Abstract
Cellulases are enzymes that normally digest cellulose; however, some are known to play essential roles in cellulose biosynthesis. Although some endogenous cellulases of plants and cellulose-producing bacteria are reportedly involved in cellulose production, their functions in cellulose production are unknown. In this study, we demonstrated that disruption of the cellulase (carboxymethylcellulase) gene causes irregular packing of de novo-synthesized fibrils in Gluconacetobacter xylinus, a cellulose-producing bacterium. Cellulose production was remarkably reduced and small amounts of particulate material were accumulated in the culture of a cmcax-disrupted G. xylinus strain (F2-2). The particulate material was shown to contain cellulose by both solid-state 13C nuclear magnetic resonance analysis and Fourier transform infrared spectroscopy analysis. Electron microscopy revealed that the cellulose fibrils produced by the F2-2 cells were highly twisted compared with those produced by control cells. This hypertwisting of the fibrils may reduce cellulose synthesis in the F2-2 strains.
INTRODUCTION
Cellulose is one of the most abundant biopolymers on earth and is produced by plants, algae, and some types of bacteria (1). It is a linear polymer of β-1,4-linked glucan chains, which are held together by hydrogen bonds to form cellulosic microfibrils (2). In cellulose I, glucan chains are arranged in a parallel fashion. Although cellulose I is the most prominent type of crystalline form in microfibrils, it can be converted to cellulose II by treatment with chemical agents, such as strong alkali (3). Plant and bacterial celluloses are synthesized on the plasma membrane, and several glucan chains assemble to form crystalline microfibrils outside the plasma membrane (4, 5). Plant cellulose molecules produced on the plasma membrane assemble into cellulose networks within cell walls, and the crystallized celluloses are Iβ-enriched cellulose. In contrast, bacterial cellulose molecules produced by the cellulose synthase complex on the plasma membrane must pass through the bacterial outer membrane to form cellulose fibrils, and most bacterial cellulose molecules after crystallization become cellulose Iα (6, 7). Although cellulose synthase complexes located on the plasma membrane play essential roles in cellulose formation, other proteins are also involved.
Plants and cellulose-producing bacteria have several cellulases. Endo-β-1,4-glucanases, the Korrigan (KOR) protein of Arabidopsis thaliana (a plant), and carboxymethylcellulase (CMCax) of Gluconacetobacter xylinus (a cellulose-producing bacterium) are endogenous cellulases that putatively digest celluloses. Several studies have shown that the KOR protein is involved in cellulose synthesis (8, 9). An insertional mutant and a mutant carrying a point mutation for an endogenous plant cellulase, known as KOR1, show typical phenotypes defined by the prevention of cellulose synthesis in primary and secondary cell walls (9, 10). Several hypotheses regarding the function of KOR have been proposed, including (i) separation of a postulated cellulose primer from the nascent polymer, (ii) separation of completed cellulose molecules from the cellulose synthase complex, and (iii) cleavage of noncrystalline glucan molecules from crystalline cellulose microfibrils to remove the tensional stress that might arise during the assembly of a large number of glucan chains into fibrils (6). However, the role of KOR in cellulose synthesis is unclear.
We used G. xylinus as a model system for investigating the role of endogenous cellulase in cellulose fibril formation. G. xylinus cellulose is produced by a cellulose synthase complex on the plasma membrane with protein components encoded by a single operon (bcsABCD or acsABCD) (11–14). The other operon constructed from two open reading frames (ORFs) is located upstream of the bcs operon (Fig. 1A), which encodes CMCax, which might be responsible for cellulose hydrolysis (15). Several findings support the possible involvement of CMCax in bacterial cellulose synthesis. The addition of endoglucanase to the culture or overproduction of cellulase by G. xylinus enhances the yield of cellulose synthesis (16, 17), whereas the addition of antibodies to recombinant glucanase inhibits the formation of the cellulose fibrils (18). In the present study, we describe a novel type of cellulose ribbon in a mutant strain of G. xylinus carrying a disrupted cellulase gene.
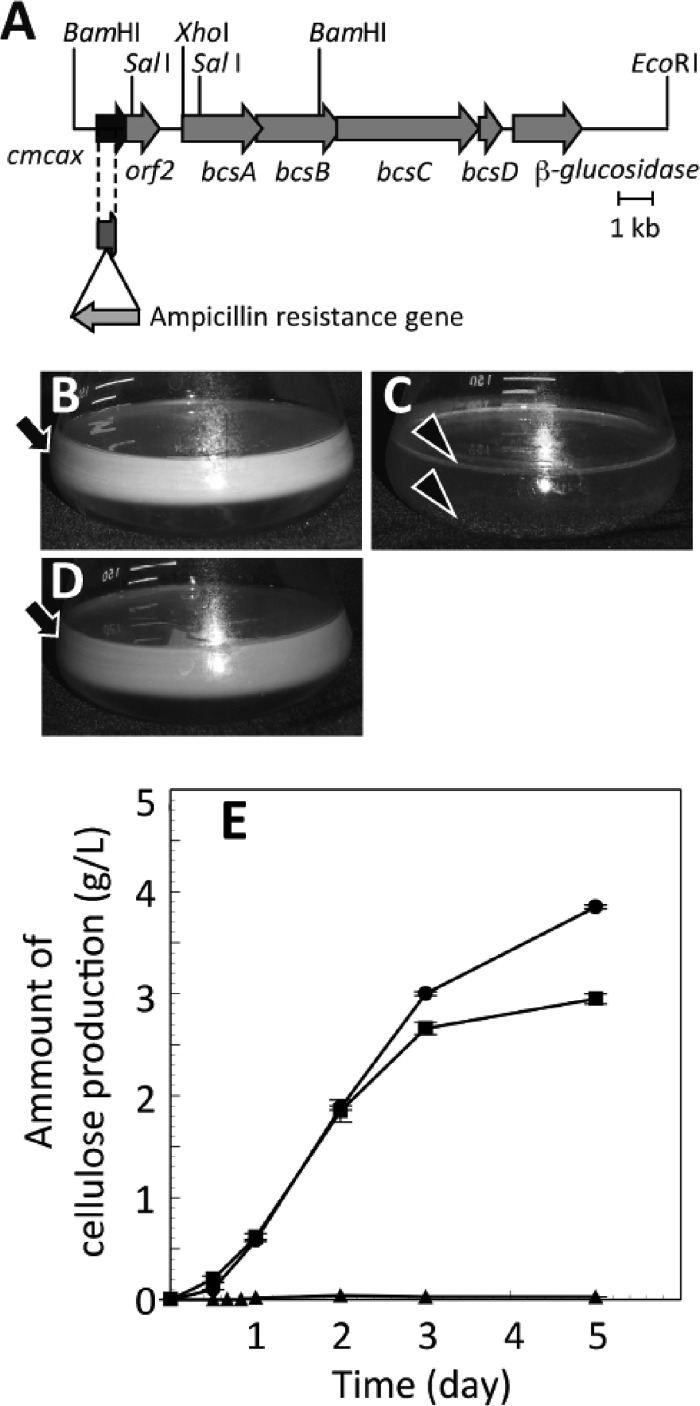
Fig 1.
