Abstract
Low oxygen tension exerts a significant effect on the replication of several DNA and RNA viruses in cultured cells. In vitro propagation of hepatitis C virus (HCV) has thus far been studied under atmospheric oxygen levels despite the fact that the liver tissue microenvironment is hypoxic. In this study, we investigated the efficiency of HCV production in actively dividing or differentiating human hepatoma cells cultured under low or atmospheric oxygen tensions. By using both HCV replicons and infection-based assays, low oxygen was found to enhance HCV RNA replication whereas virus entry and RNA translation were not affected. Hypoxia signaling pathway-focused DNA microarray and real-time quantitative reverse transcription-PCR (qRT-PCR) analyses revealed an upregulation of genes related to hypoxic stress, glycolytic metabolism, cell growth, and proliferation when cells were kept under low (3% [vol/vol]) oxygen tension, likely reflecting cell adaptation to anaerobic conditions. Interestingly, hypoxia-mediated enhancement of HCV replication correlated directly with the increase in anaerobic glycolysis and creatine kinase B (CKB) activity that leads to elevated ATP production. Surprisingly, activation of hypoxia-inducible factor alpha (HIF-α) was not involved in the elevation of HCV replication. Instead, a number of oncogenes known to be associated with glycolysis were upregulated and evidence that these oncogenes contribute to hypoxia-mediated enhancement of HCV replication was obtained. Finally, in liver biopsy specimens of HCV-infected patients, the levels of hypoxia and anaerobic metabolism markers correlated with HCV RNA levels. These results provide new insights into the impact of oxygen tension on the intricate HCV-host cell interaction.
INTRODUCTION
Hepatitis C virus (HCV) infection causes a wide range of clinical manifestations, from a healthy carrier state to acute and chronic hepatitis that can lead to fibrosis, cirrhosis, and hepatocellular carcinoma. Nearly 3% of the world's population is chronically infected with HCV (1, 2), and current therapeutic approaches are not broadly effective (3).
HCV is a positive-strand RNA virus with a 9.6-kb genome that is flanked at both termini by conserved, nontranslated regions (NTRs), required for RNA translation and replication. The 5′ NTR comprises an internal ribosome entry site (IRES) that directs the expression of a polyprotein precursor (4, 5). The polyprotein is cleaved into structural (core, E1, E2) and nonstructural (p7, NS2, NS3, NS4A, NS4B, NS5A, NS5B) proteins that, in association with cellular factors, form a membrane-associated replicase complex. This copies the viral positive-strand RNA into a negative-strand intermediate that serves as the template for the synthesis of progeny genomes. The alternative reading frame (ARFP) or core+1 and minicore proteins, with as-yet-unknown functions, appear to be synthesized from the core region by alternative translation mechanisms (6, 7).
Studies of the HCV replication cycle have first become possible in 1999 with the development of the replicon system (8). With the identification of a particular HCV genotype 2a isolate (JFH1) that replicates very efficiently in cell culture, a fully permissive HCV culture system was established (9). This system was improved upon development of intragenotypic chimeras consisting of the JFH1 replicase (NS3 to NS5B) fused to the J6 core-to-NS2 region (10, 11) as well as of cell culture-adapted variants of the wild-type (wt) JFH1 virus (JFH1/adpt) (12).
To date, HCV proliferation has been studied exclusively under atmospheric oxygen tension (20% [vol/vol] O2) (13). However, liver normoxic conditions range from 12% O2 around the portal vein to 1% O2 near the central vein (14), with a median value of 3% O2 (15). This oxygen gradient is important for a metabolic activity zonation (16, 17) that is reflected by an asymmetric distribution of key enzymes. The capacity for oxidative energy metabolism, glucose release, and oxidation protection is higher in the periportal area, whereas the capacity for glucose uptake, glutamine formation, and fatty acid synthesis is higher perivenously.
In tissue culture, low oxygen triggers an adaptive reprogramming of cellular homeostasis and bioenergetics (18). Concerning hepatocytes, low oxygen is essential for the preservation of their structure and metabolism (19). Moreover, hypoxia affects the replication of several viruses (20–26).
Based on these observations, we studied the impact of oxygen tension on HCV replication and virus production in human hepatoma (Huh7) cells. We show that low oxygen selectively enhances HCV RNA replication at an early stage and in a hypoxia-inducible factor (HIF)-independent manner. We provide evidence that oncogenes associated with increased anaerobic energy metabolism, as well as creatine kinase B (CKB), are upregulated under hypoxia and that they are responsible for the observed HCV RNA replication enhancement.
MATERIALS AND METHODS
Cell culture.
Huh7.5 (27) and Huh7-Lunet (28) cells were grown in high-glucose (25 mM) Dulbecco's modified minimal essential medium (Invitrogen) supplemented with 2 mM l-glutamine, 0.1 mM nonessential amino acids, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% (vol/vol) fetal calf serum (complete DMEM). To create oxygen tensions lower than the atmospheric one, cells were cultured in a fully humidified incubator supplied with pure nitrogen gas to reduce oxygen as well as with 5% (vol/vol) CO2 at 37°C (New Brunswick CO2 incubator; Innova) (29). For dimethyl sulfoxide (DMSO) differentiation, 8 × 104 Huh7.5 cells were seeded per well of 12-well collagen-coated plates; 24 h later (at 90% confluence), the medium was supplemented with 1% (vol/vol) DMSO, and cells were further cultured for 14 days.
Human liver biopsy specimens.
Human liver fine-needle biopsy specimens were obtained from patients with chronic HCV infection following consent. Sample collection was approved by the Ethical and Scientific Committee of the Hippokration Hospital of Athens.
Plasmid construction.
All amino acid and nucleotide numbers refer to the JFH1 genome (GenBank accession no. AB047639 [30]). Plasmids pFK-JFH1wt, pFK-JFH1/adpt1/mut4-6 (referred to as JFH1/adpt1), pFK-I389RLuc2ACore-3′-Jc1 (JcR2a), pFK-Jc1, and pFK-I389LucUbH77Core-EIJFH1NS3-3′ (LucCore-NSJFH1) have been described previously (9, 11, 12, 31, 32). pFK-I389Luc-EIJFH1NS3-3′_delGDD (Luc-NSJFH1/delGDD) has an in-frame amino acid deletion (MLVCGDDLVV) encompassing the GDD motif of NS5B. pEGFP-HIF-1α has been described previously (33). pEGFP-HIF-2α carries the human HIF-2α cDNA (obtained from pcDNA-HIF-2α; kindly provided by S. L. McKnight) fused carboxy-terminally of the enhanced green fluorescent protein (EGFP) sequence of pEGFP-C1 (Clontech). p9xHRE-Luc carries nine copies of hypoxia response element (HRE) and rat prolactin minimal promoter upstream of the firefly luciferase gene (kindly provided by R. Hernandez-Alcoceba, University of Navarra, Pamplona, Spain). The cytomegalovirus (CMV)-driven JUN-FOS expression vector and the reporter plasmid MMP1-luc, carrying a consensus AP-1 binding site, were kindly provided by L. Bakiri (Fundación Banco Bilbao Vizcaya, Madrid, Spain) (34).
In vitro transcription.
Full-length and bicistronic HCV constructs were linearized with MluI and used for in vitro transcription as described previously (12).
In vitro-transcribed RNA and plasmid DNA transfection.
Electroporation with viral RNAs into Huh7-Lunet cells was performed as described elsewhere (32). For transfection with Luc-NSJFH1/delGDD, Huh7-Lunet cells seeded at 50 to 60% confluence were treated with TransIT-mRNA transfection reagent and mRNA Boost reagent (Mirus) as recommended by the manufacturer. Plasmid DNA transfections were performed at 40 to 50% cell confluence using TransIT-LT1 transfection reagent (Mirus).
siRNA transfection.
The small interfering RNAs (siRNAs) targeting HIF-1α (5′-AGGAAGAACTATGAACATAAA-3′; NM-001530) and HIF-2α (5′-CCCGGATAGACTTATTGCCAA-3′; NM-001430) and the AllStars negative-control siRNA were obtained from Qiagen. The siRNA targeting CKB (5′-CGUCACCCUUGGUAGAGUUTT-3′; NM_001823) and the scramble negative-control siRNA to CKB (5′-GGCGUACUAGCUUAUUCGCTT-3′) were purchased from Sigma. Huh7.5 cells seeded at 40 to 50% confluence were transfected with siRNA using Lipofectamine 2000 transfection reagent (Invitrogen) as recommended by the manufacturer.
Preparation of HCV cell culture (HCVcc) virus stocks and infection assays.
Virus stocks were generated as described elsewhere (32) and used to infect naive Huh7.5 cells. Culture medium was exchanged 4 h after virus inoculation.
Virus titration in cell culture supernatants.
HCV was titrated essentially as described elsewhere (10). Infectivity titers were determined using the JFH1 NS5A-specific mouse monoclonal antibody 9E10 (kindly provided by C. Rice, The Rockefeller University, NY) (10) and expressed as the 50% tissue culture infective dose (TCID50)/ml.
Western blot analysis.
Western blotting was performed as described elsewhere (35). Dilutions of 1:1,000 for JFH1 NS5A monoclonal antibody (9E10 [10]), 1:500 for human HIF-1α mouse monoclonal antibody (BD Biosciences), 1:2,000 for human β-actin mouse monoclonal antibody, 1:100 for GFP rabbit polyclonal antibody (Santa Cruz Biotechnology), and 1:1,000 and 1:500 for human phosphorylated and total AKT rabbit monoclonal antibodies (Cell Signaling), respectively, were used.
Indirect immunofluorescence.
Indirect immunofluorescence analysis of JFH1 NS5A was performed as described elsewhere (32). DNA was stained with propidium iodide (Sigma). Images were acquired with the Leica TCS-SP four-channel confocal microscope equipped with an argon ion laser and helium-neon laser.
Luciferase assays.
Firefly and renilla luciferase (F-Luc and R-Luc) activities in cell lysates were measured using the respective chemiluminescent assay kit (Promega), as recommended by the manufacturer, in a GloMax 20/20 single-tube luminometer (Promega) for 10 s. Luciferase activities were normalized to the total protein amount determined using the Bradford assay reagent (Pierce).
Measurement of intracellular ATP levels.
ATP was measured using the ViaLight HS BioAssay kit (Lonza) according to the manufacturer's protocol in a GloMax 20/20 single-tube luminometer (Promega) for 1 s. ATP levels were normalized to total protein amounts.
RNA quantification by quantitative reverse transcription-PCR (qRT-PCR) and a branched DNA assay.
