Abstract
Actin filament (F-actin) is believed to be involved in measles virus (MV) assembly as a cellular factor, but the precise roles remain unknown. Here we show that Phe at position 50 of the MV matrix (M) protein is important for its association with F-actin, through which the function of the M protein is regulated. In plasmid-expressed or MV-infected cells, a coimmunoprecipitation study revealed that the wild-type M (M-WT) protein associated strongly with F-actin but only weakly with the cytoplasmic tail of the hemagglutinin (H) protein. Since the F50P mutation allowed the M protein the enhanced interaction with the H protein in return for the sharply declined association with F-actin, the mutant M (M-F50P) protein strongly inhibited MV cell-cell fusion and promoted the uptake of the H protein into virus particles. The abundantly incorporated H protein resulted in the increase in infectivity of the F50P virus, although the virus contained a level of genome RNA equal to that of the WT virus. When the structure of F-actin was disrupted with cytochalasin D, the M-WT protein liberated from F-actin interacted with the H protein as tightly as the M-F50P protein, suppressing cell-cell fusion and promoting virus assembly comparably efficiently as the M-F50P protein. The cell-cell fusion activity of the WT virus appeared to be upheld by F-actin, which prevents the M protein interaction with the H protein. Our results indicate that F-actin in association with the M protein alters the interaction between the M and H proteins, thereby modulating MV cell-cell fusion and assembly.
INTRODUCTION
Measles virus (MV), a member of the genus Morbillivirus in the family Paramyxoviridae, is an enveloped virus with a nonsegmented negative-sense RNA genome of about 16,000 nucleotides. The genomic RNA, encapsidated by the nucleocapsid (N) protein and associated with a virus RNA-dependent RNA polymerase composed of two subunits, the phospho (P) and large (L) proteins, forms a helical ribonucleoprotein complex (RNP) core of the virus (1). Each virus particle maintains its structure with the matrix (M) protein underlying the lipid bilayer of the envelope on which glycoprotein spikes, the hemagglutinin (H) and fusion (F) proteins, are protruding (2). The initial step of MV infection is attachment to cells via binding of the H protein to cellular receptors (3). Clinical isolates of MV utilize the signaling lymphocyte activation molecule (SLAM; also known as CD150) and recently identified nectin4 (PVRL4) as receptors to infect certain types of cells or organs of the immune system (4, 5) and epitherial cells or tissues (6, 7), respectively. Binding of the H protein to a receptor triggers fusion of the virus envelope with the plasma membrane (envelope fusion), a process mediated by the F protein (8), releasing RNP into the cytoplasm. RNP then initiates the transcription of virus mRNAs, and as synthesized virus proteins accumulate, the same RNP comes to be used as the template for synthesis of a positive-strand antigenome, from which virus genome RNA is replicated.
MV spreads in cell cultures or animal tissues in two ways, either by producing progeny virus particles that undergo multiple cycles of infection via envelope fusion or by fusing the infected cells directly with neighboring uninfected cells, forming syncytium (cell-cell fusion). Contrary to other members of Paramixoviridae that perform proper budding processes, release of the infectious progeny particles of MV is inefficient, and most of the particles remain in cell-associated form (9). Therefore, cell-cell fusion might be required for successful systemic dissemination as well as pathogenicity of MV in vivo (5, 10). Since the M protein lines the inner surface of the plasma membrane and releases viruslike particles (VLPs) when expressed independently of other virus components, it is believed that its function is intrinsically required for virus particle formation (11, 12). In addition, the M protein plays a key role in accurate virus assembly to produce infectious progeny virus by concentrating the H and F proteins, as well as the RNP (13–15), at the budding sites on the plasma membrane of the MV-infected cells. The H and F proteins are connected to the internal proteins of virus particles through interaction of their cytoplasmic tails with the M protein (16–18). On the other hand, for induction of cell-cell fusion, expression of the H and F proteins alone is sufficient (8), and the M protein inhibits cell-cell fusion by the interaction with the cytoplasmic tails of the H and F proteins (19–22). Subacute sclerosing panencephalitis (SSPE) is a neurodegenerative disease caused by persistent infection of SSPE virus, the highly mutated virus derived from MV. Abrogation of the M protein function due to the accumulated mutations (23) and shortened glycoprotein tails (24) have been postulated to account for loss of proper particle assembly and extensive cell-cell fusion of SSPE virus by limiting the interaction between the M and glycoproteins. Involvement of the interaction between the M protein and the cytoplasmic tails of envelope glycoproteins in cell-cell fusion as well as in the particle production has been reported for other paramyxoviruses (25–30).
Previous studies have demonstrated that actin, a cellular cytoskeleton component, is packaged in the completed MV particles (31, 32). Actin filaments were found to interact with the majority of MV structural proteins in the infected cells (33, 34) and appeared to be in close association with RNP in virus particles during the budding process (35). Since inhibitors of actin polymerization, such as cytochalasin B (Cyt-B) and Cyt-D and latrunculin A, restricted the growth of MV during the budding process, filamentous actin (F-actin) is considered to be involved in MV maturation (31, 36–38). However, the precise roles of F-actin in correlation with MV proteins remain to be elucidated. Actin has also been reported to be related in the maturation of other paramyxoviruses, such as Sendai virus, canine distemper virus, respiratory syncytial virus, and Newcastle disease virus (39, 40, 42–45, 58), and the direct specific binding of actin to the M protein was demonstrated for Sendai virus (46). Whether the interaction of the M protein with the cytoplasmic tails of the glycoproteins or the RNP is affected by F-actin is unknown.
Recently, we isolated an MV variant lacking syncytium formation from a clinical strain. The variant possessed a single point substitution, F50P, in the M protein, and the mutant M protein had lost the ability of the wild-type M protein to associate efficiently with F-actin. In this study, we show that F-actin interferes with the interaction of the M protein with the cytoplasmic tail of the H protein, thereby modulating the function of the M protein in virus cell-cell fusion and assembly.
MATERIALS AND METHODS
Cells and viruses.
Vero cells constitutively expressing human SLAM (Vero/hSLAM) (a gift from Y. Yanagi) (47) and HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Nissui Pharmaceutical, Tokyo, Japan) supplemented with 7% fetal bovine serum (FBS). 293T and B95a (a marmoset B-cell line transformed with Epstein-Barr virus) (9) cells were maintained in high-glucose DMEM (Wako Pure Chemical, Osaka, Japan) supplemented with 10% FBS. BHK cells constitutively expressing T7 RNA polymerase (BHK/T7-9) (a gift from N. Ito and M. Sugiyama) (48) were maintained in RPMI medium (Wako Pure Chemical) supplemented with 8% FBS and 0.6 mg/ml hygromycin B.
The kyNS strain of MV is a clinical virus of genotype H1 isolated from a measles patient and passaged once in B95a cells. The cloned viruses were obtained from the monolayer culture of Vero/hSLAM cells inoculated with the kyNS strain by plaque purification, picking either one of the syncytia (clone 1) or blindly the cells without syncytium (clone 2). The recombinant MVs (rMVs) were generated from cDNAs according to the method reported by Seki et al. (49) by transfecting BHK/T7-9 cells with the full-length genome plasmids of MV described below and three support plasmids, pCITE-IC-N, pCITE-IC-PΔC, and pCITEko9301B-L (gifts from M. Takeda). The generated rMVs were propagated in B95a cells.
Plasmid constructions.
The plasmid p(+)MV323-EGFP (a gift from Y. Yanagi) encodes the full-length antigenomic cDNA of the B95a-isolated MV IC-B strain (50) with an additional transcriptional unit of the enhanced green fluorescence protein (EGFP) (51). We eliminated the SacII site at nucleotide (nt) 2898 and the SalI site at nt 7129 of p(+)MV323-EGFP by introducing g2901a and c7134t substitutions. As the result, single amino acid substitution A72E in the C protein occurred in p(+)MV323c72-EGFP, the modified p(+)MV323-EGFP. In this paper, nucleotide position numbers are shown in accordance with the sequence of the IC-B strain genome (52) (GenBank accession number AB016162).
The cDNA of the kyNS strain mRNA was synthesized by reverse transcription of total RNA extracted from B95a cells infected with the kyNS strain followed by PCR with a primer pair amplifying the M gene (nt 4296 to 5303). The nucleotide sequence of the amplified M gene fragment was determined (GenBank accession number AB740967). The amplified cDNA was digested with BglII and BsaBI, and the released fragment was used to replace the BglII-BsaBI fragment (nt 4303 to 5147) of the p(+)MV323c72-EGFP, which generated the p(+)MV323c72-EGFP/M-WT(H1) plasmid. To introduce the substitution, Phe to Pro at residue 50 (F50P) of the M protein, nt 3957 to 5821 carrying nucleotide substitutions TT to CC at nt 4443 and 4444 was obtained and then digested with SalI and SacII. The released SalI-SacII fragment possessing the F50P substitution was used to replace the corresponding SalI-SacII region (nt 4222 to 5776) of p(+)MV323c72-EGFP/M-WT(H1), which generated the plasmid p(+)MV323c72-EGFP/M-F50P(H1).
