Abstract
Drug-resistant human immunodeficiency virus type 1 (HIV-1) minority variants increase the risk of virologic failure for first-line nonnucleoside reverse transcriptase inhibitor (NNRTI)-based regimens. We performed a pooled analysis to evaluate the relationship between NNRTI-resistant minority variants and the likelihood and types of resistance mutations detected at virologic failure. In multivariable logistic regression analysis, higher NNRTI minority variant copy numbers, non-white race, and nevirapine use were associated with a higher risk of NNRTI resistance at virologic failure. Among participants on efavirenz, K103N was the most frequently observed resistance mutation at virologic failure regardless of the baseline minority variant. However, the presence of baseline Y181C minority variant was associated with a higher probability of Y181C detection after virologic failure. NNRTI regimen choice and preexisting NNRTI-resistant minority variants were both associated with the probability and type of resistance mutations detected after virologic failure.
Keywords: HIV-1 drug resistance, minority variants, virologic failure, resistance genotyping
Human immunodeficiency virus (HIV) drug resistance mutations present below 10%–20% of the viral population are not reliably detected by genotyping techniques that use population (Sanger) sequencing [1, 2]. In a recent pooled analysis, we demonstrated that the presence of baseline drug-resistant minority variants more than doubles the risk of virologic failure in patients initiating a first-line nonnucleoside reverse transcriptase inhibitor (NNRTI)-based regimen [3]. However, it is unknown how these baseline minority variants relate to the risk and type of resistance mutations detected at virologic failure.
The mechanism by which these detectable drug-resistant minority variants mediate treatment failure would at first glance be straightforward and involve the expansion of virus harboring that resistance mutation. Such a pattern has been detected in studies of resistance developing against a CCR5-antagonist [4] and raltegravir [5]. In addition, there can be a relatively high concordance of preexisting minority variants with mutations detected after virologic failure of raltegravir-based regimens [6] and NNRTI-based regimens in treatment-experienced patients [7]. However, these studies were of limited sample size and data from treatment-naive individuals do not always support such a straightforward interpretation [8]. We used data collected during a previously described pooled analysis of drug-resistant minority variants to evaluate the relationship between baseline NNRTI-resistant minority variants, antiretroviral therapy (ART) regimen, and other factors on the likelihood and types of resistance mutations detected by population sequencing after virologic failure.
METHODS
This analysis derives from a substudy of a previously reported pooled analysis of HIV type 1 (HIV-1) drug-resistant minority variants on the risk of virologic failure for treatment-naive individuals initiating an NNRTI-based regimen [3]. Patient-level data were obtained from all studies to exclude participants with any evidence of pre-ART drug resistance and to standardize the definition of virologic failure. A total of 240 participants are included from seven studies with genotypic resistance results after virologic failure [8–14].
All of the studies evaluated baseline K103N minority variants, and 6 of the 7 studies evaluated baseline Y181C minority variants as well [8–13]. A detailed description of the assays, limits of detection, and minority variants detected can be found in the original report [3]. Mutations conferring NRTI and NNRTI resistance were defined as those with a Stanford HIV resistance DB score ≥10 for the participant's ART regimen. Medication adherence was available from 3 studies and was determined either by self-reported medication adherence over the previous 4 [8] or 7 days [9], or by a clinic-based pill count [14].
Fisher exact tests, Wilcoxon rank sum tests, and Cochran-Mantel-Haenszel statistics (stratified by study) were used to compare factors associated with either the presence of genotyping results or resistance at virologic failure. Minority variant copy number was estimated as the product of percentage of minority variant and level of HIV-1 RNA. Individuals without detectable minority variants were assigned a minority variant copy number of 0%; in a sensitivity analysis, a minority variant copy number equivalent to 10% of the assay limit of detection was imputed. A multivariable logistic regression, stratified by study, with backward elimination was performed to evaluate factors that predicted the risk of NNRTI resistance at virologic failure. Variables included in the regression model were determined a priori with the exception of ethnicity, which was found to be a significantly associated with the risk of NNRTI resistance at virologic failure on univariate analysis. Statistical analysis and the creation of figures were performed using SAS 9.2 and GraphPad Prism 5.
