Abstract
As part of a long-term investigation on the evolution of Passiflora L., we investigated the divergence ages of the genus and diversification of its subgenera, relating them with biogeographical and/or historical events, and other characteristics of this taxon. The main aim of the present work was to evaluate the biogeographic distribution of this genus to better understand its evolutionary history. This is the first time that representatives from South American and Old World Passifloraceae genera have been studied as a group comprising a total of 106 widely distributed species, with representative samples of the four suggested subgenera. Seven DNA regions were studied, comprising 7,431 nucleotides from plastidial, mitochondrial and nuclear genomes. Divergence time estimates were obtained by using a Bayesian Markov Chain Monte Carlo method and a random local clock model for each partition. Three major subgenera have been shown to be monophyletic and here we are proposing to include another subgenus in the Passiflora infrageneric classification. In general, divergence among the four subgenera in Passiflora is very ancient, ranging from ∼32 to ∼38 Mya, and Passifloraceae seems to follow a biogeographic scenario proposed for several plant groups, originating in Africa, crossing to Europe/Asia and arriving in the New World by way of land bridges. Our results indicated that Passiflora ancestors arrived in Central America and diversified quickly from there, with many long distance dispersion events.
Keywords: biogeography, molecular phylogenetics analysis, passionflowers, plant evolution, taxonomic classification
Introduction
Passiflora L. is the largest genus of the Passifloraceae family, and encompasses more than 500 wild species distributed especially in the Neotropical region (Ulmer and MacDougal, 2004). The majority of these are herbaceous, but there are also shrubs and trees among them. Killip (1938) and MacDougal (1994) asserted that among the Angiosperms no other group presents such a high foliar diversity, and its flowers display ample variation in size and color, with the corona and perianth showing diverse orientation and development. Coevolution with insect pollinators has been suggested as an explanation for these features (MacDougal, 1994). Based on morphology only (especially flower structures) Feuillet and MacDougal (2004) proposed a drastic taxonomic reevaluation of the genus that, according to them, would consist of only four subgenera (Passiflora, Decaloba, Astrophea and Deidamioides), against the 22 or 23 formerly proposed (Killip, 1938; Escobar, 1989).
The first molecular phylogeny of Passiflora, published by Muschner et al. (2003), included more than 60 species of Passiflora studied for plastidial and nuclear genome markers. They found three clearly defined major clades while the forth one remained undefined due to the small number of species classified in it. They also found that the monospecific Tetrastyllis was part of Passiflora The morphological propositions of Feuillet and MacDougal (2004) are mostly in agreement with our molecular phylogeny. Although some attempts to elucidate the phylogeny of the genus did not agree with our molecular results and the proposition by Feuillet and MacDougal (2004) with respect to the number of subgenera and their composition (e.g. Yockteng and Nadot, 2004a; Plotze et al., 2005), a study by Hansen et al. (2006) with other molecular sequences from other species in Passiflora recovered the four subgenera as monophyletic groups.
Inferences regarding the biogeographic history of tropical angiosperms based on morphology were frequently very poor, given the difficulty of formulating detailed phylogenetic hypotheses and obtaining adequate estimates of divergence times. For example, biogeographical analyses of the tropical flora attribute transtropical disjunctions at high taxonomic levels to the Gondwana breakup (Raven and Axelrod, 1974; Gentry, 1982, 1993; Barlow, 1990; Burnham and Graham, 1999). This interpretation, however, implies divergence times of 100-90 million years ago (Mya) between the African and Neotropical clades, and even higher values for taxa also found in Southeast Asia. In the absence of an adequate fossil record for key areas like South America (Burnham and Graham, 1999), the controversy between Gondwana breakup explanations and those which rely in more recent long-distance dispersion events for the interpretation of present distribution patterns remains unsettled.
Sequences of plastid, mitochondrial and nuclear DNA have been extensively utilized to study plant (especially Angiosperm) phylogenies (e.g. Qiu et al., 1999; Kuzoff and Gasser, 2000; Soltis et al., 2002; APG III 2009). The strategy of combining multiple genes with different functions from the three plant genomes should reduce the phylogenetic noise generated by gene function and/or genome specific phenomena, such as heterogeneity of rates of change, GC-content bias, RNA editing and protein structural constraints (Qiu et al., 1999). Rokas et al. (2003) showed that as the number of genes increases in a phylogenetic analysis, the better tree reflects the species’ phylogeny. The same type of relationship was examined by Rokas and Carroll (2005), who concluded that for phylo-genetic precision the number of genes considered is a more important determinant than the number of taxa examined. However, branch representativeness should also be taken into consideration, and when a large number of taxa is being studied, the ideal number of markers should be decided in cost-benefit terms.
