Abstract
The trimeric envelope glycoprotein (Env) of human immunodeficiency virus type 1 (HIV-1) mediates virus entry into host cells. CD4 engagement with the gp120 exterior envelope glycoprotein subunit represents the first step during HIV-1 entry. CD4-induced conformational changes in the gp120 inner domain involve three potentially flexible topological layers (layers 1, 2, and 3). Structural rearrangements between layer 1 and layer 2 have been shown to facilitate the transition of the envelope glycoprotein trimer from the unliganded to the CD4-bound state and to stabilize gp120-CD4 interaction. However, our understanding of CD4-induced conformational changes in the gp120 inner domain remains incomplete. Here, we report that a highly conserved element of the gp120 inner domain, layer 3, plays a pivot-like role in these allosteric changes. In the unliganded state, layer 3 modulates the association of gp120 with the Env trimer, probably by influencing the relationship of the gp120 inner and outer domains. Importantly, layer 3 governs the efficiency of the initial gp120 interaction with CD4, a function that can also be fulfilled by filling the Phe43 cavity. This work defines the functional importance of layer 3 and completes a picture detailing the role of the gp120 inner domain in CD4-induced conformational transitions in the HIV-1 Env trimer.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) entry, mediated by the viral envelope glycoproteins (Env), is a central process of the viral infectious cycle. The HIV-1 Env trimer is the only virus-specific antigen present on the virion surface; therefore, it constitutes the sole target for HIV-1-neutralizing antibodies. The mature HIV-1 Env trimer is derived from proteolytic cleavage of a trimeric gp160 precursor (1, 2) and is composed of exterior gp120 and transmembrane gp41 subunits. The gp120 exterior subunit is retained on the trimer via labile, noncovalent interactions with the gp41 ectodomain (3–5). The gp120 glycoprotein is responsible for interactions with the initial receptor, CD4 (6, 7), upon which conformational changes expose the binding site for the chemokine coreceptors (CCR5 and CXCR4) (8–12). CD4 binding induces conformational changes within the HIV-1 Env trimer that result in the transition of the gp41 ectodomain into prehairpin intermediates (13, 14). Upon chemokine receptor binding, heptad repeat regions HR1 and HR2 form a six-helix bundle, promoting fusion of the viral and target cell membranes (15, 16).
As the major viral determinant recognized by the neutralizing antibody response, the HIV-1 Env trimer represents a likely candidate for a vaccine immunogen. Binding of an antibody to any part of the functional HIV-1 Env spike has the potential to block HIV infection (17, 18). However, vaccination efforts with both monomeric and trimeric Env constructs in soluble or recombinant forms elicited responses that were minimally effective against the majority of primary HIV-1 isolates (19–21). Nonetheless, partial efficacy observed in the RV144 vaccine trial (22) renewed interest in HIV-1 Env as an immunogen candidate. However, to engineer an HIV-1 Env immunogen capable of eliciting broadly neutralizing antibodies, it may be important to understand better the different conformations sampled by the native Env trimers.
Detailed structural information has been obtained by X-ray crystallography for monomeric HIV-1 gp120 core in complex with stabilizing ligands such as soluble CD4 (sCD4) and/or antibodies (Abs) (23–27). However, crystallization has required deglycosylation and truncation of variable regions, removing structural constraints that apparently maintain the conformational integrity of the unliganded Env trimer (28). These studies have led to the hypothesis that, upon CD4 binding, the outer domain of gp120 does not change conformation, whereas three topological layers in the gp120 inner domain undergo conformational shifts (27). Until recently, other approaches such as cryo-electron tomography of virion Env spikes and cryo-electron microscopy (cryo-EM) analysis of purified Env variants have only yielded density maps at 18- to 30-Å resolution (29–34), insufficient for assessing detailed structural variations. However, the structure of the fully glycosylated, membrane-bound HIV-1 Env trimer precursor in its unliganded state, including the complete exterior and transmembrane regions, has been recently solved to ∼11-Å resolution by cryo-EM (35). This structure revealed the architectural organization of the gp120 and gp41 subunits and, consistent with our previous mutagenic and functional studies (3), the existence of conformational differences between the unliganded and CD4-bound states in the Env trimer, particularly in the inner domain (35). Whereas the role of layers 1 and 2 in the conformational transition from the unbound to the CD4-bound conformation has been previously reported (3), a role for the functionally undercharacterized layer 3 of the HIV-1 gp120 inner domain (β8 strand and α5 of the CD4-bound gp120, comprising residues 247 to 254 and 476 to 483, respectively) has not yet been addressed. Here, we characterize the importance of this element to Env integrity and to the process of viral entry and investigate its role in the transition to the CD4-bound conformation.
MATERIALS AND METHODS
Cell lines.
293T human embryonic kidney, Cf2Th canine thymocytes (American Type Culture Collection) and TZM-bl cell lines (NIH AIDS Research and Reference Reagent Program) were grown at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum (Sigma) and 100 μg/ml of penicillin-streptomycin (Mediatech). Cf2Th cells stably expressing human CD4 and CCR5 (Cf2Th-CD4-CCR5) (36) were grown in medium supplemented with 0.4 mg/ml of G418 (Invitrogen) and 0.15 mg/ml of hygromycin B (Roche Diagnostics). Cf2Th-CCR5 cells were grown in medium supplemented with 0.4 mg/ml of G418 (Invitrogen). The TZM-bl cell line is a HeLa cell line stably expressing high levels of CD4 and CCR5 and possessing an integrated copy of the luciferase gene under the control of the HIV-1 long terminal repeat (37).
Site-directed mutagenesis.
Mutations were introduced individually or in combination into the previously described pSVIIIenv vector expressing the HIV-1YU2 envelope glycoproteins (3). Site-directed mutagenesis was performed using a QuikChange II XL site-directed mutagenesis protocol (Stratagene). For cell-based enzyme-linked immunosorbent assays (ELISAs), a stop codon was introduced to replace the codon for Gly711, truncating the cytoplasmic tail (ΔCT) and enhancing cell surface expression of selected HIV-1YU2 envelope glycoproteins. The presence of the desired mutations was determined by automated DNA sequencing. The numbering of the HIV-1 envelope glycoprotein amino acid residues is based on that of the prototypic HXBc2 strain of HIV-1, where position 1 is the initial methionine (38).
Immunoprecipitation of envelope glycoproteins.
For pulse-labeling experiments, 3 × 105 293T cells were cotransfected by the calcium phosphate method with pLTR-Tat and the pSVIIIenv vector expressing the HIV-1YU2 envelope glycoproteins. One day after transfection, cells were metabolically labeled for 16 h with 100 μCi/ml [35S]methionine-cysteine ([35S] Protein Labeling Mix; Perkin-Elmer) in Dulbecco's modified Eagle's medium lacking methionine and cysteine and supplemented with 5% dialyzed fetal bovine serum. Cells were subsequently lysed in radioimmunoprecipitation assay (RIPA) buffer (140 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1% NP-40, 0.05% sodium dodecyl sulfate [SDS]). Precipitation of radiolabeled HIV-1YU2 envelope glycoproteins from cell lysates or medium was performed with a mixture of sera from HIV-1-infected individuals. Alternatively, the radiolabeled gp120 envelope glycoprotein in the medium was precipitated with various amounts of anti-gp120 monoclonal antibodies (MAbs) or the recombinant CD4-Ig protein for 1 h at 37°C in the presence of 50 μl of 10% protein A-Sepharose (American BioSciences).
Processing and association indices were determined by precipitation of radiolabeled cell lysates and supernatants with mixtures of sera from HIV-1-infected individuals. The association index is a measure of the ability of the mutant gp120 molecule to remain associated with the Env trimer complex on the expressing cell, relative to that of the wild-type (wt) Env trimers. The association index is calculated as follows: association index = ([mutant gp120]cell × [wild-type gp120]supernatant)/([mutant gp120]supernatant × [wild-type gp120]cell). The processing index is a measure of the conversion of the mutant gp160 Env precursor to mature gp120, relative to that of the wild-type Env trimers. The processing index was calculated by the following formula: processing index = ([total gp120]mutant × [gp160]wild type)/([gp160]mutant × [total gp120]wild type).
Cell-based ELISA.
Detection of trimeric Env on the surface of COS-1 cells was performed by cell-based ELISA, as described previously (39). Briefly, COS-1 cells were seeded in 96-well plates (2 × 104 cells per well) and transfected the next day with 0.15 μg of the pSVIIIenv plasmid expressing the envelope glycoproteins and 0.01 μg of a Tat-expressing plasmid per well, using the standard polyethylenimine (PEI) (Polyscience Inc., PA) transfection method. Two days later, cells were washed twice with blocking buffer (10 mg/ml nonfat dry milk, 1.8 mM CaCl2, 1 mM MgCl2, 25 mM Tris, pH 7.5, and 140 mM NaCl) and then incubated for 1 h at room temperature with a 1/2,000 dilution of heat-inactivated serum from HIV-1-infected individuals, 20 nM CD4-Ig (a fusion protein in which the N-terminal two domains of CD4 are linked to the Fc component of immunoglobulin G [40]), or anti-HIV-1 Env monoclonal antibodies. All ligands were diluted in blocking buffer. In some instances, 20 nM sCD4 or CD4-Ig was added in blocking buffer for 1 h at room temperature (RT) prior to the addition of 20 nM CD4-induced (CD4i) antibodies (17b and 48d) for another hour. A horseradish peroxidase (HRP)-conjugated antibody specific for the Fc region of human IgG (Pierce) was then incubated with the samples for 45 min at RT. In some cases CD4i antibodies directly conjugated to horseradish peroxidase (see below) were incubated with Env-expressing cells for 1 h at room temperature, removing the need to use an anti-Fc region antibody to develop the reaction. For all conditions, cells were washed five times with blocking buffer and five times with washing buffer. HRP enzyme activity was determined after the addition of 30 μl per well of a 1:1 mix of Western Lightning oxidizing and luminol reagents (PerkinElmer Life Sciences). Light emission was measured with an LB 941 TriStar luminometer (Berthold Technologies).
Horseradish peroxidase antibody conjugation.
The HRP conjugation of a CD4i antibody (17b) was achieved as follows: 5 mg of horseradish peroxidase (HRP; Sigma) was diluted in freshly prepared 100 mM NaHCO3 and incubated for 2 h in the dark at RT with agitation. At this point, the antibody was added to the mixture together with Na2CO3 (10 mM final concentration) for 4 h in the dark at RT. The HRP-antibody conjugate was stabilized with freshly prepared NaBH4 (132 mM) for 1 h at RT in the dark. Complexes were dialyzed against phosphate-buffered saline (PBS) overnight at 4°C.
