Abstract
During infection by human cytomegalovirus (HCMV), the tumor suppressor protein p53, which promotes efficient viral gene expression, is stabilized. However, the expression of numerous p53-responsive cellular genes is not upregulated. The molecular mechanism used to manipulate the transcriptional activity of p53 during infection remains unclear. The HCMV proteins IE1, IE2, pUL44, and pUL84 likely contribute to the regulation of p53. In this study, we used a discovery-based approach to identify the protein targets of the HCMV protein pUL29/28 during infection. Previous studies have demonstrated that pUL29/28 regulates viral gene expression by interacting with the chromatin remodeling complex NuRD. Here, we observed that pUL29/28 also associates with p53, an additional deacetylase complex, and several HCMV proteins, including pUL38. We confirmed the interaction between p53 and pUL29/28 in both the presence and absence of infection. HCMV pUL29/28 with pUL38 altered the activity of the 53-regulatable p21CIP1 promoter. During infection, pUL29/28 and pUL38 contributed to the inhibition of p21CIP1 as well as caspase 1 expression. The expression of several other p53-regulating genes was not altered. Infection using a UL29-deficient virus resulted in increased p53 binding and histone H3 acetylation at the responsive promoters. Furthermore, expression of pUL29/28 and its interacting partner pUL38 contributed to an increase in the steady-state protein levels of p53. This study identified two additional HCMV proteins, pUL29/28 and pUL38, which participate in the complex regulation of p53 transcriptional activity during infection.
INTRODUCTION
Human cytomegalovirus (HCMV) is a member of the beta-herpesvirus family, which also includes human herpesviruses 6 and 7. Infection by HCMV is a leading cause of birth defects and can cause severe disease upon immunosuppression (reviewed in reference 1). HCMV disease in immunosuppressed individuals is often successfully managed using the antiviral compound ganciclovir, valganciclovir, cidofovir, or foscarnet. Congenital HCMV infection, however, remains a significant problem because of limited diagnostics and treatment options as well as the lack of community awareness (2). The initial infection leads to systemic viral spread and a balance between latent and lytic replication cycles among diverse cell types within the body. These complex replication cycles result in a persistent lifelong infection.
Successful HCMV infection involves viral proteins interacting with and disconnecting cellular stress response pathways. Many of these pathways and the associated proteins are also altered in cancers and are conserved targets among diverse herpesviruses. Examples include DAXX (death domain-associated protein) (3–6), PML (promyelocytic leukemia protein) (7–11), IFI16 (interferon-inducible protein 16) (12, 13), Tip60 (Tat-interactive protein, 60 kDa) (14, 15), and p53 (16–24). Upon infection, delivery of the HCMV tegument protein pp71 (UL82) results in the degradation of cellular DAXX and disruption of an intrinsic antiviral response (3–6). The response is further influenced by the interaction between HCMV IE1 and PML (7–11). A second tegument protein, pp65 (UL83), binds the nuclear pathogen sensor and transcription factor IFI16 (25, 26), resulting in IFI16-dependent activation of the HCMV major immediate early (MIE) promoter (12, 13). Viral proteins also regulate the tumor suppressor protein Tip60 acetyltransferase (14, 15, 27). Tip60 participates in diverse pathways, including the activation of ATM (ataxia telangiectasia mutated protein) following DNA damage (28). Expression of pUL27 causes the transient degradation of Tip60 at early times of infection, resulting in increased expression of the CDK (cyclin-dependent kinase) inhibitor, p21CIP1 (15). Tip60 is also a target of several herpesvirus kinases, including HCMV pUL97 (14).
In general, the cellular responses involving PML, DAXX, IFI16, and Tip60 have all been demonstrated to influence the activities of the transcription factor and tumor suppressor protein p53 (29–32). As a central participant in stress responses, p53 is manipulated by HCMV. The steady-state amount of p53 protein but not RNA increases very early during infection (19, 20, 22). This stabilization of p53 (33) occurs, in part, by HCMV IE2-mediated repression of the E3 ubiquitin ligase protein MDM2 (20, 34). In addition, p53 is phosphorylated on serine 15 and 20 during infection (35, 36), and these modifications are typically associated with increased transcriptional activity. Expression of p53 contributes to efficient infection by influencing HCMV gene expression (16–18, 21, 23, 24). Surprisingly, however, the majority of p53-regulatable cellular genes are not induced (19). Reevaluation of expression changes in known p53-responsive genes (37) from microarray studies on HCMV infected cells (38) identified only 8 genes that increased in expression at multiple times postinfection, while 61 decreased or did not change within the first 24 h postinfection (hpi) (see Table S1 in the supplemental material). The HCMV proteins IE1, IE2, pUL44, and pUL84 participate in regulating p53 by binding to and altering p53-mediated transcription (22, 35, 39–44). In addition, regulation of p53 is partially achieved by relocalization of a subpopulation of p53 to viral replication centers within the nucleus (17). However, it is not clear whether these events are sufficient for HCMV to control p53 transcriptional activity during infection.
In this study, we observed that p53 also associates with the HCMV protein pUL29/28 during infection. This viral protein was previously identified in proteomic-based screens involving HCMV pUL38 (45) or cellular histone deacetylase 1 (HDAC1) (46) as bait. Expression of pUL29/28 is observed as early as 6 hpi and detected in sorbitol-pelleted virions, albeit at low levels (47). pUL29/28 is distributed throughout the nucleus (47) and associates with components of the HDAC1-containing nuclear remodeling and deacetylase (NuRD) complex during infection (46). Disruption of pUL29/28 (47–49) or components of NuRD result in decreased HCMV gene expression and genome replication (46). These results implicate pUL29/28 to be a transcriptional regulator.
The aim of this study was to more accurately define pUL29/28 binding partners during infection and determine the functional relevance of the interactions. Using pUL29/28 as bait, we discovered interactions with cellular p53 as well as two deacetylase complexes and several additional HCMV proteins. Expression of pUL29/28 with its binding partner pUL38 altered RNA levels for two of the six p53-regulatable cellular genes examined. In the absence of UL29, we observed changes in p53 binding as well as histone H3 acetylation at the responsive promoters during infection. Overall, our studies confirm the role of pUL29/28 as a transcriptional regulator and identify the tumor suppressor protein p53 and two p53-regulatable cellular genes as additional targets of its activity.
MATERIALS AND METHODS
Biological reagents.
The recombinant HCMV viruses ADin29FLAG, ADin28FLAG-C, and ADdel38 (ADdlUL38) were previously generated (47, 50) using AD169 BAC (ADwt virus). ADdel29 virus was constructed using BAC recombineering (51, 52) where the UL29 open reading frame (ORF) was replaced by the galK gene using BAC recombineering (5′-GCGGGTCGCCGAGGCTACTGCTGCTTCTGCTTTTTT GTCTCCTGTGGATCGTCGCGGACTGCCGGCCCTGTTGACAATTAATCATCGGCA-3′ and 5′-GTCTCAAACACGCTACTTTCGGTTATAAAAACACCGTCGCCCTATTTCTGGGCGCGTGTACACTGATGACTCAGCACTGTCCTGCTCCTT-3′). Viral stocks were prepared by either transfecting BAC DNA or infecting primary human fibroblasts. Concentrated stocks were obtained by ultracentrifugation through a sorbitol cushion (20% d-sorbitol, 50 mM Tris-HCl [pH 7.2], 1 mM MgCl2) at 55,000 × g for 1 to 2 h. Titers of viral stocks were determined using a 50% tissue culture infectious dose (TCID50) assay. Human fibroblasts were maintained in 96-well culture plates and infected with 10-fold serial dilutions of the viral stock. After an incubation period of approximately 2 weeks, the cells were fixed and stained for HCMV pUL123 (IE1). Titers were determined by counting the number of pUL123-positive wells per dilution series in the TCID50 assay and defined as infectious units (IU) per ml. The viral titers from experimental samples were determined by infecting fibroblasts with serial dilutions of the culture supernatants, fixing, and staining for pUL123 at 48 hpi.
Human foreskin fibroblasts, U-2 OS osteosarcoma cells, and 293T Phoenix cells were propagated in Dulbecco's modified Eagle medium (DMEM) (Life Technologies, Carlsbad, CA) supplemented with 7 to 10% fetal bovine serum (non-USA-qualified FBS [USDA-approved non-USA FBS]) (Life Technologies, Carlsbad, CA) and 1% penicillin-streptomycin (Life Technologies, Carlsbad, CA). For experiments where aspects of infection were compared between viruses, cells were serum starved prior to infection for approximately 24 to 48 h. For nutrient restriction experiments, fibroblasts were infected at a multiplicity of 6 IU per cell in standard medium (DMEM, 7% FBS, and 25 mM glucose) for 1 h. The cells were then washed with phosphate-buffered saline (PBS) and replenished with either standard medium or restrictive medium containing DMEM, 5.6 mM glucose, and no FBS.
