Abstract
Poly(ADP-ribose) polymerase 1 (PARP-1) is a cellular enzyme with a fundamental role in DNA repair and the regulation of chromatin structure, processes involved in the cellular response to retroviral DNA integration. However, the function of PARP-1 in retroviral DNA integration is controversial, probably due to the functional redundancy of the PARP family in mammalian cells. We evaluated the function of PARP-1 in retroviral infection using the chicken B lymphoblastoid cell line DT40. These cells lack significant PARP-1 functional redundancy and efficiently support the postentry early events of the mammalian-retrovirus replication cycle. We observed that DT40 PARP-1−/− cells were 9- and 6-fold more susceptible to infection by human immunodeficiency virus type 1 (HIV-1)- and murine leukemia virus (MLV)-derived viral vectors, respectively, than cells expressing PARP-1. Production of avian Rous-associated virus type 1 was also impaired by PARP-1. However, the susceptibilities of these cell lines to infection by the nonretrovirus vesicular stomatitis virus were indistinguishable. Real-time PCR analysis of the HIV-1 life cycle demonstrated that PARP-1 did not impair reverse transcription, nuclear import of the preintegration complex, or viral DNA integration, suggesting that PARP-1 regulates a postintegration step. In support of this hypothesis, pharmacological inhibition of the epigenetic mechanism of transcriptional silencing increased retroviral expression in PARP-1-expressing cells, suppressing the differences observed. Further analysis of the implicated molecular mechanism indicated that PARP-1-mediated retroviral silencing requires the C-terminal region, but not the enzymatic activity, of the protein. In sum, our data indicate a novel role of PARP-1 in the transcriptional repression of integrated retroviruses.
INTRODUCTION
Retroviruses have evolved a replication strategy that requires integration of the viral genome into the host genome. This event triggers a complex cellular response that is directed to preserve the integrity of the host genome, as well as its chromatin architecture. This complex cellular response includes DNA damage repair and chromatin remodeling (1–12).
Poly(ADP-ribose) polymerase 1 (PARP-1) is a key cellular enzyme in DNA repair and chromatin-remodeling processes (13, 14). In mammals, this enzyme is the founding member of a family of 18 proteins (15). PARP-1 promotes the transfer of ADP ribose molecules from NAD+ to acceptor proteins or to a previously formed poly(ADP-ribose) (PAR) chain. PARylation notoriously influences the functions of target proteins by altering their subcellular distributions, molecular interactions, and enzymatic activities. Similar to PARP-1, PARP-2 to -5 are nuclear proteins capable of catalyzing PARylation and have roles in genome stability and/or chromatin remodeling (14, 15). This functional overlap within the PARP family determines that although PARP-1 knockout (KO) mice do not exhibit major functional or structural defects (16), PARP-1/2 double-knockout mice are embryonic lethal (17). Similarly, PARP-1/3 double-knockout mice are hypersensitive to X-irradiation compared to the corresponding single-knockout mice (18). As expected, this functional redundancy imposes an additional challenge on the study of the role of PARP-1 in retroviral infection in mammalian cells.
The N-terminal region of PARP-1 (amino acids [aa] 1 to 524) contains a nuclear localization signal that determines the subcellular distribution of PARP-1, two zinc binding domains that mediate its binding to DNA, a caspase 3 cleavage site, and a breast cancer suppressor protein carboxy-terminal (BRCT) domain implicated in the interaction of PARP-1 with other proteins. The C-terminal region (amino acids 525 to 1014) of PARP-1 contains a WGR motif, proposed to mediate DNA binding, and the catalytic domain (19). These structural domains interact dynamically and coordinate different catalytic-independent and -dependent PARP-1 functions. PARP-1 is incorporated into nucleosomes in a catalytic-independent manner (20–22) and requires the DNA binding domain (20, 21, 23, 24) and the interaction of the C-terminal region (amino acids 214 to 1014) with the nucleosome core histone proteins (22). Incorporation of PARP-1 into the chromatin causes chromatin compaction and transcriptional repression (20, 21, 23–25). This repressive activity of PARP-1 on transcription mediates silencing of retrotransposable elements and the formation of heterochromatin in Drosophila (21, 25). Activation of the enzymatic activity of nucleosome-incorporated PARP-1 leads to auto-PARylation and dissociation of PARP-1 from chromatin, causing chromatin decondensation and activation of transcription (20, 22, 24). In addition, PARP-1 promotes transcription by other mechanisms, including inhibition of histone H3 demethylases and depletion of histone H1 from chromatin (26). The enzymatic activity of PARP-1 is also central in the repair of single-strand DNA breaks through the base excision repair pathway. The binding of PARP-1 to DNA breaks leads to upregulation of its enzymatic activity, resulting in PARylation of a variety of proteins, including PARP-1 itself and other regulatory and structural proteins involved in DNA repair, chromatin remodeling, transcription, and cell cycle regulation. In addition, PARylated PARP-1 recruits histone variants and chromatin-remodeling factors that produce important modifications in the chromatin structure at the DNA damage lesion, including compaction that results in transcriptional repression (13, 14).
A role for PARP-1 in HIV DNA integration has been proposed; however, this function has been a matter of intense debate (27–33). Contradictory data have been reported using either human- or mouse-derived primary or tumor cell lines in which the function of PARP-1 has been impaired by small interfering RNA (siRNA) expression (27, 29, 31, 32), gene knockout (27, 30, 32, 33), pharmacological inhibition (28, 29), or the use of a dominant mutant (29). In some of these studies, PARP-1 is reported to be required for HIV DNA integration (29, 30, 32), whereas in others it is proposed to have a dispensable role or even to impair this viral process (27, 28, 33). These contradictory results may not be surprising if we take into consideration the PARP functional redundancy found in mammalian cells (17, 18). Therefore, the study of PARP-1 functions in simpler organisms that lack this functional redundancy is advantageous, and this strategy has significantly contributed to the understanding of the cellular functions of the enzyme (21, 22, 25). In addition, PARP-1 functions identified in simpler organisms are relevant to more complex organisms due to the high evolutionary conservation of PARP-1 from Drosophila to humans.
In this study, we took advantage of the functional and structural evolutionary conservation of PARP-1. We studied the role of the enzyme in retroviral infection using the chicken B lymphoblastoid cell line DT40. These cells exhibit low PARP-1 functional redundancy and are viable after PARP-1 knockout. DT40 cells naturally lack the PARP-2 gene, the closest PARP-1 paralog, and PARylation is completely abrogated after PARP-1 knockout, indicating that the enzyme is the principal, if not the only, PARP protein with enzymatic activity in these cells (34). In addition, chicken cells can be used as a reliable model to study the molecular events that occur from entry to proviral transgene expression characteristic of the gammaretrovirus and lentivirus life cycle. Chicken cells are highly permissive to transduction with single-round infection by murine leukemia virus (MLV) and HIV reporter viruses (35–39), suggesting that the molecular mechanisms required for uncoating, reverse transcription, DNA integration, and transgene expression of these retroviruses are conserved in these cells. Further evidence supporting the conservation of the molecular mechanisms implicated in the early steps of HIV-1 infection in chicken cells indicates that, as in human cells, HIV integrates preferentially inside active transcription units in the chicken genome (35).
Using the DT40 cells as a model, we have discovered a novel role of PARP-1 in retroviral infection. Our data indicate that, in a catalytic-independent manner, PARP-1 promotes transcriptional repression of integrated retroviruses by epigenetic mechanisms that involve histone deacetylation and DNA methylation.
MATERIALS AND METHODS
Cell lines.
DT40-derived wild-type (WT) and KO cell lines and a PARP-1 knockout engineered to express human PARP-1 (h-1) and human PARP-2 (h-2) were previously described (34) and were kindly provided by Shunichi Takeda (Crest Laboratory, Department of Radiation Genetics, Faculty of Medicine, Kyoto University, Kyoto, Japan). KO cells expressing a human PARP-1 ΔC-terminal mutant were generated by electroporation of the corresponding expression plasmid in KO cells as described below. DT40-derived cells were cultured in RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum, 6% heat-inactivated chicken serum, 2 mM l-glutamine, and 1% penicillin/streptomycin.
The SupT1 and Jurkat cell lines were grown in RPMI 1640, while HEK293T-derived cells were grown in Dulbecco's modified Eagle's medium (DMEM), and both culture media were supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, and 1% penicillin/streptomycin.
Human PARP-1-deficient cells were generated by transduction of SupT1 cells with HIV-derived vectors expressing a PARP-1-specific or a control short hairpin RNA (shRNA), following procedures previously described (40). SupT1 cells were infected at different multiplicities of infection (MOIs) and 2 weeks later were sorted for high levels of enhanced green fluorescent protein (EGFP) expression (the top 10% of EGFP mean fluorescence intensity) by flow cytometry. The sorted cells were cultured for 10 days and then characterized.
Electroporation.
KO cells were cultured at a density of 106 cells/ml prior to electroporation. Cells (107) were washed once in ice-cold phosphate-buffered saline (PBS) and resuspended in 0.8 ml of ice-cold PBS that contained 25 μg of the XhoI-linearized expression plasmid. The cell suspension was then transferred to a 0.4-cm-gap electroporation cuvette (Bio-Rad 1652088) and incubated on ice for 10 min. The cells were electroporated at 550 V and 25 μF in a Bio-Rad Gene Pulser XCell electroporator. The electroporated cells were incubated on ice for 10 min and then gently transferred to a T-75-cm2 flask containing 20 ml RPMI 1640 supplemented with 20% heat-inactivated fetal calf serum, 6% heat-inactivated chicken serum, 2 mM l-glutamine, and 1% penicillin/streptomycin. Twenty-four hours later, the culture medium was removed and the cells were resuspended in fresh culture medium supplemented with puromycin (0.25 μg/ml). Robust polyclonal puromycin-resistant cultures emerged between 2 and 3 weeks after electroporation. PARP-1 expression was verified by immunoblotting with an anti-PARP-1 monoclonal antibody (MAb) 2-C-10 clone (41).
