Abstract
Polycomb-group (PcG) complex 1 acts as an E3 ubiquitin ligase both for histone H2A to silence transcription and for geminin to regulate its stability. Scmh1 is a substoichiometric component of PcG complex 1 that provides the complex with an interaction domain for geminin. Scmh1 is unstable and regulated through the ubiquitin-proteasome system, but its molecular roles are unknown, so we generated Scmh1-deficient mice to elucidate its function. Loss of Scmh1 caused derepression of Hoxb4 and Hoxa9, direct targets of PcG complex 1-mediated transcriptional silencing in hematopoietic cells. Double knockdown of Hoxb4 and Hoxa9 or transduction of a dominant-negative Hoxb4N→A mutant caused geminin accumulation. Age-related transcriptional downregulation of derepressed Hoxa9 also leads to geminin accumulation. Transduction of Scmh1 lacking a geminin-binding domain restored derepressed expression of Hoxb4 and Hoxa9 but did not downregulate geminin like full-length Scmh1. Each of Hoxb4 and Hoxa9 can form a complex with Roc1-Ddb1-Cul4a to act as an E3 ubiquitin ligase for geminin. We suggest that geminin dysregulation may be restored by derepressed Hoxb4 and Hoxa9 in Scmh1-deficient mice. These findings suggest that PcG and a subset of Hox genes compose a homeostatic regulatory system for determining expression level of geminin.
INTRODUCTION
Polycomb-group (PcG) proteins are subunits of PcG complex 1 and 2 (also designated as Polycomb repressive complexes 1 and 2, respectively) (1, 2) and other complexes and regulate the transcription of developmental regulators, including Hox genes. The hierarchical recruitment model (3) proposes that PcG complex 2 is recruited first to target loci and methylates histone H3 at lysine 27 (H3K27). PcG complex 1 is then recruited through the recognition of methylated H3K27 by the chromodomain of Cbx family members (Cbx2, Cbx4, Cbx6, Cbx7, and Cbx8) and induces mono-ubiquitination of histone H2A at lysine 119, which silences transcription (4–6). However, recruitment can occur independently of trimethylated H3K27 through molecular targeting of RYBP-PcG complex 1 (7).
Scmh1 is a mammalian homologue of the Drosophila Sex comb on midleg (Scm) gene (8). Scm and its homolog associate substoichiometrically with PcG complex 1 (9). We previously demonstrated that Scmh1 mediates a molecular interaction of PcG complex 1 with geminin (10) and that PcG complex 1 acts as an E3 ubiquitin ligase for geminin to sustain the hematopoietic stem cell (HSC) activity (11, 12).
Scmh1 encodes a protein with several characteristic domains, including the malignant brain tumor (MBT) domains, the proline-, glutamine-, serine-, and threonine-rich (PEST) domains, the N-terminal and C-terminal putative nuclear localization signals, and the Scm-polyhomeotic-l(3)mbt (SPM) domain (8, 13), also designated as the SAM domain (14). The C-terminal putative nuclear localization signal domain acts also as an interaction domain for geminin (the geminin-binding [GB] domain) (10). The MBT domains of Drosophila Scm directly interact with monomethylated H3K4, H3K9, H3K27, H3K36, and H4K20 (15). The SPM domain is conserved between Scmh1 and Rae28 (also designated Phc1), a mouse homologue of polyhomeotic, which is a member of PcG complex 1 (8). The SPM domain of Scmh1 mediates either homophilic or heterophilic molecular interaction with Rae28 (8). Molecular roles for these domains in Scmh1, however, remain insufficiently understood. In mutant mice lacking the SPM domain of Scmh1, skeletal abnormalities and male infertility were observed (16).
DNA replication licensing occurs at late M and G1 phases and may also be involved in G0-to-G1 transition (17). Geminin prevents rereplication from S phase to early M phase to ensure one round of DNA replication in a single cell cycle. Geminin forms a Cdt1-geminin complex that regulates Cdt1, which initiates DNA replication licensing (17, 18). Geminin inhibits (18) and stabilizes (19) Cdt1. The stoichiometry of the Cdt1-geminin complex controls regulation of DNA replication licensing (20).
Geminin also inhibits the chromatin remodeling factors Brahma and Brg1 to maintain an undifferentiated state (21) and acts as a transcription repressor or corepressor (10, 22). Geminin is required for maintaining pluripotency (23, 24). Thus, geminin may be a central regulator governing cellular proliferation and differentiation.
As we previously reported, either Hoxa9 or Hoxb4 can form a RDCOX complex with Roc1(Rbx1)-Ddb1-Cul4a, an E3 ubiquitin ligase core component (25, 26, 51). This Hox-containing complex downregulates geminin through the ubiquitin-proteasome system (UPS) (27) to enhance hematopoietic stem and progenitor activities. In Rae28-deficient mice, we observed geminin accumulation and resultant hematopoietic dysfunction due to defective activity of the PcG complex 1 E3 ubiquitin ligase activity for geminin (12). Therefore, there are at least two independent E3 ubiquitin ligase activities targeting geminin. In addition, the anaphase-promoting complex/cyclosome (APC/C) gives rise to the oscillating expression pattern of geminin in the cell cycle (28), but the role and relationship of these ubiquitin ligase activities has not been studied.
We show here that gross phenotypes of Scmh1-null (Scmh1−/−) mutants are relatively mild. An unexpected cell cycle-dependent association of Scmh1 with PcG bodies suggests an underlying reason for substoichiometric localization of Scmh1 with PcG complex 1 and supports a role for Scmh1 in cell cycle regulation. Interestingly, Scmh1 mutants result in decrease of geminin protein levels, which is surprising if Scmh1 contributes to the E3 ligase activity of the PcG complex 1 on geminin. The resolution to this paradox may be that derepression of Hoxa9 and Hoxb4 leads to increased activity of the RDCOX E3 ligase, which also targets geminin. We suggest that PcG complex 1 and some Hox genes provide a ubiquitin-mediated homeostatic regulatory system to control geminin levels.
MATERIALS AND METHODS
Generation of Scmh1-deficient mice.
Scmh1 genomic DNA was isolated from a 129/Sv mouse liver genomic library. We subcloned 6.0-kb XbaI-NheI and 3.7-kb NheI-NarI fragments into pBluescript and inserted them into the gene targeting vector which contains the pMC1 promoter-driven neomycin resistance (Neor) and diphtheria toxin A (DT) genes. 129/Sv RW4 embryonic stem (ES) cells (Incyte Genomics, Palo Alto, CA) were cultured on STO feeder cells (29). The targeting vector (50 μg) was separated from the plasmid vector by digesting with BstXI and was electroporated into ES cells using a BTX Electro cell manipulator 600 (Harvard Apparatus, Holliston, MA) set at 270 V and 500 μF. ES cells were plated onto G418-resistent feeder cells and were followed by selection with 175 μg of G418/ml. After 7 to 10 days, G418-resistant colonies were picked up. High-molecular-weight DNAs were isolated from the clones and subjected to Southern blot analysis. The blots were hybridized with either XbaI-BamHI 1.7-kb probe (see Fig. 2B) or XbaI-KpnI 0.5-kb probe (see Fig. 2B). Two independent ES clones with targeted disruption in Scmh1 were used to generate chimeric mice by injection of C57BL/6 blastocysts with 10 to 20 ES cells. The chimeric mice were mated with BDF1 mice and the offspring were examined for the presence of the targeted Scmh1 allele by Southern blotting. Scmh1 mutant mice were further genotyped by direct PCR with tail samples, Ampdirect plus Set (Shimadzu, Kyoto, Japan) and the following oligonucleotides: a, 5′-GCCTCTAGTCTCAAGGTAATTATTATAC-3′; b, 5′-GAACACACCATTATTCTTCAAGTTCTCA-3′; and c, 5′-GAGCTTGGCGGCGAATGGGCTGACCGC-3′. The PCR conditions consisted of 1 cycle of 95°C for 10 min (predenaturing), followed by 40 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min, followed in turn by 1 cycle of 72°C for 7 min (extension). Scmh1-deficient mice with the C57BL/6 genetic background were generated and were subjected to further analysis.
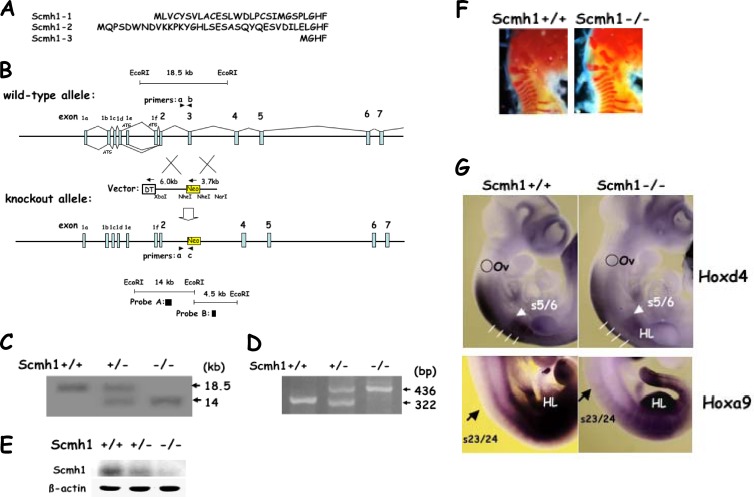
Fig 2.
Generation of Scmh1-deficient mice. (A) Amino acid sequences predicted by 5′ part of the Scmh1 cDNAs. Scmh1-1, the amino acid sequence predicted by the cDNA, which we originally reported (8). Accession numbers for the cDNA database: Scmh1-1, AB030906; Scmh1-2, AB646299; and Scmh1-3, AB646300. (B) Strategy for generating Scmh1-deficient mice. Direction of DT and Neo transcription is indicated by arrows. Either wild-type or knockout alleles were detected by Southern blot analysis with probe A or B and by PCR analysis with indicated sets of the primers. The N-terminal Scmh1-1 amino acid sequence is encoded by exon 1f, that of Scmh1-2 is encoded by exons 1b, 1c, and 1d, and that of Scmh1-3 is encoded by exon 1e, respectively. (C) Detection of the knockout allele by Southern blotting. Genomic DNA was digested with EcoRI and was subjected to Southern blot analysis with probe A. A 18.5-kb band corresponded to a wild-type allele, while 14-kb one to a knockout allele. (D) Detection of the knockout allele by PCR analysis. PCR analysis with primer set a and b detected the wild-type allele (322 bp), while that with primer set a and c detected the knockout allele (436 bp). (E) Immunoblot analysis of the testes with an anti-Scmh1 antibody. (F) Lateral views of the occipital, cervical, and thoracic regions of the skeleton. No obvious abnormality was found in the skeletons of 10 Scmh1−/− mice. Representative skeletal findings of 17.5-dpc embryos are shown. (G) Expression of Hoxd4 and Hoxa9 in embryos. Whole-mount in situ hybridization analysis of 10-dpc embryos used either Hoxd4 or Hoxa9 as the probe. Lateral views are shown. s5/6, between the 5th and 6th somites of the paraxial mesoderm; s23/24, between the 23rd and 24th somites. The anterior expression boundaries of Hoxd4 and Hoxa9 in the paraxial mesoderm were detected between the 5th and 6th somites and between the 23rd and 24th somites, respectively. HL, hind limb; Ov, otic vesicle.
Skeletal analysis.
Embryos at 17.5 days postcoitus (dpc) were fixed in Bouin's solution for 24 h. After removal of the skin, muscle, and viscera, the embryos were dehydrated in 96% ethanol and transferred to acetone for 2 days. The samples were stained in 0.001% alizarin red S and 0.003% Alcian blue in 1% acid-alcohol solution for 6 h at 37°C and, after being washed in distilled water, the samples were subjected to the clearing steps. Cleared skeletons were stored in 100% glycerol (30).
