Abstract
Smk1 is a meiosis-specific mitogen-activated protein kinase (MAPK) in Saccharomyces cerevisiae that controls spore morphogenesis. Similar to other MAPKs, it is controlled by dual phosphorylation of its T-X-Y activation motif. However, Smk1 is not phosphorylated by a prototypical MAPK kinase. Here, we show that the T residue in Smk1's activation motif is phosphorylated by the cyclin-dependent kinase (CDK)-activating kinase, Cak1. The Y residue is autophosphorylated in an independent intramolecular reaction that requires the meiosis-specific protein Ssp2. Although both SMK1 and SSP2 are expressed as middle-meiosis-specific genes, Smk1 protein starts to accumulate before Ssp2. Thus, Smk1 exists in a low-activity (pT) form early in sporulation and a high-activity (pT/pY) form later in the program. Ssp2 must be present when Smk1 is being produced to activate the autophosphorylation reaction, suggesting that Ssp2 acts through a transitional intermediate form of Smk1. These findings provide a mechanistic explanation for how Smk1 activity thresholds are generated. They demonstrate that intramolecular autophosphorylation of MAPKs can be regulated and suggest new mechanisms for coupling MAPK outputs to developmental programs.
INTRODUCTION
Mitogen-activated protein kinases (MAPKs) are ubiquitous signal-transducing enzymes that control a wide spectrum of biological processes, including cell division, differentiation, and survival. The canonical MAPK module consists of an upstream MAPK kinase kinase, a MAPK kinase, and a MAPK (1, 2). Receptor-ligand interactions at the cell surface couple to signaling molecules that activate MAPK kinase kinases, which phosphorylate residues on the activation loop of MAPK kinases, which in turn phosphorylate conserved threonine (T) and tyrosine (Y) residues in the activation loop of MAPKs. Activated MAPKs dispense phosphate to serine (S) and T residues in downstream regulatory molecules. The transfer of phosphate to the activation loop of MAPK by MAPK kinase can occur through a nonprocessive mechanism that is able to generate a switch-like increase in MAPK activity (3).
While many MAPKs are activated by canonical MAPK modules, some MAPKs are activated through noncanonical pathways that can involve autophosphorylation of their T-X-Y motifs (4, 5). For example, although p38 can be activated in many cell types in response to stress signals by a canonical MAPK module in T cells, p38 can also undergo intermolecular (trans) autophosphorylation on its activating T residue when zeta-chain-associated protein kinase 70 (Zap70) phosphorylates a distal residue on p38 following T-cell receptor activation (6). In addition, p38 undergoes intermolecular autophosphorylation of the T and Y in its T-X-Y motif when it is recruited into protein complexes by transforming growth factor β-activated protein kinase binding protein (TAB1) (7). Other examples of MAPKs that are activated by noncanonical pathways include Erk7, which is activated by intramolecular (cis) autophosphorylation of the Y in its T-X-Y activation motif (8), and Erk8, which autophosphorylates its activation motif in an apparently intramolecular fashion (9). Indeed, these findings are reminiscent of studies carried out prior to the discovery of canonical MAPK signaling in which the Erk1 and Erk2 MAPKs produced in Escherichia coli were shown to autophosphorylate their T-X-Y motifs (10–13). In these early studies, the autophosphorylation of Erk2 on its activating Y residue was shown to be intramolecular. More-recent studies have shown that the Fus3 MAPK from Saccharomyces cerevisiae can undergo intramolecular autophosphorylation of its activating Y residue when complexed with the Ste5 scaffolding protein (14). These data suggested that intramolecular autophosphorylation could play a role in regulating Erk1, Erk2, and Fus3. However, extracellular ligands couple to the activation of these MAPKs through canonical MAPK activation pathways, and the significance of these autophosphorylation reactions has not been established.
Smk1 is a meiosis-specific MAPK that controls spore formation in budding yeast (15). Similar to other MAPKs, Smk1 is activated by phosphorylation of the T-X-Y motif in its activation loop (16, 17). However, it is atypical in several respects. First, SMK1 is tightly regulated by the transcriptional cascade of sporulation, being expressed exclusively in meiotic cells around the time the nuclear divisions are taking place (15, 18). Second, while many MAPKs can be activated to yield switch-like (all-or-none) responses to small changes in the concentration of extracellular ligands, Smk1 does not appear to function as a switch. Instead, sequential increases in Smk1 activity thresholds play a role in coordinating multiple steps in the spore morphogenesis program (19). Third, MAPK kinase family members are not required for its activation (20). Taken as a whole, these observations indicate that Smk1 is activated during meiotic development by a noncanonical mechanism that generates graded catalytic outputs instead of switch-like responses.
Genetic studies have provided some insight into the regulation of Smk1. CAK1, which encodes the cyclin-dependent kinase (CDK)-activating kinase, was isolated as a multicopy suppressor of the spore formation defect of a temperature-sensitive smk1 mutant, suggesting that Cak1 functions positively in the Smk1 pathway (21). Cak1 activates Cdc28, the sole essential CDK in yeast, by phosphorylating T169 in its activation loop (22–24). Therefore, Cak1 is required for vegetative growth. However, mutants of Cdc28 that contain an acidic residue substitution for amino acid T169 and additional hyperactivating substitutions bypass the requirement of CAK1 for viability (25). Studies of one of these bypass mutants (cdc28-43244) revealed that the requirement of Cak1 for Smk1 activation is Cdc28 independent (17). Moreover, phenotypic analyses of the cdc28-43244 cak1Δ strain showed that Cak1 is also required for the activation of Ime2 (26), a meiosis-specific kinase with sequence similarity to both CDKs and MAPKs (27, 28). Despite the suggestions that Cak1 could be a direct Smk1-activating kinase (17, 21), it is unclear whether Cak1 directly phosphorylates the T and/or the Y residues of Smk1, whether Cak1 activates another kinase (such as Ime2) that in turn phosphorylates Smk1, or whether some combination of these mechanisms contributes to Smk1 activation.
Further insight into the mechanism of Smk1 activation was generated by a screen of a subset of yeast deletion mutants that are defective in spore formation. In this study, the deletion of SSP2 was found to reduce the relative abundance of the phosphorylated form of Smk1 (20). SSP2 is required after prospore membrane closure, before the spore-specific layers of the spore wall are assembled (29–31). AMA1, which encodes a meiosis-specific activator of the anaphase-promoting complex E3 ubiquitin-ligase (32–36), was also identified in the deletion mutant screen. In addition, Ime2 has been shown to increase the fraction of Smk1 that is phosphorylated (37).
The transcriptional cascade of sporulation produces early, middle, and late sets of gene transcripts as different steps in meiotic development are taking place. Almost all of the SMK1 pathway members are expressed as tightly regulated middle genes. This set of genes is expressed as cells exit meiotic prophase, enter the meiotic divisions (MI/MII), and form spores (38). Middle gene induction is controlled by the opposing activities of a transcriptional repressor named Sum1 and a meiosis-specific transcriptional activator named Ndt80 (39). Sum1 and Ndt80 bind elements in middle promoters termed middle sporulation elements (MSEs) in a mutually exclusive (competitive) fashion (40). Sum1 represses middle promoters in vegetative cells and early in meiotic development, until it is removed from MSEs late in prophase of meiosis I (around the pachytene stage) (41, 42). NDT80 is itself controlled by a Sum1-responsive MSE (43, 44), and the removal of Sum1 triggers a positive autoregulatory loop in which Ndt80 binds to its own promoter, leading to high-level NDT80 transcription (45). This is accompanied by the competitive displacement of Sum1 and activation of middle promoters. Removal of Sum1 repression is sufficient to cause moderate expression of SMK1 (independent of Ndt80) (18, 40, 41). Once the Ndt80-positive autoregulatory loop is in force, SMK1 is expressed to a higher level, and SMK1 pathway genes, including SSP2, are expressed. Although CAK1 is ubiquitously expressed, its mRNA increases as middle genes are induced (46, 47). IME2 is expressed early in sporulation, but its mRNA also increases as middle genes are induced (46, 48). Thus, Smk1 is controlled by tightly choreographed interactions between the transcriptional cascade and the signaling molecules that regulate its activation state.
In this study, we show that Cak1 is a direct Smk1-activating kinase that phosphorylates Smk1 on T207. Y209 appears to be phosphorylated by Smk1 itself in an intramolecular reaction that requires SSP2. The SSP2 requirement for Smk1 activation and the temporally offset patterns of Smk1 and Ssp2 protein production generate an early pool of Smk1 that is phosphorylated on T207 and a later pool of Smk1 that is phosphorylated on both T207 and Y209. Phenotypic analyses of strains expressing smk1 phosphosite mutants indicate that the T207-phosphorylated form of Smk1 functions to promote early events in spore morphogenesis while the dually phosphorylated form of Smk1 promotes later steps in spore wall formation. These findings indicate that intramolecular phosphorylation can be developmentally regulated and suggest a novel mechanism for coupling MAPK signaling outputs to transcriptional cascades. The transcription of both SMK1 and SSP2 is activated by Ndt80, and their mRNAs accumulate around the same time. However, while Smk1 is translated shortly after its mRNA accumulates, Ssp2 is translated only after a significant lag. These findings suggest that translational derepression of SSP2 mRNA can trigger the phosphorylation of the activating Y in the Smk1 MAPK.
MATERIALS AND METHODS
Yeast strains, plasmids, and culture conditions.
