Abstract
Paramyxovirus V proteins bind to MDA5 (melanoma differentiation-associated gene 5) and LGP2 (laboratory of genetics and physiology gene 2) but not RIG-I (retinoic acid-inducible gene I). The results demonstrate MDA5 R806 is essential for inhibition by diverse V proteins. Complementary substitution for the analogous RIG-I L714 confers V protein recognition. The analogous LGP2 R455 is required for recognition by measles V protein, but not other V proteins. These findings indicate that paramyxoviruses use a single amino acid to distinguish MDA5 from RIG-I and have evolved distinct contact sites for LGP2 interference.
TEXT
Host cell innate responses to virus infections are initiated by recognition of pathogen-associated molecular patterns, such as viral nucleic acids, by cellular pattern recognition receptor proteins. One group of intracellular responders, the retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), include proteins encoded by the RIG-I and MDA5 (melanoma differentiation-associated gene 5) genes and the LGP2 (laboratory of genetics and physiology gene 2) gene (1). These RLRs all have a DECH-box RNA helicase domain (1–4), and RIG-I and MDA5 are characterized by the presence of tandem caspase activation and recruitment domains at their N termini that are absent in LGP2.
Interaction with non-self RNAs stimulates RIG-I and MDA5 to associate with the mitochondrial adaptor protein IPS-1/MAVS (5–9), which acts as a signaling scaffold to facilitate the activation of transcription factors that induce production of beta interferon (IFN-β) and other antiviral effectors. The antiviral response is amplified by IFN-JAK-STAT signal transduction to express many effector proteins that together produce a cellular antiviral state that provides a broadly effective barrier protecting the cell against virus infections.
The importance of the RLR-IFN antiviral defense system is emphasized by the fact that many viruses have mechanisms to evade or antagonize components of this innate immune response. This phenomenon has been well documented for the Paramyxoviridae family of enveloped nonsegmented negative-strand RNA viruses, which have evolved to escape or prevent both IFN production and IFN-responsive signal transduction (10–13). In many cases, the paramyxovirus IFN evasion activities are mediated by the virus-encoded V protein. The paramyxovirus V protein is produced from the polycistronic P gene and is identifiable by a cysteine-rich C-terminal domain (CTD) that is highly conserved among diverse virus species (14).
It has been established that paramyxovirus V proteins can interfere with MDA5 and LGP2 (15, 16). The V protein specifically interacts with the MDA5 and LGP2 helicase domains through a minimal V protein-binding region that corresponds to the C-terminal lobe of the helicase domain (15). Mechanistic studies indicate that V protein binding interferes with MDA5 and LGP2 ATP hydrolysis activity that is essential for signal transduction (15). The RLR archetype, RIG-I, has lower amino acid conservation in this region, consistent with a lack of direct V protein interference (15, 17).