Plasmids.
PARP-1 expression plasmids. A human PARP-1 ΔC-terminal mutant (lacking amino acids 489 to 1014) was expressed from pPARP-1 Δ C-term. This plasmid contains the human cytomegalovirus (CMV) immediate-early gene promoter driving the transcription of PARP-1 cDNA, followed by an internal ribosome entry site and the puromycin N-acetyl-transferase gene. The PARP-1 ΔC-terminal mutant was generated by mutagenesis PCR and is C-terminally tagged with a flexible and hydrophilic 20-amino-acid peptide. The identity of pPARP-1 Δ C-term was verified by overlapping DNA sequencing of the PARP-1 cDNA.
Retroviral vector plasmids.
The plasmids used to generate retroviral vectors were described previously (40). HIV vectors were produced using pHIVluc or pTRIP (42), pCMVΔR8.91, and pMD.G (a gift from D. Trono). pHIVluc was derived from pNL4-3.Luc.R−E− (43) by introducing a deletion in the Env open reading frame. The LacZ open reading frame in pTRIP was replaced by EGFP or firefly luciferase (40). Luciferase is expressed from the HIV long terminal repeat (LTR) promoter in HIVluc, and EGFP or luciferase is expressed from an internal CMV promoter in pTRIP-derived vectors. pMLV luc was obtained by cloning firefly luciferase into pLPCX (Clontech) (40).
Human PARP-1 knockdown and control cells were generated by transduction with HIV-derived vectors produced with plasmids pTRIP EGFP shRNA PARP-1 and pTRIP EGFP shRNA control. These plasmids were constructed by cloning shRNA expression cassettes that contain PARP-1-specific or scrambled shRNA sequences at a unique PpuMI site in pTRIP EGFP. The PARP-1-specific shRNA expression cassette was generated by annealing oligonucleotides 5′-GatcccgAAGTATCCCAAAAAGTTCTttcaagagaAGAACTTTTTGGGATACTTttttttggaaa-3′ and 5′-agcttttccaaaaaaAAGTATCCCAAAAAGTTCTtctcttgaaAGAACTTTTTGGGATACTTcgG-3′ and cloning them into the pSilencer 2.1-U6 hygro expression plasmid (Ambion). Then, the U6 promoter and the shRNA sequences were amplified by PCR with primers EE5 (sense; 5′-TATAGGGACCCGTAAAACGACGGCCAGTGCC-3′) and EE6 (antisense; 5′-TATAGGGTCCCGAATTCCCCAGTGGAAAGACG-3′) and cloned into pTRIP EGFP.
Generation of viruses.
Procedures described previously (40) were followed for the production of single-round-infection retroviruses. Briefly, MLV Luc vectors were produced in Phoenix A packaging cells by calcium-phosphate cotransfection of 15 μg of pLPCXluc and 5 μg of the vesicular stomatitis virus glycoprotein G (VSV-G) expression plasmid pMD.G. Forty-eight hours after transfection, the viral supernatants were harvested and concentrated by ultracentrifugation at 124,750 × g for 2 h on a 20% sucrose cushion.
HIVluc was prepared by calcium-phosphate cotransfection of HEK293T cells with 15 μg of pHIVluc and 5 μg of pMD.G. The viral supernatant was collected 48 h after transfection, and aliquots were stored at −80°C until use.
TRIPluc, TRIPEGFP, pTRIP EGFP shRNA PARP-1, and pTRIP EGFP shRNA control vectors were produced by calcium-phosphate cotransfection of HEK293T with 15 μg of the corresponding pTRIP plasmid, 15 μg of pCMVΔR8.91, and 5 μg of pMD.G. The virus-containing supernatant was collected and concentrated by ultracentrifugation as described above.
DNase-treated single-round-infection TRIPluc viruses were prepared by incubation of the filtered viral supernatant with 40 U/ml Turbo DNase (Ambion) for 30 min at 37°C (44).
Avian sarcoma/leukosis virus (ASLV)-based vectors expressing EGFP (ASLV GP) were produced by transient transfection of DF-1 chicken embryonic fibroblasts with RCANBPM2C(797-8)PolIGFP or RCASBPM2C(797-8)GFP plasmids (45, 46) (a gift from Stephen H. Hughes, NCI-Frederick, HIV Drug Resistance Program).
Reverse transcriptase levels in the HIV- and MLV-derived viral vector preparations were measured using the EnzChek Reverse Transcriptase Assay Kit (Invitrogene), following the manufacturer's instructions.
Recombinant vesicular stomatitis virus expressing EGFP (a gift from John C. Bell) was previously described (47); it was amplified, and titers were determined on Vero cells.
Single-round infectivity assay.
Target cells were plated at 1 × 105 cells in 500 μl of RPMI 1640 culture medium in 24-well plates and infected with HIV- or MLV-derived viral supernatants containing similar reverse transcriptase levels. Four days postinfection, cells were collected by centrifugation at 1,000 × g for 6 min, and the pellet was resuspended in 200 μl of PBS. Half of the sample was mixed with 100 μl of luciferase substrate (Bright-Glow Luciferase Assay System; Promega) and the other half with 100 μl of cell viability substrate (CellTiter-Glo Assay; Promega). Cell lysates were incubated for 10 min at room temperature in the dark, and then the luminescence was measured in triplicate in 50-μl samples using a microplate luminometer reader.
Analysis of the production of RAV-1.
DT40 cells were generated by infection of a chicken with RAV-1 (48). These cells are known to continuously produce and release infectious avian Rous-associated virus type 1 (RAV-1) into the supernatant. DT40-derived WT, KO, and h-1 cells were plated at 0.4 × 106 cells/ml and cultured for 72 h. Cell culture supernatant was collected, passed through a 0.45-μm filter, and concentrated by ultracentrifugation at 124,750 × g for 2 h on a 20% sucrose cushion. The viability of the producer cells was determined at the end of the experiment by measuring ATP levels, as described above (CellTiter-Glo Assay; Promega). Viral pellets were resuspended in fresh culture medium, and reverse transcriptase was measured with the EnzChek Reverse Transcriptase Assay Kit (Invitrogen).
Immunoblotting.
Cells (3 × 106) were lysed in 100 μl of Laemmli sample buffer (12 mM Tris-Cl, pH 6.8, 0.4% SDS, 2% glycerol, 1% β-mercaptoethanol, 0.002% bromophenol blue), and 15 μl of the sample was resolved by SDS-PAGE and transferred overnight to polyvinylidene difluoride (PVDF) membranes at 100 mA at 4°C. The membranes were blocked in Tris-buffered saline (TBS) containing 10% milk for 1 h and then incubated with the corresponding primary antibody diluted in TBS-5% milk-0.05% Tween 20 (antibody dilution buffer). PARP-1 was detected with MAb 2-C-10 (41) (diluted 1/1,000), PARylation with an anti-poly(ADP-ribose) monoclonal antibody (sc-56198, diluted 1/125) and PARP-2 with a rabbit antibody (sc-133886, diluted 1/500). As a loading control, anti-alpha-tubulin MAb (clone B-5-1-2; Sigma) was used at a 1/4,000 dilution. Membranes were incubated overnight at 4°C with anti-PARP-1 or -2 or -PAR Mab, whereas anti-alpha tubulin MAb was incubated for 2 h at 25°C. Primary-antibody-bound membranes were washed in TBS-0.1% Tween 20, and bound antibodies were detected with goat anti-mouse Ig-horseradish peroxidase (HRP) (Sigma) diluted 1/2,000 in antibody dilution buffer, followed by chemiluminescence detection.
Analysis of the HIV-1 life cycle by real-time PCR.
DT40-derived cells (1 × 106) were challenged with DNase-treated single-round-infection TRIPluc viruses, and 24 h later, 90% of the cells were used for DNA extraction (High Pure PCR template preparation kit; Roche), whereas 10% were cultured for 4 days to evaluate infectivity. DNA (1 ng) was used to quantify total HIV-1 cDNA (gag DNA), two long terminal repeat (2LTR) circle DNA, or histone H4 DNA by real-time PCR in a MiniOpticon system (Bio-Rad) as described previously (49). 2LTR DNA was detected using primers MB49 (5′-AACTAGGGAACCCACTGCTTAAG) and MB50 (5′-TCCACAGATCAAGGATATCTTGTC), while total HIV cDNA (HIV gag DNA) was detected with primers MB72 (5′-CGGATCTCGACGGTATGCGCC) and MB73 (5′-TCGCCCCAAAGTGGATCTCTGC). As a loading control, histone H4 DNA was quantified in the same samples used to detect HIV DNA with primers MB74 (5′-TGCGCGACAACATCCAGGGCATCAC) and MB75 (5′-GTGACCGTCTTCCTCTTGGCGTGCTC). In other experiments, DT40 cells were infected with TRIPluc and cultured for 10 days before the DNA was extracted from 106 infected cells for real-time PCR analysis. The fold change was calculated in all these experiments using the ΔCt method, and the values were expressed relative to the values detected in the WT cells.
Pharmacological inhibition of PARP.
