Abstract
The unabated circulation of the highly pathogenic avian influenza A virus/H5N1 continues to be a serious threat to public health worldwide. Because of the high frequency of naturally occurring mutations, the emergence of H5N1 variants with high virulence has raised great concerns about the potential transmissibility of the virus in humans. Recent studies have shown that laboratory-mutated or reassortant H5N1 viruses could be efficiently transmitted among mammals, particularly ferrets, the best animal model for humans. Thus, it is critical to establish effective strategies to combat future H5N1 pandemics. In this study, we identified a broadly neutralizing monoclonal antibody (MAb), HA-7, that potently neutralized all tested strains of H5N1 covering clades 0, 1, 2.2, 2.3.4, and 2.3.2.1 and completely protected mice against lethal challenges of H5N1 viruses from clades 1 and 2.3.4. HA-7 specifically targeted the globular head of the H5N1 virus hemagglutinin (HA). Using electron microscopy technology with three-dimensional reconstruction (3D-EM), we discovered that HA-7 bound to a novel and highly conserved conformational epitope that was centered on residues 81 to 83 and 117 to 122 of HA1 (H5 numbering). We further demonstrated that HA-7 inhibited viral entry during postattachment events but not at the receptor-binding step, which is fully consistent with the 3D-EM result. Taken together, we propose that HA-7 could be humanized as an effective passive immunotherapeutic agent for antiviral stockpiling for future influenza pandemics caused by emerging unpredictable H5N1 strains. Our study also provides a sound foundation for the rational design of vaccines capable of inducing broad-spectrum immunity against H5N1.
INTRODUCTION
The unabated circulation of the highly pathogenic avian influenza A virus (IAV)/H5N1 continues to be a serious threat to public health worldwide. The transmission of H5N1 virus among humans has been rare, and no human pandemic has ever occurred as a result of this virus. Nonetheless, the first human case was reported to the WHO in 2003, and since then, there have been 608 documented human H5N1 cases with 359 deaths as of 10 August 2012, with a mortality rate approaching 60% (http://www.who.int/influenza/human_animal_interface/EN_GIP_20120810CumulativeNumberH5N1cases.pdf). Because of the high frequency of naturally occurring mutations, the lethality of H5N1 has raised great concerns about the potential transmissibility of the virus in humans.
Recently, two research groups have made significant strides in the efficient transmission of the laboratory-mutated or reassortant H5N1 virus among mammals, particularly ferrets, which is the best animal model for humans. Introducing mutations in H5N1, or creating a “reassortant” of H5N1 and H1N1 viruses causing the 2009 flu pandemic, makes it possible for the manipulated H5N1 virus to replicate efficiently in these animals (1–4). These results represent significant breakthroughs in identifying specific determinants of H5N1 transmission in ferrets but also stirred a months-long debate on global biosecurity- and biosafety-related issues (5–8). Therefore, there is a strong need to explore effective strategies to combat influenza pandemics caused by H5N1 viruses in the future.
To efficiently prevent a future influenza pandemic, a robust global surveillance system should be in place for the timely detection of novel H5N1 virus strains in animals once they arise. Such a coordinated surveillance and control effort has not always been successful (9). On the other hand, inactivated virus vaccines and live-attenuated, cold-adapted H5N1 vaccines could also be developed for the prevention of H5N1 virus infection via large-scale vaccination (10–12). Other forms of H5N1 vaccines, including those based on DNA, proteins, viral vectors, and virus-like particles as well as a number of combination vaccinations, are in the developmental stage or in preclinical or clinical trials, some of which have shown efficacy in preventing H5N1 infections (13–19). However, such vaccines are not effective enough against divergent strains of H5N1 viruses, thus limiting their ability to produce broad-spectrum protection.
As an alternative to vaccines, neutralizing monoclonal antibodies (MAbs) represent a passive therapeutic strategy to provide immediate protection against influenza virus infection. Several effective MAbs against hemagglutinins (HA) of multiple strains of IAVs from group 1 and/or group 2 have been explored, showing broad-spectrum neutralization of the viruses. It was reported previously that MAbs F10 and CR6261 were effective against all tested group 1 IAVs (20, 21), while MAb CR8020 contains broad neutralizing activity against most group 2 viruses, including H3N2 and H7N7 viruses (22). Other reports indicated that MAb F16 was able to recognize the HAs of all 16 subtypes and neutralize both group 1 and 2 IAVs (23). Thus, these identified MAbs may be used as effective passive immunotherapeutics against a broad range of IAVs during influenza pandemics or epidemics because of their ability to target a variety of IAVs in addition to H5N1 virus. However, in the case of a pandemic caused by an emerging unpredictable H5N1 strain, MAbs with broad neutralizing activity specifically targeting H5N1 strains may provide a more effective passive immunotherapeutic effect.
The HA of influenza virus plays important roles in virus infection by binding to target cells and subsequently fusing viral and cellular membranes. It is the major envelope glycoprotein of the virus for inducing neutralizing antibodies. The trimeric protein of HA is initially synthesized as a precursor polypeptide, HA0, which is then activated upon cleavage into two subunits, HA1 and HA2. The “head,” HA1, is mainly responsible for receptor binding, while the stem region, HA2, mediates fusion of the viral envelope and cellular membrane, in which the fusion peptide (FP) at the N terminus of HA2 plays an important role (24, 25). Compared with the amino acid sequences of HA2, particularly FP, the sequences from HA1 are more variable. However, HA1 still has some sequences showing high-level conservation among different strains (26, 27).
In this study, we identified a neutralizing MAb, HA-7, which targets a novel and highly conserved HA1 epitope of H5N1. The function and structural basis of this MAb were accordingly explored and are reported here in detail.
MATERIALS AND METHODS
Ethics statement.
Four- to eight-week-old female BALB/c mice were used for MAb generation and virus challenge. The animal studies were carried out in strict accordance with recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (28) and of the State Key Laboratory of Pathogen and Biosecurity at the Beijing Institute of Microbiology and Epidemiology of China. All experiments related to influenza viruses were performed in approved biosafety level 3 (BSL-3) laboratories or BSL-3 animal facilities.
Generation of HA-specific MAbs.
Mouse vaccination and HA-specific MAb generation were performed according to our previously described protocols, with some modifications (29). Briefly, five BALB/c mice were subcutaneously (s.c.) vaccinated with a recombinant protein expressing codon-optimized HA1 of A/Anhui/1/2005(H5N1) (GenBank accession no. ABD28180) fused with foldon (Fd) and Fc of human IgG1 (HA1-Fdc), as described previously (30). Vaccinated mice were sacrificed 10 days after the last boost, and the splenocytes were fused with mouse myeloma cells (SP2/0). For HA-specific MAb screening, positive hybridomas were screened by an enzyme-linked immunosorbent assay (ELISA) by coating the plates with a recombinant protein containing the above-mentioned HA1 fragment without the Fd and Fc fusion (HA1-His) as well as an inactivated homologous H5N1 virus. Positive cells were expanded, retested, and subcloned to generate stable hybridoma cell lines. The MAbs were purified from culture supernatants using protein A and G Sepharose 4 Fast Flow (GE Healthcare, Piscataway, NJ).
HA pseudovirus neutralization assay.
An assay of the neutralizing activity of MAb HA-7 against infection by divergent clades of HA pseudovirus was performed as previously described (29). Briefly, 293T cells were cotransfected with a plasmid encoding an Env-defective, luciferase-expressing HIV-1 genome (pNL4-3.luc.RE) and each of the plasmids encoding HAs of homologous A/Anhui/1/2005 (AH-HA) (clade 2.3.4) and heterologous A/Xinjiang/1/2006 (XJ-HA) (clade 2.2), A/Qinghai/59/05 (QH-HA) (clade 2.2), A/Hong Kong/156/97 (HK-HA) (clade 0), and A/Hubei/1/10 (HB-HA) (clade 2.3.2.1) viruses, respectively. The classification of the H5N1 pseudovirus was based on WHO recommendations (http://www.who.int/influenza/resources/documents/2011_09_h5_h9_vaccinevirusupdate.pdf). The full-length gene sequences of HB-HA were downloaded from the GISAID EpiFlu Database (http://www.gisaid.org/). Exogenous bacterial neuraminidase (NA) (Sigma) was added 24 and 48 h after transfection, and supernatants were harvested at 72 h posttransfection for single-cycle infection. Pseudovirus-containing supernatants were incubated with serially diluted H5N1 HA MAb at 37°C for 1 h before being added to 293T cells preplated in 96-well culture plates (104 cells/well). Twenty-four hours later, cells were refed with fresh medium, which was followed by lysing cells 72 h later with cell lysis buffer (Promega) and transferring the lysates into 96-well luminometer plates. Luciferase substrate (Promega) was added to the plates, and relative luciferase activity was determined by using an Ultra 384 luminometer (Tecan). The neutralization of HA pseudovirus was then calculated (31).
Live-virus-based neutralization assay.
Neutralizing antibody titers of MAb HA-7 were further detected by a live-virus-based neutralization assay, as previously described (30). Briefly, serially 2-fold-diluted MAb was incubated with 50% tissue culture infective doses (100 TCID50) of influenza viruses for 2 h at 37°C prior to its addition to a monolayer of Madin-Darby canine kidney (MDCK) cells. The tested influenza viruses included H5N1 viruses A/VietNam/1194/2004 (VN/1194), A/Shenzhen/406H/06 (SZ/406H), and A/Hong Kong/213/03 (HK/213); 2009 swine-origin influenza A virus (S-OIV H1N1 virus); and seasonal influenza A virus (seasonal IAV). Virus supernatant was removed and replaced with fresh medium. The virus-infected cells were incubated for 72 h at 37°C. Infectivity was identified by the presence of viral cytopathic effect (CPE) on day 4, and the titer was calculated by the Reed-Muench method.