Total cellular RNA was extracted using the NucleoSpin RNA II kit (Macherey-Nagel). cDNA synthesis was performed with Moloney murine leukemia virus reverse transcriptase (Promega) according to the manufacturer's protocol and with a mixture of the specific primers (see Table S1 in the supplemental material) JFH1-5′NTR-354R and YWHAZ-R for the HCV positive-strand RNA and the 14-3-3-zeta polypeptide (YWHAZ) mRNA, respectively, JFH1-5′NTR-276F and YWHAZ-R for the HCV negative-strand RNA (3.5 pmol/μl of each primer), or pd(N)6 random hexamer primers (GE Healthcare Life Sciences) for the cellular transcripts. Real-time quantitative PCR was performed using the SsoFast EvaGreen supermix (Bio-Rad) as well as primer pairs specific for the JFH1 5′ NTR or the cellular YWHAZ, vascular endothelial growth factor A (VEGFA), glucose transporter 1 (GLUT1), hexokinase 2 (HK2), enolase 1 (ENO1), lactate dehydrogenase A (LDH-A), CKB, FOS, MYC, SRC, α1-antitrypsin (A1AT), and hepatocyte nuclear factor 4 alpha (HNF4-α) (see Table S1 in the supplemental material). The YWHAZ housekeeping gene was selected as a normalization control, as it was confirmed that its expression was not affected under low-oxygen conditions (36). Total cellular RNA from human liver biopsy (LB) specimens was prepared using the RNeasy Plus Mini total RNA isolation kit (Qiagen), and HCV positive-strand RNA was quantified using the Versant HCV RNA 3.0 (branched DNA [bDNA]) assay as recommended by the manufacturer.
Chemicals.
AKT inhibitor VIII (AKTi-1/2) was obtained from Calbiochem.
Statistical analysis.
Only results subjected to statistical analysis using Student's t test with a P value of ≤0.05 were considered statistically significant and presented. Statistical calculations were carried out using Excel in Microsoft Office.
RESULTS
Low oxygen tension induces HCV production in hepatoma cultured cells.
HCV naturally infects hepatocytes that are exposed to an oxygen concentration ranging from 1 to 12% (14), with a median value of 3% (vol/vol) O2 (15). However, to date, the HCV replication cycle has been studied exclusively under atmospheric oxygen conditions (20% O2) in all cell culture systems (13).
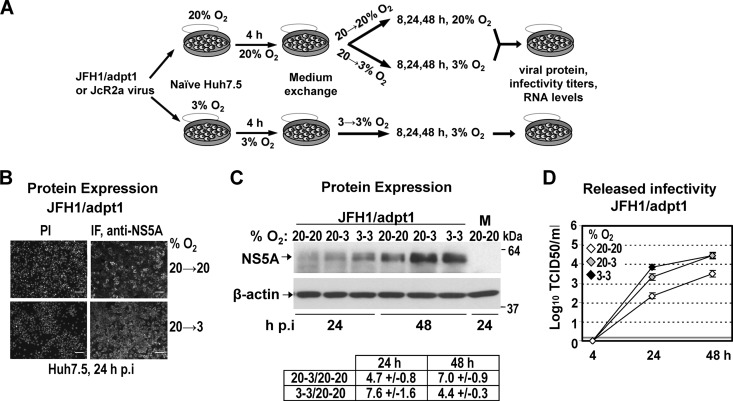
To investigate the effect of low oxygen (hypoxia and 3% O2) on the efficiency of HCV replication, Huh7.5 cells were infected with a highly assembly-competent JFH1 isolate (JFH1/adpt1) (Fig. 1), and virus production was evaluated at 3% and 20% O2 (Fig. 2A), as follows: subconfluent Huh7.5 cells preincubated for 18 h at 20% or 3% O2 were inoculated with HCV (under the oxygen conditions of preincubation), the virus inoculum was withdrawn 4 h later, and cells were further incubated at 20% O2 (a condition hereafter referred to as 20→20), transferred immediately to 3% O2 (20→3), or further incubated at 3% O2 (3→3) for the indicated time points. The last condition (3→3) was intended to induce low-oxygen-mediated cellular changes prior to infection. HCV production was assessed by evaluating viral protein expression and release of infectious HCV particles by using indirect immunofluorescence or Western blotting and TCID50 assays, respectively.
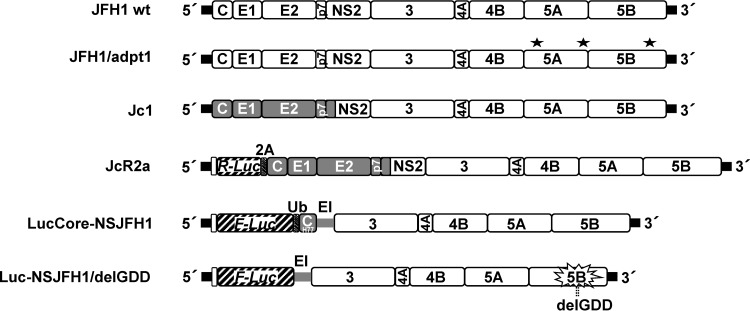
Fig 1.
Schematic representation of HCV constructs used in this study. From top to bottom: JFH1 wild-type (wt) virus genome; JFH1/adpt1, cell culture-adapted JFH1 genome containing virus titer-enhancing mutations (indicated with asterisks) (12); Jc1, chimeric virus genome composed of the J6CF structural and JFH1 nonstructural regions (gray and white boxes, respectively); JcR2a, a Jc1 derivative containing the R-Luc gene (striped box) fused N-terminally to 16 codons of the core gene (white box) and C-terminally to the FMDV 2A protease-coding sequence (gray striped box); LucCore-NSJFH1, a bicistronic replicon encoding a firefly luciferase (F-Luc)-ubiquitin (Ub)-core fusion protein (N-terminal 161 residues of the H77 isolate); Luc-NSJFH1/delGDD, containing a three-codon deletion in the NS5B region (delGDD) gene. Black bars in all panels indicate HCV NTRs. EI, IRES of the encephalomyocarditis virus.
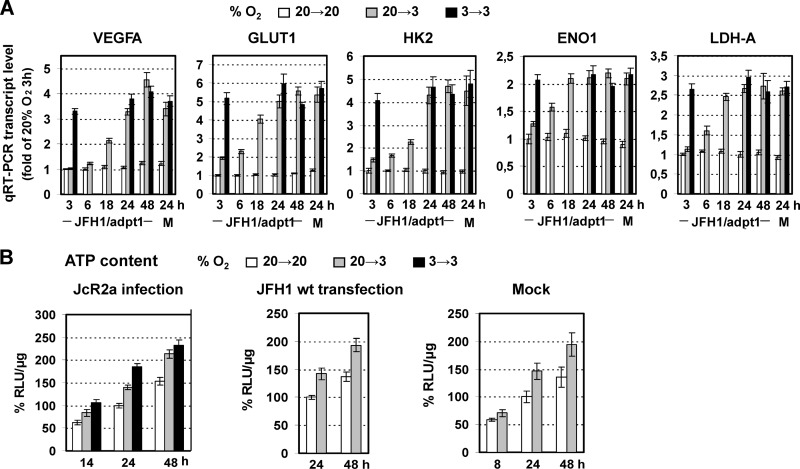
Fig 2.
Low oxygen tension enhances the production of JFH1-derived viruses in Huh7.5-infected cells. (A) Schematic representation of the experimental procedure. Supernatants of Huh7-Lunet cells electroporated with JFH1/adpt1 or JcR2a RNA (10 μg/4 × 106 cells) and incubated at 20% O2 were collected and used for infection of naive Huh7.5 cells (1 TCID50/cell) that were seeded at 30% confluence (to avoid pericellular hypoxia) and preincubated for 18 h at 20% or 3% O2, respectively. Cells preincubated at 20% O2 were cultured at either 20% or 3% O2 until harvest (20→20 or 20→3, respectively). Cells preincubated at 3% O2 were incubated at 3% O2 (3→3) until harvest. (B) Indirect immunofluorescence for NS5A in JFH1/adpt1-infected cells incubated as specified on the right of the panels. Nuclei were stained with propidium iodide (PI; left column). Bar, 80 μm. A representative experiment of two independent repetitions is shown. (C) (Top) Western blot analysis of NS5A (top) in JFH1/adpt1-infected cells incubated as specified in the top of each lane. M, mock-infected cells. (Bottom) β-Actin served as a loading control. Numbers on the right refer to the positions of molecular mass marker proteins. A representative experiment of 3 independent repetitions is shown. (Bottom) Imaging quantification of NS5A signals, normalized by using β-actin signals. Quantity I Bio-Rad software was used to quantify the respective signals from the 3 independent repetitions of the Western blot analysis. Mean values are expressed relative to that obtained at 20→20% O2. (D) Enhanced release of infectious HCV at 3% O2 from JFH1/adpt1-infected Huh7.5 cells, as determined by a TCID50 assay. The gray horizontal line indicates the cutoff of the assay (∼2 TCID50/ml). A representative experiment of three independent repetitions is shown.
As shown in Fig. 2B, at 24 h postinfection (h p.i.), the NS5A immunofluorescence staining of JFH1/adpt1-infected Huh7.5 cells was noticeably increased at 3% O2 (20→3) compared to that at 20% O2 (20→20). Accordingly, an enhanced accumulation of NS5A protein (5- to 7-fold) was also detected by Western blot analysis in lysates of JFH1/adpt1-infected Huh7.5 cells cultured under low-oxygen conditions (20→3 or 3→3) (Fig. 2C). In addition, a significant titer increase was observed with cells cultured at 3% O2 (about 10-fold for the 20→3 condition and up to 30-fold for 3→3) compared to cells cultured at 20% O2 both 24 h and 48 h p.i. (Fig. 2D), suggesting that low oxygen provides an advantage to HCV propagation in Huh7.5 cells.
Low oxygen tension induces HCV genome replication during the early stages of infection.
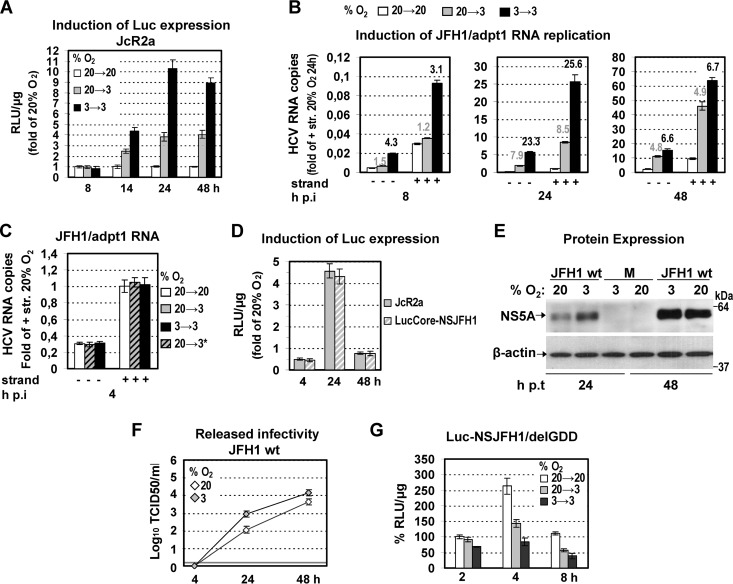
We next investigated the particular step of the viral replication cycle that is affected by low O2. We first determined viral RNA replication by using the JcR2a reporter virus (Fig. 1) containing the R-luc gene. An enhancement of luciferase activity was found with cells cultured at 3% O2, arguing for elevated viral RNA replication under hypoxia (Fig. 3A; see also Fig. S1A in the supplemental material for 20% O2). This result was supported by analysis of cells cultured under the same conditions and infected with the JFH1/adpt1 virus, thus excluding the possibility that the observed enhancement is caused by effects on luciferase (Fig. 3B). Importantly, independent of the readout (luciferase assay, negative- or plus-strand RNA by qRT-PCR), replication was stimulated most in cells cultured consistently at low O2 (3→3). We therefore concluded that low O2 has a positive effect on HCV RNA replication, best visible at early time points. The profound difference between RNA levels obtained with cells cultured at 3→3 and those obtained with cells cultured at 20→3% O2 conditions argues for a replication-promoting effect by the preexisting cellular response to low oxygen.
