Abstract
As an obligate pathogen, influenza virus requires host cell factors and compartments to mediate productive infection and to produce infectious progeny virus. Recently, several small interfering RNA (siRNA) knockdown screens revealed influenza virus host dependency proteins, all of which identified at least two subunits of the coat protein I (COPI) complex. COPI proteins oligomerize to form coated vesicles that transport contents between the Golgi apparatus and the endoplasmic reticulum, and they have also been reported to mediate endosomal trafficking. However, it remains unclear which steps in the influenza virus infection cycle rely on the COPI complex. Upon systematic dissection of the influenza virus infection cycle, from entry to progeny virion production, we found that prolonged exposure to COPI complex disruption through siRNA depletion resulted in significant defects in virus internalization and trafficking to late endosomes. Acute inhibition of COPI complex recruitment to the Golgi apparatus with pharmacological compounds failed to recapitulate the same entry defects as observed with the COPI-depleted cells but did result in specific decreases in viral membrane protein expression and assembly, leading to defects in progeny virion production. Taken together, our findings suggest that COPI complexes likely function indirectly in influenza virus entry but play direct roles in viral membrane protein expression and assembly.
INTRODUCTION
Influenza virus is a negative-sense strand RNA enveloped virus that contains eight segmented genomes which encode 13 viral proteins. The influenza virus virion envelope is derived from the infected cell's plasma membrane and contains three integral viral membrane proteins: the M2 proton channel, hemagglutinin (HA), and neuraminidase (NA). Encapsulated within the viral envelope is a coat of matrix protein 1 (M1), which forms contacts with each of the viral ribonucleoprotein (vRNP) complexes. Each of the vRNPs consists of a strand of viral RNA bound to viral nucleoprotein (NP). In addition, each vRNP is bound to an RNA-dependent RNA polymerase complex containing three proteins: PA, PB1, and PB2 (1).
Influenza A virus is the causative agent of seasonal flu and historically has led to pandemic infections, such as the 1918 Spanish influenza outbreak, which killed an estimated 50 million people worldwide (1). At a cellular level, influenza virus infection starts with virion attachment to cell surface sialylated glycoproteins or glycolipids (2). The virus particle then triggers endocytosis through clathrin-dependent and clathrin-independent pathways (3–6). Productive entry requires the virus to traffic to low-pH endosomes (pH ∼ 5.0), at which point HA mediates fusion between the viral envelope and the lipid bilayer of the endosome (1, 4, 7–9). Upon pH-mediated fusion, the vRNPs are released into the cytoplasm and subsequently transported into the nucleus to initiate viral replication and viral protein translation (10). Infected cells produce progeny virions by assembling viral proteins and vRNP complexes at the plasma membrane. Viral NA cleaves cell surface sialic acids to allow assembled virions to bud and release from the infected cell's membrane and thereby initiate another infection cycle in neighboring uninfected cells (1).
Being an obligate pathogen encoding only 13 viral proteins, influenza virus hijacks host proteins, cellular compartments/organelles in order to harbor infection and produce progeny virus. Four genome-wide knockdown screens, one in Drosophila cells and three in human cells (11–14), recently identified host proteins important for influenza virus infection. One group of host dependency proteins identified in all influenza screens included subunits of the coat protein I (COPI) complex (11–14). There are nine different COPI complex subunits: α, β1, β2, δ, ε, γ1, γ2, ζ1, and ζ2. Each COPI complex contains a single copy of the α, β1, β2, δ, and ε subunits in addition to one of the following isoform combinations: γ1/ζ1, γ1/ζ2, or γ2/ζ1 (15–17). Together, COPI complexes form a vesicle coat that traffics contents between Golgi stacks and cargoes between the Golgi apparatus and the endoplasmic reticulum (ER) (15, 17, 18). In addition, several studies reported a role for COPI complexes in the endocytic pathway, since perturbation of these complexes leads to defects in endosomal sorting, multivesicular body (MVB) formation, and/or membrane trafficking (19–24).
Previous studies indicated that COPI might play a role in influenza virus entry. König and colleagues reported that δ-COPI (ARCN1) knockdown inhibits vRNP nuclear import (14). Given the roles of COPI proteins in the endocytic pathway, König and colleagues hypothesized that the block in vRNP nuclear import might be due to defects in endosomal trafficking. Viral entry is a multistep process, however, and it remains unclear which step (if any) is ARCN1 dependent.
Recently, Cureton and colleagues reported that disruption of COPI complexes differentially affects vesicular stomatitis virus (VSV) entry versus viral gene expression (25). The authors used a temperature-sensitive CHO cell line (IdlF), which degrades ε-COPI at 40°C (23), to show that ε-COPI depletion inhibits virus binding and internalization as well as transferrin (Tfn) uptake. However, these results were recapitulated only after prolonged treatment with brefeldin A (BrefA), an inhibitor that prevents GDP-to-GTP exchange of ADP ribosylation factor 1 (ARF1), an essential step for membrane recruitment of COPI to the Golgi apparatus (26). Acute (short-term) treatment with BrefA did not lead to the same effect on viral entry as observed with the IdlF cells incubated at 40°C, but it did cause a specific decrease in VSV gene expression.
The results from Cureton and colleagues suggest that indirect effects caused by long-term inactivation of COPI lead to general defects in the clathrin-mediated endocytosis pathway (25). VSV exclusively uses clathrin-mediated endocytosis for infection (27–31). Influenza virus, on the other hand, can productively infect cells in the absence of clathrin-mediated endocytosis. Indeed, several reports have shown that depletion of key components of the clathrin complex fails to block influenza virus entry and infection because alternative productive endocytic pathways, such as macropinocytosis, can be hijacked by influenza virus (3, 5, 6, 32). Moreover, influenza virus requires lower-pH compartments for viral fusion than for VSV (4, 29, 33). Therefore, how COPI proteins are involved in each of the steps of influenza virus entry and infection remains unclear.
Here, we dissected the different steps of the influenza virus infection cycle (entry, replication, trafficking, and assembly) in order to identify the steps that COPI proteins affect. We found that the first major entry block in COPI knockdown cells occurs at virus internalization. In addition, COPI-depleted cells exhibited defects in early endosome-to-intermediate/late endosome trafficking for virus particles that did manage to enter the cells. The entry block was not limited to influenza virus only but was also observed for other cargoes (Tfn, epidermal growth factor [EGF], and dextran) that are internalized through either clathrin-mediated endocytosis or macropinocytosis. Given the large defect in viral entry upon small interfering RNA (siRNA) silencing of COPI, we used pharmacological inhibitors, namely, BrefA and golgicide A (GCA), to acutely disrupt COPI complexes. With the inhibitor treatment, we found that COPI complexes were not directly required for influenza virus entry. However, disruption of functional COPI complexes directly inhibited viral membrane protein expression, assembly of viral components at the plasma membrane, and production of infectious progeny virus.
MATERIALS AND METHODS
Antibodies for immunofluorescence assays or Western blotting.
We used the following primary antibodies for this study: rabbit anti-coatomer subunit δ antibody (1:1,000 dilution) (catalog no. ab96725; Abcam), mouse anti-influenza A virus nucleoprotein antibody (AA5H) (1:1,000) (ab20343; Abcam), mouse antinucleoprotein (1:100) (sc-57882; Santa Cruz Biotechnology), rabbit anti-β-COPI (1:1,000) (generously donated by James Rothman), rabbit anti-COPZ1 (1:1,000) (SAB4500896; Sigma-Aldrich), rabbit anti-γ1-COP1 (1:10,000) (generously donated by Felix Wieland), rabbit anti-COPE (1:1,000) (ab88824; Abcam), mouse anti-β-actin antibody (loading control) (1:1,000) (ab8227; Abcam), rabbit-anti-phospho-signal transducer and activator of transcription 1 (STAT1) Tyr701 (1:1,000) (9167; Cell Signaling Technology), mouse anti-Cy5/Alexa Fluor 647 (1:1,000) (C1117; Sigma-Aldrich), mouse anti-early endosome antigen 1 (EEA1) (1:500) (610457; BD Transduction Laboratories), mouse anti-lysobisphosphatidic acid (LBPA) antibody (50 μg, 1:500) (Z-PLBPA; Echelon), mouse anti-influenza A virus M2 protein antibody (14C2) (1:1,000) (ab5416; Abcam), rabbit antineuraminidase (1:5,000) (kind gift from Gillian Air), and mouse anti-PB1 (1:100) (sc-17601; Santa Cruz Biotechnology). We used the following secondary antibodies for this study: Alexa Fluor 647 donkey anti-mouse IgG (1:1,000 dilution) (catalog no. A31571; Molecular Probes), Alexa Fluor 555 donkey anti-mouse IgG (1:1,000) (A31570; Molecular Probes), Alexa Fluor 488 donkey anti-mouse IgG (1:1,000) (A-21202; Molecular Probes), Alexa Fluor 568 donkey anti-rabbit IgG (1:1,000) (A-100042; Molecular Probes), enhanced chemiluminescence (ECL) donkey anti-rabbit IgG, horseradish peroxidase (HRP) linked (1:5,000) (NA934; GE Healthcare), and ECL sheep anti-mouse IgG (1:5,000) (NXA931; GE Healthcare).
