Abstract
Over three-fourths of human immunodeficiency virus (HIV)–infected men who have sex with men (MSM) have at least one herpesvirus detected in their semen, and cytomegalovirus (CMV) is the most prevalent. The presence of CMV is associated with higher T-cell immune activation and with HIV disease progression in treated and untreated individuals. In this study of 113 antiretroviral (ART)–naive HIV-infected MSM, we found that CMV replication in blood and semen was associated with higher levels of HIV DNA in peripheral blood mononuclear cells. These observations suggest that interventions aimed to reduce CMV replication and, thus, systemic immune activation could decrease the size of the latent HIV reservoir.
Keywords: cytomegalovirus, HIV DNA, latent reservoir, immune activation, early HIV infection
Over three-fourths of human immunodeficiency virus (HIV)–infected men who have sex with men (MSM) have at least one actively replicating herpesvirus detected in their semen, and cytomegalovirus (CMV) is the most prevalent. While the interactions between herpesviruses and HIV are relevant to HIV transmission [1], the role of herpesviruses in HIV pathogenesis may be equally important. Presence of CMV is associated with HIV disease progression and higher T-cell immune activation [1, 2]. Such immune activation can blunt CD4+ T-cell recovery during antiretroviral therapy (ART) and lead to premature mortality [2]. Moreover, immune activation in response to CMV may be responsible for accelerated immunosenescence in HIV-infected and HIV-uninfected individuals [2]. Since activated T cells undergo extensive cell division and differentiation [3], we hypothesized that bystander proliferation of HIV-infected CD4+ T cells could be an important factor in determining the size of the HIV proviral reservoir [4].
The latent HIV reservoir is established during primary HIV infection [5], but the mechanisms that maintain this reservoir are unclear. It is therefore particularly important to understand which factors contribute to increase the immune activation and which mechanisms drive and modulate the establishment of the viral reservoir during the earliest phase of HIV infection. Here, we investigated the relationships between CMV replication in semen and paired peripheral blood mononuclear cells (PBMCs) and estimated the duration of infection, the HIV DNA level in PBMCs, the HIV RNA load in blood plasma, and CD4+ T-cell numbers.
MATERIALS AND METHODS
Participants, Samples, and Clinical Laboratory Tests
A total of 229 semen samples from 113 participants in the San Diego Primary Infection Cohort [1] were included. All subjects had recently acquired HIV infection, were naive to ART, and were CMV seropositive. Semen and blood were collected as described previously [1]. In blood, the CD4+ T-lymphocyte count was measured by flow cytometry (LabCorp), and the plasma HIV RNA load was quantified by the Amplicor HIV Monitor Test (Roche Molecular Systems). Demographic characteristics and standard laboratory values were collected, and screening for sexually transmitted infections (STIs; Neisseria gonorrhoeae infection, Chlamydia trachomatis infection, and syphilis) was performed at baseline for a subset of 95 participants. The estimated duration of infection was calculated using an established algorithm [6], and HIV subtype was determined from HIV pol sequence data (Viroseq 2.0; Applied Biosystems), using SCUEAL [7]. The study was conducted with written consent, as approved by Human Research Protections Program at University of California–San Diego.
Quantification of HIV DNA and CMV DNA
DNA was extracted from 5 million PBMCs and 200 μL of seminal plasma from each participant (QIAamp DNA Mini Kit, Qiagen, CA). Total HIV DNA was quantified by real-time polymerase chain reaction (PCR) from extracted DNA, using primers as described elsewhere [1]. Because of the large genetic variability of HIV, accurate quantification by real-time PCR may require primers and probes specific for each viral variant. This study used the conserved probe mf348 [8], which matches 81% of HIV subtype B pol sequences downloaded from the Los Alamos HIV Sequence Database (available at: http://www.hiv.lanl.gov; accessed March 2009). Since fluorescent PCR probes are particularly sensitive to mismatches [9], for each participant we determined whether the mf348 probe would match the sequence of each patient's HIV RNA population, as determined by the population-based HIV pol sequence.
Similarly, levels of CMV in DNA extracted from PBMCs and seminal plasma were measured by real-time PCR, as described previously [1]. Cellular input for both HIV and CMV DNA measurements in PBMCs was quantified by β-actin PCR, using the primers β-actin 959 F (CTGGCACCCAGCACAATG) and β-actin 1026 R (GCCGATCCACACGGAGTACT) and the detection probe β-actin 980 T (f-TCAAGATCATTGCTCCTCCTGAGCGC-q).
