n = 106) had concentrated actin patches in more than one bud (Fig. 3F, cells 1, 2, and 3), suggesting that these buds are growing simultaneously. Interestingly, among these multibudded cells with two or more actively growing buds, 76% of them (n = 58) had just a single nucleus (Fig. 3F, cell 3), suggesting that these buds are likely produced in one nuclear cycle. In addition, we found that a delay in cell separation also plays a role in the generation of the multibudded phenotype, as 33% of multibudded cells (n = 106) carried one bud still connected to its mother cell after cytokinesis was complete (judged by a discontinuity of cytoplasmic F-actin fluorescence) (Fig. 3F, cell 4, arrowhead).
These findings demonstrate that the N-terminal extension is important for Rho4 function. Further analysis revealed that the functional region within the N-terminal extension locates roughly between amino acid residues 43 and 61 as rho3Δ rho4Δ42 mutant cells that express the Rho4Δ42 mutant lacking amino acid residues 1 to 42 showed no detectable defect in either growth or cell morphology at 24°C and 37°C, compared to rho3Δ RHO4 control cells (Fig. 3C and D). In addition, rho3Δ rho4Δ42 mutant cells did not display a defect in bud-site selection (92% axial budding).
Taken together, our results indicate that the N-terminal extension of Rho4, specifically the region of aa 43 to 61, has an important role in Rho4 function. Removal of the N-terminal extension results in defects in several cellular functions, including bud-site selection, bud formation, nuclear segregation, and cell separation.
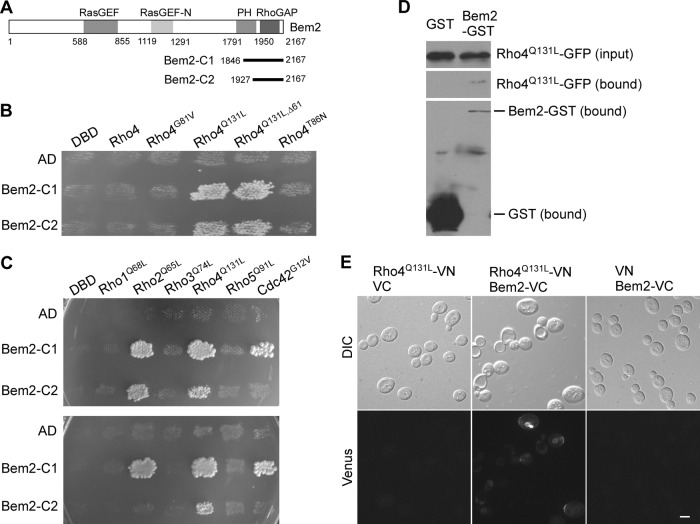
Rho4 interacts with Bem2, a RhoGAP for Cdc42 and Rho1.
As an approach to study Rho4 function and regulation in polarized growth, we performed a two-hybrid screen searching for genes whose product interacts with Rho4. Using a Rho4Q131L-Gal4 DBD (DNA-binding domain) fusion construct as bait, we isolated two short BEM2 segments that interact with Rho4Q131L from a yeast cDNA library. Bem2, a protein of 2,167 aa, contains a RasGEF and a RasGEF-N domain near the middle and a PH (pleckstrin homology) domain and a RhoGAP (GTPase-activating protein) domain at the C terminus (Fig. 4A). The two BEM2 segments encode only the C-terminal region: BEM2-C1 encodes the region of aa 1846 to 2167 (322 aa) containing the RhoGAP domain and most of the PH domain; BEM2-C2 encodes the region of aa 1927 to 2167 (241 aa) containing the RhoGAP domain and the last 24 aa of the PH domain (Fig. 4A). Thus, it appears that Bem2 interacts with Rho4 through its RhoGAP domain. The interaction of Rho4 with Bem2 segments is GTP dependent as Bem2-C1 and Bem2-C2 interacted only with the constitutively active Rho4Q131L mutant but not with wild-type Rho4 or the inactive Rho4T86N mutant in a two-hybrid assay (Fig. 4B). Removal of the first 61 aa of Rho4 did not impair the interaction of Rho4 with two Bem2 segments (Fig. 4B, RHO4Q131L,Δ61).