RESULTS
Genotypic resistance data after virologic failure were available from 240 of the 319 (75%) participants in the original pooled analysis with virologic failure [3]. The median time from virologic failure to resistance genotyping was 22 days (interquartile range [IQR], 0–49 days]. More than half of the participants had NNRTI resistance detected at the time of virologic failure (Supplementary Table 1). Those with detectable NNRTI resistance at the time of virologic failure had lower baseline CD4+ cell counts (205/mm3 vs 261/mm3, P = .008), and were more commonly receiving a nevirapine-based regimen (P = .03). The NRTI component of the ART regimen was variable with 9 different combinations represented. The most common NRTI backbone used for individuals on either efavirenz or nevirapine was zidovudine/lamivudine (AZT/3TC). Of those on AZT/3TC and efavirenz, 50% (49/99) had detectable NNRTI resistance at virologic failure compared with 73% (8/11) of those on AZT/3TC and nevirapine (P = .21). There were significant differences in the distribution of races/ethnicities (P = .003) with whites comprising a smaller proportion of those with NNRTI resistance at virologic failure than blacks. Overall mean ART adherence rates were similar between those with and without NNRTI resistance at virologic failure. No significant differences in baseline characteristics between individuals with or without resistance testing at virologic failure were detected with one exception (Supplementary Table 2). Those without genotyping data had higher rates of ART adherence (P < .01).
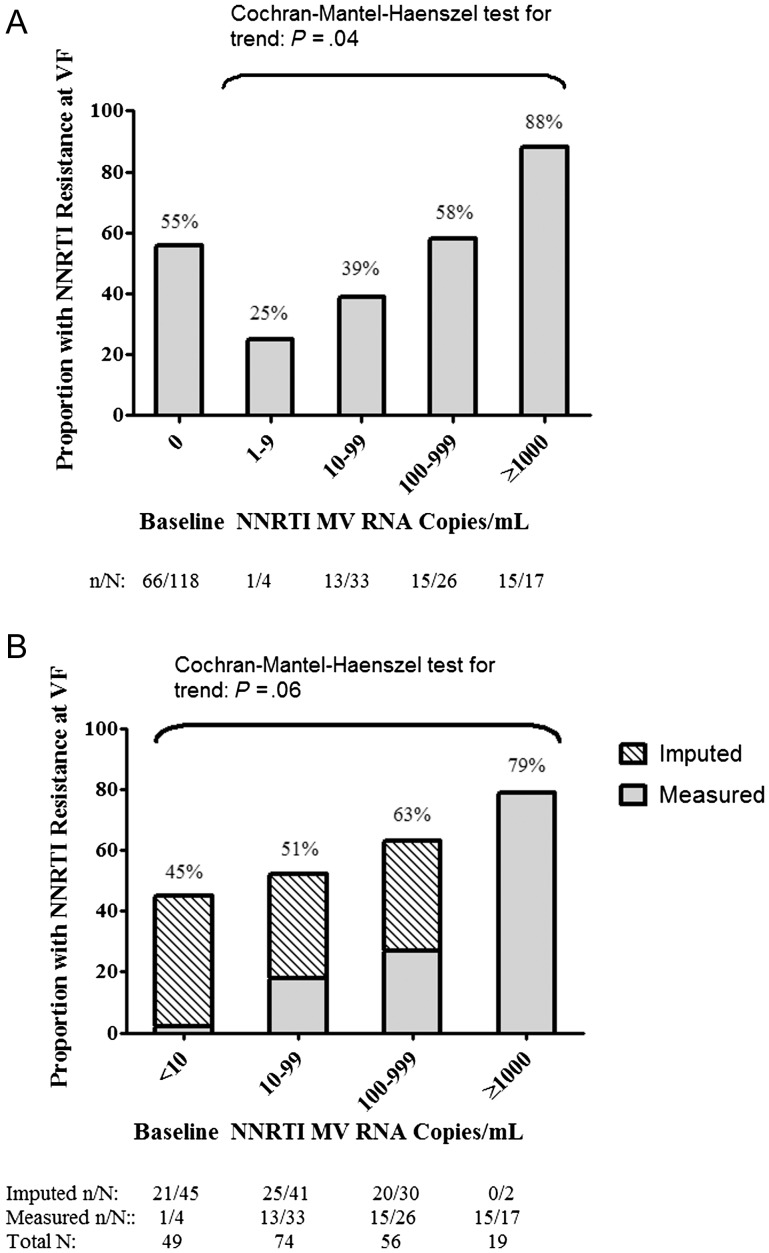
A significantly higher proportion of those with ≥1% NNRTI minority variants had detectable NNRTI resistance at virologic failure compared to either individuals harboring <1% NNRTI minority variants or no detectable minority variants (92% with ≥1% minority variants vs 49% with <1%, P = .002 and 92% ≥1% minority variants vs 58% without, P = .01). A similar outcome was seen when participants were stratified based on harboring ≥0.5% vs <0.5% minority variants. Among those with detectable minority variants at baseline, increasing copy numbers of NNRTI resistance mutations was associated with a higher probability of resistance at virologic failure (Figure 1A). Interestingly, individuals with no detectable minority variants had an intermediate outcome. This result is likely due to the varying limits of detection for