Abstract
Hepatitis C virus (HCV) is considered to have a causative role in B-cell lymphoproliferative diseases, including B-cell lymphomas, in chronic virus carriers. Previous data from in vitro HCV-infected B-cell lines and peripheral blood mononuclear cells from HCV-positive individuals suggested that HCV might have a direct mutagenic effect on B cells, inducing mutations in the tumor suppressor gene TP53 and the proto-oncogenes BCL6 and CTNNB1 (β-catenin). To clarify whether HCV indeed has a mutagenic effect on B cells in vivo, we analyzed naive and memory B cells from the peripheral blood of four chronic HCV carriers and intrahepatic B cells from the livers of two HCV-positive patients for mutations in the three reported target genes. However, no mutations were found in the TP53 and CTNNB1 genes. For BCL6, which is a physiological target of the somatic hypermutation process in germinal-center B cells, the mutation levels identified were not higher than those reported in the respective B-cell subsets in healthy individuals. Hence, we conclude that in chronic HCV carriers, the virus does not generally induce mutations in the cancer-related genes TP53, CTNNB1, and BCL6 in B cells. Based on these findings, new targets have to be investigated as potential mediators of HCV-associated B-cell lymphomagenesis.
INTRODUCTION
Epidemiological, clinical, and experimental data strongly support the idea of a causative role of hepatitis C virus (HCV) in the development of B-cell lymphoproliferative diseases in chronic virus carriers. These diseases include type II mixed cryoglobulinemia (MC), a benign B-cell proliferative disorder with monoclonal B-cell expansions, and a subgroup of B-cell non-Hodgkin lymphomas (B-NHL) (1–6). The mechanisms by which chronic HCV infection affects B-cell differentiation and predisposes patients to the occurrence of premalignant and malignant B-cell lymphoproliferations are still poorly understood. One potential mechanism is the antigen-driven proliferation of HCV-specific B cells. This is supported by the finding of restricted immunoglobulin (Ig) V gene usage in B-cell clones in MC and HCV-associated B-cell lymphomas, and the direct demonstration of B-cell receptor specificity of some HCV-associated lymphomas for HCV antigens (7–10). It is also noteworthy that in livers of chronic HCV carriers, B-cell follicles resembling in several aspects ectopic germinal centers (GC) are found, indicating chronic HCV-driven B-cell proliferation in this organ (11).
Another—not mutually exclusive—hypothesis suggests that there is a direct mutagenic effect of HCV on B cells, which might promote B-cell transformation. This idea is mainly based on in vitro studies, in which human B-cell lines were exposed to HCV virions. It was reported that in this setting, HCV activates the enzyme activation-induced cytidine deaminase (AID), which is the key factor for somatic hypermutation of Ig V genes (12). Somatic hypermutation physiologically also targets the BCL6 proto-oncogene at a low frequency in the GC reaction, so that about 30% of post-GC memory B cells carry one or more mutations in the major mutation cluster in intron 1 of BCL6 (13). In the in vitro studies, enhanced BCL6 mutations were observed in the B-cell lines (12). Besides, HCV can also induce oxidative stress, with accumulation of nitrogen and oxygen reactive species which lead to DNA damage (14–16). TP53 and β catenin (CTNNB1) were identified as target genes of the reactive oxygen species with remarkably high mutation accumulation in the in vitro studies (mutation frequencies of 5 to 11 × 10−4/bp) (12, 14). Notably, mutations in TP53, CTNNB1, and BCL6 were also identified in peripheral blood mononuclear cells (PBMC) of HCV-positive patients (12), although only about 10% of PBMC are B cells. As TP53 is a tumor suppressor gene and BCL6 and CTNNB1 are proto-oncogenes, mutations in these genes might be of pathogenetic relevance.
Regarding the reported mutagenic effect of HCV, it should be mentioned that it is still controversially discussed whether HCV indeed infects B cells and induces mutations by a hit-and-run mechanism, or whether the effects seen are due to the binding of HCV to costimulatory receptors expressed on the surface of B cells, such as CD81 (17–24).
In the present work, we aimed to clarify whether the mutagenic effect of HCV on TP53, CTNNB1, and BCL6 is detectable in B cells of chronically HCV-infected patients, and hence might represent a main factor for B-cell lymphoproliferative diseases in such patients.
MATERIALS AND METHODS
Blood and tissue samples.
Fifty ml of peripheral blood (PB) was collected from each of four HCV-positive patients (HCV1 to HCV4). These were three males and one female, with a mean age of 60 years, attending the Department of Gastroenterology and Hepatology at the University Hospital Essen. All patients were infected with genotype 1 (mean viral load, 3,278,153 ± 2,780,622 IU/ml); two patients (HCV1 and HCV2) had evidence of extrahepatic diseases (cryoglobulinemia). Liver specimens were obtained from two HCV-positive patients (2 males) attending the Liver Transplantation Unit of the University Hospital Essen for liver transplantation. They were both infected by HCV of genotype 3 with a stage 3 liver disease. The studies were approved by the local ethics committee of the University Hospital Essen.
Cell separation.
PBMC were isolated by Ficoll-Paque density centrifugation, and CD19+ B cells were enriched by magnetic cell separation using the MACS system (Miltenyi Biotec, Bergisch-Gladbach, Germany).
Cell sorting.
