Abstract
Myoblast differentiation into mature myotubes is a critical step in the development and repair of human skeletal muscle. Here we show that small interfering RNA (siRNA)-based silencing of the Ste20-like mitogen-activated protein 4 kinase 4 (Map4k4) in C2C12 myoblasts markedly enhances expression of myogenic differentiation genes, myoblast fusion, and myotube diameter. In contrast, adenovirus-mediated expression of native Map4k4 in C2C12 cells attenuates each of these processes, indicating that Map4k4 is a negative regulator of myogenic differentiation and hypertrophy. Expression of a Map4k4 kinase-inactive mutant enhances myotube formation, suggesting that the kinase activity of Map4k4 is essential for its inhibition of muscle differentiation. Map4k4 regulation of myogenesis is unlikely to be mediated by classic mitogen-activated protein kinase (MAPK) signaling pathways, because no significant difference in phosphorylation of extracellular signal-regulated kinase (ERK), p38, or c-Jun N-terminal kinase (JNK) is observed in Map4k4-silenced cells. Furthermore, silencing of these other MAPKs does not result in a hypertrophic myotube phenotype like that seen with Map4k4 depletion. Uniquely, Map4k4 silencing upregulates the expression of the myogenic regulatory factor Myf5, whose depletion inhibits myogenesis. Furthermore, Myf5 is required for enhancement of myotube formation in Map4k4-silenced cells, while Myf5 overexpression rescues Map4k4-mediated inhibition of myogenic differentiation. These results demonstrate that Map4k4 is a novel suppressor of skeletal muscle differentiation, acting through a Myf5-dependent mechanism.
INTRODUCTION
Skeletal muscle differentiation is a highly coordinated multistep process in which mononucleated myoblasts first withdraw from the cell cycle in response to extracellular cues, then differentiate into postmitotic myocytes (early differentiation), and subsequently fuse into multinucleated myotubes (late differentiation) which finally bundle to form mature muscle fibers (terminal differentiation). This process is elaborately controlled by activation of Myf5, MyoD, myogenin, and MRF4, four myogenic regulatory factors (MRFs) belonging to a family of basic helix-loop-helix transcription factors. During myogenesis, MRFs are activated and operate in concert with other transcriptional regulators, such as myocyte enhancer factor 2 (MEF2), in a coordinated manner to regulate the transcription of muscle-specific genes, including those for myosin heavy chain (MyHC) and muscle creatine kinase (MCK) (1–3). Previous studies have confirmed Myf5 and MyoD as muscle determination factors that are expressed mainly in undifferentiated myoblasts and differentiating myocytes, while myogenin is activated in early differentiation (4). MRF4 has been shown to be expressed transiently during somitogenesis and later fiber maturation (5), playing roles in myogenic lineage commitment (6) as well as myoblast fusion and differentiation (7, 8).
Mitogen-activated protein kinases (MAPKs) are components of serine/threonine protein kinase cascades that respond to extracellular stimuli and regulate essential cellular functions, such as proliferation (9), differentiation (10–12), and apoptosis (13). The MAPK family is categorized into three main groups, i.e., p38 stress-activated protein kinase, c-Jun N-terminal kinase (JNK), and extracellular signal-regulated protein kinase (ERK) (14), all of which have been demonstrated to be involved in mammalian skeletal myogenesis (15, 16). The exploration of new upstream kinases that modulate the downstream effector MAPKs identified mitogen-activated protein kinase kinase kinase kinase4 (MAP4K4), a serine/threonine protein kinase that belongs to the germinal-center kinase GCK-IV group of Saccharomyces cerevisiae sterile 20 protein (Ste20) kinases (17). Map4k4 may activate the JNK signaling pathway in some cell types and mediate cancer cell proliferation, apoptosis, and motility (18, 19). It was found to play an essential role in development (20), and it has been shown to be critical for mesoderm migration during gastrulation by acting upstream of p38 MAPK (21). In an RNA interference (RNAi)-based screen for regulators of adipocyte function, we discovered that Map4k4 downregulated expression of peroxisome proliferator-activated receptor γ (PPARγ), a transcription factor that is essential for adipocyte differentiation and function (22). Cell size, insulin-mediated glucose transport, and triglyceride content were found to be increased significantly upon Map4k4 silencing in cultured adipocytes (23), indicating that Map4k4 is a negative regulator of insulin-stimulated lipogenesis and adipose hypertrophy.
More recently, it was found that Map4k4 also functions in muscle to impair insulin sensitivity. Map4k4 gene silencing in primary human skeletal muscle cells prevented tumor necrosis factor alpha (TNF-α)-induced insulin resistance (24). However, the role of Map4k4 in skeletal myogenesis has not been addressed. In the present study, we used C2C12 murine myoblasts to investigate the function of Map4k4 in myogenic processes. We demonstrate that Map4k4 acts upstream of Myf5 as a negative regulator of skeletal muscle differentiation.
MATERIALS AND METHODS
Molecular biology.
A mouse Myf5 clone (GenBank accession number NM_008656) was purchased from Open Biosystems. The coding region of the Myf5 gene was PCR amplified and cloned into a pCMV plasmid carrying three N-terminal hemagglutinin (HA) tags (pCMV-3HA) to create an N-terminally HA-tagged Myf5 construct. Primers used to amplify PCR fragments of the Myf5 coding region were as follows: for the 5′ fragment, CGATCGCCACGCGTATCTCGAGCTATGGACATGACGGACGGCTGCCAGTTCTCCCCT; and for the 3′ fragment, GCGTACGGATCCGTCGACTCATAATACGTGATAGATAAGTCTGGAGCTGGAGGGTCC.
Cell culture and transfection.
Mouse C2C12 myoblasts (American Type Culture Collection) were cultured in growth medium (GM) consisting of Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin at 37°C with 5% CO2. To induce differentiation, 95% confluent cells were placed in a differentiation medium (DM) consisting of DMEM with 2% horse serum. Multinucleated myotubes were evident after 3 days of differentiation. For small interfering RNA (siRNA) transfection, C2C12 myoblasts cultured in growth medium were transfected with 50 pmol siRNA by use of Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions for reverse transfection. Twenty-four hours later, cells were switched to DM and cultured for the indicated times before harvest. To transfect siRNA into differentiated myotubes, siRNA-endoporter complexes were used as described previously (25). Briefly, 50 pmol of siRNA was incubated with 2.5 nmol of endoporter (Gene Tools) in phosphate-buffered saline (PBS) for 15 min and then added to cells. All siRNAs were purchased from Dharmacon (Lafayette, CO). For plasmid transfection, GM-cultured C2C12 myoblasts were transfected with 2 μg of plasmids by use of Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions for reverse transfection. Twenty-four hours later, cells were switched to DM and cultured for the indicated times before harvest.
Adenovirus infection.
C2C12 myoblasts were grown until 90% confluence and were then infected with a green fluorescent protein (GFP)-expressing control virus, wild-type Map4k4-expressing virus, or Map4k4 D152N-expressing virus at a dose of 104 virus particles per cell for 18 h in GM before differentiation. At 72 h postdifferentiation, cells were fixed for immunofluorescence or harvested for Western blotting. All adenoviruses were gifts from Diane L. Barber (Department of Cell and Tissue Biology, University of California, San Francisco, CA).
Myotube analysis.
Myotube nuclei were counted in approximately 100 randomly chosen MyHC-positive cells containing three or more nuclei. Myotubes were categorized into three groups (3 to 6 nuclei, 7 to 15 nuclei, and >15 nuclei per myotube) and were expressed as a percentage of the total myotube number. The fusion index was calculated as the ratio of nuclei in MyHC-positive myotubes to the total number of nuclei in the field for five random fields. To analyze myotube diameter, five fields were chosen randomly, and three myotubes were measured per field. The average diameter per myotube was calculated as the mean of three measurements taken along the long axis of the myotube.
Isolation of mouse satellite cells.