the assays included in this pooled analysis [3]. Thus, individuals without detectable minority variants based on a less sensitive assay may, in fact, harbor low-frequency mutations that might have been detectable by a more sensitive test. We therefore performed a sensitivity analysis using both measured and imputed minority variant copy number. Individuals without detectable minority variants were assigned an imputed minority variant copy number equivalent to 10% of the assay limit of detection. Results of this analysis closely mirrored those of the measured values alone (Figure 1B).
Figure 1.
Rates of nonnucleoside reverse transcriptase inhibitor (NNRTI) resistance detected at virologic failure increases with higher measured copy nos. of NNRTI-resistant minority variants (A) or measured and imputed copy nos. of NNRTI-resistant minority variants (B). For those without detectable minority variants (MVs) at baseline, MV copy nos. were imputed by using 10% of the assay limit of detection. Abbreviation: VF, virologic failure.
In multivariable logistic regression analysis, factors that were independently associated with higher odds of NNRTI resistance at virologic failure included having a higher baseline NNRTI minority variant copy number, nevirapine use, and nonwhite ethnicity (Supplementary Table 3). Baseline viral load, CD4+ count, and ART adherence were not found to be significant predictors of NNRTI resistance at virologic failure.
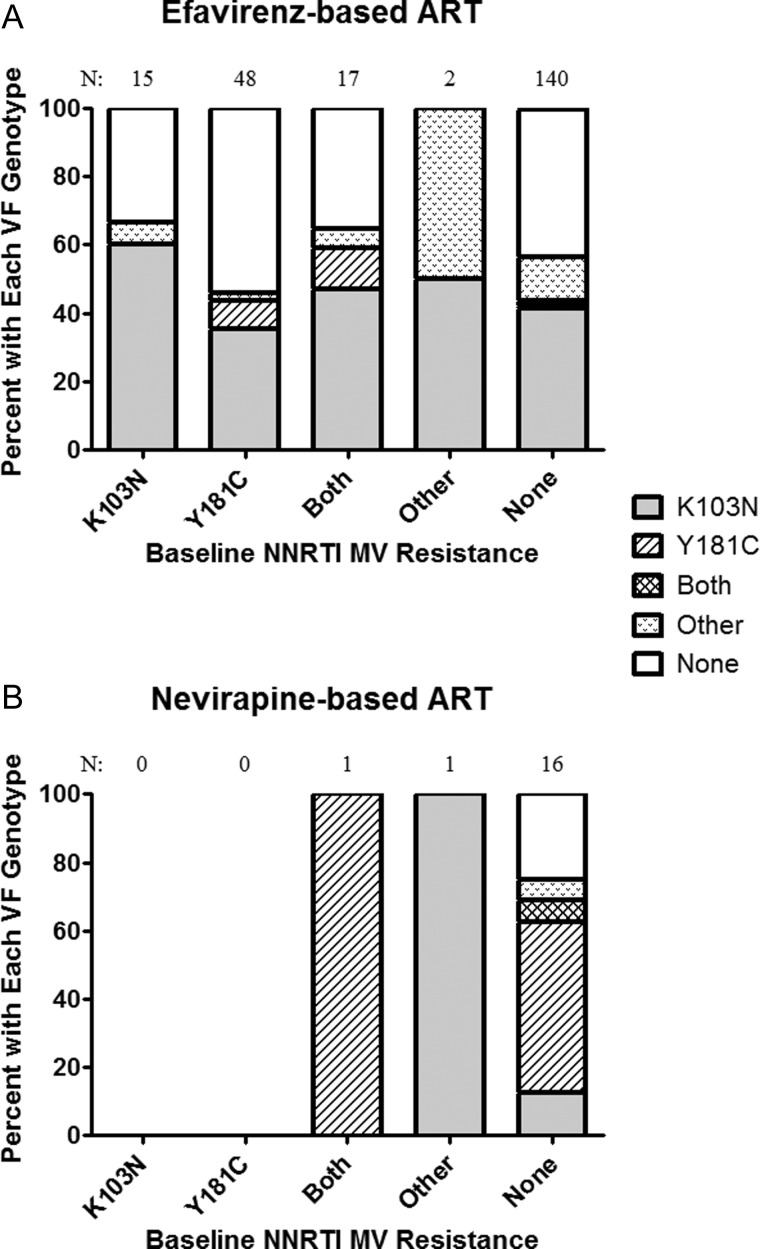
We evaluated the relationship between the NNRTI-resistant minority variants detected at baseline and the resistance mutations that emerged at virologic failure. Participants were categorized into those receiving efavirenz and those receiving a nevirapine-based regimen. Individuals receiving an efavirenz-based regimen were found to have K103N as the most common NNRTI resistance mutation detected at virologic failure regardless of the baseline resistance pattern (Figure 2A). However, the presence of baseline Y181C was associated with a higher rate of Y181C detection at virologic failure (18% vs 3%, P = .01). Y181C was the most commonly detected NNRTI resistance at virologic failure for those receiving a nevirapine-based ART regimen, although there were relatively few participants receiving nevirapine (Figure 2B). In those individuals with no baseline NNRTI resistance mutation but resistance on virologic failure, Y181C was detected in 75% (9 of 12) of participants receiving nevirapine as compared to 4% (3 of 79) of those receiving efavirenz (P < .001).