Cross-linking of envelope glycoproteins.
Analysis of cross-linked trimeric envelope glycoproteins by cell-based ELISA was performed as described above, with the exception that before the addition of blocking buffer, cells were incubated with 5 mM glutaraldehyde (GA) (Sigma) for 5 min at room temperature before the addition of 50 mM glycine to quench the unreacted GA.
Recombinant luciferase viruses.
Recombinant viruses containing the firefly luciferase gene were produced by calcium phosphate transfection of 293T cells with the HIV-1 proviral vector pNL4.3 Env− Luc and the pSVIIIenv plasmid expressing the wild-type or mutant HIV-1YU2 envelope glycoproteins at a ratio of 2:1. Two days after transfection, the cell supernatants were harvested; the reverse transcriptase activities of all viruses were measured as described previously (41). The virus-containing supernatants were stored in aliquots at −80°C.
Infection by single-round luciferase viruses.
Cf2Th-CD4-CCR5 target cells were seeded at a density of 5 × 103 cells/well in 96-well luminometer-compatible tissue culture plates (PerkinElmer) 24 h before infection. Recombinant viruses (10,000 reverse transcriptase units) in a final volume of 100 μl were then added to the target cells, followed by incubation for 48 h at 37°C; the medium was then removed from each well, and the cells were lysed by the addition of 30 μl of passive lysis buffer (Promega) and three freeze-thaw cycles. An LB 941 TriStar luminometer (Berthold Technologies) was used to measure the luciferase activity of each well after the addition of 100 μl of luciferin buffer (15 mM MgSO4, 15 mM KPO4 [pH 7.8], 1 mM ATP, and 1 mM dithiothreitol) and 50 μl of 1 mM d-luciferin potassium salt (Prolume).
Purification of recombinant HIV-1 gp120 glycoproteins.
FreeStyle 293F cells (Invitrogen) were grown in FreeStyle 293F medium (Invitrogen) to a density of 1 × 106 cells/ml at 37°C with 8% CO2 with regular agitation (125 rpm). Cells were transfected with a codon-optimized plasmid expressing His6-tagged wild-type or mutant HIV-1YU2 gp120 using the 293Fectin reagent, as directed by the manufacturer (Invitrogen). One week later, the cells were pelleted and discarded. The supernatants were filtered (0.22-μm-pore-size filter) (Corning), and the gp120 glycoproteins were purified by nickel affinity columns, as directed by the manufacturer (Invitrogen). The gp120 preparations were dialyzed against PBS and stored in aliquots at −80°C. To assess purity, recombinant proteins were loaded on SDS-PAGE polyacrylamide gels and stained with Coomassie blue.
SPR biosensor analysis.
Surface plasmon resonance (SPR) biosensor data were collected on a Biacore 3000 optical biosensor (General Electric). Four-domain soluble CD4 (sCD4) and the 17b anti-gp120 monoclonal antibody were immobilized onto separate flow cells within the same sensor chip (CM5; GE) to a surface density of ∼500 response units (RU) using standard amine coupling chemistry (42). The binding capacities of CD4 and 17b surfaces were kept low to avoid mass transport effects and steric hindrance. Flow cell 1 or 3 was left blank as a control for nonspecific binding and refractive index changes. With the instrument operating in a parallel sensing mode, soluble gp120 was injected over flow cells 1 and 2 or 3 and 4 at different concentrations ranging from 100 to 750 nM at a flow rate of 30 μl/min for 3 min. This was followed by a 10-min dissociation phase to allow an estimation of off-rates and binding affinities. Sensor data were prepared for kinetic analysis by subtracting binding responses collected from the blank reference surface. To examine the binding of the 17b antibody to gp120-CD4 complexes, a mixture of soluble gp120 and sCD4 (molar ratio of 1:1) at concentrations ranging from 100 to 750 nM was passed over the 17b chip. The association and dissociation phase data were fitted simultaneously with BIAevalution, version 3.2, RC1 software using a 1:1 Langmuir model of binding.
Statistical analysis.
Comparison between groups was performed with a Mann-Whitney rank sum test using SigmaPlot, version 6.0, software. Data were expressed as means ± standard deviations (SD), and P values of ≤0.05 were considered to be significant.
RESULTS
The HIV-1 gp120 layer 3 inner domain mutants.
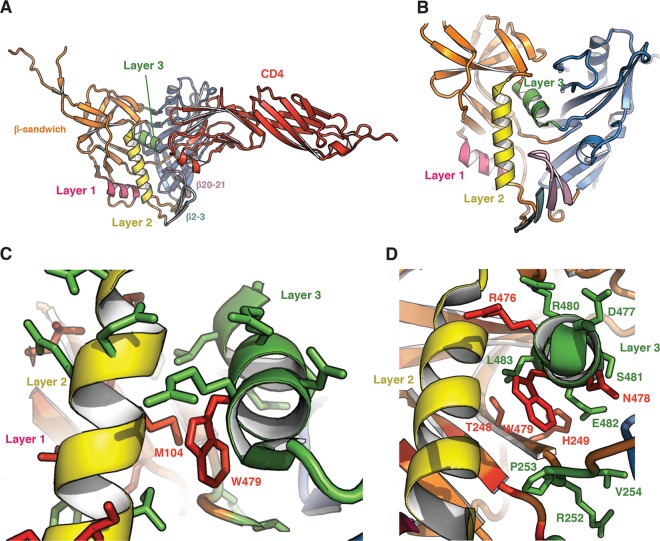
A seven-stranded β-sandwich in the HIV-1 gp120 inner domain serves as a point of departure for the gp120 N and C termini, which roam toward the viral membrane. The β-sandwich and the gp120 terminal strands have been implicated in the noncovalent association of gp120 with gp41 (3–5). Projecting from the β-sandwich toward the target cell membrane are three excursions that compose the rest of the inner domain and, in two cases, transit into the neighboring gp120 domains (Fig. 1B). These excursions form three topological layers (layers 1, 2, and 3) that have been proposed to change conformation as gp120 undergoes the transition from the unliganded to the CD4-bound state (3, 27).
Fig 1.
Structure of the inner domain of HIV-1 gp120 in the CD4-bound conformation. (A) The structure (27) of HIV-1HXBc2 gp120 (ribbon) complexed with two-domain CD4 (red) is shown from the perspective of the Env trimer axis. The β20-β21 strands of gp120 project from the outer domain and compose two of the four strands of the bridging sheet. The other two strands of the bridging sheet are derived from the distal portion of layer 2. (B) A close-up view of the conformation adopted by the inner domain layers 1, 2, and 3. (C) A close-up view of the interactions between layers 2 and 3. The side chain residues that were altered in this and a previous study (3) are colored according to the gp120-trimer association index (red, association index of <0.5; green, association index of ≥0.7). (D) The side chain residues are colored according to CD4-Ig binding ability (red, relative CD4-Ig binding of ≤0.5; green, relative CD4-Ig binding of >0.5).
In a recent study, we demonstrated the importance of layers 1 and 2 for the transition to the CD4-bound conformation (3). Our mutagenesis study indicated that after the initial contact with CD4, layer 1-layer 2 interactions strengthened gp120-CD4 binding by reducing the off-rate. This observation suggested that these layers existed in a different conformation in the unliganded state (3). However, the role of the gp120 inner domain layer 3, a highly conserved element among HIV-1 isolates (Fig. 2), remains poorly characterized. To investigate the role that layer 3 plays in Env function in the unliganded and CD4-bound states, single amino acid changes were introduced into the Env of HIV-1YU2, a primary virus directly cloned from an HIV-1-infected individual (43). The changes were focused on layer 3 although a few alterations were introduced into adjacent gp120 elements. The Env mutants were evaluated with respect to the following properties: proteolytic processing of the gp160 precursor, association of the gp120 and gp41 subunits, CD4 binding (in monomeric and trimeric context), functional ability to mediate cell-cell fusion and virus entry, and the effect of soluble CD4 (sCD4) on virus infectivity (Table 1). The conformation of gp120 in cell supernatants was assessed by precipitation with a panel of monoclonal antibodies (MAbs) that recognize conformation-dependent epitopes (Tables 1 and 2) (44, 45).
Fig 2.
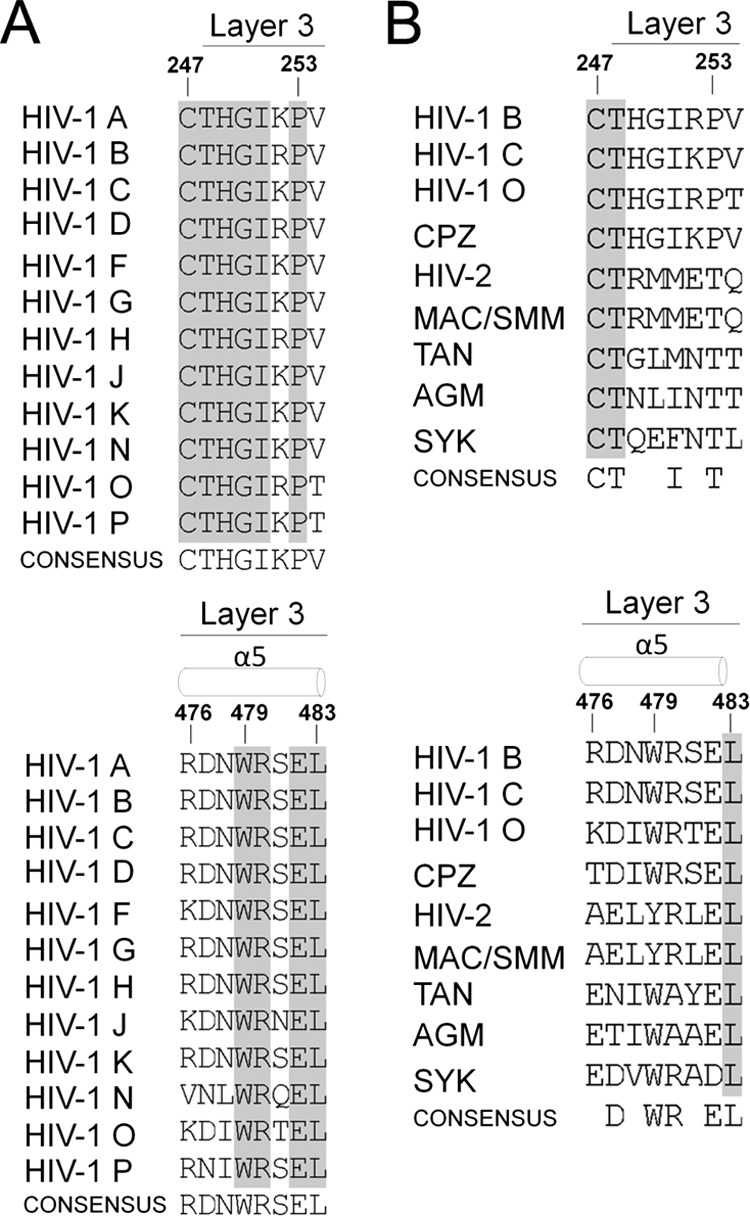
Sequences of HIV-1 and primate immunodeficiency virus Env. Primary sequence alignment of layer 3 gp120 residues from representative HIV-1 A (accession number ABB29387.1), HIV-1 B (accession number K03455), HIV-1 C (AAB36507.1), HIV-1 D (P04581.1), HIV-1 F (ACR27173.1), HIV-1 G (ACO91925.1), HIV-1 H (AAF18394.1), HIV-1 J (ABR20452.1), HIV-1 K (CAB59009.1), HIV-1 N (AAT08775.1), HIV-1 O (AAA99883.1), and HIV-1 P (ACY40659.1) viruses (A) and from primate immunodeficiency lineages SIVcpz (accession number ABD19490.1), HIV-2 (AAC95347.1), SIVmac/smm (AAA47637.1), SIVtan (AAC57057.1), SIVagm (AAA91919.1), and SIVsyk (AAA74712.1) (4, 5) (B). Secondary structure elements are shown above the sequences (4, 5). The shading highlights residues that are conserved.