To construct plasmids pcDNA-MV/M-WT and pcDNA-MV/M-F50P expressing the wild-type M (M-WT) and the mutant M (M-F50P) proteins, the M gene fragments (nt 4296 to 5303) amplified by PCR using p(+)MV323c72-EGFP/M-WT(H1) and p(+)MV323c72-EGFP/M-F50P(H1), respectively, were cloned into the pcDNA3 vector (Invitrogen Life Technologies, Carlsbad, CA). To generate plasmids expressing the H and F proteins of the IC-B strain, the H (nt 8129 to 9982) and F (nt 6316 to 7968) genes were amplified by PCR using p(+)MV323c72-EGFP as the template. The amplified H gene and F gene were cloned into the pcDNA3-myc vector and the pcDNA3 vector, which generated pcDNA-MV/H-myc and pcDNA-MV/F, respectively. The plasmid expressing the H protein with the truncated cytoplasmic tail (CT) by 20 amino acids (21), pcDNA-MV/HΔ20-myc, was generated by deleting nt 8132 to 8191. Precise experimental methods including the information of the primers are available on request.
Virus titration.
Monolayers of Vero/hSLAM cells in 24-well plates were infected with serially diluted virus samples. After 1 h of incubation at 37°C, the virus samples were removed, and the cells were overlaid with DMEM containing 7% FBS and 2.5% methylcellulose. At 3 days postinfection (p.i.), PFU was determined by counting the number of EGFP-expressing plaques under a fluorescence microscope (Axioskop; Zeiss, Goettingen, Germany).
Cell-cell fusion assay (syncytium formation of rMVs).
Vero/hSLAM cells cultured in 24-well plates were infected with the MV at a multiplicity of infection (MOI) of 0.001 and incubated for 30 h at 37°C. When cells were treated with cytochalasin D (Cyt-D) (Sigma-Aldrich, St. Louis, MO), the culture medium was replaced with medium including 2 μM Cyt-D or dimethyl sulfoxide (DMSO) only as a control at 12 h p.i. The cells were then fixed by 1% paraformaldehyde, permeabilized with 1% Triton X-100, and stained with Hoechst 33342 (Sigma-Aldrich). The number of nuclei in an EGFP-expressing syncytium was counted under a fluorescence microscope.
Quantitative cell-cell fusion assay.
Subconfluent monolayers of 293T cells in 6-well plates were transfected with 0.5 μg of an H protein-expressing plasmid (pcDNA-MV/H-myc or pcDNA-MV/HΔ20-myc), 0.5 μg of the F protein-expressing pcDNA-MV/F, 0.5 μg of an M protein-expressing plasmid (pcDNA-MV/M-WT or pcDNA-MV/M-F50P), or the pcDNA3 vector, together with 0.5 μg of T7 RNA polymerase-expressing pCAG-T7 (41) per well, using polyethyleneimine (Polysciences, Warrington, PA). At 12 h posttransfection, culture medium was replaced by medium including 2 μM Cyt-D or DMSO only. At 24 h posttransfection, the culture medium was removed and the cells were dispersed in 1 mM EDTA/phosphate-buffered saline (PBS) solution and transferred onto the 293T cells that had been transfected 24 h before with SLAM-expressing pCA7-SLAM (a gift from Y. Yanagi) (1 μg) and pTM1-EGFP (1 μg) that expresses EGFP only under the T7 RNA polymerase. At 48 h after cocultivation, the cells were fixed with 1% paraformaldehyde, and the cells expressing EGFP as the result of cell-cell fusion were counted under a fluorescence microscope.
VLP budding assay.
Subconfluent monolayer cultures of 293T cells in 6-well plates were transfected with 1 μg of an M protein-expressing plasmid (pcDNA-MV/M-WT or pcDNA-MV/M-F50P) together with 1 μg of an H protein-expressing plasmid (pcDNA-MV/H-myc or pcDNA-MV/HΔ20-myc) or pcDNA3-myc vector. At 12 h posttransfection, the culture medium was replaced with the medium including 2 μM Cyt-D or DMSO only. At 48 h posttransfection, the cell debris was removed from the culture medium by centrifugation at 3,000 × g for 10 min at 4°C, and the supernatant was loaded onto a 20% sucrose layer. The VLPs were purified as the pellets by passing through the 20% sucrose layer (134,000 × g for 2 h at 4°C); suspended in hypotonic buffer consisting of 1.5 mM MgCl2, 10 mM HEPES-NaOH (pH 7.5), 10 mM KCl, and 0.5 mM dithiothreitol; and concentrated by adding 20% trichloroacetic acid (TCA). The cells were solubilized with the lysis buffer consisting of 0.5% Triton X-100, 0.5% deoxycholate, 10 mM Tris-HCl (pH 7.5), and 5 mM NaCl (57). The VLPs and the cell extracts were resuspeded in SDS loading buffer (25 mM Tris-HCl [pH 6.8], 1% 2-mercaptoethanol, 1% SDS, 0.05% bromophenol blue, and 5% glycerol), fractionated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) in a 12% polyacrylamide gel, and blotted onto polyvinylidene disulfide (PVDF) membranes (PVDF-Plus; GE Health Care, Waukesha, WI). The membranes were incubated with mouse monoclonal antibody against MV M protein (Merck Millipore, Billerica, MA), rabbit polyclonal antibody against myc tag (Cell Signaling Technology, Danvers, MA), and mouse monoclonal antibody against β-actin (Cell Signaling Technology), followed by incubation with horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (IgG; Santa Cruz Biotechnology, Santa Cruz, CA) or HRP-conjugated goat anti-rabbit IgG (Cell Signaling Technology) for detection of the M and H proteins or cellular actin, respectively. Proteins were visualized using the ECL Plus Western blotting detection system (GE Healthcare) by exposure to an autoradiography film (Fuji Film, Tokyo, Japan).
Immunoprecipitation and Western blot analyses.
Subconfluent monolayer cultures of 293T cells in 6-well plates were transfected with pcDNA-MV/M-WT or pcDNA-MV/M-F50P (1 μg) with or without an H protein-expressing plasmid (pcDNA-MV/H-myc or pcDNA-MV/HΔ20-myc) (1 μg). At 12 h posttransfection, the culture medium was replaced by the medium including 2 μM Cyt-D or DMSO only. At 24 h posttransfection, cells were suspended in 0.1 M HEPES-NaOH (pH 7.5) including cross-linking reagent dithiobis(succinimidylpropionate) (DSP; Thermo Fisher Scientific, Waltham, MA) followed by incubation for 2 h at 4°C, and the cross-linking reaction was stopped by 10 mM Tris-HCl (pH 7.5). The cells were solubilized with 1.5 ml of the lysis buffer (composition identical to that described above), and the cell extracts were used for immunoprecipitation after centrifugation at 13,000 × g for 10 min at 4°C. To prepare the Dounce-homogenized cell extracts, 293T cells transfected and incubated equally as described above were suspended in 1 ml of hypotonic buffer (composition identical to that described above) and homogenized using a Dounce homogenizer. Cell extracts for immunoprecipitation were prepared by centrifugation of the homogenized cells at 1,000 × g for 7 min at 4°C to remove unhomogenized cells and debris. When the MV-infected cells were analyzed, monolayers of Vero/hSLAM cells in 6-well plates were infected with rMVs at an MOI of 0.001, and the culture medium was replaced with medium including 2 μM Cyt-D or DMSO only at 12 h p.i. After further incubation for 60 h at 37°C, the cells were treated with DSP for 2 h at 4°C and solubilized with lysis buffer to prepare the cell extracts as described above.
A small amount (28 μl) of each cell extract was mixed with SDS loading buffer as the sample for total cell extract. The rest of the extract was incubated for 1 h at 4°C with protein G-conjugated agarose beads (KPL, Washington, DC) which had been preincubated with an anti-MV M protein mouse monoclonal antibody overnight at 4°C. The immune complexes were obtained by centrifugation and washed with the lysis buffer, and the precipitated proteins as well as the total cell extracts were subjected to SDS-PAGE using 12% polyacrylamide gels followed by electroblotting onto PVDF membranes. The proteins were detected by incubating the membranes with one of these antibodies: mouse monoclonal antibody against MV M protein, rabbit polyclonal antibody against MV H protein (a gift from T. Kohama), rabbit polyclonal antibody against myc tag, mouse monoclonal antibody against β-actin, rabbit polyclonal antibody against β-tubulin (Thermo Fisher Scientific), and mouse monoclonal antibody against golgin97 (Molecular Probes, Eugene, OR) (54), followed by incubation with either the HRP-conjugated secondary antibody, goat anti-mouse IgG, or goat anti-rabbit IgG. Proteins were visualized using the ECL Plus Western blotting detection system by exposure to an autoradiography film.
Quantification of genome RNA in MV particle.
Virus genome RNA was extracted from virus particles, and cDNA was prepared by reverse transcription using the primer MV35F (TAAGGATAGATCAATCAA) that anneals the leader sequence of the MV genome RNA. MV genome was quantified amplifying N gene by PCR (15, 20, 25, and 30 cycles) with a set of primers, MV1099F (AATTCCTGGAGATTCCTCAA) and MV1166R (TTAGTGCCCCTGTTACTTT), followed by determination of the intensity of each DNA band using ImageJ software (http://rsbweb.nih.gov/ij/index.html).
Indirect immunofluorescence staining.
