Abstract
Following integration, HIV-1 in most cases produces active infection events; however, in some rare instances, latent infection events are established. The latter have major clinical implications, as latent infection allows the virus to persist despite antiretroviral therapy. Both the cellular factors and the viral elements that potentially determine whether HIV-1 establishes active or latent infection events remain largely elusive. We detail here the contribution of different long terminal repeat (LTR) sequences for the establishment of latent HIV-1 infection. Using a panel of full-length replication-competent virus constructs that reflect naturally occurring differences of HIV-1 subtype-specific LTRs and targeted LTR mutants, we found the primary ability of HIV-1 to establish latent infection in this system to be controlled by a four-nucleotide (nt) AP-1 element just upstream of the NF-κB element in the viral promoter. Deletion of this AP-1 site mostly deprived HIV-1 of the ability to establish latent HIV-1 infection. Extension of this site to a 7-nt AP-1 sequence massively promoted latency establishment, suggesting that this promoter region represents a latency establishment element (LEE). Given that these minimal changes in a transcription factor binding site affect latency establishment to such large extent, our data support the notion that HIV-1 latency is a transcription factor restriction phenomenon.
INTRODUCTION
Antiretroviral therapy (ART) reduces the viral load to extremely low or undetectable levels, but, following cessation of ART, viral infection rebounds within a few weeks. It is believed that the major viral reservoir driving this viral resurgence is a population of latently HIV-1-infected CD4+ memory T cells (1–6). Owing to the extremely long half-life of the memory T cells in which the latent virus resides, in the absence of any de novo infection, natural eradication would take >70 years (7, 8). The only way forward toward a cure for HIV-1 infection would thus be a therapeutic strategy that actively purges this viral reservoir.
Essential to the development of effective reactivation strategies would be a comprehensive understanding of how latency is established and controlled. Once integrated, HIV-1 can essentially be viewed as another cellular gene. In many ways, the HIV-1 promoter is highly similar to a series of promoters of cellular genes that are not active in resting T cells but are upregulated following T cell activation. Among these, the most notable are cellular promoters for the interleukin-2 (IL-2) receptor (CD25), tumor necrosis factor alpha (TNF-α), IL-2, IL-6, and IL-8 (9, 10). All of these, as in the case of HIV-1, have a CD28-responsive element (CD28RE) that is crucial for efficient gene expression, and as for HIV-1 reactivation, stimulation of CD28 is essential for the efficient activation of these genes. None of these genes is expressed in memory T cells, which constitute the majority of the latent HIV reservoir. Their expression is restricted by the absence of transcription factors, with no requirement of a special restrictive chromatin environment. For all of these genes, as for HIV-1, paused RNAP II polymerase has been found bound to the transcription start site and restriction of P-TEFb and TFIIH, two important components of the actively transcribing RNA polymerase II complex, has been shown to contribute to a latent/nonexpressing phenotype (11, 12).
As with all genomic DNA, following integration, the latent HIV-1 long terminal repeat (LTR) is expected to be embedded in a chromatin structure. The current model of latent HIV-1 infection suggests that following integration, nucleosome positioning and the formation of a restrictive chromatin environment on the latent HIV-1 LTR are the key players for the control of latent HIV-1 infection (13–15). DNA methylation patterns at the viral LTR have been suggested by some to stabilize latent infection (16, 17). Two nucleosomes have been described to be located at well-defined positions on the latent HIV-1 LTR (see Fig. 2). The two nucleosomes, nuc-0 and nuc-1, are positioned at nucleotides (nt) 40 to 200 and nt 465 to 610, respectively, and are separated by a nucleosome-free region of approximately 265 nt (18, 19). This nucleosome-free region includes the CD28RE, the enhancer element, and the core promoter. For HIV-1 subtype B viruses, the most relevant transcription factor binding sites in this area are the two NF-κB sites in the enhancer element as well as the three Sp-1 sites in the core promoter. Nuc-1 is then positioned downstream of the CATATAA box and the transcriptional start site and overlaps a series of three AP-1 sites, which also are essential for HIV-1 transcription (20–23).
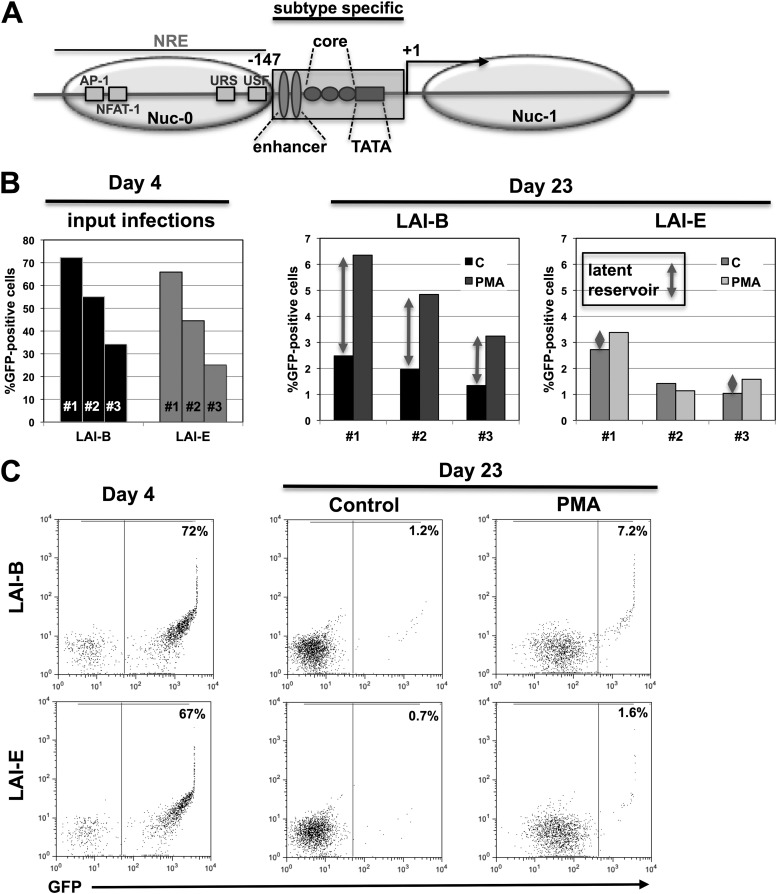
Fig 2.
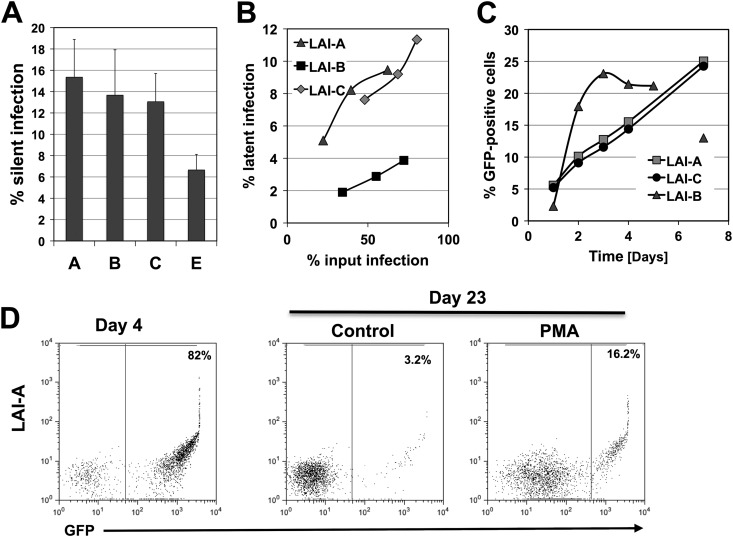
The enhancer/core promoter element of the LTR controls HIV-1 latency establishment. (A) Schematic overview of the HIV-1 LTR showing some of the key elements and transcription factor binding sites, including the positions of the nucleosomes. The 147-nt enhancer/core promoter region that is exchanged in the different viral constructs is indicated by a box (subtype specific). (B) Input infection for three independent infection experiments using LAI-B and LAI-E were measured as the level of GFP-positive cells on day 3 postinfection. The size of the latent infection reservoir was then determined on day 23 postinfection by comparing the percentages of GFP-positive cells under baseline conditions (black columns) and 24 h following PMA stimulation (gray columns). For each infection experiment, the size of the latent reservoir is indicated by gray arrows. C, control. (C) Flow cytometric data for one representative latency establishment experiment. Initial active infection levels of J2574 reporter T cells with either LAI-B or LAI-E were determined by flow cytometric analysis for GFP expression on day 4 postinfection. On day 23, when stable latent infection was established, samples of the respective cell populations were stimulated for 24 h with PMA (10 ng/ml) and active infection levels in control and PMA treated cell cultures were determined by flow cytometric analysis. The difference between the percentages of GFP-positive cells in the PMA-treated cell population and the control cell cultures represents the latent infection reservoir.
Recently, Rafati et al. reported that for latent HIV-1 infection events, the two nucleosomes that are found at the LTR would be actively repositioned away from their predicted DNA binding sites as a function of the presence of BAF or PBAF, respectively, to possibly restrict access of activating transcription factors to the LTR (24). Similar findings had been reported in 2007 for the inactive IL-2 receptor (CD25) promoter (25), again emphasizing the similarities between latent HIV-1 infection and certain inactive T cell-specific cellular genes that are controlled primarily by transcription factor restriction.
While the role of chromatin restriction for HIV-1 latency control is emphasized by some, the importance of transcription factor restriction for HIV latency establishment was suggested early on, when it was hypothesized that in primary T cells, HIV-1 latency is generated by viral shutdown during the transition of activated T cells to a memory phenotype (5). The idea was that transcription factor restriction caused by the transition of an infected T cell to a resting state would shut down viral transcription. No direct experimental evidence for this hypothesis has been provided, but restriction of P-TEFb and TFIIH, two important components of the actively transcribing RNA polymerase II complex, has been shown to contribute to a latent phenotype (11, 12). To this end, paused RNA polymerase II complex has been found associated with the latent LTR promoter (11, 26–29).