Passive immunization and virus challenge.
The protective potential of MAb HA-7 against H5N1 influenza virus infection was detected in mice. BALB/c mice were infected with 5 50% lethal doses (LD50) of VN/1194 or SZ/406H of H5N1 virus. Twenty-four hours later, the mice were intravenously (i.v.) injected with 0.5 ml (containing 1 mg) of purified MAb HA-7. The control group was given the same amount of MAb specific to the receptor-binding domain (RBD) of severe acute respiratory syndrome coronavirus (SARS-CoV). Six infected mice per group were observed daily for 14 days to calculate the survival rate and body weight change. Six mice from each group were sacrificed on day 5 postchallenge, and lung samples were collected for virological detection and histopathological evaluation.
Virus titer detection.
Virus titers were detected as previously described (30, 32–34). Briefly, lung tissues from euthanized mice were aseptically removed and homogenized in minimal essential medium (MEM) containing antibiotics to achieve 10% (wt/vol) suspensions of lungs. In 96-well cell culture plates, 10-fold serial dilutions of samples were added to the monolayer of MDCK cells in quadruplicate and allowed to absorb for 2 h at 37°C in the presence of 5% CO2. Supernatants were then removed and replaced with fresh MEM, and MDCK cells were incubated as described above for 72 h. CPE was observed daily, and virus titer was determined by the HA test. For the HA test, 50 μl of 0.5% turkey red blood cells (Lampire Biological Laboratories, Pipersville, PA) was added to 50 μl of cell culture supernatant and incubated at room temperature for 30 min. Wells containing HA were scored as positive. The virus titer was calculated by the Reed-Muench method and expressed as log10 TCID50/g of lung tissues.
Histopathological analysis.
The lung tissues of challenged mice were immediately fixed in 10% formalin and embedded in paraffin wax. Sections 4 to 6 mm in thickness were made and mounted onto slides. Histopathological changes caused by H5N1 virus infection were examined by hematoxylin and eosin (H&E) staining and viewed under a light microscope, as previously described (30, 35).
Reactivity detection and epitope mapping of MAb HA-7 by ELISA.
The reactivity detection and epitope mapping of MAb HA-7 were done by using ELISA plates coated with truncated protein fragments and synthetic peptides covering the full-length HA proteins of A/Anhui/1/2005(H5N1) as well as inactivated IAVs, as described previously (30, 36–38). Briefly, 96-well ELISA plates were respectively coated with recombinant HA proteins (1 μg/ml), overlapping peptides (10 μg/ml), or inactivated IAVs overnight at 4°C and blocked with 2% nonfat milk for 2 h at 37°C. Recombinant human IgG1-Fc2 protein (rFc), commercial human IgG Fc protein (IgG-Fc), Fd fused with HIV-1 gp41 (HIV-Fd), and SARS-CoV RBD protein were used as the controls. Serially diluted MAb was added to the plates and incubated for 1 h at 37°C. The plates were washed three times with phosphate-buffered saline (PBS)-Tween (PBST) and incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (1:2,000), IgG1 (1:2,000), IgG2a (1:5,000) (Bethyl Laboratories, Montgomery, TX), and IgG2b (1:2,000) (Invitrogen), respectively, for 1 h at 37°C. For IgG3 detection, the plates were sequentially incubated with goat anti-mouse IgG3 (1:1,000) and anti-goat HRP-conjugated antibody (1:5,000) for 1 h at 37°C. The reaction was visualized by using the substrate 3,3′,5,5′-tetramethylbenzidine (TMB) (Invitrogen) and stopped with 1 N H2SO4. The A450 was measured by using an ELISA plate reader (Tecan, San Jose, CA).
To detect the reactivity of MAb with denatured HA1 proteins, the ELISA plates were coated with recombinant HA1 proteins (1 μg/ml) overnight at 4°C, followed by treatment of the plate with dithiothreitol (DTT) (10 mM) for 1 h at 37°C. The plates were then treated with iodoacetamide (50 mM) for 1 h at 37°C to stop the reaction (39). After three washes, a regular ELISA was performed, as described above.
Virus binding assay.
A virus binding assay was performed by using QH-HA H5N1 pseudovirus as previously described, with some modifications (27). Briefly, virus was incubated with serial dilutions of MAb HA-7 or control antibodies in Dulbecco's MEM (DMEM) containing 1% bovine serum albumin (BSA) overnight at 4°C. MDCK cells were seeded into 96-well plates 24 h before infection and blocked with DMEM containing 1% BSA (100 μl/well) for 1 h at 4°C. The mixture of the virus and MAb was added to MDCK cells for 2 h at 4°C, followed by washing the cells four times with PBS containing 1% BSA to remove unbound virus. The lysed cells were quantified for HIV p24 by ELISA. Percent virus binding was expressed as a relative percentage of the p24 reading from the cells without antibodies (no Ab), which was set as 100%.
Postattachment assay.
A postattachment assay was performed by using QH-HA H5N1 pseudovirus as previously described, with some modifications (21, 27). Briefly, virus was added to MDCK cells plated 18 h before testing and incubated for 6 h at 4°C. Cells were washed three times with cold PBS, followed by the addition of serially diluted MAb HA-7 and control antibodies to the MDCK monolayer for 2 h at 4°C. After the addition of fresh DMEM, the cells were incubated at 37°C and measured for luciferase activity at 72 h postinfection. The percentage of viral entry was expressed as a relative percentage of the luciferase reading from the cells without antibodies (no Ab), which was set as 100% (% viral entry = luciferase reading of Ab/no Ab × 100%).
Cloning and sequencing of the genes encoding heavy and light chains of HA-7Fab.
The VL and VH genes of HA-7 were amplified by reverse transcriptase PCR (RT-PCR). Briefly, total RNA was isolated from 107 hybridoma cells of HA-7 using TRIzol reagent (Invitrogen). The cDNA was synthesized by using the SuperScript II RT-PCR kit (Invitrogen) according to the manufacturer's instructions. Primers mVH-f (ATGGRATGGAGCTGGATCTT) and mVH-r (ATAGACAGATGGGGGTGTCGTTTTGGC) were used to amplify the genes encoding the heavy (H) chain, and primers mVK-f (ATGGAGWCAGACACACTCCT) and mVK-r (GGATACAGTTGGTGCAGCATC) were used for the light (L) chain. The amplified DNA fragments were cloned into T vector (Invitrogen), followed by selection of positive clones for sequencing.
Structure of the HA-7Fab/H5HA complex determined by 3D-EM.
The HA-7 Fab fragment (HA-7Fab) was prepared from HA-7 IgG by papain digestion. H5HA (the HA from an H5N1 avian influenza A virus) was expressed and purified as previously reported (21). H5HA was mixed with an excess of HA-7Fab and incubated at room temperature for 30 min. The HA-7Fab/H5HA complex was then separated from HA-7Fab by gel filtration chromatography using a Superdex S200 10/300 column in a solution containing 10 mM Tris (pH 7.4), 50 mM NaCl, and 1 mM CaCl2. Negatively stained electron microscopy (EM) specimens were prepared as described previously (40). Briefly, 3 μl of the HA-7Fab/H5HA complex (at ∼0.01 mg/ml) was put onto a freshly glow-discharged carbon-coated EM grid, blotted with filter paper after 30 s, and stained with 0.75% uranyl formate. Images were acquired by using a 4,000- by 4,000-pixel TVIPS charge-coupled-device (CCD) camera (Tietz Video and Image Processing Systems, Gauting, Germany) on a Tecnai F20 electron microscope (FEI, Hillsboro, OR) equipped with a field emission electron source operated at 200 kV, at a magnification of ∼×70,000, resulting in a calibrated pixel size of 4.28 Å/pixel on the specimen scale. The defocus values were in the range of 2.0 to 3.2 μm. The electron dosage was ∼40 electrons/Å2. Image quality was monitored on the basis of power spectrum quality. Particle boxing, contrast transfer function (CTF) correction, initial model generation, three-dimensional (3D) refinement, and resolution assessment were all carried out with the EMAN2 package (41). Three-fold symmetry was imposed during the reconstruction. A data set of 3,172 particles was used for the final reconstructed map. The resolution was estimated based on the resolution criteria of Fourier shell correlation (FSC) with a cutoff of 0.5. Handedness of the map was determined on the basis of the known H5HA crystal structure.
Visualization, molecular docking, and epitope mapping of HA-7.
The H5HA trimer atomic model was obtained from its published crystal structure (Protein Data Bank [PDB] accession no. 3FKU). Mouse-origin HA-7 was found to belong to IgG1. Thus, the molecular structure of HA-7Fab was obtained by homologous modeling using Swiss-Model (http://swissmodel.expasy.org/) (42), using the mouse IgG1 crystal structure (PDB accession no. 1DBM) as a template, together with its determined primary sequences (see Fig. 4D). The resulting homology model was adjusted manually with COOT (43) to remove atom clashes and to improve geometry. Visualization and rigid-body fitting of atomic models into the 3D-EM density maps were done with UCSF Chimera (44). Models were first manually docked into their EM density using Chimera, based on prior knowledge. Their positions were then refined using the fit-in-map utility in Chimera. After fitting refinement, the positions with the highest correlation coefficient values were chosen. We first manually put the HA trimer into the center of the density map with the 3-fold axis aligned to the EM map's 3-fold axis. Once HA was docked, the density of HA-7Fab was obtained by subtracting the density of HA (calculated at 18 Å using the e2pdb2mrc.py utility in EMAN2) from the complex's EM density. The Fab molecule was manually put into its density with complementarity-determining region (CDR) loops pointing to HA. The receptor of avian influenza virus H5HA (sialic acid) was located according to the H5HA/receptor complex structure in the Protein Data Bank (accession no. 1JSN) (45). Antibody epitopes were visually identified as residues contacted by Fab's density using Chimera. Molecular figures were prepared by using Chimera.
