Abstract
The propensity for capsid disassembly and uncoating of human adenovirus is modulated by interactions with host cell molecules like integrins and alpha defensins. Here, we use atomic force microscopy (AFM) nanoindentation to elucidate, at the single-particle level, the mechanism by which binding of these host molecules affects virus particle elasticity. Our results demonstrate the direct link between integrin or defensin binding and the mechanical properties of the virus. We show that the structure and geometry of adenovirus result in an anisotropic elastic response that relates to icosahedral symmetry. This elastic response changes upon binding host molecules. Whereas integrin binding softens the vertex regions, binding of a human alpha defensin has exactly the opposite effect. Our results reveal that the ability of these host molecules to influence adenovirus disassembly correlates with a direct effect on the elastic strength of the penton region. Host factors that influence adenovirus infectivity thus modulate the elastic properties of the capsid. Our findings reveal a direct link between virus-host interactions and capsid mechanics.
INTRODUCTION
Human adenovirus (HAdV) is one of the largest known nonenveloped double-stranded DNA (dsDNA) viruses. The ∼36-kb viral genome is encapsidated by a pseudo-T=25 icosahedral capsid that is over 90 nm in diameter with a corresponding mass of ∼150 MDa (1–3). The main capsid proteins form a closed icosahedral shell that is composed of 240 trimeric hexons, 12 pentamers of the penton base, and 12 fiber trimers. In addition, there are four cement capsid proteins (IIIa, VI, VIII, and IX) and five proteins associated with the genomic core of the virion (V, VII, μ, IVa2, and terminal protein). During the final step of particle maturation, multiple capsid proteins are posttranslationally processed by a viral protease. Approximately one-third of the more than 50 adenovirus types cause acute infections in humans that are generally self-limiting except in immunocompromised patients. Replication-defective adenoviruses are also used in a significant number of gene therapy and vaccine applications (4).
Integrin αvβ5 is one of several cell surface receptors for adenovirus that mediates internalization of the virus rather than attachment (5–7). Adenovirus binding to integrin αvβ5 promotes clustering of the receptors and activates downstream signaling pathways that facilitate virus endocytosis. Integrin binds to exposed RGD motifs on the virus penton base in a maximum stoichiometry of 4:5 (8, 9). This stoichiometric mismatch between integrin αvβ5 and the penton base is the result of a steric hindrance. It is also believed that integrin binding causes a conformation change in the penton base. The spiral untwisting of the penton base protein due to integrin binding could relax the interactions of the individual penton base subunits with the neighboring peripentonal hexons as well as cause the release of the fiber protein at the cell surface, thereby facilitating capsid disassembly at a later stage of cell entry (8, 10).
Adenovirus endosome escape and infection are restricted by human alpha defensins. Defensins are a conserved family of antimicrobial proteins, two of which (HNP1 and HD5) are known to block adenovirus infection (11–14). Binding of defensins to the outer surface of adenovirus blocks endosomal escape and thermally stabilizes the virion. Whereas cell attachment and internalization are unaffected by binding of defensins, stabilization of the virus prevents release of the endosomolytic protein VI and exposure of the internal core viral DNA; this prevents subsequent steps in the virus life cycle, including endosome escape, nuclear localization, and ultimately replication of adenovirus in the host cell (15–17). Thus, binding of defensin and integrin to adenovirus has opposing effects on capsid disassembly and infectivity.
The physical properties of virus capsids are important factors at various stages of the virus life cycle, including genome packaging and capsid maturation (18). In the present study, we determined whether this is also the case for virus-host interactions that modulate adenovirus uncoating. While multiple biochemical and structural analyses also show that integrin and defensin binding to adenovirus produce opposite effects on capsid stability, we tested this idea more rigorously using a single-particle, mechanobiological approach. To do this, we used a recently developed tool in the field of physical virology: atomic force microscopy (AFM) nanoindentation (18, 19). The morphology and mechanical resilience of the virus capsid can be probed nearly simultaneously in an AFM nanoindentation experiment. The particles are analyzed in buffer solution at ambient temperature. First, an image is recorded, after which nanoindentation is performed on a single-particle basis to monitor the elastic response of the capsid. Forces can be applied beyond a threshold where capsid integrity is maintained, which allows for the study of mechanical failure of the virion particle.
In previous AFM studies of viral mechanics, it was demonstrated that the material properties of virus capsids are intimately linked with the viral life cycle. For example, genome encapsidation in cowpea chlorotic mottle virus (CCMV), minute virus of mice (MVM), and phage λ was shown to have a profoundly stabilizing effect on the virus capsid (20–22). The combined process of proteolytic maturation and genome packaging in herpes simplex virus 1 nucleocapsids was also shown to increase their mechanical resilience (23). Similarly, maturation of bacteriophage HK97 also results in capsid stabilization (24). In the HK97 study, it was shown that covalent cross-linking of capsomeres and reorganization of the capsid quaternary structure are responsible for the increase in mechanical strength of the capsid and in its resistance to material fatigue. Proteolytic maturation has the opposite effect in adenovirus, where the mature capsid is more prone to disassembly than are immature capsids, thereby priming the virion for efficient uncoating upon cell entry (25). Finally, phage φ29 and norovirus were shown to have built-in prestress in the capsid that strengthens the particles, thereby increasing their survivability in hostile environments (26, 27). Prestress refers to the fact that some capsid geometries have global nonzero lateral and shear forces acting on the shell, even in optimal structures at their free-energy minima. While accumulating evidence has demonstrated the impact of mechanical stability on the earlier stages of the viral life cycle, there is still little evidence of its importance in viral infectivity. One of the few studies that relate mechanics to infectivity was performed on a retrovirus, HIV-1, revealing that a “stiffness switch” during maturation correlates with the ability of viral particles to enter cells (28).
Here, we present AFM nanoindentation studies of intact human adenovirus type 5 (Ad5) particles equipped with the adenovirus type 35 (Ad35) fiber, designated Ad35F, to examine the effects of integrin and defensin association. We demonstrate that binding of integrin αvβ5 and human alpha defensin 5 (αHD5) have opposite effects on the elastic response of Ad35F, revealing a direct link between virus-host interactions and the mechanical properties of the capsid.
MATERIALS AND METHODS
Proteins and virus preparations.
The DAV-1 monoclonal antibody that recognizes the penton base RGD loop was purified on protein G-Sepharose as previously described (29). The purified IgG antibody was resuspended in 50 mM Tris-HCl, pH 8.0. Soluble recombinant integrin αvβ5 was produced in insect cells using baculovirus and purified by immunoaffinity chromatography as previously described (5). Chemically synthesized human alpha defensin HD5 was obtained from Peptides International, Inc. (Louisville, KY); resuspended at 100 μM in sterile water; and stored at −80°C until use. Human adenovirus type 5 (HAdV5) equipped with the Ad35 fiber, designated Ad35F, was purified as described previously (1, 30).
AFM.
Imaging and nanoindentation were performed at room temperature in solution. The buffer for Ad35F–DAV-1 complexes was 40 mM Tris (pH 8.1), 500 mM sodium chloride, 2% (wt/vol) sucrose, and 1% (wt/vol) mannitol. The buffer for Ad35F-αvβ5 integrin complexes was 20 mM Tris (pH 8.1), 150 mM sodium chloride, and 2 mM magnesium dichloride. The buffer for Ad35F-HD5 complexes was phosphate-buffered saline (PBS), 10 mM sodium phosphate (pH 7.4), 137 mM sodium chloride, 3 mM potassium chloride. The complexes were prepared by incubating either antibody DAV-1, αvβ5 integrin, or defensin HD5 with Ad35F in a 10-fold molar excess to the penton base monomer for 1 h on ice before depositing the solution on the AFM substrate. Free Ad35F was analyzed under all three buffer conditions. The final Ad35F concentration during the measurements was 6 μg/ml.
