Abstract
DivIVA proteins are curvature-sensitive membrane binding proteins that recruit other proteins to the poles and the division septum. They consist of a conserved N-terminal lipid binding domain fused to a less conserved C-terminal domain. DivIVA homologues interact with different proteins involved in cell division, chromosome segregation, genetic competence, or cell wall synthesis. It is unknown how DivIVA interacts with these proteins, and we used the interaction of Bacillus subtilis DivIVA with MinJ and RacA to investigate this. MinJ is a transmembrane protein controlling division site selection, and the DNA-binding protein RacA is crucial for chromosome segregation during sporulation. Initial bacterial two-hybrid experiments revealed that the C terminus of DivIVA appears to be important for recruiting both proteins. However, the interpretation of these results is limited since it appeared that C-terminal truncations also interfere with DivIVA oligomerization. Therefore, a chimera approach was followed, making use of the fact that Listeria monocytogenes DivIVA shows normal polar localization but is not biologically active when expressed in B. subtilis. Complementation experiments with different chimeras of B. subtilis and L. monocytogenes DivIVA suggest that MinJ and RacA bind to separate DivIVA domains. Fluorescence microscopy of green fluorescent protein-tagged RacA and MinJ corroborated this conclusion and suggests that MinJ recruitment operates via the N-terminal lipid binding domain, whereas RacA interacts with the C-terminal domain. We speculate that this difference is related to the cellular compartments in which MinJ and RacA are active: the cell membrane and the cytoplasm, respectively.
INTRODUCTION
DivIVA homologues constitute a group of highly conserved cell division proteins in Gram-positive bacteria. They bind to the cytosolic face of the cytoplasmic membrane and accumulate at membrane regions with increased negative curvature in rod-shaped bacteria (1–3). Negatively curved (i.e., concave) membrane regions occur at the cell poles and along the cytokinetic ring as soon as it starts to constrict and invaginates the cell membrane. Membrane binding and curvature sensitivity appear to be intrinsic features of DivIVA, as it was shown that DivIVA of Bacillus subtilis also localizes to curved membranes when expressed in other, nonrelated species, including yeast cells (4). DivIVA is used as a scaffold and recruits other proteins that function in cell division, cell wall biosynthesis, secretion, genetic competence, or chromosome segregation (5–13). The proteins that interact with DivIVA are therefore diverse and comprise both transmembrane and cytosolic proteins (14). The best-characterized DivIVA protein is that of B. subtilis, for which four different interaction partners are known: (i) the transmembrane protein MinJ, which acts as a molecular bridge between DivIVA and the FtsZ-inhibiting MinCD complex (11, 12); (ii) the DNA-binding protein RacA, which is required for chromosome segregation during spore formation (8, 15); (iii) the competence-specific inhibitor of cell division Maf (16); and (iv) the competence regulator ComN (17). Nothing is known about the molecular interaction between DivIVA and its interaction partners. We set out to determine DivIVA interaction domains in more detail and focused on the binding of B. subtilis DivIVA with MinJ and RacA.
The crystal structure of B. subtilis DivIVA revealed a two-domain organization: a highly conserved N-terminal domain that forms a dimeric structure with a characteristic cap structure and a less conserved C-terminal domain that is rich in coiled coils but varies in length among the different bacterial species (18). These domains are connected by a flexible ∼20-amino-acid linker (Fig. 1). The N-terminal domain is required for the lipid binding of DivIVA and for localization (1, 18, 19). The dimeric cap structure of this lipid binding domain (LBD) exposes two phenylalanine side chains (F17, one per subunit), and the insertion of these side chains into the hydrophobic core of the phospholipid bilayer is essential for lipid binding (18). This membrane interaction is stabilized by auxiliary electrostatic interactions between positively charged arginine and lysine residues (R18 and K15) in the immediate vicinity of F17 and the negatively charged phospholipid head groups (18). The crystal structure suggested that the central coiled-coil region of the C-terminal domain contributes to DivIVA dimerization (Fig. 1B) and that the end of this domain (amino acids 130 to 153) forms an antiparallel four-helix bundle constituting the tetramerization domain (TD) whereby two DivIVA dimers are linked together in an end-to-end orientation (18) (Fig. 1A and B). The C-terminal part of DivIVA is the least-conserved domain; it differs in length and can contain large insertions (Fig. 1A). It was therefore speculated that this domain is responsible for the interaction with other proteins (14).
Fig 1.
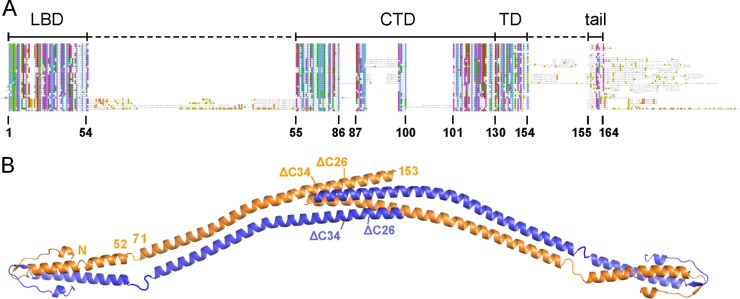
Domain arrangement of B. subtilis DivIVA. (A) Schematic sequence alignment of DivIVA proteins of different phylogenetic origins. Abbreviations above the alignment label the individual protein regions: LBD, lipid binding domain; CTD, C-terminal domain; TD, tetramerization domain; tail, C-terminal tail region. Amino acid numbering is according to the B. subtilis DivIVA sequence. (B) Model of the crystal structure of the full-length B. subtilis DivIVA tetramer which has been assembled from the individual crystal structures of the N- and the C-terminal domains (18). Crystallographic data for the linker between both domains (residues 53 to 70) are not available. Amino acid positions at the beginning and the end of the lipid binding domain as well as the C-terminal domain are indicated for one molecule. Truncation sites of DivIVAΔC26 and DivIVAΔC34 at positions 138 and 130, respectively, are also shown (compare Table 3).
To test whether the C terminus of DivIVA comprises the partner interaction domain, we tested C-terminally truncated variants of B. subtilis DivIVA for their interaction with MinJ and RacA using the bacterial two-hybrid system. These experiments proved inconclusive since removal of the tetramerization domain appeared to affect oligomerization. Therefore, we set up a complementation assay with chimeric DivIVA proteins that consist of domains from B. subtilis DivIVA and Listeria monocytogenes DivIVA. The latter protein localizes normally when expressed in B. subtilis but is biologically inactive and is unable to recruit MinJ or RacA to the cell division sites and cell poles. This experiment revealed that the sporulation activity and the cell division activity of DivIVA can be separated. It emerged that the transmembrane protein MinJ binds to the N-terminal lipid binding domain of DivIVA, whereas the C-terminal domain of DivIVA contains the binding site for the cytosolic protein RacA.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains that were used in this study are listed in Table 1. Routinely, B. subtilis strains were cultivated in LB broth or on LB agar at 37°C. If necessary, the following antibiotics were added at the indicated concentrations: tetracycline (10 μg/ml), spectinomycin (100 μg/ml), and chloramphenicol (5 μg/ml). Other supplements were IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM) and xylose (0.5%). For all cloning procedures, Escherichia coli TOP10 was used as the standard plasmid host (23).
Table 1.