Figure 2.
Association between baseline minority nonnucleoside reverse transcriptase inhibitor (NNRTI) resistance mutation and the resistance detected by standard genotyping after virologic failure (VF) for those on an efavirenz-based regimen (A) or nevirapine-based regimen (B).
In the original pooled analysis, 228 participants had pre-ART assessment of minority M184V mutations, and 10 were found to have an M184V minority variant. Of these 10, virologic failure occurred in 4 participants, and M184V was found in the virologic failure genotype of 2. By contrast, M184V was detected by viral genotyping at virologic failure in 21 of 80 participants without preexisting M18V minority variant (P = .30). The 2 participants with preexisting K65R minority variants did not have virologic failure.
DISCUSSION
In this study, we found that among treatment-naïve patients initiating an NNRTI-based regimen, the presence of NNRTI-resistant minority variants, nonwhite ethnicity, and nevirapine use were all associated with an increased risk of NNRTI resistance detected at virologic failure. Interestingly, the type of NNRTI resistance that emerged at virologic failure frequently differed from those detected as minority variants at baseline. This finding was unexpected as the dose-dependent relationship of baseline minority variants with both risk of virologic failure [3] and detectable resistance at virologic failure initially suggested a straightforward explanation for their effects. There are several potential explanations for this discrepancy. The presence of one minority variant could predispose to the development of additional resistance mutations. Despite the relatively short time from the date of virologic failure to resistance genotyping, there may have been sufficient time since the end of virologic suppression for selection of more fit resistance variants [13]. It is possible that earlier virologic sampling in patients with baseline Y181C minority variant may have detected variants containing both Y181C and K103N prior to K103N becoming the dominant species. Alternatively, the detection of baseline drug-resistant minority variant could be a marker of greater underlying viral diversity and may be associated with the presence of other undetected resistance mutations that eventually become the dominant species.
