Abstract
Hepatitis B virus immune escape mutants have been associated with vaccine failure and reinfection of grafted liver despite immune prophylaxis, but their biological properties remain largely unknown. Transfection of 20 such mutants in a human hepatoma cell line identified many with severe impairment in virion secretion, which can be rescued to various extents by coexpression of wild-type envelope proteins or introduction of a novel glycosylation site. Consistent with their role in maintaining intra- or intermolecular disulfide bonds, cysteine residues within the “a” determinant are critical for virion secretion.
TEXT
Infection by hepatitis B virus (HBV) can be prevented by vaccination with the small (S) envelope protein, the most abundant envelope protein on the virion surface and the primary component of the empty envelope protein particles, or subviral particles (1). The neutralizing antibodies thus elicited target the immunodominant loop of the S protein (residues 101 to 163), especially its “a” determinant (residues 124 to 147), with residues 141 to 146 being the most dominant epitope (2). However, breakthrough infection despite induction of neutralizing antibodies has been documented and is often attributed to single amino acid substitutions within the “a” determinant (3–8). Such immune escape mutants are also responsible for reinfection of grafted liver despite passive prophylaxis with hepatitis B immunoglobulin (HBIG) (9–14). The mutations render the S protein poorly recognizable by antibodies raised against the wild-type (WT) virus, which forms the structural basis for both immune escape and false-negative diagnostic test results. An important serological marker of HBV infection is hepatitis B surface antigen (HBsAg), the collective term for viral envelope proteins present on virions and the large excess of subviral particles. The immune escape mutations can render HBsAg undetectable or poorly detected by immunoassays based on monoclonal antibodies against wild-type virus (15–18), contributing to some cases of “occult HBV infection” (lack of detectable HBsAg despite positive HBV DNA) (8, 19–23).
There are concerns that the universal vaccination program will provide the driving force for the spread of immune escape mutants and gradually render the current vaccine ineffective in eradicating HBV infection (24, 25). The prevalence of immune escape mutants in HBV DNA-positive children in Taiwan increased from 7.8% before to 22.6% 20 years following the nationwide vaccination program, but the prevalence of chronic HBV infection among the pediatric population decreased from 9.6% to 0.5% during this same period, indicating that while immune escape does occur, the HBV vaccine continues to be highly effective (5, 26). The classic G145R immune escape mutant reverted to the wild type when HBIG was discontinued in liver transplant recipients and during follow-up of mutant-infected children (10, 26) or when the mutant was transmitted to naïve chimpanzees (27, 28). These observations strongly suggest that in the absence of immune pressure, these mutants have reduced fitness compared to the wild-type virus. In this regard, the classic G145R immune escape mutant was found to be severely impaired in virion secretion (29, 30). Similarly, we reported that the I110M and G119E mutations hamper virion secretion (29, 31). In the present study, we characterized the virion secretion impact of 20 immune escape mutations or other mutations inside the immunodominant loop (Table 1). The escape mutations chosen for the current study, including those from HIV- and hepatitis C virus (HCV)-coinfected women with alcohol abuse, have been associated with vaccine failure, breakthrough infection of transplanted liver despite HBIG prophylaxis, and occult HBV infection.
Table 1.
Mutations in the immunodominant loop for functional characterization
| S domain substitution | Nucleotide change | Polymerase substitution | Other substitution(s) of the same site | Association(s) and/or purpose(s)a |
|---|---|---|---|---|
| T114R | C495G | N122K | T (T114T) | B, C |
| T115A | A497G | N123S | I, N | C |
| T118A | A506G | Y126C | K, M, R, S, V | A, B, D |
| T118K | C507A | Y126stop | A, M, R, S, V | A, B, D |
| 120insR121 | 514insAGC515 | 128insK/M129L | B | |
| P120T | C512A | T128N | L, Q, R, S | A, B, C, D |
| C121A | T515G/G516C | M129S | E | |
| C124A | T524G/G525C | L132R | E | |
| T126N | C531A | D134E | A, I, L, S | A, B, C |
| P127S | C533T | S135F | L, T | A, B, C, |
| Q129H | A541C | Silent | L, N, P, R, S | A, B, C, D |
| G130R | G542C | R138T | E, K, N | B, C, D |
| N131S | A546G | Silent | B, C | |
| F134L | T566A | S143T | H, S, Y | A, B, C |
| C138Y | G568A | Silent | S | A, C |
| K141E | A575G | K149R | I, Q, W | A, C |
| P142S | C578T | T150I | H, L | A, B, C |
| D144G | A585G | Silent | A, E, H | A, B, C |
| C147A | T593G/G594C | L155C | F, R, W, Y | A, C, D, E |
| C147R | T593C | L155S | A, F, W, Y | A, C, D |
| C149A | T599G/G600C | L157R | R, W | A, B, E |
| C149R | T599C | L157P | A, W | A, B |
A, vaccine failure; B, graft reinfection despite passive immunoprophylaxis; C, occult infection; D, anti-HBs-positive infection; E, to test the role of cysteine residues.