Fig 4.
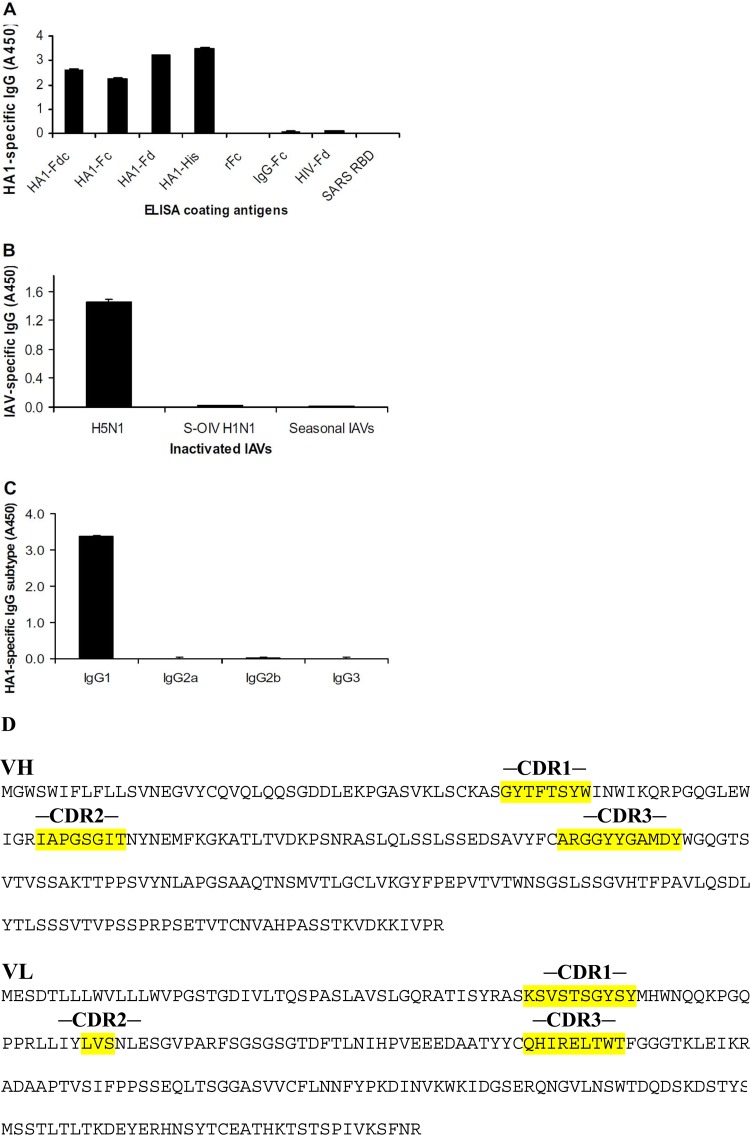
Detection of specificity and subtypes of MAb HA-7 by ELISA. (A) Reactivity of total IgG of HA-7 with fusion proteins containing full-length HA1 (residues 13 to 325 of H5 numbering [21, 25, 46], corresponding to HA1 residues +3 to 322 [30]) and truncated fragments of HA1 of A/Anhui/1/2005(H5N1) virus. ELISA plates were respectively coated with recombinant HA1 proteins fused with human Fc (HA1-Fc), Fd sequence (HA1-Fd), or Fd plus Fc (HA1-Fdc) and HA1 protein without Fd and Fc (HA1-His). Recombinant hIgG1-Fc2 protein (rFc), commercial human IgG Fc protein (IgG-Fc), Fd fused with HIV-1 gp41 (HIV-Fd), and SARS-CoV RBD protein were used as the controls. (B) Reactivity of total IgG of MAb HA-7 with inactivated IAVs containing H5N1, S-OIV H1N1, and seasonal influenza viruses (IAVs). (C) Detection of IgG subtypes of MAb HA-7 using recombinant HA1-His protein as the coating antigen. The data are presented as the mean absorbance at 450 nm (A450) ± standard deviations of data from duplicate wells of MAbs at a dilution of 1:3,200. Panels A to C show data from three independent experiments. (D) Amino acid sequences for HA-7 Fab heavy (VH) and light (VL) chains. CDR1, CDR2, and CDR3 are labeled.
Protein structure accession number.
The 3D-EM density map of the HA-7Fab/H5HA complex has been deposited in the EMDB database under accession number EMD-2074.
RESULTS
MAb HA-7 potently inhibited entry of H5N1 virus with divergent strains.
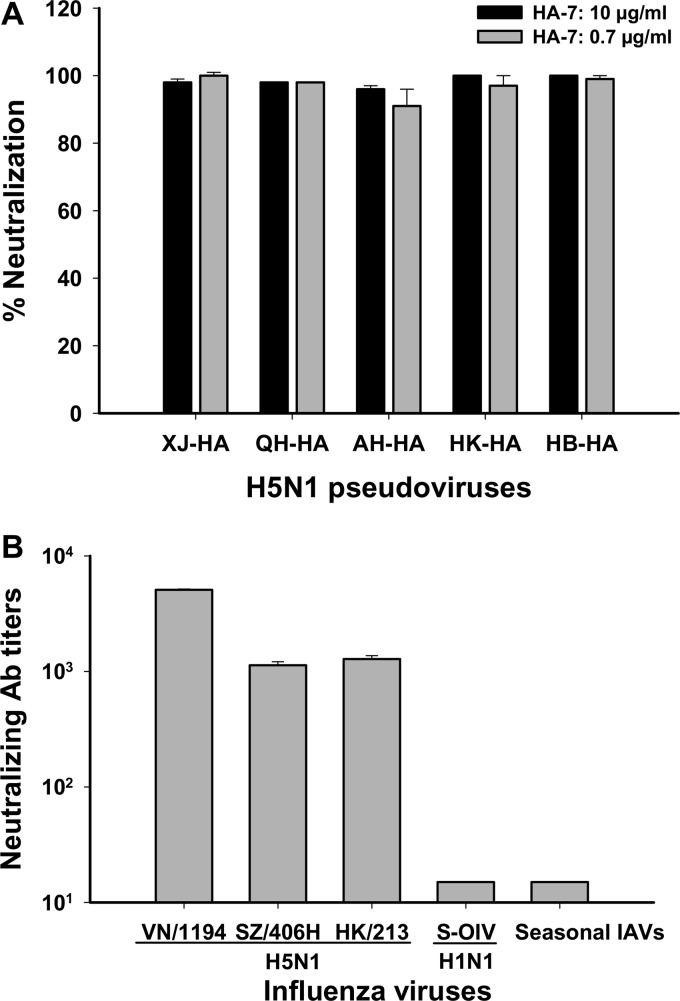
Monoclonal hybridomas were screened against the HA1 fragment (HA1-His) and inactivated H5N1 virus by ELISA, and positive clones in both ELISAs were used to generate stable hybridoma cell lines for the production of MAbs, followed by screening of the inhibition of viral entry using our established pseudovirus neutralization assay (29). Among 25 selected MAbs with high titers of antibody responses, HA-7 was shown to inhibit over 95% of viral entry for both the homologous AH-HA strain and heterologous strains XJ-HA, QH-HA, HK-HA, and HB-HA of H5N1 pseudovirus at a concentration as low as 0.7 μg/ml, reaching inhibitory activity similar to that at 10 μg/ml (Fig. 1A). Further detection of the neutralizing activity of the MAb against live influenza viruses indicated that HA-7 could strongly neutralize infection by all tested H5N1 strains, including VN/1194, HK/213, and SZ/406H. However, no significant inhibition was observed for MAb against other types of IAVs, including the S-OIV H1N1 (A/Beijing/501/2009) pandemic strain and seasonal influenza virus (Fig. 1B).
Fig 1.
Detection of inhibitory activity of MAb HA-7 on influenza virus entry by neutralization assays. (A) Pseudovirus neutralization assay. The data are presented as the mean percentages of neutralization ± standard deviations for duplicate wells of MAb HA-7 at concentrations of 0.7 and 10 μg/ml, respectively, against HAs of H5N1 pseudovirus covering four different clades, including homologous strain AH-HA (clade 2.3.4) and heterologous strains HK-HA (clade 0), XJ-HA (clade 2.2), QH-HA (clade 2.2), and HB-HA (clade 2.3.2.1). (B) Live-virus-based neutralization assay. Shown are data for neutralizing activities of HA-7 against strains VN/1194 (clade 1), SZ/406H (clade 2.3.4), and HK/213 (clade 1) of H5N1; S-OIV H1N1 (A/Beijing/501/2009 pdm strain); and seasonal IAVs. The neutralizing antibody titer was defined as the highest dilution of MAb that completely suppressed the virus-induced CPE in 50% of the wells. Panels A and B show representative data from three independent repeat experiments.