The aims of the present work were: (a) to re-examine the Passiflora phylogeny combining markers from the three genomes, thus contributing to taxonomic classification, (b) to test monophyly of the genus and its subgenera, (c) to investigate the divergence time between the main clades, and (d) to evaluate the biogeographic distribution, aiming to better understand its evolutionary history. This is the first time that representatives from South American and Old World Passifloraceae genera are included as an outgroup.
Materials and Methods
Taxon sampling
We investigated a total of 106 species distributed in the four subgenera of Feuillet and MacDougal (2004) and representatives from seven other genera of Passifloraceae (Adenia isoalensis, Adenia keramanthus, Ancystrothyrsus sp., Barteria sp., Deidamia sp., Dilkea johannesii, Mitostemma brevifilis, Paropsia brazzeana and Paropsia madagascariensis), one Malesherbiacea (Malesherbia linearifolia) and one Turneraceae (Turnera subulata). These species were utilized as outgroups, all being included in Passifloraceae by the Angiosperm Phylogeny Group APG (2003 Angiosperm Phylogeny Group APG (2009). More information about DNA sources and GenBank numbers is provided in Table S1 in Supplementary Material.
DNA extraction, amplification and sequencing
Total DNA was extracted from fresh leaves dried in silica gel or obtained from herbarium material, using the method of Roy et al. (1992). Eight DNA regions were sequenced: the rbcL and rps4 genes, trnL intron and trnL-trnF intergenic spacers from the plastid genome, nad1 b/c and nad5 d/e introns from the mitochondrial genome and a partial portion of the 26S gene from the nuclear ribosomal genome. These regions were amplified with primers 1F and 1460R (Savolainen et al., 2000), rps45 and rps43 (Souza-Chies et al., 1997), c, d, e and f (Taberlet et al., 1991), nad1/2 and nad1/3 (Duminil et al., 2002), mt3 and mt6 (Souza et al., 1991), N-nc26S1 and 1229r (Kuzoff et al., 1998). Sequencing primers were used as listed by these authors except for the nad1 b/c intron, for which we constructed an internal primer specific for Passiflora (5′-ATTCACATAGAGACAGACT).
PCR products were purified using the polyethylene glycol/NaCl precipitation method of Dunn and Blattner (1987). Sequencing was performed on a MegaBace 1000 (GE Health Care) automatic sequencer using the DYEnamicTM ET termination cycle sequencing premix kit (GE Health Care) following the manufacturer’s protocol. The sequences were deposited in Genbank (Accession numbers are given in Table S1). The sequences were aligned using Clustal W (Thompson et al., 1994) implemented on Mega5 (Tamura et al., 2011). All alignments were manually adjusted. Regions of ambiguous alignment were excluded from the analyses.
Phylogenetic analyses
The phylogenetic analyses were performed for the seven genetic markers with a Bayesian approach using BEAST 1.7.1. Less than 20% of the alignment corresponded to missing data. Two independent runs of 3 x 107 chains were performed, each with sampling at every 3,000 generations. The parameters used were as follows: a single HKY substitution model with eight gamma categories, a Yule tree prior and a random local clock model for each partition, which is highly recommended in Passiflora, since different subgenera present different DNA content (Yotoko et al., 2011). Three calibration points were used. A fossil seed with age 37 Mya assigned to the genus Passiflora (Mai, 1967), also used by Hearn (2006) in an Adenia (Passifloraceae) divergence time investigation, was used for an exponential prior for the Passiflora clade with a mean of 15 Mya and an offset of 37 Mya. A normal prior with a mean of 48 Mya and standard deviation of 10 Mya was used for the Passifloraceae/Turneraceae divergence and for the root of the phylogeny we used a uniform prior of between 70 and 110 Mya (based on Bell et al., 2010). The software Tracer v1.5 was used to check for convergence after the first 10% of generations had been discounted as burn-in. Maximum-clade-credibility trees were estimated using the program TreeAnnotator, which is part of the Beast package. Statistical support for the clades was determined by assessing Bayesian posterior probabilities.