Cell culture.
A549 lung carcinoma cells (ATCC) were cultured in Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (Serum International), 1 mM nonessential amino acids (ATCC), and antibiotics (25 U/ml penicillin and 25 μg/ml streptomycin) (ATCC) and were maintained in a humidified environment with 5% CO2 at 37°C. For siRNA knockdown experiments, A549 cells were electroporated with 100 pmol of specified COPI siGENOME SMARTpool siRNA (Dharmacon) or AllStars negative-control siRNA (Qiagen) using a Lonza Amaxa Nucleofector with kit T (catalog no. VVCA-1002; Lonza) and program X-001. Experiments were performed with the siRNA-treated A549 cells 48 h after electroporation. For plasmid expression, A549 cells were electroporated with 2 μg of either COPE-green fluorescent protein (GFP) (OriGene) or Rab7-GFP (a generous gift from Qing Zhong) (34) plasmid using kit T and Nucleofector program X-001. Experiments using these plasmids were performed within 24 h after electroporation with the plasmids.
A Live/Dead viability/cytotoxicity kit (catalog no. L-3224; Invitrogen) was used according to vendor instructions to measure cell viability, death, and growth. Stimulation of the interferon (IFN) response in A549 cells was achieved with 10,000 U/ml IFN-α (catalog no. 11101-1; PBL Biomedical Laboratories) for 3 h prior to collecting cell lysate.
Spinning disk confocal imaging.
Spinning disk confocal z-stacks were obtained with a custom-built confocal microscope which has been described in detail (35). Briefly, multicolor fluorescent fixed-cell imaging was achieved using an Yokogawa spinning disk confocal scan head attached to an Olympus IX-71 microscope (Olympus, Center Valley, PA) with an Olympus 60× oil immersion objective (numerical aperture, 1.35). A 561-nm crystal laser, 647-nm krypton ion laser, and/or 457/488/514-nm argon laser was used to image the samples with an Andor 885 electron-multiplying charge-coupled-device camera.
Cargo uptake assays and data analysis.
siRNA-treated A549 cells were rinsed extensively with phosphate-buffered saline (PBS) and cold bound with Alexa Fluor 568-transferrin (AF568-Tfn) (catalog no. T23365; Invitrogen) and Alexa Fluor 647-epidermal growth factor (AF647-EGF) (E35351; Invitrogen) for 30 min at 4°C. Unbound AF568-Tfn and AF647-EGF were removed with extensive cold PBS washes, and cells were incubated in warm DMEM with antibiotics at 37°C for different times. For fluid-phase uptake, serum-starved A549 cells were incubated with 5 mg/ml tetramethylrhodamine (TMR)-dextran (D-1868; Invitrogen) in warm DMEM with antibiotics at 37°C for different times.
At the indicated time points, surface-bound cargo was removed with acid buffer (PBS adjusted to pH 2.5 with acetic acid) for 2 min, and cells were fixed with 3.2% paraformaldehyde (PFA) (Electron Microscopy Sciences) and imaged with a spinning disk confocal microscope. Confocal z-stacks were obtained for each field of view, which contained about 9 or 10 cells per image. For the analysis of Tfn, EGF, or dextran internalization, a MatLab script based on k-clustering analysis was used to establish thresholds for the images to differentiate the background pixels from the signal pixels. Briefly, an optimized threshold value was determined for each maximal z-projection image with an iterative thresholding algorithm which found the maximal difference between the mean background pixels and the mean signal pixels. The mean signal intensities of 15 to 20 maximal z-projection images per time point were calculated and averaged.
To test the known effect of amiloride on macropinocytosis, A549 cells were treated for 60 min with different concentrations of 5-(N-ethyl-N-isopropyl)amiloride (EIPA) (50 μM; catalog no. A3085; Sigma-Aldrich) in DMEM without penicillin-streptomycin (P/S) or fetal bovine serum (FBS). The cells were then incubated with 0.2 mg/ml AF647-dextran for 30 min at 37°C prior to low-pH PBS washing, followed by PFA fixation. Additionally, inhibition of dynamin-dependent endocytosis was tested by treatment for 30 min with different doses of Dynasore (D7693; Sigma-Aldrich) in DMEM without P/S or FBS prior to cold binding of 1 μg/ml AF647-Tfn or 1:1,000 AF647-EGF for 30 min. Unbound cargo was replaced with a warmed inhibitor containing DMEM without FBS or P/S, and cells were incubated at 37°C for 10 min prior to low-pH PBS washing, followed by PFA fixation. For dextran, Tfn, and EGF uptake experiments, imaging of the samples and analyses were performed as described above.
Nonspecific amine-reactive labeling of virus.
Fifty microliters of 1 mg/ml purified influenza A virus X31, A/Aichi/68 (H3N2) (Charles River Laboratories), was mixed with 47 μl of 0.1 M freshly prepared carbonate buffer (pH ∼8 to 9) and 6 μg Alexa Fluor 647 carboxylic acid, succinimidyl ester (catalog no. A-20106; Invitrogen), dissolved in dimethyl sulfoxide (DMSO). The mixture was gently mixed in the dark for 60 min. A Nap5 size exclusion column (GE Healthcare) was used to separate free dyes from labeled virus using HEPES 145 elution buffer (50 mM HEPES [pH 7.4], 145 mM NaCl). The excess AF647-labeled X31 was aliquoted, snap-frozen, and stored at −80°C (8). No significant difference in infectivity was observed between labeled and unlabeled X31 (data not shown).
X31 membrane labeling.
One hundred microliters of 1 mg/ml purified X31 was mixed with 3 μl of 25 mM 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt (DiD solid) (catalog no. D-7757; Invitrogen), dissolved in DMSO. The mixture was gently vortexed in the dark for 2 h. A Nap5 size exclusion column was used to separate free dyes from labeled virus using HEPES 145 elution buffer. The excess DiD-labeled X31 was aliquoted, snap-frozen, and stored at −80°C. Prior to the use of DiD-labeled X31 for the experiments, the virus was filtered through a 0.2-μm filter (32). We confirmed with plaque assays that the DiD-labeled X31 remained infectious.
Bulk viral fusion assay.
For the pharmacological compound-treated cells, all steps in the fusion assay before trypsinization, except the PBS washes, included the compounds. siRNA- or pharmacological compound-treated A549 cells seeded in Lab-Tek 8-well glass dishes were rinsed extensively with PBS and cold bound with 2 × 104 PFU/ml DiD-X31 for 45 min on ice. Unbound virus was removed with extensive PBS washes, and cells were incubated with DMEM for different times at 37°C. Afterwards, the cells were washed once with PBS and trypsinized. The trypsin was neutralized with PBS supplemented with 10% FBS and 30 mM sodium azide, and the cells were collected and placed on ice to prevent any further fusion. Cells were fixed with 2% PFA for 20 min, washed once with PBS, and analyzed immediately with a flow cytometer (BD Biosciences Fortessa). The data were interpreted using FlowJo software.
Virus binding or elderberry lectin binding assays.
After 48-h siRNA knockdown, A549 cells were washed extensively with PBS and cold bound with different concentrations of AF647-X31 in DMEM for 45 min. Unbound virus was removed through PBS washes, and the cells were immediately trypsinized. The trypsin was neutralized with PBS supplemented with 10% FBS and 30 mM sodium azide. Cells were collected, fixed with 2% PFA for 20 min, washed once with PBS, and analyzed immediately with a flow cytometer.
For the elderberry lectin binding assays, siRNA-treated cells were trypsinized and fixed with 2% PFA. The cells were then treated with or without different doses of sialidase A (Prozyme) in PBS (pH ∼5) for 1 h at 37°C. The pH was raised to ∼7 to stop the sialidase A reaction, and cells were stained for 1 h with a 1:1,000 dilution of fluorescein isothiocyanate (FITC)-conjugated Sambucus nigra (elderberry) bark lectin (SNA-FITC) (Vector Laboratories). After three PBS washes, the samples were immediately analyzed by flow cytometry.
Immunofluorescence for flow cytometry.
siRNA- or pharmacological compound-treated A549 cells were infected with influenza virus at 37°C for 9 or 12 h. Infected cells were trypsinized, fixed with 2% PFA in PBS for 20 min at room temperature, and washed once to remove the fixation buffer. The cells were permeablized in buffer P (PBS containing 10% FBS and 0.075% saponin) for 5 min. The samples were incubated with mouse anti-influenza virus NP or anti-influenza virus M2 (1:1,000 dilution in buffer P) for 60 min, washed three times with buffer P, incubated with Alexa Fluor 647 donkey anti-mouse IgG (1:1,000 dilution in buffer P) for 30 min, washed three times with buffer P, washed once with PBS, and analyzed by flow cytometry.
Immunofluorescence with endosomal markers.