If transcription factor restriction were to be the driving force behind the establishment of latent HIV-1 infection, we hypothesized that it should be possible to generate virus LTR mutants that would establish different levels of latent infection in a standardized cellular environment. Similar ideas had been pursued earlier; however, in those experiments, attenuating mutations that would detrimentally affect the ability of the virus to drive gene expression and viral replication were used. Some of these attenuating mutations, e.g., one affecting the TAR element, have actually been successfully used to increase the percentage of latently infected cells in a model of HIV latency in primary T cells (12, 28, 30). However, while such mutations can be efficiently exploited to optimize experimental systems for a particular purpose, there is little evidence that such mutations are relevant for latency establishment in vivo (3, 31, 32). In fact, the ability of latent viruses to efficiently replicate upon ex vivo cell stimulation was a prerequisite for the success of the initial studies that identified and quantified the latent HIV-1 reservoir in patients (3, 33–35). In our study, only LTR mutations/alterations that were otherwise nonattenuating and that were derived from naturally occurring prototypic, subtype-specific LTR templates were considered. Using a series of subtype-specific LTR mutants that were otherwise syngeneic, we found that a 147-nt region (−1 to −147 relative to the transcriptional start site), from a prototypic HIV-1 subtype E virus, substituting for the original HIV LAI subtype B (LAI-B) sequence, would prevent latency establishment. In contrast, inserting the subtype A- or subtype C-specific 147-nt region into HIV LAI-B resulted in a massive increase in latent infection events. We narrowed down the possible actual elements responsible for these differences in latency establishment efficacy to an AP-1 binding site in the CD28RE of the LTR enhancer/core element just upstream of the NF-κB element. Extension of this 4-nt AP-1 site to a 7-nt AP-1 motif as seen in subtype A or subtype C viruses again massively increased the capacity of these viruses to establish latent infection events. Removal of this AP-1 site would practically abrogate the ability of these mutants to establish latent infection events. The results of these genetic perturbation experiments suggest that HIV-1 latency establishment and possibly latency maintenance are likely controlled at the level of transcription factor restriction. We here discuss the possible implications of our findings for future strategies to target the latent HIV-1 reservoir.
MATERIALS AND METHODS
Cell culture, plasmids, and reagents.
All T cell cultures were maintained in RPMI 1640 supplemented with 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% heat-inactivated fetal bovine serum (FBS). FBS was obtained from HyClone (Logan, UT) and was tested on a panel of latently infected cells to ensure that the utilized FBS batch did not spontaneously trigger HIV-1 reactivation (36, 37). The phorbol esters 13-phorbol-12-myristate acetate (PMA) and prostratin, the histone deacetylase (HDAC) inhibitors valproic acid, sodium butyrate (NaBu), and trichostatin A (TSA), the DNA methyltransferase inhibitor 5′-azacytidine, and the cell-differentiating agent N′-N′-hexamthylene-bisacetamide (HMBA) were purchased from Sigma. Suberoylanilide hydroxamic acid (SAHA; vorinostat) was purchased from Cayman Chemicals. Recombinant human TNF-α was obtained from R&D.
J2574 reporter T cells.
J2574 reporter T cells were generated by retrovirally transducing Jurkat T cells with an HIV-1 reporter construct (p2574) in which an HIV-1 subtype B LTR controls the expression of green fluorescent protein (GFP). The HIV-1 LTR and the GFP gene are separated by a 2,500-bp spacer element. Lentiviral particles were produced by transfecting 293T cells with p2574 and supplying gag-pol-rev-tat in trans. Vesicular stomatitis virus G (VSV-G) was used as a viral envelope protein. Following lentiviral transduction of Jurkat cells, all cells that spontaneously expressed GFP were removed by cell sorting. The GFP-negative population was then activated with PMA to identify all cells that would harbor an inducible LTR-GFP-LTR integration event. Cells that turned GFP positive following stimulation were again selected by cell sorting. GFP expression in this population ceased after a few days, leaving a population of GFP-negative reporter cells. The amount of founder cells for this population is calculated to represent >100,000 individual integration events.
HIV-1 vectors.
Some of the utilized HIV-1 clones have been previously described (38, 39). These clones are derived from the subtype B LAI molecular clone (40) but have a subtype-specific 3′ LTR from position −147 to position +63, stretching from the noncoding part of U3 to the R region, including the complete TAR hairpin. During virus production, the U3 region of the 3′ LTR is inherited by the viral progeny, and the recombinant progeny thus contains a subtype-specific −147 to −1 region. For the purpose of simplification, we refer to the viruses as LAI-B for the parental subtype B construct and LAI-A, LAI-C, and LAI-E for the viruses that hold the −147 to −1 LTR sequence of a prototypic HIV-1 strain of subtype A, subtype C, and subtype E (CRF01_AE), respectively. In all cases, viral stocks were prepared by transfection of the plasmids into 293T cells. Viral supernatants were then harvested 2 days posttransfection, divided into aliquots, and stored at −80°C.
To generate the NL43-AP-1 mutants, the NL43 viral plasmid was digested with XhoI and NcoI and the generated fragment was cloned into pGEMTeasy vector. All manipulations of the 3′ LTR were generated in this pGEM-3′LTR construct. To generate NL–ΔAP-1, forward primer ATCGAGCTTGCTACAAGGGACTTTCCGC and reverse primer GCAGTTCTTGAAGTACTCCGGATGCAGCT were used to delete the AP-1 site (TGAC) present in wild-type NL43 (NL43 wt). To generate NL-7nt/AP-1, the forward primer TCAAGAACTGCtgacacaGAGCTTGCTAC and the reverse primer AGTACTCCGGATGCAGCTCTCGG were used to generate the 7-nt AP-1 site (TGACACA; indicated with lowercase characters in the forward primer sequence). The respective PCR products were treated with DpnI to digest the methylated template DNA, the PCR products were ligated and transformed, and colonies with the correct plasmids were identified. The mutated XhoI-NcoI fragments were then transferred into the NL43 backbone.
EMSAs.
Freshly isolated primary human CD4+ T cells or CA5 T cells were stimulated with PMA (25 ng/ml) and ionomycin (1.5 μM) for 90 min. Nuclear extracts were prepared as described previously (41). An AP-1 consensus oligonucleotide was used to generate a 32P-labeled probe. Three HIV LTR oligonucleotides, reflective of the AP-1 and NF-κB sites shown (see Fig. 5), were used as unlabeled competitors. The AP-1 consensus sequence (21 nt) was CGCTTGATGACTCAGCCGGAA; the NL4-3 wild-type sequence (61 nt) was TTCAAGAACTGCTGACATCGAGCTTGCTACAAGGGACTTTCCGCTGGGGACTTTCCTAACT; the NL-7nt/AP-1 sequence (61 nt) was TTCAAGAACTGCTGACACAGAGCTTGCTACAAGGGACTTTCCGCTGGGGACTTTCCTAACT; and the NL–ΔAP-1 sequence (57 nt) was TTCAAGAACTGCATCGAGCTTGCTACAAGGGACTTTCCGCTGGGGACTTTCCTAACT. Electrophoretic mobility shift assays (EMSAs) were performed as previously described (42). Briefly, 1 μg of nuclear protein extract, 25,000 cpm of 32P-labeled probe, EMSA binding buffer (500 mM Tris-HCI, pH 7.5; 500 mM KCl; 15 mM MgCl2; 10 mg/ml bovine serum albumin [BSA]; 50% glycerol; 5 mg/ml single-stranded sperm DNA; 1 mM dithiothreitol [DTT]), and anti-c-Fos antibody or the indicated competitors were mixed and incubated for 45 min at room temperature. The mixtures were then loaded and subjected to electrophoresis on a 6% gradient using prepoured DNA retardation gels (Novex, San Diego, CA). Radioactive bands were detected by conventional autoradiography.
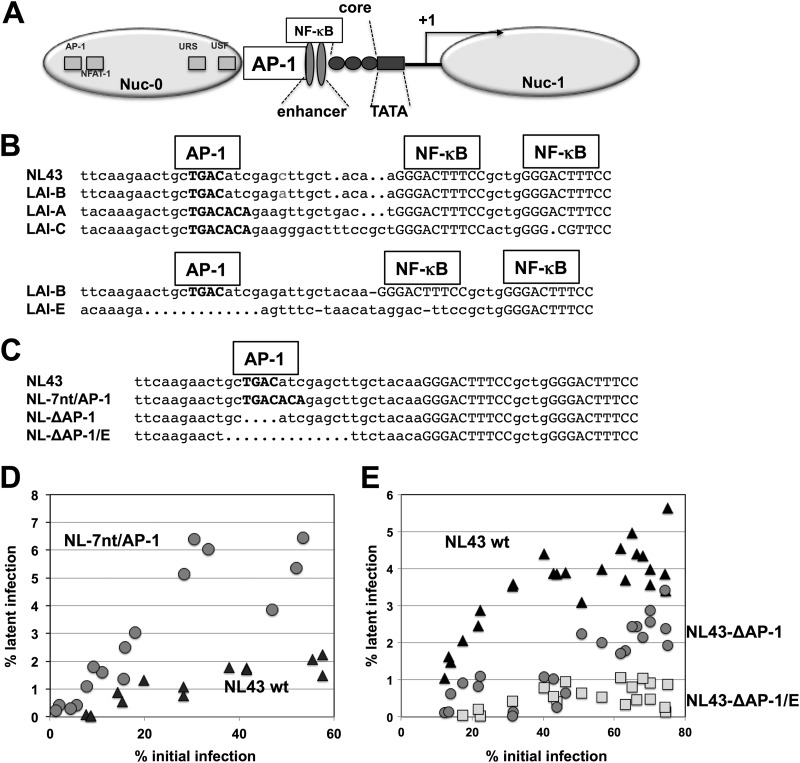
Fig 5.