Jurkat, HEK293T, SupT1, and DT40-derived cells were treated with several PARP inhibitors for 24 h and then challenged with single-round-infection HIV-derived viruses expressing luciferase. Infected cells were culture for 4 days in the presence or absence of inhibitors, and then HIV-1 infection and cell viability were measured as described above. The PARP inhibitor set (catalog no. 528820; EMD) included 3-aminobenzamide, 5-iodo-6-amino-1,2-benzopyrone, 1,5-isoquinolinediol, 8-hydroxy-2-methylquinazoline-4-one (NU 1025), and 3,4-dihydro-5[4-(1-piperindinyl)butoxy]-1(2H)-isoquinoline (DPQ).
Pharmacological inhibition of histone deacetylase and DNA CpG methylation.
DT40-derived cells were infected with TRIPluc or MLVluc and 4 days later treated or not with sodium butyrate (5 mM; B5887; Sigma) for 24 h or with 5-azacytidine (30 μM; A2385; Sigma) for 36 h. Following these treatments, the cells were analyzed for HIV infection and cell viability as described above.
RESULTS
Roles of PARP-1 and PARP-2 in retroviral infection.
In order to evaluate the roles of PARP-1 and -2 in retroviral infection, we studied DT40-derived cells. These cells efficiently support the early postentry steps of the gammaretrovirus and lentivirus life cycle (35–39). PARP-1 is the only member of the PARP family with catalytic activity in these cells, and DT40 cells are viable after PARP-1 knockout (34).
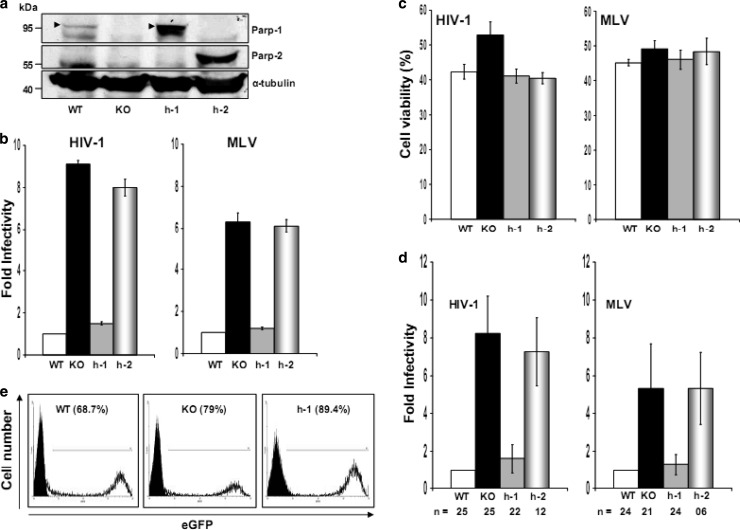
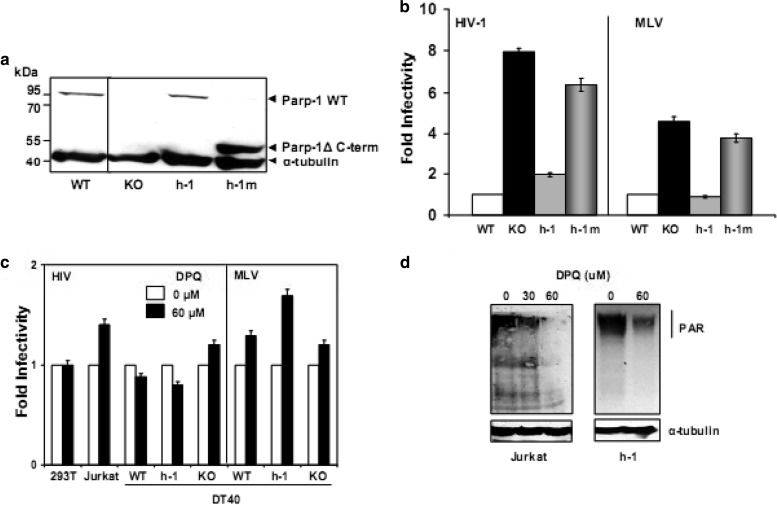
DT40 WT, KO, and h-1 and h-2 cells (Fig. 1a) were challenged with single-round-infection HIV (TRIPluc)- and MLV (MLVluc)-derived viral vectors containing similar levels of reverse transcriptase activity. These viruses express firefly luciferase from an internal CMV promoter. Four days after viral challenge, the cells were analyzed for luciferase activity and cell viability. Viability was determined by measuring intracellular levels of ATP. The data depicted in Fig. 1b indicated that KO cells were approximately nine and six times more susceptible to infection by HIV and MLV, respectively, than WT cells, suggesting that PARP-1 expression impairs MLV and HIV infection. Similar results were observed when an HIV-derived vector expressing EGFP was evaluated (data not shown).
Fig 1.
Effects of PARP-1 and PARP-2 on retroviral infection. (a) Expression of PARP proteins in DT40 WT, KO, and h-1 or h-2 cells. Alpha tubulin was detected as a loading control. (b, c, and d) Susceptibility of DT40 WT, KO, h-1, and h-2 cells to TRIPluc and MLVluc infection. Cells were challenged with TRIPluc or MLVluc, and infectivity (luciferase expression [b and d]) and cell viability (ATP levels [c]) were measured 4 days after infection. (b) Fold infectivity is expressed relative to the luciferase levels detected in WT cells, and the standard deviations (SD) (error bars) correspond to triplicate measurements from two independent experiments. (d) Fold infectivity was calculated by normalizing luciferase activity to ATP levels in the same samples and expressed relative to the values of WT cells. The standard deviations represent the variability observed in multiple experiments (n is indicated) conducted over 1 year using different viral preparations. (c) Viability of cells in panel b 4 days after HIV-1 or MLV infection. The standard deviations correspond to triplicate measurements of two independent experiments, and the percent cell viability was calculated relative to the corresponding noninfected cell lines. (e) FACS analysis of EGFP expression in DT40 WT, KO, and h-1 cells infected at a multiplicity of infection of 10 with a recombinant vesicular stomatitis virus expressing EGFP. The percentages of EGFP-positive cells are shown in parentheses.
The enhanced susceptibility of KO cells to retroviral infection was efficiently reduced to the levels observed in WT cells by stable expression of human PARP-1 (h-1 cells) (Fig. 1b). These results clearly indicated that both chicken and human PARP-1 efficiently impair HIV and MLV infection. Our observations also correlated with the high degree of evolutionary conservation of PARP-1. Comparison of the primary sequences of human and chicken PARP-1 proteins indicates 77% identity and 88% similarity.
The data in Fig. 1 indicated that, although both h-1 and WT cells were similarly susceptible to retroviral infection (Fig. 1b), h-1 cells express higher levels of PARP-1 (human protein) than WT cells (avian protein) (Fig. 1a). These results suggested that this novel activity of PARP-1 reaches saturation at the physiological levels of the protein. Alternatively, the apparent differences in PARP-1 levels observed in WT and h-1 cells could be determined by different affinities of the antibody used for the human and the avian proteins.
Unlike the effect of PARP-1, robust expression of human PARP-2 (Fig. 1a), the closest human PARP-1 paralog, did not modify the susceptibility of KO cells to retroviral infection (Fig. 1b). This observation suggested that the enzymatic activity or other PARP-1 functions shared with PARP-2 are not involved in this novel function of PARP-1.
Massive DNA damage leads to strong PARP-1 activation that triggers cellular death (50–52). Therefore, we determined whether the differences in susceptibility to retroviral infection observed in cells expressing or not expressing PARP-1 could be attributed to different cellular survival or proliferation rates following retroviral infection. To this end, we measured the viability of WT, KO, h-1, and h-2 cells 4 days after HIV-1 or MLV infection. Although infected cells exhibited a decrease in viability compared to noninfected cells, we did not observe significant differences in viability between cells expressing or not expressing PARP proteins after viral infection (Fig. 1c). Therefore, the observed differences in susceptibility to retroviral infection were not due to different cellular proliferation or survival rates of the infected cells.
In order to better characterize the range of the inhibitory effect of PARP-1 on retroviral infection, we compiled (Fig. 1d) the results of multiple experiments conducted over 1 year with different viral preparations and by different researchers in our laboratory. In the infection experiments described below, the luciferase activity of the retroviral transgene was normalized to the ATP levels detected in the studied cells.
The specificity of the effect of PARP-1 on retroviral infection was next evaluated by determining the susceptibility of cells expressing or not expressing PARP-1 to infection by a rhabdovirus, vesicular stomatitis virus, engineered to express EGFP (VSV-EGFP) (47). WT, KO, and h-1 DT40 cells were infected at multiplicities of infection (MOIs) of 100, 30, 10, and 1 and analyzed by fluorescence-activated cell sorting (FACS) 48 h later. VSV-driven EGFP expression levels were similar in these cells at all the MOIs evaluated, Fig. 1e shows the results for an MOI of 10, indicating that the susceptibility to VSV infection was not influenced by the levels of PARP-1 in the target cells. VSV-driven expression of EGFP was detected by microscopic examination of the infected cells as early as 10 h postinfection, and no differences between the infected cell lines were observed at this early time point (data not shown). Analysis of VSV infection at later time points was precluded due to the cytopathic effects caused by the replicating virus. Therefore, these data demonstrated that the decreased susceptibility to retroviral infection observed in cells expressing PARP-1 was not due to a global defect in the cells.
In summary, the data included in Fig. 1 clearly indicated that PARP-1, but not PARP-2, specifically impairs retroviral infection through an evolutionarily conserved mechanism.
Analysis of the effect of PARP-1 on the HIV life cycle.