The luciferase plasmids containing the p21Cip1 promoter was kind gifts from B. Vogelstein (The Johns Hopkins University School of Medicine) (53, 54). The pCGN-UL29/28HA plasmid has been previously described (47). The pCIcchp53FLAG plasmid was a kind gift from L. Cirillo (The Medical College of Wisconsin) (55) and pEGFP from Clontech Laboratories, Inc. (Mountain View, CA). The following antibodies were used in studies for Western blot (WB), immunoprecipitation (IP), or immunofluorescence analysis (IF): mouse anti-FLAG M2 (WB, IP, IF) (Sigma-Aldrich, St. Louis, MO), mouse anti-p53 clone DO-1 (WB, IP, IF) (Santa Cruz Biotechnology, Santa Cruz, CA; Sigma-Aldrich, St. Louis, MO), mouse anti-GAPDH clone 0411 (WB) (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-Myc clone 71D10 (WB) (Cell Signaling Technology, Danvers, MA), mouse anti-HDAC3 clone 3G6 (WB, IP) (EMD-Millipore, Billerica, MA), mouse anti-hemagglutinin (HA) clone HA-7 (WB, IP) (Sigma-Aldrich, St. Louis, MO), rabbit anti-FLAG (WB) (Sigma-Aldrich, St. Louis, MO), mouse anti-p21Cip1 clone CP74 (WB) (EMD-Millipore, Billerica, MA), mouse anti-21Cip1 clone F-5 (WB) (Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti-ICP36 clone 10D8 (WB) (Virusys Corporation, Taneytown, MD), rabbit anti-p53 acetyl (K382) (WB) (Epitomics, Burlingame, CA), mouse anti-beta actin clone AC-15 (WB) (Abcam, Cambridge, MA), and mouse control IgG (Santa Cruz Biotechnology, Santa Cruz, CA). The mouse anti-pUL38 clone 8D6 (WB) (50), and mouse anti-pUL123 clone 1B12 (WB) were generously provided by Tom Shenk (Princeton University, Princeton, NJ). Secondary antibodies for Western blot analysis were goat anti-mouse IgG–HRP (horseradish peroxidase) and donkey anti-rabbit IgG–HRP (Jackson ImmunoResearch, West Grove, PA), and those for immunofluorescences were goat anti-mouse IgG (heavy plus light chain [H+L])–Alexa Fluor 488 and donkey anti-mouse IgG (H+L)–Alexa Fluor 488 (Life Technologies, Carlsbad, CA).
Mass spectrometry analysis.
Primary foreskin fibroblasts were infected at an MOI of 3 using ADin29FLAG and harvested at 24 hpi as previously described (45). Fractionated cell powder was resuspended in lysis buffer [20 mM HEPES-KOH (pH 7.4), 110 mM potassium acetate, 2 mM MgCl2, 0.1% (vol/vol) Tween 20, 1.0% (vol/vol) Triton X-100, 250 mM NaCl, and protease inhibitors (Roche Applied Science, Indianapolis, IN)]. Protein complexes were isolated using mouse anti-FLAG M2 antibody conjugated to Dynabeads M-270 epoxy (Life Technologies, Carlsbad, CA) for approximately 1 h, eluted, dried, and resuspended in SDS-PAGE loading buffer. Samples were alkylated with iodoacetamide and separated by SDS–10% PAGE. The entire lane was cut into approximately 4 mm sections and processed as previously described (45). The isolated tryptic peptides were dried, resuspended in 5% acetonitrile and 0.1% formic acid, and identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using an ESI-LTQ XL mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Peptide identification was carried out using the search algorithm SEQUEST and a combined human cytomegalovirus and human protein database with a global false discovery rate of 5%. The results were evaluated using the free software Visualize (56). Proteins which bound to the antibody-conjugated beads were also identified following infection with wild-type HCMV as previously described (15), and this information controlled for nonspecific interactions.
Analysis of protein and nucleic acid.
For Western blot analysis of steady-state protein levels, cells were lysed in an SDS lysis buffer (1% SDS, 50 mM Tris [pH 7.4]). Protein samples were resolved using sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred by semidry transfer to Protran nitrocellulose membranes (Whatman Inc., Piscataway, NJ). The membrane was blocked with 5% milk in phosphate-buffered saline (PBS) containing 0.1% Tween 20 (PBS-T). The membrane was incubated with primary antibody diluted in PBS-T containing 3% bovine serum albumin (BSA) followed by a secondary antibody conjugated to HRP. The antibodies were detected using an enhanced chemiluminescence (ECL) or ECL Plus reagent (GE Healthcare, Piscataway, NJ) and film or FluorChem HD2 (Proteinsimple, Santa Clara, CA), and a ChemiDoc system (Bio-Rad, Hercules, CA) was used for protein quantification. For immunoprecipitation experiments, cells were lysed in an NP-40 lysis buffer (0.1 to 0.5% NP-40, 50 mM Tris, 150 mM NaCl, 1 mM EDTA [pH 7.4]). Immunoprecipitation was completed using either protein A/G Plus agarose (Santa Cruz Biotechnology, Santa Cruz, CA) or Dynabeads protein G (Life Technologies, Carlsbad, CA). Lysates were precleared for 30 min at 4°C using beads in the absence of antibody. Samples were incubated with antibody for 1 to 2 h at 4°C followed by the addition of beads for 1 h at 4°C. The beads were washed three times with lysis buffer and resuspended in Laemmli sample buffer. The samples along with lysate controls obtained prior to immunoprecipitation were evaluated by Western blotting.
For immunofluorescence microscopy, cells were grown on coverslips, infected at a multiplicity of 4, fixed using 2% paraformaldehyde, permeabilized using 0.1% Triton X-100, and blocked using BSA dissolved in PBS-T. Fixed cells were incubated for 1 h using primary antibody against p53 diluted in PBS-T and BSA, washed with PBS-T, and incubated with a fluorophore-coupled secondary antibody. Finally, coverslips were mounted using SlowFade gold antifade reagent (Life Technologies, Carlsbad, CA).
Quantification of cellular and viral RNA was completed using quantitative reverse transcriptase PCR (qRT-PCR). Total RNA was collected using TRIzol reagent (Life Technologies, Carlsbad, CA), and cDNA was synthesized using 1 to 2 μg of DNase-treated RNA, random hexamers, and SuperScript III reverse transcriptase (Life Technologies, Carlsbad, CA). Following cDNA synthesis, qPCR was performed using primers designed against the genes of interest. Primers for p21CIP1 and GAPDH were published previously (15). Additional primers (Integrated DNA Technologies, Inc., Coralville, IA) included TSC2 (5′-GGATGATAGGGCAGATTTGTGAA-3′, 5′-ACCGCCTTCCAGAGTGCTT-3′), PUMA (5′-TGGAGGGTCCTGTACAATCTCA-3′, 5′-TCTGTGGCCCCTGGGTAA-3′), CASP1 (5′-CATCACAGGCATGACAATGCTGCT-3′, 5′-TGCCTTCCCGAATACCAT GAGACA-3′), BIRC5 (5′-TCTGTCAGCCCAACCTTCACATCT-3′, 5′-TGTAACAATCCACCCTGCAGCTCT-3′), IGFBP3 (5′-AGTTCCTCAATGTGCTGAGTCCCA-3′, 5′-AGGCTGCCCATACTTATCCACACA-3′), MMP2 (5′-AGAAGGATGGCAAGTACGGCTTCT-3′, 5′-AGTGGTGCAGCTGTCATAGGATGT-3′), and MDM2 (5′-CTATTGGAAATGCACTTCATGCA-3′, 5′-CGAAGGGCCCAACATCTGT-3′). Quantification was completed using FastStart universal SYBR green master mix (Roche Applied Science, Indianapolis, IN) and a 7900HT fast real-time PCR system (Life Technologies, Carlsbad, CA). A standard curve consisting of 10-fold serial dilutions of one sample was used to determine relative abundance.
Cellular and viral DNA levels were determined by quantitative PCR (qPCR). Cells were harvested and incubated overnight in DNA lysis buffer (400 mM NaCl, 10 mM Tris [pH 8.0], 10 mM EDTA, 0.1 mg/ml of proteinase K, and 0.2% SDS) at 37°C. DNA was extracted using phenol-chloroform, treated with RNase A for 1 h at 37°C, and ethanol precipitated. qPCR was performed using primers directed against either viral (15) or cellular DNA. For these studies, we used primers targeting sequences located within the cellular gene BAX (5′-TCCCCCCGAGAGGTGTTTT-3′ and 5′-CGGCCCCAGTTGAAGTTG-3′). A standard curve consisting of 10-fold serial dilutions was used to determine relative abundance.
Luciferase reporter assays.
U-2 OS cells or U-2 OS cells expressing HCMV pUL38 growing in 12-well culture plates were transfected with reporter and expression plasmids using Fugene 6 transfection reagent (Roche Applied Science, Indianapolis, IN). Amounts of luciferase reporter vectors used ranged from 50 to 200 ng per well, and amounts of expression plasmids ranged from 1 to 100 ng per well. The cells were harvested at 2 days posttransfection, and relative light units (RLU) were measured within each lysate using the luciferase assay system (Promega Corp, Madison, WI) as previously described (46, 47). The RLUs were quantified using a Victor 3V multilabel plate reader (PerkinElmer, Inc., Waltham, MA) with a measurement time of 10 s.
Cell cycle analysis.
Quantification of cell cycle phases was completed as previously described (15) using U-2 OS cells or U-2 OS cells expressing HCMV pUL38. Both cell types were transfected with 10 μg of pCGN-pUL29/28HA expression plasmid using Fugene 6 transfection reagent (Roche Applied Science, Indianapolis, IN) following the manufacturer's instructions. The cells, culturing conditions, and plasmid were as described above (see “Biological reagents”). At 24 h posttransfection, the cells were either untreated (unsynchronized) or treated (synchronized) with 75 ng/ml nocodazole (Sigma-Aldrich, St. Louis, MO) for 16 h. The cells were then collected, fixed using 70% ice-cold ethanol, and stained using a mouse anti-HA antibody (Sigma-Aldrich, St. Louis, MO) followed by rabbit anti-mouse IgG antibody conjugated to Alexa Fluor 488 dye (Life Technologies, Carlsbad, CA). To measure DNA content, the cells were incubated for 30 min in Guava cell cycle reagent (EMD Millipore, Billerica, MA) at room temperature in the dark. Flow cytometry was performed using a Guava EasyCyte mini-flow cytometer system (EMD Millipore, Billerica, MA). The DNA content of positively and negatively HA-stained cells was determined by gating on each population using FlowJo analysis software (Tree Star, Ashland, OR).
Chromatin immunoprecipitation assay.