The B-cell-enriched cell suspensions were stained using the following antibodies: anti-IgD-phycoerythrin (PE)-Cy7 (Becton, Dickinson [BD], Heidelberg, Germany), anti-IgM-PE (BD), anti-CD27-allophycocyanin (APC) (BD), and anti-IgG- and anti-IgA-fluorescein isothiocyanate (FITC) (Dako). B-cell subsets were sorted with a FACSDiva cell sorter (BD) as naive B cells (IgM positive [IgM+] IgD+ CD27−), class-switched memory B cells (IgG/IgA+ CD27+), and non-class-switched memory B cells (IgM+ IgD+ CD27+). Each B-cell subpopulation was sorted in 200 to 500 cell aliquots into PCR tubes. PB CD3+ T cells were sorted from healthy donors and used as a control to determine the background mutations introduced by the DNA polymerases in the PCR assays.
Immunohistochemistry and laser capture microdissection.
Detection of CD20+ follicle-like structures was performed on fresh-frozen liver samples from HCV-positive patients who underwent liver transplantation. Frozen liver specimens were cut as a series of 7-μm-thick sections and mounted onto FrameSlides PET (Zeiss, Munich, Germany), air-dried, and fixed in acetone for 10 min at −20°C. Endogenous alkaline phosphatase was inhibited with 0.1 N HCl for 10 min. Sections were stained with mouse monoclonal antibodies against human CD20 (Dako, Eching, Germany) for 1 h at room temperature. The color reaction was developed using an APAAP system (Dako). Sections of human reactive tonsils were used as positive controls for anti-CD20 immunostaining. Other tonsil sections were stained with a mouse monoclonal antibody against human CD3 (Dako). Intrahepatic CD20+ B-cell follicles and tonsilar CD3+ T cells were isolated by laser capture microdissection (PALM MicroBeam; Zeiss) and catapulted into PCR caps containing 20 μl of 1× DNA polymerase buffer (either Expand High Fidelity or Phusion buffer) and 0.1% Triton X-100.
PCR amplification of the target genes.
Both sorted and microdissected cell samples were lysed with proteinase K (0.25 mg/ml) for 3 h at 55°C. The enzyme was inactivated by heating at 95°C for 10 min. A seminested PCR was performed to amplify the mutation hot spot regions of the selected target genes (BCL6 intron 1 area B; CTNNB1 exons 3 and 8; TP53 exons 4, 5 and 6, and 7 and 8) (13, 25, 26). PCR assays were performed using two different DNA polymerases: an Expand High Fidelity polymerase mixture (Roche Applied Science, Mannheim, Germany) and Phusion DNA polymerase (New England BioLabs, Frankfurt am Main, Germany) with expected error frequencies of about 3 × 10−4 (4.3 × 10−6 changes/bp/cycle error rate) and 3 × 10−5 (about 4.4 × 10−7 changes/bp/cycle error rate), respectively, after 70 cycles of 2 rounds of PCR.
The first round of amplification was performed with a single aliquot of sorted or microdissected cells as a multiplex PCR in 50-μl reaction mixtures containing the following primers, each at 0.1 μM final concentration: CTNNB1 exon 3 (for 5′-ctg gct atc att ctg ctt ttc ttg gc-3′ and rev 5′-aac tgc att ctg act ttc agt aag gc-3′) and CTNNB1 exon 8 (for 5′-cta gaa cag ata ttt agg att gat agg c-3′ and rev 5′-gtg acc aca ttt ata tca tct gaa ccc-3′); fragments of TP53 and BCL6 were amplified as previously described (13, 25). The PCRs had the following cycling conditions: a first denaturation step of 95°C for 4 min, followed by 35 cycles of 50 s at 95°C, 30 s at 56°C, and 60 s at 72°C. A final incubation was performed for 5 min at 72°C. The first-round multiplex PCR was followed by separate second-round PCRs for each amplificate, with 35 cycles using the primer-specific annealing temperatures of 56°C for BCL6, 61°C for TP53, and 50°C for CTNNB1. The second-round CTNNB1 primers had the following sequences: for ß-Cat-Ex3-ForII, 5′-ttc aat ggg tca tat cac aga ttc-3′; for ß-Cat-Ex3-RevII, 5′-aat gaa aaa taa tac tct tac cag-3′; for ß-Cat-Ex8-ForII, 5′-tct agg tgg aat gca agc ttt ag-3′; and for ß-Cat-Ex8-RevII, 5′-aag agt ccc aag gag acc ttc cat c-3′.
The PCR products were further incubated with Taq DNA polymerase (Fermentas, St. Leon-Rot, Germany) and 0.2 mM dATP for 20 min at 72°C to generate a 3′ A overhang at both ends and ligated into pGEM T-Easy vector (Promega, Mannheim, Germany). After transformation in XLB1 Blue (Stratagene, Waldbronn, Germany), plasmids were isolated. For each PCR product, 10 to 20 individual clones were sequenced by Sanger sequencing on an ABI 3010 sequencer (Applied Biosystems, Darmstadt, Germany). Sequences of PCR products were compared to the corresponding germ line sequences with SeqScape v2.5 software (Applied Biosystems). Identical base changes in different sequences (shared mutations) were counted only once.