HeLa cells were seeded on microcoverslips (Matsunami, Osaka, Japan) in a 24-well plate and transfected with 0.5 μg of an M protein-expressing plasmid (pcDNA-MV/M-WT or pcDNA-MV/M-F50P) and 0.5 μg of an H protein-expressing plasmid (pcDNA-MV/H-myc) independently or in combination. At 12 h posttransfection, culture medium was replaced by medium including 2 μM Cyt-D or DMSO only, and at 24 h or 48 h posttransfection, the cells were fixed with 1% paraformaldehyde and permeabilized with 1% Triton X-100. The cells were then incubated with mouse monoclonal antibody against MV M protein and rabbit polyclonal antibody against myc tag, followed by incubation with fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse IgG (MBL, Nagoya, Japan) and Texas Red-conjugated goat anti-rabbit IgG (Vector Laboratories, Burlingame, CA) as the secondary antibody. When F-actin was visualized with FITC-conjugated phalloidin (Sigma-Aldrich), Texas Red-conjugated goat anti-mouse IgG (Rockland, Gilbertsville, PA) was used as the secondary antibody to detect the M protein. The stained cells were observed using a confocal microscope (FV1000; Olympus, Tokyo, Japan). For detection of the kyNS strain-infected cells, monolayers of Vero/hSLAM cells in 24-well plates inoculated with the kyNS strain were fixed with 1% paraformaldehyde and permeabilized with 1% Triton X-100 at 72 h p.i. The cells were incubated with mouse monoclonal antibody against MV N protein (Merck Millipore), followed by incubation with FITC-conjugated rabbit anti-mouse IgG.
RESULTS
A single amino acid substitution in the M protein, F50P, is responsible for the defect of MV syncytium formation.
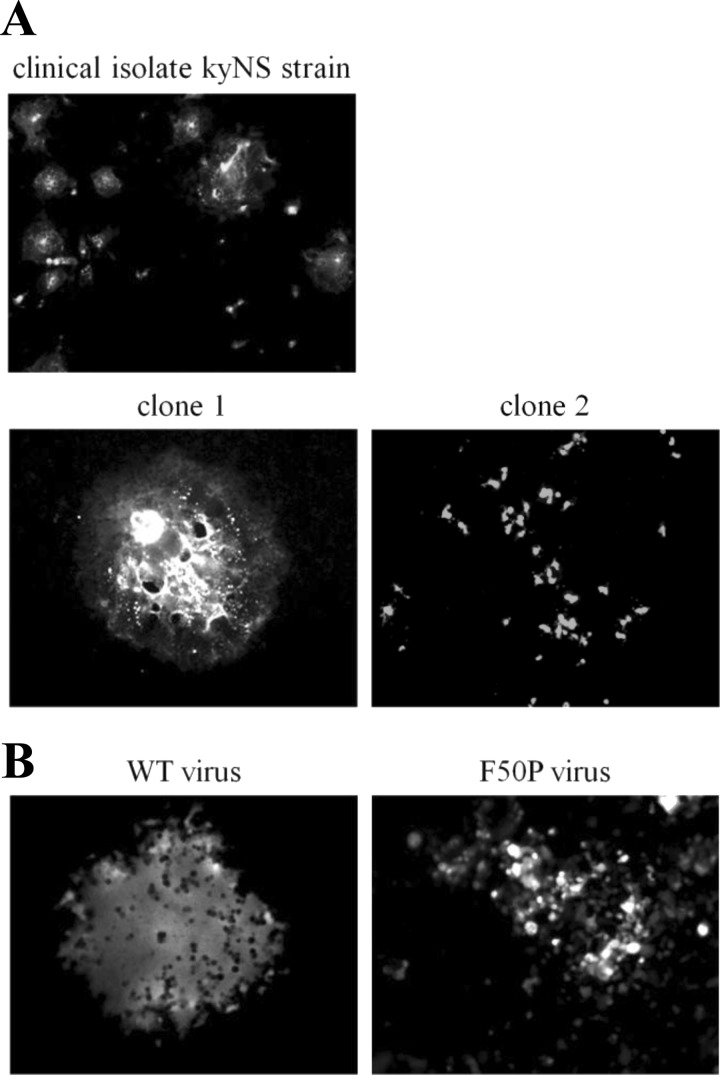
Shortly after the isolation of the kyNS strain of MV, we noticed from its cytopathic effect (CPE) that the virus was a mixture of some variants displaying different cell-cell fusion activities. By plaque purification, therefore, we obtained two clones from the kyNS strain: clone 1, forming large syncytia typical for the MV clinical viruses, and clone 2, lacking syncytium formation (Fig. 1A). Then we compared the amino acid sequences of the M, F, and H proteins that regulate cell-cell fusion of MV (20) between the two clones and found a single point substitution in the M protein. Whereas Phe at residue 50 of clone 1 was commonly observed, Pro of clone 2 was rarely found in the wild-type M protein sequences published and available in databases. To certify the effect of the F50P substitution, the recombinant MV (rMV) possessing either the wild-type or mutant M (M-WT or M-F50P) protein was generated (here called the WT virus or the F50P virus). Figure 1B demonstrates that the F50P substitution is responsible for the defect of syncytium formation of clone 2. Position 50 in the M protein had not been reported and appeared to be new as the site that regulates MV cell-cell fusion.
Fig 1.
The F50P substitution in the M protein is responsible for the defect of MV cell-cell fusion. (A) CPE of the clinical strain kyNS and clones 1 and 2 isolated from the kyNS strain. Vero/hSLAM cells inoculated with each virus were fixed and permeabilized at 72 h p.i. Virus-infected cells were visualized by incubating the cells with anti-MV N protein monoclonal antibody followed by FITC-conjugated rabbit anti-mouse IgG. Magnification of the photographs of the clones is twice (×200) of that of the kyNS strain (×100). (B) CPE of the F50P virus. B95a cells were infected with the WT or F50P virus and fixed at 72 h p.i. Infected cells expressing EGFP were observed under a fluorescence microscope. Magnification, ×200.
The M-F50P protein tightly interacts with the H protein at the Golgi apparatus.
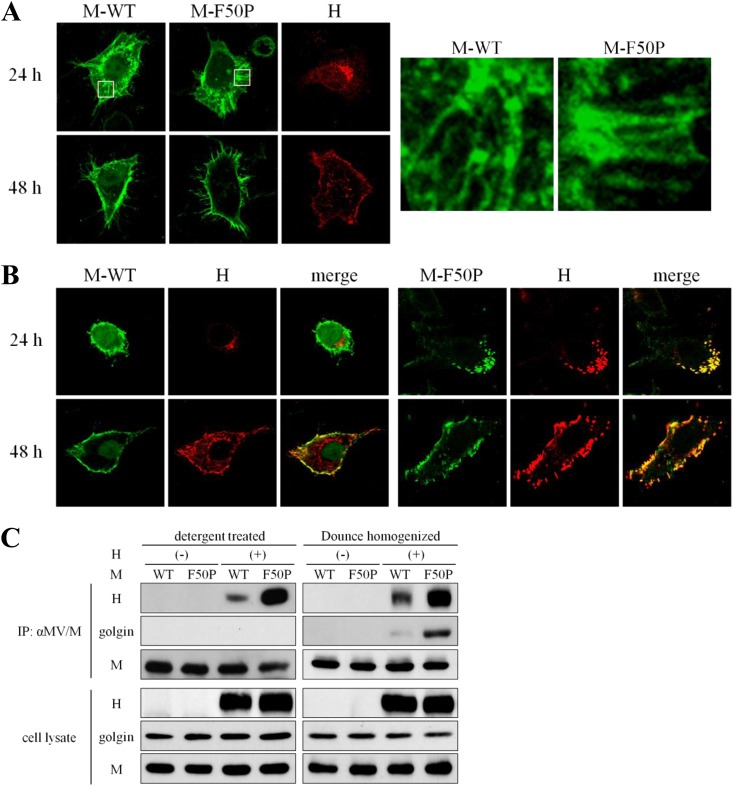
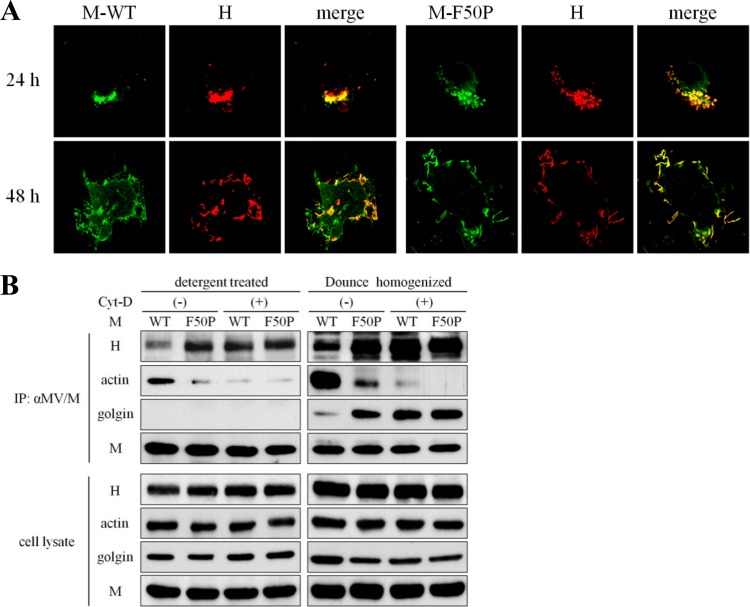
Cathomen et al. (20) showed that the interaction of the M protein with the envelope glycoproteins strongly affected cell-cell fusion of the MV Edmonston strain. Of the two glycoproteins, it was reported that the interaction of the M protein with the H protein could alter cell-cell fusion more significantly (55). Therefore, to study the mechanism by which the F50P substitution conferred the defect of cell-cell fusion on MV, we first examined the interaction of each M protein, the M-WT or M-F50P protein, with the H protein. When expressed alone, both the M-WT and M-F50P proteins were distributed, forming filamentous structure equally in the cytoplasm at 24 h posttransfection. The H glycoprotein localized in close proximity to the nuclei, presumably at the Golgi apparatus, which was quite different from the filamentous fashion of the M proteins. All the three proteins migrated to the cell surface thereafter (Fig. 2A). When coexpressed with the H protein, the M-WT protein showed the filamentous distribution, similar to that observed in the single expression, which was independent of the localization of the H protein at 24 h posttransfection. At 48 h posttransfection, the H protein spread throughout the cell and partly accumulated on the plasma membrane where the M-WT protein was concentrated predominantly, but coexpression of the H protein did not cause an apparent redistribution of the M-WT protein. In contrast, the M-F50P protein strongly colocalized with the H protein at 24 h posttranfection in the perinuclear region and seemed to move together toward the cell surface thereafter (Fig. 2B).