For chromatin immunoprecipitation (ChIP) experiments, 5 × 106 human fibroblasts were plated onto one 150-mm dish as described in “Biological reagents.” Cells were synchronized using DMEM containing 0.5% FBS for 48 h and were mock infected, infected at an MOI of 6 infectious units per cell using ADwt or ADdel29 virus, or treated with 10 μM nutlin-3 (Sigma-Aldrich, St. Louis, MO) in DMEM containing 7% FBS. Cells were fixed at 18 hpi in 1% ChIP-grade formaldehyde (Thermo Fisher Scientific, Waltham, MA) for 10 min and neutralized with 125 mM glycine for 5 min. Cells were washed three times with ice cold phosphate-buffered saline, scraped, collected, and lysed in 500 μl SDS lysis buffer (1% sodium dodecyl sulfate, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1]) containing Complete mini-EDTA-free protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN). Lysates were sonicated to an average size of 1 kbp using a microtip sonicator. Insoluble material was removed by centrifugation at 18,000 × g at 4°C for 10 min. A fraction of supernatant was kept for the input measurement and to test shearing efficiency.
The resulting supernatant was diluted with 5 volumes of ChIP buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 1.67 M Tris-HCl [pH 8.1], 176 mM NaCl) and incubated with 10 μg antibody overnight at 4°C with rotation. The antibodies used were mouse anti-p53 clone DO-1 (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-acetyl-histone 3 lys9 (Millipore, Billerica, MA), and normal mouse IgG (Santa Cruz Biotechnology, Santa Cruz, CA). Antibody complexes were precipitated with 60 μl Dynabeads protein G (Life Technologies, Carlsbad, CA) for 2 h with rotation at 4°C. The bound complexes were collected using a magnet and washed once with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 150 mM NaCl), once with high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 500 mM NaCl), once with LiCl wash (250 mM LiCl, 1% Igepal-CA630, 1% sodium deoxycholate, 1 mM EDTA), and twice with TE buffer (10 mM Tris-HCl [pH 8.1], 1 mM EDTA), then eluted twice in 250 μl elution buffer (40 mM NaHCO3, 1% SDS, 10 mM dithiothreitol) at room temperature for 15 min. The beads were removed, and the eluate was reverse cross-linked using a final concentration of 4% NaCl at 65°C for 12 h. Proteins within the eluate were digested in 10 mM EDTA and 40 mM Tris-HCl (pH 6.5) using 40 μg proteinase K (Roche Applied Science, Indianapolis, IN) at 45°C for 1 h. DNA was isolated using a gel-PCR DNA fragment extraction kit (IBI Scientific, Peosta, IA) per manufacturer protocols and eluted using 30 μl EB elution buffer provided in the kit. Cellular DNA levels were determined using quantitative PCR with sequence-specific primers. The PCR mixture was 5 μl 2× FastStart universal SYBR green master mix (Roche Applied Science, Indianapolis, IN), 3.6 μl H2O, 0.2 μl of each 100 μM primer stock, and 1 μl of sample. PCR primers detected the 3′ p53 response element (RE) in the p21CIP1 promoter (5′-CTCTCCTCCCCGAGGTCA-3′, 5′-ACATCTCACGCTGCTCACAGTCT-3′), the 5′ RE in the p21CIP1 promoter (5′-AGCAGGCTCTGGCTCTGATT-3′, 5′-CAAAATAGCCACCAGCCTCTTCT-3′), caspase 1 promoter (5′-GGCCTGTACATCTATTGG-3′, 5′-GATCTATCCAAGGGCTGGTG-3′), PUMA promoter (5′-GCGAGACTGTGGCCTTGTGT-3′, 5′-CGTTCCAGGGTCCACAAAGT-3′) (57), and exon 6 of GAPDH (5′-CCTCACAGTTGCCATGTA-3′, 5′-CATGGTACATGACAAGGTG-3′).
RESULTS
HCMV pUL29/28 interacts with cellular p53 during infection.
HCMV pUL29/28 interacts with pUL38 (45), cellular HDAC1 (histone deacetylase 1) and MTA2 (metastasis-associated protein 2) (46) during infection. These interactions were identified using pUL38 and HDAC1 as bait during immunoisolation of protein complexes and identification by mass spectrometry. To more accurately define pUL29/28 binding partners, we elected to use pUL29/28 as the bait protein. Using the ADin29FLAG virus (47), which expresses pUL29/28 in-frame with the FLAG epitope at the amino terminus, we infected primary fibroblasts at 3 infectious units (IU)/cell. At 24 hpi, protein complexes were isolated using an antibody against the FLAG epitope and further fractionated by their masses on an SDS-PAGE gel. The resulting proteins were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Proteins identified as binding nonspecifically to the reagents using lysates from AD169 wild-type (ADwt)-infected cells were removed from the results (15). In Table 1, we list putative binding proteins which were identified by at least 7 unique peptides from the combined results of three biological replicate experiments. The complete list of proteins defined by at least 3 peptides, minus the nonspecific interactions, is presented in Table S2 in the supplemental material.
Table 1.
Putative CMV pUL29/28- and pUL29-interacting proteins at 24 hpi
| Type | UniProt accession no. | HGNCa symbol | Name | Complexb | p53b | No. of peptides |
% Coverage | |
|---|---|---|---|---|---|---|---|---|
| Unique | Total | |||||||
| Viral | P16764 | UL29 | NuRD | 20 | 564 | 48.89 | ||
| P16727 | UL84 | x | 13 | 16 | 29.69 | |||
| P16761 | UL25 | 10 | 13 | 24.24 | ||||
| P16779 | UL38 | NuRD | 9 | 66 | 38.07 | |||
| P16847 | UL28 | NuRD | 9 | 60 | 36.94 | |||
| Cellular | O94776 | MTA2 | Metastasis associated 1 family, member 2 | NuRD | x | 32 | 162 | 53.89 |
| Q9BZK7 | TBL1XR1 | Transducin (beta)-like 1 X-linked receptor 1 | N-CoR | 25 | 271 | 60.89 | ||
| O75376 | NCOR1 | Nuclear receptor corepressor 1 | N-CoR | x | 22 | 56 | 10.66 | |
| Q86YP4 | GATAD2A | GATA zinc finger domain containing 2A | NuRD | 22 | 39 | 46.92 | ||
| P15924 | DSP | Desmoplakin | 19 | 22 | 10.10 | |||
| Q9BTC8 | MTA3 | Metastasis associated 1 family, member 3 | NuRD | 17 | 62 | 33.00 | ||
| O60907 | TBL1X | Transducin (beta)-like 1X-linked | N-CoR | 16 | 112 | 41.44 | ||
| Q14839 | CHD4 | Chromodomain helicase DNA binding protein 4 | NuRD | 16 | 38 | 9.88 | ||
| Q8WXI9 | GATAD2B | GATA zinc finger domain containing 2B | NuRD | x | 15 | 61 | 34.40 | |
| Q13330 | MTA1 | Metastasis associated 1 | NuRD | x | 15 | 54 | 25.59 | |
| Q92769 | HDAC2 | Histone deacetylase 2 | NuRD | x | 13 | 102 | 32.79 | |
| Q09028 | RBBP4 | Retinoblastoma binding protein 4 | NuRD | 12 | 113 | 36.71 | ||
| O15379 | HDAC3 | Histone deacetylase 3 | N-CoR | 12 | 39 | 31.54 | ||
| Q9Y618 | NCOR2 | Nuclear receptor corepressor 2 | N-CoR | 12 | 20 | 6.36 | ||
| O00505 | KPNA3 | Karyopherin alpha 3 (importin alpha 4) | x | 10 | 18 | 24.57 | ||
| Q16576 | RBBP7 | Retinoblastoma-binding protein 7 | NuRD | 9 | 90 | 21.88 | ||
| O95983 | MBD3 | Methyl-CpG binding domain protein 3 | NuRD | 9 | 20 | 28.87 | ||
| Q9BQ87 | TBL1Y | Transducin (beta)-like 1, Y-linked | N-CoR | 8 | 87 | 21.07 | ||
| Q9NP68 | TP53 | Tumor protein p53 | NuRD/NCoR | x | 8 | 11 | 23.92 | |
| P13667 | PDIA4 | Protein disulfide isomerase family A, member 4 | 7 | 16 | 15.35 | |||
| Q9UBB5 | MBD2 | Methyl-CpG binding domain protein 2 | NuRD | x | 7 | 15 | 18.25 | |
| P02786 | TFRC | Transferrin receptor (p90, CD71) | 7 | 12 | 10.92 | |||
| O00629 | KPNA4 | Karyopherin alpha 4 (importin alpha 3) | x | 7 | 10 | 17.27 | ||
| P14923 | JUP | Junction plakoglobin | 7 | 8 | 11.02 | |||
| P04181 | OAT | Ornithine aminotransferase | 7 | 7 | 21.87 | |||
HGNC, HUGO Gene Nomenclature Committee.
Published interactions.
The resulting data confirmed the previously observed interactions with the HDAC1- and MTA2-containing NuRD complex (46). However, unlike in previous studies, here we detected most of the proteins that define this chromatin remodeling complex (Table 1) (reviewed in reference 58). In addition, we observed protein components of the nuclear receptor corepressor (N-CoR) deacetylase complex (Table 1; also, see Table S2 in the supplemental material) (59). We also detected the tumor suppressor protein p53 and several p53-binding proteins, including importin α (KPNA3, KPNA4) (60) and proteins within both the NuRD (61) and N-CoR (62) complexes (Table 1). Finally, the viral proteins pUL25, pUL84, and pUL38 were observed. HCMV pUL84 has been previously demonstrated to associate with p53, the NuRD component RBBP4, and importin α (Table 1) (42, 63, 64). Our results suggest that pUL29/28 interacts with multiple viral and cellular proteins at 24 hpi, including the tumor suppressor protein p53.