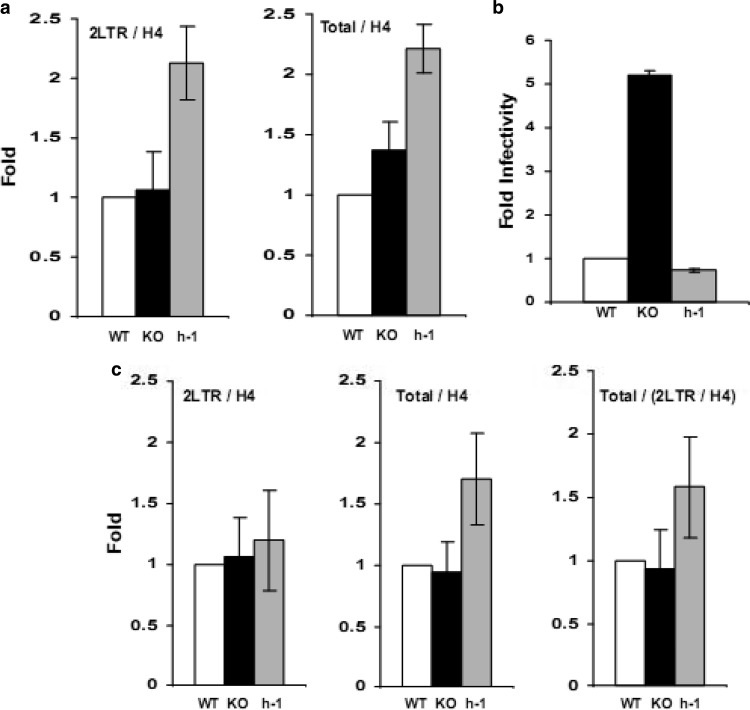
PARP-1 has been extensively studied in relation to retroviral DNA integration in mammalian cells, and contradictory data have been reported. While some studies indicated a fundamental role of PARP-1 in retroviral DNA integration and integration site selection, other investigations reported that PARP-1 is dispensable for retroviral DNA integration or infection (27–33). To evaluate the effect of PARP-1 on the HIV life cycle in DT40 cells, we quantified by real-time PCR the levels of total HIV cDNA (gag DNA), an indicator of complete and successful reverse transcription, and the levels of 2LTR circle DNA, a marker of the nuclear import of the preintegration complex that was inversely correlated with viral DNA integration. As a loading control, we detected histone H4 DNA in the same samples by real-time PCR and used these values to normalize the levels of total HIV cDNA and 2LTR circle DNA.
In these studies, we found similar amounts of histone H4-normalized total HIV cDNA and 2LTR circle DNA in WT and KO cells at 24 h postinfection, whereas the levels of both HIV-1 DNA forms were slightly higher in h-1 cells (Fig. 2a). These results established that PARP-1 impairs HIV-1 infection by a mechanism that does not involve reverse transcription, nuclear import of the preintegration complex, or viral DNA integration, therefore suggesting a postintegration effect.
Fig 2.
Effect of PARP-1 on the HIV-1 life cycle. DT40 WT, KO, and h-1 cells were infected with TRIPluc and analyzed 24 h (a) or 10 days (b and c) later. Total HIV cDNA (gag DNA), 2LTR circle DNA, and histone H4 DNA were determined by real-time PCR. Total HIV cDNA and 2LTR circle DNA levels were normalized to the levels of histone H4. The standard deviations represent the variability of triplicate real-time measurements of samples from two (a) or three (c) independent infection experiments. (b) Susceptibility of DT40-derived cells to HIV-1 infection. Infectivity (cell viability-normalized luciferase levels) was determined at 10 days postinfection. Fold infectivity is expressed relative to the infectivity of WT cells. The standard deviations represent the variability of triplicate measurements of one of the infection experiments considered in panel c.
Given the lack of techniques to directly quantify HIV-1 DNA integration in chicken cells, we could only indirectly analyze the effects of PARP-1 on viral DNA integration. However, it is well established that the nonintegrated forms of HIV cDNA exhibit a significantly shorter half-life than the integrated viral cDNA. Among other factors, the incapability of the nonintegrated HIV cDNA to undergo DNA replication determines that the amount of nonintegrated viral DNA substantially decreases in a cell after several cell divisions. In contrast, the integrated viral cDNA replicates synchronously with the host DNA and remains at constant levels throughout multiple rounds of cell division. This cell division-dependent dilution effect is especially marked in DT40 cells, which divide up to three times per day (53, 54). We took advantage of the different half-lives of integrated and nonintegrated forms of the HIV genome and the short duration of the DT40 cell cycle to quantify the relative levels of integrated HIV DNA in DT40 cells by real-time PCR. The primers used to amplify total HIV cDNA (gag DNA) detect both integrated (provirus) and nonintegrated viral cDNAs that include linear and circular forms (mainly 1LTR and 2LTR circles). Therefore, we quantified gag DNA levels (total HIV cDNA) in infected DT40 cells 10 days after infection. Due to the different half-lives of integrated and nonintegrated HIV DNA forms, we expected that the majority, if not all, of the gag DNA at this time point would correspond to integrated HIV DNA. To verify this assumption, we also measured 2LTR circle DNA levels in these cells by real-time PCR. The amounts of all these forms of HIV-1 DNA were normalized to histone H4 DNA levels to ensure equal loading.
In these experiments, WT, KO, and h-1 cells were infected with TRIPluc and cultured for 10 days, and then the luciferase and ATP levels were determined and DNA was extracted for analysis. In agreement with the results presented in Fig. 1b and d, cells expressing PARP-1 were also markedly less susceptible to HIV-1 infection than PARP-1−/− cells when infectivity was measured 10 days after viral transduction (Fig. 2b). In accordance with our predictions, in three out of six independent infection experiments, 2LTR DNA was undetectable, while very low levels were measured in the remainder of the experiments. On the other hand, total HIV cDNA levels were very robust in the six assays evaluated, suggesting that the majority of the total HIV cDNA detected with the gag primers corresponds to integrated viral cDNA. In samples with undetectable 2LTR DNA circles, similar levels of H4-normalized total HIV cDNA were found in WT, KO, and h-1 cells. KO cells contained 1.68- ± 0.72- and h-1 cells 1.71- ± 0.91-fold higher levels of histone H4-normalized total HIV cDNA than WT cells. These results demonstrated that HIV-1 DNA integration occurred with similar efficiencies in cells expressing or not expressing PARP-1.
In the other three experiments where low levels of 2LTR circle DNA could still be detected at 10 days postinfection, similar amounts of H4-normalized 2LTR circle DNA and total HIV cDNA were found in WT, KO, and h-1 cells (Fig. 2c). These data correlated with findings at 24 h after infection (Fig. 2a) and further support the conclusion that HIV-1 reverse transcription and nuclear import of the preintegration complex are not affected by PARP-1. In order to estimate the levels of integrated HIV-1 cDNA in the experiments with residual levels of 2LTR circles, we calculated the ratio of H4-normalized total HIV cDNA (containing both integrated and nonintegrated forms) to H4-normalized 2LTR circle DNA (nonintegrated form) in the different cell lines studied (Fig. 2c). Using this strategy, we observed similar quantities of integrated DNA in cells expressing or not expressing PARP-1, further supporting the data described above for cells with undetectable levels of 2LTR circle DNA at 10 days postinfection, as well as the results obtained 24 h after infection (Fig. 2b) that indicate that PARP-1 levels do not influence HIV-1 DNA integration.
Interestingly, in multiple experiments, we have observed that the levels of total HIV cDNA at 24 h postinfection and, as a consequence, the 2LTR levels at that time point and the total HIV cDNA at 10 days postinfection (more likely integrated provirus) are slightly higher (less than 2.1-fold) in h-1 than in WT or KO DT40 cells (Fig. 2a and c). Curiously, the levels of PARP-1 in h-1 cells are higher than in WT or KO cells (Fig. 1a). These data suggest that human PARP-1 could have a role in the nuclear stability of the HIV-1 preintegration complex; however, our data cannot strongly support this conclusion, and more importantly, this possibility cannot explain the observed effect of PARP-1 on retroviral infection.
In conclusion, the data in Fig. 2 demonstrated that PARP-1 impairs HIV-1 infection at a postintegration step of the viral life cycle without significantly affecting the efficiency of HIV-1 reverse transcription (total HIV cDNA at 24 h postinfection), nuclear import of the preintegration complex (2LTR circle DNA at 24 h postinfection), and HIV DNA integration (2LTR circle DNA at 24 h postinfection and total HIV cDNA and the total HIV cDNA/2LTR circle DNA ratio at 10 days postinfection). Therefore, we postulated that PARP-1 affects the transcriptional activity of the provirus.
Effect of PARP-1 on the production of RAV-1 by DT40 cells.
The HIV- and MLV-derived retroviral vectors analyzed express the luciferase transgene from an endogenous CMV promoter. In order to evaluate the effect of PARP-1 on LTR promoter-mediated transcription, we infected PARP-1-expressing (WT and h-1) and KO DT40 cells with an ASLV expressing EGFP from the viral LTR promoter. Although high-titer virus preparations were obtained by replication of the virus in DF-1 cells, ASLV replicated poorly in the infected DT40-derived cells. Thus, these low replication levels prevented us from performing a dependable analysis of the effect of PARP-1 on the replication of ASLV (data not shown). Equivalent results were obtained with an ASLV-derived vector expressing EGFP from an internal CMV promoter (45, 46). Similarly to an MLV-derived vector that we used to infect DT40 cells, these ASLV-derived viruses express the amphotropic MLV envelope, suggesting that it is unlikely that the poor ASLV replication observed in DT40 cells was due to a receptor-mediated restriction.