Fig 3.
Three percent oxygen positively regulates the HCV genome replication. (A and B) Enhanced HCV RNA replication in Huh7.5 cells infected (1 TCID50/cell) with one of two viruses. (A) JcR2a virus. Cells were cultured as specified in the upper left, and replication-derived luciferase activity is expressed as relative light units (RLU) per μg of total protein amount. Values obtained with 20→20 cells were set (each time) to one. (B) JFH1/adpt1 virus. qRT-PCR analysis of the intracellular HCV-negative (−) and -positive (+) strand RNA copies from cells incubated at 20→20, 20→3, or 3→3% O2, expressed relative to the positive-strand RNA obtained at 20→20% O2 at 24 h p.i. YWHAZ mRNA levels were used for normalization. Above the respective bars, ratios of RNA copies at 20→3 and 3→3 versus 20→20% O2 for each time point are shown. (C to F) Low oxygen tension does not affect virus entry. (C) qRT-PCR analysis of the intracellular HCV-negative (−) and -positive (+) strand RNA copies from Huh7.5 cells inoculated with JFH1/adpt1 virus (1 TCID50/cell) and incubated for 4 h as specified. 20→3* refers to cells that were preincubated at 20% O2 and transferred immediately after virus addition from 20% to 3% O2. Values (normalized as described above) are expressed relative to the positive-strand RNA obtained at 20→20% O2 at 4 h postinoculation. (D) Luciferase activities, derived from JcR2a and LucCore-NSJFH1 RNA-transfected Huh7-Lunet cells (10 and 5 μg RNA/4 × 106 cells, respectively). After transfection, the cells were further cultured at 20% or 3% O2, and luciferase activities at 3% O2 are expressed as fold inductions of those obtained at 20% O2. (E and F) Enhanced viral protein expression and release of infectivity at 3% O2 upon JFH1 RNA electroporation (10 μg RNA/4 × 106 cells) of Huh7-Lunet cells. (E) (Top) Western blot analysis of NS5A production in cells incubated at 20% or 3% O2. (Bottom) β-Actin served as a loading control. M, mock nontransfected cells. A representative experiment of two independent repetitions is shown. (F) Kinetics of release of infectivity from the viral RNA-transfected cells incubated at 20% or 3% (TCID50 assay). A representative experiment of three independent repetitions is shown. (G) Three percent oxygen does not positively affect HCV RNA expression. Huh7-Lunet cells preincubated at 20% or 3% O2 for 18 h were transfected with Luc-NSJFH1/delGDD RNA and further incubated as specified on the upper right. Luciferase values measured with 20→20% O2 cells harvested 4 h p.t. were set to 100%. In all panels, bars represent mean values from at least three independent experiments in triplicate. Error bars indicate standard deviations.
In order to examine whether the significantly higher levels of viral RNA are due to enhanced viral entry, we determined the amounts of JFH1/adpt1 RNA 4 h postinoculation in cells cultured at 20→20 or 3→3% O2 or in cells that were transferred immediately after virus addition from 20% to 3% O2 (20→3*). As shown in Fig. 3C, viral RNA levels were similar in all three conditions, suggesting that virus entry is most likely not affected by low oxygen in our system.
These results were substantiated by transfecting the bicistronic reporter replicon LucCore-NSJFH1 and the reporter virus JcR2a as well as the JFH1 wild-type (wt) genome (Fig. 1) into Huh7-Lunet cells that were subsequently incubated at 20% or 3% O2 for various time points posttransfection (p.t.). At 4 h p.t., luciferase values attained with the reporter constructs were reduced around 2-fold at 3% O2 compared to values at 20% O2, whereas at 24 h p.t., a significant rise of about 4.5-fold was detected (Fig. 3D). Similarly, at 24 h p.t. of JFH1 wt RNA, Western blot analysis revealed a higher accumulation of NS5A protein (5- to 7-fold) (Fig. 3E) and the TCID50 assay showed a significant increase (7- to 8-fold) in the production of infectious HCV (Fig. 3F) at 3% O2. However, at 48 h p.t., the 3% O2-related enhancement was significantly lower for the transfected cells than for the infected cells. We hypothesized that this may be related to the different kinetics of HCV RNA replication in transfected cells compared to infected cells (37) or to stronger exhaustion of host factors in transfected cells than infected cells (38). In fact, a more detailed kinetics analysis using different amounts of the bicistronic replicon LucCore-NSJFH1 showed that under low-oxygen conditions, RNA replication enhancement reaches maximum levels 24 h p.t. and gradually declines thereafter (see Fig. S1B in the supplemental material). Overall, these results confirm that hypoxia facilitates HCV RNA replication during early stages of viral infection independent from virus entry.
We next investigated whether low oxygen tension has a positive effect on HCV RNA stability/expression, which may contribute to the enhancement of viral replication. Indeed, it has been reported that under hypoxia, the rate of cellular protein synthesis is reduced (39). Consistently, we found that upon transfection of Huh7-Lunet cells with a capped and polyadenylated reporter mRNA, expression of F-Luc was diminished at 3% O2 compared to normoxia (data not shown). However, HCV RNA translation is IRES dependent, raising the question of whether low oxygen may favor IRES-mediated translation. For this, a nonreplicative bicistronic JFH1 reporter RNA (Luc-NSJFH1delGDD) (Fig. 1) was electroporated into Huh7-Lunet cells that were further cultured at 20% or 3% O2 for various time points p.t. Interestingly, a significant impairment of luciferase activity was detectable with cells transferred to 3% O2 right after transfection (20→3) or with cells preincubated for 18 h at 3% O2 prior to transfection (3→3) (Fig. 3G). Furthermore, no increase in the HCV IRES-dependent expression of luciferase was observed using a bicistronic reporter plasmid under infection conditions (40) at 3% O2 (see Fig S1C in the supplemental material). Thus, enhanced HCV RNA replication at low O2 concentrations is not due to elevated RNA translation.
Low-oxygen-mediated enhancement of HCV replication during early infection is not facilitated by HIF-α.
Hypoxia-inducible factors (HIFs) are important transcription factors for cell adaptation to hypoxia (18) which are stabilized and activated below 5% O2 (41). The oxygen-regulated HIF-α subunit (1α/2α/3α) and the ubiquitous HIF-1β interact as a complex with HRE-containing gene promoters. However, HIF-1α activation under normoxia is a general phenomenon in infection with bacteria, protozoa, and pathogenic viruses (29). HCV induces HIF-1α stabilization at late time points postinfection (42–44). Thus, we examined if the induction of HCV replication, observed at 3% O2, was related to HIF activation. To this end, we performed the following experiments.
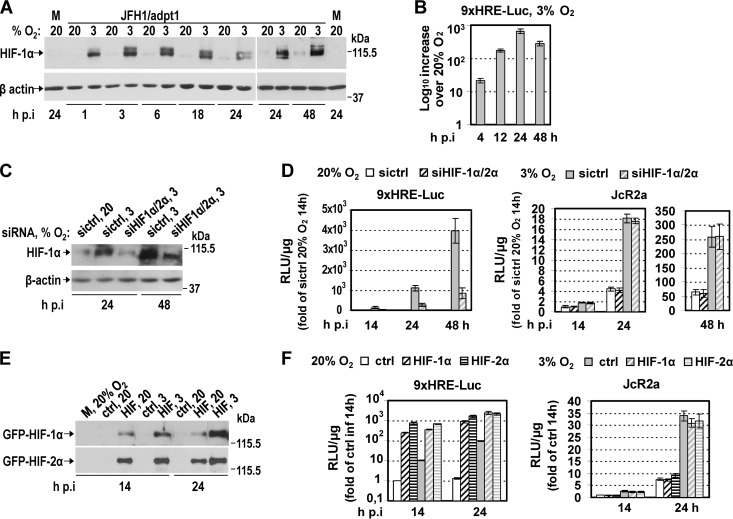
First, we confirmed that HIF-1α was stabilized already 1 h after transferring JFH1/adpt1-infected Huh7.5 cells to 3% O2 and remained at high levels up to 48 h p.i. (Fig. 4A). HIF activation at 3% O2 (∼20-fold) (Fig. 4B) was verified by measuring luciferase activity in cells transfected with the plasmid 9xHRE-Luc, infected with JFH1/adpt1, and then incubated at 20% or 3% O2 (20→20 or 20→3). Consistent with the HIF-1α protein (Fig. 4A), HRE activity (see Fig. S2 in the supplemental material) was similar between HCV- and noninfected cells at early hours p.i., whereas at 72 h p.i., an ∼2-fold increase was detected in infected cells, in agreement with studies (42–44) showing HIF-1 activation at late stages of HCV infection.
Fig 4.
Hypoxia-mediated enhancement of HCV replication is not facilitated by HIF-1α/2α. (A and B) HIF-1α is stabilized and HIFs are activated at 3% O2. (A) (Top) Western blot analysis of endogenous HIF-1α protein using lysates of JFH1/adpt1-infected Huh7.5 cells incubated at 20% or 3% O2. M, mock-infected cells. (Bottom) β-Actin served as a loading control. (B) HRE-dependent expression of F-Luc in Huh7.5 cells transfected with the 9xHRE-Luc construct, infected with JFH1/adpt1 (1 TCID50/cell) 18 h later, and subsequently cultured at 20% or 3% O2 until harvest. Values were normalized to those obtained with cells cultured at 20% O2. (C and D) HIF-1α and HIF-2α silencing does not affect low-oxygen-mediated enhancement of HCV replication. Huh7.5 cells cotransfected with the 9xHRE-Luc construct (0.05 μg/4 × 104 cells) and a mixture of siRNAs targeting HIF-1α and HIF-2α (20 nM each; siHIF1α/2α) or a control siRNA (40 nM) (sictrl) were inoculated with JcR2a (1 TCID50/cell) 18 h p.t. and further incubated at 20% or 3% O2. (C) (Top) Western blot analysis of endogenous HIF-1α protein. (Bottom) β-Actin served as a loading control. (D) Effect of HIF-1α/2α knockdown on the following. (Left) HIF activation, as determined by HRE-dependent F-Luc activity. (Right) HCV replication, as determined by R-Luc activity. Mean values were normalized to the reporter activity measured in cells that were cultured at 20% O2. (E and F) HIF-1α and HIF-2α overexpression is not sufficient to enhance HCV replication. Huh7.5 cells cotransfected with the 9xHRE-Luc construct (0.05 μg/4 × 104 cells) and one plasmid expressing GFP-HIF-1α, GFP-HIF-2α, or GFP (0.4 μg/4 × 104 cells) were inoculated with JcR2a (1 TCID50/cell) 18 h p.t. and subsequently incubated at 20% or 3% O2. ctrl, cells transfected with the plasmid expressing GFP. Transfection efficiency was 60 to 70%. (E) HIF-1α and -2α overexpression as determined by GFP-specific Western blot analysis. (F) Effect of HIF-1α or HIF-2α overexpression on the following. (Left) HIF activation, as determined by HRE-dependent luciferase expression. (Right) HCV replication, as determined by R-Luc expression. Mean values were normalized to the reporter activity detected in cells cultured at 20% O2. For Western blot analysis, at each time, a representative experiment of at least two independent repetitions is shown. In all panels, bars represent mean values from at least three independent experiments in triplicate. Error bars indicate standard deviations.