HIV-1 latency establishment is controlled by an AP-1 motif in the CD28RE of the LTR. (A) The structure of the HIV-1 LTR, including the nucleosome positions and the transcriptional start site (+1). The position of the AP-1 site is indicated. (B) Alignment of prototypic CD28RE sequences (AP-1/NF-κB region) of NL43, LAI-A, LAI-B, LAI-C, and LAI-E. (C) Alignment of the generated CD28RE regions in NL43 wt, NL-7nt/AP-1, NL–ΔAP-1, and NL–ΔAP-1/E. (D) J2574 reporter T cells were infected with increasing amounts of NL43 wt (black triangles) and NL-7nt/AP-1 (gray circles), and the pool of latent HIV-1 infection for each infection culture was determined on day 14 postinfection (46). The size of the latent reservoir for each infection was plotted over the initial level of infection as determined on day 3 postinfection. (E) Similar latency establishment experiments were performed using NL43 wt (black triangles), NL–ΔAP-1 (gray circles), and NL–ΔAP-1/E (gray squares). The size of the latently infected cell pool as determined on day 14 postinfection is plotted over the level of initial infection as determined on day 3.
FCM.
Infection levels in the cell cultures were monitored by analysis of GFP expression using flow cytometry (FCM). FCM analysis was performed on a Guava easyCyte flow cytometer (Guava Technologies, Inc.), which allows for direct sample analysis in 96-well plates. The EasyCyte flow cytometer allows for the determination of absolute cell numbers in a sample, a function that was used to ensure that the levels of cell viability in all experiments were comparable. Cell sorting experiments were performed using a FACSAria flow cytometer (Becton, Dickinson). Data analysis was performed using either CellQuest (Becton, Dickinson) or Guava Express (Guava Technologies, Inc.).
RESULTS
Generation of a GFP-reporter T cell population for HIV-1 infection.
Potential differences in the ability of HIV-1 subtypes to establish latent infection would be best studied using reporter T cells in which the transcription status of HIV-1 infection events is indicated at the single-cell level by a directly accessible surrogate marker such as GFP. Several different GFP reporter cell lines have been previously published; however, all of these reporter T cell lines are of a clonal nature (43–45). As it has been previously reported that HIV-1 subtype promoter activity would be influenced by the host cell type (39), we wanted to at least avoid a potential influence of the clonal nature of these cell lines on our studies. We thus initially generated a reporter T cell population using an HIV-1 NL43-based LTR-GFP-LTR reporter vector (p2574) that would be expected to have the same integration site profile as HIV-1 (see Materials and Methods). Based on input material calculations, this population of J2574 T cells should represent >100,000 diverse founder cells and thus limit the influence of potential clonal effects on our observations.
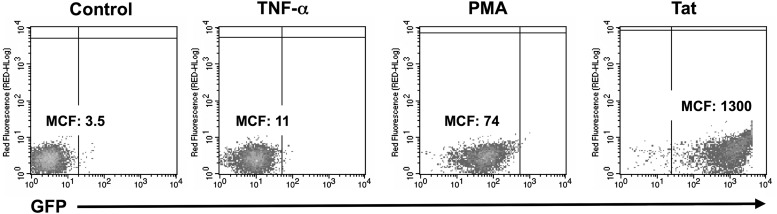
In the J2574 population, TNF-α stimulation caused a 3-fold increase in GFP expression, and PMA stimulation caused a 20-fold increase in the absence of Tat. Retroviral transduction with a Tat-expression vector triggered a >300-fold GFP shift in the transduced population and can be readily distinguished from stimulation-induced shifts in the baseline activity of the LTR-GFP-LTR reporter (Fig. 1). In this reporter T cell population, viral replication studies can be performed by directly measuring the increase in the numbers of of GFP-positive cells over time using flow cytometric analysis, without the requirement to perform other labor-intensive analyses such as p24 Gag protein determinations or reverse transcription (RT) activity assays. By the same means, changes in the viral transcriptional activity status (latent/active) in a cell population can be determined at the single-cell level. This J2574 reporter T cell population was used for the subsequent studies.
Fig 1.
GFP regulation by stimulation and Tat expression in J2574 cells. (A) To determine the responsiveness of the quantitative GFP marker to stimulation or to HIV-1 Tat, we stimulated J2574 cells with TNF-α or PMA and determined changes in GFP mean channel fluorescence (MCF) intensity after 48 h using flow cytometric analysis. To determine the effect of HIV-1 Tat, as a noncytopathic surrogate for HIV-1 infection, on the integrated LTR-GFP-LTR reporter vectors, we retrovirally transduced J2574 T cells with a Tat-expression vector and determined GFP mean channel fluorescence on day 6 postransduction (Tat).
Reduced level of latency establishment in HIV-1 LAI-E infection.
HIV-1 transcription efficacy is mostly regulated by transcription factors binding to the extended core/enhancer element of the viral LTR. Thus, we hypothesized that changes in this region would affect latency establishment efficacy if transcription factor restriction primarily governs latency establishment. To test this idea, we used a panel of fully replication-competent chimeric viruses that differed only in the sequence −147 nt to −1 nt relative to the transcriptional start site. In the various chimeric viruses, the sequence −147 nt to −1 nt was derived from prototypic, subtype-specific sequences for the HIV-1 subtypes A, B, C1, D, E, F, and G (38, 39) (Fig. 2A).
We initially tested whether the ability of HIV-1 LAI-E to drive higher levels of HIV-1 replication would result in an altered ability to establish latent infection events at the population level. For this purpose, we infected J2574 T cells with either LAI-B or LAI-E. From day 1 postinfection, the cultures were maintained in the presence of RT inhibitors to prevent the potential establishment of preintegration latency and to avert ongoing viral replication that would interfere with the establishment of a stable, latently HIV-1-infected reservoir in the respective cell populations. The results of three independent experiments performed at different infection levels are shown in Fig. 2. To allow for a meaningful comparison of the capacities of the respective viruses to establish a pool of latently infected cells, we chose infection cultures that on day 4 postinfection would produce similar levels of active infection (Fig. 2B). Active and latent infection levels in these infection cultures were then monitored until day 23, when a stable latent reservoir would have been established (46). Other than in LAI-B-infected cell populations, LAI-E infection cultures had not produced a reservoir of silently infected cells on day 3 postinfection (data not shown). HIV-1 subtype B routinely established a substantial level of silent infection events early after infection, and we previously demonstrated that the majority of latent infection events are derived from this population of silently infected cells (46). On day 23 postinfection, all infection cultures using LAI-B virus had established an appreciable reservoir of latently infected T cells, whereas LAI-E infections had produced no or only a minimal latent reservoir (Fig. 2C). Note that viability in the cell cultures on days 5 to 7 routinely dropped as a function of the infection level and then recovered over time. For example, viability in infection cultures that on day 3 exhibited active infection in 60% of the cells would drop to ∼40% on days 5 to 7 postinfection. However, we did not notice any cytopathicity difference between the various viruses used in these studies.
The inability of HIV-1 LAI-E to establish latent HIV-1 infection is correlated with an increased replication capacity.
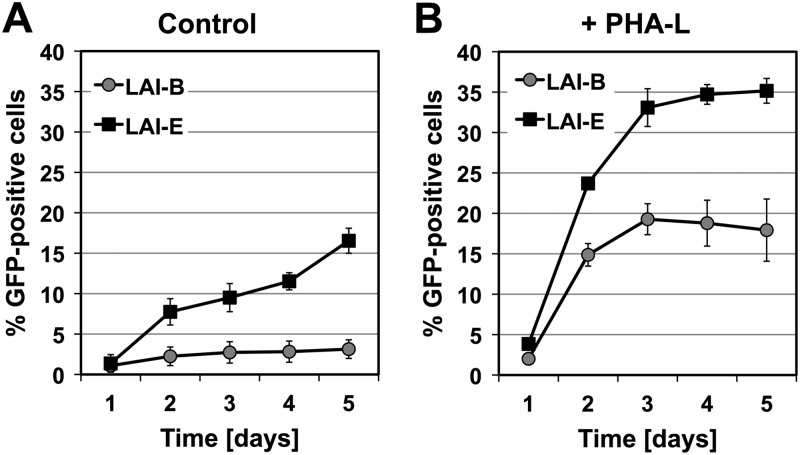
We next tested whether the ability of LAI-E to produce almost exclusively active infection events in J2574 cells would correlate with a higher level of viral replication than was seen with LAI-B. For this purpose, we infected J2574 T cells with either LAI-B or LAI-E and followed viral replication over 5 days. When adjustments for initial viral infection levels were made, we observed an ∼5-fold-higher level of viral replication until day 5 postinfection for LAI-E (Fig. 3A). Activation of the J2574 T cell population before or during infection by PHA-L with the intention to increase cellular activation levels partially ameliorated some of the replication advantage, but LAI-E replication was still more efficient (Fig. 3B). These results indicate that the observed higher replication capacity of the LAI-E LTR is an intrinsic function of the LTR. This advantage is partially retained when access to NF-κB or other transcription factors is not limiting as the result of cell activation (e.g., PHA-L). The replication advantage of LAI-E is thus somewhat independent of the presence of NF-κB and is likely mediated by another factor that increases baseline transcription. Such a scenario would explain how LAI-E prevents latency formation in low-level NF-κB activity environments that are favorable for latency formation for LAI-B. As the observed increased replication capacity is correlated to the exchange of the 147-nt enhancer/core promoter sequence, these data lend further support to the idea that establishment of latent infection is a transcription factor phenomenon and that it is inversely correlated to the transcriptional activity and replication capacity of the virus.
Fig 3.
LAI-B and LAI-E replication kinetics in J2574 cells. J2574 cells were infected with either LAI-B or LAI-E, and viral replication was monitored by determining the percentage of GFP-positive cells at the indicated time points using flow cytometric analysis. Viral replication efficacy was determined in (A) unstimulated J2574 cells or (B) J2574 cells stimulated with 1 μg/ml PHA-L, added to increase intracellular activation levels with respect to NF-κB provision. All experiments were performed in triplicate.
Subtype A and subtype C chimeric viruses generate increased levels of latent infection.
