Abstract
Coronavirus nsp1 has been shown to induce suppression of host gene expression and to interfere with the host immune response. However, the mechanism is currently unknown. The only available structural information on coronavirus nsp1 is the nuclear magnetic resonance (NMR) structure of the N-terminal domain of nsp1 from severe acute respiratory syndrome coronavirus (SARS-CoV) from the betacoronavirus genus. Here we present the first nsp1 structure from an alphacoronavirus, transmissible gastroenteritis virus (TGEV) nsp1. It displays a six-stranded β-barrel fold with a long alpha helix on the rim of the barrel, a fold shared with SARS-CoV nsp113–128. Contrary to previous speculation, the TGEV nsp1 structure suggests that coronavirus nsp1s have a common origin, despite the lack of sequence homology. However, comparisons of surface electrostatics, shape, and amino acid conservation between the alpha- and betacoronaviruses lead us to speculate that the mechanism for nsp1-induced suppression of host gene expression might be different in these two genera.
INTRODUCTION
Coronaviruses (CoVs) cause mainly respiratory and enteric disease (1). In farm animals, these viruses cause severe disease and lead to large economic losses. In humans, CoVs generally cause mild symptoms, like the common cold. However, the emergence of severe acute respiratory syndrome (SARS) in 2003 made it apparent that CoVs could also cause serious disease in the human population. CoVs contain a positive, single-stranded RNA genome of about 30 kb, which is the largest among RNA viruses (2, 3). The replicase gene, comprising two-thirds of the genome, encodes two large precursor polyproteins that are cleaved into 16 nonstructural proteins (nsp's), where nsp1 is the first to be expressed (2, 4–7).
CoVs were originally classified into three groups based on antigenic cross-reactivity (8). Subsequent phylogenetic analysis, including analysis of the replicase region, rendered the same three clusters with few exceptions. These were called groups 1, 2, and 3 (9, 10). When SARS coronavirus (SARS-CoV), the etiological agent of SARS, was discovered (11–13), it was placed as the only member in an early split-off from group 2, in subgroup 2b (10). This effectively put the viruses previously established to be members of group 2 in subgroup 2a. These groups have now been recognized as genera, where groups 1, 2, and 3 have become the genera alpha-, beta-, and gammacoronaviruses (α-CoVs, β-CoVs, and γ-CoVs), and SARS-CoV is placed in lineage B of the beta genus, β-CoVB. Since then, several SARS-like viruses have been identified, mainly in bats, and placed in β-CoVB (14, 15).
The CoV genome is generally well conserved between the genera. The largest differences in the replicase gene can be found in the 5′ end, and the most N-terminal cleavage product, nsp1, is considered one of the genus-specific markers (10, 16). This is based on both sequence comparisons and the fact that nsp1 from α-CoV, that from β-CoVA, and that from β-CoVB are different in size, ∼110, 250, and 180 amino acids, respectively. In contrast to the α-CoVs and β-CoVs, the γ-CoVs do not contain an nsp1 protein (17). The fact that no sequence homology could be inferred between the different nsp1s, or any host protein, raised the question of whether these proteins shared similar structures and functions (16). However, several studies have shown that nsp1s from both alpha- and betacoronaviruses display both differences and similarities.
It is established that nsp1 suppresses translation of host mRNA. nsp1s from human CoV-299E, murine hepatitis virus (MHV), and SARS-CoV significantly reduce reporter gene expression in HEK 293 cells (18–20). In several cell lines, SARS-CoV nsp1 suppresses host gene expression, including that of type I interferon, involved in the host immune response (21). SARS-CoV nsp1 also promotes the degradation of host mRNA (22, 23). Like SARS-CoV nsp1, transmissible gastroenteritis virus (TGEV) nsp1 can efficiently suppress host mRNA translation, although it seems to lack the ability to modify and degrade host mRNAs. There are indications that SARS-CoV nsp1 also suppresses the expression of the CoV genes (22), but recent experiments on SARS-CoV nsp1 suggest that a short sequence in the 5′ end common to all CoV mRNAs protects the viral RNA from degradation (24, 25). Deletion of nsp1 from infectious clones of MHV from β-CoVA abolishes the ability of the virus to infect cultured cells (26). A mutation in the cleavage site between nsp1 and nsp2 in the α-CoV transmissible gastroenteritis virus (TGEV), preventing the release of nsp1, leads to a drastic decrease in the viability of the virus (27).
The observed biochemical effects of nsp1 highlight the importance of this protein in the CoV life cycle and its potential role as a significant virulence factor, as well as its importance for evasion of host responses. This also indicates that nsp1 is an interesting target in the search for new antiviral drugs. The frequent detection of SARS-like CoV in mammalian hosts indicates a high risk of reintroduction into the human population (14). For development of vaccines and antivirals, it is important to understand CoV pathogenicity and its mechanism for avoiding host antiviral systems.
This article presents the first high-resolution crystal structure of nsp1 from an alphacoronavirus. To date, the only known structure of nsp1 from coronaviruses is that of SARS-CoV nsp113–128, belonging to the betacoronavirus genus, determined by nuclear magnetic resonance (NMR) (28). The structure of TGEV nsp1 reflects the structural and functional similarities and differences between α-CoV and β-CoV. It also suggests that nonstructural protein 1 was not acquired independently by the different coronavirus genera.
