Abstract
Polyomavirus origins of replication contain multiple occurrences of G(A/G)GGC, the high-affinity binding element for the viral initiator T-antigen (T-ag). The site I regulatory region of simian virus 40, involved in the repression of transcription and the enhancement of DNA replication initiation, contains two GAGGC sequences arranged head to tail and separated by a 7-bp AT-rich sequence. We have solved a 3.2-Å costructure of the SV40 origin-binding domain (OBD) bound to site I. We have also established that T-ag assembly on site I is limited to the formation of a single hexamer. These observations have enabled an analysis of the role(s) of the OBDs bound to the site I pentanucleotides in hexamer formation. Of interest, they reveal a correlation between the OBDs bound to site I and a pair of OBD subunits in the previously described hexameric spiral structure. Based on these findings, we propose that spiral assembly is promoted by pentanucleotide pairs arranged in a head-to-tail manner. Finally, the possibility that spiral assembly by OBD subunits accounts for the heterogeneous distribution of pentanucleotides found in the origins of replication of polyomaviruses is discussed.
INTRODUCTION
Polyomaviruses are small DNA tumor viruses implicated in human diseases, particularly among the immunocompromised and the elderly (1). For example, the polyomavirus JC causes progressive multifocal leukoencephalopathy (PML) (2), BK virus is correlated with renal dysfunction in kidney transplant patients (2), and Merkel cell polyomavirus (MCV) is implicated in a rare but aggressive form of skin cancer (3, 4). The recent identification of several new human polyomaviruses (e.g., KI and WU polyomaviruses [5]) have provided additional incentives to establish fundamental aspects of the polyomavirus life cycle.
The best-studied member of the polyomavirus family is simian virus 40 (SV40). Advances made with SV40 include the identification of several of the tumor suppressor proteins targeted during viral transformation (reviewed in reference 6). It has also been used to identify many of the proteins required for eukaryotic DNA replication (reviewed in references 7 to 9). In related studies, it was used to establish basic mechanisms involved in the replication process (reviewed in references 10 to 12). A current focus of research in this field is providing a molecular understanding of initiation events (reviewed in reference 13). In the long term, it is anticipated that progress made in terms of understanding SV40 replication will lead to therapies for treating polyomavirus infections.
Among the highly conserved features of the circular, double-stranded DNA genomes of polyomavirus family members are the origins of DNA replication. The minimal or “core” origin of SV40 is a 64-bp region that is necessary and sufficient for DNA replication (reference 14 and references therein) (Fig. 1A). The central region of the core origin (termed site II) contains a palindromic arrangement of four GAGGC pentanucleotides (termed P1 to P4). The pentanucleotides are the high-affinity binding sites for T antigen (T-ag), the virally encoded initiator protein (15–17). Flanking the central site II region of the core origin are the AT-rich and early palindrome (EP) regions.
Fig 1.
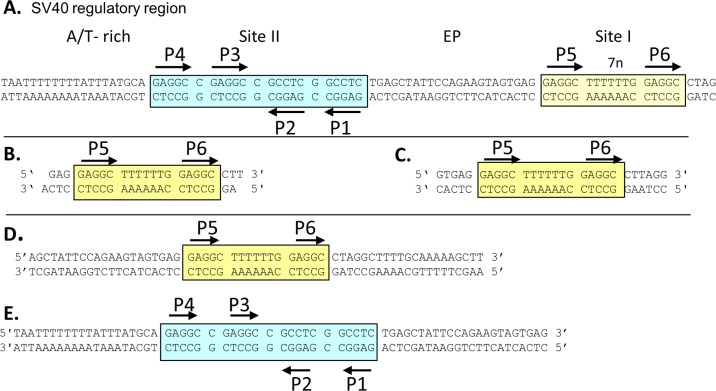
(A) DNA sequence of the core origin of replication and the site I regulatory region. Site II is indicated by a cyan box, and site I is indicated by a yellow box. Black arrows indicate both the positions and the orientations of the GAGGC pentameric sequences. The locations of the AT-rich and EP regions are also indicated. (B) Site I-containing oligonucleotide used to determine the OBD-site I costructure. Note that the oligonucleotide has a 1-bp overhang; this is commonly used to promote formation of a pseudocontinuous helix within the crystal. (C) Twenty-eight-base-pair site I-containing oligonucleotide used in the ITC studies. DNA substrates used in the EMSAs included the 59-bp site I-containing oligonucleotide (D) and the 64-bp core origin oligonucleotide (E).
The T-antigens encoded by polyomaviruses are multifunctional, modular proteins that contain an N-terminal DNA J domain, a central origin binding domain (OBD), and a C-terminal helicase domain (reviewed in references 10 to 12). The OBDs are necessary and sufficient for site-specific binding to the pentanucleotides. Once site-specific binding is completed, the helicase domains are positioned over the flanking regions (18, 19). The EP region of the origin is melted by a motif in the helicase domain termed the beta-hairpin (20–23). We have proposed that the beta-hairpin-dependent melting of the flanking sequences is coupled to the assembly of hexameric rings of T-ag around one of the newly generated single strands of DNA (21, 24). Once the oligomerization process is completed, two hexamers of T-ag are formed on the core origin (reviewed in references 13 and 25).
Recent crystallographic studies of the individual domains of T-ag have greatly advanced our understanding of its dynamic interactions with the origin of replication. For example, the costructures of the OBD bound to site II established the basis for site-specific binding to the pentanucleotides (24, 26, 74). In brief, residues in the OBD A1 and B2 motifs (27, 28) make numerous contacts with both the sugar-phosphate backbone and the major groove of individual pentanucleotides (reviewed in reference 13). Related structural studies revealed how the OBD interacts with single-stranded DNA (ssDNA) (29, 30) and with duplex DNA in a non-sequence-specific manner (26). Moreover, crystal structures of the SV40 helicase domain were solved in the presence and absence of nucleotide (20, 31). The structures of the helicase domain, and related structures of the bovine E1 helicase domain (32), established fundamental principles regarding the mechanisms employed by eukaryotic helicases (reviewed in reference 33).
The crystal structure of the SV40 T-ag OBD has also been solved in the absence of DNA. In one frequently detected crystal form, the OBDs adopt a left-handed helical filament having six molecules per turn (34, 35; reviewed in reference 13). This structure was termed the “spiral” or “lock washer” form of the T-ag OBD. It was postulated that one turn of the spiral (containing six OBDs) occurs in the context of full-length T-ag hexamers. The OBDs in the spiral structure cannot interact with double-stranded DNA (dsDNA) in a sequence-specific fashion due to conformational changes in the A1 loops, as well as steric collisions predicted to occur between the DNA and neighboring T-ag OBDs (reviewed in reference 13). These and related observations led to the proposal that the lock washer conformation is adopted by the OBDs subsequent to site-specific binding of the GAGGCs (24). An additional interesting feature of the OBD spiral is a “gap” through which ssDNA may transit (34, 36).
While the recent structures have provided many snap shots of the initiation process, it is not understood how T-ag's interactions with the origin translate into the assembly of hexamers and double-hexamers. For example, origin subfragments containing single pentanucleotides support hexamer formation (37–39) and certain pairs of pentanucleotides support double-hexamer assembly (38–40). Therefore, it is not clear why the central region of the core origin has four pentanucleotides. Moreover, it is not obvious why the pentanucleotides in polyomavirus origins are separated by spacers of different lengths (41).
To continue our investigation of T-ag's interactions with the SV40 regulatory region, we have determined the costructure of the OBD bound to a regulatory region termed site I (Fig. 1A). Site I is adjacent to the EP region and contains two GAGGC pentanucleotides (herein termed P5 and P6) arranged in a head-to-tail fashion and separated by a 7-bp spacer [containing 6 deoxythymidines and 1 deoxyguanosine and termed the poly(dT)·poly(dA) spacer]. The primary physiological function of site I is the repression of early gene transcription, although it has also been reported to facilitate viral replication (42–47). The function(s) of site I are completely dependent upon its exact sequence (see, e.g., references 48 and 49) as well as its orientation and spacing relative to site II (48).
Analyses of the costructure formed between the OBDs and site I indicated that a single hexamer of T-ag forms on this site, a possibility confirmed by additional EMSAs (see below). Therefore, the OBDs initially bound to P5 and P6 must contribute to the formation of the site I hexamer. Regarding their roles in hexamer assembly, we have observed a surprising spatial relationship between the OBDs bound to P5 and P6 and a pair of OBDs in the left-handed spiral structure. Moreover, structure-based modeling studies demonstrate that a similar correlation can be made between an additional pair of OBDs within the spiral and the OBDs bound to a pentanucleotide pair in site II that is separated by a much shorter spacer length (34). In view of these findings, we propose a unifying hypothesis regarding pentanucleotide pairs that are arranged in a head-to-tail manner in polyomavirus origins of DNA replication; namely, they promote the formation of OBD spirals.
MATERIALS AND METHODS
Molecular biology techniques. (i) Purification of SV40 T-ag and the T-ag OBD.
Protocols for the purification of the T-ag OBD, amino acids (aa) 131 to 260, were previously reported (24, 50); they were used as described previously with the exception that the Sephacryl S-100 column was replaced with a smaller Superdex 75 column. Selenomethionine (SeMet)-containing T-ag OBD was also prepared as described previously (34). To summarize the purification protocol, the T-ag OBD is expressed as a glutathione S-transferase (GST) fusion in Escherichia coli BL21(DE3), purified by affinity chromatography with glutathione Sepharose 4B resin (GE, Inc.), digested with thrombin to remove the GST, and further purified by anion and cation exchange chromatography followed by gel filtration. Upon purification, the T-ag OBD was stored in storage buffer (20 mM Tris, pH 8.0, 50 mM NaCl, 10% glycerol, 1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride [PMSF], and 0.1% beta-mercaptoethanol) at −80°C until ready for use.
Full-length SV40 T-ag (aa 1 to 708) was generated using a baculovirus expression vector containing the T-ag-encoding A gene (51). It was subsequently isolated using immunoaffinity techniques with monoclonal antibody PAb 419 as previously described (52, 53). Once purified, T-ag was dialyzed into T-ag storage buffer (see above) and stored at −80°C until ready for use.
(ii) EMSA of T-ag and DNA.
The electromobility shift assay (EMSA) reactions with T-ag and 32P-labeled doubled-stranded DNA (reference 54 and references therein) were conducted under replication conditions (55) as previously reported (38). The double-stranded DNA substrates used in these assays were prepared as described previously (38). After 20 min of incubation at 37°C, the reaction mixtures were cross-linked via the addition of glutaraldehyde (0.1% final concentration) for 5 min. Once the cross-linking was completed, the samples were applied to a 4-to-12% gradient polyacrylamide gel (19:1 acrylamide/bisacrylamide ratio) and electrophoresed in 0.5× Tris-borate-EDTA (TBE) for ∼2 h (∼600 V and 20 mA). Gels were then dried and subjected to autoradiography.
