Abstract
Background. Fluoroquinolone-resistant Escherichia coli are increasingly prevalent. Their clonal origins—potentially critical for control efforts—remain undefined.
Methods. Antimicrobial resistance profiles and fine clonal structure were determined for 236 diverse-source historical (1967–2009) E. coli isolates representing sequence type ST131 and 853 recent (2010–2011) consecutive E. coli isolates from 5 clinical laboratories in Seattle, Washington, and Minneapolis, Minnesota. Clonal structure was resolved based on fimH sequence (fimbrial adhesin gene: H subclone assignments), multilocus sequence typing, gyrA and parC sequence (fluoroquinolone resistance-determining loci), and pulsed-field gel electrophoresis.
Results. Of the recent fluoroquinolone-resistant clinical isolates, 52% represented a single ST131 subclonal lineage, H30, which expanded abruptly after 2000. This subclone had a unique and conserved gyrA/parC allele combination, supporting its tight clonality. Unlike other ST131 subclones, H30 was significantly associated with fluoroquinolone resistance and was the most prevalent subclone among current E. coli clinical isolates, overall (10.4%) and within every resistance category (11%–52%).
Conclusions. Most current fluoroquinolone-resistant E. coli clinical isolates, and the largest share of multidrug-resistant isolates, represent a highly clonal subgroup that likely originated from a single rapidly expanded and disseminated ST131 strain. Focused attention to this strain will be required to control the fluoroquinolone and multidrug-resistant E. coli epidemic.
Keywords: Escherichia coli infections, antimicrobial resistance, extended-spectrum β-lactamase, CTX-M-15, fluoroquinolone resistance, multidrug resistance, sequence type ST131, multilocus sequence typing, molecular epidemiology, FimH
Until recently, fluoroquinolones (FQs) were preferred agents for treating multiple types of bacterial infection, including urinary tract infections (UTIs), most of which are caused by Escherichia coli [1, 2]. However, FQ resistance in E. coli is increasingly prevalent, resulting in some patients progressing to severe illness or death despite receiving conventional empirical or prophylactic therapy [3, 4]. In part because of this, the authors of the 2010 Infectious Diseases Society of America guidelines recommended nitrofurantoin over FQs for empirical treatment of uncomplicated UTIs [5].
In E. coli, although upregulated efflux pumps and plasmid-encoded resistance mechanisms can reduce FQ susceptibility, high-level resistance typically requires 1–2 point mutations within the quinolone resistance-determining regions (QRDRs) of gyrA and parC, the chromosomal genes encoding the FQ targets DNA gyrase and topoisomerase IV, respectively [6]. Because of its chromosomal basis, such FQ resistance has arisen in diverse E. coli clonal lineages that presumably acquired the requisite QRDR mutations independently [7–9].
Despite the high clonal diversity of FQ-resistant (FQ-R) E. coli, the past decade has seen the rapid emergence and global spread of a specific FQ resistance-associated clonal group, ST131, which is among ≥1000 E. coli sequence types (STs), as defined by multilocus sequence typing (MLST) [9–11]. As are most STs, ST131 is both genetically and phenotypically diverse. For example, although ST131 first came to attention because of its close association with extended-spectrum β-lactamases (ESBLs), especially CTX-M-15 [12], in many locales ST131 is more frequently ESBL-negative but FQ-R [9], and these resistance traits are differentially distributed among ST131's diverse pulsed-field gel electrophoresis (PFGE) types [13]. However, whether ST131's association with FQ resistance is from frequent, independent emergence of resistance in diverse ST131 strains, vs expansion of a single strain, remains unknown [8, 9, 11, 14–17].
Thus, sub-ST analysis of ST131 isolates is critical to understanding the ongoing emergence of FQ-R E. coli. Accordingly, we analyzed FQ resistance at the sub-ST level for over 350 archived or newly obtained ST131 isolates and over 700 non-ST131 E. coli isolates, by determining fine clonal diversity based on individual gene loci and PFGE analysis. We sought thereby to define the clonal history of FQ resistance in ST131 and its impact on clinical population dynamics within E. coli generally.
METHODS
Isolates and Patients
The clonality of FQ resistance was analyzed among historical ST131 isolates (n = 236) and recent ST131 (n = 116) and non-ST131 (n = 737) human clinical isolates. The historical ST131 isolates were a convenience sample selected from multiple collections to represent diverse years of isolation (1967–2009), FQ phenotypes, locales (201 isolates from across the United States; 35 international isolates from 10 countries on 5 continents), and sources, including humans (n = 173), food/companion animals (n = 52), and food/environment (n = 10) [13]. The 853 recent clinical isolates (both ST131 and non-ST131) were consecutive, single-patient (ostensibly), human extraintestinal isolates from October 2010 and January 2011 from 5 clinical microbiology laboratories in Seattle, Washington (Group Health Cooperative, Harborview Medical Center, Seattle Children's Hospital, University of Washington Medical Center) and Minneapolis, Minnesota (Veterans Affairs Medical Center) [18]. Local institutional review boards approved the study protocol.
Sequence Analysis of Individual Loci
Isolates were assigned to an ST based on MLST allele profiles, as determined by sequencing established MLST loci (http://mlst.ucc.ie/mlst/dbs/Ecoli). Within-ST clonal variation was resolved on the basis of sequence variation in the E. coli fimbrial adhesin gene, fimH (positions 64–552) [18]. The QRDR of gyrA (6-570) and parC (1-573) also was sequenced [16]. Maximum-likelihood trees were inferred for gyrA and parC using PAUP* [19]. The ST131 gyrA and parC alleles were labeled as follows: stepwise mutational derivatives of each (numbered) main allele were indicated by using lower case letters for silent (synonymous) mutations and upper case letters for replacement (nonsynonymous) mutations.