Statistical Analysis
A linear mixed-effects model for repeated measures was used to investigate the relationship between CMV and HIV DNA levels. Fixed effects included detectable CMV in PBMCs and semen, estimated duration of infection, HIV RNA blood plasma levels, CD4+ T-cell numbers, and status of probe match, and each subject was treated as the random effect. For 59 subjects with longitudinal data, a subanalysis was performed that included a binary variable that accounted for intermittent CMV seminal shedding. Because of the low numbers of subjects with longitudinal data and detectable CMV in PBMCs (n = 12), we were not able to assess the impact of intermittent CMV shedding in those subjects.
HIV and CMV DNA levels were normalized to cellular input (expressed in copies per 106 cell equivalents) with respect to β-actin levels, and both HIV DNA and RNA levels were transformed to their base-10 logarithm values. Univariate analysis was performed using all samples and using only those samples in which HIV DNA levels were obtained with a matched probe. Associations between STIs and HIV DNA levels were evaluated using the reduced data set of 95 subjects with available STI screening information. Multivariate analysis used all samples and included the matching status of the probe as a covariate.
Associations between CMV and HIV RNA and CD4+ T-cell counts were also evaluated. Since CD4+ T-cell counts were not normally distributed, mixed-effects logistic regression modeling was used to test the association, treating the presence of CMV as the outcome variable and HIV DNA levels as the predictor. Since these relationships were tested at the univariate level only, interpretation of the relationship between CMV and CD4+ T-cell levels by using mixed-effects logistic regression is equivalent to specifying CD4+ T-cell levels as the outcome and using linear mixed-effects models.
RESULTS
Participants, Samples, and Clinical Laboratory Findings
All participants were ART-naive MSM infected with HIV. All but 3 had HIV subtype B infection; 2 had HIV subtype B/D infection, and 1 had HIV subtype B/F1 recombinant infection (Supplementary Table 1). From all individuals (n = 113), 229 paired blood and semen samples were collected. The 59 subjects (52%) with available longitudinal samples had a median follow-up duration of 67 days, ranging from 4 days to 3.15 years. The median baseline CD4+ T-cell count for all subjects was 523 cells/mL, and the median blood plasma HIV RNA level was 4.7 log10 copies/mL. The median estimated duration of infection was 70 days. Approximately 46% of semen samples were positive for CMV (median peak viral load, 4.52 log10 DNA copies/mL [range, 0.70–8.31 log10 DNA copies/mL]). CMV was found in 13% of the PBMC samples (median peak DNA level, 1.73 log10 DNA copies/106 cell-equivalents [range, 0.12–4.54 log10 DNA copies/106 cell-equivalents]). Among the 59 subjects with longitudinal samples, 11 (19%) presented intermittent CMV shedding in semen, and 10 (17%) presented intermittent CMV shedding in PBMCs. HIV DNA was detected in 91.3% of PBMC samples (median peak level, 2.19 log10 DNA copies/106 cell-equivalents [range, 0.06–4.42 log10 DNA copies/106 cell-equivalents]). Also, 159 samples (69.4%) had a complete match between HIV sequence and the detection probe mf348.
Associations Between CMV Replication and HIV RNA, HIV DNA, and CD4+ T Cells
When evaluating the entire data set (Table 1), longer estimated duration of infection (P = .08), presence of CMV in PBMCs (P = .07), and having a 100% match with the detection probe (P < .001) were positively associated with higher HIV DNA levels in PBMCs. Interestingly, blood plasma HIV RNA levels and CD4+ T-cell counts were not associated with HIV DNA levels (P > .48). To evaluate whether the lack of association between HIV RNA and HIV DNA was a consequence of the early stage of infection for most of the participants, we performed 2 subanalyses. The first analysis excluded samples with low-level HIV DNA (<1 log10 HIV DNA/106 cell-equivalents), and there was a resultant positive association between HIV DNA and HIV RNA (P = .01). The second analysis excluded samples with estimated durations of infection of <70 days and repeated excluding samples <120 days, and no association was found (P > .5). Also, the presence of STIs at baseline was not associated with higher proviral HIV DNA (data not shown; P > .6).
Table 1.