All yeast strains used in this study are in the SK1 background (Table 1). The set of strains containing the wild-type, T207A, Y209F, T207/Y209F, and K69R forms of SMK1-3×HA-HIS3MX6 were derived by standard genetic crosses from strains LH331, LH408, LH510, LH412, and LH405, respectively (16). To construct the SMK1-HH allele, 8 histidine codons and the hemagglutinin (HA) epitope coding sequence were fused in frame to the carboxy-terminal end of SMK1 using the HH-URA3 cassette in yeast strain LRY253 as described by Chen et al. (49). The ssp2Δ SMK1-HH strain JTY4 was derived by crossing a haploid derivative of LRY253 with a haploid ssp2Δ strain, PSY3-B, which contains a precise deletion of the SSP2 open reading frame (31). The SSP2-13MYC-KANMX6 allele was constructed by integration of PCR fragments as described previously (50). The ura3::PGPD1-GAL4(848).ER::URA3 PGAL1-NDT80::TRP1 alleles used in the GOY19 NDT80-inducible system were derived by standard genetic methods from strain YGS91, which was generated in the laboratory of Jacqueline Segall using strains and plasmids described by Benjamin et al. (48).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| ALY60 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 his4/his4 ho::LYS2/ho::LYS2 SMK1-3×HA-KANMX6/SMK1-3×HA-KANMX6 | 20 |
| ALY62 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 smk1::LEU2/smk1::LEU2 | 20 |
| LH416 | MATa/MATα ura3/ura3 leu2/leu2 trp1ΔFA/trp1ΔFA lys2/lys2 his3/his3 ho::hisG/ho::hisG SMK1-HIS3MX6/SMK1-3×HA-HIS3MX6 | 16 |
| LRY342 | MATa/MATα ura3/ura3 leu2/leu2 trp1ΔFA/trp1ΔFA lys2/lys2 his3/his3 ho::hisG/ho::hisG smk1-T207A-3×HA-HIS3MX6/smk1-T207A-3×HA-HIS3MX6 | This study |
| LRY343 | MATa/MATα ura3/ura3 leu2/leu2 trp1ΔFA/trp1ΔFA lys2/lys2 his3/his3 ho::hisG/ho::hisG smk1-Y209F-3×HA-HIS3MX6/smk1-Y209F-3×HA-HIS3MX6 | This study |
| LRY344 | MATa/MATα ura3/ura3 leu2/leu2 trp1ΔFA/trp1ΔFA lys2/lys2 his3/his3 ho::hisG/ho::hisG smk1-T207A,Y209F-3×HA-HIS3MX6/smk1-T207A,Y209F-3×HA-HIS3MX6 | This study |
| LRY373 | MATa/MATα ura3-SK1/ura3 leu2::hisG/leu2 trp1::hisG/trp1ΔFA lys2-SK1/lys2 his3/his4 ho::LYS2/ho::hisG smk1-Y209F-3×HA-HIS3MX6/smk1::LEU2 | This study |
| JLY3 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 CDC28-43244/CDC28-43244 | 26 |
| JLY4 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 CDC28-43244 cak1Δ::TRP1/CDC28-43244 cak1Δ::TRP1 | 26 |
| CMY95 | MATa/MATα ura3/ura3 trp1::hisG/trp1::hisG lys2/lys2 his4/his4 ho::LYS2/ho::LYS2 SSP2-13MYC-KANMX6/SSP2-13MYC-KANMX6 SMK1-3×HA-KANMX6/SMK1-3×HA-KANMX6 | This study |
| CMY58 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 ssp2Δ/ssp2Δ | This study |
| LRY274 | MATa/MATα ura3/ura3 leu2/leu2 trp1ΔFA/trp1ΔFA lys2/lys2 his3/his3 ho::hisG/ho::hisG smk1-K69R-3×HA-HIS3MX6/smk1-K69R-3×HA-HIS3MX6 | This study |
| LRY287 | MATa/MATα ura3/ura3 leu2::hisG/leu2 lys2/lys2 trp1-hisG/trpΔFA his3/his4-N ho::LYS2/ho::hisG smk1-K69R-3×HA-HIS3MX6/SMK1 | This study |
| YGS91 | MATa/ ura3 lys2 leu2::hisG trp1::hisG his3 ho::LYS2 ura3::PGPD1-GAL4(848).ER::URA3 PGAL1-NDT80::TRP1 | J. Segall |
| GOY19 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG lys2/lys2 trp1::hisG/trp1::hisG his3/HIS3 ho::LYS2/ho::LYS2 SMK1-3×HA-HIS3/SMK1-3×HA-HIS3 SSP2-13MYC-KANMX6/SSP2-13MYC-KANMX6 ura3::PGPD1-GAL4(848).ER::URA3/ura3::PGPD1-GAL4(848).ER::URA3 PGAL1-NDT80::TRP1/PGAL1-NDT80::TRP1 | This study |
| LRY253 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 his4/his4 ho::LYS2/ho::LYS2 SMK1-HH::LEU2/SMK1-HH::LEU2 | This study |
| PSY3B | MATa leu2::hisG his4-B lys2 ho::LYS2 ura3 can1 | 31 |
| JTY4 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 his4/his4 ho::LYS2/ho::LYS2 SMK1-HH::LEU2/SMK1-HH::LEU2 ssp2Δ/ssp2Δ | This study |
The plasmids used in this study are listed in Table 2. pLAK51 was constructed by inserting the KpnI/SalI fragment containing the SMK1 gene from pLAK40 (15) into the same sites of pRS314. The SMK1-HA CEN TRP plasmids pAS44 (wild-type SMK1), pAS52 (smk1-T207A), pAS40 (smk1-Y209F), and pAS48 (smk1-T207A,Y209F) were constructed by replacing the BglII-PstI fragment of pLAK51 with the BglII-PstI fragment from YCpSL1, YCpSL9, YCpSL8, and YCpSL3, respectively (17). The BglII site is located in the SMK1 open reading frame 553 bp downstream of the translational start, between codon K69 and the T-X-Y activation loop codons, while the PstI site is located 577 bp downstream of the SMK1 termination codon and is not in the SMK1 genomic clone. The mseΔ SMK1 2μm URA3 plasmids pAS16 (smk1-T207A), pAS12 (smk1-Y209F), and pAS8 (smk1-T207A,Y209F) were constructed by replacing the KpnI-BglII fragment from pSL9, pSL8, and pSL5 with the KpnI-BglII fragment insert from pSL6 (17). The endogenous KpnI site is located 219 bp upstream of the initiator ATG of SMK1. pAS1 (smk1-K69R) was generated by replacing the StyI fragment of pLAK51 with the StyI fragment insert from pLH326. The endogenous StyI sites are located 83 bp and 892 bp downstream of the SMK1 initiator ATG. The smk1-K69R CEN TRP activation loop mutant plasmids pAS29 (smk1-K69R), pAS36 (smk1-K69R,Y207A), pAS24 (smk1-K69R,Y209F), and pAS33 (smk1-K69R,T207A,Y209F) were constructed by replacing the BglII-PstI fragment of pAS1 with BglII-PstI fragment inserts from YCpSL1, YCpSL9, YCpSL8, and YCpSL3, respectively.
Table 2.
Plasmids used in this study
| Plasmid | Markers | Reference or source |
|---|---|---|
| pRS314 | CEN TRP1 | 51 |
| YEp352 | 2μm URA3 | 52 |
| pLAK51 | pRS314 + SMK1-HA | This study |
| pLH326 | pBluescript SK + smk1-K69R-3×HA | 16 |
| pAS1 | pLac51 + smk1-K69R-3×HA | This study |
| pAS44 | pLac51 + SMK1-3×HA | This study |
| pAS52 | pLac51 + smk1-T207A-3×HA | This study |
| pAS40 | pLac51 + smk1-Y209F-3×HA | This study |
| pAS48 | pLac51 + smk1-T207A,Y209F-3×HA | This study |
| pSL6 | YEp352 + SMK1-3×HA ΔMSE | 17 |
| pAS16 | pSL6 + smk1-T207A-3×HA | This study |
| pAS12 | pSL6 + smk1-Y209F-3×HA | This study |
| pAS8 | pSL6 + smk1-T207A,Y209F-3×HA | This study |
| pAS36 | pAS1 + smk1-T207A-3×HA | This study |
| pAS24 | pAS1 + smk1-Y209F-3×HA | This study |
| pAS33 | pAS1 + smk1-T207A,Y209F-3×HA | This study |
Vegetative yeast cultures were maintained in yeast extract-peptone-dextrose (YEPD) (1% yeast extract, 2% peptone, 2% glucose) containing 40 μg/ml adenine or SD medium (0.67% yeast nitrogen base without amino acids, 2% glucose, nutrients essential for auxotrophic strains). For sporulation experiments, cells were grown overnight in YEPA (1% yeast extract, 2% peptone, 2% potassium acetate) containing 40 μg/ml adenine to a density of 107 cells/ml (mid-logphase). Cells were collected by centrifugation, washed in 2% potassium acetate, and resuspended to 4 × 107 cells/ml in sporulation medium (2% potassium acetate, 10 μg/ml adenine, 5 μg/ml histidine, 30 μg/ml leucine, 7.5 μg/ml lysine, 10 μg/ml tryptophan, 5 μg/ml uracil) and placed on a roller drum at 30°C. Synchronous sporulation in the estradiol-inducible NDT80 system was carried out essentially as described previously (53). In brief, sporulation was induced as described above. At 6 h postinduction, β-estradiol was added to 2 μM to induce expression of NDT80.
Electrophoresis and immunoblot analyses.
Yeast cells harvested at different times after transfer into sporulation medium were lysed with NaOH, and proteins were prepared by trichloroacetic acid precipitation as previously described (20). Samples were electrophoretically resolved through 8% polyacrylamide (29:1 acrylamide-bis-acrylamide) gels containing 100 μM MnCl2 and 100 μM Phos-tag acrylamide at 40-mA constant current. After electrophoresis, the gel was soaked in transfer buffer with 1 mM EDTA for 10 min and then transfer buffer without EDTA for 10 min at room temperature. Proteins were transferred to an Immobilon-P membrane and probed for HA with a 1:10,000 dilution of HA.11 monoclonal antibody (Berkeley Antibody Company), for Myc with a 1:5,000 dilution of 9E10 anti-c-Myc monoclonal antibody (Berkeley Antibody Company), for Cdc28 with a 1:10,000 PSTAIR monoclonal antibody (Sigma), for Ndt80 with a 1:25,000 rabbit polyclonal antibody (a gift from Kirsten Benjamin), and for Cak1 with a 1:10,000 dilution of a 2.2-mg/ml mouse monoclonal antibody (a gift from Philip Kaldis). A donkey anti-mouse IgG conjugated to alkaline phosphatase (Jackson Immuno Research Laboratories) was used at 1:5,000 to detect immunoreactivity.
Fluorescence assays.
The fluorescence assay for the incorporation of dityrosine into insoluble material was carried out using a modification of the method of Briza et al. (54, 55). Briefly, yeast cells were collected from liquid sporulation cultures onto nitrocellulose filters using a fritted glass funnel connected to a vacuum apparatus. The filter was subsequently placed cell side up into a petri dish containing ascal wall lysis buffer (350 μl of 100 mM NaCl, 10 mM EDTA, 50 mM β-mercaptoethanol [pH 5.8], and 70 μl glusulase), incubated at 37°C for 3 h, blotted cell side up on Whatman 3MM filter paper to remove excess buffer, and placed cell side up in a petri dish containing 300 μl of concentrated NH4OH for 2 min. Fluorescence was visualized by illuminating the filter with a hand-held 304-nm UV light and photography through a blue filter.
In vitro kinase assays.
Cak1-glutathione transferase (Cak1-GST) kinase and Cdk2 were generously provided by Philip Kaldis (23). Smk1-HA and Smk1-T207A-HA were transcribed and translated in a 100-μl TNT reaction volume (Promega) as per the manufacturer's protocol, diluted 1:10 in phosphate-buffered saline supplemented with 0.05% Tween 20 and protease inhibitors at the concentration specified by Schindler and Winter (56), and immunoprecipitated using rabbit anti-HA antisera (Santa Cruz Biotechnology Inc.) and protein A/G magnetic beads (Pierce). The sample was washed six times in phosphate-buffered saline containing 0.05% Tween 20 and twice in kinase buffer (20 mM Tris-HCl, 5 mM MgCl2, 1 mM dithiothreitol [DTT]), 30% of the bead-immunoprecipitate complex was mixed with 20 ng of GST-Cak1 in 36 μl kinase buffer on ice, and the reaction was initiated by adding 4 μl of 100 mM ATP containing 10 μCi [γ-32P]ATP. The positive-control Cdk2 substrate was used at 4 ng/reaction. After incubation at 30°C for 45 min, the reaction was terminated by the addition of an equal volume of 2× gel loading buffer, and the proteins were fractionated by electrophoresis through 8% polyacrylamide gels. Proteins were then transferred to nylon membranes, and the membrane was exposed to a PhosphorImager screen. Subsequently, the filter was tested for HA and Cak1 immunoreactivity as described above, to control for levels of these proteins in the reactions.
Y209 phosphospecific antiserum.
A peptide containing residues 201 to 214 of Smk1 with phosphotyrosine at position 209 and an additional carboxy-terminal cysteine (C) (TVQPHITNYpVATRWC; the phosphorylated residue is underlined) was coupled through the C residue to keyhole limpet hemocyanin, and the complex was injected subcutaneously into two rabbits (Research Genetics). Booster injections were performed at 4 and 8 weeks after primary immunization. Animals were sacrificed at 12 weeks, and antibody was affinity purified from pooled sera by using the phosphorylated peptide conjugated to Affigel and absorbed against the nonphosphorylated peptide conjugated to Affigel. We were unable to detect a phosphospecific signal by immunoblot analyses of total cellular extracts from meiotic cells. Therefore, we used a set of SMK1-HH strains, which contain 8 histidine codons and the hemagglutinin (HA) epitope fused to the carboxy terminus of SMK1. Smk1-HH was purified from meiotic cells that had been collected at 6.5 h (when Smk1 is monophosphorylated) and 9.5 h (when Smk1 is dually phosphorylated) using nickel beads under denaturing conditions as described by Chen et al. (49). Analytical amounts of the purified protein were analyzed for HA immunoreactivity, and comparable amounts of Smk1-HH (the yield from approximately 108 cells) were resolved by electrophoresis through gels lacking Phos-tag, since the variable amounts of Ni in the Smk1-HH preparations appeared to influence sample migration when Phos-tag was present. Immunoblot analyses were carried out using a 1:1,000 dilution of the Y209p phosphospecific antisera.