Table 1.
Phenotypes of HIV-1 gp120 mutantsa
| Envelope glycoprotein | Processing index | Association index | CD4-Ig binding | Cell-cell fusion | Relative infectivity | sCD4 IC50 (nM)b |
|---|---|---|---|---|---|---|
| Wild-type YU2 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 60 |
| T248A | 0.40 | 0.94 | 0.30 | 0.98 | 0.98 | 50 |
| H249A | 0.67 | 1.18 | 0.49 | 1.32 | 0.68 | 100 |
| R252A | 0.66 | 0.65 | 0.62 | 1.04 | 1.05 | 100 |
| P253A | 0.85 | 0.82 | 1.00 | 1.48 | 0.62 | 100 |
| V254A | 0.66 | 0.51 | 0.75 | 1.22 | 1.02 | 160 |
| R476A | 1.23 | 0.90 | 0.31 | 1.48 | 1.18 | 160 |
| R476E | 0.73 | 0.95 | 0.14 | 1.52 | 0.53 | 100 |
| D477A | 0.95 | 0.95 | 0.60 | 1.40 | 0.43 | 140 |
| N478A | 0.77 | 0.75 | 0.44 | 1.24 | 0.36 | >200 |
| W479A | 0.91 | 0.12 | 0.23 | 0.97 | 0.06 | 180 |
| R480A | 0.90 | 1.60 | 0.86 | 0.91 | 0.77 | 70 |
| S481A | 1.25 | 0.74 | 1.36 | 1.17 | 0.78 | 80 |
| E482A | 0.63 | 0.94 | 0.79 | 1.17 | 0.52 | 40 |
| L483A | 0.80 | 0.91 | 0.77 | 1.36 | 0.47 | 60 |
The phenotypes of the wt and mutant Env glycoproteins were determined as described in Materials and Methods. All mutated residues are located in layer 3. Data represent the mean values derived from at least three experiments. Less than 20% deviation from the mean value was typically observed.
IC50, 50% inhibitory concentration.
Table 2.
Recognition of HIV-1YU2 gp120 variants by monoclonal antibodiesa
| Envelope glycoprotein | Binding affinity for:b |
|||||
|---|---|---|---|---|---|---|
| 17b | 412d | b12 | VRC01 | C11 | 2G12 | |
| Wild-type YU2 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| T248A | 0.09 | 0.12 | 0.20 | 0.17 | 0.14 | 0.51 |
| H249A | 0.36 | 0.49 | 0.47 | 0.39 | 0.37 | 0.64 |
| R252A | 0.27 | 0.19 | 0.86 | 0.64 | 0.54 | 0.78 |
| P253A | 0.26 | 0.29 | 0.77 | 0.71 | 0.63 | 0.93 |
| V254A | 0.48 | 0.55 | 1.16 | 0.99 | 1.07 | 1.12 |
| R476A | 0.19 | 0.30 | 0.75 | 0.43 | 0.59 | 0.84 |
| R476E | 0.11 | 0.18 | 0.32 | 0.21 | 0.22 | 0.70 |
| D477A | 0.30 | 0.57 | 0.31 | 0.48 | 0.58 | 0.87 |
| N478A | 0.14 | 0.23 | 1.08 | 0.51 | 0.50 | 0.84 |
| W479A | 0.02 | 0.04 | 0.53 | 0.29 | 0.35 | 0.60 |
| R480A | 0.50 | 0.84 | 1.21 | 0.75 | 0.81 | 1.03 |
| S481A | 0.83 | 1.35 | 1.32 | 1.32 | 1.43 | 1.06 |
| E482A | 0.53 | 1.18 | 0.89 | 0.60 | 0.81 | 0.98 |
| L483A | 0.32 | 0.40 | 1.51 | 0.71 | 0.96 | 1.33 |
293T cells transiently expressing the indicated HIV-1 Env glycoproteins were radiolabeled, and the supernatants were precipitated with a mixture of sera from HIV-1-infected individuals. The precipitates were analyzed on nonreducing and reducing SDS-polyacrylamide gels, as reported by Finzi et al. (45). All mutated residues are located in layer 3.
Comparable amounts of radiolabeled wt and mutant gp120 glycoproteins were incubated with a 13 nM concentration of the indicated antibody for 1 h at 37°C. Precipitates were analyzed by SDS-PAGE and autoradiography/densitometry. The amount of mutant gp120 precipitated by the monoclonal antibody, normalized to the amount of gp120 precipitated by a mixture of sera from HIV-1-infected individuals, is reported relative to the value obtained for wt gp120. The results shown represent the average of at least two independent experiments.
The ability of mutant Env trimers to bind CD4.
The effects of the changes introduced into layer 3 of the inner domain on the affinity of monomeric gp120 for CD4 were examined. The amounts of radiolabeled wild-type (wt) and mutant gp120 in the supernatants of Env-expressing 293T cells were normalized, and equivalent amounts of gp120 were used for precipitation by CD4-Ig. Alternatively, the ability of trimeric cell surface-expressed Env variants to interact with CD4-Ig was also evaluated by cell-based ELISA, as described in Materials and Methods. For cell-based ELISAs, the cytoplasmic tail of the Env variants was truncated to enhance cell surface expression; however, this did not affect precursor processing or gp120-gp41 association (Table 3). Finally, all cytoplasmic tail-deleted Env variants were tested for their ability to interact with the PG9 and PG16 antibodies, which preferentially bind trimeric Env (46).
Table 3.
Characterization of ligand binding to selected HIV-1YU2 gp120 variants by cell-based ELISAa
| Envelope glycoprotein | Processing index | Association index | Binding affinity for: |
|||||
|---|---|---|---|---|---|---|---|---|
| CD4-Ig | 17b | 17b + sCD4 | VRC01 | PG9 | PG16 | |||
| Wild-type YU2 | 1.00 | 1.00 | 1.00 | 1.00 | 2.77 | 1.00 | 1.00 | 1.00 |
| T248A | 0.32 | 0.75 | 0.72 | 0.92 | 1.70 | 0.72 | 0.45 | 0.44 |
| H249A | 0.38 | 1.01 | 0.83 | 1.09 | 2.15 | 0.81 | 0.68 | 0.35 |
| R252A | 0.79 | 0.75 | 0.81 | 0.48 | 2.53 | 0.91 | 1.00 | 0.62 |
| P253A | 0.61 | 0.56 | 0.67 | 0.85 | 1.69 | 0.75 | 0.63 | 0.45 |
| V254A | 1.03 | 0.73 | 0.70 | 0.68 | 1.70 | 0.86 | 1.06 | 0.49 |
| R476A | 0.83 | 0.85 | 0.59 | 0.95 | 1.72 | 0.76 | 0.60 | 0.59 |
| D477A | 0.59 | 0.81 | 0.76 | 1.06 | 2.31 | 0.65 | 0.75 | 0.78 |
| N478A | 0.75 | 0.55 | 0.64 | 0.33 | 1.41 | 0.56 | 0.38 | 0.55 |
| W479A | 0.62 | 0.19 | 0.64 | 0.35 | 1.05 | 0.38 | 0.08 | 0.16 |
| R480A | 0.50 | 0.92 | 1.15 | 0.88 | 2.51 | 0.92 | 0.48 | 0.45 |
| S481A | 0.85 | 1.02 | 1.17 | 0.89 | 2.58 | 0.72 | 0.32 | 0.43 |
| E482A | 0.59 | 1.10 | 0.79 | 0.79 | 1.78 | 0.81 | 0.50 | 0.53 |
| L483A | 0.58 | 0.44 | 0.64 | 0.44 | 1.18 | 0.45 | 0.57 | 0.60 |
The phenotypes of the wt and mutant Env glycoproteins were determined as described in Materials and Methods. All mutated residues are located in layer 3. Data represent the mean values derived from at least three experiments. Less than 20% deviation from the mean value was typically observed.
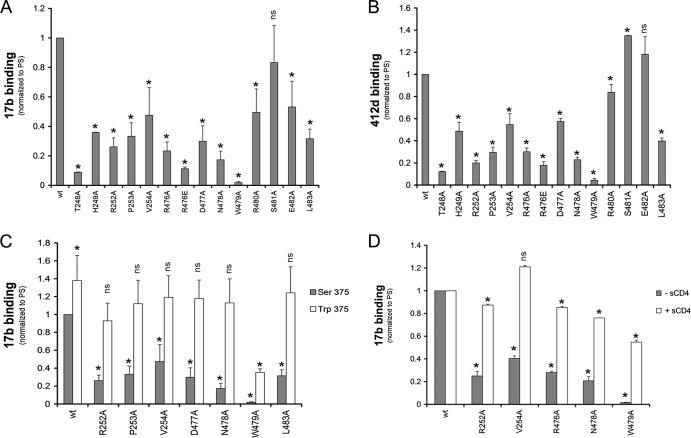
Five layer 3 changes decreased the efficiency of mutant gp120 interaction with CD4. A strong correlation between CD4-Ig binding by monomeric gp120 and trimeric Env variants was observed (Fig. 3A) although some mutants exhibited a more pronounced decrease in the context of monomeric gp120. Nevertheless, the T248A, H249A, R476A, N478A, and W479A mutants exhibited decreased CD4-Ig binding in both assays (Fig. 3A and B; Tables 1 and Table 3). The observed decreases in CD4 binding were mainly due to decreased on-rates compared with that of wt gp120 (Table 4). Thus, gp120 mutants with alterations in layer 3 fail to engage CD4 efficiently. The five layer 3 residues implicated in CD4 binding are located in the β8 strand and α5 helix of the CD4-bound gp120. A network of interactions involving some residues of the β8-α5 region in layer 3 may contribute to CD4 binding by helping to shape the nearby Phe43 cavity that directly contacts CD4 (26, 47, 48). Similarly, several layer 3 variants resulted in decreased recognition by antibodies (b12 and VRC01) directed against the CD4-binding site of gp120 (Tables 2 and 3). In summary, multiple residues in layer 3 of the gp120 inner domain that do not directly contact CD4 nonetheless contribute to the affinity of the gp120-CD4 interaction.