A loss-of-function mutation in cmcax of G. xylinus suppressed de novo cellulose synthesis. (A) Schematic diagram of the bacterial cellulose synthase (bcs) operon and its flanking region in strain PBR2001 (wild type). In a loss-of-function cmcax mutant, F2-2, an ampicillin resistance gene was inserted at the cmcax site located upstream of the bcs operon. For construction of the cmcax-complemented strain F2-2(pSA-CMCase/k), a 1.9-kb fragment including a full-length cmcax sequence was excised using the restriction enzymes BamHI and SalI. This fragment was used in subsequent experiments. (B to D) Comparison of products from static cultures of G. xylinus grown at 30°C for 1 week: wild type (B), F2-2 (C), and F2-2(pSA-CMCase/k) (D). The arrows show cellulose pellicles, and the arrowheads show particulate materials. (E) The time course of cellulose synthesis during culture on a rotary shaker. Filled circle, wild type; filled triangle, F2-2; filled square, F2-2(pSA-CMCax/k). Each point shows the mean and SEM obtained from three samples.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
G. xylinus strain BPR2001 and Escherichia coli strains DH5α and TB1 were used in this study. The strain BPR2001 was isolated by Toyosaki et al. (19) and deposited at the National Institute of Technology and Evaluation-International Patent Organism Depositary (accession number FERM BP-4545). E. coli was grown in Luria-Bertani (LB) medium (20) at 37°C. G. xylinus was grown in fructose-peptone-yeast extract (FPY) medium (2% fructose, 1% polypeptone, 0.5% yeast extract, and 0.25% K2HPO4) at 30°C on a rotary shaker (180 rpm) for 2 to 3 days. For exopolysaccharide analysis, G. xylinus was statically grown in corn steep liquor-fructose (CSL-Fru) medium (21) at 30°C for 3 days, following which the cellulose pellicle was removed by filtration. CSL-Fru medium contains 20 ml/liter corn steep liquor, 40 g/liter fructose, 3.3 g/liter (NH4)2SO4, 14.7 mg/liter CaCl2·2H2O, 1.0 g/liter KH2PO4, 3.6 mg/liter FeSO4·7H2O, 2.42 mg/liter Na2MoO4·2H2O, 250.0 mg/liter MgSO4·7H2O, 1.73 mg/liter ZnSO4·7H2O, 1.39 mg/liter MnSO4·5H2O, 0.05 mg/liter CuSO4·5H2O, 2.0 mg/liter inositol, 0.4 mg/liter niacin, 0.4 mg/liter pyridoxine HCl, 0.4 mg/liter thiamine HCl, 0.2 mg/liter calcium pantothenate, 0.2 mg/liter riboflavin, 0.2 mg/liter p-aminobenzoic acid, 0.002 mg/liter folic acid, and 0.002 mg/liter biotin (pH 5.0). The cell suspension was inoculated into fresh medium in a baffled shake flask and cultured at 30°C on a rotary shaker (180 rpm) for 5 days. To select transformants with resistance markers, the following antibiotics were used: ampicillin (50 μg/ml), chloramphenicol (50 μg/ml), and kanamycin (100 μg/ml). The E. coli plasmid vectors used in this study were pUC119, pHSG399, and pHSG299 (TaKaRa Bio Inc., Otsu, Japan). The plasmid vector pSA19 (22) was used as a shuttle vector between G. xylinus and E. coli.
Electroporation.
For preparing E. coli-competent cells, E. coli was grown in LB medium and washed with ice-cold 10% glycerol. Plasmid DNA (1 μg) was introduced into the competent E. coli cells by electroporation using the Cell-Porator apparatus (Gibco BRL, Carlsbad, CA). Electroporation was performed in 0.15-cm cuvettes at a field strength of 1.9 kV/cm, with the capacitor and resistor set at 330 μF and 4 kΩ, respectively. After electroporation, the cells were incubated in 1 ml of LB medium at 37°C for 1 h and then plated onto LB agar plates containing appropriate antibiotics. The plates were incubated at 37°C until colonies were formed.
For preparing G. xylinus-competent cells, G. xylinus was grown in FPY medium containing 1% (vol/vol) cellulase (Celluclast; Novo Nordisk A/S, Bagsvard, Denmark) and washed with ice-cold 10% glycerol. Plasmid DNA (5 μg) was introduced into G. xylinus by electroporation using the Cell-Porator apparatus. Electroporation was performed in 0.15-cm cuvettes at a field strength of 2.1 kV/cm, with the capacitor and resistor set at 330 μF and 8 kΩ, respectively. After electroporation, the cells were incubated in 1 ml of FPY medium containing 1% (vol/vol) cellulase at 30°C for 3 h and then plated onto FPY agar plates containing appropriate antibiotics. The plates were incubated at 30°C until colonies were formed.
Gene disruption and generation of recombinant strains.
The β-lactamase gene, which provides ampicillin resistance (Ampr), was amplified as a 1.1-kb PCR product from pUC119 using oligonucleotide primers Amp1 (5′-GTCAGGTGGCACTTTTCGGG-3′) and Amp2 (5′-GCTCAGTGGAACGAAAACTCACG-3′). To disrupt the gene encoding CMCax, a 0.8-kb ClaI-HincII fragment from cosmid pAM9, isolated as a plasmid including the entire region of the bcs operon from a wild-type genomic library (Fig. 1A) (23), was subcloned into pHSG399. The nucleotide sequence data of cmcax appears in the DNA Data Bank of Japan, European Molecular Biology Laboratory, and GenBank nucleotide sequence databases under the accession number AB010645. The cmcax gene corresponds to nucleotides (nt) 869 to 1994 in this nucleotide sequence. The Ampr gene was inserted at the StuI site on the 0.8-kb ClaI-HincII fragment, and the resultant plasmid was linearized and transformed into strain BPR2001 (wild type), yielding the mutant strain F2-2. To confirm the insertion of the Ampr gene into the F2-2 genomic DNA, genomic Southern hybridization was performed by the capillary transfer method (24) (see Fig. S1 in the supplemental material). The probe DNA was prepared using the digoxigenin (DIG) labeling and detection system (Roche Diagnostics, Tokyo, Japan). CMC-1 (5′-TAGCGGCGAATCCCACAGCG-3′) and CMC-2 (5′-TCGAGCGTGGGCAGTTCACC-3′) were used as primers, and the total wild-type DNA was used as a template in the PCR. Prehybridization and hybridization were performed at 68°C according to the instructions for the DIG labeling and detection kit (Roche Diagnostics). The blotting membranes were washed twice with 2× salt-sodium citrate (SSC) buffer (20× SSC contains 3 M NaCl and 0.3 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) at room temperature for 5 min and then washed twice with 0.1× SSC-0.1% SDS at 68°C for 15 min.