Miscellaneous assays and procedures.
For light microscopy, cells were fixed with ethanol and stained with DAPI (4′,6-diamidino-2-phenylindole) as previously described (15). Cells were photographed under wet mount using a Nikon Optiphot equipped for epifluorescence. For the quantitation of refractile spores, asci that had completed MII (4 DAPI-staining foci) were scored for the presence of refractile spores using phase-contrast optics. Data are presented as the fraction of MII-positive asci that contain 2 or more refractile spores. Although the 2-spore cutoff was chosen to eliminate the contribution of preferential nonsister spore packaging, which produces dyad spores in asci that have completed MII under certain nutritional conditions and in certain genetic backgrounds (57), dyad spores in MII-positive asci were rare in all samples assayed in this study (in all cases less than 5% of the MII-positive asci). Two hundred MII-positive cells were counted for each strain, and the entire set of mutants quantitated in this study were analyzed together to facilitate comparison. For electron microscopy, cells were pelleted and fixed in 2.5% glutaraldehyde in 0.13 M cacodylate buffer (pH 7.4). Cells were postfixed with 1% osmium tetroxide for 1.5 h and dehydrated through a graded series of ethanol. Cells embedded in Spurr low-viscosity resin were sliced ultrathin (600 Å), mounted on copper grids, and stained with saturated aqueous uranyl acetate and Reynold's lead citrate. Sections were photographed with a JOEL 1010 transmission electron microscope at 60 or 80 kV. For quantitating electron microscopy images, background levels of nonsporulating cells were excluded from the analyses and only asci containing more than 1 visible spore were counted. Spores were sorted into 3 phenotypic classes depending on ultrastructural features. The first class resembled the wild type. While the inner and outer layers of wild-type spore walls were readily distinguishable in our preparations, it is often not possible to identify all four layers since this requires optimal staining and this can vary even within a single sample grid. Therefore, spore walls were scored as the wild type if there was evidence of electron-lucent (inner wall) layer material surrounded by electron-dense (outer coat) material irrespective of further structural detail. The second phenotypic class of spores were surrounded only by the electron-lucent material that resembles inner spore wall layers. Spores were sorted into this class irrespective of the thickness of the layers or whether the 2 inner layers could be differentiated. The third phenotypic class was heterogeneous and contained spores that were surrounded by aberrant structures that did not resemble wild-type or inner layers. This class contained abnormal electron-dense staining structures that did not surround inner layers, structures that were devoid of recognizable structure, and some spores that appear to have been blocked at the prospore membrane stage. RNA was prepared and analyzed by Northern blotting using DNA probes that were radioactively labeled using random priming and the large (Klenow) fragment of DNA polymerase I from E. coli as previously described (58).
RESULTS
The Smk1 T-X-Y activation loop is phosphorylated on T207 and Y209 through two independent reactions.
Smk1 is produced exclusively in sporulating cells and first appears late in meiotic prophase after the Sum1 repressor has been removed from the SMK1 promoter (5 to 6 h postinduction in the rapidly sporulating SK1 genetic background) (41, 42). Subsequently, Smk1 accumulates to a high level as active Ndt80 is produced and the meiotic divisions (MI and MII) are occurring (between 6 and 8 h). As Smk1 accumulates, it is activated by phosphorylation. Smk1 protein levels remain high as spore morphogenesis takes place (between 8 and 10 h). After spore walls are formed, the level of Smk1 declines until only background levels are present at 24 h.
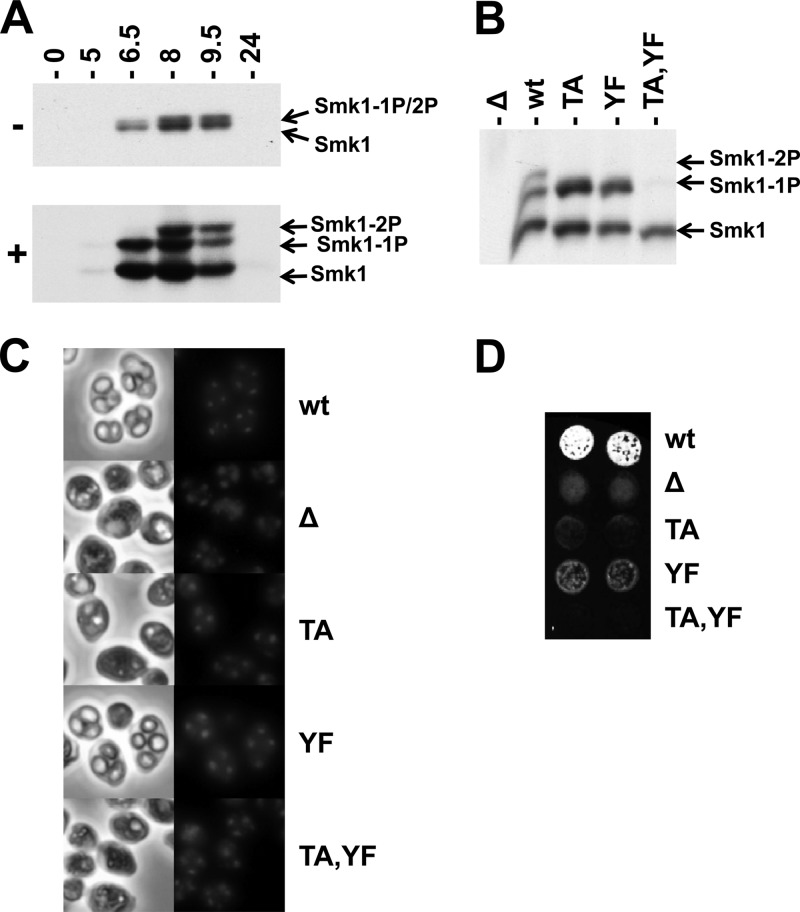
Phosphorylated Smk1 can be electrophoretically resolved from nonphosphorylated Smk1 in polyacrylamide gels, but singly and dually phosphorylated forms of Smk1 comigrate in these assays (Fig. 1A, upper panel). However, when Phos-tag acrylamide is included in the electrophoretic system, singly and dually phosphorylated forms of Smk1 can be resolved (Fig. 1A, lower panel). Smk1 from cells at 6.5 h following meiotic induction is monophosphorylated, while Smk1 at later time points is dually phosphorylated. These modifications require T207 and Y209 since they are not observed in an smk1-T207A,Y209F mutant (Fig. 1B). In addition, Smk1-T207A and Smk1-Y209F are both monophosphorylated. These findings suggest that T207 and Y209 in the activating motif of Smk1 are phosphorylated during meiotic development and that phosphorylation of the T and Y residues can occur independently.
Fig 1.
Phosphorylation of Smk1 on T207 and Y209. (A) Diploid SMK1-HA cells were transferred to sporulation media for the indicated times (in hours). Total protein extracts were fractionated by electrophoresis in the presence (+) or absence (−) of Phos-tag acrylamide as indicated and analyzed by immunoblotting using an HA antibody. (B) Meiosis was induced in an smk1Δ strain harboring the low-copy-number (CEN-based) SMK1-HA plasmids with the indicated activation loop mutations (TA, T207A; YF, Y209F). Samples were collected 9.5 h after meiotic induction and analyzed by electrophoresis through Phos-tag gels and immunoblotting using an HA antibody. (C) The indicated strains were harvested 24 h after meiotic induction. DNA was stained with DAPI, and cells were viewed by phase-contrast (left column) and fluorescence (right column) microscopy. (D) Two independent isolates of the indicated strains were harvested 24 h after meiotic induction, collected on filters, and assayed for the incorporation of dityrosine in the outer spore wall using the fluorescence assay. 1P, monophosphorylated; 2P, dually phosphorylated.
Spore formation can be monitored by direct microscopic observation and by using a fluorescence-based filter assay that measures the incorporation of dityrosine, a sporulation-specific component normally found in the outer layer of the spore wall into insoluble material (54). In previous studies, it was shown that single-copy (CEN) plasmids harboring smk1-T207A, smk1-Y209F, or smk1-T207A,Y209F did not complement the smk1Δ spore formation defect using these assays. However, smk1-Y209F, but not smk1-T207A, expressed from multicopy (2μ) plasmids complemented the smk1Δ defect (17). We tested the phenotypes of these mutations in their normal chromosomal context and found that the smk1-T207A and smk1-T207A,Y209F homozygote sporulation phenotypes were indistinguishable from the smk1Δ phenotype (<2% of the cells that completed MII formed refractile spore walls when viewed by phase-contrast microscopy, with 200 asci counted for each strain) (Fig. 1C). In addition, these strains were negative in the fluorescence assay (Fig. 1D). In contrast, 65% of the smk1-Y209F homozygotes that completed MII contained refractile spores (compared to 97% of the wild-type cells), and these cultures produced an intermediate signal in the fluorescence assay (Fig. 1C and D). Thus, T207 is required for detectable SMK1 function using these phenotypic assays, while Y209 is required for efficient completion of late steps in spore formation.
Cak1 phosphorylates Smk1 on residue T207.
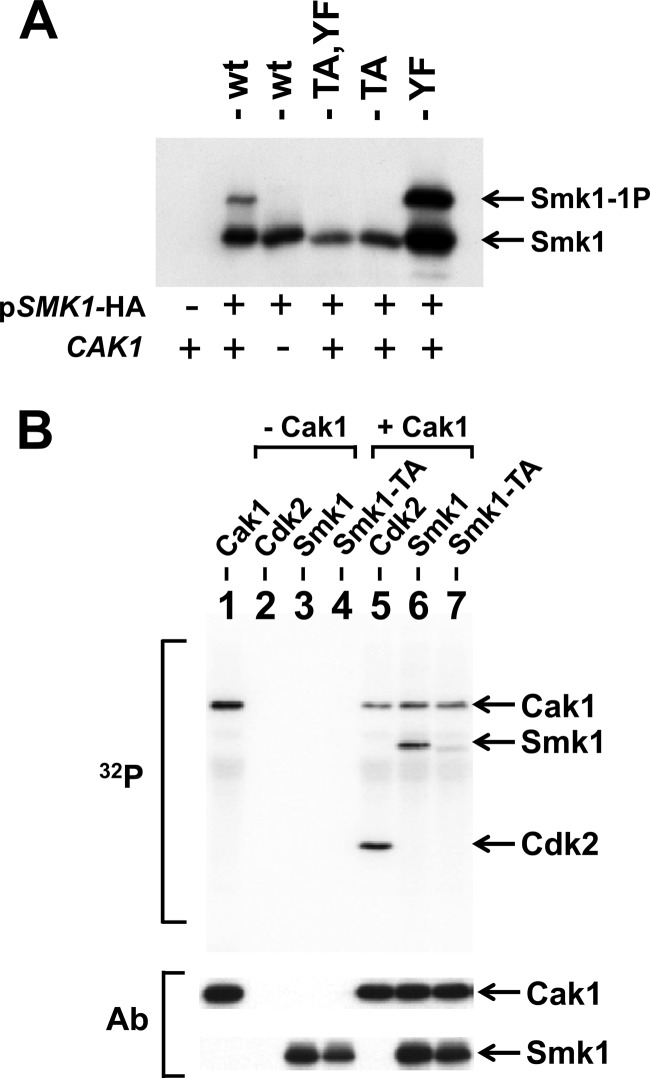
Previous studies demonstrated that CAK1 is a dosage suppressor of the sporulation defect of a temperature-sensitive smk1 hypomorph and that Cak1 promotes Smk1 activation (21). SMK1 is not expressed in mitotic cells due to the presence of the Sum1 transcriptional repression complex, which binds to the MSE in the SMK1 promoter (44). A mutation in the SMK1 MSE that eliminates Sum1 binding (referred to here as mseΔ) leads to SMK1 derepression and accumulation of Smk1 protein in vegetative cells (18, 44). This allows for analysis of Smk1 in the cdc28-43244 background, which does not require Cak1 for vegetative growth (25). To test the Cak1-dependent phosphorylation of Smk1, cdc28-43244 haploids containing or lacking CAK1 were transformed with plasmids expressing wild-type, smk1-T207A, smk1-Y209F, and smk1-T207A,Y209F from the mseΔ promoter. A singly phosphorylated form of Smk1 is produced in CAK1 cells but not in cak1Δ cells, and mutation of T207 abolishes this phosphomodification, while mutation of Y209 does not (Fig. 2A). These findings are consistent with Cak1 being required for the phosphorylation of T207 but not Y209. The absence of Y209 phosphorylation in mitotic cells implies that some other meiosis-specific factor is required for the phosphorylation of this residue.