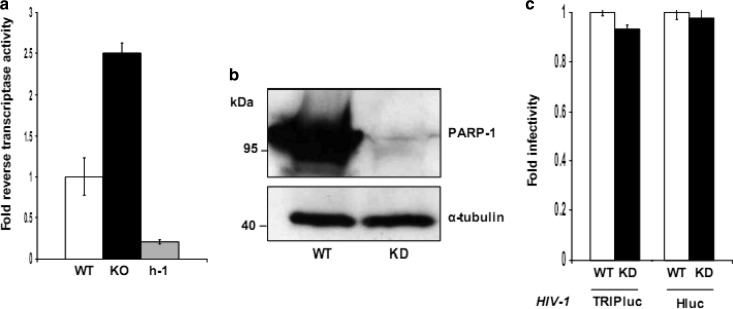
DT40 cells produce infectious RAV-1, the retroviruses used to generate the cell line (48). Therefore, we decided to evaluate the effect of PARP-1 on the activity of the LTR promoter of RAV-1 by determining the production of the virus by DT40 cells. DT40-derived cells were grown for 72 h, and the production of RAV-1 was analyzed by measuring reverse transcriptase activity in the cell culture supernatant after virus concentration on a 20% sucrose cushion. Reverse transcriptase levels produced by the different DT40-derived cell lines were normalized to the number of viable cells (ATP levels) in the cultures at the time of harvesting the viral supernatant. The data in Fig. 3 clearly indicate that the production of RAV-1 was also affected by the levels of PARP-1 in the producer cells, and the magnitude of the effect was similar to that observed with MLV- and HIV-derived viral vectors employing an internal CMV promoter (Fig. 1). RAV-1 production was 2.5-fold and 12.5-fold higher in PARP-1-null cells (KO cells) than in WT or h-1 cells, respectively. These data demonstrated that PARP-1 modulates the expression from the LTR promoter, as well as the retrovirus internal CMV promoter.
Fig 3.
Roles of PARP-1 in the expression of avian Rous-associated virus type 1 and in HIV-1 infection of human cells. (a) Reverse transcriptase activity detected in the 20% sucrose cushion-concentrated cell culture supernatant derived from DT40 WT, KO, and h-1 cells. The reverse transcriptase activity was normalized to the total number of viable producer cells. The SD represents triplicate measurements from one experiment. These results are representative of four independent experiments. (b) Levels of PARP-1 protein in control (WT) and PARP-1-knockdown (KD) human CD4+ T cells. Alpha tubulin was detected in these samples as a loading control. (c) Susceptibility of control and PARP-1 knockdown human CD4+ T cells to HIV-1 infection. The luciferase levels detected in these cells were normalized to the ATP amounts to calculate infectivity. The standard deviation values represent the variability observed in one experiment. The experiment is representative of two independent experiments.
Effect of PARP-1 on HIV-1 infection of human CD4+ T cells.
PARP-1 is expected to have significant functional redundancy in mammals. Five members of the PARP family (PARP-1, -2, -3, -4, and -5) are nuclear enzymes with a role in genome stability and/or chromatin remodeling (14, 15). This functional redundancy determines that although PARP-1 and PARP-2 single-knockout mice are viable, the double knockout is embryonic lethal (17). Similarly, the PARP-1/3 double-knockout mice exhibit higher sensitivity to X-irradiation than the corresponding single-knockout mice (18).
In order to evaluate whether this functional redundancy also affects the role of PARP-1 in repressing the expression of integrated retroviral genomes, we generated a stable PARP-1-deficient human CD4+ T cell line (Fig. 3b) and challenged these cells with two different single-round-infection HIV-derived vectors expressing luciferase, TRIPluc, and Hluc. Unlike TRIPluc, which expresses luciferase from an internal CMV promoter, the expression of this transgene is driven by the LTR promoter in HIVluc (40). Control and PARP-1-deficient cells were infected with these viruses and evaluated 4 days later for luciferase activity. The data in Fig. 3c clearly indicated that luciferase expression levels were similar in control and PARP-1-deficient cells, suggesting that other PARP family members present in human cells compensate for the loss of PARP-1 and are capable of repressing the transcription of integrated HIV-1 proviruses. Similar to our findings in human cells, lack of PARP-1 expression in mouse cells was not associated with an increase in retroviral expression upon HIV infection (27, 30, 32, 33). These findings in mammalian cells differ from our observations in avian cells, highlighting the advantages of the DT40 cell model for studying the role of PARP-1 in retroviral infection.
Role of epigenetic mechanisms in PARP-1-induced retroviral transcriptional repression.
The data in Fig. 1, 2, and 3a suggested that PARP-1 specifically impairs retroviral infection, presumably at the level of transgene expression. A decrease in retroviral transgene expression could be caused by a reduced transcription rate or by mutations of the reporter gene that affect its activity. In turn, reduced transcriptional promoter activity could be due to epigenetic or genetic (mutation) mechanisms. Since transcriptional silencing is central in HIV-1 latency in human cells (55–61), we expected that impaired transcriptional activity of the proviral internal promoter would be a major mechanism in PARP-1-induced retroviral silencing. Thus, we focused our study on the evaluation of this hypothesis.
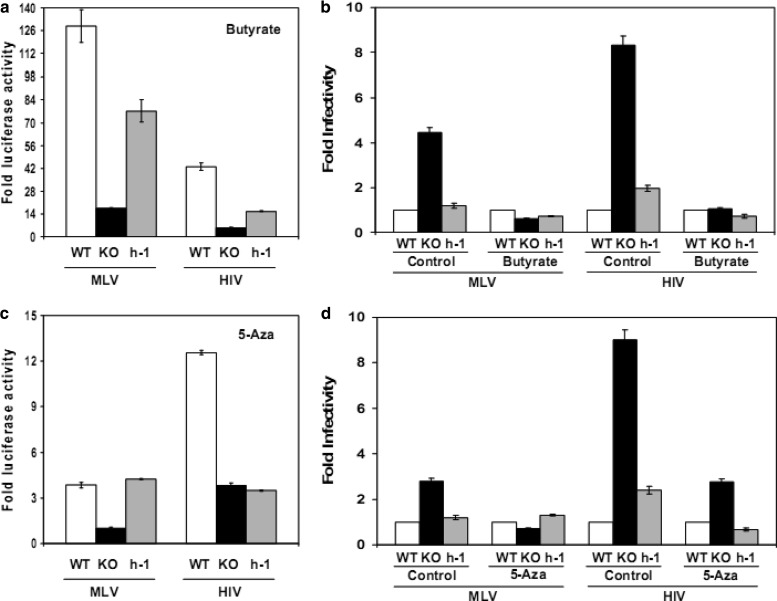
In human cells, the most common cause of HIV-1 latency is transcriptional silencing of the provirus by epigenetic mechanisms, including histone deacetylation and CpG island DNA methylation (55, 56, 60). To determine the relevance of histone deacetylation to the effect of PARP-1 on retroviral infection, we evaluated the effect of sodium butyrate (5 mM), a histone deacetylase inhibitor, on the transgene levels in HIV- or MLV-infected cells. DT40 WT, KO, and h-1 cells were infected with TRIPluc or MLVluc and 4 days later treated or not (control) with sodium butyrate for 24 h and then assessed for luciferase and ATP levels. ATP-normalized luciferase levels detected in nontreated DT40 cells were considered baseline values, and the levels observed in the corresponding treated DT40 cell lines were expressed relative to them. The data in Fig. 4a revealed that sodium butyrate treatment very effectively increased luciferase expression in DT40 cells infected with MLV or HIV. However, the increase was significantly more marked in cells expressing PARP-1. Following sodium butyrate treatment and normalization of luciferase activity using DT40 WT levels as a baseline, we observed that the difference in luciferase expression among all DT40 cell lines was abolished (Fig. 4b, butyrate-treated cells). Collectively, data in Fig. 4a and b indicated a major role of histone deacetylases in the repressive activity of PARP-1 on retroviral expression.
Fig 4.
Role of epigenetic mechanisms in the effect of PARP-1 on retroviral infection. DT40 WT, KO, and h-1 cells were infected with TRIPluc and MLVluc, and 4 days later, the cells were treated or not (control) with sodium butyrate for 24 h (a and b) or with 5-azacytidine for 36 h (c and d), and infectivity (luciferase levels) and cell viability (ATP levels) were measured. Luciferase activity was normalized to ATP levels in all of the experiments. (a and c) ATP-normalized luciferase activity is expressed relative to the values found in the corresponding infected cell lines that were not treated. (b and d) ATP-normalized luciferase activity (infectivity) expressed relative to the infectivity values detected in the WT cells in the treatment or control group. The standard deviation values represent triplicate measurements from one infection experiment. The data are representative of three independent experiments.
Recruitment of histone deacetylases to the retroviral promoter is mediated by specific transcription factors or by CpG DNA methylation of the promoter regions. To investigate the role of DNA methylation in PARP-1-induced retroviral silencing, we evaluated the effect of 5-azacytidine treatment on the levels of transgene expression in MLV- or HIV-infected DT40 cells. 5-Azacytidine substitutes cytosine residues in the cellular DNA, preventing methylation of cytosine at carbon 5. In addition, 5-azacytidine irreversibly inhibits the activity of the enzyme responsible for this modification, DNA cytosine–C5 methyltransferase, by causing enzyme-DNA adducts (62).