For statistical reasons, we cannot exclude the possibility that a few plasmid sequences derive from the same cell, in particular, as not all DNA molecules might be available for PCR (due to DNA strand breaks or incomplete release of DNA from chromatin after proteinase K digestion). However, as we observed the expected pattern of about one-third mutated BCL6 sequences in memory B cells (presented in Results), and as we observed mostly unique BCL6 mutations in the mutated sequences, this validates for BCL6 that the plasmids represent many different B cells in the sample and that there was not a preferential amplification of BCL6 from a single or very few B cells. As CTNNB1 and TP53 were coamplified from the very same DNA samples as BCL6, this argumentation can be extended to the two other genes as well, although here most sequences were unmutated.
Statistical analysis.
Statistical analysis of the data was performed by Fisher's exact test. Values of P < 0.05 were considered to be statistically significant.
RESULTS
To assess whether HCV induces a mutator phenotype in B cells in vivo, we analyzed the mutational status of the genes BCL6, TP53, and CTNNB1 in B cells of chronically HCV-infected patients. The mutation analysis included the three main PB B-cell subsets, namely, naive B cells, class-switched memory B cells, and IgM+ IgD+ memory B cells (27, 28). The analysis was performed on different B-cell subpopulations to determine whether mutations are generally present in the PB B cells, or only in post-GC, i.e., memory B cells.
The analysis was initially done with B cells from two patients (HCV1 and HCV2). Fluorescence-activated cell sorter (FACS)-sorted CD3+ T cells from a healthy donor served as a negative control. The PCR was carried out using an Expand High Fidelity DNA polymerase mix. We amplified the known mutation hot spot regions of the three genes, which are exons 4 to 8 of TP53, parts of intron 1 of BCL6, and exons 3 and 8 of CTNNB1. PCR products were cloned into plasmids, and 14 to 66 plasmids were sequenced per cell subset per PCR fragment. The sequence analysis showed mutation frequencies between 0.42 and 3.1 × 10−4 for TP53 and between 2.8 and 8.0 × 10−4 for CTNNB1 in the B cells (Table 1). However, no significant difference in the mutation frequencies of the selected genes was seen between the B cells isolated from HCV-infected patients and the T cells from the healthy donor (Table 1). These values were indeed in the range expected from the intrinsic error rate of the High Fideltiy enzyme mix. The observed mutation frequencies in the BCL6 gene were largely in concordance with previously published data for healthy individuals (28, 29), with mutations above the experimental background in both class-switched and one of the two IgM+ memory B-cell isolates from the two patients. Mutations in naive B cells were at the background level determined with T cells from healthy donor HD1. Hence, there was no increased BCL6 mutation load in B cells from HCV+ patients in comparison to B cells from healthy individuals.
Table 1.
Mutational analysis of cellular genes in PB B-cell subpopulations from HCV-positive patients and T cells from a healthy donor using an Expand High Fidelity PCR systema
| Donor | Cell type | TP53 |
CTNNB1 |
BCL6 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of clones mutated/no. of clones sequenced | No. of mutations/ total no. of nucleotides | Mutation frequency × 10−4 | No. of clones mutated/no. of clones sequenced | No. of mutations/total no. of nucleotides | Mutation frequency × 10−4 | No. of clones mutated/no. of clones sequenced | No. of mutations/total no. of nucleotides | Mutation frequency × 10−4 | ||
| HCV1 | Class switched | 4/43 | 4/17,100 | 2.3 | 4/34 | 5/7,758 | 6.4 | 10/22 | 14/15,466 | 9.0 |
| IgM+ IgD+ CD27+ | 2/38 | 2/14,212 | 1.4 | 3/29 | 2/6,544 | 3.0 | 5/14 | 5/9,842 | 5.0 | |
| Naive | 3/34 | 2/11,736 | 1.7 | 2/29 | 2/6,996 | 2.8 | 9/22 | 11/15,466 | 7.1 | |
| HCV2 | Class switched | 1/62 | 1/23,798 | 0.4 | 2/37 | 3/8,622 | 3.4 | 12/18 | 33/12,654 | 26.0 |
| IgM+ IgD+ CD27+ | 5/65 | 6/25,040 | 2.4 | 2/42 | 3/9,836 | 3.0 | 10/17 | 12/11,951 | 10.0 | |
| Naive | 7/66 | 8/25,547 | 3.1 | 8/44 | 8/10,186 | 8.0 | 8/20 | 9/14,060 | 6.4 | |
| HD1 | T | 11/96 | 12/33,718 | 3.6 | 5/65 | 5/14,765 | 3.4 | 13/29 | 13/20,387 | 6.4 |
Mutation frequencies (mutations per base pair) are calculated as the total number of single nucleotide mutations in all clones versus the total number of nucleotides sequenced. HD, healthy donor.