Free Ad35F and its complexes with either DAV-1, integrin αvβ5, or defensin HD5 were prepared for AFM nanoindentation by incubating a 100-μl droplet of virus solution for approximately 15 min at room temperature on silanized glass slides. The silanized glass substrates were prepared as described previously (19). Briefly, this procedure consisted of first rinsing the glass slides with deionized water. The slides were dried overnight and then incubated for 16 h in an ethanol-water bath and saturated with potassium hydroxide, followed by another round of thorough rinsing and drying. The cleaned glass slides were rendered hydrophobic by incubating them overnight in a hexamethyldisilazane vapor. After the 15-min incubation of the virus solution on the AFM substrate, another 100 μl of buffer solution was added before mounting the AFM head with a wetted tip onto the sample. Olympus OMCL-RC800PSA rectangular, silicon-nitride cantilevers with a nominal spring constant of 0.05 N/m and a nominal tip radius of 15 nm were used. The cantilevers were calibrated using the method of Sader et al. (31), giving an average value of 0.0524 ± 0.002 (standard deviation [SD]) N/m. Viral imaging and nanoindentation were performed on a Nanotec Electronica AFM (Tres Cantos, Spain), operated in jumping mode. Nanoindentation was performed with a probe velocity of 55 nm/s, and the data were analyzed using a home-built Labview application described previously (32). Briefly, the photodiode signal was converted to units of force based on the slope of a force distance curve (FDC) measured on the glass substrate, where distance is the z-displacement of the piezo scanner (see Fig. 1a). The spring constant of the virus particle was determined from the slope of the FDC on the virus, as determined by linear regression on the linear-like part of the FDC, using Hooke's law, treating the cantilever-virus system as two springs in series. In the case of adenovirus, the linear-like part of the FDC generally starts within 0.05 to 0.10 nN of the contact point between the tip and virus and generally ends within 0.05 nN before the breaking point. The indentation of the particles at a certain force is determined by taking the cantilever displacement at the corresponding force of an FDC measured on glass and subtracting this from the cantilever displacement of the FDC on the virus particle at the same force. Doing this for a complete nanoindentation cycle on a particular viral particle yields the force indentation curve (FIC).
Fig 1.
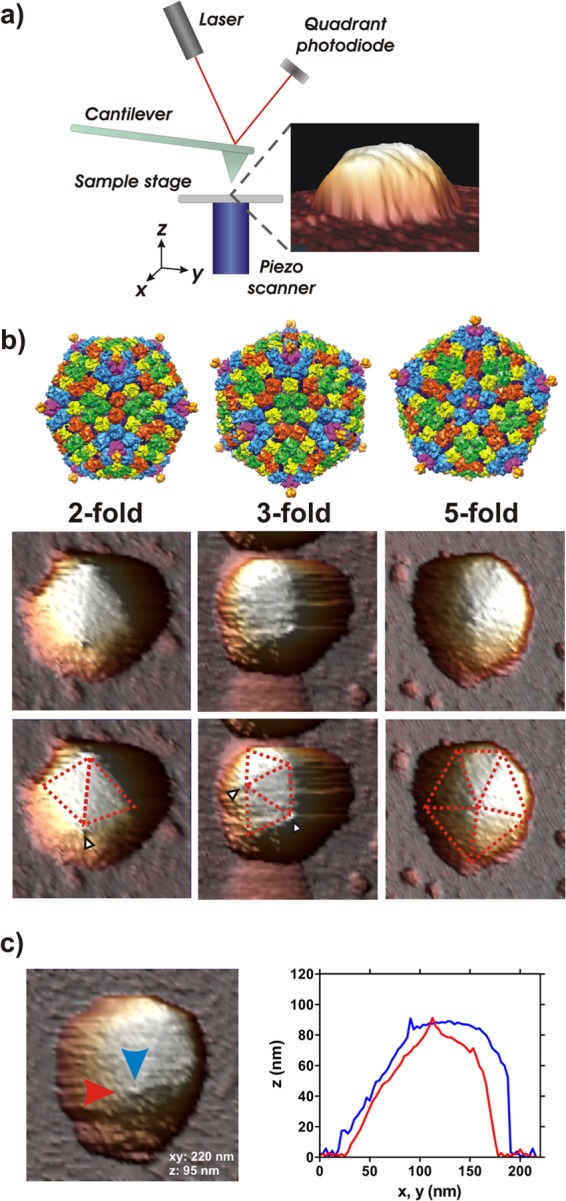
AFM imaging of intact Ad35F. (a) Principal components of the AFM setup. The piezo scanner moves the sample in all directions, allowing for the sharp tip at the end of the cantilever to sample the surface. Topography changes result in bending of the cantilever and a concomitant change in laser signal on the quadrant photodiode. The example image of an Ad35F particle lying on a coverslip is shown in three-dimensional rendering under a 77° viewing angle and shows how AFM can accurately image top surfaces but never beneath an object. (b) (Top) Surface rendering of Ad35F based on the crystal structure of the virus (Protein Data Bank [PDB] code, 1VSZ) showing the three different particle orientations of the icosahedral capsids The four unique hexon trimers are depicted in red, green, yellow, and blue (peripentonal). The penton base is shown in magenta, and the short Ad35 fiber is shown in gold. A surface-exposed cement protein that forms a helical bundle is depicted in dark blue. (Middle) Ad35F virions imaged by AFM, top-view images. Ad35F has a strongly faceted capsid which can be clearly seen from the AFM images. Typical images of Ad35F deposited on their 2-, 3-, and 5-fold axes are shown as indicated. Each image is 250 by 250 nm in the xy dimension and approximately 90 nm in the z dimension. (Bottom) The same images with morphological features annotated. Facet edges are indicated by dashed red lines. Several surface protrusions can also be distinguished, indicated by white arrowheads, and likely represent the trimeric fiber. (c) AFM image of free Ad35F with a visible surface protrusion on one of the vertices, alongside height profiles along the direction indicated by the arrows.
The average maximum imaging force was approximately 90 to 120 pN. It was recently reported that AFM imaging of HAdV5 at similar forces might result in mechanical failure of the virus (33). We observed damage during imaging on several occasions (estimated at 10 to 20%) during our measurements, and nanoindentation was not pursued for these particles. For every individual nanoindentation, the integrity of the shell was carefully checked from the AFM image. Moreover, the relatively high spring constants and breaking forces in our measurements also indicate that the mechanical integrity of the shells was preserved during imaging as performed prior to nanoindentation. Image processing and analysis were performed using WSxM software (34). The tip-sample correction for determining the width of force-induced disassembly products was performed using simple geometrical considerations, adapted from previously described procedures (35). The particles were approximated as upright cylinders with radius r and height h. Then, the broadening half-width, wh, can be estimated as wh = [h1 × (2Rt − h1)]0.5 + r, where Rt is the tip radius and h1 is the height at which wh was determined. For Rt, we used the nominal tip radius of 15 nm, and h1 was set at 0.5h (i.e., the apparent width was determined at half the maximum height of the particle, as the full width at half maximum [FWHM]). Thus, at this particular height the following relation holds: FWHM = 2wh.
Normal mode analysis of Mackay icosahedron.
To model the effects of virus nanoindentation, a Mackay icosahedron (36) was constructed, which consisted of 252 nodes and had a radius of ∼9.5 arbitrary length units (ALU). A nondimension elastic network model was constructed (37), in which any two nodes separated by no more than 4 ALU were connected by a harmonic spring. All nodes were given a uniform mass (1 arbitrary mass unit), and initially all springs were given a uniform stiffness (1 arbitrary force/length unit). This cutoff length permitted connections between both first-neighbor and second-neighbor nodes; each of the 12 nodes situated at 5-fold vertices had 15 connections, while all other nodes had 17 or 18 connections. From the Hookean potential energy function of the elastic network model, a Hessian matrix was constructed and a full diagonalization was performed to determine the system's eigenvalues and eigenvectors (normal modes) of the system. The normal modes are the principal directions and provide a natural basis for describing any deformation/conformational change. The lowest-frequency modes have been shown to associate with global/large-scale conformational changes in many biomolecules and complexes (38). As has been shown previously, for an icosahedrally symmetric structure, all modes were either nondegenerate (unique eigenvalue), indicating icosahedrally symmetric modes, or had 3-, 4-, or 5-fold degeneracy (repeated eigenvalues, same period of oscillation) (39, 40). We also constructed an elastic network model to represent αvβ5-HAdV by softening the stiffness in the region of the 5-fold vertices. The softening was accomplished by decreasing the spring constant of the springs that were directly connected to the nodes at the 5-fold vertices. In order to observe a significant shift in mode frequencies, the springs connected to the 5-fold node were reduced by a factor of 100. By softening the 5-fold node springs, there is an overall downward shift in the eigenvalue spectrum. Since the overall rigidity of the particles appears to remain intact upon integrin binding, we renormalize the softened network eigenvalues (λ), by the ratio , where the sums are performed over the lowest 200 modes, excluding the first 6 modes, which correspond to rigid body movements. With the renormalization, we conserve the sum of the eigenvalues over the low-frequency end of the spectrum between the uniform (U) and softened (S) networks.