Satellite cells were isolated from 6- to 8-week-old C57B6/J mice by fluorescence-activated cell sorting (FACS) as described previously (26). Briefly, skeletal muscles were digested with collagenase B (10 mg/ml; Roche) and dispase II (2.4 U/ml; Roche) and then filtered successively through 250-, 100, and 40-μm nylon meshes. The resulting mononuclear cells were incubated with anti-mouse integrin α7 clone 3C12 (MBL) and biotin anti-mouse CD34 (eBioscience) antibodies for 20 min and then stained with Hoechst 33258 (Sigma) and with phycoerythrin (PE)–anti-mouse CD11b (eBiosciences), PE–anti-mouse CD45 (eBiosciences), PE–anti-mouse Ly-6A-E (Sca1; BD Bioscience), PE–anti-rat CD31 (BD Bioscience), and streptavidin-allophycocyanin (APC)-Cy7 (BD Bioscience) antibodies. Integrin α7+ CD34+ CD31− CD11b− CD45− Sca1− cells were considered satellite cells and were sorted by a BD FACS Aria II flow cytometer.
Western blotting.
Cells were solubilized with ice-cold lysis buffer (20 mM HEPES, pH 7.2, 100 mM NaCl, 1 mM EDTA, 100 mM phenylmethylsulfonyl fluoride [PMSF], 0.01% Triton X-100, 1% SDS, and Halt protease and phosphatase inhibitor cocktail [Thermo Scientific]), and protein concentrations were assessed by bicinchoninic acid (BCA) assay (Thermo Scientific). Equal amounts of protein were loaded onto 8.5% SDS-polyacrylamide gels and transferred to nitrocellulose membranes. The following antibodies were used: anti-Map4k4 (Bethyl), anti-Myf5 (sc-20; Santa Cruz), anti-MyoD (BD Biosciences), anti-Mef2C (Cell Signaling), antimyogenin (F5D; Developmental Studies Hybridoma Bank [DSHB], University of Iowa), anti-sarcomeric myosin heavy chain (MHC) (MF20; DSHB, University of Iowa), anti-phospho-p38 (Cell Signaling), anti-total p38α (Cell Signaling), anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (Cell Signaling), anti-p44/42 MAPK (Erk1/2) (Santa Cruz), anti-phospho-SAPK/JNK (Thr183/Tyr185) (Cell Signaling), and anti-SAPK/JNK (Cell Signaling).
Immunofluorescence microscopy.
Cells grown on glass coverslips were fixed with 4% formaldehyde and blocked in PBS containing 2% goat serum (Invitrogen), 1% bovine serum albumin (Sigma), 0.1% Tween 20, and 0.05% Triton X-100 (American Bioanalytical) for 1 h at room temperature. The cells were then incubated with the MF20 monoclonal antibody (MAb) against MHC (1:40; DSHB) for 2.5 h and subsequently with an Alexa Fluor 488- or Alexa Fluor 594-conjugated secondary antibody (1:200; Invitrogen) for 1 h at room temperature. Cells were mounted with ProLong Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen). Images were obtained using a Zeiss Axiovert 200 inverted microscope equipped with a Zeiss AxioCam HR charge-coupled device (CCD) camera.
Creatine kinase activity assay.
Cells were lysed in ice-cold lysis buffer (20 mM HEPES, pH 7.2, 100 mM NaCl, 1 mM EDTA, 100 mM PMSF, 0.01% Triton X-100, and protease and phosphatase inhibitor cocktail). Lysates were centrifuged at 14,000 × g for 10 min at 4°C, and the supernatants were used immediately for creatine kinase (CK) activity assay. CK activity was measured using a spectrophotomety-based kit (Stanbio Laboratory, Boerne, TX) according to the manufacturer's instructions. Specific CK activity was calculated by normalization to the total protein content.
Map4k4 kinase activity assay.
Cells were solubilized with ice-cold lysis buffer (50 mM Tris, pH 7.4, 100 mM NaCl, 5 mM EDTA, 1% NP-40, 0.1% deoxycholic acid, and protease and phosphatase inhibitor cocktail). Cell lysates were immunoprecipitated with anti-Map4k4 antibody (Bethyl). Myelin basic protein (MBP) (1 μg) and 10 μCi of [γ-32P]ATP were added to the immunoprecipitates and incubated for 30 min at 30°C in kinase buffer (20 mM HEPES, 10 nM MgCl2, 1 mM dithiothreitol [DTT], and protease and phosphatase inhibitor cocktail). Samples were separated by 12% SDS-PAGE and visualized by autoradiography. Map4k4 kinase activities were determined by normalizing the radioactivity of 32P-labeled MBP to the amount of immunoprecipitated Map4k4 protein detected by Western blotting.
Isolation of RNA and RT-PCR.
RNA isolation was performed according to the TRIzol reagent protocol (Invitrogen). cDNA was synthesized from 1.5 μg of total RNA by use of an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. For real-time PCR (RT-PCR), synthesized cDNA and iQ SYBR green Supermix were run on a MyIQ real-time PCR system (Bio-Rad) with the following primer pairs: for Map4k4, 5′-CATCTCCAGGGAAATCCTCAGG-3′ and 5′-TTCTGTAGTCGTAAGTGGCGTCTG-3′; and for Myf5, 5′-TATGAAGGCTCCTGTATCCC-3′ and 5′-ACGTGCTCCTCATCGTCTG-3′. 36B4 was used as an internal loading control. Relative gene expression was determined using the ΔCT method (27).
Statistics.
The statistical significance of differences in the means of experimental groups was determined by two-tailed Student's t test, using Microsoft Excel. The data are presented as means ± standard errors of the means (SEM). A P value of <0.05 was considered significant.
RESULTS
Map4k4 expression and protein kinase activity are decreased during skeletal muscle differentiation.
To determine whether Map4k4 may play a role in muscle differentiation, we first examined Map4k4 expression in primary mouse satellite cells and mature muscle fibers. Adult satellite cells are considered to be progenitor cells of somitic origin in skeletal muscle development (28). RT-PCR analysis revealed a 4-fold decrease in Map4k4 expression in mouse quadriceps compared with that in purified satellite cells isolated from the same mice via fluorescence-activated cell sorting (Fig. 1A). We also investigated Map4k4 expression during differentiation of C2C12 cells, a well-established cell line that is derived from mouse satellite cells and faithfully mimics the skeletal muscle differentiation process in vitro. A reduction of Map4k4 protein levels during C2C12 myogenic differentiation was detected by Western blotting, concomitant with increased expression of the differentiation markers myogenin and MyHC (Fig. 1B). Furthermore, we measured Map4k4 protein kinase activity during C2C12 cell differentiation and determined that it was highest in confluent myoblasts and decreased dramatically after 24 h of differentiation (Fig. 1C). These results indicate that Map4k4 is dynamically regulated during skeletal muscle differentiation, suggesting a potential role for Map4k4 in muscle development.
Fig 1.
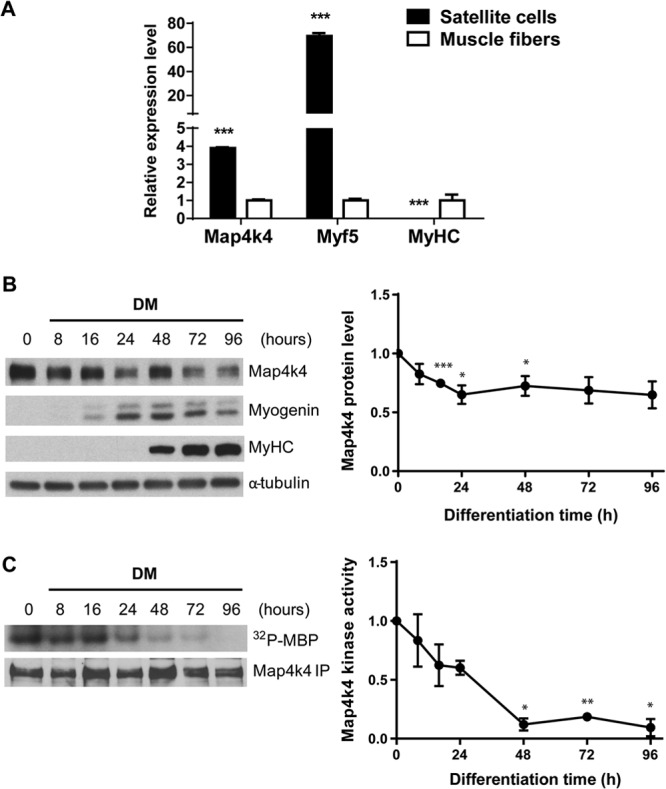
Expression and kinase catalytic activity of Map4k4 during skeletal muscle differentiation. (A) RT-PCR analysis of Map4k4 expression in isolated mouse satellite cells versus quadriceps. Data represent means ± SEM for three independent experiments. (B) Expression of Map4k4 during C2C12 myogenic differentiation. Densitometry results are representative of means ± SEM for four independent experiments. (C) C2C12 myoblasts were incubated in DM for the indicated times. Map4k4 kinase activity at each time point was assessed by kinase assay and immunoblotting. Results represent means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Map4k4 silencing promotes skeletal muscle differentiation.
