Abstract
The envelope of Bacillus anthracis encompasses a proteinaceous S-layer with two S-layer proteins (Sap and EA1). Protein assembly in the envelope of B. anthracis requires S-layer homology domains (SLH) within S-layer proteins and S-layer-associated proteins (BSLs), which associate with the secondary cell wall polysaccharide (SCWP), an acetylated carbohydrate that is tethered to peptidoglycan. Here, we investigated the contributions of two putative acetyltransferases, PatA1 and PatA2, on SCWP acetylation and S-layer assembly. We show that mutations in patA1 and patA2 affect the chain lengths of B. anthracis vegetative forms and perturb the deposition of the BslO murein hydrolase at cell division septa. The patA1 and patA2 mutants are defective for the assembly of EA1 in the envelope but retain the ability of S-layer formation with Sap. SCWP isolated from the patA1 patA2 mutant lacked acetyl moieties identified in wild-type polysaccharide and failed to associate with the SLH domains of EA1. A model is discussed whereby patA1- and patA2-mediated acetylation of SCWP enables the deposition of EA1 as well as BslO near the septal region of the B. anthracis envelope.
INTRODUCTION
Bacillus anthracis is a Gram-positive, spore-forming bacterium and the causative agent of anthrax (1). Following infection, spores germinate, and the resulting vegetative forms replicate in all tissues, eventually triggering the death of infected hosts (2). Spore formation in carcass tissue and contamination of the environment provide for the dissemination of the pathogen (2). To escape phagocytic clearance in host tissues, B. anthracis elaborates a poly-d-γ-glutamic acid (PDGA) capsule, which is tethered to cell wall peptidoglycan (3, 4). Further, bacilli form elongated chains from four or more incompletely separated cells (5). The size of B. anthracis vegetative chains precludes their engulfment by phagocytes (6).
The chain length of B. anthracis vegetative forms is controlled by proteins that are associated with the microbe's surface (S)-layer (5, 7). Two secreted proteins, surface array protein (Sap) and extractable antigen 1 (EA1), assemble into paracrystalline S-layer sheets (8, 9). Twenty-two B. anthracis S-layer-associated proteins (BSLs) are incorporated as minor constituents into the S-layer, where they fulfill specific functions during the pathogenesis of anthrax infections (10). For example, BslK promotes B. anthracis heme iron uptake (11), BslA mediates adherence of vegetative forms to host cells (12), and BslO enables separation of bacilli from elongating chains (5). Both S-layer proteins and S-layer-associated proteins are tethered to the bacterial envelope via three S-layer homology (SLH) domains, which bind to the secondary cell wall polysaccharide (SCWP) (13, 14). SLH domains fold into a three-pronged spindle structure that has been proposed to bind SCWP at the interprong grooves (15). Previous work described the SCWP of B. anthracis as a polysaccharide with the repeating structure [→4)-β-ManNAc-(1→4)-β-GlcNAc-(1→6)-α-GlcNAc-(1→]n, where α-GlcNAc is replaced with α-Gal and β-Gal at O-3 and O-4, respectively, and the β-GlcNAc is replaced with α-Gal at O-3 (16). An SCWP strand is tethered via a murein linkage unit (GlcNAc-ManNAc, where ManNAc is N-acetylmannosamine) and a phosphodiester bond to the C-6 hydroxyl of N-acetylmuramic acid (MurNAc) within the peptidoglycan (14, 17).
The SCWP of B. anthracis is modified with ketal pyruvate and acetyl groups (14, 18). Pyruvylation of the SCWP requires the csaB gene product, which is absolutely essential for B. anthracis S-layer assembly (13, 14). The ketal-pyruvyl moiety is attached to the C-4 and C-6 hydroxyls of the terminal ManNAc residue in the SCWP of B. anthracis strain CDC 684 (18). Of note, B. anthracis CDC 684 is severely attenuated in the guinea pig model for anthrax disease; however, its genome sequence is closely related to that of the virulent strain B. anthracis Vollum (19, 20). Further, the SCWP of B. anthracis CDC 684 lacks the galactosyl decorations of the SCWP found in other B. anthracis isolates (16, 18, 21).
The SCWP of B. anthracis CDC 684 is acetylated at the C-3 hydroxyl of GlcNAc, the penultimate N-acetylated sugar (18). Acetylation of SCWP appears to be a universal feature of B. anthracis and Bacillus cereus, and the sites of acetylation and pyruvylation on the polysaccharide appear similar between B. anthracis CDC 684 and B. anthracis Sterne (18, 22, 23). Nevertheless, the genetic basis for such modification is not yet known. Recently, two putative acyltransferase systems, designated PatA1/PatB1 and PatA2/PatB2, were identified in B. anthracis and proposed to function as peptidoglycan acetyltransferases (24). PatA1 and PatA2 belong to the family of membrane-bound O-acyltransferases (MBOAT; pfam 03062) (25), whose founding member, AlgI, was identified as a gene product required for Pseudomonas aeruginosa exopolysaccharide (alginate) O acetylation (26). Together with two secreted proteins, AlgF and AlgJ, AlgI is required for the transfer of acetyl groups (acetyl-coenzyme A [CoA]) from the cytoplasm, across the membrane and to the O-2 and/or O-3 positions of mannuronate residues in alginate (27). More recently, members of the MBOAT family have been proposed to function as peptidoglycan O-acetyltransferases (OAP), modifying the C-6 hydroxyl of MurNAc (28). Specifically, the gene for the AlgI homolog of Neisseria gonorrhea (patA) is located in a cluster with two other genes, encoding secreted proteins with esterase motifs (Ape1a and Ape2). patA and the Ape1a and Ape2 genes are thought to promote N. gonorrhea peptidoglycan acetylation via a pathway that utilizes acetyl-CoA (28–30).
The OatA membrane protein of Staphylococcus aureus catalyzes the O acetylation of the C-6 hydroxyl of MurNAc in peptidoglycan via its acyltransferase 3 domain (pfam 01757) (31). Homologs of oatA are expressed in other Gram-positive bacteria, including B. anthracis (24), and their products O acetylate peptidoglycan as a means to escape innate immune responses during infection (32, 33). The OatB enzyme belongs to the same family as OatA; however, OatB is thought to catalyze the acetylation of the C-6 hydroxyl of GlcNAc in peptidoglycan (34). In agreement with the general hypothesis that PatA1/PatB1, PatA2/PatB2, OatA, and OatB modify the envelope of B. anthracis, earlier work reported the saponified release of O-acetyl groups from envelope preparations in a manner requiring all of these genes (24). Nevertheless, the experimental approach used an envelope preparation comprised of peptidoglycan with the SCWP, and the possibility that PatA1/PatB1 and PatA2/PatB2 act on the SCWP, similar to AlgIJF acting on P. aeruginosa exopolysaccharide, can therefore not be excluded (24).
This possibility was explored here, and we report that the patA1 and patA2 mutant strains cannot assemble the EA1 S-layer protein in their envelopes but retain the ability of S-layer formation from Sap. Similar amounts of SCWP were isolated from patA1 patA2 mutants. Nevertheless, the SCWP harbored fewer O-acetyl modifications than wild-type (WT) B. anthracis and failed to associate with the SLH domains of EA1. We therefore propose that PatA1/PatB1- and PatA2/PatB2-mediated O acetylation of the SCWP provides for the deposition of proteins into unique locations within the S-layer, thereby controlling the chain length of B. anthracis vegetative forms.
MATERIALS AND METHODS
Bacterial strains and plasmids.
B. anthracis and Bacillus subtilis strains used in this study are listed in Table 1. Bacilli were cultured in brain heart infusion (BHI) broth or agar supplemented with antibiotics when necessary. Sodium bicarbonate (0.8%, wt/vol) was added to induce protective antigen (PA) and BslA expression when appropriate. Escherichia coli strains were grown in Luria Bertani (LB) broth. Chloramphenicol (10 μg/ml) and ampicillin (100 μg/ml) were used to maintain plasmid selection. B. anthracis strains were sporulated in modified G medium (modG) as previously described (36). Spore preparations were heat treated to kill any remaining vegetative bacilli and stored at 4°C. Spores were enumerated by CFU counts. Spores were germinated by inoculation into BHI medium and grown at 37°C.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| B. anthracis | ||
| Sterne 34F2 | (pXO1+ pXO2−) wild type | Laboratory collection |
| JDB2519 | patA1::tetL (Tetr) | 24 |
| JDB2578 | patA2::aad9 (Specr) | 24 |
| JDB1972 | patA1::aphA3::patA2 (Kanr) | 24 |
| JDB2573 | patA1::aphA3::patA2 pML360 | 24 |
| JDB2609 | patA1::aphA3::patA2 pML301 | 24 |
| JML145 | patA1::aphA3::patA2 pAD123 | This work |
| B. subtilis | ||
| PY79 | Wild type | Laboratory collection |
| E. coli | ||
| JWK440 | BL21(DE3), pJK88 | This work |
| Plasmids | ||
| pML301 | pAD123 derivative containing patA2-bas0846 on a BamHI/KpnI fragment; Cmr Ampr | 24 |
| pML360 | pAD123 derivative containing patA1-bas0843 on a BamHI/KpnI fragment; Cmr Ampr | 24 |
| pAD123 | pTA1060 ori, cat Cmr bla Ampr | 35 |
| pJK88 | pET16b plasmid with the first 180 amino acids of eag as a translational fusion with mCherry | This work |
Tetr, tetracycline resistance, Specr, spectinomycin resistance; Kanr, kanamycin resistance; Cmr, chloramphenicol resistance; Ampr, ampicillin resistance.