Plasmid construction for the complementation experiment.
We previously constructed a plasmid (25) containing the BamHI-XhoI fragment (Fig. 1A) from pAM9 (23). The plasmid was digested with SalI and Sse8387I, and the digested ends were filled by T7 DNA polymerase (TaKaRa Bio Inc.) and then self-ligated. The resultant plasmid contained a 1.9-kb fragment including the entire cmcax sequence (Fig. 1A). The constructed plasmid was digested at the SmaI site and ligated to a kanamycin resistance (Kmr) gene amplified from pHSG299 as the template DNA by PCR using the primers Km-1 (5′-GTGAAGAAGGTGTTGCTGAC-3′) and Km-R1 (5′-CTGGCGTAATAGCGAAGAGG-3′). The resultant plasmid was designated pSA-CMCase/k.
Cellulose analyses.
The amount of celluloses in the culture was measured according to the method described by Chao et al. (26).
For the analysis of solid-state 13C nuclear magnetic resonance (NMR) and Fourier transform infrared (FTIR) spectroscopy, specimens were prepared according to the following method. Using forceps, we removed sheets of cellulose film from cellulose pellicles produced by the control cells. In the mutant F2-2 culture, particulate material was collected by centrifugation. The pellet was resuspended in 10 mM K-acetate buffer (pH 5.0) and recentrifuged at 5,000 rpm for 20 min. In the F2-2 culture, cells grew rapidly to a large concentration. In order to avoid contamination of the cell fragments, we washed the resultant pellet with a sodium hydrate solution. The cellulose product (pellicle or particulate material of the mutant) was stored in 0.1 M NaOH at 80°C overnight and then washed several times with hot water. The purified cellulose and alkali-insoluble materials were dried at 60°C overnight. Avicel (microcrystalline cellulose) (Merck, Darmstadt, Germany) was treated with 4 M NaOH at 80°C and washed several times with hot water. These products were analyzed by solid-state 13C NMR and FTIR spectrometry.
High-resolution solid-state 13C NMR spectra were recorded on a Chemagnetics Infinity 400 spectrometer (13C: 100.6 MHz) (Varian, Inc., Santa Clara, CA) using cross-polarization–magic-angle sample spinning (CP-MAS). The spectral width, acquisition time, and repetition time for CP-MAS were 40 kHz, 50 ms, and 4 s, respectively. The contact time for cross polarization was 1 ms. Free induction decay signals were acquired with 2,048 data points and Fourier transformed as 32,768 data points after 30,720 data points were zero filled. The π/2 pulses for carbon and protons were 5.0 μs, and the sample spinning rate was 4 kHz. The 13C chemical shifts were referenced to the carboxyl signal of glycine (176.03 ppm from tetramethylsilane [TMS]) and expressed as relative shifts from the TMS value.
FTIR spectra were recorded on a Spectrum One spectrometer (PerkinElmer Inc., Waltham, MA) equipped with a doped triglycine sulfate (DTGS) detector in an attenuated total reflection mode. A total of 16 scans were performed at a resolution of 4 cm−1 in the range of 4,000 to 400 cm−1 and averaged. The prepared samples were dried at 60°C overnight and then analyzed.
Calcofluor white staining and light microscopy.
G. xylinus cells were cultivated for 7 days and collected by centrifugation. The resultant pellet was resuspended in 250 μl of fresh FPY medium. The suspension was incubated on polyethylenimine-coated slide glasses at 25°C for 1 h. The cells were fixed with 4% formaldehyde in 50 mM K-acetate buffer (pH 5.0) and then stained with 0.005% calcofluor white M2R dissolved in 50 mM K-acetate buffer. The stained cells were observed using an optical microscope (Optiphot-2; Nikon, Tokyo, Japan) equipped with a charge-coupled-device camera (Digital Sight DS-L1; Nikon).
Electron microscopy.
G. xylinus cells were incubated on Formvar-coated copper grids at 30°C for 1 h. The cells were kept wet. The cells on the grids were washed with a few drops of distilled water and then negatively stained with 1.5% aqueous uranium acetate. Specimens were observed with a JEOL-1010 or JEOL-2000EX electron microscope (JEOL Ltd., Tokyo, Japan) at 80 kV. Electron microscope films were digitized, and the half-pitch and width of cellulose fibrils were determined using Photoshop CS2 software (Adobe Systems, Tokyo, Japan). The F test and two-tailed Student's t test were performed using Microsoft Excel 12.0 (Microsoft Japan, Tokyo, Japan).
RESULTS
Cellulose production in the culture of the cmcax-disrupted strain.
To investigate the role of CMCax in bacterial cellulose synthesis, we prepared an artificial mutant strain, F2-2, with a disrupted cmcax sequence accompanied by insertion of the Ampr gene (Fig. 1A; see also Fig. S1 in the supplemental material). We also constructed a cmcax complement strain, F2-2(pSA-CMCase/k), in which the complementation plasmid pSA-CMCase/k (an approximately 1.9-kb fragment of G. xylinus DNA with cmcax and a Kmr gene included in the shuttle vector pSA19) was inserted into F2-2. A 5-mm-thick pellicle (Fig. 1B, arrow) was formed after 1 week of static culture of the wild-type strain. Pellicles were not formed in the 1-week static F2-2 culture, but small amounts of particulate material (Fig. 1C, arrowheads) were observed at the bottom of the flask or attached to the inner surface of the flask at the border between the medium and the air. As expected, a thick pellicle (Fig. 1D, arrow) was also formed in the static F2-2(pSA-CMCax/k) culture.
Changes in the amounts of cellulose were compared among these strains in agitation cultures prepared over 5 days on a rotary shaker. The amount of cellulose produced by F2-2(pSA-CMCax/k) cultured for 5 days was 77% of that produced by the wild-type strain; however, the amount of cellulose produced by F2-2 was very small (Fig. 1E). These results indicate that disruption of cmcax causes a remarkable reduction in cellulose production by G. xylinus.
Presence of cellulose II in the particulate material produced by the F2-2 culture.