To delineate the cell membrane, fixed A549 cells were first stained with SNA-FITC without permeabilization. For colocalization studies with influenza virus and EEA1, fixed A549 cells were blocked and permeabilized with 3% bovine serum albumin (Jackson Laboratories) and 0.1% Triton X-100 (blocking buffer) for 30 min at room temperature. The cells were incubated with mouse anti-EEA1 in blocking buffer for 1 h at room temperature, rinsed with PBS, stained with Alexa Fluor 555 donkey anti-mouse IgG in blocking buffer for 30 min, and washed with PBS.
For colocalization studies with influenza virus and LBPA, fixed A549 cells were blocked and permeabilized with 3% bovine serum albumin (Jackson Laboratories) and 0.075% (wt/vol) saponin (blocking buffer) for 30 min at room temperature. All subsequent washes and antibody incubations were performed with the blocking buffer, unless stated otherwise. The sample was incubated with mouse anti-LBPA for 1 h at room temperature, washed, stained with Alexa Fluor 555 donkey anti-mouse IgG for 30 min, washed, and rinsed with PBS.
All samples were imaged with a customized spinning disk confocal microscope, which is described briefly above and described in more detail elsewhere (35). ImageJ software was used to visualize the fluorescence confocal z-stacks, and the fraction of internalized virus particles that colocalized with the respective endosomal marker was determined.
Pharmacological compounds.
The following pharmacological compounds were used in this study: dimethyl sulfoxide (DMSO) (1:1,000) (catalog no. D2650; Sigma-Aldrich), brefeldin A (10 μg/ml) (B7651; Sigma-Aldrich), and golgicide A (10 μM) (345862; EMD). Both the brefeldin A and golgicide A were dissolved in DMSO, and incubations were performed in DMEM supplemented with 10% fetal bovine serum, 1 mM nonessential amino acids, and antibiotics. The final amount of DMSO did not exceed 0.1% (vol/vol). Specific incubation times for each of the experiments are described in the figures and legends.
Plaque assays.
We collected the supernatant from influenza-infected cells at 18 or 24 h postinfection (hpi). Serial dilutions of the virus-containing supernatant were used to inoculate an ∼80% confluent monolayer of Texas-MDCK cells (a generous gift from Robert Lamb) or Vero cells (ATCC) grown in 6-well dishes for ∼1.5 to 2 h at 37°C. The cells were washed with PBS, coated with 3 ml of 30% Noble agar (Affymetrix) in DMEM containing antibiotics and 2 μg/ml acetylated trypsin (Sigma-Aldrich), and then incubated at 37°C.
Two different detection methods were used to determine the viral titers. For immunodetection of infected cell colonies (fluorescent foci), the Noble agar disks were removed at ∼36 hpi, and the cells were fixed with 100% methanol for 8 to 10 min at −20°C. The cells were rinsed with PBS, blocked and permeabilized with 3% bovine serum albumin and 0.1% Triton X-100 in PBS for 30 min, immunostained against NP in blocking buffer for 60 min, washed three times with PBS, incubated with Alexa Fluor 488 donkey anti-mouse IgG in blocking buffer for 30 min, and washed three times with PBS. The fluorescent foci were visualized and quantified using a Typhoon fluorescence scanner (GE Healthcare).
The second detection method used to determine the viral titers was counting the numbers of plaques observed 2.5 days postinfection. To detect plaques, the agar disks were removed, and the cells were immediately fixed and stained with a solution containing 1:1,000 (vol/wt) crystal violet with 30% ethanol in water. The number of plaques for each condition was counted. The viral titer was determined as (number of foci or plaques)/(dilution × inoculation volume [in ml]). Samples were tested in quadruplicate.
Western blotting.
Cell lysates were diluted in Laemmli sample buffer (catalog no. 161-0737; Bio-Rad) containing dithiothreitol to achieve the same amounts of protein for all samples. The samples were loaded and run on a 4% to 15% Tris-HCl polyacrylamide gel (Bio-Rad). The separated proteins from the gel were transferred onto Hybond polyvinylidene difluoride membranes (GE Healthcare), blocked with 5% nonfat milk in Tris-buffered saline (TBS)-Tween, incubated with the specified primary antibodies (reported in the figure legends) overnight at 4°C, washed with TBS-Tween, incubated with HRP-conjugated secondary antibodies for 60 min at room temperature, washed with TBS-Tween, detected with Lumigen TMA-6 (TMA-100; Lumigen), and exposed/developed onto high-sensitivity Kodak film.
RESULTS
COPI depletion inhibits influenza virus infection via an entry step at or prior to viral fusion.
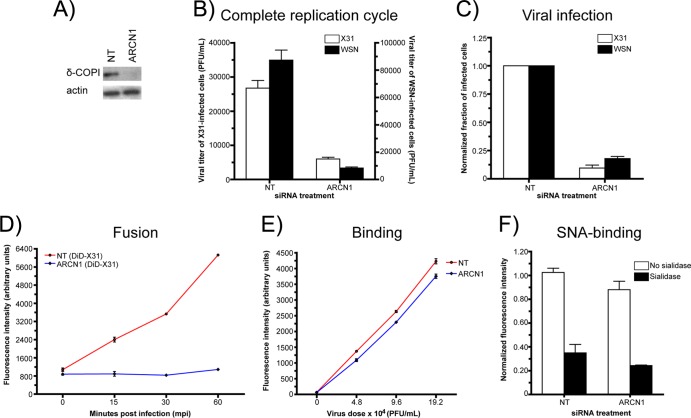
To confirm previously published results showing that COPI proteins are required for influenza virus infection, we used siRNA to deplete one of the essential subunits of the COPI complex, δ-COPI (ARCN1). We were able to achieve efficient ARCN1 depletion after 48 h, as shown with Western blotting (Fig. 1A). To test the effect of siRNA treatment on influenza infection, supernatants from influenza virus A/Aichi/1968 (H3N2) (X31)- or influenza virus A/WSN/1933 (H1N1) (WSN)-infected A549 cells were collected, and the viral titers were measured with plaque assays. Relative to the viral titers of the nontargeting siRNA control, those of the supernatants from ARCN1 knockdown cells were decreased by 78% ± 3% for X31 and 90% ± 1% for WSN infection (Fig. 1B). Furthermore, ARCN1 knockdown cells exhibited a defect in virus infection prior to viral progeny production. At 8.5 hpi with X31 or WSN, we quantified the fraction of NP-positive cells with flow cytometry. Relative to the control cells, the ARCN1 knockdown cells had 93% ± 0.01% and 82% ± 2% fewer NP-expressing cells with X31 and WSN infection, respectively (Fig. 1C). Among the cells that expressed NP, the NP expression levels were similar for the control and ARCN1 knockdown cells (data not shown).
Fig 1.
COPI depletion blocks viral entry at a step(s) prior to viral fusion. (A) Western blot probing δ-COP1 expression in nontargeting (NT) or ARCN1 siRNA-treated cells after a 48-h knockdown. (B) Plaque assay results measuring the viral titers of released virions from X31- or WSN-infected siRNA-treated cells at 24 h postinfection (hpi). The mean viral titers and standard deviations from duplicate experiments are shown. (C) Normalized fractions of NP-expressing cells in X31- or WSN-infected siRNA-treated cells at 8.5 hpi. (D) Quantification of DiD fluorescence dequenching using flow cytometry. Nontargeting siRNA-treated and ARCN1 knockdown cells were cold bound with DiD-labeled X31 (DiD-X31) and allowed to internalize virus for different times at 37°C. The fold increase in the fluorescence signal from DiD was monitored at each time point to measure the amount of viral fusion. (E) Quantification of AF647-X31 binding in siRNA-treated cells using flow cytometry. (F) Binding of FITC-conjugated elderberry lectin (SNA-FITC)—a protein that binds to α(2,6)-linked sialic acids on glycoproteins and glycolipids—to nontargeting and ARCN1 siRNA-treated cells with and without treatment with 0.01 U sialidase A. The data for each independent experiment were normalized to the non-sialidase-treated nontargeting siRNA-treated control cell value. Each condition was tested in duplicate in two independent experiments.
In addition, we checked for potential siRNA-mediated off-target effects, i.e., cytotoxicity and interferon induction. Compared with control cells, the ARCN1 knockdown cells exhibited an ∼25% decrease in cell viability, a minor increase in cytotoxicity, and a correspondingly slight decrease in cell growth after 24 and 48 h of siRNA treatment (http://zhuang.harvard.edu/Publications/Sun_et_al_supplemental_figures.pdf). Furthermore, siRNA treatment did not stimulate interferon induction (37; see also http://zhuang.harvard.edu/Publications/Sun_et_al_supplemental_figures.pdf). Given that the COPI complex is required for influenza virus infection and that the cells were largely healthy, we conducted experiments to identify which steps of the influenza virus infection cycle COPI proteins mediate.