To explore the function of Map4k4 in myogenic differentiation, we used siRNA directed against Map4k4 to deplete the protein kinase in C2C12 myoblasts and monitored morphological differences during cell differentiation. Map4k4 silencing resulted in a significant sustained reduction of Map4k4 protein throughout differentiation and formation of larger myotubes after 48 h in DM (Fig. 2A). Enhanced muscle cell fusion was observed in Map4k4-silenced cells on differentiation day 3, as there was a shift toward myotubes containing more nuclei per myotube (Fig. 2B) and having an increased fusion index (Fig. 2C). Map4k4 silencing also resulted in a 70% increase of cell diameter in day 3 myotubes (Fig. 2D), likely due to enhanced myoblast fusion. In addition, myoblast proliferation was not affected by Map4k4 depletion, as indicated by similar nuclear numbers in random microscopic fields and no change in the percentage of 5-ethynyl-2′-deoxyuridine (EdU)-positive cells following a 1-h pulse when Map4k4 was silenced in myoblasts (data not shown). These data exclude the possibility that the hypernucleated myotubes with increased size resulted from an increased number of undifferentiated myoblasts available for the fusion process.
Fig 2.
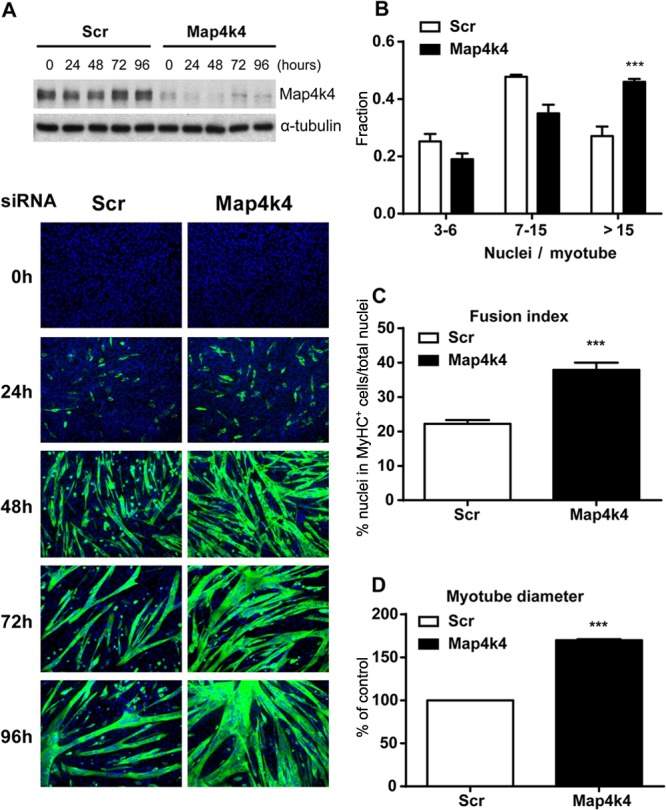
Map4k4 silencing promotes myotube formation in C2C12 cells. C2C12 myoblasts were transfected with scrambled siRNA or siRNA against Map4k4. Twenty-four hours later, the cells were transferred to DM for the indicated times. (A) (Top) Efficiency of Map4k4 knockdown as determined by immunoblotting. (Bottom) Cells were fixed and immunostained for MyHC, and myoblast differentiation was observed by fluorescence microscopy (green, MyHC; blue, DAPI). Magnification, ×100. Data are representative of at least three independent experiments. (B) Fractions of myotubes with the indicated numbers of nuclei were quantified among 100 randomly chosen myotubes after 72 h in DM. (C) The fusion index for day 3 myotubes was calculated from the ratio of the number of nuclei in MyHC-positive myotubes to the total number of nuclei in one field for five random microscopic fields. (D) Myotube diameters were measured with day 3 myotubes. Results represent means and SEM for three independent experiments. ***, P < 0.001.
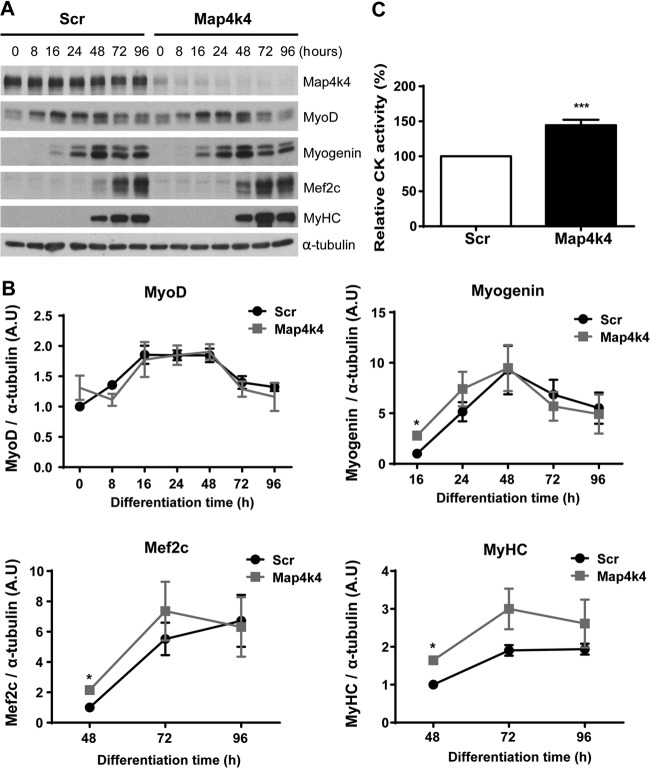
We further studied the differentiation program by examining the expression of muscle differentiation markers. No significant changes in MyoD protein levels were detected in Map4k4-silenced cells compared with scrambled siRNA-transfected controls during differentiation (Fig. 3A and B). However, significant transient increases in myogenin and Mef2C expression were detected in Map4k4-silenced cells at 16 h and 48 h of differentiation, respectively (Fig. 3A and B). MyHC expression starts in a population of mononuclear myoblasts and rapidly increases with myoblast fusion during late myogenesis (29) (Fig. 2A). Map4k4 silencing enhanced MyHC expression during late C2C12 cell differentiation, although the increase was only significant at 48 h of differentiation, with trends toward increased expression at later time points (Fig. 3A and B). MCK activity, a later marker of skeletal muscle cell differentiation, was increased by 45% in Map4k4-silenced cells at day 3 of differentiation. These results suggest that silencing of Map4k4 enhances myotube formation and promotes skeletal myogenic differentiation.
Fig 3.
Map4k4 silencing enhances C2C12 myogenic differentiation. C2C12 myoblasts were transfected with scrambled or Map4k4 siRNA. Cells were transferred to DM 24 h after transfection and differentiated for the indicated times. (A) Expression of myogenic differentiation proteins was assayed by immunoblotting with the indicated antibodies. Data are representative of three independent experiments. (B) Densitometric analysis of the Western blots in panel A. (C) Creatine kinase (CK) activities of transfected cells were measured after 3 days in DM. Data represent means ± SEM for three independent experiments. A.U, absorbance units. *, P < 0.05; ***, P < 0.001.
Inhibition of myogenic differentiation by Map4k4 requires its kinase activity.