Fig 2.
Cak1 phosphorylates Smk1 on T207. (A) Smk1-HA was constitutively produced using the mseΔ promoter in cdc28-43244 (hyperactivated) CAK1 (+) and cak1Δ (−) mitotic cells, and proteins were analyzed by electrophoresis through Phos-tag gels and immunoblotting using an HA antibody. (B) Phosphotransferase assays containing the indicated proteins were carried out in the presence of 32P-labeled ATP. The proteins were resolved on electrophoretic gels and transferred to a membrane, and radioactivity was recorded using a phosphorimager (32P). Lane 1, Cak1-GST; lane 2, mammalian Cdk2; lane 3, Smk1 immunoprecipitated from an in vitro transcription/translation (TNT) reaction; lane 4, Smk1-T207A immunoprecipitated from a TNT reaction; lane 5, mammalian Cdk2 plus Cak1-GST; lane 6, immunoprecipitated Smk1 plus Cak1-GST; lane 7, immunoprecipitated Smk1-T207A plus Cak1-GST. The positions of the proteins are indicated by arrows. The same filter was subsequently assayed for Cak1 or HA (Smk1) immunoreactivity (Ab) as indicated. The incorporation of radioactivity into the Cak1 band requires the GST moiety since untagged Cak1 does not undergo autophosphorylation in these assays.
To further establish whether Cak1 phosphorylates Smk1 on T207, Smk1 and Smk1-T207A were produced in a coupled transcription/translation in vitro system. These proteins were immunoprecipitated and incubated with purified Cak1-GST in the presence of radioactive ATP. Mammalian Cdk2 was used as a positive-control substrate in these experiments. As shown in Fig. 2B, Cak1-GST transfers phosphate to Smk1 in vitro. The T207A substitution sharply reduced the Cak1-dependent incorporation of radioactivity into the mutant Smk1-T207A protein (relative specific activity of Smk1-T207A is 18% ± 3% of Smk1, n = 3). While the radioactive, wild-type Smk1 protein migrated slightly slower than unlabeled protein (the gels used in these experiments did not contain Phos-tag), radioactive Smk1-T207A protein comigrated with unlabeled protein, suggesting that a different residue in Smk1 is being phosphorylated at a low level when T207 is not present. Cak1 and Smk1 were identified as interacting proteins in a genome-wide 2-hybrid study (59). We have confirmed that Cak1 and Smk1 interact using the yeast 2-hybrid system (data not shown). Taken as a whole, these data indicate that T207 in Smk1 is directly phosphorylated by Cak1 in vitro and in vivo. The reduced level of Cak1-dependent phosphate incorporation into Smk1-T207A may reflect the spurious transfer of phosphate onto one or more unidentified residues in Smk1 when T207 is rendered nonphosphorylatable and the local concentration of Cak1 relative to Smk1-T207A is high as a result of protein-protein interactions.
SSP2 promotes the phosphorylation of Smk1 on residue Y209.
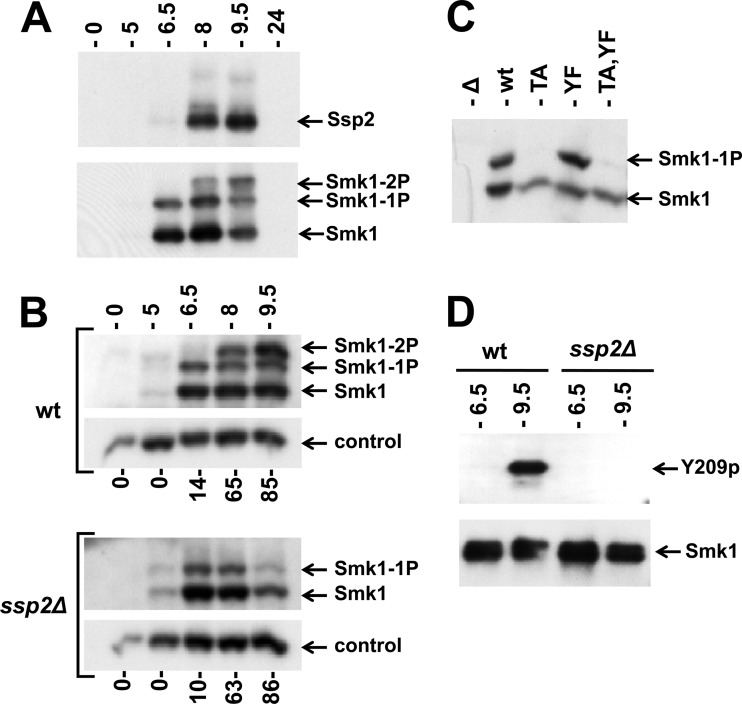
A previous screen of deletion strains that are defective in spore formation revealed that the relative amount of phosphorylated Smk1 is reduced in an ssp2Δ mutant (20). However, the mono and diphosphorylated Smk1 species were not resolved in these studies. SSP2 is controlled by a tightly regulated middle-meiotic promoter that is activated by Ndt80 (but not repressed by Sum1) (40, 46). While SSP2 is required for spore morphogenesis, the biochemical function of the Ssp2 protein is unknown (30, 31). To examine the relationship between the phosphorylated forms of Smk1 and Ssp2, cells were analyzed by immunoblot assays at different times following the induction of sporulation. As shown in Fig. 3A, high levels of Smk1 protein are present at 6.5 h following meiotic induction, while high levels of Ssp2 are not present until around 8.0 h. Importantly, Smk1 is monophosphorylated prior to the accumulation of Ssp2 and dually phosphorylated once Ssp2 is present.
Fig 3.
Phosphorylation of Smk1 on Y209 requires Ssp2. (A) Extracts from meiotic cells were prepared at the indicated times postinduction and analyzed by electrophoresis through Phos-tag gels and immunoblotting. A single filter was sequentially analyzed using anti-Myc (upper panel; Ssp2) or anti-HA (lower panel; Smk1) antibodies. (B) Sporulation was induced in wild-type (wt) and ssp2Δ strains as indicated, and Smk1-HA was analyzed as described for panel A. Cdk1 (Cdc28), which is present at a constant level throughout sporulation, was monitored using a PSTAIR antibody as a loading control. The fraction of cells that had completed MII at each time point is indicated below the immunoblots (n = 100). (C) smk1Δ ssp2Δ strains harboring low-copy-number (CEN-based) Smk1-HA plasmids with the indicated activation loop mutations (TA, T207A; YF, Y209F) were collected 9.5 h following transfer to sporulation medium, and Smk1-HA was analyzed as described for panel A. (D) Smk1-HH proteins were purified from wild-type and ssp2Δ meiotic cells collected 6.5 h and 9.5 h after transfer to sporulation medium. Comparable amounts of Smk1-HH were resolved by electrophoresis in gels lacking Phos-tag and analyzed by immunoblotting using phosphospecific Y209p peptide antisera (Y209p) or an anti-HA antibody (Smk1).
To further investigate the relationship between SSP2 and SMK1, the accumulations of singly and doubly phosphorylated Smk1 in the wild-type and ssp2Δ strains were compared at different times during meiotic development (Fig. 3B). Smk1 appears at the same time (starting around 5 h), and the fraction of Smk1 that is monophosphorylated at 5 and 6.5 h is indistinguishable in wild-type and ssp2Δ cells. However, the dually phosphorylated form of Smk1 that appears at the later time points in wild-type cells (8 and 9.5 h postinduction) is completely absent in the ssp2Δ mutant. These observations suggest that SSP2 is required for the phosphorylation of Y209.
Interestingly, the absence of dually phosphorylated Smk1 in the ssp2Δ mutant is accompanied by a reduction in the total amount of Smk1 protein (compare the 9.5-h time points in Fig. 3B). This reduction is not a consequence of differences in meiotic kinetics, since the fractions of cells that completed MI and MII are similar (Fig. 3B). These observations suggest either that Smk1 is unstable or that Smk1 translation is impaired in the ssp2Δ background (see below for further discussion).
To test whether SSP2 is required for the phosphorylation of Smk1 on Y209, the set of smk1 phosphosite mutants was examined in the ssp2Δ strain at a late time point in meiosis (9.5 h). The Smk1-T207A mutant protein, in which Y209 is available for phosphorylation, was unphosphorylated in ssp2Δ cells, while both Smk1-Y209F and wild-type Smk1 were monophosphorylated (Fig. 3C). These data suggest that SSP2 is specifically required for the phosphorylation of Smk1 on Y209. To further establish that this is the case, we used an antiserum that is specific for the Y209-phosphorylated form of Smk1. In these experiments, an allele of SMK1 that had been tagged with 8 consecutive histidine (H) residues and an HA epitope tag (SMK1-HH) was used. Smk1-HH was purified from wild-type and ssp2Δ meiotic cells collected at 6.5 h postinduction (when Smk1 is monophosphorylated) and at 9.5 h postinduction (when Smk1 is dually phosphorylated in the wild-type strain but monophosphorylated in the ssp2Δ background). Equivalent amounts of purified Smk1-HH were resolved by electrophoresis and assayed by immunoblot analyses with the Y209 phosphospecific antiserum (Fig. 3D). Y209 phosphospecific immunoreactivity was detected only in wild-type cells and only at the later (9.5 h) time point, confirming that Smk1 is phosphorylated on Y209 late in meiotic development in an SSP2-dependent fashion.
Smk1 is autophosphorylated in cis on residue Y209.
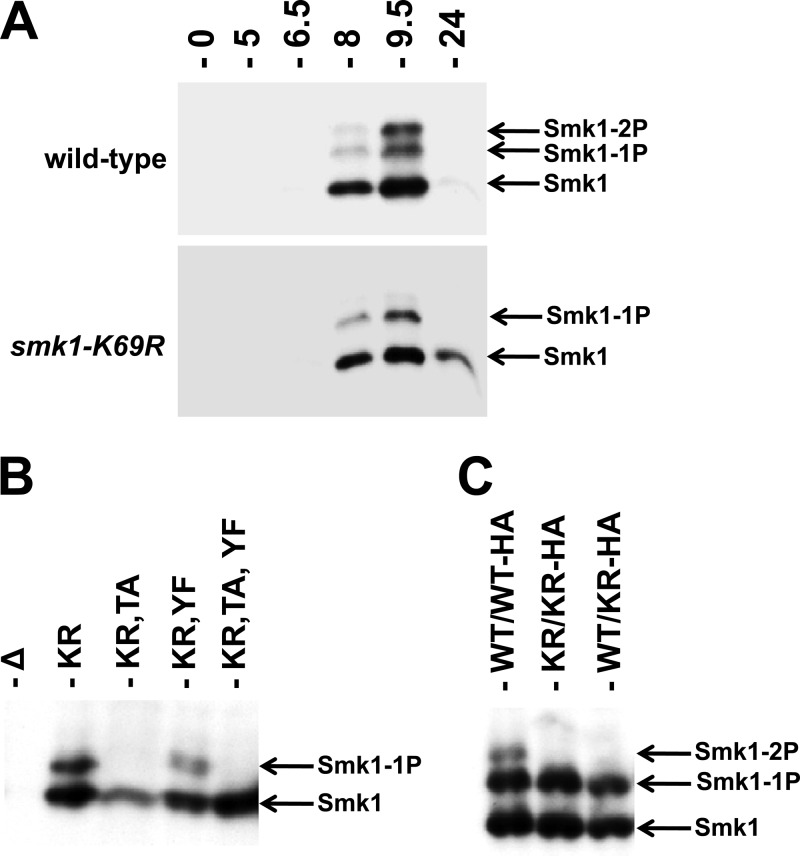
In order to test whether autocatalysis plays a role in Y209 phosphorylation, a catalytically inactive form of Smk1 containing a mutation in its ATP-binding pocket (K69R) was analyzed. Smk1-K69R is predominantly monophosphorylated (Fig. 4A). Smk1-K69R,T207A is not detectably phosphorylated (Fig. 4B). These findings are consistent with Smk1 autophosphorylating residue Y209. In contrast, Smk1-K69R and Smk1-K69R,Y209F proteins are monophosphorylated, thereby demonstrating that the Cak1-dependent phosphorylation (on T207) is unaffected by the catalytic K69R substitution in Smk1.