Plasmids and strains used in this study
| Plasmid or strain | Relevant characteristic | Source or reference |
|---|---|---|
| Plasmids | ||
| pAPNC213 | bla aprE5′ spc lacI PspacaprE3′ | 20 |
| pAPNC213cat | bla aprE5′ cat lacI PspacaprE3′ | H. Strahl |
| pDG9 | bla amyE3′ spc Pxyl-divIVA-gfp amyE5′ | 18 |
| pSG1154 | bla amyE3′ spc Pxyl-′gfp amyE5′ | 21 |
| pSH2 | bla aprE5′ spc lacI Pspac-divIVABsaprE3′ | 18 |
| pSH3 | bla amyE3′ spc Pxyl-divIVABs-gfpA206K amyE5′ | 18 |
| pKT25-racA | kan Plac-cya(T25)-racA | 1 |
| pUT18-divIVA | bla Plac-cya(T18)-divIVA | 1 |
| pUT18C-divIVA | bla Plac-cya(T18)-divIVA | 11 |
| pUT18C-minJ | bla Plac-cya(T18)-minJ | 11 |
| pUT18C-racA | bla Plac-cya(T18)-racA | 1 |
| p25-N-divIVA | kan Plac-divIVA-cya(T25) | 1 |
| p25-N-minJ | kan Plac-minJ-cya(T25) | 11 |
| pINC3 | bla amyE3′ spc Pxyl-divIVAR131A-gfp amyE5′ | This work |
| pINC12 | bla aprE5′ spc lacI Pspac-divIVAR131A aprE3′ | This work |
| pKK13 | bla amyE3′ spc Pxyl-divIVABs-57-LmamyE5′ | This work |
| pSBLH001 | bla Plac-divIVA1-459(Δ11)-cya(T18)a | This work |
| pSBLH004 | kan Plac-divIVA1-459(Δ11)-cya(T25) | This work |
| pSBLH005 | bla Plac-divIVA1-432(Δ20)-cya(T18) | This work |
| pSBLH008 | kan Plac-divIVA1-432(Δ20)-cya(T25) | This work |
| pSBLH036 | bla Plac-divIVA1-429(Δ21)-cya(T18) | This work |
| pSBLH037 | bla Plac-divIVA1-414(Δ26)-cya(T18) | This work |
| pSBLH038 | bla Plac-divIVA1-390(Δ34)-cya(T18) | This work |
| pSBLH039 | kan Plac-divIVA1-429(Δ21)-cya(T25) | This work |
| pSBLH040 | kan Plac-divIVA1-414(Δ26)-cya(T25) | This work |
| pSBLH041 | kan Plac-divIVA1-390(Δ34)-cya(T25) | This work |
| pSH19 | bla amyE3′ spc Pxyl-divIVABsamyE5′ | This work |
| pSH209 | bla amyE3′ spc Pxyl-divIVALmamyE5′ | This work |
| pSH210 | bla amyE3′ spc Pxyl-divIVALm-gfp amyE5′ | This work |
| pSH260 | bla amyE3′ spc Pxyl-divIVALm-71-BsamyE5′ | This work |
| pSH261 | bla amyE3′ spc Pxyl-divIVALm-144-BsamyE5′ | This work |
| pSH267 | bla amyE3′ spc Pxyl-divIVALm-130-BsamyE5′ | This work |
| pSH272 | bla amyE3′ spc Pxyl-divIVALm-83-BsamyE5′ | This work |
| pSH278 | bla amyE3′ spc Pxyl-divIVALm-104-BsamyE5′ | This work |
| pSH290 | bla amyE3′ spc Pxyl-divIVALm-71-Bs-gfpA206K amyE5′ | This work |
| pSH291 | bla amyE3′ spc Pxyl-divIVALm-144-Bs-gfpA206K amyE5′ | This work |
| pSH292 | bla amyE3′ spc Pxyl-divIVALm-130-Bu-gfpA206K amyE5′ | This work |
| pSH293 | bla amyE3′ spc Pxyl-divIVALm-83-Bs-gfpA206K amyE5′ | This work |
| pSH294 | bla amyE3′ spc Pxyl-divIVALm-104-Bs-gfpA206K amyE5′ | This work |
| pSH316 | bla aprE5′ cat lacI Pspac-minJ-gfp aprE3′ | This work |
| pSH317 | bla aprE5′ cat Pspac-minJ-gfp aprE3′ | This work |
| pSH320 | bla aprE5′ cat PdivIVA1-minJ-gfp aprE3′ | This work |
| pSH326 | bla amyE3′ spc Pxyl-divIVABs-57-lm-gfp amyE5′ | This work |
| pSH328 | bla aprE5′ cat PdivIVA3-minJ-gfp aprE3′ | This work |
| pSH330 | bla aprE5′ spc lacI Pspac-divIVABsR102K aprE3′ | This work |
| pSH331 | bla aprE5′ spc lacI Pspac-divIVABsR102E aprE3′ | This work |
| pSH334 | bla aprE5′ spc lacI Pspac-divIVABsΔC34 aprE3′ | This work |
| pSH335 | bla aprE5′ cat lacI Pspac-divIVABsaprE3′ | This work |
| pSH336 | bla aprE5′ cat lacI Pspac-divIVABsR102K aprE3′ | This work |
| pSH337 | bla aprE5′ cat lacI Pspac-divIVABsR102E aprE3′ | This work |
| pSH340 | bla aprE5′ cat lacI Pspac-divIVABsΔC34 aprE3′ | This work |
| pSH354 | bla amyE3′ spc Pxyl-divIVALm-gfpA206K amyE5′ | This work |
| pSH355 | bla amyE3′ spc Pxyl-divIVABs-57-Lm-gfpA206K amyE5′ | This work |
| B. subtilis strains | ||
| 168 | trpC2 | |
| 168(pSG4916) | 168 racA::pSG4916(Pxyl-gfp-racA′ cat) | 8 |
| 4041 | 168 divIVA::tet | 18 |
| BSN5 | 168 aprE::Pspac-divIVA spc lacI divIVA::tet | 18 |
| SB002 | 168 amyE::Pxyl-minJ-gfp spc minJ::tet | 22 |
| BSN51 | 168 amyE::Pxyl-divIVABsspc divIVA::tet | This work |
| BSN238 | 168 amyE::Pxyl-divIVALmspc divIVA::tet | This work |
| BSN274 | 168 amyE::Pxyl-divIVALm-144-Bsspc divIVA::tet | This work |
| BSN278 | 168 amyE::Pxyl-divIVALm-130-Bsspc divIVA::tet | This work |
| BSN287 | 168 amyE::Pxyl-divIVALm-83-Bsspc divIVA::tet | This work |
| BSN288 | 168 amyE::Pxyl-divIVALm-104-Bsspc divIVA::tet | This work |
| BSN294 | 168 amyE::Pxyl-divIVALm-71-Bs-gfpA206K spc divIVA::tet | This work |
| BSN295 | 168 amyE::Pxyl-divIVALm-144-Bs-gfpA206K spc divIVA::tet | This work |
| BSN296 | 168 amyE::Pxyl-divIVALm-130-Bs-gfpA206K spc divIVA::tet | This work |
| BSN297 | 168 amyE::Pxyl-divIVALm-83-Bs-gfpA206K spc divIVA::tet | This work |
| BSN298 | 168 amyE::Pxyl-divIVALm-104-Bs-gfpA206K spc divIVA::tet | This work |
| BSN308 | 168 aprE::Pspac-minJ-gfp cat lacI | This work |
| BSN313 | 168 aprE::Pspac-divIVAR131A spc lacI divIVA::tet | This work |
| BSN316 | 168 amyE::Pxyl-divIVALm-71-Bsspc divIVA::tet | This work |
| BSN317 | 168 aprE::Pspac-minJ-gfp cat | This work |
| BSN321 | 168 amyE::Pxyl-divIVABs-57-Lmspc divIVA::tet | This work |
| BSN332 | 168 aprE::PdivIVA3-minJ-gfp cat | This work |
| BSN333 | 168 aprE::PdivIVA3-minJ-gfp cat divIVA::tet | This work |
| BSN334 | 168 aprE::PdivIVA3-minJ-gfp cat amyE::Pxyl-divIVABsspc divIVA::tet | This work |
| BSN335 | 168 aprE::PdivIVA3-minJ-gfp cat amyE::Pxyl-divIVALmspc divIVA::tet | This work |
| BSN336 | 168 aprE::PdivIVA3-minJ-gfp cat amyE::Pxyl-divIVALm-104-Bsspc divIVA::tet | This work |
| BSN338 | 168 aprE::PdivIVA3-minJ-gfp cat amyE::Pxyl-divIVABs-57-Lmspc divIVA::tet | This work |
| BSN340 | 168 racA::pSG4916(Pxyl-gfp-racA′ cat)amyE::Pxyl-divIVABsspc divIVA::tet | This work |
| BSN341 | 168 racA::pSG4916(Pxyl-gfp-racA′ cat)amyE::Pxyl-divIVALmspc divIVA::tet | This work |
| BSN342 | 168 racA::pSG4916(Pxyl-gfp-racA′ cat)amyE::Pxyl-divIVALm-104-Bsspc divIVA::tet | This work |
| BSN344 | 168 racA::pSG4916(Pxyl-gfp-racA′ cat)amyE::Pxyl-divIVABs-57-Lmspc divIVA::tet | This work |
| BSN356 | 168 aprE::Pspac-divIVABscat lacI divIVA::tet | This work |
| BSN357 | 168 aprE::Pspac-divIVABsR102K cat lacI divIVA::tet | This work |
| BSN358 | 168 aprE::Pspac-divIVABsR102E cat lacI divIVA::tet | This work |
| BSN360 | 168 aprE::Pspac-divIVABsΔC34 cat lacI divIVA::tet | This work |
| BNS372 | 168 amyE::Pxyl-divIVABs-57-Lm-gfpA206K spc divIVA::tet | This work |
| BSN373 | 168 amyE::Pxyl-divIVALm-gfpA206K spc divIVA::tet | This work |
The designations represent, e.g., for divIVA1-459(Δ11), divIVA from nucleotide positions 1 to 459 with a deletion of the 11 C-terminal amino acid residues.