Fig 4.
Bem2 interacts with Rho4 in vivo. (A) Schematic representation of the domains of Bem2 and the position of Bem2 segments Bem2-C1 and Bem2-C2 isolated from the two-hybrid library. (B) Two-hybrid assay of the interaction of Bem2-C1 and Bem2-C2 with Rho4 and Rho4 mutants. Yeast strain pJ69-4α carrying pGBDU-C1 (DBD), pGBDU-RHO4ΔC, pGBDU-RHO4G81V,ΔC, pGBDU-RHO4Q131L,ΔC, pGBDU-RHO4Q131L,Δ61,ΔC, or pGBDU-RHO4T86N,ΔC was mated with strain pJ69-4A carrying pOAD (AD), pOAD-BEM2-C1, or pOAD-BEM2-C2. Diploids from the mating reactions were patched on SC-Leu-Ura plates and replica plated to an SC-Leu-Ura-Ade plate. Pictures were taken after incubation at 30°C for 3 days. (C) Two-hybrid assay between Bem2 and the six Rho GTPases. Yeast strain pJ69-4α carrying pODB80-S (DBD), pODB80-RHO1Q68L,ΔC, pODB80-RHO2Q65L,ΔC, pODB80-RHO3Q74L,ΔC, pODB80-RHO4Q131L,ΔC, pODB80-RHO5Q91L,ΔC, and pODB8-CDC42G12V,ΔC was mated with strain pJ69-4A carrying pOAD (AD), pOAD-BEM2-C1, and pOAD-BEM2-C2. Diploid cells were grown on SC-Leu-Trp plates and replica plated to an SC-Leu-Trp-His plus 2 mM 3-AT plate (upper panel) and an SC-Leu-Trp-Ade plate (bottom panel). Pictures were taken after 3 days. (D) GST pulldown assay. Yeast strain YEF3593 (rho4Δ) carrying the pEGKT316 (GST)/pUG34-RHO4Q131L and pEGKT316-BEM2 (Bem2-GST)/pUG34-RHO4Q131L plasmid pairs was used. Rho4Q131L-GFP was detected by anti-GFP antibody. (E) BiFC assay. Cells of strain YEF3593 (rho4Δ) carrying pVN1-RHO4Q131L/pVC1, pVN1-RHO4Q131L/pVC1-BEM2, and pVN1/pVC1-BEM2 plasmid pairs were grown on SC-Ura-His medium. Green fluorescence was examined by fluorescence microscopy. Bar, 5 μm.
Previous studies have shown that Bem2 has two in vivo targets, Cdc42 and Rho1, and the Bem2 C-terminal segment (aa 1818 to 2167) has RhoGAP activity (33–35). We tested if Bem2-C1 and Bem2-C2 segments also interact with other Rho GTPases. We observed that they both showed an interaction with Rho2Q65L, but not with Rho1Q68L, Rho3Q74L, or Rho5Q91L (Fig. 4C). Bem2-C1 also showed an interaction with Cdc42G12V. Compared to the interaction with Rho2 and Cdc42, Bem2's interaction with Rho4 appears to be stronger since Bem2-C2 showed an interaction only with Rho4Q131L but not with others, based on its ability to activate a more stringent GAL-ADE2 reporter (Fig. 4C, lower panel).
The interaction between Bem2 and Rho4 was confirmed by bimolecular fluorescence complementation (BiFC) and GST pulldown assays using Rho4Q131L and full-length Bem2 (Fig. 4D and E). The interaction could be detected on the plasma membrane and endomembranes by BiFC. Together, our study has identified Bem2 as a protein that interacts with Rho4. Bem2 interacts only with the activated GTP-bound form of Rho4, and this interaction is mediated by the RhoGAP domain of Bem2, suggesting that Rho4 could be a novel target of Bem2.