We found that the type of minority variant mutation at baseline clearly influenced which resistance mutations detected at virologic failure as participants on efavirenz were more likely to have Y181C detected at virologic failure when that mutation was present as a baseline minority variant. Which resistance mutation emerged at virologic failure also was strongly correlated with the NNRTI regimen. Our results support the observation both in vitro and in vivo that the Y181C mutation offers relatively high levels of resistance and fitness preservation in the setting of nevirapine exposure [15–17]. The same association has been found of the K103N resistance mutation in the setting of efavirenz use [17, 18].
A number of studies have now shown that nonwhite ethnicity is associated with increased risk of virologic failure [19, 20]. We found that nonwhite ethnicity was also associated with a higher risk of NNRTI resistance at the time of virologic failure. One potential explanation may lie in ethnic-specific distributions of genetic polymorphisms (eg, in CYP2B6) that affect antiretroviral medication (ARV) metabolism and drug concentrations. These genotypes have been shown to affect the risk of virologic failure and likely affect the risk of resistance emergence given that slow-metabolizer genotypes are more frequent in nonwhite participants and allow for longer periods of functional monotherapy after treatment discontinuation [21].
Due to its low cost, nevirapine continues to be one of the most commonly used ARVs, especially in developing countries. In this study, nevirapine use was also independently associated with a higher risk of NNRTI resistance at the time of virologic failure. However, this finding should be interpreted with caution given the variation in the NRTI backbones, which could modify the risk of treatment failure and resistance emergence.
This study has several limitations. First, we combined data from 7 studies using assays with varying limits of minority variant detection [3]. We performed a sensitivity analysis using an imputed proportion of minority variants for those without detectable minority variants using 10% of the assay limit of detection. The results were consistent with the analysis performed using measured minority variant proportion alone. In addition, one of the studies evaluating an efavirenz-based regimen did not test for the presence of the Y181C minority variant [14]. This omission may have led to an underestimation of the association between baseline Y181C minority variant presence and Y181C detection by standard genotyping at virologic failure. Finally, only a small proportion of the total study population was tested for the presence of NRTI resistance, which limits our ability to evaluate the impact of baseline NRTI-resistant minority variants on their emergence during treatment failure.
We have now demonstrated that for individuals initiating a first-line NNRTI-based regimen, NNRTI-resistant minority variants increase both the risk of virologic failure and NNRTI resistance detection at the time of treatment failure. Our results also show that the minority variants detected at baseline frequently differ from the resistance mutations observed at virologic failure. Additional studies of viral diversity, linkage analysis of low-frequency resistance mutations, and longitudinal observations during early virologic failure would provide further insights on how drug resistance mutations emerge and evolve during antiretroviral treatment failure.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank the study participants of all studies for their contributions to this analysis. We thank Melanie Balduin (University of Cologne), Rodolphe Thiebaut (Bordeaux School of Public Health), and Bernard Masquelier (Bordeaux University Hospital) for their invaluable contributions to the original systematic review and pooled analysis. We thank Paul Bain, PhD (Countway Library of Medicine, Harvard Medical School), for his assistance with the systematic review and Christian Pou (irsiCaixa AIDS Research Institute) for his help in the data collection. We thank Christina Lalama, MS (Harvard School of Public Health), and the AIDS Clinical Trials Group for contributing data from ACTG A5095. The findings and conclusions of this article are those of the authors and do not necessarily represent the official view of the Centers for Disease Control and Prevention.