Construction of bacterial two-hybrid plasmids.
In order to construct C-terminal truncations of divIVA for use in the bacterial two-hybrid assay, plasmids p25N-divIVA and pUT18-divIVA were used as the templates in a PCR with oligonucleotide 25_N_18_F as the forward primer and the oligonucleotides divIVA_11_B2H_R (DivIVA which lacks the last 11 C-terminal amino acids [DivIVAΔ11]), divIVA_19_B2H_R (DivIVAΔ20), divIVA_21_R (DivIVAΔ21), divIVA_26_R (DivIVAΔ26), and divIVA_34_R (DivIVAΔ34) as the complementary primers (for all primer sequences, see Table 2). The PCR products were KpnI digested, self-ligated, and transformed to E. coli. The appropriate clones were identified using restriction analysis and DNA sequencing.
Table 2.
Oligonucleotides used in this study
| Name | Sequence (5′ → 3′) |
|---|---|
| 25_N_18_F | CCGGGTACCGAGCTCGAATTCA |
| divIVA_11_B2H_R | GCGGGTACCTCAAGGAGATGATCCCA |
| divIVA_19_B2H_R | GCGGGTACCAATTTCAGAAGATCAAG |
| divIVA_21_R | GCGGGTACCAGAAGATCAAGCTGAGCT |
| divIVA_26_R | GCGGGTACCGCTTCAATCAGCATTTGG |
| divIVA_34_R | GCGGGTACCGTTCTGAACACTTTAGAC |
| SHW109 | CTTAGGTACCAAGCTAGTAACTATGGTAGAATG |
| SHW110 | GCGCTCGAGTTAACGTTCTTCAGATTCAGCTG |
| SHW111 | GCGCTCGAGACGTTCTTCAGATTCAGCTG |
| SHW180 | CGAAGTAAATGACTTCCTCGATC |
| SHW184 | CTTAACTAGTTTTGTATAGTTCATCCATGCC |
| SHW237 | GAACGTTTAGGTCATTTTACAAACATTGAGGAGACATTGAATAAATC |
| SHW238 | GTGGAAGCACAAATGGATTTAATTAAAAATGACGATTGGGATCATC |
| SHW247 | CGTCAATCCAAAGTATTCCGTACACGTTTCCAAATGCTGATTG |
| SHW265 | CAAACAGCTGCCGAAGAAGTGAAACGCAATTCTCAAAAAGAAGCAAAG |
| SHW266 | GCAGAAAAAAATGCTGACCGAATTATCAACGAATCGTTATCAAAATC |
| SHW304 | AAAGGAAGGACTTGATATCGAATTCC |
| SHW305 | TATCAAGTCCTTCCTTTTCCTCAAATAC |
| SHW342 | ATATGTCGACACATAAAATGCATCTAGAAAGGAG |
| SHW343 | GCGCGGATCCTTATTTGTATAGTTCATCCATGCC |
| SHW349 | TTGCGCTCACATCAAATCGTCTCCCTCCG |
| SHW350 | GATTTGATGTGAGCGCAACGCAAGCTTC |
| SHW354 | GTCGTATGGAGGTGCTAGATATGCCATTAACGCCAAATGATATTC |
| SHW355 | GTTTCTTCAATGTTTGTAAAATGACCGATTCTTTCATCAAGCTCATTG |
| SHW356 | GACACCTCGAGCATGATGCCACCTCCATTTTTAC |
| SHW357 | TGATGCCACCTCCATTTTTACATTTC |
| SHW358 | AAATGGAGGTGGCATCATGCTCGAGG |
| SHW366 | GAAGAACGTGGACTCGAGGTCGACGGTATC |
| SHW367 | GACCTCGAGTCCACGTTCTTCAGATTCAGCTG |
| SHW378 | CGCTGATAAAATTATCAACGAATCG |
| SHW379 | GATAATTTTATCAGCGTTTTTCTCC |
| SHW380 | CGCTGATGAAATTATCAACGAATC |
| SHW381 | GATAATTTCATCAGCGTTTTTCTCC |
| SHW386 | TCAGAACATAATTCCAAATGCTGATTGAAG |
| SHW387 | ATTTGGAATTATGTTCTGAACACTTTAGAC |
| SHW425 | CACAATCTAAACTTTCCAAAGATCCCAACG |
| SHW426 | GGAAAGTTTAGATTGTGTGGACAGGTAATG |
| SV23 | GAAAAGGAATAACTTGATATCGAATTC |
| SV24 | GATATCAAGTTATTCCTTTTCCTCAAATAC |
| SV81 | CGCGCGAGCTCTTATTCCTTTTCCTCAAATACAGC |
| SV98 | CTTAGGTACCTTGGCCGGTGCAGCTTAAC |
| SV123 | CGGGATCCAAAATGGAGGTGGCATCATGCCATTAACGCCAAATG |
| R131A_fw | TCAGAACAGCTTTCCAAATGCTGATTG |
| R131A_rev | TTTGGAAAGCTGTTCTGAACACTTTAG |
Construction of plasmids containing divIVA from B. subtilis and L. monocytogenes.
For xylose-inducible expression of B. subtilis divIVA (divIVABs), we constructed plasmid pSH19. This plasmid was obtained by introducing a stop codon between divIVA and gfp using plasmid pSH3 as the template and the oligonucleotides SV23/SV24 as the primers in a QuikChange mutagenesis reaction. In order to express the L. monocytogenes divIVA (divIVALm) gene in B. subtilis cells, plasmid pSH209 was constructed. This plasmid contains the complete lmo2020 open reading frame of L. monocytogenes under the control of the Pxyl promoter. It was obtained by amplification of the L. monocytogenes divIVA DNA fragment with the oligonucleotides SHW109/SHW110 and subsequent cloning of the obtained fragment into plasmid pSG1154 using KpnI/XhoI. Plasmid pSH210 was constructed in the same way to allow the expression of L. monocytogenes DivIVA-green fluorescent protein (GFP) in cells of B. subtilis. However, for this cloning, the divIVA DNA fragment was amplified with primers SHW109/SHW111 to fuse the divIVA gene in frame to the gfp gene of the vector backbone. The A206K mutation, which prevents dimerization of GFP (24, 25), was introduced into the gfp part of plasmid pSH210 using QuikChange mutagenesis with SHW425/SHW426 as the mutagenic primers. The resulting plasmid was sequenced and named pSH354.
Construction of divIVA chimeras.
For the construction of chimeric divIVA genes consisting of N-terminal parts from L. monocytogenes and C-terminal parts from B. subtilis, a PCR-based restriction-free cloning strategy was used (26). C-terminal fragments of the B. subtilis divIVA gene were amplified from plasmid pSH19 with SHW237 (pSH260), SHW238 (pSH261), SHW247 (pSH267), SHW265 (pSH272), and SHW266 (pSH278) as the respective forward primers and SHW184 as the reverse primer in a first step. All forward primers were identical to the desired fusion sites in the L. monocytogenes divIVA gene in their 5′ regions, whereas the reverse primer SHW184 annealed outside the B. subtilis divIVA gene in the pSH19 plasmid backbone. For the construction of the divIVABs-57-Lm chimera, a DNA fragment corresponding to the first 57 amino acids of the B. subtilis divIVA gene was amplified in a PCR with pSH19 as the template and SHW354 and SHW355 as the primers. All PCR products were purified using a PCR purification kit from Qiagen and used as megaprimers in a second PCR with plasmid pSH209 as the template in order to fuse the N- and C-terminal fragments of B. subtilis divIVA to the corresponding portions of the L. monocytogenes divIVA gene. For the construction of the divIVABs-57-Lm chimera (pKK13), primer SHW180 was added as a reverse primer to the PCR mixture. The PCR mixtures were then DpnI digested and transformed, and the correct clones were identified using restriction analysis and DNA sequencing.
GFP was fused to all DivIVA chimeras by replacing the stop codon of the chimeric divIVA genes by a glycine codon in a way in that the divIVA genes were fused to the downstream gfp open reading frame that was already present in these plasmids. For this purpose, we used the oligonucleotides SHW304/SHW305 to replace the divIVA stop codons in plasmids pSH260, pSH261, pSH267, pSH272, and pSH278. The replacement of the divIVA stop codon in plasmid pKK13 was performed using the primer pair SHW366/SHW367. The DNA sequences of all plasmid clones were verified, and the plasmids were named pSH290 (divIVALm-71-Bs-gfpA206K, where gfpA206K represents the A-to-K change at position 206 encoded by gfp), pSH291 (divIVALm-144-Bs-gfpA206K), pSH292 (divIVALm-130-Bs-gfpA206K), pSH293 (divIVALm-83-Bs-gfpA206K), pSH294 (divIVALm-104-Bs-gfpA206K), and pSH326 (divIVABs-57-Lm-gfp). Finally, the gfpA206K mutation was also introduced into plasmid pSH326 as described above to result in plasmid pSH355.