Second, we evaluated the role of HIFs in the enhancement of HCV replication under low-oxygen conditions by silencing the two most well-studied HIF-α isoforms, 1α and 2α (45). A mixture of two siRNAs (siHIF1α/2α) efficiently reduced HIF-1α levels at 3% O2, as shown by Western blotting of JcR2a-infected cells (Fig. 4C), without affecting cell viability (see Fig. S3D in the supplemental material). Also, silencing of HIF1α/2α was confirmed upon cotransfection of the 9xHRE-Luc plasmid, as HRE activity was downregulated ∼5-fold (Fig. 4D, left). Surprisingly, silencing had no effect on the enhancement of HCV RNA replication at 3% O2 (Fig. 4D, right). Third, we overexpressed HIF-1α and -2α (Fig. 4E), leading to upregulation of HRE comparable to that observed at 3% O2 (Fig. 4F, left) and induction of HIF-regulated genes (see Fig. S3A in the supplemental material), albeit lower than that observed at 3% O2; however, this had no effect on HCV replication (Fig. 4F, right) or particle production (see Fig. S3B in the supplemental material). Finally, we pharmacologically induced HIF stability and/or activity (46) by treating Huh7.5 cells with CoCl2, desferrioxamine (DFO), or dimethyloxallyl glycine (DMOG). This enhanced HRE activity (see Fig. S3E, left, in the supplemental material) again had no effect on viral replication (see Fig. S3E, right, in the supplemental material). These results suggest that hypoxia-related enhancement of HCV replication, observed at the early stages of infection, occurs independently from HIF-1 and HIF-2 activation.
Low-oxygen-mediated enhancement of HCV replication is linked to hypoxia-induced reprogramming of cellular energetics.
Since the activation of HIFs did not contribute to the detected upregulation of HCV replication at 3% O2, we examined global cellular gene expression changes in mock- and JFH1/adpt1-infected Huh7.5 cells using hypoxia signaling pathway-focused DNA microarrays. Cells cultured for 24 h at 3% O2 showed (18, 47) an upregulation (by at least 2-fold) of several hypoxia-related genes (see Table S2 in the supplemental material), such as HIFs, directing transcriptional targets (VEGFA, EPO), genes encoding stress-related proteins, and genes involved in oxidative stress and reactive oxygen species (ROS) production, glucose transport (GLUT1, GLUT8) and metabolism (HK2, ENO1), cell growth, proliferation, and cell cycle regulation, including the FOS, MYCN, and KIT oncogenes. Notably, the respective transcript levels in uninfected and infected cells were similar. Moreover, qRT-PCR analysis of selected genes revealed an early response of transcriptional reprogramming to low oxygen (Fig. 5A) and confirmed a metabolic shift toward a higher rate of anaerobic glycolysis. Cell cycle distribution experiments using hydroxyurea (HU) treatment and fluorescence-activated cell sorting (FACS) analysis (see Table S3 in the supplemental material) also supported an acceleration of the cell cycle kinetics at 3% O2, consistent with previous studies (48, 49). Interestingly, HCV replication enhancement at 3% O2 was also observed in the HU-arrested cells, suggesting that the effect on HCV is related neither to cell cycle acceleration nor to a specific cell cycle phase (data not shown).
Fig 5.
Early induced reprogramming of cellular energetics by hypoxia. (A) Upregulation of VEGFA and GLUT1, HK2, ENO1, and LDH-A genes in JFH1/adpt1-infected Huh7.5 cells that were cultured at 20→3 or 3→3% O2 relative to 20→20% O2. Values were normalized to those obtained with cells kept at 20% O2 and harvested 3 h p.i. For 3→3 infected cells, values were measured only at 3, 24, and 48 h p.i. J, JFH1/adpt1-infected cells; M, mock-infected cells. (B) Three percent oxygen increases the ATP content of hepatoma cells. Intracellular ATP levels in JcR2a-infected Huh7.5 cells (1 TCID50/cell) (right), JFH1 RNA-electroporated Huh7-Lunet cells (10 μg RNA/4 × 106 cells) (middle), or mock-transfected Huh7-Lunet cells (left). Cells were incubated as specified at the top and harvested at given time points. Mean values were normalized to the ones detected in cells cultured at 20→20% O2 and harvested 24 h postinfection/posttransfection. In all panels, bars represent mean values from at least three independent experiments in triplicate or quintuplicate (ATP content). Error bars indicate standard deviations.
Given that viral replication processes depend strongly on cell energetics, we assessed the effect of 3% O2 on the intracellular levels of ATP in JcR2a-infected (Fig. 5B, left), JFH1 RNA-transfected (Fig. 5B, middle), and mock-transfected (Fig. 5B, right) Huh7 cells. Incubation of JcR2a-infected Huh7.5 cells at 3% O2 resulted in an increase of ATP levels by 40 to 45% (20→3) or 50 to 80% (3→3) at 24 and 48 h p.i. No significant difference in the ATP content was observed between infected and control cells up to 48 h p.i. (data not shown). Notably, the ATP ratio at 3% versus 20% O2 was cell confluence dependent, most likely due to the development of pericellular hypoxia (50) in high cell densities (see Fig. S4 in the supplemental material).
To test the relationship between cell energetics and HCV replication, we modulated ATP production of cells by feeding with high or low glucose, galactose, or glutamine, followed by incubation of cells at different O2 concentrations. As shown in Fig. 6A and B, glucose reduction or substitution resulted in a gradual attenuation of the increase of both the intracellular ATP content and HCV replication observed under low oxygen conditions. qRT-PCR analysis confirmed a concomitant strong reduction of HK2 gene expression at 3% O2 (Fig. 6C), suggesting an impairment of glycolysis. We therefore concluded that HCV replication enhancement at 3% O2 correlates primarily with an ATP increase due to the induction of anaerobic glycolysis.
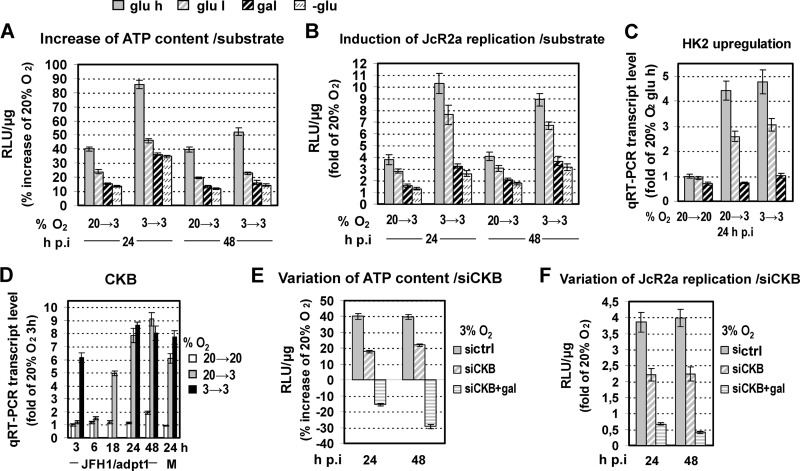
Fig 6.
Replacing the glycolytic substrate with oxidative ones, as well as CKB silencing, reduces low-oxygen-mediated HCV enhancement. (A to C) Huh7.5 cells preincubated at 20% or 3% O2 were inoculated with JcR2a (1 TCID50/cell) and cultured at the given conditions in complete DMEM containing high glucose (glu h; 25 mM), low glucose (glu l; 5.56 mM), galactose instead of glucose (gal; 10 mM), or no glucose (-glu). (A) Bioluminescent measurement of intracellular ATP levels. Mean values (%) were normalized to ATP levels detected in cells cultured at 20→20% O2. (B) JcR2a-derived luciferase activity expressed as fold induction relative to values obtained with cells cultured at 20→20% O2. (C) qRT-PCR-based quantification of HK2 mRNA in cells treated as described above. Results are expressed as fold induction of gene expression relative to that in cells cultured at 20→20% O2 in DMEM with high glucose. (D) Upregulation of CKB expression at 3% O2. qRT-PCR analysis of CKB mRNA amounts in JFH1/adpt1-infected Huh7.5 cells cultured as specified on the right. Data were normalized to values obtained with cells cultured at 20→20% O2 and harvested 3 h p.i. For 3→3 infected cells, values were measured only at 3, 24, and 48 h p.i. J, JFH1/adpt1-infected cells. M, mock-infected cells. (E and F) Huh7.5 cells transfected with an siRNA targeting CKB (siCKB) or a control siRNA (sictrl) (100 nM) were inoculated 18 h p.t. with JcR2a (1 TCID50/cell), supplied with DMEM containing either high glucose or galactose (gal), and incubated at 20% O2 or transferred to 3% O2. (E) Intracellular ATP levels as determined by a bioluminescent assay. (F) JcR2a-derived luciferase activity. Values were normalized to those obtained with cells cultured at 20→20% O2 and transfected with the same siRNA. In all panels, bars represent mean values from at least three independent experiments in triplicate or quintuplicate (ATP content). Error bars indicate standard deviations.
Low-oxygen-mediated HCV replication enhancement is also linked to an increase in CKB-dependent ATP synthesis.
Based on previous studies (51–53), another important metabolic route that can still produce ATP under low-oxygen conditions in Huh7 cells is catalyzed by creatine kinase (CK) (54). The brain-type isoform of this enzyme (CKB) has been shown to promote HCV replication (55). Interestingly, qRT-PCR analysis demonstrated a significant (8- to 9-fold) increase in CKB mRNA at 3% O2, indicating directly that CKB is a hypoxia-induced gene (Fig. 6D). To determine the possible effect of this kinase on HCV replication induction at 3% O2, we silenced CKB expression. This reduced the ATP increase observed at 3% O2 by 50% (Fig. 6E) and resulted in a lower enhancement of JcR2a RNA replication (Fig. 6F). Replacement of glucose by galactose simultaneously with CKB silencing resulted in lower intracellular ATP levels and viral replication at 3% than at 20% O2. These data suggest that under low-oxygen conditions, glycolysis and CKB are the main contributors to ATP production and play a key role in the observed enhancement of HCV replication.
At 3% O2, virus replication is induced in DMSO-treated Huh7.5 cells.