Whole-mount in situ hybridization.
Embryos were fixed in phosphate buffered-saline (PBS) containing 4% paraformaldehyde, bleached, and treated with 10 μg of proteinase K (Sigma-Aldrich, St. Louis, MO)/ml. After additional fixation in 0.2% glutaraldehyde and 4% paraformaldehyde in PBS, the embryos were soaked in prewarmed prehybridization buffer for 1 h at 70°C and hybridized overnight with a digoxigenin-labeled Hox riboprobe. Hybridization was detected by treatment of embryos with a preabsorbed alkaline phosphatase-conjugated anti-digoxigenin antibody (Roche Diagnostics GmbH, Mannheim, Germany), followed by treatment with 4-nitroblue tetrazolium chloride and BCIP (5-bromo-4-chloro-3-indolylphosphate; Roche Diagnostics GmbH). The samples were investigated under a stereoscopic microscope (31).
Real-time PCR.
Total cellular RNA was extracted from cells by using a Quick-RNA MicroPrep kit (ZYMO Research, Orange, CA) and reverse transcribed using TaqMan reverse transcription reagents (Life Technologies, Carlsbad, LA), and the product was subjected to real-time quantitative PCR analysis using TaqMan gene expression assays and an ABI 7500 real-time PCR system (Life Technologies). The relative expression levels for the specific transcripts were detected by normalization with transcripts for GAPDH (glyceraldehyde-3-phosphate dehydrogenase).
Indirect immunofluorescence labeling.
Cells were fixed in 3% paraformaldehyde PBS for 10 min, permeabilized with 0.5% NP-40 PBS for 10 min, and stained with primary and fluorescence-labeled secondary antibodies and further concurrently with Hoechst 33258 (1 μg/ml; Calbiochem, La Jolla, CA). Images were captured using an epifluorescence optics equipped with a charge-coupled device camera (BX60 and DP70; Olympus, Tokyo, Japan) or an inverted confocal laser scanning LSM5 Pascal microscope (Carl Zeiss, Oberkochen, Germany). The cells were synchronized by treatment with roscovitine (100 μM), hydroxyurea (5 mM), colchicine (1 μg/ml; Calbiochem), aphidicolin (5 μg/ml; Wako Pure Chemical, Osaka, Japan), or thymidine (2.5 mM; Sigma-Aldrich) for 24 h.
Hematopoiesis and cell cycle analyses.
Clonogenic activity was assayed as follows. The fetal liver (FL) cells were cultured in Dulbecco modified Eagle medium (DMEM; Life Technologies) supplemented with 15% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA), 100 ng of mouse stem cell factor (SCF)/ml, 100 ng of human thrombopoietin (TPO)/ml, and 100 ng of mouse Flt3 ligand (R&D Systems, Minneapolis, MN)/ml for 24 h. The cells were then infected with retroviruses. After being washed in minimal essential medium alpha medium (Life Technologies), 104 cells were resuspended in methylcellulose semisolid medium (Methocult M3231; Stem Cell Technologies, Vancouver, British Columbia, Canada) and plated in 35-mm culture dishes in the presence of 10 ng of mouse SCF/ml, 10 ng of mouse granulocyte-macrophage colony-stimulating factor/ml, 10 ng of mouse interleukin-3 (R&D Systems)/ml, and 3 U of human erythropoietin (EPO) (Chugai Pharmaceutical, Tokyo, Japan)/ml. Colonies were counted after 7 to 9 days of culture under an inverted microscope. The long-term-culture-initiating cell (LTC-IC) activity was examined by detecting myeloid CFU in culture (CFU-Cs) in FL cells, which were cultured on an S17 feeder layer in the myeloid long-term culture medium (Myelocult M5300; Stem Cell Technologies) containing 10−6 M hydrocortisone sodium hemisuccinate (Sigma-Aldrich), changing half of the medium every week for 6 weeks. Long-term repopulating (LTR) activity was assayed by injecting 106 retrovirally transduced FL cells and 2 × 105 competitors into C57BL/6 congenic mice lethally irradiated with 9.0 Gy administered in a single dose from a 60Co gamma ray source. Enhanced yellow fluorescent protein-positive (EYFP+) multilineage cells were examined 1 and 4 months after the transplantation.
Cell cycle analysis was performed with the APC BrdU flow kit (BD Pharmingen, San Diego, CA). In vitro labeling with bromodeoxyuridine (BrdU) was performed at a final concentration of 10 μM for 45 min, and in vivo labeling was performed as follows. BrdU (5 mg) was intraperitoneally injected into mice. At 20 h after the injection, mice were sacrificed, and bone marrow (BM) cells were subjected to further analyses. Geminin protein expression in each phase of the cell cycle was detected by additional immunostaining with a rabbit polyclonal antibody raised against glutathione S-transferase (GST)–geminin. Cell sorting analysis was performed on the FACSCalibur flow cytometer and FACSAria II cell sorter (BD Biosciences Immunocytometry Systems, San Jose, CA). More than three independent experiments were performed, and the data were subjected to statistical analyses (11, 12, 25).
DNA transfection.
cDNAs or Flag-tagged cDNAs were subcloned into the downstream of the cytomegalovirus promoter of pcDNA3.1 expression vector (Life Technologies). HEK-293 or U-2 OS cells were grown in DMEM supplemented with 10% FBS. The plasmid DNAs were transfected by the calcium phosphate coprecipitation method (25), and the resultant transfectants were subjected to further analyses.
siRNA transfection.
Freshly prepared mouse BM were cultured in DMEM supplemented with 15% FBS, 100 ng of mouse SCF/ml, 100 ng of human TPO/ml, and 100 ng of mouse Flt3 ligand/ml for 24 h. Cells (5 × 105) were harvested and resuspended in 1 ml of Accell small interfering RNA (siRNA) delivery media (Thermo Fisher Scientific) supplemented with 100 ng of mouse SCF/ml, 100 ng of human TPO/ml, and 100 ng of mouse Flt3 ligand/ml and cultured with 0.5 mM (each) Accell SMART pool of four double-stranded siRNAs for mouse Hoxa9 (CGCUUGACACUCACACUUU, UGACUAUGCUUGUGGUUCU, CUUGCAGCUUCCAGUCCAA, and UGGGCAACUACUAUGUGGA) and for mouse Hoxb4 (GCAUCGUUUAUUAUUAUUA, CCAUCUGUCUUGUUUACUC, CUCUUAGGUUACAAAUGAA, and CCAGAUAGCACAGUGUUUU) or the negative control (Accell nontargeting pool [Thermo Fisher Scientific]) for 72 h. The cells were then subjected to further analysis. The efficiency of the cotransfection was monitored by using fluorescent dye-labeled nontargeting siRNA (Accell green or red nontargeting siRNA; Thermo Fisher Scientific) as indicators.
Retrovirus-mediated gene transduction.
The murine stem cell virus vector with the EYFP gene driven by the pgk promoter as a selection marker (MEP) was cotransfected with Gag, Pol, and vesicular stomatitis virus glycoprotein (VSV-G) envelope expression plasmids into HEK-293 cells with Lipofectamine 2000 (Life Technologies). The ecotropic packaging cells line, PlatE, was infected 3 to 10 times with a virus, and the supernatants were concentrated by centrifugation at 6,000 × g for 16 h to produce a high-titer helper-free retrovirus. FL cells were extracted from 15.5-dpc embryos and cultured for 48 h in DMEM supplemented with 15% FBS and three cytokines (100 ng of mouse SCF/ml, 100 ng of human TPO/ml, 100 ng of mouse Flt3 ligand/ml). The cells were then cultured with retrovirus in retronectin-coated dishes (TaKaRa Bio, Otsu, Japan) for 72 h in the same medium with the addition of 5 μg protamine sulfate (Sigma-Aldrich)/ml. Retrovirally transduced cells were detected by flow cytometry based on their EYFP expression (12, 25).
Immunoprecipitation and immunoblot analysis.
Cell extracts were obtained by resuspending cell pellets in radioimmunoprecipitation assay buffer consisting of 10% glycerol, 0.5% Triton X-100, 20 mM HEPES (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1.5 mM MgCl2, and a protease inhibitor cocktail (Complete Mini; Roche Diagnostics GmbH), sonicated for 30 s on ice, and centrifuged for 15 min at 15,000 × g. The supernatant of the lysate was subjected to immunoprecipitation experiments, and the lysate was subjected to immunoprecipitation with GammaBind G Sepharose (GE Healthcare, Milwaukee, WI). Proteins were separated by SDS-PAGE, transferred to Immobilon-P (Millipore, Billerica, MA), immunoblotted with primary antibodies, and visualized with horseradish peroxidase-conjugated anti-rabbit IgG and SuperSignal West Femto maximum sensitivity substrate (Thermo Fisher Scientific). To examine protein stability and ubiquitination in vivo, cells were treated with MG132 (20 μM; Peptide Institute, Osaka, Japan).
Reconstitution of PcG complex 1 in Spodoptera frugiperda insect cells (Sf9) and purification.
Sf9 were cultured in Grace's insect cell culture medium (Life Technologies) supplemented with 10% FBS and 0.06% tryptose phosphate broth-Bacto (BD Pharmingen) in the presence of 0.1 mg of streptomycin/ml and 100 U of penicillin/ml (Wako Pure Chemical). cDNAs were inserted into either pV-IKS to produce the GST fusion product or pVL1392, and the vectors were cotransfected into Sf9 with a linearized BaculoGold baculovirus DNA (BD Pharmingen) for viral particle formation by using Cellfectin (Life Technologies). Recombinant baculoviruses were amplified by repeating infection. Sf9 were then infected with high-titer viruses, and at 72 h postinfection the cells were washed with cold PBS and suspended in homogenizing buffer (20 mM Tris [pH 7.9], 4 mM MgCl2, 0.4 mM EDTA, 2 mM dithiothreitol [DTT], 20% glycerol, 0.1% NP-40, 1 mM ZnCl2, with Complete Mini). The suspension was homogenized and centrifuged at 15,000 × g for 10 min, and the supernatant was subjected to a glutathione affinity column chromatography (Glutathione-Sepharose 4 Fast Flow; GE Healthcare) (12, 25).
In vitro ubiquitination assay.
Recombinant geminin, which was tagged with His6 and myc in the N- and C-terminal portions, respectively, was produced in Escherichia coli BL2, and was purified from supernatant of the extracts by means of cobalt affinity chromatography (Co-agarose; Wako Pure Chemical). Purified recombinant geminin or nucleosomal histone H2A (Calbiochem) was incubated in a 20-μl reaction mixture containing 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 2 mM DTT, 100 μM ZnCl2, 3 mM ATP, 0.1 μg of ubiquitin-activating enzyme E1 (Wako Pure Chemical), 1.0 μg of ubiquitin-conjugating enzyme UbcH5c (Enzo Life Science, Plymouth Meeting, PA), 10 μg of ubiquitin or myc-ubiquitin (Biochem, Cambridge, MA), and the affinity-purified complex (1 μg). After incubation at 37°C for 1 h, the reaction was terminated with protein sample buffer, run on SDS-PAGE, and was subjected to immunoblot analysis (12, 25).
ChIP assay.
