Abstract
MicroRNAs are key regulators of the immune response, but their role in CD8 T cell differentiation in vivo is not known. We show that miR-155 is important in both effector and memory antiviral CD8 T cell responses. Without miR-155, there was a weaker effector response and a skewing toward memory precursor cells. At the memory stage, miR-155-deficient CD8 T cells preferentially differentiated into central memory cells and were capable of mounting a potent secondary response.
TEXT
For intracellular pathogens, a crucial part of long-term immunity is the generation of memory CD8 T cells. Naïve T cells can be directed toward at least two different fate-determining pathways after antigen (Ag) exposure. One pathway, driven by IL-12 and IL-2 signaling, upregulates T box expressed in T cell (T-bet) expression, resulting in terminal differentiation of short-lived effector cells (SLECs). Alternatively, a pathway driven by lower levels of IL-12 but stimulated by IL-21 or IL-10 leads to production of memory precursor effector cells (MPECs), which display less effector activity than SLECs (1). The ultimate repository for long-term memory is the central memory T cell (Tcm), and understanding how T cells can be directed toward this fate has profound implications for vaccine design. Given the distinct transcriptional profiles of effector and memory cells (2), it is an open question if there are master regulators of gene expression in these cells. MicroRNAs (miRNAs) are important posttranscriptional regulators in many developmental processes. Several miRNAs have been shown to be critical in many stages of hematopoietic development (3). While miRNAs have been shown to influence CD4 T cell and regulatory T cell differentiation, it is currently unclear which miRNAs influence CD8 T cell differentiation.
Studies using CD8 T cells lacking dicer, an enzyme essential for the processing of all miRNAs, showed deficits in CD8 T cell accumulation and survival (4). Other studies profiled miRNA expression following CD8 T cell activation in vitro (5, 6). Upregulation of miR-21 and miR-155 was observed, which was interesting due to previous studies implicating roles for them in immune cell function (7, 8). In another study, CD8 T cells cultured in vitro to resemble central memory cells downregulated miR-155 relative to effector CD8 T cells (9). While these studies implied miR-155 involvement in CD8 T cell differentiation and memory development, it is still unclear which processes are affected by miR-155 during T cell differentiation in vivo. Here we report that miR-155 plays a critical role in the generation of memory CD8 T cells during virus infection in vivo.
Discussion.
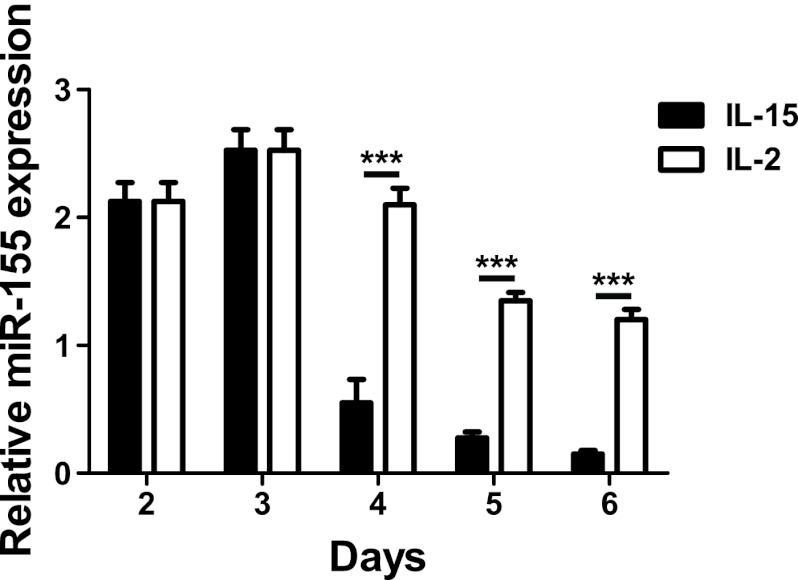
To determine the kinetics of miR-155 expression after CD8 T cell activation, we simulated OT-I cells with antigen in vitro and measured miR-155 expression by PCR. miR-155 expression peaked at 3 days poststimulation and then declined at all subsequent time points (Fig. 1). The decline in miR-155 was much more marked in cells cultured with IL-15 than in cultures supplemented with IL-2, resulting in significantly lower levels of miR-155 at days 4, 5, and 6 after activation. Culture with IL-2 generates effector/effector memory phenotype CD8 T cells, whereas IL-15 generates cells with central memory characteristics (10). These data therefore implied that sustained miR-155 expression was associated with effector/effector memory cells, whereas central memory cells were associated with lower miR-155 expression.
