Abstract
Rhabdomyosarcoma is a pediatric tumor of skeletal muscle that expresses the myogenic basic helix-loop-helix protein MyoD but fails to undergo terminal differentiation. Prior work has determined that DNA binding by MyoD occurs in the tumor cells, but myogenic targets fail to activate. Using MyoD chromatin immunoprecipitation coupled to high-throughput sequencing and gene expression analysis in both primary human muscle cells and RD rhabdomyosarcoma cells, we demonstrate that MyoD binds in a similar genome-wide pattern in both tumor and normal cells but binds poorly at a subset of myogenic genes that fail to activate in the tumor cells. Binding differences are found both across genomic regions and locally at specific sites that are associated with binding motifs for RUNX1, MEF2C, JDP2, and NFIC. These factors are expressed at lower levels in RD cells than muscle cells and rescue myogenesis when expressed in RD cells. MEF2C is located in a genomic region that exhibits poor MyoD binding in RD cells, whereas JDP2 exhibits local DNA hypermethylation in its promoter in both RD cells and primary tumor samples. These results demonstrate that regional and local silencing of differentiation factors contributes to the differentiation defect in rhabdomyosarcomas.
INTRODUCTION
We have recently performed chromatin immunoprecipitation coupled to high-throughput sequencing (ChIP-seq) for the myogenic regulatory factor MyoD in murine cells of the skeletal muscle lineage and described widespread binding of MyoD in both intra- and intergenic regions of the genome (1). MyoD is a member of the basic helix-loop-helix (bHLH) family of transcription factors, a large group of factors that bind DNA through a basic region and use amphipathic helices to heterodimerize with other bHLH proteins (2, 3). In myogenic cells, MyoD heterodimerizes with members of the E-protein bHLH family, binds DNA in a sequence-specific fashion, and transactivates gene targets (4). We found that MyoD bound extensively both in undifferentiated, proliferating myoblasts and in terminally differentiated myotubes. Genes that had increased expression with differentiation were associated with an increased MyoD ChIP-seq signal, and genes that decreased expression were associated with a decreased signal. Analysis of the areas neighboring MyoD-bound sites revealed potential binding sites for a variety of other factors that are known or believed to play roles during myogenesis (e.g., Ap-1, Meis, Runx, and Sp1).
Rhabdomyosarcoma (RMS) is a pediatric tumor of skeletal muscle that resembles undifferentiated myogenic cells (5, 6). Puzzlingly, the tumors typically express MyoD, even though MyoD expression is normally capable of driving terminal skeletal muscle differentiation in not only myogenic cells but those of other lineages as well (7). Previous work identified no defect in the ability of MyoD in RMS to bind to DNA but, rather, identified a defect in its ability to activate myogenic target genes (8), but the binding of MyoD in these tumors has never been investigated in a genome-wide fashion. More recently, our work in rhabdomyosarcoma cells has suggested that they are actually representative of an arrested state of development in normal skeletal muscle, offering the possibility of providing information on a specific point in the myoblast-myotube transition shortly preceding terminal differentiation (9, 10).
To further investigate both the normal molecular mechanisms of MyoD-mediated myogenesis in human cells and the basis for the impaired myogenesis in rhabdomyosarcomas, we have performed ChIP-seq for MyoD in primary human myoblasts and myotubes, as well as in an embryonal cell culture model of RMS, RD cells, alongside gene expression analysis in the same cells. RMS cells exhibit widespread binding of MyoD throughout the genome, with a striking level of similarity to the binding found in primary cells, but possess differences in MyoD binding at a relatively small subset of locations. Differential MyoD binding and impaired MyoD gene target activation implicate numerous transcription factors that are expressed at lower levels in RD cells than primary cells, including MEF2C, RUNX1, JDP2, and NFIC, in impaired myogenesis, and all of the factors are capable of rescuing myogenesis to various extents. We find evidence of differential DNA accessibility across large-scale regions of the genome in RD cells, one of which contains MEF2C, suggesting a role for regional suppression of genes associated with the final stages of myogenesis, in addition to more local effects. Finally, we identify DNA hypermethylation of the promoter of JDP2 in both RD cells and multiple primary human tumor samples compared to normal human cells, implicating DNA methylation-mediated silencing of myogenic cofactors as a potential event in tumor progression and/or formation.
MATERIALS AND METHODS
Cell culture.
RD cells were obtained from the American Type Culture Collection (ATCC), and all analyses were performed on cells that originated from low-passage-number frozen aliquots. RD cells were maintained in Dulbecco modified Eagle medium (DMEM) with 10% bovine calf serum and 1% penicillin-streptomycin (Gibco). Primary myoblasts were collected and cultured as has been previously described (11). They were maintained in F-10 medium supplemented with 20% fetal bovine serum, 1% penicillin-streptomycin, dexamethasone (1 μM), and human basic fibroblast growth factor (final concentration, 10 ng/ml; Promega). Low-serum differentiation medium consisted of either DMEM with 1% horse serum, 1% penicillin-streptomycin, and 10 μg/ml insulin and transferrin (RD cells) or an F-10-based version with the same additives (primary cells).
Chromatin immunoprecipitation.
Primary myoblast samples were collected in growth medium at low density, and both primary myotubes and RD cells were collected after being cultured in differentiation medium (72 h for myotubes, 24 h for RD cells). ChIP and ChIP-seq were performed as has been described previously (1), using a previously characterized MyoD antibody (1, 12). The ChIP primers used for loci A to D were the same as those used for locus A, locus C, locus H, and locus D, respectively, in the PvuII accessibility studies. Locus E primer sequences were as follows: CATGCAGGGCACTCTAGTCTC and TAAGGGTTTAGCCTGCCACA.
Cell staining and Western blotting.
For 5-ethynyl-2′-deoxyuridine (EdU) stains, after 24 h in low-serum differentiation medium, cells were shifted to differentiation medium supplemented with EdU at a final concentration of 50 μM (Invitrogen) and incubated for a further 24 h. Cells were then fixed and stained according to the manufacturer's protocols using a Click-iT kit, and total nuclei and EdU-positive nuclei were counted by hand. Three independent biological replicates were performed for each condition, and the nuclei in five microscope fields were quantified for each replicate to determine the percentage of EdU-positive nuclei. On the basis of a previously established protocol, the highest- and lowest-value measurements were removed, and the remaining three values were averaged; however, similar data were obtained without excluding potential outlier samples. Those averages were used to calculate the mean and standard error of the mean (SEM) of each condition.
For other cell stains, cells were fixed with 2% paraformaldehyde for 6 min at room temperature before permeabilization with Triton X-100. Myosin heavy chain (MHC) was detected with the MF-20 antibody, and nuclei were detected with DAPI (4′,6-diamidino-2-phenylindole).
Western blots were performed on whole-cell lysates collected in Laemmli buffer containing 10% beta-mercaptoethanol. All blots were blocked in 3% (wt/vol) milk in 0.5% Tween 20-containing phosphate-buffered saline before incubation with primary antibody (MHC with antibody MF-20 and MEF2C with antibody 5030 [Cell Signaling]; α-tubulin with antibody T-9026 [Sigma]), a horseradish peroxidase-conjugated secondary antibody (Jackson), and chemiluminescent detection (Pierce).
Peak calling.
Sequences were extracted by the GApipeline program (version 0.3.0). Reads mapping to the X and Y chromosomes were excluded from our analysis. Reads were aligned using BWA to the human genome (hg19). Duplicate sequences were discarded to minimize the effects of PCR amplification. Each read was extended in the sequencing orientation to a total of 200 bases to infer the coverage at each genomic position. Peak calling was performed by an in-house-developed R package, which models background reads by a negative binomial distribution as previously described (13).
Motif analysis.
We applied a house-developed Bioconductor package, motifRG, for discriminative de novo motif discovery as previously described (1, 14). To find discriminative motifs between MyoD peaks in myotubes versus myoblasts and MyoD peaks in myotubes versus RD cells, we selected cell-type-specific peaks (present in one cell type with a P-value cutoff of 10−10 and absent in the other cell type at a P-value cutoff of 10−5).