Results of Univariate and Multivariate Analyses of the Association Between Various Factors and Higher Human Immunodeficiency Virus (HIV) DNA Levels in Peripheral Blood Mononuclear Cells (PBMCs)
| Univariate Regression |
Multivariate Regression, All Samples |
|||||
|---|---|---|---|---|---|---|
| All Samplesa |
Matched Samplesb |
|||||
| Variable | Estimate (95% CI) | P | Estimate (95% CI) | P | Estimate (95% CI) | P |
| Presence of CMV in PBMCs | .26 (−.02 to .54) | .07 | .30 (−.01 to .61) | .06 | .28 (.001–.56) | .049 |
| Presence of CMV in semen | .17 (−.05 to .39) | .135 | .27 (.003–.55) | .048 | … | |
| Longer interval since HIV infection (mo) | .01 (−.001 to .02) | .083 | .004 (−.01 to .02) | .453 | .01 (−.00003 to .02) | .051 |
| HIV RNA load (log10 copies/mL) | .05 (−.09 to .20) | .481 | −.01 (−.19 to .16) | .873 | … | |
| CD4+ T-cell count (cells/mm3) | −.00 (.00–.00) | .482 | −.00 (.00–.00) | .529 | … | |
| 100% match with detection probe | .80 (.36–1.23) | <.001 | … | .83 (.39–1.27) | <.001 | |
Estimated slopes and 2-sided P values (α = 0.05) were calculated using linear mixed-effects models for repeated measures univariate and multivariate analysis. Univariate analysis was performed on all samples and on a subset of 159 samples with a complete match between the detection probe and the main viral population. All covariates with P values of < .10 in the univariate models were considered for multivariate modeling. The presence of CMV in semen was eliminated from our multivariate analysis since it was highly correlated with the presence of CMV in the cell (P < .0001).
Abbreviations: CI, confidence interval; CMV, cytomegalovirus.
a Data are for 113 subjects and 229 samples.
b Data are for 80 subjects and 159 samples.
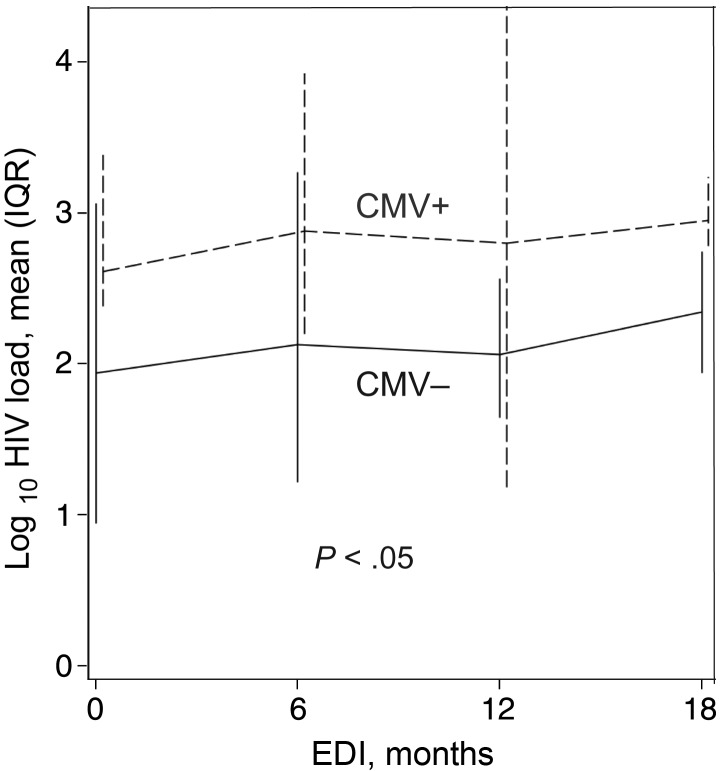
When evaluating only the samples with a complete match between HIV sequence and the detection probe (n = 159), detectable CMV levels in semen (P = .05) and in PBMCs (P = .06) were each associated with higher levels of HIV DNA in PBMCs (Figure 1). The association between CMV in PBMCs and higher levels of HIV DNA in PBMCs remained significant in a multivariate analysis (P = .05) that included estimated duration of infection and presence of a mismatch between probe and target sequence as covariates. The presence of CMV in semen was not included in the multivariate analysis since it was highly correlated with the presence of CMV in PBMCs (P < .0001) and had weaker association with HIV DNA levels than CMV in PBMCs. Overall, subjects with detectable CMV in both semen and PBMCs were more likely to have higher HIV DNA levels in PBMCs than participants with undetectable CMV in both semen and PBMCs (P = .02 for the entire data set; P = .008 for the matched subset) and those with detectable CMV in only semen or PBMCs (P = .07 for the entire data set; P = .05 for the matched subset). After including a binary variable in the model that indicated intermittent CMV shedding, the associations between HIV DNA levels and seminal CMV remained unchanged, and the presence of intermittent shedding versus consistent shedding was not significantly different than 0 (P = .47).
Figure 1.