In contrast to LAI-E, which yielded almost exclusively productive infection events, we found that LAI-A and LAI-C produced greatly increased levels of latent infection compared to HIV-1 LAI-B. Similar to LAI-B, but different from LAI-E, LAI-A and LAI-C both established a pool of silently infected cells early after infection (day 3) (Fig. 4A). We demonstrated earlier that this silent infection reservoir serves as the primary source for latent subtype B infection (46), but we could not produce any evidence that the silent infection reservoir was more pronounced in LAI-A or LAI-C infections than in LAI-B infections. On day 23 postinfection, latency levels determined as a function of the initial infection level in these cultures were increased by a factor of 3 to 5 relative to LAI-B infection levels (Fig. 4B). Again, the only difference between these two chimeric viruses and the parental LAI-B was a 147-nt region that would represent a prototypic subtype A or subtype C enhancer/core promoter sequence (38, 39), further emphasizing the governing role of this sequence for latency formation. The increased ability of LAI-A and LAI-C to establish latent infection was not associated with a decrease in replication capacity (Fig. 4C). This is different from what we observed for LAI-E, where we found the decreased ability of LAI-E to establish latent infection events to be associated with an increased ability to replicate relative to LAI-B (Fig. 3). Taken together, the results suggest a dominant role for the enhancer/core promoter element of the LTR (−147 nt to −1 nt) for latency establishment.
Fig 4.
Increased level of latency establishment in LAI-A and LAI-C infections. (A) J2574 reporter T cells were infected with LAI-A, -B, -C, or -E. On day 2 postinfection, samples from each culture were stimulated with PMA to test for the presence of transcriptional silent infection events. Adjusted for comparable input infection levels, the size of the silent infection reservoir is indicated for each virus. (B) On day 23 postinfection, we determined the size of the latent reservoir established in each infection culture. To be able to compare the actual ability of each virus to establish latent infection, the size of the established latent infection is plotted over the input infection level as determined on day 3 postinfection. (C) Replication kinetics of LAI-A, LAI-B, and LAI-C in PHA-L-stimulated J2574 T cells. (D) Flow cytometric data for one representative latency establishment experiment. Initial active infection levels of J2574 reporter T cells with LAI-A were determined by flow cytometric analysis of GFP expression on day 4 postinfection. On day 23, when stable latent infection was established, samples of the respective cell populations were stimulated for 24 h with PMA (10 ng/ml) and active infection levels in control and PMA-treated cell cultures were determined by flow cytometric analysis. The difference between the percentages of GFP-positive cells in the PMA-treated cell population and the control cell cultures represents the latent infection reservoir.
An AP-1 transcription factor binding site essentially controls HIV-1 latency establishment.
To detail the exact transcription factor binding site(s) that would be responsible for the observed differences in the capacity of these LAI viruses to establish latent infection events, we used Transcription Element Search Software (TESS) (47) as a pattern recognition software that would guide our mutational studies. Among several candidate sites, TESS indicated the presence of a 4-bp AP-1 site in the subtype B-specific LTR sequence, just upstream of the NF-κB element, that was extended to a 7-nt AP-1 element in the corresponding subtype A or subtype C LTR sequence. To test the relevance of this AP-1 element for HIV-1 latency establishment, we initially created two HIV-1 NL43 AP-1 mutants. In HIV-1 NL–ΔAP-1, we removed the 4 nt constituting the naturally occurring AP-1 site, whereas in NL-7nt/AP-1, two nucleotides were exchanged (TC > CA), to generate a 7-nt AP-1 site as seen in HIV-1 subtype A (or subtype C). It is important to appreciate that the mutations were limited to this site and did not include any of the naturally occurring subtype-specific changes of the NF-κB element sequence that are present in LAI-A, LAI-B, or LAI-E. With the exception of the AP-1, mutations NL43 wt, NL–ΔAP-1, and NL-7nt/AP-1 were syngeneic based on an HIV-1 NL43 backbone. The relevant original subtype-specific LTR sequences and the NL43 mutations are shown in Fig. 5.
To compare the abilities of the various LTR mutant viruses to establish latent infection at the highest experimentally possible resolution, we infected J2574 reporter T cells with each virus at various multiplicities of infection (MOIs), resulting in a total of 15 to 25 infections within a range of 5% to 70% active infection on day 3 postinfection. Again, RT or integrase (INT) inhibitors were added 24 h following infection to each culture, to prevent the establishment of preintegration latency and to inhibit ongoing HIV-1 replication, which would affect the formation of a latent HIV-1 reservoir (46). As previously reported, a stable latent infection reservoir is established about 10 days postinfection (46). To determine whether the LTR mutants would establish different levels of latent infection reservoirs, we initially determined active infection levels on day 3 postinfection. On day 14 postinfection, we again took samples from each infection culture and stimulated these with the phorbol ester PMA, likely the most potent HIV-1-reactivating agent in this system. At 24 h post-PMA stimulation, the levels of active HIV-1 infection in the parental unstimulated populations were compared with the level of induced HIV-1 infection in the corresponding PMA-treated cultures. Both baseline active HIV-1 expression levels and PMA-induced HIV-1 expression levels were directly quantified by determining the percentage of GFP-positive T cells using flow cytometry. In Fig. 5D and E, the size of this latent viral infection reservoir as measured on day 14 postinfection is plotted over the active infection level determined on day 3 postinfection. These data indicate that, relative to that of the parental NL43, the extension of the AP-1 site in NL-7nt/AP-1 using the respective subtype A or C sequence as the template created a virus that produced greatly increased levels of latent infection, similar to those seen with the viral template viruses LAI-A and LAI-C (Fig. 5D). Conversely, NL–ΔAP-1, in which the AP-1 site was deleted, produced significantly less latent infection, emphasizing the importance of the AP-1 motif for latency establishment (Fig. 5E). NL–ΔAP-1/E, a slightly extended deletion of the AP-1 motif which was designed using LAI-E as the template, totally abrogated the ability of this LTR mutant virus to establish any latent infection (Fig. 5E). The inability of NL–ΔAP-1/E to establish latent infection events reproduces our finding for LAI-E, albeit NL–ΔAP-1/E is syngeneic with NL4-3wt outside the ΔAP-1/E deletion, including the NF-κB sites. These data suggest that the extended AP-1 motif likely represents a latency establishment element (LEE) and should constitute a therapeutic target to trigger HIV-1 reactivation.
Differential binding affinity of AP-1 to the HIV CD28-responsive element (AP-1/NF-κB).
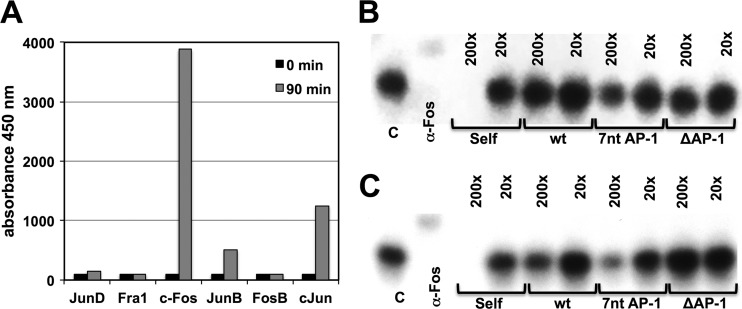
The finding that a 2-nt mutation or a 4-nt deletion in the vicinity of the NF-κB element triggered a vastly different ability of HIV-1 NL4-3 to establish latent infection would be suggestive that these mutations locally affect transcription factor binding, rather than having global effects on the interaction of the promoter sequence with other higher-level transcriptional control mechanisms. Given identical cellular environments, this would provide the viruses with differential abilities to transition to a fully expressing state. In the absence of any knowledge of what factors bind to this LTR region, it is difficult to test this hypothesis, particularly as it is even unclear whether the differences in the capacity to establish latent infection events are caused by gain-of-function or loss-of-function effects. Also, we would likely have to consider the interaction of the AP-1 motif with adjacent transcription factor binding sites that define the respective CD28-responsive element, in particular, the NF-κB binding sites. In this situation, the most promising way forward would be to test whether oligonucleotides that represent the various AP-1 motifs combined with the NF-κB element would actually differ in their ability to bind to AP-1 factors. For this purpose, we initially determined which AP-1 factors would be present in nuclear extracts from relevant T cells (CA5 T cells and primary CD4+ T cells) and found that c-Fos was highly active after induction (Fig. 6A). c-Fos also seemed to be the primary AP-1 in the nuclear extracts, as an anti-c-Fos antibody would completely shift the AP-1 consensus band. When we used oligonucleotides reflecting the AP-1/NF-κB site combinations of NL43 wt, NL-7nt AP-1, and NL–ΔAP-1 to compete at different concentrations for binding of c-Fos to the 32P-labeled AP-1 consensus oligonucleotide, we found that, relative to the AP-1/NF-κB NL43 wt oligonucleotide, the NL-7nt AP-1 oligonucleotide was much more efficient at competing for binding of AP-1 to the consensus oligonucleotide. In contrast, the NL–ΔAP-1 oligonucleotide did not compete for binding at either competitor concentration. The experiments produced identical results when we used nuclear extracts from primary CD4+ T cells (Fig. 6B) or from the latently HIV-1-infected CA5 T cells (Fig. 6C). While these data do not suggest that AP-1 or, more specifically, c-Fos is functionally relevant for the establishment of latent HIV-1 infection, the findings are consistent with the idea that the various AP-1/NF-κB sites exhibit differential levels of affinity for transcription factors. Differential affinity for select transcription factors, including c-Fos, would thus be sufficient to explain the differential ability of these LTR mutants to establish latent HIV-1 infection.
Fig 6.