MATERIALS AND METHODS
Cloning, protein expression, and purification.
A full-length construct of TGEV nsp1, including an N-terminal His6 tag, was cloned into the expression plasmid pDEST14 (Invitrogen). The protein was expressed in Escherichia coli BL21-AI cells (Invitrogen) grown in LB medium at 37°C. When the optical density at 600 nm (OD600) reached 0.6, the culture was transferred to 25°C and protein expression was induced with l-arabinose (2 g/liter). After 3 to 5 h, the cells were harvested by centrifugation. The cells were washed in 1× SSP buffer (150 mM NaCl, 250 mM NaH2PO4, pH 7.4) prior to storage at −20°C. For protein purification, the cells from a 1-liter culture were thawed and resuspended in 20 ml lysis buffer (50 mM Na2HPO4, 50 mM Na2SO4, 100 mM HEPES, 200 mM NaCl, 10 mM imidazole, 0.5% Triton X-100, 14 mM β-mercaptoethanol, pH 8.0) supplemented with 0.01 mg/ml RNase, 0.02 mg/ml DNase, and 0.25 mg/ml lysozyme. The cells were subsequently lysed under 2 × 105 kPa of pressure using a Constant cell disrupter (Constant Systems Ltd.), and the lysate was centrifuged at 8°C and 45,000 × g (SS-34 rotor; Sorvall) for 20 min. The cleared cell lysate was incubated with 0.5 ml Ni-Sepharose (6 Fast Flow; GE Healthcare) for 30 min at 8°C on a shaker. The Ni matrix was washed on a column with 20 ml wash buffer (50 mM Na2HPO4, 50 mM Na2SO4, 100 mM HEPES, 200 mM NaCl, 20 mM imidazole, 14 mM β-mercaptoethanol, pH 8.0), and the protein was eluted with 2.5 ml elution buffer (same as wash buffer but with 250 mM imidazole). Directly after elution, the buffer was exchanged on a PD-10 column (Bio-Rad) and eluted with 20 mM Tris-HCl, 300 mM NaCl, pH 8.0, 14 mM β-mercaptoethanol. Ni-Sepharose purification and buffer exchange were performed at 8°C. The protein was further purified by size exclusion chromatography (HiLoad 16/60 Superdex-75; GE Healthcare). The fractions from the peak corresponding to a monomer of the TGEV nsp1 protein were pooled and diluted four times with 20 mM Tris-HCl, pH 8.0, to a NaCl concentration of 75 mM. The protein was then applied to a 1-ml HiTrapQ anion exchange column (GE Healthcare), which was washed with 20 ml of buffer A (20 mM Tris-HCl, 75 mM NaCl, pH 8.0, and 14 mM β-mercaptoethanol) and eluted with a gradient to buffer B (20 mM Tris-HCl, 1 M NaCl, pH 8.0, and 14 mM β-mercaptoethanol) over a volume of 20 ml. Both size exclusion chromatography and anion-exchange chromatography were carried out at 25°C. The TGEV nsp1 eluted at 500 mM NaCl, and the purity of the sample was >98% as judged by analysis using SDS-PAGE. The protein sample was diluted with 20 mM Tris-HCl to a NaCl concentration of 150 mM and thereafter concentrated to between 3 to 10 μg/μl in a Vivaspin concentrator (Vivascience).
A second construct with a 5-amino-acid C-terminal truncation and an N-terminal His6 tag was cloned into the expression vector pEXP5 (Invitrogen). Expression and purification were performed as for the full-length construct.
Crystallization.
For crystallization screening, drops containing 0.5 μl protein solution and 0.5 μl reservoir solution were set up as sitting-drop vapor diffusion experiments using an Oryx 4 crystallization robot (Douglas Instruments Ltd.). Initial crystal hits were obtained at 20°C under two conditions in the JCSG+ suite (Qiagen): A9 (200 mM ammonium chloride and 20% [wt/vol] polyethylene glycol [PEG] 3350) and H7 (200 mM ammonium sulfate, 100 mM Bis-Tris, pH 5.5, and 25% [wt/vol] PEG 3350). The crystallization conditions were optimized in terms of precipitant, buffer, pH, and protein concentration. Optimal concentrations of protein and PEG 4000 were batch dependent. Drops, in volumes varying between 3 and 20 μl, containing protein and reservoir solution in a 2:1 ratio, were set up. The drops were seeded with previously obtained crystals 30 min after setup. After several rounds of optimization, the best crystals were obtained in 5% (wt/vol) PEG 4000, 200 mM ammonium chloride, 30 mM HEPES, and 30 mM morpholineethanesulfonic acid (MES), pH 6.2, with a protein concentration of 5 μg/μl. Native crystals were dipped for a few seconds in reservoir solution supplemented with 15% glycerol before vitrification in liquid nitrogen. Crystals for phasing were soaked for 2 h in reservoir solution supplemented with 10 mM K2PtCl4 and thereafter back soaked for 30 min in the same solution without K2PtCl4. The Pt-soaked crystals were cryoprotected and vitrified as described above.
Data collection, phasing, and refinement.