To examine whether the F2-2 culture produced cellulose, the chemical nature of the particulate material in the static culture was investigated. The sample was prepared from the static culture because the amount of particulate material was greater than that found in the agitated culture. Solid-state 13C NMR spectra were obtained with proton-carbon CP-MAS. The 13C CP-MAS NMR spectra of alkali-insoluble materials from wild-type (Fig. 2Aa) and F2-2(pSA-CMCax/k) (Fig. 2Ab) pellicles were found to be very similar to previously published spectra of cellulose I (7). In contrast, the C6 signal in the spectrum of the alkali-insoluble material obtained from the F2-2 culture was shifted upfield (Fig. 2Ac). Furthermore, the C4 signal (Fig. 2Ac, dagger) was also shifted slightly upfield, and the C1 signal became a doublet at 104.5 and 105.6 ppm. The spectrum of the particulate material obtained from the F2-2 culture was similar to that of NaOH-treated Avicel (see Materials and Methods), which represents typical cellulose II (Fig. 2Ad). These observations (Fig. 2A) suggest that the particulate material from the F2-2 culture contained cellulose with a crystalline structure similar to that of cellulose II.
Fig 2.
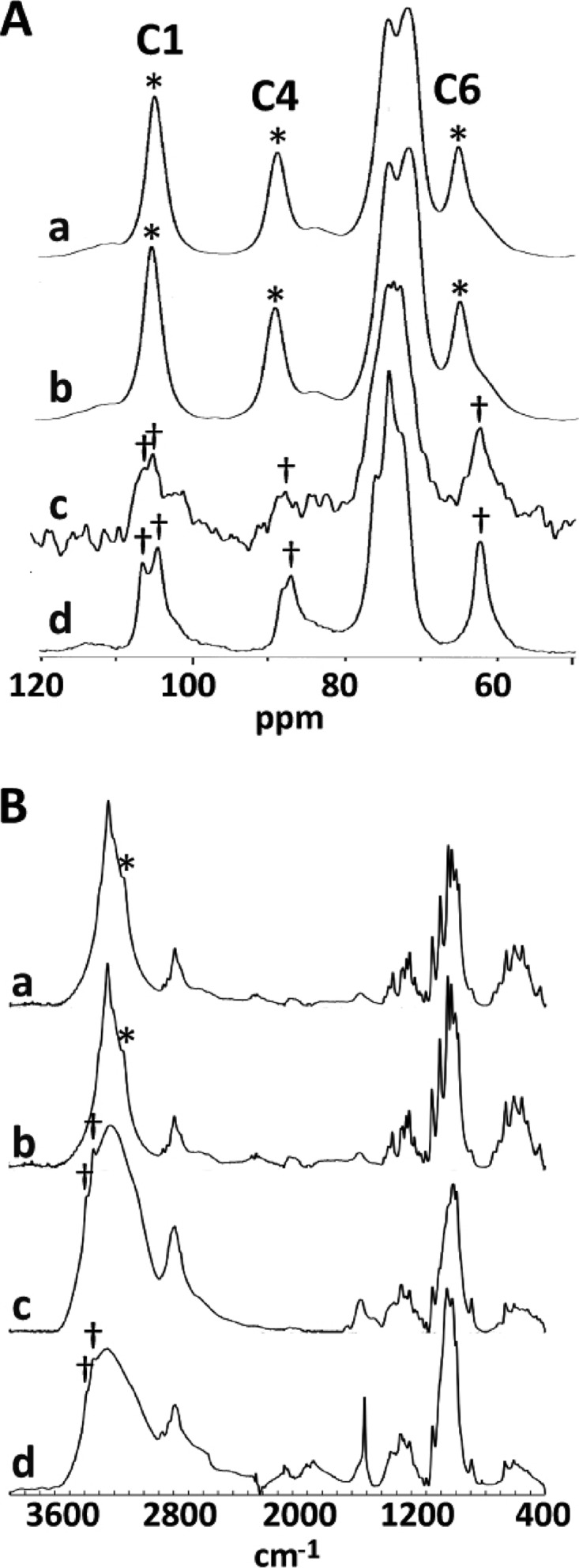
An alkali-insoluble fraction obtained from the culture of the F2-2 strain containing cellulose II. (A) 13C CP-MAS NMR spectra of alkali-insoluble materials. (B) FTIR spectra of alkali-insoluble materials. Alkali-insoluble materials were prepared from cellulose pellicles produced by wild-type cells (a) (cellulose I), cellulose pellicles produced by F2-2(pSA-CMCax/k) cells (b), and particulate material produced by F2-2 (c). (d) A cellulose II spectrum (Avicel washed with 4 M NaOH). Asterisks and daggers in panel A indicate the positions of peaks in C1, C4, and C6 of cellulose I and cellulose II, respectively; those in panel B indicate the positions of distinctive peaks in cellulose I (3,240 cm−1) and cellulose II (3,440 and 3,480 cm−1), respectively.
FTIR spectrometry also revealed a significant difference in the spectra of the cellulose of the wild-type and F2-2 cultures (Fig. 2B). In the OH-stretching frequency region from 3,600 to 3,000 cm−1, the spectrum of the cellulose specimen from the F2-2 culture contained two bands near 3,440 and 3,480 cm−1 (Fig. 2Bc, dagger). These bands were not present in the spectrum of the wild-type strain (Fig. 2Ba) and were assigned to the intramolecular hydrogen bonds of O3 H···O5 in cellulose II (27). Similar bands were also detected in the NaOH-treated cellulose (Fig. 2Bd, dagger), suggesting that the main component of the alkali-insoluble fraction from F2-2 is cellulose II. In contrast, these bands were absent in the spectrum of F2-2(pSA-CMCax/k). Instead, a band was present near 3,240 cm−1 (Fig. 2Bb, asterisk). This absorption band was observed only for cellulose Iα (28).
Highly twisted cellulose fibrils associated with F2-2.
To investigate the morphology of the cellulose fibrils produced by strain F2-2, negatively stained bacteria were examined by transmission electron microscopy. Approximately 10% of the examined cells had cellulose fibrils in the wild-type and F2-2(pSA-CMCax/k) cultures (Fig. 3A and C). Although the frequencies were very low (approximately 1% of all cells), cellulose fibrils associated with the F2-2 cells were observed (Fig. 3B). The existence of cellulose fibrils in F2-2 cells was also confirmed by fluorescence microscopy of cells stained with 0.005% calcofluor white M2R (see Fig. S2 in the supplemental material). The cellulose fibrils produced by F2-2 cells (Fig. 3E to G) were more twisted than those produced by the wild-type (Fig. 3D) or F2-2(pSA-CMCax/k) (Fig. 3H) cells.
Fig 3.