Financial Support. J. Z. L. has received research support from Bristol-Myers Squibb and Tobira. R. P. has received consulting fees from Pfizer, Merck, Roche Diagnostics; the IrsiCaixa AIDS Research Institute and the Lluita contra la SIDA Foundation have received grant support from Pfizer, ViiV Healthcare, Siemens, Merck, and Boehringer Ingelheim for studies that R. P. serves as principal investigator. E. S. S. and M. D. M. are employees and stock-holders of Gilead Sciences, Inc. Yale University receives grant support from Merck, Pfizer, Gilead, Abbott, ViiV, Vertex, and Bristol-Myers Squibb for studies that M. D. K. serves as the principal investigator. M. D. K. receives royalties from patents owned by Stanford University for some HIV diagnostic tests. A. M. G. has served as a consultant to and/or has received research and travel grants from Bristol-Myers Squibb, Gilead, GlaxoSmithKline, Merck, Jenssen/Tibotec, and ViiV Healthcare. K. J. M. has received travel grants and honoraria from Gilead, Roche Diagnostics, GlaxoSmithKline, Bristol-Myers Squibb, Tibotec, and Abbott; and the University of Zurich received research grants from Gilead, Roche, and Merck Sharp & Dohme for studies that K. J. M. serves as principal investigator. D. R. K. has served as a consultant to and/or has received research grant support from Abbott, Avexa, Bristol-Myers Squibb, Boehringer-Ingelheim, Bristol-Myers Squibb, Gilead, GlaxoSmithKline, Merck, Roche, Tobira, Vertex, ViroStatistics, and ViiV Healthcare; he has also received speaking honoraria from Gilead and Roche. J. Z. L. is the recipient of a Clinical Investigator Training Program Fellowship: Harvard/MIT Health Sciences and Technology—Beth Israel Deaconess Medical Center, in collaboration with Pfizer Inc. and Merck & Co. R. P. was supported in part by the CHAIN (Collaborative HIV and Anti-HIV Drug Resistance Network), Integrated Project no. 223131, funded by the European Commission Framework 7 Program. H. J. R. is supported in part by grants from the National Institutes of Health (NIH; Statistical and Data Management Center of the AIDS Clinical Trials Group U01 AI068634 and Harvard University CFAR P30 AI060354). D. K. is supported by a VA Merit Award. K. H. H. was supported in part by a grant from the NIH (U01 AI042170). K. J. M. is supported by the Swiss National Science Foundation (SNF), grants 324700-120793 and CR32I2_127017, and the European Community's Seventh Framework Programme (FP7/2007-2013) under the project "Collaborative HIV and Anti-HIV Drug Resistance Network (CHAIN)," grant agreement no. 223131. R. K. is supported in part by grants from the NIH (U01 AI 068636, K24 RR016482) and a Virology Specialty Laboratory subcontract from the ACTG.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Larder BA, Kohli A, Kellam P, Kemp SD, Kronick M, Henfrey RD. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature. 1993;365:671–3. doi: 10.1038/365671a0. [DOI] [PubMed] [Google Scholar]
- 2.Church JD, Jones D, Flys T, et al. Sensitivity of the ViroSeq HIV-1 genotyping system for detection of the K103N resistance mutation in HIV-1 subtypes A, C, and D. J Mol Diagn. 2006;8:430–2. doi: 10.2353/jmoldx.2006.050148. quiz 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li JZ, Paredes R, Ribaudo HJ, et al. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA. 2011;305:1327–35. doi: 10.1001/jama.2011.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsibris AM, Korber B, Arnaout R, et al. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS ONE. 2009;4:e5683. doi: 10.1371/journal.pone.0005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Codoner FM, Pou C, Thielen A, et al. Dynamic escape of pre-existing raltegravir-resistant HIV-1 from raltegravir selection pressure. Antiviral Res. 2010;88:281–6. doi: 10.1016/j.antiviral.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Miller MD, Danovich RM, et al. Analysis of low-frequency mutations associated with drug resistance to raltegravir before antiretroviral treatment. Antimicrob Agents Chemother. 