Results and Discussion
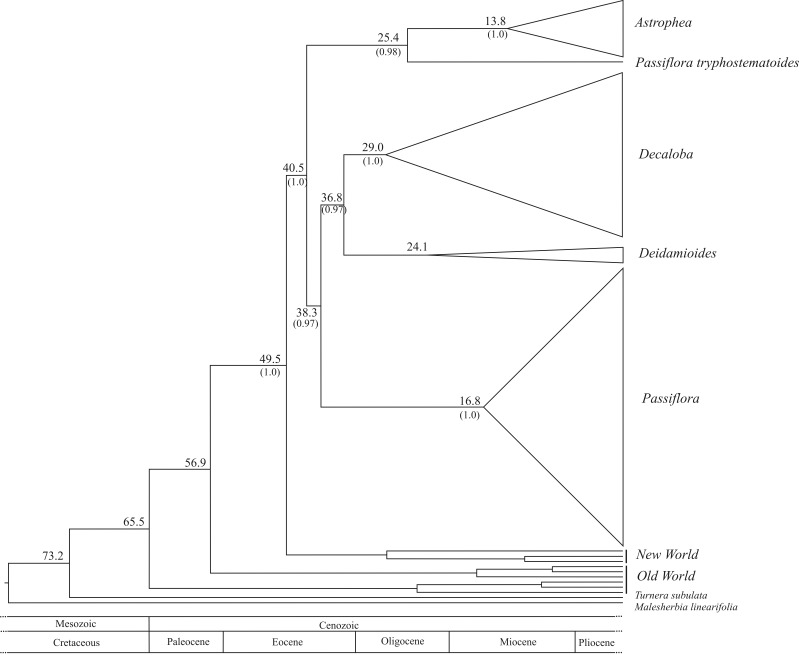
The alignment for all loci totaled 7,431 nucleotides. The numbers of variable and parsimony informative sites for each marker are shown in Table 1. In Figure 1 we present the phylogenetic relationship and divergence times obtained by the Bayesian approach for the main clades. Diversification ages, geological periods and outstanding events for these clades are shown in Table 2. The full non-collapsed branches of the Bayesian tree can be obtained by request from the corresponding author.
Table 1.
Sequences characterization, alignment size, variable and parsimony informative site numbers.
| Marker | Total sites | Variable sites | Parsimony informative |
|---|---|---|---|
| rbcL | 1345 | 354 | 218 |
| rps4 | 615 | 231 | 146 |
| trnL-trnF | 411 | 158 | 78 |
| trnL intron | 681 | 204 | 111 |
| nad1 | 1704 | 323 | 120 |
| nad5 | 1550 | 210 | 75 |
| 26S | 1125 | 228 | 122 |
| All | 7431 | 1708 | 870 |
Figure 1.
Phylogenetic relationship and divergence times obtained by the Bayesian approach using seven genetic markers from three plant genomes of Passiflora species and related taxa.
Table 2.
Divergence times, geological periods and outstanding events for the clades presented in Figure 1.
| Group | Age | Period | Outstanding events |
|---|---|---|---|
| Malesherbiaceae (Passifloraceae + Turneraceae) | 73.2 | Cretaceous | Migration from Old to New World through land bridges |
| Passifloraceae | 65.5 | Paleocene | |
| Old and New World genera | 49.5 | Eocene (E) | Land bridges linking North-Central-South Americas |
| Passiflora genus | 40.5 | Eocene (M) | Andes uplifting first stage |
| (Astrophea + Tryphostematoides) | 38.3 | Eocene (L) | Andes uplifting second stage |
| (Passiflora+Decaloba+Deidamioides) | |||
| (Decaloba+Deidamioides) (Passiflora) | 36.8 | Eocene (L) | Andes uplifting completed |
| Decaloba diversification | 29.0 | Oligocene (L) | Paleo-Orinoco fluvial system |
| Deidamioides diversification | 24.1 | Oligocene (L) | |
| Astrophea diversification | 13.8 | Miocene (M) | Lake Pebas environment; internal migration/diversification |
| Passifora diversification | 16.8 | Miocene (M) | Lake Pebas environment; internal migration/diversification |
In the present analysis, Malesherbiaceae appeared as a sister group of Passifloraceae and Turneraceae, as already proposed by Davis et al. (2005) and Krosnick et al. (2006), and the divergence time found here (73.2 Mya) is also in agreement with Davis et al. (2002). Wikström et al. (2001) estimated divergence between Passifloraceae s.s and Turneracea between 32-36 Mya. We found an older date (65.5 Mya), but note that other authors (i.e., Bremer et al., 2004) also obtained dates older than those reported by Wikström et al. (2001) for different groups. A possible explanation for this difference may be that Wikström et al. (2001) focused their work on higher taxonomic groups, with a very sparse sample density at lower (below family) taxonomic levels and used a different method to estimate divergence.
Considering that sampling at the molecular level in the present study is similar to other biogeographical analyses (Renner, 2004; Richardson et al., 2004; Bell and Donoghue, 2005; Yuan et al., 2005), the above listed divergence time suggests a post-Gondwanic origin of the Passifloraceae. According to Raven and Axelrod (1974) migration between South America and Africa could have occurred even after the Gondwana breakup at 90-105 Mya. Morley (2003) reviewed the potential world migration routes for the megathermal angiosperms, suggesting that connections between South America and Africa may have existed up to the Oligocene (around 35 Mya). These connections may have been used for stepping stone dispersal across islands of the Rio Grande Rise and the Walvis Ridge, which according to Parrish (1993), were above water southwest of the coast of Africa up until that time, as well as through the Sierra Leone Rise.
