Abstract
Animal influenza viruses (AIVs) are a major threat to human health and the source of pandemic influenza. A reliable small-mammal model to study the pathogenesis of infection and for testing vaccines and therapeutics against multiple strains of influenza virus is highly desirable. We show that cotton rats (Sigmodon hispidus) are susceptible to avian and swine influenza viruses. Cotton rats express α2,3-linked sialic acid (SA) and α2,6-linked SA residues in the trachea and α2,6-linked SA residues in the lung parenchyma. Prototypic avian influenza viruses (H3N2, H9N2, and H5N1) and swine-origin 2009 pandemic H1N1 viruses replicated in the nose and in the respiratory tract of cotton rats without prior adaptation and produced strong lung pathology that was characterized by early lung neutrophilia, followed by subsequent pneumonia. Consistent with other natural and animal models of influenza, only the H5N1 virus was lethal for cotton rats. More importantly, we show that the different avian and pandemic H1N1 strains tested are strong activators of the type I interferon (IFN)-inducible MX-1 gene both locally and systemically. Our data indicate that the cotton rat is a suitable small-mammal model to study the infection of animal influenza viruses and for validation of vaccines and therapeutics against these viruses.
INTRODUCTION
The pathogenesis and interspecies transmission of avian- and swine-origin influenza viruses remain major research topics. The 1997 outbreak of H5N1 avian influenza virus, which was transmitted to individuals with close contact with birds, propelled efforts to understand the pathogenesis of human infections with the potential to cause pandemics (1, 2). Additional human infections with H7N7, H7N3, and H9N2 avian influenza viruses in Canada, Europe, and Asia have further highlighted the necessity for the investigation of factors controlling avian influenza virus-host interactions (3–5).
More recently, the emergence of the swine-origin 2009 H1N1 pandemic influenza (H1N1pdm) virus highlighted the need for an improved understanding of the viral and host factors that lead to efficient replication of influenza viruses in new hosts. Since its emergence in Mexico in 2009 (6), H1N1pdm has become established in the human population and appears to have displaced the seasonal H1N1 viruses but not the seasonal H3N2 and influenza B viruses. Although it was perceived as generally mild, approximately 18,000 deaths were attributed to the H1N1pdm during the pandemic period (7). The illness in hospitalized patients with H1N1pdm was associated with an early onset that included fever, cough, pharyngalgia, shortness of breath, headache, myalgia, diarrhea, and vomiting, which progressed to respiratory failure or multiple organ failure in fatal cases (8).
Increasing evidence suggests that the host immune response plays a critical role in determining various outcomes of influenza virus infection (9). The inflammatory response induced during infection may favor viral clearance (10, 11). However, when this response is exacerbated, it may be highly detrimental to the host (12, 13). It is accepted that the high mortality rates seen in infections with highly pathogenic avian influenza viruses are associated with an overactive inflammatory response, and the severity of the disease is closely related to the magnitude of the virus-induced cytokine storm (13).
The mechanism of how animal influenza viruses (AIVs) directly infect humans and other mammals is multifactorial, although it is considered that the viral surface hemagglutinin (HA) protein plays a pivotal role due to its specificity for sialic acid (SA) receptors. Animal models are critical for studies on this mechanism, as findings in animal models are more reflective of these intricacies than those in cell cultures. Receptor specificity and the pattern of expression and distribution of influenza virus SA receptors dictate, at least in part, the fate of influenza virus infection. For example, the airway epithelium of mice, which is a small-animal model of influenza, predominantly express receptors with SA linked to cell surface glycoproteins or glycolipids in an α2,3-linkage (α2,3-linked SA receptors) (14). Most human influenza viruses preferentially bind α2,6-linked SA receptors. Thus, it is not surprising that the majority of human influenza viruses require prior adaptation before being able to replicate efficiently in the mouse.
The cotton rat (Sigmodon hispidus) has been established as a model for influenza research as it is highly permissive for human influenza A and B viruses without prior adaptation (15, 16). It has been shown that viral titers in nasal and lung tissues in cotton rats infected with human influenza viruses were proportional to the infectious dose, and this model replicated many respiratory features observed in humans (16). In addition, cotton rats have also been used in evaluation of cross-protective or heterosubtypic immunity (17, 18), vaccines (19), and antiviral treatments against influenza (20). Upon challenge with influenza viruses, cotton rats develop signs of disease that can be evaluated quantitatively, allowing the assessment of pathogenicity and its reduction through treatment. However, these studies have been limited to only human influenza virus strains. In this study, we expanded on the characterization of cotton rats as a potential small-animal model of animal influenza infections.
MATERIALS AND METHODS
Animals.
Young adult (4- to 8-week-old) cotton rats (Sigmodon hispidus) of both genders were obtained from the inbred colony maintained at Sigmovir Bioystems, Inc. (SBI). The animals were housed in large polycarbonate cages and fed a diet of standard rodent chow and water. Cotton rats were seronegative for adventitious respiratory viruses and other common rodent pathogens. All experiments were performed using protocols that followed federal guidelines and were approved by SBI's and University of Maryland School of Veterinary Medicine's Institutional Animal Care and Use Committees (IACUC). Cotton rats were infected intranasally (i.n.) with the various influenza virus strains under isoflurane anesthesia by application of 0.1 ml of virus solution. Animals were sacrificed by carbon dioxide inhalation.
Lectin-based staining.
Trachea and lung were freshly collected, embedded in OCT (Sakura Finetek USA Inc., Torrance, CA), and cut into 5-mm-thick sections. After air drying overnight, the sections were rinsed with tap water for 1 h, followed by incubation with 3% H2O2 in methanol for 20 min and fixing with cold acetone for 15 min. Subsequently, the sections were blocked with 1% bovine serum albumin (BSA) in Tris-buffered saline (TBS) for 1 h. For detection of receptors, the sections were incubated with digoxigenin (DIG)-labeled Maackia amurensis agglutinin (MAA) (specific for SAα2,3-gal; Boehringer Mannheim Biochemicals, Germany) or Sambucus nigra agglutinin (SNA) (specific for SAα2,6-gal; Boehringer Mannheim Biochemicals, Germany) in buffer I (TBS containing Mg2+, Mn2+, Ca2, and 1% BSA) for 1 h. After washing with TBS, the sections were incubated with peroxidase-conjugated anti-DIG Fab fragments (Boehringer Mannheim Biochemicals, Germany) in TBS containing 1% BSA for 1 h. After washing with 0.05 M Tris (pH 7.6), the sections were developed in a solution of diaminobenzidine (DAB) (for SAα2,3-gal) or aminoethylcarbazole (AEC) (for SAα2,6-gal) containing 3% H2O2 for 10 min, counterstained with hematoxylin, mounted, and observed under a light microscope. All incubation steps were done at room temperature unless otherwise specified. Photos were taken using SPORT ADVANCED software.