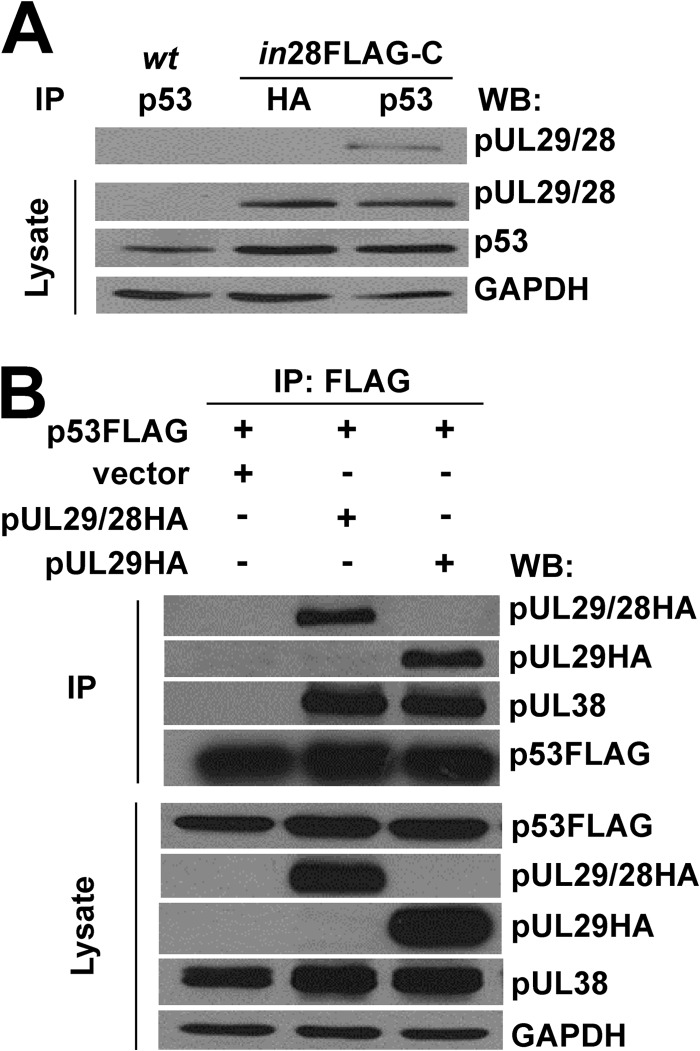
Because several of the identified proteins are known regulators of p53 (Table 1), and p53 is important for HCMV replication, we focused the remaining studies on determining the relationship between pUL29/28 and p53. First, to confirm the interaction, we infected fibroblasts with ADin28FLAG-C, which contains a FLAG epitope at the carboxyl terminus of pUL29/28 (47). At 24 hpi, we immunoprecipitated protein complexes using an antibody against p53 and completed a Western blot using an antibody against the FLAG epitope placed in pUL29/28. As seen in Fig. 1A, we detected a protein at the correct molecular weight of pUL29/28 using lysates from ADin28FLAG-C- but not ADwt-infected cells. We did not observe this protein using a control antibody against the HA (hemagglutinin) epitope (Fig. 1A). Based upon these results, we conclude that pUL29/28 does associate with p53 during infection.
Fig 1.
HCMV pUL29/28 and pUL38 interact with cellular p53. (A) Fibroblasts were infected at an MOI of 6 with ADwt (wt) or ADin28FLAG-C (in28FLAG-C). Immunoprecipitation was performed at 24 hpi from whole-cell lysates using an antibody against p53 or the HA epitope. Western blotting was performed using antibodies directed against the indicated proteins. (B) U2OS cells stably expressing pUL38 (U2OS-pUL38) were transfected with p53FLAG and either empty pCGN, pUL29/28HA, or pUL29HA. Immunoprecipitation was performed at 48 h posttransfection using whole-cell lysates and an antibody against the FLAG epitope. Western blot analysis was completed using antibodies against the indicated proteins.
To determine whether the interaction occurs in the absence of infection, we transfected cells with a p53FLAG expression plasmid and either an empty or a pUL29/28HA expression plasmid. We elected to use p53FLAG because the antibody heavy chain prevented detection of endogenous p53. Constructs were introduced into U-2 OS osteosarcoma cells which express pUL38 (U2OS-UL38 cells) (46, 47). We evaluated interactions by immunoprecipitating p53FLAG with an antibody against the FLAG epitope and Western blot analysis using an antibody against HA. We observed an interaction between p53 and pUL29/28 (Fig. 1B) as well as the smaller pUL29 protein (47). Furthermore, we detected pUL38 within these complexes (Fig. 1B). Interestingly, pUL38 failed to associate with p53 in the absence of expression of either UL29 isoforms (Fig. 1B). Our data indicate that pUL29/28 and pUL38 likely interact with p53 and suggest that no additional viral proteins are necessary for the event.
Expression of pUL29/28 and pUL38 alters expression of the p53-regulatable p21CIP1 promoter.
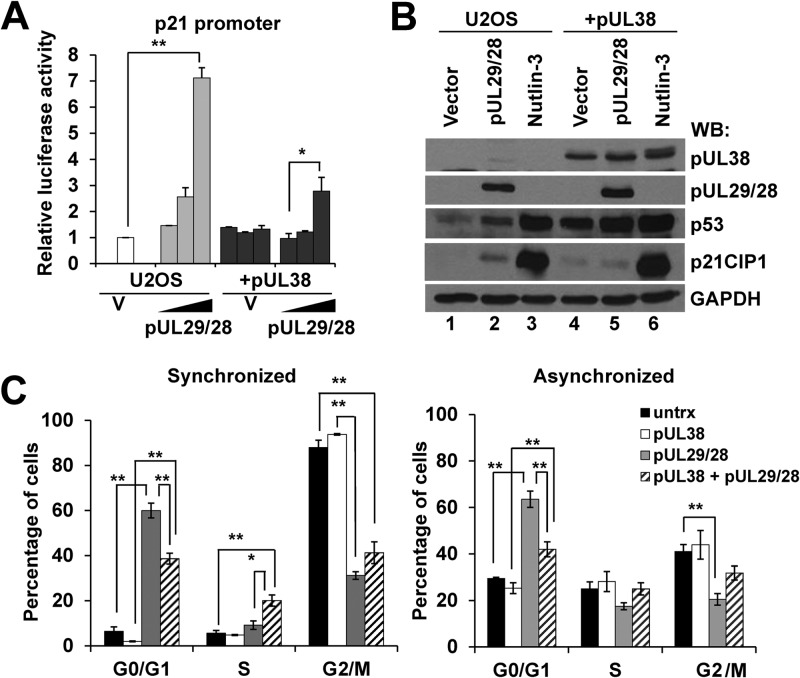
We next asked whether pUL29/28 could influence p53-regulated gene expression. We initiated these studies by determining changes in promoter activity using a luciferase reporter. Increasing amounts of pUL29/28HA expression plasmid were transfected into U2OS cells along with a reporter regulated by the p21CIP1 promoter (CDK-interacting protein 1) (53) (Fig. 2A). Luciferase activity was determined for each sample and presented relative to the empty plasmid control as previously described (46, 47). Following transfection, similar levels of luciferase DNA relative to cellular DNA were observed by qPCR (data not shown). At the largest amount of pUL29/28HA plasmid used, we observed an average 7.1-fold increase in luciferase activity (Fig. 2A). To determine the contribution of pUL38, we also completed the studies using U2OS-UL38 cells. We have previously demonstrated that pUL38 does not affect pUL29/28 protein expression levels (46). Furthermore, we did not observe differences in luciferase activity between U2OS and U2OS-UL38 cells receiving empty vector (Fig. 2A). Transfection of the pUL29/28 expression plasmid into U2OS-UL38 cells resulted in only a 2.8-fold increase in p21CIP1 promoter activity (Fig. 2A). These observations suggest that pUL29/28 does influence p21CIP1promoter activity but that pUL38 inhibits maximal pUL29/28-mediated activation of the promoter.
Fig 2.
Expression of HCMV pUL29/28 and pUL38 alters p53-regulatable gene expression in the absence of infection. (A) Control U2OS or U2OS-pUL38 (+pUL38) were transfected with a plasmid containing the p21CIP1 promoter driving luciferase along with 1.0, 10.0, and 100.0 ng of either empty pCGN or pUL29/28 plasmid DNA. Relative luciferase activity was measured at 48 h after transfection. The data are means for two replicate samples ± standard deviations. Statistical analysis was performed using Student's t test (**, P ≤ 0.01). (B) The U2OS and U2OS-pUL38 (+pUL38) cells were either transfected with empty vector or pUL29/28, or treated with nutlin-3 for 24 h. Cells were harvested 48 h posttransfection. Western blot analysis was performed using antibodies against the indicated proteins. (C) U2OS cells were transfected with pUL29/28HA expression vector. At 40 h posttransfection, the cells were treated with or without nocodazole to synchronize the population. Cells were stained using antibody against the HA epitope and propidium iodide, and DNA content was measured by flow cytometry. The data are displayed as percentages of cells in each phase of cell cycle and are the means from two biological experiments ± standard errors of the means. Statistical analysis was performed using Student's t test (*, P ≤ 0.05; **, P ≤ 0.01).
In addition to promoter activity, we evaluated changes in endogenous p21CIP1 levels by Western blotting in U2OS cells. We elected to evaluate protein expression in order to detect p53 levels in the experiments. Transfection of the pUL29/28 plasmid resulted in a consistent increase in both p53 and p21CIP1 expression levels compared to vector control (Fig. 2B, lanes 1 and 2). For a positive control, we treated U2OS cells with nutlin-3 which inhibits the MDM2 E3 ligase and stabilizes p53 (65). We observed robust increases in p53 and p21CIP1 protein levels following treatment with nutlin-3 (Fig. 2B, lanes 1 and 3). In the U2OS-UL38 cells alone, we observed elevated levels of p53 compared to U2OS cells (Fig. 2B, lane 1 and 4). Following expression of pUL29/28, we detected an additional increase in p53 which approached the levels observed in the nutlin-treated control (Fig. 2B, lanes 5 and 6). Surprisingly, however, the level of p21CIP1 remained low in spite of the increase in p53. These changes follow the pattern observed using the reporter plasmids (Fig. 2A). Furthermore, our data suggest that expression of pUL38 and pUL29/28 causes an increase in a population of p53 which is not capable of inducing p21CIP1 expression.