Fig 4.
Smk1 is autophosphorylated on residue Y209. (A) Extracts from wild-type SMK1-HA (upper panel) and smk1-K69R-HA (lower panel) strains prepared at the indicated times after transfer to sporulation medium were analyzed by electrophoresis through Phos-tag gels and immunoblotting using an HA antibody. (B) Samples from an smk1Δ/smk1Δ strain harboring the indicated low-copy-number (CEN-based) Smk1-HA plasmids were harvested 9.5 h after meiotic induction and assayed as described for panel A. (C) SMK1/SMK1-HA (WT/WT-HA), smk1-K169R/smk1-K169R-HA (KR/KR-HA), and SMK1/smk1-K169R-HA (WT/KR-HA) cells were harvested 9.5 h following meiotic induction and assayed as described for panel A.
To further test the mechanism of Smk1 autocatalysis, the phosphorylation of a heterozygous strain expressing an epitope-tagged (HA), catalytically inactive (K69R) copy of SMK1 and an untagged wild-type copy of SMK1 was analyzed. If autocatalysis takes place in an intermolecular reaction, the nontagged, wild-type protein is expected to phosphorylate the tagged, catalytically inactive protein. In an intramolecular reaction, the wild-type Smk1 would be unable to phosphorylate the catalytically inactive form of the protein, resulting in monophosphorylated HA-tagged Smk1. Immunoblot analysis of the heterozygous smk1-K69R-HA/SMK1 strain showed only a singly phosphorylated form of Smk1-K69R-HA (Fig. 4C). These data are consistent with Smk1 autophosphorylating Y209 in an intramolecular reaction. Nevertheless, the hypothesized autophosphorylation reaction must be tightly controlled, since autophosphorylation is undetectable when Smk1 isolated from meiotic cells (16) or Smk1 produced in the in vitro transcription/translation system (Fig. 2) is assayed in vitro.
The smk1-Y209F and ssp2Δ phenotypes are similar.
If the sole function of SSP2 is to promote the autophosphorylation of Smk1 on Y209, then the ssp2Δ and smk1-Y209F sporulation phenotypes should be similar. ssp2Δ and smk1-Y209F cells were compared by light microscopy (Fig. 5A, columns 1 and 2) and by using the fluorescence assay (Fig. 5B). Consistent with previously published reports, we find that less than 1% of the ssp2Δ cells that have completed MII form refractile spore walls when viewed by light microscopy (n = 200 asci in all cases). We also find that ssp2Δ cells are negative in the fluorescence assay. While Sarkar et al. observed that nuclei in the ssp2Δ strain became fragmented after meiosis had been completed (31), we did not observe extensive nuclear fragmentation at the 24 h time point (Fig. 5A, column 2). This may reflect differences in the sporulation media used in the two studies (the 2% acetate used in this study may osmotically stabilize nuclear structures, in contrast to the 1% acetate used by Sarkar et al.). In contrast to the ssp2Δ strain, 65% of the smk1-Y209F/smk1-Y209F homozygotes that completed MII formed refractile spores, and these spores generated a weak signal in the fluorescence assay (Fig. 5; see also Fig. 1C and D). These data show that the ssp2Δ homozygote has a more severe sporulation defect than the smk1-Y209F homozygote. We also tested the sporulation phenotype of an smk1-Y209F/smk1Δ heterozygote. In contrast to the smk1-Y209F/smk1-Y209F homozygote, a fluorescence signal was undetectable in the smk1-Y209F/smk1Δ heterozygote, and <2% of the smk1-Y209F/smk1Δ asci contained refractile spores. Thus, a 50% reduction in the copy number of smk1-Y209F dramatically reduces spore formation, and the smk1-Y209F/smk1Δ and ssp2Δ homozygote sporulation phenotypes are indistinguishable in these assays.
Fig 5.
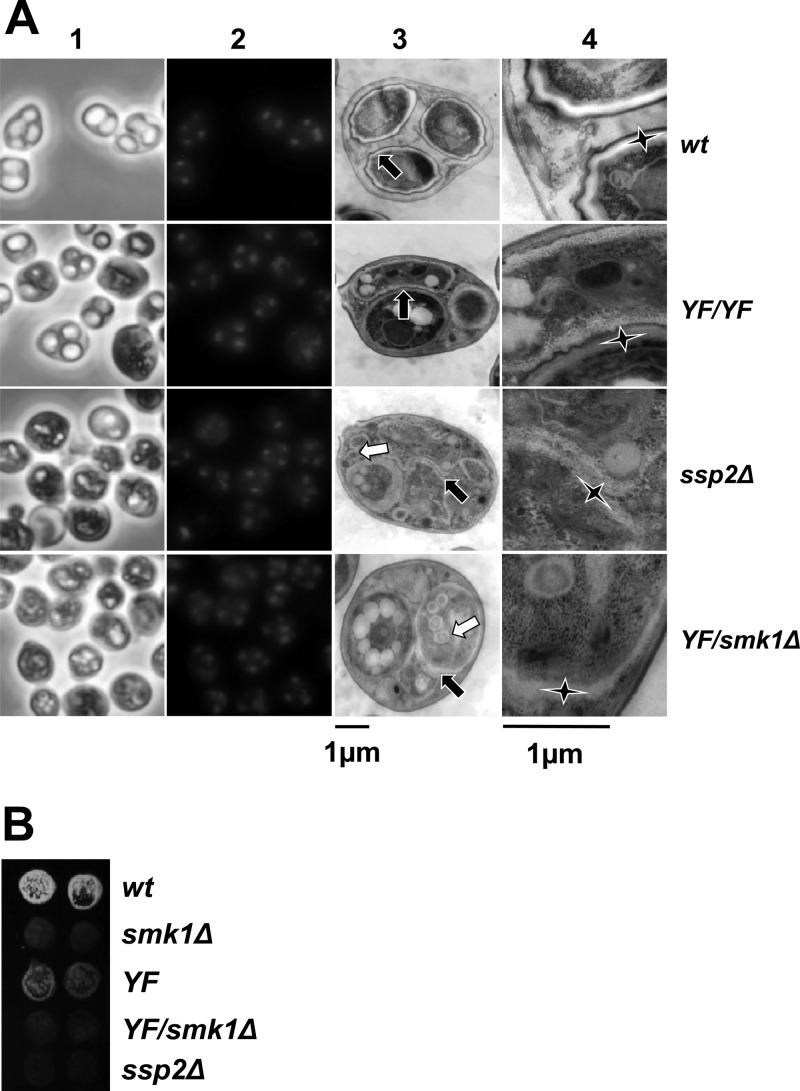
The smk1-Y209F/smk1Δ phenotype is indistinguishable from the ssp2Δ/ssp2Δ phenotype. (A) Cells with the indicated genotypes were harvested 24 h after meiotic induction. Samples were stained with DAPI and images recorded using phase-contrast (column 1) and fluorescence (column 2) microscopy. Aliquots of these samples were stained with uranyl acetate and examined by thin-section electron microscopy. A representative low-magnification electron micrograph (column 3) and corresponding high-magnification electron micrograph (column 4) are shown. Each black arrow in column 3 points to the center of a magnified area shown in column 4 for reference. The elongated vertices of the stars in column 4 point to the inner layers of the spore wall. Note that in the wild type (wt), these inner layers are surrounded by the chitin/chitosan and the electron-dense dityrosine coat (outer layers). In the smk1-Y209F/smk1-Y209F (YF/YF) spores, the indicated inner layers (star) are surrounded by the chitin/chitosan layer and dityrosine coat resembling wild-type spore walls, while the inner layers surrounding the adjacent spore (indicated by the short upward-pointing vertex of the star) are absent. While the inner layers are present in ssp2Δ and smk1-Y209F/smk1Δ (YF/smk1Δ) spores, the outer layers are missing in almost all spores. The white arrows in the ssp2Δ and YF/smk1Δ samples in column 3 point to the supernumerary vesicles that appear to be surrounded by inner spore wall layers. (B) Sporulating cells of the indicated genotypes were harvested 24 h after meiotic induction in duplicate, equivalent numbers of cells were collected on filters, and the incorporation of dityrosine into insoluble material was assayed using the fluorescence assay.
To gain additional insight into the relationship between the ssp2Δ and smk1-Y209F phenotypes, cells that had been incubated in sporulation medium for 24 h were examined using thin-section electron microscopy (Fig. 5A, columns 3 and 4). In the wild-type control sample, 98% of the spores were surrounded by multilayered spore walls characteristic of mature spores (n = 71 spores counted). In contrast to the wild-type spore walls, 95% of the meiotic products in the ssp2Δ cells were surrounded by only inner electron-lucent spore wall layers (n = 71). In many cases the ssp2Δ spore wall layers appear thicker than normal, and as previously described, membranous structures/remnants not present in wild-type cells are seen within the ssp2Δ spores (30). Moreover, spherical structures that are smaller than spores but that appear to be surrounded by inner spore wall-like layers were present (white arrows in column 3). We observed a strikingly similar spore wall defect in the smk1-Y209F/smk1Δ heterozygote (89% of spores were surrounded by only the inner layers, 4% resembled wild-type, and 7% were blocked at other stages; n = 85). In contrast to the phenotype of the smk1-Y209F heterozygote, most of the spores in the smk1-Y209F/smk1-Y209F homozygote were surrounded by spore walls that resembled those of the wild type (62%). Of the remaining spores in the homozygote, 35% appear to be surrounded only by inner layers, and 4% appear to have blocked spore morphogenesis at an early stage prior to the deposition of the inner layers (n = 69). As a control, we also analyzed the smk1-T207A,Y209F homozygote, which had the most severe phenotype (47% of the spores were surrounded by only inner layers, and 53% of the spores appeared to have been blocked at an early stage in spore morphogenesis; n = 93). Taken as a whole, these data show that the ssp2Δ and single-copy smk1-Y209F strains have strong sporulation-defective phenotypes that are indistinguishable, while the two-copy smk1-Y209F strain undergoes some steps in spore wall formation, leading to partially defective spore walls. These findings suggest that in addition to promoting autophosphorylation, SSP2 modestly increases SMK1 activity by an additional mechanism. This mechanism might involve the stabilization or translational activation of Smk1, since as described above, a decreased amount of Smk1 is present at the later stages of the program in the ssp2Δ mutant (Fig. 3B). It is therefore possible that the phenotype of ssp2Δ is entirely due to a deficiency in SMK1 activity (see Discussion).
Ssp2 is unable to promote autophosphorylation of preexistent Smk1.