A trans-complementation assay to study virion secretion.
Experiments were performed on a genotype A clone. As previously reported (32), we used a subgenomic HBV DNA fragment to express the large (L), middle (M), and S envelope proteins. The 2.3-kb (0.7mer) fragment (nucleotides 2721 to 3221/1 to 1770) was inserted upstream of the SV40 polyadenylation signal and cloned into the pBluescript vector. Mutations were introduced to the 0.7mer construct by replacement of its AvrII-EcoRV restriction fragment with PCR products. To examine the impact of immune escape mutations on virion secretion, the 0.7mer construct was cotransfected with a 1.5mer (4.8-kb) HBV genome (nucleotides 1044 to 3221/1 to 2600) cloned into the pBluescript vector (32). The latter construct, which was rendered unable to express the envelope proteins through a G261A nonsense mutation at the 5′ end of the S gene, served as the source for core and polymerase protein translation and genome replication. Since the genome thus packaged inside virions is unable to express envelope proteins, this approach minimizes biological hazard. Using a 1.5mer construct as the common source for all functions other than envelope protein expression also simplifies data interpretation, because some immune escape mutations induce missense or even nonsense mutations in the overlapping polymerase gene (Table 1) that might affect HBV DNA replication and, consequently, the amount of virions secreted.
Envelope protein expression and HBsAg secretion.
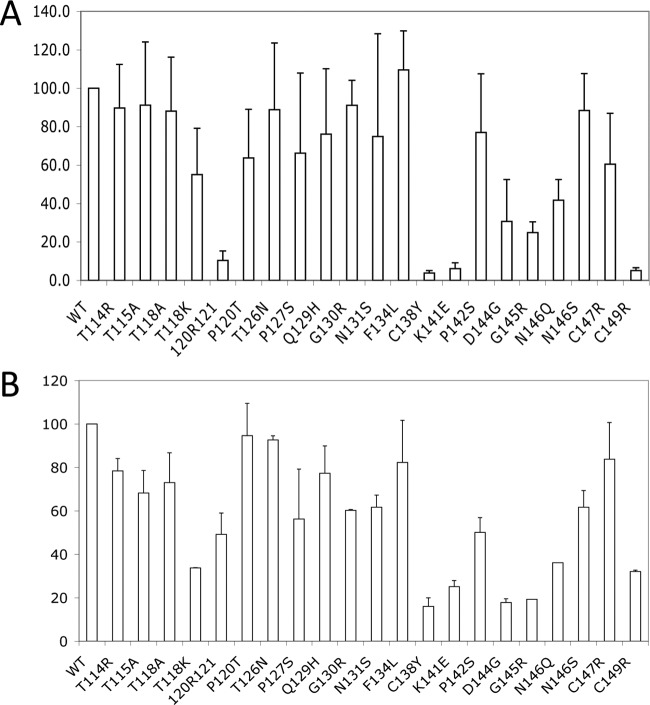
The Huh7 human hepatoma cell line grown in 6-well plates was cotransfected with the 1.5mer replication construct (1.5 μg) and the 0.7mer expression construct (0.5 μg). A Western blot analysis of cell lysate revealed expression of L, M, and S proteins by all the mutants (Fig. 1A and B). HBsAg secreted to culture supernatant was quantified by an enzyme-linked immunosorbent assay (ELISA) using three commercial kits. The Auszyme Monoclonal kit (Abbott Laboratories) revealed a negligible amount of HBsAg for the 120R121 insertion mutant as well as the C138Y, K141E, and C149R substitution mutants and significantly reduced HBsAg for the D144G and G145R mutants (Fig. 2A). According to the ETI-MAK-2plus kit (DiaSorin), HBsAg was lowest for the C138Y, D144G, G145R, and K141E mutants (Fig. 2B). Another kit (Abazyme) found a negligible amount of secreted HBsAg for a larger number of mutants, including 120R121, P120T, C138Y, K141E, P142S, D144G, G145R, C147R, and C149R (data not shown). Further studies are needed to clarify whether the low values of HBsAg in culture supernatant are a consequence of impaired HBsAg detection, secretion, or stability.