Crystallographic data were collected at the European Synchrotron Radiation Facility (ESRF), Grenoble, France. Native data were collected at beamline ID23eh2, at a wavelength of 0.873 Å to 1.6 Å resolution. Anomalous data were collected at beamline ID14eh4. Two 360-degree data sets were collected at the Pt edge (λ = 1.072 Å) at different κ angles, with an oscillation angle of 3 degrees, to a resolution of 2.5 Å. The images were indexed and integrated in the software program MOSFLM (29) and scaled in the program Scala (30, 31). The space group was determined to be P1, with two molecules in the asymmetric unit, related by a noncrystallographic 2-fold axis as revealed by a self-rotation function calculated by the software program Molrep (31, 32). The solvent content was estimated to be 40%, with a Matthews coefficient of 2.08 (31, 33). Four platinum sites were identified by single isomorphous replacement with anomalous scattering (SIRAS) using the software program ShelxD (34). The sites were further refined in SHARP (35). Subsequent solvent flattening and histogram matching using the program DM (36) resulted in a significantly improved electron density map. The Buccaneer software program (37) was used to create a first trace of the polypeptide backbone. This initial model was further improved by alternate cycles of model rebuilding in O (38) and refinement in the Buster-TNT program (39, 40). Final refinement was performed using translation, libration, skew (TLS) refinement with the two chains as separate groups. In the N-terminal region of the A chain, an additional four residues from the His tag could be modeled. In the B chain, density for the two first residues in the N-terminal region was missing. For full phasing and refinement statistics, see Table 1.
Table 1.
Data collection and refinement statisticsa
| Statistic | Value for TGEV nsp1 |
|
|---|---|---|
| Data collection | Native | Pt |
| Beamline | ID23eh2 (ESRF) | ID14eh4 (ESRF) |
| Wavelength (Å) | 0.873 | 1.072 |
| Space group | P1 | P1 |
| Cell axial lengths (Å) | 35.4, 36.0, 42.2 | 35.6 36.1 42.7 |
| Cell angles (°) | 91.3, 109.1, 94.2 | 90.9, 109.4, 93.9 |
| Resolution range (Å) | 27.4–1.5 (1.58–1.50) | 40.3–2.5 (2.64–2.5) |
| No. of reflections measured | 122,299 (17,500) | 167,992 (24,887) |
| No. of unique reflections | 30,811 (4,441) | 6,888 (9,98) |
| Avg multiplicity | 4.0 (3.9) | 24.4 (24.9) |
| Anomalous avg multiplicity | 12.0 (12.3) | |
| Completeness (%) | 96.6 (95.4) | 99.6 (99.8) |
| Anomalous completeness (%) | 99.4 (99.8) | |
| Rmerge | 0.056 (0.476) | 0.079 (0.263) |
| 〈I/σI〉 | 14.3 (3.0) | 45.5 (19.7) |
| Refinement | ||
| Resolution range (Å) | 26.1–1.5 | |
| No. of reflections used in working set | 31,262 | |
| No. of reflections for Rfree calculation | 1,535 | |
| R (%) | 17.5 | |
| Rfree (%) | 20.9 | |
| No. of nonhydrogen atoms | 1,718 | |
| No. of solvent waters | 207 | |
| Mean B factor (Å2) | 23.3 | |
| Ramachandran plot outliers (%)b | 0.0 | |
| RMSD from ideal bond length (Å)c | 0.01 | |
| RMSD from ideal bond angle (°)b | 1.06 | |
Values in parentheses refer to the outer resolution shell. Data collection statistics were calculated using Scala, part of the CCP4 software program suite (30, 31). Refinement statistics, except for Ramachandran outliers, were calculated using Buster-TNT (39).
Calculated using a strict-boundary Ramachandran plot definition (51).
Root mean square deviation; ideal values from Engh and Huber (52).
Protein structure accession number.
The TGEV nsp1 structure has been deposited in the Protein Data Bank under PDB ID 3ZBD.
RESULTS
The TGEV nsp1 structure exhibits an irregular β-barrel fold.
Initial crystallization trials were performed with protein from a construct expressing full-length TGEV nsp1. Crystals were obtained and tested for diffraction, but no data of sufficient quality could be collected. The sequence from TGEV nsp1 was analyzed using the secondary structure prediction software programs Phyre (41) and I-Tasser (42), and a new construct was produced, containing a 5-amino-acid C-terminal truncation. This new construct yielded crystals under the same crystallization conditions as the full-length protein, and native data were collected to 1.5 Å. Anomalous data from platinum soaks were collected to 2.4 Å. The structure was subsequently solved by single isomorphous replacement with anomalous scattering using both data sets. Details of phasing and refinement are in Table 1.
The TGEV nsp1 structure is characterized by an irregular six-stranded β-barrel, flanked by a small β-sheet connected to a short 310 helix (Fig. 1). A 15-amino-acid-long α-helix is placed on the rim of the barrel. Four antiparallel strands, β3, β7, β5, and β6, make up one side of the barrel, with β3 and β6 loosely connected to strands β1 and β8, which create the other side of the barrel. β2 and β4 form a small parallel sheet flanking the barrel adjacent to β3 and β7. Some of the strands are irregular and have breaks in the hydrogen bonding pattern. This is due to a β-bulge in β7 involving the carbonyl oxygen from Val64 and amide nitrogens from Gln85 and Gly86, as well as Pro74, which makes a kink in the end of the strand β6.