Association of differential peaks with differentially expressed genes.
To detect differential peaks between two samples, we first took the square root of the peak heights in both samples and fit a local regression curve on the difference of the two versus the mean. The residual is defined as the difference subtracted by the fitted value, which adjusts for mean dependent bias. We fit another local regression model on the squared residual versus mean, to estimate mean dependent variance. The z value is defined by the residual divided by the standard deviation (square root of the estimated variance). This approach draws some similarity with the mean dependent dispersion estimate by the DESeq package, except that we used a nonparametric approach instead of modeling by a negative binomial distribution. The defined z value fits the normal distribution well, so we used a cutoff of 1.28, which corresponds to the 90% confidence interval of the normal distribution to define differential peaks. This definition of differential peaks is used in Fig. 3D, which is different from the cell type-specific peaks used for de novo motif analysis described above.
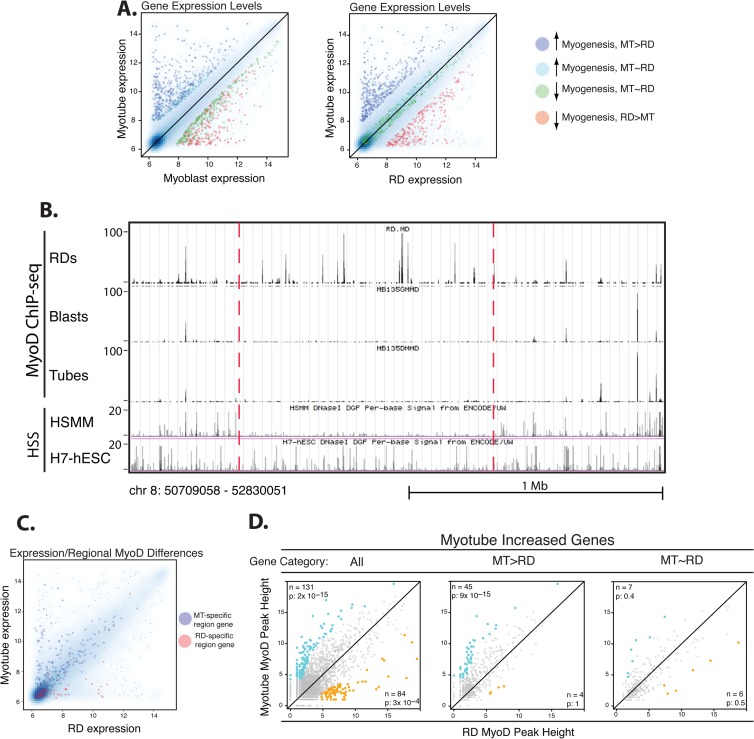
Fig 3.
MyoD binding in RD cells is impaired relative to that in human myotubes at genes that are poorly expressed in RD cells. (A) Scatterplots of gene expression determined by expression microarrays comparing either myotube gene expression to myoblast expression or myotube gene expression to RD cell gene expression. Genes are given the following colors on the basis of their categories: blue, genes that normally have increased expression during myogenesis but are expressed at a substantially lower level in RD cells; cyan, genes that normally have increased expression during myogenesis and are expressed at comparable levels between RD and primary cells; red, genes that normally decrease expression during myogenesis and continue to be expressed in RD cells; green, genes that normally decrease during myogenesis and also are expressed at low levels in RD cells. See below for definitions of MT > RD and MT ∼ RD. (B) A screenshot from the UCSC genome browser shows a region that has low MyoD ChIP-seq peaks in primary cells but substantial peaks in RD cells and that agrees with HSS data from human myoblasts. Track identity is indicated along the side (HSMM is the ENCODE human myoblast data, and H7-hESC is from human embryonic stem cells). The scale of peak heights is indicated along the left for each track, and the scale and genomic coordinates are indicated below. (C) There is a positive association between cell type-specific regional increases in MyoD ChIP-seq peak heights and expression of genes in those regions. The scatterplot of gene expression is as described for panel A, but with genes colored on the basis of their association with regional changes in MyoD ChIP-seq: blue dots, genes physically located (defined as either some portion of a transcript itself or 5 kb up- or downstream from either end of a transcript overlapping with the region) in regions determined to have higher MyoD peaks in myotubes; red dots, genes in regions with higher MyoD peaks in RD cells. (D) Scatterplots of MyoD peak height associated with genes significantly upregulated during myogenic differentiation in the primary cells, further grouped on the basis of gene expression, as indicated by gene category. All, all upregulated genes are plotted; MT > RD, the plot contains only those genes that are expressed at levels at least 3 times higher in myotubes (MT) than RD cells; MT ∼ RD, plots of those genes expressed comparably (expression within 50%) between myotubes and RD cells. Similar to Fig. 1B, the x and y axes correspond to the square root of peak heights in two cell types. Increased peaks along the y axis (myotubes) relative to the x axis (RD cells) are colored blue, and decreased peaks are colored gold. Numbers (n) and P values (p) for enrichment of differential peaks are indicated in the appropriate corner. See Materials and Methods for the definition of differential peaks and calculation of the enrichment P value. The myotube/RD cell comparison shows an enrichment for higher MyoD peaks in myotubes only at genes that are poorly expressed in RD cells (MT > RD plot) and not at those expressed at comparable levels in myotubes and RD cells (MT ∼ RD plot).
To detect the enrichment of differential peaks in the given gene sets, we calculated the enrichment of differential peaks (up, z value of >1.28; down, z value of <−1.28) that associated with the given gene sets within the promoter regions among all promoter peaks based on a hypergeometric distribution.
ChIP-seq binding site regional differences.
To identify large-scale regions with peaks that are consistently higher or lower in a two-sample comparison, we designed an algorithm to partition the genome into regions that maximize the sum of the absolute differential scores for peaks within the regions. The differential score is defined as the log odds ratio of the peak height corrected by the log odds ratio of the number of background reads within 100-kb flanking regions. The correction is necessary, as we have identified by another method large regions with noticeable copy number variation, which would bias our method. We also required that all peaks within the region show differences in the same direction, uninterrupted by strong peaks (peak height > 60) with comparable heights or peaks with a difference in the opposite direction. The boundaries of regions needed to be peaks with an absolute differential score greater than 1. We reported the computed regions with a sum of the differential score of greater than 30 and a width of greater than 100 kb. To estimate the false discovery rate (FDR) for the number of reported regions, we partitioned the genome by 5-kb windows to conserve locally correlated peaks and permutated windows with the peaks inside 100 times. We applied the above-described algorithm on each permutated genome and reported the regions using the same criteria. The empirical FDR is estimated by the ratio of the average number of reported regions in each permutation over the number of actual reported regions. Hypersensitivity (HSS) data were acquired from publically available data sets at http://genome.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeUwDgf (15, 16)
PvuII accessibility assays.
PvuII accessibility studies were performed as has been described previously (13), using nuclei isolated from RD cells and primary myotubes. The genomic coordinates and PCR primers for loci tested by quantitative PCR (qPCR) are as follows: for A-locus (chromosome 11 [chr11]) positions 5242257 to 5242328, primers GCTGCGTTACTTTGGAGGAG and GGGAAAAAGCAGAGCAGAAA; for B-locus (chr3) positions 195335273 to 195335637, GCGCACACACACTCACTCA and CGTGCCAGGATGTTTTATTG; for C-locus (chr5) positions 88249331 to 88249405, CATGCATTTTCAGGTCACCA and CCCCTCCACTTTGATTCGTA; for D-locus (chr5) positions 88365439 to 88365512, TTTTTGAAGGTTGCTTAGATTTCTG and CCCAAAGACATTCCTTAGTGTAAAA; for E-locus (chr5) positions 88276783 to 88276872, TGGTACATTTTCAAACCAGCTATG and TGCAAGACTGCAATTTACAAAA; for F-locus (chr5) positions 88401722 to 88401806, TCTTGATCCTTGGACCCATT and ATGGGGCTGTATGTGGTCTG; for G-locus (chr11) positions 1942741 to 1942824, AGCCGTGGCAGTTACAAGAG and ACGGGGGTAGCTTTTTCCTA; for H-locus (chr16) positions 31439840 to 31439927, TTGGCAAGGAGGCATTTAAG and GCCACCAGGCATAGAATAGG; for I-locus (chr16) positions 31444755 to 31444840, ATCATCGAATGACAGGCACA and TTGGAAAGTTAGCCAAGGTGA; and for J-locus (chr2) positions 71167375 to 71167452, GGAAGGGAGGAAGTTGAAAGA and CCAAGGACCAGAAAAGTCCA.