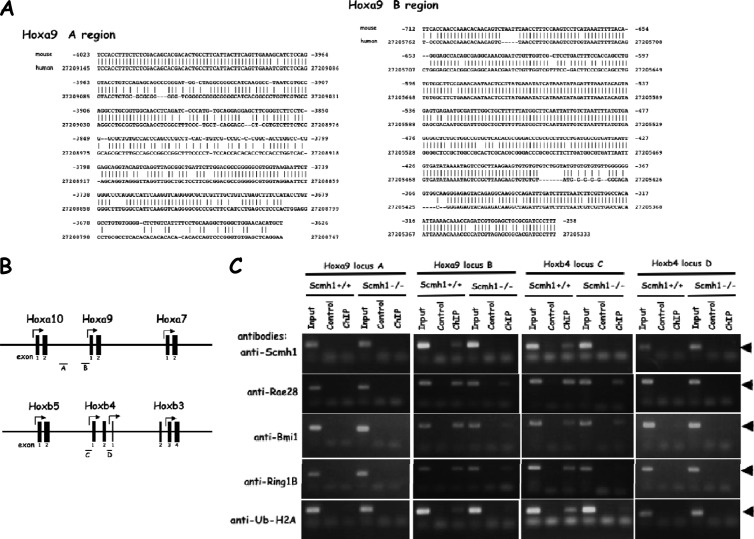
A chromatin immunoprecipitation (ChIP) assay was performed by using a LowCell ChIP kit (Diagenode, Liege, Belgium) according to the manufacturer's instructions. Freshly prepared mouse FL (∼105) were fixed with 0.01% formaldehyde for 8 min at room temperature, which was terminated by the addition of 125 mM glycine. DNA-protein cross-linked cells were washed twice with cold PBS, and were treated with lysis buffer supplemented with 20 mM sodium butylate for 5 min on ice. The samples were then subjected to sonication to shear the chromatin using the Bioruptor (Diagenode) for 12 cycles (30 s on, 30 s off). The average size of the DNA fragments was confirmed to be approximately 500 bp, ranging 200 to 1,000 bp. The sheared chromatin was incubated with protein A- or G-coated paramagnetic beads bound with the antibody of interest (anti-Ub1-histone H2A, anti-Scmh1, anti-Ring1B, anti-Bmi1, and anti-Rae28 antibodies) overnight at 4°C. The samples were then washed and immunoprecipitated. DNA was isolated from the immunoprecipitates by boiling for 10 min and was purified by using supplied DNA purifying slurry. ChIP DNA was detected by standard PCR for genomic locus A (from −4023 to −3626, 398 bp; the nucleotide numbers are designated by counting the adenine in each of the initiation codons as +1) and locus B (from bp −712 to −258, 455 bp) in the Hoxa9 gene and for locus C (from bp −268 to +88, 356 bp) and locus D (from bp +4266 to +4549, 284 bp) in the Hoxb4 gene. The PCR primer pairs used were as follows: locus A, 5′-TCCACCTTTCTCTCGACAGCAC-3′ and 5′-GAGCATGTGTTCCAGCCCAGC-3′; locus B, 5′-TTCACCAACCAAACACAACAGTCT-3′ and 5′-AAAGGGATCGCGCAGCTCCAC-3′; locus C, 5′-CCCCGCGGGAGCCCTATGTA-3′ and 5′-GGTAGGTAATCGCTCTGTGAATA-3′; and locus D, 5′-AATCA AGTCAAAGCTGCTCCTTG-3′ and 5′-ATCCGGAGAGACGGCTAACACT-3′.
Statistical analysis.
More than three independent experiments were performed, and the data were analyzed using the Student t test. The results are shown with the standard errors of the mean. Correlation was analyzed using the Spearman's rank correlation coefficient, and the trend line was estimated by the least-squares method.
Antibodies.
The primary and secondary antibodies used were listed in Table 1.
Table 1.
Antibodies used in this study
| Antibody (clone or product no.) | Species (antibody type)b | Manufacturerc |
|---|---|---|
| Probe-conjugated antibody | ||
| Anti-CD34 + APC (RAM34) | Rat (m) | eBioscience |
| Anti-Sca-1/Ly-6A/E + PE-Cy7 (D7) | Rat (m) | eBioscience |
| Anti-c-Kit/CD117 + APC-eFluor780 (2B8) | Rat (m) | eBioscience |
| Anti-B220/CD45R + biotin (RA3-6B2)a | Rat (m) | eBioscience |
| Anti-CD3ε + biotin (145-2C11)a | Hamster (m) | eBioscience |
| Anti-Gr-1/Ly-6G + biotin (RB6-8C5)a | Rat (m) | eBioscience |
| Anti-Mac-1α/CD11b + biotin (M1/70)a | Rat (m) | eBioscience |
| Anti-TER-119 + biotin (TER-119)a | Rat (m) | eBioscience |
| Primary antibody | ||
| Anti-Flag (anti-Flag M2) | Mouse (m) | Sigma |
| Anti-myc (sc-789) | Rabbit (p) | Santa Cruz |
| Anti-myc (9E10) | Mouse (m) | Santa Cruz |
| Anti-HA (12CA5) | Mouse (m) | Roche |
| Antigeminin | Rabbit (p) | — |
| Anti-Scmh1 | Rabbit (p) | — |
| Anti-Rae28 | Rabbit (p) | — |
| Anti-Bmi1 (05-637) | Mouse (m) | Merck |
| Anti-Ring1B (H0000604-M01) | Mouse (m) | Abnova |
| Anti-β-actin (AC-74) | Mouse (m) | Sigma |
| Anti-Ub1-histone H2A | Mouse (m) | Merck |
| Anti-histone H2A | Rabbit (p) | Merck |
| Secondary antibodyd | ||
| Anti-rabbit IgG + PE (611-108-122) | Goat (p) | Rockland |
| Anti-rabbit IgG + HRP (1858415) | Goat (p) | Thermo Fisher |
| Anti-mouse IgG + HRP (1858413) | Goat (p) | Thermo Fisher |
| Anti-goat IgG + HRP (sc-2020) | Donkey (p) | Santa Cruz |
| Anti-rabbit IgG + Alexa Fluor 488 (A11034) | Goat (p) | Life Technologies |
| Anti-mouse IgG + Alexa Fluor 549 (A11032) | Goat (p) | Life Technologies |
Lin− cells were purified by using the indicated biotin-conjugated antibodies with streptavidin plus PerCP-Cy5.5 (BD Pharmingen).
m or p, monoclonal or polyclonal antibody, respectively.
Thermo Fisher, Thermo Fisher Scientific; Roche, Roche Diagnostics GmbH; Sigma, Sigma-Aldrich. —, polyclonal antibodies raised against the GST fusion recombinant molecules (12, 30).
Secondary antibodies were conjugated with labels.
RESULTS
UPS-mediated regulation of Scmh1.
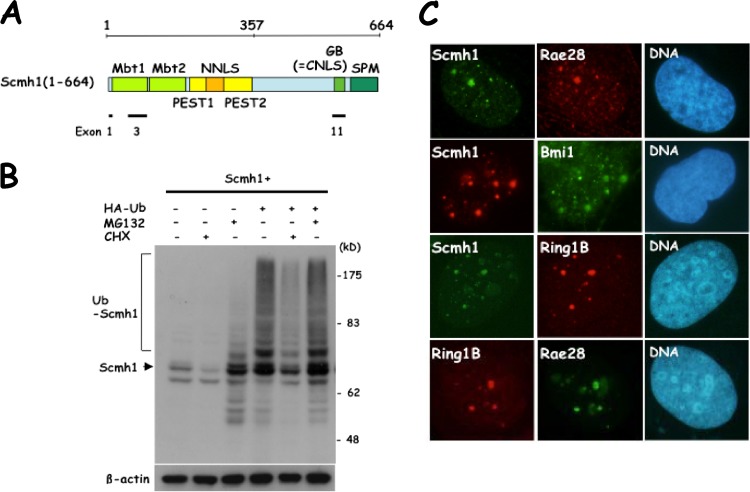
Scmh1 encodes a protein with several characteristic domains as described above (Fig. 1A) (8, 10). We previously showed that Scmh1 was highly sensitive to MG132, a proteasome inhibitor (12), so we transfected Scmh1 into a human embryonic kidney cell line, HEK-293 cells, and examined stability and ubiquitination of Scmh1 (Fig. 1B). Scmh1 levels were reduced after 6 h of cycloheximide treatment, suggesting that Scmh1 is unstable. However, MG132 treatment stabilized the Scmh1 protein and allowed detection of weak mobility-shifted bands of the same mobility as those detected in ubiquitin-cotransfected cells, suggesting that Scmh1 is ubiquitinated and is under the regulation of UPS.
Fig 1.
Regulation of the protein's stability and the subcellular localization of Scmh1. (A) Schematic representation of structure of Scmh1. Characteristic domains are indicated by colors. The parts corresponding to the representative exons are indicated in the lower part of the panel. The third exon, which was deleted in the knockout allele, encodes Mbt1. The GB domain is encoded by the eleventh exon. (B) Immunoblot analysis with an anti-Scmh1 antibody. HEK-293 cells were transfected with expression vectors for Scmh1 and HA-Ub and were treated with MG132 or cycloheximide (CHX) for 6 h. (C) Subcellular colocalization of each member of PcG complexes in polycomb bodies. Flag-Scmh1, Ring1B, Bmi1, and Rae28 were detected by indirect immunofluorescence labeling in Flag-Scmh1-transfected U-2 OS cells synchronized with hydroxyurea. Nuclear DNA was stained with DAPI.
Expression of endogenous Scmh1 was weakly detectable but unstable, a finding consistent with the previous finding that Scmh1 was substoichiometrically detected in PcG complex 1 (9), so we stably transfected U-2 OS cells with an expression plasmid for Flag-Scmh1 to improve the sensitivity of our assays. These transfected cells were treated with five different drugs (roscovitine, aphidicolin, hydroxyurea, thymidine, or colchicine) to synchronize the cell cycle, and the cells were double stained with an anti-Flag monoclonal antibody and rabbit polyclonal antibodies against each member of PcG complex 1 (Fig. 1C). In cells treated with hydroxyurea and thymidine, which synchronize cells at early S phase, Flag-Scmh1 colocalizes with Ring1B, Rae28, and Bmi1 in polycomb bodies (hydroxyurea, Fig. 1C; thymidine, data not shown). In contrast, Scmh1 was undetectable in cells synchronized at M phase by treatment with colchicine (data not shown). These findings suggest that Scmh1 is detectable in polycomb bodies in a specific phase of the cell cycle.
Generation and phenotype analysis of Scmh1-deficient mice.
Intensive analysis of mouse brain mRNAs by rapid amplification of 5′ cDNA ends (5′-RACE) discovered two additional cDNAs with alternative 5′ sequences (8). The alignment of the predicted amino acid sequences is shown in Fig. 2A. These N-terminal parts of Scmh1 are derived from multiple alternative exons (exons 1a to 1f) (Fig. 2B). To determine the roles of Scmh1 in mammals, we disrupted Scmh1 by homologous recombination (Fig. 2B). The targeting vector replaces the third exon, the second common protein-coding exon, with the Neor gene. The linearized targeting vector was introduced into ES cells. Genomic DNA from G418-resistant ES clones was examined by Southern blotting. The wild-type Scmh1 allele (Scmh1+) displayed an 18.5-kb single EcoRI band, while the knockout allele displayed a 14-kb band with probe A, and the result was confirmed with an independent probe (data not shown).
Scmh1-deficient mice were generated from two independent ES clones with the targeted allele. Generation of the offspring at the Mendelian ratios was confirmed by the Southern blot and PCR analyses (Fig. 2C and D). The testis, which expresses Scmh1 at high levels, was analyzed by immunoblot analysis with an anti-Scmh1 antibody. Scmh1 protein was not detectable in Scmh1−/− mice but was detectable in a gene dosage-dependent manner in Scmh1+/− animals (Fig. 2E).
Scmh1−/− mice were fertile and had normal average life span. We did not observe any obvious developmental abnormalities in Scmh1−/− mice. Although mice deficient for PcG genes display skeletal transformations that may result from altered Hox gene expression boundaries along the anteroposterior axis (30, 32, 33), no abnormality was observed in 10 Scmh1−/− embryos (Fig. 2F). The anterior expression boundary of Hoxd4 was shifted anteriorly in the paraxial mesoderm in Rae28-deficient mice (30) and expression of Hoxa9 was increased in hematopoietic cells from Scmh1−/− mice as described below. However, we did not observe any alteration in the expression domains of Hoxa9 and Hoxd4 in the paraxial mesoderm of 10.5-dpc Scmh1−/− embryos (Fig. 2G).