Fig 3.
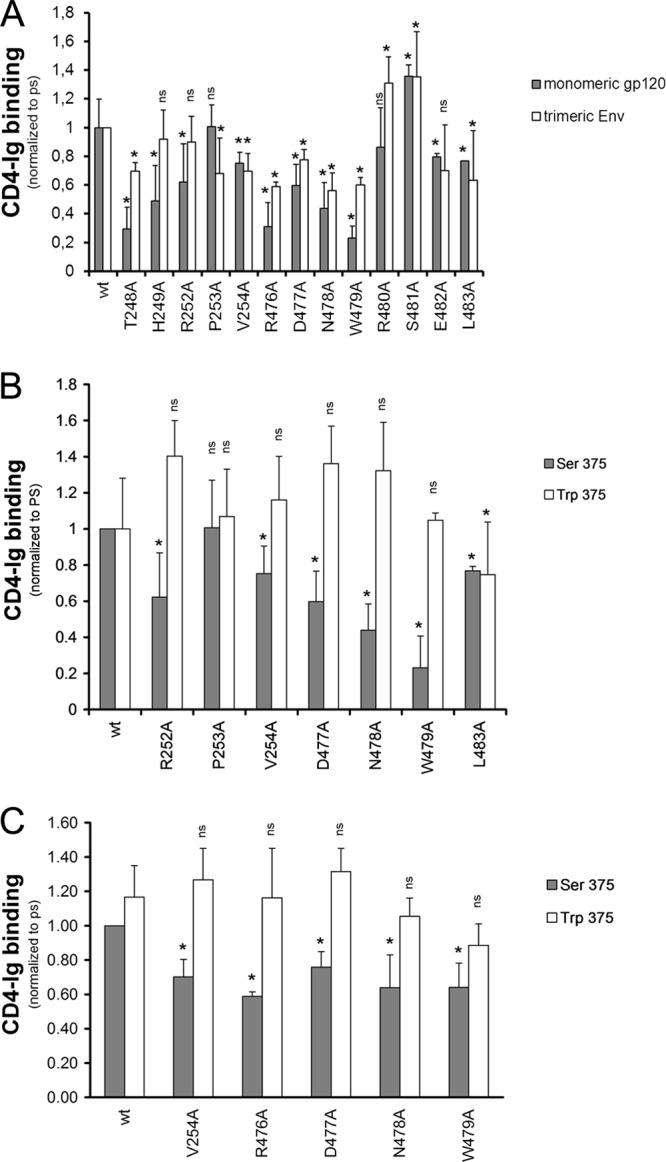
Recognition of layer 3 gp120 variants by CD4. The effects of alterations in layer 3 on gp120 recognition by CD4-Ig were examined for monomeric soluble gp120 by immunoprecipitation or for cellular-expressed trimeric Env by cell-based ELISA (A). In both contexts, the gp120 variants were normalized by patient serum (PS) and to the signal obtained for their wt counterparts. The effect of filling the Phe43 cavity with a tryptophan (S375W) on layer 3 variants was analyzed by immunoprecipitation (B) or by cell-based-ELISA (C) with CD4-Ig. Data shown represent the means ± standard errors of the means of at least two independent experiments. *, P < 0.05 by a Mann-Whitney rank sum test. Differences with P values of ≥0.05 were not considered significant (ns).
Table 4.
Characterization of ligand binding to selected HIV-1YU2 gp120 mutants by surface plasmon resonance
| gp120 protein | Binding affinity for:a |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| sCD4 |
17b |
17b + sCD4 |
|||||||
| On-rate (M−1 s−1) | Off-rate (s−1) | KD (M [fold change]) | On-rate (M−1 s−1) | Off-rate (s−1) | KD (M [fold change]) | On-rate (M−1 s−1) | Off-rate (s−1) | KD (M [fold change]) | |
| Wild type | 2.23 × 104 | 2.21 × 10−4 | 9.90 × 10−9 (1.00) | 5.54 × 104 | 5.98 × 10−5 | 1.08 × 10−9 (1.00) | 4.20 × 104 | 5.77 × 10−5 | 1.38 × 10−9 (1.00) |
| T248A | 7.74 ×103 | 3.59 × 10−4 | 4.64 × 10−8 (4.69) | 6.61 × 104 | 6.81 × 10−3 | 1.03 × 10−7 (95.54) | 1.88 × 104 | 1.32 × 10−4 | 6.99 × 10−9 (5.08) |
| V254A | 1.40 × 104 | 2.88 × 10−4 | 2.05 × 10−8 (2.08) | 1.08 × 104 | 1.11 × 10−4 | 1.02 × 10−8 (9.49) | 3.08 × 104 | 8.48 × 10−5 | 2.75 × 10−9 (2.00) |
| R476A | 1.07 × 104 | 2.61 × 10−4 | 2.45 × 10−8 (2.46) | 4.40 × 103 | 5.75 × 10−5 | 1.31 × 10−8 (12.14) | 2.70 × 104 | 5.19 × 10−5 | 1.92 × 10−9 (1.40) |
| D477A | No binding | 9.62 × 103 | 5.14 × 10−5 | 5.34 × 109 (4.96) | 9.30 × 103 | 8.90 × 10−5 | 9.57 × 10−9 (6.96) | ||
| N478A | 4.84 × 103 | 3.32 × 10−4 | 6.86 × 10−8 (6.93) | 2.99 × 103 | 1.88 × 10−4 | 6.29 × 10−8 (58.33) | 2.13 × 104 | 6.93 × 10−5 | 3.26 × 10−9 (2.37) |
| W479A | No binding | No binding | 7.42 × 103 | 8.56 × 10−5 | 1.15 × 10−8 (8.39) | ||||
| S375W | 1.43 × 104 | 1.94 × 10−6 | 1.33 × 10−10 (0.01) | 7.67 × 104 | 4.96 × 10−5 | 6.47 × 10−10 (0.60) | 2.94 × 104 | 6.90 × 10−5 | 2.35 × 10−9 (1.70) |
| V254A S375W | 4.37 × 104 | 4.97 × 10−5 | 1.14 × 10−9 (0.12) | 6.94 × 104 | 5.19 × 10−5 | 7.47 × 10−10 (0.69) | 3.83 × 104 | 7.81 × 10−5 | 2.04 × 10−9 (1.48) |
| R476A S375W | 2.88 × 104 | 5.64 × 10−5 | 1.96 × 10−9 (0.19) | 5.11 × 104 | 4.84 × 10−5 | 9.47 × 10−10 (0.88) | 2.64 × 104 | 8.49 × 10−5 | 3.21 × 10−9 (2.34) |
| D477A S375W | 5.67 × 104 | 4.39 × 10−5 | 7.74 × 10−10 (0.08) | 5.78 × 104 | 4.10 × 10−5 | 7.11 × 10−10 (0.66) | 2.80 × 104 | 7.27 × 10−5 | 2.60 × 10−9 (1.89) |
| W479A S375W | 1.55 × 104 | 4.00 × 10−4 | 2.59 × 10−8 (2.61) | 4.29 × 104 | 3.13 × 10−4 | 7.29 × 10−9 (6.76) | 1.29 × 104 | 7.24 × 10−5 | 5.62 × 10−9 (4.08) |
The gp120-reactive ligand (sCD4 or 17b) was immobilized directly onto a CM5 sensor chip and the binding of the indicated gp120 protein was evaluated as described in Materials and Methods. No binding, detected signal below accurate analysis of kinetics parameters; KD, equilibrium dissociation constant.
Effect of filling the Phe43 cavity on the phenotypes of inner domain mutants.
Serine 375 flanks the Phe43 cavity; substitution of a tryptophan residue for serine 375 fills the Phe43 cavity with the indole ring (49). As a result, the S375W mutant favors conformation(s) closer to that of the CD4-bound state (50). To determine whether such a change in gp120 conformation would influence the phenotypes of the inner domain mutants, we introduced the S375W change into some of the gp120 variants described above. Remarkably, the S375W change completely restored the CD4-binding abilities of gp120 mutants with alterations in R252, V254, D477, N478, and W479 (Fig. 3B). The compensatory effect of S375W was observed in the context of monomeric and trimeric Env, as shown by cell-based ELISA (Fig. 3C and Table 3). In the context of monomeric gp120, introduction of the S375W change restored CD4 binding affinity by increasing the on-rate and decreasing the off-rate (Table 4). Thus, some changes in layer 3 disrupt the conformational transition of gp120 from the unliganded state to the CD4-bound state, and these detrimental effects on CD4 binding can be compensated by filling the Phe43 cavity with a hydrophobic tryptophan side chain.
Recognition of mutants by ligands that prefer the CD4-bound conformation.
We examined the binding of a panel of layer 3 gp120 mutants to CD4-induced (CD4i) Abs (17b and 412d) that preferentially recognize the CD4-bound conformation (51, 52). These ligands bind overlapping, conserved regions on gp120 (26, 53).
In the monomeric context and in the absence of sCD4, all layer 3 changes tested, with the exception of S481A, moderately (H249A, V254A, R480A, and E482A) or substantially (T248A, R252A, P253A, R476A, R476E, D477A, N478A, W479A, and L483A) decreased CD4i binding (17b and 412d) compared with the binding of wt gp120 (Fig. 4A and B; Table 2). As expected (50), the S375W change that fills the Phe43 cavity increased 17b binding of wt gp120 in the absence of sCD4. Importantly, with the exception of W479A, the S375W change restored 17b binding to wt levels for the rest of the layer 3 variants tested (Fig. 4C and Table 4). Moreover, addition of sCD4 also reestablished the ability of layer 3 mutants, including W479A, to interact efficiently with 17b (Fig. 4D and Table 4). Thus, alteration of layer 3 residues in the gp120 inner domain decreases CD4i antibody binding to monomeric gp120 in a manner that can be compensated by the S375W change or by sCD4 binding. These results suggest that the layer 3 gp120 residues do not directly interact with the 17b antibody, consistent with the crystal structure of the gp120 core-CD4-17b complex (26, 53).