The above-described results demonstrate that HA-7 effectively inhibited the entry of divergent strains of H5N1 virus covering five different clades, including clade 0 (HK-HA), clade 1 (VN/1194 and HK/213), clade 2.2 (XJ-HA and QH-HA), clade 2.3.4 (AH-HA and SZ/406H), and clade 2.3.2.1 (HB-HA). The inability of the MAb to neutralize other types of IAVs suggested that MAb HA-7 is specific to H5N1 virus.
Passive immunotherapeutic effects of MAb HA-7 in vivo.
We then investigated the passive immunotherapeutic effects of MAb HA-7 in protecting mice against H5N1 virus infection. Mice were passively immunized i.v. with purified MAb HA-7, and protective efficacy was evaluated by calculating the survival rate and weight loss for a period of 14 days. The mice were sacrificed 5 days after challenge to detect viral titers and pathological changes in the lungs.
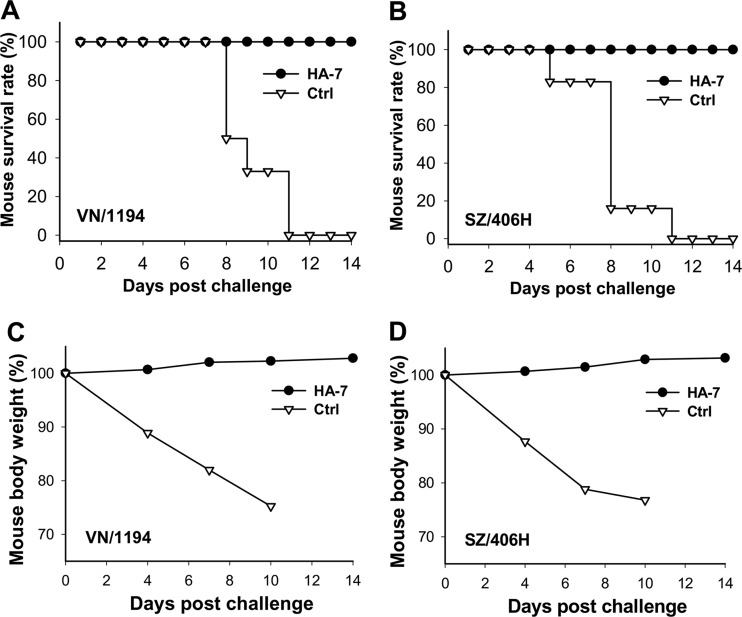
All mice injected with MAb HA-7 survived challenge with VN/1194 (clade 1) (Fig. 2A) and SZ/406H (clade 2.3.4) (Fig. 2B) H5N1 viruses, while no mice from the control group injected with the MAb against the RBD of SARS-CoV survived challenge by these two viruses. In addition, no obvious body weight loss was observed for the mice immunized with MAb HA-7 after challenge with a lethal infection by the VN/1194 (Fig. 2C) or SZ/406H (Fig. 2D) strain of H5N1 virus, while the mice in the SARS-CoV RBD control group exhibited a continuous decrease of body weight, and none of them survived more than 10 days after virus challenge.
Fig 2.
Detection of cross-protection of HA-7 neutralizing MAb against lethal H5N1 virus challenge. (A and B) Survival rate of HA-7-injected mice after challenge with 5 LD50 of strain VN/1194 (clade 1) (A) or SZ/406H (clade 2.3.4) (B) of H5N1 virus. (C and D) Percent body weight change of HA-7-treated mice after challenge with 5 LD50 of the VN/1194 (C) and SZ/406H (D) strains of H5N1 virus. A MAb targeting the RBD of SARS-CoV was used as the control (Ctrl). Panels A to D show representative data from two independent repeat experiments with six mice per group.
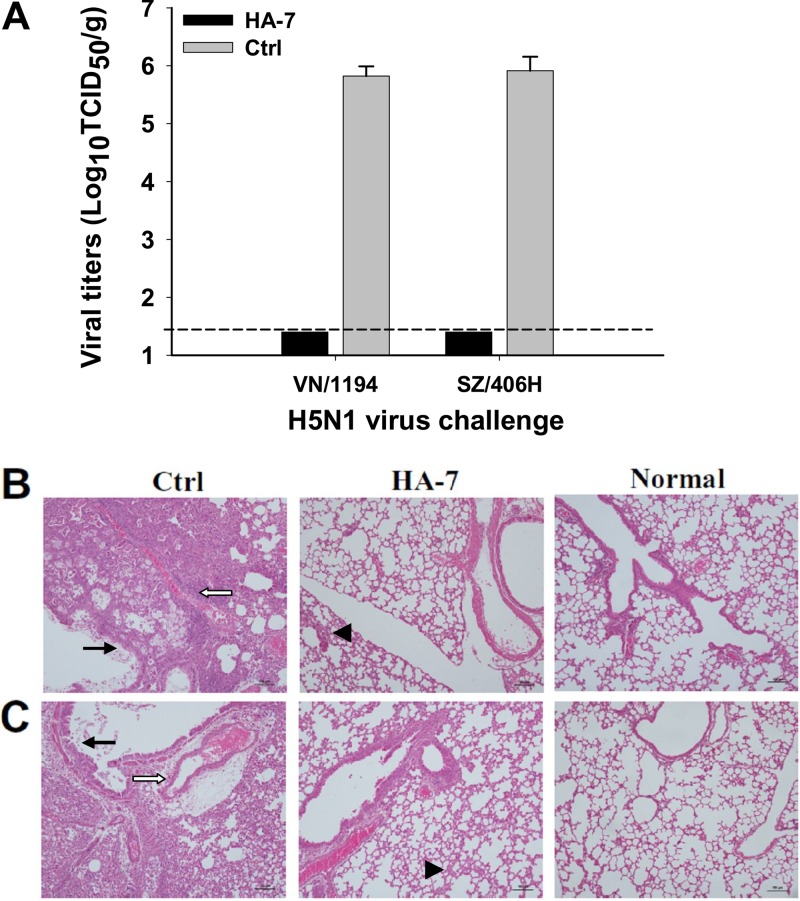
Observation of the viral titers in the lung tissues of challenged mice indicated that no virus titer was detectable in HA-7-immunized mice challenged with VN/1194 and SZ/406H viruses, while a high virus titer was detected in the control mice injected with the MAb specific to the RBD of SARS-CoV (Fig. 3A). Observation of the histopathological change from the H&E-stained lung tissues further revealed that the control mice receiving the SARS-CoV RBD-specific MAb developed a high degree of histopathological damage in their lungs after challenge with both the VN/1194 and SZ/406H strains of H5N1 virus. Serious interstitial pneumonia and inflammation were observed, with diffused alveolar collapse, significant lymphocyte infiltration, pulmonary vascular dilatation and congestion, as well as epithelial cell degeneration. On the contrary, mice receiving the HA-7 treatment had significantly diminished pathological changes caused by VN/1194 (clade 1) and SZ/406H (clade 2.3.4) virus challenges (Fig. 3B and C). The above-described results suggest that the identified neutralizing HA-7 MAb could completely protect immunized mice against lethal challenges with tested strains of clade 1 and clade 2.3.4 of H5N1 virus, indicating their potential passive immunotherapeutic effects against H5N1 virus infection.
Fig 3.
Virus titers and histopathological changes in the lungs of MAb-treated mice challenged with H5N1 viruses. HA-7-injected mice were challenged with 5 LD50 of clade 1 strain VN/1194 and clade 2.3.4 strain SZ/406H of H5N1, and lung tissues were collected at day 5 postchallenge. (A) Detection of viral titers of VN/1194 and SZ/406H H5N1 viruses in collected lung tissues. Mice injected with SARS-CoV RBD-specific MAb were used as the negative control (Ctrl). The data are expressed as means ± standard deviations of virus titers (log10 TCID50/g) of lung tissues from six mice per group. The limit of detection is 1.5 log10 TCID50/g of tissues, as shown by the dotted line. The figure shows representative data from two independent repeat experiments. (B and C) Evaluation of histopathological changes in collected lung tissues. Lung tissues from the mice injected with a MAb to the RBD of SARS-CoV and those from uninfected mice were used as the negative control (Ctrl) and normal control (Normal), respectively. Lung tissues were stained with H&E and observed by light microscopy (magnification, ×100). Shown are representative images of histopathological changes from two independent experiments with six mice per group challenged with VN/1194 (B) and SZ/406H (C) H5N1 viruses, respectively. Serious histopathological damage was seen in the control mice, with bronchial epithelial cell degeneration, necrosis, and desquamation (solid arrows). Large amounts of exudates and severe edema with infiltration of lymphocytes and mononuclear cells were also seen around blood vessels (open arrows). However, the HA-7-immunized mice showed only mild interstitial pneumonia changes with focal broadening interstitial spaces and lymphocytic infiltration (arrowheads).
MAb HA-7 is highly specific to HA1 of H5N1 virus.