The cyclin-dependent kinase inhibitor p21CIP1 plays a downstream role in p53-dependent cell cycle arrest. To provide additional support for the idea that pUL29/28 and pUL38 influence p53 activities and p21CIP1 expression, we evaluated relative changes in the cell cycle. We transfected either U2OS or U2OS-UL38 cells with the pUL29/28 expression plasmid. Following synchronization in G2/M by nocodazole treatment, 88% of the untransfected control cells were in G2/M, as determined by flow cytometry (Fig. 2C). In contrast, only 31% of the pUL29/28-positive cells were found in G2/M, while 60% were in G0/G1 (Fig. 2C). In an unsynchronized population, we again observed 63% of pUL29/28-expressing U2OS cells were in G0/G1 (Fig. 2C). These data suggest that expression of pUL29/28 promotes cell cycle arrest in G0/G1, which is consistent with elevated levels of p21CIP1. In pUL38- and pUL29/28-expressing cells, however, we detected 39% of cells in G0/G1 (Fig. 2C). Expression of pUL38 alone had no influence on the cell cycle (Fig. 2C). Although cell cycle control is complex, these results correlate with the observed reduction in p21CIP1 upon expression of both pUL29/28 and pUL38 (Fig. 2B, lanes 2 and 5) and elevated levels of p53 (Fig. 2B, lane 5).
Our studies show that in the absence of HCMV infection and other viral proteins, expression of pUL29/28 can induce the p53-regulatable p21CIP1 promoter and p21CIP1 protein expression. However, the response is significantly dampened upon coexpression of pUL38 with pUL29/28. Furthermore, expression of both viral proteins together appears to increase the levels p53 without a subsequent increase in p21CIP1 expression.
HCMV pUL29/28 and pUL38 are necessary to inhibit expression of a subset of p53-regulatable genes during infection.
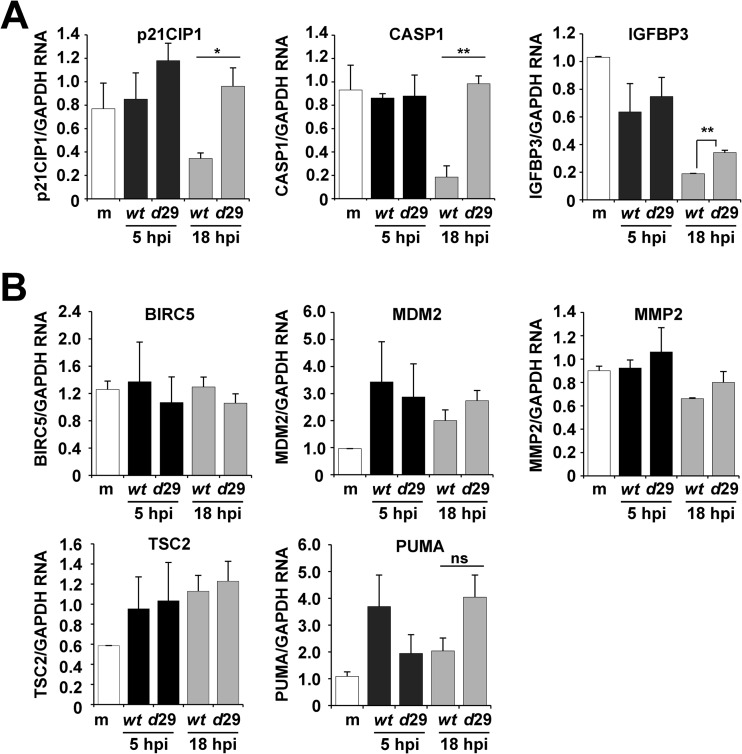
The cellular environment during HCMV infection is substantially more complex than that of uninfected cells involving the manipulation of diverse cellular processes. Therefore, we quantified changes in a subset of p53-regulatable genes following infection by either ADwt or a UL29-deficient virus, ADdel29. In addition to p21CIP1, we evaluated expression of CASP1 (caspase 1), BIRC5 (baculoviral IAP repeat containing 5), MDM2 (mdm2 p53 binding protein homolog), TSC2 (tuberous sclerosis 2), PUMA (p53 upregulated modulator of apoptosis), IGFBP3 (insulin-like growth factor binding protein 3), and MMP2 (matrix metallopeptidase 2) (37) using sequence-specific primers and quantitative RT-PCR (Fig. 3). The experiments were completed by infecting fibroblasts that had been synchronized by serum starvation with either ADwt or ADdel29 virus at a multiplicity of 6 IU/cell. We harvested total RNA at 5 and 18 hpi and determined expression changes using relative quantification with a standard curve for each primer pair and normalizing to GAPDH expression. Based upon the results of our studies in U2OS cells, we had predicted that a UL29-deficient viral infection would express reduced levels of p53-regulatable genes compared to an infection with wild-type virus. However, in contrast to our prediction, we observed increased expression at 18 hpi (Fig. 3A). Expression of p21CIP1 as well as CASP1 was on average 2.8- and 5.3-fold higher, respectively, during infection with ADdel29 than during infection with ADwt (Fig. 3A). Although statistically significant, the change in IGFBP3 was less than 2-fold (Fig. 3A). Also, surprisingly, we did not observe statistically significant differences in gene expression for the other genes (Fig. 3B) between the two infections. Our data demonstrate that infection in the absence of the UL29 ORF results in >2-fold-increased expression of only two of the eight p53-regulatable genes examined.
Fig 3.
Disruption of pUL29/28 during infection results in elevated expression of a subset of p53-regulatable genes. (A) Fibroblasts were either mock infected or infected with ADwt or ADdel29 at an MOI of 6 and harvested at the indicated times postinfection. Relative RNA was measured by qRT-PCR for the indicated genes and normalized to cellular GAPDH levels. The data are the means for two technical replicates ± standard deviations, with the results being validated in a separate independent biological replicate. Statistical analysis was performed using Student's t test (*, P ≤ 0.05; **, P ≤ 0.01). (B) Experiments were completed as described above, but no statistical difference in expression were observed.
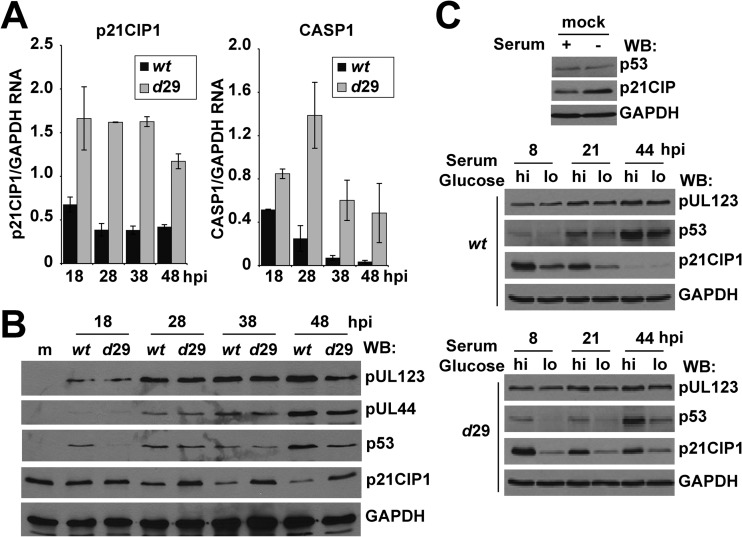
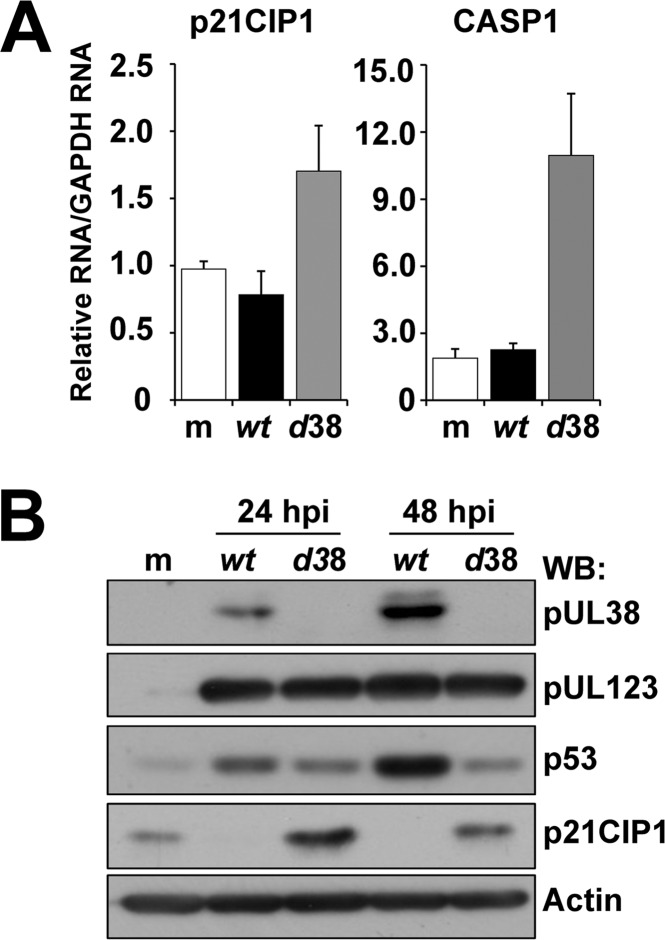
Disruption of the UL29 gene results in both altered viral gene expression and DNA replication using a low MOI (47). To determine whether the changes in p21CIP1 and CASP1 were possibly due to a lag in the replication cycle, we quantified expression over a time course using a high MOI. As shown in Fig. 4A, expression levels of these genes remained elevated in ADdel29-infected cells compared to ADwt as late as 48 hpi. At the protein level, p21CIP1 was higher during ADdel29 infection compared to wild-type starting at 28 hpi (Fig. 4B). Dependent on the specific time point, we also observed reduced levels of p53 in the absence of UL29 (Fig. 4B). We did detect similar levels of expression for the viral proteins pUL123 and pUL44 between viruses under high-MOI conditions (Fig. 4B). These data suggest that the changes are not the result of a lag in the replication cycle.