Antimicrobial Susceptibility, ESBL Status, and blaCTX-M-15
Susceptibility to ciprofloxacin and 11 other antimicrobial agents was determined by disk diffusion, as specified by the Clinical and Laboratory Standards Institute [20]. Intermediate was considered resistant. Penicillins and cephalosporins were counted as separate antimicrobial classes. Historic isolates were previously characterized for ESBL production and presence of blaCTX-M-15 (encoding CTX-M-15) [13].
Pulsed-field Gel Electrophoresis Analysis
The 352 historical and recent ST131 isolates underwent standardized XbaI pulsed-field gel electrophoresis (PFGE) analysis, with pulsotypes defined at ≥94% PFGE profile similarity to index strains for each pulsotype [13]. For dendrogram construction, a 24% subsample (n = 85) was used to allow single-page readability. The 85 ST131 isolates were selected randomly after deliberate inclusion of the earliest (plus a second, as available) representative of each fimH-gyrA-parC combination. The dendrogram was inferred within BioNumerics (Applied Maths) according to the unweighted pair group method based on Dice similarity coefficients.
Statistical Analysis
Fisher exact test (2-tailed) was used to test comparisons of proportions. The significance criterion was P < .05.
RESULTS
FQ Resistance Within ST131 Is Associated With a Specific Subclone
To explore the subclonal structure of ST131, the 352 historical and recent ST131 isolates (1967–2011) underwent fimH sequencing and PFGE profiling. This analysis identified 185 unique PFGE pulsotypes and 7 distinct fimH-based putative clonal lineages (H15, H22, H27, H30, H35, H41, H94).
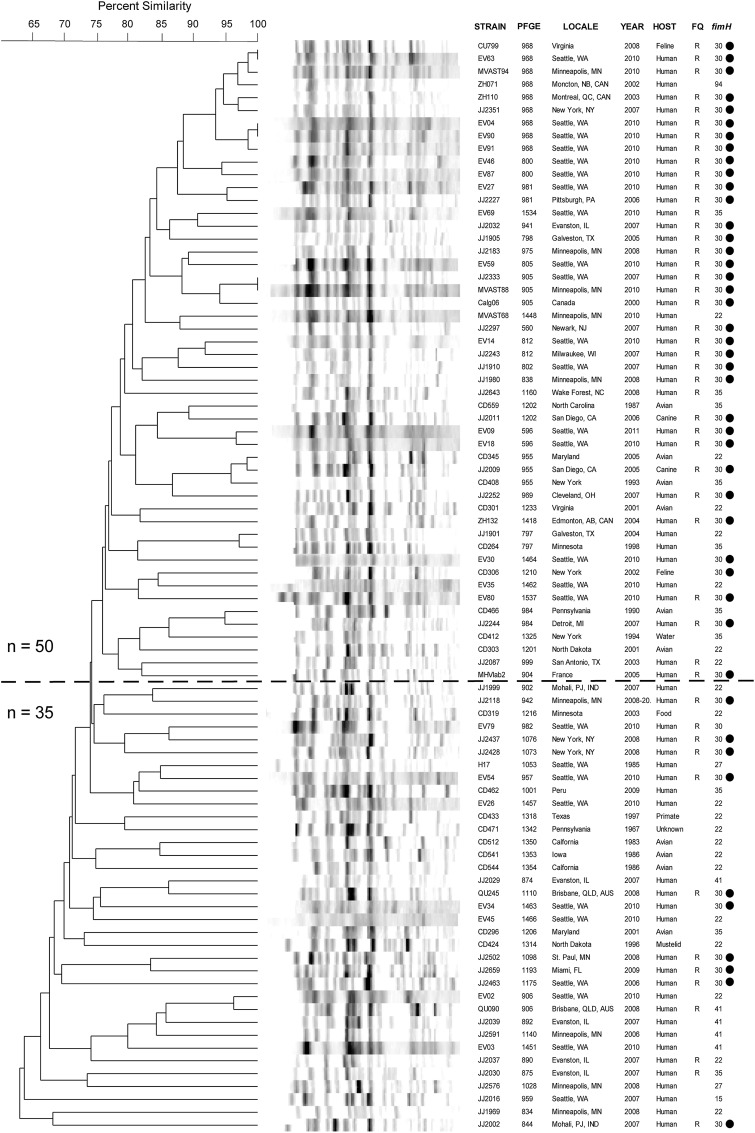
The fimH-based typing corresponded broadly with PFGE profiles, as reflected in the statistically significant segregation of fimH alleles within the PFGE dendrogram (Figure 1). For example, the dominant fimH30 allele was concentrated within the upper dendrogram region (≥75% profile similarity), as compared with the lower region (<75% profile similarity; 35 of 50 [70%] vs 10 of 35 [29%]; P < .001). Conversely, the secondmost-common fimH22 allele was concentrated within the lower dendrogram region (13 of 35 [37%] vs 7 of 50 [14%]; P = .02). However, none of the fimH-based lineages were confined to a single clade on the PFGE tree (Figure 1), reflecting certain level of phylogenetic incongruence between the 2 typing methods.
Figure 1.
XbaI pulsed-field gel electrophoresis-based dendrogram for 85 sequence type ST131 Escherichia coli isolates (1967–2011). FQ, fluoroquinolone phenotype (R, resistant; S, susceptible); fimH, allele of fimH (type 1 fimbrial adhesin); PFGE, pulsotype; Year, year of isolation or submission to reference laboratory. Bullets (to right) mark isolates with the fimH30 allele. Horizontal line separates isolates with ≥75% overall profile similarity (top, n = 50) from less-similar isolates (bottom, n = 35).