Mean human immunodeficiency virus (HIV) DNA levels, by presence of cytomegalovirus (CMV) in peripheral blood mononuclear cells (PBMCs). The unbroken line shows mean values and interquartile ranges for proviral HIV DNA for samples with undetectable CMV DNA in PBMCs, whereas the dashed line represents mean values and interquartile ranges for proviral HIV DNA for samples with detectable CMV DNA in PBMCs. Data shown for matched samples only. Abbreviation: EDI, estimated duration of infection.
Of note, detectable CMV in semen and/or PBMCs were not significantly associated with HIV RNA levels or with CD4+ T-cell counts (P > .5).
DISCUSSION
The impact of low-level HIV replication on the replenishment and size of the latent reservoir is controversial [10, 11]. Recent studies have identified immune activation and homeostatic cell proliferation as a major mechanism to sustain the persistence of the HIV reservoir [4]. Since HIV DNA levels in blood predict the rate of CD4+ T-cell loss, time to AIDS, virologic failure of ART, and dementia [12–14], reducing these proviral levels alone may have substantial clinical benefits.
This study demonstrated that presence of CMV in PBMCs and seminal plasma of HIV-infected ART-naive MSM was associated with higher levels of proviral HIV DNA. It also found that simultaneous detection of CMV in semen and PBMCs was associated with the highest levels of HIV DNA in PBMCs. These findings were most robust when evaluating samples that harbored an HIV population whose sequences completely matched that of the real-time PCR detection probe. Interestingly, levels of HIV DNA in PBMCs and CMV replication were not associated with CD4+ T-cell numbers or HIV RNA levels. A significant association between HIV RNA and HIV DNA levels was found after excluding samples with low-level HIV DNA, suggesting that the HIV DNA reservoir had to be at a sufficient size to correlate with measured HIV RNA levels, independent of the estimated duration of infection.
This study has several limitations. First, it was conducted on ART-naive individuals, so the measured HIV DNA levels included both integrated and unintegrated viral forms and might represent the extent of HIV replication, in addition to the size of the stable HIV reservoir. Since the latent viral reservoir includes only integrated HIV DNA, our total HIV DNA levels cannot represent solely the latent reservoir, although other studies found that >90% of HIV-infected cells in both untreated and treated HIV-infected individuals have a latent viral transcription pattern [15]. Future studies should evaluate the relationships between continuous and intermittent CMV reactivation and HIV DNA levels in the setting of ART and suppressed HIV RNA (which is representative of the latent reservoir).
Second, although we used a conserved set of primers and probe to quantify HIV DNA [8], we observed that samples with mismatches in the probe-binding site in their major HIV population presented significantly lower HIV DNA levels. We evaluated this by introducing a “mismatch variable” in our statistical model. We recognize that accounting for the mismatches as a binary variable is not ideal, since each primer mismatch could affect the efficiency of the PCR in a different way. Future studies should account for this potential technological artifact by perhaps using newer techniques, like digital droplet PCR, which may be less sensitive to single-base mismatches in primer and probe sequences. Moreover screening for STIs was performed only on a subset of participants at baseline, and unrecognized STIs might have confounded our findings. Last, because this was a retrospective, observational study, we cannot establish a causal relationship between CMV reactivation and HIV DNA levels, and the extent of immune activation during untreated HIV infection could be a determinant of CMV shedding and HIV DNA levels. Alternatively, subjects with larger HIV reservoirs might be more immunosuppressed and thus have more CMV reactivation. Despite those limitations, this study provides important insights in regard to one possible mechanism contributing to the establishment of the viral reservoir during early HIV infection. Future studies should determine whether persistent CMV replication could be targeted as a strategy to reduce the size of the latent HIV reservoir.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We are grateful to all of the participants in the San Diego Primary Infection Cohort; the CFAR Genomic and Translational Virology Cores; and Nadir Weibel, for his support and inspiring discussions. We also thank and commemorate our dear friend and outstanding research colleague, Marek Fischer, for all his contributions and support to our research over many years. The HIV RNA quantification standard was obtained through the NIH AIDS Research and Reference Reagent Program, DAIDS, NIAID. The HIV VQA RNA quantification standard was obtained from the DAIDS Virology Quality Assurance Program. Primer and probe for quantification of herpesviruses, as well as the plasmids and quantification standards, were kindly provided by Fred Lakeman.
S. G. participated in the study design, performed the laboratory experiments, participated in the data analyses for this study, and wrote the primary version of the manuscript; C. M. A. performed the statistical analysis and wrote the primary version of the manuscript; S. R. M. participated in the data analyses and revised the manuscript; M. V. V. performed the laboratory experiments; S. J. L. and D. M. S. enrolled participants; D. D. R., S. J. L., and D. M. S. designed the present study, participated in data analysis, and revised the manuscript; and all authors read and approved the final manuscript.