Since suppression of Map4k4 expression enhanced skeletal muscle differentiation, we hypothesized that Map4k4 overexpression would have the opposite effect. To test this, adenoviruses expressing a GFP control (AdGFP) or wild-type (wt) Map4k4 (AdMap4k4 wt) (30) were used to infect C2C12 myoblasts for 18 h prior to differentiation. Overexpression of wt Map4k4 impeded MyHC-positive myotube formation (Fig. 4A) and myoblast fusion (Fig. 4B) within 72 h of serum deprivation. Western blot analysis confirmed that the expression of Mef2c and the late myogenic differentiation marker MyHC was inhibited in wt Map4k4-overexpressing cells (Fig. 4C). We also assessed the effect of a Map4k4 kinase-inactive mutant on myogenic differentiation. C2C12 myoblasts were infected with adenoviruses expressing Map4k4 D152N, a kinase-inactive mutant of Map4k4 (AdMap4k4 D152N) (30), and were induced to differentiate into myotubes for 72 h. Interestingly, Map4k4 D152N overexpression caused the formation of larger myotubes and a substantial increase in myoblast fusion (Fig. 4A and B), similar to the results of the Map4k4 knockdown experiments (Fig. 2A and C). An increase in Mef2c and MyHC expression was also observed in Map4k4 D152N-overexpressing cells (Fig. 4C). These data suggest that the Map4k4 kinase-inactive mutant functions as a dominant-negative inhibitor, possibly by competing with functional endogenous Map4k4 in C2C12 cells, and that Map4k4 kinase activity is required to repress skeletal muscle differentiation.
Fig 4.
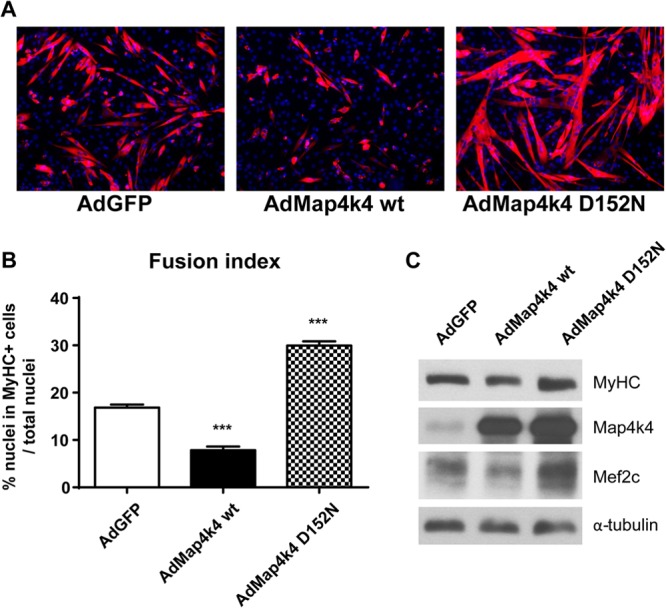
Map4k4 kinase activity is required for its inhibition of C2C12 myogenic differentiation. C2C12 myoblasts were infected with adenoviruses expressing GFP, wild-type Map4k4, or a Map4k4 kinase-inactive D152N mutant and differentiated for 72 h. (A) Cells were immunostained with anti-MyHC antibody. Images were photographed by fluorescence microscopy (red, MyHC; blue, DAPI). Magnification, ×100. (B) Quantitative analysis of myogenic conversion as scored by the fusion index. Data represent means and SEM for two independent experiments. ***, P < 0.001. (C) Immunoblot analysis of Map4k4 and myogenic markers. Data are representative of three independent experiments.
Map4k4 does not regulate myogenic differentiation through canonical MAPK signaling pathways.
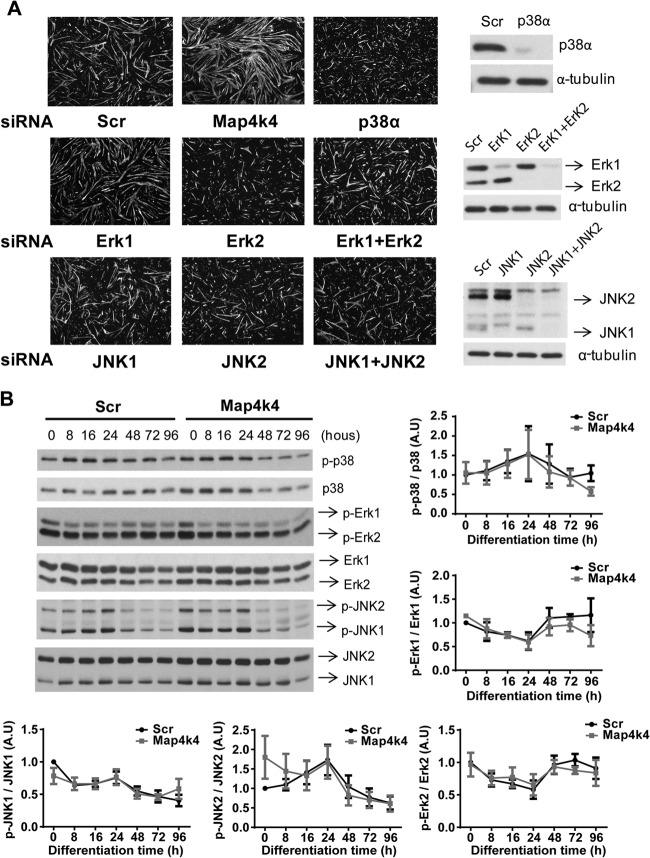
In other systems, Map4k4 has been described as an upstream effector in JNK, ERK, and p38 signaling pathways. These pathways are also reportedly involved in skeletal muscle differentiation, and thus it seemed possible that Map4k4 regulates myogenic differentiation through these canonical MAPK pathways. To assess this hypothesis, we used siRNA to suppress MAPK expression separately or in combination in C2C12 myoblasts and monitored myogenic differentiation by visualizing MyHC-positive myotube formation. We posited that if Map4k4 functions upstream in the respective signaling pathway to regulate myogenesis, then silencing of the downstream effectors would result in a phenotype similar to that with Map4k4 silencing. However, depletion of p38α abolished myogenic differentiation because few p38α-silenced cells fused into multinuclear myotubes (Fig. 5A), consistent with the conclusion derived from previous studies that p38α is critical for skeletal myogenesis (31–33). Other reports have shown that basal JNK activity is essential for regulation of skeletal muscle differentiation and that inhibition of JNK activation inhibits myogenesis by inducing myoblast apoptosis (34). We suppressed JNK1 expression in myoblasts and observed a minimal effect of JNK1 silencing on myotube formation under our experimental conditions. However, JNK2 or JNK1/2 silencing in combination inhibited myogenic differentiation, as shown by reduced myotube formation (Fig. 5A). Erk1/2 is essential for myoblast proliferation and is inhibitory to early differentiation, but it is also required for myocyte fusion. Inhibition of ERK activity early in myogenesis promotes differentiation, whereas later inhibition impedes differentiation (32). In our study, Erk1 silencing in C2C12 myoblasts promoted myotube formation, while knockdown of Erk2 resulted in the formation of smaller myotubes. Myotubes differentiated from Erk1 and Erk2 double-knockdown myoblasts had a modestly decreased size compared to those differentiated from the scrambled siRNA-transfected control cells (Fig. 5A). These results revealed that Map4k4 functions differently on myogenic differentiation than the canonical MAPK pathways. This conclusion was further confirmed by measurement of phosphorylation levels of the MAPKs during differentiation. No significant changes in p38α, ERK1/2, or JNK1/2 phosphorylation were observed in Map4k4-silenced cells (Fig. 5B) or C2C12 cells overexpressing kinase-inactive Map4k4 D152N (data not shown), indicating that Map4k4 failed to regulate their activities during myogenic differentiation.
Fig 5.