Fig 4.
pUL29/28 influences p21CIP1 and CASP1 expression during infection. (A) Fibroblasts were infected with ADwt or ADdel29 at an MOI of 6 and harvested at the indicated times postinfection. Relative RNA was measured using qRT-PCR. (B) Samples were collected at the indicated times postinfection and processed for Western blot analysis using antibodies against the indicated proteins. (C) For nutrient stress, fibroblasts were serum starved for 24 h and infected with ADwt or ADdel29 at an MOI of 6 in 7% FBS, 25 mM glucose (hi) for 1 h. Cells were then washed and replenished with either normal medium (hi) or medium containing 5.56 mM glucose and no serum (lo). Western blot analysis was performed using whole-cell lysates harvested at the indicated times postinfection as well as from mock-infected samples and antibodies against the indicated proteins.
Cellular p53 and p53-regulated gene expression are influenced by diverse forms of stress, including nutrient stress (66). Furthermore, pUL38 has been demonstrated to regulate the nutrient-sensitive mTOR signaling pathway (45). We next determined whether pUL29/28 could regulate p53 and p21CIP1 expression following perturbation using medium lacking serum and containing reduced levels of glucose. Fibroblasts were synchronized by removing serum for 24 h prior to infection, which resulted in elevated levels of p21CIP1 at the time of infection (Fig. 4C). Following infection for 1 h in 25 mM glucose and 7% serum (high-nutrient conditions), cultures were placed in either the same high-nutrient conditions or moved to stressed conditions defined by 5.6 mM glucose and no serum (low-nutrient conditions). Using Western blot analysis, we observed reduced levels of both p53 and p21CIP1 under low-nutrient conditions at 8 and 21 hpi for both ADwt and ADdel29 viral infections (Fig. 4C). However, expression of p21CIP1 decreased in ADwt- but not in ADdel29-infected cells at 44 hpi regardless of medium conditions (Fig. 4C). HCMV pUL123 protein levels appeared to be unaffected (Fig. 4C). Our data suggest that pUL29/28 negatively regulates p21CIP1 expression independently of perturbation using nutrient stress.
To determine the impact of the pUL29/28-binding protein pUL38 on gene expression, we quantified expression of p21CIP1 and CASP1 expression from infected cells at 18 hpi using either ADwt or a UL38-deficient virus, ADdlUL38 (50). Expression of both genes was elevated during infection with ADdlUL38 compared to ADwt (Fig. 5A). Similar to ADdel29, we detected increased p21CIP1 and decreased p53 protein expression using Western blot analysis of lysates from ADdlUL38-infected cells (Fig. 5B). The levels of expression for pUL123 were similar between viruses (Fig. 5B). Cumulatively, our data demonstrate that pUL38 and pUL29/28 inhibit the expression of the p53-regulatable genes p21CIP1 and CASP1. Furthermore, elevated levels of p21CIP1 in both the UL29- and UL38-deficient viral infections correlated with reduced levels of p53.
Fig 5.
HCMV pUL38 contributes to the regulation of p53 during infection. (A) Fibroblasts were either mock infected or infected with ADwt or ADdel38 at an MOI of 5. At 18 hpi, RNA was collected, and qRT-PCR was used to measure relative RNA levels normalized to GAPDH for the indicated genes. (B) Fibroblasts were either mock infected or infected with ADwt or ADdel38 and evaluated by Western blotting at 24 and 48 hpi using the indicated antibodies.
pUL29/28 expression alters p53 binding and histone acetylation at the p21CIP1 and CASP1 promoters.
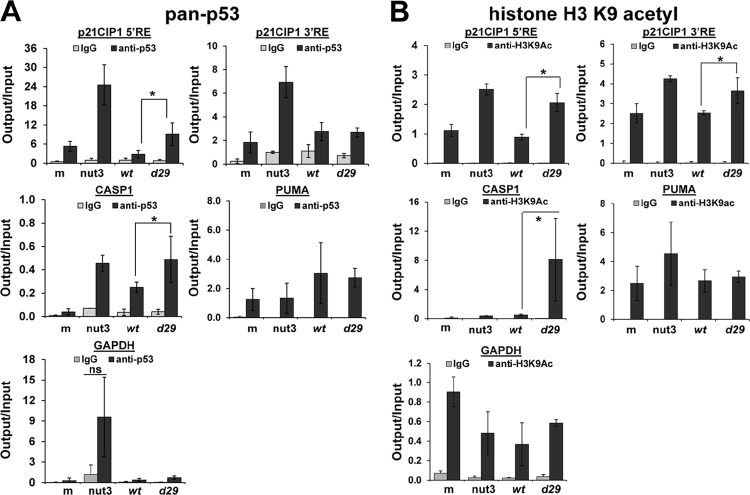
The regulation of p53 in cells involves diverse mechanisms. To determine how pUL29/28 might influence p53, we explored several possibilities, which included changes in acetylation of p53, relocalization within the infected cells (17, 67), and modifications at p53-regulatable promoters (reviewed in reference 68). We did not detect changes either in p53 acetylation at lysine 382 between 6 and 24 hpi or in p53 localization between ADwt and ADdel29 infections (data not shown). As previously seen for the adenovirus E4-ORF3 protein (69), an alternative possibility for p53 regulation involves changes occurring at selected p53-responsive promoters. Using chromatin immunoprecipitation (ChIP), we determined the levels of p21CIP1 and CASP1 promoter sequences associating with p53 and acetylated histone H3 during infection. Two p53 response elements (RE) have been identified within the p21CIP1 promoter (Fig. 6A) (53) and one in the CASP1 promoter (70). The 5′ RE in the p21CIP1 promoter binds to p53 relatively strongly compared to the 3′ RE. To evaluate p53 binding, fibroblasts were serum starved and then infected using either ADwt or ADdel29 virus at 6 IU/cell. We harvested the cells at 18 hpi, completed ChIP using an antibody against p53 or control IgG, and determined changes in the associating DNA by quantitative PCR. As a positive control, we treated uninfected cells with nutlin-3 and observed increased DNA associated with p53 (Fig. 6A). Following infection by ADdel29, we detected an average 3-fold increase in DNA binding of p53 to the p21CIP1 5′ RE compared to ADwt (Fig. 6A). However, no changes were detected at the 3′ RE (Fig. 6A). For the CASP1 promoter, we observed an average 2-fold increase between viruses (Fig. 6A). Although modest, these changes were detected in four replicate experiments. In contrast, we did not observe differences in binding to PUMA promoter sequence or an exonic region of GAPDH (Fig. 6A). For GAPDH, we did see an average increase in binding upon nutlin-3 treatment (Fig. 6A). However, these changes were highly variable and were not statistically significant. Our data suggest that p53 binding to the 5′ RE of p21CIP1 promoter and the CASP1 promoter is negatively influenced by the expression of pUL29/28.
Fig 6.
pUL29/28 influences p53 and histone H3 K9 acetyl binding to responsive promoters. (A) Fibroblasts were mock infected, treated with 10 μM nutlin-3 for 18 h, or infected with ADwt or ADdel29 at an MOI of 6 for 18 h. Chromatin immunoprecipitation (ChIP) was performed using an antibody against pan-p53 or control IgG. Quantitative PCR was used to determine the relative output DNA levels normalized to input. The data are means from four replicate experiments ± standard deviations and are presented as percentage of input. The PUMA data set represents two experiments. (B) ChIP analysis was performed as described above but using an antibody against histone 3 lysine 9 acetylation. The data are means for four replicates ± standard deviation and are presented as percentages of input. Statistical analysis was performed using Student's t test (*, P ≤ 0.05).
Acetylation of histone H3 on lysine 9 (H3K9Ac) is associated with actively transcribed genes. We next quantified changes of H3K9Ac at the p53-regulatable promoters during infection. Serum-starved fibroblasts were infected with either ADwt or ADdel29 virus at 5 IU/cell, and we completed ChIP at 18 hpi using an antibody against H3K9Ac. For both p21CIP1 and CASP1 promoters, we detected increased DNA associated with H3K9Ac following infection with the UL29-deficient virus, compared to ADwt (Fig. 6B). Specifically, we observed an average 2.3-fold increase for the p21CIP1 5′ RE, 1.4-fold for the 3′ RE, and 16-fold for the CASP1 promoter sequence (Fig. 6B). We did not observe changes when using primers against PUMA and GAPDH (Fig. 6B). Although correlative, our data demonstrate increased p53 and H3K9Ac associating with p21CIP1 and CASP1 promoter sequences during HCMV infection in the absence of the UL29 ORF. These data are consistent with the observations that p21CIP1 and CASP1 RNA expression are elevated during an ADdel29 viral infection compared to infection with the wild type.
DISCUSSION
Immediately upon HCMV infection, the steady-state amount of cellular p53 protein, but not RNA, increases (19, 20, 22). Surprisingly, however, the majority of p53-regulatable cellular genes are not induced (19) (see Table S1 in the supplemental material). In this study, we identified an interaction between p53 and the HCMV protein pUL29/28 using a proteomics-based approach (Table 1 and Fig. 1). With this discovery, we have demonstrated that HCMV pUL29/28 and pUL38 contribute to negatively regulating p53-dependent gene expression during infection for two of eight genes examined more than 2-fold (Fig. 3). Recombinant viruses containing disruptions in either UL29 or UL38 resulted in elevated RNA levels for p21CIP1 (CDK-interacting protein 1) and CASP1 (caspase 1) (Fig. 3A and 4A). Although our studies do not comprehensively evaluate all p53-regulated genes, we observed that additional genes were not significantly influenced by pUL29/28 and pUL38 (Fig. 3B), suggesting that the regulation is limited.