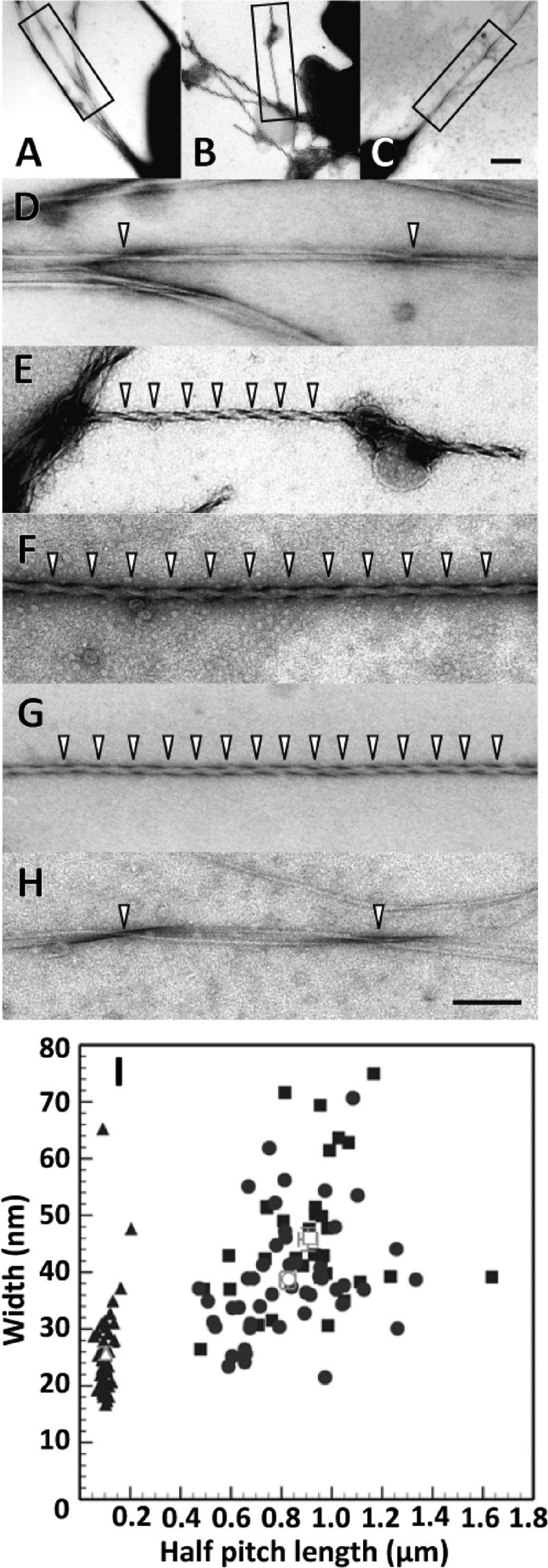
Cellulose fibrils produced by the F2-2 cells were highly twisted. (A to H) Electron micrographs of negatively stained cellulose fibrils. Images A and D show cellulose fibrils produced by a wild-type cell. Images B, E, F, and G show cellulose fibrils produced by the F2-2 cells. Images C and H show the cellulose fibrils produced by a F2-2(pSA-CMCax/k) cell. Images D, E, and H are enlargements of the regions within the rectangle in the images A, B, and C, respectively. The arrowheads indicate twisted points in each fibril. Images A to C are at the same magnification, and images D to H are at the same magnification. The scale bar in image C is 400 nm, and that in H is 200 nm. (I) Relationships between the half-pitch and width of the cellulose fibrils produced by wild-type cells (filled squares) (n = 50), F2-2 (filled triangles) (n = 34), and F2-2(pSA-CMCax/k) cells (filled circles) (n = 47). Open symbols and bars denote the means and SEM, respectively. In the wild-type cells, observations were carried out for 6 independent experiments, and the half-pitches and widths of 34 fibrils produced by 33 cells were determined. In the F2-2 cells, the 9 independent experiments were performed, and the half-pitches and widths of 50 fibrils produced by 24 cells were determined. In the F2-2(pSA-CMCax/k) cells, the half-pitches and widths of 47 fibrils produced by 46 cells were determined from 4 independent experiments.
To characterize the morphology of the fibrils, we further quantified their half-pitches and widths (Fig. 3I). The half-pitches of the fibrils produced by F2-2 cells were 103.0 ± 3.5 nm (mean ± standard error of the mean, n = 50), which was significantly different from those of the fibrils produced by the wild-type (906.1 ± 37.4 nm, n = 34, P < 0.0001 by two-tailed t test) and F2-2(pSA-CMCax/k) (825.4 ± 30.2 nm, n = 47, P < 0.0001) strains. The widths of the fibrils produced by the F2-2 strain (25.0 ± 1.2 nm, n = 50) were approximately half of those of the fibrils produced by the wild-type strain (45.8 ± 2.0 nm, n = 34, P < 0.0001) and two-thirds of those of the fibrils produced by the F2-2(pSA-CMCax/k) strain (38.6 ± 1.5, n = 47, P < 0.0001) (Fig. 3G). In contrast, the widths of the fibrils produced by the complement strain were slightly less than those of the fibrils produced by the wild-type strain (P = 0.005), although no significant differences were observed between the half-pitches of the two fibrils (P = 0.094). These results indicate that fibrils produced by the F2-2 strain were highly twisted compared with those produced by the control cells.
DISCUSSION
Cellulose ribbons produced by wild-type G. xylinus strains are gently twisted, with a pitch of approximately 900 nm (Fig. 3D). In contrast, de novo-synthesized ribbons became entangled and highly twisted with a pitch of approximately 103 nm in the F2-2 strain (Fig. 3E to G). Morphological alterations of cellulose ribbons by the addition of cellulose-binding molecules have been reported. Calcofluor white (29) is a fluorescent agent that binds to cellulose and induces fibrils with broad bands that do not condense laterally in the ribbon. The cellulose-binding domain (CBD) (30) is a polypeptide that binds to cellulose and interferes with its synthesis. Electron microscopic observations have shown that the fibrils produced in the presence of the CBD are splayed ribbons composed of separate fibrillar subunits. Similar splayed ribbons have also been reported in wild-type bacteria cultured in the presence of CMCax (16). The highly twisted ribbon in the cmcax-disrupted strain represents a morphology that is markedly different from that of the cellulose ribbons altered by these cellulose-binding molecules; thus, the ribbons presented in this study may represent a novel type of alternation of cellulose ribbons. The addition of CMCax (16) and loss of function of CMCax (in this study) affected the morphologies of the cellulose ribbons; the addition produced a splayed ribbon, and the loss of function formed a highly twisted ribbon. These observations suggest an essential role of CMCax in cellulose assembly.