Strain JWK440 is the E. coli BL21(DE3) strain transformed with the pJK88 plasmid containing the first 180 amino acids of EA1 as a translational fusion with mCherry. This vector was constructed by amplifying mCherry with P287 (5′-TTTCTCGAGAGCAAGGGCGAGGAGG-3′) and P288 (5′-TTTGGATCCTTACTTGTACAGCTCGTCCATG-3′) and digested with XhoI-BamHI. Cut DNA was ligated with pET16b vector DNA, also cleaved with XhoI-BamHI, and transformed into DH5α cells. The recombinant product was then digested with NdeI-XhoI and ligated with the NdeI-XhoI-cut, PCR-amplified portion of EA1 encoding the SLH domains (amino acids 30 to 210) (P289, 5′-GGAATTCCATATGGGTAAATCATTCCCAGACGTTC-3′; P288, 5′-TTTGGATCCTTACTTGTACAGCTCGTCCATG-3′). The ligation product was transformed into E. coli BL21(DE3). Strain JML145 was generated by purifying pAD123 plasmid DNA from E. coli K1077 (dcm dam mutant) and transforming JDB1972 (patA1 patA2) bacilli by electroporation.
Chain length of vegetative bacilli.
B. anthracis spores were germinated by suspension in 2 ml of BHI broth at 37°C for 4 h. Cells were fixed using 4% paraformaldehyde and analyzed by microscopy. Images were obtained with a charge-coupled-device (CCD) camera on an Olympus IX81 microscope using a 100× or 40× objective. The chain lengths of bacilli were measured from acquired differential interference contrast (DIC) images using ImageJ and converted to lengths in micrometers using reference images of an objective micrometer. The statistical significance of chain length phenotypes was analyzed using an unpaired, two-tailed Student's t test.
S-layer fractionation.
B. anthracis overnight cultures, grown in BHI broth supplemented with 0.8% sodium bicarbonate (wt/vol), were diluted 1:100 into fresh medium and grown to an A600 of 2.5. One milliliter of culture was centrifuged at 16,000 × g and separated into medium (supernatant) and pellet fractions. Proteins in the medium were precipitated with 10% (vol/vol) trichloroacetic acid (TCA) for 30 min on ice and centrifuged at 16,000 × g for 10 min. Bacterial sediments (pellets) were washed twice with 1× phosphate-buffered saline (PBS) and boiled at 95°C for 10 min in 100 μl of PBS–3 M urea to extract S-layer and S-layer-associated proteins. Extracts were centrifuged at 16,000 × g, and S-layer extract was separated with the supernatant from the bacterial sediment. S-layer extract was added to an equal volume of sample buffer (4% SDS, 1% β-mercaptoethanol, 10% glycerol, 50 mM Tris-HCl [pH 7.5], bromophenol blue). The bacterial sediment was washed twice with PBS and mechanically lysed by silica bead beating for 3 min (Fastprep-24; MP Biomedical). After sedimentation of the beads, proteins in the cell lysates were precipitated with TCA and sedimented by centrifugation at 16,000 × g for 10 min. All TCA precipitates were washed with ice-cold acetone and dried. Samples were suspended in 100 μl of 1 M Tris-HCl (pH 8.0)–4% SDS and mixed with an equal volume of sample buffer. Aliquots (10 μl) of each sample were separated by 10% SDS-PAGE and analyzed by Coomassie staining or electro-transferred to polyvinylidene difluoride (PVDF) membranes for immunoblot analysis. Proteins were detected with rabbit antiserum raised against purified antigens. Immunoreactive products were revealed by chemiluminescent detection after incubation with horseradish peroxidase (HRP)-conjugated secondary antibody (Cell Signaling Technology). The percent amount of protein in subcellular fractions was calculated by averaging the ratio of immunoblot signals from one fraction to the sum of the signal in all fractions using three independent experimental determinations.
Purification of EA1SLH-mCherry.
One liter of LB culture of E. coli BL21(DE3)(pJK88) was grown at 37°C with rotation to an A600 of 0.6 and induced for expression of the SLH domain of EA1 fused to the fluorescent protein mCherry (EA1SLH-mCherry) with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h. Cells were sedimented by centrifugation, suspended in 50 mM Tris-HCl (pH 7.5) with 150 mM NaCl and disrupted by two passages at 14,000 lb/in2 in a French press. Lysate was cleared by centrifugation at 17,000 × g for 10 min to remove unbroken cells and then by ultracentrifugation at 100,000 × g for 30 min to sediment cell membranes. Soluble supernatant was subjected to Ni-nitrilotriacetic acid (NTA) affinity chromatography according to the manufacturer's recommendations (Qiagen). The His-tagged EA1SLH-mCherry protein was eluted with 0.1 M imidazole, dialyzed against PBS overnight, and used for binding studies.
EA1SLH-mCherry binding to B. anthracis vegetative forms.
B. anthracis Sterne wild-type or mutant strains (csaB, patA1, patA2, and patA1 patA2 strains) were grown in BHI broth to an A600 of 2.5, and vegetative bacilli were sedimented by centrifugation. Bacilli were suspended in PBS supplemented with 3 M urea and heated at 95°C for 10 min to solubilize the S-layer (14). Cells were again sedimented by centrifugation, washed extensively with water, and suspended in PBS. Cell density was normalized to an A600 of 1. These cells (100-μl samples) were then incubated with 4 μl of 15 mM EA1SLH-mCherry for 10 min at room temperature in the dark. After incubation, cells were sedimented by centrifugation, washed extensively with PBS, and imaged by fluorescence or phase-contrast microscopy on an Olympus IX81 microscope using a 100× objective. Images were captured via a CCD camera. Alternatively, fluorescence was quantified using a Biotek Synergy HT microplate reader and 96-well format. The excitation wavelength was 590 (±20) nm, and the emission was read at 645 (±40) nm. In the SCWP inhibition assay, the EA1SLH-mCherry protein solution was preincubated with reverse-phase high-performance liquid chromatography (rpHPLC) fractions that had been taken to dryness in a Speed-Vac and solubilized in 1 M HEPES-KOH (pH 7.5). Subsequently, 100-μl suspensions of S-layer-stripped B. anthracis Sterne cells (A600 of 1) were added. The suspension was sedimented by centrifugation, washed extensively with PBS, and quantified by fluorescence measurements as described in the EA1SLH-mCherry binding assay. Fluorescence measurements were compared using an unpaired, two-tailed Student's t test.
Purification of B. anthracis SCWP.
Bacilli were grown overnight at 37°C on BHI agar, scraped off the surface, and washed in water. Bacilli were boiled for 30 min in a 4% SDS solution, washed, and suspended in water. The bacteria were mechanically lysed with 0.1-mm glass beads. The resulting murein sacculi were sedimented by centrifugation at 17,000 × g for 15 min, suspended in 100 mM Tris-HCl (pH 7.5), and incubated for 4 h at 37°C with 10 μg/ml DNase and 10 μg/ml RNase supplemented with 20 mM MgSO4. Samples were then incubated for 16 h at 37°C with 10 μM trypsin supplemented with 10 mM CaCl2. Enzymes were inactivated by boiling for 30 min in a water bath in 1% SDS. The SDS was removed by repeated cycles of centrifugation and washing in water. The murein sacculi were then washed with water, 100 mM Tris-HCl (pH 8.0), water, 0.1 M EDTA (pH 8.0), water, acetone, and finally twice with water. Murein sacculi were suspended in 5 to 7 ml of water, and 25 ml of 48% hydrofluoric acid (HF) was added. Samples were incubated overnight on ice with shaking. The acid-extracted murein sacculi were sedimented by centrifugation at 17,000 × g for 15 min. SCWP-containing supernatant was mixed with ice-cold ethanol in a 1:5 ratio, causing SCWP precipitation. The polysaccharide was sedimented by centrifugation at 17,000 × g and 4°C for 15 min, and the sediment was washed extensively with ice-cold ethanol. The SCWP was suspended in water at a concentration of 100 mg/ml, and 100 μl of this solution was subjected to rpHPLC analysis.
rpHPLC analysis was performed over a 250-mm by 4.6-mm C18-ODS Hypersile column with a 3-μm-particle-size guard column (Thermos) using a water-acetonitrile gradient where the mobile phase was supplemented with 0.1% trifluoroacetic acid (TFA). The gradient was derived from buffer A (water–0.1% TFA) and buffer B (acetonitrile–0.1% TFA), with a continuous flow rate of 0.5 ml per minute and the following parameters: 0 to 10 min, 0.1% buffer B; 11 to 15 min, linear gradient of 0.1 to 10% buffer B; 16 to 35 min, linear gradient of 10 to 20% buffer B; 36 to 100 min, linear gradient of 20 to 99.9% buffer B; 101 to 110 min, 99.9% buffer B. The eluate was monitored for absorbance at 206 nm, and 0.5-ml fractions were collected.
Measurement of O acetylation.
Ethanol-precipitated SCWP was suspended in 25 mM sodium phosphate buffer (pH 6.5). The SCWP was treated with either 1 M or 0.01 M NaOH (final concentration) and, after 1 h at room temperature or 10 h at 4°C, samples were neutralized with the appropriate volume of 1 M HCl. For quantification of released acetate, a Megazyme Acetic Acid Kit (Megazyme International Ireland, Ltd., Wicklow, Ireland) was used according to the manufacturer's recommendations. For normalization of wild-type and patA1 patA2 mutant SCWP concentrations, the SCWP solutions were subjected to high-performance size exclusion chromatography (HPSEC) using a 300-mm by 7.8-mm BioBasic SEC300 (Thermos) column equilibrated with 50 mM sodium phosphate buffer (pH 7.0) and a 0.1 ml/min flow rate. Chromatographs were analyzed by measuring absorbance at 206 nm. The singular peak for the SCWP was used to normalize the released acetate concentration relative to the abundance of SCWP.