We first probed the effect of COPI depletion on viral fusion using a fluorescence dequenching assay. To detect viral fusion, we incorporated a saturating amount of a lipophilic dye, DiD (36), into enveloped viruses, such that the detected fluorescence signal from the virus particles became partially quenched due to intermolecular dye interactions. For DiD-labeled viruses, pH-mediated fusion results in a dramatic increase in the fluorescence signal (dequenching) stemming from the diffusion of DiD molecules from the viral envelope into the endosomal membrane (8, 38). To measure the amount of viral fusion on a population level, we incubated siRNA-treated cells with DiD-labeled X31 on ice, allowed the virus to infect the cells at 37°C for different times, and quantified the DiD fluorescence intensity using a flow cytometer. As shown in Fig. 1D, we observed an enhancement in fluorescence intensity with increasing incubation times at 37°C for the nontargeting siRNA-treated control cells. By the 60-min time point, the control cells exhibited a 5.70- ± 0.02-fold increase in the DiD signal. In stark contrast, the signal enhancement in ARCN1 knockdown cells was only 24% ± 0.03%. In addition, we tested COPE, COPG1, and COPZ1 knockdown cells with the bulk DiD fusion assay to determine whether depletion of other COPI subunits resulted in similar viral fusion defects. Indeed, knockdown of those COPI subunits also yielded significant defects in viral fusion (http://zhuang.harvard.edu/Publications/Sun_et_al_supplemental_figures.pdf), although the effect due to COPE depletion appeared to be weaker. Collectively, these results show that COPI subunit knockdown blocks a viral entry step(s) at or before viral fusion.
We next tested the amounts of Alexa Fluor 647-conjugated X31 (AF647-X31) virus bound to control and ARCN1 knockdown cells by using flow cytometry. The largest difference in binding was observed with the lowest virus dose tested, at which ARCN1 knockdown cells bound 20% ± 3% less virus than the nontargeting siRNA-treated control cells (Fig. 1E). We also quantified the level of α(2,6)-linked sialic acids presented on the surface of the siRNA-treated cells by measuring the amount of FITC-conjugated elderberry lectin (SNA-FITC) bound to nonpermeabilized fixed cells. This assay was validated with A549 cells treated with different sialidase A units, followed by SNA-FITC staining; exposure to higher levels of sialidase A resulted in a dose-dependent decrease in SNA-FITC binding (http://zhuang.harvard.edu/Publications/Sun_et_al_supplemental_figures.pdf). Applying this assay to the nontargeting and ARCN1 siRNA-treated cells, we found that the latter bound 17% ± 1% less elderberry lectin than the control cells (Fig. 1F). Because X31 and SNA can still bind to ARCN1-depleted cells, it is unlikely that siRNA-mediated COPI complex disruption leads to a general defect in sialylation or presentation of glycoproteins or glycolipids at the cell surface. Taken together, the data in Fig. 1 show that COPI depletion with siRNA knockdown inhibits viral infection and replication through an entry block after virus binding but prior to or at viral fusion.
siRNA knockdown of COPI abrogates the internalization of influenza virus, as well as other clathrin-mediated endocytosis or macropinocytosis cargos.
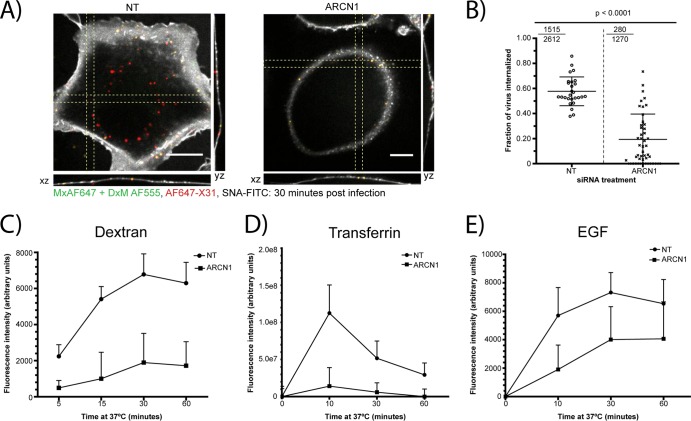
The small defects in virus binding could not explain the dramatic decrease in viral fusion in the COPI-depleted cells. Therefore, we subsequently tested the effect of COPI depletion on influenza virus internalization by quantifying the fraction of internalized AF647-X31 in the siRNA-treated cells at 30 min postinfection (mpi). The cells were fixed and labeled with SNA-FITC to delineate the plasma membrane. In addition, to confidently identify the virus particles close to the plasma membrane as being internalized or noninternalized, we performed nonpermeabilizing immunofluorescence staining to stain surface-bound noninternalized virus with an antibody specific for AF647 (Fig. 2A). The ARCN1 knockdown cells exhibited a significant (P < 0.0001) defect in virus uptake compared to that of the control cells. Specifically, the mean internalization fractions in the nontargeting siRNA-treated control cells and ARCN1 knockdown cells were 0.58 and 0.19, respectively (Fig. 2B). In addition, we report a ratio that represents the sum of internalized particles divided by the total number of virus particles among all cells. The ratios for the control and ARCN1 knockdown cells were 0.58 and 0.22, respectively (Fig. 2B, top left corners).
Fig 2.
ARCN1 knockdown cells exhibit defective internalization of influenza virus and other cargoes. (A) Representative confocal xy, yz, and xz cross-sectional images of AF647-X31-infected siRNA-treated cells (red) at 30 min postinfection (mpi). The plasma membrane was labeled using SNA-FITC (white). Nonpermeabilizing immunofluorescence staining with an antibody specific for AF647 (green) was performed to differentiate plasma membrane-proximal particles as being noninternalized (yellow) or internalized (red). Scale bars, 10 μm. (B) Fraction of virus internalized, of the total number of virus particles per cell. The internalized virus fraction was counted for more than 30 cells per siRNA treatment condition, and values are graphed in the form of a dot plot. Statistical analysis was performed using a one-sided Student t test. Furthermore, for each siRNA treatment, a ratio is presented in the top left corner (the numerator denotes the total particles internalized, and the denominator represents the total number of particles counted among all cells). (C to E) Quantification of TMR-dextran (C), AF568-Tfn (D), and AF647-EGF (E) uptake in nontargeting and ARCN1 siRNA-treated cells from at least 18 different confocal z-stacks per time point.
Given the substantial defect in influenza virus internalization in the ARCN1 knockdown cells, we tested whether other cargoes of endocytic pathways previously described as productive influenza virus entry pathways, i.e., clathrin-mediated endocytosis and macropinocytosis, were also inhibited. Tfn, EGF, and dextran were used as representative markers of clathrin-mediated endocytosis and macropinocytosis. The cargoes were allowed to be internalized for different amounts of time at 37°C, and surface-bound cargoes were stripped from the cells using a low-pH buffer (pH 2.5) prior to fixation and imaging. We validated our analysis method by quantifying the amount of cargo internalized upon treatment with well-established inhibitors of macropinocytosis and dynamin-dependent endocytosis. To verify our algorithm for macropinocytosis, we treated A549 cells with different concentrations of EIPA prior to allowing for fluid phase uptake; for dynamin-dependent endocytosis, we treated A549 cells with different doses of Dynasore prior to Tfn or EGF uptake (39, 40). For all three cargoes tested, we observed dose-dependent decreases in measured cargo uptake with increasing concentrations of inhibitor treatment (http://zhuang.harvard.edu/Publications/Sun_et_al_supplemental_figures.pdf). We applied the assay to the siRNA-treated cells and found that the uptake of not only dextran but also Tfn and EGF was significantly impaired (Fig. 2C to E). The largest observed defects in internalization were at the earlier time points tested, i.e., 15 min for dextran and 10 min for Tfn and EGF. EGF uptake was relatively less perturbed than Tfn uptake, because it has been shown that EGF might be internalized through a clathrin-independent pathway at high concentrations (36, 41, 42).
Collectively, our results show that COPI depletion blocks influenza virus internalization, which in part contributes to the defect in viral fusion shown in Fig. 1D. However, representative cargoes for two different productive entry pathways for influenza virus—clathrin-mediated endocytosis and macropinocytosis (3–6)—also exhibited defects in internalization in ARCN1-depleted cells.
ARCN1 knockdown cells exhibit impaired endosomal trafficking upon virus internalization.
We next tested whether ARCN1 knockdown leads to any postinternalization defects. Upon internalization, influenza virus enters Rab5+ endosomes near the cell periphery and then is sorted into Rab7+ endosomes toward the perinuclear region, in which late endosomes/lysosomes are dense (43). Upon exposure to a pH of ∼5.0, the influenza virus envelope fuses with the endosomal membrane, releasing the vRNP into the cytoplasm for nuclear import (44, 45).
To test whether ARCN1 knockdown cells exhibit any defects in trafficking to intermediate/late endosomes, we cold bound AF647-X31 to siRNA-treated cells and incubated the samples at 37°C for 90 min. The cells were then fixed and immunostained for the intermediate/late endosome marker lysobisphosphatidic acid (LBPA) (46; see also http://zhuang.harvard.edu/Publications/Sun_et_al_supplemental_figures.pdf). We found a significant (P < 0.0001) decrease in AF647-X31 colocalization with LBPA+ vesicles, with a mean colocalization fraction for individual cells of 0.42 for the nontargeting control cells versus 0.10 for the ARCN1 knockdown cells. In addition, we report the ratios of the sum of colocalized particles to the total number of internalized particles among all analyzed cells. The ratios for the nontargeting and ARCN1 siRNA-treated cells were 0.62 and 0.11, respectively (Fig. 3A and B, upper left corners). Regardless of the type of measurement, these findings strongly suggest that virus particles exhibit a defect in trafficking to intermediate/late endosomes in ARCN1 siRNA-treated cells.