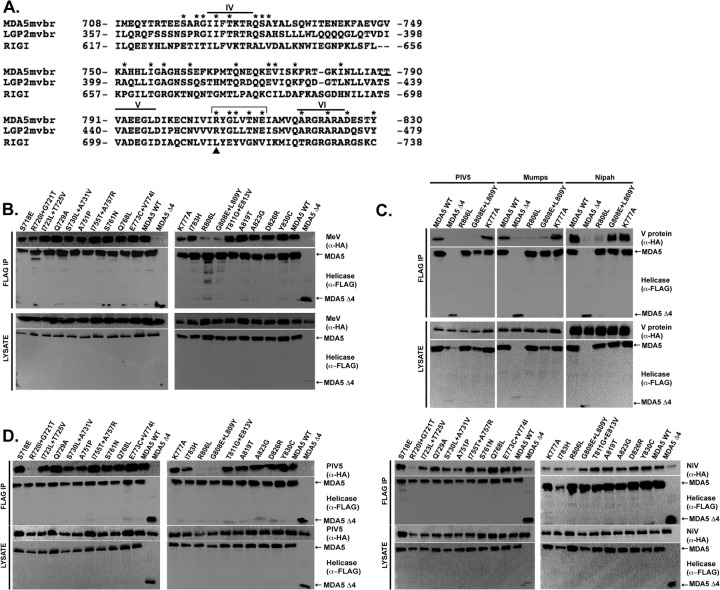
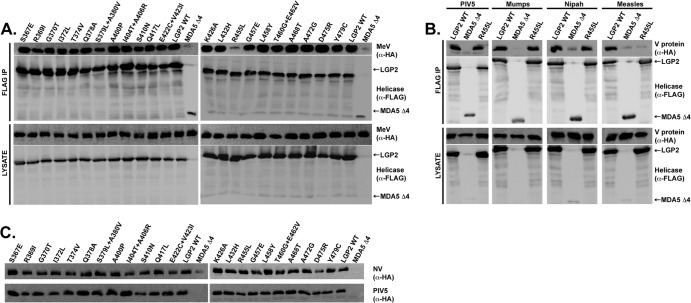
Alignment of the amino acid sequences in the minimal V protein-binding region reveals 26 residues that are identical in MDA5 and LGP2 but different in RIG-I (Fig. 1A). To test if these amino acids contribute to V protein targeting specificity, site-directed mutagenesis (Quick Change; Agilent) was used to substitute the conserved MDA5 or LGP2 amino acids for the RIG-I correlate. For MDA5, 19 mutants were generated containing single or double adjacent substitutions. The FLAG epitope-tagged MDA5 proteins were expressed in HEK293T cells along with hemagglutinin (HA) epitope-tagged measles virus V protein. Cell lysates were subjected to FLAG immunoaffinity purification, and FLAG peptide-eluted proteins were separated by SDS-PAGE and subjected to HA immunoblotting to determine if the mutation disrupted the MDA5-V protein interaction (Fig. 1B). The wild-type (WT) MDA5 and most of the mutants robustly coprecipitated with measles virus V protein, and a previously characterized negative control, MDA5 fragment Δ4, consisting of residues 747 to 1025, was used to establish baseline sensitivity (15). Two variant MDA5 proteins, with the single mutation R806L and double mutation G808E L809Y, were defective in V protein coprecipitation (Fig. 1B). This analysis defines a small epitope that is important for measles virus recognition of MDA5 and reveals that few other individual conserved amino acid residues are required.
Fig 1.
MDA5 mutagenesis reveals R806 is essential for interaction with paramyxovirus V proteins. (A) Multiple-sequence alignment of the MDA5, LGP2, and RIG-I proteins within the minimal V protein-binding region (MVBR). Residues that are the same in MDA5 and LGP2 but different in RIG-I are marked by an asterisk and were subjected to substitution with the RIG-I residue. Roman numerals represent the conserved helicase motifs. The arrowhead indicates MDA5 R806, LGP2 R455, and RIG-I L714; the bracket indicates the multimutant changes. (B) Coimmunoprecipitation analysis of MDA5 and MDA5 mutants with measles virus V protein (MeV). HEK293T cells were transfected with HA-tagged MeV and FLAG-tagged MDA5 (wild type [WT] or with the indicated substitutions), and cell lysates were subjected to FLAG immunoprecipitation (IP) prior to immunoblotting with either anti-HA (α-HA) or anti-FLAG (α-FLAG) antibody. MDA5 Δ4 refers to a previously characterized negative control. (C) Coimmunoprecipitation analysis of MDA5 or the MDA5 R806L and G808E L809Y mutants with the V proteins of parainfluenza virus 5 (PIV5), mumps virus, and Nipah virus. The experimental procedures were identical to those described for panel B. (D) Coimmunoprecipitation analysis of MDA5 and MDA5 mutants with PIV5 and Nipah virus V protein (NiV). The experimental procedures were identical to those for panel B.