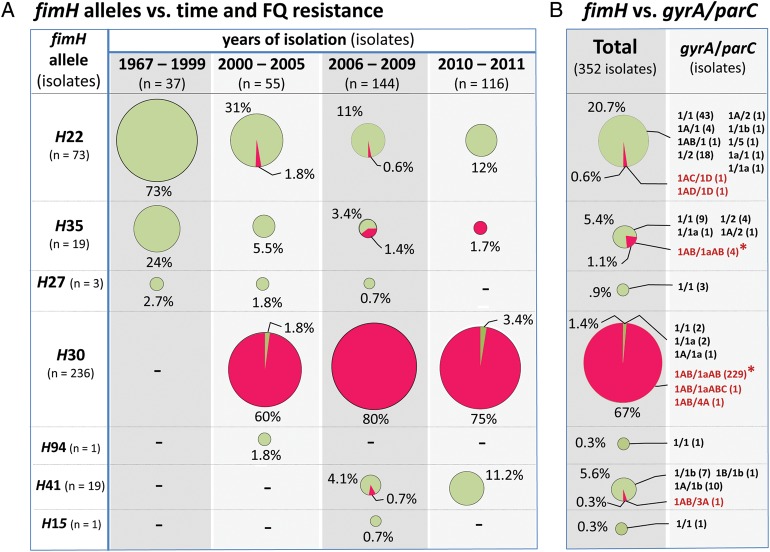
The H subclones were next analyzed for associations with FQ resistance and prevalence during the study period (1967–2011; Figure 2A). During the earliest period (1967–1999), only FQ-susceptible (FQ-S) subclones were encountered, predominantly H22 and H35. FQ-R isolates appeared first during 2000–2005, associated almost exclusively with the (newly detected) H30 subclone. Thereafter, the H30 subclone continued to account for >97% of FQ-R ST131 isolates and constituted an increasing proportion of ST131 isolates overall.
Figure 2.
Distribution of fluoroquinolone-susceptible (FQ-S) and resistant (FQ-R) isolates among the 7 fimH-based (H) ST131 subclones. Area of circle is proportional to the relative abundance of the particular H subclone within the particular time period. Percentage values are shown relative to the total no. of isolates within the time period. (A) Clonal distribution by time period. (B) Overall clonal distribution. The gyrA and parC allele combinations observed among FQ-S (in green) and FQ-R (in red) isolates are labeled according to the nomenclature shown in Figure 3. Asterisks identify the ST131 isolates’ principal gyrA/parC allele combination (ie, gyrA1AB/parC1aAB).
The FQ-R ST131 Clonal Expansion Involved Almost Exclusively a Single gyrA/parC Combination
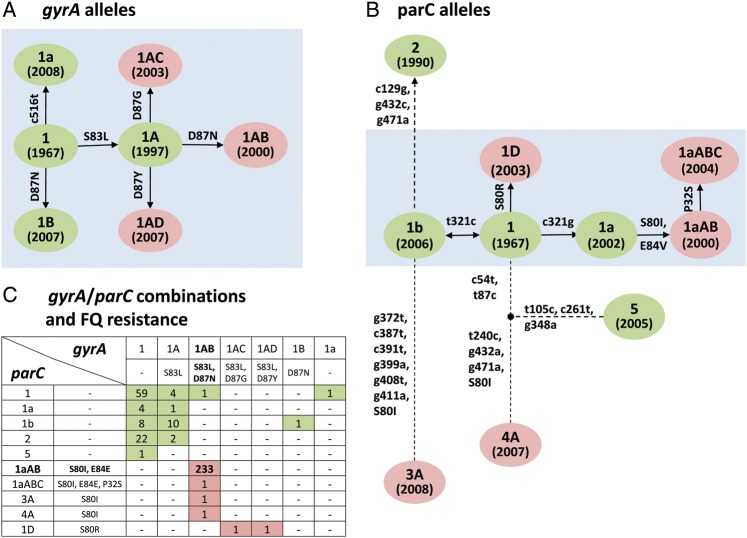
Among the 352 E. coli ST131 study isolates, sequence analysis of gyrA and parC identified 7 gyrA alleles and 10 parC alleles (Tables 1 and 2; Figure 3). The gyrA alleles differed by no more than 1 single-nucleotide polymorphism (SNP), suggesting evolution by point mutation (Table 1; Figure 3A). All FQ-S isolates possessed either the putative ancestor allele gyrA1 or a derivative thereof containing either one silent SNP (gyrA1a) or one amino acid replacement mutation, ie, Ser-83-Leu (gyrA1A) or Asp-87-Asn (gyrA1B). In contrast, all FQ-R isolates possessed gyrA alleles that differed from gyrA1A by distinct secondary replacement mutations at Asp-87: gyrA1AB (Asp-87-Asn), gyrA1AC (Asp-87-Gly), and gyrA1AD (Asp-87-Tyr).
Table 1.
Single-Nucleotide Polymorphisms in gyrA Among 352 Recent and Historical Escherichia coli Isolates of Sequence Type ST131
| gyrA allele | Nucleotide at Indicated Positiona (Associated Amino Acid Shift, if Any)b |
|||
|---|---|---|---|---|
| bp 248 | bp 259 | bp 260 | bp 516 | |
| 1 | c | g | a | t |
| 1a | c | |||
| 1A | t (S83L) | |||
| 1AB | t (S83L) | a (D87N) | ||
| 1AC | t (S83L) | g (D87G) | ||
| 1B | a (D87N) | |||
| 1AD | t (S83L) | t (D87Y) | ||
a Only polymorphic sites are shown. Bases are shown for ancestral allele (allele 1) and for other alleles, only if different from allele 1. Blank cells, identity with allele 1.
b Amino acid code: D, aspartate; G, glycine; L, leucine; N, asparagine; S, serine; Y, tyrosine.