Viruses and cells.
The following low-pathogenic avian influenza viruses were used in this study: influenza A/duck/Hong Kong/375/1975 (H3N2) and A/guinea fowl/Hong Kong/WF10/1999 (H9N2) viruses (here referred to as 375/H3N2 and WF10/H9N2, respectively). These viruses were obtained from the influenza virus repository at St. Jude Children's Hospital, Memphis, TN. The highly pathogenic avian influenza H5N1 (A/Vietnam/1203/2004) virus and the pandemic H1N1 (A/California/04/2009) virus were kindly provided by the Centers for Disease Control and Prevention (CDC), Atlanta, GA. A/Netherland/602/09 (H1N1) virus (21, 22), mouse-adapted A/California/04/2009 (H1N1) virus (ma-ca/04) (22), and human H1N1 (A/Brisbane/59/07) and H3N2 (A/Wuhan/359/95) viruses (16) were previously described. Avian viruses were propagated in 10-day-old embryonated, specific-pathogen-free chicken eggs, and all pandemic and seasonal influenza viruses were propagated in Madin-Darby canine kidney (MDCK) cells. Virus stocks were maintained at −80°C until use. Virus stocks generated in eggs were titrated by 50% egg infectious dose (EID50) to equalize the dose of infection. MDCK cells were maintained in modified Eagle's medium (MEM) containing 5% fetal calf bovine serum.
Virus titration.
All viruses from tissue samples were titrated on MDCK cells grown to confluence in 96-well plates by 50% tissue culture infectious dose (TCID50) using HA assay as a readout as described previously (22). All titrations were performed in the presence of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin (Worthington) at a concentration of 1 μg/ml. The tissues analyzed were lung and nose, which were homogenized in 10 parts (wt/vol) of Earle's modified Eagle's medium supplemented with 0.218 M sucrose, 4.4 mM glutamate, 3.8 mM KH2HPO4, and 7.2 mM K2HPO4 (to stabilize virus during freeze-thawing). After centrifugation, supernatants were removed and stored at −80°C until assayed. The lowest level of detection of this assay was 102.5 TCID50 of influenza virus per gram of lung and nose tissue.
RNA isolation and reverse transcription-PCR (RT-PCR) analysis.
Lung tissue (lingular lobe) was flash-frozen in liquid nitrogen and homogenized in 0.5 ml RLT buffer with β-mercaptoethanol using a TissueLyzer LT (two 5-mm beads per sample, 50 Hz, 2 min) (Qiagen). Lung RNA was isolated using an RNeasy kit (Qiagen, catalog number 74106). Blood (∼1 ml) was collected in EDTA-containing tubes and lysed with EL buffer within 1 h of collection, and RNA was isolated using the QIAamp RNA Blood Minikit (Qiagen, catalog number 52304).
Reverse transcription was performed using aQuantiTect reverse transcription kit (Qiagen, catalog number 205314). cDNA was diluted in water to give a ratio of 1 µg RNA of the original RNA per 100 ml final volume. Three microliters of cDNA per reaction was used for real-time PCR using the SYBR green PCR kit (Qiagen catalog number 204057). All reactions were done in duplicates. Primers and conditions for MX-1 were described previously (23). MX-1 mRNA expression was normalized to the β-actin gene as a housekeeping gene using the Pfaffi method. Primers for β-actin in this study were 5′-CCCATTGAACACGGCATTGTC-3′ (forward) and 5′-TGTCACGCACGATTTCCCTCTC-3′ (reverse).
Lung pathology.
Lungs were dissected with the lower 1/3 of the trachea. They were inflated to their normal volume and immersed in 10% neutral buffered formalin. Lungs were evaluated for four indices of pulmonary inflammatory changes: peribronchiolitis (inflammatory cells clustered around the periphery of small airways), perivasculitis (inflammatory cell infiltration around blood vessels), interstitial pneumonia (inflammatory cell infiltration and thickening of alveoli walls), and alveolitis (cells within the alveolar spaces). Slides were scored blind, with validation of scoring by two pathologists experienced in respiratory viral pathogenesis, on a severity scale of 0 to 4 as previously reported (24).
Statistical analysis.
Viral titers were calculated as arithmetic means ± standard errors for all animals in a group at a given time. Pulmonary scores were expressed as the mean ± standard error for all animals in a group. Differences among groups in weight, temperature, and MX-1 expression were evaluated by Student's t test.
RESULTS
Characterization of influenza virus receptors in cotton rats.
Influenza virus infections are mediated by specific interactions between the viral HA and cell oligosaccharides that contain SA residues. Most avian and equine influenza viruses bind to α2,3-linked SA receptors, whereas human viruses, including the earliest available isolates from the 1957 and 1968 pandemics, bind to α2,6-linked SA receptors (25–27). Cell surface receptors for both human (α2,6) and avian (α2,3) influenza viruses have been described in humans (14, 28–30), pigs (31), chickens (32), and quail and other minor poultry species (33, 34).
Since the cotton rat is susceptible to infection with strains of human influenza virus without prior adaptation (16), the receptor distribution in the upper and lower respiratory tracts was analyzed. By performing lectin-based staining, the trachea and minor airways of cotton rats showed strong reactivity with Maackia amurensis agglutinin (MAA), a lectin with specificity for α2,3-linked SA, and Sambucus nigra agglutinin (SNA), a lectin specific for α2,6-linked SA (Fig. 1). By comparing similar sections stained for either α2,3-linked SA or α2,6-linked SA or β-tubulin (a marker of ciliated epithelial cells), we established that α2,6-linked SA receptors are colocalized with ciliated cells, whereas α2,3-linked SA receptors are more associated with mucin-producing cells in the trachea of cotton rats. In addition, staining of the lung parenchyma showed a consistent staining of type I and type II pneumocytes with α2,6-linked SA receptors, whereas α2,3-linked SA receptors were rarely expressed (Fig. 1, lung). These results indicate that both α2,3-linked and α2,6-linked SA receptors are present in the airways of cotton rats, in contrast to the case for mice, where only α2,6-linked SA receptors predominate in the lung tissue. These results explain the susceptibility of cotton rats to infection with human strains of influenza virus and suggest that the cotton rat is potentially susceptible to infection with avian influenza viruses.