Differential affinities of the HIV-1 CD28-responsive elements to c-Fos/AP-1. (A) Induction of AP-1 factor activity in CA5 T cells following stimulation with 10 ng/ml PMA. (B) EMSA to determine the affinities of the different HIV-1 CD28-responsive elements representing NL wt, NL-7nt/AP-1, and NL–ΔAP-1 (as seen in Fig. 5) to c-Fos/AP-1 using nuclear extracts from primary CD4+ T cells. A 32P-labeled consensus AP-1 site oligonucleotide (C) was either shifted with an anti-c-Fos MAb or competed with a 200× or 20× excess of cold AP-1 consensus oligonucleotide (self) or oligonucleotides representing the respective CD28-responsive elements of NL43 wt, NL-7nt/AP-1, and NL–ΔAP-1. The reduction in band intensities is inversely correlated to the affinity of the respective oligonucleotide to c-Fos/AP-1. (C) EMSA experiments were performed as described for panel B using nuclear extracts from CA5 T cells.
LTR sequences comprising the extended negative regulatory element are not required for the formation of latent HIV-1 infection.
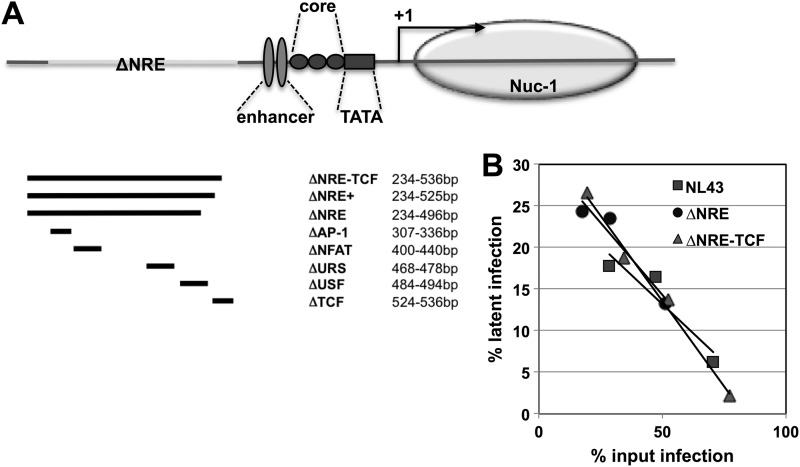
To confirm the importance of the identified AP-1 motif-based latency establishment element, we investigated whether nonattenuating mutations in other reported regulatory regions of the HIV LTR would affect HIV-1 latency establishment. For this purpose, we generated a panel of LTR deletion mutants comprising targeted mutations of individual transcription factor binding sites within the negative regulatory element (NRE) or complete and extended NRE deletion mutants (Fig. 7).
Fig 7.
The negative regulatory element of the LTR is not important for HIV-1 latency establishment. (A) To test whether relative nuc-0/nuc-1 positioning would influence latency establishment, we generated a series of LTR mutants in which we removed parts of the LTR DNA sequence that, following viral integration into the cellular genome, would be associated with nuc-0. The position of the maximal deletion in the LTR and its effect on nucleosome positioning are depicted. (B) The various LTR-deletion viruses were used to infect GFP-reporter T cells at different MOIs. On day 2 postinfection, a sample of each culture was stimulated with PMA to activate transcriptionally silent infection events. The percentage of silently infected cells was then plotted over the percentage of actively infected cells to obtain latency establishment profiles. Comparing the level of latency establishment over a range of infectivity levels is crucial, as the relative level of latent infection decreases with increasing infection levels. The latency establishment profiles of three representative experiments comparing wt NL43 with the ΔNRE-LTR mutant and with the ΔNRE-TCF-LTR mutant are depicted.
In our hands, none of these mutations resulted in attenuated viral replication capacity in our Jurkat T cell-based experimental systems (data not shown). In the subsequently performed latency establishment experiments, neither targeted mutations of individual transcription factor binding sites within the NRE nor complete or extended NRE deletion mutants (Fig. 7A) exhibited any change in the latency establishment phenotype relative to the parental virus. This was particularly striking with the largest LTR deletions, which remove more than one-third of the LTR (e.g., ΔNRE-TCF; 302 of 789 nt). In Fig. 7B, the latency phenotypes for the parental NL43 and two of the largest deletion mutants (ΔNRE and ΔNRE-TCF) are shown. Taken together, these findings suggest that the transcription factor binding sites in this region are not important for HIV-1 latency establishment, adding further support to the idea that the AP-1 site identified above is crucial for the establishment of latent HIV-1 infection.
A similar knockout approach cannot be pursued for the LTR regions downstream of the NF-κB element. This LTR region holds essential transcription factor binding sites (e.g., Sp1, AP-1) (22, 48) or essential functional elements such as the TAR element (49). Elimination of this region would be lethal to the ability of the LTR to promote transcription or viral replication. Even smaller manipulations in this area have been repeatedly reported to greatly attenuate the capacity of a virus to promote transcription or to replicate (22, 48). In fact, cells of the ACH-2 cell line, one of the latent cell lines reported early on, hold an integrated provirus that is driven into a transcriptional inactive state by an attenuating mutation in the TAR sequence (30), a finding that is now exploited by some to establish latent infection in primary T cell (12, 28).
Kinetic reactivation profiles of latently LAI-A- and LAI-B-infected T cells.
The observed differences in the abilities of the utilized HIV-1 LTR mutants to establish latent infection raised the issue of whether these would also translate into differential levels of responsiveness to reactivation. Within the panels of latently infected T cell clones established for each of the viruses, we observed great variation in their responsiveness to a defined stimulus. For example, reactivation by PMA could result in >95% reactivation for one cell clone or was limited to less than 40% in a second cell clone. Such variability has also been seen in studies from other laboratories. For example, reactivation in the various J-LAT clones from the Verdin laboratory ranged between 30% and 95% (50). While we made similar observations, we published only latently infected T cell clones in which reactivation levels generally exceed the 80% range (37, 46). In our hands, these differences are independent of the integration type, referring to the possibility that latent HIV-1 can integrate in the same orientation or the converse orientation relative to the transcription direction of the host gene or even in an intergenic region (51). Due to these large variations in responsiveness to the different stimuli in the clonal cell lines, we decided to test the ability of the different known HIV-1-reactivating agents to trigger reactivation in latently HIV-1-infected T cell populations, in which up to 20% of the cell population was actually latently HIV-1 infected. Based on input calculations, these T cell populations hold a minimum of >10,000 independent latently HIV-1-infected T cell clones. Given our results showing that LAI-A (or LAI-C) produced more latent infection events relative to the initially active infection level than LAI-B, we were particularly interested in using the latent LAI-A population to address the issue of whether latent LAI-A infection would be less responsive to known HIV-1-reactivating agents than latent LAI-B infection.
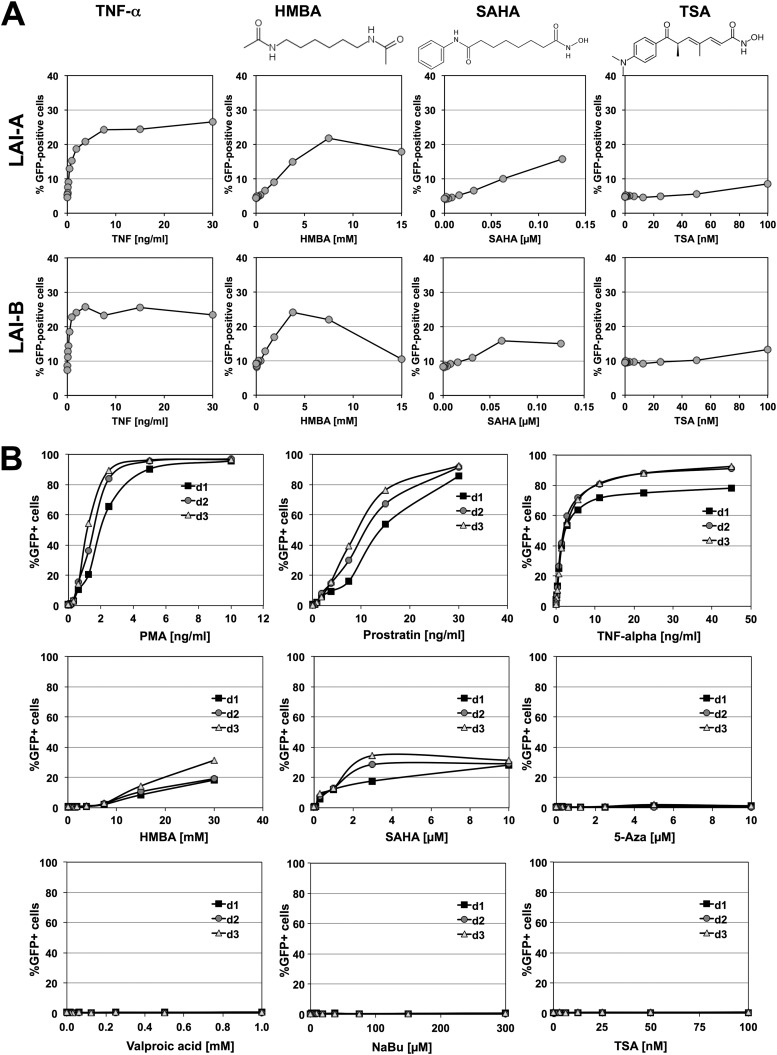
As shown in Fig. 8A, TNF-α, a strong NF-κB activator, acted equally well to reactivate latent LAI-A and LAI-B infections at the population level. Similar results were obtained for the phorbol esters PMA and prostratin (data not shown). At the population level, the cell-differentiating agent N′-N′-hexamthylene-bisacetamide (HMBA) would trigger relatively high levels of reactivation in both T cell populations. Suberoylanilide hydroxamic acid (SAHA; vorinostat), a bimodal drug that triggers cell differentiation and acts as an HDAC inhibitor, was also effective in either population. SAHA was initially developed as a cell-differentiating agent to improve HMBA (52) and 2 years later was discovered to act also as a HDAC inhibitor (53). SAHA was designed using HMBA and the HDAC inhibitor trichostatin A as structural templates (52). As reported previously (26, 31, 46), specific HDAC inhibitors such as trichostatin A, sodium butyrate, and valproic acid did not trigger relevant levels of reactivation in either cell population. Similar data were obtained for a population of latently LAI-C-infected T cells.
Fig 8.