Functional interaction between RHO4 and BEM2 in vivo.
Bem2 is a RhoGAP whose in vivo targets are thought to be Cdc42 and Rho1. Our results suggest that Rho4 could be a new potential target of Bem2 in the cells. To examine the functional relationship between Bem2 and Rho4, we evaluated the effect of BEM2 overexpression in several rho3Δ rho4 mutant strains. We observed that GAL1-driven overexpression of BEM2 impaired growth in rho3Δ RHO4, rho3Δ rho4Δ61, and rho3Δ rho4-3 cells at 24°C. However, the inhibitory effect appeared to be more pronounced in rho3Δ rho4Δ61 and rho3Δ rho4-3 cells (Fig. 5A). Upon BEM2 overexpression, rho3Δ RHO4 cells showed a slight increase in cell size but could form small colonies in 6 days. In contrast, rho3Δ rho4Δ61 and rho3Δ rho4-3 cells became large and round and failed to grow into visible colonies in 6 days (Fig. 5A and B). This result indicates that an excess of Bem2 aggravated the defect of rho3Δ rho4Δ61 and rho3Δ rho4-3 mutants. Given that Bem2 interacts with Rho4 via its RhoGAP domain, this finding suggests that Bem2 may negatively regulate Rho4 function as a Rho4 GAP in yeast cells.
Fig 5.
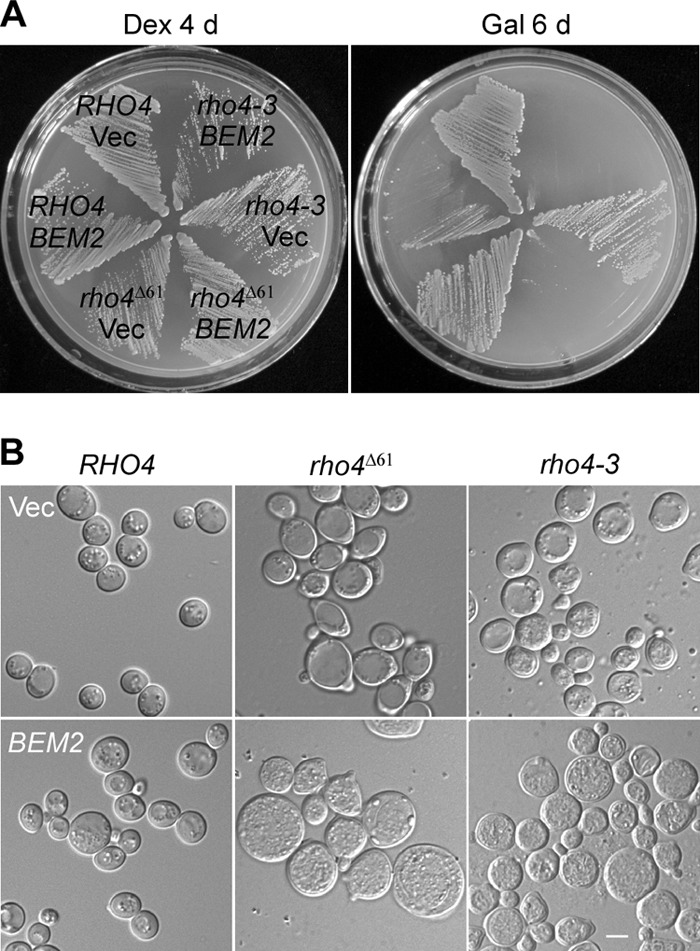
Functional interaction between BEM2 and RHO4 in vivo. (A) Overexpression of BEM2 in rho3Δ rho4 mutants aggravates the growth defect. Plasmid pEGKT316 (Vec) or pEGKT316-BEM2 was transformed into yeast strains JGY2243 (rho3Δ RHO4), JGY2242 (rho3Δ rho4Δ61), and JGY2093 (rho3Δ rho4-3). The cells were streaked on SC-Leu-Ura (containing glucose [Dex]) and SRG-Leu-Ura (containing galactose [Gal]) and incubated at 24°C for 4 and 6 days, respectively. (B) The morphology of cells overexpressing BEM2. Cells as shown in panel A after growth on an SRG-Leu-Ura plate for 4 days were visualized by DIC microscopy. Bar, 5 μm.