To investigate the effect of hypoxia on HCV replication in a cellular system that better mimics the in vivo situation and to exclude effects caused by cell proliferation, we produced highly differentiated, growth-arrested Huh7.5 cells (Huh7.5dif) by DMSO treatment (see Fig. S5A and C, left, in the supplemental material) (56). These cells were readily infectible with HCV, albeit with lower efficiency than Huh7.5 cells (see Fig. S5B and C in the supplemental material).
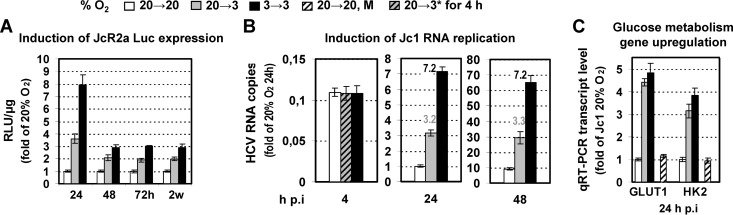
Under hypoxia, HCV replication enhancement was also observed in Huh7.5dif cells, detected early after infection (Fig. 7A and B), and sustained throughout the 2-week observation period (Fig. 7A; see also Fig. S5D in the supplemental material). Consistently, an upregulation of the glycolysis-related genes GLUT1 and HK2 was detected at 20→3 and 3→3% O2 (Fig. 7C).
Fig 7.
Three percent oxygen enhances HCV replication in Huh7.5dif cells. (A) Effect of low oxygen tension on JcR2a-infected Huh7.5dif cells that were incubated as specified at the top and harvested at various time points. Mean values are expressed relative to that obtained with cells cultured at 20→20% O2. (B) Effect of low oxygen on Jc1 RNA replication as determined by quantification of intracellular positive-strand RNA copies. Values are expressed relative to those obtained at 20→20% O2 24 h p.i. Numbers above the bars refer to ratios of RNA copies. 20→3*, cells that were transferred immediately after virus addition from 20% to 3% O2. (C) Effect of 3% O2 on GLUT1 and HK2 genes in Jc1-infected Huh7.5dif cells. mRNA amounts were quantified by qRT-PCR and expressed relative to values obtained with cells cultured at 20→20% O2 and harvested 24 h p.i. M, mock-infected cells. In all panels, bars represent mean values from at least three independent experiments in triplicate. Error bars indicate standard deviations.
Correlation of VEGF, HK2, LDH-A, and CKB with HCV RNA levels in liver biopsy specimens from chronically infected patients.
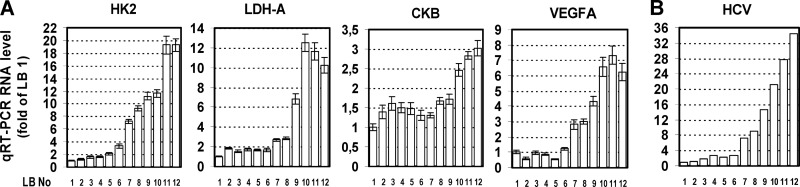
In an attempt to correlate results obtained in cell culture with the in vivo situation, we obtained liver fine-needle biopsy specimens from 12 patients with liver fibrosis due to chronic HCV infection. Total RNA was extracted and subjected to qRT-PCR analysis for quantification of mRNA levels of the energy production-related genes HK2, LDH-A, and CKB as well as the hypoxic stress-responsive gene VEGFA. HCV positive-strand RNA was quantified by a branched DNA assay. Importantly, liver biopsy (LB) samples 10 to 12, containing the largest amounts of the cellular transcripts (Fig. 8A), also contained the largest HCV RNA amounts (Fig. 8B). Moreover, LB samples 1 to 6, containing the smallest amounts of the selected cellular genes, also contained the smallest HCV RNA amounts. Intermediate values of RNAs were detected in LB samples 7 to 9. Despite the limited number of clinical samples, these results argue for an in vivo correlation between the degree of metabolic activity and response to hypoxic stress and the HCV replication level.
Fig 8.
Analysis of human liver biopsy specimens reveals a correlation between elevated HCV RNA amounts and increased expression of genes involved in anaerobic metabolism. (A) qRT-PCR quantification of HK2, LDH-A, CKB, and VEGF mRNA amounts in total RNA isolated from 12 liver samples (LB 1 to 12) from patients with chronic hepatitis C. Bars represent mean values from three independent repetitions, and error bars indicate standard deviations. YWHAZ mRNA levels were used for normalization. (B) HCV positive-strand RNA copy levels from the same samples, as quantified using the branched DNA assay. Values were normalized to those obtained with sample LB 1.
Possible role of oncogene induction for hypoxia-related enhancement of HCV replication in Huh7.5 cells.
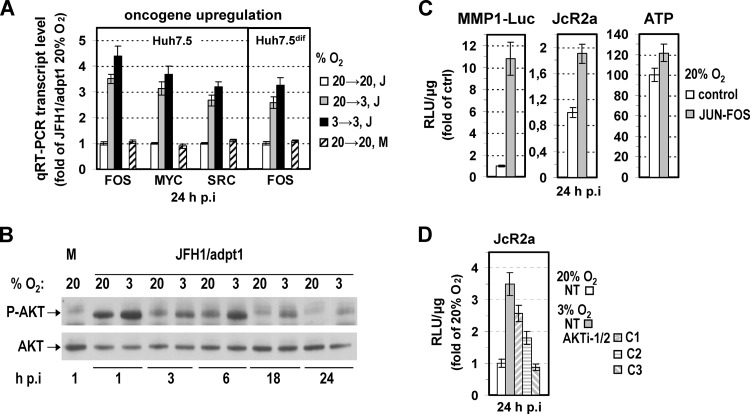
ln addition to HIF, reprogramming of bioenergetics during hypoxia is also known to be affected by certain oncogenes and tumor suppressor genes (47, 57–62). Hypoxia-mediated upregulation of AKT1, MYC, FOS, H-RAS, SRC, BCR/ABL, and p53 has been reported to enhance glucose uptake and glycolysis through direct activation of related genes, including GLUT1, HK2, ENO1, and LDH-A. Since our results (see Table S2 in the supplemental material) suggested the upregulation of certain oncogenes (FOS, MYCN, and KIT) in HCV-infected cells under hypoxia, we investigated further the induction of selected oncogenes by using qRT-PCR. As shown in Fig. 9A, at 24 h of cell incubation at 20→3 or 3→3% O2, FOS, MYC, and SRC were upregulated 3- to 4-fold compared to levels after incubation at 20→20% O2. Notably, mRNA levels in noninfected (mock) cells were similar to those in infected cells. Expression of AKT1, H-RAS, and BCR/ABL genes was unaffected (data not shown). However, consistent with previous studies (63), Western blot analysis revealed a significant increase of AKT phosphorylation levels in HCV-infected Huh7.5 cells cultured at 3% O2 compared to cells at 20% O2 (Fig. 9B). Notably, AKT phosphorylation was also induced early during infection of Huh7.5 cells at 20% O2 (compare mock- and JFH1-infected cells), as previously described for other flaviviruses (64, 65), and this increase was higher at 3% O2, even after elimination of the infection-related enhancement of AKT (63). Interestingly, in the nondividing Huh7.5dif cells, FOS was also upregulated at 20→3 and 3→3% O2 24 h p.i. (Fig. 9A) whereas MYC and SRC were not changed (data not shown). Moreover, FOS and SRC were not induced upon HIF-1α/2α overexpression (see Fig. S6A in the supplemental material). Therefore, we hypothesized that certain hypoxia-regulated oncogenes can operate separately from HIFs to modulate cell energetics favoring HCV RNA replication.
Fig 9.
Low-oxygen-mediated oncogene upregulation is linked to enhancement of HCV replication. (A) Upregulation of FOS, MYC, and SRC genes in JFH1/adpt1 (J)-infected Huh7.5 or Huh7.5dif cells. M, mock-infected cells. (B) Elevated levels of AKT phosphorylation in infected Huh7.5 cells cultured under hypoxia. Western blot analysis of endogenous levels of phosphorylated (top) and total (bottom) AKT. M, mock-infected cells. A representative experiment of two independent repetitions is shown. (C) JUN-FOS overexpression enhances intracellular ATP levels and HCV replication. Huh7.5 cells cotransfected with the MMP1-Luc (0.05 μg/4 × 104 cells)- and the JUN-FOS (0.4 μg/4 × 104 cells)-encoding plasmids or the respective vector (control) were inoculated 18 h p.t. with JcR2a (1 TCID50/cell) and further incubated at 20% O2 for 24 h. JUN-FOS overexpression enhances TRE-dependent expression (left), increases HCV RNA replication (middle), and results in a detectable upregulation of the intracellular ATP (right). For all panels, values were normalized to those obtained with control-transfected cells. (D) AKT inhibition reduces hypoxia-mediated HCV enhancement. Huh7.5 cells were inoculated with JcR2a and 4 h later were untreated or treated with the AKT inhibitor VIII (Akti-1/2) at the following concentrations: C1, 2.5 μC; C2, 5 μC; C3, 10 μC. After 24 h of incubation of cells at 20% or 3% O2, luciferase activities were determined. Values were normalized to those obtained with untreated cells cultured at 20% O2. In all panels, bars represent mean values from at least three independent experiments in triplicate or quintuplicate (ATP content). Error bars indicate standard deviations.
To challenge this hypothesis, we evaluated the role of FOS and AKT in HCV replication. FOS was chosen because its expression was upregulated in both normal and DMSO-treated Huh7.5 cells. As shown in Fig. 9C, overexpression of a JUN-FOS tethered dimer in cells cultured at 20% O2 enhanced virus replication ∼2-fold (middle panel), concomitant with an ∼20% elevation of intracellular ATP levels (right panel). As expected, the consensus tissue plasminogen-activator response element (TRE) within the human matrix metalloproteinase 1 (MMP1) promoter, known to be recognized by JUN-FOS, was strongly activated (∼10-fold) (left panel). Concerning AKT, its role in the induction of glycolysis is well documented. Treating Huh7.5 cells with the AKT inhibitor VIII (AKTi-1/2) reduced JcR2a replication enhancement at 3% O2 (Fig. 9D), even at an inhibitor concentration (C1 in Fig. 9D) that does not significantly alter cellular ATP content (data not shown). These results suggest that activator protein 1 (AP-1) (FOS/JUN or JUN/JUN dimers) and AKT are factors responsible for the enhancement of HCV replication under low-oxygen conditions.
In support of this assumption, incubation of cells at 12% O2, a condition prevailing in the liver, did not stabilize or activate HIF in Huh7.5 cells (see Fig. S6B and C, respectively, in the supplemental material), consistent with HIF activation at oxygen levels lower than 5% (vol/vol) (41). However, transcription of glycolysis-related genes and CKB (see Fig. S6D, left, in the supplemental material) and expression of the FOS oncogene (see Fig. S6D, right, in the supplemental material) were enhanced. Consistent with these results, JcR2a replication (see Fig. S6E in the supplemental material) and intracellular ATP levels (see Fig. S6F in the supplemental material) were also enhanced at 12% O2, albeit with delayed kinetics and to a lower extent compared to levels at 3% O2.