In the absence of infection, pUL29/28 alone induced p21CIP1 expression in U2OS cells (Fig. 2A). However, we did not observe any evidence of pUL29/28-mediated induction during infection (Fig. 3A). When pUL29/28 was expressed with pUL38 again in the absence of infection, maximal induction did not occur, suggesting that pUL38 may disrupt pUL29/28-p53 activities (Fig. 2A). Under these conditions, both pUL29/28 and pUL38 likely interact with p53 (Fig. 1B). From our studies using U2OS cells, we can conclude that pUL29/28 and pUL38 influence p53. However, because of the artificial nature of the experiments and the absence of other viral proteins, interpretation of the results as they pertain to HCMV infection is not clear-cut. It is conceivable that pUL29/28 and pUL38 function cooperatively as repressors of p53. In support of this possibility, the interaction of p53 with pUL38 required expression of pUL29/28 (Fig. 1B). We have previously demonstrated that the interaction between the pUL38 and NuRD complex also requires pUL29/28 (46). Further support for a negative regulatory role comes from the observation that infection in the absence of UL29 resulted in increased association of p53 and acetylated histone H3 K9 with the p21CIP1 and CASP1 promoters (Fig. 6A and B). These results support the overall conclusion that pUL29/28 in the context of pUL38 expression functions to inhibit two p53-regulatable promoters during infection.
Expression of p21CIP1 was previously demonstrated to be tightly regulated during infection (71). p21CIP1 contributes to HCMV IE gene expression during infection of cells in G2/M (24). Furthermore, HCMV pUL27 promotes p21CIP1 expression by disrupting the Tip60 acetyltransferase (15). Our data demonstrate that pUL29/28 and pUL38 also participate in regulating p21CIP1. Initially, p21CIP1 protein levels are maintained within infected cells up to 9 hpi (42, 71) and then begin to decrease. Regulation occurs at the levels of both transcriptional control and protein degradation (71). Here, we demonstrate that steady-state p21CIP1 protein expression is maintained in the absence of pUL29/28 (Fig. 4B and C) or pUL38 (Fig. 5B). Since these viral proteins influence expression at the RNA level, our studies suggest that transcriptional regulation plays a critical role in controlling p21CIP1 during HCMV infection.
We have demonstrated that a subset of p53-regulatable genes is influenced by pUL29/28 and pUL38. This indicates a role for other HCMV proteins in regulating p53. Studies completed by several laboratories have demonstrated the involvement of the immediate early proteins IE1 (pUL123) and IE2 (pUL122) in regulating p53 activities (22, 35, 39–42, 44). Furthermore, pUL84 and pUL44 have also been implicated (42, 43). One challenge that we faced in these studies was distinguishing between the role of pUL29/28 in influencing HCMV IE expression (46, 47) and direct regulation of p53. In support of a direct role, we detected similar levels of IE1 protein expression in ADwt and both UL29- and UL38-deficient viral infections using a high multiplicity of infection (Fig. 4B and 5B). We also demonstrated changes in in vitro promoter studies in the absence of other HCMV proteins (Fig. 2A), and these changes were consistent with the responses observed during infection (Fig. 3A). Furthermore, we showed that pUL29/28 interacts with p53 (Fig. 1A). However, we were unable to show direct association of pUL29/28 with the responsive promoters due to high background in ChIP experiments using the antibody against the FLAG epitope (data not shown). Furthermore, the pUL29/28 binding partner pUL38 regulates an endoplasmic reticulum stress response (72, 73) and mTOR activity (45), which may influence p53. Based upon our data and the complexity of infection as well as p53 regulation, it is likely that our observations are the result of both direct and indirect involvement with p53.
In addition to changes in gene expression, we observed increased steady-state levels of p53 protein upon expression of pUL29/28 and pUL38 following transfection (Fig. 2B). During infection in the absence these protein, we detected reduced levels of p53 protein (Fig. 4B and 5B). These changes coincided with opposing changes in cellular gene expression in vitro (Fig. 2) and during infection (Fig. 4 and 5). Therefore, we hypothesize that pUL29/28 and pUL38 stabilize a population of p53 which is no longer capable of binding to select promoters. A similar phenotype has been observed for adenovirus protein E4-ORF3 (69). Expression of E4-ORF3 results in stabilization of p53, whose activity is dominantly suppressed. E4-ORF3 expression prevents p53 DNA binding by inducing heterochromatin formation at select p53-regulatable promoters, including p21CIP1.
Numerous stress responses involving p53 are activated during infection and have the potential to disrupt HCMV replication by inducing response such as apoptosis, cell cycle arrest, or cellular senescence. However, p53 is required for efficient HCMV gene expression and viral replication (16). Therefore, regulating p53-transcriptional responses during infection is critical. Our studies indicate that pUL29/28 and pUL38 participate in repression of a limited number of p53-regulated cellular genes during HCMV infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Katherine Stabelfeldt for her excellent technical assistance and Andrew Vallejos for reevaluating microarray data. We thank Lee Fortunato and Eain Murphy for their input on these studies. We also thank Tom Shenk for the generous gift of CMV-specific antibodies and for giving us permission to include a list of p53-regulatable genes extracted from previously published microarray data.
This work was supported by MCW grant 9305200 and NIH/NIAID grant 5R01AI083281, awarded to S.S.T., and by NIH/NCI grant 5R01CA120768, awarded to D.Y.
Footnotes
Published ahead of print 12 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01926-12.
REFERENCES
- 1. Mocarski E, Shenk T, Pass RF. 2007. Cytomegaloviruses, p 2702–2772 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2. Cannon MJ. 2009. Congenital cytomegalovirus (CMV) epidemiology and awareness. J. Clin. Virol. 46(Suppl 4):S6–S10 [DOI] [PubMed] [Google Scholar]
- 3. Cantrell SR, Bresnahan WA. 2005. Interaction between the human cytomegalovirus UL82 gene product (pp71) and hDaxx regulates immediate-early gene expression and viral replication. J. Virol. 79:7792–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hofmann H, Sindre H, Stamminger T. 2002. Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J. Virol. 76:5769–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ishov AM, Vladimirova OV, Maul GG. 2002. Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71 at ND10 facilitates initiation of viral infection at these nuclear domains. J. Virol. 76:7705–7712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saffert RT, Kalejta RF. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 80:3863–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ahn JH, Brignole EJ, III, Hayward GS. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol. 18:4899–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ahn JH, Hayward GS. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J. Virol. 71:4599–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Korioth F, Maul GG, Plachter B, Stamminger T, Frey J. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 229:155–158 [DOI] [PubMed] [Google Scholar]
- 10. Muller S, Dejean A. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wilkinson GW, Kelly C, Sinclair JH, Rickards C. 1998. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J. Gen. Virol. 79(Pt 5):1233–1245 [DOI] [PubMed] [Google Scholar]
- 12. Cristea IM, Moorman NJ, Terhune SS, Cuevas CD, O'Keefe ES, Rout MP, Chait BT, Shenk T. 2010. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J. Virol. 84:7803–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gariano GR, Dell'oste V, Bronzini M, Gatti D, Luganini A, De Andrea M, Gribaudo G, Gariglio M, Landolfo S. 2012. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog. 8:e1002498 doi:10.1371/journal.ppat.1002498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li R, Zhu J, Xie Z, Liao G, Liu J, Chen MR, Hu S, Woodard C, Lin J, Taverna SD, Desai P, Ambinder RF, Hayward GS, Qian J, Zhu H, Hayward SD. 2011. Conserved herpesvirus kinases target the DNA damage response pathway and TIP60 histone acetyltransferase to promote virus replication. Cell Host Microbe 10:390–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reitsma JM, Savaryn JP, Faust K, Sato H, Halligan BD, Terhune SS. 2011. Antiviral inhibition targeting the HCMV kinase pUL97 requires pUL27-dependent degradation of Tip60 acetyltransferase and cell-cycle arrest. Cell Host Microbe 9:103–114 [DOI] [PubMed] [Google Scholar]
- 16. Casavant NC, Luo MH, Rosenke K, Winegardner T, Zurawska A, Fortunato EA. 2006. Potential role for p53 in the permissive life cycle of human cytomegalovirus. J. Virol. 80:8390–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fortunato EA, Spector DH. 1998. p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus. J. Virol. 72:2033–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hannemann H, Rosenke K, O'Dowd JM, Fortunato EA. 2009. The presence of p53 influences the expression of multiple human cytomegalovirus genes at early times postinfection. J. Virol. 83:4316–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jault FM, Jault JM, Ruchti F, Fortunato EA, Clark C, Corbeil J, Richman DD, Spector DH. 1995. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J. Virol. 69:6697–6704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muganda P, Mendoza O, Hernandez J, Qian Q. 1994. Human cytomegalovirus elevates levels of the cellular protein p53 in infected fibroblasts. J. Virol. 68:8028–8034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rosenke K, Samuel MA, McDowell ET, Toerne MA, Fortunato EA. 2006. An intact sequence-specific DNA-binding domain is required for human cytomegalovirus-mediated sequestration of p53 and may promote in vivo binding to the viral genome during infection. Virology 348:19–34 [DOI] [PubMed] [Google Scholar]