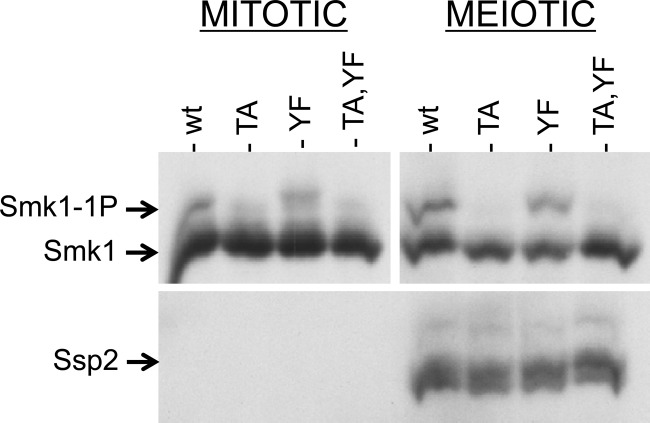
We next asked whether Smk1 translated in vegetative cells can be phosphorylated on Y209 once these cells are induced to sporulate. For this purpose, we used the set of plasmids that express SMK1 from the mseΔ promoter. Because mseΔ eliminates binding of the Sum1 repressor and the Ndt80 activator, SMK1 is not only derepressed in vegetative cells but also uninducible in meiotic cells (18). Most of the Smk1 that is present in sporulating cells harboring these plasmids was therefore translated prior to meiotic induction. The wild-type, T207A, Y209F, and T207A Y209F forms of SMK1 were expressed from mseΔ promoters in vegetative diploid cells, and the proteins were compared with the proteins present in sporulating cells 9.5 h postinduction, after Ssp2 had been translated (Fig. 6). Although the fraction of Smk1 that is monophosphorylated increased during sporulation, this increase is a consequence of residue T207 phosphorylation since it was completely absent in the smk1-T207A and smk1-T207A,Y209F strains. The increase in Cak1-dependent T207 phosphorylation that takes place in meiotic cells is consistent with the transcriptional induction of CAK1 during sporulation (CAK1 mRNA and Cak1 protein increase first as early genes are induced and more substantially as middle genes are induced) (46, 47). Notably, in the meiotic samples, wild-type Smk1 was monophosphorylated and Smk1-T207A was not detectably phosphorylated. Thus, Smk1 that is produced in vegetative cells cannot undergo the SSP2-dependent autophosphorylation of Y209 despite the fact that the same pool of protein can be phosphorylated on T207 by Cak1. While the experiment whose results are shown in Fig. 6 was carried out in an smk1Δ background harboring the indicated mseΔ-SMK1-HA plasmids, identical results were obtained when a wild-type SMK1 (untagged) background harboring the same plasmids was assayed (data not shown). The absence of SSP2-dependent autophosphorylation of Y209 in these experiments is therefore not an indirect consequence of an SMK1 deficiency. One explanation for the lack of Y209 phosphorylation in these experiments is that Ssp2 acts on a transient intermediate form of Smk1 (present during translation or protein folding).
Fig 6.
Smk1 produced prior to meiotic induction is not autophosphorylated on Y209 in sporulating cells. Vegetative smk1Δ SSP2-MYC cells harboring plasmids expressing the indicated SMK1-HA mutant from the mseΔ promoter were transferred to sporulation medium. Samples were taken at 0 h and at 9.5 h and analyzed by electrophoresis through Phos-tag gels and immunoblotting using an HA (Smk1) or a MYC (Ssp2) antibody. There is more Smk1 in the mitotic cells (left) than in the meiotic cells (right) due to degradation of Smk1 that takes place during the 9.5-h interval. The mitotic and meiotic Smk1 signals were analyzed in the same blot, but the exposure of the meiotic samples was approximately 4 times as long as the mitotic exposure to facilitate the comparison of the relative levels of phosphorylated Smk1 in mitotic and meiotic cells. TA, T207A; YF, Y209F; wt, wild type.
Ssp2 is translated just prior to the accumulation of the doubly phosphorylated form of Smk1.
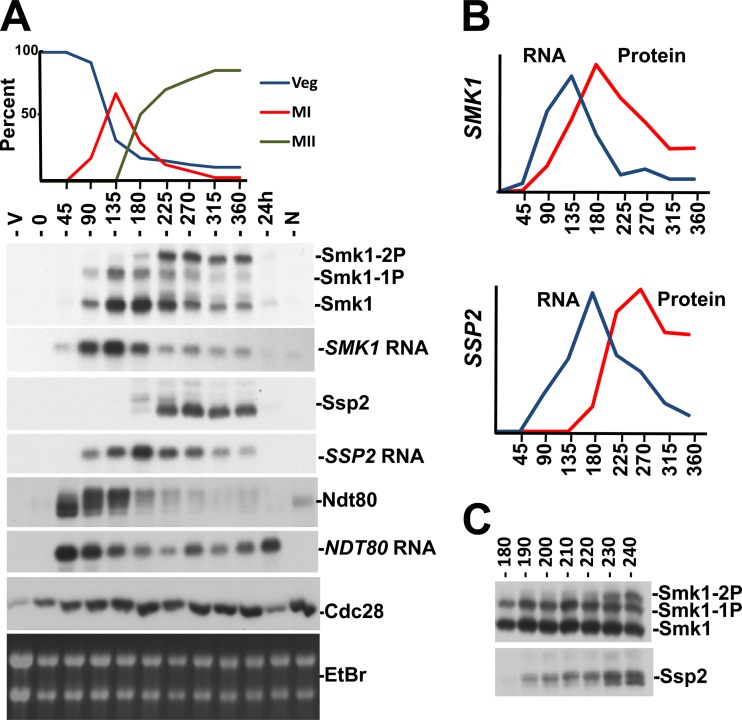
To further investigate the connection between Smk1 autophosphorylation and SSP2, we utilized a strain in which NDT80 is controlled by an estradiol-inducible promoter (48, 53). In this genetic background, cells that have been transferred to sporulation medium stall in the pachytene stage of MI prophase due to NDT80 insufficiency. NDT80 induction causes exit from pachytene and the synchronous completion of MI, MII, and spore formation. We monitored Ndt80, Smk1, and Ssp2 proteins and mRNAs from cells at different times following estradiol addition (Fig. 7). NDT80 mRNA and Ndt80 protein accumulate at the earliest postinduction time point tested (45 min following estradiol addition). The series of slowly migrating forms of Ndt80 whose relative abundance increases during the first 2 h after estradiol addition are consistent with Ndt80 being activated by phosphorylation (48, 60–62).
Fig 7.
Smk1 phosphorylation in synchronized meiotic cells. (A) A strain expressing NDT80 from an estradiol-inducible promoter was transferred to sporulation medium for 6 h to enrich for pachytene-arrested cells (0 hours). β-Estradiol was added, and samples were withdrawn every 45 min. A vegetatively growing sample (V), a sample without any β-estradiol (at a time corresponding to the 360 min time point) (N), and a sample 24 h after β-estradiol addition (greater than 90% spores) were also analyzed. Progression through meiosis was assayed by counting DAPI-stained cells (upper panel). Veg, nuclei from vegetatively growing cells; MI, binucleated cells that had completed MI; MII, the sum of cells that were tri- or tetranucleated. Samples were analyzed by immunoblotting of proteins resolved on Phos-tag gels using the indicated antibodies or by Northern blot analysis of total RNA using probes specific for the indicated mRNAs. Cdc28, whose levels are constant throughout meiosis, and the ethidium bromide (EtBr)-stained rRNA from the gel used in the Northern blot analysis are shown to control for total protein and RNA, respectively. (B) Quantitation of SMK1 and SSP2 mRNA and proteins from data shown in panel A. (C) Smk1 and Ssp2 were analyzed by immunoblot analyses of Phos-tag gel-resolved proteins taken at 10-min intervals after the addition of β-estradiol. Only the time points when Ssp2 accumulates and Smk1 autophosphorylation is taking place are shown for simplicity. Note the correlation of the lower electrophoretic form of Ssp2 (not resolved when proteins are analyzed on gels lacking Phos-tag and therefore likely to be a dephosphorylated form of Ssp2) and Smk1 autophosphorylation (Smk1-2P).
SMK1 mRNA levels increase as the slowly migrating forms of Ndt80 accumulate. An increase in Smk1 protein closely follows the increase in SMK1 mRNA, and Smk1 protein levels peak around the 180 min time point. Importantly, dually phosphorylated Smk1 protein is undetectable prior to the 180 min time point, while much of the Smk1 after 180 min is dually phosphorylated. Significantly, Ssp2 protein starts to accumulate at this 180 min time point. The near-simultaneous appearance of Ssp2 and dually phosphorylated Smk1 is consistent with the coincident appearance of Ssp2 and dually phosphorylated Smk1 in cells that express NDT80 from its endogenous promoter (Fig. 3A).
To further examine the temporal relationship between Ssp2 production and Smk1 phosphorylation, the experiment was repeated and samples were withdrawn every 10 min around the interval when the dually phosphorylated form of Smk1 starts to accumulate (as Smk1 is autophosphorylating Y209). In this experiment, the dually phosphorylated form of Smk1 increased rapidly between 220 and 230 min. Ssp2 is present prior to this transition at 220 min, yet its mobility changes as the doubly phosphorylated form of Smk1 appears.
Interestingly, the translation of SSP2 mRNA is delayed relative to the translation of NDT80 or SMK1 mRNAs. Even though SSP2 mRNA is present at relatively high levels at the 90 and 135 min time points in Fig. 7A, Ssp2 protein is undetectable in these samples and quantitation of protein and mRNA levels shows that there is a significant delay in the translation of Ssp2 compared to Smk1 (Fig. 7B). These observations are consistent with ribosome-profiling studies, which demonstrate that SSP2 mRNA is a member of a set of meiotic mRNAs that are present as the nuclear divisions are taking place but not engaged by ribosomes until relatively late in meiotic development (63). These findings suggest that the apparent translational derepression of SSP2 that occurs as cells complete MII controls the appearance of Ssp2 and Smk1 autophosphorylation.
DISCUSSION
This study demonstrates that the meiosis-specific Smk1 MAPK is activated by two mechanistically distinct phosphotransferase reactions that take place at different stages in meiotic development. In the first reaction, the newly synthesized Smk1 that is translated after middle genes are induced is phosphorylated on residue T207 by Cak1. This reaction generates a pool of monophosphorylated Smk1 as MI is taking place. Cak1 is present in a constitutively active form in vegetative cells (22–24, 64). However, CAK1 expression increases above its vegetative level as middle genes are induced and Smk1 levels are rising (46, 47). Taken together, these findings suggest that the accumulation of partially activated Smk1-T207p is largely controlled by NDT80. Despite the tight connection between the transcriptional cascade of sporulation and the accumulation of the monophosphorylated form of Smk1, it should be pointed out that all of the experiments in this study were carried out in the absence of environmental glucose and nitrogen, which are known to negatively regulate other protein kinases that control meiotic processes (39). Whether the phosphorylation of Smk1 on T207 is controlled by nutritionally regulated signaling pathways independent of the transcriptional cascade remains to be established.