Table 2.
Single-Nucleotide Polymorphisms in parC Among 352 Recent and Historical Escherichia coli Isolates of Sequence Type ST131
| parC allele | Nucleotide at Indicated Positiona (Associated Amino Acid Shift, if Any)b |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bp 54 | bp 87 | bp 94 | bp 105 | bp 129 | bp 238 | bp 239 | bp 240 | bp 251 | bp 261 | bp 321 | bp 348 | bp 372 | bp 387 | bp 391 | bp 399 | bp 408 | bp 411 | bp 432 | bp 471 | |
| 1 | c | t | c | t | c | a | g | t | a | c | c | g | g | c | c | g | g | g | g | g |
| 1a | g | |||||||||||||||||||
| 1b | t | |||||||||||||||||||
| 1aAB | t (S80I) | t (E84V) | g | |||||||||||||||||
| 1aABC | t (P32S) | t (S80I) | t (E84V) | g | ||||||||||||||||
| 1D | c (S80R) | |||||||||||||||||||
| 2 | g | t | c | a | ||||||||||||||||
| 3A | t (S80I) | t | t | t | t | a | t | a | ||||||||||||
| 4A | t | c | t (S80I) | c | a | a | ||||||||||||||
| 5 | t | c | c | t | a | |||||||||||||||
a Only polymorphic sites are shown. Bases are shown for ancestral allele (allele 1) and, for other alleles, only if different from allele 1. Blank cells, identity with allele 1.
b Amino acid codes: E, glutamate; I, isoleucine; P, proline; R, arginine; S, serine; V, valine.
Figure 3.
ST131-associated gyrA and parC alleles: gene phylogeny and combinations. Green, alleles (or combinations) associated with fluoroquinolone-susceptible isolates. Red, alleles (or combinations) associated with fluoroquinolone-resistant isolates. Single letter amino acid code: G (Gly), D (Asp), E (Glu), I (Ile), L (Lys), N (Asn), P (Pro), R (Arg), S (Ser), V (Val), and Y (Tyr). (A and B) Phylogeny of the ST131-associated gyrA and parC alleles. Labels inside circles: allele designations. In parentheses: earliest known year of isolation for the allele. Along the branches: lower-case numbers are nucleotide positions with silent mutations, upper-case numbers are amino acid positions with amino acid replacement mutations. Arrows: putative evolutionary order of mutations (double arrow between allele 1 and 1′ indicates uncertainty of the order). Gray boxes: phylogenetic clades within which nearest alleles differ by no more than one silent nucleotide change. (C) gyrA and parC allele combinations. Numbers inside cells: number of ST131 isolates with the corresponding allele combination. The predominant allele combination among FQ-R isolates is shown in boldface.
Among the 10 parC alleles, 6 closely related alleles likewise appeared to have evolved by point mutation (Table 2; Figure 3B, gray box). Most FQ-S isolates possessed the putative ancestor allele, parC1, or a variant containing 1 silent SNP (parC1a and parC1b). In contrast, most FQ-R isolates possessed parC1aAB (parC1a plus replacement mutations Ser-80-Ile and Glu-84-Val), parC1aABC (parC1aAB plus replacement mutation Pro-32-Ser), or parC1D (parC1 plus replacement mutation Ser-80-Arg). The 4 remaining parC alleles instead differed by multiple (≥3) silent SNPs, suggesting horizontal gene transfer. Two such alleles, parC4A and parC3A, occurred in FQ-R isolates and shared replacement mutation Ser-80-Ile.
The 7 gyrA and 10 parC alleles occurred in ST131 in 18 combinations (Figure 3C). Among FQ-S isolates the gyrA1/parC1 ancestral allele combination (Figure 3C) occurred in slightly more than half of isolates and in most H subclones (Figure 2B). In contrast, among FQ-R isolates the gyrA1AB/parC1aAB combination predominated overwhelmingly (98% of FQ-R isolates; Figure 3C), associated almost exclusively with the H30 subclone (Figure 2B).
The H30 ST131 Subclone Is Associated With FQ Resistance Globally, Regardless of Source, and With CTX-M-15
Among the 236 historical ST131 isolates, the H30 ST131 subclone was closely associated with FQ resistance regardless of locale and source, and with ESBL production and blaCTX-M-15. Specifically, among US isolates the H30 subclone accounted for 122 of 126 (97%) FQ-R isolates, vs 1 of 75 (1%) FQ-S isolates (P < .001), and among international isolates for 26 of 27 (96%) FQ-R isolates, vs 0 of 8 (0%) FQ-S isolates (P < .001). Similarly, among human-source isolates it accounted for 136 of 140 (97%) FQ-R isolates, vs 0 of 32 (0%) FQ-S isolates (P < .001), and among non-human-source isolates for 13 of 13 (100%) FQ-R isolates, vs 1 of 51 (2%) FQ-S isolates (P < .001). The 13 non-human-source FQ-R H30 subclone isolates, all with the gyrA1AB/parC1aAB combination, represented diverse animal hosts, including dogs, cats, a primate, and a dolphin (data not shown). Regarding cephalosporin resistance, the H30 subclone accounted for 92 of 108 (85%) ESBL-positive isolates, vs 57 of 128 (45%) ESBL-negative isolates (P < .001), and among ESBL-positive isolates for 63 of 68 (93%) blaCTX-M-15-positive isolates, vs 29 of 40 (73%) blaCTX-M-15-negative isolates (P < .01).