MALDI-TOF mass spectrometry.
rpHPLC fractions were spotted onto a Prespotted AnchorChip II, PAC II, plate (Bruker) that contained an α-cyano-4-hydroxycinnamic acid (HCCA) matrix. Calibrants were used directly from the PAC II plate. The samples were subjected to matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry using an Autoflex Speed Bruker MALDI instrument in positive-reflectron mode. The Bruker data analysis package was used for analysis of the spectra, and peaks were defined as having a signal-to-noise (S/N) ratio of >3.
Immunofluorescence microscopy.
Spores of B. anthracis strains were germinated in 2 ml of BHI broth at 37°C for 3 h. Cells were sedimented by centrifugation and fixed with 4% buffered formaldehyde. Fixed samples were incubated with diluted rabbit antiserum raised against purified recombinant Sap, EA1, or BslO, and bound antibody was detected with fluorophore-conjugated secondary antibody and boron dipyrromethene (BODIPY)-vancomycin. A Leica SP5 Tandem Scanner Spectral 2-Photon confocal microscope with a 63× objective was used to observe B. anthracis cells.
RESULTS
Mutations in patA1 and patA2 cause chain length phenotypes of B. anthracis mutants.
An important cue for the function of the patA1 and patB1 and the patA2 and patB2 genes can be derived from their location on the B. anthracis chromosome (Fig. 1A). The patA1-patB1-patA2-patB2 cluster is located immediately adjacent to three genes, csaB-sap-eag, encoding the two S-layer proteins (Sap and EA1) and the ketal pyruvyl transferase that modifies the SCWP (CsaB). Earlier work reported that B. anthracis patA1, patA2, and patA1 patA2 mutants, compared to wild-type bacilli, display increased chain lengths of their vegetative forms (24). Nevertheless, the chain length phenotypes have not yet been quantified. We sought to address the questions of whether the patA1, patA2, and patA1 patA2 mutants produce variable chain lengths and how these phenotypes compare with those of B. anthracis csaB and bslO variants. Spore preparations of B. anthracis wild-type, csaB, patA1, patA2, and patA1 patA2 strains were inoculated in BHI broth at 37°C and incubated for 4 h. The resulting vegetative forms were fixed and visualized by light microscopy, and the lengths of 100 chains were quantified (Fig. 1B and C). Wild-type bacilli displayed an average chain length of 16.9 μm. As expected, the average chain length of csaB and bslO mutant bacilli was increased to 109.0 μm and 95.2 μm, respectively (Fig. 1C) (5). The average chain lengths of strains with a deletion of the patA1 or the patA2 gene was moderately increased to 35.9 μm or 29.5 μm, respectively (WT versus the patA1 strain, P < 0.0001; WT versus the patA2 strain, P < 0.0001). The B. anthracis patA1 patA2 mutant lacks both MBOAT acetyltransferases and displayed a further increase in average chain length (64.5 μm) (WT versus patA1 patA2 mutant, P < 0.0001; patA1 strain versus patA1 patA2 mutant, P < 0.0001; patA2 strain versus patA1 patA2 mutant, P < 0.0001). Transformation of the patA1 patA2 strain with pML360, a plasmid containing patA1 and patB1, or pML301, harboring patA2 and patB2, diminished the average chain length relative to transformation with the empty pAD123 vector alone (strain carrying pAD123 versus pML360, P < 0.0001; strain carrying pAD123 versus pML301, P < 0.0001) (Fig. 1D). These data reveal a chain length phenotype of the patA1 patA2 mutant strain, which is, however, not as severe as the exaggerated chain length phenotypes of the bslO and csaB mutant strains (5). Further, the patA1 and patA2 genes fulfill complementary but not redundant roles in B. anthracis S-layer assembly as the mutant lacking both genes displays an additive chain length phenotype over mutants with a deletion of only one gene.
Fig 1.
Mutations in the patA1 and patA2 genes cause chain length phenotypes in Bacillus anthracis vegetative forms. (A) Genes for the PatA1 and PatA2 acetyltransferases are located on the Bacillus anthracis chromosome immediately adjacent to genes for the major S-layer proteins (sap and eag) and to csaB, which encodes a ketal pyruvyl transferase essential for S-layer formation. (B) DIC microscopy images of vegetative forms germinated from B. anthracis wild-type and csaB, bslO, patA1, patA2, and patA1 patA2 mutant spores. Scale bar, 10 μm. (C) The chain lengths of 100 vegetative bacilli were quantified for each strain; the data are presented as a box-and-whiskers plot, where the size of each box is defined by the boundaries of the 25th and 75th percentiles, the whiskers are the maximum and minimum measured lengths, and the center bar indicates the median length. Data were examined with an unpaired two-tailed Student's t test. A significant difference (P < 0.0001) relative to wild-type is indicated with an asterisk (*). The patA1 patA2 mutant was also compared to mutants with singular deletions of patA1 and patA2 genes. (D) The B. anthracis patA1 patA2 mutant strain was transformed with pAD123, i.e., the empty vector, pML360, a plasmid expressing patA1-patB1, or pML301, a plasmid expressing patA2-patB2. A significant difference (P < 0.0001) between the empty vector and the pML360 and pML301 strains was observed after data sets were compared with a Student t test.
B. anthracis patA1 patA2 mutants cannot retain EA1 in their S-layers.
Earlier work used lithium chloride extraction of B. anthracis S-layer proteins and analysis via Coomassie-stained SDS-PAGE gels to characterize patA1 patA2 mutants (24). The results suggested that the patA1 patA2 mutant, but not mutants with a deletion of only patA1 or patA2, are defective in retaining S-layer proteins, presumably Sap, in the bacterial envelope (24). We used Coomassie-stained SDS-PAGE gels and immunoblotting with rabbit polyclonal antibodies, which had been raised against purified proteins, to analyze the subcellular location of S-layer proteins and BSLs in B. anthracis cultures fractionated into extracellular medium (M), S-layer (S), and cells (C) (Fig. 2). Unlike the csaB mutant, which does not assemble an S-layer, the patA1, patA2, and patA1 patA2 mutants all assembled S-layers (Fig. 2A). Quantifying the distribution of S-layer proteins in wild-type and variant strains, we noted that the patA1, patA2, and patA1 patA2 mutants form S-layers via Sap assembly but incorporated reduced amounts of EA1 into these structures (Table 2). As a control, wild-type and mutant bacilli secreted protective antigen (PA) into the extracellular medium and deposited PrsA1 peptidyl-prolyl-isomerase into the cytoplasmic membrane (Fig. 2B). As expected, the csaB mutant was unable to deposit BslA and BslO into its envelope. The patA1 and patA2 mutants incorporated BslO, but not BslA, into their Sap S-layers. In contrast, the patA1 patA2 mutant was defective not only for the deposition of BslA but also for the incorporation of BslO into its S-layer. These results indicate that patA1, patA2, and patA1 patA2 mutants form S-layers predominantly via the assembly of Sap, which suggests further that the corresponding chain length phenotypes cannot be explained as defects in the envelope assembly of Sap (37).
Fig 2.
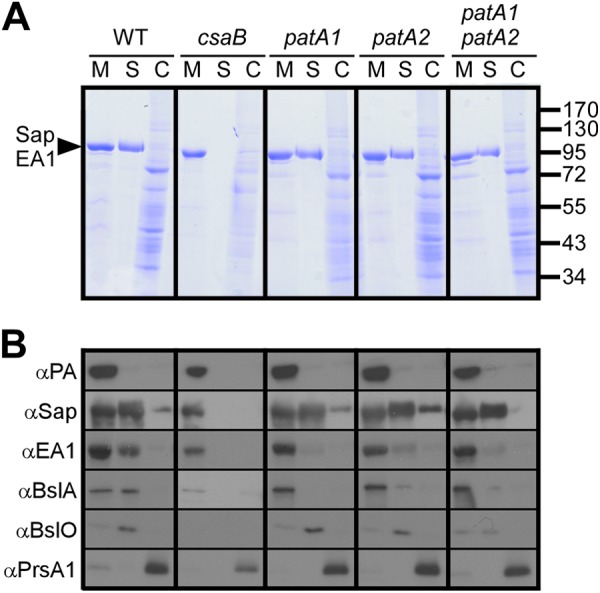
Impact of patA1 and patA2 mutations on S-layer protein and S-layer-associated protein trafficking in B. anthracis. (A) B. anthracis vegetative cultures were fractionated into medium (M), S-layer (S), and cell (C) fractions, and proteins in each sample were analyzed by Coomassie-stained SDS-PAGE gels. The arrowhead denotes the migratory position of the S-layer proteins Sap and EA1, and the numbers indicate the position of molecular mass markers (in kDa). (B) Samples generated in the experiment shown in panel A were examined by immunoblotting with antibodies raised against protective antigen (PA), a secreted protein and component of anthrax toxins (42), and PrsA1, a peptidylprolyl isomerase and membrane lipoprotein (43), as well as S-layer proteins (Sap and EA1) and S-layer-associated proteins (BslA and BslO). α, anti.
Table 2.