Map4k4 silencing does not promote skeletal muscle differentiation through canonical MAPK signaling pathways. C2C12 myoblasts were transfected with scrambled siRNA or siRNA against Map4k4, p38α, JNK1, JNK2, JNK1 and JNK2, Erk1, Erk2, or Erk1 and Erk2. Cells were transferred to DM 24 h after differentiation and then incubated for 72 h. (A) (Left) Myoblast differentiation was observed by immunostaining for MyHC expression. Magnification, ×50. (Right) Efficiency of MAP kinase knockdown as determined by Western blotting. (B) C2C12 myoblasts were transfected with scrambled or Map4k4 siRNA. Cells were transferred to DM 24 h after transfection and then incubated for the indicated times. Lysates were immunoblotted with the indicated antibodies. Densitometric analysis data represent means ± SEM for three independent experiments.
Map4k4 functions mainly at the early stage of myogenic differentiation.
To investigate the stages of myogenic differentiation in which Map4k4 functions, C2C12 cells were transfected with scrambled siRNA or siRNA targeting Map4k4 at multiple stages of differentiation and for various periods (Fig. 6A, right panels), and myotube formation was assessed on day 4 after initiation of differentiation by measuring the fusion index. In these experiments, 90% of Map4k4 proteins were depleted in myotubes on day 4 in response to transfections of Map4k4 siRNA at the different time points shown (Fig. 6B). Map4k4 silencing in myoblasts (day −1) provoked the most robust myotube formation (Fig. 6A, upper left panels), as the fusion index in Map4k4-silenced cells was 60% higher than that in the control cells on day 4 (Fig. 6C). Map4k4 depletion on day 1 in myocytes that were about to enter the late stage of differentiation still resulted in larger myotubes and increased myoblast fusion compared to those in the control cells. However, the promotion of myotube formation was less than that resulting from Map4k4 silencing at earlier stages in myoblast differentiation (Fig. 6A, middle left panels, and C). When siRNA against Map4k4 was transfected into day 2 myotubes, coincident with the onset of terminal differentiation, the myotubes showed even smaller changes in size or fusion than the results obtained from Map4k4 suppression in myoblasts and day 1 myocytes (Fig. 6A, lower left panels, and C). These results indicate that Map4k4 functions in multiple stages of muscle differentiation but that the enhanced myotube formation observed in Map4k4-depleted cells at later stages of differentiation results mainly from a role that Map4k4 plays at the onset of myogenic differentiation. The observation that Map4k4 apparently plays an early role in the process is also consistent with the higher Map4k4 kinase activity at the early stage of muscle differentiation (Fig. 1B).
Fig 6.
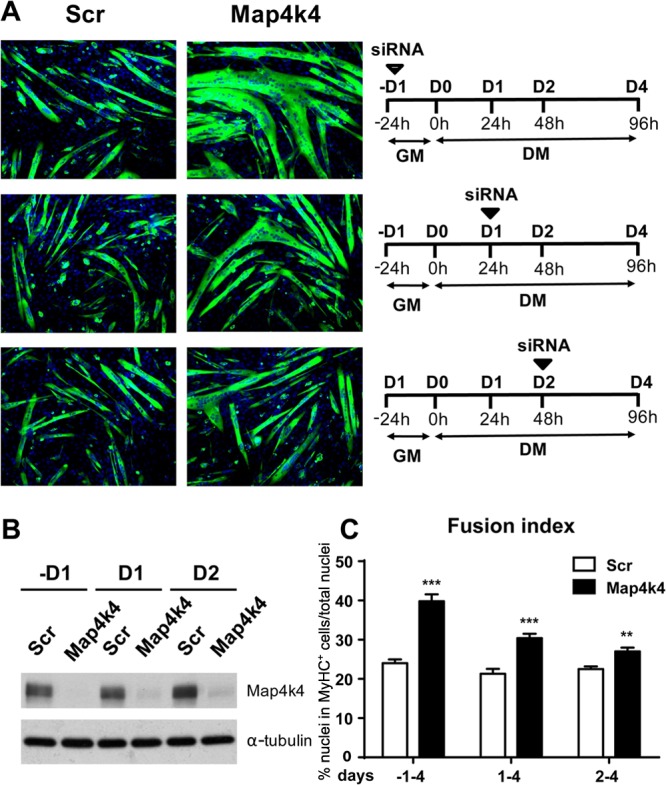
Map4k4 functions mainly at the early stage of myogenic differentiation. C2C12 myoblasts were transfected with scrambled or Map4k4 siRNA at different stages of differentiation for the indicated times. (A) (Left) Cells were fixed and immunostained for MyHC (green) after 4 days in DM. Magnification, ×100. (Right) Schematics of time courses of siRNA application. (B) Efficiency of Map4k4 knockdown as determined by immunoblotting. (C) The fusion index was calculated by dividing the number of nuclei in MHC-positive cells by the total number of nuclei in that field. Data represent means and SEM for two independent experiments. **, P < 0.01; ***, P < 0.001.
Map4k4 regulates myogenic differentiation in a Myf5-dependent manner.
Among the four myogenic regulatory factors, Myf5 and MyoD regulate the early stage of skeletal muscle differentiation. Because no change in MyoD expression was detected in Map4k4-silenced cells during differentiation (Fig. 3A and B), we examined Myf5 expression by RT-PCR and Western blotting. In cells treated with scrambled siRNA, Myf5 expression increased in early differentiation, peaked at 24 h, and then decreased (Fig. 7A and B). Map4k4 silencing increased Myf5 mRNA transcripts 1.4-fold within 16 h of myogenic differentiation (Fig. 7A). More dramatically, a 3-fold increase in Myf5 protein levels was detected in Map4k4-depleted undifferentiated myoblasts and myocytes at the early stage of differentiation (Fig. 7B).
Fig 7.
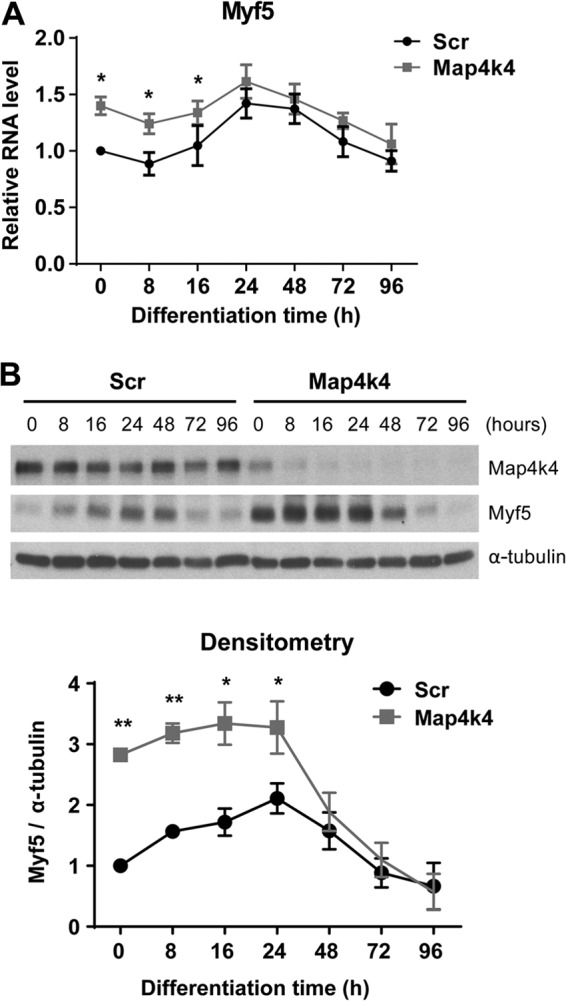
Map4k4 silencing increases protein levels of Myf5. (A) C2C12 myoblasts were transfected with scrambled or Map4k4 siRNA and then transferred to DM for the indicated times at 24 h posttransfection. (B) (Top) Myf5 expression determined by immunoblotting. (Bottom) Densitometry of Myf5 normalized to α-tubulin. Data are presented as means ± SEM for three independent experiments. *, P < 0.05; **, P < 0.01.