- 22. Speir E, Modali R, Huang ES, Leon MB, Shawl F, Finkel T, Epstein SE. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265:391–394 [DOI] [PubMed] [Google Scholar]
- 23. Wing BA, Johnson RA, Huang ES. 1998. Identification of positive and negative regulatory regions involved in regulating expression of the human cytomegalovirus UL94 late promoter: role of IE2-86 and cellular p53 in mediating negative regulatory function. J. Virol. 72:1814–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zydek M, Hagemeier C, Wiebusch L. 2010. Cyclin-dependent kinase activity controls the onset of the HCMV lytic cycle. PLoS Pathog. 6:e1001096 doi:10.1371/journal.ppat.1001096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. 2011. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe 9:363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11:997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, Confalonieri S, Cesaroni M, Marchesi F, Gasco M, Scanziani E, Capra M, Mai S, Nuciforo P, Crook T, Lough J, Amati B. 2007. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature 448:1063–1067 [DOI] [PubMed] [Google Scholar]
- 28. Sun Y, Jiang X, Chen S, Fernandes N, Price BD. 2005. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. U. S. A. 102:13182–13187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gostissa M, Morelli M, Mantovani F, Guida E, Piazza S, Collavin L, Brancolini C, Schneider C, Del Sal G. 2004. The transcriptional repressor hDaxx potentiates p53-dependent apoptosis. J. Biol. Chem. 279:48013–48023 [DOI] [PubMed] [Google Scholar]
- 30. Johnstone RW, Wei W, Greenway A, Trapani JA. 2000. Functional interaction between p53 and the interferon-inducible nucleoprotein IFI 16. Oncogene 19:6033–6042 [DOI] [PubMed] [Google Scholar]
- 31. Legube G, Linares LK, Tyteca S, Caron C, Scheffner M, Chevillard-Briet M, Trouche D. 2004. Role of the histone acetyl transferase Tip60 in the p53 pathway. J. Biol. Chem. 279:44825–44833 [DOI] [PubMed] [Google Scholar]
- 32. Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, Pelicci PG. 2000. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406:207–210 [DOI] [PubMed] [Google Scholar]
- 33. Chen Z, Knutson E, Wang S, Martinez LA, Albrecht T. 2007. Stabilization of p53 in human cytomegalovirus-initiated cells is associated with sequestration of HDM2 and decreased p53 ubiquitination. J. Biol. Chem. 282:29284–29295 [DOI] [PubMed] [Google Scholar]
- 34. Zhang Z, Evers DL, McCarville JF, Dantonel JC, Huong SM, Huang ES. 2006. Evidence that the human cytomegalovirus IE2-86 protein binds mdm2 and facilitates mdm2 degradation. J. Virol. 80:3833–3843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Castillo JP, Frame FM, Rogoff HA, Pickering MT, Yurochko AD, Kowalik TF. 2005. Human cytomegalovirus IE1-72 activates ataxia telangiectasia mutated kinase and a p53/p21-mediated growth arrest response. J. Virol. 79:11467–11475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Luo MH, Rosenke K, Czornak K, Fortunato EA. 2007. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J. Virol. 81:1934–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Riley T, Sontag E, Chen P, Levine A. 2008. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9:402–412 [DOI] [PubMed] [Google Scholar]
- 38. Browne EP, Wing B, Coleman D, Shenk T. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bonin LR, McDougall JK. 1997. Human cytomegalovirus IE2 86-kilodalton protein binds p53 but does not abrogate G1 checkpoint function. J. Virol. 71:5861–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Castillo JP, Yurochko AD, Kowalik TF. 2000. Role of human cytomegalovirus immediate-early proteins in cell growth control. J. Virol. 74:8028–8037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hsu CH, Chang MD, Tai KY, Yang YT, Wang PS, Chen CJ, Wang YH, Lee SC, Wu CW, Juan LJ. 2004. HCMV IE2-mediated inhibition of HAT activity downregulates p53 function. EMBO J. 23:2269–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hwang ES, Zhang Z, Cai H, Huang DY, Huong SM, Cha CY, Huang ES. 2009. Human cytomegalovirus IE1-72 protein interacts with p53 and inhibits p53-dependent transactivation by a mechanism different from that of IE2-86 protein. J. Virol. 83:12388–12398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kwon Y, Kim MN, Young Choi E, Heon Kim J, Hwang ES, Cha CY. 2012. Inhibition of p53 transcriptional activity by human cytomegalovirus UL44. Microbiol. Immunol. 56:324–331 [DOI] [PubMed] [Google Scholar]
- 44. Tsai HL, Kou GH, Chen SC, Wu CW, Lin YS. 1996. Human cytomegalovirus immediate-early protein IE2 tethers a transcriptional repression domain to p53. J. Biol. Chem. 271:3534–3540 [PubMed] [Google Scholar]
- 45. Moorman NJ, Cristea IM, Terhune SS, Rout MP, Chait BT, Shenk T. 2008. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3:253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Terhune SS, Moorman NJ, Cristea IM, Savaryn JP, Cuevas-Bennett C, Rout MP, Chait BT, Shenk T. 2010. Human cytomegalovirus UL29/28 protein interacts with components of the NuRD complex which promote accumulation of immediate-early RNA. PLoS Pathog. 6:e1000965 doi:10.1371/journal.ppat.1000965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mitchell DP, Savaryn JP, Moorman NJ, Shenk T, Terhune SS. 2009. Human cytomegalovirus UL28 and UL29 open reading frames encode a spliced mRNA and stimulate accumulation of immediate-early RNAs. J. Virol. 83:10187–10197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223–14228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U. S. A. 100:12396–12401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Terhune S, Torigoi E, Moorman N, Silva M, Qian Z, Shenk T, Yu D. 2007. Human cytomegalovirus UL38 protein blocks apoptosis. J. Virol. 81:3109–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ. 2008. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: Implications for latency. Proc. Natl. Acad. Sci. U. S. A. 105:5453–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817–825 [DOI] [PubMed] [Google Scholar]
- 54. Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. 2001. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol. Cell 7:673–682 [DOI] [PubMed] [Google Scholar]
- 55. Miyagishi M, Fujii R, Hatta M, Yoshida E, Araya N, Nagafuchi A, Ishihara S, Nakajima T, Fukamizu A. 2000. Regulation of Lef-mediated transcription and p53-dependent pathway by associating beta-catenin with CBP/p300. J. Biol. Chem. 275:35170–35175 [DOI] [PubMed] [Google Scholar]
- 56. Halligan BD, Greene AS. 2011. Visualize: a free and open source multifunction tool for proteomics data analysis. Proteomics 11:1058–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schlereth K, Beinoraviciute-Kellner R, Zeitlinger MK, Bretz AC, Sauer M, Charles JP, Vogiatzi F, Leich E, Samans B, Eilers M, Kisker C, Rosenwald A, Stiewe T. 2010. DNA binding cooperativity of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 38:356–368 [DOI] [PubMed] [Google Scholar]
- 58. Lai AY, Wade PA. 2011. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat. Rev. Cancer. 11:588–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang J, Kalkum M, Chait BT, Roeder RG. 2002. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9:611–623 [DOI] [PubMed] [Google Scholar]
- 60. Marchenko ND, Hanel W, Li D, Becker K, Reich N, Moll UM. 2010. Stress-mediated nuclear stabilization of p53 is regulated by ubiquitination and importin-alpha3 binding. Cell Death Differ. 17:255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Luo J, Su F, Chen D, Shiloh A, Gu W. 2000. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408:377–381 [DOI] [PubMed] [Google Scholar]
- 62. Dohi Y, Ikura T, Hoshikawa Y, Katoh Y, Ota K, Nakanome A, Muto A, Omura S, Ohta T, Ito A, Yoshida M, Noda T, Igarashi K. 2008. Bach1 inhibits oxidative stress-induced cellular senescence by impeding p53 function on chromatin. Nat. Struct. Mol. Biol. 15:1246–1254 [DOI] [PubMed] [Google Scholar]
- 63. Gao Y, Colletti K, Pari GS. 2008. Identification of human cytomegalovirus UL84 virus- and cell-encoded binding partners by using proteomics analysis. J. Virol. 82:96–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lischka P, Sorg G, Kann M, Winkler M, Stamminger T. 2003. A nonconventional nuclear localization signal within the UL84 protein of human cytomegalovirus mediates nuclear import via the importin alpha/beta pathway. J. Virol. 77:3734–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303:844–848 [DOI] [PubMed] [Google Scholar]
- 66. Feng Z, Zhang H, Levine AJ, Jin S. 2005. The coordinate regulation of the p53 and mTOR pathways in cells. Proc. Natl. Acad. Sci. U. S. A. 102:8204–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Utama B, Shen YH, Mitchell BM, Makagiansar IT, Gan Y, Muthuswamy R, Duraisamy S, Martin D, Wang X, Zhang MX, Wang J, Vercellotti GM, Gu W, Wang XL. 2006. Mechanisms for human cytomegalovirus-induced cytoplasmic p53 sequestration in endothelial cells. J. Cell Sci. 119:2457–2467 [DOI] [PubMed] [Google Scholar]
- 68. Brooks CL, Gu W. 2011. The impact of acetylation and deacetylation on the p53 pathway. Protein Cell 2:456–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Soria C, Estermann FE, Espantman KC, O'Shea CC. 2010. Heterochromatin silencing of p53 target genes by a small viral protein. Nature 466:1076–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gupta S, Radha V, Furukawa Y, Swarup G. 2001. Direct transcriptional activation of human caspase-1 by tumor suppressor p53. J. Biol. Chem. 276:10585–10588 [DOI] [PubMed] [Google Scholar]
- 71. Chen Z, Knutson E, Kurosky A, Albrecht T. 2001. Degradation of p21cip1 in cells productively infected with human cytomegalovirus. J. Virol. 75:3613–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Qian Z, Xuan B, Gualberto N, Yu D. 2011. The human cytomegalovirus protein pUL38 suppresses endoplasmic reticulum stress-mediated cell death independently of its ability to induce mTORC1 activation. J. Virol. 85:9103–9113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Xuan B, Qian Z, Torigoi E, Yu D. 2009. Human cytomegalovirus protein pUL38 induces ATF4 expression, inhibits persistent JNK phosphorylation, and suppresses endoplasmic reticulum stress-induced cell death. J. Virol. 83:3463–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.