S-layer and S-layer-associated protein distribution in B. anthracis wild-type and mutant strains
| Protein and fraction | Amt of protein in the indicated strain (%)a |
||||
|---|---|---|---|---|---|
| Wild type | csaB mutant | patA1 mutant | patA2 mutant | patA1 patA2 mutant | |
| Sap | |||||
| Medium | 45 (±5) | 99 (±1) | 38 (±9) | 37 (±4) | 47 (±2) |
| S-layer | 41 (±4) | 0 (±0) | 44 (±3) | 41 (±5) | 44 (±8) |
| Cell | 13 (±7) | 1 (±1) | 17 (±9) | 22 (±3) | 9 (±10) |
| EA1 | |||||
| Medium | 55 (±12) | 100 (±0) | 85 (±5) | 71 (±7) | 88 (±9) |
| S-layer | 43 (±12) | 0 (±0) | 12 (±4) | 24 (±7) | 7 (±4) |
| Cell | 2 (±1) | 0 (±0) | 3 (±0) | 5 (±2) | 5 (±7) |
| BslA | |||||
| Medium | 55 (±1) | 100 (±0) | 95 (±6) | 83 (±6) | 90 (±7) |
| S-layer | 44 (±1) | 0 (±0) | 3 (±4) | 15 (±8) | 9 (±6) |
| Cell | 1 (±1) | 0 (±0) | 1 (±2) | 2 (±2) | 1 (±1) |
| BslO | |||||
| Medium | 19 (±6) | ND | 22 (±13) | 24 (±16) | 55 (±10) |
| S-layer | 77 (±9) | ND | 72 (±2) | 67 (±26) | 45 (±9) |
| Cell | 4 (±3) | ND | 7 (±6) | 10 (±10) | 1 (±1) |
The immunoblot signals of two S-layer proteins (Sap, surface array protein, and EA1 extractable antigen 1) and two S-layer-associated proteins (BslA and BslO, S-layer-associated proteins A and O, respectively) in the medium, S-layer, and cell fraction of B. anthracis cultures were quantified in triplicate (Fig. 2). The mean and associated standard deviation of the percent amount in each fraction is presented here. The percentage of Sap that was retained in the S-layer did not change in strains with mutations in patA1 or patA2. These strains showed defects in the S-layer retention of EA1, BslA, and BslO. ND, not detected.
Mutations in csaB, patA1, and patA2 do not affect B. anthracis synthesis of the SCWP.
To address the question of whether there is a defect in the synthesis of the SCWP in csaB, patA1, patA2, and patA1 patA2 mutant B. anthracis strains, we utilized a polyclonal anti-SCWP antibody to investigate the abundance and localization of SCWP in vegetative bacilli. B. anthracis cultures were grown in BHI broth to an A600 of 1, stripped of their S-layers by incubation at 95°C in phosphate-buffered 3 M urea, and fixed for immunofluorescence microscopy experiments. Rabbit polyclonal antibodies raised against SCWP-bovine serum albumin (BSA) detected the presence of SCWP in the envelope of wild-type bacilli (Fig. 3A). Neither the abundance nor the distribution of the SCWP was affected in B. anthracis mutants lacking the csaB, patA1, and patA2 genes (Fig. 3A). As a control, antibodies raised against SCWP did not stain the envelope of Bacillus subtilis, a related spore-forming microbe that does not synthesize SCWP or S-layers (Fig. 3B).
Fig 3.
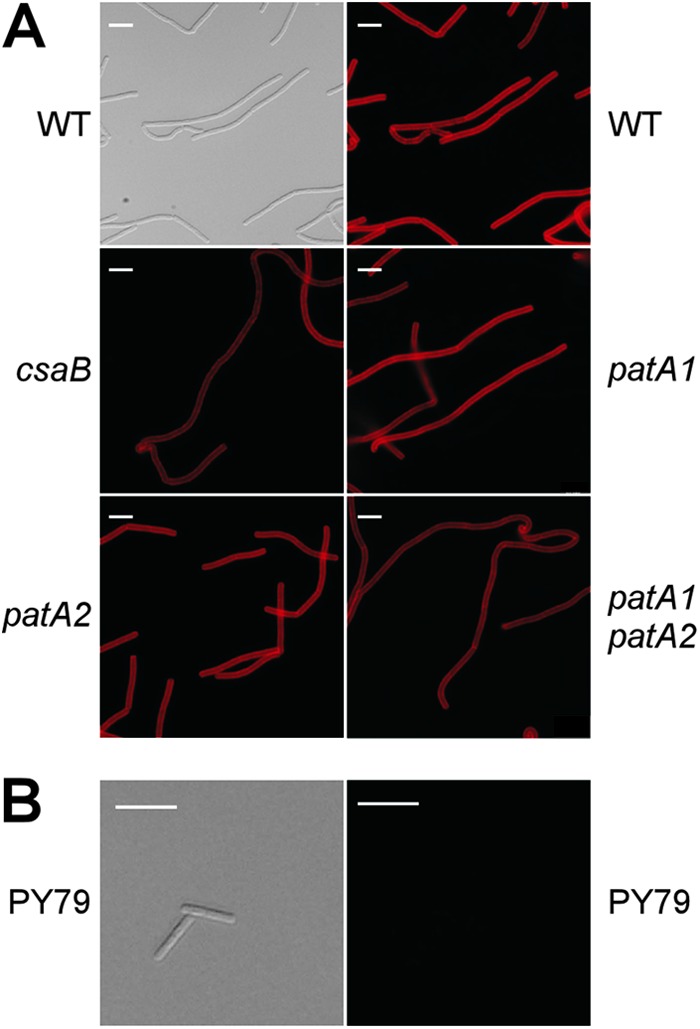
Mutation in csaB, patA1, or patA2 does not alter the abundance of the SCWP in B. anthracis. (A) Vegetative forms derived from wild-type, csaB, patA1, patA2, and patA1 patA2 mutant B. anthracis spores were stripped of their S-layers via treatment with 3 M urea. Bacilli were subsequently fixed with 4% paraformaldehyde and subjected either to DIC microscopy or to immunofluorescence microscopy with rabbit polyclonal antibodies raised against purified SCWP-BSA conjugate followed by secondary antibody conjugated to DyLight594. Scale bar, 10 μm. (B) B. subtilis strain PY79, a strain that does not elaborate an SCWP, does not show immunoreactivity toward the SCWP antibody. A refreshed overnight growth was fixed with 4% paraformaldehyde and subjected to DIC microscopy and immunofluorescence microscopy in the same manner as the B. anthracis strains. Scale bar, 10 μm.
EA1SLH-mCherry association with the SCWP of B. anthracis requires patA1 and patA2.
To dissect the biochemical defect of patA1 patA2 mutants, we used a previously established in vitro assay for S-layer assembly (14). The EA1SLH-mCherry hybrid, which encompasses the SLH domain of EA1 fused to the fluorescent protein mCherry, was purified from the cytoplasm of E. coli via affinity chromatography and analyzed by SDS-PAGE (Fig. 4A). When incubated with vegetative bacilli that had been stripped of their S-layers, EA1SLH-mCherry bound to ketal-pyruvylated SCWP in the envelope of B. anthracis, as visualized by fluorescence microscopy and quantified in fluorescence plate reader experiments (Fig. 4B and C). As a control, no binding of EA1SLH-mCherry to SCWP was observed for csaB mutant bacilli (Fig. 4B and C). The SCWP of patA1 but not patA2 bacilli displayed a moderate reduction in the ability to bind EA1SLH-mCherry (Fig. 3B and C) (WT versus the patA1 mutant, P < 0.001; WT versus the patA2 mutant, P > 0.05). However, the defect in EA1SLH-mCherry binding was more pronounced in the patA1 patA2 mutant strain than in the patA1 mutant alone (Fig. 4B and C) (patA1 mutant versus patA1 patA2 mutant, P < 0.001).
Fig 4.
The SLH domain of EA1 displays reduced binding to the SCWP from patA1 patA2 mutant B. anthracis. (A) Coomassie-stained SDS-PAGE gel of the purified fusion protein EA1SLH-mCherry. Numbers indicate the positions of molecular mass markers (in kDa). (B) Vegetative forms of B. anthracis strains were stripped of their S-layers via treatment with 3 M urea. Differential interference contrast (DIC) and fluorescence microscopy images of the wild type and fluorescence microscopy images of csaB, patA1, patA2, and patA1 patA2 mutant B. anthracis strains incubated with EA1SLH-mCherry are shown. Scale bar, 15 μm. (C) The fluorescence intensity of EA1SLH-mCherry binding to the envelope of S-layer-stripped B. anthracis strains was quantified in three independent experimental determinations. Data were averaged and examined with an unpaired two-tailed Student's t test. *, P < 0.01; **, P < 0.001.
SCWP from the B. anthracis patA1 patA2 mutant does not prevent EA1SLH-mCherry binding to wild-type bacilli.
The SCWP was released from the murein sacculi of wild-type and patA1 patA2 mutant B. anthracis cells via hydrofluoric acid (HF)-mediated hydrolysis of the phosphodiester bond between the C-6 hydroxyl of MurNAc in peptidoglycan and the GlcNAc-ManNAc moiety of the murein linkage units for the SCWP. Acid-extracted polysaccharide was precipitated with ethanol and subjected to rpHPLC. Absorbance was monitored at 206 nm, and a single, large, compound peak eluted at 48 to 52 min during the chromatography of HF-extracted wild-type SCWP (Fig. 5A). Elution fractions corresponding to this absorbance peak, but not any other rpHPLC fraction, inhibited the binding of EA1SLH-mCherry to ketal-pyruvylated SCWP in B. anthracis cells that were stripped of their S-layers (Fig. 5B). A similar absorbance peak at 48 to 52 min was observed during rpHPLC of HF-released SCWP from the patA1 patA2 mutant strain (Fig. 5A). Unlike wild-type SCWP, the patA1 patA2 mutant SCWP did not inhibit binding of EA1SLH-mCherry to ketal-pyruvylated SCWP in vegetative bacilli (Fig. 5B).