(iii) 1,10-Phenanthroline–copper footprinting.
A single-stranded oligonucleotide (∼25 pmol), derived from the previously described 47-bp “site I + wt 30” oligonucleotide (50), was 32P labeled at its 5′ terminus using standard methods (56). It was then hybridized to its unlabeled complementary strand, and the asymmetrically labeled substrates were purified by electrophoresis (38).
The gel retardation 1,10-phenanthroline–copper reactions were performed as described by Kuwabara and Sigman (57); T-ag-specific protocols are presented in references 38 and 58. In brief, the products of standard band shift reactions with the 47-bp site I + wt 30 oligonucleotide and T-ag (54, 59) or the T-ag OBD (50) were separated on 4-to-12% gradient polyacrylamide gels in 0.5× TBE for ∼2 h (∼500 V and 20 mA). Following electrophoresis, the gel was rinsed in 50 mM Tris-HCl (pH 8.0); cleavage of the DNA was catalyzed by soaking the gel at room temperature in 1,10-phenanthroline–CuSO4–3-mercaptopropionic acid (final concentrations, 0.17 mM, 38 μM, and 5 mM, respectively) for 20 min. The DNA cleavage reaction was quenched by the addition of 2,9-dimethyl-1,10-phenanthroline (final concentration, 2.2 mM) for ∼2 min. The gel was then rinsed in H2O, wrapped in plastic wrap, and subjected to autoradiography for ∼1.5 to 2 h. Polyacrylamide slices containing complexes formed between the 47-bp site I + wt 30 oligonucleotide and either T-ag or the T-ag OBD or containing free DNA were excised and eluted overnight at 37°C in ∼0.5 ml of elution buffer (0.3 M sodium acetate [pH 5.2], 0.2% sodium dodecyl sulfate [SDS], 10 mM magnesium acetate, 10 μg of proteinase K per ml [56]). Polyacrylamide fragments were removed by centrifugation, and the DNA-containing supernatants were subjected to phenol-chloroform-isoamyl alcohol (25:24:1) and chloroform-isoamyl alcohol (24:1) extractions; the DNA fragments were then precipitated with 100% (vol/vol) ethanol. The DNA pellets were washed with 70% (vol/vol) ethanol, dried, and resuspended to ∼2,000 cpm/μl in loading dye (80% [vol/vol] formamide, 10 mM NaOH, 1 mM EDTA, 0.1% bromophenol blue, and 0.1% xylene cyanol). Aliquots were boiled for ∼3 min and applied to a 14% polyacrylamide-8 M urea gel in 1× TBE at ∼2,000 V and 28 mA. Sequencing markers were obtained after performance of Maxam-Gilbert (60) G and G+A reactions on the asymmetrically labeled 47-bp site I + wt 30 oligonucleotide.
Biophysical techniques. (i) Crystallography.
(a) DNA purification. Synthetic oligonucleotides were ordered from Integrated DNA Technologies (IDT). The sequence of the double-stranded site I-containing oligonucleotide used in crystallization is shown in Fig. 1B. The two strands used to form this oligonucleotide were purified by anion exchange chromatography (DNAPak; Vydac Inc.). The eluted DNA was lyophilized, desalted on a PD-10 column (GE Inc.), and lyophilized to dryness. The oligonucleotides were resuspended in annealing buffer (10 mM Tris, pH 7.5, 50 mM NaCl), and the complementary strands were mixed in a 1:1 molar ratio based on calculated molar extinction coefficients. The DNA was annealed by heating it to 95°C for 3 min, followed by slow cooling to 4°C. The duplex DNA was stored at −20°C.
(b) Crystallization. The complexes of site I DNA with either the selenomethione-containing or the nonlabeled T-ag OBD were prepared in a 1.2:2 DNA/protein molar ratio. The complex was concentrated via ultrafiltration using a Vivaspin concentrator (10,000 molecular weight cutoff) to a protein concentration of ∼10 mg/ml, aliquoted, quick-frozen in liquid nitrogen, and then stored at −80°C.
Crystals of the T-ag OBD in complex with the site I-containing oligonucleotide were grown by vapor diffusion over a 1-ml reservoir using a Linbro plate at 18°C by mixing 1 μl of the complex with 1 μl of the reservoir solution (16.875% [wt/vol] polyethylene glycol 3350 [PEG 3350], 0.18 M CaCl2, and 3% [wt/vol] sucrose). Crystals of the SeMet-T-ag OBD–site I DNA complex were grown by vapor diffusion over a 1-ml reservoir using a Linbro plate at 18°C by mixing 2 μl of the complex with 2 μl of the reservoir solution (0.1 M sodium acetate, pH 4.6, 0.2 M ammonium sulfate, 30% [wt/vol] PEG 4000, and 10 mM CaCl2).
(c) X-ray data collection and structure solution. The crystals were transferred to a cryogenic solution (25.5% [wt/vol] PEG 4000, 15% ethylene glycol, 0.085 M sodium acetate, pH 4.6, 0.17 M ammonium sulfate, and 8.5 mM CaCl2) using a cryoloop (Hampton Research Inc.) and then frozen in liquid nitrogen. X-ray data were collected at 100 K at NSLS Beamline X29 (Brookhaven National Laboratory, NY). The X-ray data were processed with HKL2000 (61). The costructure was solved by using single anomalous diffraction and molecular replacement (SAD and MR) and T-ag OBD coordinates bound to GAGGC as a search model (RCSB Protein Data Bank [PDB] identification number [ID] 2NTC) and the program PHASER-EP (62). The structure was refined using the program REFMAC5 (63), within the CCP4 suite of programs (64), and the PHENIX suite of programs (65). The resulting maps and models were visualized and improved with the molecular graphics program COOT (66).
(ii) ITC.
Isothermal titration calorimetry (ITC) data were collected with a VP-ITC system (MicroCal, Northampton, MA); the data were analyzed with Origin software provided by the manufacturer. The site I-containing dsDNA used in the ITC experiments is presented in Fig. 1C. Prior to our conducting the experiments, the dsDNA oligonucleotides and proteins were buffer exchanged into 10 mM sodium phosphate buffer, pH 7.0, and 50 mM NaCl, using PD-10 columns (GE Healthcare). Protein and DNA concentrations were determined spectrophotometrically, using calculated extinction coefficients from the ProtParam Web server and the IDT website, respectively.
(iii) Molecular modeling.
Unless otherwise indicated, all molecular graphic images were generated using the program PyMOL (67). Superpositions were generated using the program LSQMAN (68). DNA parameters were analyzed using either the program CURVES+ (69) or 3DNA (70). Protein-DNA interactions were analyzed using the program PDBSUM (71). The previously solved structures used in the molecular modeling discussed here are the apo-T-ag OBD and T-ag helicase domain (RCSB PDB IDs 2FUF and 1SVM, respectively).
RESULTS
Our previous structures of the SV40 OBD revealed that it is highly versatile in terms of the complexes it forms (reviewed in references 10 and 13). Given this property and the unique features of site I, we elected to investigate the structural basis for the interaction of the OBD with this region.
(i) The T-ag OBD–site I costructure.
(a) Overview of T-ag OBD bound to site I. The T-ag OBD–site I crystal structure was refined to 3.2 Å, with an Rwork of 18.76 and an Rfree of 26.74. The refinement statistics are presented in Table 1. The complex crystallized in space group C2221, with two T-ag OBD molecules and one 23-mer site I-containing oligonucleotide in the asymmetric unit. The DNA substrate used in this study, shown in Fig. 1B, contains two GAGGC sequences (i.e., P5 and P6) arranged as head-to-tail direct repeats and separated by the previously described 7-bp spacer (molecule A is bound to P5, and molecule B is bound to P6). The N-terminal residues (i.e., 131 to 133 within molecule A and 131 to 134 within molecule B) and the C-terminal residues (i.e., 258 to 260 within molecule A and 257 to 260 within molecule B) were not visible in the electron density, and therefore they are not included in the final model. Side chain electron density was also missing from several lysines (i.e., K136 in molecule A and K136, K167, K214, and K228 in molecule B). Therefore, in the final model (Fig. 2A), the side chains of these residues were truncated. The bound T-ag OBDs are on the same face of the site I DNA and are oriented in a head-to-tail fashion. This orientation is in contrast to that of our previous structure of a T-ag OBD pair on a DNA derivative of site II (i.e., on P1 and P3), wherein the GAGGC sequences were oriented in a head-to-head fashion (RCSB PDB ID 2NTC). As a result, when OBDs are bound to site I, the N terminus of each OBD is distal to the core origin, while the C terminus is proximal.
Table 1.
X-ray data collection and refinement statistics for the T-ag OBD–site I costructure
| Parameter | Value(s) for SV40 OBD–site Ia |
|---|---|
| Data collection statistics (λ = 0.9792 Å) | |
| Space group | C2221 |
| Cell dimensions | |
| a, b, c (Å) | 58.604, 139.672, 132.112 |
| α, β, γ (°) | 90.000, 90.000, 90.000 |
| Resolution (Å) | 47.99–3.2 (3.28–3.2) |
| Rsym or Rmerge | 10.60 (41.2) |
| I /σI | 31.49 (7.36) |
| Completeness (%) | 100.0 (100.0) |
| Redundancy | 16.1 (14.5) |
| Refinement statistics | |
| Resolution (Å) | 3.2 |
| No. of reflections | 8,841 |
| Rwork/Rfree | 18.76/26.74 |
| No. of: | |
| All atoms | 2,966 |
| Protein molecules | 2,963 |
| Water molecules | 3 |
| B-factors (all) | 137.1 |
| RMSDs | |
| Bond length (Å) | 0.011 |
| Bond angle (°) | 1.767 |
Values in parentheses are for the highest-resolution shell.
Fig 2.