Since the observed mutation frequencies were quite variable and in the range of the experimental background introduced by the DNA polymerase mixture, we performed the analysis on B cells from two additional HCV-positive patients (HCV3 and HCV4) and T cells from a further healthy donor (HD2) using Phusion DNA polymerase, which has about a 10-fold-lower error rate than the High Fidelity system. Indeed, the experimental mutation background level as seen in the T cells was clearly lower with the Phusion polymerase than with the Expand High Fidelity system (0.5 to 1.1 × 10−4/bp and 3.4 to 6.4 × 10−4/bp, respectively). Using Phusion polymerase, the mutation level for CTNNB1 in all B-cell subsets was in the range of the experimental background (Table 2). The TP53 gene showed a slightly but statistically significantly higher mutation frequency in the naive B-cell subset from patient HCV3 and in the non-class-switched B cells from patient HCV4 than in the control T cells (P < 0.05). However, the four other B-cell samples did not show mutations above the background. For BCL6, naive B cells did not show a significantly higher mutation frequency than the control T cells (Table 2). The IgM+ IgD+ memory B cells harbored BCL6 mutations with frequencies of 6.4 × 10−4 and 8.7 × 10−4 for patients HCV3 and HCV4, respectively, and the class-switched memory B cells of these two patients showed BCL6 mutation frequencies of 6.1 × 10−4 and 14 × 10−4. These mutation frequencies are similar to the respective frequencies in memory B cells of healthy individuals (ca. 4 to 8 × 10−4 for IgM+ IgD+ and ca. 8 to 9 × 10−4 for class-switched memory B cells) as previously published (28). This is particularly true when one considers that the experimental background mutation level of 1.1 × 10−4 (value for control T cells) should be subtracted from the values determined in our study, whereas the published data (28) are devoid of polymerase errors.
Table 2.
Mutational analysis of cellular genes in PB B-cell subpopulations from HCV-positive patients and T cells from a healthy donor using Phusion DNA polymerasea
| Donor | Cell type | TP53 |
CTNNB1 |
BCL6 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of clones mutated/no. of clones sequenced | No. of mutations/total no. of nucleotides | Mutation frequency × 10−4 | No. of clones mutated/no. of clones sequenced | No. of mutations/total no. of nucleotides | Mutation frequency × 10−4 | No. of clones mutated/no. of clones sequenced | No. of mutations/total no. of nucleotides | Mutation frequency × 10−4 | ||
| HCV3 | Class switched | 3/86 | 4/32,295 | 1.2 | 1/45 | 1/10,361 | 0.9 | 8/23 | 24/16,169 | 14 |
| IgM+ IgD+ CD27+ | 5/76 | 5/27,062 | 1.8 | 1/44 | 1/10,186 | 0.9 | 8/22 | 10/15,466 | 6.4 | |
| Naive | 7/74 | 10/30,769b | 3.2 | 2/46 | 2/10,762 | 1.8 | 1/23 | 1/16,169 | 0.6 | |
| HCV4 | Class switched | 1/63 | 1/25,394 | 0.4 | 1/38 | 1/8,797 | 1.1 | 6/23 | 10/16,169 | 6.1 |
| IgM+ IgD+ CD27+ | 9/60 | 10/22,879b | 4.3 | 1/42 | 1/9,723 | 1.0 | 7/18 | 11/12,654 | 8.7 | |
| Naive | 1/59 | 1/21,918 | 0.4 | 0/39 | 0/8,972 | 0 | 4/23 | 4/16,169 | 2.5 | |
| HD2 | T | 3/146 | 3/56,177 | 0.5 | 2/90 | 2/21,174 | 0.9 | 3/38 | 3/26,714 | 1.1 |
Mutation frequencies (mutations per base pair) are calculated as the total number of single nucleotide mutations in all clones versus the total number of nucleotides sequenced. HD, healthy donor.
For B cells versus T cells, P < 0.05.
As there were hardly any HCV-associated mutations in the PB B cells, we wondered whether B cells in the liver, being in more direct contact with HCV, might be targeted by mutations. Since proliferating B-cell populations within follicle-like structures are frequently detected in the liver of chronically HCV-infected patients (11, 30), we analyzed B cells isolated from the liver of two HCV-positive patients, using the Phusion polymerase. Microdissected CD3-positive T cells from a reactive tonsil of an HCV-negative patient were used as a negative control. CD20-positive follicle-like structures were isolated by laser microdissection (Fig. 1). The mutation analysis was performed on 3 distinct follicles per liver, of about 100 isolated B cells each (Table 3). The analysis showed no statistically significant difference in the mutation frequency of TP53 and CTNNB1 in the intrahepatic B cells compared to the tonsilar T cells. For BCL6, one donor did not show mutations above the experimental background level, whereas the other patient showed a BCL6 mutation frequency (4 × 10−4/bp) not higher than that seen with post-GC B cells in healthy individuals. These results strongly suggest that HCV does not induce mutations of the selected cellular genes, even in B cells from the liver microenvironment.