We then detected the specificity of MAb HA-7 by ELISA, by coating the ELISA plates with different antigens consisting of HA1 proteins of H5N1 with or without fusion with Fd and/or Fc or inactivated IAVs. The HA-7 neutralizing MAb reacted strongly with recombinant HA1 proteins respectively fused with the Fc immunoenhancer (HA1-Fc), the trimeric Fd sequences (HA1-Fd), or Fd plus Fc (HA1-Fdc) and HA1 protein without Fd and Fc (HA1-His), but only background levels of immunoreactivity were seen with rFc, IgG-Fc, and the recombinant control proteins HIV-Fd and SARS-CoV RBD (Fig. 4A). In addition, this MAb had strong reactivity with inactivated H5N1 virus but low to no reactivity with 2009 S-OIV H1N1 and 2009-2010 seasonal IAVs consisting of H1N1 and H3N2 influenza A viruses (Fig. 4B). The above-described results indicate that the selected MAb is highly specific to the HA1 protein of H5N1 virus. This neutralizing MAb was found to belong to the IgG1 subtype after coating of the ELISA plate with a HA1-specific recombinant protein (HA1-His) (Fig. 4C). The amino acid sequences of the heavy (VH) and light (VL) chains of HA-7Fab were analyzed, and CDR1, CDR2, and CDR3 were labeled (Fig. 4D).
MAb HA-7 recognized a highly conserved region in HA1 of H5N1.
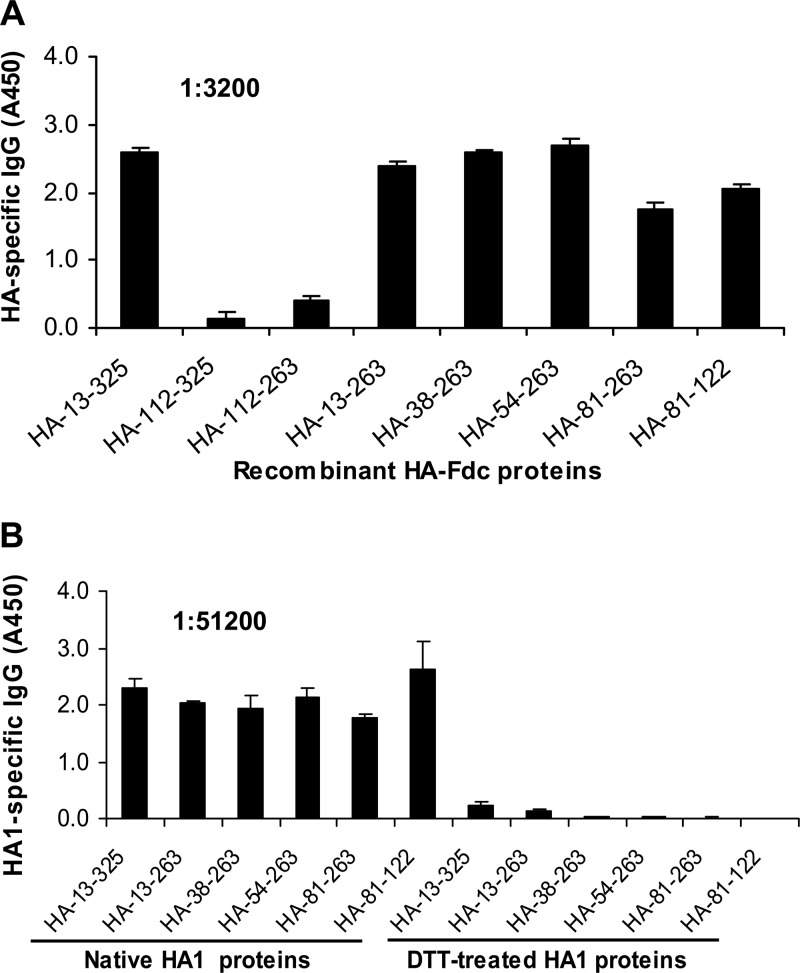
The epitope mapping of HA-7 was initially done by using ELISA plates coated with truncated recombinant HA fragments covering the full length of the HA1 protein. The results indicated that HA-7 reacted strongly with the proteins covering full-length HA1, residues 13 to 325, as well as truncated HA1 fragments containing residues 13 to 263, 38 to 263, 54 to 263, 81 to 263, and 81 to 122 (the sequences were numbered according to the H5 numbering system, as previously described [21, 25, 46]). However, the HA-7 MAb had low to no responses with proteins containing HA residues 112 to 325 (HA-112–325) and HA-112–263 (Fig. 5A). These results suggest that the neutralizing HA-7 MAb is specific to an epitope mapped to residues 81 to 122 (NVPEWSYIVEKANPANDLCYPGNFNDYEELKHLLSRINHFEKIQ) of the HA1 region of H5N1, suggesting that the sequences at residues 81 to 122 of HA1 were essential for MAb binding. The identified epitopes of HA-7 were then aligned by comparing a series of representative sequences covering clades 0 to 9 and their subclades of human and nonhuman H5N1 viruses, including clades 0, 1, 1.1, 2.2, 2.2.1, 2.2.1.1, 2.2.2, 2.3.2, 2.3.2.1, 2.3.4, 2.3.4.1, 2.3.4.2, 2.3.4.3, 3, 4, 5, 6, 7, 8, and 9. Analysis of the amino acid sequence corresponding to this epitope indicated that residues 81 to 122 of HA1 are highly conserved among analyzed strains of H5N1 viruses causing human and nonhuman infections (see Table S1 in the supplemental material).
Fig 5.
Epitope mapping and detection of reactivity of MAb HA-7 with denatured recombinant proteins by ELISA. (A) ELISA plates were coated with truncated recombinant HA1 protein fragments covering the full-length HA1 region, followed by the addition of MAb HA-7 to detect the reactivity of total IgG. The data are presented as mean A450 values ± standard deviations of data from duplicate wells of MAbs at a dilution of 1:3,200. Shown are representative data from three independent repeat experiments. (B) Reactivity of MAb HA-7 with DTT-denatured recombinant HA1 protein fragments, using corresponding proteins without DTT treatment as a comparison. The data are presented as mean A450 values ± standard deviations of data from duplicate wells of MAb at a dilution of 1:51,200.
MAb HA-7 recognized conformational structures of HA.
In order to identify the effects of protein conformation on the ability of the MAb to bind to the specific antigens, ELISA plates were coated with recombinant HA1 proteins and then treated with DTT to break the disulfide bonds of these proteins, followed by the addition of HA-7 to detect reactivity. As shown in Fig. 5B, the reactivity of MAb HA-7 with DTT-treated HA1 fusion proteins decreased dramatically, or to an undetectable level, although HA-7 had strong reactivity with native forms of these HA1 proteins fused with the trimeric motif Fd and immunoenhancer Fc. Further detection of the reactivity of HA-7 with synthetic peptides covering the full-length HA protein of H5N1 indicated that this MAb had very low reactivity with the overlapping peptides containing linear structures of HA (see Fig. S1 in the supplemental material). The fact that HA-7 had a strong reaction with recombinant proteins encompassing different fragments (covering residues 81 to 122) of H5N1 HA1 but had little to no reactivity with DTT-denatured HA1 proteins or overlapping peptides covering this region indicates that the identified neutralizing HA-7 MAb recognizes conformational structures similar to the structure of the native HA1 protein rather than linear structures of HA1.
Mechanism of MAb HA-7 inhibition of viral entry.
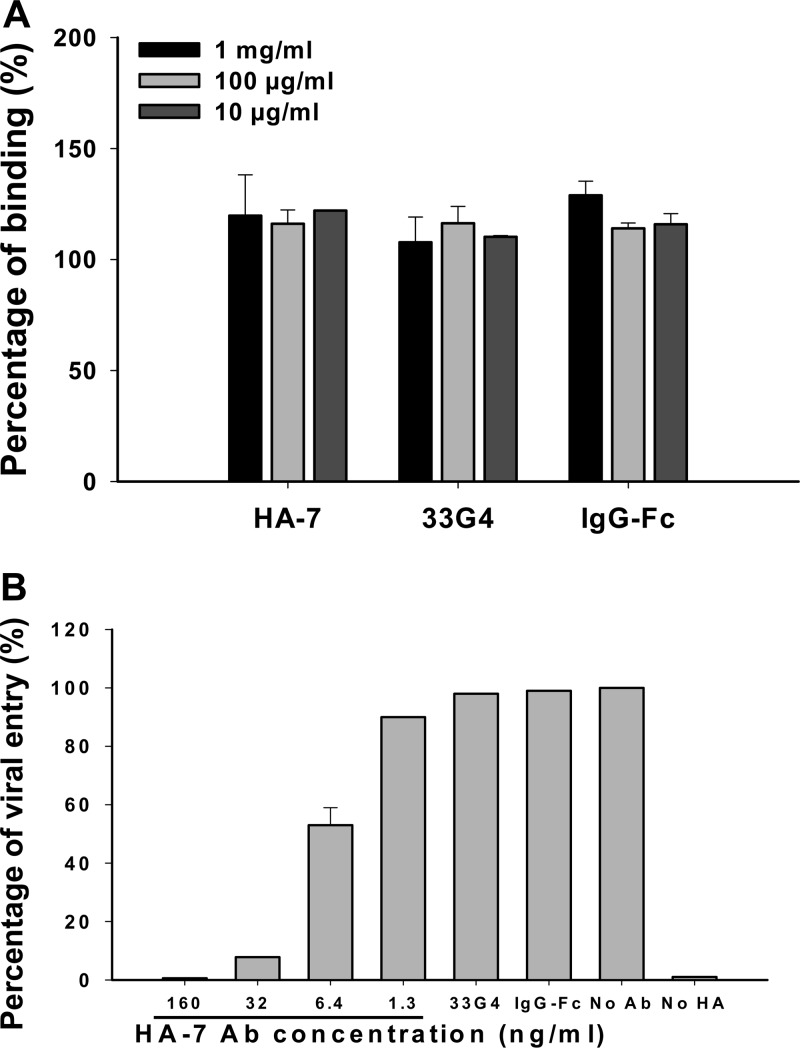
Virus binding assays and postattachment assays were performed using QH-HA H5N1 pseudovirus to detect the mechanism of inhibition. Results from the virus binding assay showed that neither the presence of MAb HA-7 nor the increase of the HA-7 concentration decreased the ability of this MAb to bind to the virus, which are similar to those of the negative-control MAb to SARS-CoV RBD (33G4) and human IgG-Fc (Fig. 6A), suggesting that the inhibition of binding of MAb HA-7 to H5N1 viruses might not be attributed to a blockade of receptor binding. On the contrary, results from the postattachment process indicated that HA-7 inhibited virus entry in a dose-dependent manner, while the negative-control antibodies 33G4 and human IgG-Fc did not neutralize virus entry, even at the highest concentration of 10 μg/ml (Fig. 6B), confirming that MAb HA-7 inhibited virus entry at the postattachment stage rather than through the receptor-binding process.