Fig 2.
Interaction of the M-F50P protein with the H protein. (A) Filamentous distribution of the M protein expressed alone. HeLa cells were transfected with a plasmid expressing the M-WT, M-F50P, or the H-myc protein separately and were fixed and permeabilized at 24 or 48 h posttransfection. Each protein was visualized by incubating the cells with the mouse monoclonal antibody against the M protein followed by the FITC-conjugated rabbit anti-mouse IgG or with the rabbit anti-myc tag antibody followed by the Texas Red-conjugated goat anti-rabbit IgG. Observation was performed under a confocal microscope, and images of 13 compiled sections are shown. Filamentous appearance of the M protein in the boxed area is enlarged on the right side. (B) Colocalization of the M-F50P protein with the H protein. HeLa cells were transfected with a plasmid expressing the M-WT or M-F50P protein together with the H-myc protein-expressing plasmid. At 24 or 48 h posttransfection, the cells were fixed, permeabilized, and incubated with the mouse monoclonal antibody against the M protein and the rabbit polyclonal antibody against the myc tag as the first antibodies and then FITC-conjugated rabbit anti-mouse IgG and Texas Red-conjugated goat anti-rabbit IgG as the second antibodies. Observation was performed under a confocal microscope. (C) Efficient coprecipitation of the H protein with the M-F50P protein. 293T cells were transfected with a plasmid expressing the M-WT or M-F50P protein without (−) or together with (+) a plasmid expressing the H-myc protein. At 24 h posttransfection, cells were solubilized with detergent solution after the incubation with cross-linking reagent DSP for 2 h (detergent treated) or homogenized using a Dounce homogenizer (Dounce homogenized). The M proteins were precipitated with anti-M antibody, and coprecipitated proteins were analyzed by immunoblotting. The M and H proteins and golgin were detected using mouse monoclonal antibody against MV M protein, rabbit polyclonal antibody against myc tag, and mouse monoclonal antibody against golgin97 as the first antibodies, respectively.
Then immunoprecipitation using the extract of the cells coexpressing either the M-WT or M-F50P protein with the H protein was performed. As shown in Fig. 2C, anti-M antibody coprecipitated much more of the H protein with the M-F50P protein than with the M-WT protein. In an immunoprecipitation study using the cell extracts prepared by homogenizing the cells mechanically with a Dounce homogenizer to preserve the structure of the Golgi apparatus, the same antibody coprecipitated golgin, a marker protein of the Golgi body (54), as well as the H protein with the M-F50P protein. The results demonstrated that the M-F50P protein tightly interacts with the H protein at the Golgi body.
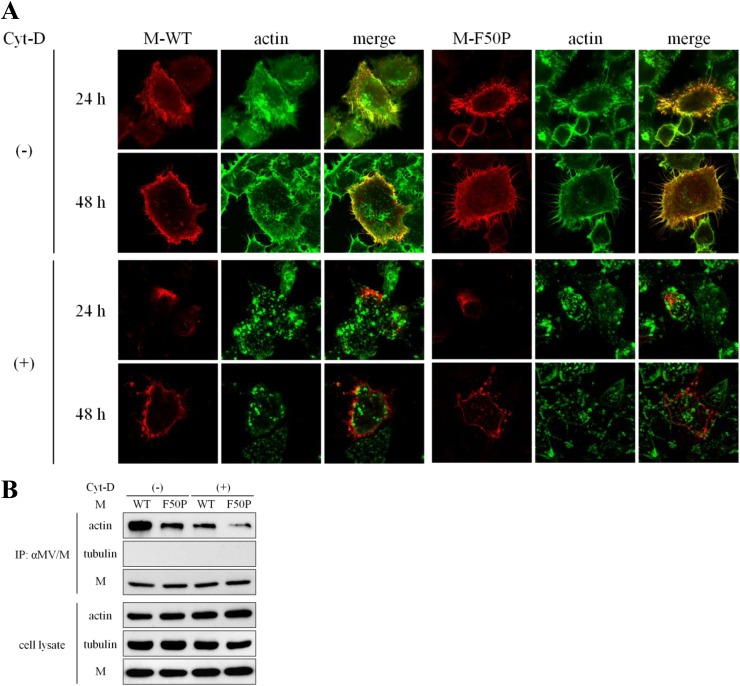
The M-WT protein associates firmly with F-actin.
Since each M protein expressed alone distributed forming filamentous structure, we then examined whether the M proteins associate with some cellular cytoskeleton proteins. As shown in Fig. 3A, both of the M-WT and M-F50P proteins expressed in HeLa cells colocalized with F-actin but not with tubulin (data not shown). When the cells were treated with Cyt-D, the M proteins distributed apart from the patches of the disrupted F-actin. To verify the observation, the M protein was immunoprecipitated using anti-M antibody. Actin was coprecipitated with either of the M proteins, but the amount precipitated with the M-WT protein was clearly much more. When F-actin was disrupted by the treatment with Cyt-D, the amount of coprecipitated actin with either M protein was reduced significantly. Tubulin was not detected in the precipitates at all. Through the coprecipitation study, the M-WT protein appeared to associate with F-actin more firmly than the M-F50P protein.
Fig 3.
Tight association of the M-WT protein with F-actin. (A) Colocalization of the M proteins with F-actin. HeLa cells were transfected with a plasmid expressing the M-WT or M-F50P protein. At 12 h posttransfection, culture medium was replaced by medium including Cyt-D or DMSO. At 24 or 48 h posttransfection, the cells were fixed, permeabilized, and incubated with the mouse monoclonal antibody against the M protein followed by Texas Red-conjugated goat anti-mouse IgG. F-actin was visualized with FITC-conjugated phalloidin. Observation was performed under a confocal microscope. (B) Efficient coprecipitation of actin with the M-WT protein. 293T cells were transfected with a plasmid expressing M-WT or M-F50P protein. At 12 h posttransfection, culture medium was replaced by medium including Cyt-D or DMSO. The cells were incubated for a further 12 h and then solubilized with detergent solution after the incubation with cross-linking reagent DSP for 2 h. Proteins immunoprecipitated with the anti-M antibody (IP) as well as whole-cell extracts (cell lysate) were subjected to immunoblot analysis. The M protein, actin, and tubulin were detected using mouse monoclonal antibody against MV M protein, mouse monoclonal antibody against β-actin, and rabbit polyclonal antibody against β-tubulin as the first antibodies, respectively.
Interaction of the M-WT protein with the H protein is prevented by the associated F-actin.
We next wanted to know whether F-actin in association with the M protein affects the interaction between the M and H proteins. When the structure of F-actin was disrupted with Cyt-D, no peculiar change in the distribution of the M-F50P protein was observed; the M-F50P protein strongly colocalized with the H protein as in the cells without Cyt-D treatment (compare Fig. 4A with Fig. 2B). On the other hand, the distribution of the M-WT protein drastically changed; the M-WT protein predominantly colocalized with the H protein in the perinuclear region at 24 h posttransfection and moved to the plasma membrane together, which was almost the same behavior as that shown by the M-F50P protein (Fig. 4A).
Fig 4.
Stimulated interaction of the M-WT protein with the H protein after the disruption of F-actin. (A) Colocalization of the M proteins with the H protein after Cyt-D treatment. The M-WT or M-F50P protein was coexpressed with the H-myc protein. At 12 h posttransfection, the culture medium was replaced by medium including Cyt-D. At 24 or 48 h posttransfection, the cells were treated as described for Fig. 2B, and the distribution of the M and H proteins was observed under a confocal microscope. (B) Enhanced coprecipitation of the H protein with the M-WT protein after Cyt-D treatment. The M-WT or M-F50P protein was coexpressed with the H-myc protein in 293T cells. At 12 h posttransfection, culture medium was replaced by medium including Cyt-D or DMSO. At 24 h posttransfection, cell extracts were prepared by 2 ways with detergent solution and a Dounce homogenizer and subjected to immunoprecipitation as described in Fig. 2C. Proteins precipitated with anti-M antibody were analyzed by immunoblotting with anti-M protein mouse monoclonal antibody, anti-myc tag rabbit polyclonal antibody, anti-β-actin mouse monoclonal antibody, and anti-golgin97 mouse monoclonal antibody as the first antibodies.
The mutual interactions of the M and H proteins with F-actin were studied by immunoprecipitation using the extract of the cells coexpressing the M and H proteins. In the experiment without Cyt-D treatment, while the anti-M antibody precipitated the H protein and a significant amount of actin with the M-WT protein, the clearly larger amount of the H protein and only trace amounts of actin were coprecipitated with the M-F50P protein. When the cells were treated with Cyt-D, however, elimination of actin and augmentation of the H protein in the complex coprecipitated with the M-WT protein were observed, which suggested that Cyt-D treatment abrogated the association of the M-WT protein with F-actin, resulting in a simultaneous increase of the interaction with the H protein up to the level equivalent to that of the M-F50P protein (Fig. 4B). When the cell extract prepared by a Dounce homogenizer was used, the F-actin disrupted by the treatment with Cyt-D could not associate with the M-WT protein, as observed before in the experiment where the detergent-treated cell extract was used, and the M-WT protein free from F-actin interacted with the H protein tightly at the Golgi body just like the M-F50P protein. The data obtained here indicated that F-actin in firm association prevents the M-WT protein from interacting with the H protein. Contrarily, the M-F50P protein is allowed to interact with the H protein because it associates with F-actin only very slightly. Thus, Cyt-D treatment caused no effect on the interaction between the M-F50P and H proteins.