P value peak overlap analysis.
We adopted a nonparametric rank-based paradigm to compare two ChIP-seq samples as previously described (13). We ranked all peaks by their P values and group ranks into bins of 3,000 (i.e., the top 3,000 peaks, then the top 6,000 peaks, etc). Then, we computed the fraction of top x peaks in one sample that overlap the top y peaks in another sample, where x and y vary from 3,000 to 30,000 and y is equal to or greater than x.
RT-PCR and qPCR.
All qPCRs were performed using Sybr green from Bio-Rad on an Applied Biosystems 7900HT apparatus. Relative expression levels were calculated using cDNA dilution standard curves or delta-delta threshold cycle (CT) calculations. All values are reported as the mean ± SEM of at least three independent biological experiments, and significance was tested with t tests, unless otherwise noted. All reverse transcription-PCRs (RT-PCRs) were performed simultaneously with minus reverse transcriptase controls to check the absence of a signal. The primer sequences used are as follows: for CKM, forward (F) primer CCAAGTTCGAGGAGATCCTC and reverse (R) primer AGCTGCACCTGTTCTACTTCG; for TIMM17b, F primer GGAGCCTTCACTATGGGTGT and R primer CACAGCATTGGCACTACCTC; for RUNX1, F primer GGGAACTGTCAAGCTGGTGT and R primer GCCTGCTCTCCTGTGCTATC; for MEF2C, F primer CCAAGGACTAATCTGATCGGG and R primer TTTCCTGTTTCCTCCAAACAA; for JDP2, F primer AGCCCGTGAAAAGTGAGCTA and R primer CTTCTTCTTGTTCCGGCATC; for NFIC, F primer CCCGCTCAAAGATCTTGTCT and R primer ATCCCACAAAGGAACGGTTT; for COX6A2, F primer GAGTTCCGTCCCTACCAACA and R primer CAGAGGGTTCACGTGGCTAT; for PGM5, F primer CATATTTCCGTCAGATGGGG and R primer AGGGACCTTCATTGATTTGG; for SOHLH2, F primer GCAGAGGTCCTACAGCGAAC and R primer CGAACTCTGACAACGAAGCA; for TNNT3, F primer CAAGTTCGAGTTTGGGGAGA and R primer CTGGCCTCTCTACTTCCAGC; for CACNB1, F primer TGACACCATCAATCACCCAG and R primer GGGACTTGATGAGCCTTTGA; and for DUSP22, F primer GCCTGTACATCGGCAACTTC and R primer GCTGGGATGCACAGGTATTT.
Microarrays.
The microarray analysis of green fluorescent protein-infected RD cells in low-serum differentiation medium has been previously reported (9). For primary cells, three independent biological samples were collected both in growth medium at a low cell density for the myoblast condition and after 72 h at complete cellular confluence in low-serum differentiation medium for the myotube condition. All samples had RNA isolated using an RNeasy kit (Qiagen). RNA was hybridized to Illumina human HT-12 (version 4) BeadChips. Analysis was performed in R/Bioconductor software using the lumi and limma packages with the annotations found in the lumiHumanAll.db package. P values were adjusted to account for multiple testing using the Benjamini and Hochberg method, and cutoffs for significant changes were a FDR of <0.05 and a fold change of >2.
Primary human samples.
Primary rhabdomyosarcoma patient samples were obtained from Seattle Children's Hospital in accordance with an Institutional Review Board protocol. Adult skeletal muscle genomic DNA was obtained from BioChain and was from two different individuals. Genomic DNA was isolated from patient samples using a DNeasy blood and tissue kit (Qiagen).
RRBS.
Reduced-representation bisulfite sequencing (RRBS) was performed to assess DNA methylation at single-nucleotide resolution, with analysis restricted to the promoter of JDP2 (17). RRBS was performed using the EpiQuest Genomic Service of ZymoResearch. Five hundred nanograms of genomic DNA was used as input for RRBS, and library preparation and bioinformatic analysis were performed as described previously (18).
RESULTS
MyoD binding in primary human myoblasts and myotubes.
ChIP-seq was performed for endogenous MyoD in primary human myoblasts and myotubes. Similar to our findings in murine muscle cells, MyoD peaks were detected at ∼30,000 to 60,000 locations throughout the genome in both human myoblasts and myotubes (Table 1). Most peaks were present in both myoblasts and myotubes, although the relative height of some peaks changed with differentiation (Fig. 1A and data not shown).
Table 1.
Number and genomic location of MyoD ChIP-seq peaks in RD cells
| Cell type | No. of MyoD peaks at the following P value cutoffb: |
Fraction of peaks at the following genomic locationa: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 10−5 | 10−7 | 10−10 | Promc | Prox promd | 3 primee | Exon | Intron | Upstreamf | Downg | Intergenich | |
| Myoblast | 44,364 | 32,313 | 23,158 | 0.144 | 0.222 | 0.032 | 0.166 | 0.553 | 0.212 | 0.192 | 0.418 |
| Myotube | 58,449 | 43,334 | 31,630 | 0.117 | 0.195 | 0.034 | 0.147 | 0.570 | 0.208 | 0.200 | 0.396 |
| RD | 52,762 | 36,267 | 25,231 | 0.120 | 0.195 | 0.031 | 0.147 | 0.557 | 0.211 | 0.191 | 0.415 |
The fraction of MyoD peaks found in each listed type of genomic region is given. Note that categories are not mutually exclusive, and a single peak may be included in multiple categories.
Three P value cutoffs were used to evaluate whether ChIP-seq reads are considered a peak and included in the count of the total number of peaks.
Prom, promoter, 500 bp up- or downstream from the TSS.
Prox prom, proximal promoter, 2 kb up- or downstream from the TSS.
3 prime, 500 nt up- or downstream from the end of the transcript.
Upstream, 2 kb to 10 kb upstream of the TSS.
Down, 2 kb to 10 kb downstream from the end of the transcript.
Intergenic, >10 kb from any annotated gene.
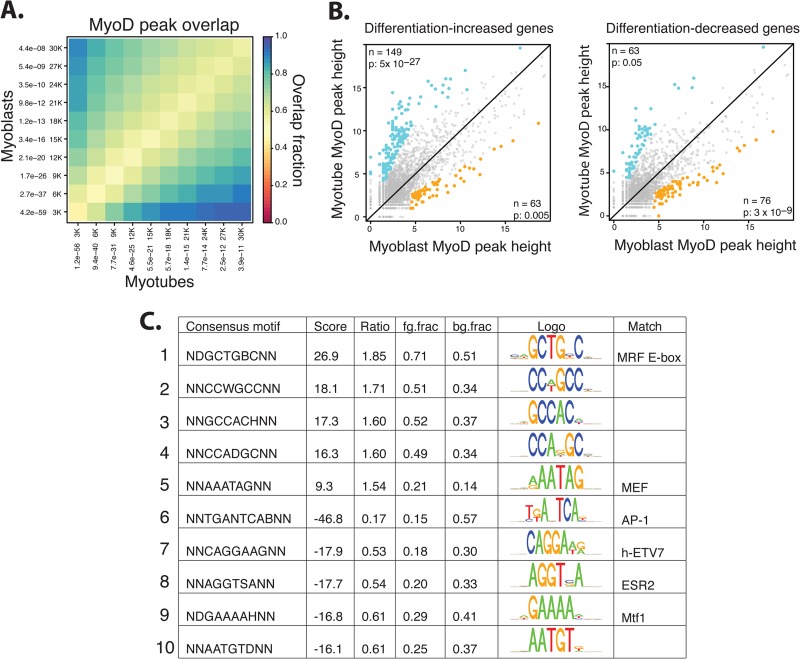
Fig 1.