To summarize, certain oncogenes, including FOS and AKT1, in combination with other cellular factors, such as CKB, modulate cell energetics and, thus, HCV RNA replication.
DISCUSSION
Recent studies have shown that oxygen tension exerts a significant effect on the replication of several viruses. In general, hypoxia restricts the replication of viruses that naturally infect tissues exposed to ambient oxygen (simian virus 40, adenovirus) (25, 26) and induces the propagation of viruses that naturally target tissues exposed to low oxygen concentrations (vesicular stomatitis virus, herpesviruses, human immunodeficiency virus, parvovirus B19) (20–24).
To date, HCV in vitro infection has been studied exclusively under atmospheric oxygen conditions using mostly Huh7 and derivatives, although the liver microenvironment is hypoxic (14, 15) and hepatocytes are highly differentiated (66). Here, we investigated HCV replication efficiency at 3% (vol/vol) O2 to mimic the oxygen tension sensed by most hepatocytes in the liver or at 12% O2, which is present around the liver portal vein. Our results reveal the following.
Low oxygen tension (3% O2) enhances HCV replication in cultured cells.
This was most pronounced by preincubation of cells at 3% O2 prior to infection (3→3% O2), arguing that a preexisting hypoxia-induced cellular factor(s) promotes HCV replication. This enhancement was due to an RNA replication increase at early hours of infection, whereas virus entry, RNA translation, assembly, and release were not detectably affected (Fig. 2 and 3). Several mechanisms responsible for replication enhancement can be envisaged.
Low-oxygen-mediated increase of HCV replication is independent of HIF-1α/2α activation.
HIFs are fundamental control transcription factors of the cellular metabolic state under low-oxygen conditions (18). Although low-oxygen-dependent enhancement of HCV replication may reflect HIF-1-mediated alterations of cellular homeostasis, overexpression and silencing studies revealed that HIF-1α/2α activation did not correlate with the hypoxia-related increase of HCV replication observed at the early stages of infection (Fig. 4; see also Fig. S3 in the supplemental material). In agreement, chemically induced hypoxia failed to elevate HCV replication, whereas 12% O2, which fails to activate HIF-1α (41) (as well as HIF-2α), positively affected HCV replication (see Fig. S6 in the supplemental material). Notably, HIF-1α activation from HCV at late stages of viral infection has been reported under normoxia and has been linked to mitochondrial dysfunction and oxidative stress caused by HCV proteins (42–44). In agreement with these studies, we also detected an increase in HIF-1α activation, but only 72 h p.i. (see Fig. S2 in the supplemental material). Interestingly, a HIF-1-independent increase of genome replication under hypoxic conditions has been found also for B19V (67).
Low-oxygen-mediated increase in HCV replication in cultured cells is directly linked to cellular energetic changes.
Hypoxia is associated with an adaptive cell metabolic reprogramming, including a shift in glucose metabolism from oxidative phosphorylation to anaerobic glycolysis and lactic acid production (18, 47). Huh7.5 cells, like most cancer cells, exhibit high rates of glycolysis and lactate production under normoxia (Warburg effect) (58, 62) and even higher rates under hypoxia conditions (Pasteur effect) (68). For highly proliferating cells, the advantage of such a metabolic switch is to combine energy (ATP) from enhanced glycolysis with production of nutrients/intermediates for cell growth and division (69). Oncogenes and tumor suppressor proteins are known to regulate this phenomenon in addition to HIF activation (60).
Indeed, transcriptome analysis of Huh7.5 cells confirmed that several oncogenes, as well as genes involved in glucose uptake (GLUT1) and anaerobic glycolysis (LDH-A, HK2, ENO1), were upregulated at 3% O2 (Fig. 5A; see Table S2 in the supplemental material). A comparison of HCV-infected and mock cells up to 24 h p.i. suggested that during the early stages of infection, HCV does not affect the cellular transcription profile, as previously shown (70). As expected, enhanced glucose uptake and use compensated for the ATP levels under low-oxygen conditions (Fig. 5B) and promoted cell proliferation (see Table S3 in the supplemental material), consistent with previous reports (48, 49).
Interestingly, the ATP gain correlated well with the low-oxygen-mediated HCV replication enhancement and was directly linked to hypoxia-induced glycolysis, as revealed by experiments comprising the use of different energy substrates in the culture medium of infected Huh7.5 cells (Fig. 6A to C). Consistently, previous studies have directly implicated two glycolytic enzymes in the regulation of HCV replication (71, 72), hexokinase III, which is transcriptionally activated by both HIF and MYC under hypoxia, and pyruvate kinase, which is enhanced in a HIF-independent manner (73–75). Thus, hypoxia-mediated upregulation of glycolysis represents an HCV replication-promoting metabolic modification. Moreover, oxidative phosphorylation, although active at 3% O2, had no role in the observed gain of energy, as detected by oligomycin treatment (mitochondrial ATP synthase inhibitor) of the infected cells (data not shown).
Oncogene induction may be implicated in hypoxia-related HCV replication enhancement.
Oncogenes and tumor suppressor proteins, such as AKT, AP-1 (FOS/JUN or JUN dimers), MYC, SRC, and p53, are known to coordinate glycolysis under both anaerobic and aerobic (Warburg) conditions (47, 58–62). Furthermore, induction of oxidative stress- and ROS-related genes (76) or other HIF-1-independent pathways, such as that of RAS and intracellular Ca2+ effector pathways (77, 78), may modulate oncogene expression under certain cellular conditions. Inasmuch as our results indicated a HIF-independent hypoxia-induced HCV replication, we hypothesized that certain oncogenes and/or tumor suppressor genes may play a critical role in cellular changes that favor virus replication enhancement under low-oxygen conditions.
Indeed, several oncogenes were found to be upregulated (Fig. 9A; see Table S2 in the supplemental material) or activated (Fig. 9B) at 3% O2, together with oxidative stress- and ROS-related genes (see Table S2 in the supplemental material) that may contribute to oncogene induction. Among the induced oncogenes, FOS, MYC, SRC, and AKT have been already shown to directly activate glucose-related transporter and glycolytic gene promoters independently of HIFs (47, 57–62) and, in addition, have been reported to stimulate HCV replication through either association with viral nonstructural proteins or other ways of action not completely defined (71, 79–83). In fact, our results confirmed the HIF-independent transcriptional activation of FOS and SRC (see Fig. S6A and D in the supplemental material) under hypoxia and showed that JUN-FOS overexpression was sufficient to increase HCV replication and intracellular ATP (Fig. 9C). In contrast, no ATP increase was detected during HIF overexpression, even though the expression of certain glycolytic enzymes was affected (see Fig. S3A and C in the supplemental material), or upon chemical-induced hypoxia (data not shown), whereas no decrease was observed after HIF-1α/2α silencing (see Fig. S3D in the supplemental material). Moreover, the enhanced FOS expression in the DMSO-differentiated Huh7.5 cells at 3% O2 (Fig. 9A) argues for a cellular proliferation-unrelated role of this oncogene in HCV replication enhancement. In addition, although expression of the AKT1 gene was found unaffected at 3% O2 in infected cells, AKT phosphorylation/activation was increased under hypoxia (Fig. 9B). Interestingly, the addition of the AKT inhibitor VIII (Akti-1/2) had a negative effect on the HCV replication enhancement at 3% O2 (Fig. 9D), even at concentrations that do not significantly alter intracellular ATP, implicating AKT activity in the hypoxia-mediated increase of HCV propagation.
Finally, supportive of our hypothesis that oncogenes are crucial for the hypoxia-mediated HCV enhancement, our results from 12% O2 show that although, as expected (41), 12% O2 does not support HIF stabilization/activation, it enhances HCV replication and, concomitantly, the expression of glycolytic enzymes and oncogenes (see Fig. S6B to E in the supplemental material).
Thus, at least in hepatoma cells, oncogenes appear to be important players in the hypoxia-driven cellular metabolic shift, enhancing glycolysis and ATP production and, concomitantly HCV replication in a HIF-independent manner. Interestingly, hypoxia-mediated induction of oncogenes has been reported also for primary liver cells (49).
CKB induction contributes to hypoxia-related HCV replication enhancement.
Creatine kinase covers the immediate energetic needs of the cell by catalyzing the reversible reaction of phosphocreatine (PCr) and ADP to creatine and ATP (54). To date, CKB upregulation under hypoxia has been reported in a three-dimensional hepatocellular culture model (53) as well as in the rat liver (51), and CKB overexpression in the liver of transgenic mice has been shown to stabilize the energy metabolism protecting against low-oxygen stress (52). Moreover, CKB, the main isotype expressed in the liver (84), is known to regulate HCV replication by forming a complex with NS3-4A and locally delivering the energy required for the helicase activity (55).
In our study, the CKB mRNA levels were strongly induced at 3% O2 (Fig. 6D), and this contributed significantly to both HCV replication enhancement and higher intracellular ATP levels at 3% O2 (Fig. 6E and F), as revealed by silencing experiments. Interestingly, CKB upregulation observed at 12% O2 argues for a HIF-independent transcriptional activation under low-oxygen conditions (see Fig. S6D in the supplemental material). Oncogene-mediated regulation of the CKB promoter has been suggested only for the case of p53 (repression) (54). However, the details of CKB modulation by oxygen tension remain elusive. Sp1 and cyclic AMP (cAMP)/protein kinase A (PKA) response elements of CKB promoter may be involved, as shown for glucose metabolism genes during low-oxygen conditions (73, 85).
Overall, our results indicate that, in addition to glycolysis, CKB may act to enhance the intracellular ATP levels under low-oxygen conditions in a HIF-independent manner. Thus, oncogene-mediated glycolysis and CKB activation are the main contributors to ATP production, at least in Huh7.5 cells, and the observed hypoxia-related enhancement of the HCV replication is directly linked to the activity of those processes. Therefore, it is tempting to speculate that HCV-infected patients may benefit by combining the current therapy with the use of glycolysis or CKB inhibitors, used already in cancer treatment (62, 86, 87).
Low oxygen tension promotes a sustained induction of HCV infection in Huh7dif cells.
A low-oxygen-mediated increase in HCV replication was also observed in Huh7.5dif cells (differentiated by DMSO) that better mimic the conditions of HCV infection in vivo (56). Interestingly, HCV replication enhancement at 3% O2 was maintained up to 2 weeks in this system (Fig. 7), consistent with its ability to support long-term HCV replication. Moreover, this system confirmed that hypoxia selectively affects the establishment of HCV replication, since preincubation of cells at 3% O2 determined the level of viral RNA throughout a 2-week observation period.
Importantly, bioenergetics reprogramming and HCV replication enhancement in the nondividing Huh7.5dif cells at 3% O2 can be exclusively linked to hypoxia adaptation, excluding confounding effects caused by increased cell proliferation (47, 58, 62, 69, 88), similarly to the HU-arrested Huh7.5 cells.
Increased HCV RNA levels correlate with increased activity of anaerobic metabolism in liver biopsy specimens.
Liver cells regulate their metabolism and structure in response to oxygen tension (16, 19). A correlation between low oxygen and increased glycolysis prevails in the liver (16, 17). Also, at least in mouse liver, CKB overexpression stabilizes energy metabolism upon hypoxic stress (51, 52).