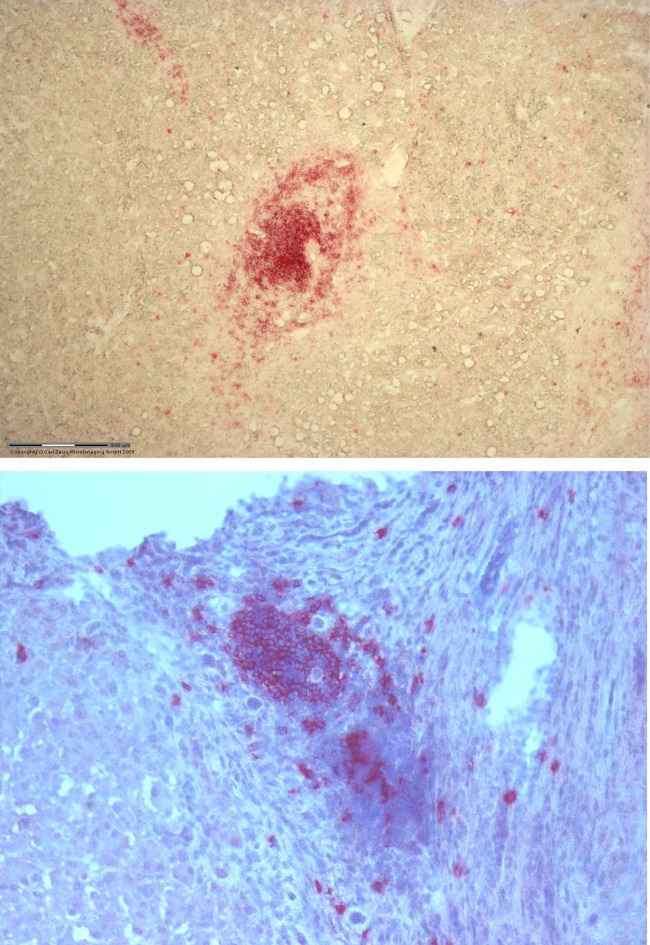
Fig 1.
Representative immunohistochemical staining for CD20-positive B cells in two different HCV-positive liver specimens. The staining shows the presence of B-cell clusters around the portal areas.
Table 3.
Mutational analysis of cellular genes in intrahepatic B cells from HCV-positive patients and tonsillar CD3+ T cellsa
| Donor | Cell type | TP53 |
CTNNB1 |
BCL6 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of clones mutated/no. of clones sequenced | No. of mutations/total no. of nucleotides | Mutation frequency × 10−4 | No. of clones mutated/no. of clones sequenced | No. of mutations/total no. of nucleotides | Mutation frequency × 10−4 | No. of clones mutated/no. of clones sequenced | No. of mutations/total no. of nucleotides | Mutation frequency × 10−4 | ||
| HCV5b | Intrahepatic CD20+ | 6/115 | 6/42,419 | 1.4 | 1/80 | 1/18,407 | 0.5 | 9/46 | 16/32,338c | 4.9 |
| HCV6b | Intrahepatic CD20+ | 3/117 | 4/45,875 | 0.9 | 0/82 | 0/18,757 | 0 | 6/47 | 6/33,041 | 1.8 |
| Tonsil 1 | Tonsil CD3+ | 3/73 | 3/28,087 | 1.0 | 2/51 | 2/11,976 | 1.7 | 2/27 | 2/18,981 | 1.1 |
Mutation frequencies (mutations per base pair) are calculated as the total number of single nucleotide mutations in all clones versus the total number of nucleotides sequenced.
All data represent the mean values of the analysis performed on three distinct CD20+ follicles per liver.
For B cells versus T cells, P < 0.05.
DISCUSSION
Comprehensive studies with B-cell lymphoma lines have shown that HCV can induce a mutator phenotype in B cells by causing mutations in proto-oncogenes (BCL6, CTNNB1) and tumor suppressor genes (TP53) in in vitro HCV-infected B cells (12, 14). Moreover, increased mutation frequencies of these genes were also seen in PBMC of chronically HCV-infected individuals (12). As B cells typically account for only about 10% of PBMC (the other 90% being composed of T cells, monocytes, and natural killer cells), the previous study did not clarify whether B cells are indeed a target for HCV-induced mutations in vivo. Since expanded B-cell clones in HCV-associated lymphoproliferations carry somatically mutated Ig V genes, suggesting a post-GC derivation (7), we wanted to define in which B-cell subpopulations, pre- or post-GC B cells, virus-induced mutations might occur. Therefore, to further assess the impact of HCV infection on the mutational status of cellular genes in vivo, we analyzed the mutation frequencies of TP53, CTNNB1, and BCL6 in naive and memory PB B cells isolated from chronically HCV-infected individuals. Unexpectedly, the analysis of PB B cells of four patients revealed that TP53 and CTNNB1 did not acquire mutations in naive or memory B cells, as shown by the low number of nucleotide differences observed, which were in the same range as those of the control T cells from healthy individuals. Hence, they represent the experimental background. Slightly higher values were seen for TP53 in naive B cells from one and IgM+ IgD+ CD27+ B cells from another of the four patients, but these may represent outliers, considering the overall rarity of the mutations. Moreover, even these values (3.2 and 4.3 × 10−4) were only one-third of those reported for HCV-exposed B-cell lymphoma lines (12). Mutations were consistently identified in the BCL6 gene in both subsets of PB memory B cells of all four patients. However, BCL6 is a physiological target of somatic hypermutation, and the mutation frequencies seen here are in the same range as those previously reported for memory B cells in healthy individuals (13). No BCL6 mutations were found in naive B cells. Hence, there is also no increased mutation targeting of BCL6 in PB B cells of HCV-positive patients.