Fig 1.
Impact of amino acid substitutions in the immunodominant loop on virion secretion. Huh7 cells grown in 6-well plates were cotransfected with 1.5 μg of the 1.5mer replication construct and 0.5 μg of the 0.7mer WT or mutant envelope protein construct or 0.5 μg of vector DNA. Alternatively, cells were transfected with 1.5 μg of N16, a 1.5mer construct capable of envelope protein expression. Cells and culture supernatant were harvested at day 5 posttransfection. (A and B) One aliquot of cell lysates was used for a Western blot analysis with a monoclonal anti-preS2 antibody (S26; Virogen) (A) and, following stripping, with a horse polyclonal anti-S antibody (anti-Ad/Ay; Abcam) (B). The two bands for L, M, and S proteins are gp42/p39, gp36/gp33, and gp27/p24, respectively. Please notice the lack of gp42, gp36, and gp27 bands in the N146S mutant. (C) Another aliquot was used for a Western blot analysis of core protein. (D) A third aliquot was used for a Southern blot analysis of replicative DNA. (E) Virions were immunoprecipitated from culture supernatant using the monoclonal anti-preS2 antibody (Virogen), and DNA extracted was subjected to a Southern blot analysis with a full-length HBV DNA probe.
Fig 2.
Detection of secreted HBsAg by two commercial ELISA kits. An aliquot of culture supernatant (a 1:10 diluted sample corresponding to the original volume of 2 to 10 μl) was used for the assay to avoid signal saturation. Values for the WT construct are set at 100%. Results are based on three independent experiments. (A) Abbott kit. (B) DiaSorin kit.
Impairment in virion secretion.
Analysis of intracellular core protein (Fig. 1C), replicative DNA (Fig. 1D), and secreted HBeAg (data not shown) revealed similar transfection efficiencies of the 1.5mer construct, as anticipated. Virions were concentrated from culture supernatant by immunoprecipitation with a monoclonal anti-preS2 antibody (Virogen), followed by DNA extraction and Southern blot analysis. We did not use anti-S antibodies for immunoprecipitation in case immune escape mutations weaken antibody binding to mutant envelope proteins. The G130R, C138Y, D144G, N146S, and C149R mutants were completely defective in virion secretion (Fig. 1E), while the T114R, T115A, P127S, Q129H, C147R, and K141E mutants secreted very few virions. In contrast, the T118A, P120T, and N131S mutants displayed efficient virion secretion. Very similar results were obtained when virions were immunoprecipitated with R254, a rabbit polyclonal antibody targeting the preS1 domain of the L protein (data not shown) (33).
Most mutants with low HBsAg values in culture supernatant (suggestive of impaired HBsAg secretion or detection or reduced HBsAg stability) were also impaired in virion secretion. On the other hand, several mutants with severely impaired virion secretion had no major defect in HBsAg recognition, at least according to the three kits employed in this study (G130R, N146S). As we already reported, the N146S mutant is deficient in virion secretion due to ablation of the N-linked glycosylation site in the S protein (29). As for the G130R mutant, its S protein was easily detectable by Western blotting if preconcentrated from culture supernatant by immunoprecipitation with the same anti-S polyclonal antibody but not if precipitated by polyethylene glycol (PEG) (data not shown). Thus, this mutation might disrupt particle formation or trigger its disassembly.
The observation that the C138Y, C147R, and C149R mutants were all severely hampered in virion secretion is consistent with the importance of cysteine residues in maintaining the proper intramolecular or intermolecular disulfide bonds in the immunodominant loop (34, 35). Analysis of additional mutations (C121A, C124A, C147A, and C149A) revealed that they severely reduce virion secretion as well (Fig. 3C), although HBsAg secretion was unaltered by the C147A mutation and only moderately reduced by the C149A mutation when analyzed by the Abbott kit (Fig. 3D).