Hematopoietic abnormalities in Scmh1-deficient mice.
The cellularity of FL or BM mildly increased in Scmh1−/− mice relative to wild-type mice (FL, Fig. 3A; BM, Fig. 4A). The number of cells in lineage subpopulations of the hematopoietic cells was not affected in FL (Fig. 3B), but the numbers of B220+ and CD3+ lymphoid cells were reduced, and myeloid-lineage cells mildly increased in BM from Scmh1−/− animals (Fig. 4B). The clonogenic and LTC-IC activities were augmented in Scmh1−/− FL (Fig. 3C and D).
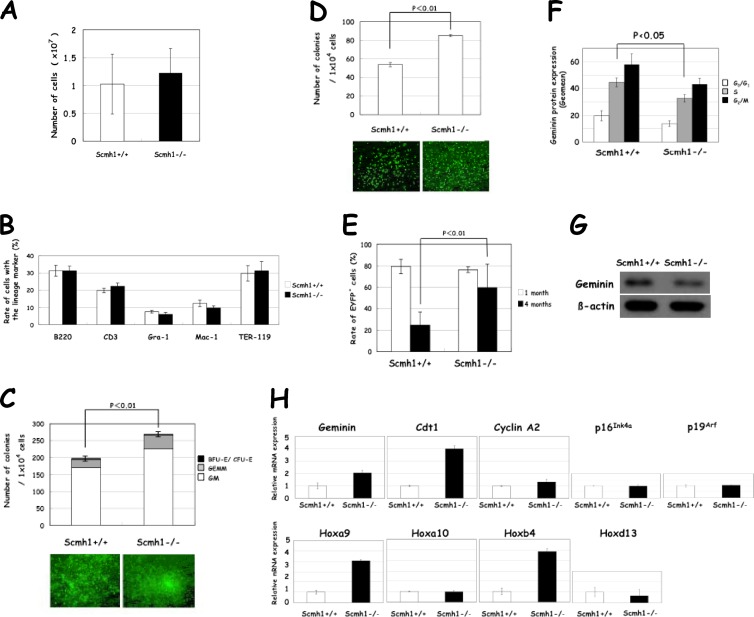
Fig 3.
Hematopoietic and molecular analyses of FL. (A) Total number of FL cells. (B) Percentage of each of the lineage-committed cells. Cells were analyzed by flow cytometry with the indicated lineage markers. (C) Clonogenic activity. Microscopic views of representative colonies are shown in the lower part of the panel. (D) LTC-IC activity. (E) LTR activity. The percentage of EYFP+ cells in the peripheral blood was examined 1 month and 4 months after the injection. (F) Geminin protein expression in each phase of the cell cycle. FL cells were pulse-labeled with BrdU and was examined by flow cytometry. The geometric mean (Geomean) of fluorescence intensity for geminin protein expression is shown. (G) Immunoblot analysis with an antigeminin antibody. (H) Real-time PCR analysis of mRNAs for geminin, Cdt1, cyclin A2, p16Ink4a, p19Arf, Hoxa9, Hoxa10, Hoxb4, and Hoxd13.
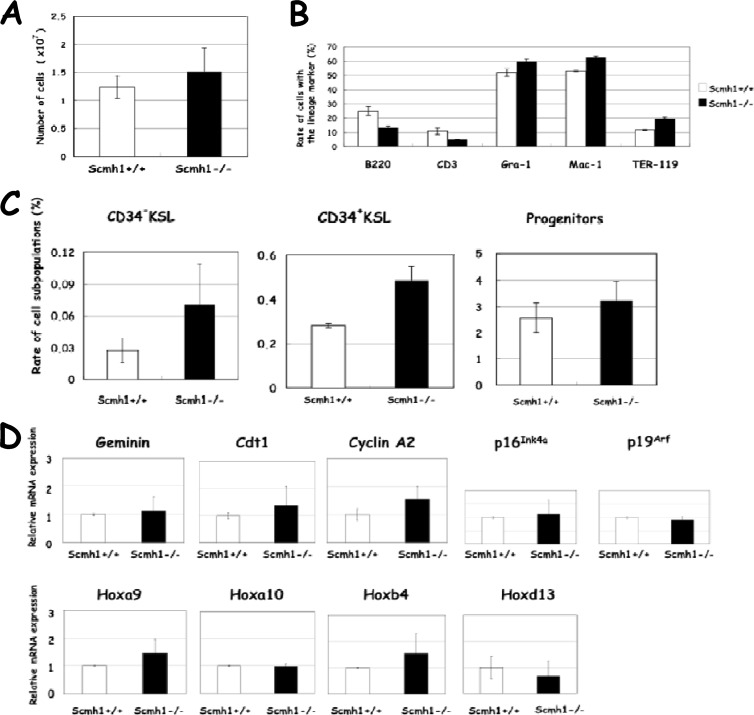
Fig 4.
Hematopoietic and molecular analyses of BM. (A) Total numbers of BM cells in Scmh1+/+ and Scmh1−/− mice. (B) Proportion of lineage-committed cells in BM. (C) Proportion of CD34− c-Kit+ Sca1+ Lin− (KSL), CD34+ KSL, and progenitor subpopulations in BM. (D) Real-time PCR analysis of mRNAs for geminin, Cdt1, cyclin A2, p16Ink4a, p19Arf, Hoxa9, Hoxa10, Hoxb4, and Hoxc13 in BM.
We then examined the LTR activity. FL cells were retrovirally labeled with EYFP, injected into lethally irradiated congenic mice, and EYFP+ cells in the peripheral blood were examined 1 and 4 months after the injection. The numbers of EYFP+ Scmh1−/− and control cells were almost equal after 1 month, but Scmh1−/− cells were preferentially maintained after 4 months, indicating that activity of the hematopoietic stem cells was augmented in Scmh1−/− FL (Fig. 3E).
Hematopoietic stem and progenitor subpopulations are poorly enriched by cell sorting of FL because these immature cells are CD11b(Mac-1)low or CD11b(Mac-1)+ in FL (34). Therefore, we analyzed the frequency of HSC (CD34− c-Kit+ Sca1+ Lin−), multipotent progenitor cell (CD34+ c-Kit+ Sca1+ Lin−), and hematopoietic progenitor cell (c-Kit+ Sca1− Lin− [Progenitors in Fig. 4C]) subpopulations in BM. All of these cell types were increased in BM from Scmh1−/− mice (Fig. 4C), a finding consistent with the findings above that the hematopoietic stem and progenitor activities are promoted in Scmh1−/− FL.
Role for derepressed Hoxb4 and Hoxa9 in the regulation of geminin protein in Scmh1−/− FL.
PcG complex 1 that includes Scmh1 acts as an E3 ubiquitin ligase for geminin (12). Therefore, we expected that deficiency of Scmh1 would impair the E3 ubiquitin ligase activity, thus stabilizing geminin and leading to the accumulation of geminin in Scmh1−/− mutants relative to control mice. Surprisingly, expression of geminin protein in FL cells was decreased in each phase of the cell cycle (Fig. 3F). This finding was confirmed by immunoblot analysis (Fig. 3G). Real-time PCR analysis indicated that expression of mRNA for geminin, Cdt1, and cyclin A2, which is under the regulation of E2F (35), was increased in Scmh1−/− FL. However, expression of mRNA for p16Ink4a and p19Arf encoded by Cdkn2a, a well-known downstream target for PcG complex 1 (36), was not altered (Fig. 3H). The expression of Hoxa9 and Hoxb4 was markedly enhanced in Scmh1−/− FL, although the expression of Hoxa10 and Hoxd13 was not affected (Fig. 3H). Similar but milder alterations in Hoxa9 and Hoxb4 expression were also observed in BM from 3-month-old Scmh1−/− mice (Fig. 4D).
To determine whether Scmh1 directly represses Hoxa9 and Hoxb4, we assayed Scmh1 binding and histone H2A monoubiquitination at Hoxa9 and Hoxb4 loci using ChIP assays. We compared Scmh1+/+ and Scmh1−/− FL in two evolutionary conserved intergenic regions between the Hoxa10 and Hoxa9 genes (Fig. 5A and B), the promoter region of the Hoxb4 genes (Fig. 5C) (37), and an intergenic region between the Hoxb4 and Hoxb3 genes (Fig. 5D), that includes a neural regulatory element, CR3 (38) (Fig. 5B). An anti-Scmh1 antibody efficiently immunoprecipitated the chromatin in both of the promoter regions (Fig. 5B and C) in FL from Scmh1+/+ but not from Scmh1−/− embryos. Similarly, anti-Ring1B, anti-Bmi1, and anti-Rae28 antibodies immunoprecipitated these regions in Scmh1+/+ FL but did so less efficiently in Scmh1−/−. As expected, an anti-monoubiquitinated histone H2A (anti-Ub1-histone H2A) antibody gave similar results (Fig. 5C). Little binding of PcG complex 1 members, Ring1B, Bmi1, Rae28, Scmh1, and Ub1-histone H2A was detected in the A and D regions (Fig. 5C). Together, these results indicate that PcG complex 1 members bind less efficiently in the absence of Scmh1 and that Scmh1 directly represses Hoxa9 and Hoxb4.
Fig 5.
ChIP analysis of Hoxa9 and Hoxb4 loci. (A) Evolutionally conserved regions in the Hoxa9 locus. We compared nucleotide sequence of the mouse Hoxa9 locus from the 4-kb upstream region to the 4-kb downstream region of Hoxa9 (NC_000072) against that of the human (NC_000007) by using the NCBI BLAST program to search for a conserved region in the intergenic region of the mouse Hoxa9 locus. Two well-conserved regions were designated A and B, which reside from bp −4023 to bp −3626 and from bp −712 to bp −258 upstream to the initiation codon, respectively. The nucleotide sequences of these regions are aligned between those of the mouse and those of the human. (B) Schematic representation of the ChIP analysis. Primer sets specific for the Hoxa9 and Hoxb4 regions were used for PCR amplification. (C) ChIP analysis of Hoxa9 and Hoxb4 loci with anti-Scmh1, anti-Ring1B, anti-Bmi1, anti-Rae28, and anti-Ub1-histone H2A antibodies. FL cells were subjected to the ChIP analysis, and PCR-amplified specific DNA fragments are indicated by arrows.
We recently demonstrated that either Hoxb4 (25) or Hoxa9 (51) can form a RDCOX complex to act as an E3 ubiquitin ligase for geminin. The derepression of Hoxa9 and Hoxb4 described above would lead to increased activity of an alternative E3 ligase for geminin and thus might compensate for defective activity of the PcG complex 1 E3 ligase for geminin caused by Scmh1 deficiency.