Fig 3.
Defective endosomal trafficking of internalized influenza virus particles in ARCN1-depleted cells. (A) Representative spinning disk confocal images of nontargeting and ARCN1 siRNA-treated cells infected with AF647-X31 for 90 min. The plasma membrane was stained with SNA-FITC, and the late endosomes were immunostained with LBPA. Scale bars, 10 μm. (B) The fractions of internalized particles that colocalized with LBPA, a late endosome marker, were counted for at least 20 cells per siRNA treatment. The dot plot average line represents the mean colocalization fraction of the analyzed cells; each dot within the plot is the colocalized fraction from an individual cell. Statistical analysis was performed using a one-sided Student t test. In addition, for each siRNA treatment, a ratio is presented in the top left corner (the numerator denotes the total number of internalized particles colocalized with LBPA, and the denominator represents the total number of internalized virus particles among all cells). The dot plot mean and ratio in the top left corner for each respective treatment may not necessarily be equal, especially in the presence of substantial cell-to-cell variation. (C) Representative spinning disk confocal images of nontargeting and ARCN1 siRNA-treated cells infected with AF647-X31 for 90 min, plasma membrane stained with SNA-FITC, and immunostained with EEA1, an early endosome marker. Scale bars, 10 μm. (D) Quantification of the internalized particles that colocalize with EEA1 at 90 mpi was performed as described for panel B for more than 35 cells per siRNA treatment. Statistical analysis was performed using a one-sided Student t test.
Given the large observed defect in trafficking to LBPA+ vesicles, we tested whether the ARCN1 knockdown cells were accumulating the AF647-X31 in early endosomes. Indeed, at 90 mpi, the ARCN1 knockdown cells exhibited significant (P < 0.0001) increases in the mean colocalization fractions, which were 0.24 for the ARCN1 knockdown cells and 0.07 for the nontargeting control cells (Fig. 3C and D). The values for the sum of colocalized particles divided by the total number of internalized particles for all nontargeting and ARCN1 siRNA-treated cells were 0.1 and 0.36, respectively.
Previous studies reported that COPI proteins might be important for endosomal trafficking (19, 22). However, it remains unclear whether COPI vesicles directly mediate sorting of influenza virus between different types of endosomes. To test whether AF647-X31 traffics within COPI vesicles, we transiently expressed GFP-tagged ε-COPI (COPE-GFP) in A549 cells. Of the GFP-COPI subunit constructs tested, ε-COP-GFP minimized disruption of the Golgi architecture. After several rounds of live cell-imaging experiments with A549 cells expressing COPE-GFP and infected with AF647-X31, we found only one example of COPE-GFP colocalized with AF647-X31 out of many virus particles tracked (http://zhuang.harvard.edu/Publications/Sun_et_al_supplemental_figures.pdf). Although this does not completely negate the possibility that COPI proteins play a specific role in endosomal trafficking between MVBs and late endosomes, our data suggest that the effect of COPI in transporting the influenza virus from early to late endosomes is likely indirect.
Rapid disruption of COPI vesicles with pharmacological inhibitors does not block viral entry.
The results described thus far help explain why several of the COPI subunits emerged as important host proteins in influenza virus infection in the siRNA genome-wide knockdown screens (11, 13, 14). However, efficient siRNA depletion can take a few days, potentially leading to the accumulation of siRNA-mediated indirect effects due to long-term depletion of critical host proteins. In order to test whether we could recapitulate the observations in the siRNA-treated cells and to determine whether there is a direct role for COPI during the influenza virus infection cycle, we conducted experiments using pharmacological inhibitors that rapidly disrupt COPI vesicle formation.
The conversion of inactive cytosolic ADP ribosylation factor 1 (ARF1)-GDP to the activated ARF1-GTP form triggers its attachment to the Golgi membrane and subsequent recruitment of COPI complexes (15–17). Two pharmacological inhibitors—BrefA and GCA—prevent the exchange of ARF1-GDP and ARF1-GTP via different mechanisms. BrefA binds to the protein-protein interface of ARF1 and several different BrefA-sensitive guanine nucleotide exchange factors (GEFs) to prevent the exchange of GDP and GTP (26, 47). Therefore, treatment with BrefA inhibits not only COPI membrane recruitment but also other ARF1-GTP-dependent vesicular trafficking processes, such as activator protein 1-dependent trans-Golgi network trafficking to endosomal compartments (15, 16). A more specific inhibitor of COPI complex vesicle formation is GCA, which binds to Golgi apparatus-specific brefeldin A resistance guanine nucleotide exchange factor 1 (GBF1), a COPI-specific ARF GEF (48).
We confirmed that the DMSO-treated control cells exhibited compact COPI perinuclear staining reminiscent of the Golgi apparatus. In stark contrast, the BrefA- or GCA-treated cells exhibited dispersed β-COPI staining, indicative of an inability to recruit the COPI complex to the Golgi apparatus (48; see also http://zhuang.harvard.edu/Publications/Sun_et_al_supplemental_figures.pdf). To test whether the drug treatment also inhibits viral fusion, as observed for siRNA COPI knockdown, we performed a viral fusion assay as described for Fig. 1D. Viral fusion was measured in A549 cells pretreated and incubated with 10 μg/ml BrefA, 10 μM GCA, or 0.1% (vol/vol) DMSO (as a control for the total amount of DMSO added to the BrefA- or GCA-treated cells) during the entire experiment. In contrast to the results shown in Fig. 1D, we found that the inhibition of COPI complex vesicle formation does not affect viral fusion (Fig. 4A).
Fig 4.
Pharmacological disruption of COPI complexes does not block virus entry. (A) Representative flow cytometry-based quantification of DiD dequenching from three independent experiments. The DiD signal was measured at different time points after DiD-X31 infection in A549 cells treated with 0.1% (vol/vol) DMSO, 10 μg/ml BrefA, or 10 μM GCA. Standard deviations represent duplicate samples for each time point. (B) Quantification of the fraction of internalized AF647-X31 particles at 30 mpi upon treatment with inhibitors or DMSO, using the same immunofluorescence-based internalization assay in Fig. 2A. At least 17 different cells were quantified for virus internalization. Statistical analysis was performed using one-way analysis of variance (ANOVA). (C) Quantification of the fraction of AF647-X31 particles colocalized with Rab7-GFP+ endosomes at 90 mpi in A549 cells treated with 0.1% (vol/vol) DMSO, 10 μg/ml BrefA, or 10 μM GCA. At least 16 different cells were quantified. In the top left corner of each panel, we report the ratio that represents the sum of all AF647-X31 particles colocalized with Rab7 versus the total number of internalized virus particles for all cells quantified. Statistical analysis was performed using one-way ANOVA. (D) Flow cytometry-based quantification of the percentage of NP+ cells and the NP expression levels of the NP+ cells for A549 cells treated with 0.1% (vol/vol) DMSO, 10 μg/ml BrefA, or 10 μM GCA during the entire infection with X31 (MOI, ∼1). (E) Flow cytometry-based quantification of the percentage of NP+ cells and the NP expression levels of the NP+ cells for A549 cells infected with X31 (MOI, ∼1.5) and exposed to 0.1% (vol/vol) DMSO, 10 μg/ml BrefA, or 10 μM GCA only during entry. (F) Flow cytometry-based quantification of the percentage of NP+ cells and the NP expression levels of the NP+ cells for A549 cells infected with X31 (MOI, ∼1.5) and exposed to 0.1% (vol/vol) DMSO, 10 μg/ml BrefA, or 10 μM GCA only after 90 mpi. In D to F, the bar chart data represent the averages from two independent experiments (consisting of three replicate samples for each inhibitor or DMSO treatment), with the corresponding standard errors of the mean (SEM).
Because internalization and transport to late endosomes constituted the two major blocks in viral entry for ARCN1 knockdown cells, we tested whether BrefA and GCA treatment could recapitulate those same defects. Using the same immunofluorescence-based internalization assay as described for Fig. 2A and B, we found that the mean internalization fractions were 0.58, 0.56, and 0.57 for DMSO-, BrefA-, and GCA-treated cells, respectively (Fig. 4B). Furthermore, we tested whether inhibitor treatment affects trafficking to intermediate/late endosomes by measuring the fraction of internalized AF647-X31 within Rab7-GFP+ endosomes. The mean colocalization fractions for the virus and Rab7 in the DMSO-, BrefA-, and GCA-treated cells were 0.74, 0.67, and 0.61, respectively (Fig. 4C). Our AF647-X31 internalization and Rab7-GFP colocalization results were consistent with the viral fusion results; only minor differences were observed upon BrefA and GCA treatment, indicating that it is unlikely that rapid disruption of COPI vesicles results in an entry defect.