Previous studies have revealed both universal and virus-specific requirements for V protein interactions with MDA5 (10). To test if the mutated MDA5 proteins are insensitive to a broad range of paramyxovirus V proteins, additional coimmunoprecipitation assays were carried out with V proteins from parainfluenza virus 5 (PIV5), mumps virus, and Nipah virus (Fig. 1C). The results indicate that all of the tested V proteins were defective in interaction with MDA5 R806L. The effect of the MDA5 G808E L809Y mutations on interaction with PIV5 and mumps virus V proteins is similar to that in measles virus V protein, but Nipah virus V protein remained capable of precipitating the G808E L809Y double mutant. These results indicate virus-specific differences in MDA5 recognition. To test for additional idiosyncrasies, all MDA5 mutants were tested in a coimmunoprecipitation assay with PIV5 and Nipah virus V proteins (Fig. 1D). Only one other mutant, MDA5 R720I T725V, resulted in defective interactions, but due to variability among biological replicates, this double mutant was not analyzed further. These results demonstrate that MDA5 R806 is widely important for interaction with all of the paramyxovirus V proteins tested and that additional residues contribute to virus-specific associations.
Expression of MDA5 by plasmid transfection can stimulate downstream signaling to potently induce IFN-β gene expression, and V protein coexpression can prevent this antiviral response (4, 10, 15). To validate the coimmunoprecipitation results with a biologically relevant endpoint, an IFN-β-promoter luciferase reporter gene assay was conducted with wild-type MDA5, MDA5 R806L, and a mutant that retains V protein association, MDA5 K777A (Fig. 2A). Both of these mutant proteins were able to activate the IFN-β promoter and also exhibited 3- to 5-fold-higher signaling activity than wild-type MDA5. Importantly, MDA5- and MDA5 K777A-dependent antiviral signaling was antagonized by measles virus V protein, but MDA5 R806L-dependent signaling was insensitive to V protein expression.
Fig 2.
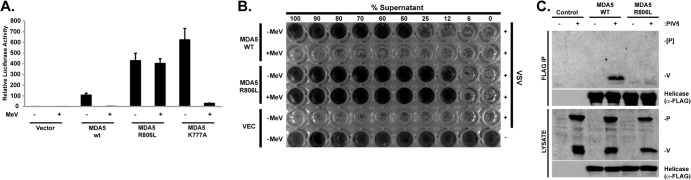
MDA5 mutant R806L retains antiviral signaling activity but is insensitive to V protein interference. (A) R806L activates IFN-β transcription and is insensitive to V protein interference. The MDA5 WT and K777A and R806L mutants were subjected to antiviral signaling assays. HEK293T cells were transfected with the MDA5 protein indicated (or empty vector), the −110 IFN-β promoter luciferase reporter gene, and a Renilla luciferase vector for normalization, with (+) or without (−) measles virus V protein (MeV). The cells were harvested and assayed for firefly and Renilla luciferase activities. Data are plotted as average values (n = 3), with error bars representing standard deviations. Data are presented relative to those from vector controls for MDA5 and MDA5 R806L. (B) R806L protects cells from virus-induced cytopathic effects and is insensitive to V protein interference. HEK293T cells were transfected with empty vector or expression vectors for MDA5 (WT) or MDA5 R806L in the presence (+) or absence (−) of expressed measles virus V protein (MeV). Conditioned medium was harvested 24 h posttransfection, diluted (% Supernatant), and applied to freshly plated 2fTGH cells in 96-well dishes. Following 8 h of incubation, cells were infected with 6 × 103 PFU of vesicular stomatitis virus (VSV; Indiana strain). Cells were fixed and stained with methylene blue in 50% ethanol 16 h later. (C) Differential MDA5 coprecipitation with V protein from paramyxovirus-infected cells. HEK293T cells expressing the indicated MDA5 protein (or empty vector) were infected with 1 PFU/cell of PIV5 (+) or mock infected (−). Cell lysates were FLAG immunoaffinity purified (IP) and immunoblotted with antiserum that recognizes the PIV5 P and V proteins. Only V protein coprecipitated with MDA5, but not the R806L mutant.