ST131's Principal FQ-R gyrA/parC Combination Is Confined to ST131
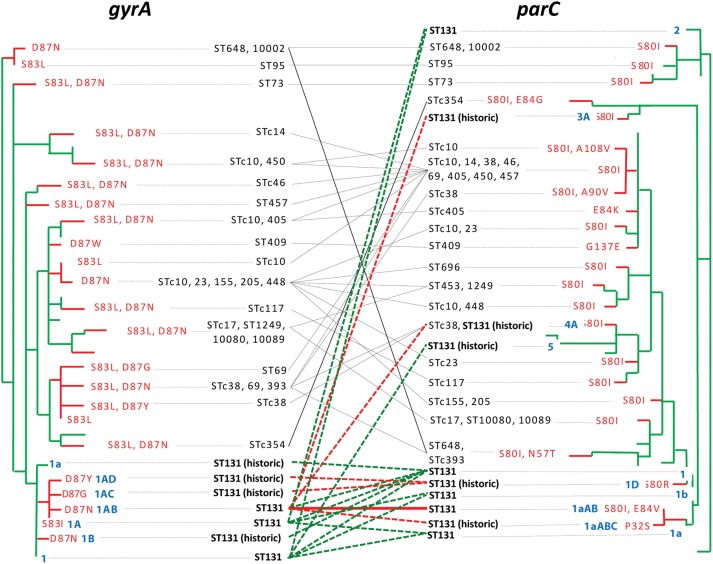
The isolates gyrA and parC were sequenced also from selected non-ST131 recent clinical isolates, including all 78 non-ST131 FQ-R isolates, plus 63 FQ-S isolates from the same STs. To avoid false inferences of evolutionary relatedness due to repeated occurrence of the same resistance-associated replacement mutations in different lineages, phylogenetic trees were inferred for gyrA and parC based only on silent SNPs (green: Figure 4), after which the replacement mutations were added to the corresponding tree branches (red: Figure 4).
Figure 4.
gyrA and parC phylogenetic trees for ST131 isolates and diverse fluoroquinolone (FQ)-resistant non-ST131 Escherichia coli isolates. Trees were built based on silent variation only (green branches); FQ-resistance-determining amino acid replacement changes were then added manually (red branches and labels). STc: ST complexes (groups of closely related STs) within which the indicated alleles are found. Diagonal lines indicate gyrA/parC combinations (dashed green, in FQ-susceptible ST131 isolates; dashed red, in FQ-R ST131 isolates; solid black, in FQ-R non-ST131 isolates). Blue/boldface labels: the 17 alleles found in ST131, named according to the nomenclature used in Figure 3. “ST131 (historic)” marks ST131-associated alleles found only in historic ST131 isolates.
In contrast to ST131's mutation-evolved gyrA and parC alleles, the 4 parC alleles suspected of being horizontally transferred into ST131 (Figure 3B) clearly derived from non-ST131 regions of the parC tree (Figure 4). Moreover, horizontal transfer of gyrA and parC occurred extensively throughout the species, with most alleles appearing in multiple STs (as indicated by the diverse STs listed for certain alleles) and/or in diverse combinations (as indicated by the cross-links connecting certain gyrA alleles to multiple parC alleles, and vice versa; Figure 4). In contrast, ST131's signature gyrA1AB and parC1aAB alleles, and their combination (heavy cross-link), occurred only within ST131 (Figure 4).
The H30 ST131 Subclone Represents the Largest Clonal Expansion in E. coli and Is Associated With Extensively Antimicrobial-resistant Infections
When analyzed across the 853 recent clinical E. coli isolates, the H30 ST131 subclone was strongly associated with FQ resistance, accounting for 86 of 166 (52%) FQ-R isolates but only 4 of 687 (0.6%) FQ-S isolates (P < .001). In contrast, the combined non-H30 ST131 subclones exhibited no such association, accounting for 2 of 166 (1.2%) FQ-R isolates and 29 of 687 (4.2%) FQ-S isolates (P > .10).
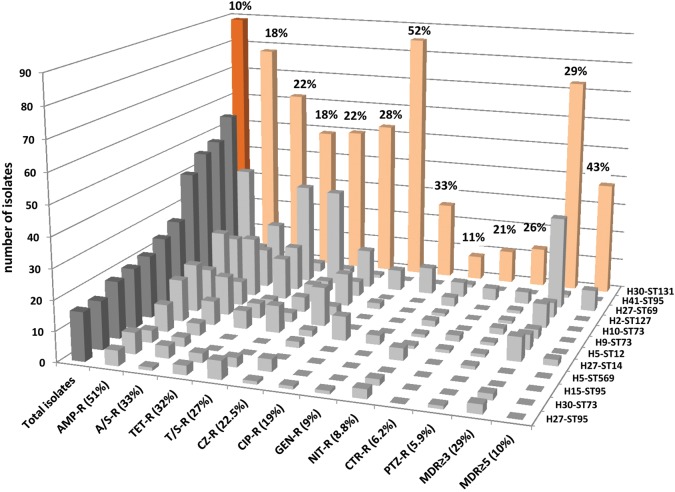
Overall, based on fimH typing (combined with MSLT), 185 total H subclones were identified among the 853 recent clinical E. coli isolates. The H30 ST131 subclone was by far the most prevalent (10.4% overall), followed only distantly by H subclones from historically dominant lineages such as ST95, ST69, ST127, and ST73 (Figure 5). The H30 subclone also dominated for resistance to each studied antimicrobial, for example, FQs (52% H30) and ceftriaxone (21% H30), and for multidrug resistance (Figure 5).
Figure 5.