Fig 3.
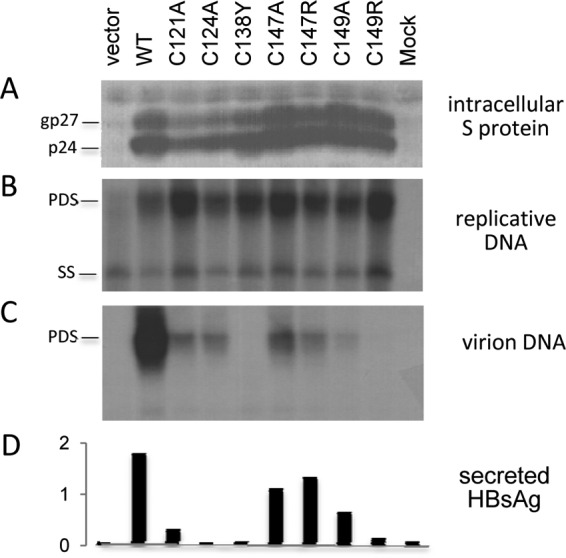
Critical role of cysteine residues in the immunodominant loop on virion secretion. Huh7 cells grown in 6-well plates were cotransfected with 1.5 μg of the 1.5mer replication construct and 0.5 μg of the 0.7mer expression construct as indicated or with vector DNA. Cells and culture supernatant were harvested at day 5 posttransfection. (A and B) An aliquot of cell lysate was used for a Western blot analysis of S protein (A), and another aliquot was used for a Southern blot analysis of replicative DNA (B). (C) Virions were immunoprecipitated from culture supernatant using an anti-preS2 monoclonal antibody, followed by Southern blotting. (D) Secreted HBsAg was determined by the Abbott ELISA kit.
Rescue of virion secretion by coexpression of wild-type envelope proteins.
Immune escape mutants often coexist with a small amount of wild-type virus. We cotransfected a mutant 0.7mer construct with a wild-type 0.7mer construct at a 5:1 (0.5 μg/0.1 μg) or 1:1 (0.3 μg/0.3 μg) ratio to examine whether the wild-type envelope proteins can rescue virion secretion through phenotypic mixing. At a 5:1 ratio, virion secretion was rescued efficiently only for the P127S and K141E mutants and moderately for the G145R mutant (Fig. 4A). It is interesting to note that the K141E mutant has been associated with vaccine failure in two Gambian children despite protective levels of anti-HBs antibody (7). Coexpressing mutant and wild-type envelope proteins at a 1:1 ratio led to efficient rescue of virion secretion for all the mutants except C138Y (Fig. 4C). However, at such a ratio, a high level of HBsAg was detectable in culture supernatant, especially according to PEG precipitation followed by Western blotting (Fig. 4E), suggesting a reversal of the immune escape phenotype or restoration in HBsAg secretion. It remains to be determined whether virions produced by coexpression of mutant and wild-type envelope proteins contain a mixture of the two types of proteins or just the wild-type proteins (if the mutant proteins have a dominant negative effect on virion formation or secretion). In the latter case, viral infectivity will still be neutralized by anti-S antibodies raised against wild-type envelope proteins.
Fig 4.
Rescue of virion secretion by coexpression of wild-type envelope proteins at two different ratios. Huh7 cells grown in 6-well plates were transfected with 1.4 μg of the 1.5mer replication construct, together with 0.5 μg of the mutant 0.7mer expression construct plus 0.1 μg of vector DNA or the WT 0.7mer expression construct (A and B) or 0.3 μg each of the mutant 0.7mer expression construct and vector DNA or the WT expression construct (C to F). Cells and culture supernatant were harvested at day 5 posttransfection. (F) Cell lysate was used for a Western blot analysis of S protein. (A and C) The bulk of the culture supernatant was used for immunoprecipitation by an anti-preS2 antibody, followed by Southern blotting. (E) Another aliquot was used for PEG precipitation, followed by Western blotting with an anti-S antibody. A small volume was used for HBsAg detection by the Abbott kit (10 μl for panel B and 5 μl for panel D).
Rescue of virion secretion by an M133T mutation.