In these experiments, WT, KO, and h-1 DT40 cells were infected with TRIPluc or MLVluc. Four days later, the cells were treated or not (control) with 5-azacytidine (30 μM) for 36 h, approximately four to five DT40 cell cycles (53, 54), and then the luciferase and ATP levels were measured. Similar to the effect of sodium butyrate (Fig. 4a and b), 5-azacytidine treatment suppressed the differences in retroviral transgene levels observed in the infected cells (Fig. 4c) by increasing the luciferase expression, preferentially, in PARP-1-expressing cells (Fig. 4d). Unlike sodium butyrate, 5-azacytidine caused cytopathic effects, and the absolute increase in luciferase activity in the treated cells was significantly lower than in the cells exposed to sodium butyrate. In summary, the results in Fig. 4c and d demonstrated a central role of CpG DNA methylation in PARP-1-induced retroviral silencing.
The data in Fig. 4 revealed that PARP-1 induces retroviral transcriptional repression by epigenetic mechanisms involving histone deacetylation and DNA methylation. These are also the main mechanisms of HIV-1 silencing in human cells, highlighting the relevance of our findings.
Role of the catalytic activity of PARP-1 in retroviral infection.
PARP-1 has a major role in transcriptional repression of retrotransposons in Drosophila (21, 25). PARP-1 represses transcription by direct incorporation into the chromatin, a process that requires both the DNA binding domain (20, 21, 23, 24) and the C-terminal region of the protein (22). Chromatin incorporation of PARP-1 and transcriptional repression are independent of its enzymatic activity (20, 21, 23, 24). In order to determine the mechanism of PARP-1-induced retroviral silencing, we evaluated the role of the C-terminal region of PARP-1 and its enzymatic activity in retroviral infection.
We generated a truncated PARP-1 mutant (PARP-1 ΔC-terminal) missing the C-terminal region of the protein (amino acids 489 to 1014). This mutant is expected to completely lack the enzymatic activity and the ability to interact with the nucleosome core histones (22) but to preserve the nuclear localization (amino acids 202 to 233), binding to DNA (aa 12 to 90 and 116 to 200), and the BRCT domain (aa 388 to 461)-mediated protein interaction functions of the full-length protein.
PARP-1 ΔC-terminal was stably expressed in DT40 PARP KO cells, and the expression was verified by immunoblotting analysis with an anti-PARP-1 specific antibody (Fig. 5a). Then, DT40 WT, KO, and h-1 cells and KO cells expressing the PARP-1 ΔC-terminal mutant (h-1m cells) were infected with TRIPluc and MLVluc viruses, and luciferase activity and cellular ATP levels were determined 4 days later. The data shown in Fig. 5b indicated that, contrary to the effect of full-length PARP-1, expression of a C-terminally truncated PARP-1 mutant (h-1m cells) failed to reduce the susceptibility of KO cells to retroviral infection. Similar results were observed with two other independently generated cell lines expressing the mutant (data not shown). These data indicated that the C-terminal region of PARP-1 is required for its effect on retroviral infection.
Fig 5.
Roles of the C-terminal region and the enzymatic activity of PARP-1 in retroviral infection. (a) Expression of PARP-1 wild type and the ΔC-terminal mutant in DT40 cells as determined by immunoblotting with an anti-PARP-1 specific monoclonal antibody. Alpha tubulin was detected as a loading control. (b) Susceptibility of DT40-derived cells to HIV and MLV infection. DT40 WT, KO, h-1, and h-1m cells were challenged with TRIPluc or MLVluc viral vectors, and 4 days later, infectivity (luciferase activity) and cell viability (ATP levels) were measured. Luciferase activity was normalized to ATP levels to calculate infectivity, and the fold infectivity was expressed relative to WT cells. The standard deviations represent the variability of triplicate measurements from two independent experiments. (c) Effect of DPQ, a PARP inhibitor, on retroviral infection. Different cell lines were treated or not (control) with DPQ and then infected with TRIPluc or MLVluc viral vectors. Cell viability-normalized infectivity was calculated as described above, and the fold infectivity is expressed relative to the infectivity values found in the control cells. The standard deviations represent the variability of triplicate measurements from three independent experiments. (d) Effect of DPQ treatment on PARP activity in Jurkat and h-1 cells. Cells were treated or not with DPQ for 24 h, and the levels of PAR were measured by immunoblotting with a specific antibody. Alpha tubulin was detected as a loading control.
The C-terminal region of PARP-1 contains the active site of the enzyme and mediates the binding of the protein to the nucleosome core histones (22). PARP-1 mediates transcriptional repression in a catalytic-independent manner (20–22); therefore, the requirement for the C-terminal region of PARP-1 to repress retroviral expression is unlikely to be related to the enzymatic activity of the protein. Nonetheless, to directly analyze the role of the enzymatic activity of PARP-1 in retroviral infection, we evaluated the effects of a panel of PARP inhibitors on the susceptibility of the human CD4+ T cell line Supt1 to HIV-1 infection. Target cells were treated for 24 h with different PARP inhibitors and then infected with TRIPluc or HIVluc (40). Then, the infected cells were cultured for 4 days in the presence of the corresponding PARP inhibitors until infectivity and cell viability were measured. The PARP inhibitors assayed included 3-aminobenzamide (5 μM and 10 μM), 5-iodo-6-amino-1,2-benzopyrene (100 μM and 600 μM), 1,5-isoquinolinediol (20 μM and 200 μM), NU 1025 (500 μM and 1,000 μM), and DPQ (30 μM, 60 μM, and 120 μM).
In three independent experiments, we observed significant cellular toxicity at the highest doses of inhibitors used, preventing further analysis of the infection. On the other hand, toxicity was minimal at the lower doses; however, we did not observe any increase in the susceptibility of Supt1 cells to infection at these doses (data not shown). In an attempt to reduce the cellular toxicity observed, we treated Supt1 cells with these compounds at the doses indicated above for only 18 h prior to infection, and then the cells were cultured for 4 days in the absence of the drugs until infectivity and cell viability were measured. Even in this experimental setting, the higher doses of the inhibitors were toxic to the cells and the lower doses, although nontoxic, did not increase HIV-1 infection (data not shown).
Among the assayed PARP inhibitors, DPQ (30 μM and 60 μM) exhibited the lowest degree of cellular toxicity, and therefore, it was used in further experiments. The human cell lines Jurkat and 293T and the DT40-derived WT, KO, and h-1 cells were treated with DPQ (30 μM and 60 μM) 24 h before infection with HIVluc or TRIPluc (human cells) and MLVluc or TRIPluc (DT40 cells), and 4 h after infection, the same doses of DPQ were added to the cultures until analysis of infection and cell viability was conducted 4 days later. In these experiments, DPQ treatment did not significantly alter the susceptibility of the target cells to retroviral infection at any of the doses evaluated (Fig. 5c). Similar results were found with 30 μM DPQ (data not shown).
In order to determine whether DPQ treatment caused efficient PARP inhibition, we determined by immunoblotting the basal levels of poly(ADP-ribose) in Jurkat and h-1 cells treated with the inhibitor for 24 h. The data in Fig. 5d demonstrated that DPQ (60 μM) markedly reduced the catalytic activity of PARP enzymes in the treated cells, and the effect was more potent in Jurkat than in h-1 cells. Therefore, these results demonstrated a lack of correlation between susceptibility to retroviral infection and the enzymatic activity of PARP-1.
In summary, the results in Fig. 5 indicated that although the C-terminal region of PARP-1 is necessary to promote retroviral silencing (Fig. 5b), it is unlikely that the enzymatic activity of PARP-1 is involved in this effect (Fig. 5c and d). Therefore, these findings suggest that PARP-1 causes retroviral silencing by a previously described transcriptional repressive mechanism that requires the C-terminal region of the protein but is independent of its enzymatic activity (22). This conclusion is also indirectly supported by our observations that human PARP-2, which is also catalytically active, did not have any effect on retroviral infection (Fig. 1b and d).
DISCUSSION
We have identified a novel role of PARP-1 in the cellular response to integrated retroviruses. Our data indicate that PARP-1 promotes viral transcriptional repression by epigenetic mechanisms that involve the activity of histone deacetylases and DNA methylation. These epigenetic mechanisms also have a central role in HIV-1 latency in human cells (55, 56, 60), highlighting the relevance of our findings. As expected, these epigenetic processes are also implicated in PARP-1-induced transcriptional repression of cellular genes. In normal cells, PARP-1 silences the thrombomodulin promoter by inducing promoter DNA methylation (63), while in cardiomyocytes, the enzyme transcriptionally represses the α-myosin heavy-chain promoter through interaction with histone deacetylases (64). It is not surprising that both DNA methylation and histone deacetylation are implicated in the PARP-1 effect on retroviral expression, since these two processes functionally interact to cause transcriptional repression. DNA methyl transferase 1 methylates cytosine residues in the DNA that attract DNA methyl-binding proteins, which in turn recruit histone deacetylases. These enzymes remove acetyl groups from the tails of histones, causing chromatin condensation that results in repression of gene transcription (65–69).
Previous reports indicated that PARP-1 regulates HIV-1 LTR promoter activity (31, 70, 71). However, contradictory conclusions were derived from these studies. Pharmacological inhibition of PARP-1 enzymatic activity or the reduction of PARP-1 cellular levels was associated with impaired HIV-1 LTR-driven transcription, indicating a positive role of PARP-1 in LTR promoter activity (31, 70). However, more recently, a repressive role of PARP-1 in Tat-regulated HIV-1 LTR-driven transcription was demonstrated, and the mechanism involved the competition of PARP-1 with Tat for binding to TAR RNA (71). Unlike the previously reported inhibitory activity of PARP-1, the function that we described in this study is Tat independent and involves the activity of histone deacetylases and DNA methylation, indicating a novel type of PARP-1 regulation of retroviral expression.