Characteristics of MyoD binding in primary human myoblasts and myotubes. (A) A comparison of the top 30,000 (30K) MyoD peaks in myoblasts and myotubes demonstrates a high proportion of overlap in the sites occupied by MyoD, regardless of differentiation status. Peaks were ranked by P value and grouped into bins that increase by 3,000 peaks each time (i.e., first the 3,000 most significant peaks are considered, then the 6,000 most significant are considered, etc.). The overlap fraction scale is indicated. (B) Scatterplots showing the square root of the height of MyoD peaks within 2 kb of the TSS of the genes that either increase or decrease with myogenic differentiation (as indicated above the panels) demonstrate that upregulated genes are associated with increasing peaks and that the converse is true for downregulated genes. Differential peaks are colored blue if increased during differentiation and colored gold if decreased, and the number (n) and P value (p) are indicated in the appropriate corner. P-value statistics are calculated on the basis of the enrichment of differential peaks associated with the given gene set out of all differential peaks present within 2 kb up- or downstream of promoters based on a hypergeometric distribution (see Materials and Methods). (C) Myotube-specific MyoD peaks were compared to myoblast-specific MyoD peaks, and a de novo motif analysis was performed on the DNA immediately surrounding (100 bp up- or downstream) the peaks to determine over- and underrepresented DNA motifs. Positive scores indicate those motifs that are associated with MyoD binding in myotubes, while negative scores indicate those that are associated with MyoD in myoblasts. Motifs were compared to those in a transcription factor database to determine the closest annotated match. ratio, enriched/depleted ratio of motifs; fg.frac and bg.frac, fractions of foreground and background sequences that contain at least one motif occurrence, respectively.
To correlate MyoD binding with gene regulation, gene expression arrays were performed on primary human myoblasts and myotubes under the same conditions used for the ChIP-seq experiments. Using a 2-fold change and an FDR of less than 0.05, the expression of 758 genes increased and that of 705 genes decreased with differentiation (see Table S1 in the supplemental material). Increased gene expression was associated with an increase in MyoD binding in the gene promoter-proximal region (2 kb up- or downstream from the annotated transcription start site [TSS]) and decreased expression with decreased binding (Fig. 1B).
De novo motif analysis was performed to identify DNA motifs associated with either myotube-increased or myoblast-increased peaks. Compared to myoblast-increased peaks, myotube-increased peaks were enriched for additional E boxes (Fig. 1C, row 1) and the motif for MEF protein family members (19, 20) (Fig. 1C, row 5), whereas myoblast-increased peaks were enriched for AP-1, ETS (human ETV7), estrogen receptor 2 (ESR2), and metal-regulatory family 1 (Mtf1) motifs (Fig. 1C, rows 6 to 9).
MyoD binding in rhabdomyosarcoma cells.
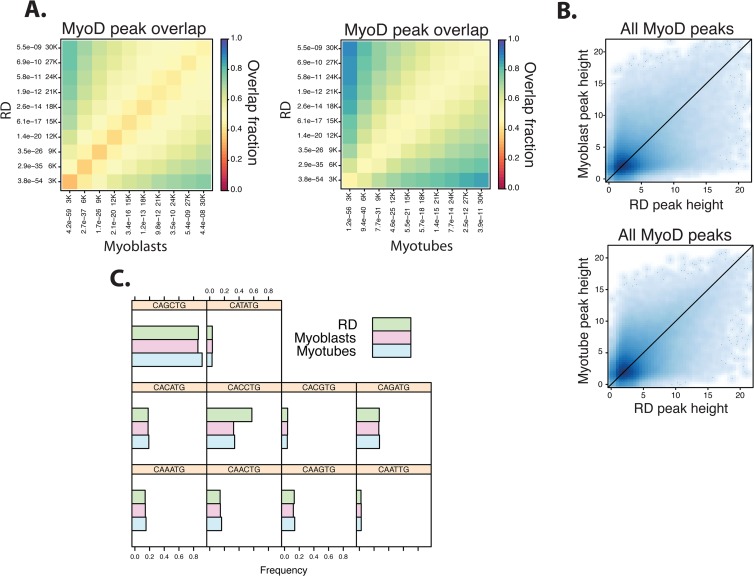
ChIP-seq for MyoD was performed in RD cells, a human embryonal cell culture model of rhabdomyosarcomas, and the results were compared to the results from the primary human muscle cells. The overall number of sites bound by MyoD was comparable between RD and primary human cells, with a similar genome-wide distribution (Table 1). Comparisons of relative peak location showed that most of the peaks were present in both RD and primary muscle cells, with an ∼50 to 90% overlap between RD and differentiated myotubes and an ∼50 to 80% overlap between RD and myoblasts (Fig. 2A), and the distribution of peak heights was also comparable between cell types (Fig. 2B). In addition, MyoD peaks in both RD cells and primary muscle cells showed a similar distribution of E-box sequences, including the MyoD-preferred CAGCTG and CACCTG E boxes, although peaks in RD cells had more CACCTG E boxes (Fig. 2C). Therefore, the overall pattern of MyoD binding is highly similar in RD cells and primary muscle cells, but a subset of peaks is differentially bound.
Fig 2.
The DNA binding characteristics of MyoD in RD rhabdomyosarcoma cells resemble MyoD binding in primary human myotubes. (A) As in Fig. 1A, the top 30,000 MyoD peaks were compared between primary human cells and RD cells. Peaks were ranked by P value and grouped into bins that increase by 3,000 peaks each time (i.e., first the 3,000 most significant peaks are considered, then the 6,000 most significant are considered, etc.). While MyoD peaks in RD cells overlap to a substantial degree with those in both myoblasts and myotubes, the overlap is higher with the RD/myotube comparison. The overlap fraction scale is indicated. (B) Scatterplots show a relatively equal distribution of the square root of the MyoD peak heights of all peaks found in myotubes, myoblasts, and RD cells. (C) Histograms of the proportion of E boxes bound by MyoD in myoblasts, myotubes, and RD cells, grouped by the central dinucleotide core of the E-box sequence, show no difference in sequence preference, apart from an increase in CC E boxes in RD cells.
Poor expression of genes in RD cells correlates with both regional and local decreases in MyoD binding.
RMS cells are characterized by a failure to properly execute the expression of a subset of the myogenic gene program. We had previously performed gene expression array analysis on RD cells cultured under the same conditions as the primary cells (9) and used those data to compare expression between the RD and primary cells. Genes were grouped into four categories on the basis of their expression patterns (Fig. 3A): those genes that (i) normally increase expression during myogenesis but are expressed at substantially lower levels (≤3-fold compared to myotube expression) in RD cells (n = 308 genes) (see Table S2 in the supplemental material), (ii) normally increase expression during myogenesis and are expressed at comparable levels (<50% difference) between RD and primary cells (n = 160 genes) (see Table S3 in the supplemental material), (iii) normally decrease expression during myogenesis and continue to be expressed in RD cells (≥3-fold compared to myotube expression) (n = 176 genes) (see Table S4 in the supplemental material), and (iv) normally decrease expression during myogenesis and also are expressed at low levels in RD cells (<50% difference) (n = 224 genes) (see Table S5 in the supplemental material).