Deformations to the model were performed by orienting the model with the axis of deformation aligned with the positive Z axis. The deformation was performed in spherical coordinates, where the radial positions, ri, were displaced a distance Δri = 2.44cos(ϴi), where ϴi is the polar angle node i makes between the positive Z axis and its radial vector; only those atoms in the upper hemisphere of the shell were deformed. The cosine form of the deformation is analogous to an l10 spherical harmonic deformation, which has previously been shown to be a good descriptor of the nanoindentation deformation shape (41). Projections were computed between the 200 lowest-frequency normal modes and the displacements, calculated using only the upper hemisphere nodes, by computing normalized dot products. The modes were then sorted to identify those modes which had the largest-magnitude projection onto the displacements. Those modes which project onto the displacement share components of their direction with the direction of the displacement; in essence, those are the modes which characterize (partially) the motion/displacement of interest. We found that the top five modes had nontrivial projections (>0.2), and we used these modes to interpret the stiffness. For a harmonic oscillator, the frequency of oscillation is given by , where k is the spring constant and m is the mass of the bead. For a given mode, λ = ω2, so we can take λ ∼ k, and we then sum the λs over the top five modes to calculate the effective spring constants, which are presented in Results.
RESULTS
AFM imaging of intact Ad35F virions.
Adenovirus particles (Ad35F) were investigated by AFM on a single-particle basis. First, images were recorded in order to determine the centers of the particles, the target for the nanoindentation experiments (Fig. 1a). The Ad35F particles average ∼90 nm in height, which is in agreement with previously published cryo-electron microscopy (cryo-EM) and crystal structure data of the Ad35F capsid (Fig. 1b; see also Table A1 in the Appendix) (1, 2). The icosahedral symmetry of the Ad35F particles can be clearly distinguished from the AFM images, both from the hexagonal/pentagonal contour and from the triangular facets of the capsid surface (Fig. 1b). One key morphological feature of adenovirus capsids is the presence of fibers that protrude radially outwards from the vertex. The adenovirus strain of the present study, Ad35F, is based on Ad5, equipped with the short fiber protein of Ad35. As the fiber is a relatively flexible component of the capsid (42), it is hard to distinguish this as a clear morphological feature due to the inherent force-application of AFM imaging. This is consistent with previous reports on AFM imaging of Ad5, which possesses the longer Ad5 wild-type (WT) fiber, in which no visible fibers were reported (25, 33). In the AFM images of Ad35F, a small number of particles did exhibit small surface protrusions that are located on each of the vertices of the icosahedral shell (Fig. 1b, white arrowheads, and Fig. 1c). These likely represent the trimeric fiber of Ad35F. The spikes appear relatively short compared to the expected length of the Ad35F fiber protein (expected, ∼9 nm from the base emerging from the penton to the tip of the fiber knob; mean height in imaging, 6.6 ± 2.2 nm [SD]; n = 13). On the one hand, this will be accounted for by the fact that the fiber is unlikely to be perfectly perpendicular to the imaging plane and will thus appear shorter based on simple trigonometric considerations, and on the other hand, this could indicate that the fibers are partially pushed away by the AFM tip.
The Ad35F particles are randomly adsorbed onto the substrate with respect to their icosahedral symmetry. As a result, the individual particles are probed along different icosahedral symmetry axes, i.e., either on the edge between two triangular facets (2-fold), at the center of a facet (3-fold), or on a vertex (5-fold). Hence, by correlating AFM imaging with the nanoindentation results, the mechanical response of Ad35F can be distinguished with respect to the icosahedral structure of the capsid. The contact area between the tip and sample is estimated to be on the scale of single capsomeres (43). It was recently demonstrated that the elastic response as probed by AFM correlates with localized structural dynamics, hence allowing the mechanical response of adenovirus to be discussed in terms of localized structural dynamics with respect to the icosahedral symmetry of the capsid (44).
Mechanical failure of the virion.
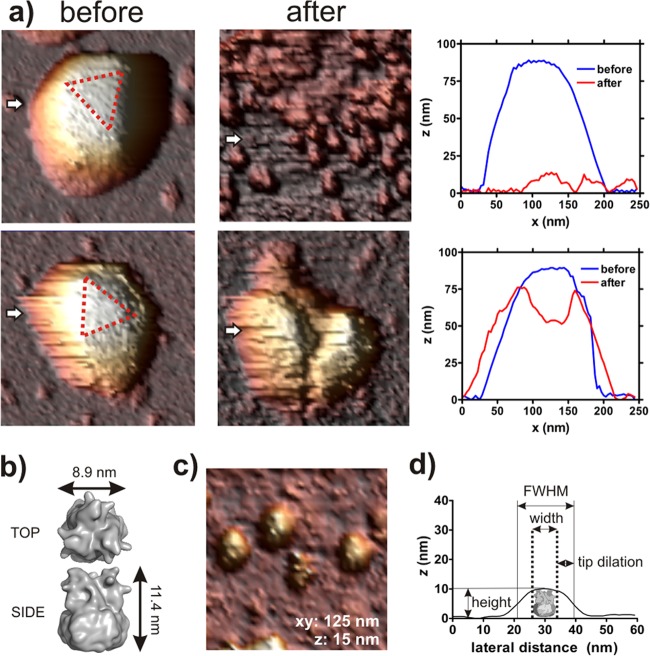
Regardless of particle orientation, the virions exhibit an initial linear-like force response, and the spring constant of the particle can be determined from the slope of the FDC. With indentations of ∼5 to 20 nm, the particles are no longer able to withstand the relatively high forces, and they suddenly break (Fig. 2a and 3). This is reflected by sharp transitions to lower forces in the curve (Fig. 3a). Imaging of Ad35F after nanoindentation reveals that the high load that was applied to the capsid results in severe capsid damage (Fig. 2a). Most particles appear to have completely disintegrated into a large number of smaller particles. A subset of particles (5 to 10%) display a hole where the AFM tip broke through, sometimes accompanied by cracks running along the remainder of the shell. We did not find a correlation between particle orientation or mechanical response and the two observed mechanically induced disassembly pathways.
Fig 2.
AFM imaging of Ad35F reveals virion disassembly upon mechanical failure. (a) Ad35F particles subjected to mechanical failure. The top row shows a particle that completely disintegrated into individual smaller particles in response to mechanical failure. Left, particle before nanoindentation; right, particle after nanoindentation. The corresponding height profiles are taken along the white arrows indicated in the images. The complete breakage of virions into their component hexon subunits is observed in the vast majority of cases. The bottom row shows a particle that partially maintained capsid integrity upon mechanical failure. There is clearly a hole where the AFM tip broke through the shell, as well as a crack running along the remainder of the capsid. This type of response is observed in approximately 5 to 10% of all particles. Images are 250 by 250 nm in xy and z dimensions as indicated in the corresponding height profile. (b) Surface rendering of an individual hexon trimer as described in the crystal structure of Ad35F (PDB code, 1VSZ). (c) AFM images of the smaller particles that arise from mechanical failure of Ad35F. The image is 125 by 125 nm in xy dimensions and 15 nm in the z dimension as indicated. (d) A typical height profile of force-induced disassembly products. A schematic drawing of the hexon trimer is presented to scale, and the imaging parameters that were used to assess the particle dimensions are indicated on the height profile.