Construction of a minJ-gfp fusion.
In order to express a minJ-gfp fusion in the divIVA chimera strains, the minJ-gfp allele of strain SB002 was PCR amplified using the oligonucleotides SHW342/SHW343, the resulting PCR fragment was cut with BamHI/SalI and ligated to pAPNC213cat digested with the same enzymes, and the resulting plasmid was named pSH316 after DNA sequencing. However, there was only marginal MinJ-GFP fluorescence, when pSH316 was inserted into the B. subtilis chromosome under inducing conditions (strain BSN308; data not shown). Therefore, plasmid pSH317 was constructed, in which the lacI gene of pSH316 was deleted by PCR using the primer pair SHW349/SHW350 in order to enhance the fluorescence signal. MinJ-GFP fluorescence was still not sufficient with this allele (strain BSN317; data not shown), so the promoter region of the B. subtilis divIVA gene, including the ribosomal binding site (RBS), was amplified with primers SV98/SHW356, and the resulting PCR fragment was cut with KpnI and XhoI and ligated to pSH317, which had been cut with the same enzymes. Two clones that contained a divIVA promoter insert of the right size were isolated, but DNA sequencing revealed single mutations in the RBS (PdivIVA1 on pSH320). In order to correct this, QuikChange mutagenesis with primers SHW357/SHW358 was employed on pSH320, and several plasmid clones were isolated and transformed to B. subtilis. From these transformations, only three plasmid clones conferred the typical fluorescence pattern of MinJ-GFP on cells of B. subtilis. When sequenced, one of these clones had a corrected RBS but also an unintended G deletion between the RBS and the MinJ-GFP start codon (PdivIVA3). This clone was named pSH328 and used for all further studies.
Construction of point mutations and C-terminal truncations in divIVA.
For the construction of plasmid pINC12 encoding the divIVAR131A gene (where divIVAR131A represents the R-to-A change at position 131 encoded by divIVA) under the control of the Pspac promoter, we made use of plasmid pINC3, which already contained the divIVAR131A-gfp allele. pINC3 was originally obtained by QuikChange mutagenesis using the mutagenesis primers R131A_fw/R131A_rev on plasmid pDG9. divIVAR131A of pINC3 was then amplified using the primers SV123/SV81, and the resulting PCR fragment was cut with BamHI/SalI and ligated to the BamHI/SalI-cut vector backbone of plasmid pAPNC213. The DNA sequence of one clone was verified, and this clone was named pINC12. The R102K, R102E, and ΔC34 (deletion of the C-terminal 34 amino acid residues) mutations were introduced into plasmid pSH2 by QuikChange mutagenesis using the oligonucleotides SHW378/SHW379 (pSH330), SHW380/SHW381 (pSH331), and SHW386/SHW387 (pSH334), respectively. In order to exchange the spc marker for a cat cassette in these plasmids, the KpnI/SacI Pspac-divIVA fragments of pSH2, pSH330, pSH331, and pSH334 were then subcloned into the KpnI/SacI-cut backbone of pAPNC213cat in a second step. The resulting plasmids were sequenced and named pSH335 (wild type [wt]), pSH336 (R102K), pSH337 (R102E), and pSH340 (ΔC34).
Strain construction.
Plasmids designed for the expression of divIVA alleles in B. subtilis were inserted into the amyE gene of B. subtilis 168, and amylase-negative transformants were selected on the basis of iodine staining of starch-containing agar plates. Alternatively, the aprE locus was also used for chromosomal integrations. Insertions at aprE were generally confirmed by PCR. Combinations of alleles were generated by transformation (27).
Bacterial two-hybrid analysis.
In order to investigate the interaction of the DivIVA and C-terminally truncated DivIVA proteins with MinJ and RacA, the bacterial two-hybrid system was used (28). Plasmids encoding divIVA alleles fused to the T18 or the T25 fragment of the Bordetella pertussis adenylate cyclase were cotransformed in E. coli BTH101 along with plasmids encoding T25 and T18 fusions to RacA and MinJ. Transformants were selected on nutrient agar plates containing ampicillin (100 μg/ml), kanamycin (50 μg/ml), X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 0.004%), and IPTG (0.1 mM), and photographs were taken after 40 h of growth at 30°C.
Microscopic techniques.
For microscopy of bacterial cells, a small volume (0.3 μl) of an exponentially growing culture was mounted on a microscope slide covered with a thin film of 1.5% agarose (dissolved in distilled water). Membranes were stained using FM5-95. Images were taken with a Nikon Eclipse Ti microscope coupled to a Nikon DS-MBWc charge-coupled-device camera and processed using the NIS elements AR software package (Nikon).
Sporulation assays.
B. subtilis strains were streaked on Schaeffer′s sporulation agar (29) containing 0.5% xylose or 1 mM IPTG and incubated for up to 7 days at 37°C, until lysis of the translucent sporulation-deficient strains could be comfortably discriminated from the optically dense appearance of sporulation-proficient strains. Plates were photographed against a black background.
Isolation of cellular proteins, PAGE techniques, and Western blotting.
Exponentially growing cells of B. subtilis were harvested by centrifugation (13,000 rpm for 1 min in an Eppendorf 5415R tabletop centrifuge), and the cell pellet was washed once in ZAP buffer (10 mM Tris-HCl, pH 7.5, 200 mM NaCl). Cells were disrupted by sonication in ZAP buffer containing 1 mM phenylmethylsulfonyl fluoride, and cell debris was pelleted in another centrifuge run. Aliquots of the resulting supernatant were separated either by SDS-PAGE or by blue native PAGE, which was performed using NativePAGE Novex 4 to 16% bis-Tris gels (Invitrogen) and carried out according to the instructions given by the manufacturer. Subsequently, gels were blotted onto a polyvinylidene difluoride (PVDF) membrane employing a semidry electroblotting unit. Proteins of interest were visualized using polyclonal rabbit antisera recognizing DivIVA (5) or GFP (lab stock) as the primary antibodies and an anti-rabbit immunoglobulin G conjugated to horseradish peroxidase as the secondary one. An ECL chemiluminescence detection kit (Thermo Scientific) was then used for the detection of the peroxidase conjugates on the PVDF membranes.
RESULTS
C-terminal DivIVA truncations interfere with MinJ and RacA binding.
The tetramerization domain of B. subtilis DivIVA is followed by 11 nonconserved amino acid residues. The atomic structure of this C-terminal stretch could not be solved using crystallography, suggesting that it is a flexible tail. To determine whether this C-terminal tail is involved in the binding of MinJ and/or RacA, we made use of the bacterial two-hybrid assay and cloned two DivIVA truncations in this system: DivIVAΔ11, which lacks the last 11 C-terminal amino acids, and DivIVAΔ20, which lacks the last 20 C-terminal amino acids, including a part of the tetramerization domain (Table 3). Both truncations were still able to interact with full-length DivIVA, indicating that both mutants are expressed normally and can form dimers (Table 3; a colored image of the bacterial two-hybrid plate is available in Fig. S1 in the supplemental material). MinJ interacted strongly with full-length DivIVA and with both DivIVA truncations in the bacterial two-hybrid assay, whereas RacA showed a weak interaction with full-length DivIVA that was abolished when the last 11 amino acids of DivIVA were removed (Table 3; see Fig. S1 in the supplemental material). It seems that the RacA-DivIVA interaction depends on the 11 C-terminal amino acids of DivIVA, whereas the MinJ contact site is located more to the N terminus of DivIVA. To test this, additional DivIVA truncations were constructed: DivIVAΔ21, DivIVAΔ26, and DivIVAΔ34. These truncations successively removed the complete tetramerization domain. The last two truncations, DivIVAΔ26 and DivIVAΔ34, were severely impaired in their ability to interact with MinJ (Table 3; see Fig. S1 in the supplemental material), suggesting that the tetramerization domain contains residues required for MinJ binding.
Table 3.