We further extended the analysis to B cells isolated from HCV-positive livers. The liver is the primary target for HCV replication, and one main histologic characteristic of chronic HCV infection is the presence of lymphoid aggregates in the liver, which resemble GC structures where B cells proliferate and are clonally restricted (21, 30). The distribution of HCV within the liver is patchy (31), with infected cells generally appearing in clusters consistent with cell-to-cell spread of virus. Moreover, the retention of B cells in the liver parenchyma has been shown to be promoted by the viral infection which stabilizes B-cell/hepatocyte interactions, perhaps allowing viral transmission between the two cell types (32). Based on these observations, it is conceivable that in the B-cell aggregates in the liver, the interaction between HCV and B cells is more direct than in other biological compartments. Our analysis performed on the CD20+ follicle-like structures isolated from the liver of two HCV-positive patients, however, showed an absence of mutations in TP53 and CTNNB1 (Table 3). For BCL6, the mutation loads were even lower than those seen in PB memory B cells. This may be due to the presence of a mixed population of pre- and post-GC B cells within the intrahepatic follicles, even though the composition of these B-cell clusters in the liver has not yet been investigated. Moreover, among the microdissected areas, some non-B cells with an unmutated BCL6 gene may be present.
We do not have direct information about the clonality of the B cells we studied. However, there is no clear indication in the literature that naive or class-switched PB B cells in HCV patients are clonally restricted. Moreover, our BCL6 mutation analysis revealed, in addition to unmutated cells, many B cells with distinct (physiological) BCL6 mutations among IgM+ IgD+ CD27+ or class-switched B cells, indicating that they were polyclonal. In any case, our observation of a lack of mutations in the genes analyzed in B cells exposed to HCV in vivo is relevant and conclusive regardless of the clonality of the B cells.
Taking these data from the PB and the liver of six HCV-infected individuals together, it clearly emerges that B cells in these patients are not acquiring HCV-driven mutations in the three genes analyzed. This is an unexpected finding with respect to a previous report (12). Regarding the in vitro studies with B-cell lines, it has to be considered that these were Burkitt lymphoma cell lines (12). It is conceivable that these transformed B cells, which also have a GC B-cell phenotype, are particularly susceptible to mutation induction by HCV. It may also be that in the in vitro cultures, the viral titers are much higher than in vivo, causing a stronger mutagenic effect of HCV on the B cells.
Whether HCV directly infects the B cells analyzed was not studied either in the previous in vitro study or in the present work. However, our aim was to clarify the consequences of the exposure of human B cells to HCV independently of the controversial issue about HCV lymphotropism.
The discrepancy between the previously reported data on PBMC of HCV-infected individuals (12) and our study of B cells from chronically HCV-infected patients is difficult to reconcile. The previous work did not identify in which cell type among the PBMC the mutations were present. Hence, the mutations might potentially be present only in T cells, monocytes, or NK cells, which we did not analyze. However, this scenario appears unlikely. The reported mutagenic effect of HCV on PBMC should affect not only T cells, monocytes, and/or NK cells but also B cells, considering the mutagenic effect of HCV seen with B-cell lines (12). In addition, as in PB only B cells express low levels of BCL6 transcripts (transcription of a gene is essential for somatic hypermutation of the gene) (33, 34), it is unlikely that a presumed AID induction in non-B cells would cause BCL6 mutations in these cells. In line with our findings, another study of BCL6 mutations in PBMC (not isolated B cells) of HCV-positive patients with or without lymphoproliferative disorders or B-NHL also did not find BCL6 mutations (35). In any case, even if mutations occur in PB non-B cells in vivo, they would not be relevant for B-cell transformation in HCV carriers.
Taken together, although genetic alterations of TP53 and CTNNB1 genes are implicated in the development of hepatocellular carcinoma associated with HCV infection (26, 36), the results of the present work revealed that these genes, as well as the BCL6 proto-oncogene, are not (frequent) targets of HCV-induced mutagenesis in PB or intrahepatic B cells of HCV-infected patients. In this regard, it is also notable that aberrant hypermutation of the proto-oncogenes PIM1, PAX5, RHOH, and MYC, which is seen in diffuse large B-cell lymphomas (37), has not been detected in HCV-associated B-cell lymphomas (38), in line with our finding that HCV does not induce a general mutator phenotype in B cells. Hence, it remains to be clarified how HCV contributes to the development of B-cell lymphoproliferative diseases in chronic virus carriers, and whether the contribution of the virus to the pathogenesis of these diseases includes direct mutagenic effects.
ACKNOWLEDGMENTS
We thank Kerstin Heise, Andrea Kopplin, and Gwen Lorenz for expert technical assistance, Marc Seifert for help with cell sorting, and Berit Jungnickel for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft through TRR60. P.J. was supported by the IFORES program of the Medical Faculty of the University Duisburg-Essen.
We declare that no competing interests exist.