Fig 4.
Recognition of soluble gp120 variants by CD4i monoclonal antibodies. Comparable amounts of radiolabeled wt and mutant gp120 were incubated with 13 nm of 17b (A) or 412d (B) CD4i Abs for 1 h at 37°C. Alternatively, the effect of filling the Phe43 cavity with a tryptophan (S375W) (C) or of adding sCD4 (D) to layer 3 on 17b recognition was assessed by immunoprecipitation. gp120 variants were normalized by PS and to the signal obtained for their wt counterparts. Precipitates were analyzed by SDS-PAGE and densitometry. Incubation with sCD4 increased the binding of wt gp120 to 17b 1.5-fold. Data shown represent the means ± standard errors of the means of at least two independent experiments. *, P < 0.05 by a Mann-Whitney rank sum test. Differences with P values of ≥0.05 were not considered significant (ns).
Reductions mainly in on-rates, with some acceleration of off-rates, contributed to the decreased affinity of layer 3 variants for 17b (Table 4). These results are consistent with a model in which these mutants do not spontaneously sample the conformation recognized by the 17b antibody, and when they do, they have difficulty in retaining the antibody as efficiently as wt gp120. Thus, layer 3 in the inner domain contributes to the ability of HIV-1 gp120 to assume the conformation preferred by CD4i Abs.
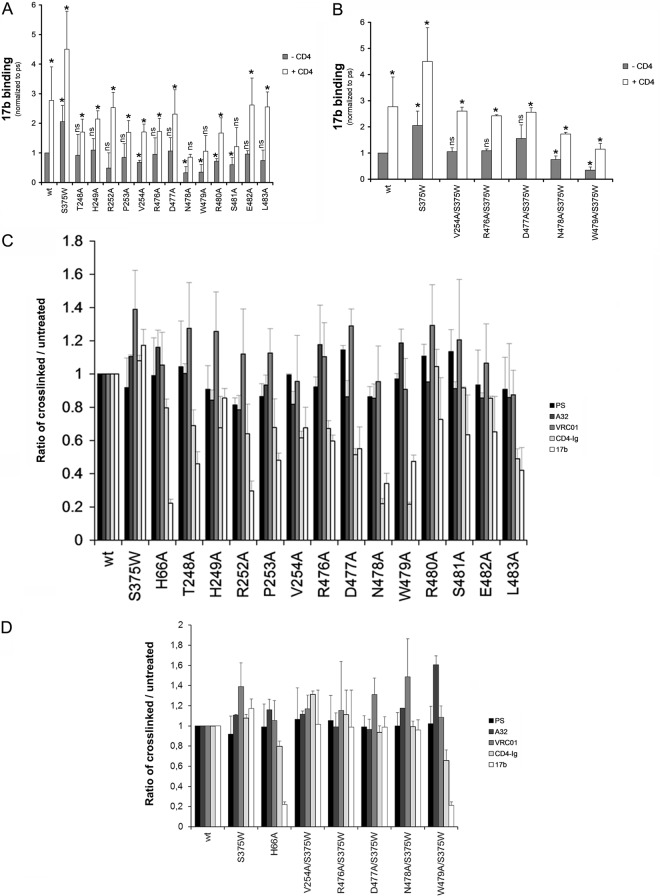
Similar observations on decreased CD4i interaction, albeit less pronounced, were made when layer 3 variants were expressed in the trimeric context and assessed for CD4i binding by cell-based ELISA (Fig. 5A). Significantly, 17b recognition of most Env mutants in the trimeric context could be restored by addition of sCD4 (Fig. 5A), as was seen for the monomeric gp120 variants. As described above, interaction with sCD4 forces the Env trimers to assume the CD4-bound conformation; in our assay this is measured by a 2- to 3-fold enhancement of 17b recognition, upon sCD4 addition, for the wt Env (Fig. 5A and B). For Envs expressed on the cell surface, introduction of the S375W change restored 17b binding to wt levels for the majority of layer 3 mutants, with the exception of W479A (Fig. 5B). Association with sCD4 further enhanced 17b binding of all S375W-containing layer 3 variants (including W479A/S375W).
Fig 5.
Recognition of cellular-expressed trimeric HIV-1 Env variants by a CD4i monoclonal antibody. The effects of alterations in layer 3 on gp120 recognition by 17b, a CD4i antibody, were examined in the absence or presence of CD4 (13 nM CD4-Ig) by cell-based ELISA (A). To avoid recognition of the Ig portion of the CD4-Ig fusion protein by a secondary antibody, 17b was directly conjugated to HRP, as described in Materials and Methods. The effect of filling the Phe43 cavity with a tryptophan (S375W) on layer 3 variants was also analyzed by cell-based-ELISA (B). HIV-1 Env variants were normalized by PS and to the signal obtained for the wt. The S375W mutant, known to interact better with CD4i Abs (50), was introduced for comparison. Data shown represent the means ± standard errors of the means of at least four independent experiments. (C and D) Conformational fixation followed by ligand selection. The ratio of ligand binding to cross-linked versus untreated HIV-1 Env trimer is shown for wt and layer 3 variants without (C) or with (D) filling of the Phe43 cavity. Signals were normalized to those obtained for the wt. Data shown are representative of at least three independent experiments performed in duplicate. *, P < 0.05 by a Mann-Whitney rank sum test. Differences with P values of ≥0.05 were not considered significant (ns).
To investigate further the contribution of layer 3 to the conformation sampled by HIV-1 Env trimers in the unliganded state, we conformationally fixed cell surface Envs prior to probing with specific ligands in the cell-based ELISA (28, 54). In this procedure, glutaraldehyde (GA) cross-linking is used to fix the conformation of Env, and then the relative occupancy of gp120 conformations is assessed by quantifying the binding of ligands with different requirements for specific gp120 conformations. GA cross-links lysine residues and thereby fixes the conformation of the protein component of the envelope glycoproteins (55). Cells were left untreated or cross-linked (5 mM GA) and incubated with a polyclonal mixture of sera (PS) from HIV-1-infected individuals, CD4-Ig, or monoclonal Abs that recognize different conformations of the HIV-1 Env trimers. The polyclonal mixture of sera recognizes multiple conformations sampled by HIV-1 Env trimers; therefore, the signal obtained with PS can be used to normalize mutant Env expression levels at the cell surface (1). The CD4i 17b antibody recognizes gp120 epitopes that overlap the chemokine receptor-binding site and that are normally formed and exposed after CD4 binding (51). Therefore, it recognizes conformations closer to the CD4-bound state. In contrast, the CD4-binding site VRC01 antibody recognizes multiple gp120 conformations (56), and the inner-domain-recognizing antibody A32 has only modest conformational preference (28). We also included two HIV-1 Env trimer variants known to preferentially sample distinct unliganded conformations: the S375W change favors the spontaneous sampling of a CD4-bound-like conformation (50), whereas a change in layer 1, H66A, has been shown to block the transition to the CD4-bound conformation (3, 57, 58). Accordingly, the ratio of cross-linked to untreated HIV-1 Env trimers recognized by 17b was enhanced for the S375W mutant and dramatically reduced for the H66A mutant, compared to this ratio for wt HIV-1 Env trimers. Similar to the H66A mutant, the majority of layer 3 variants exhibited greater-than-wt decreases in 17b recognition upon cross-linking, with the more pronounced phenotypes observed for the T248A, R252A, P253A, N478A, W479A, and L483A mutants (Fig. 5C). VRC01 and A32 recognized the cross-linked wt and layer 3 variants similarly. Importantly, filling the Phe43 cavity restored 17b recognition to wt levels for all the layer 3 variants tested, with the exception of W479A (Fig. 5D). Altogether, these data support the hypothesis that layer 3 integrity is important for the spontaneous sampling of the CD4-bound conformation by HIV-1 Env trimers.
Proteolytic processing and subunit association of the Env glycoprotein mutants.
Proteolytic processing of the gp160 precursor and association of the gp120 and gp41 subunits of each mutant Env trimer were evaluated. All of the mutants were expressed efficiently, and only mutant T248A exhibited a marked decrease in proteolytic processing of the gp160 Env precursor (Table 1), perhaps due to its proximity to cysteine 247. In striking contrast to the major contribution of inner domain layer 1 and layer 2 residues to subunit association within the Env trimer (3), changes in only one layer 3 residue (W479) significantly disrupted the noncovalent association of gp120 with the Env trimer (Table 1). Apparently, layer 3 as a whole is not directly responsible for gp120 association with the unliganded Env glycoprotein trimer. Residue W479 locates at the center of a hydrophobic interface between layers 2 and 3 (Fig. 1C and D). Changes in Trp479 might affect trimer stability through effects on layer 2-layer 3 association, which could indirectly affect gp120-trimer association by altering either the gp120-trimer association domain or the gp120-gp41 interface. Consistent with the relevance of layer 2-layer 3 interaction to this phenotype, M104 in layer 2, which contacts W479 in the CD4-bound conformation (26), has been previously shown to be involved in gp120-gp41 association (3). Indeed, substitution of a tryptophan residue for methionine 104 enhanced gp120-trimer association on its own; more importantly, the M104W change was sufficient to restore gp120-trimer association of W479A to a wt level (Fig. 6). This observation supports the hypothesis that the phenotypic effect of the W479A change on gp120-trimer association results from a disruption of an interaction between layers 2 and 3.
Fig 6.