The newly described inhibitory function of PARP-1 presented here suggests that the other members of the PARP family perform a broad range of overlapping functions in mammals, including the capacity to repress retroviral transcription. Human and mouse cells express several members of the PARP family with demonstrated functional redundancy with PARP-1 in genome integrity preservation (17, 18). However, DT40 cells express only PARP-1 (34). Thus, PARP-1-deficient chicken cells consist of a valuable and advantageous system that is able to highlight the particular functions of PARP-1 regardless of the activities of other members of the PARP family.
Although PARP-1 represses the transcription of the retroviruses integrated into the host genome, cells expressing the enzyme are still susceptible to retroviral infection. The ability of some proviruses to escape PARP-1-mediated transcriptional repression is very unlikely to be due to insufficient amounts of PARP-1 in the cell, since it is a very abundant protein with more than 106 molecules per cell (72), whereas only a few preintegration complexes integrate into infected cells. Therefore, these observations suggest that not all PARP-1 molecules in the cell are competent to silence the integrated retroviruses. This functional disparity could be the result of different interactions of PARP-1 with cofactors required for retroviral silencing or the existence of posttranslational modifications regulating this PARP-1 function.
PARP-1 could also cause retroviral silencing by promoting integration of retroviruses into transcriptionally disfavored areas of the chromatin. In support of this possibility, it has been shown that PARP-1 promotes integration of HIV-1 near alphoid DNA in the centromeric regions in mouse and human cells (32). HIV-1 DNA integration at these genome locations results in transcriptional silencing of the provirus in human cells (60).
Based on our observations, PARP-1 represses integrated retroviruses by a mechanism that requires the C-terminal region, but not the enzymatic activity, of the protein. A similar mechanism has been described for PARP-1 in the repression of transcription (22). In this case, PARP-1 is incorporated into nucleosomes through the interaction of its C-terminal region with the nucleosome core histone proteins and chromatin incorporation of PARP-1, causing chromatin condensation and transcriptional silencing.
A role of PARP-1 in transcriptional silencing of retrotransposons that are integrated in heterochromatin regions has been described in Drosophila (21, 25). Cells lacking PARP-1 have a reduced amount of heterochromatin and increased transcription of retrotransposons. These reports and our findings urge us to propose that PARP-1 plays a fundamental role in transcriptional silencing of genomes that invade the host genome. This transcriptional repression activity could be part of the mechanisms employed by PARP-1 to preserve the integrity of the host genome in response to the assault of exogenous genomes.
ACKNOWLEDGMENTS
This work was supported by NIH grant number 5 SC1 AI098238-02 to M.L. C.M. was funded through a fellowship from the NIGMS RISE grant 5R25GM069621-09. UTEP core facilities are funded by BBRC grant 5G12RR008124.
We thank Shunichi Takeda (Department of Radiation Genetics, Kyoto University, Kyoto, Japan) for kindly providing us with the cell lines used in this study; Jose Yelamos (Institut Municipal d'Investigació Mèdica, Barcelona, Spain) for a PARP-1 expression plasmid; Vanessa Rodríguez (UTEP) for the generation of PARP-1 expression plasmids; German Rosas-Acosta (UTEP) for providing us with a preparation of the recombinant VSV-EGFP; Erik Edlund (UTEP) for help with the design of the PARP-1 shRNA used; and Alice M. Nyakeriga (Texas Tech University, Health Sciences Center) for help with the flow cytometry cell sorting of the human PARP-1-deficient cells.
Footnotes
Published ahead of print 19 December 2012
REFERENCES
- 1. Allouch A, Di Primio C, Alpi E, Lusic M, Arosio D, Giacca M, Cereseto A. 2011. The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host Microbe 9:484–495 [DOI] [PubMed] [Google Scholar]
- 2. Colin L, Dekoninck A, Reichert M, Calao M, Merimi M, Van den Broeke A, Vierendeel V, Cleuter Y, Burny A, Rohr O, Van Lint C. 2011. Chromatin disruption in the promoter of bovine leukemia virus during transcriptional activation. Nucleic Acids Res. 39:9559–9573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cosnefroy O, Tocco A, Lesbats P, Thierry S, Calmels C, Wiktorowicz T, Reigadas S, Kwon Y, De Cian A, Desfarges S, Bonot P, San Filippo J, Litvak S, Cam EL, Rethwilm A, Fleury H, Connell PP, Sung P, Delelis O, Andreola ML, Parissi V. 2012. Stimulation of the human RAD51 nucleofilament restricts HIV-1 integration in vitro and in infected cells. J. Virol. 86:513–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Espeseth AS, Fishel R, Hazuda D, Huang Q, Xu M, Yoder K, Zhou H. 2011. siRNA screening of a targeted library of DNA repair factors in HIV infection reveals a role for base excision repair in HIV integration. PLoS One 6:e17612 doi:10.1371/journal.pone.0017612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lesbats P, Botbol Y, Chevereau G, Vaillant C, Calmels C, Arneodo A, Andreola ML, Lavigne M, Parissi V. 2011. Functional coupling between HIV-1 integrase and the SWI/SNF chromatin remodeling complex for efficient in vitro integration into stable nucleosomes. PLoS Pathog. 7:e1001280 doi:10.1371/journal.ppat.1001280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Quivy V, De Walque S, Van Lint C. 2007. Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies. Subcell Biochem. 41:371–396 [PubMed] [Google Scholar]
- 7. Rafati H, Parra M, Hakre S, Moshkin Y, Verdin E, Mahmoudi T. 2011. Repressive LTR nucleosome positioning by the BAF complex is required for HIV latency. PLoS Biol. 9:e1001206 doi:10.1371/journal.pbio.1001206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smith JA, Yeung J, Kao GD, Daniel R. 2010. A role for the histone deacetylase HDAC4 in the life-cycle of HIV-1-based vectors. Virol. J. 7:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Studamire B, Goff SP. 2010. Interactions of host proteins with the murine leukemia virus integrase. Viruses 2:1110–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Duyne R., Guendel I., Narayanan A., Gregg E., Shafagati N., Tyagi M., Easley R., Klase Z., Nekhai S., Kehn-Hall K., Kashanchi F. 2011. Varying modulation of HIV-1 LTR activity by Baf complexes. J. Mol. Biol. 411:581–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang YX, Guen V, Richard J, Cohen EA, Berthoux L. 2010. Cell context-dependent involvement of ATR in early stages of retroviral replication. Virology 396:272–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoder KE, Espeseth A, Wang XH, Fang Q, Russo MT, Lloyd RS, Hazuda D, Sobol RW, Fishel R. 2011. The base excision repair pathway is required for efficient lentivirus integration. PLoS One 6:e17862 doi:10.1371/journal.pone.0017862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ji Y, Tulin AV. 2010. The roles of PARP1 in gene control and cell differentiation. Curr. Opin. Genet. Dev. 20:512–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krishnakumar R, Kraus WL. 2010. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell 39:8–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ame JC, Spenlehauer C, de Murcia G. 2004. The PARP superfamily. Bioessays 26:882–893 [DOI] [PubMed] [Google Scholar]
- 16. Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner EF. 1995. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 9:509–520 [DOI] [PubMed] [Google Scholar]
- 17. Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, Sabatier L, Chambon P, de Murcia G. 2003. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 22:2255–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boehler C, Gauthier LR, Mortusewicz O, Biard DS, Saliou JM, Bresson A, Sanglier-Cianferani S, Smith S, Schreiber V, Boussin F, Dantzer F. 2011. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc. Natl. Acad. Sci. U. S. A. 108:2783–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim MY, Zhang T, Kraus WL. 2005. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 19:1951–1967 [DOI] [PubMed] [Google Scholar]
- 20. Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. 2004. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 119:803–814 [DOI] [PubMed] [Google Scholar]
- 21. Kotova E, Jarnik M, Tulin AV. 2010. Uncoupling of the transactivation and transrepression functions of PARP1 protein. Proc. Natl. Acad. Sci. U. S. A. 107:6406–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pinnola A, Naumova N, Shah M, Tulin AV. 2007. Nucleosomal core histones mediate dynamic regulation of poly(ADP-ribose) polymerase 1 protein binding to chromatin and induction of its enzymatic activity. J. Biol. Chem. 282:32511–32519 [DOI] [PubMed] [Google Scholar]
- 23. Langelier MF, Ruhl DD, Planck JL, Kraus WL, Pascal JM. 2010. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J. Biol. Chem. 285:18877–18887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wacker DA, Ruhl DD, Balagamwala EH, Hope KM, Zhang T, Kraus WL. 2007. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol. Cell. Biol. 27:7475–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tulin A, Stewart D, Spradling AC. 2002. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 16:2108–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kraus WL, Lis JT. 2003. PARP goes transcription. Cell 113:677–683 [DOI] [PubMed] [Google Scholar]