DISCUSSION
Functions of Rho4 during yeast budding.
As rho4Δ cells do not display a detectable defect, the exact functions of Rho4 during yeast budding are not readily identified. Rho4 is thought to play a role in the maintenance, but not in the initiation, of bud growth based on the finding that rho3Δ rho4Δ cells often died at the small-budded stage at 30°C (6). In this study, we showed that overexpression of the constitutively active rho4Q131L mutant in rdi1Δ cells caused the accumulation of large, round, unbudded cells with a depolarized actin cytoskeleton, indicating that an excess of Rho4 activity could interfere with bud emergence. In S. pombe, overexpression of wild-type rho4+ as well as the constitutively active rho4G23V or rho4Q74L mutant in wild-type cells induced large, round cells with depolarized actin organization (12, 13). This phenotype is similar to what we observed in Rho4Q131L-overproducing cells. These results suggest that S. cerevisiae Rho4 shares similar functions with S. pombe SpRho4 in the control of polarized growth.
The phenotype shown by cells overproducing Rho4Q131L mutant is similar to that of cells overproducing the constitutively active Bni1ΔN or Bnr1ΔRBD mutant lacking the Rho-binding domain (7, 36). Bni1 and Bnr1 are two formins responsible for the polymerization of G-actin into actin cables. They are thought to be normally kept in a catalytically inactive state by intramolecular inhibition. The binding of Rho GTPase to their N-terminal Rho-binding domains relieves the inhibition, leading to the activation of their actin-nucleating activity (37). Because Rho4 binds to Bni1 and Bnr1 (7, 8) and Rho4 is known to play an important role in the activation of the formins (9), we speculate that the loss-of-cell-polarity phenotype shown by Rho4Q131L-overproducing cells likely results from a hyperactivation of Bni1 and Bnr1. Alternatively, overproduced Rho4Q131L may disrupt actin organization by affecting the functions of the type I myosins and the Arp2/3 complex, which regulate the dynamics of actin patches. This might be achieved by the binding of Rho4Q131L to Exo70. The latter interacts with the myosins and Arp2/3 complex via a protein complex containing Rvs167 (amphiphysin), Vrp1 (verprolin), Las17 (WASP), and G-actin (38).
Apart from a role in regulating actin organization, Rho4 homologs in the yeast S. pombe and C. albicans are required for cell separation since deletion of SpRHO4 (at high temperature) or CaRHO4 caused the formation of short cell chains (12, 13, 15). In contrast, in S. cerevisiae, both rho4Δ and rho3Δ rho4Δ cells did not show a cell separation defect. However, in this study, we found that rho3Δ rho4Δ61 cells expressing an N-terminally truncated version of Rho4 displayed a mild cell separation defect. Some rho3Δ rho4Δ61 cells carry more than one bud, and a fraction of multibudded cells showed a bud whose cytoplasm already separated from the mother cells. This finding suggests that Rho4 may normally have a role in cell separation. Rho4 is known to mediate the interaction between Bnr1 and Hof1, two proteins involved in cytokinesis (39). Like rho3Δ rho4Δ61 cells, hof1Δ cells also displayed a growth defect and multiple buds at high temperature (39). It will be interesting to investigate if Hof1 function is affected in rho3Δ rho4Δ61 cells. The S. pombe SpRho4 and C. albicans CaRho4 are thought to be involved in cell separation by controlling polarized secretion of cell-wall-degrading enzymes such as endo-1,3-glucanases SpEng1, SpAgn1, and CaEng1 to the septation sites (14, 15). In S. cerevisiae, cell-wall-degrading enzymes such as Eng1 also play a critical role in cell separation (40). Further investigation will be required to address whether Rho4 is involved in the polarized secretion of these lytic enzymes.