Fig 1.
Immunologic detection of SAα2,3-gal- and SAα2,6-gal-linked receptors in cotton rat trachea (left panels) and alveoli (right panels). The brown color (developed from DAB) indicates the presence of SAα2,3-gal or SAα2,6-gal as indicated. The original magnification is ×400. Arrows emphasize the positively stained cells. Note that in cotton rat trachea the staining for SAα2,3-gal is located mainly in mucin-producing cells (arrowheads), whereas the staining for SAα2,6-gal is seen mainly in ciliated cells (arrows). In lung, there is low to nondetectable staining for SAα2,3-gal but a strong staining for SAα2,6-gal, associated with type II (arrows) and some type I (arrowheads) pneumocytes.
Infection of cotton rats with low-pathogenic avian influenza virus strains.
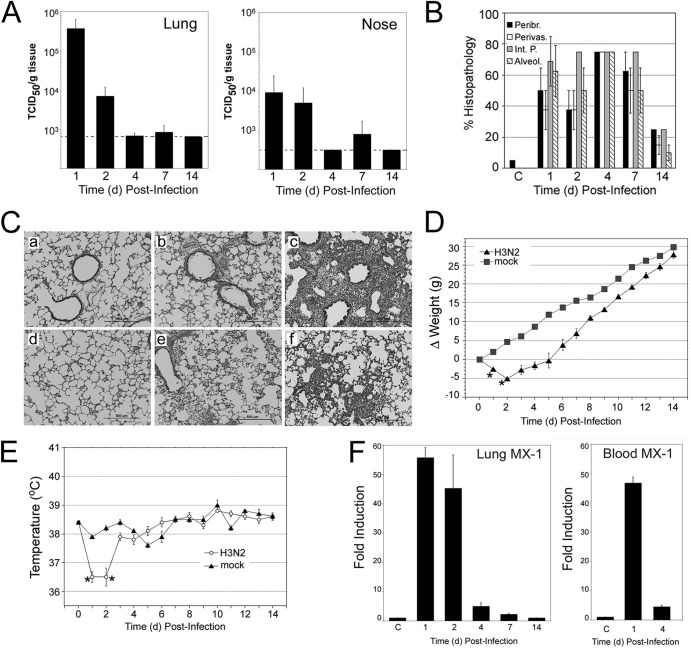
To assess the susceptibility of cotton rats to avian influenza viruses, infections were performed using two low-pathogenic strains, A/duck/Hong Kong/375/1975 (H3N2) and A/guinea fowl/Hong Kong/WF10/1999 virus (H9N2) (here referred to as 375/H3N2 and WF10/H9N2, respectively). Cotton rats were infected with a wide range of 375/H3N2 virus inocula (from 102 to 107 EID50/rat), and virus shedding was analyzed at different days postinfection (dpi). When cotton rats were inoculated i.n. with a dose of 107 EID50, virus was isolated from noses and lungs of all animals at 1 and 2 dpi and from a few animals at 7 dpi (Fig. 2A). Lung and nose titers reached ∼5 × 105 and ∼104 TCID5050/g of tissue, respectively, at 1 dpi (Fig. 2A). Animals inoculated with the lowest dose of virus (102 EID50) still yielded replicative virus in lungs (detected only at 1 dpi) and noses (detected at 1, 2, and 4 dpi) (data not shown). Strong pathology was detected in the lungs of animals infected with 107 EID50 (Fig. 2B and C). The lung pathology was slightly reduced when a dose of 106 EID50 was used, and there was no detectable pathology in animals infected with 105 EID50 (data not shown). At 1 dpi, all four lung pathological parameters measured, i.e., peribronchiolitis, perivasculitis, interstitial pneumonia, and alveolitis, were dramatically enhanced. These parameters peaked at 4 dpi and remained high through 7 dpi. Signs of inflammation remained detectable at 14 dpi (Fig. 2B). The pathology at 1 dpi was characterized by a rapid increase in exfoliation and degeneration of bronchiolar ciliated epithelial cells (Fig. 2C, panel b). Alveolitis was also present, with infiltration of inflammatory cells (mainly neutrophils and macrophages) of the alveolar space (Fig. 2C, panel e). By 4 dpi, there was consolidation of the pathology, with patches of pneumonia as the result of massive cellular infiltration (Fig. 2C, panels c and f). Animals infected with 107 EID50 showed significant decreases in body weight and temperature at 1 and 2 dpi (Fig. 2D and E).
Fig 2.
Characterization of infection of cotton rat with H3N2 A/Duck/HK/375/75. Cotton rats were infected with 107 EID50/rat. The control group (mock) was inoculated with allantoid fluid diluted 1:10 in phosphate-buffered saline (PBS). (A) Viral titers in the lung and nose. (B) Lung pathology scores evaluated for 4 relevant parameters: peribronchiolitis (Peribr.), perivasculitis (Perivas.), interstitial pneumonia (Int. P.), and alveolitis (Alveol.). (C) Representative photographs of hematoxylin and eosin (H&E)-stained lung sections from uninfected rats (panels a and d) and rats at 1 dpi (panels b and e) and 4 dpi (panels c and f). Panels b and c best exemplify the progressive increases in epithelial damage, peribronchiolitis, and perivasculitis. Panels e and f demonstrate the progressive increase in pneumonia and alveolitis. Magnification, is ×100. (D and E) Weight (D) and temperature (E) changes were recorded at different days postinfection. *, P < 0.05. (F) mRNA expression of the type I interferon-inducible gene MX-1 in lung (left panel) and blood (right panel) measured at the indicated dpi. C, group of mock (allantoid)-inoculated animals.
Mx proteins are powerful inhibitors of influenza virus replication and sensitive markers of in vivo expression of type I interferons (IFNs) (35). We have previously characterized MX-1 and MX-2 genes in the cotton rat and corroborated the induction of both genes by type I IFN (23), influenza virus (23), and respiratory syncytial virus (36), as well as the capability of MX-1 to inhibit influenza virus infection (37). To further characterize the pathogenesis of these influenza viruses, we measured the expression of the cotton rat MX-1 gene in lungs and blood samples of infected animals (Fig. 2F). We found that infection was followed by strong induction of MX-1 mRNA in the lung, peaking at 1 and 2 dpi, and in the blood, peaking at 1 dpi. MX-1 expression remained significantly higher than that for the control on day 4 and day 7 in the lung and on day 4 in blood (P < 0.05).