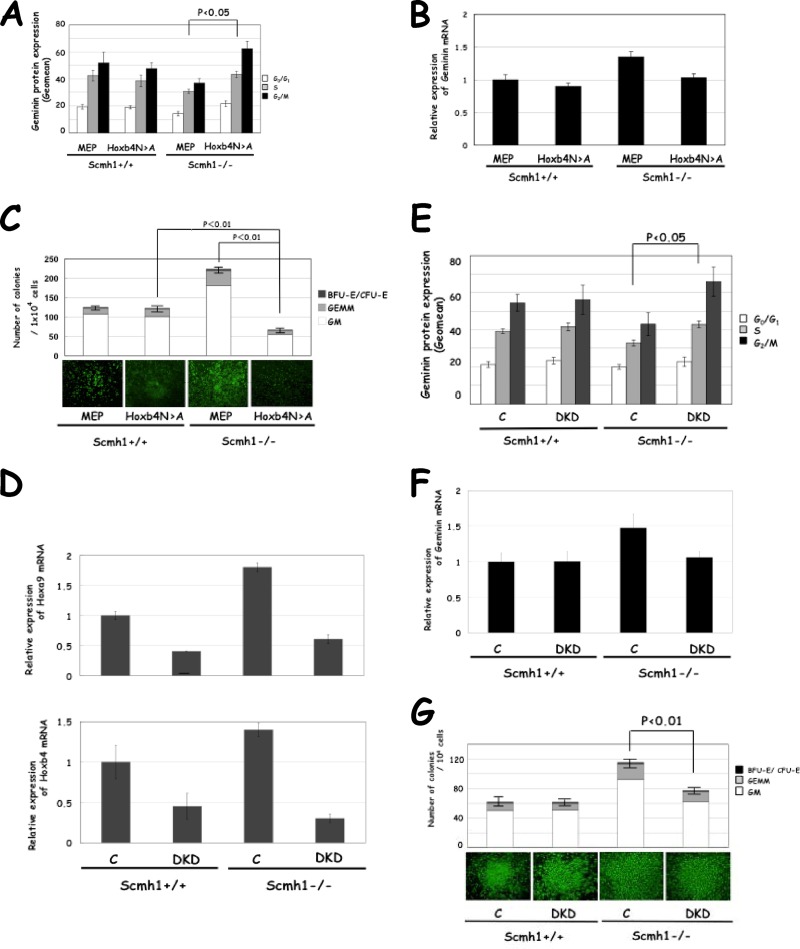
To confirm this hypothesis, we performed three experiments. The Hoxb4N→A mutant forms RDCOXB4 complex more stably than Hoxb4 but, because Hoxb4N→A does not interact with geminin, the resulting complex does not display the E3 ubiquitin ligase activity for geminin, and Hoxb4N→A may affect the E3 ubiquitin ligase activity of the RDCOX complex in the cells (25). First, we showed that transduction of Hoxb4N→A increased geminin protein expression (Fig. 6A) through the dominant-negative effect on the RDCOX complex, even though geminin mRNA levels decreased (Fig. 6B). Augmented clonogenic activity was effectively downregulated by Hoxb4N→A transduction in Scmh1−/− FL cells but had little effect on Scmh1+/+ FL cells (Fig. 6C).
Fig 6.
Effect of Hoxa9 and Hoxb4 on geminin expression and clonogenic activity. (A to C) Effect of Hoxb4N→A transduction on FL cells. (A) Geminin protein expression. Geomean for the geminin protein expression level in each phase of the cell cycle are shown. MEP, an empty vector. (B) Geminin mRNA. (C) Clonogenic activity. (D to G) Effect of DKD on FL cells. DKD, mixture of siRNA for Hoxa9 and Hoxb4; C, nontargeting control siRNA. (D) mRNA for Hoxa9 and Hoxb4. (E) Geminin protein expression in each phase of the cell cycle. (F) mRNA for geminin. (G) Clonogenic activity.
Second, we simultaneously knocked down both Hoxb4 and Hoxa9 (double knockdown [DKD]) using the transfection of siRNA. We controlled for the efficiency of transfection of siRNA into FL cells using flow cytometric analysis and showed that the majority of FL cells were efficiently transfected with the fluorescence-labeled siRNA (data not shown). DKD effectively downregulated Hoxa9 and Hoxb4 in both Scmh1+/+ and Scmh1−/− FL (Fig. 6D). As expected, DKD increased the amount of geminin protein in every cell cycle phase (Fig. 6E), even though geminin mRNA levels decreased in Scmh1−/− FL (Fig. 6F). DKD exerted less effect on geminin expression in Scmh1+/+ FL (Fig. 6E and F). DKD downregulated the augmented clonogenic activity in Scmh1−/− FL but had less effect on Scmh1+/+ FL (Fig. 6G).
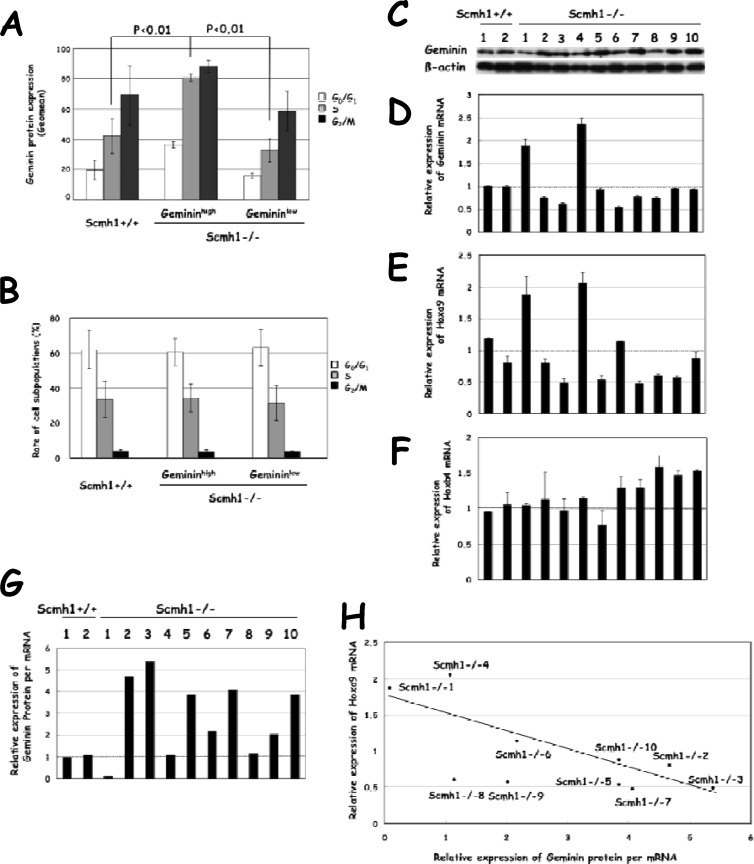
Third, we examined geminin expression in BM from mice that were older than 20 months. In vivo labeling experiments with BrdU showed that geminin accumulation occurred in each phase of the cell cycle in approximately half of aged Scmh1−/− mice (gemininhigh Scmh1−/− mice) (Fig. 7A), although the proportion of cells in each phase of the cell cycle was not significantly altered (Fig. 7B). Overt geminin protein accumulation was also detected by immunoblot analysis in BM from 5 of 10 Scmh1−/− mice (Fig. 7C). Geminin protein accumulation occurred in Schm1−/− BM, with lower expression of Hoxa9 than controls (Fig. 7C and E). Hoxa9 is the most abundantly expressed Hoxa cluster gene in BM (39), and its expression is essential for normal function of HSCs (40). Curiously, the expression levels of Hoxa9 mRNA (Fig. 7E) and those of geminin protein relative to mRNA (Fig. 7C, D, and G) were negatively correlated (Fig. 7H) at a statistically significant level (Spearman's r = 0.697 > 0.4, P < 0.05). We suggest that in aged Scmh1−/− mice, decreased expression levels of Hoxa9 lead to the inability to prevent geminin accumulation caused by Scmh1 deficiency. The expression levels of geminin protein relative to mRNA level were low despite a low Hoxa9 mRNA expression in Scmh1−/− mice (Fig. 7E and G), in which Hoxb4 mRNA expression was the highest (Fig. 7F).
Fig 7.
Expression of geminin, Hoxa9, and Hoxb4 in BM from aged mice. (A and B) Cell sorting analysis of geminin protein expression and proportion of cell subpopulations in each phase of the cell cycle. Three aged mice of each genotype (Scmh1+/+, gemininhigh and gemininlow Scmh1−/−) were subjected to in vivo BrdU-labeling experiments. (A) Geminin protein expression. (B) Proportion of the cell subpopulation in each phase of the cell cycle. Gemininhigh, mice with geminin accumulation; gemininlow, mice without geminin accumulation. (C) Immunoblot analysis of geminin protein expression in aged mice. (D to F) Real-time PCR analysis. (D) Geminin mRNA. (E) Hoxa9 mRNA. (F) Hoxb4 mRNA. Individual mice were numbered 1 and 2 for Scmh1+/+ animals and 1 to 10 for Scmh1−/− animals. (G) Relative expression level of geminin protein to mRNA. The expression levels of geminin protein were normalized to expression levels of β-actin and were divided by the mRNA levels to calculate the ratios. (H) Correlation of expression levels of Hoxa9 mRNA to geminin protein or mRNA in BM from aged mice. Bands for geminin in immunoblot analysis were scanned using the ImageJ program (NIH). Geminin protein expression levels were normalized to β-actin and divided by the level of geminin mRNA to determine the ratio of protein to mRNA as shown above. Correlation of Hoxa9 expression with the geminin protein/RNA ratio was estimated with the Spearman rank correlation coefficient and the least-squares method.
Molecular role for Scmh1 in the E3 ubiquitin ligase activity.
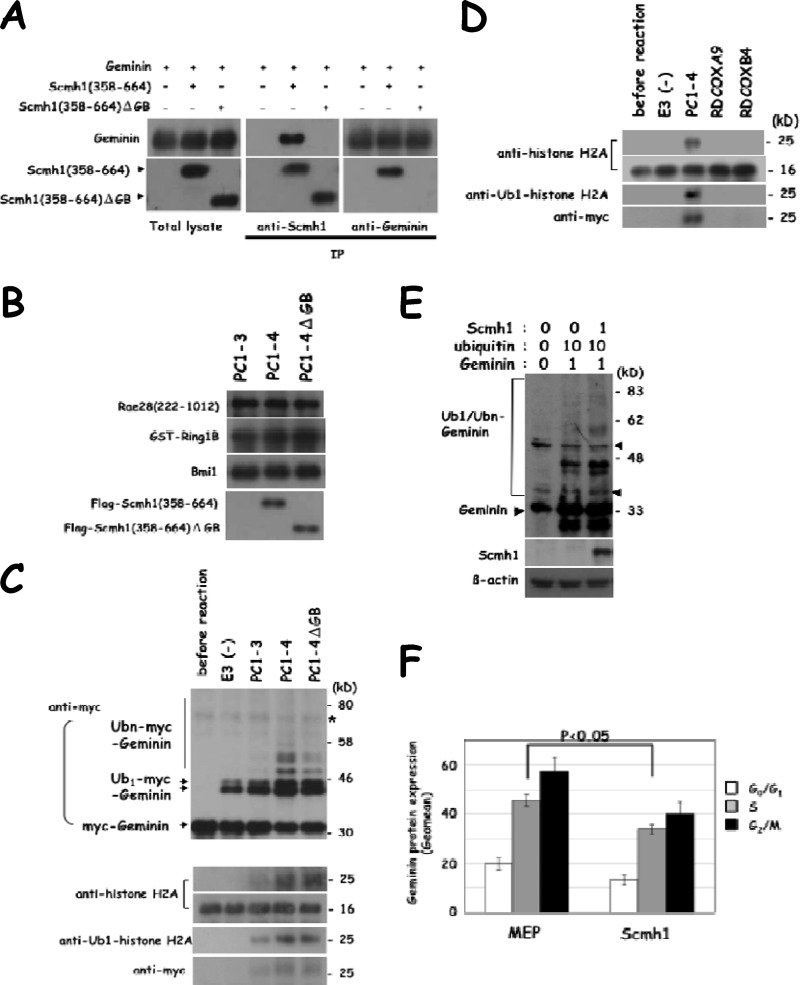
We transfected Scmh1(358-664) and geminin in HEK-293 cells and confirmed the molecular interaction of Scmh1(358-664) with geminin by immunoprecipitation analysis. This molecular interaction was impaired by deletion of the GB domain (540 to 568 amino acids) from Scmh1(358-664) (Fig. 8A).
Fig 8.