Next, to test whether BrefA or GCA treatment affects influenza infection, we pretreated A549 cells with 0.1% DMSO (as a control), 10 μg/ml BrefA, or 10 μM GCA for 60 min, allowed X31 to infect the cells at a multiplicity of infection (MOI) of ∼1 for 9 h, and measured NP expression with a flow cytometer. In stark contrast to the results shown in Fig. 1C, we found that the disruption of COPI complexes resulted in a moderate decrease in infection; relative to the DMSO control treatment, BrefA or GCA treatment reduced the percentage of NP-expressing cells by 21.3% ± 7.9% or 40.6% ± 1.3%, respectively. In addition, we found a moderate decrease in the NP expression level for NP+ cells relative to that for the DMSO control, i.e., 31.1% ± 1.2% and 35.3% ± 3.6% in the BrefA- and GCA-treated cells, respectively (Fig. 4D). However, it remains unclear whether these defects resulted from an entry or postentry exposure to BrefA or GCA.
To assess whether exposure to BrefA or GCA during entry affects productive influenza virus infection, we pretreated cells with the inhibitors for 60 min, followed by X31 infection in the presence of the compounds for 90 min. Afterward, the compounds were removed, and the infection was continued for an additional 7.5 h before processing for flow cytometry. We found that treatment with BrefA or GCA during entry alone did not affect the percentage of NP+ cells or the NP expression levels (Fig. 4E). However, when BrefA and GCA were added and maintained only after 90 mpi (Fig. 4F), a moderate decrease in NP expression was observed again (Fig. 4F), indicating that the effects of BrefA or GCA treatment on viral gene expression originate from postentry steps. Collectively, these results indicate that functional COPI vesicle recruitment to cellular membranes is not required for influenza virus internalization, endosomal trafficking, or viral fusion but has a moderate effect on viral NP expression.
Rapid disruption of COPI complex function decreases viral membrane protein expression and progeny virus production.
To test whether COPI complex disruption inhibits viral membrane protein expression, we infected A549 cells with X31 (MOI, ∼1.5). At 90 mpi, we added 0.1% DMSO (as a control), 10 μg/ml BrefA, or 10 μM GCA to the virus-infected cells. After 9 hpi, we collected the cells, fixed them, immunostained them for M2, and analyzed the samples with a flow cytometer. Immunostaining of the X31-infected cells for M2 in the presence of detergents allowed detection of total M2 expression. Compared with the results of postentry effects of BrefA and GCA on NP expression (Fig. 4F), M2 was more substantially perturbed upon treatment with BrefA or GCA than was the DMSO control (Fig. 5A).
Fig 5.
Functional COPI complex recruitment to the cis-Golgi is required for viral membrane protein expression and assembly of progeny virions. (A) Flow cytometry-based quantification of the percentage of M2-positive cells and the M2 expression levels in the M2-positive cells for A549 cells infected with X31 (MOI, ∼1.5) and exposed to 0.1% (vol/vol) DMSO, 10 μg/ml BrefA, or 10 μM GCA only after 90 mpi. Immunofluorescence staining was performed either in the presence of saponin, to detect total M2 expression, or in the absence of detergent, to detect surface M2. The bar chart data represent the averages from two independent experiments (consisting of three replicate samples for each inhibitor or DMSO treatment) with the corresponding SEM. (B) Quantification of the relative surface or total NA expression based on the acquired confocal images. Error bars indicate standard deviations for at least 18 images per treatment condition. (C) Quantification of the fraction of infected cells treated with 0.1% (vol/vol) DMSO, 10 μg/ml BrefA, or 10 μM GCA that contain budding zones at the cell periphery. The fraction in the top left corner of each panel represents the number of cells with a budding zone (numerator) relative to the total number of virus-infected cells (denominator) for all cells counted. (D) Viral titers of supernatants collected from X31-infected A549 cells treated with 0.1% (vol/vol) DMSO, 10 μg/ml BrefA, or 10 μM GCA at 18 hpi, as measured with plaque assays (see Materials and Methods). The bar chart includes the standard deviation from samples tested in quadruplicate.
To determine whether the disruption of COPI vesicle formation was essential to M2 trafficking to the plasma membrane, we performed the experiment as described above but immunostained cells for M2 under nonpermeabilizing conditions. M2 cell surface expression was decreased even more for the BrefA-treated cells than the total M2 level, indicating the presence of a trafficking defect. Surprisingly, we observed only a minor defect in M2 trafficking in GCA-treated cells (Fig. 5A). Additionally, we confirmed our results through sequential immunofluorescence assays for M2 without and with permeabilization. By 9 hpi, M2 readily trafficked to the plasma membrane in the DMSO-treated control cells. In stark contrast, no M2 was detected on the plasma membrane of BrefA-treated cells, while some M2 was trafficked to the cell surface and some was trapped within some intracellular vesicles in GCA-treated cells (http://zhuang.harvard.edu/Publications/Sun_et_al_supplemental_figures.pdf). We confirmed with β-COPI staining that COPI vesicle formation and concentration within the Golgi apparatus were still perturbed in both BrefA- and GCA-treated cells.
To test whether BrefA or GCA treatment also impaired the expression or plasma membrane targeting of another influenza virus membrane protein, we stained for surface and total NA (http://zhuang.harvard.edu/Publications/Sun_et_al_supplemental_figures.pdf). BrefA, and to a lesser extent GCA, impaired NA protein expression. Furthermore, consistent with the M2 transport experiments, BrefA treatment also inhibited NA trafficking to the plasma membrane. However, in contrast to the minor trafficking effects on M2, GCA treatment strongly inhibited NA transport to the cell surface (Fig. 5B). The discrepancy between M2 and NA plasma membrane trafficking in GCA-treated cells suggests that the two membrane proteins exhibit different transport requirements.
It remains unclear whether BrefA and GCA treatment leads to additional perturbations in the assembly of viral components at the plasma membrane. Influenza virus assembly is initiated by HA and NA recruitment to cholesterol-enriched plasma membrane domains. M1 is believed to interact with the cytoplasmic tails of HA and NA, and M1 interactions with vRNPs facilitate the incorporation of the viral genome into the budding virions (49). The vRNPs can be detected with PB1-specific antibodies. The PB1 patches are sites enriched with assembling virus particles, or budding zones. To determine whether functional COPI vesicle formation is essential for virus assembly, we immunostained X31-infected cells treated with DMSO, BrefA, or GCA for M2 (to detect infected cells) and PB1 (http://zhuang.harvard.edu/Publications/Sun_et_al_supplemental_figures.pdf). We found that 38% of the infected DMSO-treated cells contained budding zones enriched with PB1 at 18 hpi. In stark contrast, only 1% and 2% of BrefA- and GCA-treated cells, respectively, infected with X-31 had budding zones (Fig. 5C). The inability to traffic NA (and, to some extent, M2) to the plasma membrane in either the BrefA- or GCA-treated cells might help to explain the large defect in assembly and the failure to concentrate progeny virions at budding zones within infected cells.
To assess whether the defects in assembly were important for progeny virus production, we performed the experiment depicted in Fig. 5D. The supernatant of the virus-infected cells was collected at 18 hpi, and the viral titers were measured with standard plaque assays. We found that the presence of either BrefA or GCA reduced viral titers by about 2 orders of magnitude relative to those of the DMSO control (Fig. 5D). In summary, the results obtained from using the pharmacological inhibitors strongly suggest that the inactivation of COPI vesicle formation does not result in an entry defect but does specifically inhibit viral membrane protein expression, plasma membrane transport, assembly, and infectious progeny virion production.
DISCUSSION
As an obligate pathogen encoding 13 viral proteins, influenza virus requires host factors and compartments to mediate productive infection. Recently, genome-wide knockdown screens identified host dependency proteins important for influenza virus infection. Despite relatively little overlap in identified host dependency proteins among the screens, all of the screens identified subunits of the COPI complex as critical host proteins mediating productive virus infection. COPI proteins form complexes which oligomerize to coat vesicles. These coated vesicles mediate retrograde trafficking between the Golgi apparatus and the ER, as well as between the Golgi stacks. Previous studies also identified a role for COPI proteins in endosomal trafficking. Given that COPI proteins mediate trafficking between different cellular compartments in both the synthesis and endosomal trafficking pathways, it remained unclear which steps in the influenza virus infection cycle COPI proteins regulate.
Identification of host dependency proteins through siRNA depletion can result from four general outcomes: (i) siRNA-induced cytotoxicity, (ii) off-target effects (e.g., interferon induction or siRNA sequence overlap with other mRNA sequences), (iii) direct effects of siRNA-mediated depletion of the gene of interest, or (iv) indirect effects mediated by long-term depletion of the target of interest. We confirmed that, within our experimental time frame, siRNA-mediated cytotoxicity did not play a dominant role in our studies. No differences in cell viabilities were observed, with minor changes in cytotoxicities and thus cell growth rates. Based on the control experiments, we believe that the defects in influenza virus infection upon ARCN1 depletion result in either direct or indirect effects specific to siRNA-mediated silencing. In order to determine whether COPI depletion results in direct or indirect effects, we used both siRNA silencing and rapid COPI vesicle disruption via BrefA or GCA treatment to analyze how these two methods of perturbation affect different steps in the influenza virus infection cycle, i.e., binding, internalization, transport into late endosomes, fusion, viral protein expression, trafficking, assembly, and progeny virion production.