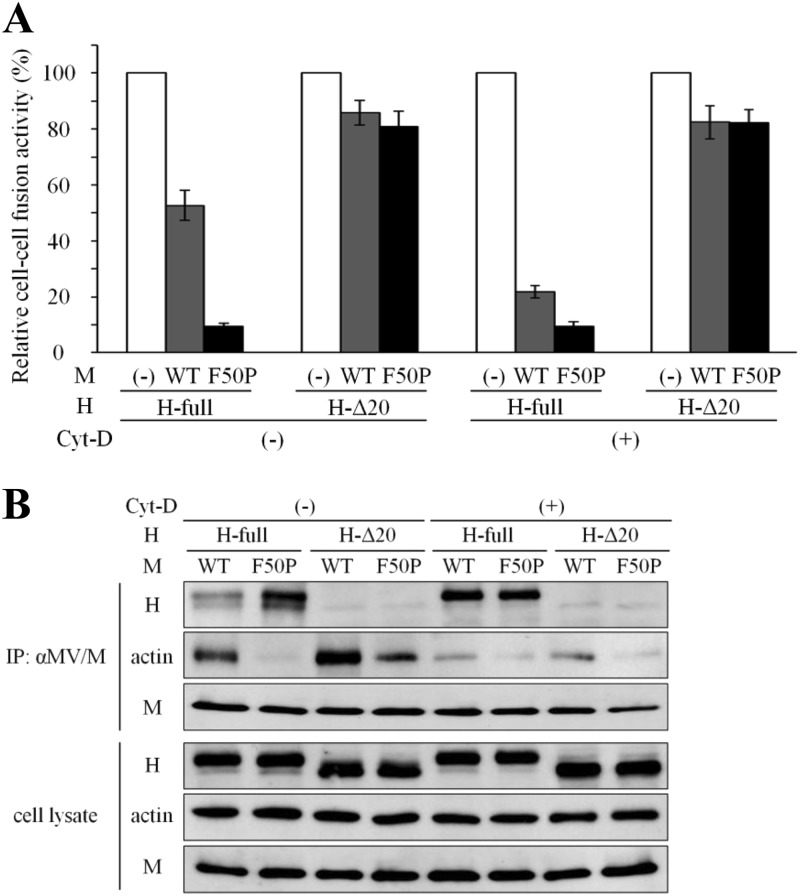
Cell-cell fusion-inhibiting activity of the M-WT protein is disturbed by the associated F-actin.
We next studied whether the associated F-actin could affect cell-cell fusion-inhibiting activity of the M protein. Cell-cell fusion induced by the plasmid-expressed H and F proteins was inhibited by 50% and more sharply by 90% in the presence of the M-WT and M-F50P proteins, respectively (Fig. 5A). When the cytoplasmic tail of the H protein was truncated by 20 amino acids from the N terminus (H-Δ20 protein) (21), the inhibition of cell-cell fusion by each M protein was greatly relieved to 15%. After the treatment with Cyt-D, the inhibition by the M-WT protein increased up to 80%, while that by the M-F50P protein remained unchanged. Cyt-D treatment exhibited no effect on the activity of either M protein to inhibit cell-cell fusion induced by the H-Δ20 protein. Immunoprecipitation analysis directly proved that the M-F50P protein interacted with the cytoplasmic tail of the H protein encompassing the 20 amino acids more strongly than the M-WT protein and that the M-WT protein improved its interacting ability with the same domain of the H protein when F-actin was disrupted by Cyt-D treatment and nearly failed to associate with the M-WT protein (Fig. 5B). Neither of the M proteins could interact with the H-Δ20 protein. These data showed that inhibition of cell-cell fusion by the M protein is closely related to its interaction with the cytoplasmic tail of the H protein, which is disturbed by F-actin in firm association with the M protein.
Fig 5.
Enhanced cell-cell fusion-inhibiting activity of the M-WT protein after the disruption of F-actin. (A) Effect of the Cyt-D treatment on cell-cell fusion inhibition by the M proteins. 293T cells were transfected with a plasmid expressing the H-myc protein (H-full) or the one lacking the cytoplasmic tail (H-Δ20), the F protein-expressing plasmid, and one of the M protein (M-WT or M-F50P)-expressing plasmids or empty pcDNA vector together with the T7 RNA polymerase-expressing plasmid, pCAG-T7. At 12 h posttransfection, the culture medium was replaced by medium including Cyt-D or DMSO. At 24 h posttransfection, cells were overlaid onto 293T cells pretransfected with SLAM-expressing plasmid, pCA7-SLAM, and T7 RNA polymerase-driven reporter plasmid, pTM1-EGFP, and were incubated for a further 48 h. EGFP-expressing cells were counted under a fluorescence microscope, and the relative cell-cell fusion activity was displayed, setting the count of the cells in the absence of the M protein as 100%. White bar, without the M protein (100%); gray bar, the M-WT protein; black bar, the M-F50P protein. (B) Effect of the Cyt-D treatment on the M protein interaction with the H protein and F-actin. The M-WT or M-F50P protein was coexpressed with the H-myc protein (H-full) or the one lacking the cytoplasmic tail (H-Δ20) in 293T cells as described for Fig. 4B. At 24 h posttransfection, the cells were treated with cross-linking reagent DSP for 2 h and solubilized with detergent solution. The M proteins were then precipitated with anti-M antibody, and the coprecipitated proteins were analyzed by immunoblotting with anti-M protein mouse monoclonal antibody, anti-myc tag rabbit polyclonal antibody, and anti-β-actin mouse monoclonal antibody as the first antibodies.
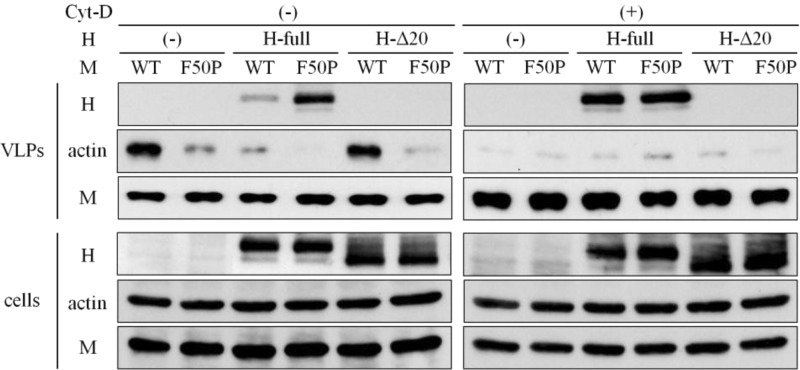
Uptake of the H protein by the M-WT protein into the VLP is prevented by the associated F-actin.
To examine whether the virus assembly operated by the M protein could be affected by the associated F-actin, the VLPs released from the M protein-expressing cells were analyzed. When expressed alone, there was no difference between the amount of the M-WT and M-F50P proteins in the culture medium released as VLPs, though much more actin was taken into the VLP released by the M-WT protein (Fig. 6). Supposing the amount of the M protein in the VLP reflects the number of VLPs, the data suggest that each M protein possesses the equivalent activity to release VLP. When the H protein was coexpressed, the markedly large amount of the H protein was detected in the VLP released by the M-F50P protein. In case F-actin was disrupted with Cyt-D, nearly the equal amount of the H protein was carried into the VLP produced by each M-WT or M-F50P protein. We then characterized VLPs released from the cells coexpressing the H-Δ20 protein. Neither the M-WT nor the M-F50P protein took the H-Δ20 protein into the VLPs at all, even when the cells were treated with Cyt-D. The protein composition of the VLPs released under the coexpression of the H-Δ20 protein was quite similar to that of the VLPs produced in the absence of the H protein. Both the M proteins behaved as if they were in the absence of the H protein when the M protein-interacting domain on the H protein was depleted. The striking difference between the two M proteins in the ability to carry the H protein into the VLPs was in good correlation with the efficiency of the M protein to interact with the cytoplasmic tail of the H protein (Fig. 5B). The data indicated that the M-H protein interaction plays a key role in the uptake of the H protein into the VLP, which is interrupted by F-actin in tight association with the M protein.
Fig 6.
Enhanced incorporation of the H protein into VLPs by the M-WT protein after the disruption of F-actin. 293T cells were transfected with a plasmid expressing the M-WT or M-F50P protein without (−) or together with a plasmid expressing the H-myc protein (H-full) or the one lacking the cytoplasmic tail (H-Δ20). At 12 h posttransfection, culture medium was replaced by medium including Cyt-D or DMSO, and at 48 h, VLPs released into the culture medium were collected as pellets by centrifugation passing through 20% sucrose layer (VLPs). The cells were solubilized with detergent solution as whole-cell extract (cells). These samples were subjected to the immunoblotting analysis, and the M and H proteins and actin were detected as described in the legend to Fig. 5B.