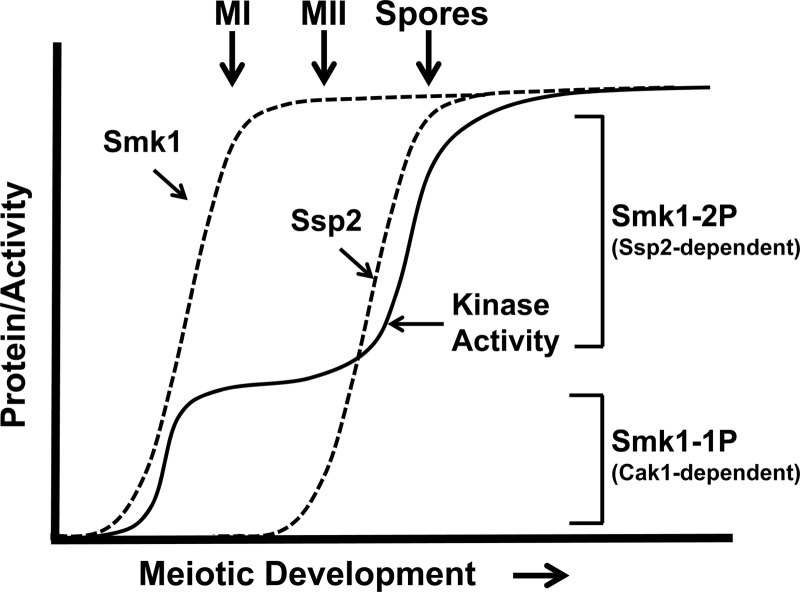
The second reaction that activates Smk1 is the SSP2-dependent phosphorylation of residue Y209, which takes place around the time that cells complete MII. While the phosphorylation of T207 appears to be sufficient to partially activate Smk1 (as evidenced by the partial sporulation deficiency of smk1-Y209F), the phosphorylation of Y209 appears to be unable to activate Smk1 unless T207 is also phosphorylated (as evidenced by the null-like smk1-T207A phenotype). These findings suggest that the low (T207p) form of Smk1 promotes early steps in spore morphogenesis that take place as cells are undergoing the meiotic divisions, while the high T207p/Y209p state triggers events that occur after cells have completed MII (see model in Fig. 8).
Fig 8.
Model for the establishment of Smk1 catalytic thresholds by Cak1-dependent phosphorylation of T207 and Ssp2-dependent autophosphorylation of Y209. Dashed lines indicate proteins, and the solid line indicates Smk1 catalytic activity. The Ssp2-dependent changes in Smk1 protein level are not shown for simplicity. See the text for details.
Wagner et al. previously described a collection of temperature-sensitive smk1 hypomorphs that block meiotic development at different stages in spore morphogenesis. In that study, increases in smk1 allelic dosage were shown to promote stepwise advancement of the spore morphogenesis program (19). For example, at a semipermissive temperature (27.5°C), incorporation of dityrosine into insoluble material was nearly undetectable in cells containing 1 copy of smk1-4, while cells containing 2 copies of smk1-4 incorporated wild-type levels of dityrosine. Moreover, a 2-copy smk1-4 strain produced <5% refractile spores, while a 3-copy smk1-4 strain produced >90% refractile spores. The switch-like transitions caused by changes in smk1 allelic dosage suggest that quantitative changes in Smk1 MAPK activity play a role in coordinating spore morphogenesis. The phenotypic analyses of the ssp2Δ and smk1 phosphosite mutant phenotypes reported in this study are consistent with the Wagner et al. report in several respects. First, the ssp2Δ/ssp2Δ and smk1-Y209F/smk1Δ sporulation phenotypes (which are indistinguishable) resemble the smk1-4/smk1Δ semipermissive-temperature phenotype (all of these mutants produce background levels of refractile spores that are fluorescent negative and contain inner spore but not outer spore wall layers as assayed by electron microscopy). Second, similar to smk1-4, small increases in smk1-Y209F dosage dramatically increased the fraction of cells that formed refractile spores (a 1-copy smk1-Y209F strain produces <2% refractile spores, and a 2-copy strain produces 65%). These observations are consistent with the phosphorylation of T207 partially activating Smk1 and with the quantitative model for Smk1 phosphoregulation depicted in Fig. 8.
While our genetic data indicate that Smk1-Y209F is partially active, a previously published study reported that Smk1-Y209F protein isolated from meiotic cells, in contrast to wild-type Smk1, showed only background levels of catalytic activity against a mammalian MAPK (Erk1/2) substrate (Elk1) in vitro (16). One explanation for the apparent discrepancy between the genetic and biochemical experiments is that the phosphorylation of Smk1 on T207 increases Smk1 catalytic activity only modestly. Indeed, Pierce et al. showed that one can reduce SMK1 expression by more than 90% without causing detectable sporulation defects (18). These observations raise the possibility that a relatively low level of Smk1 catalytic activity (which might have been missed in the biochemical assay) can have significant effects on spore morphogenesis. Alternatively, the phosphorylation of T207 might increase the activity of Smk1 substantially, but only for a restricted subset of phosphoconsensus sites that are poorly represented in the apparently high-Km surrogate substrate used in the in vitro study. Enzymatic analyses of purified Smk1 that is singly and doubly phosphorylated are needed to establish whether the low/high T-X-Y activity toggle hypothesis illustrated in Fig. 8 is accompanied by significant changes in substrate specificity.
A key finding in this study is that Y209 phosphorylation requires SSP2. However, our attempts to influence the phosphorylation of Smk1 by ectopically expressing the SSP2 open reading frame (using the GAL1 promoter) in cells harboring the mseΔ-SMK1-HA plasmid have been unsuccessful. Although there are many possible explanations for these negative data, they do raise the possibility that additional regulatory features that are present only in meiotic cells promote the SSP2-dependent phosphorylation of Smk1. AMA1 and IME2 have been shown to promote Smk1 activation (20, 37), and the role of these meiosis-specific genes in controlling the SSP2-dependent activation of Smk1 requires further investigation.
Although SSP2 mRNA is present in cells that are undergoing the meiotic divisions, Ssp2 protein is not translated until cells are completing MII (compare the kinetics of MII with the Smk1 phosphorylation pattern in Fig. 7A). These observations are consistent with the ribosome profiling experiments of Brar et al., which showed that SSP2 translational efficiency (ribosomal occupancy/kb of mRNA) increases by about 3 orders of magnitude around the time that cells are completing MII (63). Interestingly, the earliest detectable forms of Ssp2 are electrophoretically distinguishable from the later forms of the protein, and the later Ssp2 electrophoretic forms temporally correlate with the appearance of dually phosphorylated Smk1 (Fig. 7A and C). These observations suggest that regulatory interactions operating at the level of Ssp2 translation and modification of the Ssp2 protein control the Smk1 autophosphorylation reaction. We speculate that these regulatory interactions couple the autocatalytic Y209 reaction to exit from MII.
Smk1 produced in vegetative cells using the mseΔ promoter system is unable to be modified on Y209 when these cells are transferred to sporulation medium. However, the vegetatively produced Smk1 in these experiments is phosphorylated on T207, suggesting that it is appropriately folded. There are several possible explanations for the inability of Ssp2 to promote the Y209 reaction on a preexisting pool of Smk1, including sequestration of Smk1 that is translated in vegetative cells in a compartment incompatible with the reaction, or inactivation of the vegetatively produced Smk1 by a modification that we have not detected. Nevertheless, our favored class of model to explain these data is that Ssp2 promotes the intramolecular autophosphorylation of a transitional intermediate form of Smk1.
Two different families of S/T kinases have been shown to autophosphorylate activation loop residues via reactions that require transitory intermediates. The best-characterized example is the dual-specificity tyrosine phosphorylation-regulated protein kinase (DYRK) from Drosophila, which has been shown to autophosphorylate a Y residue in its activation loop as it is being translated (65, 66). Once DYRK is released from the ribosome, it is unable to carry out this reaction. Another example is GSK3β, which autophosphorylates a Y residue in an intramolecular reaction that requires the Hsp90 chaperone (67). Similar to the DYRKs, mature GSK3β is unable to carry out this activating reaction. We suggest that the autophosphorylation of Smk1 on Y209 occurs either as the protein is emerging from the ribosome (cotranslationally, as is seen with DYRK) or shortly after it is released from the ribosome (during folding of the Smk1 protein, as is seen with GSK3β). In this class of model, the inability of folded Smk1 produced in vegetative cells to autophosphorylate on Y209 would be due to structural barriers in the mature protein that prevent it from adopting the transitional intermediate structure.
How does Ssp2 promote the intramolecular transfer of phosphate to Y209? One possibility is that Ssp2 directly interacts with Smk1 to stabilize the hypothesized Smk1 intermediate or to force it into a structure that promotes autophosphorylation prior to the final stages of protein folding. In the case of class 2 DYRKs, a conserved region from the amino terminus (termed the NAPA domain), is required for autophosphorylation of the activating Y residue, and this domain can complement the cotranslational autophosphorylation defect of an amino-terminally truncated DYRK1 mutant in trans (68). Although protein segments resembling the DYRK NAPA domain are not present in Ssp2, it is conceivable that regions of Ssp2 function in a NAPA-like fashion. Another class of model to explain the role of SSP2 in promoting Smk1 autophosphorylation is that Ssp2 stalls protein translation when the hypothesized Smk1 intermediate has emerged from the ribosome, thus increasing transitional-form dwell time. However, translational stalling is rare in eukaryotes and stalled ribosomes on SMK1 mRNA were not detected in ribosome profiling experiments (63). Another possibility is that Ssp2 promotes Smk1 autophosphorylation by directing Smk1 translation to a subcellular location where a protein with a NAPA-like function resides or by influencing NAPA-like activities associated with ribosomes. Further work is required to elucidate how Ssp2 promotes Smk1 autophosphorylation. It will be interesting to learn whether related pathways activate MAPK autophosphorylation during developmental programs in animals.
ACKNOWLEDGMENTS
We thank Kirsten Benjamin, Philip Kaldis, Linda Huang, Dilip Nag, Jacqueline Segal, and Ghadeer Shubassi for generously providing biochemical reagents, antibodies, plasmids, and yeast strains. We thank Sham Sunder and Daniel Corbi for comments on the manuscript.
This work was supported by a grant from the National Science Foundation (MCB-0950009).