Fig 5.
SCWP isolated from wild-type and patA1 patA2 mutant B. anthracis strains. (A) The SCWP was released via hydrofluoric acid (HF) treatment from murein sacculi that had been isolated from either wild-type or patA1 patA2 mutant vegetative forms. Ethanol-precipitated SCWP was subjected to rpHPLC, and the eluate's absorbance at 206 nm (A206) was recorded in milli-arbitrary units (mAu). (B) Collected rpHPLC fractions from panel A (A1), were tested for competitive inhibition of EA1SLH-mCherry binding to the SCWP of wild-type bacilli. SCWPs from wild-type but not SCWPs from patA1 patA2 mutant bacilli displayed competitive inhibitor activities toward the binding of bacilli to EA1SLH-mCherry.
Mass spectrometry of the SCWP from wild-type and patA1 patA2 mutant B. anthracis.
The SCWP of wild-type and patA1 patA2 mutant B. anthracis, eluting at 48 to 52 min during rpHPLC, was subjected to MALDI-TOF mass spectrometry (Fig. 6A and B). For example, m/z 1,136.8 and m/z 2,232.5 were abundant ion signals derived from each SCWP preparation (Fig. 6). These compounds represent the sodiated adducts of one and two SCWP repeating units, respectively (Fig. 6). Table 3 summarizes ion signals from wild-type and patA1 patA2 mutant SCWPs that were found in both samples. Sodiated ion signals conforming to variable lengths of the SCWP repeating units were abundantly present in each spectrum. Less abundant ions differed from the repeating units, in agreement with the general hypothesis of SCWP heterogeneity in galactosyl modification and acetylation. For example, m/z 1,500.55 conforms to a compound structure of four acetylated amino sugars and three decorating galactosyl sugars with one deacetylated amino sugar in the repeating sequence of the SCWP (Table 3).
Fig 6.
MALDI-TOF mass spectrometry of SCWP isolated from wild-type and patA1 patA2 mutant B. anthracis strains. (A) rpHPLC-purified SCWP from wild-type B. anthracis was subjected to MALDI-TOF mass spectrometry, and ion spectra were recorded. m/z, mass-to-charge ratio. The ion signal marked with an asterisk (*) identifies the sodiated adduct of a compound with the predicted structure [β-ManNAc-(1→4)-β-GlcNAc(Gal)-(1→6)-α-GlcNAc(Gal)2]. (B) rpHPLC-purified SCWP from patA1 patA2 mutant B. anthracis was analyzed by MALDI-TOF mass spectrometry, and ion spectra were recorded. (C) The SCWP is comprised of the repeating unit [→4)-β-ManNAc-(1→4)-β-GlcNAc-(1→6)-α-GlcNAc-(1→]n (printed in black), where α-GlcNAc is replaced with α-Gal and β-Gal at O-3 and O-4, respectively, and the β-GlcNAc is replaced with α-Gal at O-3 (galactosyl substitutions printed in blue) (16).
Table 3.
Ions present in MALDI-TOF spectra of hydrofluoric acid-released SCWP from wild-type and patA1 patA2 mutant murein sacculi of B. anthracis Sternea
| Observed m/z in: |
Proposed compositionb | Theoretical m/z (monoisotopic) | WT Δm/zd | patA1 patA2 mutant Δm/ze | |
|---|---|---|---|---|---|
| WT strainc | patA1 patA2 mutant | ||||
| 1,094.792 | 1,094.636 | HexNAc2-GlcN-Gal3 | 1,094.39 | −0.41 | −0.25 |
| 1,136.849** | 1,136.826 | HexNAc3-Gal3 | 1,136.40 | −0.45 | −0.43 |
| 1,177.869 | 1,177.868 | HexNAc4-Gal2 | 1,177.42 | −0.45 | −0.44 |
| 1,218.926 | 1,218.920 | HexNAc5-Gal | 1,218.45 | −0.48 | −0.47 |
| 1,338.926 | 1,338.926 | HexNAc4-Gal3 | 1,339.48 | 0.55 | 0.55 |
| 1,380.005 | 1,379.966 | HexNAc5-Gal2 | 1,380.50 | 0.50 | 0.54 |
| 1,460.034 | 1,460.014 | HexNAc3-GlcN-Gal4 | 1,459.52 | −0.52 | −0.50 |
| 1,500.091* | 1,500.061 | HexNAc4-GlcN-Gal3 | 1,500.55 | 0.45 | 0.48 |
| 1,542.101 | 1,542.114 | HexNAc5-Gal3 | 1,542.56 | 0.45 | 0.44 |
| 1,584.103* | 1,583.995 | HexNAc6-Gal2 | 1,583.58 | −0.52 | −0.41 |
| 1,662.177 | 1,662.183 | HexNAc4-GlcN-Gal4 | 1,662.60 | 0.42 | 0.41 |
| 1,704.213 | 1,704.212 | HexNAc5- Gal4 | 1,704.61 | 0.40 | 0.40 |
| 1,746.253 | 1,746.257 | HexNAc6- Gal3 | 1,745.63 | −0.62 | −0.62 |
| 1,824.292 | 1,824.292 | HexNAc4-GlcN-Gal5 | 1,824.65 | 0.36 | 0.36 |
| 1,866.287 | 1,866.282 | HexNAc5-Gal5 | 1,866.66 | 0.37 | 0.38 |
| 1,908.304* | 1,908.318 | HexNAc6-Gal4 | 1,907.69 | −0.62 | −0.63 |
| 1,986.422 | 1,986.356 | HexNAc5-GlcN-Gal6 | 1,986.70 | 0.28 | 0.35 |
| 2,028.405 | 2,028.377 | HexNAc5-Gal6 | 2,028.71 | 0.31 | 0.34 |
| 2,070.431* | 2,070.403 | HexNAc6-Gal5 | 2,069.74 | −0.69 | −0.66 |
| 2,232.542* | 2,232.519 | HexNAc6-Gal6 | 2,231.79 | −0.75 | −0.73 |
| 2,392.756 | 2,392.413 | HexNAc5-GlcN- Gal6 | 2,392.86 | 0.11 | 0.45 |
| 2,434.629 | 2,434.629 | HexNAc6-Gal6 | 2,434.87 | 0.24 | 0.24 |
| 2,476.694 | 2,476.520 | HexNAc8-Gal5 | 2,475.90 | −0.79 | −0.62 |
| 2,554.825 | 2,554.663 | HexNAc6-GlcN-Gal7 | 2,554.91 | 0.09 | 0.25 |
| 2,596.851 | 2,596.683 | HexNAc7-Gal7 | 2,596.93 | 0.07 | 0.24 |
| 2,638.864 | 2,638.688 | HexNAc8-Gal6 | 2,637.95 | −0.91 | −0.74 |
| 2,758.834 | 2,758.651 | HexNAc7-GlcN-Gal7 | 2,757.99 | −0.84 | −0.83 |
| 2,800.836 | 2,800.651 | HexNAc8-Gal7 | 2,800.00 | −0.83 | −0.65 |
| 2,920.999 | 2,920.999 | HexNAc7-GlcN-Gal8 | 2,920.05 | −0.95 | −0.95 |
| 2,963.070 | 2,963.070 | HexNAc8-Gal8 | 2,962.06 | −1.01 | −1.01 |
Ion signals and proposed composition of compounds from MALDI-TOF mass spectra of rpHPLC-purified SCWPs from wild-type B. anthracis Sterne and the patA1 patA2 strain.
HexNAc, ManNAc (N-acetylmannosaminyl) and GlcNAc (N-acetylglucosaminyl); GlcN, glucosaminyl; Gal, galactosyl. The SCWP isolated from wild-type and the patA1 patA2 mutant strains generated nearly identical sets of ions, similar to spectra characterizing the SCWP structure. The proposed composition was derived from sodiated, positively charged ions. Of note, additional compositional explanations for the observed masses exist; however, they are not listed here.
Observed ions noted with a single asterisk (*) were accompanied by less intense ions with an additional m/z 15.999 and were interpreted as oxygen adducts of the parent ion. The ion noted with a double asterisk (**) was accompanied by a less intense ion with reduced m/z 2.011; this accompanying ion was interpreted as an oxygen adduct (+15.999) and dehydration product (−18.01) of the parent ion.
Theoretical m/z − observed WT m/z.
Theoretical m/z − observed patA1 patA2 mutant m/z.
Both wild-type and patA1 patA2 mutant SCWPs generated the ion signal m/z 2,800.7, which can be interpreted as the sodiated ion [ManNAc-GlcNAc2(Gal3)]2-ManNac-GlcNAc(Gal) (Fig. 7A and B). Wild-type SCWP generated also m/z 2,842.8, which can be interpreted as the acetylated form of [ManNAc-GlcNAc2(Gal3)]2-ManNac-GlcNAc(Gal) (Fig. 7A). On the other hand, the SCWP isolated from the patA1 patA2 mutant did not generate m/z 2,842.8, suggesting that the appearance of acetylated SCWP species may require the expression of patA1 and patA2. Of note, although the mass spectrometry data in Fig. 7A and B are in agreement with the proposed compound structures, they do not constitute experimental proof for the structural assignments. Similar observations were made for the ion signals m/z 2,680.849 and m/z 2,638.864 from wild-type SCWP; m/z 2,638.688 but not m/z 2,680.849 was identified in the SCWP from the patA1 patA2 mutant strain (Table 3).