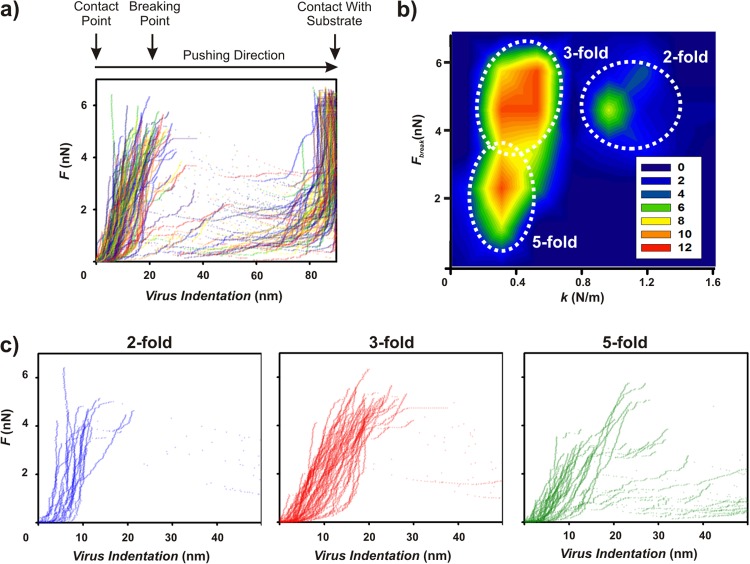
Fig 3.
AFM nanoindentation of free Ad35F reveals distinct elastic responses along each of the icosahedral symmetry axes. (a) FICs for all individual free Ad35F virions. As the tip approaches and pushes into the virion, there is a steady increase in force. As the indentation increases to between approximately 5 and 20 nm, the particles break as revealed by sharp transitions toward lower forces. (b) Two-dimensional density analysis of spring constant versus breaking force reveals the presence of three distinct populations in the mechanical response of Ad35F. (c) FICs as presented in panel a, binned according to the orientation along which the particles were probed. The force response is distinct along each of the three icosahedral symmetry axes, each accounting for one of the maxima observed in panel b.
We further investigated the identity of the force-induced disassembly products that appear with the majority of virions. Due to their large number, they are likely hexon trimers. This was confirmed by analyzing the height and apparent width of 50 of these disassembly products from the AFM images. The hexon trimer as described in the crystal structure of Ad35F is approximately 11.4 nm in height and averages 8.9 nm in width (1) (Fig. 2b). The apparent height in AFM is not distorted by tip-sample dilation and can be directly extracted from the AFM image (Fig. 2c and d). The 50 analyzed particles average 11.0 ± 0.8 nm in height, which is consistent with the height of the hexon trimer. The apparent width of the particles is heavily dilated by the relatively large tip (nominal tip-radius of 15 nm). The expected, calculated FWHM of the disassembly products is 32 nm as follows from the relations described in Materials and Methods. However, the measured FWHM will be lower due to the fact that the hexon will be pushed sideways when imaging off-center and due to the fact that the hexons are not perfect cylinders but rather possess slightly curved corners on top. On average, the FWHM of the 50 particles is 25 nm, which is a reasonable figure to expect in light of above considerations for the hexon trimers (Fig. 2c and d). Hence, the number of observed particles and their dimensions provide major evidence that mechanical failure of HAdV results in disassembly into individual capsomeres, in particular hexons. It is possible that some of these hexons also contain associated cement proteins that are too small (relative to the hexon) to be detected using this method.
Icosahedral symmetry and the mechanical response of the virion.
We observed a rather large spread in the mechanical response of Ad35F (Fig. 3a). In a two-dimensional analysis of spring constant versus breaking force, three mechanically distinct populations were detected (Fig. 3b). These populations are distinguished based on the mechanical response according to icosahedral symmetry. In particular, the force response of Ad35F is substantially different when probed on an edge, facet, or vertex (Fig. 3b and c and Table 1). The spring constant and breaking force are lowest along the 5-fold icosahedral symmetry axis. The stiffness along the 3-fold symmetry axis is 1.5-fold higher, and that along the 2-fold axis is 3.6-fold higher than that on the 5-fold axis. The breaking forces are approximately equal along the 2- and 3-fold axes; both are 1.9-fold higher than that on the 5-fold axis. Figure 3c shows all force indentation curves for each of the three icosahedral axes to illustrate that the mechanical response is strongly influenced by the icosahedral symmetry of the capsid. Notably, the spring constant obtained along the icosahedral 3-fold symmetry axis (0.43 N/m) in our studies is in good agreement with that reported for wild-type Ad5 in an independent study using a similar approach (0.46 N/m) (25).
Table 1.
Elastic response of HAdV and virus-host cell molecular complexes
| Axis | Sample |
kvira (N/m) |
||
|---|---|---|---|---|
| Avg | SEM | n | ||
| 2 (edge) | Ad35F | 1.03 | 0.04 | 25 |
| Ad35F + integrin | 1.25 | 0.04 | 15 | |
| Ad35F + HD5 | 0.98 | 0.06 | 12 | |
| 3 (facet) | Ad35F | 0.43 | 0.01 | 49 |
| Ad35F + integrin | 0.41 | 0.02 | 23 | |
| Ad35F + HD5 | 0.49 | 0.03 | 27 | |
| 5 (vertex) | Ad35F | 0.31 | 0.02 | 36 |
| Ad35F + integrin | 0.15 | 0.03 | 8 | |
| Ad35F + HD5 | 0.48 | 0.04 | 13 | |
kvir, virion spring constant.
The anisotropic mechanical response of Ad35F, i.e., the different elastic responses along the three icosahedral axes, can be explained by the structure of the capsid. The anisotropic response of icosahedral shells scales with shell thickness, and the stiffness along the 5-fold symmetry axis becomes proportionally lower with thicker shells (20). The contiguous part of the Ad35F capsid is approximately 10 nm thick, roughly 25% of the average shell radius. In shells of these dimensions, the force response is expected to be lowest along the 5-fold symmetry axis, consistent with our results. Around the 2-fold axis of Ad35F, there are several structural features that could be related to its relatively high spring constant. First, the hexons at the 2-fold axis have a unique “handshake” association comprised of extended hypervariable region loop associations containing amino residues 184 to 194. This interaction is not present between 3-fold related hexons (1). The capsid also contains an extensive network of a cement protein (IX) that connects the triangular facets via tetrameric helix bundles that are located along the ribs of the shell (1, 2). Figure 4 illustrates how the 2-fold axis is reinforced by hexon-hexon and cement protein associations. The extended conformation of protein VIII mainly stabilizes quasi-3-fold junctions on the capsid interior. However, it also cements the hexons (red) at the 2-fold symmetry axes with the 3-fold related hexons (green).
Fig 4.
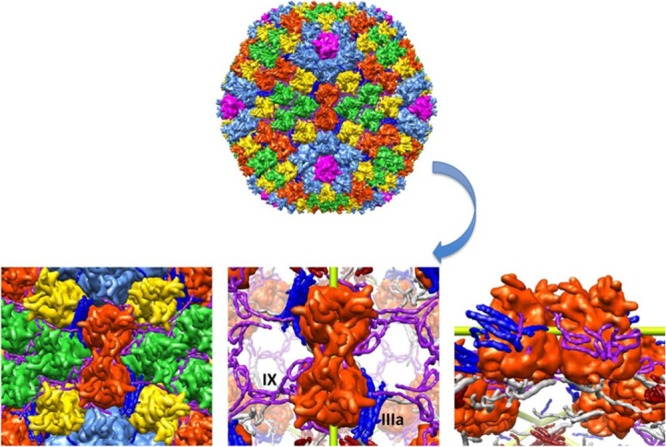
Surface rendering of Ad35F oriented at its 2-fold axis based on the crystal structure. The individual proteins are color coded as described for Fig. 1. Two lower panels (left and center) show a close-up view of the two hexons that are in tight association and in contact with multiple cement proteins. Another lower panel (right) shows a side view of the interacting cement proteins that bolster hexon associations.
Elastic properties of Ad35F in a complex with host cell factors.