Fig 6.
Mechanism study of MAb HA-7 against H5N1 pseudovirus infection. (A) Virus binding assay with QH-HA pseudovirus-infected MDCK cells. The antibodies were diluted to 10 μg/ml, 100 μg/ml, and 1 mg/ml, respectively, and binding ability was measured by quantification of HIV p24 content by ELISA. A MAb to the RBD of SARS-CoV (33G4) and human IgG-Fc were used as controls. The data are presented as mean A450 values ± standard deviations of data from duplicate wells. (B) Inhibition by HA-7 of the postattachment process in QH-HA pseudovirus-infected MDCK cells. A MAb to the RBD of SARS-CoV (33G4) and human IgG-Fc were used as the negative controls. No-Ab and no-HA wells were included as the system control. The data are presented as mean percentages ± standard deviations for duplicated wells. Panels A and B show representative data from three independent repeat experiments.
Structural characterization of HA-7Fab/H5HA by 3D-EM.
The HA-7Fab/H5HA complex was well separated from excessive HA-7Fab by gel filtration chromatography (see Fig. S2A in the supplemental material). The 2-dimensional EM images of the negatively stained HA-7Fab/H5HA complex were of high contrast and immediately showed the characteristic HA1 head binding (see Fig. S2B and S2C in the supplemental material). Particle boxing was done semiautomatically, using the EMAN2 package, and then visually checked. CTF correction was done automatically, followed by manual fitting for each micrograph. The initial 3D density model was unambiguously generated using the reference-free class-averaged images. Iterative 3D refinement with a total of 3,172 particle images was done by using EMAN2 until it converged with no significant visual changes checked by Chimera. The effective resolution of this final 3D reconstruction is 18.0 Å based on the criterion of an FSC coefficient of 0.5 (see Fig. S2D in the supplemental material). The final 3D-EM map contoured at 2 times the standard deviation is shown in Fig. S2E in the supplemental material.
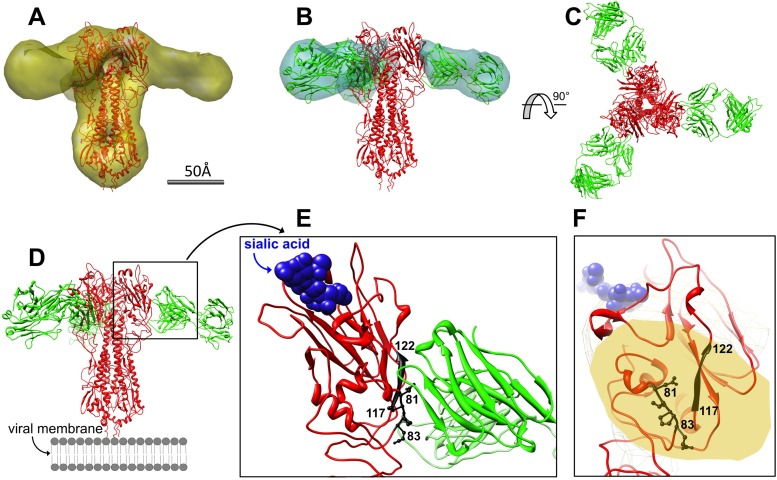
Docking of atomic models of H5HA (X ray) and HA-7Fab (homology built) was straightforward and unambiguous after using the fit-in-map utility in Chimera (Fig. 7A and B), based on correlation coefficient values. From the resulting complex model (Fig. 7C and D; see also Movie S1 the supplemental material), we made two major discoveries. First, HA-7Fab binds laterally to the sides of the H5HA head. It does not contact the receptor-binding site or potentially interact with the receptor itself (Fig. 7E and F; see also Movie S1 in the supplemental material), indicating that HA-7 would not interfere with virus attachment to the host cell surface. Second, we found that residues 81 to 83 (Asn81, Val82, Pro82A, and Glu83) and 117 to 122 (His117, Phe118, Glu119, Lys120, Ile121, and Gln122) constitute the entire core epitope map for HA-7 (Fig. 7E and F). These two segments are in the center of Fab's “footprint” on HA1 and are spatially the closest to Fab. The two segments are 38 amino acid residues away from each other. Residues 81 to 83 exist as a loop, while residues 117 to 122 adopt a β-strand conformation. These features manifest the conformational nature of the epitope. In addition to the core epitope, other scattered HA1 residues that are peripheral to Fab's density and might come into contact with HA-7 include residues Asp77, Glu78, Ile80, Tyr141, Gln142, Arg149, Tyr256, Lys259, and Val261 of HA1.
Fig 7.
3D-EM-based model docking and epitope analysis of HA-7. (A) Docking of H5HA trimer (red) into the EM density map (yellow). (B) Docking of Fab (green) into its density map (cyan). (C) Threefold top view of the trimeric HA-7Fab/H5HA complex. (D) Side view of the trimeric HA-7Fab/H5HA complex. (E) Zoom-in side view of the HA/Fab interface. Residues 81 to 83 and 117 to 122 of HA1 (black) are in contact with Fab, while the receptor (sialic acid [blue spheres]) is not. (F) Zoom-in view from Fab showing that residues 81 to 83 and 117 to 122 of HA1 (black) lie at the center of Fab's “footprint” (yellow) on HA and comprise the core epitope. The receptor (sialic acid [light blue spheres]) is in the background.
DISCUSSION
H5N1 infection has become an increasing threat to public health worldwide (47–50). If the H5N1 virus, with either naturally occurring mutations or laboratory-generated mutants and/or reassortants, acquires the ability to transmit from human to human and maintains its current virulence, a potential influenza epidemic could emerge, for which the world is inadequately prepared (6, 51). Thus, it seems prudent to evaluate a series of comprehensive prevention and immunotherapy strategies based on neutralizing MAbs specifically targeting the HA of H5N1 virus, a key viral protein capable of inducing potent neutralizing antibodies against virus infection.
In this study, we generated such a neutralizing MAb, HA-7, that strongly inhibited the entry of H5N1 virus with all tested strains covering clades 0, 1, 2.2, 2.3.4, and 2.3.2.1 as well as completely protected immunized mice against lethal challenge by tested clade 1 and 2.3.4 strains of H5N1 virus. Interestingly, unlike other previously reported neutralizing MAbs targeting group 1 and/or group 2 IAVs (21–23, 52), our identified MAb neutralized only H5N1 virus, rather than other types of IAVs, such as S-OIV(H1N1) and seasonal influenza virus, indicating that this MAb is highly specific to the H5N1 virus.
To further characterize the HA-7 neutralizing MAb and map its epitope(s), we constructed and expressed a series of recombinant HA protein fragments covering the full-length HA1 protein (residues 13 to 325) fused with the Fd trimeric motif and Fc immunoenhancer, expecting them to maintain the conformational structures of native HA (30). The detection of MAb with these recombinant proteins indicated that HA-7 had strong reactivity with proteins covering residues 81 to 122 of HA1 but little or no reactivity with the same proteins denatured by DTT or synthetic overlapping peptides covering this region, further suggesting the ability of MAb HA-7 to recognize conformational, rather than linear, structures of HA. This is consistent with the results from the 3D-EM study of the HA-7Fab/H5HA complex. By aligning the epitope residue sequences, we confirmed that this newly discovered neutralizing epitope covering residues 81 to 122 of the HA1 subunit, part of the RBD region, particularly its core residues (residues 81 to 83 and 117 to 122), is highly conserved among clades 0 to 9 and their subclades of H5N1 viruses, according to the WHO and CDC (see Table S1 in the supplemental material). Thus, the identification of this novel conformation-dependent neutralizing epitope may guide us to develop effective innovative vaccines, in consideration of the RBD of HA as the primary target of neutralizing antibodies.
The anti-HA antibodies function in two ways to neutralize influenza virus infection, either by blocking binding of the viral HA to its cellular receptor (the first step) or by interfering with the subsequent HA-mediated membrane fusion (the second step) (21, 53). Unlike some of the other HA-neutralizing MAbs which block the step of virus attachment and receptor binding (54, 55), this mechanistic study showed that HA-7 did not inhibit virus binding to the cells (receptor-binding step) but affected the postattachment stage (Fig. 6A and B), indicating that HA-7 inhibits virus entry, possibly by blocking HA-mediated membrane fusion. Distinct from the MAb targeting the stalk region of the fusion peptide of HA2, which usually conferred broad cross-protection against group 1 and/or group 2 IAVs (21–23), MAbs targeting the head region have the liability to induce strain-specific immunity against a particular subgroup of IAVs (26, 27). Here, however, we showed that the HA1-targeting HA-7 MAb maintains broad neutralization and is specific to the H5N1 virus.