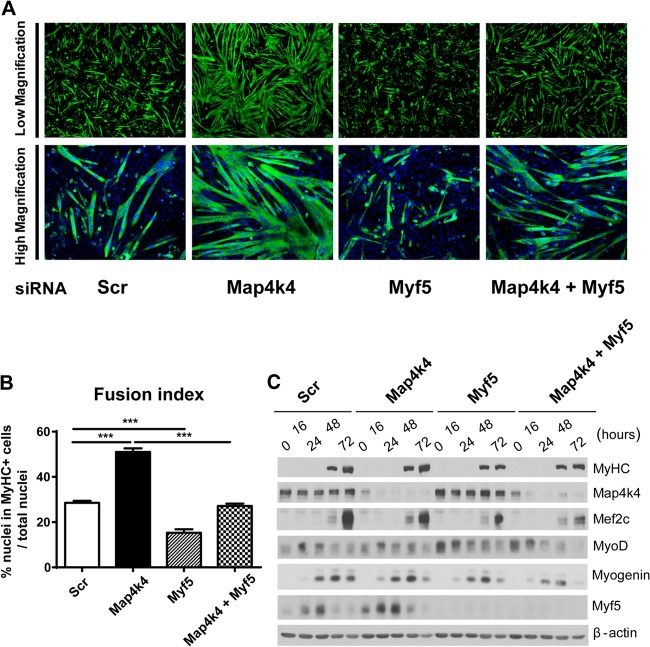
To determine whether the increase in Myf5 protein levels is essential for the enhanced myogenic differentiation that is found after Map4k4 depletion, we performed double-knockdown experiments to suppress Map4k4 and Myf5 expression simultaneously in C2C12 myoblasts and examined differentiation by microscopic analysis and Western blotting. As expected, Map4k4 knockdown promoted myogenic differentiation and Myf5 expression (Fig. 8). In contrast, Myf5 silencing impeded myogenic differentiation, as shown by reduced myotube formation (Fig. 8A), decreased myoblast fusion (Fig. 8B), and less expression of myogenin, Mef2C, and MyHC during differentiation (Fig. 8C). Importantly, compared with Map4k4 suppression alone, smaller myotubes with less fusion and myogenic differentiation factor expression were observed when Map4k4 and Myf5 were silenced simultaneously (Fig. 8), indicating that reduced levels of Myf5 expression partially inhibit the effects of Map4k4 silencing on myogenic differentiation.
Fig 8.
Suppression of Myf5 impairs Map4k4 silencing-enhanced myogenic differentiation. C2C12 myoblasts were transfected with scrambled siRNA or with siRNA against Map4k4, Myf5, or a combination of both. (A) Cells were fixed and immunostained for MyHC (green) and DAPI (blue) at day 3 after differentiation, and myogenic conversion was observed by fluorescence microscopy. Magnification, ×50 (top) and ×100 (bottom). (B) The fusion index was calculated for transfected cells after 3 days in DM. Results represent means and SEM for three independent experiments. ***, P < 0.001. (C) Immunoblots of myogenic differentiation proteins. Data are representative of three independent experiments.
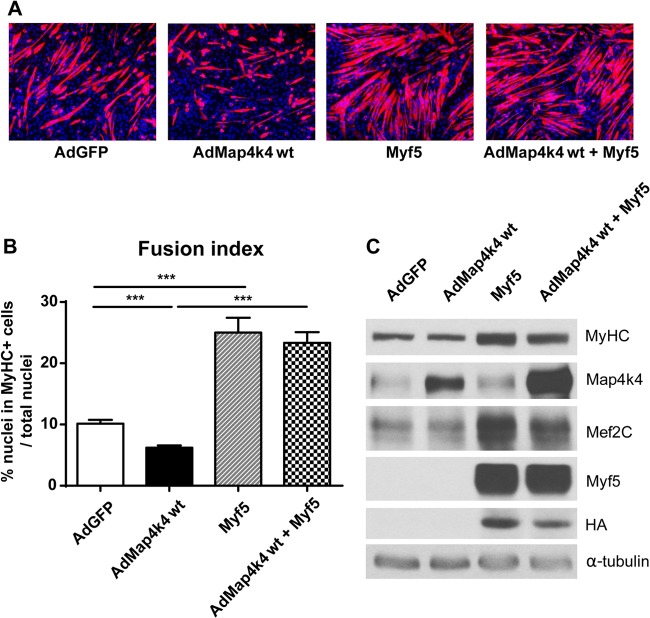
To further demonstrate that Myf5 is a downstream effector of Map4k4 signaling in the regulation of skeletal muscle differentiation, we overexpressed Map4k4 and Myf5 simultaneously in C2C12 myoblasts and examined muscle differentiation by microscopy and immunoblotting. Expression of native Map4k4 protein kinase inhibited C2C12 myogenic differentiation, while Myf5 expression robustly enhanced myotube formation (Fig. 9A and B) and late differentiation marker expression (Fig. 9C). Interestingly, Myf5 expression significantly reversed the inhibitory effect of Map4k4 expression in myogenic differentiation (Fig. 9). Based on these results, we concluded that Map4k4 regulates skeletal myogenesis at least partially by regulating Myf5 expression.
Fig 9.
Myf5 expression reverses the impaired myogenic differentiation caused by Map4k4. C2C12 myoblasts were transfected with empty vector or a construct expressing Myf5. Six hours later, the cells were infected with adenoviruses expressing GFP or Map4k4. (A) Cells were fixed and immunostained for MyHC (red) and DAPI (blue) on day 3 after differentiation, and myogenic conversion was observed by fluorescence microscopy. Magnification, ×100. (B) The fusion index was calculated for transfected cells after 3 days in DM. Results represent means and SEM for three independent experiments. ***, P < 0.001. (C) Immunoblots of myogenic differentiation proteins. Data are representative of three independent experiments.
DISCUSSION
In this study, we demonstrated that Map4k4 negatively regulates C2C12 myogenic differentiation through a Myf5-dependent mechanism. RNAi-mediated gene-specific silencing of Map4k4 expression or expression of a Map4k4 kinase-inactive mutant enhanced the differentiation of C2C12 myoblasts (Fig. 2, 3, and 4), while Map4k4 expression inhibited this process (Fig. 4). Map4k4 suppression resulted in a significant increase in Myf5 expression at the early stage of differentiation (Fig. 7). Furthermore, silencing of Myf5 inhibited C2C12 myoblast differentiation and also suppressed the ability of Map4k4 silencing to enhance myotube formation (Fig. 8). In contrast, Myf5 expression promoted myotube formation and reversed Map4k4-mediated myogenic differentiation inhibition (Fig. 9), suggesting that Map4k4 regulates C2C12 myogenesis at least in part through Myf5.
We found that Map4k4 does not likely suppress myogenic differentiation via activation of the canonical MAPK signaling pathways, although it has been reported that Map4k4 can act upstream of p38, JNK, and ERK. In contrast to the dramatic enhancement of myotube formation upon Map4k4 silencing, a slight increase of myotube formation was observed in Erk1-silenced cells, while suppression of other MAPKs resulted in either no morphological difference or smaller myotubes. In addition, no change in levels of phosphorylated MAPKs was observed upon Map4k4 knockdown, which further suggests that the Map4k4 silencing-mediated increase in myogenesis is independent of MAPKs (Fig. 5). These findings implicating signaling pathways distinct from canonical MAP kinase cascades are consistent with our studies of Map4k4 signaling in other cellular contexts. We have previously shown that Map4k4 functions to mediate lipopolysaccharide (LPS)-induced TNF-α expression in macrophages, in a MAP kinase-independent manner. In that study, silencing of macrophage Map4k4 had no effect on the phosphorylation of p38, ERK1/2, JNK1/2, or their substrate ATF2 or c-Jun in response to LPS (35). These findings strongly suggest that Map4k4 may regulate cellular processes independently of MAPK signaling pathways.
In the current study, Myf5 expression was enhanced at the early stage of myogenic differentiation in Map4k4-depleted myocytes (Fig. 7). We further found that Myf5 is critical for myogenic differentiation, because siRNA-mediated Myf5 suppression significantly inhibited C2C12 myoblast differentiation (Fig. 8), consistent with the observations reported in a recently published study (36). Furthermore, we found that silencing of Myf5 reduced the enhanced myotube formation that ensued upon Map4k4 suppression to the same level as that with the scrambled siRNA control (Fig. 8), indicating that Map4k4 regulates myogenic differentiation in a Myf5-dependent manner. The marked rescue by Myf5 overexpression of myogenic differentiation inhibition because of Map4k4 in myoblasts demonstrates that Map4k4 regulates skeletal muscle differentiation by acting upstream of Myf5. Other regulators may also be involved in the regulation of myogenesis by Map4k4, such as proteins related to the myoblast fusion that occurs in late differentiation. This concept is supported by the observation that Map4k4 suppression during late differentiation still significantly increased myotube formation (Fig. 6A, middle panels).