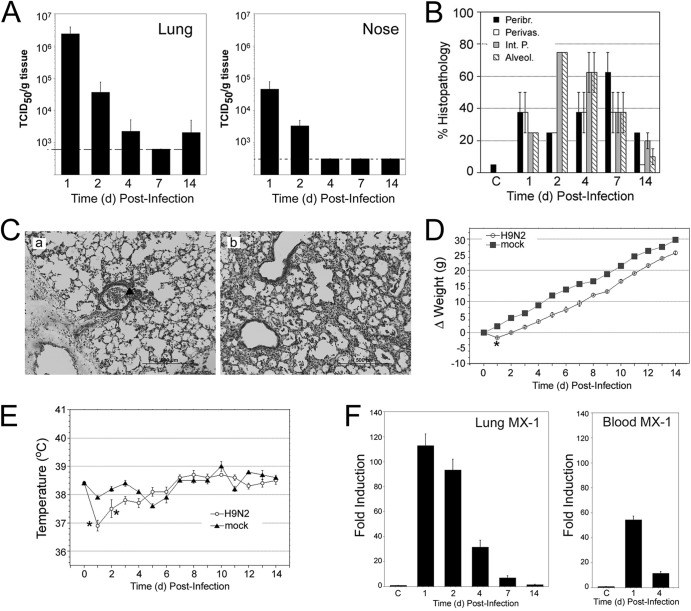
Infection of cotton rats with WF10/H9N2 virus showed a pattern of replication similar to that seen for 375/H3N2 virus. When cotton rats were infected with 107 EID50, nasal replication of WF10/H9N2 peaked at 1 dpi (∼7 × 104 TCID50/g tissue) and remained relatively high at 2 dpi (Fig. 3A). Higher titers in the lungs than in the nose were observed also for this virus at 1 and 2 dpi. Infection with decreasing doses of WF10/H9N2 (from 107 to 102 EID50) showed virus in the lung even at the lowest dose of 102 EID50 at 1 dpi and in the nose at doses of 103 EID50 or greater (data not shown). The most characteristic lung pathology features detected during infection of cotton rats with the WF10/99 strain was the strong defoliation of the ciliated epithelia and extravasations of neutrophils into bronchioles at 1 dpi (Fig. 3B and C, panel a). Peak histopathology was detected between 2 and 7 dpi, with strong interstitial pneumonia and alveolitis at 2 dpi and strong peribronchiolitis at day 7 and patches of lesion consolidation (Fig. 3C, panel b). Subtle but significant changes in body weight and temperature were detected in cotton rats infected with the WF10/H9N2 virus. Weight loss and temperature drop were evident at 1 dpi (Fig. 3D and E). Strong induction of MX-1 mRNA was detected in lung tissue and blood samples (Fig. 3F), peaking at 1 dpi and remaining significantly higher until 7 dpi in the lung or until 4 dpi in blood (P < 0.05).
Fig 3.
Characterization of infection of cotton rat with H9N2 A/guinea fowl/Hong Kong/WF10/1999. (A) Lung and nose viral replication after infection with 107 EID50/rat. (B) Quantification of lung pathology for 4 relevant parameters of damage: peribronchiolitis, perivasculitis, interstitial pneumonia, and alveolitis. Pathology peaked at 2 to 7 dpi and was still detectable at 14 dpi. (C) Lung pathology was characterized by strong accumulation of cell debris and granulocytes in the airways, forming plugs at 1 dpi (panel a, arrow). The major airway infiltration is resolved by day 4 (panel b), and the pathology progresses to a strong interstitial pneumonia accompanied by alveolitis. Magnification, is ×100. (D and E) Weight (D) and temperature (E) changes were recorded at different days postinfection. *, P < 0.05. (F) mRNA expression of the type I interferon-inducible gene MX-1 in lung and blood measured at the indicated dpi. C, group of mock (allantoid)-inoculated animals.
Infection of cotton rats with highly pathogenic H5N1 virus.
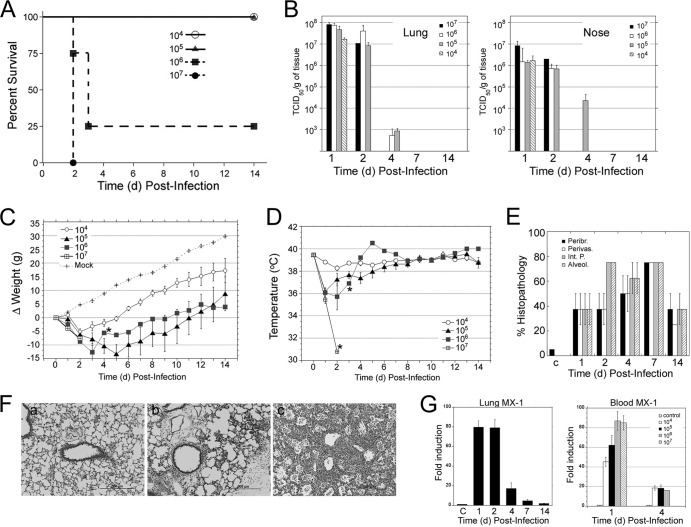
To further test the suitability of the cotton rat model for the study of avian influenza, we tested the replication and associated pathology after infection with A/Vietnam/1203/2004 (H5N1), a highly pathogenic virus isolate from a lethal human case (here referred to as 1203/H5N1). Infection of cotton rats with a high dose of 1203/H5N1 virus (107 EID50) given i.n. caused 100% mortality by 2 dpi (Fig. 4A). When animals were infected with106 EID50, 3 out 4 animals died by 3 dpi, while all animals infected with a dose of 105 EID50 or lower survived (Fig. 4A). Our data are consistent with the notion that in humans, a high dose of virus is required to produce a life-threatening infection (38). Furthermore, we found that cotton rats are highly permissive to H5N1 replication. Viral titers in the lungs of animals infected with as low as 104 EID50 reached >107 TCID50/g and were not very different from viral titers in the noses of animals infected with 104 EID50. The same was true for 1203/H5N1 replication in the nose, although titers reached at this site were in general 1 log TCID50/g lower. Virus was isolated from both lungs and noses of all animals infected with a dose of 105 TCID50/g at 4 dpi. Animals infected with 1203/H5N1 showed evident signs of disease, including hunching and piloerection. In addition, all infected animals showed dramatic weight loss (up to 15%) between 1 and 5 dpi, and hypothermia (>3°C drop in body temperature) at day 1, when infected at doses of 105 and 106 EID50 (Fig. 4C and D). Lung pathology scores of cotton rats infected with the 1203/H5N1 virus were plotted after infection of animals with 105 EID50, since either all or 3 out of 4 animals died before 4 dpi in groups infected with 107 and 106 TCID50, respectively. Lung pathology in these animals was pronounced by 1 dpi and further increased, peaking by day 7 postinfection, in animals infected with 105 EID50 (Fig. 4E). Some of the characteristic features observed included rapid infiltration of the alveolar space by neutrophils (alveolitis) (Fig. 4F, panel b) that subsequently developed into a consolidated pneumonia by 4 dpi (Fig. 4F, panel c). Analysis of lung expression of mRNA for the MX-1 gene showed a peak of expression at 1 to 2 dpi, with levels of induction remaining significant at 7 dpi (P < 0.05). Analysis of the expression of MX-1 in blood samples was dose dependent at 1 dpi, reaching a plateau at a dose of virus of 106 to 107 EID50 (Fig. 4G). The expression of mRNA for MX-1 remained strong at 4 dpi in those surviving animals, with levels of activation of MX-1 mRNA expression close to 20-fold above the levels in uninfected control animals (P < 0.05).