To verify the reporter gene assays with endogenous antiviral responses, the V protein sensitivity of MDA5 and MDA5 R806L was assessed in an antiviral cytopathic effect interference assay (Fig. 2B). The wild-type and mutant MDA5 proteins were expressed in HEK293T cells in the presence or absence of measles virus V protein, and conditioned growth media were collected 24 h later. The conditioned media were diluted and applied to freshly plated 2fTGH cells for 8 h prior to infection with vesicular stomatitis virus (VSV) for 16 h. Medium from control cells does not provide protection from VSV-induced cytopathicity, but medium from MDA5-expressing cells, which contains secreted IFN, protects the cells. Again, the R806L protein was found to be hyperactive compared to WT MDA5. Antiviral protection conferred by wild-type MDA5 is sensitive to V protein interference, but protection conferred by MDA5 mutant R806L is insensitive to V protein-mediated interference (Fig. 2B).
To further test the differential ability of the MDA5 proteins to support interaction with V proteins expressed physiologically, coprecipitation was tested in the context of native PIV5 infection. Cells expressing tagged wild-type MDA5 or MDA5 R806L were infected with PIV5, and whole-cell lysates were subjected to FLAG immunoprecipitation. Eluates were probed with antiserum that recognizes the PIV5 P and V proteins (Fig. 2C). The PIV5 V protein, but not the P protein, was specifically coprecipitated by wild-type MDA5, but V protein did not coprecipitate with MDA5 R806L. This supports the importance of MDA5 R806 as crucial for association with the PIV5 V protein under the natural conditions of virus infection.
To verify the importance of R806 for V protein recognition, two complementary mutations were designed to replace the analogous leucine of RIG-I (residue 714) with the arginine of MDA5, either as a single point mutation (creating RIG-I L714R) or by substituting MDA5 residues 806, 808, 809, 811, and 813 (creating RIG-I Multi-Mut; bracket in Fig. 1A). Both RIG-I mutants acquired the ability to be recognized by measles virus and Nipah virus V proteins in coimmunoprecipitation assays, but the single mutation was insufficient for PIV5. RIG-I Multi-Mut was recognized by all of the V proteins (Fig. 3). These results demonstrate that this arginine residue is both necessary and sufficient for V protein recognition and interference for most V proteins, but PIV5 requires additional MDA5 residues in the context of RIG-I.
Fig 3.
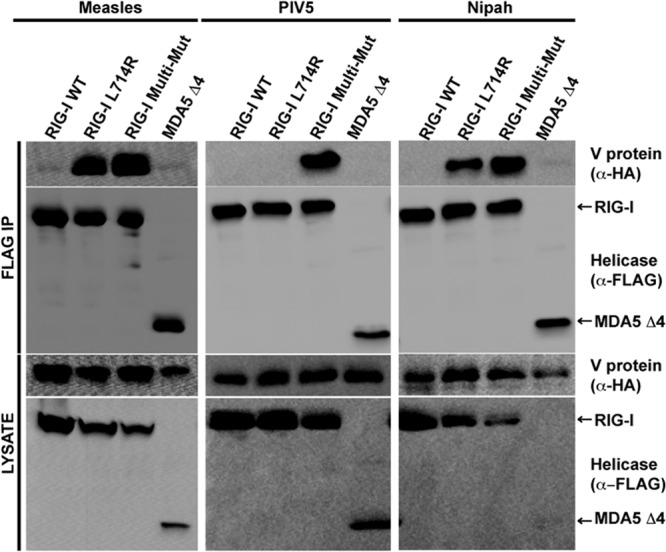
Arginine substitution for RIG-I L714 confers V protein recognition. (A) Coimmunoprecipitation analysis of RIG-I with V proteins. FLAG-tagged RIG-I or RIG-I L714R or RIG-I Multi-Mut mutants were expressed with V proteins from measles virus, PIV5, or Nipah virus and subjected to analysis as described in the legend to Fig. 1B. RIG-I Multi-Mut composition is indicated by a bracket in Fig. 1A.