We recently found that a virion secretion defect associated with two missense mutations in the viral envelope proteins (I110M and G119E) as well as the classic G145R immune escape mutation can be overcome by another naturally occurring mutation, M133T, which creates a novel N-linked glycosylation site (29, 31). In fact, M133T itself is frequently associated with occult HBV infection or failed HBIG prophylaxis and often associates with “a” determinant mutations such as G130N, F134L, D144A, D144G, G145A, G145K, and G145R (12, 36–40). Introduction of the M133T mutation rescued virion secretion efficiently for the T118K, P127S, and N146S mutants (Fig. 5B). It also rescued virion secretion for the G130R, C138Y, C147R, and C149R mutants but much less efficiently. As expected, the M133T mutation is associated with an additional protein band consistent with the doubly glycosylated S protein (Fig. 5A).
Fig 5.

Rescue of virion secretion by the M133T mutation. Huh7 cells were cotransfected with 1.5 μg of the 1.5mer replication construct and 0.5 μg of the 0.7mer mutant envelope protein construct with or without the extra M133T mutation. (A) Western blot analysis of intracellular S protein. (B) Southern blot analysis of virion DNA following immunoprecipitation with the anti-preS2 antibody. (C) Secreted HBsAg as measured by the Abbott kit.
Are S protein residues critical for virion secretion also important for infectivity?
The S protein is involved not only in HBV virion secretion but also in viral infectivity. Interestingly, many immune escape mutations also severely impaired infectivity of hepatitis delta virus (41), which hijacks HBV envelope proteins for release from and entry into hepatocytes. The immune escape mutations were detrimental to infectivity when present on the S but not the L protein (33). Similarly, we found that introducing the I110M, G119E, and R169P mutations into the S protein impaired HBV virion secretion but that virion secretion was unaltered when the same mutations were introduced to L and M proteins only (29). Moreover, I110, G119, C124, K141, P142, C147, and C149 are critical for both virion formation and viral infectivity, with the notable exception that the G145R mutation did not compromise infectivity despite severely impairing virion formation (29, 33, 41). This maintenance of infectivity might explain its higher prevalence compared to other immune escape mutations. Further investigation is warranted to establish whether the same set of amino acid residues in the S protein controls both HBV release from and entry into hepatocytes, if entry (uncoating) is the reverse of envelopment.
The threat posed by immune escape mutants depends on the degree of immune escape as well as their biological fitness. Besides mutations in the “a” determinant, which may affect virion release from and entry into hepatocytes, missense mutations in the overlapping P gene (Table 1) might influence viral replication capacity, a topic which remains to be explored in the future. Antibodies made in response to the HBV vaccine mostly target an epitope composed of residues 141 to 146 (2), and mutations at this site will have the highest degree of immune escape. Nevertheless, as shown before and here, K141E, P142S, D144G, G145R, and loss of the N-linked glycosylation site at N146 severely impair virion secretion and are thus predicted to delay virus spread (29, 30). As these mutations (except G145R) probably also impair HBV infectivity (33, 41), they might not spread infection in the new host quickly enough to avoid clearance by the immune response. The deficiency in virion formation can be rescued by the WT virus at a 1:1 ratio, but that will most likely reverse the immune escape phenotype of the mosaic virus particle. A more worrisome scenario is the acquisition of a second-site mutation, such as M133T, which efficiently restores virion secretion of some immune escape mutants without reversing the immune escape phenotype (Fig. 5C). It should be pointed out that most non-A HBV genotypes contain T131 instead of N131, and for these genotypes, the M133T mutation alone is insufficient to create a novel N-linked glycosylation site. Nevertheless, a T131N/M133T double mutation creating such a glycosylation site in non-A genotypes is frequently observed (9, 42) (X. Zhang and J. Zhang, personal communications) and can be associated with G145R (38).
ACKNOWLEDGMENTS
This work was supported by NIH grants R21-CA-133976 and P01-AA-019072.