Fig 1.
miR-155 expression in CD8 T cells following activation. OT-I T cells were cultured with SIINFEKL peptide (1 ng/ml) for 3 days and then cultured with either IL-2 (10 U/ml) or IL-15 (10 ng/ml) for an additional 3 days. Total RNA was extracted from cultures on days 2, 3, 4, 5, and 6. We then performed microRNA-specific reverse transcription followed by quantitative PCR (qPCR) for miR-155, using PCR for small nucleolar RNA 202 (snoRNA202) as an internal standard. Graphs show means ± standard deviations. ***, P < 0.01.
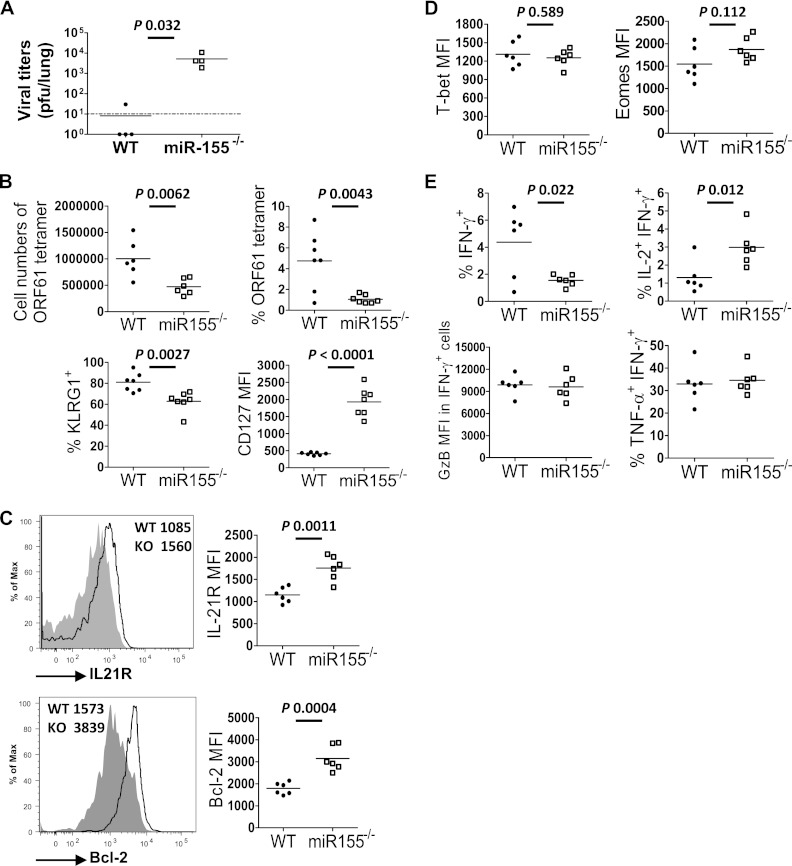
Next we assessed the role of miR-155 in the antiviral CD8 T cell response in vivo. As miR-155 has been shown to affect the biology of multiple hematopoietic cell types (11–13), we chose a system where CD8 T cell-intrinsic effects could be measured. To measure the relevance of miR-155 for viral control, we constructed mixed-bone marrow chimeric mice such that miR-155 was absent from the entire CD8 T cell compartment. Following infection with murid herpesvirus 68 (MHV-68), viral titers in the lung were determined by standard plaque assay (13) at day 14 postinfection. Viral titers were significantly higher in chimeric mice lacking miR-155 in CD8 T cells than in mice with intact miR-155 in CD8 T cells (Fig. 2A). In order to study the effect of miR-155 deficiency on the biology of the CD8 T cell compartment, without the potentially confounding factor of differences in viral load, mixed-bone marrow chimeric mice were generated such that both wild-type (WT) and miR-155−/− CD8 T cells were present in the same infected mouse (14). The proportions and surface phenotypes (CD127, KLRG-1, CD122, and CD62L) were comparable between the miR-155−/− and WT CD8 T cells before infection in these mice (data not shown). Upon infection with MHV-68, the epitope-specific primary CD8 T cell response from each population was measured. We observed an attenuated effector response from miR-155−/− CD8 T cells at the acute phase (Fig. 2B, top). Phenotypic analysis showed markedly higher CD127 expression by miR-155−/− cells and lower KLRG1 expression (Fig. 2B, bottom), indicative of a skewing toward MPECs. Similar data were obtained from the lung and mediastinal lymph nodes. These changes were also observed for the subdominant ORF6 epitope, showing that this was not an epitope-dependent phenomenon (data not shown).