Role for Scmh1 in the E3 ubiquitin ligase activity. (A) Immunoprecipitation analysis of Scmh1 and geminin. HEK-293 cells were transfected with Scmh1(358-664) and geminin. Molecular interaction of Scmh1(358-664) and geminin is detected but was impaired by deletion of the GB domain in Scmh1(358-664). Immunoprecipitation analysis with the indicated antibodies. (B) Immunoblot analysis of affinity-purified recombinant complexes, PC1-3, PC1-4, and PC1-4ΔGB, which were reconstituted in Sf9. PC1-3, GST-Ring1B+Bmi1+Rae28(222-1012); PC1-4, GST-Ring1B+Bmi1+Rae28(222-1012)+Flag-Scmh1(358-664); PC1-4ΔGB, GST-Ring1B+Bmi1+Rae28(222-1012)+Flag-Scmh1(358-664)ΔGB. (C and D) In vitro ubiquitination reaction with the recombinant complexes. The purified recombinant PcG complex 1 and RDCOX complexes were subjected to an in vitro ubiquitination assay (+E1+E2+ubiquitin+recombinant myc-geminin or +E1+E2+myc-ubiquitin+nuclesomal histones). Ub-myc-geminin and myc-Ub1-histone H2A were detected in the reaction products. Ubn-myc-geminin was almost undetectable in PC1-3 and was weakly detectable in PC1-4ΔGB. E3(−), the ubiquitination reaction was done without an E3 ubiquitin ligase. (E) Effect of Scmh1 on geminin ubiquitination. HEK-293 cells were transfected with Scmh1, ubiquitin, and geminin and were subjected to immunoblot analysis. Amount of the transfected plasmids (μg) is indicated in the upper part of the panel. The mobility-shifted bands were assumed to be Ub-geminin due to close similarity to those detected in the in vitro ubiquitination analysis. Triangle, unspecified bands. (F) Effect of Scmh1 transduction on geminin protein expression in FL cells. Geomean for geminin protein expression in each phase of the cell cycle is shown.
To further characterize the molecular role for Scmh1 and its GB domain, in the E3 ubiquitin ligase activity in vitro, we reconstituted PcG complex 1, which is composed of Ring1B, Bmi1, Rae28, and Scmh1, in Sf9. The insect cells were coinfected with baculoviruses encoding GST-Ring1B, Bmi1, Rae28, and either Flag-Scmh1 or Flag-Scmh1 lacking the GB domain (Flag-Scmh1ΔGB). Since full-length Rae28 and Scmh1 were unstable in Sf9, a truncated form of Rae28 lacking an N-terminal region including serine threonine-rich and glutamine-rich domains [Rae28(222-1012)] and that of Scmh1 lacking an N-terminal region, including the MBT and PEST domains preceded by a Flag tag in the N-terminal portion [Flag-Scmh1(358-664)], were expressed together with GST-Ring1B and Bmi1 to obtain the recombinant PcG complex 1 (PC1-4). We also prepared for PC1-4 with Flag-Scmh1(358-664)ΔGB (PC1-4ΔGB), and a complex designated PC1-3, which is composed of GST-Ring1B, Bmi1, and Rae28(222-1012) but lacks Scmh1. Cell extracts were prepared from Sf9 expressing PC1-3, PC1-4, and PC1-4ΔGB, and the complexes were purified by means of a glutathione affinity chromatography. A pulldown assay showed that GST-Ring1B, Bmi1, and Rae28(222-1012) formed a complex with either Flag-Scmh1(358-664) or Flag-Scmh1(358-664)ΔGB (Fig. 8B).
Equal amounts of the affinity-purified recombinant PcG complex 1 were analyzed using an in vitro ubiquitination assay with myc-tagged geminin as a substrate. The reaction product was analyzed by immunoblot analysis with an anti-myc monoclonal antibody. As described previously, PC1-4 mediated the formation of polyubiquitin chains in geminin. The two faster-migrating mobility-shifted bands correspond to monoubiquitinated (Ub1) geminin and the other more mobility-shifted bands correspond to polyubiquitinated (Ubn) geminin (12). The polyubiquitination activity was impaired but not abolished by deletion of the GB domain in Scmh1 (Fig. 8C). We suggest that the E3 ubiquitin ligase activity of PC1-4 for geminin mediated by the GB domain provides PC1-4 with a higher-affinity interaction domain with geminin.
Next, we examined activity of the E3 ubiquitin ligase for chromatin histone H2A by using myc-tagged ubiquitin. As shown in the lower panels of Fig. 8C, a mobility-shifted band with the molecular mass of 25 kDa was detected in the reaction product with PC1-4 or PC1-4ΔGB by either of anti-histone H2A, anti-Ub1-histone H2A or anti-myc antibodies. The intensity of the mobility-shifted bands was not significantly affected by deletion of the GB domain in Scmh1, but a deficiency of Scmh1 (PC1-3) reduced labeling. Thus, Scmh1 augments the activity of the E3 ubiquitin ligase for histone H2A as well as for geminin, but the activity of the E3 ubiquitin ligase for histone H2A is not mediated by the GB domain. We could not detect the E3 ubiquitin ligase activity for histone H2A in the recombinant RDCOXA9 and RDCOXB4 complex (Fig. 8D), showing that the PcG complex 1 and RDCOX complex have different substrate specificities.
We further examined in vivo whether exogenous expression of Scmh1 exerted an effect on the ubiquitination and expression of geminin. We transiently expressed full-length Scmh1, geminin, and ubiquitin in HEK-293 cells and examined the ubiquitination of geminin. The transfection of exogenous Scmh1 induced ubiquitination of geminin (Fig. 8E) and reduced the endogenous geminin expression levels in each phase of the cell cycle in FL (Fig. 8F), probably through the increased ubiquitination of geminin, as shown in the in vitro and in vivo assay systems described above.
Effect of Scmh1, Scmh1ΔGB, and Scmh1ΔMBT transduction on Hox and geminin expression in Scmh1−/− FL.
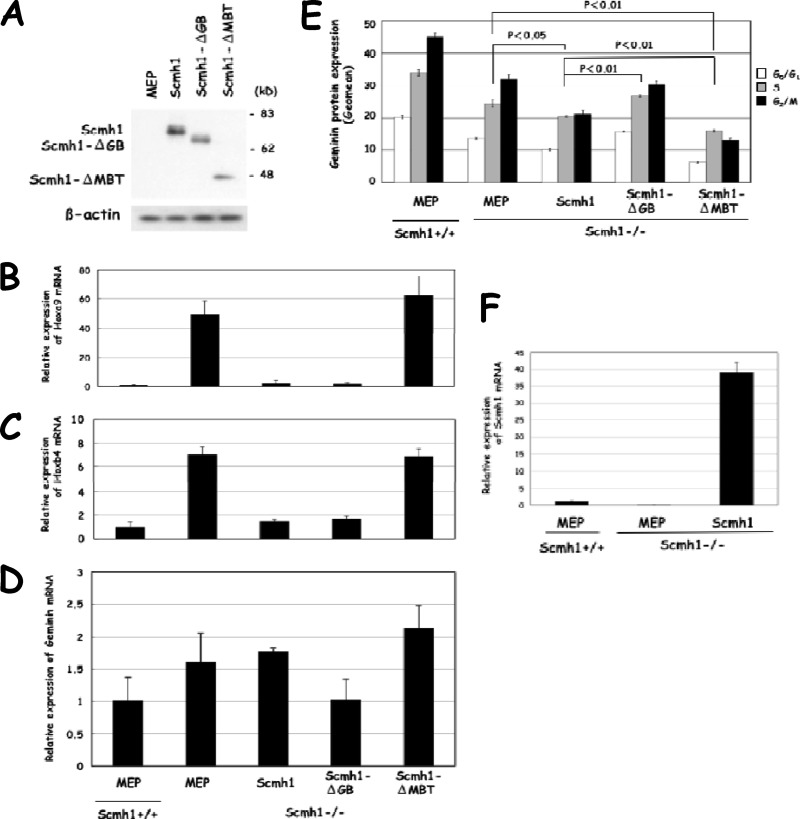
We prepared several murine stem cell virus vectors with EYFP: MEP, Scmh1, Scmh1ΔGB, and Scmh1ΔMBT, in which amino acids 28 to 287 of the MBT domain were deleted. After we confirmed the expression of these constructs in HEK-293 cells (Fig. 9A), we retrovirally transduced each of the mutants into Scmh1−/− FL cells. As expected, the Scmh1 deficiency by itself was responsible for the derepression of the Hox genes, because full-length Scmh1 reduced expression of Hoxa9 and Hoxb4 in Scmh1−/− FL cells (Fig. 9B and C).
Fig 9.
Transduction of Scmh1, Scmh1ΔGB, and Scmh1ΔMBT and the effect on expression of Hoxa9, Hoxb4, and geminin in Scmh1−/− FL cells. (A) Expression of Scmh1 derivatives in HEK-293 cells. HEK-293 cells were retrovirally transduced with Scmh1, Scmh1ΔGB, and Scmh1ΔMBT. After treatment with MG132 for 6 h, the cells were subjected to immunoblot analysis with a polyclonal anti-Scmh1 antibodies. MEP, an empty vector. (B to E) Effect of transduced Scmh1 derivatives on Scmh1−/− FL cells. (B) Hoxa9 mRNA. (C) Hoxb4 mRNA. (D) Geminin mRNA. (E) Geminin protein. Relative expression of Hoxa9, Hoxb4, and geminin was detected by real-time PCR analysis and Geomean for geminin protein expression in each phase of the cell cycle was determined after flow cytometry. MEP, an empty vector. (F) Relative expression of Scmh1 mRNA in FL cells retrovirally transduced with Scmh1. MEP, an empty vector.
Transduction of Scmh1ΔGB decreased geminin mRNA levels and reduced the expression of Hoxa9 and Hoxb4 in Scmh1−/− FL cells (Fig. 9B to D) but, unlike full-length Scmh1, did not downregulate the geminin protein (Fig. 9E). This observation is consistent with our observations that the GB domain is required for the efficient E3 ubiquitin ligase activity of PcG complex 1 for geminin (Fig. 8C). The MBT domain of Drosophila Scm aids transcriptional repression of PcG complex 1 through direct interaction with monomethylated histones (15). We transduced Scmh1ΔMBT into Scmh1−/− FL cells to determine whether the MBT domains of Scmh1 are required to repress the transcription of Hox genes. The Scmh1ΔMBT transduction failed to lower expression levels of Hoxa9 and Hoxb4 in Scmh1−/− cells (Fig. 9B and C). We conclude that the MBT domains are required for efficient transcriptional repression of the target genes by PcG complex 1 in mice.
Transduction of full-length Scmh1 into Scmh1−/− FL cells could alter geminin protein levels, either by increasing the E3 ligase activity of PcG complex 1 or indirectly by decreasing expression of Hox genes, which in turn would decrease the E3 ligase activity of the RDCOX complex. Scmh1 overexpression induced ubiquitination of geminin in HEK-293 cells (Fig. 8E) and retroviral Scmh1 transduction reduced geminin expression in each phase of the cell cycle in FL cells (Fig. 8F). The transduction of Scmh1 downregulated geminin protein levels without significantly affecting the mRNA level, probably as a consequence of enhanced activity of the PcG complex 1 E3 ubiquitin ligase for geminin by overdose of Scmh1 (Fig. 9F).
Expression levels of geminin protein were more strongly reduced by transduction of Scmh1ΔMBT than by full-length Scmh1. We suggest that Scmh1ΔMBT was not able to downregulate Hoxa9 and Hoxb4 but retained polyubiquitination activity for geminin. As described above, the E3 ubiquitin ligase activity for histone H2A was detected in vitro in PC1-4 which lacked the MBT domains in Scmh1. Therefore, we suggest that the MBT domains are required for the recruitment of PcG complex 1 in the Hoxa9 and Hoxb4 loci, and this leads to the derepression of Hox loci.