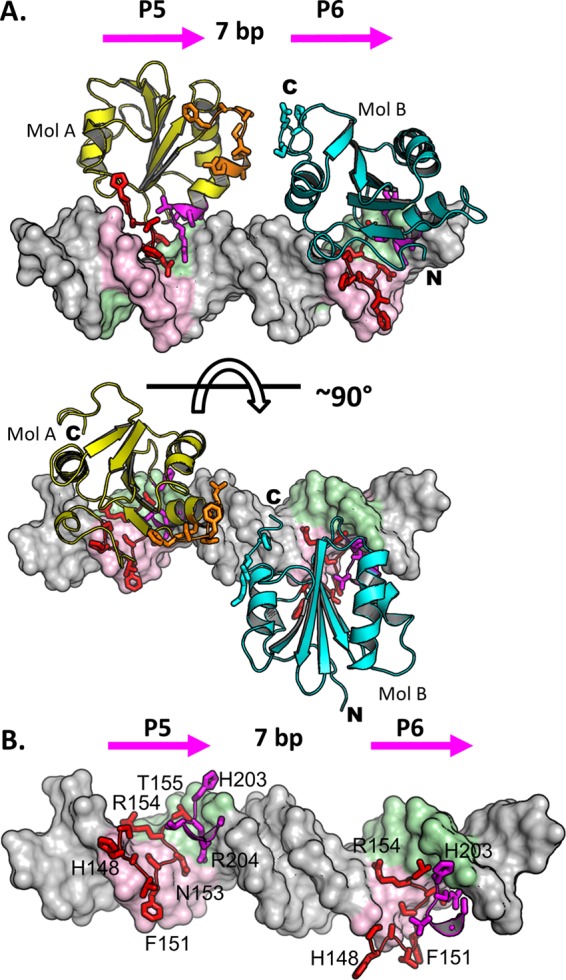
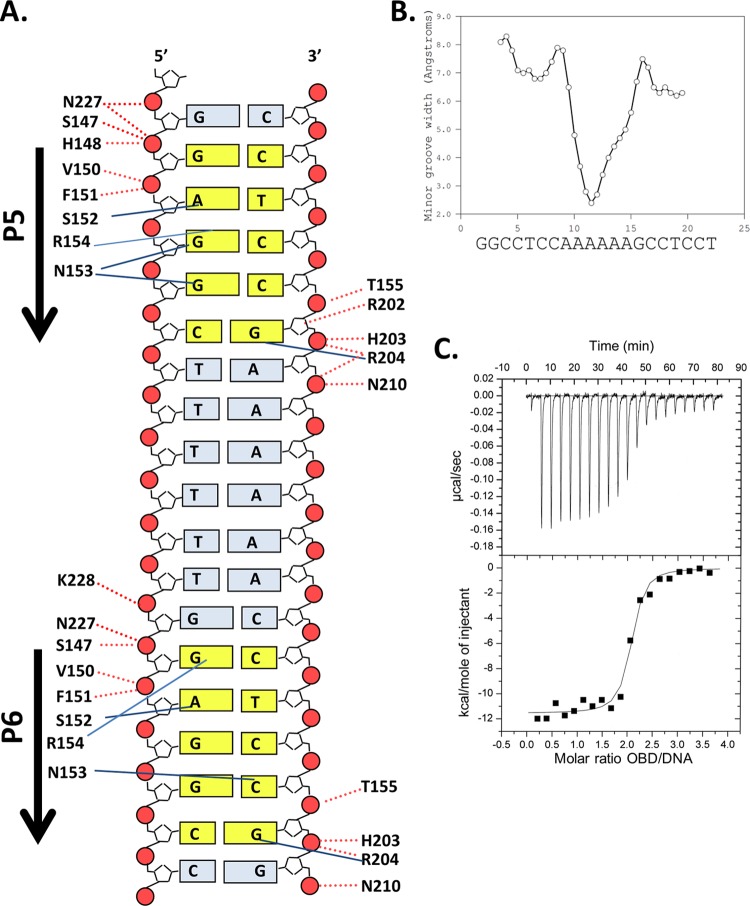
Costructure of the T-ag OBD bound to site I. (A, top) Ribbon diagrams of the two T-ag OBDs (yellow and cyan) bound to the site I oligonucleotide (shown as a surface representation). Pink arrows indicate the orientations of the GAGGC sequences; P5 and P6 are labeled. The GAGGC sequences are pink, and their complements are light green. The DNA-binding A1 (aa 147 to 159) and B2 (aa 203 to 207) motifs are shown in red. The N and C termini of the OBDs are labeled when visible. The B3 residues (aa 213 to 220) of the OBD bound to P5 are orange and shown as sticks. The C-terminal residues of the OBD bound to P6 are light blue and shown as sticks. (Bottom) Same as in panel A, but the view is rotated 90 degrees. Mol A and Mol B, molecules A and B, respectively. (B) Closeup of the protein-DNA interactions in which residues in the A1 and B2 motifs are selectively depicted. Residues in A1 making site-specific contacts with the GAGGCs include N153, R154, and T155. Residues in B2 making site-specific contacts include H203 and R204.
In the site I costructure, superposition of T-ag OBD molecule A upon molecule B requires a rotation of ∼60° and a translation of approximately 30 Å (data not shown). The superposition of the two T-ag OBDs revealed that they have nearly identical structures, as indicated by a low root mean square deviation (RMSD) (0.72 Å over 115 Cαs). Additional notable features of the costructure include the following.
(b) Protein-protein interactions. The crystal structure indicates that the OBDs bound to site I do not interact (Fig. 2A). However, a region of the OBD bound to P5 (i.e., residues 214 to 218) is juxtaposed next to the C-terminal portion of the OBD bound to pentamer P6 (i.e., residues 253 to 256). (The closest point of interaction [∼4.2 Å] is between Asp 256 and Lys 214). Of interest, Thr 217 and Phe 218 are among the residues comprising the B3 motif (72, 73) (aa 213 to 218). The B3 motif is a flexible region of the T-ag OBD, and we previously proposed that this flexibility contributes to the plasticity of T-ag as it transits to larger complexes during oligomerization (34). Consistent with this proposal, T-ags containing mutations in the B3 regions are known to be defective in oligomerization (reviewed in reference 30). Thus, in the context of full-length T-ag, or under solution conditions, the OBDs bound to P5 and P6 may interact.
(c) Protein-DNA interactions. As in the previously described T-ag OBD–site II costructures (24, 26), the DNA-binding motifs (A1 and B2) interact deep in the major groove. A closeup view of the residues in the A1 and B2 motifs interacting with the major groove is presented in Fig. 2B. A schematic of the set of protein-DNA interactions involved in DNA binding, calculated using the program PDBSUM (71), is shown in Fig. 3A. A superposition of either OBD bound to a GAGGC on site I upon either OBD bound to a GAGGC in site II (PDB ID 2NTC [24]) established that they are virtually identical (RMSDs = 0.49 to 0.73 over 115 Cαs for all possible combinations of OBD pairs). Thus, as one might predict, the network of protein-DNA interactions between the OBD and the GAGGCs are nearly identical to those observed in the previous T-ag OBD–site II DNA structure (e.g., RCSB PDB ID 2NTC). However, differences are observed in the contacts, most of which can be attributed to the lower resolution of this structure. Nevertheless, it is apparent that the A1 motif makes preferential interaction with one strand and that the B2 motif interacts with the opposite strand.
Fig 3.
Set of interactions between the T-ag OBD and site I and related biophysical data. (A) Schematic of the protein-DNA interactions. Dashed red lines indicate interactions with the phosphate backbone; solid blue lines indicate base-specific interactions. Residues from the A1 motif extend between residues 147 and 159, while those from the B2 motif extend between residues 203 and 207. (B) Analysis of DNA structure within the complex. The minor groove width is shown on the y axis; the DNA sequence is presented on the x axis. (C) ITC-based measurement of the thermodynamic parameters for the interaction of the T-ag OBD with site I. The 28-bp duplex DNA used in these experiments, used at a concentration of ∼1.5 μM, is presented in Fig. 1C. Titration of the T-ag OBD into the site I-containing oligonucleotide took place at 25°C in reaction buffer (Materials and Methods). The protein concentration in the syringe was ∼37.5 μM. The actual calorimetric trace is shown in the top panel. The y axis of the isotherm is power in μcal/s; the x axis is time in minutes. The stoichiometry and association constants were determined from curve fitting the integrated calorimetric trace presented in the bottom panel.
(d) DNA distortions. The duplex DNA forms a continuous pseudohelix in the crystal lattice. The conformation of the DNA is basically B-form. Analysis of each of the GAGGC duplexes, measured using the program CURVES+ (69), revealed consistent structural distortions (data not shown), such as a low helical twist and a low helical rise in the center of the sequence (20° and 2.6 Å, respectively). These structural distortions were also observed in previous OBD-GAGGC-containing structures (see, e.g., reference 24). There is also a small kink on either side of the A-tract. Furthermore, there is an overall curvature of the helical axis of 10.2 degrees; this bend decreases to 4.3 degrees in the A-tract region. A compression of the minor groove between the T-ag OBDs is also observed (Fig. 3B). Owing to these distortions, the T-ag OBDs are positioned closer together than they would be on perfectly straight DNA. A compression of the minor groove was also observed in the costructure of the OBD bound to pentanucleotides 1 and 3 in site II (24).
(e) Comparison with apo T-ag OBD. Structures of the T-ag OBD in the absence of DNA are also available (34, 74). Comparison of the apo form (RCSB PDB ID 2FUF) to the OBD structure when it is bound to the GAGGCs in site I revealed that the main conformational shift occurs in the DNA-binding region termed the A1 loop. In the present structure, the DNA-binding A1 motif is in the bound state (i.e., extended away from the protein core), a conformation seen previously in other T-ag OBD–GAGGC costructures (24; reviewed in reference 13).
(ii) ITC studies of the interaction of the T-ag OBD with site I.
It was previously reported that full-length T-ag binds site I approximately 10 times more tightly than site II [e.g., site I dissociation constant (Kd) = 0.7 nM; site II Kd = 5 nM (75)]. Therefore, we investigated whether this relationship extends to the isolated T-ag OBD. To conduct these studies, we used the technique of isothermal titration calorimetry (ITC), which allows for determination of both the stoichiometry of binding and the binding affinity of the components. The data (Fig. 3C) show that two T-ag OBDs bind to the 2 pentamers in site I with a Kd of ∼12.3 nM. This Kd is ∼8-fold tighter than that determined from the recently reported ITC data of the T-ag OBD bound to a 4-GAGGC-containing site II DNA target (Kd = 93.5 nM) (76). Thus, the tighter affinity of full-length T-ag for site I than for site II is largely a function of the OBD-DNA interactions.
(iii) A single hexamer of T-ag assembles on site I.