Kinetic reactivation profiles for latent LAI-A and LAI-B infections. (A) A latently HIV-1 LAI-A-infected J2574 T cell population and a latently HIV-1 LAI-B-infected T cell population were stimulated with the NF-κB-activating cytokine TNF-α, the cell-differentiating agent HMBA, the HDAC inhibitor TSA, and the bimodal agent SAHA, which acts as both cell-differentiating agent and HDAC inhibitor. SAHA was conceived as a second-generation cell-differentiating agent using HMBA and TSA as structural templates (52). The level of HIV-1 reactivation over a low-level active background infection was determined 24 h poststimulation by determining the increase in the numbers of GFP-expressing cells. All compounds were titrated over a concentration range that did not decrease cell viability below 60%. (B) The latently HIV-1 LAI-B-infected J-BH12 T cell clone was stimulated with increasing amounts of the phorbol esters PMA and prostratin, TNF-α, the cell-differentiating agent HMBA, the bimodal (cell-differentiating/HDAC-inhibiting) SAHA, the DNA methyltransferase inhibitor 5-azacytidine, and the HDAC inhibitors valproic acid, sodium butyrate (NaBu), and trichostatin A (TSA). The HIV-1-reactivating compound effect was determined on days 1 to 3 by determinig the percentage of GFP-positive cells using flow cytometric analysis. For data inclusion, cell viability had to be >30% at the time point of analysis.
Responsiveness to a particular stimulus or the absence thereof could be confirmed using a clonal latently infected T cell clone that was selected based on the ability of several activating stimuli to trigger full reactivation at the population level. Using this clonal latently NL43 wt-infected T cell line, we found that PMA, prostratin, and TNF-α all could trigger full reactivation at the population level in a concentration-dependent manner. The cell-differentiating agents HMBA and SAHA triggered low levels of reactivation. The DNA methyltransferase inhibitor 5-azacytidine and the specific HDAC inhibitors trichostatin A, valproic acid, and sodium butyrate had no HIV-1-reactivating effect at subtoxic concentrations (Fig. 8B).
Thus, while subtype-specific mutations of the LTR greatly influence latency establishment in a given cellular environment, our data do not suggest that the molecular requirements to trigger HIV-1 reactivation greatly differ between the tested HIV-1 subtypes.
DISCUSSION
Once integrated, HIV-1 acts merely as another gene controlled by a promoter that contains a CD28-responsive element (CD28RE), very much like some T cell activation markers or cytokines (CD25, IL-2, IL-6, IL-8, TNF-α) (9, 54). Interestingly, none of these genes is actually expressed in (resting) CD4-positive memory T cells, which constitute the primary cell type known to harbor latent HIV-1 (4, 33). Transcription factor restriction is considered the primary control mechanism for the expression of these cellular genes, and in vitro stimulation with PHA-L or CD3/CD28 monoclonal antibody (MAb) combinations efficiently triggers their expression, without the requirement for any pharmacological inhibitors that would reverse a restrictive, epigenetically altered chromatin environment. Nevertheless, despite these apparent similarities, there seems to be a general assumption that latent HIV-1 infection requires a special restrictive chromatin environment that would differ from the chromatin environment found on inducible cellular genes (12, 13, 15, 55–57).
In line with the idea that, given a defined cellular environment, transcription factor restriction would be key to HIV-1 latency establishment and control, we here demonstrate that minimal mutations of a single AP-1 site within the CD28RE just upstream of the NF-κB element can dramatically alter the capacity of HIV-1 to establish latent infection events.
In the initial experiments, we found that substitution of the core/enhancer element of a subtype B virus with the corresponding region of a prototypic subtype A or subtype C virus would greatly enhance the capacity of these chimeric viruses to establish latent infection, while the subtype E core/enhancer element would deprive the chimeric virus of the ability to establish latent infection events. All chimeric viruses were syngeneic in the LTR regions outside the subtype-specific 147-nt enhancer/core promoter sequences, suggesting that any observed differences in the capacity of these viruses to establish latent infection have to be correlated with differences in the transcription factor binding site pattern of the respective 147-nt enhancer/core promoter sequences. HIV-1 latency establishment should thus primarily be a function of transcription factor availability at the time point of infection.
The increase in latent infection observed in LAI-A- and LAI-C-infected cells relative to LAI-B infection is interesting in the context of a previous observation from our laboratory. In the study by Duverger et al., we demonstrated that for subtype B viruses, the majority of latent infection events would be derived from infections that integrated in a nonactive, silent state (46). The size of this silent infection reservoir that can be observed early after infection (day 3 or 4) is substantially larger than that of the stable latent reservoir established at a later time point in these infected cultures. For LAI-A or LAI-C, which were found to produce higher levels of latent infection, we would have assumed that the silent infection reservoir on day 3 postinfection would also be larger. However, this was not the case. The larger latent reservoir in these T cell populations could thus be explained only by a decreased rate at which silent LAI-A or LAI-C infection events spontaneously transition to an active state. This notion is supported by the finding that LAI-A and LAI-C exhibit slower replication kinetics in Jurkat T cells than LAI-B (Fig. 4). Conversely, the observed increased replication capacity of the subtype E chimeric virus LAI-E relative to the subtype B reference virus LAI-B (Fig. 3) indicates that the 147-nt subtype E enhancer/core element provides the virus with a higher transcriptional baseline activity. This may then facilitate the initial production of Tat that, once it is made, feeds back to improve transcriptional elongation and possibly occlude the transcriptional stop signal that is located downstream of the TAR element (58). As a result, the subtype E chimeric virus can avoid latent infection. A possible explanation for this effect could be the growth-associated binding protein (GABP) site that has replaced the distal NF-κB element in LAI-E and is unique to HIV-1 subtype E (59). As a member of the family of Ets factors, GABP is a unique transcription factor that is an obligate multimeric complex and has originally been described to control the immediate early gene activation of herpes simplex virus 1 cytokine promoters in T cells (60). GABP is also known to enhance IL-2 promoter activity (61, 62). It is thus conceivable that GABP binding could increase baseline promoter expression of the LAI-E LTR and thus prevent the establishment of latent infection events.
The ability of a virus to establish latent infection would thus be a function of the capacity of a given LTR sequence or transcription factor binding site composition in the context of a given transcription factor profile to drive baseline LTR expression in the absence of Tat.
If this idea were correct, we hypothesized that latency establishment should be governed by a distinct transcription factor/binding site interaction within the 147-nt enhancer/core promoter sequence. As we detailed the transcription factor binding sites that mediate the observed effects on latency establishment, we identified a single AP-1 site just upstream of the NF-κB element as key for the control of latent infection. The 4-nt AP-1 site in the subtype B-specific LTR sequence is extended to a 7-nt AP-1 element in the corresponding subtype A or subtype C LTR sequence. Mutation of the 4-nt AP-1 site in HIV NL43 (subtype B) to the subtype A (or C) sequence required changes of two nucleotide positions and provided HIV NL-7nt/AP-1 with the same increased relative capacity to establish latent infection events that is found for LAI-A or LAI-C infections, compared to the parental subtype B viruses. Conversely, deletion of the 4-nt AP-1 site in HIV NL–ΔAP-1 reduced the capacity of the virus to establish latent infection events, demonstrating the importance of this transcription factor binding site for the control of latent HIV-1 infection. An extended deletion of the AP-1 motif modeled after the respective LTR sequence found in LAI-E abrogated the ability of this virus (HIV NL–ΔAP-1/E) to establish any latent infection. The different AP-1 motifs of the otherwise syngeneic CD28RE elements, indeed, exhibited differential levels of binding affinity for c-Fos. While this finding does not suggest a causal relationship with the latency establishment profile of the respective full-length viruses, it provides a proof of principle that the introduced minor mutations actually alter the composition of transcription factors binding to the CD28RE, which is in line with the idea that transcription factor restriction is key to the control of latent HIV-1 infection.
It is important to appreciate that in these mutants, other than in HIV LAI-A or HIV LAI-E, even the NF-κB elements in all three viruses are identical, supporting the notion that NF-κB may not be the limiting factor that triggers establishment of latent HIV-1 infection. Also, the finding may suggest that high-level NF-κB activation may not be required to trigger HIV-1 reactivation if the transcription factor composition at the AP-1 site can be pharmacologically altered to allow the virus to transition from a silent to an active expression state.
To this end, it is interesting that, as we recently reported, AS601245, a Jun N-terminal protein kinase (JNK) inhibitor, would be able prevent HIV-1 reactivation in both primary T cells and T cell lines, despite high levels of NF-κB activation (63). As AP-1 is a major molecular substrate of JNK, our data support the idea that AP-1 may play a central role in HIV-1 latency establishment and maintenance. Again, whether these findings allow for the argumentum e contrario that it may be possible to selectively trigger HIV-1 reactivation remains to be seen, but the data strongly suggest that HIV-1 latency is a transcriptional regulation phenomenon. Our data suggest that latency establishment is likely controlled by the transcription factor composition available at the time point of infection or, more specifically, transcription factor restriction (11). Following integration, the LTR eventually becomes active as a function of the transcription factor binding site composition and the cellular transcription factor profile. Latency is then maintained due to continuous transcription factor restriction. At that time, nucleosomes form on the LTR and are positioned by interaction with other cellular factors such as the SWI/SNF complex BAF or PBAF (24), similar to reports for cellular genes, such as CD25, that are also not actively expressed in resting T cells but can be induced following T cell activation (10, 25, 64).
The results clearly indicate that the capacity of HIV-1 to establish latent infection events can be modulated by altering the transcription factor binding site composition in a short stretch of the viral LTR that represents the CD28RE, a key site through which the cellular environment can affect HIV-1 expression status by transcription factor restriction (65–68). Again, such regulation would not be unlike that of cellular genes such as IL-2, IL-8, and TNF-α. The promoters of these genes also contain a CD28-responsive element; the genes are not expressed in resting T cells (or Jurkat T cells) and are differentially regulated through transcription factor restriction (9).