Next, the effect of integrin αvβ5 and defensin HD5 binding to the Ad35F vertex region was investigated. The components of the buffers used to produce stable virus-host cell molecular complexes were different from the components of the buffer used for the virus alone. Therefore, the mechanical properties of free Ad35F were tested in all the different buffer systems used in this study, and no significant effects of buffer composition were observed (see Table A1 in the Appendix). Ad35F was incubated with an excess of the soluble extracellular domain of human integrin αvβ5 to form a complex and then subjected to nanoindentation. As with the free particles, integrin αvβ5-HAdV complexes exhibited an initial linear force response that was followed by a sudden breaking event (Fig. 5). Differentiating the force response along the different icosahedral symmetry axes revealed that integrin αvβ5 binding has a significant effect on the mechanical response of the capsid (Fig. 6). The stiffness of the vertex region in integrin αvβ5-HAdV is 55% lower than that of free particles (P < 0.0001 in a one-way analysis of variance [ANOVA], including the particles in complex with defensin described below, and P < 0.01 in a direct Bonferroni-corrected two-tailed t test between free and integrin-bound particles). The spring constant along the 3-fold axis remains unchanged but is accompanied by a moderate 25% increase in the stiffness of the 2-fold axis (P = 0.0007 in the one-way ANOVA and P < 0.01 in a direct comparison between free and integrin-bound particles in the Bonferroni-corrected two-tailed t test).
Fig 5.
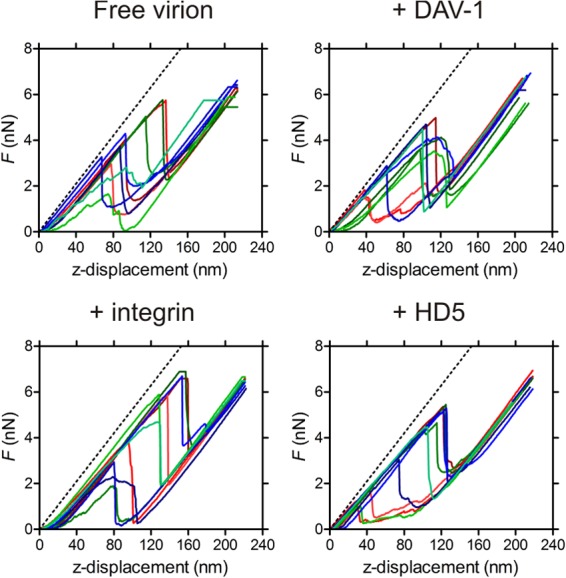
FDCs of free Ad35F and Ad35F complexed with DAV-1, integrin, and HD5. For each experiment, 10 representative curves are shown in different colors. The reference FDC on glass is shown as a dashed black line.
Fig 6.
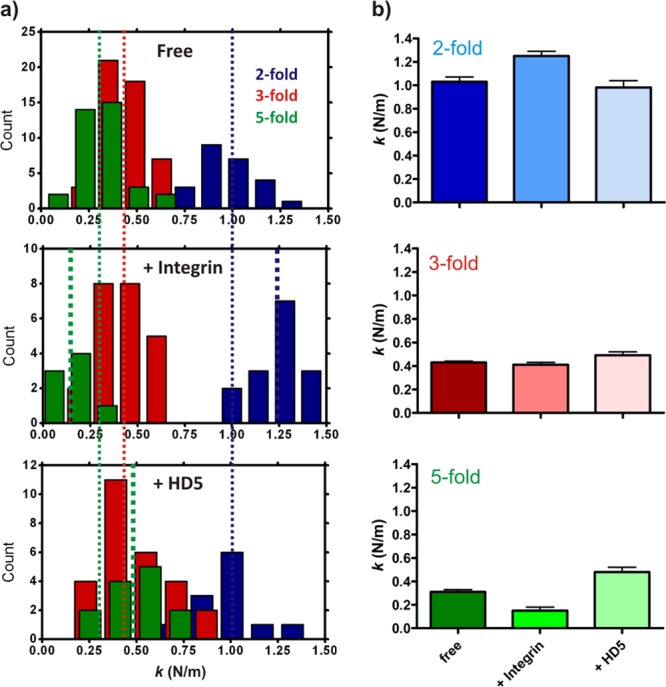
The anisotropic elastic response of Ad35F following host cell molecular interactions that modulate capsid disassembly. (a) Frequency distributions of spring constants determined for Ad35F alone, Ad35F-integrin, or Ad35F-HD5 complexes. Thin dashed lines indicate the averages determined for Ad35F alone (free); thick dashed lines indicate the averages for Ad35F-host cell molecular complexes. (b) Average spring constants determined for Ad35F, Ad35F-integrin, or Ad35F-HD5 complexes, as determined along each of the icosahedral symmetry axes. Error bars represent standard errors of the means; source data are presented in Table 1.
To further investigate the seemingly allosteric effects of integrin binding, specifically, the increase in stiffness at the 2-fold axis concomitant with decreased stiffness at the 5-fold vertex, we performed a modeling study. The complexity and large size of adenovirus make it computationally prohibitive for performing structure-based calculations. Therefore, we performed calculations employing a toy-model, a Mackay icosahedron (36, 40) (Fig. 7a), to investigate if the icosahedral symmetry alone encodes a link between the lower stiffness at the 5-fold axis and greater stiffness at the 2-fold axis. A simple bead-spring elastic network model was constructed, and the normal modes of the structure were determined. The normal modes were determined for a network with a uniform spring constant (representing free Ad35F) and for a network in which the springs directly connected to the bead at the 5-fold vertex are softened (representing the integrin αvβ5-HAdV complex). We then proceeded to deform the capsid along either the 5-fold or the 2-fold axis (Fig. 7b and c) and determined which normal modes contribute (project onto) the deformation mode. As a control calculation, we also performed the deformation along the 3-fold axis (not shown).
Fig 7.
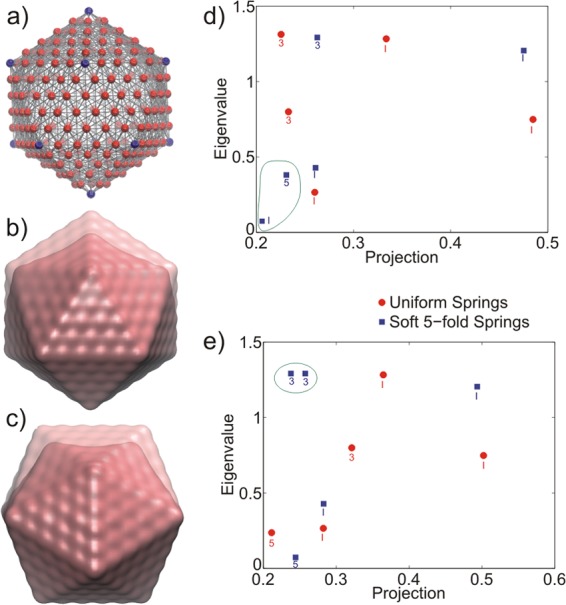
Normal mode analysis of Mackay icosahedron. (a) Elastic network representation of the icosahedron. The 5-fold nodes are colored blue; all other nodes are red. (b and c) Deformations along the 5-fold (b) and 2-fold (c) axes. The undeformed configuration is shown by the transparent structure. (d and e) Normal mode projections onto the 5-fold displacement (d) and 2-fold displacement (e) are shown against the mode eigenvalues; the 5 modes with the largest projections are shown. Each mode is identified by its degeneracy (icosahedral [I], 5-fold degenerate, or 3-fold degenerate) and whether the network contained uniform springs (red circles) or softened springs at the 5-fold vertices (blue squares). The modes that are circled in green are the modes most responsible for shifting the overall stress response between the uniform and softened networks.
When the deformation is performed along either the 5-fold or the 2-fold axis, the dominant mode in both the uniform and softened networks is an icosahedral mode, which is consistent with the previously observed robustness of icosahedral modes in capsid motions (45, 46). However, there are differences when examining the secondary modes, which still contribute significantly (projection > 0.2) to the deformation mode. For the 5-fold deformation (Fig. 7d) on the softened network, low-frequency modes (a 5-fold degenerate and an icosahedral mode) which are not present in the uniform network contribute to the deformation. Conversely, when the deformation is performed along the 2-fold axis (Fig. 7e), two higher-frequency modes (both 3-fold degenerate) contribute to the deformation mode but are not contributing to the deformation on the network with uniform springs. By summing the eigenvalues of the top 5 modes that project onto the deformation mode (those shown in Fig. 7d and e), we can estimate an effective spring constant for the deformation. Let us denote these different spring constants as k5U, k5S, k2U, and k2S, where the U and S subscripts represent the uniform or softened network, respectively, and the “2” and “5” subscripts represent deformation along the 5-fold axis or 2-fold axis, respectively. From this analysis, we obtain the ratios k5U//k5S = 1.30 and k2U//k2S = 0.78.