Role of the N-terminal extension of Rho4.
Rho4 has an unusually long N-terminal extension. Our study on the rho3Δ rho4Δ61 mutant harboring the N-terminally truncated rho4Δ61 allele showed that the N-terminal extension of Rho4 plays an important role in Rho4 function. In contrast to rho3Δ rho4Δ cells that displayed a large, round cell morphology with a small bud (5), the phenotypes of rho3Δ rho4Δ61 cells are quite complex and only partially resemble those of rho3Δ rho4Δ cells. rho3Δ rho4Δ61 cells had a growth defect at high temperature. Moreover, a high percentage of rho3Δ rho4Δ61 cells carried two buds, instead of just one. Furthermore, some of the cells may also have a defect in bud emergence and nuclear segregation.
Regarding the multibudded phenotype, rho3Δ rho4Δ61 cells resemble cdc42G60D mutant cells, except that the phenotype is milder than that of cdc42G60D cells (41). Deletion of BEM2 in a certain strain background is also known to produce a small percentage of cells carrying multiple buds (42). All three mutants are able to produce two or more buds that are growing simultaneously. It was believed that the phenotype of bem2Δ and cdc42G60D cells resulted from increased levels of activated Cdc42 in the cells (41, 42). The mechanism underlying multiple budding in rho3Δ rho4Δ61 cells is not clear.
Rho4 is not the only Rho GTPase that carries a long N-terminal extension. Rho3 also has an N-terminal region that is 13 aa longer than that of Cdc42 but is significantly shorter than that of Rho4 (Fig. 3A). Interestingly, like Rho4, the N-terminal region (amino acid residues 1 to 18) of Rho3 was also critical for its function (43). The N-terminal region of Rho3 contains a cysteine residue at position 5 whose posttranslational modification by palmitoylation is important for Rho3's localization to the plasma membrane. In addition, the N-terminal region is thought to participate in the binding with Exo70, a downstream effector of Rho3, since mutations of two basic residues, R17 and K18, in the Rho3Q74L mutant greatly reduced the binding with Exo70 (43). In the case of Rho4, we found that the N-terminal extension is not required for the normal localization of Rho4 since the Rho4Δ61 mutant localized to bud tip and bud neck normally, as wild-type Rho4 did (data not shown). So far, we do not know the mechanism underlying the observed phenotypes shown by rho3Δ rho4Δ61 cells. The rho4Δ61 allele could be a hyperactive allele, given that it could produce multibudded cells, resembling the cdc42G60D mutation. The yeast Ras2 provides an example that shows that an insert in the molecule of a Ras-family GTPase could modulate its activity. Ras2 contains an unusually long insert about 130 aa in length located near the C terminus that is absent in mammalian H-ras. It was shown that this insert has a negative regulatory function on Ras2 since the expression of a Ras2 mutant lacking the C-terminal insert, but not wild-type Ras2, can rescue lethal mutations in CDC25, a gene encoding a GEF for Ras2 (44). It is possible that the removal of amino acid residues 1 to 61 in Rho4 may affect its intrinsic GTPase activity. Alternatively, the removal of the N-terminal extension may alter the binding affinity of Rho4 with downstream effectors or regulatory proteins. Our two-hybrid assay with Rho4Q131L,Δ61 showed that it interacts with Bem2's RhoGAP domain, similarly to Rho4Q131L. This suggests that Bem2 may not be one of these proteins.
Bem2, a novel GAP for Rho4.