Comparison of the MyoD peaks in myotubes and RD cells revealed two types of differences: (i) regional differences, where an area of greater than 100 kb had consistently higher or lower peaks, suggesting a regional alteration in DNA accessibility, and (ii) local differences, where one peak in a region would show differential binding, whereas surrounding peaks would not. To investigate whether large-scale regional differences might contribute to differential gene expression, the MyoD ChIP-seq data were compared to detect regions of ≥100 kb with a consistently higher MyoD signal in one of the cell types. At an FDR of <0.05, 107 regions of difference were detected, with 31 regions being higher in RD cells and 76 being higher in the primary muscle cells (Fig. 3B; see Table S6 in the supplemental material).
The regional differences coincide with a subset of genes differentially expressed between RD cells and myotubes (Fig. 3C). Forty-nine genes that are expressed at higher levels in myotubes than RD cells are found in or immediately adjacent to (±5 kb) regions with higher peaks in the primary cells, and 13 genes that are expressed at higher levels in RD cells are found in regions with higher peaks in RD cells (see Table S7 in the supplemental material).
To determine whether local peak-specific alterations in binding might also contribute to differential gene expression, we focused on the set of MyoD peaks in the promoter-proximal regions (±2 kb from the TSS) of genes grouped on the basis of the previously mentioned expression profiles. We identified a statistically significant enrichment for higher MyoD peaks in myotubes at 45 genes expressed poorly in RD cells but no such enrichment at those genes expressed comparably between cells (Fig. 3D). Only 17 of the genes identified as having differential MyoD peaks by the proximal promoter analysis were also found in the regional difference analysis at the chosen statistical criteria (see Table S7 in the supplemental material), suggesting that effects leading to differential MyoD binding operate at a more local scale for a subset of genes. Examples of peak-specific differences in MyoD binding are shown in Fig. 4A.
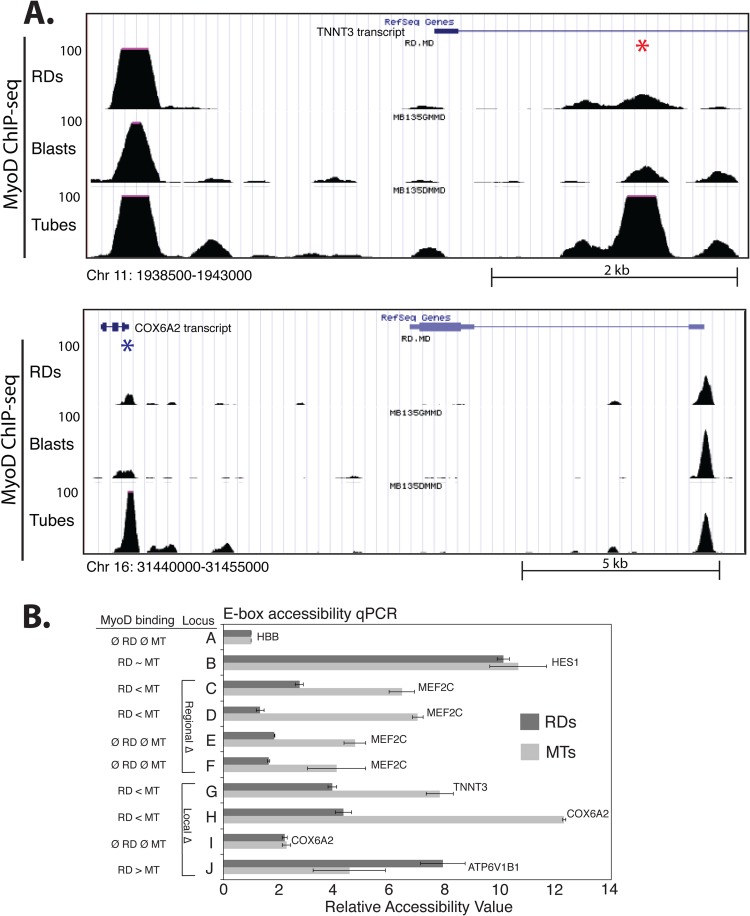
Fig 4.
Differential MyoD binding in RD cells and myotubes is due to regional and local differences in chromatin accessibility. (A) UCSC genome browser screenshots of local MyoD binding differences between RD cells and myotubes at peaks located in the region of two of the genes tested for accessibility differences. Track identity is indicated along the side. The scale of peak heights is indicated along the left for each track, and the scale and genomic coordinates are indicated below. Individual differential peaks are indicated with an asterisk. (B) PvuII endonuclease accessibility was assessed in RD cells and human primary myotubes at several loci with regional (rows C to F) or local (rows G to J) differences (Δ) in MyoD binding patterns. The genes located closest to the tested loci are indicated at the end of the bars. The status of MyoD binding, as determined by ChIP-seq, is indicated in the left-hand column. Locus G corresponds to the area indicated by the red asterisk in the top of panel A, and locus H corresponds to the blue asterisk in the bottom of panel A. Locus I is approximately 5 kb from locus H in an area with no MyoD binding. Relative accessibility was determined by qPCR and calculated compared to the control locus (A), a GC E box ∼5 kb upstream of the HBB gene. Data are represented as the means ± standard deviations of two technical replicates. Genomic coordinates and PCR primers for each locus are listed in Materials and Methods. Ø, not present.
Analysis of publicly available DNase I accessibility data from ENCODE shows lower cleavage signals in human myoblasts at regions with higher MyoD peaks in the RD cells (data not shown), suggesting that a relatively inaccessible chromatin context might prevent MyoD binding in these regions in primary muscle cells. We assessed relative DNA accessibility in RD cells and primary muscle cells by exposing nuclei to the PvuII restriction endonuclease, which cleaves the CAGCTG E-box sequence, as previously described (13). E boxes located in a region of reduced MyoD binding in RD cells exhibited reduced accessibility in RD cells, regardless of whether or not a MyoD peak was identified at the E box (Fig. 4B, compare rows C to D and E to F), indicating that these entire regions were in relatively inaccessible chromatin in the RD cells compared to the primary muscle cells. Regions with peak-specific differences showed a relatively lower accessibility at the site of the peak decreased in RD cells but not in the adjacent region (Fig. 4B, compare rows H and I), further supporting a model in which both regional and local effects operate on MyoD binding.
Differences in MyoD binding between RMS and primary cells identify binding motifs for potential myogenic cofactors that rescue RMS myogenesis.
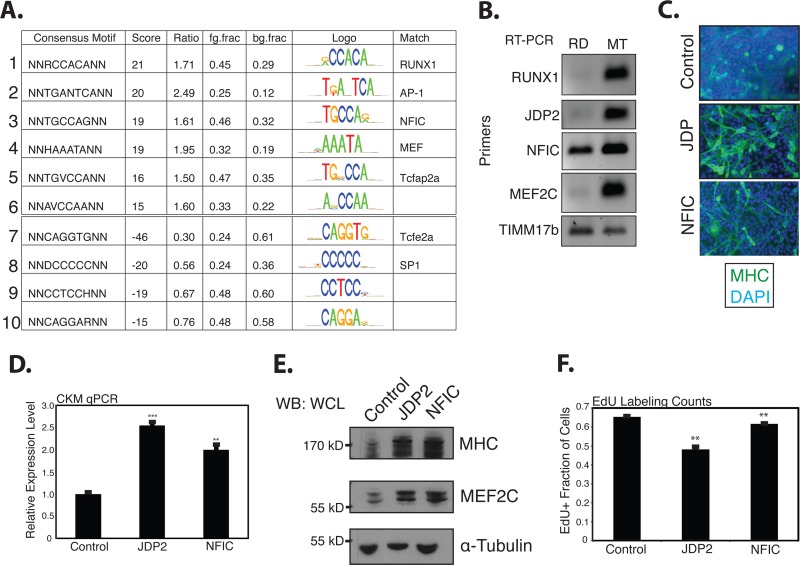
A de novo motif analysis was performed to analyze sequences enriched, either positively or negatively, under the MyoD peaks found in the myotubes that were absent or substantially reduced in RD cells. The strongest-scoring motifs were for RUNX1, AP-1, NFIC, and MEF2 (Fig. 5A). RUNX1 and JDP2, a potential component of AP-1 complexes, have previously been shown to be sufficient to drive myogenic differentiation in RD cells (9, 21); NFIC has previously been shown to cooperate with bHLH proteins in myogenic cells (22); and MEF2 factors cooperate with MyoD at many muscle promoters. Our expression array analysis identified, and RT-PCR confirmed, that RD cells express fewer RUNX1, JDP2, NFIC, and MEF2C transcripts than primary myotubes (Fig. 5B).