Fig 2.
Effect of miR-155 on the primary antiviral CD8 T cell response to MHV-68. (A) Bone marrow from miR-155−/− and CD8α−/− mice were mixed in a 1:1 ratio and then injected intravenously into lethally irradiated RAG−/− mice (miR-155 group) to generate mice lacking miR-155 in all CD8 T cells. As a control, wild-type bone marrow was mixed with CD8α−/− bone marrow (WT group), such that all CD8 T cells were wild type. After reconstitution of the immune system, mice were infected with MHV-68 and the lung viral titer was measured on day 10 postinfection. (B to E) Mixed-bone marrow chimeric mice were generated by injecting lethally irradiated B6 mice with a 1:1 ratio of miR-155 and WT bone marrow. Therefore, both miR-155−/− and WT CD8 T cells were present in the same mouse but expressed different congenic markers, allowing us to distinguish the two populations. Following immune reconstitution, mice were infected intranasally with 400 PFU of MHV-68, and various organs were removed at 14 days postinfection. The antigen-specific CD8 T response was measured by ORF61 tetramer staining. MFI, mean fluorescence intensity. (B, top) Numbers of total ORF61 tetramer-positive CD8 T cells and percentages of ORF61-specific CD8 T cells among total CD8 T cells. (Bottom) Percentages of KLRG1+ cells and the fluorescence intensity of CD127 staining in the ORF61-specific CD8 T cell population. (C) Representative flow cytometric plots for IL-21R and Bcl-2 staining gated on ORF61-specific CD8 T cell populations. Graphs show values obtained, with each point representing a single mouse. KO, knockout. (D) Mean fluorescence intensity of T-bet and eomesodermin staining in the ORF61-specific CD8 populations. (E) CD8 T cells were stained to measure the production of IFN-γ, IL-2, TNF-α, and granzyme B after brief antigen simulation in vitro. Plots were gated on IFN-γ-producing CD8 T cell populations. Each point represents data from an individual mouse. Data are representative of three experiments. The limit of detection was 10 PFU per lung (dotted line). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The miR-155 population also exhibited elevated expression of the IL-2 receptor (IL-21R) and Bcl-2, markers indicative of memory cells (Fig. 2C). Levels of expression of transcription factors associated with the activation of effector-associated genes, T-bet and eomesodermin, were equivalent between WT and miR-155−/− cells (Fig. 2D). As expected, the frequency of CD8 T cells producing gamma interferon (IFN-γ) was reduced in the absence of miR-155 (Fig. 2E). No difference was detected in the proportions of cells producing tumor necrosis factor alpha (TNF-α) or the levels of granzyme B produced (Fig. 2E). However, we detected a significantly elevated production of IL-2 (Fig. 2E), consistent with phenotypic skewing toward the MPEC phenotype.
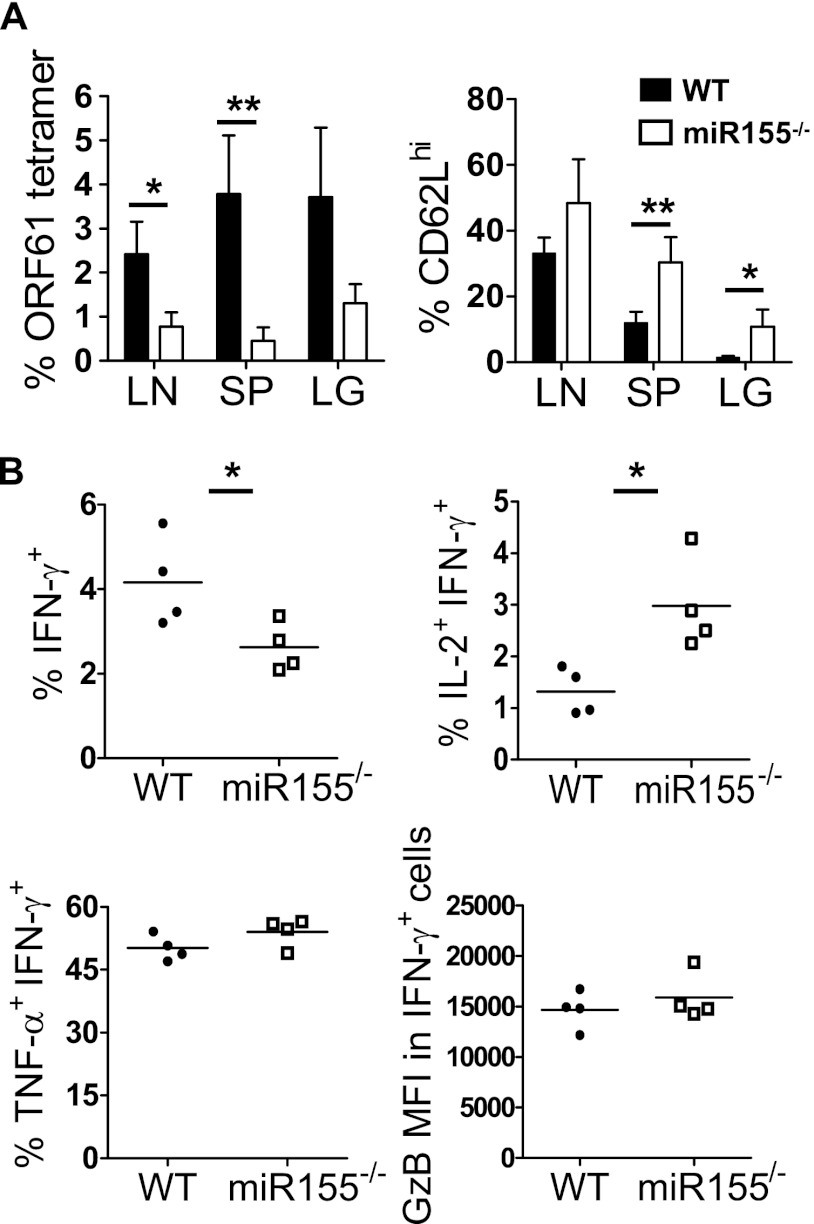
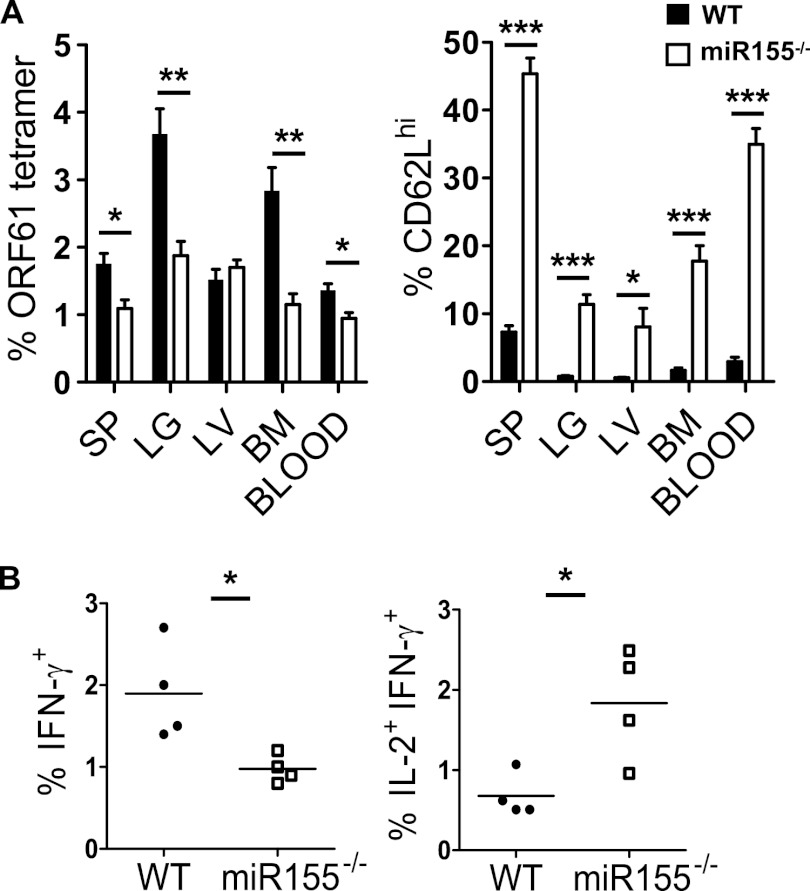
As our data indicated preferential generation of an MPEC phenotype, we tested a later time postinfection (day 28), after the resolution of the acute infection. The overall magnitude of the memory response was decreased in the miR-155−/− population (Fig. 3A). Consistently with our previous research (15), the memory CD8 T cell response was dominated by effector memory (low-expression CD62L [CD62Llo]) cells in the WT population. However, a significantly larger proportion of miR-155−/− cells were CD62Lhi, consistent between several different organs (Fig. 3A). Analysis of effector functions revealed a decreased proportion of IFN-γ, but TNF-α and granzyme B production was unaffected (Fig. 3B). However, miR-155−/− cells produced markedly more IL-2 than the equivalent WT cells (Fig. 3B). These phenotypic and functional changes were also observed at day 60 postinfection but with a larger difference between the experimental groups with regard to CD62L expression (Fig. 4A and B). Collectively, these data indicated a significantly larger proportion of central memory cells within the miR-155−/− CD8 T population. This was consistent with our in vitro data, which associated lower miR-155 expression with central memory differentiation.