Fig 7.
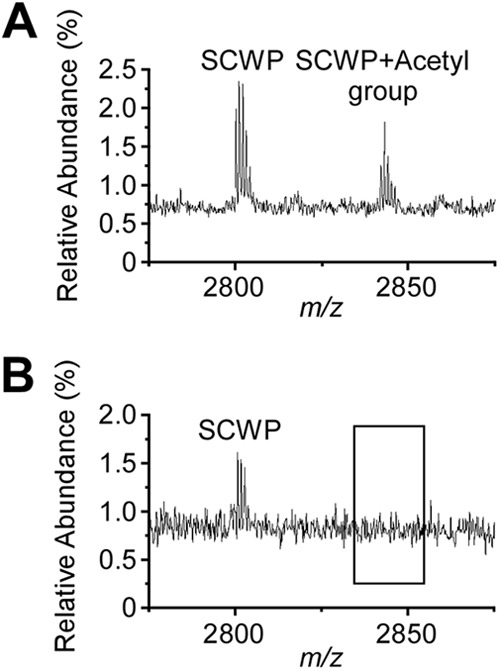
Ion signals for acetylated SCWP in wild-type but not in patA1 patA2 mutant B. anthracis cells. (A) Ion signals from the wild-type SCWP were detected with MALDI-TOF mass spectrometry; m/z 2,800.7 was interpreted as the sodiated ion [ManNAc-GlcNAc2(Gal3)]2-ManNac-GlcNAc(Gal), and m/z 2,842.8 was interpreted as its acetylated variant. (B) Ion signals from the patA1 patA2 mutant SCWP were detected with MALDI-TOF mass spectrometry; m/z 2,800.7 was interpreted as the sodiated ion [ManNAc-GlcNAc2(Gal3)]2-ManNac-GlcNAc(Gal). m/z 2,842.7 was not detected in the spectrum. Of note, although mass spectrometry data are in agreement with the proposed compound structures, they do not constitute experimental proof for the structural assignments.
Quantification of sodium hydroxide-released acetate from wild-type and patA1 patA2 mutant SCWP.
To examine whether the SCWP of patA1 patA2 mutant bacilli harbors fewer O-acetyl groups than the SCWP of wild-type B. anthracis cells, we extracted polysaccharides with hydrofluoric acid from the envelope of bacilli and subjected HPSEC-purified SCWP to base-catalyzed hydrolysis of O-acetyl groups. Released acetate was quantified via enzyme assay and normalized to the SCWP peak area during HPSEC chromatography (see Fig. S1 in the supplemental material). Compared to acetate groups released from wild-type SCWP, which were set as 100% (±3%), patA1 patA2 mutant SCWP harbored 60% (±2%) of the 1 M sodium hydroxide-released acetate. Using a milder alkaline treatment, 0.01 M NaOH at 4°C, the patA1 patA2 mutant SCWP harbored 45% (±0%) of the acetate released by wild-type SCWP; the wild type was again set as 100% (±1%). These data indicate that acetylation of the SCWP indeed requires the patA1 and patA2 genes.
Localizing S-layer proteins and BslO in the envelope of wild-type and patA1 patA2 mutant B. anthracis strains.
Data presented here suggest that the patA1 and patA2 genes promote acetylation of the SCWP, a modification that impacts the deposition of some, but not all, S-layer and S-layer-associated proteins in the envelope of B. anthracis. To visualize protein deposition into the S-layer, we used polyclonal rabbit antibodies raised against purified recombinant proteins for fluorescence microscopy of B. anthracis vegetative forms. Immunofluorescence microscopy experiments revealed incorporation of Sap into the S-layer of wild-type B. anthracis (see Fig. S2 in the supplemental material). Although Sap was deposited throughout the envelope of B. anthracis, fluorescence microscopy experiments detected areas of abundant Sap deposition near the poles but not at the cell division septa of vegetative chains (see Fig. S2). Division septa are defined as sites of intense BODIPY-vancomycin staining, a compound that binds to the precursors of peptidoglycan synthesis (see Fig S2). Deposition of Sap into the S-layer was abolished in csaB mutant B. anthracis (see Fig. S2). In contrast, deletion of patA1 or patA2 or of patA1 and patA2 did not prevent the incorporation of Sap into the S-layer. Of note, the patA1 patA2 mutant, but not variants with a single deletion of patA1 or patA2, lacked the abundant deposition of Sap near the poles of vegetative chains (see Fig. S2). This defect was rescued when plasmid pML360 (patA2) was transformed into the patA1 patA2 mutant, whereas pML301 (patA1) had no effect.
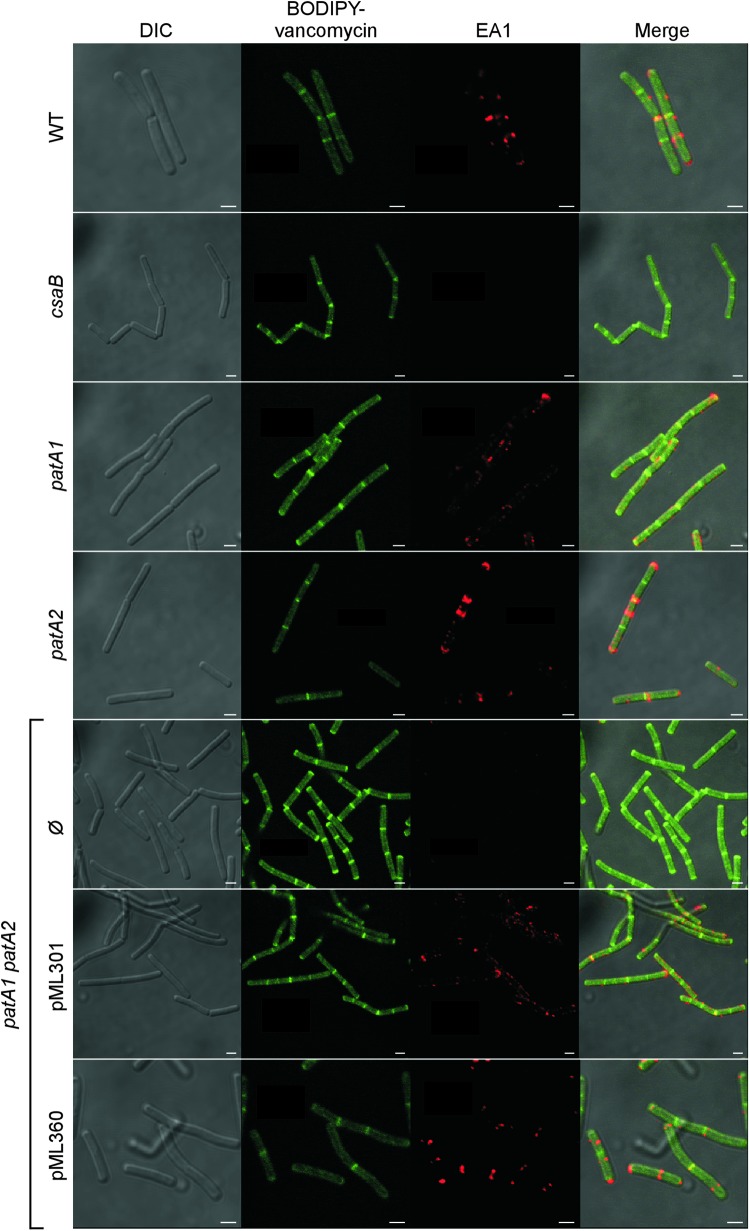
Immunofluorescence microscopy experiments revealed the deposition of EA1 near the cell division septa of wild-type vegetative chains, similar to recently reported data (Fig. 8) (7). As expected, the csaB mutant did not deposit EA1 into the B. anthracis envelope (Fig. 8). The patA1 and patA2 mutant strains incorporated EA1 into the B. anthracis envelope; however, the fluorescent signals did not coincide with BODIPY-vancomycin staining of cell division septa (Fig. 8). In contrast, the patA1 patA2 mutant strain failed to deposit EA1 into the S-layer of vegetative chains (Fig. 8). This defect was rescued by plasmids providing for the expression of either patA1 or patA2 (Fig. 8).
Fig 8.
Localization of the S-layer protein EA1 in the envelope of wild-type, csaB, patA1, patA2, and patA1 patA2 B. anthracis strains. Spores from wild-type and mutant B. anthracis strains were diluted into BHI broth, and germinated bacilli were incubated for 3 h. Vegetative forms were fixed in 4% buffered formalin. Differential interference contrast (DIC) and fluorescence microscopy with BODIPY-vancomycin or rabbit polyclonal antibodies staining against the S-layer protein EA1 followed by secondary antibody DyLight594 conjugate were used to acquire images. Data sets were merged to reveal the location of cell wall septa (BODIPY-vancomycin) and S-layer protein EA1 in wild-type and mutant bacilli. The patA1 patA2 mutant strain was transformed with plasmid pML301, expressing patA1 and patB1, or pML360, expressing patA2 and patB2, or left untransformed (Ø). Scale bar, 1 μm.