Other studies (Wolfe, 1978; Renner et al., 2001; Davis et al., 2002) suggested boreo-tropical migration into southern areas during the Oligocene and Miocene, which could explain the distribution of plants including Passifloraceae. A possible route to dispersion through Laurasia during the Eocene climatic optimum, which may have supported tropical vegetation, could be the best explanation for many organisms that now have a disjunct distribution in the South American, African, and southeastern Asian tropics (Richardson et al., 2004), such as the Passifloraceae. In this hypothesis, the North Atlantic region was at a thermal maximum between the Eocene/Oligocene (see Wolfe, 1978) and the North Hemisphere was at its warmest period during the Paleocene/Eocene (according to Davies et al., 2004). The land bridges could thus have been warm enough to support plants like Passifloraceae. Given the estimated age of the family, this is a viable route for its migration. As global temperatures dropped during the Oligocene, species might have become extinct in colder regions and expanded their ranges into the warmer south.
Molecular phylogenetic studies have also demonstrated that the role of long-distance dispersals to explain modern distribution patterns may have been underestimated (Renner et al., 2001; Renner, 2004; Yuan et al., 2005). Especially in Passifloraceae, the time frame postulated in the land bridge hypothesis is more plausible than a Gondwana hypothesis because the former allows a larger time window for family evolution and expansion, which would be more favorable for multiple radiations and migrations from South America to Africa and Australia.
Another explanation to the family distribution range is offered by the climate changes that occurred during the late Cretaceous, when the opening of the Tethys Seaway caused a global warming between five and eight degrees (Fluteau, 2003), that would have allowed tropical plants to expand northward. During the following climate cooling, extinctions occurred and many species were restricted to warmer regions in Asia, Africa and the Neotropics. This is congruent with both the Boreo-Tropical and the land bridges hypothesis.
The genus Passiflora was monophyletic with high support (PP = 1) in this analysis. Three subgenera were equally well supported, but Passiflora subg. Deidamioides as described by Feuillet and MacDougal (2004) emerged as paraphyletic because P. tryphostemmatoides appeared with high support as sister to the Passiflora subg. Astrophea. Passiflora tryphostemmatoides is the type species of a session in the subgenus Deidamioides (Feuillet and MacDougal, 2004), but presents unique morphological traits in that group, such that Killip (1938) and Escobar (1989) considered it as the type species of a new subgenus. Although the positioning of P. tryphostemmatoides in a separate group was also obtained by Yockteng and Nadot (2004b), our results must be considered with caution because we included only one species of Session Tryphostemmatoides. Yockteng and Nadot (2004b) proposed a different infrageneric classification to Passiflora, including eight subgenera. Our results did not support the three extra subgenera (in addition to the four subgenera above plus a clade with P. tryphostemmatoides). We therefore suggest a review of the infrageneric classification, including the well supported Tryphostemmatoides as a new subgenus.
In general, divergence among the four subgenera in Passiflora is very ancient, ranging from ∼33 to ∼38 Mya (Table 1 and Figure 1). The first divergence event in the Passiflora occurred as a split up between the clade Tryphostematoides+Astrophea and the clade Passiflora+Decaloba+Deidamioides (38.3 Mya). In the latter, the two major subclades (Deidamioides+Decaloba and Passiflora) split 36.8 Mya. The subgenera Deidamioides and Decaloba diverged around 33.5 Mya and Tryphostematoides and Astrophea 25.4 Mya.
The very ancient (∼40 Mya) separation of Astrophea from the clade Passiflora+Decaloba+Deidamioides could help to explain why the former encompasses species that present the most unusual morphological traits within Passiflora, some do not even look much like passionflowers. In Astrophea there are species that present tree, shrub or woody vine habits. Similarly, the older divergence of the Passiflora subgenus in relation to Decaloba+Deidamioides is intriguing since many authors have suggested that within the genus the former present ancestral traits in relation to morphology (see Ulmer and MacDougal, 2004, for a review of species description and characteristics) and genetics (Melo and Guerra, 2003 for cytogenetics; Muschner et al., 2006 for organelar inheritance; Yotoko et al., 2011 for genome size evolution). However, the diversification age within Passiflora (∼16.8 Mya) was much more recent than diversification in Decaloba (∼29 Mya).
Differences in evolutionary rates between taxa are widespread in plants (Muse, 2000), and can be ascribed to factors intrinsic to each genome type (plastidial, mitochondrial and nuclear) and to extrinsic factors like speciation dynamics, population size and life history (Bousquet et al., 1992; Muse, 2000; Andreasen and Baldwin, 2001; Barraclough and Savolainen, 2001; Smith and Donoghue, 2009). The Decaloba and Deidamioides subgenera have longer branches (see Fig. S1) than those of the Passiflora and Astrophea subgenera, indicating a pattern of accelerated molecular evolution. The mechanisms that could lead to high evolutionary rates in the former subgenera are a generation time that is shorter in Decaloba and Deidamioides than those in the others, and that they comprise most of the self-compatible species described so far in Passiflora (Benson et al., 1975; Ulmer and MacDougal, 2004).