Fig 4.
Infection of cotton rats with A/Vietnam/1203/04 H5N1. (A) Cotton rats were infected i.n. with the indicated doses of 1203/H5N1. (A) Survival graph of cotton rats after infection with different doses of 1203/H5N1. (B) Viral replication of 1203/H5N1 in the lungs and noses of infected cotton rats at the indicated dpi. Nose and lung viral titers were plotted as follows: from 4 animals/group at 1 dpi; from 1 animal infected with 107 EID50, 3 animals infected with 106 EID50, and 4 animals infected with 105 EID50 at 2 dpi; and from 2 animals infected with 106 EID50 and 4 animals infected with 105 EID50 at 4 dpi. (C and D) Weight (C) and temperature (D) changes were recorded at different days postinfection. *, weight and temperature were measured for 1 out of 4 infected animals. (E) Quantification of lung pathology in animals infected with 105 EID50. (F) Representative microscopic, H&E-stained images of lung sections from uninfected animals (panel a) and animals at 1 dpi (panel b) and 4 dpi (panel c) with a dose of virus of 105 EID50. Panels b and c exemplify the progressive increase of peribronchiolitis and perivasculitis. Magnification, ×100. (G) mRNA expression of the type I interferon-inducible gene MX-1 in lung (left panel) and blood (right panel) measured at the indicated dpi. MX1 expression in lung was measured in animals infected with 105 EID50/rat. MX-1 in blood at 4 dpi was measured from 1 animal infected with 106 EID50, whereas no animals infected with 107 EID50 survived until day 4. Uninfected animals were used as a control (C).
Infection of cotton rats with 2009 H1N1pdm strains.
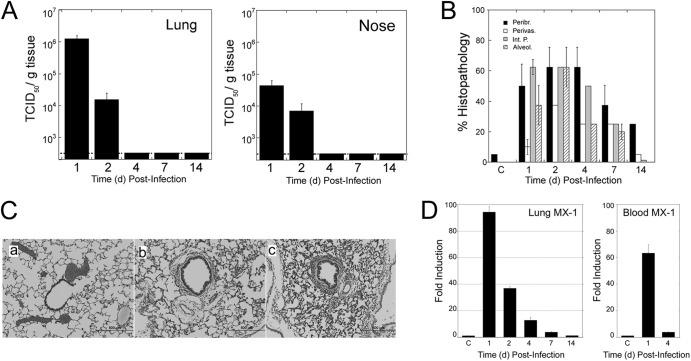
We evaluated the infectivity of the prototype pandemic isolate A/California/07/09 (Ca/07) in the cotton rat using two different doses of inocula, 107 and 106 EID50/animal. Virus was isolated from lung and nose at 1 and 2 dpi at both concentrations of viral inocula, with peak viral titers at 1 dpi in both tissues. No virus was isolated after 4 dpi (Fig. 5A). Peak lung histopathology was seen on day 2 postinfection (Fig. 5B), and its intensity was dose dependent, with stronger pathology seen in the lungs of animals infected with the highest dose of virus (Fig. 5C, panels b and c). Detectable lung pathology was observed out to 14 dpi. Infection of cotton rats with Ca/07 did not cause changes in clinical parameters as detected with avian viruses (i.e., in body weight and temperature) at any dose used for infection (data not shown). However, infection with Ca/07 produced a strong increase in the expression of the MX-1 gene in lung and blood (Fig. 5D), which preceded the peak in histopathology.
Fig 5.
Infection of cotton rats with 2009 pandemic H1N1 influenza strain. Animals (5 per group) were inoculated i.n. with 100 μl of PBS (control [C]) or infected with a dose of 107 EID50 of A/California/07/09 H1N1. (A) Viral replication of H1N1pdm in lungs and noses of cotton rats at the indicated dpi. (B) Quantification of lung histopathology for the 4 relevant parameters described in the legend to Fig. 2. Pathology peaked at 2 dpi, remained moderate at 7 dpi, and was still detectable at 14 dpi. (C) Peak lung pathology was dependent on the dose of virus used for infection (106 and 107 EID50 in panels b and c, respectively) and characterized by increases in all the histopathology parameters measured. Panel a represents an uninfected control animal. Magnification, ×100. (D) mRNA expression of the type I interferon-inducible gene MX-1 in lung and blood measured at the indicated dpi with 107 EID50 of H1N1pdm. C, group of uninfected animals.
Differential type I IFN production in cotton rats infected with H1N1pdm corroborated by MX gene expression.
To characterize the pathogenesis of pandemic H1N1 isolates further, we measured the expression of cotton rat MX-1 gene expression in lung and blood samples of animals infected with two additional H1N1pdm strains and compared its expression in cotton rats infected with seasonal H1N1 and H3N2 viruses (Fig. 6). Infected animals showed measurable levels of MX-1 expression in the lung that peaked by 1 dpi and decreased by 4 dpi, following the pattern previously observed with seasonal influenza virus strains (Fig. 6A) (23). However, the level of expression of MX-1 in the lungs of animals infected with the H1N1pdm strains surpassed that induced by seasonal H1N1 or H3N2. This increase correlated with a stronger induction of MX-1 expression in blood samples of animals infected with H1N1pdm strains (Fig. 6B). Our data indicate that in cotton rats, H1N1pdm is a more potent inducer of type I IFN than nonpandemic strains. Moreover, the data also suggest that at least the H1N1pdm strains tested in this work, and in contrast to seasonal strains, have the unique ability to induce MX-1 antiviral gene expression that can be detected in blood.
Fig 6.