To further support the applicability of our toy-model to understand elastic properties of virus capsids, we performed the deformation along a 3-fold axis in both the uniform and softened networks. By performing the same analysis as was done for the 2- and 5-fold deformations, we found that the spectrum of eigenvalues, corresponding to the top modes which project onto the displacement, showed little change between the uniform and softened networks. We computed effective spring constants for the 3-fold deformation and found the ratio k3U/k3S = 0.98, indicating that the stiffness at the 3-fold axis is nearly unchanged upon softening of the 5-fold vertex. These results show that when the spring constant along the 5-fold axis decreases (due to, for instance, integrin binding), the spring constant along the 2-fold axis increases and the spring constant along the 3-fold axis is constant. These modeling results qualitatively support the experimental findings on αvβ5-HAdV compared to free Ad35F and, given the simplicity of the model, raise the intriguing possibility that these features could be observed in other icosahedrally symmetric structures (e.g., other viral capsids).
Next, the mechanical properties of the complex of Ad35F with human alpha defensin 5 (αHD5) were studied by nanoindentation to reveal whether the virion's elasticity is affected upon binding this host cell molecule. We observed a force response in αHD5-HAdV complexes similar to that in the free virion (i.e., linear force with indentation followed by sudden breaking of the shell [Fig. 5]). There was no effect of defensin binding on the mechanical response of the facets and edges of the capsids. However, there was a pronounced increase (66%) in the stiffness of the vertex (P < 0.0001 in a one-way ANOVA, P < 0.001 in the Bonferroni-corrected t test) (Fig. 6). These results show that the ability of alpha defensins to block HAdV disassembly is correlated with mechanical reinforcement of the penton region and suggest that defensins prevent release of the penton via a mechanism that relates to diminished structural dynamics in this region.
To confirm that the observed effect of integrin αvβ5 and defensin binding to adenovirus is due to specific interactions with the penton base, an analysis of adenovirus in complex with the monoclonal antibody DAV-1 was included as a control. DAV-1 also interacts with the penton base RGD loop, but the intact antibody has no effect on the infectivity of adenovirus (29). We observed no changes in the mechanical response of the DAV-1–Ad35F complex compared to the free particles (Fig. 5; see also Table A1 in the Appendix). This confirms that the mechanical responses in the integrin αvβ5- and defensin-adenovirus complexes are uniquely related to their ability to impact both uncoating and infectivity.
DISCUSSION
The mechanical properties of Ad35F were probed with AFM nanoindentation, and we revealed that the elasticity of the capsid is correlated with the early events in the virus life cycle, in particular with virus-host interactions that modulate adenovirus capsid disassembly. A striking mechanical feature of Ad35F is the strong anisotropic response with regard to the icosahedral symmetry of the capsid. Integrin αvβ5 binding to Ad35F pentons has been suggested to precede capsid disassembly and release of the DNA from the core. Furthermore, it was recently shown that the vertex regions of large, faceted viruses are under an internal compressive prestress (47). Due to the prestress at the 5-fold vertices, the energy penalty of creating a hole for genome release is likely to be lower at those sites than around it, making the pentons a favorable candidate for a genome release site. Here we used a nanoindentation approach to show that integrin binding increases the flexibility of the vertex region, and an increase in flexibility generally precedes structural events such as the breaking of protein-protein interactions. The mechanical consequences of a strongly faceted icosahedral shell with a high T number thus appear to promote the infectivity of adenovirus through the interaction with its cell surface receptor at the 5-fold axis. In contrast, the host defense system intervenes through the action of defensin HD5, which “cements” the penton into the capsid. This reinforces and stiffens the vertex of the icosahedral capsid, thus preventing the most crucial steps of capsid disassembly and genome uncoating. The anisotropic mechanical properties of adenovirus alone and in association with the host factors thus play a crucial role in the infectivity of the virus, and several structural features of the capsid are likely responsible for the anisotropic force response.
The mechanism of how disassembly of the capsid and uncoating of the adenovirus genome are triggered is still under debate. Cryo-EM data for adenovirus without integrin and complexed with integrin indicate that the penton is inserted in a slightly twisted way in the virion and that integrin binding leads to an untwisting of the penton (8, 9). So, the binding of integrin to a penton causes a conformational change relaxing the interactions of the penton base with the peripentonal hexons. This loosening of the penton base would then facilitate uncoating. In fact, the electron density for the penton base region appears to be more diffuse in the complexed particle than in the free particle, suggesting that a subset of the pentons may have already been released from the complex. As it was recently demonstrated that the elastic strength of virus capsids correlates with localized structural dynamics (44), the current nanoindentation findings, showing a clear elastic softening along the vertex regions, represent a substantial support for the theory that integrin binding loosens the penton base in the adenovirus capsid. Hence, our AFM nanoindentation experiments support the notion that the conformational change in the penton base of HAdV upon integrin binding facilitates uncoating. Together with the recent results of Pérez-Berná et al., this indicates that there are at least two consecutive and distinct mechanical events that lead to uncoating (25). The first step occurs during assembly and maturation as the capsid organization is loosened by the proteolytic maturation. The second and final step occurs during infection by complex formation of the virions with integrin, leading to the further, necessary destabilization of the penton regions and to subsequent genome release.
In addition to the large, relative decrease in stiffness along the 5-fold symmetry axis, we also observed a moderate increase in stiffness along the 2-fold axis upon integrin binding. This is possibly related to the localization of stress at the vertices of the capsid that is the result of the icosahedral structure of the shell. Our modeling results support this notion, by showing that deformation along a 2-fold axis is characterized by a stiffer effective spring constant, when the network has softer interactions at the 5-fold vertex, compared to uniform interactions throughout. Furthermore, theoretical studies of stress distribution in viral capsids show that for high-T-number structures, there is considerable positive lateral and shear stress on the vertices (48, 49). The stress is apparently relieved from the vertices upon integrin binding: it appears from our comparison of free Ad35F with the integrin-Ad35F complex that the built-in stress accounts for at least half of the elastic strength of the vertex (it decreases from 0.31 N/m to 0.15 N/m after integrin binding).
The mechanism by which HD5 prevents capsid disassembly and uncoating appears to be quite distinct from how integrin binding facilitates this process. It was shown in heat-stability assays of adenovirus that defensin HD5 binding prevents release of the penton from the capsid (16). The subsequent release of the interior capsid protein VI, which is required for endosomal escape, is prevented, thus restricting infection (17). Whereas HD5 acts on the same crucial step in capsid disassembly as does integrin, it does not appear to modulate the stress distribution in the icosahedral shell but simply cements the penton into capsid. The mechanical strength of the vertex region, in terms of the elasticity, is increased by almost 70%, thereby presumably preventing capsid disassembly and subsequent replication.
Another striking feature in the mechanical response of Ad35F is the complete disassembly of the capsid upon mechanical failure. We observed only a small fraction of particles where a recognizable capsid remained after nanoindentation, and most particles completely disintegrated into individual capsomeres. We observed no evidence for preservation of Group-Of-Nine/Six hexons, indicating that force-induced disassembly is more complete than that achieved by treatments with organic solvents such as pyridine. Markedly different effects of mechanical failure on capsid integrity have been reported for other virus capsids. In herpes simplex virus 1, the AFM tip was shown to penetrate the capsid upon mechanical failure, displacing several capsomeres but leaving the rest of the shell mostly intact (23). A similar pattern of just several displaced capsomeres was reported for phage φ29 (35). Phage λ and HK97 proheads also exhibit discrete fracture patterns (21, 24). In hepatitis B virus and HK97 head II, it was shown that capsid integrity is almost completely maintained upon mechanical failure, apart from the observation that capsid height is not fully restored (24, 32, 50). Even though clear differences in mechanical failure can be distinguished between different virus capsids, a common theme for adenovirus, herpes simplex virus 1, and φ29 is also apparent. In all three systems, it is evident that mechanically induced disassembly follows a path that is delineated by penton-hexon or hexon-hexon boundaries. This confirms that these capsomeres are fundamental structural units of virus (dis)assembly.