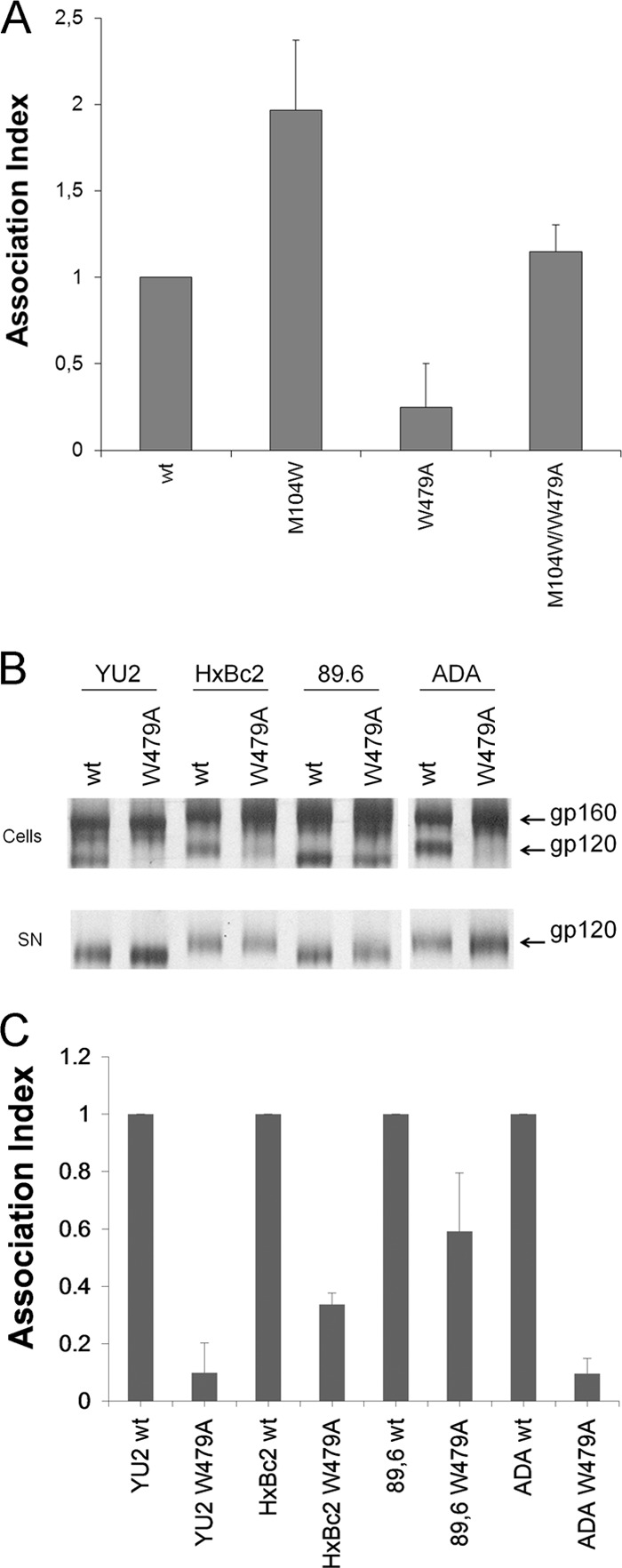
Inner domain residues important for gp120-trimer association. Cell lysates and supernatants (SN) of 35S-labeled cells transiently expressing the HIV-1YU2 (A), HIV-1HxBc2, HIV-189.6, and HIV-1ADA (B) and indicated mutant envelope glycoproteins were precipitated with serum from HIV-1-infected patients. The precipitated proteins were loaded onto SDS-PAGE polyacrylamide gels and analyzed by autoradiography and densitometry. (C) A quantification of the data shown in panel B. The association index is a measure of the ability of the mutant gp120 molecule to remain associated with the envelope glycoprotein complex on the expressing cell relative to that of the wild-type envelope glycoproteins. The association index was calculated as described in Materials and Methods. Data shown represent the average ± standard deviation of at least two independent experiments.
In the unliganded state, layer 2-layer 3 interactions could potentially influence the spatial relationship of the gp120 inner and outer domains. Elements of both domains (V1/V2 in the inner domain and V3 in the outer domain) contribute to the trimer association domain (TAD) of gp120 (35). Unlike the well-conserved gp120 regions that directly interact with gp41, changes in the gp120 TAD result in more dramatic phenotypes in R5 HIV-1 Envs than in R5 or R5X4 HIV-1 Envs (59). To examine whether W479A exhibited this property, we introduced this change into Envs derived from the R5 HIV-1ADA, the dual-tropic (R5X4) HIV-189.6 (60, 61), and the CXCR4-using (X4) HIV-1HXBc2 (62) viruses. The W479A change resulted in massive shedding of gp120 in the two R5 strains tested (YU2 and ADA) and less extensive shedding in the X4 (HXBc2) and R5X4 (89.6) Envs (Fig. 6B and C), as predicted for an effect mediated through the gp120 TAD. In this sense, the W479A change almost completely abrogated recognition of trimeric Envs by PG9/PG16 antibodies (Table 3) that recognize epitopes present in conserved portions of the gp120 variable regions (46).
Function of the mutant HIV-1 Env trimers.
The mutant HIV-1 Env trimers were assessed for the ability to mediate cell-cell fusion and to support virus entry into cells expressing CD4 and CCR5. In both assays, most of the mutant Env trimers exhibited detectable activity (Table 1). The mutant W479A, with the lowest gp120-gp41 association index, also exhibited the lowest infectivity. However, it mediated cell-cell fusion much more efficiently than cell-free virus infection. This phenotype has previously been observed for alterations in the inner domain layers 1 and 2 and the β-sandwich that affect gp120-trimer association (3, 5). Because the time between Env synthesis and engagement of the target cell is much longer in the virion infectivity assay than in the cell-cell fusion assay, the former assay is more sensitive to decreases in the stability of gp120-trimer association.
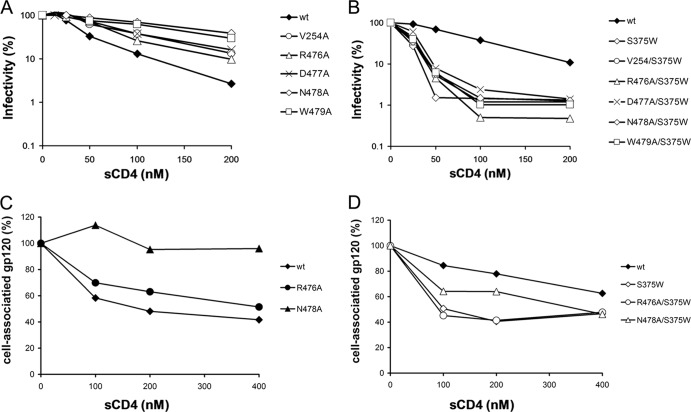
Sensitivity of the mutant viruses to neutralization by sCD4.
Changes in CD4-binding affinity can result in altered HIV-1 sensitivity to sCD4-mediated neutralization (3, 63). Several viruses bearing Env trimers with alterations in layer 3 (V254A, R476A, D477A, N478A, and W479A) were more resistant to sCD4 than viruses with wt Env trimers (Fig. 7A and Table 1). The introduction of the S375W change, which fills the Phe43 cavity (49, 50), into these sCD4-resistant mutant Env trimers resulted in viruses that were neutralized very efficiently by sCD4. Indeed, they were neutralized to the same extent as a virus with the S375W change alone, known to be neutralized more efficiently by sCD4 than viruses with the wt Env (3, 50) (Fig. 7 B). Thus, changes in layer 3 of the gp120 inner domain specifically affect sCD4 binding and virus neutralization.
Fig 7.
Sensitivity of HIV-1 Env gp120 variants to soluble CD4. (A and B) Recombinant HIV-1 expressing luciferase and bearing wt or mutant HIV-1 Env trimer were normalized by reverse transcriptase activity. Equal amounts of viruses were incubated with serial dilutions of sCD4 at 37°C for 1 h prior to infection of Cf2Th-CD4/CCR5 cells. Infectivity at each dilution of sCD4 tested is shown as the percentage of infection without sCD4 for each particular mutant. Quadruplicate samples were analyzed; data shown are representative of values obtained in at least three independent experiments. (C and D) The sCD4-induced shedding of gp120 from HIV-1 Env expressed on the cell surface. Transfected 293T cells were metabolically labeled with [35S]methionine-cysteine for 16 h with increasing concentrations of sCD4 (0 to 400 nM). Cell lysates were precipitated with PS. Precipitates were analyzed by SDS-PAGE and densitometry. Data shown are representative of values obtained in at least two independent experiments.
A prominent consequence of sCD4 binding is the shedding of gp120 from the Env trimer (3, 64, 65). Mutants R476A and N478A exhibited degrees of sCD4-induced gp120 shedding proportional to their degrees of sCD4-mediated neutralization. Indeed, R476A, which is partially resistant to sCD4 neutralization, showed a slight decrease in shedding upon sCD4 addition, whereas N478A (highly resistant to sCD4-mediated neutralization) was completely resistant to sCD4-induced shedding (Fig. 7C). Importantly, the S375W change restored the efficiency of sCD4-induced gp120 shedding in the R476A and N478A mutant Env trimers to levels comparable to the wt Env level (Fig. 7D). Thus, for this group of layer 3 mutants, sCD4 binding to gp120 and sCD4 induction of gp120 shedding correlate, in agreement with previously observed phenotypes of layer 1 and layer 2 mutants involved in modulating CD4 binding indirectly (3).
Soluble CD4 is also known to induce a transiently activated state in the HIV-1 Env trimers (39). We previously reported that layer 1 and layer 2 played a role in this process as changes in the interface between these layers resulted in a long-lived activated state (3). However, none of our layer 3 mutants were activated by sCD4 and were therefore unable to infect CD4-negative, CCR5-expressing cells when incubated with different concentrations of sCD4 (not shown).
DISCUSSION
Current structural and functional studies have led to the hypothesis that, in the process of moving from the unliganded state to the CD4-bound state, the gp120 inner domain undergoes layered movement, whereas the outer domain maintains its conformation (35). Here, we extended our comprehension of the network of interactions involving the gp120 inner domain that modulate gp120 association with the Env trimer, CD4 binding affinity, and susceptibility to inactivation by sCD4 (3).
Before engaging the CD4 receptor, gp120 must maintain its noncovalent association with gp41 and prevent gp41 from prematurely undergoing transitions to lower-energy conformations. In addition to the role of the inner domain β-sandwich and the N and C termini of gp120 in mediating gp120-gp41 association (4, 5), layers 1 and 2 of the gp120 inner domain are also involved in maintaining the association of gp120 with the Env trimer (3). An additional element of the gp120 inner domain, layer 3, apparently contributes to the association of gp120 with the unliganded Env trimer, likely through an indirect mechanism. Indeed, various layer 2 residues, including M104, were previously shown to be important for gp120-trimer association (3). Interestingly, the only layer 3 residue implicated in trimer stability, W479, directly contacts M104 in the CD4-bound conformation (Fig. 1C). Introduction of a compensatory mutation in layer 2 (M104W) (Fig. 6) is sufficient to restore gp120-trimer association to the layer 3 W479A mutant. This observation supports a model in which the effect of the W479A change on trimer stability is mediated by modulating layer 2-layer 3 interactions required for proper gp120-trimer association. Shifts between the inner domain and outer domain that occur as a result of the W479A change in the layer 2-layer 3 interface might affect the orientation of V3 (which has an outer-domain base) and V1/V2 (with an inner-domain base). These effects on the V3 and V1/V2 relationship could affect the conformation of the trimer association domain (TAD), which comprises the V1, V2, and V3 variable regions and appears to be a key element in maintaining interactions among the gp120 subunits in the Env trimer (35). Consistent with this model, the W479A change resulted in greater gp120 shedding in the context of R5 HIV-1 isolates (thought to represent a more “compact” conformation) than in X4 or R5X4 isolates; Envs of the latter viruses are thought to have an open conformation where the contribution of the TAD to trimer stability might be less important (31, 35, 59, 66). Previous work has demonstrated a more prominent role of V3 in trimer stability for primary R5 isolates than for X4 or R5X4 isolates (59).