Cellulose production decreased remarkably in the F2-2 strain (Fig. 1); however, highly twisted ribbons associated with a bacterium were observed by electron microscopy (Fig. 3). Solid-state 13C NMR and FTIR spectroscopy experiments on particulate materials from the F2-2 culture (Fig. 2) showed the existence of cellulose in the F2-2 strains. These results suggest that the highly twisted ribbons reported in this study were cellulose ribbons. The low productivity of cellulose in the mutant can be explained if the formation of highly twisted ribbons causes reduction of cellulose synthesis or if the synthesis stops prematurely in the mutant when highly twisted ribbons are formed.
G. xylinus strain AY201 has two genes coding for cellulose synthetase: acsAB of the acs operon and acsAII (31, 32). In mutants with disrupted acsAB, in vivo cellulose production was not detected, but in vitro cellulose synthase activity was observed (11). The disruption of both acsAB and acsAII was necessary to inhibit in vitro cellulose synthase activity (31). A comparative genomic analysis of G. xylinus NBRC 3288, a cellulose-nonproducing strain, with other cellulose-producing strains showed that a nonsense mutation in the catalytic subunit of cellulose synthase (BcsB1) caused the cellulose-nonproducing phenotype (33). This evidence indicates that the loss of function of the cellulose synthase genes completely inhibited cellulose synthesis. In this study, residual cellulose was detected in the culture of the cmcax-disrupted mutant, suggesting the functional differences between cellulose synthase and CMCax. Complementation experiments using the point-mutated CMCax based on available information about its crystal structure (34) and hydrolysis activity (15) should be helpful for elucidating the function of CMCax in cellulose production.
It is well established that cellulose polymerization and crystallization are coupled reactions during cellulose synthesis in G. xylinus and that crystallization limits the rate of polymerization (2, 11, 35). CMCax digests amorphous cellulose; however, it cannot digest the crystalline form (15). One possible explanation for the hypertwisted ribbons is that disruption of cmcax causes irregular packing of the de novo-synthesized fibrils (Fig. 4). The cellulose produced by the wild-type G. xylinus strain is rich in cellulose Iα (7). The present results show that the cellulose produced by the F2-2 cells was cellulose II (Fig. 2). Because cellulose II is thermodynamically more stable than cellulose I (36), inhibition of cellulose I crystallization in the mutant strain may cause accumulation of cellulose II in the particulate cellulose of the mutant culture. In this study, sodium hydrate solution was used to prepare samples for solid-state 13C NMR and FTIR analysis from the F2-2 culture. A high concentration of sodium hydrate solution (>4 M) digested the hydrogen bonds between β-1,4-d-glucan chains and caused changes in the cellulose allomorph from cellulose I to the more stable form, cellulose II. However, the concentration of sodium hydrate solution used in this study was very low (0.1 M [0.4%]), and cellulose I may not have been converted to cellulose II (3). Of course, there still remains the possibility that cellulose I can be converted to cellulose II even in low concentrations of sodium hydrate solution under conditions where the cellulose fibrils do not crystallize completely.
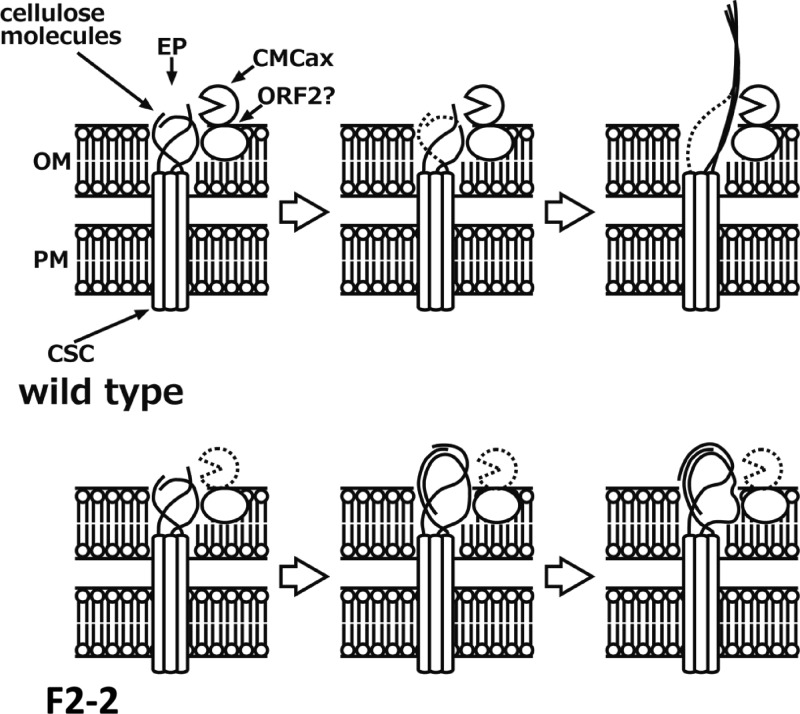
Fig 4.
Schematic representation of the possible mechanism of cellulose fibril formation in wild-type and F2-2 cells. The cellulose synthase complex (CSC) is a transmembrane protein complex that spans the plasma membrane (PM) and outer membrane (OM). Cellulose molecules synthesized by CSCs tend to tangle in the OM. In wild-type cells, CMCax near the extrusion pore (EP) cleaved the tangled cellulose molecules to allow normal crystallization. ORF2 likely assists the crystallization. In F2-2 cells, the irregularly packed cellulose molecules cannot detangle because of the loss of the cellulase activity of CMCax. These irregularly packed cellulose molecules reduce the rate of cellulose synthesis. In some F2-2 cells, the tangled cellulose fibrils transversed the EP, although the cellulose fibrils tended to twist.
In our previous study, we showed that an orf2-disrupted strain produced cellulose I, cellulose II, and amorphous cellulose (25). Together, orf2 (located downstream of cmcax) and cmcax comprise an operon (Fig. 1A) (15, 25). In an orf2-disrupted strain, cmcax may be normally expressed because it locates upstream of orf2. Therefore, cellulose I is produced by the function of CMCax in orf2-disrupted strains. The facts that cellulose production in the F2-2 culture was lower than that in the orf2-disrupted culture (Fig. 1E) (25) and that the F2-2 strain mainly produces cellulose II (Fig. 2) suggest that CMCax and ORF2 polypeptides collaborate in cellulose crystallization and that cellulose I crystallization is severely interrupted without the function of CMCax.