This is the first study that considers Passiflora diversification times in detail. Just a few species grow in North America, mainly in Mexico, which could be attributed to the presently unfavorable climate for these species that prefer warmer and moister conditions. Passifloraceae in general seems to follow a biogeographic scenario proposed by other authors for several plant groups (see Antonelli et al., 2009 and Antonelli and Sanmartín, 2011, for more details), with an origin in Africa, crossing to Europe/Asia and arrival in the New World by way of land bridges. Our results indicate that Passiflora ancestors arrived in Central America and diversified quickly from there. Passiflora subgenera divergence times show the gradual colonization of Americas from north to south. Below we present a more detailed account of this biogeographic scenario.
An alternative hypothesis explaining the disjunct distribution of Passiflora (see below) could be by Trans-Pacific dispersion, as suggested for other groups (Sanmartín and Ronquist, 2004). The relationship [(South America, New Zealand) Australia] is the most frequently observed in the flora and fauna of the South Hemisphere and is in conflict with the geologically predicted vicariance patterns (Renner et al., 2000; Winkworth et al., 2002). Sanmartín and Ronquist (2004) documented highly asymmetrical, westward long-distance plant dispersal from South America to New Zealand, against the prevailing wind and oceanic currents (Winkworth et al., 2002). Instead of direct jumps, the dispersal could have occurred in a stepping stone manner along the Antarctic coastline (Renner et al., 2000). This hypothesis is supported by the presence of temperate forests in this area until at least the Pliocene (Swenson and Bremer, 1997; Sanmartín and Ronquist, 2004). This dispersal could have been mediated by the west-flowing East Wind Drift, which runs close to the Antarctic coast, or could have followed the West Wind Drift around Antarctica, involving dispersal first to the sub-Antarctic islands (and/or Australia) and from there to New Zealand (Swenson and Bremer, 1997). Large birds could have contributed to this dispersion, as suggested by Winkworth et al. (2002) and Ulmer and MacDougal (2004). Such processes have been proposed for species with characteristics very similar to those of the Decaloba subgenus (Renner et al., 2001; Knapp et al., 2005). This type of dispersion could therefore explain the presence in southeast Asia and Australia of species of a monophyletic session of the Decaloba subgenus, as found by Krosnick and Freudenstein (2005).
Acknowledgments
We thank Alba Lins, Alessandra Selbach, Aline P. Lorenz-Lemke, Armando C. Cervi, Cássio van den Berg, Cláudio Mondin, Fernando Campos Neto, Karla Gengler, Luis Carlos Bernacci, Marcelo C. Dornelas, Marcelo S. Guerra-Filho, Mark Chase, Maurizio Vecchia, Natoniel Franklin de Melo, Roxana Yockteng, Shawn Krosnick and Teonildes S. Nunes for specimen donations, and two anonymous reviewers for comments and suggestions that have improved this manuscript. This research was financially supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento do Pessoal de Nível Superior (CAPES), Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS), and Programa de Pós-Graduação em Genética e Biologia Molecular (PPGBM-UFRGS).
Supplementary Material
The following online material is available for this article:
- Table S1 - DNA sources and GenBank numbers of the Passiflora species included in the analysis.
This material is available as part of the online article from http://www.scielo.br/gmb.