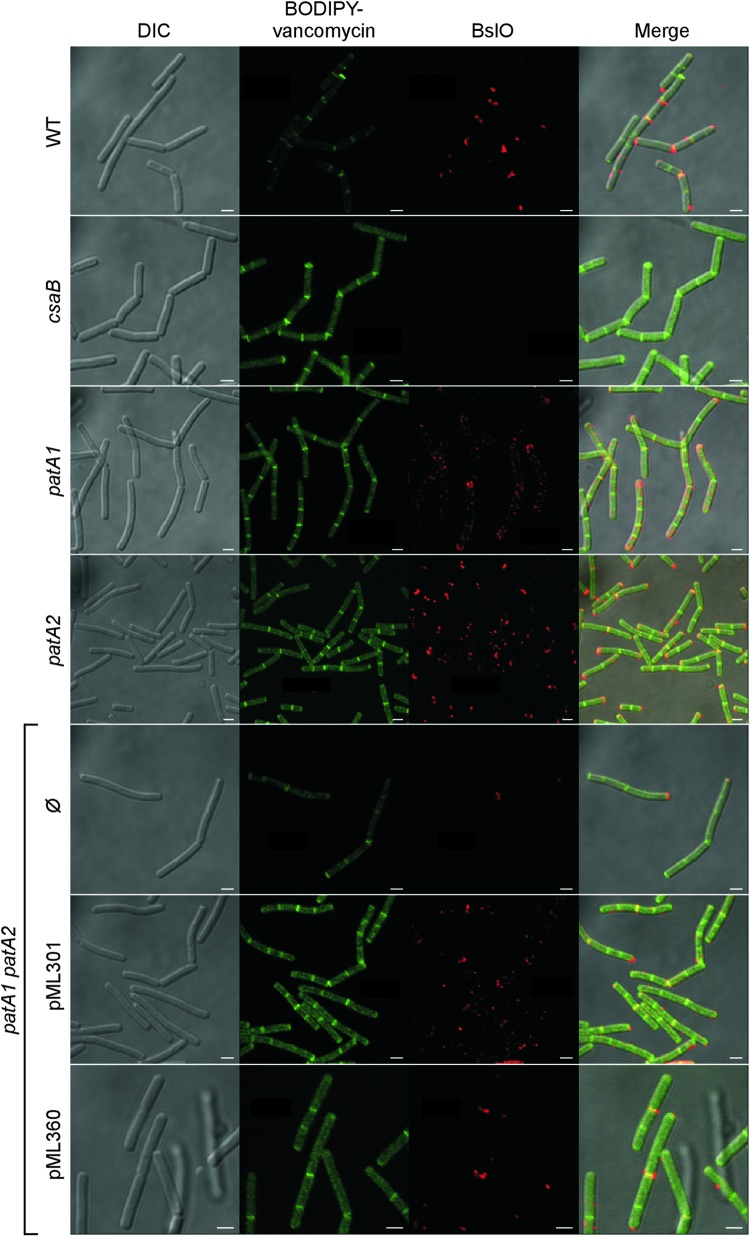
In agreement with earlier reports, immunofluorescence microscopy revealed the deposition of BslO near the cell division septa of wild-type B. anthracis vegetative chains (Fig. 9) (5). As a control, BslO deposition into the S-layer did not occur in vegetative forms of the csaB mutant strain (7) (Fig. 9). Both the patA1 and the patA2 mutant strains displayed an aberrant pattern of BslO deposition, in which the protein was detected in about half of the cell division septa of vegetative chains but was absent from the remaining septa (Fig. 9). Further, the patA1 patA2 mutant strain deposited very little, if any, BslO near division septa (Fig. 9). This defect was rescued in part by plasmids expressing either patA1 or patA2 in the patA1 patA2 mutant strain (Fig. 9).
Fig 9.
Localization of the S-layer protein EA1 in the envelope of wild-type, csaB, patA1, patA2, and patA1 patA2 B. anthracis strains. Spores from wild-type and mutant B. anthracis strains were diluted into BHI broth, and germinated bacilli were incubated for 3 h. Vegetative forms were fixed in 4% buffered formalin. Differential interference contrast (DIC) and fluorescence microscopy with BODIPY-vancomycin or rabbit polyclonal antibodies staining against the S-layer-associated protein BslO followed by secondary antibody DyLight594 conjugate were used to acquire images. Data sets were merged to reveal the location of cell wall septa (BODIPY-vancomycin) and the S-layer-associated protein BslO in wild-type and mutant bacilli. The patA1 patA2 mutant strain was transformed with plasmid pML301, expressing patA1 and patB1, or pML360, expressing patA2 and patB2, or left untransformed (Ø). Scale bar, 1 μm.
DISCUSSION
The goal of our study was to elucidate the role of patA1 and patA2 in S-layer assembly and function in B. anthracis. Several lines of evidence suggest that patA1 and patA2 affect S-layer assembly. First, patA1 and patA2 as well as the genes for their cognate secreted acetyltransferases are located immediately adjacent to the S-layer genes (csaB-sap-eag) on the B. anthracis chromosome. Second, mutations in patA1 and patA2 cause a chain length phenotype, which has previously been attributed to the improper assembly of the S-layer protein Sap and of the S-layer-associated protein BslO in the bacterial envelope (7, 37). Third, in agreement with this model, patA1, patA2, and patA1 patA2 mutants display defects in the deposition of BslO, a murein hydrolase that separates cells from the chains of B. anthracis vegetative forms (7). patA1 and patA2 likely fulfill nonoverlapping, complementary roles, as the deletion of individual genes causes partial phenotypes for both BslO deposition and chain lengths and as the phenotypic defects are additive for the double mutant strain. Fourth, the patA1 patA2 mutant strain releases the S-layer protein EA1 as well as BslA and BslO into the extracellular medium. Fifth, the patA1 patA2 mutant strain is defective in binding EA1SLH-mCherry, a reporter harboring the B. anthracis SLH domain. Sixth, the SCWP of the patA1 patA2 mutant strain cannot associate with the SLH domain of the EA1 S-layer protein, and, seventh, the SCWP of the patA1 patA2 mutant lacks acetyl modifications otherwise found in the SCWP from wild-type bacilli. Taken together, these data suggest that patA1 and patA2 function as acetyltransferases that modify the B. anthracis cell wall polysaccharide, similar to AlgI of P. aeruginosa (26).
Our experimental approach did not investigate the question of whether PatA1 and PatA2 also act to acetylate the peptidoglycan of B. anthracis. In this regard, it seems noteworthy that B. anthracis harbors homologs of oatA and oatB (31, 32), acetyltransferases that modify the C-6 hydroxyl of MurNAc or GlcNAc moieties in the repeating disaccharide of peptidoglycan strands (38). Mutants lacking oatA or oatB harbor defects in cell envelope acetylation but do not display chain length phenotypes (24; also data not shown). These data suggest that the O-6 acetylation of amino sugars in peptidoglycan may not impact S-layer assembly in B. anthracis.
Data presented here suggest that B. anthracis S-layer assembly does not occur via a mechanism of random deposition for either S-layer proteins or S-layer-associated proteins. The acetylation of the SCWP promotes the deposition of the EA1 S-layer protein and of the BslO murein hydrolase near the cell division septa of vegetative forms. If one entertained a model whereby SCWP acetylation were a prerequisite for association with the SLH domains of EA1 and BslO, but not for the SLH domain of Sap, this would imply that SCWP acetylation must be restricted to the cell division septa. If so, one could assume that PatA1 and PatA2 may also be deposited near cell division septa. Alternative possibilities exist when we take into account the recent discovery that the S-layer proteins (Sap and EA1), but not the BSLs, travel via a dedicated secretory pathway across the bacterial envelope to promote S-layer assembly (37). The subcellular location of the secretion genes, SecA2 and SlaP, is not yet known; however, these translocases could contribute to the overall mechanism of S-layer assembly that also takes into account the mechanisms of S-layer acetylation described here.
Mutations in B. anthracis csaB, patA1, patA2, sap, secA2, and slaP affect the deposition of BslO at cell division septa and the chain length of vegetative bacilli (5, 7, 14, 37); however, these mutations do not affect B. anthracis autolysis, as has been reported for the cell separation active murein hydrolases of the Gram-positive microbe Staphylococcus aureus (39–41).
Supplementary Material
ACKNOWLEDGMENTS
We thank members of our laboratory for discussion and comments on the manuscript.
J.M.L. is a trainee of the National Institutes of Health Medical Scientist Training Program at the University of Chicago (grant GM07281). This work was supported by a grant from the National Institute of Allergy and Infectious Diseases (NIAID), Infectious Disease Branch (AI069227 to O.S. and D.M.M.). We acknowledge membership within and support from the Region V Great Lakes Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (NIAID Award 1-U54-AI-057153).
Footnotes
Published ahead of print 14 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01274-12.