Prevalence of the H30 ST131 subclone among 853 recent clinical Escherichia coli isolates. Orange, H30 ST131 subclone. Gray, 11 most prevalent non-ST131 H subclones. Each H subclone is labeled along the Z-axis (right side of chart) with its fimH allele and ST number. “Total isolates” columns (at left) are darker than the rest. Antimicrobials are listed along the X-axis (front of chart) in descending order of resistance prevalence (shown in parentheses). Y-axis (vertical) gives number of total or resistant isolates. Percentage numbers above the pink H30 columns indicate the H30 subclone's relative prevalence within each category (total, or specific resistance phenotypes). Abbreviations: AMP, ampicillin; A/S, ampicillin/sulbactam; CIP, ciprofloxacin; CZ, cefazolin; GM, gentamicin; NIT, nitrofurantoin; PTZ, piperacillin/tazobactam; TET, tetracycline; T/S, trimethoprim/sulfamethoxazole. No imipenem resistance was detected. MDR, multidrug resistant (ie, to ≥3 or ≥5 classes, with penicillins and cephalosporins counted separately).
DISCUSSION
Here we show that, despite the repeated independent emergence of FQ resistance within E. coli, most current FQ-R E. coli clinical isolates appear to have originated about a decade ago from a single strain that emerged within ST131 as the fimH-based H30 subclone. This subclone expanded rapidly to become what is now the dominant and most extensively multidrug-resistant lineage of extraintestinal pathogenic E. coli worldwide. Horizontal transfer and recombination involving gyrA and parC were newly identified as a widespread mechanism for acquisition of chromosomal FQ resistance in E. coli, including ST131. However, these genetic mechanisms have not affected the dominating H30 subclone of ST131.
Although MLST has become the preferred method for clonal typing of bacterial pathogens, MLST-based clonal lineages (ie, STs) of E. coli commonly comprise highly heterogeneous strains. For example, ST73, a major ST among extraintestinal pathogenic E. coli, also includes nonpathogenic strains Nissle1917 and ABU83972, which are used as probiotics [18, 21]. Recently, we showed that STs can be divided into ecologically distinct subclones by sequencing an internal region of fimH [18]. This gene, encoding the type 1 fimbrial adhesin, is subject to more rapid evolution and horizontal gene transfer than the traditional MLST loci, which are expected to evolve neutrally [22]. Our results suggest that FQ resistance, a hallmark phenotype of (clonally diverse) ST131, is associated almost exclusively with a single fimH-based subclone, H30, carrying a distinct gyrA and parC allele combination. The close relatedness of H30 isolates strongly suggests that, remarkably, the H30 subclone—and, thus, most current FQ-R E. coli—arose from a single strain as recently as 1 decade ago. Notably, the single-strain origin of H30 subclone was more apparent from sequence analysis of gyrA and parC (which are widely separated on the chromosome) than from PFGE analysis, consistent with the limited phylogenetic validity of PFGE [7].
The finding of a predominantly single-strain origin of FQ resistance within ST131 and E. coli generally is quite surprising. Indeed, FQ resistance can potentially emerge in any E. coli strain by appropriate, analogous point mutations in the QRDR of the ubiquitous housekeeping genes gyrA and parC [6, 23]. Our study confirms this and newly documents that FQ resistance-conferring gyrA and parC alleles also exhibit extensive horizontal mobility, thereby disseminating FQ resistance rapidly among different E. coli STs. Even within ST131, FQ resistance-conferring parC alleles have entered multiple times via horizontal transfer, as illustrated in Figure 4. Nevertheless, a single allelic combination—gyrA1AB/parC1aAB – has achieved predominance within ST131 and, consequently, the species overall.
The observed tight linkage between a single gyrA/parC combination and the H30 ST131 subclone conceivably could be due to this lineage's superior FQ resistance, commensal fitness, and/or pathogenicity, compared with other E. coli. Greater FQ resistance, if present, could represent an effect of the distinctive Glu-84-Val parC replacement mutation, which occurs in ST131's hallmark parC1aAB allele along with the widespread Ser-80-Leu mutation. Further, although plasmid-borne FQ resistance and up-regulated efflux pumps are uncommon in ST131 [15, 16], other traits of the H30 subclone conceivably could augment its resistance, for example, by blocking FQ entry or increasing intracellular FQ inactivation [6]. Comparative assessment of FQ minimum inhibitory concentrations among FQ-R ST131 vs non-ST131 E. coli isolates is needed.
Regarding commensalism, resistance-conferring mutations in housekeeping genes may decrease overall bacterial fitness when antimicrobial selective pressure is absent, unless compensatory mutations have coevolved with the resistance [24], possibly resulting in a competitive advantage in the commensal niche over other E. coli. In this regard, the H30 subclone's broad host range may also promote its dissemination through the population, both within and among locales [25].
Finally, the H30 subclone might be more fit in the pathogenic niche, and this, in combination with FQ resistance, could underlie its rapid rise to clinical dominance over other E. coli. The current dominance of the H30 subclone clearly surpasses such common lineages as ST69 (“clonal group A”; associated with trimethoprim-sulfamethoxazole resistance [7]) and classic extraintestinal pathogenic lineages ST95, ST73, and ST127. We are currently exploring the successful features of the H30 subclone through comparative genomics and molecular epidemiological surveys.
Recognition of the H30 ST131 subclone has direct clinical implications. First, rapid diagnostics that detect this single-strain lineage, analogous to those currently used for rapid detection of methicillin-resistant Staphylococcus aureus, could allow better targeted selection of empirical antimicrobial therapy for patients in whom FQ-R E. coli are of concern [5]. Second, identification of relevant reservoirs and transmission pathways of the H30 subclone, and development of effective interventions against them, could limit its spread [3]. Third, development of an effective vaccine directed toward H30 subclone-associated antigens (eg, surface-exposed virulence factors, and/or the O25 somatic antigen or H4 flagellar antigen) could help protect at-risk hosts [11, 26]. Thus, focused attention to the highly successful H30 ST131 subclone can potentially lead to substantially improved prevention and management of FQ-R E. coli infections.