Fig 1.
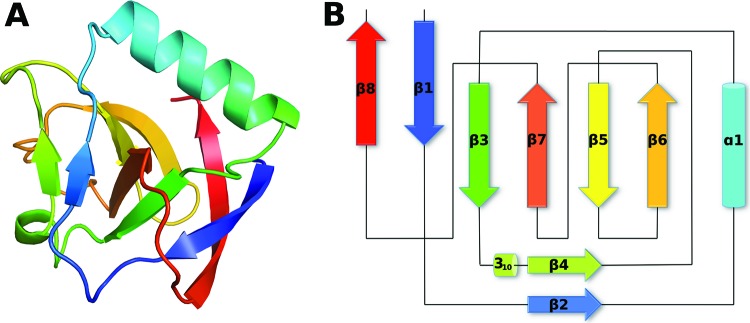
(A) Overall structure of TGEV nsp1 in rainbow colors from blue in the N-terminal region to red in the C-terminal region. (B) Topology diagram with coloring corresponding to that in panel A. Strands β1, β3, β5, β6, β7, and β8 make up the barrel. Strands β2 and β4 are unique to TGEV nsp1 and are not found in SARS-CoV nsp113–128.
A small cavity is located at the top of the barrel between β5 and β7, next to the α-helix. The cavity is lined by residues Val18, Pro19, Leu21, Val26, Glu26, Tyr41, Val61, Ile62, and Val89 and the aliphatic stem of Arg90. Glu29 has a different conformation in the A chain compared to that in the B chain, and the position of the side chain determines the size of the opening of the cavity to the solvent.
The TGEV nsp1 sequence shares no significant similarity to any known structures in the Protein Data Bank. However, a search with the crystal structure in the PDBeFold database resulted in a single significant hit, the NMR structure of the N-terminal domain of nsp1 from SARS-CoV (residues 13 to 128; PDB ID 2HSX/2GDT), with a q score of 0.37 and a z score of 6.3. The q score indicates the quality of the alignment, where 1 corresponds to an identical hit. The z score measures statistical significance of the match, where a higher number corresponds to a higher statistical significance (43).
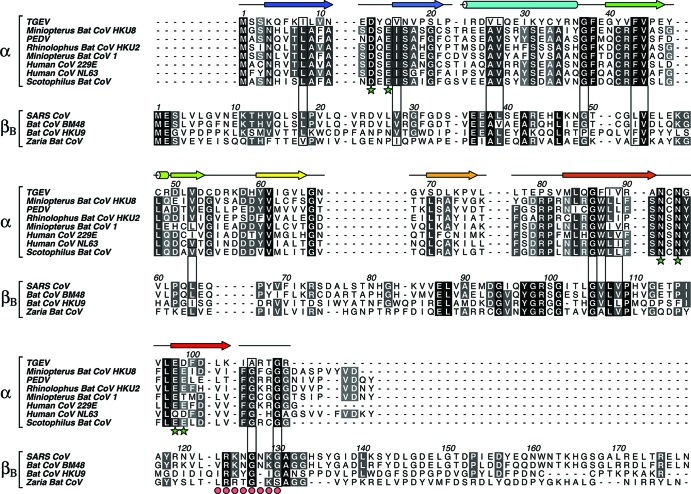
To explore the relationship between the nsp1 proteins in α-CoV and β-CoVB, sequences from both groups were gathered and aligned separately using the software program Clustal W (44). A careful structure-based sequence alignment between TGEV nsp1 and SARS-CoV nsp113–128 was used to merge the alignments of α-CoV and β-CoVB.
Conservation within the alphacoronavirus genus.
The α-CoV alignment shows a number of highly conserved areas (Fig. 2). A large portion of the conserved residues in α-CoV make up the hydrophobic core of the β-barrel fold: these include Val44, Val52, Val61, Leu77, Leu84, Phe87, Ile88, and Val89. This cluster of residues is connected to a conserved solvent-exposed hydrophobic patch consisting of Phe43 and Phe100, via Gly86, which is highly conserved due to space restraints. The Ile23-Arg35 helix shows little conservation. However, it is anchored to the hydrophobic core by the highly conserved residues Gly37 and Phe38.
Fig 2.
Eight nsp1 sequences from α-CoV and four nsp1 sequences from β-CoVB were aligned separately. The two alignments were subsequently merged by using the three-dimensional structure alignment of TGEV nsp1 and SARS-CoV nsp113–128. The level of sequence conservation within each genus is highlighted with dark background color and white letters. A darker background color indicates a higher level of conservation. Residues conserved between the genera are marked with boxes. Residues likely to be important for α-CoV function are marked by stars. These residues are further highlighted in Fig. 3 and 4. The β-CoVB consensus sequence suggested by Almeida et al. (28) is marked by circles. Secondary structure elements from the TGEV nsp1 structure are displayed above the sequence and colored according to the scheme in Fig. 1. The figure was prepared using the software program Aline (47).
The TGEV nsp1 structure contains two salt bridges: Lys7-Asp99, connecting β1 to β8, and Lys103-Asp71, connecting β8 to β6. Although Lys7-Asp99 also can be found in e.g. mink CoV in the α-CoV genus, none of these electrostatic interactions seem to be well retained throughout the CoV family.