Disclaimer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial support. This work was supported by the Department of Veterans Affairs, the James Pendleton Charitable Trust, the National Institutes of Health (awards AI69432, AI043638, MH62512, MH083552, DA034987, AI077304, AI36214 [Center for AIDS Research {CFAR}], AI047745, AI74621, GM093939 and AI080353, and AI306214 [CFAR]), the Swiss National Science Foundation (grant PASMP3_136983), the California HIV Research Program (grant RN07-SD-702), and the National Institute of General Medical Sciences (grant GM093939).
Potential conflicts of interest. D. D. R. has served as a consultant for Bristol-Myers Squibb, Gilead Sciences, Merck, Monogram Biosciences, Biota, Chimerix, Tobira, and Gen-Probe. D. M. S. has received grant support from ViiV Pharmaceuticals and consultant fees from Gen-Probe. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Gianella S, Strain MC, Rought SE, et al. Associations between virologic and immunologic dynamics in blood and in the male genital tract. J Virol. 2012;86:1307–15. doi: 10.1128/JVI.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunt PW. HIV and inflammation: mechanisms and consequences. Curr HIV/AIDS Rep. 2012;9:139–47. doi: 10.1007/s11904-012-0118-8. [DOI] [PubMed] [Google Scholar]
- 3.Okoye A, Meier-Schellersheim M, Brenchley JM, et al. Progressive CD4+ central memory T cell decline results in CD4+ effector memory insufficiency and overt disease in chronic SIV infection. J Exp Med. 2007;204:2171–85. doi: 10.1084/jem.20070567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chomont N, El-Far M, Ancuta P, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strain MC, Little SJ, Daar ES, et al. Effect of treatment, during primary infection, on establishment and clearance of cellular reservoirs of HIV-1. J Infect Dis. 2005;191:1410–8. doi: 10.1086/428777. [DOI] [PubMed] [Google Scholar]
- 6.Little SJ, Frost SD, Wong JK, et al. Persistence of transmitted drug resistance among subjects with primary human immunodeficiency virus infection. J Virol. 2008;82:5510–8. doi: 10.1128/JVI.02579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kosakovsky Pond SL, Posada D, Stawiski E, et al. An evolutionary model-based algorithm for accurate phylogenetic breakpoint mapping and subtype prediction in HIV-1. PLoS Comput Biol. 2009;5:e1000581. doi: 10.1371/journal.pcbi.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Althaus CF, Gianella S, Rieder P, et al. Rational design of HIV-1 fluorescent hydrolysis probes considering phylogenetic variation and probe performance. J Virol Methods. 2010;165:151–60. doi: 10.1016/j.jviromet.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Livak KJ. Allelic discrimination using fluorogenic probes and the 5’ nuclease assay. Genet Anal. 1999;14:143–9. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 10.Tobin NH, Learn GH, Holte SE, et al. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: expression of archival virus and replication of virus. J Virol. 2005;79:9625–34. doi: 10.1128/JVI.79.15.9625-9634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joos B, Fischer M, Kuster H, et al. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc Natl Acad Sci U S A. 2008;105:16725–30. doi: 10.1073/pnas.0804192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pellegrin I, Caumont A, Garrigue I, et al. Predictive value of provirus load and DNA human immunodeficiency virus genotype for successful abacavir-based simplified therapy. J Infect Dis. 2003;187:38–46. doi: 10.1086/345860. [DOI] [PubMed] [Google Scholar]
- 13.Rouzioux C, Hubert JB, Burgard M, et al. Early levels of HIV-1 DNA in peripheral blood mononuclear cells are predictive of disease progression independently of HIV-1 RNA levels and CD4+ T cell counts. J Infect Dis. 2005;192:46–55. doi: 10.1086/430610. [DOI] [PubMed] [Google Scholar]
- 14.Valcour VG, Shiramizu BT, Sithinamsuwan P, et al. HIV DNA and cognition in a Thai longitudinal HAART initiation cohort: the SEARCH 001 Cohort Study. Neurology. 2009;72:992–8. doi: 10.1212/01.wnl.0000344404.12759.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer M, Joos B, Niederost B, et al. Biphasic decay kinetics suggest progressive slowing in turnover of latently HIV-1 infected cells during antiretroviral therapy. Retrovirology. 2008;5:107. doi: 10.1186/1742-4690-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.