(a) EMSAs establish that T-ag forms single hexamers on site I. To continue the characterization of T-ag assembly on site I, EMSAs were conducted using full-length T-ag and adenylyl imidodiphosphate (AMP-PNP) (Fig. 4A). The site I-containing 59-bp oligonucleotide used in these experiments is shown in Fig. 1D. As a positive control, experiments were conducted with an oligonucleotide containing the 64-bp core origin (Fig. 1E). T-ag forms both single and double hexamers on this substrate (Fig. 4A, lanes 5 to 6). In contrast, T-ag oligomerization on the 59-bp site I DNA-containing substrate is limited to hexamer formation (lanes 2 to 3). (There is a trace amount of a lower-molecular-weight species [lanes 2 and 3] that is presumed to be an assembly intermediate). These studies establish that T-ag assembly on this site I-containing oligonucleotide is limited to formation of a single hexamer.
Fig 4.
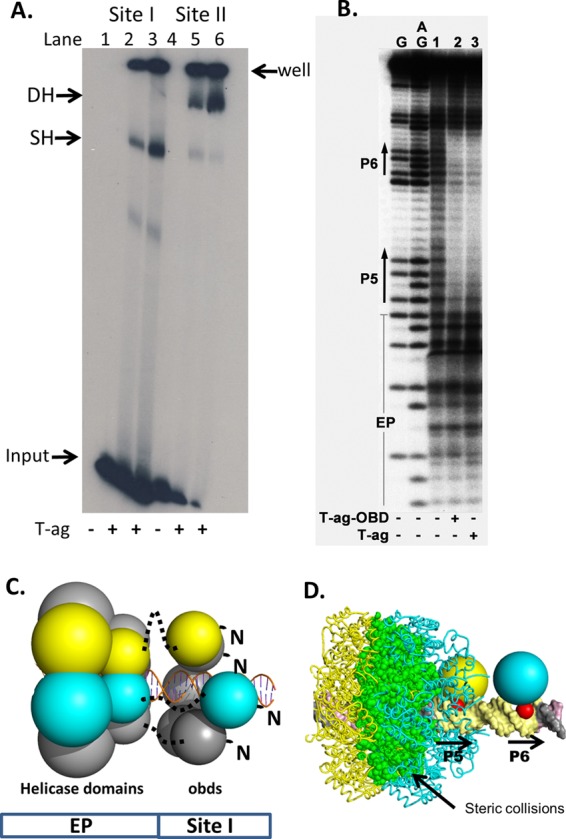
Full-length T-ag forms single hexamers on site I-containing DNA. (A) EMSA of full-length SV40 T-ag bound to a 59-bp oligonucleotide containing site I (lanes 2 and 3) or the 64-bp core origin oligonucleotide (lanes 5 and 6). The presence (+) or absence (−) of T-ag is indicated at the bottom. Lanes 2 and 5, 1.5 pmol T-ag; lanes 3 and 6, 3 pmol T-ag. The reaction products include single hexamers (SH) and double hexamers (DH). The DNA that did not enter the gel is labeled “well,” and the unbound DNA substrates are labeled “input.” (B) In situ footprinting of full-length T-ag and the OBD, when complexed to site I. The footprints were obtained using the gel retardation 1,10-phenanthroline–copper ion footprinting technique (57). The initial EMSAs were conducted with the previously described 47-bp site I + wt 30 oligonucleotide (50). Free DNA (i.e., DNA obtained from reactions conducted in the absence of protein and used as a control) is presented in lane 1. The locations of sequence features, including P5 and P6, are indicated. (C) Structure-based modeling of hexamers of the T-ag helicase and OBD domains on site I. The OBDs initially bound to P5 and P6 are shown as yellow and cyan, respectively. The OBDs are represented by single spheres, which are centered at the geometric center of mass, and the radius is approximately the radius of gyration of the domain (i.e., ∼17.5 Å). The helicase domains are represented by 2 spheres; those helicase domains connected to the initially bound OBDs are also shown as yellow and cyan. Dotted black lines represent the flexible linkers connecting the N terminus of the helicase domains to the C terminus of the OBDs. Finally, an idealized three-dimensional model of site I DNA, positioned along the 6-fold screw axis of the OBD spiral, is shown as a ribbon representation. (D) Molecular modeling studies indicating that two independent T-ag hexamers cannot form on P5 and P6. Two models of T-ag hexamers were constructed; one nucleated at P5 and one at P6. In the resulting model, significant collisions occur between the helicase domains (the collisions are shown in green).
EMSAs were also previously conducted with the shorter 47-bp site I + wt 30 oligonucleotide and both T-ag and the OBD (50). The products formed in these EMSA reactions were footprinted using the 1,10-phenanthroline–copper footprinting technique (57). These data were not previously published; therefore, a representative footprint is shown in Fig. 4B. The product formed in the reaction conducted with full-length T-ag and the site I + wt 30 oligonucleotide is presented in lane 3. The protected region starts at the 5′ end of P5, extends through the poly(dT)·poly(dA) spacer, and terminates at the 3′ end of P6. The footprint that resulted when the experiment was repeated with the purified T-ag OBD is presented in lane 2. It is apparent that the same ∼18-nucleotide (nt) footprint is formed by both T-ag and the OBD. Of interest, the same-sized footprint was generated when T-ag or the OBD was footprinted on a site II-containing oligonucleotide (38, 50). The significance of these observations is considered in Discussion. Finally, it is noted that the T-ag footprint does not extend into the flanking regions. We previously attributed this pattern to the routing of ssDNA over the helicase domain, where it would be subject to cleavage by oxygen radicals (21, 24).
(b) Molecular modeling of hexamer formation. It is possible to generate structural models for full-length T-antigen assembled on site I using the available crystal structures (i.e., the structures of the T-ag OBD [aa 131 to 260] bound to site I and the crystal structure of the T-ag helicase domain [aa 266 to 627] [31]). A schematic of a hexamer bound to site I, based on these studies, is shown in Fig. 4C. (The N-terminal J domain is omitted from this model since it is not necessary for replication in vitro [77, 78].) The OBD and helicase domains are connected via a flexible hinge (approximately aa 250 to 266, shown as a dotted line). Additional molecular modeling studies indicate that extensive molecular clashes would prevent two independent hexamers from forming on P5 and P6 (Fig. 4D). Thus, the modeling studies support the conclusion that T-ag binding to site I results in the formation of a single hexamer. Finally, the C termini of the OBDs bound to site I are proximal to the EP region (Fig. 2A); this indicates that the helicase domains are positioned over the EP region.
(iv) Crystal-structure-based modeling suggests that during hexamer formation on site I, the T-ag OBDs form a spiral structure.
Given that T-ag assembly on site I is limited to the formation of single hexamers, the OBDs bound to P5 and P6 must participate in the assembly of this oligomer. Therefore, we considered how the OBDs bound to these pentanucleotides might participate in hexamer formation. In particular, we compared the site I costructure with an additional structure that is adopted by the SV40 T-ag OBD, the apo OBD lock washer structure (RCSB PDB ID 2FUF).
A depiction of the OBD-site I costructure is presented in Fig. 5 (top). Initially, we examined the spatial relationship between the OBDs bound to site I. Superimposition of monomer A at P5 upon monomer B at P6 requires a linear translation of approximately 31 Å and a rotation of 60°. The previously reported left-handed OBD spiral (34) is presented at the bottom of Fig. 5. Of interest, OBD subunits a and f, the OBD pair proximal to the spiral gap, are separated by 29.6 Å and offset by an angle of 60°. Thus, the OBDs bound to P5 and P6 have a spatial relationship that is very similar to that of subunits a and f of the left-handed spiral structure.
Fig 5.
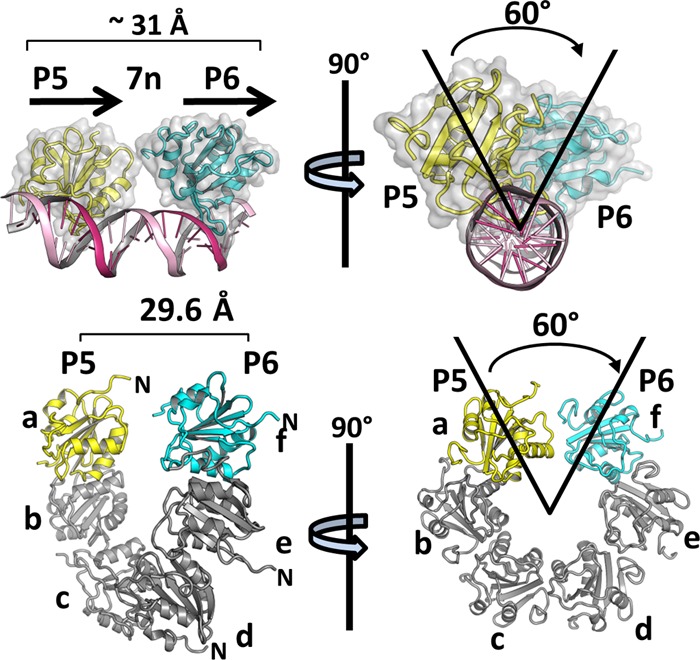
Spatial correlations between the T-ag OBDs site-specifically bound to pairs of GAGGC pentanucleotides, with OBD pairs within the left-handed spiral. (Top) Two views (rotated by ∼90°) of the OBDs bound to site I are presented as translucent surface representations; the DNA is shown as a ribbon diagram. Values describing the relationships between the OBDs (i.e., the translation and angular rotation) are indicated. (Bottom) Two views of the left-handed spiral structure of the T-ag OBD rotated by ∼90° (the subunits are labeled a to f). Spiral subunit a is yellow, while subunit f is cyan; they are offset by the same angle that separates the OBDs bound to P5 and P6 (i.e., spiral subunits a and f are also 60 degrees apart).
A simplified rendering of the OBD-site I costructure, which further highlights its relationship to the spiral, is presented in Fig. 6A. In this figure, the DNA-binding A1 and B2 loops are depicted as red spheres. Inspection of the two structures presented in Fig. 6A (top and bottom) reveals that the positions of the A1 and B2 loops are significantly different. Indeed, in order for the OBDs bound to P5 and P6 to become subunits a and f of the spiral, the bound OBDs must rotate as well as translate away from the DNA (movements modeled in Fig. 6B). Nevertheless, it is apparent that a spatial relationship exists between the OBDs bound to P5 and P6 and spiral subunits a and f.
Fig 6.