As NL-7nt/AP-1 and NL–ΔAP-1 differ from NL43 wt in only two and four nucleotides, respectively, at a defined transcription factor binding site in the LTR, our data suggest that drugs that would actually alter the transcription factor profile in latently infected cells could be most useful to trigger HIV-1 reactivation. In particular, cell-differentiating agents could have this capacity. N′-N′-Hexamthylene-bisacetamide (HMBA), a polar cell-differentiating agent, has been reported to trigger HIV-1 reactivation (69–71). HMBA, among many transcription factors, also alters AP-1 fos/jun complex availability (72). Extensive effects of suberoylanilide hydroxamic acid (SAHA) on gene expression and transcription factor regulation, including effects on c-myc or c-Jun, have also been described (73, 74). SAHA, another HIV-1-reactivating agent (75–77), was initially developed as an extremely potent cell-differentiating agent using HMBA as a structural template (52). It is now marketed as the HDAC inhibitor vorinostat, representing a capacity of SAHA that was discovered 2 years after it was published as a cell-differentiating agent (53). It is noteworthy that there is no formal demonstration of whether the bimodal drug vorinostat acts as an HDAC inhibitor or as a cell-differentiating agent to trigger HIV-1 reactivation. Another HIV-1-reactivating drug that is currently being evaluated is disulfiram, also known as Antabuse (78). Interestingly, disufiram, too, was reported to act as a cell-differentiating agent (79). In a recent publication, we added to this list of cell-differentiating agents that would alter the responsiveness of latent HIV-1 infection, when we demonstrated that FDA-approved drugs such as dactinomycin, aclacinomycin, and cytarabine, all drugs with reported cell-differentiating capacity, would prime latent HIV-1 infection for reactivation (51). Whether any of these drugs will actually be successful in triggering system-wide HIV-1 reactivation as a monotherapy in patients remains to be seen. Recent studies by Burnett et al. and by our group suggest that combinatorial use of drugs/compounds that target latent HIV-1 infection at different levels of molecular control may be more promising than the currently pursued monotherapeutic approaches (51, 80). In the context of our finding that an AP-1 site and not the NF-κB element is key to latency control, the ability of these cell-differentiating drugs to trigger HIV-1 reactivation could suggest that drugs with the ability to alter the transcription factor composition in resting memory T cells could be the key to HIV-1 reactivation in a therapeutic setting.
ACKNOWLEDGMENTS
This work was funded in part by NIH grant R01AI064012 to O.K. Parts of the work were performed in the UAB CFAR BSL-3 facilities and by the UAB CFAR Flow Cytometry Core/Joint UAB Flow Cytometry Core, which are funded in part by NIH/NIAID P30 AI027767 and by NIH 5P30 AR048311.
Footnotes
Published ahead of print 12 December 2012
REFERENCES
- 1. Blankson JN, Finzi D, Pierson TC, Sabundayo BP, Chadwick K, Margolick JB, Quinn TC, Siliciano RF. 2000. Biphasic decay of latently infected CD4+ T cells in acute human immunodeficiency virus type 1 infection. J. Infect. Dis. 182:1636–1642 [DOI] [PubMed] [Google Scholar]
- 2. Bukrinsky MI, Stanwick TL, Dempsey MP, Stevenson M. 1991. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science 254:423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. 1995. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat. Med. 1:1284–1290 [DOI] [PubMed] [Google Scholar]
- 4. Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300 [DOI] [PubMed] [Google Scholar]
- 5. Stevenson M. 1997. Molecular mechanisms for the regulation of HIV replication, persistence and latency. AIDS 11(Suppl A):S25–S33 [PubMed] [Google Scholar]
- 6. Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213–222 [DOI] [PubMed] [Google Scholar]
- 7. Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512–517 [DOI] [PubMed] [Google Scholar]
- 8. Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9:727–728 [DOI] [PubMed] [Google Scholar]
- 9. Civil A, Rensink I, Aarden LA, Verweij CL. 1999. Functional disparity of distinct CD28 response elements toward mitogenic responses. J. Biol. Chem. 274:34369–34374 [DOI] [PubMed] [Google Scholar]
- 10. Yeh JH, Spicuglia S, Kumar S, Sanchez-Sevilla A, Ferrier P, Imbert J. 2002. Control of IL-2Ralpha gene expression: structural changes within the proximal enhancer/core promoter during T-cell development. Nucleic Acids Res. 30:1944–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim YK, Bourgeois CF, Pearson R, Tyagi M, West MJ, Wong J, Wu SY, Chiang CM, Karn J. 2006. Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J. 25:3596–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tyagi M, Pearson RJ, Karn J. 2010. Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J. Virol. 84:6425–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pearson R, Kim YK, Hokello J, Lassen K, Friedman J, Tyagi M, Karn J. 2008. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J. Virol. 82:12291–12303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pion M, Jordan A, Biancotto A, Dequiedt F, Gondois-Rey F, Rondeau S, Vigne R, Hejnar J, Verdin E, Hirsch I. 2003. Transcriptional suppression of in vitro-integrated human immunodeficiency virus type 1 does not correlate with proviral DNA methylation. J. Virol. 77:4025–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Van Lint C, Emiliani S, Ott M, Verdin E. 1996. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15:1112–1120 [PMC free article] [PubMed] [Google Scholar]
- 16. Bednarik DP, Cook JA, Pitha PM. 1990. Inactivation of the HIV LTR by DNA CpG methylation: evidence for a role in latency. EMBO J. 9:1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bednarik DP, Mosca JD, Raj NB. 1987. Methylation as a modulator of expression of human immunodeficiency virus. J. Virol. 61:1253–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Verdin E. 1991. DNase I-hypersensitive sites are associated with both long terminal repeats and with the intragenic enhancer of integrated human immunodeficiency virus type 1. J. Virol. 65:6790–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Verdin E, Paras P, Jr, Van Lint C. 1993. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 12:3249–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roebuck KA, Gu DS, Kagnoff MF. 1996. Activating protein-1 cooperates with phorbol ester activation signals to increase HIV-1 expression. AIDS 10:819–826 [DOI] [PubMed] [Google Scholar]
- 21. Roebuck KA, Rabbi MF, Kagnoff MF. 1997. HIV-1 Tat protein can transactivate a heterologous TATAA element independent of viral promoter sequences and the trans-activation response element. AIDS 11:139–146 [DOI] [PubMed] [Google Scholar]
- 22. Van Lint C, Amella CA, Emiliani S, John M, Jie T, Verdin E. 1997. Transcription factor binding sites downstream of the human immunodeficiency virus type 1 transcription start site are important for virus infectivity. J. Virol. 71:6113–6127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang X, Chen Y, Gabuzda D. 1999. ERK MAP kinase links cytokine signals to activation of latent HIV-1 infection by stimulating a cooperative interaction of AP-1 and NF-kappaB. J. Biol. Chem. 274:27981–27988 [DOI] [PubMed] [Google Scholar]
- 24. Rafati H, Parra M, Hakre S, Moshkin Y, Verdin E, Mahmoudi T. 2011. Repressive LTR nucleosome positioning by the BAF complex is required for HIV latency. PLoS Biol. 9:e1001206 doi:10.1371/journal.pbio.1001206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Letimier FA, Passini N, Gasparian S, Bianchi E, Rogge L. 2007. Chromatin remodeling by the SWI/SNF-like BAF complex and STAT4 activation synergistically induce IL-12Rbeta2 expression during human Th1 cell differentiation. EMBO J. 26:1292–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blazkova J, Chun TW, Belay BW, Murray D, Justement JS, Funk EK, Nelson A, Hallahan CW, Moir S, Wender PA, Fauci AS. 2012. Effect of histone deacetylase inhibitors on HIV production in latently infected, resting CD4+ T cells from infected individuals receiving effective antiretroviral therapy. J. Infect. Dis. 206:765–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klatt A, Zhang Z, Kalantari P, Hankey PA, Gilmour DS, Henderson AJ. 2008. The receptor tyrosine kinase RON represses HIV-1 transcription by targeting RNA polymerase II processivity. J. Immunol. 180:1670–1677 [DOI] [PubMed] [Google Scholar]
- 28. Tyagi M, Karn J. 2007. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 26:4985–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Z, Klatt A, Gilmour DS, Henderson AJ. 2007. Negative elongation factor NELF represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. J. Biol. Chem. 282:16981–16988 [DOI] [PubMed] [Google Scholar]
- 30. Emiliani S, Van Lint C, Fischle W, Paras P, Jr, Ott M, Brady J, Verdin E. 1996. A point mutation in the HIV-1 Tat responsive element is associated with postintegration latency. Proc. Natl. Acad. Sci. U. S. A. 93:6377–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bosque A, Planelles V. 2009. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 113:58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang HC, Xing S, Shan L, O'Connell K, Dinoso J, Shen A, Zhou Y, Shrum CK, Han Y, Liu JO, Zhang H, Margolick JB, Siliciano RF. 2009. Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J. Clin. Invest. 119:3473–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188 [DOI] [PubMed] [Google Scholar]
- 34. Chun TW, Engel D, Mizell SB, Ehler LA, Fauci AS. 1998. Induction of HIV-1 replication in latently infected CD4+ T cells using a combination of cytokines. J. Exp. Med. 188:83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chun TW, Engel D, Mizell SB, Hallahan CW, Fischette M, Park S, Davey RT, Jr, Dybul M, Kovacs JA, Metcalf JA, Mican JM, Berrey MM, Corey L, Lane HC, Fauci AS. 1999. Effect of interleukin-2 on the pool of latently infected, resting CD4+ T cells in HIV-1-infected patients receiving highly active anti-retroviral therapy. Nat. Med. 5:651–655 [DOI] [PubMed] [Google Scholar]