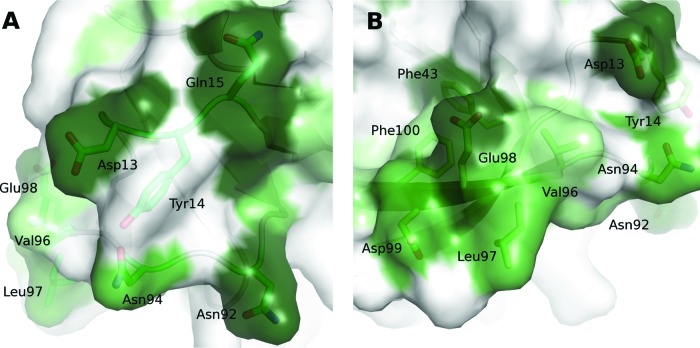
The surface of the TGEV nsp1 structure exhibits two highly conserved areas. The first is located on a ridge formed by the loops between strands β1-β2 and β7-β8 together with strand β2 (Fig. 3 and 4). On the ridge, Asp13, Gln15, Asn92, and Asn94, all conserved, are positioned in a ring around Tyr14, which is not. Gln15 is consistently replaced by a Glu in the other α-CoVs. The second conserved patch is mainly made up from side chains from strand β8, where the highest level of conservation is found in the N-terminal part. The cluster also includes one residue from β3, Phe43. Together with Phe100, this residue forms a small exposed hydrophobic patch. In this conserved area, two more hydrophobic surface-exposed residues are found: Val96 and Leu97.
Fig 3.
The surface of TGEV nsp1 displays two areas with high sequence conservation. Two loops make up the first area (A), where Asp13, Gln15, Asn92, and Asn94 make up a conserved circle. The second area (B), centered on strand β8, displays both exposed hydrophobic residues and charged residues that potentially could interact with a partner molecule or partner protein.
Fig 4.
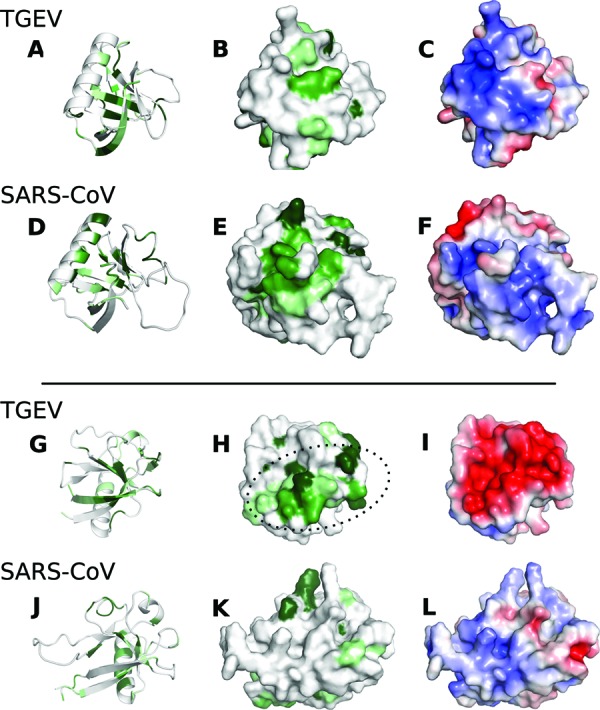
Superposed structures of TGEV nsp1 and SARS-CoV nsp113–128 are presented separately in two different rotations: rotation 1 (A to F) and rotation 2 (G to L). Conserved residues within each genus are shown in green, where darker green corresponds to a higher level of conservation. In each rotation, TGEV nsp1 and SARS-CoV nsp113–128 are shown as cartoons and surface representation displaying the conserved areas. Subfigures C, F, I, and L show the electrostatic surface potentials of the two structures from −4/+4 mV. The electrostatic potential was calculated using the software programs APBS and PDB2PQR using the PARSE force field (48–50). The area within the dotted oval in subfigure H corresponds to the two conserved areas shown in Fig 3.
Overall comparison between α-CoV and β-CoVB.
A comparison of TGEV nsp1 with the structure of SARS-CoV nsp113–128 clearly shows that the two structures share the same fold, with a characteristic six-stranded β-barrel with a long alpha helix on one side of the barrel. However, a three-dimensional alignment of TGEV nsp1 with SARS-CoV nsp113–128 reveals that there are large differences between the structures. The location of the strands in the barrel is shifted, along with an outward shift of the α-helix in TGEV nsp1 compared to SARS-CoV nsp113–128, where the helix is positioned closer to the barrel. The loop between β5 and β6 is significantly shorter in the TGEV nsp1 structure. In addition, the small β-sheet, comprising β2 and β4, flanking the barrel next to strands β3 and β7, is found only in the TGEV nsp1 structure.
The small cavity with Glu29 as a gatekeeper is not conserved. Instead, there is a narrow tunnel in the SARS-CoV nsp113–128 structure, not found in TGEV nsp1, that stretches through the center of the barrel. It appears too large to be an artifact from poor packing of the protein core. However, it is not likely to be conserved throughout the β-CoVB lineage, given the low conservation of the neighboring side chains.