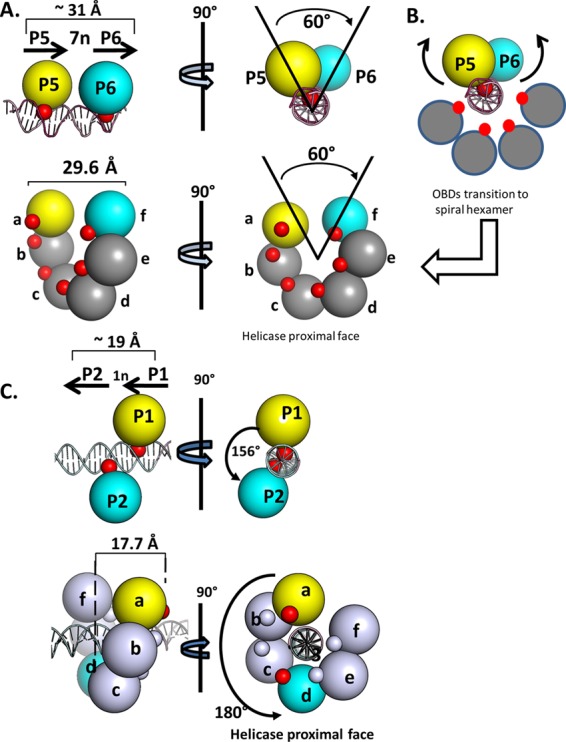
Simplified renderings of the OBD interactions with pairs of pentanucleotides. (A, top) Two views of the OBD interactions with site I. The OBDs are represented by spheres centered at the geometric center of mass. The T-ag OBDs are yellow on P5 and cyan on P6. The smaller red spheres represent the DNA-binding A1 and B2 motifs. (Bottom) Rendering of the left-handed spiral in which the OBDs proximal to the gap are represented by spheres colored as described above. This depiction serves to further illustrate that spiral subunits a and f have the same spatial relationship as the OBDs bound to P5 and P6. As in the top images, the red spheres indicate the relative locations of the A1 and B2 motifs. (B) Modeling studies indicate that the OBDs initially bound to P5 (yellow) and P6 (cyan) undergo a significant transition during spiral formation. This involves rearrangement of the A1 loop from the bound to the free form, subsequent rotation and translation away from the major groove of the dsDNA, and interaction(s) with other OBDs (reviewed in reference 13). (C, top) Two views of the OBD interactions with site II, rotated by ∼90°, used to illustrate the generality of the relationship (24, 26) (RCSB PDB ID 2ITL). P1 and P2 are separated by a 1-bp spacer; therefore, the OBDs bound to P1 (yellow) and P2 (cyan) are ∼156 degrees apart and separated by a translation of ∼19 Å. (As in previous examples, the DNA-binding A1 and B2 motifs are red and magenta, respectively). (Bottom left) Side view of the left-handed OBD spiral hexamer with DNA modeled within the central channel (the 6 subunits are labeled a to f). It is apparent that in this instance, spiral subunits a and d (yellow and cyan, respectively) have a spatial relationship that is analogous to that of the OBD subunits bound to P1 and P2. (Bottom right) Helicase-proximal view of the left-handed spiral structure with duplex DNA modeled in the central channel.
(v) Modeling spiral formation on a pentanucleotide pair having a 1-bp spacer.
We previously proposed (24) that the OBDs bound to pentanucleotides 1 and 2 in site II, a GAGGC pair separated by 1 base pair, could also assemble via a spiral. In this instance, superimposition of the monomer bound to P1 upon the monomer bound to P2 requires a linear translation of approximately 19 Å and a rotation of 156° (reproduced in Fig. 6C, top). As shown in Fig. 6C (bottom), spiral subunits a and d have a very similar spatial relationship. Therefore, we suggested that the OBDs bound to P1 and P2 become subunits a and d of the spiral in a T-ag hexamer (24). Moreover, as with spiral formation on site I (Fig. 6A), the relative positions of the A1 and B2 loops (symbolized by the red spheres) indicate that the initially bound OBDs must rotate as well as translate away from the pentanucleotides during spiral formation.
Based on these structure-based modeling studies, it is concluded that the left-handed spiral can accommodate OBD pairs bound to GAGGC pentanucleotides separated by either 1 or 7 base pairs. This indicates that by simply utilizing different pairs of OBD subunits, the left-handed OBD spiral can accommodate all of the GAGGC pairs, arranged in a head-to-tail manner, present in the SV40 regulatory region. Thus, the helical properties of the DNA appear to be correlated to the helical properties of the OBD spiral.
DISCUSSION
The cocrystal structure presented herein has significantly increased our understanding of SV40 T-ag's interactions with site I. As predicted by methylation interference studies (see, e.g., references 79 and 80), the two T-ag OBDs contact the major groove of the GAGGC sequences. Moreover, the specific interactions between the OBDs and the pentanucleotides in site I are very similar to those seen in previous T-ag OBD–GAGGC crystal structures (24, 26). Limited contacts with the phosphate backbone of the poly(dT)·poly(dA) spacer region were also observed (Fig. 3A, via K228, N210, and R204). These backbone interactions were, however, also observed in previous studies of the T-ag OBD–GAGGC interaction (24, 81). Therefore, these findings are consistent with the proposal that the site I spacer does not function with the pentanucleotides to form an extended recognition motif (49) and that T-ag has limited interactions with the bases in the poly(dT)·poly(dA) spacer (17, 79, 82).
A curve with a single binding constant fits the ITC data for the T-ag OBD–site I interaction; therefore, this result is consistent with those of previous studies indicating that P5 and P6 contribute to T-ag binding to roughly the same extent (82). The affinity of the T-ag OBD for site I is ∼8-fold greater than that for site II. Regarding the molecular basis for this tighter interaction, DNA bending is known to occur at adenine-thymine tracts (83). Indeed, our costructure demonstrates that there is a slight bend in the site I-containing DNA (∼10 degrees) and that the minor groove in the adenine-thymine tract is compressed. It is noted, however, that A-tract-containing DNA crystal structures (PDB IDs 1BDN, 1D98, and 1D89), solved in the absence of binding proteins, exhibit related features (e.g., little bending of the A-tract [2.4 to 3.0 degrees] and a narrowing of the minor groove in the A-tract region [3.4 to 3.5 Å]) (reference 84 and references therein). In the current costructure, the DNA bends 4.3 degrees in the A-tract region, and the minor groove compresses to a minimum value of 2.4 Å in the center of the A-tract. Although the free A-tract DNA structures exhibit similar distortions, they are more pronounced in the OBD-site I costructure. Thus, the intrinsic structural features of the A-tract may contribute to the high-affinity binding of the OBD to site I, and conversely, OBD binding may increase the structural perturbations.
A model for hexamer-dependent spiral formation.
Based on our EMSAs, it was concluded that only a single hexamer forms on site I, a hypothesis that is supported by our structure-based modeling studies. Therefore, since T-ag assembly is limited to hexamer formation, it was of interest to consider how the OBDs bound to P5 and P6 contribute to hexamer formation. As shown in Fig. 5, the OBDs docked to P5 and P6 have a spatial relationship similar to that of subunits a and f in the left-handed spiral structure. These and related observations have led to a model that accounts for the roles of pentanucleotide pairs, arranged in a head-to-tail manner, in T-ag assembly (Fig. 7, with site I as the example).
Fig 7.
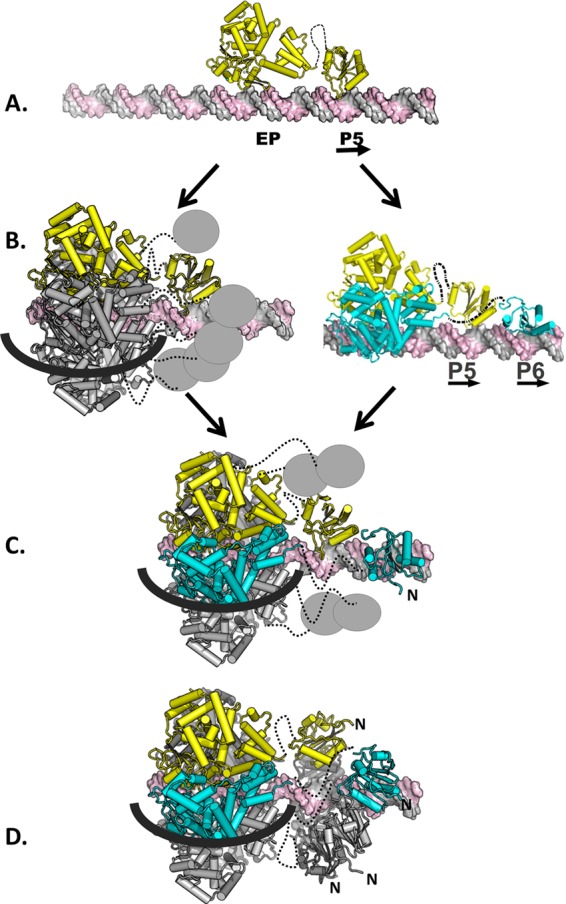
Model depicting hexamer-dependent OBD spiral formation on site I. (A) Initially, a monomer of T-ag, shown in yellow, binds to site I (the J-domain is not shown). The flexible linker that connects the two domains is shown as a dashed line. (B) Intermediates in hexamer formation. (Left) Following the binding of the initial monomer, additional T-ag molecules are recruited to site I owing to the high affinity of the helicase domains for each other. Upon recruitment, the helicase domains assemble into a hexamer (18). The five newly recruited monomers are shown in gray; the OBDs are symbolized by the small gray spheres. The black line represents one strand of DNA going over the outer surface of the helicase domain; the second strand is routed through the central channel. (Right) An alternative possibility is that a second monomer of T-ag binds to the second pentanucleotide in site I. (C) Formation of an immature hexamer. In this intermediate, the helicase domains have formed a hexamer over the flanking region, and P5 and P6 are bound by two of the OBDs. The four unbound OBDs are shown as gray spheres. The routing of the DNA is as described above. (D) Maturation of the hexamer to form the spiral structure. It is proposed that the four free OBDs shown in panel C interact with the OBDs bound to P5 and P6. As a result of this interaction, the OBDs adopt the left-handed spiral structure. (A left-handed helix is defined as turning in a counterclockwise fashion as it progresses forward. The N termini of the OBDs in the spiral are indicated.) Once formed, the spiral has a gap though which ssDNA may pass. Finally, it is noted that the isolated T-ag OBDs are monomeric in solution even at high concentrations (74). This is one indication that the protein-protein interactions that occur in the OBD spiral are not strong; therefore, the spiral structure is presumably dynamic in nature.