Analyses of liver biopsy specimens derived from HCV patients with liver fibrosis due to chronic infection showed a significant association between the expression levels of anaerobic energy metabolism (HK2, LDH-A, CKB)- and hypoxia (VEGFA)-related genes and the HCV RNA load (Fig. 8). These results are in perfect agreement with our cell culture data, indicating a strong link among hypoxia, increased glycolysis rate and CKB activity and higher HCV RNA replication.
In summary, we report that HCV infection of Huh7-derived cell lines cultured under low-oxygen conditions (3% or 12%) promotes the early stages of HCV infection/replication compared to standard cell culture conditions (20% O2). This enhancement directly correlated with hypoxia-induced changes in cell energetics, including an increase in anaerobic glycolysis and CKB expression that leads to elevated ATP production. Surprisingly, activation of HIF-1α/2α was dispensable for this enhancement, whereas activation of certain oncogenes appears to be the central event in the HCV RNA replication induction. This study opens new possibilities in optimizing conditions for in vitro propagation of HCV and defining novel therapeutic targets.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to T. Wakita for the JFH1 isolate, C. Rice for Huh7.5 cells and the 9E10 antibody, R. Hernandez-Alcoceba for the p9xHRE-Luc plasmid, and L. Bakiri for the JUN-FOS and MMP1-luc vectors. We acknowledge H. Boleti for scientific advice and editing as well as E. Xingi for technical assistance in the microscopy experiments.
This work was funded by the Hellenic General Secretariat of Research and Technology and partially by Institute Pasteur, Paris, France. Work in the laboratory of R.B. was supported by grants from the Deutsche Forschungsgemeinschaft (FOR1202, TP1, and SFB/TRR77, TP A1).
Footnotes
Published ahead of print 26 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02534-12.
REFERENCES
- 1. Levrero M. 2006. Viral hepatitis and liver cancer: the case of hepatitis C. Oncogene 25:3834–3847 [DOI] [PubMed] [Google Scholar]
- 2. Seeff LB. 2002. Natural history of chronic hepatitis C. Hepatology 36:S35–S46 [DOI] [PubMed] [Google Scholar]
- 3. Feld JJ, Hoofnagle JH. 2005. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436:967–972 [DOI] [PubMed] [Google Scholar]
- 4. Moradpour D, Penin F, Rice CM. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453–463 [DOI] [PubMed] [Google Scholar]
- 5. Tang H, Grise H. 2009. Cellular and molecular biology of HCV infection and hepatitis. Clin. Sci. (Lond.) 117:49–65 [DOI] [PubMed] [Google Scholar]
- 6. Eng FJ, Walewski JL, Klepper AL, Fishman SL, Desai SM, McMullan LK, Evans MJ, Rice CM, Branch AD. 2009. Internal initiation stimulates production of p8 minicore, a member of a newly discovered family of hepatitis C virus core protein isoforms. J. Virol. 83:3104–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vassilaki N, Mavromara P. 2009. The HCV ARFP/F/core+1 protein: production and functional analysis of an unconventional viral product. IUBMB Life 61:739–752 [DOI] [PubMed] [Google Scholar]
- 8. Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed] [Google Scholar]
- 9. Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 11. Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, Abid K, Negro F, Dreux M, Cosset FL, Bartenschlager R. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. U. S. A. 103:7408–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaul A, Woerz I, Meuleman P, Leroux-Roels G, Bartenschlager R. 2007. Cell culture adaptation of hepatitis C virus and in vivo viability of an adapted variant. J. Virol. 81:13168–13179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ashfaq UA, Khan SN, Nawaz Z, Riazuddin S. 2011. In-vitro model systems to study hepatitis C virus. Genet. Vaccines Ther. 9:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Groot H, Noll T. 1987. Oxygen gradients: the problem of hypoxia. Biochem. Soc. Trans. 15:363–365 [DOI] [PubMed] [Google Scholar]
- 15. Fleckenstein W, Weiss C. 1984. Ein neues Gewebe-pO2-Messverfahren zum Nachweis von Mikrozirkulationsstorungen. Focus Med. Hochschule Lubeck. 1:74–84 [Google Scholar]
- 16. Jungermann K, Kietzmann T. 2000. Oxygen: modulator of metabolic zonation and disease of the liver. Hepatology 31:255–260 [DOI] [PubMed] [Google Scholar]
- 17. Torre C, Perret C, Colnot S. 2010. Molecular determinants of liver zonation. Prog. Mol. Biol. Transl. Sci. 97:127–150 [DOI] [PubMed] [Google Scholar]
- 18. Semenza GL. 2011. Oxygen sensing, homeostasis, and disease. N. Engl. J. Med. 365:537–547 [DOI] [PubMed] [Google Scholar]
- 19. Martinez I, Nedredal GI, Oie CI, Warren A, Johansen O, Le Couteur DG, Smedsrod B. 2008. The influence of oxygen tension on the structure and function of isolated liver sinusoidal endothelial cells. Comp. Hepatol. 7:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aghi MK, Liu TC, Rabkin S, Martuza RL. 2009. Hypoxia enhances the replication of oncolytic herpes simplex virus. Mol. Ther. 17:51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Connor JH, Naczki C, Koumenis C, Lyles DS. 2004. Replication and cytopathic effect of oncolytic vesicular stomatitis virus in hypoxic tumor cells in vitro and in vivo. J. Virol. 78:8960–8970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haque M, Davis DA, Wang V, Widmer I, Yarchoan R. 2003. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains hypoxia response elements: relevance to lytic induction by hypoxia. J. Virol. 77:6761–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pillet S, Le Guyader N, Hofer T, NguyenKhac F, Koken M, Aubin JT, Fichelson S, Gassmann M, Morinet F. 2004. Hypoxia enhances human B19 erythrovirus gene expression in primary erythroid cells. Virology 327:1–7 [DOI] [PubMed] [Google Scholar]
- 24. Polonis VR, Anderson GR, Vahey MT, Morrow PJ, Stoler D, Redfield RR. 1991. Anoxia induces human immunodeficiency virus expression in infected T cell lines. J. Biol. Chem. 266:11421–11424 [PubMed] [Google Scholar]
- 25. Riedinger HJ, van Betteraey M, Probst H. 1999. Hypoxia blocks in vivo initiation of simian virus 40 replication at a stage preceding origin unwinding. J. Virol. 73:2243–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shen BH, Hermiston TW. 2005. Effect of hypoxia on Ad5 infection, transgene expression and replication. Gene Ther. 12:902–910 [DOI] [PubMed] [Google Scholar]
- 27. Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Friebe P, Boudet J, Simorre JP, Bartenschlager R. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Werth N, Beerlage C, Rosenberger C, Yazdi AS, Edelmann M, Amr A, Bernhardt W, von Eiff C, Becker K, Schafer A, Peschel A, Kempf VA. 2010. Activation of hypoxia inducible factor 1 is a general phenomenon in infections with human pathogens. PLoS One 5:e11576 doi:10.1371/journal.pone.0011576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kato T, Furusaka A, Miyamoto M, Date T, Yasui K, Hiramoto J, Nagayama K, Tanaka T, Wakita T. 2001. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J. Med. Virol. 64:334–339 [DOI] [PubMed] [Google Scholar]
- 31. Schmitt M, Scrima N, Radujkovic D, Caillet-Saguy C, Simister PC, Friebe P, Wicht O, Klein R, Bartenschlager R, Lohmann V, Bressanelli S. 2011. A comprehensive structure-function comparison of hepatitis C virus strain JFH1 and J6 polymerases reveals a key residue stimulating replication in cell culture across genotypes. J. Virol. 85:2565–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vassilaki N, Friebe P, Meuleman P, Kallis S, Kaul A, Paranhos-Baccala G, Leroux-Roels G, Mavromara P, Bartenschlager R. 2008. Role of the hepatitis C virus core+1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J. Virol. 82:11503–11515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mylonis I, Chachami G, Samiotaki M, Panayotou G, Paraskeva E, Kalousi A, Georgatsou E, Bonanou S, Simos G. 2006. Identification of MAPK phosphorylation sites and their role in the localization and activity of hypoxia-inducible factor-1α. J. Biol. Chem. 281:33095–33106 [DOI] [PubMed] [Google Scholar]
- 34. Bakiri L, Matsuo K, Wisniewska M, Wagner EF, Yaniv M. 2002. Promoter specificity and biological activity of tethered AP-1 dimers. Mol. Cell. Biol. 22:4952–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vassilaki N, Boleti H, Mavromara P. 2008. Expression studies of the HCV-1a core+1 open reading frame in mammalian cells. Virus Res. 133:123–135 [DOI] [PubMed] [Google Scholar]
- 36. Zhou W, Dosey TL, Biechele T, Moon RT, Horwitz MS, Ruohola-Baker H. 2011. Assessment of hypoxia inducible factor levels in cancer cell lines upon hypoxic induction using a novel reporter construct. PLoS One 6:e27460 doi:10.1371/journal.pone.0027460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Woerz I, Lohmann V, Bartenschlager R. 2009. Hepatitis C virus replicons: dinosaurs still in business? J. Viral Hepat. 16:1–9 [Google Scholar]
- 38. Binder M, Quinkert D, Bochkarova O, Klein R, Kezmic N, Bartenschlager R, Lohmann V. 2007. Identification of determinants involved in initiation of hepatitis C virus RNA synthesis by using intergenotypic replicase chimeras. J. Virol. 81:5270–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. 2006. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol. Cell 21:521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Arnaud N, Dabo S, Maillard P, Budkowska A, Kalliampakou KI, Mavromara P, Garcin D, Hugon J, Gatignol A, Akazawa D, Wakita T, Meurs EF. 2010. Hepatitis C virus controls interferon production through PKR activation. PLoS One 5:e10575 doi:10.1371/journal.pone.0010575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bracken CP, Fedele AO, Linke S, Balrak W, Lisy K, Whitelaw ML, Peet DJ. 2006. Cell-specific regulation of hypoxia-inducible factor (HIF)-1α and HIF-2α stabilization and transactivation in a graded oxygen environment. J. Biol. Chem. 281:22575–22585 [DOI] [PubMed] [Google Scholar]