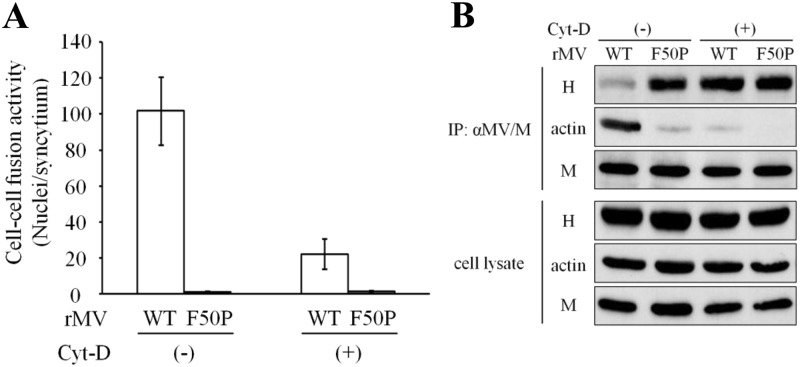
F-actin supports syncytium formation of MV by interfering with the interaction of the M protein with the H protein.
To study the involvement of F-actin in syncytium formation by MV, cell-cell fusion assay was done in cells infected with the WT or F50P virus shown in Fig. 1. While the F50P virus did not exhibit cell-cell fusion irrespective of Cyt-D treatment, the WT virus formed notable syncytia, the size of which enlarged only limitedly after the Cyt-D treatment (Fig. 7A). The immunoprecipitation study with the anti-M antibody revealed that the interaction of the M protein with the H protein was closely related to the inhibition of cell-cell fusion by the M protein (Fig. 7B). These data show that the WT virus demonstrates significantly high cell-cell fusion activity because F-actin in firm association prevents the M-WT protein from interacting with the H protein and that the M-WT protein free from the disrupted F-actin inhibits cell-cell fusion via effective interaction with the H protein. F-actin supports syncytium formation of MV by interfering with the interaction between the M and H proteins.
Fig 7.
MV cell-cell fusion affected by the association of the M protein with F-actin. (A) Effect of the Cyt-D treatment on cell-cell fusion of the WT and F50P viruses. Vero/hSLAM cells were infected with WT or F50P virus at an MOI of 0.001, and at 12 h p.i., culture medium was replaced by medium including Cyt-D or DMSO. At 30 h p.i., cells were fixed, permeabilized, and incubated with Hoechst 33342. Nuclei in the EGFP-expressing cells were counted under a fluorescence microscope, and cell-cell fusion activity was displayed as the number of nuclei in a syncytium (n = 3). White bar, the WT virus; gray bar, the F50P virus. (B) Effect of Cyt-D treatment on the interaction of the M protein with the H protein and F-actin. Vero/hSLAM cells were inoculated with the WT or F50P virus at an MOI of 0.01, and at 12 h p.i., culture medium was replaced by medium including Cyt-D or DMSO. At 24 h p.i., the cell extracts were prepared and subjected to immunoprecipitation analysis as described for Fig. 5B. The M and H proteins and actin were detected with anti-M protein mouse monoclonal antibody, anti-H protein rabbit polyclonal antibody, and anti-β-actin mouse monoclonal antibody as the first antibodies, respectively.
F-actin suppresses MV assembly by restricting the uptake of the H protein into the particle.
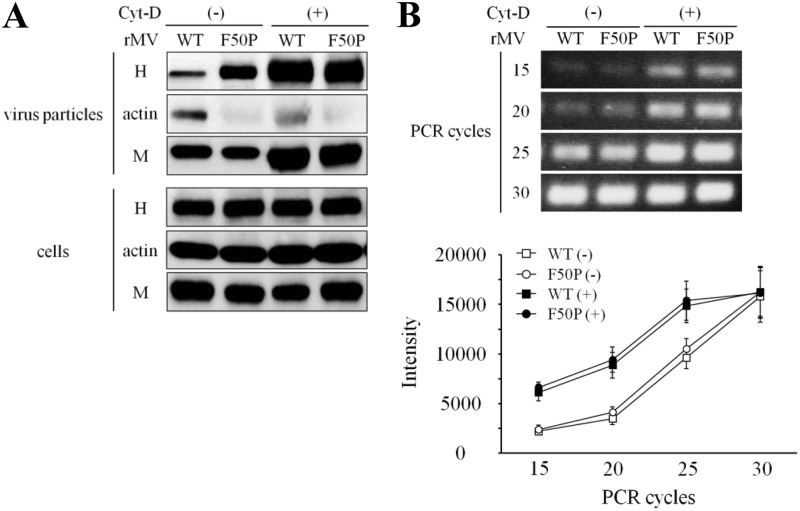
The role of F-actin in the virus assembly was studied. Figure 8A shows the protein components of the virus particle released from the cells infected with the WT virus or F50P virus. The amount of each protein, determined by measuring the intensity of the protein band, and the ratio of the H protein to the M protein (H/M) are presented in Table 1. There was no difference between the two viruses in the amount of the M protein detected in the culture medium as virus particles, indicating that these two M proteins possess nearly equal abilities to release virus particles, as suggested in Fig. 6. The H protein taken into the WT virus was strongly restricted; instead, a significant amount of actin was carried in. The H/M ratio showed that the H protein present in a particle of the WT virus was one-third of the H protein in the F50P virus. When the infected cells were treated with Cyt-D, the M protein detected as virus particles increased, suggesting that the stimulation of virus release occurred. Cyt-D treatment should have some common effects on virus release of the two viruses, with which the F50P substitution in the M protein has no concern. While the H protein in the F50P virus increased in parallel with the M protein and the H/M ratio remained unchanged, strikingly augmented H protein in the WT virus resulted in the indistinguishable H/M ratio between the two viruses. Figure 8B demonstrates that there was no difference in the level of virus genome RNA packaged in the released particles between the WT and F50P viruses irrespective of Cyt-D treatment, which indicates that the F50P mutation does not change the ability of the M protein to take the virus RNP into the particle.
Fig 8.
Restricted incorporation of the H protein into the WT virus particle. (A) Uptake of the H and M proteins. Vero/hSLAM cells were inoculated with the WT or F50P virus at an MOI of 0.01, and at 12 h p.i., the culture medium was replaced by medium including Cyt-D or DMSO. At 72 h p.i., virus particles in the culture medium were collected as pellets by centrifugation passing through a 20% sucrose layer (virus particles), and the cells were solubilized with detergent solution (cells). The M and H proteins as well as actin in the samples were analyzed by immunoblotting with the same antibodies described in the legend for Fig. 7B. (B) Amount of the genome RNA packaged in MV particles. cDNA of virus genome RNA was prepared by reverse transcription using the primer annealing the leader sequence, and then the N gene was amplified by PCR as described in Materials and Methods. □, WT virus without Cyt-D; ○, F50P virus without Cyt-D; ■, WT virus with Cyt-D; ●, F50P virus with Cyt-D.
Table 1.
Effect of the Cyt-D treatment on virus infectivity and the amount of the H and M proteins in virus particles
| Cyt-D | Virus | Infectivitya (PFU/ml) | Intensityb |
Ratio of H/Mc | |
|---|---|---|---|---|---|
| H | M | ||||
| Negative | WT | 8.55 × 102 | 5,390 ± 1,210 | 16,590 ± 720 | 0.324 ± 0.029 |
| F50P | 2.24 × 103 | 14,860 ± 1,890 | 15,160 ± 1,940 | 0.980 ± 0.034 | |
| Positive | WT | 3.18 × 103 | 27,790 ± 2,290 | 27,880 ± 1,690 | 0.997 ± 0.024 |
| F50P | 4.42 × 103 | 27,650 ± 1,420 | 26,720 ± 1,050 | 1.035 ± 0.006 | |
Infectivity of the viruses in the culture medium of which protein components were analyzed in Fig. 8.
Amount of the protein in the particles determined by quantifying the intensity of each protein band in Fig. 8 using ImageJ software (http://rsbweb.nih.gov/ij/index.html).
Relative amount of the H protein to that of the M protein.
We then assayed the infectivity of the released viruses. The virus titer of the F50P virus was about 3 times higher than that of the WT virus (Table 1). Therefore, the F50P substitution actually caused the stimulation in virus infectivity, although the amount of the virus particle released into the culture medium, judging from the amount of the M protein, was equivalent to that of the WT virus, and the level of genome RNA contained in each virus particle was indistinguishable. After Cyt-D treatment, the infectivity of the WT virus and F50P virus went up by nearly 4- and 2-fold, respectively. The increase of the virus titer of the F50P virus might be explained by the enhanced virus release after Cyt-D treatment. On the other hand, the highly improved titer of the WT virus could be due to the increased uptake of the H protein in addition to the enhanced virus release. Combined with the results of the immunoprecipitation study (Fig. 7B), it is clear that the interaction between the M and H proteins is responsible for the efficient uptake of the H protein into the virus particle, accompanied by the highly regulated virus infectivity. F-actin in association with the M protein possesses the ability to interfere with the proper virus assembly by interrupting the interaction of the M protein with the H protein.
DISCUSSION
The M protein of MV operates accurate assembly of virus components by combining RNP with envelope H and F proteins via interaction with their cytoplasmic tails (13–15, 17, 18, 55, 56). Since the association of the M protein with these proteins is also known to modulate cell-cell fusion (19–22), the M protein plays the key role in determining the way by which MV spreads its infection, particle production or cell-cell fusion. Tahara et al. (55) reported that P64S/E89K substitutions conferred on MV M protein the increased ability to interact with the H protein, to produce infectious virus particles, and to suppress cell-cell fusion. They, however, did not present the mechanism of how the M protein acquired the stimulated affinity to the H protein. In this study, we identified the amino acid at position 50 in the M protein as a new determinant for the interaction of the M protein with the H protein. Here, we demonstrated that the interaction of the M-WT protein with the cytoplasmic tail of the H protein was restricted because of the tight association with F-actin and that the F50P substitution disturbed the association, permitting the M protein interaction with the H protein, which conferred on the M-F50P protein the enhanced ability to inhibit cell-cell fusion and to take the H protein into the virus particle. Tight interaction with the M protein was required for the successful incorporation of the H protein into the virus particle. We concluded that the M-F50P protein produces a virus particle carrying a larger amount of the H protein accompanied by the enhanced infectivity. Contrarily, the M-WT protein leaves the H protein behind at budding, and the H protein is retained on the infected-cell surface, which might play some role in promoting the cell fusion with the neighboring uninfected cells. However, to observe the spikes of the H protein on each virus particle, electron microscope analysis was unsuccessful, because virus particles sufficient for the study could not be recovered. In this report, we first demonstrated that F-actin modulates MV assembly as well as syncytium formation via altering the M protein interaction with the H protein.