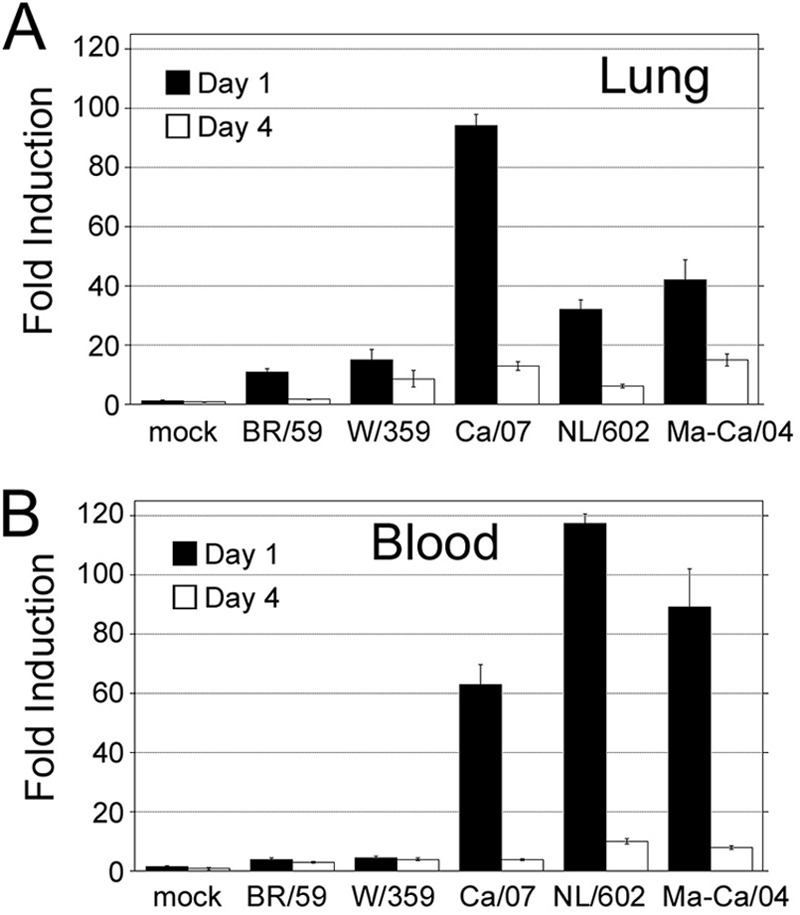
Analysis of the steady-state expression of the cotton rat MX-1 mRNA in lung (A) and blood (B) samples of animals infected with A/Brisbane/59/07 H1N1 (Br/59), A/Wuhan/359/95 H3N2 (Wu/359), A/California/07/09 (Ca/07), A/Netherlands/602/09 (NL/602), and mouse-adapted A/California/04/09 H1N1 (Ma-Ca) H1N1pdm viruses. The mock group was inoculated with PBS i.n. Mock represents a group of animals inoculated with PBS.
DISCUSSION
In this work we demonstrated that cotton rats are permissive to infection with 2009 pandemic H1N1 strains of influenza virus and several strains of avian influenza virus. All viruses replicated in the respiratory tract of cotton rats and were able to cause moderate to severe lung pathology. In addition, we observed that infection with H1N1pdm and avian influenza virus strains generated stronger type I IFN expression than that with other circulating human influenza virus strains. The high production of type I IFN was evidenced by the simultaneous induction of expression of MX-1, a sensitive marker of type I IFN (35). Importantly, we have shown that the effect of type I IFN became systemic, as high expression of MX-1 mRNA in blood cells was evidenced in animals infected only with H1N1pdm strains.
We hypothesized that the susceptibility of cotton rats to infection with nonadapted strains of avian and human influenza viruses may reside in their sialic acid distribution within the respiratory tract. Although receptors carrying α2,3- and α2,6-SA linkages are found in humans, their distribution is controversial. Initially, a selective localization of α2,3-linked SA in goblet cells and of α2,6-linked SA in ciliated cells of the epithelia of the trachea was reported (28, 39). Mastrosovich and collaborators have shown contrasting results using differentiated cultures of human tracheobronchial epithelium (29). In this study, we found that the localization of α2,6-linked SA in the cotton rat airway was restricted to ciliated cells, whereas α2,3-linked SA was found in nonciliated cells. Our results suggest that the distribution of receptors in cotton rats is similar to the pattern observed in human trachea by Couceiro et al. (28). In addition, we found that the cotton rat lung stained strongly for α2,6-linked SA but not for α2,3-linked SA. The staining for α2,6-linked SA was confined to the base of type II pneumocytes, with some scattered staining in type I pneumocytes (Fig. 1B). The presence of abundant α2,6-linked SA in lungs and tracheae of cotton rats explains, at least in part, the susceptibility of cotton rats to infection with field influenza virus isolates.
The three avian influenza viruses tested in this study replicated and induced strong pathology in the lungs of infected cotton rats, characterized by defoliation of the ciliated epithelium, alveolitis produced by the infiltration of inflammatory cells, and the final development of pneumonia. These viruses included the low-pathogenicity strains H9N2 and H3N2 and a highly pathogenic strain of H5N1. The replication of the low-pathogenic H9N2 virus was likely facilitated by the abundance of α2,3-linked SA receptors in the cotton rat airway. However, our previous data have indicated that the WF10/H9N2 virus used in this study possesses human-like receptor specificity that favors the binding of the α2,6-linked SA moiety (34). Importantly, the H5N1 virus, which preferentially binds to α2,3-linked SA structures (40), infected cotton rats and induced 100% mortality when inoculated at a high dose. This is similar to what has been observed in the ferret model (41, 42). It also correlates with the lethal outcome of this virus in humans, which has been proposed to be the result of infection with a large viral dose (38). Taken together, our infection studies demonstrated that the cotton rat model can be used in the evaluation of avian influenza viruses of different receptor specificities and virulences.
For all avian viruses tested in this study, peak viral replication coincided with the peak of changes in clinical parameters but preceded the peak of the lung pathology. This has been previously seen also for seasonal influenza virus strains in the cotton rat (16). However, this effect was not seen in H1N1pdm infection, which did not show overt changes in the clinical parameters measured and where the peak in lung pathology coincided with the peak in viral titers. This could reflect a different mechanism of engaging the host innate response by H1N1pdm virus.