The G. xylinus cellulose synthase complex is composed of transmembrane proteins that span the plasma and outer membranes and are arranged in a row on the membrane along the longitudinal axis of the cell (35, 37). A row of pores (Fig. 4, EP) with 3.5-nm diameters has been found on the outer surface of the outer membrane (38). These pores are considered to be extrusion pores for cellulose fibrils and are somehow involved in the correct crystallization of cellulose fibrils. There is a possibility that CMCax and/or ORF2 near the pore may assist or function as components of the “extrusion machine” (Fig. 4, upper row). CMCax can cleave the tangled celluloses to crystallize correctly when de novo-synthesized cellulose molecules fail to arrange themselves correctly before or during extrusion, or the CMCax sorts out tangles in the cellulose fibrils near the pore in case tensional stress arises during the assembly of cellulose fibrils. In the F2-2 strain, without inhibited CMCax cellulase activity, the cellulose molecules produced by the cellulose synthase complex tend to tangle, and the cellulose fibrils that accidentally crystallize are composed of cellulose II molecules (Fig. 4, lower row).
The AcsD subunit, a product of acsD comprising the cellulose synthase operon of G. xylinus ATCC 53582, is reportedly involved in cellulose crystallization, as the acsD mutant reduced the amounts of two cellulose allomorphs (cellulose I and cellulose II) (11). Wild-type bcsD is a homolog of acsD of G. xylinus ATCC 53582 (11, 23). Cellulose productivity in the N-terminal deletion mutant of bcsD also decreased, although cellulose crystallization produced by the mutant was not investigated (39). These observations suggested that CMCax worked together with BcsD in cellulose crystallization in the wild-type strain.
Individual β-1,4-d-glucan chains are synthesized by cellulose synthase complexes, which are arranged in a row on the cell membrane and associate to form crystalline fibrils. The structure of BcsD (AxCeSD), a component of the cellulose synthase complex, was found to be a homo-octamer comprised of four dimer subunits with a central pore and cylindrical structure (39). The interfaces between the dimers form four spiral interstices on the wall of the molecular cylinder at a 50° angle from the cylinder (vertical) axis (39). There is a possibility that the spiral interstices may affect the twist formation in cellulose microfibrils without cell rotation. It is also possible that when microfibrils are secreted by bacterial cells, the synthesized cellulose fibrils may become distorted without the action of CMCax, thereby producing highly twisted ribbons. CMCax helps to remove the tensional stress of the glucan molecules within the crystalline cellulose microfibrils that may arise during the assembly of a large number of glucan chains into fibrils. Further studies will be necessary to elucidate the twisting mechanism of bacterial cellulose fibrils.
Data obtained from the present study on bacteria show the possibility of hypertwisting of cellulose fibrils upon disruption of the cellulase gene. This may also apply to plant-based cellulases such as KOR, although there are structural differences, such as the lack of the plasma membrane-binding domain of KOR in CMCax (40). In future studies, in order to further elucidate the functions of cellulases, we will examine whether KOR or other cellulases, such as the celC product in Agrobacterium, contribute to their own cellulose synthesis (41) and whether celC can compensate for CMCax in G. xylinus cellulose production.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Hayashi (Tokyo University of Agriculture) for useful discussions.
This work was partly supported by grants from the University of Hyogo (to T.N.) and KAKENHI on Priority Areas (17049019) (from MEXT to Y.M.).
T.N., D.Y., and Y.M. designed the experiments; T.N., Y.S., and M.S. produced and characterized the mutants; S.T. performed the NMR experiments; T.I. and J.S. performed the FTIR experiments; and T.N., H.S., K.O., M.T., and Y.M. made the EM observations and performed analysis. Each author has discussed the results and commented on the manuscript.
We declare no conflicts of interest.
Footnotes
Published ahead of print 14 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01473-12.
REFERENCES
- 1. Delmer DP, Amor Y. 1995. Cellulose biosynthesis. Plant Cell 7:987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Haigler CH, Benziman M. 1982. Biogenesis of cellulose I microfibrils occurs by cell-directed self-assembly in Acetobacter xylinum, p 273–297 In Brown RM., Jr. (ed), Cellulose and other natural polymer systems. Plenum Press, New York, NY [Google Scholar]
- 3. Shibazaki H, Kuga S, Okano T. 1997. Mercerization and acid hydrolysis of bacterial cellulose. Cellulose 4:75–87 [Google Scholar]
- 4. Ross P, Mayer R, Benziman M. 1991. Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 55:35–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Somerville C. 2006. Cellulose synthesis in higher plants. Annu. Rev. Cell and Dev. Biol. 22:53–78 [DOI] [PubMed] [Google Scholar]
- 6. Albersheim P, Darvill A, Roberts K, Sederoff R, Staehelin A. 2010. Plant cell walls: from chemistry to biology. Garland Science, New York, NY [Google Scholar]
- 7. VanderHart DL, Atalla RH. 1984. Studies of microstructure in native celluloses using solid-state carbon-13 NMR. Macromolecules. 17:1465–1472 [Google Scholar]
- 8. Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Hofte H. 1998. A plasma membrane-bound putative endo-1,4-β-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17:5563–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sato S, Kato T, Kakegawa K, Ishii T, Liu YG, Awano T, Takabe K, Nishiyama Y, Kuga S, Nakamura Y, Tabata S, Shibata D. 2001. Role of the putative membrane-bound endo-1,4-β-glucanase KORRIGAN in cell elongation and cellulose synthesis in Arabidopsis thaliana. Plant Cell Physiol. 42:251–263 [DOI] [PubMed] [Google Scholar]
- 10. Turner SR, Somerville CR. 1997. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9:689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saxena IM, Lin FC, Brown RM., Jr 1990. Cloning and sequencing of the cellulose synthase catalytic subunit gene of Acetobacter xylinum. Plant Mol. Biol. 15:673–683 [DOI] [PubMed] [Google Scholar]
- 12. Saxena IM, Lin FC, Brown RM., Jr 1991. Identification of a new gene in an operon for cellulose biosynthesis in Acetobacter xylinum. Plant Mol. Biol. 16:947–954 [DOI] [PubMed] [Google Scholar]