Hexamer formation is initiated (Fig. 7A) when the OBD on a T-ag monomer interacts with a single pentanucleotide (P5 used in this example), using the repertoire of bonds reported herein (Fig. 3A). One consequence of site-specific binding is that the helicase domain is positioned over the adjacent EP region (e.g., Fig. 4C). Regarding the next step, recall that single pentanucleotides are known to support the formation of full-length T-ag hexamers (see, e.g., references 37, 38, and 40). Therefore, it is proposed that, following monomer binding, five additional T-ag molecules are recruited, a reflection of the high affinity of helicase domains for each other (31). The product formed in this step would contain the helicase domains assembled into a hexamer and five unbound OBDs (Fig. 7B, left). Based on previous studies, it is predicted that one strand of DNA goes through the central channel, while the second traverses the outer surface of the helicase domain (21, 24, 85). An alternative possibility is, however, that a second OBD binds to the second pentanucleotide in site I, resulting in the formation of a T-ag dimer (Fig. 7B, right). In either case, it is proposed that the next intermediate has the helicase domains oligomerized into a hexamer, with the DNA routed as described above, and two OBDs bound to pentanucleotides P5 and P6 (Fig. 7C). This intermediate would have four unbound OBDs; their local concentration would, however, be increased. According to the spiral hypothesis, left-handed spiral formation is nucleated by subsequent interactions between the four unbound OBDs with the OBDs associated with P5 and P6 (Fig. 7D). The protein-protein interactions needed for spiral formation may include those in the previously described OBD-OBD interface (34).
Spiral formation is incompatible with OBDs that are site-specifically bound to DNA (13) (Fig. 6B). Therefore, it is proposed that spiral assembly is coupled to the disruption of site-specific binding to P5 and P6, perhaps as a result of the structural distortions of the flanking sequences stemming from helicase assembly. Consistent with this possibility, Gutierrez et al. reported that T-ag binding to site I stimulated DNA unwinding (86). Once hexamer assembly is completed, the OBDs assemble into the previously discussed left-handed spiral. The left-handed direction of this quaternary structure is opposite to that of right-handed B-form DNA. Therefore, one consequence of left-handed spiral formation by the OBD may be to promote DNA melting, a process in which the OBDs are known to play a role (87). Characterization of the left-handed conformation of the MCM2-MCM7 complex also led to the suggestion that this conformation is adopted to promote DNA melting (88).
In support of the spiral hypothesis, Cuesta et al. (36) analyzed the T-ag fragment from aa 108 to 627 assembled on the SV40 origin via electron microscopy (EM) and concluded that the OBD forms a spiral. In addition, the footprint formed by either T-ag or the OBD on the 47-bp site I + wt 30 oligonucleotide was determined; these studies revealed that both T-ag and the OBD form a footprint that extends over ∼18 nt (∼ 61.2 Å) (Fig. 4B). The diameter of an OBD subunit is ∼35 Å (see, e.g., reference 24); thus, if the OBDs in T-ag form a single closed or planar ring on site I, the complex would form a footprint of approximately 11 bp (∼35.2 Å). Therefore, the footprinting studies are consistent with the hypothesis that upon oligomerization on site I, the OBDs within T-ag adopt a structure that extends between P5 and P6.
Implications of the spiral assembly hypothesis to OBD interactions with site II.
A single OBD spiral can accommodate the OBDs bound to pentanucleotides P1 and P2 (or P3 and P4) in site II. Of further interest, the pentanucleotide containing regulatory regions of polyomaviruses are highly conserved (see, e.g., reference 89). Therefore, the spiral assembly model may be generally applicable to the assembly of hexamers on polyomavirus origins of replication. However, since the spiral formed by the SV40 OBD is the only one reported to date (34, 35), this hypothesis remains to be verified.
Furthermore, given that site II contains four pentanucleotides, an interesting question is, in the context of a double hexamer assembled on the core origin, are two OBD spirals formed? Molecular modeling demonstrates that a double spiral of the OBD fits into the central region of electron density derived from EM images of T-ag double hexamers (Fig. 8A). Indeed, we previously proposed (34) the relative orientations of the double spiral formed on site II based on earlier mutagenesis studies (73). Moreover, the model presented in Fig. 8B demonstrates that a double OBD spiral may explain the ∼18-nt footprint generated by double-hexamer formation on site II (38). However, phenanthroline-copper footprinting studies of T-ag double hexamers revealed that pentanucleotide 4 is not protected from cleavage by oxygen radicals (38). Thus, in the context of a T-ag double hexamer, all four pentanucleotides may not be simultaneously bound by OBDs. Therefore, it is uncertain if site II supports the concurrent formation of two OBD spirals. Consistent with double-spiral plasticity on site II, EM studies indicate that wild-type T-ag bound to the core origin is present in two conformations; in one, the central module is an open ring with a gap, while in the second, the central module is a compact closed ring (36). Furthermore, the formation of both open and closed OBD rings may be related to previous biochemical experiments indicating that a feature of the two hexamers formed on site II is asymmetry. (For instance, one hexamer forms in a manner that is independent of the phosphorylation of Thr 124, while formation of the second hexamer is enhanced by Thr 124 phosphorylation (reviewed in reference 37); additional examples of asymmetric assembly of T-ag hexamers are presented in references 22 and 25). Finally, asymmetric assembly events involving the OBD (i.e., formation of single spirals) may help to explain related observations, such as the recent finding that only three pentanucleotides are needed for the assembly of the Merkel cell T-antigen (76).
Fig 8.
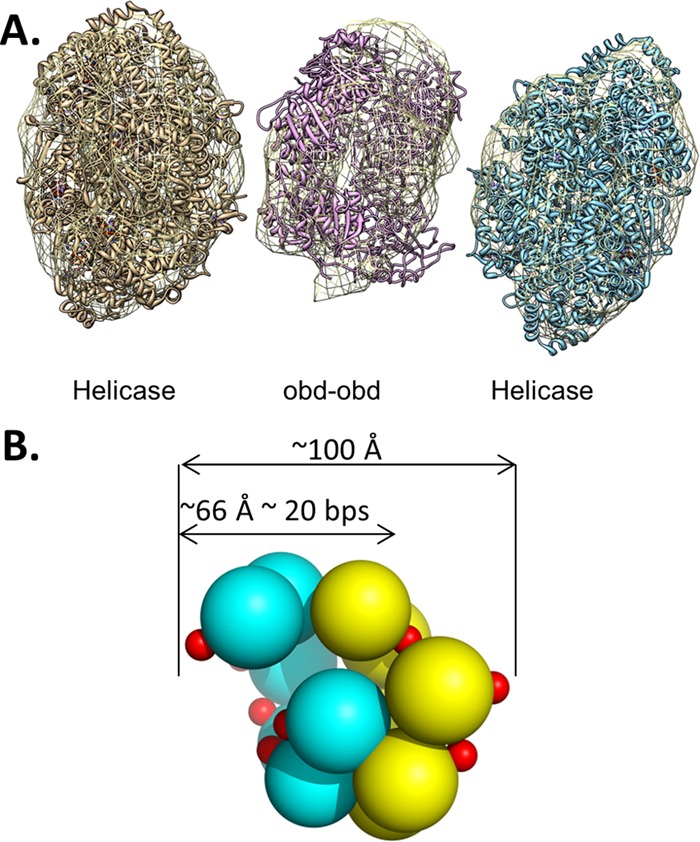
OBD oligomerization in the context of T-ag double hexamers formed on the core origin. (A) The PDB coordinates for the helicase domain (1SVM) and the OBD (2FUF) were fit into the EM density map (EMD-1681 [36]). The figure was generated using the program CHIMERA (103); the helicase domains were fit using the CHIMERA fit-in-map option. A double-hexamer spiral of the OBDs, shown as a ribbon representation, was generated and manually positioned into the central density. (B) Model of a T-ag OBD double spiral. The individual spirals are shown in yellow and cyan. The smaller arrow illustrates that its diameter (∼66 Å) is similar to the area protected in previous footprinting studies (see, e.g., reference 38).
Considerations regarding T-antigen's dynamic interactions with site I and site II.
The shifting of T-ag hexamers from site I to site II is coupled to the repression of transcription of the early region and the initiation of DNA replication (see, e.g., references 45, 46, 90, and 91). Given the importance of these events, an interesting question is what causes the transition from binding to site I to site II? Previous studies using full-length T-ag established that one of the key determinants is phosphorylation of T-ag, particularly at two clusters located at the N and C termini of the molecule (92, 93). How these phosphorylation events regulate the interaction(s) between the OBD and the pentanucleotides in site I and site II has yet to be determined. However, a prediction of the assembly events described herein is that a hexamer formed on site I would block hexamer formation on the proximal side of site II. As shown in Fig. 9, collisions would occur between the opposing helicase domains. This prediction is consistent with the results of previous studies that demonstrated that T-ag assembly on site I precedes its assembly on site II (see, e.g., reference 45).
Fig 9.
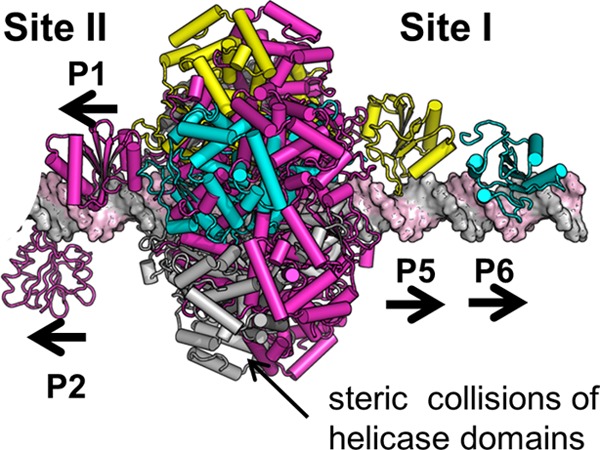
T-ag hexamers cannot exist simultaneously on site I and the proximal side of site II. As reported herein, a T-ag hexamer nucleated through the OBDs bound at P5 and P6 positions the helicase domains over the EP region. Furthermore, when T-ag assembles into a double hexamer on site II, the helicase domains span both the AT-rich and EP regions (19). Therefore, if both site I and the EP-proximal side of site II were occupied by T-ag hexamers, the helicase domains would completely overlap and suffer steric clashes.
Initiator proteins and spiral formation.
Helical protein filaments are known to be formed by other replication initiators (reviewed in reference 94). For instance, EM studies provided evidence for a helical filament in the Drosophila melanogaster origin recognition complex (ORC) (95). As previously noted, the left-handed lock washer structure is also a feature of certain MCM complexes (88) and MCM complexes contain gaps through which DNA may pass (96–98). Moreover, the prokaryotic initiator DnaA forms a right-handed superhelix that orients DNA on the outside of the helical assembly (99, 100). Importantly, the protein filament formed by DnaA provides a simple explanation for the heterogeneous arrangement of DnaA boxes found in bacterial origins (100). Reasons for suggesting that the spiral formed by the T-ag OBD may also explain the heterogeneous distribution of pentanucleotides in polyomavirus origins of replication were previously discussed. An important difference between DnaA and T-ag is, however, that the AAA+ domain is the primary determinant of filament formation in DnaA, whereas the available evidence indicates that the OBD is the primary unit of spiral formation in T-ag. Thus, a universal feature of all cellular initiator proteins may be spiral formation (100), but spiral formation may be a property of different initiator domains. It is also important to note that the hexameric helicase DnaB adopts a closed spiral staircase quaternary structure during translocation (101). This observation has led to the proposal of the hand-over-hand mechanism for DNA helicase activity. If the spiral structure of the T-ag OBD is shown to be a feature of the replication fork, it will be of interest to determine if the OBDs are engaged in a similar hand-over-hand mechanism.