We observed a modest increase in Myf5 mRNA (∼1.4-fold) but an abundant increase (∼3-fold) in Myf5 protein in Map4k4-silenced myocytes. The discrepancy in alteration levels between Myf5 mRNA and protein suggests the possibility of a regulation by Map4k4 of Myf5 at the translational or posttranslational level. Mammalian target of rapamycin (mTOR) is a major regulator of cell proliferation, survival, and growth via modulating protein synthesis (37). It was reported that Map4k4 silencing in cultured adipocytes promoted PPARγ translation and global protein synthesis via the activation of the mTOR signaling pathway (23). However, Map4k4 silencing in C2C12 myoblasts had no detectable effect on the mTOR signaling pathway during myogenic differentiation, as determined by unaltered phosphorylation of the mTOR downstream effectors p70S6K and 4EBP1 (data not shown). Moreover, Map4k4 suppression did not enhance insulin-like growth factor (IGF)/Akt/mTOR signaling in IGF1-treated C2C12 myoblasts compared with scrambled siRNA controls (data not shown). These results suggest that Map4k4 may not be a regulator of the mTOR signaling pathway in skeletal muscle. However, Map4k4 may regulate Myf5 translation in a protein-specific manner during early myogenic differentiation, a concept that requires further investigation. Besides enhanced translation, the increased Myf5 protein content could also be because of reduced protein degradation. However, we could not detect a difference in Myf5 protein stability in Map4k4-silenced myoblasts compared with controls (data not shown). In addition, Myf5 has been shown to be activated by phosphorylation at serine49 and serine133 by casein kinase 2 (CK2) in vitro (38). Thus, Map4k4 may regulate Myf5 transcriptional activity through modulation of CK2 signaling or direct Myf5 phosphorylation at other sites to regulate skeletal muscle differentiation. Further studies are required to assess these hypotheses.
In human skeletal muscle, Map4K4 silencing protected against TNF-α-mediated insulin resistance by preventing activation of JNK1/2 and ERK1/2 (24). TNF-α has been demonstrated to inhibit skeletal muscle differentiation in several studies (39–41). Therefore, Map4k4 may mediate in part the inhibitory effect of TNF-α on C2C12 differentiation. When C2C12 myoblasts were differentiated with DM containing 5 ng/ml of TNF-α for 3 days, we observed a marked inhibition of myotube formation, dramatically reduced myogenic MyHC gene expression (∼10-fold), and increased phosphorylation of JNK1/2. However, silencing of Map4k4 in TNF-α-treated cells only marginally rescued the inhibitory effect of TNF-α on C2C12 cell differentiation, as demonstrated by a slight increase in myotube formation and MyHC expression (∼1.9-fold), with no significant alteration in JNK1/2 phosphorylation (data not shown). The fact that the silencing was not complete in these studies makes data interpretation difficult, and cells from Map4k4 knockout mice will be required to address whether Map4k4 plays a role in TNF-α-mediated inhibition of myogenic differentiation.
The negative effect of Map4k4 on myoblast fusion suggests a potential involvement of Map4k4 in skeletal muscle regeneration. Muscle regeneration is a rapid and extensive self-renewal process relying on the presence of satellite cells, a population of quiescent, mononucleated stem cells that are resident in adult skeletal muscle (42, 43). Upon work overload or injury, satellite cells are activated and then proliferate and differentiate into myoblasts that either fuse to each other to create new myofibers or fuse to existing damaged myofibers for repair. The fusion process in regeneration shares similar features to muscle cell fusion in myogenic differentiation. Because Map4k4 is abundantly present in satellite cells (Fig. 1) and is able to regulate myoblast fusion in C2C12 differentiation, it may also suppress the fusion process in muscle regeneration. Moreover, we showed here that Map4k4 silencing in C2C12 myoblasts substantially increased Myf5 expression during early myogenic differentiation (Fig. 6). In muscle regeneration, satellite cell activation is associated with Myf5 upregulation (44), and Myf5 null mice had a significant delay in the regenerative process (45). These studies support Map4k4 as a potential regulator in skeletal muscle regeneration by regulating Myf5 expression and fusion in satellite cells.
In summary, the results of this study reveal a novel role for Map4k4 in skeletal myogenesis and identify Myf5 as a protein that is regulated by Map4k4 to mediate myogenic differentiation. Given the effects that Map4k4 exerts on fusion and Myf5 expression, Map4k4 inhibition may enhance the regenerative capacity of damaged muscles in trauma and degenerative diseases such as muscular dystrophies. The hypertrophic myotube formation induced by Map4k4 suppression also suggests that Map4k4 may be an attractive therapeutic target for the treatment of cachexia or sarcopenia. Further investigation into other Map4k4 effectors that are involved in myogenesis, especially muscle-specific Map4k4 substrates, will be necessary to fully understand the role of Map4k4 as a new signaling node in muscle development and function.
ACKNOWLEDGMENTS
We thank Diane L. Barber for the generous gifts of adenoviruses expressing GFP, wt Map4k4, and the kinase-inactive Map4k4 mutant. We thank Anthony N. Imbalzano and Joseph Virbasius for critical comments and helpful discussions.
This work was supported by grants from the NIH to M.P.C. (DK030898) and the Genomics Core Facility of the University of Massachusetts Diabetes and Endocrinology Research Center (DK032520).
Footnotes
Published ahead of print 3 December 2012
REFERENCES
- 1. Braun T, Gautel M. 2011. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 12:349–361 [DOI] [PubMed] [Google Scholar]
- 2. Molkentin JD, Black BL, Martin JF, Olson EN. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83:1125–1136 [DOI] [PubMed] [Google Scholar]
- 3. Olson EN, Klein WH. 1994. bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes Dev. 8:1–8 [DOI] [PubMed] [Google Scholar]
- 4. Berkes CA, Tapscott SJ. 2005. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 16:585–595 [DOI] [PubMed] [Google Scholar]
- 5. Hinterberger TJ, Sassoon DA, Rhodes SJ, Konieczny SF. 1991. Expression of the muscle regulatory factor MRF4 during somite and skeletal myofiber development. Dev. Biol. 147:144–156 [DOI] [PubMed] [Google Scholar]
- 6. Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. 2004. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 431:466–471 [DOI] [PubMed] [Google Scholar]
- 7. Suelves M, Lluis F, Ruiz V, Nebreda AR, Munoz-Canoves P. 2004. Phosphorylation of MRF4 transactivation domain by p38 mediates repression of specific myogenic genes. EMBO J. 23:365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sumariwalla VM, Klein WH. 2001. Similar myogenic functions for myogenin and MRF4 but not MyoD in differentiated murine embryonic stem cells. Genesis 30:239–249 [DOI] [PubMed] [Google Scholar]
- 9. Zhang W, Liu HT. 2002. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 12:9–18 [DOI] [PubMed] [Google Scholar]
- 10. Aouadi M, Bost F, Caron L, Laurent K, Le Marchand Brustel Y, Binetruy B. 2006. p38 mitogen-activated protein kinase activity commits embryonic stem cells to either neurogenesis or cardiomyogenesis. Stem Cells 24:1399–1406 [DOI] [PubMed] [Google Scholar]