Our studies using H1N1pdm strains and comparing them with seasonal human strains of influenza virus show that cotton rats recapitulate some of the features observed in other animal models (mice and ferrets) (22). Type I IFN is produced at the site of the viral infection. It is expected that this type of response would impact the clinical presentation of the disease, and thus our study provides clues as to the effectors of H1N1pdm virulence. In fact, expression of Mx-A, the human counterpart of MX-1 in blood, has been used as marker to differentiate between viral and bacterial infection in febrile children (43, 44). However, at this time, it is not clear how different respiratory viruses (including different human strains of influenza virus) regulate the expression of systemic IFN, since its augmentation in blood has been detected for some (e.g., coxsackievirus, adenovirus, respiratory syncytial virus, and cytomegalovirus) (45) but was not found for others (e.g., rhinovirus) (46). The ability of the cotton rat to serve as an animal model for multiple respiratory viruses will likely be instrumental in defining some of the clinical consequences associated with systemic IFN production during infection with certain influenza virus strains.
In short, our studies provided significant evidence that the cotton rat is a viable mammalian model for evaluation of AIVs, particularly those of public health concern. More isolates of AIV will need to be tested to accumulate knowledge of the pathogenesis, immune response, antiviral treatment, and vaccine efficacy against AIVs in this model. The cotton rat model has been demonstrated to be highly susceptible to infection with many respiratory viruses, including respiratory syncytial virus (24), and recently to human rhinovirus serotype 16 (J. C. G. Blanco and A. Kajon, unpublished results), all of which may be able to coinfect a given individual. Therefore, the cotton rat model could become pivotal to decipher the intricacies of mixed respiratory infections.
ACKNOWLEDGMENTS
We are indebted to Gregory Prince for his guidance. We thank Marina Boukhvalova for her suggestions throughout this work. We thank Charles Smith and Freddy and Ana Rivera for their excellent assistance with animal husbandry and Andrea Ferrero for assistance in the biosafety level 3 (BSL3) facility at the University of Maryland, College Park.
This work was supported by Virion Systems, Inc., and Sigmovir Biosystems, Inc., corporate funds to J.C.G.B. and by NIAID contract HHSN266200700010C and CDC-HHS 1U01CI000355-01 to D.R.P.
J.C.G.B. is President of Sigmovir Biosystems, Inc., a basic and contract research organization that uses the cotton rat as an animal model for the study of respiratory viruses.
Footnotes
Published ahead of print 28 November 2012
REFERENCES
- 1. de Jong JC, Claas EC, Osterhaus AD, Webster RG, Lim WL. 1997. A pandemic warning? Nature 389:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Watanabe Y, Ibrahim MS, Suzuki Y, Ikuta K. 2012. The changing nature of avian influenza A virus (H5N1). Trends Microbiol. 20:11–20 [DOI] [PubMed] [Google Scholar]
- 3. Aamir UB, Naeem K, Ahmed Z, Obert CA, Franks J, Krauss S, Seiler P, Webster RG. 2009. Zoonotic potential of highly pathogenic avian H7N3 influenza viruses from Pakistan. Virology 390:212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, Koch G, Bosman A, Koopmans M, Osterhaus AD. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. U. S. A. 101:1356–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF. 1999. Human infection with influenza H9N2. Lancet 354:916–917 [DOI] [PubMed] [Google Scholar]
- 6. Anonymous 2009. Outbreak of swine-origin influenza A (H1N1) virus infection—Mexico, March-April 2009. MMWR Morb. Mortal. Wkly. Rep. 58:467–470 [PubMed] [Google Scholar]
- 7. World Health Organization 2010. Pandemic H1N1 112. World Health Organization, Geneva, Switzerland [Google Scholar]
- 8. Jain S, Kamimoto L, Bramley AM, Schmitz AM, Benoit SR, Louie J, Sugerman DE, Druckenmiller JK, Ritger KA, Chugh R, Jasuja S, Deutscher M, Chen S, Walker JD, Duchin JS, Lett S, Soliva S, Wells EV, Swerdlow D, Uyeki TM, Fiore AE, Olsen SJ, Fry AM, Bridges CB, Finelli L. 2009. Hospitalized patients with 2009 H1N1 influenza in the United States, April-June 2009. N. Engl. J. Med. 361:1935–1944 [DOI] [PubMed] [Google Scholar]
- 9. Taubenberger JK, Morens DM. 2008. The pathology of influenza virus infections. Annu. Rev. Pathol. 3:499–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seo SH, Webster RG. 2002. Tumor necrosis factor alpha exerts powerful anti-influenza virus effects in lung epithelial cells. J. Virol. 76:1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tumpey TM, Szretter KJ, Van Hoeven N, Katz JM, Kochs G, Haller O, Garcia-Sastre A, Staeheli P. 2007. The Mx1 gene protects mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses. J. Virol. 81:10818–10821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheung CY, Poon LL, Lau AS, Luk W, Lau YL, Shortridge KF, Gordon S, Guan Y, Peiris JS. 2002. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet 360:1831–1837 [DOI] [PubMed] [Google Scholar]
- 13. Perrone LA, Plowden JK, Garcia-Sastre A, Katz JM, Tumpey TM. 2008. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 4:e1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ibricevic A, Pekosz A, Walter MJ, Newby C, Battaile JT, Brown EG, Holtzman MJ, Brody SL. 2006. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J. Virol. 80:7469–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eichelberger MC. 2007. The cotton rat as a model to study influenza pathogenesis and immunity. Viral Immunol. 20:243–249 [DOI] [PubMed] [Google Scholar]
- 16. Ottolini MG, Blanco JC, Eichelberger MC, Porter DD, Pletneva L, Richardson JY, Prince GA. 2005. The cotton rat provides a useful small-animal model for the study of influenza virus pathogenesis. J. Gen. Virol. 86:2823–2830 [DOI] [PubMed] [Google Scholar]
- 17. Straight TM, Ottolini MG, Prince GA, Eichelberger MC. 2008. Antibody contributes to heterosubtypic protection against influenza A-induced tachypnea in cotton rats. Virol. J. 5:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Straight TM, Ottolini MG, Prince GA, Eichelberger MC. 2006. Evidence of a cross-protective immune response to influenza A in the cotton rat model. Vaccine 24:6264–6271 [DOI] [PubMed] [Google Scholar]