- 27. Ariumi Y, Turelli P, Masutani M, Trono D. 2005. DNA damage sensors ATM, ATR, DNA-PKcs, and PARP-1 are dispensable for human immunodeficiency virus type 1 integration. J. Virol. 79:2973–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baekelandt V, Claeys A, Cherepanov P, De Clercq E, De Strooper B, Nuttin B, Debyser Z. 2000. DNA-dependent protein kinase is not required for efficient lentivirus integration. J. Virol. 74:11278–11285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gaken JA, Tavassoli M, Gan SU, Vallian S, Giddings I, Darling DC, Galea-Lauri J, Thomas MG, Abedi H, Schreiber V, Menissier-de Murcia J, Collins MK, Shall S, Farzaneh F. 1996. Efficient retroviral infection of mammalian cells is blocked by inhibition of poly(ADP-ribose) polymerase activity. J. Virol. 70:3992–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ha HC, Juluri K, Zhou Y, Leung S, Hermankova M, Snyder SH. 2001. Poly(ADP-ribose) polymerase-1 is required for efficient HIV-1 integration. Proc. Natl. Acad. Sci. U. S. A. 98:3364–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kameoka M, Nukuzuma S, Itaya A, Tanaka Y, Ota K, Ikuta K, Yoshihara K. 2004. RNA interference directed against poly(ADP-ribose) polymerase 1 efficiently suppresses human immunodeficiency virus type 1 replication in human cells. J. Virol. 78:8931–8934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kameoka M, Nukuzuma S, Itaya A, Tanaka Y, Ota K, Inada Y, Ikuta K, Yoshihara K. 2005. Poly(ADP-ribose)polymerase-1 is required for integration of the human immunodeficiency virus type 1 genome near centromeric alphoid DNA in human and murine cells. Biochem. Biophys. Res. Commun. 334:412–417 [DOI] [PubMed] [Google Scholar]
- 33. Siva AC, Bushman F. 2002. Poly(ADP-ribose) polymerase 1 is not strictly required for infection of murine cells by retroviruses. J. Virol. 76:11904–11910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hochegger H, Dejsuphong D, Fukushima T, Morrison C, Sonoda E, Schreiber V, Zhao GY, Saberi A, Masutani M, Adachi N, Koyama H, de Murcia G, Takeda S. 2006. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 25:1305–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barr SD, Leipzig J, Shinn P, Ecker JR, Bushman FD. 2005. Integration targeting by avian sarcoma-leukosis virus and human immunodeficiency virus in the chicken genome. J. Virol. 79:12035–12044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Beitzel B, Bushman F. 2003. Construction and analysis of cells lacking the HMGA gene family. Nucleic Acids Res. 31:5025–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mitta B, Weber CC, Fussenegger M. 2005. In vivo transduction of HIV-1-derived lentiviral particles engineered for macrolide-adjustable transgene expression. J. Gene Med. 7:1400–1408 [DOI] [PubMed] [Google Scholar]
- 38. Mitta B, Weber CC, Rimann M, Fussenegger M. 2004. Design and in vivo characterization of self-inactivating human and non-human lentiviral expression vectors engineered for streptogramin-adjustable transgene expression. Nucleic Acids Res. 32:e106 doi:10.1093/nar/gnh104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Randow F, Sale JE. 2006. Retroviral transduction of DT40. Subcell Biochem. 40:383–386 [DOI] [PubMed] [Google Scholar]
- 40. Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. 2006. An essential role for LEDGF/p75 in HIV integration. Science 314:461–464 [DOI] [PubMed] [Google Scholar]
- 41. von Kobbe C, Harrigan JA, May A, Opresko PL, Dawut L, Cheng WH, Bohr VA. 2003. Central role for the Werner syndrome protein/poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol. Cell. Biol. 23:8601–8613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zennou V, Serguera C, Sarkis C, Colin P, Perret E, Mallet J, Charneau P. 2001. The HIV-1 DNA flap stimulates HIV vector-mediated cell transduction in the brain. Nat. Biotechnol. 19:446–450 [DOI] [PubMed] [Google Scholar]
- 43. He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lu R, Limon A, Devroe E, Silver PA, Cherepanov P, Engelman A. 2004. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J. Virol. 78:12735–12746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hu J, Renaud G, Gomes TJ, Ferris A, Hendrie PC, Donahue RE, Hughes SH, Wolfsberg TG, Russell DW, Dunbar CE. 2008. Reduced genotoxicity of avian sarcoma leukosis virus vectors in rhesus long-term repopulating cells compared to standard murine retrovirus vectors. Mol. Ther. 16:1617–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hughes SH, Greenhouse JJ, Petropoulos CJ, Sutrave P. 1987. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J. Virol. 61:3004–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stojdl DF, Lichty BD, ten Oever BT, Paterson JM, Power AT, Knowles S, Marius R, Reynard J, Poliquin L, Atkins H, Brown EG, Durbin RK, Durbin JE, Hiscott J, Bell JC. 2003. VSV strains with defects in their ability to shut down innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263–275 [DOI] [PubMed] [Google Scholar]
- 48. Baba TW, Giroir BP, Humphries EH. 1985. Cell lines derived from avian lymphomas exhibit two distinct phenotypes. Virology 144:139–151 [DOI] [PubMed] [Google Scholar]
- 49. Garcia-Rivera JA, Bueno MT, Morales E, Kugelman JR, Rodriguez DF, Llano M. 2010. Implication of serine residues 271, 273, and 275 in the human immunodeficiency virus type 1 cofactor activity of lens epithelium-derived growth factor/p75. J. Virol. 84:740–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Berger NA, Berger SJ. 1986. Metabolic consequences of DNA damage: the role of poly(ADP-ribose) polymerase as mediator of the suicide response. Basic Life Sci. 38:357–363 [DOI] [PubMed] [Google Scholar]
- 51. Berger SJ, Sudar DC, Berger NA. 1986. Metabolic consequences of DNA damage: DNA damage induces alterations in glucose metabolism by activation of poly(ADP-ribose) polymerase. Biochem. Biophys. Res. Commun. 134:227–232 [DOI] [PubMed] [Google Scholar]
- 52. Luo X, Kraus WL. 2012. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 26:417–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, Cheeseman IM, Fukagawa T. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135:1039–1052 [DOI] [PubMed] [Google Scholar]
- 54. Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, Ikura T, Wang X, Kobayashi M, Yamamoto K, Boulton SJ, Takeda S. 2007. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol. Cell 25:663–675 [DOI] [PubMed] [Google Scholar]
- 55. Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A, Verdin E, Olive D, Van Lint C, Hejnar J, Hirsch I. 2009. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 5:e1000554 doi:10.1371/journal.ppat.1000554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Coiras M, Lopez-Huertas MR, Perez-Olmeda M, Alcami J. 2009. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat. Rev. Microbiol. 7:798–812 [DOI] [PubMed] [Google Scholar]
- 57. Emiliani S, Fischle W, Ott M, Van Lint C, Amella CA, Verdin E. 1998. Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J. Virol. 72:1666–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Emiliani S, Van Lint C, Fischle W, Paras P, Jr, Ott M, Brady J, Verdin E. 1996. A point mutation in the HIV-1 Tat responsive element is associated with postintegration latency. Proc. Natl. Acad. Sci. U. S. A. 93:6377–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Imai K, Ochiai K, Okamoto T. 2009. Reactivation of latent HIV-1 infection by the periodontopathic bacterium Porphyromonas gingivalis involves histone modification. J. Immunol. 182:3688–3695 [DOI] [PubMed] [Google Scholar]
- 60. Jordan A, Bisgrove D, Verdin E. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22:1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. 2009. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5:e1000495 doi:10.1371/journal.ppat.1000495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kuo HK, Griffith JD, Kreuzer KN. 2007. 5-Azacytidine induced methyltransferase-DNA adducts block DNA replication in vivo. Cancer Res. 67:8248–8254 [DOI] [PubMed] [Google Scholar]
- 63. Nocchi L, Tomasetti M, Amati M, Neuzil J, Santarelli L, Saccucci F. 2011. Thrombomodulin is silenced in malignant mesothelioma by a poly(ADP-ribose) polymerase-1-mediated epigenetic mechanism. J. Biol. Chem. 286:19478–19488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, Zhou B, Chang CP. 2010. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 466:62–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Berger SL. 2007. The complex language of chromatin regulation during transcription. Nature 447:407–412 [DOI] [PubMed] [Google Scholar]
- 66. Imai K, Ochiai K. 2011. Role of histone modification on transcriptional regulation and HIV-1 gene expression: possible mechanisms of periodontal diseases in AIDS progression. J. Oral Sci. 53:1–13 [DOI] [PubMed] [Google Scholar]
- 67. Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293:1074–1080 [DOI] [PubMed] [Google Scholar]
- 68. Li B, Carey M, Workman JL. 2007. The role of chromatin during transcription. Cell 128:707–719 [DOI] [PubMed] [Google Scholar]
- 69. Pedersen MT, Helin K. 2010. Histone demethylases in development and disease. Trends Cell Biol. 20:662–671 [DOI] [PubMed] [Google Scholar]
- 70. Kameoka M, Tanaka Y, Ota K, Itaya A, Yoshihara K. 1999. Poly(ADP-ribose) polymerase is involved in PMA-induced activation of HIV-1 in U1 cells by modulating the LTR function. Biochem. Biophys. Res. Commun. 262:285–289 [DOI] [PubMed] [Google Scholar]
- 71. Parent M, Yung TM, Rancourt A, Ho EL, Vispe S, Suzuki-Matsuda F, Uehara A, Wada T, Handa H, Satoh MS. 2005. Poly(ADP-ribose) polymerase-1 is a negative regulator of HIV-1 transcription through competitive binding to TAR RNA with Tat-positive transcription elongation factor b (p-TEFb) complex. J. Biol. Chem. 280:448–457 [DOI] [PubMed] [Google Scholar]
- 72. D'Amours D, Desnoyers S, D'Silva I, Poirier GG. 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342:249–268 [PMC free article] [PubMed] [Google Scholar]