Cdc42 and Rho1 each have several GAPs (4). In contrast, there is only one GAP, Rgd1, identified so far for Rho4. Rgd1 is also a GAP for Rho3 (19). In this study, we identified Bem2 as a potential new GAP for Rho4. We found that Bem2 interacts with Rho4 in a GTP-dependent manner. In addition, the interaction with Rho4 is mediated by the RhoGAP domain of Bem2. Furthermore, overexpression of Bem2 caused stronger growth impairment and cell enlargement in rho3Δ rho4Δ61 and rho3Δ rho4-3 cells than in rho3Δ RHO4 cells. These results suggest that Bem2 plays a negative role in the regulation of Rho4 in vivo.
Bem2 is known to be a GAP for Cdc42 and Rho1 (34, 45). It plays a critical role in cell polarity establishment, as bem2Δ cells are inviable at 37°C and the cells often died as large, round, unbudded, and multinucleate cells with defective actin organization (33). Bem2's function in polarity establishment requires its RhoGAP activity (33). Bem2 is also known to play a GAP-independent role in the morphogenesis checkpoint that delays cell cycle progression at G2 upon perturbation of actin organization (34). Given that Rho4 shares a higher amino acid sequence identity with Rho1 (47% identity) and Cdc42 (46% identity) than with Rho3 (37% identity), it is reasonable that Bem2 interacts with Rho4 in addition to Rho1 and Cdc42.
In this study, we showed that Bem2 also interacted with Rho2. Why does Bem2 have several Rho GTPase targets? Since Cdc42, Rho1, Rho2, and Rho4 play partially redundant roles during bud emergence, we speculate that, by using Bem2 as a global regulator of these Rho proteins, it is convenient for the cells to integrate various signals to the control of Rho GTPases to ensure proper bud assembly. On the other hand, why does Rho4 need Bem2 in addition to Rgd1? We speculate that Bem2 and Rgd1 may play differential roles in the control of Rho4 activity. These two GAPs may coordinately regulate Rho4 activity at different cell cycle stages or locations. Future studies will be required to address the specific roles of each GAP in Rho4 function.
ACKNOWLEDGMENTS
We thank Anthony Bretscher, Erfei Bi, François Doignon, Marc Crouzet, Patrick Brennwald, Thomas Höfken, and Won-Ki Huh for kindly providing plasmids and yeast strains.
This work was supported by grants 30770017 and 30871347 from the National Natural Science Foundation of China and the Chinese 111 Project grant B06018.
Footnotes
Published ahead of print 21 December 2012
REFERENCES
- 1. Ellenbroek SI, Collard JG. 2007. Rho GTPases: functions and association with cancer. Clin. Exp. Metastasis 24:652–672 [DOI] [PubMed] [Google Scholar]
- 2. Ridley AJ. 2006. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 16:522–529 [DOI] [PubMed] [Google Scholar]
- 3. Pruyne D, Bretscher A. 2000. Polarization of cell growth in yeast. II. The role of the cortical actin cytoskeleton. J. Cell Sci. 113:571–585 [DOI] [PubMed] [Google Scholar]
- 4. Park HO, Bi E. 2007. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 71:48–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matsui Y, Toh-e A. 1992. Isolation and characterization of two novel ras superfamily genes in Saccharomyces cerevisiae. Gene 114:43–49 [DOI] [PubMed] [Google Scholar]
- 6. Matsui Y, Toh-e A. 1992. Yeast RHO3 and RHO4 ras superfamily genes are necessary for bud growth, and their defect is suppressed by a high dose of bud formation genes CDC42 and BEM1. Mol. Cell. Biol. 12:5690–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. 1997. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276:118–122 [DOI] [PubMed] [Google Scholar]