Thus far, the viruses that belong to β-CoVB show lower diversity than those in α-CoV. An alignment of nsp1 from four viruses in β-CoVB, including SARS-CoV, shows three conserved areas. The mapping of these onto the SARS-CoV nsp113–128 structure is illustrated in Fig. 4. The two separate alignments of α-CoV and β-CoVB were merged using the structure-based alignment of TGEV nsp1 and SARS-CoV nsp113–128 (Fig. 2). Interestingly, the conserved regions within each group show very little overlap in the combined α-CoV and β-CoVB alignment. For example, the β-CoVB nsp1 proteins show a high level of conservation in helix α1, absent in α-CoV. The conservation pattern in the barrel is also different between the two groups. The consensus sequence LRKxGxKG, referred to by Almeida et al. (28), is roughly conserved within β-CoVB. Compared with the α-CoV sequences, only the two glycines are conserved, both of which seem to be located in the linker region between nsp1 and nsp2. However, a few residues seem to be retained across the α-CoV genus and the β-CoVB lineage. Most of these, like Ile8, Ile88, Phe43, Val44, Val52, Ile88, and Val89, are part of the conserved hydrophobic cluster, extending from the core of the barrel to the surface. Val44 is the center of a less well conserved cluster on the other side of β3. The β-bulge located by Gly86 seems to be absolutely conserved throughout α-CoV and β-CoVB and might be a characteristic feature of the nsp1 β-barrel fold.
The poor sequence conservation between α-CoV and β-CoVB is also reflected in the surface electrostatics. The open side of the TGEV nsp1 barrel exhibits a strong negative electrostatic potential, whereas the long helix features mainly positive electrostatics (Fig. 4C and I). The SARS-CoV nsp113–128 structure reveals a significantly different pattern. The electrostatic potential also seems to be slightly more conserved in β-CoVB than in α-CoV (Fig. 4).
DISCUSSION
The high-resolution crystal structure of TGEV nsp1 reveals that nsp1s from α-CoV share a fold with the N-terminal domain of the nsp1s in β-CoVB, despite their lack of sequence similarity. At the same time, the structure also highlights that there are important structural differences between the two lineages, potentially explaining their differences in function. SARS-CoV nsp1 inhibits interferon (IFN) expression in infected cells (19) and interferes with antiviral signaling pathways of the host (21). TGEV nsp1, together with SARS-CoV nsp1 and several other CoV nsp1s, can also efficiently inhibit expression of host mRNA. However, little is known about the mechanism behind this function.
The structure of TGEV nsp1 is characterized by an irregular six-stranded β-barrel flanked by an α-helix. In order to identify the evolutionarily conserved areas, an alignment was made from various nsp1 sequences from viruses in the alpha genus. The conserved residues were plotted onto the surface of the TGEV nsp1 protein. The conservation pattern within α-CoV does not give any immediate clues about the function or mechanism of α-CoV nsp1. A large portion of the conserved residues, centered on the highly conserved strand β7, make up the core of the protein and are more likely to be involved in the structural stability of the protein than to be important for the function. However, the TGEV nsp1 surface features two highly conserved patches. From these, a few residues stand out as candidates for potential interaction with a partner or target molecule. The patch made up from the two loops between strands β1-β2 and β7-β8 together with strand β2 has four residues that are of special interest. These are Asp13, which is completely conserved, Gln15, which is a conserved Glu residue in all α-CoVs except TGEV, and two asparagines on the neighboring loop, Asn92 and Asn94. These conserved residues are all placed on a protruding, ridge formation. The highest conservation of the second patch is found mainly on the edge of the ridge and going down on one side (Fig. 3B), including residues Leu97, Glu98, and Asp99. Both of these patches are potential surfaces for interaction with another molecule. The protruding shape of the ridge, as opposed to a cavity or a bowl shape, suggests that the partner molecule may be another protein. There are indications that TGEV nsp1 may need a host factor for its function. Experiments performed with cell-free HeLa extracts and rabbit reticulocyte lysate (RRL) reveal that TGEV nsp1 suppresses protein translation in the first experiment but not the second, suggesting that a host factor that exists in the HeLa extracts but not in RRL is needed for TGEV nsp1 function (22).
Intriguingly, the combined alignment of α-CoV and β-CoVB nsp1s shows that there is not much overlap between the conservation patterns of the two groups. The lack of conservation is also reflected in the shape and the electrostatics of the TGEV and SARS-CoV nsp113–128 structures, resulting in different three-dimensional volumes despite the similar β-barrel fold (Fig. 4).
It has been previously speculated that the SARS-CoV nsp1 might be a unique SARS protein and that its ability to suppress host gene expression potentially could account for its elevated pathogenicity relative to that of other coronaviruses (16). It is now established that nsp1 from α-CoV, as well as that from β-CoVA and β-CoVB, can induce suppression of host mRNA (18, 19, 20, 22, 23, 45). It is also established that SARS-CoV nsp1 binds the 40S subunit of the ribosome to make it translationally inactive. The nsp1-40S complex can modify the 5′ end of capped mRNA and induce cleavage in certain mRNAs containing the internal ribosome entry site (IRES). However, this activity cannot alone account for the substantially reduced expression of the reporter protein, suggesting that there is an additional mechanism for the suppression of host gene expression (23).