Study limitations included the exclusive focus on gyrA/parC-mediated FQ resistance, the use of convenience sample historical isolates (albeit broadly distributed by year, locale, and source/host), and the current isolates’ geographical restriction to 2 US cities (albeit from 5 separate institutions serving distinct patient populations). Strengths include the large and diverse study population for comparison of ST131 and non-ST131 E. coli, the use of multiple advanced molecular modalities to define sub-ST131 clonal structure, the clonal phylogenetic analysis of mutations in gyrA and parC, and the clonal assessment of resistance.
In summary, we provide evidence suggesting that the major source of the current FQ resistance epidemic in E. coli is the rapid clonal expansion of a single strain within ST131, which over the past decade has become the most successful lineage of extraintestinal E. coli overall, particularly of multidrug-resistant E. coli. Although confirmation of these findings in other locales and host populations is needed, this discovery provides a novel perspective on the E. coli antimicrobial resistance epidemic, with profound implications and exciting opportunities.
Notes
Acknowledgments. Ruth Anway (Minneapolis VA Medical Center) performed medical record abstraction. Dave Prentiss prepared the dendrogram image. Alena Gileva and Elena Linardopoulou provided excellent technical assistance. Sandip Paul provided computational assistance. Drs. Steve Moseley and Oliwia Zurek provided critical reading and discussion of the manuscript. Walter E. Stamm, MD (deceased) provided formative inspiration, guidance, and support.
Additional providers of historic ST131 isolates and associated data were the RESIST (“Resistance in E. coli ST131 as Investigated Using Sequence Typing”) Study Group members, who included: Jo Ellen Abraham (Abbott Northwestern Hospital, Minneapolis, MN), Javier Adachi (The University of Texas M. D. Anderson Cancer Center, Houston, TX), Aristides Assimacopoulos (University of South Dakota, Sioux Falls, SD), Robert L. Bergsbaken (Health Partners and Regions Medical Center, St. Paul, MN), Mariana Castanheira (JMI Laboratories, North Liberty, IA), Timothy Cleary (University of Miami, Miami, FL), Michael Cooperstock (University of Missouri School of Medicine, Columbia, MO), Yohei Doi (University of Pittsburgh, Pittsburgh, PA), Paul Edelstein (Hospital of the University of Pennsylvania, Philadelphia, PA), Nancy Hanson (Creighton University, Omaha, NE), John Holter (Veterans Affairs Medical Center, Minneapolis, MN), Thomas M. Hooton (University of Miami, Miami, FL), Michelle Hulse (Children's Hospital and Clinics of Minnesota, Minneapolis, MN; current affiliation 3M, St. Paul, MN), James H. Jorgensen (University Health System, San Antonio, TX), James S. Lewis II (University Health System, San Antonio, TX), Sybille Miller (Leesburg Veterinary Internal Medicine, Leesburg, VA), Rob Owens (Cubist Pharmaceuticals, Falmouth, ME), Elizabeth Palavecino (Wake Forest University Baptist Medical Center, Winston-Salem, NC), David L. Paterson (University of Queensland Centre for Clinical Research, Royal Brisbane and Women's Hospital Campus, Brisbane, Australia), Johann Pitout (Calgary Laboratory Services, Calgary, AB), John Quinn (Pfizer Global Research, Groton, CT), James Rice (The Scripps Research Institute, La Jolla, CA), Daniel F. Sahm (Eurofins-Medinet, Chantilly, VA), and Karen Vigil (The University of Texas Health Science Center at Houston—Medical School, Houston, TX).
Financial support. This material is based upon work supported by Office of Research and Development, Medical Research Service, Department of Veterans Affairs, Merit Review grant 1 I01 CX000192 01 (J. R. J.), and NIH ARRA award 1RC4AI092828 (E. V. S.).