It might be surprising that the WT virus possesses the M protein with the restricted activity in virus assembly and produces virus particles with limited infectivity due to insufficient uptake of the H protein. The WT virus, however, grows faster than the F50P virus in Vero/hSLAM cells due to the intense cell-cell fusion (data not shown). Tahara et al. demonstrated that the increased particle production is not readily advantageous to replicate SLAM dependently in the cell cultures and probably in the human body (55). Since release of the infectious progeny particles of MV is inefficient and most of the particles remain in cell-associated form (9), syncytium formation should contribute significantly to the efficient spread of the MV infection. Therefore, F-actin plays an important role in WT virus infection, interfering with the interaction of the M protein with the H protein to support cell-cell fusion induced by the H and F proteins. On the other hand, it should be noted that the MV with the M-F50P protein was obtained as a variant from the clinical virus shortly after isolation. Even though MV spreads mainly in the cells of the immune system by using SLAM as the receptor (4, 5), it also infects epithelial cells using nectin4 (PVRL4) as the receptor in the human body (6, 7). It might be possible that MV shows quasispecies in our body if the characteristics of the virus advantageous for replication depended on the types of the host cells. In fact, the F50P virus lacking cell-cell fusion replicates more efficiently in epithelial HT29 cells than the WT virus (data not shown). There is a report showing that one out of 12 MV strains of which M genes were analyzed by RT-PCR directly from RNA extracted from clinical samples possessed the F50V mutation in the M protein (53), and the substitution of Pro, Ser, or Leu is found in the databases at the same position of several SSPE viruses isolated from human brain. It might be possible that escape of the M protein from F-actin is one of the mechanisms to generate MV variation.
It is clear that Phe at position 50 in the M protein is important for the association with F-actin. The site involved in the interaction with the H protein is unknown, but it could be quite close to or overlap position 50 on the M protein molecule, since the association of both the M proteins with F-actin seemed to be improved in the absence of the interaction with the H protein (Fig. 5B). The mutual interaction of the proteins might be explained as follows. In the presence of the H protein, the M-WT protein with strong affinity to F-actin associates with F-actin first, which interrupts the M protein interaction with the H protein because it is blocked by F-actin. On the other hand, the M-F50P protein with the reduced affinity to F-actin interacts preferentially with the H protein that interferes with the association with F-actin. When F-actin was disrupted with Cyt-D, there was no difference between the amounts of the H protein coprecipitated with the M-WT and M-F50P proteins (Fig. 4B and 5B), suggesting that the affinities of the two M proteins to the H protein in the absence of F-actin are almost equivalent. It is unknown whether the M protein directly associates with the actin molecule on F-actin. The M protein might not associate with globular actin, because the amount of actin coprecipitated with the M protein clearly decreased after its polymerization was inhibited with Cyt-D. Therefore, it is possible that the M protein associates with some proteins combined with F-actin other than the actin molecule itself.
There have been several reports on the role of actin in assembly of MV, most of which presented the conclusion that actin is indispensable in virus assembly because the treatment with Cyt-D suppresses the infectious particle formation (31, 36–38). In the present paper, we obtained the quite different results showing that Cyt-D treatment enhanced MV assembly. In the preliminary experiments we have done using the Edmonston strain, which is genetically close to the MV strains used in most of the previous experiments, infectious virus titers clearly decreased after Cyt-D treatment (data not shown). Therefore, the discrepancy could have arisen from the difference of the MV strains used in the experiments. There are 4 amino acid substitutions in the M protein of the H1 strain we used in this study compared with the M protein of the Edmonston strain, which should be responsible for the reversed behavior of the M protein upon Cyt-D treatment. A study on the mechanism of how the amino acid substitutions change the role of F-actin in MV assembly is now going on.
ACKNOWLEDGMENTS
We thank Y. Yangi for providing Vero/hSLAM cells, p(+)MV323-EGFP, pCA7, and pCA7-SLAM; M. Takeda for providing pCITE-IC-N, pCITE-IC-PΔC, and pCITEko9301B-L; N. Ito and M. Sugiyama for providing the BHK/T7-9 cells; and T. Kohama for providing antiserum. We also thank J. Miyazaki and B. Moss for the permissions to use the CAG promoter of pCA7 and pTM1, respectively.
This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print 5 December 2012
REFERENCES
- 1. Lamb RA, Parks GD. 2007. Paramyxoviridae: the viruses and their replication, p 1449–1496 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2. Choppin PW. 1984. Membrane proteins and virus virulence. Trans. Am. Clin. Climatol. Assoc. 95:138–156 [PMC free article] [PubMed] [Google Scholar]
- 3. Griffin DE. 2007. Measles virus, p 1551–1585 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 4. Tatsuo H, Ono N, Tanaka K, Yanagi Y. 2000. SLAM (CDw150) is a cellular receptor for measles virus. Nature 406:893–897 [DOI] [PubMed] [Google Scholar]
- 5. Yanagi Y, Takeda M, Ohno S. 2006. Measles virus: cellular receptors, tropism and pathogenesis. J. Gen. Virol. 87:2767–2779 [DOI] [PubMed] [Google Scholar]
- 6. Mühlebach MD, Mateo M, Sinn PL, Prüfer S, Uhlig KM, Leonard VHJ, Navaratnarajah CK, Frenzke M, Wong XX, Sawatsky B, Ramachandran S, McCray PB, Cichutek K, von Messling V, Lopez M, Cattaneo R. 2011. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature 480:530–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Noyce RS, Bondre DG, Ha MN, Lin L-T, Sisson G, Tsao M-S, Richardson CD. 2011. Tumor cell marker PVRL4 (Nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 7:e1002240 doi:10.1371/journal.ppat.1002240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wild TF, Malvoisin E, Buckland R. 1991. Measles virus: both the haemagglutinin and fusion glycoproteins are required for fusion. J. Gen. Virol. 72:439–442 [DOI] [PubMed] [Google Scholar]
- 9. Kobune F, Sakata H, Sugiura A. 1990. Marmoset lymphoblastoid cells as a sensitive host for isolation of measles virus. J. Virol. 64:700–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moll M, Pfeuffer J, Klenk H-D, Niewiesk S, Maisner A. 2004. Polarized glycoprotein targeting affects the spread of measles virus in vitro and in vivo. J. Gen. Virol. 85:1019–1027 [DOI] [PubMed] [Google Scholar]
- 11. Pohl C, Duprex WP, Krohne G, Rima BK, Schneider-Schaulies S. 2007. Measles virus M and F proteins associate with detergent-resistant membrane fractions and promote formation of virus-like particles. J. Gen. Virol. 88:1243–1250 [DOI] [PubMed] [Google Scholar]
- 12. Salditt A, Koethe S, Pohl C, Harms H, Kolesnikova L, Becker S, Schneider-Schaulies S. 2010. Measles virus M protein-driven particle production does not involve the endosomal sorting complex required for transport (ESCRT) system. J. Gen. Virol. 91:1464–1472 [DOI] [PubMed] [Google Scholar]
- 13. Hirano A, Ayata M, Wang AH, Wong TC. 1993. Functional analysis of matrix proteins expressed from cloned genes of measles virus variants that cause subacute sclerosing panencephalitis reveals a common defect in nucleocapsid binding. J. Virol. 67:1848–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iwasaki M, Takeda M, Shirogane Y, Nakatsu Y, Nakamura T, Yanagi Y. 2009. The matrix protein of measles virus regulates viral RNA synthesis and assembly by interacting with the nucleocapsid protein. J. Virol. 83:10374–10383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Runkler N, Pohl C, Schneider-Schaulies S, Klenk H-D, Maisner A. 2007. Measles virus nucleocapsid transport to the plasma membrane requires stable expression and surface accumulation of the viral matrix protein. Cell. Microbiol. 9:1203–1214 [DOI] [PubMed] [Google Scholar]
- 16. Blau DM, Compas RW. 1995. Entry and release of measles virus are polarized in epithelial cells. Virology 210:91–99 [DOI] [PubMed] [Google Scholar]