References
- Andreasen K, Baldwin B. Unequal evolutionary rates between annual and perennial lineages of checker mallows (Sidalcea, Malvaceae): Evidence from 18S-26S rDNA internal and external transcribed spacers. Mol Biol Evol. 2001;18:936–944. doi: 10.1093/oxfordjournals.molbev.a003894. [DOI] [PubMed] [Google Scholar]
- Antonelli A, Sanmartín I. Why are there so many plant species in the Neotropics? Taxon. 2011;60:403–414. [Google Scholar]
- Antonelli A, Nylander JAA, Persson C, Sanmartín I. Tracing the impact of the Andean uplift on Neotropical plant evolution. Proc Natl Acad Sci USA. 2009;106:9749–9754. doi: 10.1073/pnas.0811421106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiosperm Phylogeny Group An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc. 2003;141:399–436. [Google Scholar]
- The Angiosperm Phylogeny Group An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 2009;161:105–121. [Google Scholar]
- Barlow BA. Biogeographical relationships of Australia and Malasia: Loranthaceae as a model. In: Baas P, Kalkman K, Geesink R, editors. The Plant Diversity of Malasia. Kluwer; Dordrecht: 1990. pp. 273–292. [Google Scholar]
- Barraclough TG, Savolainen V. Evolutionary rates and species diversity in flowering plants. Evolution. 2001;55:677–683. doi: 10.1554/0014-3820(2001)055[0677:erasdi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Bell CD, Donoghue MJ. Dating the Dipsacales: Comparing models, genes and evolutionary implications. Am J Bot. 2005;92:284–296. doi: 10.3732/ajb.92.2.284. [DOI] [PubMed] [Google Scholar]
- Bell CD, Soltis DE, Soltis PS. The age and diversification of the angiosperms re-revisited. Am J Bot. 2010;97:1296–1303. doi: 10.3732/ajb.0900346. [DOI] [PubMed] [Google Scholar]
- Benson WW, Brown KS, Gilbert LE. Coevolution of plants and herbivores: Passionflower butterflies. Evolution. 1975;29:659–680. doi: 10.1111/j.1558-5646.1975.tb00861.x. [DOI] [PubMed] [Google Scholar]
- Bousquet J, Strauss SH, Doerksen AH, Price RA. Extensive variation in evolutionary rate of rbcL gene sequences among seed plants. Proc Natl Acad Sci USA. 1992;89:7844–7848. doi: 10.1073/pnas.89.16.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer K, Friis EM, Bremer B. Molecular phylogenetic dating of Asterids flowering plants shows early Cretaceous diversification. Syst Biol. 2004;53:496–505. doi: 10.1080/10635150490445913. [DOI] [PubMed] [Google Scholar]
- Burnham RJ, Graham A. The history of neotropical vegetation: New developments and status. Ann Mis Bot Gard. 1999;86:546–589. [Google Scholar]
- Davies TJ, Savolainen V, Chase MW, Moat J, Barraclough TG. Environmental energy and evolutionary rates in flowering plants. Proc R Soc B. 2004;271:2195–2200. doi: 10.1098/rspb.2004.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CC, Bell CD, Fritsch PW, Mathews S. Phylogeny of Acridocarpus-Brachylophon (Malpighiaceae): Implications for tertiary tropical floras and Afroasian biogeography. Evolution. 2002;56:2395–2405. doi: 10.1111/j.0014-3820.2002.tb00165.x. [DOI] [PubMed] [Google Scholar]
- Davis CC, Webb CO, Wurdack KJ, Jaramillo CA, Donoghue MJ. Explosive radiation of Malpighiales supports a mid Cretaceous origin of modern tropical rain forests. Am Nat. 2005;165:36–65. doi: 10.1086/428296. [DOI] [PubMed] [Google Scholar]
- Duminil J, Pemonge M-H, Petit RJ. A set of 35 consensus primer pairs amplifying genes and introns of plant mitochondrial DNA. Mol Ecol Notes. 2002;2:428–430. [Google Scholar]
- Dunn IS, Blattner FR. Charons 36 to 40: Multi-enzyme, high capacity, recombination deficient replacement vectors with polylinkers and polystuffers. Nucleic Acid Res. 1987;15:2677–2698. doi: 10.1093/nar/15.6.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar LK. A new subgenus and five new species in Passiflora (Passifloraceae) from South America. Ann Mis Bot Gard. 1989;76:877–885. [Google Scholar]
- Feuillet C, MacDougal JM. A new infrageneric classification of Passiflora. Passiflora. 2004;14:1–4. [Google Scholar]
- Fluteau F. Earth dynamics and climate changes. C R Geosci. 2003;335:157–174. [Google Scholar]
- Gentry AH. Neotropical floristic diversity: Phytogeographical connections between Central and South America, Pleistocene climatic fluctuations, or an accident of the Andean orogeny? Ann Mis Bot Gard. 1982;69:557–593. [Google Scholar]
- Gentry AH. Diversity and floristic composition of lowland tropical forest in Africa and South America. In: Goldblatt P, editor. Biological Relationships between Africa and South America. Yale University Press; New Haven: 1993. pp. 500–547. [Google Scholar]