In contrast to these results, although TGEV nsp1 has been shown to effectively suppress host gene expression, no binding to the 40S ribosomal subunit has been observed (22). TGEV nsp1 also failed to promote host mRNA degradation (22). In SARS-CoV nsp1, it seems that the ability to bind to the 40S subunit is related to the second domain (residues 129 to 180), since SARS-CoV nsp1 carrying the K164A and H165A mutations was inactive in terms of 40S binding and consequently unable to degrade mRNA (23). However, nsp1 proteins from two other alphacoronaviruses, HCoV-229E and HCoV-NL63, have been shown to immunoprecipitate together with the S6 protein, which is part of the 40S subunit (46). Interestingly, these two nsp1 proteins share all of the conserved regions in the α-CoV group (Fig. 2). Thus, it cannot be ruled out that α-CoV TGEV nsp1 might interact with parts of the 40S ribosomal subunit under certain conditions.
It is tempting to speculate that the β-barrel domain of SARS-CoV nsp113–128 and TGEV nsp1 share a similar mechanism for the additional suppression of host mRNA, not accounted for by the SARS-CoV nsp1-induced modification of mRNA. However, there is no experimental data to support this. On the contrary, the K164A H165A double mutation harbored in the second domain renders SARS-CoV nsp1 completely inactive in experiments where TGEV nsp1 effectively suppressed host translation (22).
The structural differences in TGEV nsp1 and SARS-CoV nsp1, together with the available biochemical data, lead us to speculate that the nsp1 proteins from α-CoV and β-CoVB have different mechanisms for 40S-independent suppression of host mRNA. However, since the TGEV nsp1 structure has the same fold as SARS-CoV nsp113–128, it is very unlikely that the nsp1s were acquired independently by the different genera. This suggests that the coronavirus nsp1s are evolutionarily related and that the different mechanisms are a result of divergent evolution.
ACKNOWLEDGMENTS
I thank Linda Boomaars-van der Zanden and Eric J. Snijder (Leiden University Medical Center, The Netherlands) for providing the initial TGEV nsp1 full-length expression clone and Luis Enjuanes (Centro Nacional de Biotecnologia, Madrid, Spain) for providing the cloned cDNA that was used as a template. I thank Bruno Coutard and Bruno Canard (AFMB, Marseille, France) for assistance and Terese Bergfors for critical reading of the manuscript.
This work was supported by the EU IP Project VIZIER (CT 2004-511960) and Uppsala University Faculty Support to T. Alwyn Jones.
Footnotes
Published ahead of print 26 December 2012
REFERENCES
- 1. Weiss SR, Navas-Martin S. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69:635–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee HJ, Shieh CK, Gorbalenya AE, Koonin EV, Lamonica N, Tuler J, Bagdzhadzhyan A, Lai MMC. 1991. The complete sequence (22 kilobases) of murine coronavirus gene-1 encoding the putative proteases and RNA-polymerase. Virology 180:567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lomniczi B. 1977. Biological properties of avian coronavirus RNA. J. Gen. Virol. 36:531–533 [DOI] [PubMed] [Google Scholar]
- 4. Bredenbeek PJ, Pachuk CJ, Noten AFH, Charite J, Luytjes W, Weiss SR, Spaan WJM. 1990. The primary structure and expression of the 2nd open reading frame of the polymerase gene of the coronavirus MHV-A59—a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 18:1825–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brian DA, Baric RS. 2005. Coronavirus genome structure and replication, p 1–30 In Enjuanes L. (ed), Coronavirus replication and reverse genetics, vol 287 Springer, Berlin, Germany: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gorbalenya AE. 2001. Big nidovirus genome—when count and order of domains matter. Adv. Exp. Med. Biol. 494:1–17 [PubMed] [Google Scholar]
- 7. Ziebuhr J. 2005. The coronavirus replicase, p 57–94 In Enjuanes L. (ed), Coronavirus replication and reverse genetics, vol 287 Springer, Berlin, Germany [Google Scholar]
- 8. Lai MMC, Holmes KV. 2001. Coronaviruses, p 1163–1185 In Knipe DM, Howley PM. (ed), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 9. Gorbalenya AE, Snijder EJ, Spaan WJM. 2004. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 78:7863–7866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LLM, Guan Y, Rozanov M, Spaan WJM, Gorbalenya AE. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967–1976 [DOI] [PubMed] [Google Scholar]
- 12. Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953–1966 [DOI] [PubMed] [Google Scholar]
- 13. Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, Cheung MT, Cheng VC, Chan KH, Tsang DN, Yung RW, Ng TK, Yuen KY. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li WD, Shi ZL, Yu M, Ren WZ, Smith C, Epstein JH, Wang HZ, Crameri G, Hu ZH, Zhang HJ, Zhang JH, McEachern J, Field H, Daszak P, Eaton BT, Zhang SY, Wang LF. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679 [DOI] [PubMed] [Google Scholar]