Footnotes
Published ahead of print 5 December 2012
REFERENCES
- 1. Zanetti AR, Van Damme P, Shouval D. 2008. The global impact of vaccination against hepatitis B: a historical overview. Vaccine 26:6266–6273 [DOI] [PubMed] [Google Scholar]
- 2. Steward MW, Partidos CD, D'Mello F, Howard CR. 1993. Specificity of antibodies reactive with hepatitis B surface antigen following immunization with synthetic peptides. Vaccine 11:1405–1414 [DOI] [PubMed] [Google Scholar]
- 3. Carman WF, Zanetti AR, Karayiannis P, Waters J, Manzillo G, Tanzi E, Zuckerman AJ, Thomas HC. 1990. Vaccine-induced escape mutant of hepatitis B virus. Lancet 336:325–329 [DOI] [PubMed] [Google Scholar]
- 4. Harrison TJ, Hopes EA, Oon CJ, Zanetti AR, Zuckerman AJ. 1991. Independent emergence of a vaccine-induced escape mutant of hepatitis B virus. J. Hepatol. 13(Suppl 4):S105–S107 [DOI] [PubMed] [Google Scholar]
- 5. Hsu HY, Chang MH, Ni YH, Chen HL. 2004. Survey of hepatitis B surface variant infection in children 15 years after a nationwide vaccination programme in Taiwan. Gut 53:1499–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hsu HY, Chang MH, Ni YH, Lin HH, Wang SM, Chen DS. 1997. Surface gene mutants of hepatitis B virus in infants who develop acute or chronic infections despite immunoprophylaxis. Hepatology 26:786–791 [DOI] [PubMed] [Google Scholar]
- 7. Karthigesu VD, Allison LM, Fortuin M, Mendy M, Whittle HC, Howard CR. 1994. A novel hepatitis B virus variant in the sera of immunized children. J. Gen. Virol. 75(Part 2):443–448 [DOI] [PubMed] [Google Scholar]
- 8. Shahmoradi S, Yahyapour Y, Mahmoodi M, Alavian SM, Fazeli Z, Jazayeri SM. 2012. High prevalence of occult hepatitis B virus infection in children born to HBsAg-positive mothers despite prophylaxis with hepatitis B vaccination and HBIG. J. Hepatol. 57:515–521 [DOI] [PubMed] [Google Scholar]
- 9. Carman WF, Trautwein C, van Deursen FJ, Colman K, Dornan E, McIntyre G, Waters J, Kliem V, Muller R, Thomas HC, Manns MP. 1996. Hepatitis B virus envelope variation after transplantation with and without hepatitis B immune globulin prophylaxis. Hepatology 24:489–493 [DOI] [PubMed] [Google Scholar]
- 10. Ghany MG, Ayola B, Villamil FG, Gish RG, Rojter S, Vierling JM, Lok AS. 1998. Hepatitis B virus S mutants in liver transplant recipients who were reinfected despite hepatitis B immune globulin prophylaxis. Hepatology 27:213–222 [DOI] [PubMed] [Google Scholar]
- 11. McMahon G, Ehrlich PH, Moustafa ZA, McCarthy LA, Dottavio D, Tolpin MD, Nadler PI, Ostberg L. 1992. Genetic alterations in the gene encoding the major HBsAg: DNA and immunological analysis of recurrent HBsAg derived from monoclonal antibody-treated liver transplant patients. Hepatology 15:757–766 [DOI] [PubMed] [Google Scholar]
- 12. Protzer-Knolle U, Naumann U, Bartenschlager R, Berg T, Hopf U, Meyer zum Buschenfelde KH, Neuhaus P, Gerken G. 1998. Hepatitis B virus with antigenically altered hepatitis B surface antigen is selected by high-dose hepatitis B immune globulin after liver transplantation. Hepatology 27:254–263 [DOI] [PubMed] [Google Scholar]
- 13. Shields PL, Owsianka A, Carman WF, Boxall E, Hubscher SG, Shaw J, O'Donnell K, Elias E, Mutimer DJ. 1999. Selection of hepatitis B surface “escape” mutants during passive immune prophylaxis following liver transplantation: potential impact of genetic changes on polymerase protein function. Gut 45:306–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Terrault NA, Zhou S, McCory RW, Pruett TL, Lake JR, Roberts JP, Ascher NL, Wright TL. 1998. Incidence and clinical consequences of surface and polymerase gene mutations in liver transplant recipients on hepatitis B immunoglobulin. Hepatology 28:555–561 [DOI] [PubMed] [Google Scholar]