- Hansen AK, Gilbert LE, Simpson BB, Downie SR, Cervi AC, Jansen RK. Phylogenetic relationships and chromosome number evolution in Passiflora. Syst Bot. 2006;31:138–150. [Google Scholar]
- Hearn DJ. Adenia (Passifloraceae) and its adaptive radiation: Phylogeny and growth form diversification. Syst Bot. 2006;31:805–821. [Google Scholar]
- Killip EP. The American species of Passifloraceae. Field Mus Nat Hist, Bot Series. 1938;19:1–613. [Google Scholar]
- Knapp M, Stöckler K, Havell D, Delsuc F, Sebastiani F, Lockhart PJ. Relaxed molecular clock provides evidence for long-distance dispersal of Notophagus (Southern Beech) PLoS Biology. 2005;3:e14. doi: 10.1371/journal.pbio.0030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krosnick SE, Freudenstein JV. Monophyly and floral character homology of Old World Passiflora (Subgenus Decaloba, Supersection Disemma) Syst Bot. 2005;30:139–152. [Google Scholar]
- Krosnick SE, Harris EM, Freudenstein JV. Patterns of anomalous floral development in the Asian Passiflora (subgenus Decaloba, Supersection Disemma) Am J Bot. 2006;93:620–636. doi: 10.3732/ajb.93.4.620. [DOI] [PubMed] [Google Scholar]
- Kuzoff RK, Gasser CS. Recent progress in reconstructing Angiosperm phylogeny. Trends Plant Sci. 2000;5:330–336. doi: 10.1016/s1360-1385(00)01685-x. [DOI] [PubMed] [Google Scholar]
- Kuzoff RK, Sweere JA, Soltis DE, Soltis PS, Zimmer EA. The phylogenetic potential of entire 26S rDNA sequences in plants. Mol Biol Evol. 1998;15:251–263. doi: 10.1093/oxfordjournals.molbev.a025922. [DOI] [PubMed] [Google Scholar]
- MacDougal JM. Revision of Passiflora subgenus Decaloba section Pseudodysosmia (Passifloraceae) Syst Bot Monogr. 1994;41:1–46. [Google Scholar]
- Mai DH. Die Florenzonen, der Florenwechsel, und die Vorstellungen über den Kilmaablauf im Jungtertiär der Deutschen Demokratischen Republik. Abh Zent Geol Inst. 1967;10:55–81. [Google Scholar]
- Melo NF, Guerra FMS. Variability of the 5S and 45S rDNA sites in Passiflora L. species with distinct base chromosome numbers. Ann Bot. 2003;92:309–316. doi: 10.1093/aob/mcg138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley RJ. Interplate dispersal paths for megathermal angiosperms. Persp Plant Ecol Evol Syst. 2003;6:5–20. [Google Scholar]
- Muschner VC, Lorenz AP, Cervi AC, Bonatto SL, Souza-Chies TT, Salzano FM, Freitas LB. A first molecular phylogenetic analysis of Passiflora (Passifloracae) Am J Bot. 2003;90:1229–1238. doi: 10.3732/ajb.90.8.1229. [DOI] [PubMed] [Google Scholar]
- Muschner VC, Lorenz-Lemke AP, Vecchia M, Bonatto SL, Salzano FM, Freitas LB. Differential organellar inheritance in Passiflora’s subgenera. Genetica. 2006;128:449–453. doi: 10.1007/s10709-006-7726-4. [DOI] [PubMed] [Google Scholar]
- Muse SV. Examining rates and patterns of nucleotide substitution in plants. Plant Mol Biol. 2000;42:25–43. [PubMed] [Google Scholar]
- Parrish JT. Climate of the supercontinent Pangea. J Geol. 1993;101:215–233. [Google Scholar]
- Plotze RD, Falvo M, Padua JG, Bernacci LC, Vieira MLC, Oliveira GCX, Bruno OM. Leaf shape analysis using the multiscale Minkowski fractal dimension, a new morpho-metric method: A study with Passiflora (Passifloraceae) Can J Bot. 2005;83:287–301. [Google Scholar]
- Qiu Y-L, Lee J, Bernasconi-Quadroni F, Soltis DE, Soltis PS, Zanis M, Zimmer EA, Chen Z, Savolainen V, Chase MW. The earliest Angiosperms: Evidence from mitochondrial, plastid and nuclear genomes. Nature. 1999;402:404–407. doi: 10.1038/46536. [DOI] [PubMed] [Google Scholar]
- Raven pH, Axelrod DI. Angiosperm biogeography and past continental movements. Ann Mis Bot Gard. 1974;61:39–39. [Google Scholar]
- Renner SS. Bayesian analysis of combined chloroplast loci, using multiple calibrations, supports the recent arrival of Melastomataceae in Africa and Madagascar. Am J Bot. 2004;91:1427–1435. doi: 10.3732/ajb.91.9.1427. [DOI] [PubMed] [Google Scholar]
- Renner SS, Murray D, Foreman D. Timing transantarctic disjunctions in the Atherospermataceae (Laurales): Evidence from coding and noncoding chloroplast sequences. Syst Biol. 2000;49:579–591. doi: 10.1080/10635159950127402. [DOI] [PubMed] [Google Scholar]
- Renner SS, Clausing G, Meyer K. Historical biogeography of Melastomataceae: The roles of Tertiary migration and long-distance dispersal. Am J Bot. 2001;88:1290–1300. [PubMed] [Google Scholar]