Potential conflicts of interest. The following authors report these conflicts of interest: F. C. F.: Consultancy, research grants, and educational presentations with Cepheid. D. C. H.: Research grant from Rib-X Pharmaceuticals, consultancies with Fab Pharma and Pfizer, speakers bureau with Pfizer. J. R. J.: Research grants and contracts from Merck, Rochester Medical, and Syntiron; patent pending for detection of the H30 ST131 subclone. L. B. P.: Patent pending for detection of the H30 ST131 subclone. M.-H. N.-C.: Expert testimony for Region Wallon; research grant from Direction de la Recherche Clinique AP-HP; lectures for Novartis; travel reimbursement for 2012 ICAAC speaker. E. V. S.: Research grant from Group Health; patent pending for detection of the H30 ST131 subclone. D. T.: Research grant from ARC Linkage; consultancy from Pfizer and Bayer; speaker for Bayer Animal Health. C. U.: Consultancy with Pfizer, lecture/speaker for Forest Pharmaceuticals, Cubist, and Pfizer. G. Z.: Research grants from Achaogen, Merck, Pfizer, Cubist, Affinium, and Sunovion. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Gilbert DN, Moellering RCJ, Eliopoulos GM, Chambers HF, Saag MS. The Sanford Guide to Antimicrobial Therapy 2010. 40th ed. Vienna, VA: Antimicrobial Therapy, Inc; 2010. pp. 4–64. [Google Scholar]
- 2.Russo TA, Johnson JR. Medical and economic impact of extraintestinal infections due to Escherichia coli: an overlooked epidemic. Microbes Infect. 2003;5:449–56. doi: 10.1016/s1286-4579(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 3.Owens RC, Johnson JR, Stogstill P, Yarmus L, Lolans K, Quinn J. Community transmission in the United States of a CTX-M-15-producing sequence type ST131 Escherichia coli strain resulting in death. J Clin Microbiol. 2011;49:3406–8. doi: 10.1128/JCM.00993-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tufan Z, Bulut C, Yazan T, et al. A life-threatening Escherichia coli meningitis after prostate biopsy. Urol J. 2011;8:69–71. [PubMed] [Google Scholar]
- 5.Gupta K, Hooton TM, Naber KG, et al. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: a 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis. 2011;52:e103–20. doi: 10.1093/cid/ciq257. [DOI] [PubMed] [Google Scholar]
- 6.Hooper DC. Emerging mechanisms of fluoroquinolone resistance. Emerg Infect Dis. 2001;7:337–41. doi: 10.3201/eid0702.010239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson JR, Menard M, Johnston B, Kuskowski MA, Nichol K, Zhanel GG. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002–2004. Antimicrob Agents Chemother. 2009;53:2733–9. doi: 10.1128/AAC.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cagnacci S, Gualco L, Debbia E, Schito GC, Marchese A. European emergence of ciprofloxacin-resistant Escherichia coli clonal groups O25:H4-ST 131 and O15:K52:H1 causing community-acquired uncomplicated cystitis. J Clin Microbiol. 2008;46:2605–12. doi: 10.1128/JCM.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States (2007) Clin Infect Dis. 2010;51:286–94. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 10.Peirano G, Pitout JDD. Molecular epidemiology of Escherichia coli producing CTX-M β-lactamases: the worldwide emergence of clone ST131 O25:H4. Int J Antimicrob Agents. 2010;35:316–21. doi: 10.1016/j.ijantimicag.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother. 2011;66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 12.Nicolas-Chanoine M-H, Blanco J, Leflon-Guibout V, et al. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother. 2008;61:273–81. doi: 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 13.Johnson JR, Nicolas-Chanoine M, Debroy C, et al. Comparison of Escherichia coli sequence type ST131 pulsotypes by epidemiologic traits, 1967–2009. Emerg Infect Dis. 2012;18:598–607. doi: 10.3201/eid1804.111627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee MY, Choi HJ, Choi JY, et al. Dissemination of ST131 and ST393 community-onset, ciprofloxacin-resistant Escherichia coli clones causing urinary tract infections in Korea. J Infection. 2010;60:146–53. doi: 10.1016/j.jinf.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Cerquetti M, Guifre M, Garcia-Fernandez A, et al. Ciprofloxacin-resistant, CTX-M-15-producing Escherichia coli ST131 clone in extraintestinal infections in Italy. Clin Microbiol Infect. 2010;16:1555–8. doi: 10.1111/j.1469-0691.2010.03162.x. [DOI] [PubMed] [Google Scholar]
- 16.Leflon-Guibout V, Jurand C, Bonacorsi S, et al. Emergence and spread of three clonally related virulent isolates of CTX-M-15-producing Escherichia coli with variable resistance to aminoglycosides and tetracycline in a French geriatric hospital. Antimicrob Agents Chemother. 2004;48:3736–42. doi: 10.1128/AAC.48.10.3736-3742.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Platell JL, Cobbold RN, Johnson JR, et al. Commonality among fluoroquinolone-resistant sequence type ST131 extraintestinal Escherichia coli isolates from humans and companion animals in Australia. Antimicrob Agents Chemother. 2011;55:3782–7. doi: 10.1128/AAC.00306-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weissman SJ, Johnson J, Tchesnokova V, et al. High-resolution two-locus clonal typing of extraintestinal Escherichia coli. Appl Environ Microbiol. 2012;78:1353–60. doi: 10.1128/AEM.06663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swofford DL. PAUP* phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- 20.Clinical and Laboratory Standards Institute. M100-S20: Performance standards for antimicrobial susceptibility testing. Wayne, PA: Clinical and Laboratory Standards Institute; 2010. 20th informational supplement. [Google Scholar]
- 21.Salvador E, Wagenlehner F, Köhler CD, et al. Comparison of asymptomatic bacteriuria Escherichia coli isolates from healthy individuals versus those from hospital patients shows that long-term bladder colonization selects for attenuated virulence phenotypes. Infect Immun. 2012;80:668–78. doi: 10.1128/IAI.06191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weissman SJ, Chattopadhyay S, Aprikian P, et al. Clonal analysis reveals high rate of structural mutations in fimbrial adhesins of extraintestinal pathogenic Escherichia coli. Mol Microbiol. 2006;59:975–88. doi: 10.1111/j.1365-2958.2005.04985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hooper DC. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin Infect Dis. 2000;31(Suppl 2):S24–8. doi: 10.1086/314056. [DOI] [PubMed] [Google Scholar]
- 24.Schrag SJ, Perrot V, Levin BR. Adaptation to the fitness costs of antibiotic resistance in Escherichia coli. Proc Biol Sci. 1997;264:1287–91. doi: 10.1098/rspb.1997.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Platell JL, Johnson JR, Cobbold RN, Trott DJ. Multidrug-resistant extraintestinal pathogenic Escherichia coli of sequence type ST131 in animals and foods. Vet Microbiol. 2011;153:99–108. doi: 10.1016/j.vetmic.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Russo TA, Johnson JR. Extraintestinal isolates of Escherichia coli: identification and prospects for vaccine development. Expert Rev Vaccines. 2006;5:45–54. doi: 10.1586/14760584.5.1.45. [DOI] [PubMed] [Google Scholar]