- 36. Jones J, Rodgers J, Heil M, May J, White L, Maddry JA, Fletcher TM, III, Shaw GM, Hartman JL, IV, Kutsch O. 2007. High throughput drug screening for human immunodeficiency virus type 1 reactivating compounds. Assay Drug Dev. Technol. 5:181–189 [DOI] [PubMed] [Google Scholar]
- 37. Kutsch O, Benveniste EN, Shaw GM, Levy DN. 2002. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J. Virol. 76:8776–8786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jeeninga RE, Hoogenkamp M, Armand-Ugon M, de Baar M, Verhoef K, Berkhout B. 2000. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J. Virol. 74:3740–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Opijnen T, Jeeninga RE, Boerlijst MC, Pollakis GP, Zetterberg V, Salminen M, Berkhout B. 2004. Human immunodeficiency virus type 1 subtypes have a distinct long terminal repeat that determines the replication rate in a host-cell-specific manner. J. Virol. 78:3675–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peden K, Emerman M, Montagnier L. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185:661–672 [DOI] [PubMed] [Google Scholar]
- 41. Schreiber E, Matthias P, Muller MM, Schaffner W. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17:6419 doi:10.1093/nar/17.15.6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang M, Genin A, Cron RQ. 2004. Overexpression of octamer transcription factors 1 or 2 alone has no effect on HIV-1 transcription in primary human CD4 T cells. Virology 321:323–331 [DOI] [PubMed] [Google Scholar]
- 43. Gervaix A, West D, Leoni LM, Richman DD, Wong-Staal F, Corbeil J. 1997. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc. Natl. Acad. Sci. U. S. A. 94:4653–4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jones J, Whitford W, Wagner F, Kutsch O. 2007. Optimization of HIV-1 infectivity assays. Biotechniques 43:589–590, 592,, 594 [DOI] [PubMed] [Google Scholar]
- 45. Ochsenbauer-Jambor C, Jones J, Heil M, Zammit KP, Kutsch O. 2006. T-cell line for HIV drug screening using EGFP as a quantitative marker of HIV-1 replication. Biotechniques 40:91–100 [DOI] [PubMed] [Google Scholar]
- 46. Duverger A, Jones J, May J, Bibollet-Ruche F, Wagner FA, Cron RQ, Kutsch O. 2009. Determinants of the establishment of human immunodeficiency virus type 1 latency. J. Virol. 83:3078–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schug J. 2003. Using TESS to predict transcription factor binding sites in DNA sequence. Curr. Protoc. Bioinformatics 21:2.6.1–2.6.15 [DOI] [PubMed] [Google Scholar]
- 48. Roebuck KA, Brenner DA, Kagnoff MF. 1993. Identification of c-fos-responsive elements downstream of TAR in the long terminal repeat of human immunodeficiency virus type-1. J. Clin. Invest. 92:1336–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Feng S, Holland EC. 1988. HIV-1 tat trans-activation requires the loop sequence within tar. Nature 334:165–167 [DOI] [PubMed] [Google Scholar]
- 50. Jordan A, Bisgrove D, Verdin E. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22:1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shishido T, Wolschedorf F, Duverger F, Wagner F, Kappes J, Jones J, Kutsch O. 2012. Selected drugs with reported secondary cell-differentiating capacity prime latent HIV-1 infection for reactivation. J. Virol. 86:9055–9069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Richon VM, Webb Y, Merger R, Sheppard T, Jursic B, Ngo L, Civoli F, Breslow R, Rifkind RA, Marks PA. 1996. Second generation hybrid polar compounds are potent inducers of transformed cell differentiation. Proc. Natl. Acad. Sci. U. S. A. 93:5705–5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Richon VM, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind RA, Marks PA. 1998. A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc. Natl. Acad. Sci. U. S. A. 95:3003–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shapiro VS, Truitt KE, Imboden JB, Weiss A. 1997. CD28 mediates transcriptional upregulation of the interleukin-2 (IL-2) promoter through a composite element containing the CD28RE and NF-IL-2B AP-1 sites. Mol. Cell. Biol. 17:4051–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. He G, Ylisastigui L, Margolis DM. 2002. The regulation of HIV-1 gene expression: the emerging role of chromatin. DNA Cell Biol. 21:697–705 [DOI] [PubMed] [Google Scholar]
- 56. Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. 2009. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5:e1000495 doi:10.1371/journal.ppat.1000495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ylisastigui L, Archin NM, Lehrman G, Bosch RJ, Margolis DM. 2004. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS 18:1101–1108 [DOI] [PubMed] [Google Scholar]
- 58. Weichs an der Glon C, Ashe M, Eggermont J, Proudfoot NJ. 1993. Tat-dependent occlusion of the HIV poly(A) site. EMBO J. 12:2119–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Verhoef K, Sanders RW, Fontaine V, Kitajima S, Berkhout B. 1999. Evolution of the human immunodeficiency virus type 1 long terminal repeat promoter by conversion of an NF-kappaB enhancer element into a GABP binding site. J. Virol. 73:1331–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. LaMarco K, Thompson CC, Byers BP, Walton EM, McKnight SL. 1991. Identification of Ets- and notch-related subunits in GA binding protein. Science 253:789–792 [DOI] [PubMed] [Google Scholar]
- 61. Avots A, Hoffmeyer A, Flory E, Cimanis A, Rapp UR, Serfling E. 1997. GABP factors bind to a distal interleukin 2 (IL-2) enhancer and contribute to c-Raf-mediated increase in IL-2 induction. Mol. Cell. Biol. 17:4381–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hoffmeyer A, Avots A, Flory E, Weber CK, Serfling E, Rapp UR. 1998. The GABP-responsive element of the interleukin-2 enhancer is regulated by JNK/SAPK-activating pathways in T lymphocytes. J. Biol. Chem. 273:10112–10119 [DOI] [PubMed] [Google Scholar]
- 63. Wolschendorf F, Bosque A, Shishido T, Duverger A, Jones J, Planelles V, Kutsch O. 2012. Kinase control prevents HIV-1 reactivation in spite of high levels of induced NF-kappaB activity. J. Virol. 86:4548–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cullen SJ, Ponnappan S, Ponnappan U. 2009. Catalytic activity of the proteasome fine-tunes Brg1-mediated chromatin remodeling to regulate the expression of inflammatory genes. Mol. Immunol. 47:600–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Coudronniere N, Villalba M, Englund N, Altman A. 2000. NF-kappa B activation induced by T cell receptor/CD28 costimulation is mediated by protein kinase C-theta. Proc. Natl. Acad. Sci. U. S. A. 97:3394–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ott M, Emiliani S, Van Lint C, Herbein G, Lovett J, Chirmule N, McCloskey T, Pahwa S, Verdin E. 1997. Immune hyperactivation of HIV-1-infected T cells mediated by Tat and the CD28 pathway. Science 275:1481–1485 [DOI] [PubMed] [Google Scholar]
- 67. Ott M, Lovett JL, Mueller L, Verdin E. 1998. Superinduction of IL-8 in T cells by HIV-1 Tat protein is mediated through NF-kappaB factors. J. Immunol. 160:2872–2880 [PubMed] [Google Scholar]
- 68. Verweij CL, Geerts M, Aarden LA. 1991. Activation of interleukin-2 gene transcription via the T-cell surface molecule CD28 is mediated through an NF-kB-like response element. J. Biol. Chem. 266:14179–14182 [PubMed] [Google Scholar]
- 69. Choudhary SK, Archin NM, Margolis DM. 2008. Hexamethylbisacetamide and disruption of human immunodeficiency virus type 1 latency in CD4(+) T cells. J. Infect. Dis. 197:1162–1170 [DOI] [PubMed] [Google Scholar]
- 70. Contreras X, Barboric M, Lenasi T, Peterlin BM. 2007. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 3:1459–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vlach J, Pitha PM. 1993. Hexamethylene bisacetamide activates the human immunodeficiency virus type 1 provirus by an NF-kappa B-independent mechanism. J. Gen. Virol. 74(Part 11):2401–2408 [DOI] [PubMed] [Google Scholar]
- 72. Eliopoulos A, Spandidos D. 1993. Changes in fos/jun binding-activity on a negative regulatory element of C-myc during differentiation of mouse erythroleukemic cells. Int. J. Oncol. 2:883–888 [DOI] [PubMed] [Google Scholar]
- 73. Beliakova-Bethell N, Zhang J, Terry V, Spina C, Richman D, Woelk C. 2012. Off-target effects of suberoylanilide hydroxamic acid (SAHA) in resting CD4 T cells. Abstr. 19th Conf. Retroviruses Opportunistic Infect., poster 380 [Google Scholar]
- 74. Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Hideshima T, Akiyama M, Chauhan D, Munshi N, Gu X, Bailey C, Joseph M, Libermann TA, Richon VM, Marks PA, Anderson KC. 2004. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc. Natl. Acad. Sci. U. S. A. 101:540–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, Margolis DM. 2009. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res. Hum. Retroviruses 25:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Contreras X, Schweneker M, Chen CS, McCune JM, Deeks SG, Martin J, Peterlin BM. 2009. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J. Biol. Chem. 284:6782–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Edelstein LC, Micheva-Viteva S, Phelan BD, Dougherty JP. 2009. Short communication: activation of latent HIV type 1 gene expression by suberoylanilide hydroxamic acid (SAHA), an HDAC inhibitor approved for use to treat cutaneous T cell lymphoma. AIDS Res. Hum. Retroviruses 25:883–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Xing S, Bullen CK, Shroff NS, Shan L, Yang HC, Manucci JL, Bhat S, Zhang H, Margolick JB, Quinn TC, Margolis DM, Siliciano JD, Siliciano RF. 2011. Disulfiram reactivates latent HIV-1 in a Bcl-2-transduced primary CD4+ T cell model without inducing global T cell activation. J. Virol. 85:6060–6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Rovira M, Huang W, Yusuff S, Shim JS, Ferrante AA, Liu JO, Parsons MJ. 2011. Chemical screen identifies FDA-approved drugs and target pathways that induce precocious pancreatic endocrine differentiation. Proc. Natl. Acad. Sci. U. S. A. 108:19264–19269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Burnett JC, Lim KI, Calafi A, Rossi JJ, Schaffer DV, Arkin AP. 2010. Combinatorial latency reactivation for HIV-1 subtypes and variants. J. Virol. 84:5958–5974 [DOI] [PMC free article] [PubMed] [Google Scholar]