Conformational metastability in the unliganded HIV-1 Env trimer (28, 54, 67, 68) is expected to provide advantages for immune evasion (69, 70). By mechanisms that are not yet well understood, the gp120 TAD can apparently modulate this property. Removing the gp120 TAD, composed of the V1, V2, and V3 variable regions, allows the gp120 core to assume the CD4-bound conformation (28). In the transition from the unliganded state to the CD4-bound state, the inner domain of gp120, but not the outer domain, experiences dramatic conformational rearrangements (35). Located at the interface between the inner domain and outer domain, layer 3 is structurally expected to play a pivot-like role in the layered allosteric changes in the inner domain. Changes in layer 3 apparently decrease the spontaneous sampling of the CD4-bound conformation by HIV-1 gp120, in several instances lowering the on-rate and binding affinity of CD4i Abs and dramatically decreasing the recognition of cross-linked, cell surface-expressed Env trimers by CD4i Abs.
In contrast to what was observed for alteration of the layer 1 and layer 2 interaction, where decreased CD4 binding was mainly due to an accelerated off-rate (3), the layer 3 variants tested by SPR in this study (T248A, V254A, R476A, D477A, N478A, and W479A) mainly affected CD4 binding by decreasing the initial contact with CD4 (on-rate). The integrity of layer 3 thus appears to be necessary for the optimal presentation and exposure of the CD4-binding site. The S375W change, which fills the Phe43 cavity, completely compensates for the inability of most layer 3 mutants to initiate CD4 binding. Furthermore, the S375W change is sufficient to allow spontaneous sampling of the conformation required for efficient recognition by CD4i Abs, even after cross-linking. Thus, in addition to layers 1 and 2 in the inner domain, layer 3 contributes to the ability of gp120 to make the transition from the unliganded to the CD4-bound state.
Alteration of some gp120 residues in layer 3 (V254A, R476A, D477A, N478A, and W479A) resulted in increased resistance to neutralization by sCD4. Differences in the affinity of monomeric gp120 or trimeric Env for CD4 likely contribute to the sCD4 resistance of these mutants, as has been observed for other HIV-1 Env variants with decreases in CD4 binding (3, 63). Consistent with this, the S375W change restores the CD4-binding affinity of all these mutants and reverts the phenotypes of resistance to both sCD4 neutralization and sCD4 induction of gp120 shedding.
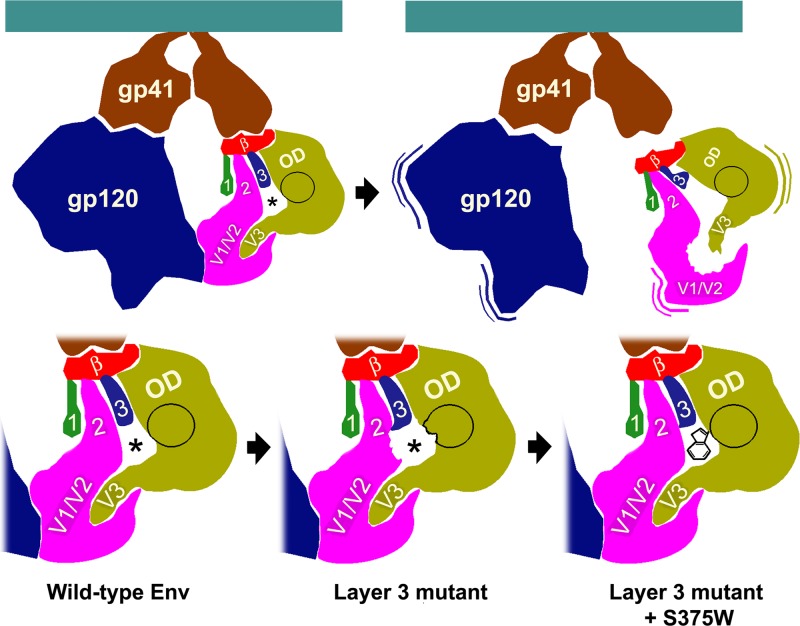
The involvement of layer 3 residues in CD4 and CD4i antibody binding and the particular influence of W479 in gp120 association with the Env trimer suggest a model for the triggering of the conformational changes required for HIV-1 entry. According to this model (Fig. 8), in the unliganded state, layer 3 helps expose the initial site of contact with CD4 and helps to maintain a relationship between the gp120 inner and outer domains that is necessary for TAD-mediated gp120 association with the Env trimer. Upon CD4 binding, layer 3 interacts with layer 2, contributing to the rearrangement of the gp120 inner domain that slows the off-rate of CD4. CD4 binding also leads to TAD conformational changes contributing to the “opening” of the Env trimer at its apex.
Fig 8.
Model for the role of layer 3 in HIV-1 Env trimer stability and CD4 binding. The image on the top left depicts the unliganded HIV-1 Env trimer, with two gp120 subunits and two gp41 subunits visible from the perspective shown. The gp120 subunit on the right is subdivided into the outer domain (OD), inner domain (β-sandwich [red] and layers 1, 2, and 3), and the TAD (V1/V2, and V3 regions) (35). The initial site of CD4 binding is circled in black, and the location of the Phe43 cavity is shown by an asterisk. Layer 3 modulates the interaction of the outer domain and layer 2 in the inner domain. The upper right image shows the consequences of changes in Trp479, which is located in layer 3, in the gp120 interdomain interface. Changes in Trp479 significantly disrupt the relationship of the inner and outer domains. Because the V1/V2 and V3 regions are anchored in the inner and outer domains, respectively, the gp120 TAD, which comprises the V1/V2 and V3 regions, is disrupted. Thus, changes in Trp479 lead to trimer instability and gp120 shedding. The images in the bottom row illustrate the phenotypic consequences of changes in layer 3 that are less disruptive than the alteration of Trp479. These more subtle changes in layer 3 cause local alterations of gp120 conformation around the Phe-43 cavity that affect CD4 binding, with effects on both on-rate and off-rate (bottom panels). The more subtle changes in layer 3 can be compensated by the S375W change, which fills the Phe43 cavity with the indole ring of tryptophan.
Importantly, the residues in layer 3 implicated in the CD4-induced conformational transition are well conserved among the HIV-1 lineage (Fig. 2A). Of interest, some residues (such as W479) are also conserved among the HIV-2/simian immunodeficiency virus SIVsm lineage although differences between the HIV-1/SIVcpz and HIV-2/SIVsm lineages are evident (Fig. 2B). For example, residue 375 is naturally a tryptophan in the HIV-2/SIVsm viruses (71); in HIV-1, substitution of tryptophan for serine 375 fills the Phe43 cavity and diminishes the impact of alterations in layers 1, 2, and 3 on CD4 and CD4i antibody binding. Thus, while both immunodeficiency virus lineages preserve the potential to form a network of inner domain interactions involving layers 1, 2, and 3, some lineage-specific differences exist. Indeed, subtle modifications in the organization of layers 1 and 2 of the inner domain of HIV-1 and SIV gp120 were recently reported to modulate trimer stability and CD4 binding differentially (72). Therefore, fine differences in the regulation of this network of inner domain interactions might help explain how these Env trimers adapted to their respective hosts; additional work is required to assess the role of layer 3 in the HIV-2/SIVsm lineage. Work aimed at understanding the underlying molecular mechanism of the transition from the unbound to the CD4-bound conformation may greatly expedite the development of interventions.
ACKNOWLEDGMENTS
We thank J. Robinson for kindly sharing antibodies (17b, 48d, 412d, A32, and C11). We thank Dennis Burton and Pascal Poignard and IAVI for their generous gift of PG9 and PG16 MAbs, M. Hancock from the McGill Sheldon Biotechnology Centre for helping with SPR analysis, M. Pancera and P. D. Kwong for comments, and Y. McLaughlin and E. Carpelan for helping in manuscript preparation.
This work was supported by an amfAR Mathilde Krim Fellowship in Basic Biomedical Research (Phase II, no. 108092-50-RKVA), by a Canada Foundation for Innovation Program Leader grant (no. 29866), by a CIHR operating grant (no. 257792), and by an FRQS Establishment of Young Scientist grant (no. 24639) to A.F. A.F. is the recipient of an FRSQ Chercheur Boursier Junior 1 Fellowship (no. 24639). This work was also supported by grants from the National Institutes of Health (AI24755 and AI67854), by the International AIDS Vaccine Initiative, and by the late William F. McCarty-Cooper.
The authors have no conflicts of interest to report.