- 19. Yim K, Miles B, Zinsou R, Prince G, Boukhvalova M. 2012. Efficacy of trivalent inactivated influenza vaccines in the cotton rat Sigmodon hispidus model. Vaccine 30:1291–1296 [DOI] [PubMed] [Google Scholar]
- 20. Ottolini M, Blanco J, Porter D, Peterson L, Curtis S, Prince G. 2003. Combination anti-inflammatory and antiviral therapy of influenza in a cotton rat model. Pediatr. Pulmonol. 36:290–294 [DOI] [PubMed] [Google Scholar]
- 21. Munster VJ, de Wit E, van den Brand JM, Herfst S, Schrauwen EJ, Bestebroer TM, van de Vijver D, Boucher CA, Koopmans M, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA. 2009. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science 325:481–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ye J, Sorrell EM, Cai Y, Shao H, Xu K, Pena L, Hickman D, Song H, Angel M, Medina RA, Manicassamy B, Garcia-Sastre A, Perez DR. 2010. Variations in the hemagglutinin of the H1N1 pandemic virus: potential for strains with altered virulence phenotype? PLoS Pathog. 6:e1001145 doi:10.1371/journal.ppat.1001145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pletneva LM, Haller O, Porter DD, Prince GA, Blanco JC. 2006. Interferon-inducible Mx gene expression in cotton rats: cloning, characterization, and expression during influenza viral infection. J. Interferon Cytokine Res. 26:914–921 [DOI] [PubMed] [Google Scholar]
- 24. Prince GA, Curtis SJ, Yim KC, Porter DD. 2001. Vaccine-enhanced respiratory syncytial virus disease in cotton rats following immunization with Lot 100 or a newly prepared reference vaccine. J. Gen. Virol. 82:2881–2888 [DOI] [PubMed] [Google Scholar]
- 25. Connor RJ, Kawaoka Y, Webster RG, Paulson JC. 1994. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 205:17–23 [DOI] [PubMed] [Google Scholar]
- 26. Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. 2000. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J. Virol. 74:8502–8512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rogers GN, Paulson JC. 1983. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 127:361–373 [DOI] [PubMed] [Google Scholar]
- 28. Couceiro JN, Paulson JC, Baum LG. 1993. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 29:155–165 [DOI] [PubMed] [Google Scholar]
- 29. Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. 2004. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. U. S. A. 101:4620–4624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. 2006. Avian flu: influenza virus receptors in the human airway. Nature 440:435–436 [DOI] [PubMed] [Google Scholar]
- 31. Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, Donatelli I, Kida H, Paulson JC, Webster RG, Kawaoka Y. 1998. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 72:7367–7373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gambaryan A, Webster R, Matrosovich M. 2002. Differences between influenza virus receptors on target cells of duck and chicken. Arch. Virol. 147:1197–1208 [DOI] [PubMed] [Google Scholar]
- 33. Kimble B, Nieto GR, Perez DR. 2010. Characterization of influenza virus sialic acid receptors in minor poultry species. Virol. J. 7:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wan H, Perez DR. 2006. Quail carry sialic acid receptors compatible with binding of avian and human influenza viruses. Virology 346:278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haller O, Kochs G. 2002. Interferon-induced mx proteins: dynamin-like GTPases with antiviral activity. Traffic 3:710–717 [DOI] [PubMed] [Google Scholar]
- 36. Pletneva LM, Haller O, Porter DD, Prince GA, Blanco JC. 2008. Induction of type I interferons and interferon-inducible Mx genes during respiratory syncytial virus infection and reinfection in cotton rats. J. Gen. Virol. 89:261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stertz S, Dittmann J, Blanco JC, Pletneva LM, Haller O, Kochs G. 2007. The antiviral potential of interferon-induced cotton rat Mx proteins against orthomyxovirus (influenza), rhabdovirus, and bunyavirus. J. Interferon Cytokine Res. 27:847–855 [DOI] [PubMed] [Google Scholar]
- 38. Tran TH, Nguyen TL, Nguyen TD, Luong TS, Pham PM, Nguyen VC, Pham TS, Vo CD, Le TQ, Ngo TT, Dao BK, Le PP, Nguyen TT, Hoang TL, Cao VT, Le TG, Nguyen DT, Le HN, Nguyen KT, Le HS, Le VT, Christiane D, Tran TT, de Menno J, Schultsz C, Cheng P, Lim W, Horby P, Farrar J. 2004. Avian influenza A (H5N1) in 10 patients in Vietnam. N. Engl. J. Med. 350:1179–1188 [DOI] [PubMed] [Google Scholar]
- 39. Baum LG, Paulson JC. 1990. Sialyloligosaccharides of the respiratory epithelium in the selection of human influenza virus receptor specificity. Acta Histochem. Suppl. 40:35–38 [PubMed] [Google Scholar]
- 40. Matrosovich M, Zhou N, Kawaoka Y, Webster R. 1999. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J. Virol. 73:1146–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Govorkova EA, Rehg JE, Krauss S, Yen HL, Guan Y, Peiris M, Nguyen TD, Hanh TH, Puthavathana P, Long HT, Buranathai C, Lim W, Webster RG, Hoffmann E. 2005. Lethality to ferrets of H5N1 influenza viruses isolated from humans and poultry in 2004. J. Virol. 79:2191–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schmolke M, Manicassamy B, Pena L, Sutton T, Hai R, Varga ZT, Hale BG, Steel J, Perez DR, Garcia-Sastre A. 2011. Differential contribution of PB1-F2 to the virulence of highly pathogenic H5N1 influenza A virus in mammalian and avian species. PLoS Pathog. 7:e1002186 doi:10.1371/journal.ppat.1002186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Halminen M, Ilonen J, Julkunen I, Ruuskanen O, Simell O, Makela MJ. 1997. Expression of MxA protein in blood lymphocytes discriminates between viral and bacterial infections in febrile children. Pediatr. Res. 41:647–650 [DOI] [PubMed] [Google Scholar]
- 44. Nakabayashi M, Adachi Y, Itazawa T, Okabe Y, Kanegane H, Kawamura M, Tomita A, Miyawaki T. 2006. MxA-based recognition of viral illness in febrile children by a whole blood assay. Pediatr. Res. 60:770–774 [DOI] [PubMed] [Google Scholar]
- 45. Chieux V, Hober D, Harvey J, Lion G, Lucidarme D, Forzy G, Duhamel M, Cousin J, Ducoulombier H, Wattre P. 1998. The MxA protein levels in whole blood lysates of patients with various viral infections. J. Virol. Methods 70:183–191 [DOI] [PubMed] [Google Scholar]
- 46. Makela MJ, Halminen M, Ruuskanen O, Puhakka T, Pirhonen J, Julkunen I, Ilonen J. 1999. Lack of induction by rhinoviruses of systemic type I interferon production or enhanced MxA protein expression during the common cold. Eur. J. Clin. Microbiol. Infect. Dis. 18:665–668 [DOI] [PMC free article] [PubMed] [Google Scholar]