Footnotes
Published ahead of print 3 December 2012
REFERENCES
- 1. Chen RE, Thorner J. 2007. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1773:1311–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Raman M, Chen W, Cobb MH. 2007. Differential regulation and properties of MAPKs. Oncogene 26:3100–3112 [DOI] [PubMed] [Google Scholar]
- 3. Ferrell JE, Jr, Machleder EM. 1998. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science 280:895–898 [DOI] [PubMed] [Google Scholar]
- 4. Coulombe P, Meloche S. 2007. Atypical mitogen-activated protein kinases: structure, regulation and functions. Biochim. Biophys. Acta 1773:1376–1387 [DOI] [PubMed] [Google Scholar]
- 5. Pimienta G, Pascual J. 2007. Canonical and alternative MAPK signaling. Cell Cycle 6:2628–2632 [DOI] [PubMed] [Google Scholar]
- 6. Salvador JM, Mittelstadt PR, Guszczynski T, Copeland TD, Yamaguchi H, Appella E, Fornace AJ, Jr, Ashwell JD. 2005. Alternative p38 activation pathway mediated by T cell receptor-proximal tyrosine kinases. Nat. Immunol. 6:390–395 [DOI] [PubMed] [Google Scholar]
- 7. Ge B, Gram H, Di Padova F, Huang B, New L, Ulevitch RJ, Luo Y, Han J. 2002. MAPKK-independent activation of p38alpha mediated by TAB1-dependent autophosphorylation of p38alpha. Science 295:1291–1294 [DOI] [PubMed] [Google Scholar]
- 8. Abe MK, Kahle KT, Saelzler MP, Orth K, Dixon JE, Rosner MR. 2001. ERK7 is an autoactivated member of the MAPK family. J. Biol. Chem. 276:21272–21279 [DOI] [PubMed] [Google Scholar]
- 9. Klevernic IV, Stafford MJ, Morrice N, Peggie M, Morton S, Cohen P. 2006. Characterization of the reversible phosphorylation and activation of ERK8. Biochem. J. 394:365–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Crews CM, Alessandrini AA, Erikson RL. 1991. Mouse Erk-1 gene product is a serine/threonine protein kinase that has the potential to phosphorylate tyrosine. Proc. Natl. Acad. Sci. U. S. A. 88:8845–8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rossomando AJ, Wu J, Michel H, Shabanowitz J, Hunt DF, Weber MJ, Sturgill TW. 1992. Identification of Tyr-185 as the site of tyrosine autophosphorylation of recombinant mitogen-activated protein kinase p42mapk. Proc. Natl. Acad. Sci. U. S. A. 89:5779–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seger R, Ahn NG, Boulton TG, Yancopoulos GD, Panayotatos N, Radziejewska E, Ericsson L, Bratlien RL, Cobb MH, Krebs EG. 1991. Microtubule-associated protein 2 kinases, ERK1 and ERK2, undergo autophosphorylation on both tyrosine and threonine residues: implications for their mechanism of activation. Proc. Natl. Acad. Sci. U. S. A. 88:6142–6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu J, Rossomando AJ, Her JH, Del Vecchio R, Weber MJ, Sturgill TW. 1991. Autophosphorylation in vitro of recombinant 42-kilodalton mitogen-activated protein kinase on tyrosine. Proc. Natl. Acad. Sci. U. S. A. 88:9508–9512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bhattacharyya RP, Remenyi A, Good MC, Bashor CJ, Falick AM, Lim WA. 2006. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science 311:822–826 [DOI] [PubMed] [Google Scholar]
- 15. Krisak L, Strich R, Winters RS, Hall JP, Mallory MJ, Kreitzer D, Tuan RS, Winter E. 1994. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev. 8:2151–2161 [DOI] [PubMed] [Google Scholar]
- 16. Huang LS, Doherty HK, Herskowitz I. 2005. The Smk1p MAP kinase negatively regulates Gsc2p, a 1,3-beta-glucan synthase, during spore wall morphogenesis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 102:12431–12436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schaber M, Lindgren A, Schindler K, Bungard D, Kaldis P, Winter E. 2002. CAK1 promotes meiosis and spore formation in Saccharomyces cerevisiae in a CDC28-independent fashion. Mol. Cell. Biol. 22:57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pierce M, Wagner M, Xie J, Gailus-Durner V, Six J, Vershon AK, Winter E. 1998. Transcriptional regulation of the SMK1 mitogen-activated protein kinase gene during meiotic development in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5970–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wagner M, Briza P, Pierce M, Winter E. 1999. Distinct steps in yeast spore morphogenesis require distinct SMK1 MAP kinase thresholds. Genetics 151:1327–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McDonald CM, Cooper KF, Winter E. 2005. The Ama1-directed APC/C ubiquitin ligase regulates the Smk1 MAPK during meiosis in yeast. Genetics 171:901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wagner M, Pierce M, Winter E. 1997. The CDK-activating kinase CAK1 can dosage suppress sporulation defects of smk1 MAP kinase mutants and is required for spore wall morphogenesis in Saccharomyces cerevisiae. EMBO J. 16:1305–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Espinoza FH, Farrell A, Erdjumentbromage H, Tempst P, Morgan DO. 1996. A cyclin-dependent kinase-activating kinase (Cak) in budding yeast unrelated to vertebrate Cak. Science 273:1714–1717 [DOI] [PubMed] [Google Scholar]
- 23. Kaldis P, Sutton A, Solomon MJ. 1996. The cdk-activating kinase (CAK) from budding yeast. Cell 86:553–564 [DOI] [PubMed] [Google Scholar]
- 24. Thuret J-V, Valay J-G, Faye G, Mann C. 1996. Civ1 (CAK in vivo), a novel Cdk-activating kinase. Cell 86:565–576 [DOI] [PubMed] [Google Scholar]
- 25. Cross FR, Levine K. 1998. Molecular evolution allows bypass of the requirement for activation loop phosphorylation of the Cdc28 cyclin-dependent kinase. Mol. Cell. Biol. 18:2923–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schindler K, Benjamin KR, Martin A, Boglioli A, Herskowitz I, Winter E. 2003. The Cdk-activating kinase Cak1p promotes meiotic S phase through Ime2p. Mol. Cell. Biol. 23:8718–8728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Honigberg SM. 2004. Ime2p and Cdc28p: co-pilots driving meiotic development. J. Cell. Biochem. 92:1025–1033 [DOI] [PubMed] [Google Scholar]
- 28. Irniger S. 2011. The Ime2 protein kinase family in fungi: more duties than just meiosis. Mol. Microbiol. 80:1–13 [DOI] [PubMed] [Google Scholar]
- 29. Coluccio A, Bogengruber E, Conrad MN, Dresser ME, Briza P, Neiman AM. 2004. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell 3:1464–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li J, Agarwal S, Roeder GS. 2007. SSP2 and OSW1, two sporulation-specific genes involved in spore morphogenesis in Saccharomyces cerevisiae. Genetics 175:143–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sarkar PK, Florczyk MA, McDonough KA, Nag DK. 2002. SSP2, a sporulation-specific gene necessary for outer spore wall assembly in the yeast Saccharomyces cerevisiae. Mol. Genet. Genomics 267:348–358 [DOI] [PubMed] [Google Scholar]
- 32. Cooper KF, Mallory MJ, Egeland DB, Jarnik M, Strich R. 2000. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc. Natl. Acad. Sci. U. S. A. 97:14548–14553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Diamond AE, Park JS, Inoue I, Tachikawa H, Neiman AM. 2009. The anaphase promoting complex targeting subunit Ama1 links meiotic exit to cytokinesis during sporulation in Saccharomyces cerevisiae. Mol. Biol. Cell 20:134–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oelschlaegel T, Schwickart M, Matos J, Bogdanova A, Camasses A, Havlis J, Shevchenko A, Zachariae W. 2005. The yeast APC/C subunit Mnd2 prevents premature sister chromatid separation triggered by the meiosis-specific APC/C-Ama1. Cell 120:773–788 [DOI] [PubMed] [Google Scholar]
- 35. Okaz E, Arguello-Miranda O, Bogdanova A, Vinod PK, Lipp JJ, Markova Z, Zagoriy I, Novak B, Zachariae W. 2012. Meiotic prophase requires proteolysis of M phase regulators mediated by the meiosis-specific APC/C(Ama1). Cell 151:603–618 [DOI] [PubMed] [Google Scholar]
- 36. Penkner AM, Prinz S, Ferscha S, Klein F. 2005. Mnd2, an essential antagonist of the anaphase-promoting complex during meiotic prophase. Cell 120:789–801 [DOI] [PubMed] [Google Scholar]
- 37. McDonald CM, Wagner M, Dunham MJ, Shin ME, Ahmed NT, Winter E. 2009. The Ras/cAMP pathway and the CDK-like kinase Ime2 regulate the MAPK Smk1 and spore morphogenesis in Saccharomyces cerevisiae. Genetics 181:511–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Neiman AM. 2011. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189:737–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Winter E. 2012. The Sum1/Ndt80 transcriptional switch and commitment to meiosis in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 76:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pierce M, Benjamin KR, Montano SP, Georgiadis MM, Winter E, Vershon AK. 2003. Sum1 and Ndt80 proteins compete for binding to middle sporulation element sequences that control meiotic gene expression. Mol. Cell. Biol. 23:4814–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ahmed NT, Bungard D, Shin ME, Moore M, Winter E. 2009. The Ime2 CDK-like kinase enhances the disassociation of the Sum1 repressor from middle meiotic promoters. Mol. Cell. Biol. 29:4352–4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shin ME, Skokotas A, Winter E. 2010. The Cdk1 and Ime2 protein kinases trigger exit from meiotic prophase in Saccharomyces cerevisiae by inhibiting the Sum1 transcriptional repressor. Mol. Cell. Biol. 30:2996–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pak J, Segall J. 2002. Regulation of the premiddle and middle phases of expression of the NDT80 gene during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 22:6417–6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xie J, Pierce M, Gailus-Durner V, Wagner M, Winter E, Vershon AK. 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18:6448–6454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chu S, Herskowitz I. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1:685–696 [DOI] [PubMed] [Google Scholar]
- 46. Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699–705 [DOI] [PubMed] [Google Scholar]
- 47. Kaldis P, Pitluk ZW, Bany IA, Enke DA, Wagner M, Winter E, Solomon MJ. 1998. Localization and regulation of the cdk-activating kinase (Cak1p) from budding yeast. J. Cell Sci. 111:3585–3596 [DOI] [PubMed] [Google Scholar]
- 48. Benjamin KR, Zhang C, Shokat KM, Herskowitz I. 2003. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 17:1524–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen XL, Reindle A, Johnson ES. 2005. Misregulation of 2 micron circle copy number in a SUMO pathway mutant. Mol. Cell. Biol. 25:4311–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961 [DOI] [PubMed] [Google Scholar]
- 51. Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hill JE, Myers AM, Koerner TJ, Tzagoloff A. 1986. Yeast E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163–167 [DOI] [PubMed] [Google Scholar]
- 53. Carlile TM, Amon A. 2008. Meiosis I is established through division-specific translational control of a cyclin. Cell 133:280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Briza P, Winkler G, Kalchhauser H, Breitenbach M. 1986. Dityrosine is a prominent component of the yeast ascospore wall. A proof of its structure. J. Biol. Chem. 261:4288–4294 [PubMed] [Google Scholar]
- 55. Esposito RE, Dresser M, Breitenbach M. 1991. Identifying sporulation genes, visualizing synaptonemal complexes, and large-scale spore and spore wall purification. Methods Enzymol. 194:110–131 [DOI] [PubMed] [Google Scholar]
- 56. Schindler K, Winter E. 2006. Phosphorylation of Ime2 regulates meiotic progression in Saccharomyces cerevisiae. J. Biol. Chem. 281:18307–18316 [DOI] [PubMed] [Google Scholar]
- 57. Nickas ME, Diamond AE, Yang MJ, Neiman AM. 2004. Regulation of spindle pole function by an intermediary metabolite. Mol. Biol. Cell 15:2606–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lindgren A, Bungard D, Pierce M, Xie J, Vershon A, Winter E. 2000. The pachytene checkpoint in Saccharomyces cerevisiae requires the Sum1 transcriptional repressor. EMBO J. 19:6489–6497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623–627 [DOI] [PubMed] [Google Scholar]
- 60. Shubassi G, Luca N, Pak J, Segall J. 2003. Activity of phosphoforms and truncated versions of Ndt80, a checkpoint-regulated sporulation-specific transcription factor of Saccharomyces cerevisiae. Mol. Genet. Genomics 270:324–336 [DOI] [PubMed] [Google Scholar]
- 61. Sopko R, Raithatha S, Stuart D. 2002. Phosphorylation and maximal activity of Saccharomyces cerevisiae meiosis-specific transcription factor Ndt80 is dependent on Ime2. Mol. Cell. Biol. 22:7024–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tung KS, Hong EJ, Roeder GS. 2000. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc. Natl. Acad. Sci. U. S. A. 97:12187–12192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. 2012. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335:552–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kaldis P. 1999. The cdk-activating kinase (CAK): from yeast to mammals. Cell. Mol. Life Sci. 55:284–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Han J, Miranda-Saavedra D, Luebbering N, Singh A, Sibbet G, Ferguson MA, Cleghon V. 2012. Deep evolutionary conservation of an intramolecular protein kinase activation mechanism. PLoS One 7:e29702 doi:10.1371/journal.pone.0029702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lochhead PA, Sibbet G, Morrice N, Cleghon V. 2005. Activation-loop autophosphorylation is mediated by a novel transitional intermediate form of DYRKs. Cell 121:925–936 [DOI] [PubMed] [Google Scholar]
- 67. Lochhead PA, Kinstrie R, Sibbet G, Rawjee T, Morrice N, Cleghon V. 2006. A chaperone-dependent GSK3β transitional intermediate mediates activation-loop autophosphorylation. Mol. Cell 24:627–633 [DOI] [PubMed] [Google Scholar]
- 68. Kinstrie R, Luebbering N, Miranda-Saavedra D, Sibbet G, Han J, Lochhead PA, Cleghon V. 2010. Characterization of a domain that transiently converts class 2 DYRKs into intramolecular tyrosine kinases. Sci. Signal. 3:ra16 doi:10.1126/scisignal.2000579 [DOI] [PubMed] [Google Scholar]