Footnotes
Published ahead of print 19 December 2012
REFERENCES
- 1. Allan JS, Coligan JE, Barin F, McLane MF, Sodroski JG, Rosen CA, Haseltine WA, Lee TH, Essex M. 1985. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science 228:1091–1094 [DOI] [PubMed] [Google Scholar]
- 2. Robey WG, Safai B, Oroszlan S, Arthur LO, Gonda MA, Gallo RC, Fischinger PJ. 1985. Characterization of envelope and core structural gene products of HTLV-III with sera from AIDS patients. Science 228:593–595 [DOI] [PubMed] [Google Scholar]
- 3. Finzi A, Xiang SH, Pacheco B, Wang L, Haight J, Kassa A, Danek B, Pancera M, Kwong PD, Sodroski J. 2010. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol. Cell 37:656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Helseth E, Olshevsky U, Furman C, Sodroski J. 1991. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J. Virol. 65:2119–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang X, Mahony E, Holm GH, Kassa A, Sodroski J. 2003. Role of the gp120 inner domain beta-sandwich in the interaction between the human immunodeficiency virus envelope glycoprotein subunits. Virology 313:117–125 [DOI] [PubMed] [Google Scholar]
- 6. Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767 [DOI] [PubMed] [Google Scholar]
- 7. Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, Montagnier L. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767–768 [DOI] [PubMed] [Google Scholar]
- 8. Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. 1996. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955–1958 [DOI] [PubMed] [Google Scholar]
- 9. Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148 [DOI] [PubMed] [Google Scholar]
- 10. Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661–666 [DOI] [PubMed] [Google Scholar]
- 11. Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667–673 [DOI] [PubMed] [Google Scholar]
- 12. Feng Y, Broder CC, Kennedy PE, Berger EA. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872–877 [DOI] [PubMed] [Google Scholar]
- 13. Furuta RA, Wild CT, Weng Y, Weiss CD. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276–279 [DOI] [PubMed] [Google Scholar]
- 14. He Y, Vassell R, Zaitseva M, Nguyen N, Yang Z, Weng Y, Weiss CD. 2003. Peptides trap the human immunodeficiency virus type 1 envelope glycoprotein fusion intermediate at two sites. J. Virol. 77:1666–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu M, Blacklow SC, Kim PS. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075–1082 [DOI] [PubMed] [Google Scholar]
- 16. Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430 [DOI] [PubMed] [Google Scholar]
- 17. Yang X, Kurteva S, Lee S, Sodroski J. 2005. Stoichiometry of antibody neutralization of human immunodeficiency virus type 1. J. Virol. 79:3500–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang X, Lipchina I, Cocklin S, Chaiken I, Sodroski J. 2006. Antibody binding is a dominant determinant of the efficiency of human immunodeficiency virus type 1 neutralization. J. Virol. 80:11404–11408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bures R, Gaitan A, Zhu T, Graziosi C, McGrath KM, Tartaglia J, Caudrelier P, El Habib R, Klein M, Lazzarin A, Stablein DM, Deers M, Corey L, Greenberg ML, Schwartz DH, Montefiori DC. 2000. Immunization with recombinant canarypox vectors expressing membrane-anchored glycoprotein 120 followed by glycoprotein 160 boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 16:2019–2035 [DOI] [PubMed] [Google Scholar]
- 20. Koff WC. 2012. HIV vaccine development: Challenges and opportunities towards solving the HIV vaccine-neutralizing antibody problem. Vaccine 30:4310–4315 [DOI] [PubMed] [Google Scholar]
- 21. Mascola JR, Snyder SW, Weislow OS, Belay SM, Belshe RB, Schwartz DH, Clements ML, Dolin R, Graham BS, Gorse GJ, Keefer MC, McElrath MJ, Walker MC, Wagner KF, McNeil JG, McCutchan FE, Burke DS. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. J. Infect. Dis. 173:340–348 [DOI] [PubMed] [Google Scholar]
- 22. Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220 [DOI] [PubMed] [Google Scholar]
- 23. Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, Stanfield RL, Robinson J, Sodroski J, Wilson IA, Wyatt R, Bewley CA, Kwong PD. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317:1930–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure 8:1329–1339 [DOI] [PubMed] [Google Scholar]
- 26. Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. U. S. A. 107:1166–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, Zhu J, Vicic DA, Debnath AK, Shapiro L, Bewley CA, Mascola JR, Sodroski JG, Kwong PD. 2012. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc. Natl. Acad. Sci. U. S. A. 109:5663–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu G, Liu J, Taylor KA, Roux KH. 2011. Structural comparison of HIV-1 envelope spikes with and without the V1/V2 loop. J. Virol. 85:2741–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz MJ, Bess JW, Nandwani R, Hoxie JA, Lifson JD, Milne JL, Subramaniam S. 2010. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 6:e1001249 doi:10.1371/journal.ppat.1001249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu SR, Loving R, Lindqvist B, Hebert H, Koeck PJ, Sjoberg M, Garoff H. 2010. Single-particle cryoelectron microscopy analysis reveals the HIV-1 spike as a tripod structure. Proc. Natl. Acad. Sci. U. S. A. 107:18844–18849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. 2006. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2:e83 doi:10.1371/journal.ppat.0020083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441:847–852 [DOI] [PubMed] [Google Scholar]
- 35. Mao Y, Wang L, Gu C, Herschhorn A, Xiang SH, Haim H, Yang X, Sodroski J. 2012. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nat. Struct. Mol. Biol. 19:893–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. LaBonte JA, Patel T, Hofmann W, Sodroski J. 2000. Importance of membrane fusion mediated by human immunodeficiency virus envelope glycoproteins for lysis of primary CD4-positive T cells. J. Virol. 74:10690–10698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Korber B, Foley BT, Kuiken C, Pillai SK, Sodroski JG. 1998. Numbering positions in HIV relative to HXB2CG. Hum. Retroviruses AIDS III:102–111 [Google Scholar]
- 39. Haim H, Si Z, Madani N, Wang L, Courter JR, Princiotto A, Kassa A, DeGrace M, McGee-Estrada K, Mefford M, Gabuzda D, Smith AB, III, Sodroski J. 2009. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 5:e1000360 doi:10.1371/journal.ppat.1000360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chowdhury IH, Koyanagi Y, Takamatsu K, Yoshida O, Kobayashi S, Yamamoto N. 1991. Evaluation of anti-human immunodeficiency virus effect of recombinant CD4-immunoglobulin in vitro: a good candidate for AIDS treatment. Med. Microbiol. Immunol. 180:183–192 [DOI] [PubMed] [Google Scholar]
- 41. Rho HM, Poiesz B, Ruscetti FW, Gallo RC. 1981. Characterization of the reverse transcriptase from a new retrovirus (HTLV) produced by a human cutaneous T-cell lymphoma cell line. Virology 112:355–360 [DOI] [PubMed] [Google Scholar]
- 42. Johnsson B, Lofas S, Lindquist G. 1991. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal. Biochem. 198:268–277 [DOI] [PubMed] [Google Scholar]
- 43. Li Y, Hui H, Burgess CJ, Price RW, Sharp PM, Hahn BH, Shaw GM. 1992. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J. Virol. 66:6587–6600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moore JP, Sodroski J. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Finzi A, Pacheco B, Zeng X, Kwon YD, Kwong PD, Sodroski J. 2010. Conformational characterization of aberrant disulfide-linked HIV-1 gp120 dimers secreted from overexpressing cells. J. Virol. Methods 168:155–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lasky LA, Nakamura G, Smith DH, Fennie C, Shimasaki C, Patzer E, Berman P, Gregory T, Capon DJ. 1987. Delineation of a region of the human immunodeficiency virus type 1 gp120 glycoprotein critical for interaction with the CD4 receptor. Cell 50:975–985 [DOI] [PubMed] [Google Scholar]
- 48. Olshevsky U, Helseth E, Furman C, Li J, Haseltine W, Sodroski J. 1990. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J. Virol. 64:5701–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xiang SH, Kwong PD, Gupta R, Rizzuto CD, Casper DJ, Wyatt R, Wang L, Hendrickson WA, Doyle ML, Sodroski J. 2002. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 76:9888–9899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179–183 [DOI] [PubMed] [Google Scholar]
- 53. Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705–711 [DOI] [PubMed] [Google Scholar]
- 54. Yuan W, Bazick J, Sodroski J. 2006. Characterization of the multiple conformational States of free monomeric and trimeric human immunodeficiency virus envelope glycoproteins after fixation by cross-linker. J. Virol. 80:6725–6737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Das M, Fox CF. 1979. Chemical cross-linking in biology. Annu. Rev. Biophys. Bioeng. 8:165–193 [DOI] [PubMed] [Google Scholar]
- 56. Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Do Kwon Y, Scheid J, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kassa A, Finzi A, Pancera M, Courter JR, Smith AB, III, Sodroski J. 2009. Identification of a human immunodeficiency virus (HIV-1) envelope glycoprotein variant resistant to cold inactivation. J. Virol. 83:4476–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kassa A, Madani N, Schon A, Haim H, Finzi A, Xiang SH, Wang L, Princiotto A, Pancera M, Courter J, Smith AB, III, Freire E, Kwong PD, Sodroski J. 2009. Transitions to and from the CD4-bound conformation are modulated by a single-residue change in the human immunodeficiency virus type 1 gp120 inner domain. J. Virol. 83:8364–8378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xiang SH, Finzi A, Pacheco B, Alexander K, Yuan W, Rizzuto C, Huang CC, Kwong PD, Sodroski J. 2010. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J. Virol. 84:3147–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Collman R, Balliet JW, Gregory SA, Friedman H, Kolson DL, Nathanson N, Srinivasan A. 1992. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J. Virol. 66:7517–7521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li Y, Kappes JC, Conway JA, Price RW, Shaw GM, Hahn BH. 1991. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J. Virol. 65:3973–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fisher AG, Collalti E, Ratner L, Gallo RC, Wong-Staal F. 1985. A molecular clone of HTLV-III with biological activity. Nature 316:262–265 [DOI] [PubMed] [Google Scholar]
- 63. Thali M, Olshevsky U, Furman C, Gabuzda D, Li J, Sodroski J. 1991. Effects of changes in gp120-CD4 binding affinity on human immunodeficiency virus type 1 envelope glycoprotein function and soluble CD4 sensitivity. J. Virol. 65:5007–5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hart TK, Kirsh R, Ellens H, Sweet RW, Lambert DM, Petteway SR, Jr, Leary J, Bugelski PJ. 1991. Binding of soluble CD4 proteins to human immunodeficiency virus type 1 and infected cells induces release of envelope glycoprotein gp120. Proc. Natl. Acad. Sci. U. S. A. 88:2189–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Moore JP, McKeating JA, Weiss RA, Sattentau QJ. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139–1142 [DOI] [PubMed] [Google Scholar]
- 66. Zhuang K, Finzi A, Tasca S, Shakirzyanova M, Knight H, Westmoreland S, Sodroski J, Cheng-Mayer C. 2011. Adoption of an “open” envelope conformation facilitating CD4 binding and structural remodeling precedes coreceptor switch in R5 SHIV-infected macaques. PLoS One 6:e21350 doi:10.1371/journal.pone.0021350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kong L, Huang CC, Coales SJ, Molnar KS, Skinner J, Hamuro Y, Kwong PD. 2010. Local conformational stability of HIV-1 gp120 in unliganded and CD4-bound states as defined by amide hydrogen/deuterium exchange. J. Virol. 84:10311–10321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle ML. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. U. S. A. 97:9026–9031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682 [DOI] [PubMed] [Google Scholar]
- 70. Kwong PD, Wilson IA. 2009. HIV-1 and influenza antibodies: seeing antigens in new ways. Nat. Immunol. 10:573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kuiken C, Marx FBP, Wolinsky S, Leitner T, Hahn B, McCutchan F, Korber B. 2008. HIV sequence compendium 2008. Los Alamos National Laboratory, Los Alamos, NM [Google Scholar]
- 72. Finzi A, Pacheco B, Xiang SH, Pancera M, Herschhorn A, Wang L, Zeng X, Desormeaux A, Kwong PD, Sodroski J. 2012. Lineage-specific differences between human and simian immunodeficiency virus regulation of gp120 trimer association and CD4 binding. J. Virol. 86:8974–8986 [DOI] [PMC free article] [PubMed] [Google Scholar]