These findings further support the model that Scmh1 regulates geminin protein directly via PcG complex 1 and indirectly via repression of Hoxa9 and Hoxb4, leading to decreased levels of RDCOX E3 ligase activity. The GB domain of Scmh1 is crucial to the E3 ubiquitin ligase activity for geminin, and the MBT domains are required for the transcriptional repression of a subset of Hox genes.
DISCUSSION
Overall, the developmental and hematopoietic phenotypes observed in Scmh1−/− mice are very mild. Notably, we did not observe posterior transformations of paraxial mesoderm, or anterior derepression of Hox loci similar to those reported for mutations in other PcG complex 1 subunits (30, 32, 33). This may be because Scmh1 homologues such as L3MBTL1-3 (41), Scml1 (42), Scml2 (43), and Sfmbt1 (44, 45) compensated for deficiency of Scmh1. A second possibility is that Scmh1, which is a substoichiometric member of the PcG complex 1, has a minor effect on the activity of PcG complex 1. On the other hand, mice lacking SPM domain of Scmh1 display skeletal abnormalities (16), which we do not observe in our mutants. We speculate that the stronger phenotypes observed in mutants lacking the SPM domain may result from a dominant-negative effect of the N-terminal truncated form of Scmh1 on the activity of PcG complex 1, consistent with the observation that truncated transcripts were observed in homozygous mutants (16). It would be interesting to monitor Scmh1 protein expression in mice lacking the SPM domain to confirm this hypothesis.
Scmh1 may have a specific role in target selection or in cell cycle-dependent activity of PcG complex 1. In support of this idea, our analysis shows that the MBT and GB domains contribute to Scmh1 recruitment and specificity. MBT domains are required for repression of Hoxa9 and Hoxb4 in FL cells. The MBT domains were originally found in Scm, Sfmbt, and l(3)mbt in Drosophila (13) and are evolutionarily related to the chromatin-binding Tudor domain “Royal Family,” including the Tudor, plant Agenet, chromo, and PWWP domains (46). The MBT domains of Scmh1 may be required for recruitment of PcG complex 1 to Hoxa9 and Hoxb4. This finding argues against the model that the chromodomain of Cbx proteins is sufficient for recruitment of PcG complex 1 to target loci (3). We could not, however, address the contribution of the MBT domain to E3 ligase activity, because we could not prepare enough recombinant PcG complex 1 in vitro to compare full-length Scmh1 with the MBT deletion in our in vitro E3 ligase assay (Fig. 8A to D).
The expression of most PcG genes is ubiquitous. Nevertheless, there is evidence for cell cycle-specific PcG binding in mitosis (47, 48). Bmi1 binding to heterochromatin is high in early S phase and is not detectable in the late S through the G2/M phases. This cell cycle-dependent heterochromatin binding is inversely correlated with the phosphorylation of Bmi1 (49). It is very striking that Scmh1 associates with PcG bodies in cells synchronized at the G1/S boundary (Fig. 1C) but not in cells synchronized at the G2/M boundary. The NLS and PEST sequences are required to regulate Scmh1 stability through UPS and the subcellular localization (Ohtsubo et al., unpublished). Since PEST sequences can be phosphorylated, we speculate that cell cycle-dependent phosphorylation by cyclin-dependent kinases may indirectly regulate presence of Scmh1 in PcG bodies by controlling Scmh1 stability. Cell cycle-dependent association of Scmh1 with PcG bodies is consistent with the observation that Scmh1 is a substoichiometric component of the PcG complex 1 (9). Given our observation that the MBT domain of Scmh1 is required for recruitment to Hox loci, these observations together suggest that PcG complex 1 may bind targets or act as an E3 ligase for histone H2A in a cell cycle-dependent manner. One intriguing possibility is that Scmh1 recruits PcG complex 1 to nascent DNA after replication, and its E3 ligase activity reestablishes posttranslational modifications of histones necessary for epigenetic inheritance of gene expression patterns.
Scmh1 contributes to the E3 ligase activity toward histone H2A and geminin itself (Fig. 8C). Scmh1 directly regulates Hoxa9 and Hoxb4 (Fig. 3H, 4D, and 5C) and is necessary for the recruitment of the PcG complex 1 (Fig. 5C) and thus for the ubiquitination of histone H2A (Fig. 5C). The failure of this E3 ligase activity on histone H2A presumably leads to the derepression of the Hox genes. It would be interesting to determine whether the form of Scmh1 that binds Hox genes lacks the GB domain, since the GB domain confers target specificity for the E3 ligase activity of PcG complex 1 to geminin, but not to histone H2A. Such experiments could distinguish whether Scmh1 exerts transcriptional repression and geminin downregulation coordinately through the MBT and GB domains in the specific chromatin loci.
We expected that deficiency for Scmh1 would prevent the E3 ligase activity of PcG complex 1 and thus lead to increased stability of geminin. So, we were surprised to observe decreased stability of geminin in Scmh1−/− mice. The simplest explanation for this paradox is provided by our recent observations that Hox proteins themselves are members of a protein complex with E3 ligase activity for geminin (25). Derepression of Hoxa9 and Hoxb4 will lead to increased levels of RDCOX E3 ligase activity, which will compensate for the loss of the PcG complex 1 E3 ligase activity. Several experiments support this conclusion. DKD of Hoxa9 and Hoxb4 in Scmh1−/− mice prevents the decrease in geminin levels (Fig. 6E). Aged mice have lower levels of Hoxa9 and Hoxb4 expression and, correspondingly, in old Scmh1−/− mice there are increased levels of geminin (Fig. 7A and C). These results are not an indirect effect of RDCOX activity on ubiquitination of H2A because this complex lacks activity on H2A (Fig. 8D).
Most phenotypes of PcG mutants in embryos and in stem cells are assumed to arise from defects in transcriptional repression of targets, including Hox genes. The findings presented here suggests that more attention should be paid to nontranscriptional effects of PcG complex 1 in generation of phenotypes. This proposal is consistent with a recent study showing a role for posterior sex combs in the destruction of cyclin B (50).
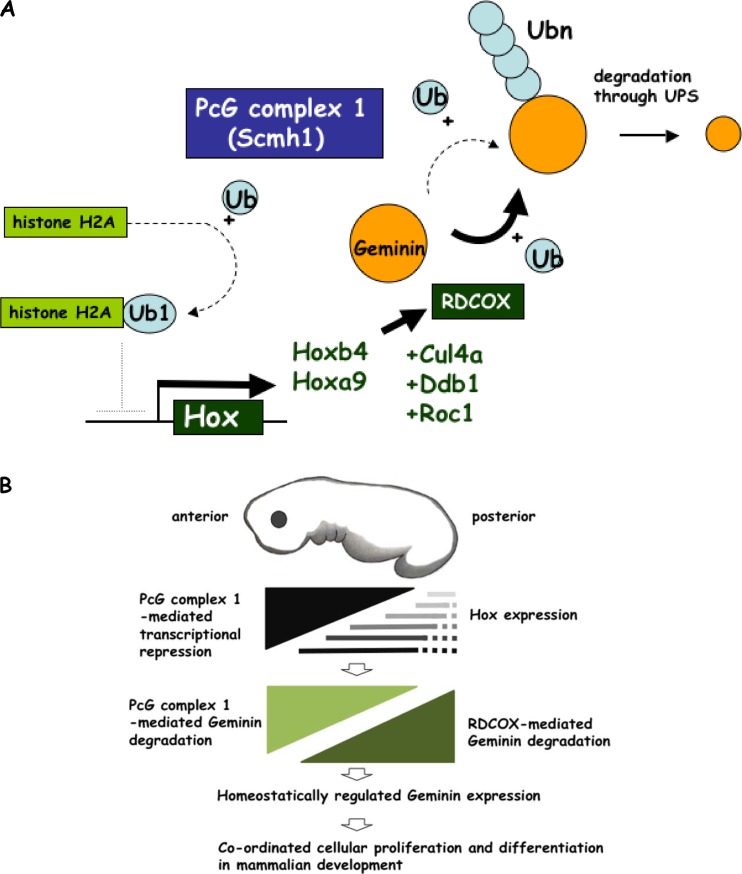
We previously showed that Hoxb4 transduction suppressed the accumulation of geminin and rescued impaired hematopoietic stem cell activity in Rae28−/− mice, suggesting that Hox and PcG complex 1 proteins work in concert to regulate geminin levels via modulation of E3 ubiquitin ligase activity (25). Thus, we suggest that derepressed Hoxa9 and Hoxb4 compensate for impaired activity of the E3 ubiquitin ligase for geminin and accelerated hematopoietic stem and progenitor activities through the downregulation of geminin in Scmh1−/− (Fig. 10A). Here, we propose that PcG complex 1 and a subset of the downstream Hox target genes form a regulatory homeostatic network for tuning the expression level of geminin protein (Fig. 10A). Unlike the APC/C that mediates polyubiquitination of geminin from the late M to the G1 phase, the PcG complex 1 and RDCOX complex could regulate geminin in other phases of the cell cycle. This model provides a way to couple gene regulation and the control of DNA replication in stem cell regulation and development. PcG complex 1 may repress Hox genes in the anterior region of embryos through the epigenetic regulation, suggesting the possibility that Hox-mediated regulation of transcription and geminin protein stability takes part in the posterior expression domains and that the PcG complex 1-mediated effect is limited to the anterior part of embryos (Fig. 10B).
Fig 10.
Proposed molecular role for Scmh1 in geminin regulation. (A) Scmh1 provides PcG complex 1 with the GB domain, which acts as an E3 ubiquitin ligase for geminin. Scmh1 also induces the activity of an E3 ubiquitin ligase for histone H2A to repress transcription of Hox genes. Some Hox proteins form an RDCOX complex, which also acts as the E3 ubiquitin ligase for geminin. The cascades suppressed or induced by Scmh1 deficiency are indicated by dotted and bold lines, respectively. Degradation of geminin may be homeostatically tuned up by a molecular network mediated by Scmh1. (B) Model for a role of PcG complex 1- and RDCOX-mediated geminin degradation in mammalian development. Hox gene transcription is segmentally regulated along the anteroposterior axis of the body. Hox genes express from a specific anterior border to more posterior part of embryos. The PcG complex 1 represses the transcription of Hox genes in the anterior part. Thus, PcG complex 1 may determine the expression level of geminin in the anterior part, while the RDCOX complex in the posterior part, which might coordinately regulate cellular proliferation and differentiation during mammalian development.
ACKNOWLEDGMENTS
This study was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, the Japan Society for the Promotion of Science Summer Program, the Ube Foundation, the Takeda Science Foundation, the Tsuchiya Medical Foundation, the Children's Cancer Foundation of Japan, the Mitsubishi Pharma Research Foundation, the Daiwa Securities Health Foundation, Novartis Pharma Japan, Okinaka Memorial Institute for Medical Research, and the Mother and Child Health Foundation.
We thank the Analysis Center of Life Science and Radiation Research Facility in Hiroshima University and M. Tsumura, Y. Kageyama, S. Mori, A. Kawashima, and Y. Nakashima, as well as H. Tetsuguchi, and T. Furutani for technical assistance and H. Shimamoto and N. Aritani for secretarial assistance.