Fig 5.
A genome-wide comparison of differential MyoD binding between myotubes and RD cells identifies binding motifs for promyogenic transcription factors enriched at MyoD-bound sites in myotubes. (A) A de novo motif analysis performed as described in the legend to Fig. 1C to determine motifs enriched adjacent to either myotube- or RD cell-specific MyoD peaks. Scores and motif matches were determined as described in the legend to Fig. 1C. (B) RT-PCR for RUNX1, JDP2, NFIC, and MEF2C transcripts, as indicated, in either RD cells (72 h in differentiation medium) or primary human myotubes (MT; 72 h in differentiation medium). TIMM17b RT-PCR serves as the internal control. (C) Immunostains of RD cells infected with an empty retrovirus or a retrovirus expressing either JDP2 or NFIC and put in low-serum differentiation medium for 72 h. MHC is a marker of terminal myogenic differentiation, and DAPI was used to stain all nuclei. (D) qPCR for muscle-specific creatine kinase, a marker of terminal myogenic differentiation on RD cells treated as described for panel C. (E) Western blots (WB) on whole-cell lysates (WCL) from RD cells treated as described for panel C, as indicated along the top of the panels. The protein blotted for is indicated to the right of each panel, and protein size is indicated along the left. (F) Cell counts indicating the percentage of RD cells that, after 24 h in differentiation medium, incorporated EdU during a 24-h pulse. For qPCR, minus reverse transcriptase controls were monitored for the absence of product, and TIMM17b served as the internal control. Both the qPCR and EdU data are reported as the means ± SEMs of three independent biological replicates. **, P < 0.01; ***, P < 0.001.
In agreement with the previous report (21), the expression of JDP2 in RD cells through retroviral introduction rescued myogenesis, leading to the formation of myotubes that stained for MHC (Fig. 5C), and RD cells exhibited an increase in expression of the differentiation marker muscle-specific creatine kinase (CKM) (Fig. 5D). The cells were found by Western blotting to express substantially more MHC and MEF2C protein (Fig. 5E) and show decreased incorporation of the thymidine analog EdU, consistent with cell cycle withdrawal (Fig. 5F). Similar results were seen with NFIC (Fig. 5C to F), though its expression did not result in substantial cell cycle withdrawal. Given the fact that both factors upregulated MEF2C protein levels and MEF2 can act directly at muscle gene promoters, we hypothesized that increased MEF2 levels would also rescue myogenesis and upregulate genes poorly expressed in RD cells.
MEF2C was introduced into RD cells by retroviral transduction and tested for its ability to increase the expression of genes identified as being poorly expressed and exhibiting reduced MyoD binding in RD cells. The genes contained in the data set for genes that normally have increased expression during myogenesis but are expressed at a substantially lower level in RD cells (MT > RD) were ranked on the basis of the MEF2C position weight matrix score and the expression of the top-six-scoring genes tested with and without increased MEF2C expression. Five of the six genes showed a statistically significant increase in expression in response to MEF2C expression, with four of the five exhibiting a substantial (>4-fold) change (Fig. 6A). In addition, MEF2C-transduced RD cells formed substantially more myotubes that stained for the differentiation marker MHC (Fig. 6B), expressed substantially more CKM, as assessed by qPCR (Fig. 6C), and were found by Western blotting to express more MHC and MEF2C (Fig. 6D). MEF2C is one of the genes identified as being poorly expressed in RD cells and is located in a region of reduced MyoD binding in RD cells (Fig. 6E), as was confirmed by PvuII studies (Fig. 4B). Notably, MEF2C expression did not induce the transcription of either the MyoD-regulated promyogenic microRNA 206 (miR-206) (9, 23–26) or the transcription factor JDP2, though it did increase the expression of NFIC (see Fig. S1 in the supplemental material), suggesting that it is acting directly at target genes to upregulate expression. Site-specific MyoD ChIP in MEF2C-transduced RD cells demonstrated an increase in MyoD binding at a subset of sites with a decreased MyoD peak in RD cells and an adjacent high position weight matrix (PWM) MEF2C motif (Fig. 6F), suggesting that MEF2C might facilitate MyoD binding at a subset of targets. Together with our previous study showing that RUNX1 expression induces myogenesis in RD rhabdomyosarcomas, our current results indicate that a relative deficiency of a set of transcription factors accounts for the inability of MyoD to fully execute the differentiation program in RD rhabdomyosarcoma cells.
Fig 6.
MEF2C acts at myogenic targets in RD cells to rescue expression and myogenesis and is located in a genomic region with low levels of MyoD binding in RD cells. (A) qPCR for potential MEF2C/MyoD targets. The subset of MT > RD genes from Fig. 3D was restricted to those genes with higher MyoD peaks in myotubes (blue dots), ranked on the basis of MEF2C PWM scores, and then the top six genes were chosen for qPCR analysis. All qPCRs were performed on three independent biological samples of RD cells transduced with either an empty control retrovirus or one expressing an Mef2C isoform and placed in differentiation medium for 36 h. The qPCR data are reported as the means ± SEMs of the three replicates, and TIMM17b served as the internal control. (B) Immunostains of RD cells treated as described for panel A. MHC is a marker of myogenesis, and DAPI stains all cell nuclei. (C) qPCR for muscle-specific creatine kinase in RD cells treated as described for panel A. The qPCR data are reported as the means ± SEMs of the three replicates, and TIMM17b served as the internal control. (D) Western blots on whole-cell lysates from RD cells treated as described for panel A and immunoblotted for the proteins indicated along the right. (E) A screenshot from the UCSC genome browser shows that MEF2C is in a genomic region that has low MyoD ChIP-seq peaks compared to primary myoblasts and myotubes. Track identity is indicated along the side. HSS indicates hypersensitivity data (HSMM is ENCODE human myoblast data, and H7-hESC is from human embryonic stem cells). The scale of peak heights is indicated along the left for each track, and the scale and genomic coordinates are indicated below. (F) RD cells expressing MEF2C have increased MyoD binding at a subset of E boxes located near MEF2 binding sites. MyoD ChIP was performed in RD cells transduced with control virus or after differentiation with MEF2C retrovirus. Relative enrichment for each locus was determined by qPCR and normalized to the control locus (A). Data are indicated as the means ± standard deviations of two technical replicates. The genes located closest to the tested loci are indicated at the top of the bars. *, P < 0.05; **, P < 0.01.
The CpG shore upstream of JDP2 is hypermethylated in the rhabdomyosarcoma cell line RD and primary rhabdomyosarcoma tumors.