Finally, it is noted that other proteins known to form open-ring spirals have been suggested to recruit protein substrates in a spiral-dependent manner (e.g., DnaA [100] and the ParR protein involved in DNA segregation [reviewed in reference 102]). Therefore, it will be of interest to determine if spiral assembly by the T-ag OBDs is related to the recruitment of the cellular factors needed to initiate viral DNA replication.
ACKNOWLEDGMENTS
We thank Howard Robinson at the National Synchrotron Light Source, Brookhaven National Laboratory, for X-ray data collection at Beamline X29. We thank Jacques Archambault and Andrew Bohm for helpful discussions.
This work was supported by a grant from the National Institutes of Health to P.A.B. (9R01GM55397).
Footnotes
Published ahead of print 26 December 2012
REFERENCES
- 1. Jiang M, Abend JR, Johson SF, Imperiale MJ. 2009. The role of polyomaviruses in human disease. Virology 384:266–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eash S, Manley K, Gasparovic M, Querbes W, Atwood WJ. 2006. The human polyomaviruses. Cell. Mol. Life Sci. 63:865–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. 2008. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl. Acad. Sci. U. S. A. 105:16272–16277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Babakir-Mina M, Ciccozzi M, Bonifacio D, Bergeallo M, Costa C, Cavallo R, DiBonito L, Perno CF, Ciotti M. 2009. Identification of the novel KI and WU polyomaviruses in human tonsils. J. Clin. Virol. 46:75–79 [DOI] [PubMed] [Google Scholar]
- 6. Ahuja D, Saenz-Robles MT, Pipas JM. 2005. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 24:7729–7745 [DOI] [PubMed] [Google Scholar]
- 7. Hurwitz J, Dean FB, Kwong AD, Lee S-H. 1990. The in vitro replication of DNA containing the SV40 origin. J. Biol. Chem. 265:18043–18046 [PubMed] [Google Scholar]
- 8. Kelly TJ. 1988. SV40 DNA replication. J. Biol. Chem. 263:17889–17892 [PubMed] [Google Scholar]
- 9. Waga S, Stillman B. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721–751 [DOI] [PubMed] [Google Scholar]
- 10. Bullock PA. 1997. The initiation of simian virus 40 DNA replication in vitro. Crit. Rev. Biochem. Mol. Biol. 32:503–568 [DOI] [PubMed] [Google Scholar]
- 11. Fanning E, Knippers R. 1992. Structure and function of simian virus 40 large tumor antigen. Annu. Rev. Biochem. 61:55–85 [DOI] [PubMed] [Google Scholar]
- 12. Simmons DT. 2000. SV40 large T antigen functions in DNA replication and transformation. Adv. Virus Res. 55:75–134 [DOI] [PubMed] [Google Scholar]
- 13. Meinke G, Bullock PA. 2012. Structural “snap-shots” of the initiation of SV40 replication, p 195–215 In Gaston K. (ed), Small DNA tumor viruses. Horizon Scientific Press, Norwich, United Kingdom [Google Scholar]
- 14. Deb S, DeLucia AL, Baur C-P, Koff A, Tegtmeyer P. 1986. Domain structure of the simian virus 40 core origin of replication. Mol. Cell. Biol. 6:1663–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DeLucia AL, Lewton BA, Tjian R, Tegtmeyer P. 1983. Topography of simian virus 40 A protein-DNA complexes: arrangement of pentanucleotide interaction sites at the origin of replication. J. Virol. 46:143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Titolo S, Welchner E, White PW, Archambault J. 2003. Characterization of the DNA-binding properties of the origin-binding domain of SV40 large T antigen by fluorescence anisotropy. J. Virol. 77:5512–5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tjian R. 1978. Protein-DNA interactions at the origin of simian virus 40 DNA replication. Cold Spring Harbor Symp. Quant. Biol. 43:655–662 [DOI] [PubMed] [Google Scholar]
- 18. Mastrangelo IA, Hough PVC, Wall JS, Dodson M, Dean FB, Hurwitz J. 1989. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature 338:658–662 [DOI] [PubMed] [Google Scholar]
- 19. Valle M, Gruss C, Halmer L, Carazo JM, Donate LE. 2000. Large T-antigen double hexamers imaged at the simian virus 40 origin of replication. Mol. Cell. Biol. 20:34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gai D, Zhao R, Li D, Finkielstein CV, Chen XS. 2004. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119:47–60 [DOI] [PubMed] [Google Scholar]
- 21. Kumar A, Meinke G, Reese DK, Moine S, Phelan PJ, Fradet-Turcotte A, Archambault J, Bohm A, Bullock PA. 2007. Model for T-antigen-dependent melting of the simian virus 40 core origin based on studies of the interaction of the beta-hairpin with DNA. J. Virol. 81:4808–4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reese DK, Sreekumar KR, Bullock PA. 2004. Interactions required for binding of simian virus 40 T antigen to the viral origin and molecular modeling of initial assembly events. J. Virol. 78:2921–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shen J, Gai D, Patrick A, Greenleaf WB, Chen XS. 2005. The roles of the residues on the channel β-hairpin and loop structures of simian virus 40 hexameric helicase. Proc. Natl. Acad. Sci. U. S. A. 102:11248–11253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meinke G, Phelan PJ, Moine S, Bochkareva E, Bochkarev A, Bullock PA, Bohm A. 2007. The crystal structure of the SV40 T-antigen origin binding domain in complex with DNA. PLoS Biol. 5:e23 doi:10.1371/journal.pbio.0050023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Borowiec JA, Dean FB, Bullock PA, Hurwitz J. 1990. Binding and unwinding-how T antigen engages the SV40 origin of DNA replication. Cell 60:181–184 [DOI] [PubMed] [Google Scholar]
- 26. Bochkareva E, Martynowski D, Seitova A, Bochkarev A. 2006. Structure of the origin-binding domain of simian virus 40 large T antigen bound to DNA. EMBO J. 25:5961–5969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Simmons DT, Wun-Kim K, Young W. 1990. Identification of simian virus 40 T-antigen residues important for specific and nonspecific binding to DNA and for helicase activity. J. Virol. 64:4858–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wun-Kim K, Upson R, Young W, Melendy T, Stillman B, Simmons DT. 1993. The DNA-binding domain of simian virus 40 tumor antigen has multiple functions. J. Virol. 67:7608–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meinke G, Phelan PJ, Fradet-Turcotte A, Bohm A, Archambault J, Bullock PA. 2011. Structure-based analysis of the interaction between the simian virus 40 T-antigen origin binding domain and single-stranded DNA. J. Virol. 85:818–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reese DK, Meinke G, Kumar A, Moine S, Chen K, Sudmeier JL, Bachovchin W, Bohm A, Bullock PA. 2006. Analyses of the interaction between the origin binding domain from simian virus 40 T-antigen and single stranded DNA provides insights into DNA unwinding and initiation of DNA replication. J. Virol. 80:12248–12259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li D, Zhao R, Lilyestrom W, Gai D, Zhang R, DeCaprio JA, Fanning E, Jochimiak A, Szakonyi G, Chen XS. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423:512–518 [DOI] [PubMed] [Google Scholar]
- 32. Enemark EJ, Joshua-Tor L. 2006. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442:270–275 [DOI] [PubMed] [Google Scholar]
- 33. Brewster AS, Chen XS. 2010. Insights into the MCM functional mechanism: lessons learned from the archaeal MCM complex. Crit. Rev. Biochem. Mol. Biol. 45:243–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meinke G, Bullock PA, Bohm A. 2006. The crystal structure of the T-ag origin binding domain. J. Virol. 80:4304–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meinke G, Phelan P, Fradet-Turcotte A, Archambault J, Bullock PA. 2011. Structure-based design of a disulfide-linked oligomeric form of the simian virus 40 (SV40) large T antigen DNA-binding domain. Acta Crystallogr. D Biol. Crystallogr. 67:560–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cuesta I, Nunez-Ramirez R, Scheres SHW, Gai D, Chen XS, Fanning E, Carazo JM. 2010. Conformational rearrangements of SV40 large T antigen during early replication events. J. Mol. Biol. 397:1276–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barbaro BA, Sreekumar KR, Winters DR, Prack AE, Bullock PA. 2000. Phosphorylation of simian virus 40 T-antigen on Thr 124 selectively promotes double-hexamer formation on subfragments of the viral core origin. J. Virol. 74:8601–8613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Joo WS, Kim HY, Purviance JD, Sreekumar KR, Bullock PA. 1998. Assembly of T-antigen double hexamers on the simian virus 40 core origin requires only a subset of the available binding sites. Mol. Cell. Biol. 18:2677–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim HY, Barbaro BA, Joo WS, Prack A, Sreekumar KR, Bullock PA. 1999. Sequence requirements for the assembly of simian virus 40 T-antigen and T-antigen origin binding domain on the viral core origin of replication. J. Virol. 73:7543–7555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sreekumar KR, Prack AE, Winters DR, Barbaro BA, Bullock PA. 2000. The simian virus 40 core origin contains two separate sequence modules that support T-antigen double-hexamer assembly. J. Virol. 74:8589–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MAA, Dalianis T, Ramqvist T, Andersson B. 2007. Identification of a third human polyomavirus. J. Virol. 81:4130–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alwine JC, Reed SI, Stark GR. 1977. Characterization of the autoregulation of simian virus 40 gene A. J. Virol. 24:22–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hansen U, Tenen DG, Livingston DM, Sharp PA. 1981. T antigen repression of SV40 early transcription from two promoters. Cell 27:603–612 [DOI] [PubMed] [Google Scholar]
- 44. Khoury G, May E. 1977. Regulation of early and late simian virus 40 transcription: overproduction of early viral RNA in the absence of a functional T-antigen. J. Virol. 23:167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Myers RM, Rio DC, Robbins AK, Tjian R. 1981. SV40 gene expression is modulated by the cooperative binding of T antigen to DNA. Cell 25:373–384 [DOI] [PubMed] [Google Scholar]
- 46. Rio D, Robbins A, Myers R, Tjian R. 1980. Regulation of simian virus 40 early transcription in vitro by a purified tumor antigen. Proc. Natl. Acad. Sci. U. S. A. 77:5706–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tegtmeyer P, Schwartz M, Collins JK, Rundell K. 1975. Regulation of tumor antigen synthesis by simian virus 40 gene A. J. Virol. 16:168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guo ZS, Heine U, DePamphilis ML. 1991. T-antigen binding to site I facilitates initiation of SV40 DNA replication but does not affect bidirectionality. Nucleic Acids Res. 19:7081–7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ryder K, Silver S, DeLucia AL, Fanning E, Tegtmeyer P. 1986. An altered DNA conformation in origin region 1 is a determinant for the binding of SV40 large T antigen. Cell 44:719–725 [DOI] [PubMed] [Google Scholar]