Footnotes
Published ahead of print 3 December 2012
REFERENCES
- 1. Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. 2007. Genome regulation by polycomb and trithorax proteins. Cell 128:735–745 [DOI] [PubMed] [Google Scholar]
- 2. Takihara Y, Hara J. 2000. Polycomb-group genes and hematopoiesis. Int. J. Hematol. 72:165–172 [PubMed] [Google Scholar]
- 3. Wang L, Brown J, Cao R, Zhang Y, Kassis JA, Jones RS. 2004. Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell 14:637–646 [DOI] [PubMed] [Google Scholar]
- 4. Nakagawa T, Kajitani T, Togo S, Masuko N, Ohdan H, Hishikawa Y, Koji T, Matsuyama T, Ikura T, Muramatsu M, Ito T. 2008. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 22:37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. 2004. Role of histone H2A ubiquitination in polycomb silencing. Nature 431:873–878 [DOI] [PubMed] [Google Scholar]
- 6. Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG. 2008. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol. Cell 29:69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, Wutz A, Vidal M, Elderkin S, Brockdorff N. 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tomotsune D, Takihara Y, Berger J, Duhl D, Joo S, Kyba M, Shirai M, Ohta H, Matsuda Y, Honda BM, Simon J, Shimada K, Brock HW, Randazzo F. 1999. A novel member of murine polycomb-group proteins, Sex comb on midleg homolog protein, is highly conserved, and interacts with RAE28/mph1 in vitro. Differentiation 65:229–239 [DOI] [PubMed] [Google Scholar]
- 9. Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. 2002. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 22:6070–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Luo L, Yang X, Takihara Y, Knoetgen H, Kessel M. 2004. The cell-cycle regulator geminin inhibits Hox function through direct and polycomb-mediated interactions. Nature 427:749–753 [DOI] [PubMed] [Google Scholar]
- 11. Ohta H, Sawada A, Kim JY, Tokimasa S, Nishiguchi S, Humphries RK, Hara J, Takihara Y. 2002. Polycomb group gene rae28 is required for sustaining activity of hematopoietic stem cells. J. Exp. Med. 195:59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohtsubo M, Yasunaga S, Ohno Y, Tsumura M, Okada S, Ishikawa N, Shirao K, Kikuchi A, Nishitani H, Kobayashi M, Takihara Y. 2008. Polycomb-group complex 1 acts as an E3 ubiquitin ligase for geminin to sustain hematopoietic stem cell activity. Proc. Natl. Acad. Sci. U. S. A. 105:10396–10401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bornemann D, Miller E, Simon J. 1996. The Drosophila Polycomb group gene Sex comb on midleg (Scm) encodes a zinc finger protein with similarity to polyhomeotic protein. Development 122:1621–1630 [DOI] [PubMed] [Google Scholar]
- 14. Kim CA, Gingery M, Pilpa RM, Bowie JU. 2002. The SAM domain of polyhomeotic forms a helical polymer. Nat. Struct. Biol. 9:453–457 [DOI] [PubMed] [Google Scholar]
- 15. Grimm C, de Ayala Alonso AG, Rybin V, Steuerwald U, Ly-Hartig N, Fischle W, Müller J, Müller CW. 2007. Structural and functional analyses of methyl-lysine binding by the malignant brain tumour repeat protein Sex comb on midleg. EMBO Rep. 8:1031–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takada Y, Isono K, Shinga J, Turner JM, Kitamura H, Ohara O, Watanabe G, Singh PB, Kamijo T, Jenuwein T, Burgoyne PS, Koseki H. 2007. Mammalian polycomb Scmh1 mediates exclusion of polycomb complexes from the XY body in the pachytene spermatocytes. Development 134:579–590 [DOI] [PubMed] [Google Scholar]
- 17. Blow JJ, Hodgson B. 2002. Replication licensing–defining the proliferative state? Trends Cell Biol. 12:72–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. 2000. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290:2309–2312 [DOI] [PubMed] [Google Scholar]
- 19. Ballabeni A, Melixetian M, Zamponi R, Masiero L, Marinoni F, Helin K. 2004. Human geminin promotes pre-RC formation and DNA replication by stabilizing CDT1 in mitosis. EMBO J. 23:3122–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lutzmann M, Maiorano D, Méchali M. 2006. A Cdt1-geminin complex licenses chromatin for DNA replication and prevents rereplication during S phase in Xenopus. EMBO J. 25:5764–5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seo S, Herr A, Lim JW, Richardson GA, Richardson H, Kroll KL. 2005. Geminin regulates neuronal differentiation by antagonizing Brg1 activity. Genes Dev. 19:1723–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Del Bene F, Tessmar-Raible K, Wittbrodt J. 2004. Direct interaction of geminin and Six3 in eye development. Nature 427:745–749 [DOI] [PubMed] [Google Scholar]
- 23. Gonzalez MA, Tachibana KE, Adams DJ, van der Weyden L, Hemberger M, Coleman N, Bradley A, Laskey RA. 2006. Geminin is essential to prevent endoreduplication and to form pluripotent cells during mammalian development. Genes Dev. 20:1880–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang VS, Carter SA, Hyland SJ, Tachibana-Konwalski K, Laskey RA, Gonzalez MA. 2011. Geminin escapes degradation in G1 of mouse pluripotent cells and mediates the expression of Oct4, Sox2, and Nanog. Curr. Biol. 21:692–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ohno Y, Yasunaga S, Ohtsubo M, Mori S, Tsumura M, Okada S, Ohta T, Ohtani K, Kobayashi M, Takihara Y. 2010. Hoxb4 transduction downregulates geminin protein, providing hematopoietic stem and progenitor cells with proliferation potential. Proc. Natl. Acad. Sci. U. S. A. 107:21529–21534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Petroski MD, Deshaies RJ. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell. Biol. 6:9–20 [DOI] [PubMed] [Google Scholar]
- 27. Ciechanover A. 1998. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17:7151–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McGarry TJ, Kirschner MW. 1998. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93:1043–1053 [DOI] [PubMed] [Google Scholar]
- 29. Ikeda W, Nakanishi H, Miyoshi J, Mandai K, Ishizaki H, Tanaka M, Togawa A, Takahashi K, Nishioka H, Yoshida H, Mizoguchi A, Nishikawa S, Takai Y. 1999. Afadin: a key molecule essential for structural organization of cell-cell junctions of polarized epithelia during embryogenesis. J. Cell Biol. 146:1117–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takihara Y, Tomotsune D, Shirai M, Katoh-Fukui Y, Nishii K, Motaleb MA, Nomura M, Tsuchiya R, Fujita Y, Shibata Y, Higashinakagawa T, Shimada K. 1997. Targeted disruption of the mouse homologue of the Drosophila polyhomeotic gene leads to altered anteroposterior patterning and neural crest defects. Development 124:3673–3682 [DOI] [PubMed] [Google Scholar]
- 31. Shirai M, Osugi T, Koga H, Kaji Y, Takimoto E, Komuro I, Hara J, Miwa T, Yamauchi-Takihara K, Takihara Y. 2002. The Polycomb-group gene Rae28 sustains Nkx2.5/Csx expression and is essential for cardiac morphogenesis. J. Clin. Invest. 110:177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van der Lugt NM, Alkema M, Berns A, Deschamps J. 1996. The Polycomb-group homolog Bmi-1 is a regulator of murine Hox gene expression. Mech. Dev. 58:153–164 [DOI] [PubMed] [Google Scholar]
- 33. van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, van der Valk M, Deschamps J, Sofroniew M, van Lohuizen M, Berns A. 1994. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 8:757–769 [DOI] [PubMed] [Google Scholar]
- 34. Morrison SJ, Hemmati HD, Wandycz AM, Weissman IL. 1995. The purification and characterization of fetal liver hematopoietic stem cells. Proc. Natl. Acad. Sci. U. S. A. 92:10302–10306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yoshida K, Inoue I. 2004. Regulation of geminin and Cdt1 expression by E2F transcription factors. Oncogene 23:3802–3812 [DOI] [PubMed] [Google Scholar]
- 36. Takihara Y. 2008. Role of Polycomb-group genes in normal and malignant stem cells. Int. J. Hematol. 87:25–34 [DOI] [PubMed] [Google Scholar]
- 37. Morrison A, Chaudhuri C, Ariza-McNaughton L, Muchamore I, Kuroiwa A, Krumlauf R. 1995. Comparative analysis of chicken Hoxb-4 regulation in transgenic mice. Mech. Dev. 53:47–59 [DOI] [PubMed] [Google Scholar]
- 38. Gould A, Morrison A, Sproat G, White RA, Krumlauf R. 1997. Positive cross-regulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 11:900–913 [DOI] [PubMed] [Google Scholar]
- 39. Lebert-Ghali CE, Fournier M, Dickson GJ, Thompson A, Sauvageau G, Bijl JJ. 2010. HoxA cluster is haploinsufficient for activity of hematopoietic stem and progenitor cells. Exp. Hematol. 38:1074–1086 [DOI] [PubMed] [Google Scholar]
- 40. Lawrence HJ, Christensen J, Fong S, Hu YL, Weissman I, Sauvageau G, Humphries RK, Largman C. 2005. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood 106:3988–3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Trojer P, Cao AR, Gao Z, Li Y, Zhang J, Xu X, Li G, Losson R, Erdjument-Bromage H, Tempst P, Farnham PJ, Reinberg D. 2011. L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol. Cell 42:438–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van de Vosse E, Walpole SM, Nicolaou A, van der Bent P, Cahn A, Vaudin M, Ross MT, Durham J, Pavitt R, Wilkinson J, Grafham D, Bergen AA, van Ommen GJ, Yates JR, den Dunnen JT, Trump D. 1998. Characterization of SCML1, a new gene in Xp22, with homology to developmental polycomb genes. Genomics 49:96–102 [DOI] [PubMed] [Google Scholar]
- 43. Montini E, Buchner G, Spalluto C, Andolfi G, Caruso A, den Dunnen JT, Trump D, Rocchi M, Ballabio A, Franco B. 1999. Identification of SCML2, a second human gene homologous to the Drosophila sex comb on midleg (Scm): a new gene cluster on Xp22. Genomics 58:65–72 [DOI] [PubMed] [Google Scholar]
- 44. Grimm C, Matos R, Ly-Hartig N, Steuerwald U, Lindner D, Rybin V, Müller J, Müller CW. 2009. Molecular recognition of histone lysine methylation by the polycomb group repressor dSfmbt. EMBO J. 28:1965–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Usui H, Ichikawa T, Kobayashi K, Kumanishi T. 2000. Cloning of a novel murine gene Sfmbt, Scm-related gene containing four mbt domains, structurally belonging to the polycomb group of genes. Gene 248:127–135 [DOI] [PubMed] [Google Scholar]
- 46. Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. 2003. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP, and MBT domains. Trends Biochem. Sci. 28:69–74 [DOI] [PubMed] [Google Scholar]
- 47. Buchenau P, Hodgson J, Strutt H, Arndt-Jovin DJ. 1998. The distribution of polycomb-group proteins during cell division and development in Drosophila embryos: impact on models for silencing. J. Cell Biol. 1998:469–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fanti L, Perrini B, Piacentini L, Berloco M, Marchetti E, Palumbo G, Pimpinelli S. 2008. The trithorax group and Pc group proteins are differentially involved in heterochromatin formation in Drosophila. Chromosoma 117:25–39 [DOI] [PubMed] [Google Scholar]
- 49. Voncken JW, Schweizer D, Aagaard L, Sattler L, Jantsch MF, van Lohuizen M. 1999. Chromatin-association of the polycomb group protein BMI1 is cell cycle-regulated and correlates with its phosphorylation status. J. Cell Sci. 112:4627–4639 [DOI] [PubMed] [Google Scholar]
- 50. Mohd-Sarip A, Lagarou A, Doyen CM, van der Knaap JA, Aslan Ü Bezstarosti K, Yassin Y, Brock HW, Demmers JA, Verrijzer CP. 2012. Transcription-independent function of polycomb group protein PSC in cell cycle control. Science 336:744–747 [DOI] [PubMed] [Google Scholar]
- 51. Ohno Y, Yasunaga S, Janmohamed S, Ohtsubo M, Saeki K, Kurogi T, Mihara K, Iscove NN, Takihara Y. Hoxa9 transduction induces hematopoietic stem and progenitor cell activity through direct down-regulation of geminin protein. PLoS One, in press [DOI] [PMC free article] [PubMed] [Google Scholar]