- 15. Chiou HL, Lee TS, Kuo J, Mau YC, Ho MS. 1997. Altered antigenicity of ‘a’ determinant variants of hepatitis B virus. J. Gen. Virol. 78(Part 10):2639–2645 [DOI] [PubMed] [Google Scholar]
- 16. Cooreman MP, van Roosmalen MH, te Morsche R, Sunnen CMG, de Ven EM, Jansen JB, Tytgat GN, de Wit PL, Paulij WP. 1999. Characterization of the reactivity pattern of murine monoclonal antibodies against wild-type hepatitis B surface antigen to G145R and other naturally occurring “a” loop escape mutations. Hepatology 30:1287–1292 [DOI] [PubMed] [Google Scholar]
- 17. Seddigh-Tonekaboni S, Waters JA, Jeffers S, Gehrke R, Ofenloch B, Horsch A, Hess G, Thomas HC, Karayiannis P. 2000. Effect of variation in the common “a” determinant on the antigenicity of hepatitis B surface antigen. J. Med. Virol. 60:113–121 [DOI] [PubMed] [Google Scholar]
- 18. Waters JA, Kennedy M, Voet P, Hauser P, Petre J, Carman W, Thomas HC. 1992. Loss of the common “A” determinant of hepatitis B surface antigen by a vaccine-induced escape mutant. J. Clin. Invest. 90:2543–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jeantet D, Chemin I, Mandrand B, Tran A, Zoulim F, Merle P, Trepo C, Kay A. 2004. Cloning and expression of surface antigens from occult chronic hepatitis B virus infections and their recognition by commercial detection assays. J. Med. Virol. 73:508–515 [DOI] [PubMed] [Google Scholar]
- 20. Minuk GY, Sun DF, Uhanova J, Zhang M, Caouette S, Nicolle LE, Gutkin A, Doucette K, Martin B, Giulivi A. 2005. Occult hepatitis B virus infection in a North American community-based population. J. Hepatol. 42:480–485 [DOI] [PubMed] [Google Scholar]
- 21. Wands JR, Marciniak RA, Isselbacher KJ, Varghese M, Don G, Halliday JW, Powell LW. 1982. Demonstration of previously undetected hepatitis B viral determinants in an Australian Aboriginal population by monoclonal anti-hbs antibody radioimmunoassays. Lancet i:977–980 [DOI] [PubMed] [Google Scholar]
- 22. Weinberger KM, Bauer T, Bohm S, Jilg W. 2000. High genetic variability of the group-specific a-determinant of hepatitis B virus surface antigen (HBsAg) and the corresponding fragment of the viral polymerase in chronic virus carriers lacking detectable HBsAg in serum. J. Gen. Virol. 81:1165–1174 [DOI] [PubMed] [Google Scholar]
- 23. Zaaijer HL, Torres P, Ontanon A, Ponte LG, Koppelman MH, Lelie PN, Hemert FJ, Boot HJ. 2008. Multiple surface antigen mutations in five blood donors with occult hepatitis B virus infection. J. Med. Virol. 80:1344–1349 [DOI] [PubMed] [Google Scholar]
- 24. Wilson JN, Nokes DJ, Carman WF. 1998. Current status of HBV vaccine escape variants—a mathematical model of their epidemiology. J. Viral Hepat. 5(Suppl 2):25–30 [DOI] [PubMed] [Google Scholar]
- 25. Zuckerman AJ. 2000. Effect of hepatitis B virus mutants on efficacy of vaccination. Lancet 355:1382–1384 [DOI] [PubMed] [Google Scholar]
- 26. Hsu HY, Chang MH, Ni YH, Chiang CL, Chen HL, Wu JF, Chen PJ. 2010. No increase in prevalence of hepatitis B surface antigen mutant in a population of children and adolescents who were fully covered by universal infant immunization. J. Infect. Dis. 201:1192–1200 [DOI] [PubMed] [Google Scholar]
- 27. Kamili S, Araujo A, Fields H, Khudyakov Y, Krawczynski K, Locarinini S, Spelbring J, Trautwein C, Yokosawa J. 2002. Experimental infection of chimpanzees with genetically engineered S-gene mutants of hepatitis B virus. Hepatology 36:313A [Google Scholar]
- 28. Ogata N, Zanetti AR, Yu M, Miller RH, Purcell RH. 1997. Infectivity and pathogenicity in chimpanzees of a surface gene mutant of hepatitis B virus that emerged in a vaccinated infant. J. Infect. Dis. 175:511–523 [DOI] [PubMed] [Google Scholar]