A number of neutralizing antibodies were previously reported to target HA1 of H5N1. It has been shown that neutralizing antibody AVFluIgG01 recognized the sequence IIPKSSWS at residues 123 to 128 of HA1, with the key residues I123, I124, P125, W127, S128, Y168, and T171 (conversion to H5 numbering) (56, 57). Similarly, MAb 65C6 binds to a conformational epitope comprising residues P125, S126, K165, Y168, and T171 (conversion to H5 numbering) at the globular domain of HA (26), with one key overlapping residue with AVFluIgG01 at P125. Previous reports also showed that neutralizing antibody mAb12-1G6 bound to residues at positions 140 to 145 of H5N1 HA, with amino acids Q142, K144, and S145 being critical for binding (58), while others demonstrated that H5N1 HA1-specific neutralizing MAbs, such as 9F4, bind the epitope below the globular head of the HA1 subunit (27). Different from the above-described HA1-binding neutralizing MAbs, HA-7 targets a neutralizing epitope at residues 81 to 122 of the HA1 globular head domain, with N81, V82, P82A, E83, H117, F118, E119, K120, I121, and Q122 as the core epitope, which was not previously described.
Previous cryo-EM studies of influenza virus HA at neutral and fusogenic pHs indicated that the HA head domain is required to undergo a conformational change in order for HA to acquire a fusion-competent status (59). The binding of HA-7 to the head region may well prevent, or retard, this head domain change and thus interfere with subsequent membrane fusion needed for viral infection. Therefore, our 3D-EM results are consistent with the results of the mechanistic study using a virus binding assay and a postattachment assay. These results all point to the conclusion that HA-7 inhibits virus entry at the postattachment membrane fusion stage rather than the receptor-binding step. We propose that it is the conservation of the core epitope (residues 81 to 83 and 117 to 122) among avian influenza virus strains that accounts for the broad neutralization ability of HA-7 and that confers the specificity of HA-7 for avian influenza viruses. At this point, limited by the current resolution of the 3D-EM approach, we cannot pinpoint the residues of HA-7 and H5HA which are involved in antibody-antigen binding. More detailed HA1/HA-7 interactions at atomic resolution can be obtained by further X-ray crystallography studies on the HA-7Fab/H5HA complex.
To summarize, this report described the identified neutralizing MAb, HA-7, which specifically targets a novel and highly conserved conformational region at the HA1 head of H5N1 virus. This antibody binds laterally to the “head” of viral HA but does not interfere with its receptor binding. The core epitope residues 81 to 83 and 117 to 122 of HA1 are conserved at the protein primary structure level in almost all of the H5N1 stains identified to date. Furthermore, we have shown that HA-7 specifically targets and neutralizes H5N1 virus rather than other types of IAVs, suggesting that HA-7 can be humanized as an effective passive immunotherapeutic agent for antiviral stockpiling for future influenza pandemics caused by emerging highly pathogenic avian H5N1 viruses with unpredictable HA sequences. The identified epitope would also provide a significant foundation for the design of novel H5N1 vaccines covering this particular region, which in turn offers the promise of inducing broad-spectrum immunity for the prevention of infections caused by divergent H5N1 influenza virus strains.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the NIH (grant R03AI088449 to L.D.), the Department of Defense (grant W81XWH-10-1-0093 to R.C.L.), and the National 973 Basic Research Program of China (grant 2011CB504706 to Y.Z. and grant 2012CB519001 to S.J.). We acknowledge the use of instruments at the Electron Imaging Center for NanoMachines, supported by the NIH (grant 1S10RR23057 to Z. Hong Zhou), and CNSI at UCLA. We also acknowledge the use of the UCSD Cryo-Electron Microscopy Facility, which is supported by NIH grants to Timothy S. Baker and a gift from the Agouron Institute to UCSD. We have no conflicting financial interests.
We greatly acknowledge the contributions of the originating laboratories that have provided the samples and/or submitted sequence data to the GenBank and GISAID EpiFlu databases. We are grateful to Ivo Atanasov (UCLA) and Norm Olson (UCSD) for assistance with EM imaging.
Footnotes
Published ahead of print 5 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02344-12.
REFERENCES
- 1. Fouchier RA, Garcia-Sastre A, Kawaoka Y. 2012. Pause on avian flu transmission studies. Nature 481:443 doi:10.1038/481443a [DOI] [PubMed] [Google Scholar]
- 2. Fouchier RA, Herfst S, Osterhaus AD. 2012. Public health and biosecurity. Restricted data on influenza H5N1 virus transmission. Science 335:662–663 [DOI] [PubMed] [Google Scholar]
- 3. Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berns KI, Casadevall A, Cohen ML, Ehrlich SA, Enquist LW, Fitch JP, Franz DR, Fraser-Liggett CM, Grant CM, Imperiale MJ, Kanabrocki J, Keim PS, Lemon SM, Levy SB, Lumpkin JR, Miller JF, Murch R, Nance ME, Osterholm MT, Relman DA, Roth JA, Vidaver AK. 2012. Policy: adaptations of avian flu virus are a cause for concern. Nature 482:153–154 [DOI] [PubMed] [Google Scholar]
- 6. Berns KI, Casadevall A, Cohen ML, Ehrlich SA, Enquist LW, Fitch JP, Franz DR, Fraser-Liggett CM, Grant CM, Imperiale MJ, Kanabrocki J, Keim PS, Lemon SM, Levy SB, Lumpkin JR, Miller JF, Murch R, Nance ME, Osterholm MT, Relman DA, Roth JA, Vidaver AK. 2012. Public health and biosecurity. Adaptations of avian flu virus are a cause for concern. Science 335:660–661 [DOI] [PubMed] [Google Scholar]
- 7. Fouchier RA, Garcia-Sastre A, Kawaoka Y, Barclay WS, Bouvier NM, Brown IH, Capua I, Chen H, Compans RW, Couch RB, Cox NJ, Doherty PC, Donis RO, Feldmann H, Guan Y, Katz J, Klenk HD, Kobinger G, Liu J, Liu X, Lowen A, Mettenleiter TC, Osterhaus AD, Palese P, Peiris JS, Perez DR, Richt JA, Schultz-Cherry S, Steel J, Subbarao K, Swayne DE, Takimoto T, Tashiro M, Taubenberger JK, Thomas PG, Tripp RA, Tumpey TM, Webby RJ, Webster RG. 2012. Pause on avian flu transmission research. Science 335:400–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Le Duc JW, Franz DR. 2012. Genetically engineered transmissible influenza A/H5N1: a call for laboratory safety and security. Biosecur. Bioterror. 10:153–154 [DOI] [PubMed] [Google Scholar]
- 9. Cohen J. 2012. Bird flu controversy. Does forewarned = forearmed with lab-made avian influenza strains? Science 335:785–787 [DOI] [PubMed] [Google Scholar]
- 10. Harada Y, Ninomiya-Mori A, Takahashi Y, Shirakura M, Kishida N, Kageyama T, Tada Y, Tashiro M, Odagiri T. 2011. Inactivated and adjuvanted whole-virion clade 2.3.4 H5N1 pre-pandemic influenza vaccine possesses broad protective efficacy against infection by heterologous clades of highly pathogenic H5N1 avian influenza virus in mice. Vaccine 29:8330–8337 [DOI] [PubMed] [Google Scholar]
- 11. Huang MH, Lin SC, Hsiao CH, Chao HJ, Yang HR, Liao CC, Chuang PW, Wu HP, Huang CY, Leng CH, Liu SJ, Chen HW, Chou AH, Hu AY, Chong P. 2010. Emulsified nanoparticles containing inactivated influenza virus and CpG oligodeoxynucleotides critically influences the host immune responses in mice. PLoS One 5:e12279 doi:10.1371/journal.pone.0012279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Steel J. 2011. New strategies for the development of H5N1 subtype influenza vaccines: progress and challenges. BioDrugs 25:285–298 [DOI] [PubMed] [Google Scholar]
- 13. Ding H, Tsai C, Gutierrez RA, Zhou F, Buchy P, Deubel V, Zhou P. 2011. Superior neutralizing antibody response and protection in mice vaccinated with heterologous DNA prime and virus like particle boost against HPAI H5N1 virus. PLoS One 6:e16563 doi:10.1371/journal.pone.0016563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hessel A, Schwendinger M, Holzer GW, Orlinger KK, Coulibaly S, Savidis-Dacho H, Zips ML, Crowe BA, Kreil TR, Ehrlich HJ, Barrett PN, Falkner FG. 2011. Vectors based on modified vaccinia Ankara expressing influenza H5N1 hemagglutinin induce substantial cross-clade protective immunity. PLoS One 6:e16247 doi:10.1371/journal.pone.0016247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Khurana S, Wu J, Verma N, Verma S, Raghunandan R, Manischewitz J, King LR, Kpamegan E, Pincus S, Smith G, Glenn G, Golding H. 2011. H5N1 virus-like particle vaccine elicits cross-reactive neutralizing antibodies that preferentially bind to the oligomeric form of influenza virus hemagglutinin in humans. J. Virol. 85:10945–10954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ledgerwood JE, Wei CJ, Hu Z, Gordon IJ, Enama ME, Hendel CS, McTamney PM, Pearce MB, Yassine HM, Boyington JC, Bailer R, Tumpey TM, Koup RA, Mascola JR, Nabel GJ, Graham BS. 2011. DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. Lancet Infect. Dis. 11:916–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin SC, Huang MH, Tsou PC, Huang LM, Chong P, Wu SC. 2011. Recombinant trimeric HA protein immunogenicity of H5N1 avian influenza viruses and their combined use with inactivated or adenovirus vaccines. PLoS One 6:e20052 doi:10.1371/journal.pone.0020052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Suguitan AL, Jr, Cheng X, Wang W, Wang S, Jin H, Lu S. 2011. Influenza H5 hemagglutinin DNA primes the antibody response elicited by the live attenuated influenza A/Vietnam/1203/2004 vaccine in ferrets. PLoS One 6:e21942 doi:10.1371/journal.pone.0021942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Torrieri-Dramard L, Lambrecht B, Ferreira HL, Van den Berg T, Klatzmann D, Bellier B. 2011. Intranasal DNA vaccination induces potent mucosal and systemic immune responses and cross-protective immunity against influenza viruses. Mol. Ther. 19:602–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16:265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856 [DOI] [PubMed] [Google Scholar]