Footnotes
Published ahead of print 26 December 2012
REFERENCES
- 1. Agnello V, Chung RT, Kaplan LM. 1992. A role for hepatitis C virus infection in type II cryoglobulinemia. N. Engl. J. Med. 327:1490–1495 [DOI] [PubMed] [Google Scholar]
- 2. Zuckerman E, Zuckerman T, Levine AM, Douer D, Gutekunst K, Mizokami M, Qian DG, Velankar M, Nathwani BN, Fong TL. 1997. Hepatitis C virus infection in patients with B-cell non-Hodgkin lymphoma. Ann. Intern. Med. 127:423–428 [DOI] [PubMed] [Google Scholar]
- 3. Turner NC, Dusheiko G, Jones A. 2003. Hepatitis C and B-cell lymphoma. Ann. Oncol. 14:1341–1345 [DOI] [PubMed] [Google Scholar]
- 4. Gisbert JP, Garcia-Buey L, Pajares JM, Moreno-Otero R. 2003. Prevalence of hepatitis C virus infection in B-cell non-Hodgkin's lymphoma: systematic review and meta-analysis. Gastroenterology 125:1723–1732 [DOI] [PubMed] [Google Scholar]
- 5. Ferri C, La Civita L, Longombardo G, Greco F, Bombardieri S. 1993. Hepatitis C virus and mixed cryoglobulinaemia. Eur. J. Clin. Invest. 23:399–405 [DOI] [PubMed] [Google Scholar]
- 6. Dammacco F, Sansonno D, Piccoli C, Racanelli V, D'Amore FP, Lauletta G. 2000. The lymphoid system in hepatitis C virus infection: autoimmunity, mixed cryoglobulinemia, and overt B-cell malignancy. Semin. Liver Dis. 20:143–157 [DOI] [PubMed] [Google Scholar]
- 7. Ivanovski M, Silvestri F, Pozzato G, Anand S, Mazzaro C, Burrone OR, Efremov DG. 1998. Somatic hypermutation, clonal diversity, and preferential expression of the VH 51p1/VL kv325 immunoglobulin gene combination in hepatitis C virus-associated immunocytomas. Blood 91:2433–2442 [PubMed] [Google Scholar]
- 8. De Re V, De Vita S, Marzotto A, Rupolo M, Gloghini A, Pivetta B, Gasparotto D, Carbone A, Boiocchi M. 2000. Sequence analysis of the immunoglobulin antigen receptor of hepatitis C virus-associated non-Hodgkin lymphomas suggests that the malignant cells are derived from the rheumatoid factor-producing cells that occur mainly in type II cryoglobulinemia. Blood 96:3578–3584 [PubMed] [Google Scholar]
- 9. Carbonari M, Caprini E, Tedesco T, Mazzetta F, Tocco V, Casato M, Russo G, Fiorilli M. 2005. Hepatitis C virus drives the unconstrained monoclonal expansion of VH1-69-expressing memory B cells in type II cryoglobulinemia: a model of infection-driven lymphomagenesis. J. Immunol. 174:6532–6539 [DOI] [PubMed] [Google Scholar]
- 10. Quinn ER, Chan CH, Hadlock KG, Foung SK, Flint M, Levy S. 2001. The B-cell receptor of a hepatitis C virus (HCV)-associated non-Hodgkin lymphoma binds the viral E2 envelope protein, implicating HCV in lymphomagenesis. Blood 98:3745–3749 [DOI] [PubMed] [Google Scholar]
- 11. Sansonno D, De Vita S, Iacobelli AR, Cornacchiulo V, Boiocchi M, Dammacco F. 1998. Clonal analysis of intrahepatic B cells from HCV-infected patients with and without mixed cryoglobulinemia. J. Immunol. 160:3594–3601 [PubMed] [Google Scholar]
- 12. Machida K, Cheng KT, Sung VM, Shimodaira S, Lindsay KL, Levine AM, Lai MY, Lai MM. 2004. Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes. Proc. Natl. Acad. Sci. U. S. A. 101:4262–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pasqualucci L, Migliazza A, Fracchiolla N, William C, Neri A, Baldini L, Chaganti RS, Klein U, Küppers R, Rajewsky K, Dalla-Favera R. 1998. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc. Natl. Acad. Sci. U. S. A. 95:11816–11821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Machida K, Cheng KT, Sung VM, Lee KJ, Levine AM, Lai MM. 2004. Hepatitis C virus infection activates the immunologic (type II) isoform of nitric oxide synthase and thereby enhances DNA damage and mutations of cellular genes. J. Virol. 78:8835–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Machida K, McNamara G, Cheng KT, Huang J, Wang CH, Comai L, Ou JH, Lai MM. 2010. Hepatitis C virus inhibits DNA damage repair through reactive oxygen and nitrogen species and by interfering with the ATM-NBS1/Mre11/Rad50 DNA repair pathway in monocytes and hepatocytes. J. Immunol. 185:6985–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lai MM. 2002. Hepatitis C virus proteins: direct link to hepatic oxidative stress, steatosis, carcinogenesis and more. Gastroenterology 122:568–571 [DOI] [PubMed] [Google Scholar]
- 17. Lerat H, Rumin S, Habersetzer F, Berby F, Trabaud MA, Trepo C, Inchauspe G. 1998. In vivo tropism of hepatitis C virus genomic sequences in hematopoietic cells: influence of viral load, viral genotype, and cell phenotype. Blood 91:3841–3849 [PubMed] [Google Scholar]
- 18. Karavattathayyil SJ, Kalkeri G, Liu HJ, Gaglio P, Garry RF, Krause JR, Dash S. 2000. Detection of hepatitis C virus RNA sequences in B-cell non-Hodgkin lymphoma. Am. J. Clin. Pathol. 113:391–398 [DOI] [PubMed] [Google Scholar]