- 11. Aouadi M, Laurent K, Prot M, Le Marchand-Brustel Y, Binetruy B, Bost F. 2006. Inhibition of p38MAPK increases adipogenesis from embryonic to adult stages. Diabetes 55:281–289 [DOI] [PubMed] [Google Scholar]
- 12. Bost F, Aouadi M, Caron L, Even P, Belmonte N, Prot M, Dani C, Hofman P, Pages G, Pouyssegur J, Le Marchand-Brustel Y, Binetruy B. 2005. The extracellular signal-regulated kinase isoform ERK1 is specifically required for in vitro and in vivo adipogenesis. Diabetes 54:402–411 [DOI] [PubMed] [Google Scholar]
- 13. Wada T, Penninger JM. 2004. Mitogen-activated protein kinases in apoptosis regulation. Oncogene 23:2838–2849 [DOI] [PubMed] [Google Scholar]
- 14. Johnson GL, Lapadat R. 2002. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298:1911–1912 [DOI] [PubMed] [Google Scholar]
- 15. Knight JD, Kothary R. 2011. The myogenic kinome: protein kinases critical to mammalian skeletal myogenesis. Skelet. Muscle 1:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lluis F, Perdiguero E, Nebreda AR, Munoz-Canoves P. 2006. Regulation of skeletal muscle gene expression by p38 MAP kinases. Trends Cell Biol. 16:36–44 [DOI] [PubMed] [Google Scholar]
- 17. Dan I, Watanabe NM, Kusumi A. 2001. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11:220–230 [DOI] [PubMed] [Google Scholar]
- 18. Collins CS, Hong J, Sapinoso L, Zhou Y, Liu Z, Micklash K, Schultz PG, Hampton GM. 2006. A small interfering RNA screen for modulators of tumor cell motility identifies MAP4K4 as a promigratory kinase. Proc. Natl. Acad. Sci. U. S. A. 103:3775–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu AW, Cai J, Zhao XL, Jiang TH, He TF, Fu HQ, Zhu MH, Zhang SH. 2011. shRNA-targeted MAP4K4 inhibits hepatocellular carcinoma growth. Clin. Cancer Res. 17:710–720 [DOI] [PubMed] [Google Scholar]
- 20. Xue Y, Wang X, Li Z, Gotoh N, Chapman D, Skolnik EY. 2001. Mesodermal patterning defect in mice lacking the Ste20 NCK interacting kinase (NIK). Development 128:1559–1572 [DOI] [PubMed] [Google Scholar]
- 21. Zohn IE, Li Y, Skolnik EY, Anderson KV, Han J, Niswander L. 2006. p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell 125:957–969 [DOI] [PubMed] [Google Scholar]
- 22. Tang X, Guilherme A, Chakladar A, Powelka AM, Konda S, Virbasius JV, Nicoloro SM, Straubhaar J, Czech MP. 2006. An RNA interference-based screen identifies MAP4K4/NIK as a negative regulator of PPARgamma, adipogenesis, and insulin-responsive hexose transport. Proc. Natl. Acad. Sci. U. S. A. 103:2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guntur KV, Guilherme A, Xue L, Chawla A, Czech MP. 2010. Map4k4 negatively regulates peroxisome proliferator-activated receptor (PPAR) gamma protein translation by suppressing the mammalian target of rapamycin (mTOR) signaling pathway in cultured adipocytes. J. Biol. Chem. 285:6595–6603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bouzakri K, Zierath JR. 2007. MAP4K4 gene silencing in human skeletal muscle prevents tumor necrosis factor-alpha-induced insulin resistance. J. Biol. Chem. 282:7783–7789 [DOI] [PubMed] [Google Scholar]
- 25. Tesz GJ, Aouadi M, Prot M, Nicoloro SM, Boutet E, Amano SU, Goller A, Wang M, Guo CA, Salomon WE, Virbasius JV, Baum RA, O'Connor MJ, Jr, Soto E, Ostroff GR, Czech MP. 2011. Glucan particles for selective delivery of siRNA to phagocytic cells in mice. Biochem. J. 436:351–362 [DOI] [PubMed] [Google Scholar]
- 26. Pasut A, Oleynik P, Rudnicki MA. 2012. Isolation of muscle stem cells by fluorescence activated cell sorting cytometry. Methods Mol. Biol. 798:53–64 [DOI] [PubMed] [Google Scholar]
- 27. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 28. Gros J, Manceau M, Thome V, Marcelle C. 2005. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature 435:954–958 [DOI] [PubMed] [Google Scholar]
- 29. Nishiyama T, Kii I, Kudo A. 2004. Inactivation of Rho/ROCK signaling is crucial for the nuclear accumulation of FKHR and myoblast fusion. J. Biol. Chem. 279:47311–47319 [DOI] [PubMed] [Google Scholar]
- 30. Baumgartner M, Sillman AL, Blackwood EM, Srivastava J, Madson N, Schilling JW, Wright JH, Barber DL. 2006. The Nck-interacting kinase phosphorylates ERM proteins for formation of lamellipodium by growth factors. Proc. Natl. Acad. Sci. U. S. A. 103:13391–13396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cuenda A, Cohen P. 1999. Stress-activated protein kinase-2/p38 and a rapamycin-sensitive pathway are required for C2C12 myogenesis. J. Biol. Chem. 274:4341–4346 [DOI] [PubMed] [Google Scholar]
- 32. Wu Z, Woodring PJ, Bhakta KS, Tamura K, Wen F, Feramisco JR, Karin M, Wang JY, Puri PL. 2000. p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol. Cell. Biol. 20:3951–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zetser A, Gredinger E, Bengal E. 1999. p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the Mef2c transcription factor. J. Biol. Chem. 274:5193–5200 [DOI] [PubMed] [Google Scholar]
- 34. Khurana A, Dey CS. 2004. Involvement of c-Jun N-terminal kinase activities in skeletal muscle differentiation. J. Muscle Res. Cell Motil. 25:645–655 [DOI] [PubMed] [Google Scholar]
- 35. Aouadi M, Tesz GJ, Nicoloro SM, Wang M, Chouinard M, Soto E, Ostroff GR, Czech MP. 2009. Orally delivered siRNA targeting macrophage Map4k4 suppresses systemic inflammation. Nature 458:1180–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rajan S, Chu Pham Dang H, Djambazian H, Zuzan H, Fedyshyn Y, Ketela T, Moffat J, Hudson TJ, Sladek R. 2012. Analysis of early C2C12 myogenesis identifies stably and differentially expressed transcriptional regulators whose knock-down inhibits myoblast differentiation. Physiol. Genomics 44:183–197 [DOI] [PubMed] [Google Scholar]
- 37. Hay N, Sonenberg N. 2004. Upstream and downstream of mTOR. Genes Dev. 18:1926–1945 [DOI] [PubMed] [Google Scholar]
- 38. Winter B, Kautzner I, Issinger OG, Arnold HH. 1997. Two putative protein kinase CK2 phosphorylation sites are important for Myf-5 activity. Biol. Chem. 378:1445–1456 [DOI] [PubMed] [Google Scholar]
- 39. Alter J, Rozentzweig D, Bengal E. 2008. Inhibition of myoblast differentiation by tumor necrosis factor alpha is mediated by c-Jun N-terminal kinase 1 and leukemia inhibitory factor. J. Biol. Chem. 283:23224–23234 [DOI] [PubMed] [Google Scholar]
- 40. Chen SE, Jin B, Li YP. 2007. TNF-alpha regulates myogenesis and muscle regeneration by activating p38 MAPK. Am. J. Physiol. Cell Physiol. 292:C1660–C1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Coletti D, Yang E, Marazzi G, Sassoon D. 2002. TNFalpha inhibits skeletal myogenesis through a PW1-dependent pathway by recruitment of caspase pathways. EMBO J. 21:631–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Charge SB, Rudnicki MA. 2004. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 84:209–238 [DOI] [PubMed] [Google Scholar]
- 43. Tedesco FS, Dellavalle A, Diaz-Manera J, Messina G, Cossu G. 2010. Repairing skeletal muscle: regenerative potential of skeletal muscle stem cells. J. Clin. Invest. 120:11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cooper RN, Tajbakhsh S, Mouly V, Cossu G, Buckingham M, Butler-Browne GS. 1999. In vivo satellite cell activation via Myf5 and MyoD in regenerating mouse skeletal muscle. J. Cell Sci. 112:2895–2901 [DOI] [PubMed] [Google Scholar]
- 45. Gayraud-Morel B, Chretien F, Flamant P, Gomes D, Zammit PS, Tajbakhsh S. 2007. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev. Biol. 312:13–28 [DOI] [PubMed] [Google Scholar]