Bacterial two-hybrid analysis of C-terminal DivIVA truncation mutants regarding their ability to interact with full-length DivIVA, MinJ, and RacA
| DivIVA | C-terminal protein sequencea | Reactivityb |
||
|---|---|---|---|---|
| DivIVA | MinJ | RacA | ||
| wt | LKKQSKVFRTRFQMLIEAQLDLLKNDDWDHLLEYEVDAVFEEKE-164 | + | + | ± |
| Δ11 | LKKQSKVFRTRFQMLIEAQLDLLKNDDWDHLLE-153 | + | + | − |
| Δ20 | LKKQSKVFRTRFQMLIEAQLDLLK-144 | + | + | − |
| Δ21 | LKKQSKVFRTRFQMLIEAQLDLL-143 | + | + | − |
| Δ26 | LKKQSKVFRTRFQMLIEA-138 | ± | ± | − |
| Δ34 | LKKQSKVFRT-130 | ± | ± | − |
Starting from position 121. The shadowed sequence stretch corresponds to the DivIVA tetramerization domain. The position of the last amino acid in the truncated DivIVA proteins is given at the end of the sequence.
Symbols coincide with the colors in Fig. S1 in the supplemental material: +, dark blue; ±, light blue; −, white.
Importance of the tetramerization domain for DivIVA activity.
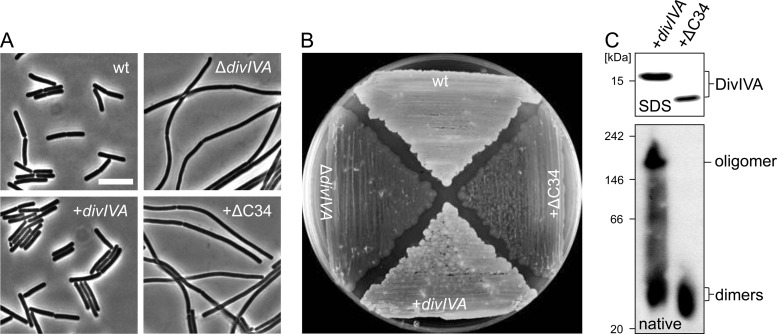
The bacterial two-hybrid assay also revealed a weakened interaction of the DivIVAΔ26 (corresponding to DivIVA amino acids 1 to 138) and DivIVAΔ34 (corresponding to DivIVA amino acids 1 to 130) truncations with full-length DivIVA, whereas the DivIVAΔ21 truncation (corresponding to DivIVA amino acids 1 to 143) still behaved normally (Table 3; see Fig. S1 in the supplemental material). So far there is no biochemical corroboration that amino acids 130 to 143 are involved in tetramerization in vivo. Own preliminary alanine mutagenesis experiments in this region identified R131 as an essential residue for DivIVA activity (see Fig. S3A to C in the supplemental material), suggesting a special importance of this region for DivIVA function. Thus, DivIVAΔC34 was expressed in a ΔdivIVA mutant (strain BSN360), and phenotypic analysis of this strain clearly demonstrated the inability of the divIVAΔC34 allele to complement the cell division and the sporulation phenotype of the ΔdivIVA mutation (Fig. 2A and B), even though DivIVAΔC34 was clearly expressed (Fig. 2C, top). Blue native PAGE of strain BSN356 expressing wild-type DivIVA showed that DivIVA exists in two different oligomeric states since two signals of different molecular masses were detected by the DivIVA antiserum (Fig. 2C, bottom). The molecular masses were calculated to be 159 ± 8 kDa for the upper signal and 41 ± 13 kDa for the lower signal in the wild-type cell extract. Given the molecular mass of B. subtilis DivIVA (19.34 kDa), these molecular masses correspond to an octamer and a dimer, respectively. Blue native PAGE of strain BSN360 revealed the existence of a dimer signal (Fig. 2C, bottom). Previous gel filtration analyses have indicated that purified DivIVA forms octamers and higher-order structures (18, 30). Therefore, it seems plausible that the region beyond residue 130 indeed contains the tetramerization domain and that tetramerization is a prerequisite step for octamerization.
Fig 2.
Complementation activity and oligomerization of a C-terminally truncated DivIVA protein devoid of the tetramerization domain. (A) Phase-contrast micrographs showing cellular morphology of strain BSN360 expressing the DivIVAΔC34 protein. Cultures of strain 168 (wt), strain 4041 (ΔdivIVA), and the complemented ΔdivIVA mutant strain BSN356 (+divIVA) were included as controls. Cells were cultivated in LB broth (containing 1 mM IPTG where necessary) to mid-logarithmic growth phase at 37°C before images were taken. Bar, 5 μm. (B) Sporulation of the same set of strains on Schaeffer's sporulation agar containing 1 mM IPTG. Cells were kept for 3 days at 37°C until lysis of the Spo− strains became apparent. (C) Western blots after SDS-PAGE and blue native PAGE to analyze expression and oligomerization of DivIVAΔC34. Strains BSN356 (+divIVA) and BSN360 (+ΔC34) were cultivated as described above, and cellular protein extracts were subjected to SDS-PAGE (top) or blue native PAGE (bottom) and subsequent Western blotting. DivIVA was detected using the polyclonal anti-DivIVA antiserum. The NativeMark standard (Invitrogen) was used as a molecular mass marker for blue native PAGE.
Domain swapping to identify DivIVA interaction domains.
It is possible that the C-terminal truncations used in the bacterial two-hybrid system result in misfolded and/or unstable DivIVA variants. This complicates the interpretation of the bacterial two-hybrid data. Because of this, we changed tactics and explored the possibility to swap domains between B. subtilis DivIVA and the closely homologous DivIVA from L. monocytogenes. In a previous study, we have shown that L. monocytogenes DivIVA displays the same localization pattern as B. subtilis DivIVA and is involved in SecA2-dependent protein secretion (10). L. monocytogenes does not sporulate and does not contain a RacA homologue. It is also unlikely that L. monocytogenes DivIVA interacts with MinJ, since deletion of the divIVA gene does not result in a minicell phenotype in L. monocytogenes, indicating that the listerial division site control system is DivIVA independent (10). This would enable us to separate the DivIVA domains required for localization and for RacA and MinJ interaction. First, it was necessary to confirm that L. monocytogenes DivIVA is normally localized when expressed in B. subtilis. Indeed, L. monocytogenes DivIVA fused to GFP and when expressed in a divIVA-knockout background (strain BSN373) showed a localization pattern that was similar to that of B. subtilis DivIVA (Fig. 3), even though L. monocytogenes DivIVA predominately exists as a dimer and just to a minor extent in an oligomeric form when expressed in B. subtilis (see Fig. S2 in the supplemental material). More importantly, however, L. monocytogenes DivIVA does not complement the cell division and sporulation defects of a B. subtilis ΔdivIVA mutant (strain BSN238; see Fig. 5A and B). Thus, L. monocytogenes DivIVA seems to be unable to bind MinJ and RacA and is therefore well suited for domain swapping.
Fig 3.
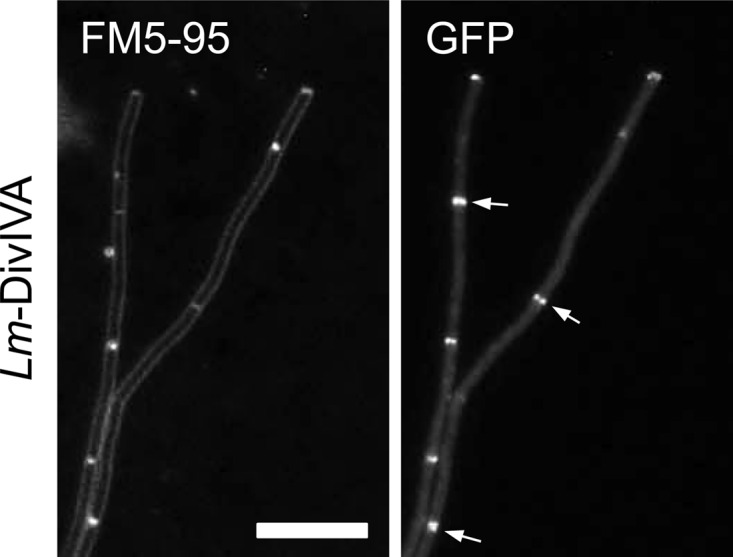
Localization of L. monocytogenes DivIVA-GFP in a B. subtilis ΔdivIVA background. Strain BSN373 (expressing L. monocytogenes DivIVA-GFPA206K) was grown in LB medium supplemented with 0.5% xylose. The localization pattern of L. monocytogenes DivIVA-GFP was analyzed by epifluorescence microscopy (right), and for orientation, a FM5-95-stained image (left) was taken in parallel. Several septal DivIVA-GFP signals are indicated by arrows. Bar, 5 μm.