- 8. Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. 1997. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 16:2745–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dong Y, Pruyne D, Bretscher A. 2003. Formin-dependent actin assembly is regulated by distinct modes of Rho signaling in yeast. J. Cell Biol. 161:1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Adamo JE, Rossi G, Brennwald P. 1999. The Rho GTPase Rho3 has a direct role in exocytosis that is distinct from its role in actin polarity. Mol. Biol. Cell 10:4121–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robinson NG, Guo L, Imai J, Toh-e A, Matsui Y, Tamanoi F. 1999. Rho3 of Saccharomyces cerevisiae, which regulates the actin cytoskeleton and exocytosis, is a GTPase which interacts with Myo2 and Exo70. Mol. Cell. Biol. 19:3580–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakano K, Mutoh T, Arai R, Mabuchi I. 2003. The small GTPase Rho4 is involved in controlling cell morphology and septation in fission yeast. Genes Cells 8:357–370 [DOI] [PubMed] [Google Scholar]
- 13. Santos B, Gutiérrez J, Calonge TM, Pérez P. 2003. Novel Rho GTPase involved in cytokinesis and cell wall integrity in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 2:521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Santos B, Martín-Cuadrado AB, Vázquez de Aldana CR, del Rey F, Pérez P. 2005. Rho4 GTPase is involved in secretion of glucanases during fission yeast cytokinesis. Eukaryot. Cell 4:1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dunkler A, Wendland J. 2007. Candida albicans Rho-type GTPase-encoding genes required for polarized cell growth and cell separation. Eukaryot. Cell 6:844–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rasmussen CG, Glass NL. 2005. A Rho-type GTPase, rho-4, is required for septation in Neurospora crassa. Eukaryot. Cell 4:1913–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Si H, Justa-Schuch D, Seiler S, Harris SD. 2010. Regulation of septum formation by the Bud3-Rho4 GTPase module in Aspergillus nidulans. Genetics 185:165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tiedje C, Sakwa I, Just U, Hofken T. 2008. The Rho GDI Rdi1 regulates Rho GTPases by distinct mechanisms. Mol. Biol. Cell 19:2885–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Doignon F, Weinachter C, Roumanie O, Crouzet M. 1999. The yeast Rgd1p is a GTPase activating protein of the Rho3 and Rho4 proteins. FEBS Lett. 459:458–462 [DOI] [PubMed] [Google Scholar]
- 20. Bi E, Pringle JR. 1996. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:5264–5675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. James P, Halladay J, Craig EA. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guthrie C, Fink GR. (ed). 1991. Methods in enzymology, vol 194 Guide to yeast genetics and molecular biology. Academic Press, San Diego, CA: [PubMed] [Google Scholar]
- 23. Johnston M, Davis RW. 1984. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol. Cell. Biol. 4:1440–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roumanie O, Peypouquet MF, Bonneu M, Thoraval D, Doignon F, Crouzet M. 2000. Evidence for the genetic interaction between the actin-binding protein Vrp1 and the RhoGAP Rgd1 mediated through Rho3p and Rho4p in Saccharomyces cerevisiae. Mol. Microbiol. 36:1403–1414 [DOI] [PubMed] [Google Scholar]
- 25. Guo J, Gong T, Gao X-D. 2011. Identification of an amphipathic helix important for the formation of ectopic septin spirals and axial budding in yeast axial landmark protein Bud3p. PLoS One 6:e16744 doi:10.1371/journal.pone.0016744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao X-D, Sperber LM, Kane SA, Tong Z, Tong AH, Boone C, Bi E. 2007. Sequential and distinct roles of the cadherin domain-containing protein Axl2p in cell polarization in yeast cell cycle. Mol. Biol. Cell 18:2542–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sung M-K, Huh W-K. 2007. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast 24:767–775 [DOI] [PubMed] [Google Scholar]