- 13. Saxena IM, Brown RM., Jr 1995. Identification of a second cellulose synthase gene (acsAII) in Acetobacter xylinum. J. Bacteriol. 177:5276–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wong HC, Fear AL, Calhoon RD, Eichinger GH, Mayer R, Amikam D, Benziman M, Gelfand DH, Meade JH, Emerick AW, Bruner R, Ben-Bassat A, Tal R. 1990. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc. Natl. Acad. Sci. U. S. A. 87:8130–8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Standal R, Iversen TG, Coucheron DH, Fjaervik E, Blatny JM, Valla S. 1994. A new gene required for cellulose production and a gene encoding cellulolytic activity in Acetobacter xylinum are colocalized with the bcs operon. J. Bacteriol. 176:665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kawano S, Tajima K, Kono H, Erata T, Munekata M, Takai M. 2002. Effects of endogenous endo-β-1,4-glucanase on cellulose biosynthesis in Acetobacter xylinum ATCC23769. J. Biosci. Bioeng. 94:275–281 [DOI] [PubMed] [Google Scholar]
- 17. Tonouchi N, Tahara N, Tsuchida T, Yoshinaga F, Beppu T, Horinouchi S. 1995. Addition of a small amount of an endoglucanase enhances cellulose production by Acetobacter xylinum. Biosci. Biotechnol. Biochem. 59:805–808 [Google Scholar]
- 18. Koo HM, Song SH, Pyun YR, Kim YS. 1998. Evidence that a β-1,4-endoglucanase secreted by Acetobacter xylinum plays an essential role for the formation of cellulose fiber. Biosci. Biotechnol. Biochem. 62:2257–2259 [DOI] [PubMed] [Google Scholar]
- 19. Toyosaki H, Naritomi T, Seto A, Matsuoka M, Tsuchida T, Yoshinaga F. 1995. Screening of bacterial cellulose-producing Acetobacter strains suitable for agitated culture. Biosci. Biotechnol. Biochem. 59:1498–1502 [Google Scholar]
- 20. Maniatis T, Fritsch EF, Sambrook J. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, New York [Google Scholar]
- 21. Matsuoka M, Tsuchida T, Matsushita K, Adachi O, Yoshinaga F. 1996. A synthetic medium for bacterial cellulose production by Acetobacter xylinum subsp. sucrofermentans. Biosci. Biotechnol. Biochem. 60:575–579 [Google Scholar]
- 22. Tonouchi N, Tsuchida T, Yoshinaga F, Horinouchi S, Beppu T. 1994. A host-vector system for a cellulose-producing Acetobacter strain. Biosci. Biotechnol. Biochem. 58:1899–1901 [DOI] [PubMed] [Google Scholar]
- 23. Nakai T, Moriya A, Tonouchi N, Tsuchida T, Yoshinaga F, Horinouchi S, Sone Y, Mori H, Sakai F, Hayashi T. 1998. Control of expression by the cellulose synthase (bcsA) promoter region from Acetobacter xylinum BPR 2001. Gene 213:93–100 [DOI] [PubMed] [Google Scholar]
- 24. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1988. Current protocols in molecular biology, p 487–491, vol 239 Wiley, New York, NY [Google Scholar]
- 25. Nakai T, Nishiyama Y, Kuga S, Sugano Y, Shoda M. 2002. ORF2 gene involves in the construction of high-order structure of bacterial cellulose. Biochem. Biophys. Res. Commun. 295:458–462 [DOI] [PubMed] [Google Scholar]
- 26. Chao Y, Ishida T, Sugano Y, Shoda M. 2000. Bacterial cellulose production by Acetobacter xylinum in a 50-L internal-loop airlift reactor. Biotechnol. Bioeng. 68:345–352 [DOI] [PubMed] [Google Scholar]
- 27. Marchessault RH, Pearson FG, Liang CY. 1960. Infrared spectra of crystalline polysaccharides. VI. Effect of orientation on the tilting spectra of chitin films. Biochim. Biophys. Acta 45:499–507 [DOI] [PubMed] [Google Scholar]
- 28. Sugiyama J, Persson J, Chanzy H. 1991. Combined infrared and electron diffraction study of the polymorphism of native celluloses. Macromolecules 24:2461–2466 [Google Scholar]
- 29. Benziman M, Haigler CH, Brown RM, Jr, White AR, Cooper KM. 1980. Cellulose biogenesis: polymerization and crystallization are coupled processes in Acetobacter xylinum. Proc. Natl. Acad. Sci. U. S. A. 77:6678–6682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shpigel E, Roiz L, Goren R, Shoseyov O. 1998. Bacterial cellulose-binding domain modulates in vitro elongation of different plant cells. Plant Physiol. 117:1185–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saxena IM, Kudlicka K, Okuda K, Brown RM., Jr 1994. Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. J. Bacteriol. 176:5735–5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Umeda Y, AHirno Ishibashi M, Hideo A, Onizuka T, Ikeuchi M, Inoue Y. 1999. Cloning of cellulose synthase genes from Acetobater xylinum JCM 7664: implication of a novel set of cellulose synthase genes. DNA Res. 6:109–115 [DOI] [PubMed] [Google Scholar]
- 33. Ogino H, Azuma Y, Hosoyama A, Nakazawa H, Matsutani M, Hasegawa A, Otsuyama K, Matsushita K, Fujita N, Shirai M. 2011. Complete genome sequence of NBRC 3288, a unique cellulose-nonproducing strain of Gluconasevtobacter xylinus isolated from vinegar. J. Bacteriol. 193:6997–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yasutake Y, Kawano S, Tajima K, Yao M, Satoh Y, Munekata M, Tanaka I. 2006. Structural characterization of the Acetobacter xylinum endo-β-1,4-glucanase CMCax required for cellulose biosynthesis. Proteins 64:1069–1077 [DOI] [PubMed] [Google Scholar]
- 35. Brown RM, Jr, Willison JHM, Richardson CL. 1976. Cellulose biosynthesis in Acetobacter xylinum: visualization of the site of synthesis and direct measurement of the in vivo process. Proc. Natl. Acad. Sci. U. S. A. 73:4565–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gardner KH, Blackwell J. 1974. The structure of native cellulose. Biopolymers 13:1975–2001 [Google Scholar]
- 37. Kimura S, Chen HP, Saxena IM, Brown RM, Jr., Itho T. 2001. Localization of c-di-GMP-binding protein with the linear terminal complexes of Acetobacter xylinum. J. Bacteriol. 183:5668–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zaar K. 1979. Visualization of pores (export sites) correlated with cellulose production in the envelope of the gram-negative bacterium Acetobacter xylinum. J. Cell Biol. 80:773–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hu SQ, Gao YG, Tajima K, Sunagawa N, Zhou Y, Kawano S, Fujiwara T, Yoda T, Shimura D, Satoh Y, Munekata M, Tanaka I, Yao M. 2010. Structure of bacterial cellulose synthase subunit D octamer with four inner passageways. Proc. Natl. Acad. Sci. U. S. A. 107:17957–17961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mølhøj M, Pagant S, Höfte H. 2002. Towards understanding the role of membrane-bound endo-β-1,4-glucanases in cellulose biosynthesis. Plant Cell Physiol. 43:1399–1406 [DOI] [PubMed] [Google Scholar]
- 41. Matthysse AG, White S, Lightfoot R. 1995. Genes required for cellulose synthesis in Agrobacterium tumefaciens. J. Bacteriol. 177:1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.