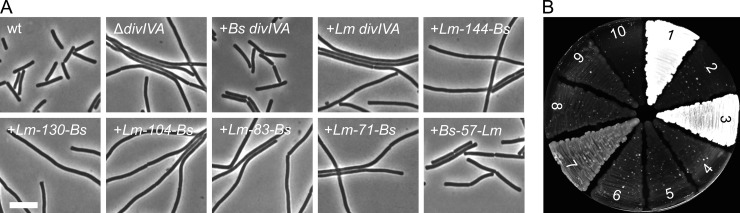
Fig 5.
Complementation activity of L. monocytogenes and B. subtilis DivIVA chimeras in the B. subtilis ΔdivIVA background. (A) Phase-contrast micrographs showing the ability of the tested L. monocytogenes and B. subtilis DivIVA chimeras to complement the filamentous ΔdivIVA phenotype. Cells were cultivated in LB broth containing 0.5% xylose until mid-log growth phase at 37°C before cell morphology was assessed microscopically. Bar, 5 μm. (B) Sporulation plate assay to test the activity of the L. monocytogenes and B. subtilis DivIVA chimeras to complement the sporulation defect of the B. subtilis ΔdivIVA mutant. Strains expressing the DivIVA chimeras were streaked on Schaeffer′s sporulation agar containing 0.5% xylose and kept at 37°C until lysis of nonsporulating strains was comfortably distinguishable from the brownish Spo+ strains. The wild type, the ΔdivIVA mutant, and strains complemented with either the B. subtilis (BSN51) or the L. monocytogenes (BSN238) divIVA gene were used as controls. Sections: 1, strain 168 (wt); 2, strain 4041 (ΔdivIVA); 3, strain BSN51 (B. subtilis divIVA); 4, strain BSN238 (L. monocytogenes divIVA); 5, strain BSN274 (Lm-144-Bs divIVA); 6, strain BSN278 (Lm-130-Bs divIVA); 7, strain BSN288 (Lm-104-Bs divIVA); 8, strain BSN287 (Lm-83-Bs divIVA); 9, strain BSN316 (Lm-71-Bs divIVA); 10, strain BSN321 (Bs-57-Lm divIVA). Note that sporulation of strain BSN288 (Lm-104-Bs divIVA) did not reach the wild-type level. This might be explained by the lack of MinCD activity in this strain, which is required for full sporulation (37).
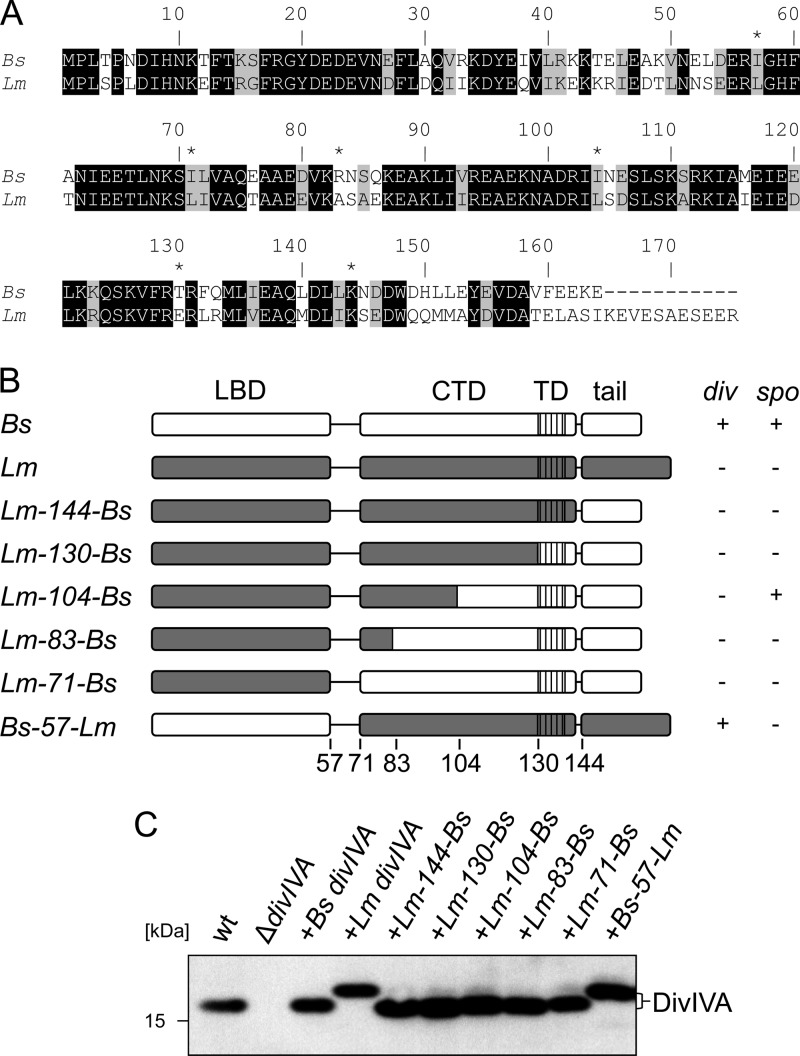
The most prominent difference between L. monocytogenes and B. subtilis DivIVA is found in the C-terminal tail (Fig. 4A), which is 11 amino acids longer in the L. monocytogenes protein and which has been shown to be important for binding RacA, according to the bacterial two-hybrid data. To test this, a DivIVA chimera was constructed by replacing the last 32 amino acids of L. monocytogenes DivIVA with the last 21 amino acids of B. subtilis DivIVA (Lm-144-Bs DivIVA [144 N-terminal amino acid residues of L. monocytogenes DivIVA fused to the corresponding C-terminal part of B. subtilis DivIVA; see Fig. 4A]), so that the C-terminal tails were exchanged between both proteins, whereas the core tetramerization domain (amino acids 130 to 143) was left intact (Fig. 4B). Expression of this chimera in a B. subtilis ΔdivIVA background (strain BSN274) did not restore either cell division or sporulation (Fig. 5A and B). Western blotting showed that Lm-144-Bs DivIVA was stably expressed and not degraded (Fig. 4C). The chimeric protein localized normally, as a GFP fusion indicated (strain BSN295; see Fig. S4 in the supplemental material) and formed a stable oligomer (see Fig. S2 in the supplemental material), suggesting that the last 21 amino acids of B. subtilis DivIVA alone are insufficient for binding of MinJ or RacA.
Fig 4.
Expression of L. monocytogenes and B. subtilis DivIVA chimeras in B. subtilis. (A) Sequence alignment of the DivIVA proteins from B. subtilis (Bs) and L. monocytogenes (Lm). Identical amino acid positions are indicated by a black background, and similar amino acid positions are indicated by a gray background. The exchange sites in the different chimeras are labeled by asterisks. (B) Schematic illustration of the domain organization of the B. subtilis and L. monocytogenes DivIVA proteins and compositions of all L. monocytogenes and B. subtilis DivIVA chimeras. The abbreviations are defined in the legend to Fig. 1A. The complementation activity of the DivIVA chimeras in the complementation assays for division (div) and sporulation (spo) is indicated in the table on the right (compare Fig. 5). (C) Western blot showing expression of the DivIVA chimeras in a B. subtilis ΔdivIVA background. The wild-type strain 168 and the ΔdivIVA mutant (strain 4041), as well as strains expressing B. subtilis divIVA (BSN51) or L. monocytogenes divIVA (BSN238), were included as controls, and the DivIVA proteins were detected with an antiserum that had been raised against B. subtilis DivIVA (5).
Systematic domain swapping.