- 28. Zhang X, Bi E, Novick P, Du L, Kozminski KG, Lipschutz JH, Guo W. 2001. Cdc42 interacts with the exocyst and regulates polarized secretion. J. Biol. Chem. 276:46745–46750 [DOI] [PubMed] [Google Scholar]
- 29. Fernandes H, Roumanie O, Claret S, Gatti X, Thoraval D, Doignon F, Crouzet M. 2006. The Rho3 and Rho4 small GTPases interact functionally with Wsc1p, a cell surface sensor of the protein kinase C cell-integrity pathway in Saccharomyces cerevisiae. Microbiology 152:695–708 [DOI] [PubMed] [Google Scholar]
- 30. Omholt K, Karsberg S, Platz A, Kanter L, Ringborg U, Hansson J. 2002. Screening of N-ras codon 61 mutations in paired primary and metastatic cutaneous melanomas: mutations occur early and persist throughout tumor progression. Clin. Cancer Res. 8:3468–3474 [PubMed] [Google Scholar]
- 31. Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- 32. Rechsteiner M, Rogers SW. 1996. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21:267–271 [PubMed] [Google Scholar]
- 33. Kim Y-J, Francisco L, Chen G-C, Marcotte E, Chan CSM. 1994. Control of cellular morphogenesis by the Ipl2/Bem2 GTPase-activating protein: possible role of protein phosphorylation. J. Cell Biol. 127:1381–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marquitz AR, Harrison JC, Bose I, Zyla TR, McMillan JN, Lew DJ. 2002. The Rho-GAP Bem2p plays a GAP-independent role in the morphogenesis checkpoint. EMBO J. 21:4012–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang T, Bretscher A. 1995. The rho-GAP encoded by BEM2 regulates cytoskeletal structure in budding yeast. Mol. Biol. Cell 6:1011–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gao L, Bretscher A. 2008. Analysis of unregulated formin activity reveals how yeast can balance F-actin assembly between different microfilament-based organizations. Mol. Biol. Cell 19:1474–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. 2002. Role of formins in actin assembly: nucleation and barbed-end association. Science 297:612–615 [DOI] [PubMed] [Google Scholar]
- 38. Roumanie O, Peypouquet MF, Thoraval D, Doignon F, Crouzet M. 2002. Functional interactions between the VRP1-LAS17 and RHO3-RHO4 genes involved in actin cytoskeleton organization in Saccharomyces cerevisiae. Curr. Genet. 40:317–325 [DOI] [PubMed] [Google Scholar]
- 39. Kamei T, Tanaka K, Hihara T, Umikawa M, Imamura H, Kikyo M, Ozaki K, Takai Y. 1998. Interaction of Bnr1p with a novel Src Homology 3 domain-containing Hof1p. Implication in cytokinesis in Saccharomyces cerevisiae. J. Biol. Chem. 273:28341–28345 [DOI] [PubMed] [Google Scholar]
- 40. Baladrón V, Ufano S, Duenas E, Martín-Cuadrado AB, del Rey F, Vázquez de Aldana CR. 2002. Eng1p, an endo-1,3-beta-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot. Cell 1:774–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Caviston JP, Tcheperegine SE, Bi E. 2002. Singularity in budding: a role for the evolutionarily conserved small GTPase Cdc42p. Proc. Natl. Acad. Sci. U. S. A. 99:12185–12190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Knaus M, Pelli-Gulli MP, van Drogen F, Springer S, Jaquenoud M, Peter M. 2007. Phosphorylation of Bem2p and Bem3p may contribute to local activation of Cdc42p at bud emergence. EMBO J. 26:4501–4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu H, Brennwald P. 2010. The function of two Rho family GTPases is determined by distinct patterns of cell surface localization. Mol. Cell. Biol. 30:5207–5217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marshall MS, Gibbs JB, Scolnick EM, Sigal IS. 1987. Regulatory function of the Saccharomyces cerevisiae RAS C-terminus. Mol. Cell. Biol. 7:2309–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peterson J, Zheng Y, Bender L, Myers A, Cerione R, Bender A. 1994. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J. Cell Biol. 127:1395–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]