- 29. Ito K, Qin Y, Guarnieri M, Garcia T, Kwei K, Mizokami M, Zhang J, Li J, Wands JR, Tong S. 2010. Impairment of hepatitis B virus virion secretion by single-amino-acid substitutions in the small envelope protein and rescue by a novel glycosylation site. J. Virol. 84:12850–12861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kalinina T, Iwanski A, Will H, Sterneck M. 2003. Deficiency in virion secretion and decreased stability of the hepatitis B virus immune escape mutant G145R. Hepatology 38:1274–1281 [DOI] [PubMed] [Google Scholar]
- 31. Khan N, Guarnieri M, Ahn SH, Li J, Zhou Y, Bang G, Kim KH, Wands JR, Tong S. 2004. Modulation of hepatitis B virus secretion by naturally occurring mutations in the S gene. J. Virol. 78:3262–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garcia T, Li J, Sureau C, Ito K, Qin Y, Wands J, Tong S. 2009. Drastic reduction in the production of subviral particles does not impair hepatitis B virus virion secretion. J. Virol. 83:11152–11165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Le Duff Y, Blanchet M, Sureau C. 2009. The pre-S1 and antigenic loop infectivity determinants of the hepatitis B virus envelope proteins are functionally independent. J. Virol. 83:12443–12451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. El Chaar M, Candotti D, Crowther RA, Allain JP. 2010. Impact of hepatitis B virus surface protein mutations on the diagnosis of occult hepatitis B virus infection. Hepatology 52:1600–1610 [DOI] [PubMed] [Google Scholar]
- 35. Mangold CM, Unckell F, Werr M, Streeck RE. 1995. Secretion and antigenicity of hepatitis B virus small envelope proteins lacking cysteines in the major antigenic region. Virology 211:535–543 [DOI] [PubMed] [Google Scholar]
- 36. Beckebaum S, Malago M, Dirsch O, Cicinnati VR, Trippler M, Lampertico P, Lama N, Treichel U, Gerken G, Broelsch CE. 2003. Efficacy of combined lamivudine and adefovir dipivoxil treatment for severe HBV graft reinfection after living donor liver transplantation. Clin. Transplant. 17:554–559 [DOI] [PubMed] [Google Scholar]
- 37. Cheung WI, Chan HL, Leung VK, Tse CH, Fung K, Lin SY, Wong A, Wong VW, Chau TN. 2010. Reactivation of hepatitis B virus infection with persistently negative HBsAg on three HBsAg assays in a lymphoma patient undergoing chemotherapy. J. Clin. Virol. 47:193–195 [DOI] [PubMed] [Google Scholar]
- 38. Hou J, Wang Z, Cheng J, Lin Y, Lau GK, Sun J, Zhou F, Waters J, Karayiannis P, Luo K. 2001. Prevalence of naturally occurring surface gene variants of hepatitis B virus in nonimmunized surface antigen-negative Chinese carriers. Hepatology 34:1027–1034 [DOI] [PubMed] [Google Scholar]
- 39. Jongerius JM, Wester M, Cuypers HT, van Oostendorp WR, Lelie PN, van der Poel CL, van Leeuwen EF. 1998. New hepatitis B virus mutant form in a blood donor that is undetectable in several hepatitis B surface antigen screening assays. Transfusion 38:56–59 [DOI] [PubMed] [Google Scholar]
- 40. Kato J, Hasegawa K, Torii N, Yamauchi K, Hayashi N. 1996. A molecular analysis of viral persistence in surface antigen-negative chronic hepatitis B. Hepatology 23:389–395 [DOI] [PubMed] [Google Scholar]
- 41. Salisse J, Sureau C. 2009. A function essential to viral entry underlies the hepatitis B virus “a” determinant. J. Virol. 83:9321–9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Song BC, Kim SH, Kim H, Ying YH, Kim HJ, Kim YJ, Yoon JH, Lee HS, Cha CY, Kook YH, Kim BJ. 2005. Prevalence of naturally occurring surface antigen variants of hepatitis B virus in Korean patients infected chronically. J. Med. Virol. 76:194–202 [DOI] [PubMed] [Google Scholar]