- 19. Inokuchi M, Ito T, Uchikoshi M, Shimozuma Y, Morikawa K, Nozawa H, Shimazaki T, Hiroishi K, Miyakawa Y, Imawari M. 2009. Infection of B cells with hepatitis C virus for the development of lymphoproliferative disorders in patients with chronic hepatitis C. J. Med. Virol. 81:619–627 [DOI] [PubMed] [Google Scholar]
- 20. Zignego AL, Giannini C, Monti M, Gragnani L. 2007. Hepatitis C virus lymphotropism: lessons from a decade of studies. Dig. Liver Dis. 39(Suppl 1):S38–S45 [DOI] [PubMed] [Google Scholar]
- 21. Sansonno D, Lauletta G, De Re V, Tucci FA, Gatti P, Racanelli V, Boiocchi M, Dammacco F. 2004. Intrahepatic B cell clonal expansions and extrahepatic manifestations of chronic HCV infection. Eur. J. Immunol. 34:126–136 [DOI] [PubMed] [Google Scholar]
- 22. Ducoulombier D, Roque-Afonso AM, Di Liberto G, Penin F, Kara R, Richard Y, Dussaix E, Feray C. 2004. Frequent compartmentalization of hepatitis C virus variants in circulating B cells and monocytes. Hepatology 39:817–825 [DOI] [PubMed] [Google Scholar]
- 23. Blackard JT, Kemmer N, Sherman KE. 2006. Extrahepatic replication of HCV: insights into clinical manifestations and biological consequences. Hepatology 44:15–22 [DOI] [PubMed] [Google Scholar]
- 24. Pal S, Sullivan DG, Kim S, Lai KK, Kae J, Cotler SJ, Carithers RL, Jr, Wood BL, Perkins JD, Gretch DR. 2006. Productive replication of hepatitis C virus in perihepatic lymph nodes in vivo: implications of HCV lymphotropism. Gastroenterology 130:1107–1116 [DOI] [PubMed] [Google Scholar]
- 25. Montesinos-Rongen M, Roers A, Küppers R, Rajewsky K, Hansmann ML. 1999. Mutation of the p53 gene is not a typical feature of Hodgkin and Reed-Sternberg cells in Hodgkin's disease. Blood 94:1755–1760 [PubMed] [Google Scholar]
- 26. Huang H, Fujii H, Sankila A, Mahler-Araujo BM, Matsuda M, Cathomas G, Ohgaki H. 1999. Beta-catenin mutations are frequent in human hepatocellular carcinomas associated with hepatitis C virus infection. Am. J. Pathol. 155:1795–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klein U, Rajewsky K, Küppers R. 1998. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J. Exp. Med. 188:1679–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seifert M, Küppers R. 2009. Molecular footprints of a germinal center derivation of human IgM+(IgD+)CD27+ B cells and the dynamics of memory B cell generation. J. Exp. Med. 206:2659–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shen HM, Peters A, Baron B, Zhu X, Storb U. 1998. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science 280:1750–1752 [DOI] [PubMed] [Google Scholar]
- 30. Racanelli V, Sansonno D, Piccoli C, D'Amore FP, Tucci FA, Dammacco F. 2001. Molecular characterization of B cell clonal expansions in the liver of chronically hepatitis C virus-infected patients. J. Immunol. 167:21–29 [DOI] [PubMed] [Google Scholar]
- 31. Liang Y, Shilagard T, Xiao SY, Snyder N, Lau D, Cicalese L, Weiss H, Vargas G, Lemon SM. 2009. Visualizing hepatitis C virus infections in human liver by two-photon microscopy. Gastroenterology 137:1448–1458 [DOI] [PubMed] [Google Scholar]
- 32. Stamataki Z, Shannon-Lowe C, Shaw J, Mutimer D, Rickinson AB, Gordon J, Adams DH, Balfe P, McKeating JA. 2009. Hepatitis C virus association with peripheral blood B lymphocytes potentiates viral infection of liver-derived hepatoma cells. Blood 113:585–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bachl J, Carlson C, Gray-Schopfer V, Dessing M, Olsson C. 2001. Increased transcription levels induce higher mutation rates in a hypermutating cell line. J. Immunol. 166:5051–5057 [DOI] [PubMed] [Google Scholar]
- 34. Fukita Y, Jacobs H, Rajewsky K. 1998. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity 9:105–114 [DOI] [PubMed] [Google Scholar]
- 35. Hofmann WP, Fernandez B, Herrmann E, Welsch C, Mihm U, Kronenberger B, Feldmann G, Spengler U, Zeuzem S, Sarrazin C. 2007. Somatic hypermutation and mRNA expression levels of the BCL-6 gene in patients with hepatitis C virus-associated lymphoproliferative diseases. J. Viral Hepat. 14:484–491 [DOI] [PubMed] [Google Scholar]
- 36. Edamoto Y, Hara A, Biernat W, Terracciano L, Cathomas G, Riehle HM, Matsuda M, Fujii H, Scoazec JY, Ohgaki H. 2003. Alterations of RB1, p53 and Wnt pathways in hepatocellular carcinomas associated with hepatitis C, hepatitis B and alcoholic liver cirrhosis. Int. J. Cancer 106:334–341 [DOI] [PubMed] [Google Scholar]
- 37. Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Küppers R, Dalla-Favera R. 2001. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature 412:341–346 [DOI] [PubMed] [Google Scholar]
- 38. Libra M, Capello D, Gloghini A, Laura P, Berra E, Cerri M, Gasparotto D, Franca S, De Re V, Gaidano G, Carbone A. 2005. Analysis of aberrant somatic hypermutation (SHM) in non-Hodgkin's lymphomas of patients with chronic HCV infection. J. Pathol. 206:87–91 [DOI] [PubMed] [Google Scholar]