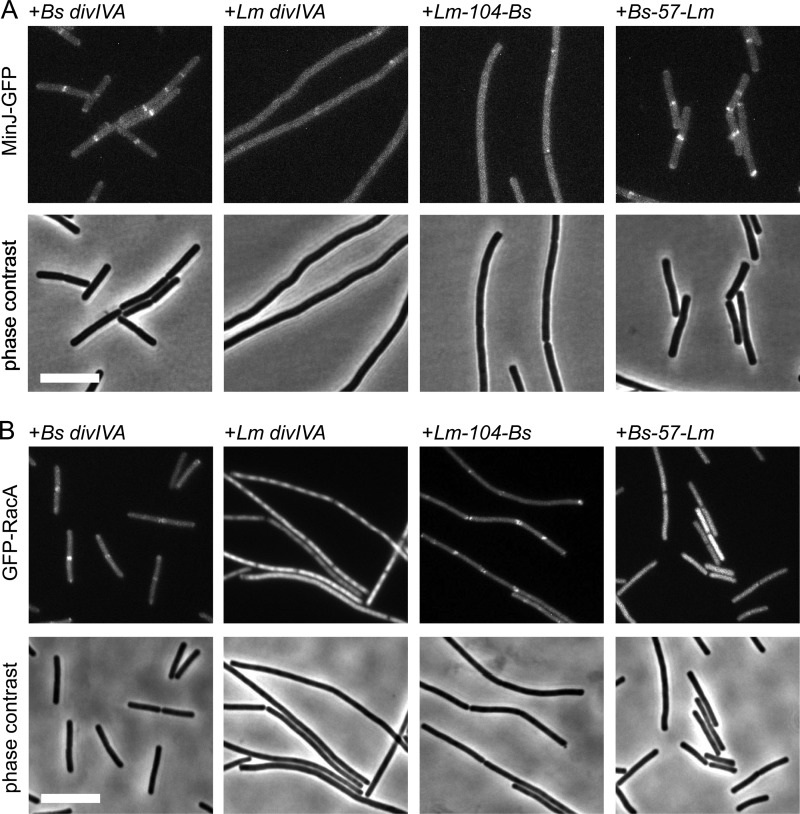
Since MinJ and RacA binding seems to require a larger part of B. subtilis DivIVA, we constructed a set of chimeric DivIVA proteins in which the fusion point between the N-terminal L. monocytogenes and the C-terminal B. subtilis parts was shifted from the tail region toward the N terminus of the C-terminal domain in a stepwise fashion, as schematically indicated in Fig. 4B. Position 130 exchanged the complete C terminus beginning from the TD, positions 104 and 83 mark the beginning of short stretches in the coiled-coil region at which both proteins differ at three to four consecutive amino acid positions, whereas the domain swap at position 71 exchanged the complete C-terminal domain behind the flexible linker (Fig. 4A and B). Stability and oligomerization of the chimeras was checked by Western blotting, indicating that all the chimeric DivIVA proteins were expressed at comparable levels (Fig. 4C) and were oligomeric (see Fig. S2 in the supplemental material). Expression of these chimeras in the B. subtilis ΔdivIVA background did not restore normal vegetative cell division (Fig. 5A), suggesting that they were unable to recruit MinJ. Of the different chimeras, only the Lm-104-Bs DivIVA chimera was able to restore spore formation. To confirm that the Lm-104-Bs DivIVA chimera was indeed able to recruit RacA and not MinJ, the chimera was expressed in ΔdivIVA mutant strains containing either the GFP-RacA or the MinJ-GFP reporter. As shown in Fig. 6A and B, the Lm-104-Bs DivIVA chimera can recruit RacA but not MinJ. In conclusion, the RacA interaction domain resides in the last 60 amino acids of DivIVA and requires residues in the coiled-coil region beyond amino acid 104.
Fig 6.
Localization of MinJ and RacA in B. subtilis strains expressing selected L. monocytogenes and B. subtilis divIVA chimeras. (A) Fluorescence micrographs showing the subcellular localization of MinJ-GFP in L. monocytogenes and B. subtilis divIVA chimera strains during mid-logarithmic growth in LB broth supplemented with 0.5% xylose at 37°C (top). MinJ-GFP was imaged in strains expressing the Lm-104-Bs DivIVA (strain BSN336) and the Bs-57-Lm DivIVA (strain BSN338) chimeras. As a control, MinJ-GFP was also visualized in ΔdivIVA strains which express B. subtilis divIVA (strain BSN334) or L. monocytogenes divIVA (strain BSN335). Phase-contrast images were included for better orientation (bottom). (B) Localization of RacA in B. subtilis strains expressing the same L. monocytogenes and B. subtilis divIVA chimeras as in panel A. Fluorescence images were obtained on cells during growth in LB broth containing 0.5% xylose at 37°C (top). GFP-RacA was visualized in strains expressing the Lm-104-Bs DivIVA (strain BSN342) and Bs-57-Lm DivIVA (strain BSN344) chimeras. As controls, GFP-RacA was also imaged in strain BSN340, which expresses B. subtilis divIVA, and in strain BSN341, which expresses L. monocytogenes divIVA. Phase-contrast images were included for better orientation (bottom). Bar, 5 μm.
It is surprising that larger replacements of the C terminus (as in Lm-71-Bs and Lm-83-Bs) were again unable to restore sporulation. A possibility is that these chimeras did not localize properly anymore. To test this, we expressed C-terminal GFP fusions to the chimeric DivIVA constructs and analyzed their localization in B. subtilis ΔdivIVA cells. Expression of all DivIVA-GFP proteins gave rise to polar and septal fluorescence signals; however, these signals occurred to different degrees (see Fig. S4A in the supplemental material). While DivIVALm-144-Bs-GFP and DivIVALm-130-Bs-GFP clearly accumulated at the division septa, the septal fluorescence signals of DivIVALm-104-Bs-GFP, DivIVALm-83-Bs-GFP, and DivIVALm-71-Bs-GFP were less intense but still visible (see Fig. S4A in the supplemental material). This suggests that all DivIVA chimeras are functional in terms of lipid binding and membrane curvature sensing.
The lipid binding domain recruits MinJ.
As none of the chimeras was able to complement the cell division phenotype, it may be that regions in the N-terminal domain are critical for the interaction of B. subtilis DivIVA with MinJ. To test this, we fused the N-terminal 57 amino acids of B. subtilis DivIVA spanning the entire lipid binding domain to the complete C-terminal domain of L. monocytogenes DivIVA (Fig. 4B). When this DivIVA chimera was expressed in a ΔdivIVA background (strain BSN321), short cells and no minicells were observed (Fig. 5A), indicating that the Bs-57-Lm DivIVA protein recruits MinJ. The localization as well as oligomerization of this chimera is comparable to that of wild-type DivIVA (see Fig. S4 and Fig. S2 in the supplemental material, respectively) and indeed restores normal septal and polar localization of GFP-MinJ (Fig. 6A). Strikingly, sporulation was still defective in strain BSN321 (Fig. 5B) and the Bs-57-Lm chimera was unable to recruit RacA (Fig. 6B). In conclusion, the lipid binding N-terminal domain of DivIVA contains the MinJ binding site, whereas the C-terminal coiled-coil domain contains the binding site for RacA.
DISCUSSION
Here we show that two of the DivIVA interaction partners from B. subtilis, MinJ and RacA, bind to mutually exclusive surface regions of DivIVA. This was concluded from complementation assays with DivIVA chimeras constructed from B. subtilis and L. monocytogenes DivIVA. Analysis of a set of such DivIVA chimeras in complementation experiments surprisingly revealed that the N-terminal lipid binding domain provides the MinJ interaction module, whereas RacA binds to the C-terminal domain of DivIVA. This was unexpected since it was assumed that the C-terminal domain would constitute the protein recruitment module for both proteins, with the LBD being important only for dimerization and lipid binding. However, a dual function of the lipid binding domain is in good agreement with the two-domain nature of DivIVA proteins. The lipid binding domain is in close contact with the cytoplasmic membrane and even partially inserts into it, which makes it a good candidate for interacting with transmembrane proteins like MinJ. Since most sequence differences between the LBDs of B. subtilis and L. monocytogenes DivIVA cluster between residues E28 and I57 (Fig. 4A), this region most likely represents the MinJ binding surface of DivIVA. Support for this assumption comes from the observation that a replacement of the region from amino acids 1 to 16 of Corynebacterium glutamicum DivIVA by the corresponding region from B. subtilis DivIVA was without effect (19). Lipid binding of DivIVA via its N-terminal domain would in turn leave the C terminus free to reach into the cytoplasm. Fitting with this, our experiments indicated that the C-terminal domain is the interaction module for RacA, which is a soluble cytoplasmic protein. It was recently reported that the interaction of C. glutamicum ParB, which is a chromosome-binding protein like RacA, with its cognate DivIVA requires central regions of the C-terminal domain as well (9). Our results thus confirm earlier speculations that the sporulation and the division functions of DivIVA can be separated. This had been concluded from the observation that a divIVA mutation consisting of an N-to-D change at position 99 severely affected sporulation but not division (31). Another classical divIVA point mutation is the divIVA1 mutation in which the alanine at position 78 is replaced by a threonine. This mutation causes a Div− Spo− phenotype (32), even though it lies outside the RacA and MinJ binding regions. Neither expression nor oligomerization of DivIVA is impaired by this mutation (33). Thus, the A78T exchange might possibly affect subcellular localization of DivIVA or induce structural changes in the protein that reduce its activity but do not influence formation of oligomers.