König and colleagues identified ARCN1, an essential subunit of the COPI complex, as a critical host protein for influenza virus infection (14). Upon further analysis, the authors also provided evidence that ARCN1 depletion affects influenza virus entry. Because of the data suggesting a role for ARCN1 in mediating influenza virus entry and because ARCN1 is known to be an essential component of the COPI vesicle complex, we decided to focus our studies on understanding the role of ARCN1 in influenza virus infection as a representative subunit of the COPI complex. After conducting a systematic dissection of the different entry steps, we found that ARCN1-depleted cells exhibit a significant defect in viral fusion. The abrogation of viral fusion in ARCN1-depleted cells likely explains why König and colleagues observed a significant defect in vRNP accumulation in the nucleus compared to that in the nontargeting control cells. Consistent with the viral fusion data, we found that ARCN1 knockdown cells exhibited an ∼66% decrease in internalization, an ∼76% decrease in colocalization of the virus with the late endosome marker LBPA, and an ∼90% decrease in virus infection relative to the those of the nontargeting siRNA-treated control cells. Taken together, our results show that COPI depletion leads to defects in virus internalization, transport to late endosomes, and viral fusion; viral fusion is likely a consequence of the first two effects.
Cureton and colleagues showed that COPI depletion also leads to defects in VSV entry (25), although there are differences between the two viruses in which particular steps are perturbed. Cureton and colleagues found that COPI depletion results in a decrease in viral fusion due to an ∼40% decrease in VSV binding and a decreased rate of VSV internalization. However, the VSV particles that manage to be internalized in COPI-depleted cells can still traffic to early endosomes and undergo vRNP release, indicating that the endosome trafficking of VSV is not perturbed. In contrast, our findings show that COPI depletion did not perturb influenza virus binding but did inhibit virus internalization and transport to late endosomes.
Treatment of cells with two pharmacological compounds, BrefA and GCA, that rapidly prevent the recruitment of COPI complexes to the Golgi apparatus resulted in strikingly different results from the siRNA experiments. Specifically, we did not observe any significant difference in influenza virus fusion or colocalization with a late endosome marker when cells were treated with DMSO, BrefA, or GCA. Substantiating the results from BrefA and GCA treatment, no extensive colocalization was observed between GFP-tagged ε-COPI, another essential component of the COPI complex, and AF647-labeled influenza virus in live cells during entry. Combining the siRNA, pharmacological inhibitor, and live cell imaging results, we conclude that functional COPI complex vesicle formation is not directly required for influenza virus entry, although ARCN1 depletion leads to indirect effects that result in defects in influenza virus internalization, trafficking to late endosomes, and, consequently, viral fusion.
Functional COPI complex formation and COPI vesicles mediate trafficking between the Golgi apparatus and the ER, as well as between different Golgi stacks. The Golgi apparatus-derived vesicles then traffic contents from the Golgi apparatus to other locations within the cell, including late endosomes, lysosomes, and the plasma membrane (16, 17). Defects in Golgi vesicular trafficking over an extended period of time, such as during siRNA depletion, could result in improper trafficking of membrane proteins or lipids. Changes in protein or lipid compositions (50) at the plasma membrane and endosomes may provide an explanation for the observed defects in viral internalization, trafficking, and fusion. The potential changes in protein and lipid compositions are, however, not reflected in the amount of α(2,6)-linked sialic acids (the attachment factor for influenza virus) presented on the surfaces of cells, consistent with the observation that viral binding is not substantially affected by ARCN1 knockdown.
The severely compromised uptake of general clathrin-mediated endocytosis and macropinocytosis cargoes in the ARCN1 knockdown cells suggests that protein and/or lipid compositions at the cell surface may be altered during the course of siRNA depletion. ARCN1 knockdown cells internalized ∼60% less virus by 30 mpi than the nontargeting control. Other cargoes of clathrin-mediated endocytosis (Tfn and EGF) and macropinocytosis (dextran) also exhibited significant internalization defects in ARCN1 knockdown cells relative to the nontargeting siRNA-treated cells. Consistent with our findings, Cureton and colleagues found that the uptake of VSV and Tfn was perturbed only after prolonged exposure to COPI disruption inhibitors (25).
Another phenomenon that might be related to the defect in trafficking of influenza virus to late endosomes is the anomalous LBPA+ late endosome staining pattern observed in Fig. 3A. In contrast to the dense perinuclear LBPA+ late endosome staining observed with the nontargeting control cells, we observed a reproducible dispersion of late endosomes—with no apparent aberrant changes to the early endosome organization—in ARCN1 knockdown cells. This observation is consistent with previous reports showing that COPI subunit depletion alters the late endosome trafficking pattern (51). It has also been found that the inhibition of endosomal trafficking through blockade of the uptake pathways leads to the losses of lysosomal acidification and perinuclear localization (52). We also noted that the shape of the ARCN1 knockdown cells sometimes appeared different from that of the nontargeting control cells, although the underlying mechanism for the change in cell morphology remains unclear. It is possible that the changes in the spatial distribution of late endosomes and the cell morphology are also related to the inhibition of viral entry.
Together, these results show that prolonged exposure to siRNA depletion of an essential COPI subunit results in an indirect effect of compromised cargo endocytosis. In contrast, the inhibition of COPI complex recruitment to the Golgi apparatus through acute pharmacological treatment directly affects viral protein expression and infectious progeny virus production. Disruption of COPI complexes by BrefA and GCA had only moderate effects on NP expression. However, BrefA and GCA treatment inhibited M2 and NA expression, suggesting that COPI complex formation is specifically required for at least some membrane protein expression. The inhibition might be due to retention of proteins within the ER from defective trafficking between the ER and the Golgi apparatus. Alternatively, treatment might cause rerouting of Golgi apparatus contents to alternative endosomal populations rather than intended target sites, such as the plasma membrane (53).
We also observed that BrefA- and GCA-treated cells exhibited defects in progeny virion assembly and production. Previous studies found that NA, but not M2, associates with lipid rafts (49). For GCA-treated cells, transport of M2 to the plasma membrane was modestly affected, while both the plasma membrane targeting of NA and progeny virus assembly were strongly impaired. These results suggest that the large assembly defect in GCA-treated cells might stem from inhibition of the trafficking of lipid raft-associated membrane proteins (which are critical for initiating virus budding and subsequent assembly steps) to the plasma membrane.
In summary, through dissection of the influenza virus infection cycle with both siRNA silencing and pharmacological compound perturbation, we have identified which steps of influenza virus infection are directly or indirectly dependent on functional COPI complex formation. Long-term siRNA COPI depletion in cells leads to indirect effects on viral entry through defective cargo uptake and vesicular trafficking to late endosomes, while acute treatment with COPI complex inhibitors impairs viral membrane protein expression and assembly, suggesting a direct role of COPI complexes in these later steps of infection. It was found recently that COPI proteins mediate many additional effects beyond the well-studied trafficking roles in the synthesis and secretory pathways, including lipid droplet formation and lipid metabolism, autophagosome formation, and endocytosis of EGF and Tfn (50, 51, 54, 55), which might be related to the observed indirect effects of COPI depletion on viral entry. Future studies on how COPI siRNA treatment affects protein or lipid expression, modification, and transport to different cellular compartments should be helpful for understanding how COPI proteins regulate such diverse cellular processes, as well as how these effects can lead to the observed defects of viral infection in COPI-depleted cells.
ACKNOWLEDGMENTS
We thank John Young for initial discussions that inspired this work, and we thank James Hogle, Sean Whelan, Raymond Chung, David Cureton, and Joshua Vaughan for useful discussions. We also thank Patricia Rogers and Brian Tilton from the Bauer Core Facility for technical advice on flow cytometry, as well as Robert Lamb, James Rothman, Felix Wieland, Gillian Air, and Qing Zhong for reagents.
This work was funded by NIH grant R01GM068518 (to X.Z.). E.S. is supported by a National Science Foundation graduate research fellowship. X.Z. is a Howard Hughes Medical Institute investigator.