- Richardson JE, Chatrou LW, Mols JB, Erkens RHJ, Pirie MD. Historical biogeography of two cosmopolitan families of flowering plants: Annonaceae and Rhamnaceae. Phil Trans R Soc Lond B. 2004;359:1495–1508. doi: 10.1098/rstb.2004.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Carroll SB. More genes or more taxa? The relative contribution of gene number and taxon number to phylogenetic accuracy. Mol Biol Evol. 2005;22:1337–1344. doi: 10.1093/molbev/msi121. [DOI] [PubMed] [Google Scholar]
- Rokas A, Williams BL, King N, Carroll SB. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- Roy A, Frascaria N, MacKay J, Bousquet J. Segregating random amplified polymorphic DNAs (RAPDs) in Betula alleghaniensis. Theor App Genet. 1992;85:173–180. doi: 10.1007/BF00222856. [DOI] [PubMed] [Google Scholar]
- Sanmartín I, Ronquist F. Southern Hemisphere bio-geography inferred by event-based models: Plant vs. animal patterns. Syst Biol. 2004;53:216–243. doi: 10.1080/10635150490423430. [DOI] [PubMed] [Google Scholar]
- Savolainen V, Chase MW, Hoot SB, Morton CM, Soltis DE, Bayer C, Fay MF, Bruijn AY, Sullivan S, Qiu YL. Phylogenetics of flowering plants based on combined analysis of plastid atpB and rbcL gene sequences. Syst Biol. 2000;49:306–362. doi: 10.1093/sysbio/49.2.306. [DOI] [PubMed] [Google Scholar]
- Smith SA, Donoghue MJ. Rates of molecular evolution are linked to life history in flowering plants. Science. 2009;322:86–89. doi: 10.1126/science.1163197. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE, Savolainen V, Crane PR, Barraclough TG. Rate heterogeneity among lineages of tracheophytes: Integration of molecular and fossil data and evidence for molecular living fossils. Proc Natl Acad Sci USA. 2002;99:4430–4435. doi: 10.1073/pnas.032087199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza AP, Jubier M-F, Delcher E, Lancelin D, Lejeune B. A trans-splicing model for the expression of the tripartite nad5 gene in wheat and maize mitochondria. Plant Cell. 1991;3:1363–1378. doi: 10.1105/tpc.3.12.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza-Chies TT, Bittar G, Nadot S, Carter L, Besin E, Lejeune B. Phylogenetic analysis of Iridaceae with parsimony and distance methods using the plastid gene rps4. Plant Syst Evol. 1997;204:109–123. [Google Scholar]
- Swenson U, Bremer K. Patterns of floral evolution of four Asteraceae genera (Senecioneae-Blennospermatinae) and the origin of white flowers in New Zealand. Syst Biol. 1997;46:407–425. [Google Scholar]
- Taberlet P, Gielly L, Patou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. Mega5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer T, MacDougal JM. Passiflora: Passionflowers of the World. Timber Press; Portland: 2004. p. 430. [Google Scholar]
- Wikström N, Savolainen V, Chase MW. Evolution of the angiosperms: Calibrating the family tree. Proc R Soc Lond. 2001;268:2211–220. doi: 10.1098/rspb.2001.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkworth RC, Wagstaff SJ, Glenny D, Lockhart P. Plant dispersal N.E.W.S. from New Zealand. Trends Ecol Evol. 2002;17:514–520. [Google Scholar]
- Wolfe JA. A paleobotanical interpretation of Tertiary climates in the northern hemisphere. Am Sci. 1978;66:694–703. [Google Scholar]
- Yockteng R, Nadot S. Phylogenetic relationships among Passiflora species based on the glutamine synthetase nuclear gene expressed in chloroplast (ncpGS) Mol Phylogenet Evol. 2004a;31:379–396. doi: 10.1016/S1055-7903(03)00277-X. [DOI] [PubMed] [Google Scholar]
- Yockteng R, Nadot S. Infrageneric phylogenies: A comparison of chloroplast-expressed glutamine synthetase, cytosol-expressed glutamine synthetase and cpDNA maturase K in Passiflora. Mol Phylogenet Evol. 2004b;31:397–402. doi: 10.1016/S1055-7903(03)00276-8. [DOI] [PubMed] [Google Scholar]
- Yotoko KSC, Dornelas MC, Togni PD, Fonseca TC, Salzano FM, Bonatto SL, Freitas LB. Does variation in genome sizes reflect adaptive or neutral processes? New clues from Passiflora. PLoS One. 2011;6:e18212. doi: 10.1371/journal.pone.0018212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y-M, Wohlhauser S, Möller M, Klackenberg J, Callmander MW, Küpfer P. Phylogeny and biogeography of Exacum (Gentianaceae): A disjunctive distribution in the Indian Ocean Basin resulted from long distance dispersal and extensive radiation. Syst Biol. 2005;54:21–34. doi: 10.1080/10635150590905867. [DOI] [PubMed] [Google Scholar]
Internet Resources
- Tracer v1.5 software, http://beast.bio.ed.ac.uk/Tracer (accessed in October 22, 2012)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
- Table S1 - DNA sources and GenBank numbers of the Passiflora species included in the analysis.