- 15. Shi ZL, Hu ZH. 2008. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 133:74–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Connor RF, Roper RL. 2007. Unique SARS-CoV protein nsp1: bioinformatics, biochemistry and potential effects on virulence. Trends Microbiol. 15:51–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ziebuhr J, Schelle B, Karl N, Minskaia E, Bayer S, Siddell SG, Gorbalenya AE, Thiel V. 2007. Human coronavirus 229E papain-like proteases have overlapping specificities but distinct functions in viral replication. J. Virol. 81:3922–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kamitani W, Narayanan K, Huang C, Lokugamage K, Ikegami T, Ito N, Kubo H, Makino S. 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. U. S. A. 103:12885–12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng Makino C-TKS. 2008. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 82:4471–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zust R, Cervantes-Barragan L, Kuri T, Blakqori G, Weber F, Ludewig B, Thiel V. 2007. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 3:1062–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wathelet MG, Orr M, Frieman MB, Baric RS. 2007. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 81:11620–11633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. 2011. Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate. J. Virol. 85:638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kamitani W, Huang C, Narayanan K, Lokugamage KG, Makino S. 2009. A novel two-pronged strategy to suppress host protein synthesis by SARS coronavirus nsp1 protein. Nat. Struct. Mol. Biol. 16:1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tanaka T, Kamitani W, Dediego ML, Enjuanes L, Matsuura Y. 2012. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J. Virol. 86:11128–11137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thiel V, Ivanov KA, Putics A, Hertzig T, Schelle B, Bayer S, Weissbrich B, Snijder EJ, Rabenau H, Doerr HW, Gorbalenya AE, Ziebuhr J. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305–2315 [DOI] [PubMed] [Google Scholar]
- 26. Brockway SM, Denison MR. 2005. Mutagenesis of the murine hepatitis virus nsp1-coding region identifies residues important for protein processing, viral RNA synthesis, and viral replication. Virology 340:209–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Galan C, Enjuanes L, Almazan F. 2005. A point mutation within the replicase gene differentially affects coronavirus genome versus minigenome replication. J. Virol. 79:15016–15026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Almeida MS, Johnson MA, Herrmann T, Geralt M, Wuthrich K. 2007. Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus. J. Virol. 81:3151–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leslie AGW, Powell HR. 2007. Processing diffraction data with MOSFLM. Evolving Methods Macromol. Crystallogr. 245:41–51 [Google Scholar]
- 30. Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr. D 62:72–82 [DOI] [PubMed] [Google Scholar]
- 31. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vagin A, Teplyakov A. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30:1022–1025 [Google Scholar]
- 33. Matthews BW. 1968. Solvent content of protein crystals. J. Mol. Biol. 33:491–497 [DOI] [PubMed] [Google Scholar]
- 34. Sheldrick GM. 2008. A short history of SHELX. Acta Crystallogr. A. 64:112–122 [DOI] [PubMed] [Google Scholar]
- 35. de La Fortelle E, Bricogne G. 1997. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods, p 472–494 In Carter CW. (ed), Methods in enzymology, vol 276 Academic Press, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- 36. Cowtan K, Main P. 1998. Miscellaneous algorithms for density modification. Acta Crystallogr. D 54:487–493 [DOI] [PubMed] [Google Scholar]
- 37. Cowtan K. 2006. The Buccaneer software for automated model building. Acta Crystallogr. D 62:1002–1011 [DOI] [PubMed] [Google Scholar]
- 38. Jones TA, Zou JY, Cowan SW, Kjeldgaard M. 1991. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A 47:110–119 [DOI] [PubMed] [Google Scholar]
- 39. Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Sharff A, Smart O, Vonrhein C, Womack T. 2011. AutoBuster, version 1.10.0. Global Phasing Ltd., Cambridge, United Kingdom [Google Scholar]
- 40. Smart OS, Brandl M, Flensburg C, Keller P, Paciorek W, Vonrhein C, Womack TO, Bricogne G. 2008. Refinement with local structure similarity restraints (LSSR) enables exploitation of information from related structures and facilitates use of NCS, abstr. TP139. Abstr. Annu. Meet. Am. Crystallogr. Assoc [Google Scholar]
- 41. Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 42. Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40 doi:10.1186/1471-2105-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krissinel E, Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60:2256–2268 [DOI] [PubMed] [Google Scholar]
- 44. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 45. Tohya Y, Narayanan K, Kamitani W, Huang C, Lokugamage K, Makino S. 2009. Suppression of host gene expression by nsp1 proteins of group 2 bat coronaviruses. J. Virol. 83:5282–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang YJ, Shi HL, Rigolet P, Wu NN, Zhu LC, Xi XG, Vabret A, Wang XM, Wang TH. 2010. Nsp1 proteins of group I and SARS coronaviruses share structural and functional similarities. Infect. Genet. Evol. 10:919–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bond CS, Schuttelkopf AW. 2009. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr. D 65:510–512 [DOI] [PubMed] [Google Scholar]
- 48. Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 98:10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. 2007. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35:W522–W525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. 2004. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32:W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kleywegt GJ, Jones TA. 1996. Phi/psi-chology: Ramachandran revisited. Structure 4:1395–1400 [DOI] [PubMed] [Google Scholar]
- 52. Engh RA, Huber R. 2006. Structure quality and target parameters, p 382–392 In Rossmann MG, Arnold E. (ed), International tables for crystallography, vol F Wiley, New York, NY [Google Scholar]