2011;55:1114–9. doi: 10.1128/AAC.01492-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halvas EK, Wiegand A, Boltz VF, et al. Low frequency nonnucleoside reverse-transcriptase inhibitor-resistant variants contribute to failure of efavirenz-containing regimens in treatment- experienced patients. J Infect Dis. 2010;201:672–80. doi: 10.1086/650542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paredes R, Lalama CM, Ribaudo HJ, et al. Preexisting minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J Infect Dis. 2010;201:662–71. doi: 10.1086/650543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simen BB, Simons JF, Hullsiek KH, et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis. 2009;199:693–701. doi: 10.1086/596736. [DOI] [PubMed] [Google Scholar]
- 10.Jakobsen MR, Tolstrup M, Sogaard OS, et al. Transmission of HIV-1 drug-resistant variants: prevalence and effect on treatment outcome. Clin Infect Dis. 2010;50:566–73. doi: 10.1086/650001. [DOI] [PubMed] [Google Scholar]
- 11.Johnson JA, Li JF, Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 2008;5:e158. doi: 10.1371/journal.pmed.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geretti AM, Fox ZV, Booth CL, et al. Low-frequency K103N strengthens the impact of transmitted drug resistance on virologic responses to first-line efavirenz or nevirapine-based highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2009;52:569–73. doi: 10.1097/QAI.0b013e3181ba11e8. [DOI] [PubMed] [Google Scholar]
- 13.Metzner KJ, Giulieri SG, Knoepfel SA, et al. Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and -adherent patients. Clin Infect Dis. 2009;48:239–47. doi: 10.1086/595703. [DOI] [PubMed] [Google Scholar]
- 14.Goodman D, Zhou Y, Margot NA, et al. Low level of the K103N HIV-1 above a threshold is associated with virological failure in treatment-naive individuals undergoing efavirenz-containing therapy. AIDS. 2011;25:325–33. doi: 10.1097/QAD.0b013e3283427dcb. [DOI] [PubMed] [Google Scholar]
- 15.Iglesias-Ussel MD, Casado C, Yuste E, Olivares I, Lopez-Galindez C. In vitro analysis of human immunodeficiency virus type 1 resistance to nevirapine and fitness determination of resistant variants. J Gen Virol. 2002;83:93–101. doi: 10.1099/0022-1317-83-1-93. [DOI] [PubMed] [Google Scholar]
- 16.Hanna GJ, Johnson VA, Kuritzkes DR, et al. Patterns of resistance mutations selected by treatment of human immunodeficiency virus type 1 infection with zidovudine, didanosine, and nevirapine. J Infect Dis. 2000;181:904–11. doi: 10.1086/315329. [DOI] [PubMed] [Google Scholar]
- 17.Ngo-Giang-Huong N, Jourdain G, Amzal B, et al. Resistance patterns selected by nevirapine vs. efavirenz in HIV-infected patients failing first-line antiretroviral treatment: a bayesian analysis. PLoS One. 2011;6:e27427. doi: 10.1371/journal.pone.0027427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuritzkes DR, Lalama CM, Ribaudo HJ, et al. Preexisting resistance to nonnucleoside reverse-transcriptase inhibitors predicts virologic failure of an efavirenz-based regimen in treatment-naive HIV-1-infected subjects. J Infect Dis. 2008;197:867–70. doi: 10.1086/528802. [DOI] [PubMed] [Google Scholar]
- 19.Weintrob AC, Grandits GA, Agan BK, et al. Virologic response differences between African Americans and European Americans initiating highly active antiretroviral therapy with equal access to care. J Acquir Immune Defic Syndr. 2009;52:574–80. doi: 10.1097/QAI.0b013e3181b98537. [DOI] [PubMed] [Google Scholar]
- 20.Ribaudo H, Smith K, Robbins GK, et al. Race differences in the efficacy of initial ART on HIV infection in randomized trials undertaken by ACTG. 2011 18th Conference on Retroviruses and Opportunistic Infections. Boston, Massachusetts, Feb 27–Mar 2 (abstract 50) [Google Scholar]
- 21.Ribaudo HJ, Haas DW, Tierney C, et al. Pharmacogenetics of plasma efavirenz exposure after treatment discontinuation: An Adult AIDS Clinical Trials Group Study. Clin Infect Dis. 2006;42:401–7. doi: 10.1086/499364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.