In conclusion, we demonstrated with a direct experimental approach that virus-host cell molecular interactions impact the mechanical properties of the virus capsid. Whereas previous mechanical studies focused on the overall stability of virion particles, we were able to reveal the actual location and mechanism of stabilization and destabilization. Host cell molecules with opposite effects on capsid disassembly modulate the elastic response of the HAdV virion accordingly. The infectivity of HAdV appears crucially dependent on the icosahedral geometry of the capsid, mainly through the anisotropic stiffness and stress distribution that arise as a consequence of specific structural features of the virion. These findings represent significant advances in our understanding of viral infectivity, as we have uncovered a direct link between virus-host interactions that critically influence cell entry processes and the mechanical properties of virus capsids.
ACKNOWLEDGMENTS
G.J.L.W. is supported by an STW-administered NanoSci E+ grant, by Fundamenteel Onderzoek der Materie (FOM) through the “Physics of the genome” program, and by the NanoNextNL consortium of the Dutch Government and partners. G.R.N. received support from NIH grants RO1 HL054352 and EY011431. V.S.R. received support from NIH grant AI070771.
APPENDIX
The complexes of Ad35F with DAV-1, integrin, or defensin were incubated and measured by AFM in different buffer solutions. Hence, to ensure that none of the observed effects of host-factor binding on the elastic response were due to the accompanying differences in buffer composition, we performed control experiments on free Ad35F in all corresponding buffers. Table A1 lists these results together with the average height and breaking forces found in our experiments.
Table A1.
Summary of Ad35F nanoindentationa
| Axis | Preparation | Buffer |
k (N/m) |
Fbreak (nN) |
h (nm) |
n | |||
|---|---|---|---|---|---|---|---|---|---|
| Avg | SEM | Avg | SEM | Avg | SEM | ||||
| 2 | Ad35F | DX | 1.03 | 0.04 | 5.4 | 0.3 | 91.3 | 0.6 | 10 |
| TMN | 1.00 | 0.07 | 4.0 | 0.2 | 92.4 | 0.5 | 10 | ||
| PBS | 1.03 | 0.08 | 4.5 | 0.2 | 91.4 | 0.4 | 5 | ||
| Ad35F + DAV-1 | DX | 0.96 | 0.06 | 4.8 | 0.3 | 93.1 | 0.3 | 10 | |
| Ad35F + integrin | TMN | 1.25 | 0.04 | 5.3 | 0.3 | 92.2 | 0.3 | 15 | |
| Ad35F + HD5 | PBS | 0.98 | 0.06 | 5.4 | 0.2 | 92.6 | 0.6 | 12 | |
| 3 | Ad35F | DX | 0.42 | 0.03 | 4.6 | 0.2 | 91.5 | 0.3 | 21 |
| TMN | 0.44 | 0.02 | 4.6 | 0.2 | 92.1 | 0.3 | 20 | ||
| PBS | 0.41 | 0.02 | 4.2 | 0.3 | 92.1 | 0.3 | 8 | ||
| Ad35F + DAV-1 | DX | 0.40 | 0.04 | 4.9 | 0.2 | 91.2 | 0.5 | 8 | |
| Ad35F + integrin | TMN | 0.41 | 0.02 | 5.1 | 0.2 | 92.9 | 0.3 | 23 | |
| Ad35F + HD5 | PBS | 0.49 | 0.03 | 4.2 | 0.2 | 91.5 | 0.2 | 27 | |
| 5 | Ad35F | DX | 0.29 | 0.01 | 3.6 | 0.5 | 95.4 | 0.6 | 10 |
| TMN | 0.33 | 0.03 | 2.4 | 0.2 | 93.7 | 0.4 | 16 | ||
| PBS | 0.29 | 0.04 | 2.0 | 0.4 | 94.5 | 0.5 | 10 | ||
| Ad35F + DAV-1 | DX | 0.27 | 0.04 | 2.7 | 0.4 | 93.8 | 0.7 | 9 | |
| Ad35F + integrin | TMN | 0.15 | 0.03 | 2.4 | 0.3 | 95.4 | 0.6 | 8 | |
| Ad35F + HD5 | PBS | 0.48 | 0.04 | 3.6 | 0.5 | 94.8 | 0.3 | 13 | |
Reported are the average virion spring constant k, breaking force Fbreak, and maximum particle height h. Buffer conditions as follows: DX, 40 mM Tris (pH 8.1), 500 mM sodium chloride, 2% (wt/vol) sucrose, and 1% (wt/vol) mannitol; TMN, 20 mM Tris (pH 8.1), 150 mM sodium chloride, and 2 mM magnesium dichloride; PBS, 10 mM sodium phosphate (pH 7.4), 137 mM sodium chloride, and 3 mM potassium chloride.
Footnotes
Published ahead of print 26 December 2012
REFERENCES
- 1. Reddy VS, Natchiar SK, Stewart PL, Nemerow GR. 2010. Crystal structure of human adenovirus at 3.5 Å resolution. Science 329:1071–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu H, Jin L, Koh SBS, Atanasov I, Schein S, Wu L, Zhou ZH. 2010. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329:1038–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nemerow GR, Stewart PL, Reddy VS. 2012. Structure of human adenovirus. Curr. Opin. Virol. 2:115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kay MA. 2011. State-of-the-art gene-based therapies: the road ahead. Nat. Rev. Genet. 12:316–328 [DOI] [PubMed] [Google Scholar]
- 5. Mathias P, Galleno M, Nemerow GR. 1998. Interactions of soluble recombinant integrin αvβ5 with human adenoviruses. J. Virol. 72:8669–8675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stewart PL, Nemerow GR. 2007. Cell integrins: commonly used receptors for diverse viral pathogens. Trends Microbiol. 15:500–507 [DOI] [PubMed] [Google Scholar]
- 7. Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. 1993. Integrins α(v)β3 and α(v) β5 promote adenovirus internalization but not virus attachment. Cell 73:309–319 [DOI] [PubMed] [Google Scholar]
- 8. Lindert S, Silvestry M, Mullen TM, Nemerow GR, Stewart PL. 2009. Cryo-electron microscopy structure of an adenovirus-integrin complex indicates conformational changes in both penton base and integrin. J. Virol. 83:11491–11501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiu CY, Mathias P, Nemerow GR, Stewart PL. 1999. Structure of adenovirus complexed with its internalization receptor, α(v)β5 integrin. J. Virol. 73:6759–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nakano MY, Boucke K, Suomalainen M, Stidwill RP, Greber UF. 2000. The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. J. Virol. 74:7085–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bastian A, Schäfer H. 2001. Human α-defensin 1 (HNP-1) inhibits adenoviral infection in vitro. Regul. Pept. 101:157–161 [DOI] [PubMed] [Google Scholar]
- 12. Gropp R, Frye M, Wagner TOF, Bargon J. 1999. Epithelial defensins impair adenoviral infection: implication for adenovirus-mediated gene therapy. Hum. Gene Ther. 10:957–964 [DOI] [PubMed] [Google Scholar]
- 13. Klotman ME, Chang TL. 2006. Defensins in innate antiviral immunity. Nat. Rev. Immunol. 6:447–456 [DOI] [PubMed] [Google Scholar]
- 14. Selsted ME, Ouellette AJ. 2005. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6:551–557 [DOI] [PubMed] [Google Scholar]
- 15. Smith JG, Silvestry M, Lindert S, Lu W, Nemerow GR, Stewart PL. 2010. Insight into the mechanisms of adenovirus capsid disassembly from studies of defensin neutralization. PLoS Pathog. 6:e1000959 doi:10.1371/journal.ppat.1000959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smith JG, Nemerow GR. 2008. Mechanism of adenovirus neutralization by human α-defensins. Cell Host Microbe 3:11–19 [DOI] [PubMed] [Google Scholar]
- 17. Nguyen EK, Nemerow GR, Smith JG. 2010. Direct evidence from single-cell analysis that human α-defensins block adenovirus uncoating to neutralize infection. J. Virol. 84:4041–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roos WH, Bruinsma R, Wuite GJL. 2010. Physical virology. Nat. Phys. 6:733–743 [Google Scholar]
- 19. Roos WH. 2011. How to perform a nanoindentation experiment on a virus. Methods Mol. Biol. 783:251–264 [DOI] [PubMed] [Google Scholar]