- 17. Naim HY, Ehler E, Billeter MA. 2000. Measles virus matrix protein specifies apical virus release and glycoprotein sorting in epithelial cells. EMBO J. 19:3576–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Spielhofer P, Bächi T, Fehr T, Christiansen G, Cattaneo R, Kaelin K, Billeter MA, Naim HY. 1998. Chimeric measles viruses with a foreign envelope. J. Virol. 72:2150–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cathomen T, Mrkic B, Spehner D, Drillien R, Naef R, Pavlovic J, Aguzzi A, Billeter MA, Cattaneo R. 1998. A matrix-less measles virus is infectious and elicits extensive cell fusion: consequences for propagation in the brain. EMBO J. 17:3899–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cathomen T, Naim HY, Cattaneo R. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moll M, Klenk H-D, Maisner A, Moll M, Klenk H, Maisner A. 2002. Importance of the cytoplasmic tails of the measles virus glycoproteins for fusogenic activity and the generation of recombinant measles viruses. J. Virol. 76:7174–7186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reuter T, Weissbrich B, Schneider-Schaulies S, Schneider-Schaulies J. 2006. RNA interference with measles virus N, P, and L mRNAs efficiently prevents and with matrix protein mRNA enhances viral transcription. J. Virol. 80:5951–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cattaneo R, Schmid A, Eschle D, Baczko K, ter Meulen V, Billeter MA. 1988. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell 55:255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmid A, Spielhofer P, Cattaneo R, Baczko K, ter Meulen V, Billeter MA. 1992. Subacute sclerosing panencephalitis is typically characterized by alterations in the fusion protein cytoplasmic domain of the persisting measles virus. Virology 188:910–915 [DOI] [PubMed] [Google Scholar]
- 25. Ali A, Nayak DP. 2000. Assembly of Sendai virus: M protein interacts with F and HN proteins and with the cytoplasmic tail and transmembrane domain of F protein. Virology 276:289–303 [DOI] [PubMed] [Google Scholar]
- 26. Ghildyal R, Li D, Peroulis I, Shields B, Bardin PG, Teng MN, Collins PL, Meanger J, Mills J. 2005. Interaction between the respiratory syncytial virus G glycoprotein cytoplasmic domain and the matrix protein. J. Gen. Virol. 86:1879–1884 [DOI] [PubMed] [Google Scholar]
- 27. Schmitt AP, He B, Lamb RA. 1999. Involvement of the cytoplasmic domain of the hemagglutinin-neuraminidase protein in assembly of the paramyxovirus simian virus 5. J. Virol. 73:8703–8712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shaikh FY, Cox RG, Lifland AW, Hotard AL, Williams JV, Moore ML, Santangelo PJ, Crowe JE., Jr 2012. A critical phenylalanine residue in the respiratory syncytial virus fusion protein cytoplasmic tail mediates assembly of internal viral proteins into viral filaments and particles. mBio 3:e00270–11 doi:10.1128/mBio.00270-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tong S, Li M, Vincent A, Compans RW, Fritsch E, Beier R, Klenk C, Ohuchi M, Klenk H-D. 2002. Regulation of fusion activity by the cytoplasmic domain of a paramyxovirus F protein. Virology 301:322–333 [DOI] [PubMed] [Google Scholar]
- 30. Waning DL, Schmitt AP, Leser GP, Lamb RA. 2002. Roles for the cytoplasmic tails of the fusion and hemagglutinin-neuraminidase proteins in budding of the paramyxovirus simian virus 5. J. Virol. 76:9284–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stallcup CK, Raine CS, Fields BN. 1983. Cytochalasin B inhibits the maturation of measles virus. Virology 124:59–74 [DOI] [PubMed] [Google Scholar]
- 32. Tyrrell DL, Norrby E. 1978. Structural polypeptides of measles virus. J. Gen. Virol. 39:219–229 [DOI] [PubMed] [Google Scholar]
- 33. Bohn W, Rutter G, Hohenberg H, Mannweiler K, Nobis P. 1986. Involvement of actin filaments in budding of measles virus: studies on cytoskeletons of infected cells. Virology 149:91–106 [DOI] [PubMed] [Google Scholar]
- 34. Moyer SA, Baker SC, Horikami SM. 1990. Host cell proteins required for measles virus reproduction. J. Gen. Virol. 71:775–783 [DOI] [PubMed] [Google Scholar]
- 35. Bohn W, Mannweiler K, Hohenberg H, Rutter G. 1987. Replica-immunogold technique applied to studies on measles virus morphogenesis. Scanning Microsc. 1:319–330 [PubMed] [Google Scholar]
- 36. Bedows E, Rao KM, Welsh MJ. 1983. Fate of microfilaments in Vero cells infected with measles virus and herpes simplex virus type 1. Mol. Cell. Biol. 3:712–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Berghäll H, Wallén C, Hyypiä T, Vainionpää R. 2004. Role of cytoskeleton components in measles virus replication. Arch. Virol. 149:891–901 [DOI] [PubMed] [Google Scholar]
- 38. Bohn W, Rutter G, Hohenberg H, Mannweiler K. 1983. Inhibition of measles virus budding by phenothiazines. Virology 130:44–55 [DOI] [PubMed] [Google Scholar]
- 39. Jeffree CE, Brown G, Aitken J, Su-Yin DY, Tan BH, Sugrue RJ. 2007. Ultrastructural analysis of the interaction between F-actin and respiratory syncytial virus during virus assembly. Virology 369:309–323 [DOI] [PubMed] [Google Scholar]
- 40. Kallewaard NL, Bowen AL, Crowe JE., Jr 2005. Cooperativity of actin and microtubule elements during replication of respiratory syncytial virus. Virology 331:73–81 [DOI] [PubMed] [Google Scholar]
- 41. Kitagawa Y, Yamaguchi M, Zhou M, Komatsu T, Nishio M, Sugiyama T, Takeuchi K, Itoh M, Gotoh B. 2011. A tryptophan-rich motif in the human parainfluenza virus type 2 V protein is critical for the blockade of Toll-like receptor 7 (TLR7)- and TLR9-dependent signaling. J. Virol. 85:4606–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miazza V, Mottet-Osman G, Startchick S, Chaponnier C, Roux L. 2011. Sendai virus induced cytoplasmic actin remodeling correlates with efficient virus particle production. Virology 410:7–16 [DOI] [PubMed] [Google Scholar]
- 43. Morrison TG, McGinnes LJ. 1985. Cytochalasin D accelerates the release of Newcastle disease virus from infected cells. Virus Res. 4:93–106 [DOI] [PubMed] [Google Scholar]
- 44. Orvell C. 1980. Structural polypeptides of canine distemper virus. Arch. Virol. 66:193–206 [DOI] [PubMed] [Google Scholar]
- 45. Takimoto T, Murti KG, Bousse T, Scroggs RA, Portner A. 1984. Role of matrix and fusion proteins in budding of Sendai virus. J. Virol. 75:11384–11391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Giuffre RM, Tovell DR, Kay CM, Tyrrell DLJ. 1982. Evidence for an interaction between the membrane protein of a paramyxovirus and actin. J. Virol. 42:963–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ono N, Tatsuo H, Hidaka Y, Aoki T, Minagawa H, Yanagi Y. 2001. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J. Virol. 75:4399–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ito N, Takayama-Ito M, Yamada K, Hosokawa J, Sugiyama M, Minamoto N. 2003. Improved recovery of rabies virus from cloned cDNA using a vaccinia virus-free reverse genetics system. Microbiol. Immunol. 47:613–617 [DOI] [PubMed] [Google Scholar]
- 49. Seki F, Yamada K, Nakatsu Y, Okamura K, Yanagi Y, Nakayama T, Komase K, Takeda M. 2011. The SI strain of measles virus derived from a patient with subacute sclerosing panencephalitis possesses typical genome alterations and unique amino acid changes that modulate receptor specificity and reduce membrane fusion activity. J. Virol. 85:11871–11882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Takeda M, Takeuchi K, Miyajima N, Kobune F, Ami Y, Nagata N, Suzaki Y, Nagai Y, Tashiro M. 2000. Recovery of pathogenic measles virus from cloned cDNA. J. Virol. 74:6643–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hashimoto K, Ono N, Tatsuo H, Takeda M, Takeuchi K, Minagawa H, Yanagi Y. 2002. SLAM (CD150)-independent measles virus entry as revealed by recombinant virus expressing green fluorescent protein. J. Virol. 76:6743–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Takeuchi K, Miyajima N, Kobune F, Tashiro M. 2000. Comparative nucleotide sequence analyses of the entire genomes of B95a cell-isolated and Vero cell-isolated measles viruses from the same patient. Virus Genes 20:253–257 [DOI] [PubMed] [Google Scholar]
- 53. Jin L, Richards A, Brown DW. 1996. Development of a dual target-PCR for detection and characterization of measles virus in clinical specimen. Mol. Cell. Probes 10:191–200 [DOI] [PubMed] [Google Scholar]
- 54. Griffith KJ, Chan EK, Lung CC, Hamel JC, Guo X, Miyachi K, Fritzler MJ. 1997. Molecular cloning of a novel 97-kd Golgi complex autoantigen associated with Sjögren's syndrome. Arthritis Rheum. 40:1693–1702 [DOI] [PubMed] [Google Scholar]
- 55. Tahara M, Takeda M, Yanagi Y. 2007. Altered interaction of the matrix protein with the cytoplasmic tail of hemagglutinin modulates measles virus growth by affecting virus assembly and cell-cell fusion. J. Virol. 81:6827–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Runkler N, Dietzel E, Moll M, Klenk H-D, Maisner A. 2008. Glycoprotein targeting signals influence the distribution of measles virus envelope proteins and virus spread in lymphocytes. J. Gen. Virol. 89:687–696 [DOI] [PubMed] [Google Scholar]
- 57. Jiang D-P, Ide Y-H, Nagano-Fujii M, Shoji I, Hotta H. 2009. Single-point mutations of the M protein of a measles virus variant obtained from a patient with subacute sclerosing panencephalitis critically affect solubility and subcellular localization of the M protein and cell-free virus production. Microbes Infect. 11:467–475 [DOI] [PubMed] [Google Scholar]
- 58. Katayama H, Hori M, Sato K, Kajita M, Ozaki H, Karaki H, Ohashi K, Kai C. 2004. Role of actin microfilaments in canine distemper virus replication in Vero cells. J. Vet. Med. Sci. 66:409–415 [DOI] [PubMed] [Google Scholar]