- 50. Joo WS, Luo X, Denis D, Kim HY, Rainey GJ, Jones C, Sreekumar KR, Bullock PA. 1997. Purification of the simian virus 40 (SV40) T-antigen DNA binding domain and characterization of its interactions with the SV40 origin. J. Virol. 71:3972–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. O'Reilly DR, Miller LK. 1988. Expression and complex formation of simian virus 40 large T antigen and mouse p53 in insect cells. J. Virol. 62:3109–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dixon RAF, Nathans D. 1985. Purification of simian virus 40 large T antigen by immunoaffinity chromatography. J. Virol. 53:1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Simanis V, Lane DP. 1985. An immunoaffinity purification procedure for SV40 large T antigen. Virology 144:88–100 [DOI] [PubMed] [Google Scholar]
- 54. Dean FB, Dodson M, Echols H, Hurwitz J. 1987. ATP-dependent formation of a specialized nucleoprotein structure by simian virus 40 (SV40) large tumor antigen at the SV40 replication origin. Proc. Natl. Acad. Sci. U. S. A. 84:8981–8985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wobbe CR, Dean F, Weissbach L, Hurwitz J. 1985. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc. Natl. Acad. Sci. U. S. A. 82:5710–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 57. Kuwabara MD, Sigman DS. 1987. Footprinting DNA-protein complexes in situ following gel retardation assays using 1,10-phenanthroline-copper ion: Escherichia coli RNA polymerase-lac promoter complexes. Biochemistry 26:7234–7238 [DOI] [PubMed] [Google Scholar]
- 58. Sreekumar KR, Barbaro BA, Prack A, Bullock PA. 2000. Methods for studying interactions between simian virus 40 T-antigen and the viral origin of replication. Methods Mol. Biol. 165:49–67 [DOI] [PubMed] [Google Scholar]
- 59. Murakami Y, Hurwitz J. 1993. DNA polymerase α stimulates the ATP-dependent binding of simian virus tumor T antigen to the SV40 origin of replication. J. Biol. Chem. 268:11018–11027 [PubMed] [Google Scholar]
- 60. Maxam AM, Gilbert W. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499–560 [DOI] [PubMed] [Google Scholar]
- 61. Otwinowski Z, MW 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 62. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winnn MD, Storoni LC, Read RJ. 2007. Phaser crystalographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 64. Collaborative Computational Project Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 65. Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 67. DeLano WL. 2002. The PyMOL molecular graphics system. Delano Scientific, Palo Alto, CA [Google Scholar]
- 68. Kleywegt GJ. 1996. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D Biol. Crystallogr. 52:842–857 [DOI] [PubMed] [Google Scholar]
- 69. Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. 2009. Conformational analysis of nucleic acids revisited: Curves+. Nucleic Acids Res. 37:5917–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zheng G, Lu XJ, Olson WK. 2009. Web 3DNA—a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 37:W240–W246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Laskowski RA. 2009. PDBsum new things. Nucleic Acids Res. 37:D355–D359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Simmons DT, Loeber G, Tegtmeyer P. 1990. Four major sequence elements of simian virus 40 large T antigen coordinate its specific and nonspecific DNA binding. J. Virol. 64:1973–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Weisshart K, Taneja P, Jenne A, Herbig U, Simmons DT, Fanning E. 1999. Two regions of simian virus 40 T antigen determine cooperativity of double-hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional origin DNA unwinding. J. Virol. 73:2201–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Luo X, Sanford DG, Bullock PA, Bachovchin WW. 1996. Structure of the origin specific DNA binding domain from simian virus 40 T-antigen. Nat. Struct. Biol. 3:1034–1039 [DOI] [PubMed] [Google Scholar]
- 75. Müller D, Ugi I, Ballas K, Reiser P, Henning R, Montenarh M. 1987. The AT-rich sequence of the SV40 control region influences the binding of SV40 T antigen to binding sites II and III. Virology 161:81–90 [DOI] [PubMed] [Google Scholar]
- 76. Harrison CJ, Meinke G, Kwun HJ, Rogalin H, Phelan PJ, Bullock PA, Chang Y, Moore PS, Bohm A. 2011. Asymmetric assembly of Merkel cell polyomavirus large T-antigen origin binding domain at the viral origin. J. Mol. Biol. 409:529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Collins BS, Pipas JM. 1995. T antigens encoded by replication-defective simian virus 40 mutants dl1135 and 5080. J. Biol. Chem. 270:15377–15384 [DOI] [PubMed] [Google Scholar]
- 78. Weisshart K, Bradley MK, Weiner BM, Schneider C, Moarefi I, Fanning E, Arthur AK. 1996. An N-terminal deletion mutant of simian virus 40 (SV40) large T antigen oligomerizes incorrectly on SV40 DNA but retains the ability to bind to DNA polymerase A and replicate SV40 DNA in vitro. J. Virol. 70:3509–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jones KA, Tjian R. 1984. Essential contact residues within SV40 large T antigen binding sites I and II identified by alkylation-interference. Cell 36:155–162 [DOI] [PubMed] [Google Scholar]
- 80. Tegtmeyer P, Lewton BA, DeLucia AL, Wilson VG, Ryder K. 1983. Topography of simian virus 40 A protein-DNA complexes: arrangement of protein bound to the origin of replication. J. Virol. 46:151–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Fradet-Turcotte A, Vincent C, Joubert S, Bullock PA, Archambault J. 2007. Quantitative analysis of the binding of simian virus 40 large T antigen to DNA. J. Virol. 81:9162–9174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ryder K, Vakalopoulou E, Mertz R, Mastrangelo I, Hough P, Tegtmeyer P, Fanning E. 1985. Seventeen base pairs of region I encode a novel tripartite binding signal for SV40 T antigen. Cell 42:539–548 [DOI] [PubMed] [Google Scholar]
- 83. Koo H-S, Wu H-M, Crothers DM. 1986. DNA bending at adenine-thymine tracts. Nature 320:501–506 [DOI] [PubMed] [Google Scholar]
- 84. DiGabriele AD, Steitz TA. 1993. A DNA dodecamer containing an adenine tract crystallizes in a unique lattice and exhibits a new bend. J. Mol. Biol. 231:1024–1039 [DOI] [PubMed] [Google Scholar]
- 85. Yardimici H, Wang X, Loveland AB, Tappin I, Rudner DZ, Hurwitz J, van Oijen AM, Walter JC. 2012. Bypass of a protein barrier by a replicative DNA helicase. Nature 492:205–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gutierrez C, Guo Z-S, Roberts J, DePamphilis ML. 1990. Simian virus 40 origin auxiliary sequences weakly facilitate T-antigen binding but strongly facilitate DNA unwinding. Mol. Cell. Biol. 10:1719–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Foster EC, Simmons DT. 2010. The SV40 large T-antigen origin binding domain directly participates in DNA unwinding. Biochemistry 49:2087–2096 [DOI] [PubMed] [Google Scholar]
- 88. Lyubimov AY, Costa A, Bleichert F, Botchan MR, Berger JM. 2012. ATP-dependent conformational dynamics underlie the functional asymmetry of the replicative helicase form a minimalist eukaryote. Proc. Natl. Acad. Sci. U. S. A. 109:11999–12004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Li L, Li BL, Hock M, Wang E, Folk WR. 1995. Sequences flanking the pentanucleotide T-antigen binding sites in the polyomavirus core origin help determine selectivity of DNA replication. J. Virol. 69:7570–7578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. DiMaio D, Nathans D. 1982. Regulatory mutants of simian virus 40: effect of mutations at a T antigen binding site on DNA replication and expression of viral genes. J. Mol. Biol. 156:531–548 [DOI] [PubMed] [Google Scholar]
- 91. Stetter G, Montenarh M. 1989. Complex interaction of SV40 large T antigen with the control region on the SV40 DNA. Oncogene 4:1353–1357 [PubMed] [Google Scholar]
- 92. Mohr IJ, Stillman B, Gluzman Y. 1987. Regulation of SV40 DNA replication by phosphorylation of T antigen. EMBO J. 6:153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Schneider J, Fanning E. 1988. Mutations in the phosphorylation sites of simian virus 40 (SV40) T antigen alter its origin DNA-binding specificity for sites I or II and affect SV40 DNA replication activity. J. Virol. 62:1598–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. O'Donnell M, Jeruzalmi D. 2006. Helical proteins initiate replication of DNA helices. Nat. Struct. Mol. Biol. 13:665–667 [DOI] [PubMed] [Google Scholar]
- 95. Clarey MG, Erzberger JP, Grob P, Leschziner AE, Berger JM, Nogales E, Botchan MR. 2006. Nucleotide-dependent conformational changes in the DnaA-like core of the origin recognition complex. Nat. Struct. Mol. Biol. 13:684–690 [DOI] [PubMed] [Google Scholar]
- 96. Bochman ML, Schwacha A. 2008. The Mcm2-7 complex has in vitro helicase activity. Mol. Cell 31:287–293 [DOI] [PubMed] [Google Scholar]
- 97. Bochman ML, Schwacha A. 2010. The Saccharomyces cerevisiae Mcm6/2 and Mcm5/3 ATPase active sites contribute to the function of the putative Mcm2-7 ‘gate.’ Nucleic Acids Res. 38:6078–6088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. 2011. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 18:471–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Duderstadt KE, Chuang K, Berger JM. 2011. DNA stretching by bacterial initiators promotes replication origin opening. Nature 478:209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Erzberger JP, Mott ML, Berger JM. 2006. Structural basis for ATP-dependent DNA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 13:676–683 [DOI] [PubMed] [Google Scholar]
- 101. Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA. 2012. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell 1512:267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Salje J, Gayathri P, Lowe J. 2010. The Par MRC system: molecular mechanisms of plasmid segregation by actin-like filaments. Nat. Rev. Microbiol. 8:683–692 [DOI] [PubMed] [Google Scholar]
- 103. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 13:1605–1612 [DOI] [PubMed] [Google Scholar]