Fig 3.
Early skewing toward a central memory phenotype. Cells from the organs indicated were harvested from MHV-68-infected mixed-bone marrow chimeric mice at day 28 postinfection. Cells were stained with ORF61 tetramer plus antibody to CD62L. (A) Percentages of ORF61-specific cells among the CD8 T cell populations and percentages of CD62Lhi cells among the ORF61-specific CD8 T cell population. (B) CD8 T cells were stained to measure the production of IFN-γ, IL-2, TNF-α, and granzyme B. Plots were gated on IFN-γ-producing CD8 T cell populations. Each point represents data from an individual mouse. Data are representative of three experiments. *, P < 0.05; **, P < 0.01. LN, mediastinal lymph node; SP, spleen; LG, lung.
Fig 4.
Memory Ag-specific miR155−/− CD8 T cells that are present at a low frequency have a central memory phenotype. Mixed-bone marrow chimeric mice were intranasally infected with 400 PFU of MHV-68. (A) Sixty days postinfection, Ag-specific CD8 T cells were identified using ORF61 tetramer in the organs shown, and the percentages of CD62Lhi were measured. (B) IFN-γ or IL-2 production in the IFN-γ+ CD8 T cells was measured with intracellular cytokine staining. Each point represents data from an individual mouse. Data are representative of three experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. SP, spleen; LG, lung; LV, liver; BM, bone marrow.
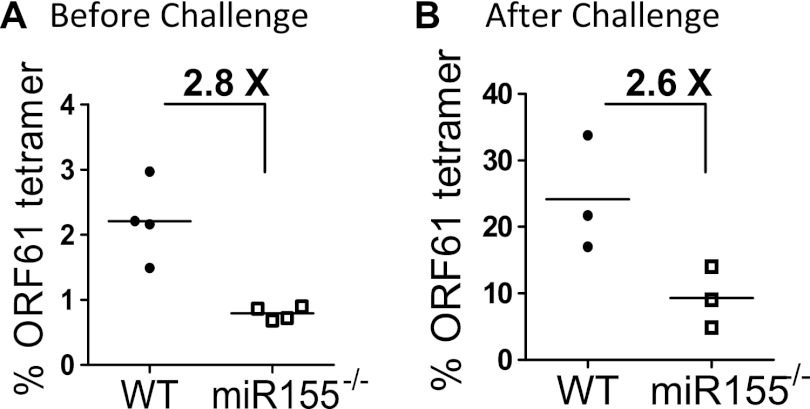
To test whether memory CD8 T cells remained functional in the absence of miR-155, chimeric mice were challenged with a recombinant vaccinia virus encoding the ORF61 epitope from MHV-68, 60 days after MHV-68 infection. Both the WT and miR-155−/− populations expanded equivalently (Fig. 5), indicating that the secondary proliferative capacity of central memory-skewed miR-155−/− memory CD8 T cells was intact.
Fig 5.
The secondary proliferative response was intact in the absence of miR-155. To measure the magnitude of the recall response, MHV-68-infected mice at day 60 postinfection were challenged with 2 × 10e7 vaccinia virus encoding ORF61 intraperitoneally and monitored at 6 days postchallenge. Ag-specific responses were measured before rechallenge (A) and after rechallenge (B). The numbers shown indicate the average fold differences in numbers of ORF-61-specific CD8 T cells for each group. Each dot represented an individual recipient. Note the different scales in panels A and B, reflecting an increased proportion of ORF61-specific cells after rechallenge. Representative plots from two independent experiments using three to four mice per group are shown.