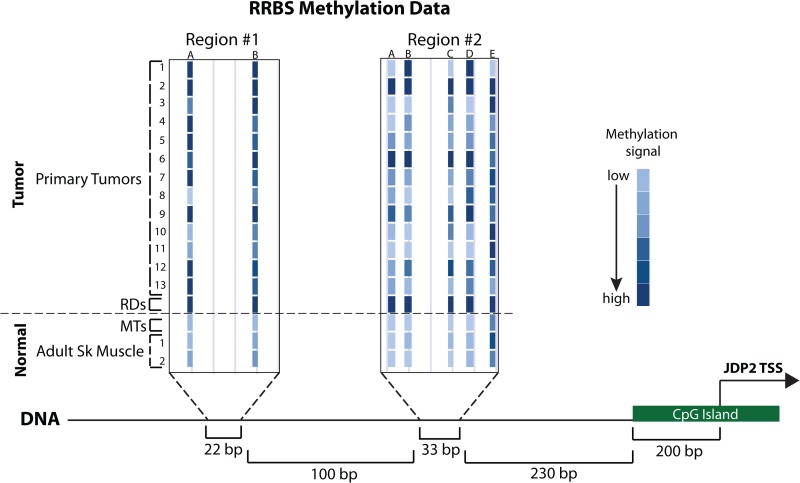
In contrast to MEF2C, JDP2 was not in a region of decreased MyoD binding (data not shown). To determine whether local epigenetic modifications might suppress JDP2 expression, we performed bisulfite sequencing of the promoter region of JDP2 in RD cells and primary myotubes. Regions of hypermethylation were found in the promoter adjacent to the CpG island present at the TSS of JDP2 (Fig. 7). The CpG island itself showed no differential methylation (see Fig. S2 in the supplemental material). Hypermethylation of the JDP2 promoter was also found in primary tumors of both the embryonal and alveolar subtypes (Fig. 7; see Table S8 in the supplemental material).
Fig 7.
The RD cell line and primary RMS tumors are DNA hypermethylated in the promoter region of the promyogenic factor JDP2. RRBS data from the promoter region of JDP2 for 13 primary tumors (for diagnostic details, see Table S8 in the supplemental material), RD cells, primary myotubes (MTs), and two different samples of normal adult skeletal muscle (Adult Sk Muscle). Higher levels of methylation at a given genomic position are indicated by darker coloration, as indicated in the key. JDP2 TSS indicates the position of the transcription start site of the gene, and the annotated CpG island at the JDP2 promoter is also indicated (also see Fig. S2 in the supplemental material). Genomic distances are indicated at the bottom.
DISCUSSION
Our experiments comparing MyoD binding between normal human myotubes and rhabdomyosarcoma cells have identified (i) regions of decreased MyoD binding that include critical myogenic cofactors, such as MEF2C, (ii) local impairment of MyoD binding at peaks associated with specific cofactors (MEF2C, RUNX, NFIC, and JDP2), and (iii) local DNA hypermethylation at the promoter of the transcription factor JDP2, a factor capable of inducing myogenic differentiation in RMS, in numerous primary tumors, as well as the RD cell line. Together, these findings indicate both local and regional suppression of cofactors that cooperate with MyoD to regulate muscle gene expression and result in a decreased expression of a subset of MyoD-regulated genes.
Widespread genome-wide binding of MyoD in RD cells agrees with our prior findings that MyoD activity is compromised in RMS but that DNA binding is not globally affected (8, 10). Given the global similarity in MyoD binding between myotubes and RD cells, the observed differences in subsets of MyoD-bound sites, such as decreased MyoD binding adjacent to myogenic genes that fail to activate in RMS, begin to provide mechanistic insight into how RMSs fail to differentiate. Rather than a wholesale failure of MyoD activity, MyoD function fails at a discrete subset of its targets.
We identified two causes for decreased MyoD binding at subsets of sites in RD cells. Differences in binding over large regions spanning 100 or more kilobases correlated with differential nuclease access and indicated that large regional differences in chromatin compaction might restrict MyoD access to some critical genes, e.g., the MEF2C gene (Fig. 4B and 6E). In addition to regionally decreased MyoD peaks, many peaks were locally decreased (i.e., in a region where the surrounding peaks were not different between RD and primary muscle cells). The de novo motif analysis of the DNA sequences under these peaks identified potential cofactors that are expressed at lower levels in RD cells than primary muscle cells and can induce the expression of muscle genes when expressed in RD cells. Our demonstration that MEF2C expression also increases MyoD binding at some peaks with an adjacent MEF2 binding motif suggests that MEF2C and, possibly, the other cofactors increase the accessibility of these sites to MyoD and/or stabilize the binding of MyoD at some of these sites. We should note, however, that site-specific ChIPs for MyoD with increased RUNX1 expression did not detect an increase in MyoD binding (K. L. MacQuarrie, unpublished data), but it is possible either that the appropriate sites were not interrogated or that some myogenic cofactors identified by our analysis increase MyoD binding, while others do not.
In normal myogenesis, MyoD causes histone acetylation on a genome-wide scale (1). We have previously demonstrated the presence of histone acetylation at MyoD targets in RD cells (9), demonstrating that MyoD can still function in that capacity in RMS cells. While a low level of histone acetylation would be expected across the regions that we identify to be binding less MyoD in RD cells than in primary cells, it is not clear what occurs in the situation of local changes, such as individual low peaks. It is possible that local acetylation, such as on the scale of a single nucleosome, might be impaired, but it is also possible that acetylation occurs at some or all such sites. Such possibilities will need to be tested directly to determine how specifically targeted MyoD-mediated histone acetylation is at such sites.
We have previously proposed that rhabdomyosarcoma cells are poised on the verge of differentiation and require only a final push for the process of myogenesis to proceed to completion, after the activation of nested feed-forward circuits that center on MyoD and utilize cooperative factors such as RUNX1. The data presented in this paper not only offer further support for that model but also suggest that myogenesis may be controlled by a dosage of myogenic factors, both the myogenic regulatory factors and cooperating factors. RMS differentiation can be induced by JDP2, NFIC, RUNX1, ZNF238, and MEF2C, and other groups have demonstrated similar effects with other components of the normal myogenic gene network (9, 21, 26–29), all of which interact with and/or potentiate MyoD-mediated activity in some manner. The ability of a variety of MyoD-cooperating factors to complete the final steps of differentiation in RMS cells and complete the process of terminal differentiation suggests that myogenic cells have evolved to respond to a cumulative dosage of promyogenic factors. A similar model has previously been proposed for B-lymphocyte development, with the relative dosage of three of the E proteins being critical for development of the cells in vivo (30).
Using a genome-wide DNA methylation assay developed in our lab, we have recently shown hypermethylation of CpG islands in rhabdomyosarcoma cells relative to normal skeletal muscle (31). While this assay robustly identifies differential DNA methylation in large CpG islands, it does not detect more subtle differences in DNA methylation outside these regions (32). Methylation of CpG shores, areas adjacent to but outside CpG islands, has been implicated in both developmental processes and tumor biology and linked to reduced gene expression (33, 34). The identification of hypermethylation in the CpG shore in the promoter region of JDP2 in primary tumors as well as the RD cell line demonstrates that the differences seen in myogenic pathways in cell culture models can be relevant to primary tumor biology and suggests that the silencing of promyogenic genes apart from the myogenic regulatory factors may be a mechanism that these tumors use to escape terminal differentiation. It is of interest to note that the AP-1 motif was identified as being enriched with myoblast-specific peaks in the primary cell comparison (Fig. 1C), while it was identified as being enriched with myotube-specific peaks in the later comparison (Fig. 5A); but JDP2 can homodimerize as well as form heterodimers with a variety of partners, including ATF-2, c-Jun, and C/EBPγ (35–37), and the primary cell comparison may represent an AP-1 complex with functional differences from the activity that occurs with increased JDP2 expression in RMS cells. JDP2 activity has been implicated in the control of cellular senescence (38), and HES1, a bHLH protein involved in the control of the quiescence-senescence decision in cells, contributes to the failure of terminal differentiation in RMS cells (39). The relation of both factors to each other and MyoD activity during normal myogenesis or in RMSs is unclear at this time.
The hypermethylation of the JDP2 promoter will need to be further investigated to determine if it is capable of tumor induction or is related to continued tumor growth after formation. Regardless of whether JDP2 hypermethylation is a causative factor in the genesis of rhabdomyosarcoma, it suggests that the development of prodifferentiation-based therapies that will impair or halt tumor growth by screening for their ability to affect specific cellular targets inducing terminal differentiation may be a possibility.