- 20. Carrasco C, Carreira A, Schaap IAT, Serena PA, Gómez-Herrero J, Mateu MG, De Pablo PJ. 2006. DNA-mediated anisotropic mechanical reinforcement of a virus. Proc. Natl. Acad. Sci. U. S. A. 103:13706–13711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ivanovska I, Wuite G, Jönsson B, Evilevitch A. 2007. Internal DNA pressure modifies stability of WT phage. Proc. Natl. Acad. Sci. U. S. A. 104:9603–9608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Michel JP, Ivanovska IL, Gibbons MM, Klug WS, Knobler CM, Wuite GJL, Schmidt CF. 2006. Nanoindentation studies of full and empty viral capsids and the effects of capsid protein mutations on elasticity and strength. Proc. Natl. Acad. Sci. U. S. A. 103:6184–6189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roos WH, Radtke K, Kniesmeijer E, Geertsema H, Sodeik B, Wuite GJL. 2009. Scaffold expulsion and genome packaging trigger stabilization of herpes simplex virus capsids. Proc. Natl. Acad. Sci. U. S. A. 106:9673–9678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roos WH, Gertsman I, May ER, Brooks CL, III, Johnson JE, Wuite GJL. 2012. Mechanics of bacteriophage maturation. Proc. Natl. Acad. Sci. U. S. A. 109:2342–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pérez-BernÁ AJ, Ortega-Esteban A, Menendez-Conejero R, Winkler DC, Menendez M, Steven AC, Flint SJ, de Pablo PJ, San Martin C. 2012. The role of capsid maturation on adenovirus priming for sequential uncoating. J. Biol. Chem. 287:31582–31595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carrasco C, Luque A, Hernando-Pérez M, Miranda R, Carrascosa JL, Serena PA, De Ridder M, Raman A, Gómez-Herrero J, Schaap IAT, Reguera D, De Pablo PJ. 2011. Built-in mechanical stress in viral shells. Biophys. J. 100:1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baclayon M, Shoemaker GK, Uetrecht C, Crawford SE, Estes MK, Prasad BVV, Heck AJR, Wuite GJL, Roos WH. 2011. Prestress strengthens the shell of Norwalk virus nanoparticles. Nano Lett. 11:4865–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kol N, Shi Y, Tsvitov M, Barlam D, Shneck RZ, Kay MS, Rousso I. 2007. A stiffness switch in human immunodeficiency virus. Biophys. J. 92:1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stewart PL, Chiu CY, Huang S, Muir T, Zhao Y, Chait B, Mathias P, Nemerow GR. 1997. Cryo-EM visualization of an exposed RGD epitope on adenovirus that escapes antibody neutralization. EMBO J. 16:1189–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith TAG, Idamakanti N, Rollence ML, Marshall-Neff J, Kim J, Mulgrew K, Nemerow GR, Kaleko M, Stevenson SC. 2003. Adenovirus serotype 5 fiber shaft influences in vivo gene transfer in mice. Hum. Gene Ther. 14:777–787 [DOI] [PubMed] [Google Scholar]
- 31. Sader JE, Chon JWM, Mulvaney P. 1999. Calibration of rectangular atomic force microscope cantilevers. Rev. Sci. Instrum. 70:3967–3969 [Google Scholar]
- 32. Snijder J, Ivanovska IL, Baclayon M, Roos WH, Wuite GJL. 2012. Probing the impact of loading rate on the mechanical properties of viral nanoparticles. Micron 43:1343–1350 [DOI] [PubMed] [Google Scholar]
- 33. Ortega-Esteban A, Horcas I, Hernando-Pérez M, Ares P, Pérez-BernÁ AJ, San Martín C, Carrascosa JL, De Pablo PJ, Gómez-Herrero J. 2012. Minimizing tip-sample forces in jumping mode atomic force microscopy in liquid. Ultramicroscopy 114:56–61 [DOI] [PubMed] [Google Scholar]
- 34. Horcas I, Fernández R, Gómez-Rodríguez JM, Colchero J, Gómez-Herrero J, Baro AM. 2007. WSXM: a software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 78:013705 [DOI] [PubMed] [Google Scholar]
- 35. Ivanovska IL, Miranda R, Carrascosa JL, Wuite GJL, Schmidt CF. 2011. Discrete fracture patterns of virus shells reveal mechanical building blocks. Proc. Natl. Acad. Sci. U. S. A. 108:12611–12616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mackay AL. 1962. A dense non-crystallographic packing of equal spheres. Acta Crystallogr. 15:916–918 [Google Scholar]
- 37. Tirion MM. 1996. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys. Rev. Lett. 77:1905–1908 [DOI] [PubMed] [Google Scholar]
- 38. Tama F, Brooks CL., III 2006. Symmetry, form, and shape: guiding principles for robustness in macromolecular machines. Annu. Rev. Biophys. Biomol. Struct. 35:115–133 [DOI] [PubMed] [Google Scholar]
- 39. Van Vlijmen HWT, Karplus M. 2005. Normal mode calculations of icosahedral viruses with full dihedral flexibility by use of molecular symmetry. J. Mol. Biol. 350:528–542 [DOI] [PubMed] [Google Scholar]
- 40. Yang Z, Bahar I, Widom M. 2009. Vibrational dynamics of icosahedrally symmetric biomolecular assemblies compared with predictions based on continuum elasticity. Biophys. J. 96:4438–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. May ER, Aggarwal A, Klug WS, Brooks CL. 2011. Viral capsid equilibrium dynamics reveals nonuniform elastic properties. Biophys. J. 100:L59–L61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu E, Pache L, Von Seggern DJ, Mullen TM, Mikyas Y, Stewart PL, Nemerow GR. 2003. Flexibility of the adenovirus fiber is required for efficient receptor interaction. J. Virol. 77:7225–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gibbons MM, Klug WS. 2008. Influence of nonuniform geometry on nanoindentation of viral capsids. Biophys. J. 95:3640–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Castellanos M, Perez R, Carrasco C, Hernando-Perez M, Gomez-Herrero J, De Pablo PJ, Mateu MG. 2012. Mechanical elasticity as a physical signature of conformational dynamics in a virus particle. Proc. Natl. Acad. Sci. U. S. A. 109:12028–12033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tama F, Brooks CL., III 2005. Diversity and identity of mechanical properties of icosahedral viral capsids studied with elastic network normal mode analysis. J. Mol. Biol. 345:299–314 [DOI] [PubMed] [Google Scholar]
- 46. May ER, Feng J, Brooks CL., III 2012. Exploring the symmetry and mechanism of virus capsid maturation via an ensemble of pathways. Biophys. J. 102:606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Klug WS, Roos WH, Wuite GJL. 2012. Unlocking internal prestress from protein nanoshells. Phys. Rev. Lett. 109:168104. [DOI] [PubMed] [Google Scholar]
- 48. Zandi R, Reguera D. 2005. Mechanical properties of viral capsids. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 72:1–12 [DOI] [PubMed] [Google Scholar]
- 49. Aznar M, Luque A, Reguera D. 2012. Relevance of capsid structure in the buckling and maturation of spherical viruses. Phys. Biol. 9:036003. doi:10.1088/1478-3975/9/3/036003 [DOI] [PubMed] [Google Scholar]
- 50. Roos WH, Gibbons MM, Arkhipov A, Uetrecht C, Watts NR, Wingfield PT, Steven AC, Heck AJR, Schulten K, Klug WS, Wuite GJL. 2010. Squeezing protein shells: how continuum elastic models, molecular dynamics simulations, and experiments coalesce at the nanoscale. Biophys. J. 99:1175–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]