Footnotes
Published ahead of print 19 December 2012
REFERENCES
- 1. Palese P, Shaw ML. 2007. Orthomyxoviridae: the viruses and their replication, p 1647–1689 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2. Wiley DC, Skehel JJ. 1987. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 56:365–394 [DOI] [PubMed] [Google Scholar]
- 3. de Vries E, Tscherne DM, Wienholts MJ, Cobos-Jimenez V, Scholte F, Garcia-Sastre A, Rottier PJ, de Haan CA. 2011. Dissection of the influenza A virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLoS Pathog. 7:e1001329 doi:10.1371/journal.ppat.1001329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Matlin KS, Reggio H, Helenius A, Simons K. 1981. Infectious entry pathway of influenza virus in a canine kidney cell line. J. Cell Biol. 91:601–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rust MJ, Lakadamyali M, Zhang F, Zhuang X. 2004. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat. Struct. Mol. Biol. 11:567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sieczkarski SB, Whittaker GR. 2002. Influenza virus can enter and infect cells in the absence of clathrin-mediated endocytosis. J. Virol. 76:10455–10464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doms RW, Helenius A, White J. 1985. Membrane-fusion activity of the influenza-virus hemagglutinin: the low pH-induced conformational change. J. Biol. Chem. 260:2973–2981 [PubMed] [Google Scholar]
- 8. Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. 2003. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 100:9280–9285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Skehel JJ, Bayley PM, Brown EB, Martin SR, Waterfield MD, White JM, Wilson IA, Wiley DC. 1982. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 79:968–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Whittaker GR, Kann M, Helenius A. 2000. Viral entry into the nucleus. Annu. Rev. Cell Dev. Biol. 16:627–651 [DOI] [PubMed] [Google Scholar]
- 11. Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454:890–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Maurer AP, Muller E, Wolff T, Rudel T, Meyer TF. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822 [DOI] [PubMed] [Google Scholar]
- 14. König R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, Garcia-Sastre A, Young JA, Palese P, Shaw ML, Chanda SK. 2010. Human host factors required for influenza virus replication. Nature 463:813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beck R, Rawet M, Wieland FT, Cassel D. 2009. The COPI system: molecular mechanisms and function. FEBS Lett. 583:2701–2709 [DOI] [PubMed] [Google Scholar]
- 16. Hsu VW, Yang JS. 2009. Mechanisms of COPI vesicle formation. FEBS Lett. 583:3758–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Popoff V, Adolf F, Brugger B, Wieland F. 2011. COPI budding within the Golgi stack. Cold Spring Harb. Perspect. Biol. 3:a005231 doi:10.1101/cshperspect.a005231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gu F, Gruenberg J. 1999. Biogenesis of transport intermediates in the endocytic pathway. FEBS Lett. 452:61–66 [DOI] [PubMed] [Google Scholar]
- 19. Aniento F, Gu F, Parton RG, Gruenberg J. 1996. An endosomal beta COP is involved in the pH-dependent formation of transport vesicles destined for late endosomes. J. Cell Biol. 133:29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daro E, Sheff D, Gomez M, Kreis T, Mellman I. 1997. Inhibition of endosome function in CHO cells bearing a temperature-sensitive defect in the coatomer (COPI) component ϵ-COP. J. Cell Biol. 139:1747–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gabriely G, Kama R, Gerst JE. 2007. Involvement of specific COPI subunits in protein sorting from the late endosome to the vacuole in yeast. Mol. Cell. Biol. 27:526–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gu F, Aniento F, Parton RG, Gruenberg J. 1997. Functional dissection of COP-I subunits in the biogenesis of multivesicular endosomes. J. Cell Biol. 139:1183–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo Q, Vasile E, Krieger M. 1994. Disruptions in Golgi structure and membrane traffic in a conditional lethal mammalian cell mutant are corrected by ϵ-COP. J. Cell Biol. 125:1213–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Whitney JA, Gomez M, Sheff D, Kreis TE, Mellman I. 1995. Cytoplasmic coat proteins involved in endosome function. Cell 83:703–713 [DOI] [PubMed] [Google Scholar]
- 25. Cureton DK, Burdeinick-Kerr R, Whelan SP. 2012. Genetic inactivation of COPI coatomer separately inhibits vesicular stomatitis virus entry and gene expression. J. Virol. 86:655–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lippincott-Schwartz J, Liu W. 2006. Insights into COPI coat assembly and function in living cells. Trends Cell Biol. 16:e1–e4 [DOI] [PubMed] [Google Scholar]
- 27. Cureton DK, Massol RH, Saffarian S, Kirchhausen TL, Whelan SPJ. 2009. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 5:e1000394 doi:10.1371/journal.ppat.1000394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johannsdottir HK, Mancini R, Kartenbeck J, Amato L, Helenius A. 2009. Host cell factors and functions involved in vesicular stomatitis virus entry. J. Virol. 83:440–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matlin KS, Reggio H, Helenius A, Simons K. 1982. Pathway of vesicular stomatitis virus entry leading to infection. J. Mol. Biol. 156:609–631 [DOI] [PubMed] [Google Scholar]
- 30. Sun XJ, Yau VK, Briggs BJ, Whittaker GR. 2005. Role of clathrin-mediated endocytosis during vesicular stomatitis virus entry into host cells. Virology 338:53–60 [DOI] [PubMed] [Google Scholar]
- 31. Superti F, Seganti L, Ruggeri FM, Tinari A, Donelli G, Orsi N. 1987. Entry pathway of vesicular stomatitis virus into different host cells. J. Gen. Virol. 68:387–399 [DOI] [PubMed] [Google Scholar]
- 32. Chen C, Zhuang XW. 2008. Epsin 1 is a cargo-specific adaptor for the clathrin-mediated endocytosis of the influenza virus. Proc. Natl. Acad. Sci. U. S. A. 105:11790–11795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. White J, Matlin K, Helenius A. 1981. Cell fusion by Semliki Forest, influenza, and vesicular stomatitis viruses. J. Cell Biol. 89:674–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun QM, Westphal W, Wong KN, Tan I, Zhong Q. 2010. Rubicon controls endosome maturation as a Rab7 effector. Proc. Natl. Acad. Sci. U. S. A. 107:19338–19343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vaughan JC, Brandenburg B, Hogle JM, Zhuang X. 2009. Rapid actin-dependent viral motility in live cells. Biophys. J. 97:1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen H, De Camilli P. 2005. The association of epsin with ubiquitinated cargo along the endocytic pathway is negatively regulated by its interaction with clathrin. Proc. Natl. Acad. Sci. U. S. A. 102:2766–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47 [DOI] [PubMed] [Google Scholar]
- 38. Stegmann T, Morselt HW, Scholma J, Wilschut J. 1987. Fusion of influenza virus in an intracellular acidic compartment measured by fluorescence dequenching. Biochim. Biophys. Acta 904:165–170 [DOI] [PubMed] [Google Scholar]
- 39. Doherty GJ, McMahon HT. 2009. Mechanisms of endocytosis. Annu. Rev. Biochem. 78:857–902 [DOI] [PubMed] [Google Scholar]
- 40. Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. 2006. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 10:839–850 [DOI] [PubMed] [Google Scholar]
- 41. Jiang X, Sorkin A. 2003. Epidermal growth factor receptor internalization through clathrin-coated pits requires Cbl RING finger and proline-rich domains but not receptor polyubiquitylation. Traffic 4:529–543 [DOI] [PubMed] [Google Scholar]
- 42. Sigismund S, Woelk T, Puri C, Maspero E, Tacchetti C, Transidico P, Di Fiore PP, Polo S. 2005. Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl. Acad. Sci. U. S. A. 102:2760–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lakadamyali M, Rust MJ, Zhuang X. 2006. Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell 124:997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lakadamyali M, Rust MJ, Zhuang X. 2004. Endocytosis of influenza viruses. Microbes Infect. 6:929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mercer J, Schelhaas M, Helenius A. 2010. Virus entry by endocytosis. Annu. Rev. Biochem. 79:803–833 [DOI] [PubMed] [Google Scholar]
- 46. Kobayashi T, Stang E, Fang KS, de Moerloose P, Parton RG, Gruenberg J. 1998. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature 392:193–197 [DOI] [PubMed] [Google Scholar]
- 47. Mossessova E, Corpina RA, Goldberg J. 2003. Crystal structure of ARF1*Sec7 complexed with brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol. Cell 12:1403–1411 [DOI] [PubMed] [Google Scholar]
- 48. Sáenz JB, Sun WJ, Chang JW, Li J, Bursulaya B, Gray NS, Haslam DB. 2009. Golgicide A reveals essential roles for GBF1 in Golgi assembly and function. Nat. Chem. Biol. 5:157–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rossman JS, Lamb RA. 2011. Influenza virus assembly and budding. Virology 411:229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Misselwitz B, Dilling S, Vonaesch P, Sacher R, Snijder B, Schlumberger M, Rout S, Stark M, von Mering C, Pelkmans L, Hardt WD. 2011. RNAi screen of Salmonella invasion shows role of COPI in membrane targeting of cholesterol and Cdc42. Mol. Syst. Biol. 7:474 doi:10.1038/msb.2011.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Razi M, Chan EYW, Tooze SA. 2009. Early endosomes and endosomal coatomer are required for autophagy. J. Cell Biol. 185:305–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huotari J, Helenius A. 2011. Endosome maturation. EMBO J. 30:3481–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang CJ, Rosenwald AG, Willingham MC, Skuntz S, Clark J, Kahn RA. 1994. Expression of a dominant allele of human ARF1 inhibits membrane traffic in vivo. J. Cell Biol. 124:289–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Collinet C, Stoter M, Bradshaw CR, Samusik N, Rink JC, Kenski D, Habermann B, Buchholz F, Henschel R, Mueller MS, Nagel WE, Fava E, Kalaidzidis Y, Zerial M. 2010. Systems survey of endocytosis by multiparametric image analysis. Nature 464:243–249 [DOI] [PubMed] [Google Scholar]
- 55. Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453:657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]