- 24. Prabhu N, Prabakaran M, Ho HT, Velumani S, Qiang J, Goutama M, Kwang J. 2009. Monoclonal antibodies against the fusion peptide of hemagglutinin protect mice from lethal influenza A virus H5N1 infection. J. Virol. 83:2553–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. 2006. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312:404–410 [DOI] [PubMed] [Google Scholar]
- 26. Hu H, Voss J, Zhang G, Buchy P, Zuo T, Wang L, Wang F, Zhou F, Wang G, Tsai C, Calder L, Gamblin SJ, Zhang L, Deubel V, Zhou B, Skehel JJ, Zhou P. 2012. A human antibody recognizing a conserved epitope of H5 hemagglutinin broadly neutralizes highly pathogenic avian influenza H5N1 viruses. J. Virol. 86:2978–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oh HL, Akerstrom S, Shen S, Bereczky S, Karlberg H, Klingstrom J, Lal SK, Mirazimi A, Tan YJ. 2010. An antibody against a novel and conserved epitope in the hemagglutinin 1 subunit neutralizes numerous H5N1 influenza viruses. J. Virol. 84:8275–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. National Research Council 1996. Guide for the care and use of laboratory animals. National Academies Press, Washington, DC [Google Scholar]
- 29. Du L, Zhao G, Zhang X, Liu Z, Yu H, Zheng BJ, Zhou Y, Jiang S. 2010. Development of a safe and convenient neutralization assay for rapid screening of influenza HA-specific neutralizing monoclonal antibodies. Biochem. Biophys. Res. Commun. 397:580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Du L, Leung VH, Zhang X, Zhou J, Chen M, He W, Zhang HY, Chan CC, Poon VK, Zhao G, Sun S, Cai L, Zhou Y, Zheng BJ, Jiang S. 2011. A recombinant vaccine of H5N1 HA1 fused with foldon and human IgG Fc induced complete cross-clade protection against divergent H5N1 viruses. PLoS One 6:e16555 doi:10.1371/journal.pone.0016555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chou TC. 2006. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 58:621–681 [DOI] [PubMed] [Google Scholar]
- 32. Zhao G, Du L, Xiao W, Sun S, Lin Y, Chen M, Kou Z, He Y, Lustigman S, Jiang S, Zheng BJ, Zhou Y. 2010. Induction of protection against divergent H5N1 influenza viruses using a recombinant fusion protein linking influenza M2e to Onchocerca volvulus activation associated protein-1 (ASP-1) adjuvant. Vaccine 28:7233–7240 [DOI] [PubMed] [Google Scholar]
- 33. Zhao G, Lin Y, Du L, Guan J, Sun S, Sui H, Kou Z, Chan CC, Guo Y, Jiang S, Zheng BJ, Zhou Y. 2010. An M2e-based multiple antigenic peptide vaccine protects mice from lethal challenge with divergent H5N1 influenza viruses. Virol. J. 7:9 doi:10.1186/1743-422X-7-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao G, Sun S, Du L, Xiao W, Ru Z, Kou Z, Guo Y, Yu H, Jiang S, Lone Y, Zheng BJ, Zhou Y. 2010. An H5N1 M2e-based multiple antigenic peptide vaccine confers heterosubtypic protection from lethal infection with pandemic 2009 H1N1 virus. Virol. J. 7:151 doi:10.1186/1743-422X-7-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Du L, Zhao G, Lin Y, Sui H, Chan C, Ma S, He Y, Jiang S, Wu C, Yuen KY, Jin DY, Zhou Y, Zheng BJ. 2008. Intranasal vaccination of recombinant adeno-associated virus encoding receptor-binding domain of severe acute respiratory syndrome coronavirus (SARS-CoV) spike protein induces strong mucosal immune responses and provides long-term protection against SARS-CoV infection. J. Immunol. 180:948–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Du L, Zhao G, Chan CC, Sun S, Chen M, Liu Z, Guo H, He Y, Zhou Y, Zheng BJ, Jiang S. 2009. Recombinant receptor-binding domain of SARS-CoV spike protein expressed in mammalian, insect and E. coli cells elicits potent neutralizing antibody and protective immunity. Virology 393:144–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Du L, Zhao G, Li L, He Y, Zhou Y, Zheng BJ, Jiang S. 2009. Antigenicity and immunogenicity of SARS-CoV S protein receptor-binding domain stably expressed in CHO cells. Biochem. Biophys. Res. Commun. 384:486–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Du L, Zhao G, Chan CC, Li L, He Y, Zhou Y, Zheng BJ, Jiang S. 2010. A 219-mer CHO-expressing receptor-binding domain of SARS-CoV S protein induces potent immune responses and protective immunity. Viral Immunol. 23:211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. He Y, Li J, Heck S, Lustigman S, Jiang S. 2006. Antigenic and immunogenic characterization of recombinant baculovirus-expressed severe acute respiratory syndrome coronavirus spike protein: implication for vaccine design. J. Virol. 80:5757–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ohi M, Li Y, Cheng Y, Walz T. 2004. Negative staining and image classification—powerful tools in modern electron microscopy. Biol. Proced. Online 6:23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157:38–46 [DOI] [PubMed] [Google Scholar]
- 42. Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a Web-based environment for protein structure homology modelling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- 43. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 44. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 45. Ha Y, Stevens DJ, Skehel JJ, Wiley DC. 2001. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc. Natl. Acad. Sci. U. S. A. 98:11181–11186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weis WI, Brunger AT, Skehel JJ, Wiley DC. 1990. Refinement of the influenza virus hemagglutinin by simulated annealing. J. Mol. Biol. 212:737–761 [DOI] [PubMed] [Google Scholar]
- 47. Butler D. 2012. Caution urged for mutant flu work. Nature 481:417–418 [DOI] [PubMed] [Google Scholar]
- 48. Kawaoka Y. 2012. H5N1: flu transmission work is urgent. Nature 482:155 doi:10.1038/nature10884 [DOI] [PubMed] [Google Scholar]
- 49. Perez DR. 2012. Public health and biosecurity. H5N1 debates: hung up on the wrong questions. Science 335:799–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Garcia-Sastre A. 2012. Working safely with H5N1 viruses. mBio 3(2):e00049–12 doi:10.1128/mBio.00049-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cohen J. 2012. Bird flu controversy. Dead reckoning the lethality of bird flu. Science 335:786 doi:10.1126/science.335.6070.786 [DOI] [PubMed] [Google Scholar]
- 52. Yoshida R, Igarashi M, Ozaki H, Kishida N, Tomabechi D, Kida H, Ito K, Takada A. 2009. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog. 5:e1000350 doi:10.1371/journal.ppat.1000350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Skehel JJ, Wiley DC. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531–569 [DOI] [PubMed] [Google Scholar]
- 54. Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao HX, Harrison SC. 2011. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U. S. A. 108:14216–14221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Han T, Sui J, Bennett AS, Liddington RC, Donis RO, Zhu Q, Marasco WA. 2011. Fine epitope mapping of monoclonal antibodies against hemagglutinin of a highly pathogenic H5N1 influenza virus using yeast surface display. Biochem. Biophys. Res. Commun. 409:253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sun L, Lu X, Li C, Wang M, Liu Q, Li Z, Hu X, Li J, Liu F, Li Q, Belser JA, Hancock K, Shu Y, Katz JM, Liang M, Li D. 2009. Generation, characterization and epitope mapping of two neutralizing and protective human recombinant antibodies against influenza A H5N1 viruses. PLoS One 4:e5476 doi:10.1371/journal.pone.0005476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cao Z, Meng J, Li X, Wu R, Huang Y, He Y. 2012. The epitope and neutralization mechanism of AVFluIgG01, a broad-reactive human monoclonal antibody against H5N1 influenza virus. PLoS One 7:e38126 doi:10.1371/journal.pone.0038126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ohkura T, Kikuchi Y, Kono N, Itamura S, Komase K, Momose F, Morikawa Y. 2012. Epitope mapping of neutralizing monoclonal antibody in avian influenza A H5N1 virus hemagglutinin. Biochem. Biophys. Res. Commun. 418:38–43 [DOI] [PubMed] [Google Scholar]
- 59. Bottcher C, Ludwig K, Herrmann A, van Heel M, Stark H. 1999. Structure of influenza haemagglutinin at neutral and at fusogenic pH by electron cryo-microscopy. FEBS Lett. 463:255–259 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.