- 42. Nasimuzzaman M, Waris G, Mikolon D, Stupack DG, Siddiqui A. 2007. Hepatitis C virus stabilizes hypoxia-inducible factor 1α and stimulates the synthesis of vascular endothelial growth factor. J. Virol. 81:10249–10257 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43. Ripoli M, D'Aprile A, Quarato G, Sarasin-Filipowicz M, Gouttenoire J, Scrima R, Cela O, Boffoli D, Heim MH, Moradpour D, Capitanio N, Piccoli C. 2010. Hepatitis C virus-linked mitochondrial dysfunction promotes hypoxia-inducible factor 1α-mediated glycolytic adaptation. J. Virol. 84:647–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wilson GK, Brimacombe CL, Rowe IA, Reynolds GM, Fletcher NF, Stamataki Z, Bhogal RH, Simoes ML, Ashcroft M, Afford SC, Mitry RR, Dhawan A, Mee CJ, Hubscher SG, Balfe P, McKeating JA. 2012. A dual role for hypoxia inducible factor-1α in the hepatitis C virus lifecycle and hepatoma migration. J. Hepatol. 56:803–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Loboda A, Jozkowicz A, Dulak J. 2010. HIF-1 and HIF-2 transcription factors—similar but not identical. Mol. Cells 29:435–442 [DOI] [PubMed] [Google Scholar]
- 46. Nagel S, Talbot NP, Mecinovic J, Smith TG, Buchan AM, Schofield CJ. 2010. Therapeutic manipulation of the HIF hydroxylases. Antioxid. Redox Signal. 12:481–501 [DOI] [PubMed] [Google Scholar]
- 47. Webster KA. 2003. Evolution of the coordinate regulation of glycolytic enzyme genes by hypoxia. J. Exp. Biol. 206:2911–2922 [DOI] [PubMed] [Google Scholar]
- 48. Gwak GY, Yoon JH, Kim KM, Lee HS, Chung JW, Gores GJ. 2005. Hypoxia stimulates proliferation of human hepatoma cells through the induction of hexokinase II expression. J. Hepatol. 42:358–364 [DOI] [PubMed] [Google Scholar]
- 49. Lee SH, Lee MY, Lee JH, Han HJ. 2008. A potential mechanism for short time exposure to hypoxia-induced DNA synthesis in primary cultured chicken hepatocytes: correlation between Ca(2+)/PKC/MAPKs and PI3K/Akt/mTOR. J. Cell. Biochem. 104:1598–1611 [DOI] [PubMed] [Google Scholar]
- 50. Sheta EA, Trout H, Gildea JJ, Harding MA, Theodorescu D. 2001. Cell density mediated pericellular hypoxia leads to induction of HIF-1alpha via nitric oxide and Ras/MAP kinase mediated signaling pathways. Oncogene 20:7624–7634 [DOI] [PubMed] [Google Scholar]
- 51. Govorova LV, Teplov SI. 1976. Changes in the creatine kinase activity of the brain, heart, liver and plasma of rats subjected to oxygen starvation. Biull. Eksp. Biol. Med. 81:177–179 (In Russian.) [PubMed] [Google Scholar]
- 52. Miller K, Halow J, Koretsky AP. 1993. Phosphocreatine protects transgenic mouse liver expressing creatine kinase from hypoxia and ischemia. Am. J. Physiol. 265:C1544–C1551 [DOI] [PubMed] [Google Scholar]
- 53. Pruksakorn D, Lirdprapamongkol K, Chokchaichamnankit D, Subhasitanont P, Chiablaem K, Svasti J, Srisomsap C. 2010. Metabolic alteration of HepG2 in scaffold-based 3-D culture: proteomic approach. Proteomics 10:3896–3904 [DOI] [PubMed] [Google Scholar]
- 54. Wyss M, Kaddurah-Daouk R. 2000. Creatine and creatinine metabolism. Physiol. Rev. 80:1107–1213 [DOI] [PubMed] [Google Scholar]
- 55. Hara H, Aizaki H, Matsuda M, Shinkai-Ouchi F, Inoue Y, Murakami K, Shoji I, Kawakami H, Matsuura Y, Lai MM, Miyamura T, Wakita T, Suzuki T. 2009. Involvement of creatine kinase B in hepatitis C virus genome replication through interaction with the viral NS4A protein. J. Virol. 83:5137–5147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bauhofer O, Ruggieri A, Schmid B, Schirmacher P, Bartenschlager R. 2012. Persistence of HCV in quiescent hepatic cells under conditions of an interferon-induced antiviral response. Gastroenterology 143:429–438.e8 doi:10.1053/j.gastro.2012.04.018 [DOI] [PubMed] [Google Scholar]
- 57. Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, Thompson CB. 2004. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 64:3892–3899 [DOI] [PubMed] [Google Scholar]
- 58. Kim JW, Dang CV. 2006. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 66:8927–8930 [DOI] [PubMed] [Google Scholar]
- 59. Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. 2007. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol. Cell. Biol. 27:7381–7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Levine AJ, Puzio-Kuter AM. 2010. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 330:1340–1344 [DOI] [PubMed] [Google Scholar]
- 61. Michiels C, Minet E, Michel G, Mottet D, Piret JP, Raes M. 2001. HIF-1 and AP-1 cooperate to increase gene expression in hypoxia: role of MAP kinases. IUBMB Life 52:49–53 [DOI] [PubMed] [Google Scholar]
- 62. Pelicano H, Martin DS, Xu RH, Huang P. 2006. Glycolysis inhibition for anticancer treatment. Oncogene 25:4633–4646 [DOI] [PubMed] [Google Scholar]
- 63. Polytarchou C, Iliopoulos D, Hatziapostolou M, Kottakis F, Maroulakou I, Struhl K, Tsichlis PN. 2011. Akt2 regulates all Akt isoforms and promotes resistance to hypoxia through induction of miR-21 upon oxygen deprivation. Cancer Res. 71:4720–4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lee CJ, Liao CL, Lin YL. 2005. Flavivirus activates phosphatidylinositol 3-kinase signaling to block caspase-dependent apoptotic cell death at the early stage of virus infection. J. Virol. 79:8388–8399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhu YZ, Cao MM, Wang WB, Wang W, Ren H, Zhao P, Qi ZT. 2012. Association of heat-shock protein 70 with lipid rafts is required for Japanese encephalitis virus infection in Huh7 cells. J. Gen. Virol. 93:61–71 [DOI] [PubMed] [Google Scholar]
- 66. Michalopoulos GK, DeFrances MC. 1997. Liver regeneration. Science 276:60–66 [DOI] [PubMed] [Google Scholar]
- 67. Chen AY, Kleiboeker S, Qiu J. 2011. Productive parvovirus B19 infection of primary human erythroid progenitor cells at hypoxia is regulated by STAT5A and MEK signaling but not HIFα. PLoS Pathog. 7:e1002088 doi:10.1371/journal.ppat.1002088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Racker E. 1974. History of the Pasteur effect and its pathobiology. Mol. Cell. Biochem. 5:17–23 [DOI] [PubMed] [Google Scholar]
- 69. Vander Heiden MG, Cantley LC, Thompson CB. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324:1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Walters KA, Syder AJ, Lederer SL, Diamond DL, Paeper B, Rice CM, Katze MG. 2009. Genomic analysis reveals a potential role for cell cycle perturbation in HCV-mediated apoptosis of cultured hepatocytes. PLoS Pathog. 5:e1000269 doi:10.1371/journal.ppat.1000269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, Longerich T, Diehl S, Ramirez F, Balla T, Rohr K, Kaul A, Buhler S, Pepperkok R, Lengauer T, Albrecht M, Eils R, Schirmacher P, Lohmann V, Bartenschlager R. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9:32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wu X, Zhou Y, Zhang K, Liu Q, Guo D. 2008. Isoform-specific interaction of pyruvate kinase with hepatitis C virus NS5B. FEBS Lett. 582:2155–2160 [DOI] [PubMed] [Google Scholar]
- 73. Discher DJ, Bishopric NH, Wu X, Peterson CA, Webster KA. 1998. Hypoxia regulates beta-enolase and pyruvate kinase-M promoters by modulating Sp1/Sp3 binding to a conserved GC element. J. Biol. Chem. 273:26087–26093 [DOI] [PubMed] [Google Scholar]
- 74. Hoque M, Mathews MB, Pe'ery T. 2010. Progranulin (granulin/epithelin precursor) and its constituent granulin repeats repress transcription from cellular promoters. J. Cell. Physiol. 223:224–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wyatt E, Wu R, Rabeh W, Park HW, Ghanefar M, Ardehali H. 2010. Regulation and cytoprotective role of hexokinase III. PLoS One 5:e13823 doi:10.1371/journal.pone.0013823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fruehauf JP, Meyskens FL., Jr 2007. Reactive oxygen species: a breath of life or death? Clin. Cancer Res. 13:789–794 [DOI] [PubMed] [Google Scholar]
- 77. Mizukami Y, Kohgo Y, Chung DC. 2007. Hypoxia inducible factor-1 independent pathways in tumor angiogenesis. Clin. Cancer Res. 13:5670–5674 [DOI] [PubMed] [Google Scholar]
- 78. Zeng M, Kikuchi H, Pino MS, Chung DC. 2010. Hypoxia activates the K-ras proto-oncogene to stimulate angiogenesis and inhibit apoptosis in colon cancer cells. PLoS One 5:e10966 doi:10.1371/journal.pone.0010966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ishida H, Tatsumi T, Hosui A, Nawa T, Kodama T, Shimizu S, Hikita H, Hiramatsu N, Kanto T, Hayashi N, Takehara T. 2011. Alterations in microRNA expression profile in HCV-infected hepatoma cells: involvement of miR-491 in regulation of HCV replication via the PI3 kinase/Akt pathway. Biochem. Biophys. Res. Commun. 412:92–97 [DOI] [PubMed] [Google Scholar]
- 80. Kang SM, Lim S, Won SJ, Shin YJ, Lim YS, Ahn BY, Hwang SB. 2011. c-Fos regulates hepatitis C virus propagation. FEBS Lett. 585:3236–3244 [DOI] [PubMed] [Google Scholar]
- 81. Kim K, Kim KH, Kim HY, Cho HK, Sakamoto N, Cheong J. 2010. Curcumin inhibits hepatitis C virus replication via suppressing the Akt-SREBP-1 pathway. FEBS Lett. 584:707–712 [DOI] [PubMed] [Google Scholar]
- 82. Pfannkuche A, Buther K, Karthe J, Poenisch M, Bartenschlager R, Trilling M, Hengel H, Willbold D, Haussinger D, Bode JG. 2011. C-Src is required for complex formation between the hepatitis C virus-encoded proteins NS5A and NS5B: a prerequisite for replication. Hepatology 53:1127–1136 [DOI] [PubMed] [Google Scholar]
- 83. Zhang Z, Harris D, Pandey VN. 2008. The FUSE binding protein is a cellular factor required for efficient replication of hepatitis C virus. J. Virol. 82:5761–5773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Meffert G, Gellerich FN, Margreiter R, Wyss M. 2005. Elevated creatine kinase activity in primary hepatocellular carcinoma. BMC Gastroenterol. 5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Firth JD, Ebert BL, Ratcliffe PJ. 1995. Hypoxic regulation of lactate dehydrogenase A. Interaction between hypoxia-inducible factor 1 and cAMP response elements. J. Biol. Chem. 270:21021–21027 [DOI] [PubMed] [Google Scholar]
- 86. Teicher BA, Menon K, Northey D, Liu J, Kufe DW, Kaddurah-Daouk R. 1995. Cyclocreatine in cancer chemotherapy. Cancer Chemother. Pharmacol. 35:411–416 [DOI] [PubMed] [Google Scholar]
- 87. Tennant DA, Duran RV, Gottlieb E. 2010. Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer. 10:267–277 [DOI] [PubMed] [Google Scholar]
- 88. DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. 2008. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7:11–20 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.