Similar mutagenic analysis was conducted with LGP2, the second V protein target. To analyze the 26 residues in LGP2, 22 LGP2 constructs containing single or double adjacent substitutions were screened for interaction with measles virus V protein (Fig. 4A). Similar to MDA5, few individual mutations resulted in interaction defects, but replacement of LGP2 R455 with the leucine of RIG-I (R455L) severely impaired its ability to associate with measles virus V protein. LGP2 R455 is the paralog of MDA5 R806, indicating a conserved mode of measles virus V protein interaction with both MDA5 and LGP2.
Fig 4.
LGP2 mutagenesis reveals R455 is essential for interaction with measles virus V protein, but not other paramyxovirus V proteins. (A) Coimmunoprecipitation analysis of the LGP2 WT and mutants with measles virus V protein (MeV). Analysis was carried out exactly as in Fig. 1B, but using LGP2 instead of MDA5. (B) LGP2 R455L is defective for interaction with measles V protein, but not V proteins of PIV5, mumps virus, or Nipah virus. Analysis was carried out as in Fig. 1C, but using LGP2. (C) Coprecipitation of LGP2 mutants with PIV5 and Nipah V proteins. Analysis was carried out identically to the method used for Fig. 1B. Only coprecipitation panels are shown.
The ability of the LGP2 R455L mutant to interact with different V proteins was also tested by coimmunoprecipitation (Fig. 4B). All V proteins interacted with wild-type LGP2. Although defective for interaction with measles virus V protein, LGP2 R455L retained the ability to coprecipitate with V proteins from PIV5, mumps virus, and Nipah virus. Screening of all of the LGP2 mutants for associations with PIV5 and Nipah V proteins indicated that no specific mutation resulted in defective coprecipitation (Fig. 4C). Thus, while measles virus requires LGP2 R455 for interaction, this is not a universal property shared by all of the V proteins tested. Moreover, as the importance of LGP2 R455 differs from that of the widely important MDA5 R806, we conclude that paramyxoviruses have evolved distinct molecular interactions for interference with MDA5 and LGP2.
Prior investigations of V protein interactions with MDA5 and LGP2 delineated a conserved minimal V protein-binding region of approximately 130 amino acids that is necessary and sufficient for targeting by all paramyxoviruses tested (15). Based on the high degree of amino acid identity between MDA5 and LGP2 in this region, it was postulated that individual conserved amino acids constitute a V protein contact surface within the 130 amino acids. The present data support this hypothesis and furthermore reveal both general and virus-specific mechanisms for V protein suppression of MDA5 and LGP2. To suppress MDA5, V proteins from the Rubulavirus genus (PIV5 and mumps virus), the Morbillivirus genus (measles virus), and the Henipavirus genus (Nipah virus) universally require MDA5 R806. This residue alone accounts for the ability of V proteins to suppress MDA5 but not RIG-I. A subset of the V proteins (PIV5, mumps, and measles) were unable to bind to a G808E L809Y double mutant, but Nipah virus V protein remained able to coprecipitate with the double mutant. This indicates a distinct adaptation by Nipah virus for MDA5 interaction.
Remarkably, a single amino acid substitution in RIG-I, L714R, renders the protein sensitive to measles and Nipah virus V protein recognition and interference. It was observed that V protein interacts with the RIG-I protein at steady state without the requirement for virus infection or RNA-mediated RIG-I activation. The fact that paramyxoviruses have not adapted to RIG-I interference underscores the conclusion that these viruses effectively avoid recognition by RIG-I unless defective RNA genomes are present (18).