Supplementary Material
ACKNOWLEDGMENTS
K.L.M. was supported by a Developmental Biology Predoctoral Training Grant (T32HD007183). Z.Y. was supported by an NIH Interdisciplinary Training Grant in Cancer Research (T32CA080416). A.P.F. was supported by a grant from the University of Washington Child Health Research Center (NIH U5K12HD043376-10) and Hyundai Hope on Wheels. S.J.D. was supported as a St. Baldrick's Foundation Scholar and by Hyundai Hope on Wheels. S.J.T. was supported by NIH NIAMS (R01AR045113).
Footnotes
Published ahead of print 10 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00916-12.
REFERENCES
- 1. Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, Gentleman RC, Tapscott SJ. 2010. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell 18:662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lassar AB, Buskin JN, Lockshon D, Davis RL, Apone S, Hauschka SD, Weintraub H. 1989. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell 58:823–831 [DOI] [PubMed] [Google Scholar]
- 3. Murre C, McCaw PS, Baltimore D. 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56:777–783 [DOI] [PubMed] [Google Scholar]
- 4. Lassar AB, Davis RL, Wright WE, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. 1991. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 66:305–315 [DOI] [PubMed] [Google Scholar]
- 5. Merlino G, Helman L. 1999. Rhabdomyosarcoma—working out the pathways. Oncogene 18:5340–5348 [DOI] [PubMed] [Google Scholar]
- 6. Sebire NJ, Malone M. 2003. Myogenin and MyoD1 expression in paediatric rhabdomyosarcomas. J. Clin. Pathol. 56:412–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davis RL, Weintraub H, Lassar AB. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51:987–1000 [DOI] [PubMed] [Google Scholar]
- 8. Tapscott S, Thayer MJ, Weintraub H. 1993. Deficiency in rhabdomyosarcomas of a factor required for MyoD activity and myogenesis. Science 259:1450–1453 [DOI] [PubMed] [Google Scholar]
- 9. MacQuarrie KL, Yao Z, Young JM, Cao Y, Tapscott SJ. 2012. miR-206 integrates multiple components of differentiation pathways to control the transition from growth to differentiation in rhabdomyosarcoma cells. Skeletal Muscle 2:7 doi:10.1186/2044-5040-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang Z, MacQuarrie KL, Analau E, Tyler AE, Dilworth FJ, Cao Y, Diede SJ, Tapscott SJ. 2009. MyoD and E-protein heterodimers switch rhabdomyosarcoma cells from an arrested myoblast phase to a differentiated state. Genes Dev. 23:694–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Snider L, Geng LN, Lemmers RJ, Kyba M, Ware CB, Nelson AM, Tawil R, Filippova GN, van der Maarel SM, Tapscott SJ, Miller DG. 2010. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 6:e1001181 doi:10.1371/journal.pgen.1001181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. 1988. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242:405–411 [DOI] [PubMed] [Google Scholar]
- 13. Fong AP, Yao Z, Zhong JW, Cao Y, Ruzzo WL, Gentleman RC, Tapscott SJ. 2012. Genetic and epigenetic determinants of neurogenesis and myogenesis. Dev. Cell 22:721–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Palii CG, Perez-Iratxeta C, Yao Z, Cao Y, Dai F, Davison J, Atkins H, Allan D, Dilworth FJ, Gentleman R, Tapscott SJ, Brand M. 2011. Differential genomic targeting of the transcription factor TAL1 in alternate haematopoietic lineages. EMBO J. 30:494–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sabo PJ, Hawrylycz M, Wallace JC, Humbert R, Yu M, Shafer A, Kawamoto J, Hall R, Mack J, Dorschner MO, McArthur M, Stamatoyannopoulos JA. 2004. Discovery of functional noncoding elements by digital analysis of chromatin structure. Proc. Natl. Acad. Sci. U. S. A. 101:16837–16842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, Weaver M, Shafer A, Lee K, Neri F, Humbert R, Singer MA, Richmond TA, Dorschner MO, McArthur M, Hawrylycz M, Green RD, Navas PA, Noble WS, Stamatoyannopoulos JA. 2006. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods 3:511–518 [DOI] [PubMed] [Google Scholar]
- 17. Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454:766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. 2011. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protoc. 6:468–481 [DOI] [PubMed] [Google Scholar]
- 19. Molkentin JD, Black BL, Martin JF, Olson EN. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83:1125–1136 [DOI] [PubMed] [Google Scholar]
- 20. Yu YT, Breitbart RE, Smoot LB, Lee Y, Mahdavi V, Nadal-Ginard B. 1992. Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev. 6:1783–1798 [DOI] [PubMed] [Google Scholar]
- 21. Ostrovsky O, Bengal E, Aronheim A. 2002. Induction of terminal differentiation by the c-Jun dimerization protein JDP2 in C2 myoblasts and rhabdomyosarcoma cells. J. Biol. Chem. 277:40043–40054 [DOI] [PubMed] [Google Scholar]
- 22. Hebert SL, Simmons C, Thompson AL, Zorc CS, Blalock EM, Kraner SD. 2007. Basic helix-loop-helix factors recruit nuclear factor I to enhance expression of the NaV 1.4 Na+ channel gene. Biochim. Biophys. Acta 1769:649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. 2006. Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 174:677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. 2006. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. U. S. A. 103:8721–8726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. 2006. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J. Cell Biol. 175:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, Tuschl T, Ponzetto C. 2009. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J. Clin. Invest. 119:2366–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Puri P, Wu Z, Zhang P, Wood L, Bhakta K, Han J, Feramisco J, Karin M, Wang J. 2000. Induction of terminal differentiation by constitutive activation of p38 MAP kinase in human rhabdomyosarcoma cells. Genes Dev. 14:574–584 [PMC free article] [PubMed] [Google Scholar]
- 28. Sirri V, Leibovitch M, Leibovitch S. 2003. Muscle regulatory factor MRF4 activates differentiation in rhabdomyosarcoma RD cells through a positive-acting C-terminal protein domain. Oncogene 22:5658–5666 [DOI] [PubMed] [Google Scholar]
- 29. Wang H, Garzon R, Sun H, Ladner K, Singh R, Dahlman J, Cheng A, Hall B, Qualman S, Chandler D, Croce C, Guttridge D. 2008. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 14:369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhuang Y, Cheng P, Weintraub H. 1996. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2, and HEB. Mol. Cell. Biol. 16:2898–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mahoney SE, Yao Z, Keyes CC, Tapscott SJ, Diede SJ. 2012. Genome-wide DNA methylation studies suggest distinct DNA methylation patterns in pediatric embryonal and alveolar rhabdomyosarcomas. Epigenetics 7:400–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Diede SJ, Guenthoer J, Geng LN, Mahoney SE, Marotta M, Olson JM, Tanaka H, Tapscott SJ. 2010. DNA methylation of developmental genes in pediatric medulloblastomas identified by denaturation analysis of methylation differences. Proc. Natl. Acad. Sci. U. S. A. 107:234–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, Miller J, Schlaeger T, Daley GQ, Feinberg AP. 2009. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 41:1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP. 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 41:178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aronheim A, Zandi E, Hennemann H, Elledge SJ, Karin M. 1997. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol. 17:3094–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Broder YC, Katz S, Aronheim A. 1998. The ras recruitment system, a novel approach to the study of protein-protein interactions. Curr. Biol. 8:1121–1124 [DOI] [PubMed] [Google Scholar]
- 37. Jin C, Ugai H, Song J, Murata T, Nili F, Sun K, Horikoshi M, Yokoyama KK. 2001. Identification of mouse Jun dimerization protein 2 as a novel repressor of ATF-2. FEBS Lett. 489:34–41 [DOI] [PubMed] [Google Scholar]
- 38. Nakade K, Pan J, Yamasaki T, Murata T, Wasylyk B, Yokoyama KK. 2009. JDP2 (Jun dimerization protein 2)-deficient mouse embryonic fibroblasts are resistant to replicative senescence. J. Biol. Chem. 284:10808–10817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sang L, Coller HA, Roberts JM. 2008. Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science 321:1095–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.