The question that we cannot answer conclusively is, why do the two chimeras with the more N-terminally located fusion points (Lm-83-Bs and Lm-71-Bs) not behave similarly to the Lm-104-Bs DivIVA protein? Initially, this conflicted with the idea that the C-terminal domain is the protein recruitment module for RacA, since C-terminal exchanges longer than those in Lm-104-Bs should result at least in the same degree of complementation activity. We do not think that this is explained by misfolding of the respective chimeric proteins, since they still oligomerize (see Fig. S2 in the supplemental material) and because GFP-tagged versions of these chimeras still localized to the septum to the same degree as the Lm-104-Bs GFP fusion protein (see Fig. S4A in the supplemental material) and therefore appeared to be folded properly. Moreover, a strain expressing an Lm-57-Bs DivIVA protein showed the same sporulation defect as strains expressing Lm-83-Bs and Lm-71-Bs DivIVA chimeras (data not shown). Possibly, longer C-terminal exchanges do not function in the context of an unrelated lipid binding domain. With regard to this issue, the fact that the arginine 102 residue of B. subtilis DivIVA is phosphorylated might be of special interest here (34). This phosphorylation could be critical for RacA recruitment and may add an extra dimension of activity control on the different DivIVA chimeras. Therefore, we constructed a phosphoablative (R102K) and a phosphomimetic (R102E) mutant allele of divIVA and tested their activity in our complementation system. Both of these mutations cause a Div+ Spo− phenotype (see Fig. S5 in the supplemental material). Hence, arginine 102 might indeed have a crucial function in RacA binding, but it is not relevant for the interaction with MinJ. Phosphorylations at the C-terminal domains are well described for DivIVA from Mycobacterium tuberculosis (named Wag31 in mycobacteria) and Streptococcus pneumoniae, even though they both occurred at threonine side chains (T73 and T201, respectively). Phenotypic analysis of phosphomimetic and phosphoablative divIVA mutant strains in these organisms also revealed that these phosphorylations are indeed involved in cell shape control (35, 36). In the future it will be interesting to address the regulatory impact of such phosphorylations for DivIVA binding partner recruitment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ling Juan Wu (Newcastle University) for the kind gift of the gfp-racA strain and Henrik Strahl (Newcastle University) for sharing plasmid pAPNC213cat.
This work was financially supported by a DAAD fellowship to K.G.K., a DFG grant (BR-2915/2-1) to M.B., and a Wellcome Trust Research Career Development Fellowship as well as a Biological Sciences Research Council Grant to L.W.H.
Footnotes
Published ahead of print 21 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02171-12.
REFERENCES
- 1. Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ, Errington J, Marenduzzo D, Hamoen LW. 2009. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 28:2272–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ramamurthi KS, Losick R. 2009. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc. Natl. Acad. Sci. U. S. A. 106:13541–13545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eswaramoorthy P, Erb ML, Gregory JA, Silverman J, Pogliano K, Pogliano J, Ramamurthi KS. 2011. Cellular architecture mediates DivIVA ultrastructure and regulates Min activity in Bacillus subtilis. mBio 2:e00257–11. 10.1128/mBio.00257-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Edwards DH, Thomaides HB, Errington J. 2000. Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J. 19:2719–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 12:3419–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Letek M, Ordonez E, Vaquera J, Margolin W, Flärdh K, Mateos LM, Gil JA. 2008. DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum. J. Bacteriol. 190:3283–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu H, Chater KF, Deng Z, Tao M. 2008. A cellulose synthase-like protein involved in hyphal tip growth and morphological differentiation in Streptomyces. J. Bacteriol. 190:4971–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu LJ, Errington J. 2003. RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol. Microbiol. 49:1463–1475 [DOI] [PubMed] [Google Scholar]
- 9. Donovan C, Sieger B, Krämer R, Bramkamp M. 2012. A synthetic Escherichia coli system identifies a conserved origin tethering factor in Actinobacteria. Mol. Microbiol. 84:105–116 [DOI] [PubMed] [Google Scholar]
- 10. Halbedel S, Hahn B, Daniel RA, Flieger A. 2012. DivIVA affects secretion of virulence-related autolysins in Listeria monocytogenes. Mol. Microbiol. 83:821–839 [DOI] [PubMed] [Google Scholar]
- 11. Bramkamp M, Emmins R, Weston L, Donovan C, Daniel RA, Errington J. 2008. A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD. Mol. Microbiol. 70:1556–1569 [DOI] [PubMed] [Google Scholar]
- 12. Patrick JE, Kearns DB. 2008. MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol. Microbiol. 70:1166–1179 [DOI] [PubMed] [Google Scholar]
- 13. Flärdh K. 2003. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol. Microbiol. 49:1523–1536 [DOI] [PubMed] [Google Scholar]
- 14. Kaval KG, Halbedel S. 2012. Architecturally the same, but playing a different game: the diverse species-specific roles of DivIVA proteins. Virulence 3:406–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ben-Yehuda S, Rudner DZ, Losick R. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299:532–536 [DOI] [PubMed] [Google Scholar]
- 16. Briley K, Jr, Prepiak P, Dias MJ, Hahn J, Dubnau D. 2011. Maf acts downstream of ComGA to arrest cell division in competent cells of B. subtilis. Mol. Microbiol. 81:23–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dos Santos VT, Bisson-Filho AW, Gueiros-Filho FJ. 2012. DivIVA-mediated polar localization of ComN, a post-transcriptional regulator of B. subtilis. J. Bacteriol. 194:3661–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oliva MA, Halbedel S, Freund SM, Dutow P, Leonard TA, Veprintsev DB, Hamoen LW, Löwe J. 2010. Features critical for membrane binding revealed by DivIVA crystal structure. EMBO J. 29:1988–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Letek M, Fiuza M, Ordonez E, Villadangos AF, Flärdh K, Mateos LM, Gil JA. 2009. DivIVA uses an N-terminal conserved region and two coiled-coil domains to localize and sustain the polar growth in Corynebacterium glutamicum. FEMS Microbiol. Lett. 297:110–116 [DOI] [PubMed] [Google Scholar]
- 20. Morimoto T, Loh PC, Hirai T, Asai K, Kobayashi K, Moriya S, Ogasawara N. 2002. Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology 148:3539–3552 [DOI] [PubMed] [Google Scholar]
- 21. Lewis PJ, Marston AL. 1999. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene 227:101–110 [DOI] [PubMed] [Google Scholar]
- 22. van Baarle S, Bramkamp M. 2010. The MinCDJ system in Bacillus subtilis prevents minicell formation by promoting divisome disassembly. PLoS One 5:e9850 doi:10.1371/journal.pone.0009850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24. Zacharias DA, Violin JD, Newton AC, Tsien RY. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296:913–916 [DOI] [PubMed] [Google Scholar]
- 25. Landgraf D, Okumus B, Chien P, Baker TA, Paulsson J. 2012. Segregation of molecules at cell division reveals native protein localization. Nat. Methods 9:480–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van den Ent F, Löwe J. 2006. RF cloning: a restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys. Methods 67:67–74 [DOI] [PubMed] [Google Scholar]
- 27. Hamoen LW, Smits WK, de Jong A, Holsappel S, Kuipers OP. 2002. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 30:5517–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95:5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harwood CR, Coxon RD, Hancock IC. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 30. Stahlberg H, Kutejova E, MuchovÁ K, Gregorini M, Lustig A, Müller SA, Olivieri V, Engel A, Wilkinson AJ, Barák I. 2004. Oligomeric structure of the Bacillus subtilis cell division protein DivIVA determined by transmission electron microscopy. Mol. Microbiol. 52:1281–1290 [DOI] [PubMed] [Google Scholar]
- 31. Thomaides HB, Freeman M, El Karoui M, Errington J. 2001. Division site selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation. Genes Dev. 15:1662–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cha JH, Stewart GC. 1997. The divIVA minicell locus of Bacillus subtilis. J. Bacteriol. 179:1671–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Muchová K, Kutejova E, Scott DJ, Brannigan JA, Lewis RJ, Wilkinson AJ, Barák I. 2002. Oligomerization of the Bacillus subtilis division protein DivIVA. Microbiology 148:807–813 [DOI] [PubMed] [Google Scholar]
- 34. Elsholz AK, Turgay K, Michalik S, Hessling B, Gronau K, Oertel D, Mäder U, Bernhardt J, Becher D, Hecker M, Gerth U. 2012. Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 109:7451–7456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kang CM, Abbott DW, Park ST, Dascher CC, Cantley LC, Husson RN. 2005. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 19:1692–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fleurie A, Cluzel C, Guiral S, Freton C, Galisson F, Zanella-Cleon I, Di Guilmi AM, Grangeasse C. 2012. Mutational dissection of the S/T-kinase StkP reveals crucial roles in cell division of Streptococcus pneumoniae. Mol. Microbiol. 83:746–758 [DOI] [PubMed] [Google Scholar]
- 37. Barák I, Prepiak P, Schmeisser F. 1998. MinCD proteins control the septation process during sporulation of Bacillus subtilis. J. Bacteriol. 180:5327–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.