For interaction with LGP2, the analogous R455 was found to be required for measles virus V protein association. Despite its importance for measles V protein binding, the LGP2 R455L mutant did not disrupt LGP2 association with mumps virus, PIV5, or Nipah virus V proteins. Again, V proteins have diversified to use distinct means to bind to a common target, LGP2. However, no point mutations were identified to have a deleterious effect on LGP2 interaction with PIV5 or Nipah virus V proteins.
These universal attributes and variations on the common theme of MDA5 and LGP2 interference are reminiscent of the great diversity in mechanisms of V protein interference with STAT proteins (12). All of the paramyxovirus V proteins have been found to disrupt IFN signaling by direct interference with STAT1 and/or STAT2, but each genus has a specialized mechanism for STAT suppression. The present study indicates that RLR antagonism by V proteins is similarly accomplished by diversified virus-specific adaptations.
ACKNOWLEDGMENTS
We are grateful to Jean-Patrick Parisien for general advice and technical expertise and members of the Horvath laboratory for helpful comments.
This work was supported by NIH grant R01AI50707 to C.M.H. and by support from an Initiative for Maximizing Student Development grant R25GM079300 to K.R.R.
Footnotes
Published ahead of print 26 December 2012
REFERENCES
- 1. Bruns AM, Horvath CM. 2012. Activation of RIG-I-like receptor signal transduction. Crit. Rev. Biochem. Mol. Biol. 47:194–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cordin O, Banroques J, Tanner NK, Linder P. 2006. The DEAD-box protein family of RNA helicases. Gene 367:17–37 [DOI] [PubMed] [Google Scholar]
- 3. Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo Gale Y-MM, Akira S, Yonehara S, Kato A, Fujita T. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851–2858 [DOI] [PubMed] [Google Scholar]
- 4. Bamming D, Horvath CM. 2009. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J. Biol. Chem. 284:9700–9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988 [DOI] [PubMed] [Google Scholar]
- 6. Xu L-G, Wang Y-Y, Han K-J, Li L-Y, Zhai Z, Shu H-B. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727–740 [DOI] [PubMed] [Google Scholar]
- 7. Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172 [DOI] [PubMed] [Google Scholar]
- 8. Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682 [DOI] [PubMed] [Google Scholar]
- 9. Hiscott J, Lin R, Nakhaei P, Paz S. 2006. MasterCARD: a priceless link to innate immunity. Trends Mol. Med. 12:53–56 [DOI] [PubMed] [Google Scholar]
- 10. Ramachandran A, Horvath CM. 2010. Dissociation of paramyxovirus interferon evasion activities: universal and virus-specific requirements for conserved V protein amino acids in MDA5 interference. J. Virol. 84:11152–11163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goodbourn S, Randall RE. 2009. The regulation of type I interferon production by paramyxoviruses. J. Interferon Cytokine Res. 29:539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramachandran A, Horvath CM. 2009. Paramyxovirus disruption of interferon signal transduction: STATus report. J. Interferon Cytokine Res. 29:531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Childs K, Randall R, Goodbourn S. 2012. Paramyxovirus V proteins interact with the RNA helicase LGP2 to inhibit RIG-I-dependent interferon induction. J. Virol. 86:3411–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thomas SM, Lamb RA, Paterson RG. 1988. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell 54:891–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parisien J-P, Bamming D, Komuro A, Ramachandran A, Rodriguez JJ, Barber G, Wojahn RD, Horvath CM. 2009. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J. Virol. 83:7252–7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Andrejeva J, Childs K, Young D, Carlos T, Stock N, Goodbourn S, Randall R. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. U. S. A. 101:17264–17269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Childs K, Stock N, Ross C, Andrejeva J, Hilton L, Skinner M, Randall R, Goodbourn S. 2007. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359:190–200 [DOI] [PubMed] [Google Scholar]
- 18. Yount JS, Kraus TA, Horvath CM, Moran TM, López CB. 2006. A novel role for viral-defective interfering particles in enhancing dendritic cell maturation. J. Immunol. 177:4503–4513 [DOI] [PubMed] [Google Scholar]