REFERENCES
- 1. Koch R. 1876. Die Ätiologie der Milzbrand-Krankheit, begründet auf die Entwicklungsgeschichte des Bacillus anthracis. Beitr. Biol. Pflanz. 2: 277– 310 [Google Scholar]
- 2. Marraffini LA, Schneewind O. 2006. Targeting proteins to the cell wall of sporulating Bacillus anthracis. Mol. Microbiol. 62: 1402– 1417 [DOI] [PubMed] [Google Scholar]
- 3. Candela T, Fouet A. 2006. Poly-gamma-glutamate in bacteria. Mol. Microbiol. 60: 1091– 1098 [DOI] [PubMed] [Google Scholar]
- 4. Richter GS, Anderson VJ, Garufi G, Lu L, Joachimiak A, He C, Schneewind O, Missiakas D. 2009. Capsule anchoring in Bacillus anthracis occurs by a transpeptidation mechanism that is inhibited by capsidin. Mol. Microbiol. 71: 404– 420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anderson VJ, Kern JW, McCool JW, Schneewind O, Missiakas DM. 2011. The SLH domain protein BslO is a determinant of Bacillus anthracis chain length. Mol. Microbiol. 81: 192– 205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ruthel G, Ribot WJ, Bavari S, Hoover T. 2004. Time-lapse confocal imaging of development of Bacillus anthracis in macrophages. J. Infect. Dis. 189: 1313– 1316 [DOI] [PubMed] [Google Scholar]
- 7. Kern VJ, Kern JW, Theriot JA, Schneewind O, Missiakas DM. 2012. Surface (S)-layer proteins Sap and EA1 govern the binding of the S-layer associated protein BslO at the cell septa of Bacillus anthracis. J. Bacteriol. 194: 3833– 3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mesnage S, Tosi-Couture E, Mock M, Gounon P, Fouet A. 1997. Molecular characterization of the Bacillus anthracis main S-layer component: evidence that it is the major cell-associated antigen. Mol. Microbiol. 23: 1147– 1155 [DOI] [PubMed] [Google Scholar]
- 9. Etienne-Toumelin I, Sirard JC, Duflot E, Mock M, Fouet A. 1995. Characterization of the Bacillus anthracis S-layer: cloning and sequencing of the structural gene. J. Bacteriol. 177: 614– 620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kern JW, Schneewind O. 2008. BslA, a pXO1-encoded adhesin of Bacillus anthracis. Mol. Microbiol. 68: 504– 515 [DOI] [PubMed] [Google Scholar]
- 11. Tarlovsky Y, Fabian M, Solomaha E, Honsa E, Olson JS, Maresso AW. 2010. A Bacillus anthracis S-layer homology protein that binds heme and mediates heme delivery to IsdC. J. Bacteriol. 192: 3503– 3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kern JW, Schneewind O. 2010. BslA, the S-layer adhesin of Bacillus anthracis, is a virulence factor for anthrax pathogenesis. Mol. Microbiol. 75: 324– 332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mesnage S, Fontaine T, Mignot T, Delepierre M, Mock M, Fouet A. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19: 4473– 4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kern J, Ryan C, Faull K, Schneewind O. 2010. Bacillus anthracis surface-layer proteins assemble by binding to the secondary cell wall polysaccharide in a manner that requires csaB and tagO. J. Mol. Biol. 401: 757– 775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kern JW, Wilton R, Zhang R, Binkowski A, Joachimiak A, Schneewind O. 2011. Structure of the SLH domains from Bacillus anthracis surface array protein. J. Biol. Chem. 286: 26042– 26049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Choudhury B, Leoff C, Saile E, Wilkins P, Quinn CP, Kannenberg EL, Carlson RW. 2006. The structure of the major cell wall polysaccharide of Bacillus anthracis is species specific. J. Biol. Chem. 281: 27932– 27941 [DOI] [PubMed] [Google Scholar]
- 17. Kojima N, Arakai Y, Ito E. 1985. Structure of the linkage units between ribitol teichoic acids and peptidoglycan. J. Bacteriol. 161: 299– 306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Forsberg LS, Abshire TG, Friedlander A, Quinn CP, Kannenberg EL, Carlson RW. 2012. Localization and structural analysis of a conserved pyruvylated epitope in Bacillus anthracis secondary cell wall polysaccharides and characterization of the galactose deficient wall polysaccharide from avirulent B. anthracis CDC 684. Glycobiology 22: 1103– 1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Okinaka RT, Price EP, Wolken SR, Gruendike JM, Chung WK, Pearson T, Xie G, Munk C, Hill K, Challacombe J, Ivins BE, Schupp JM, Beckstrom-Sternberg SM, Friedlander A, Keim P. 2011. An attenuated strain of Bacillus anthracis (CDC 684) has a large chromosomal inversion and altered growth kinetics. BMC Genomics 12: 477– 490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ezzell JWJ, Abshire TG, Little SF, Lidgerding BC, Brown C. 1990. Identification of Bacillus anthracis by using monoclonal antibody to cell wall galactose-N-acetylglucosamine polysaccharide. J. Clin. Microbiol. 28: 223– 231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Forsberg LS, Choudhury B, Leoff C, Marston CK, Hoffmaster AR, Saile E, Quinn CP, Kannenberg EL, Carlson RW. 2011. Secondary cell wall polysaccharides from Bacillus cereus strains G9241, 03BB87 and 03BB102 causing fatal pneumonia share similar glycosyl structures with the polysaccharides from Bacillus anthracis. Glycobiology 21: 934– 948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leoff C, Saile E, Sue D, Wilkins PP, Quinn CP, Carlson RW, Kannenberg EL. 2008. Cell wall carbohydrate compositions of strains from Bacillus cereus group of species correlate with phylogenetic relatedness. J. Bacteriol. 190: 112– 121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leoff C, Saile E, Rauvolfova J, Quinn C, Hoffmaster AR, Zhong W, Mehta AS, Boons GJ, Carlson RW, Kannenberg EL. 2009. Secondary cell wall polysaccharides of Bacillus anthracis are antigens that contain specific epitopes which cross-react with three pathogenic Bacillus cereus strains that caused severe disease, and other epitopes common to all the Bacillus cereus strains tested. Glycobiology 19: 665– 673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Laaberki M-H, Pfeffer J, Clarke AJ, Dworkin J. 2011. O-Acetylation of peptidoglycan is required for proper cell separation and S-layer anchoring in Bacillus anthracis. J. Biol. Chem. 286: 5278– 5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hofmann K. 2000. A superfamily of membrane-bound O-acyltransferases with implications for Wnt signaling. Trends Biochem. Sci. 25: 111– 112 [DOI] [PubMed] [Google Scholar]
- 26. Franklin MJ, Ohman DE. 1996. Identification of algI and algJ in the Pseudomonas aeruginosa alginate biosynthetic gene cluster which are required for alginate O acetylation. J. Bacteriol. 178: 2186– 2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Franklin MJ, Ohman DE. 2002. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J. Bacteriol. 184: 3000– 3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weadge JT, Pfeffer JM, Clarke AJ. 2005. Identification of a new family of enzymes with potential O-acetylpeptidoglycan esterase activity in both Gram-positive and Gram-negative bacteria. BMC Microbiol. 5: 49 doi:10.1186/1471-2180-5-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weadge JT, Clarke AJ. 2006. Identification and characterization of O-acetylpeptidoglycan esterase: a novel enzyme discovered in Neisseria gonorrhoeae. Biochemistry 45:839– 851 [DOI] [PubMed] [Google Scholar]
- 30. Dillard JP, Hackett KT. 2005. Mutations affecting peptidoglycan acetylation in Neisseria gonorrhoeae and Neisseria meningitidis. Infect. Immun. 73: 5697– 5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bera A, Herbert S, Jakob A, Vollmer W, Götz F. 2005. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55: 778– 787 [DOI] [PubMed] [Google Scholar]
- 32. Bera A, Biswas R, Herbert S, Götz F. 2006. The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect. Immun. 74: 4598– 4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aubry C, Goulard C, Nahori M, Cayet N, Decalf J, Sachse M, Boneca I, Cossart P, Dussurget O. 2011. OatA, a peptidoglycan O-acetyltransferase involved in Listeria monocytogenes immune escape, is critical for virulence. J. Infect. Dis. 204: 731– 740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bernard E, Rolain T, Courtin P, Guillot A, Langella P, Hols P, Chapot-Chartier MP. 2011. Characterization of O-acetylation of N-acetylglucosamine: a novel structural variation of bacterial peptidoglycan. J. Biol. Chem. 286: 23950– 23958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dunn AK, Handelsman J. 1999. A vector for promoter trapping in Bacillus cereus. Gene 226: 297– 305 [DOI] [PubMed] [Google Scholar]
- 36. Kim HU, Goepfert JM. 1974. A sporulation medium for Bacillus anthracis. J. Appl. Bacteriol. 37: 265– 267 [DOI] [PubMed] [Google Scholar]
- 37. Nguyen-Mau S-M, Oh SY, Kern V, Missiakas D, Schneewind O. 2012. Secretion genes as determinants of Bacillus anthracis chain length. J. Bacteriol. 194: 3841– 3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ghuysen JM, Strominger JL. 1963. Structure of the cell wall of Staphylococcus aureus, strain Copenhagen. II. Separation and structure of the disaccharides. Biochemistry 2: 1119– 1125 [DOI] [PubMed] [Google Scholar]
- 39. Kajimura J, Fujiwara T, Yamada S, Suzawa Y, Nishida T, Oyamada Y, Hayashi I, Yamagishi J, Komatsuzawa H, Sugai M. 2005. Identification and molecular characterization of an N-acetylmuramyl-l-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol. Microbiol. 58: 1087– 1101 [DOI] [PubMed] [Google Scholar]
- 40. Yamada S, Sugai M, Komatsuzawa H, Nakashima S, Oshida T, Matsumoto A, Suginaka H. 1996. An autolysin ring associated with cell separation of Staphylococcus aureus. J. Bacteriol. 178: 1565– 1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oshida T, Sugai M, Komatsuzawa H, Hong YM, Suginaka H, Tomasz A. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-β-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. U. S. A. 92: 285– 289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vodkin MH, Leppla SH. 1983. Cloning of the protective antigen gene of Bacillus anthracis. Cell 34: 693– 697 [DOI] [PubMed] [Google Scholar]
- 43. Williams RC, Rees ML, Jacobs MF, Praqai Z, Thwaite JE, Baillie LW, Emmerson PT, Harwood CR. 2003. Production of Bacillus anthracis protective antigen is dependent on extracellular chaperone, PrsA. J. Biol. Chem. 278: 18056– 18062 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.