Little is known about the role played by specific miRNAs in CD8 T cell immunity to viruses in vivo, and this study highlights a critical role for miR-155 in this process. These data indicate that miR-155 plays an important role in the generation and/or maintenance of both effector and effector memory CD8 T cells during virus infection. Our data show that the absence of miR-155 results in a marked reduction in the effector response during the acute stage of infection, but the remaining cells have a higher proportion of MPECs. During the memory phase of the response, in the absence of miR-155, more cells expressed CD62L and IL-2, indicative of a central memory phenotype. This was observed in several organs, suggesting that this represents a systemic change in the CD8 T cell phenotype rather than a redistribution of central or effector memory cells between the WT and miR-155−/− populations. Interestingly, memory cells expanded similarly upon secondary viral challenge, indicating that the defect observed during the primary response was not manifest during the recall response. As central memory CD8 T cells have been shown to confer superior immunity both in vivo and in adoptive-cell transfer studies, suppressing miR-155 may prove a useful way to preferentially generate these cells and lead to better immune therapies.
ACKNOWLEDGMENTS
This work was supported by NIH grants R01CA069943 and R01AI076274.
Footnotes
Published ahead of print 5 December 2012
REFERENCES
- 1. Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. 2011. An interleukin-21–interleukin-10–STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity 35:792–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Willinger T, Freeman T, Hasegawa H, McMichael AJ, Callan MFC. 2005. Molecular signatures distinguish human central memory from effector memory CD8 T cell subsets. J. Immunol. 175:5895–5903 [DOI] [PubMed] [Google Scholar]
- 3. Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. 2008. MicroRNAs: new regulators of immune cell development and function. Nat. Immunol. 9:839–845 [DOI] [PubMed] [Google Scholar]
- 4. Zhang N, Bevan MJ. 2010. Dicer controls CD8+ T-cell activation, migration, and survival. Proc. Natl. Acad. Sci. U. S. A. 107:21629–21634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N. miRNA profiling of naïve, effector and memory CD8 T cells. PLoS One 2(10):e1020 doi:10.1371/journal.pone.0001020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Salaun B, Yamamoto T, Badran B, Tsunetsugu-Yokota Y, Roux A, Baitsch L, Rouas R, Fayyad-Kazan H, Baumgaertner P, Devevre E, Ramesh A, Braun M, Speiser D, Autran B, Martiat P, Appay V, Romero P. 2011. Differentiation associated regulation of microRNA expression in vivo in human CD8+ T cell subsets. J. Transl. Med. 9:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. 2007. Requirement of bic/microRNA-155 for normal immune function. Science 316:608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu TX, Hartner J, Lim E-J, Fabry V, Mingler MK, Cole ET, Orkin SH, Aronow BJ, Rothenberg ME. 2011. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-γ pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J. Immunol. 187:3362–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Almanza G, Fernandez A, Volinia S, Cortez-Gonzalez X, Croce CM, Zanetti M. Selected microRNAs define cell fate determination of murine central memory CD8 T cells. PLoS One 5(6):e11243 doi:10.1371/journal.pone.0011243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Manjunath N, Shankar P, Wan J, Weninger W, Crowley MA, Hieshima K, Springer TA, Fan X, Shen H, Lieberman J, von Andrian UH. 2001. Effector differentiation is not prerequisite for generation of memory cytotoxic T lymphocytes. J. Clin. Invest. 108:871–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu C, Huang X, Zhang X, Roensch K, Cao Q, Nakayama KI, Blazar BR, Zeng Y, Zhou X. 2011. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of P27kip1, KPC1, and SOCS-1. Blood 117:4293–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Forrest ARR, Kanamori-Katayama M, Tomaru Y, Lassmann T, Ninomiya N, Takahashi Y, de Hoon MJL, Kubosaki A, Kaiho A, Suzuki M, Yasuda J, Kawai J, Hayashizaki Y, Hume DA, Suzuki H. 2010. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia 24:460–466 [DOI] [PubMed] [Google Scholar]
- 13. He M, Xu Z, Ding T, Kuang D-M, Zheng L. 2009. MicroRNA-155 regulates inflammatory cytokine production in tumor-associated macrophages via targeting C/EBPβ. Cell. Mol. Immunol. 6:343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allie SR, Zhang W, Fuse S, Usherwood EJ. 2011. Programmed death 1 regulates development of central memory CD8 T cells after acute viral infection. J. Immunol. 186:6280–6286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Obar JJ, Crist SG, Gondek DC, Usherwood EJ. 2004. Different functional capacities of latent and lytic antigen-specific CD8 T cells in murine gammaherpesvirus infection. J. Immunol. 172:1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]