Abstract
Integrated retroviral DNA is subject to epigenetic transcriptional silencing at different frequencies. This process is mediated by repressive DNA methylation and histone modifications on viral chromatin. However, the detailed mechanisms by which retroviral silencing is initiated and maintained are not well understood. Using a model system in which avian sarcoma virus (ASV) DNA is epigenetically repressed in mammalian cells, we previously found that a cellular scaffolding protein, Daxx, acts as an antiretroviral factor that promotes epigenetic repression through recruitment of histone deacetylases (HDACs). Here we show that human Daxx protein levels are increased in response to retroviral infection and that Daxx acts at the time of infection to initiate epigenetic repression. Consistent with a rapid and active antiviral epigenetic response, we found that repressive histone marks and long terminal repeat (LTR) DNA methylation could be detected within 12 h to 3 days postinfection, respectively. Daxx was also found to be required for long-term ASV silencing maintenance and full viral DNA methylation, and it was physically associated with both viral DNA and DNA methyltransferases (DNMTs). These findings support a model in which incoming retroviral protein-DNA complexes are detected by Daxx, and the integrated provirus is rapidly chromatinized and repressed by DNA methylation and histone modification as part of an antiviral response. These results uncover a possible direct and active antiviral mechanism by which DNMTs can be recruited to retroviral DNA.
INTRODUCTION
Retroviruses are important agents of disease and serve as valuable vectors for gene delivery, and their study has provided seminal insights into cellular functions. A defining feature of retroviral replication is the integration of a DNA copy of the retroviral RNA genome into host chromatin, a process that establishes the DNA provirus. Integration provides a permanent association of viral DNA with the host cell and all of its progeny, and it also allows the provirus to efficiently mobilize the cellular transcriptional machinery for synthesis of viral mRNAs and viral RNA genomes.
DNA integration is an essential step in retroviral replication, and it is catalyzed by the virus-encoded integrase (IN) protein. However, establishment of the provirus does not guarantee its expression; transcriptional repression by epigenetic mechanisms (epigenetic silencing) is often observed in both natural and interspecies retroviral infections. Examples include the silencing of retroviruses in embryonic stem cells (1, 2), the progressive silencing of expression of genes transduced by retroviral vectors during long-term cell propagation (3), and HIV latency (4). Epigenetic mechanisms also repress the expression of endogenous retroviruses (5–9).
Retroviral epigenetic silencing is mediated by the enzymatic placement, and subsequent reading, of DNA methylation marks (addition of a methyl group to position 5 of the cytosine pyrimidine [5MeCpG]) and repressive nucleosomal histone modifications. These epigenetic mechanisms also play a key role in the silencing of cellular genes during development and differentiation (10). In both cases, the enzymes that place these repressive modifications on DNA and histones must be targeted appropriately. DNA methylation is catalyzed by DNA methyltransferases (DNMTs) (11, 12). Three DNMTs, DNMT1, DNMT3A, and DNMT3B, account for all 5MeCpG methylation in mammalian cells. DNMT1 has been viewed as a maintenance DNMT, with its activity being coupled to DNA replication. DNMT3A and DNMT3B are categorized as de novo DNMTs, although the distinction between de novo and maintenance DNMTs has recently been reevaluated (11). The histone modifications are placed or removed by large families of enzymes (e.g., histone deacetylases [HDACs], histone methyltransferases). These histone “marks” (including ca. 100 unique lysine and arginine modifications) encompass acetylation, phosphorylation, and mono-, di-, or trimethylation. Repressive histone modifications include histone H3 lysine 9 trimethylation (H3K9me3) and histone H4 lysine 20 trimethylation (H4K20me3). Both histone modifications and 5MeCpG DNA marks are recognized by protein “readers” through modular recognition domains. The readers then guide additional effector proteins that ultimately control epigenetic gene silencing or gene activation. When initially inserted into the host chromatin, retroviral DNA is epigenetically naive. The earliest events that contribute to the epigenetic fate of the provirus are largely unknown and may include the passive influence of the chromatin environment around the integration site (13–16) or the initiation of epigenetic repression through specific recognition of viral DNA sequence or protein components (2, 17–21). In particular, very little is known about how the cellular DNA methylation machinery is recruited to the integrated retroviral DNA.
Early studies showed that infection of mammalian cells with avian sarcoma viruses (ASV) could result in proviral epigenetic silencing, while infection of natural avian host cells appeared to be fully permissive for virus expression (22). Recent studies indicated that the ASV DNA integration site patterns are similar in human and avian cells (23–25), suggesting that the chromosomal position of the provirus is likely not the major determinant for epigenetic silencing. Our previous findings have led us to focus on a model whereby mammalian host cell factors initiate ASV proviral epigenetic silencing as part of an antiviral response in a largely position-independent manner (17, 18). The initial finding that led us in this direction was our identification of the human, ubiquitously expressed, cellular protein Daxx, as a binding partner of ASV integrase, as detected by the yeast two-hybrid system (17). Daxx was first discovered as a cytoplasmic Fas death domain-associated protein (26). However, Daxx localization in promyelocytic nuclear bodies (PML-NBs) (27, 28) and its association with numerous nuclear binding partners, including DNA-binding transcription factors and repressive epigenetic regulators (29–32) and viral proteins (17, 33, 34), have suggested additional roles for Daxx (35).
Our studies on retroviral epigenetic silencing have utilized full-length ASV-green fluorescent protein (GFP) reporter viruses that encode the murine leukemia virus (MLV) amphotropic (ampho) envelope gene, allowing efficient single-round infection of mammalian cells, with further spread being restricted by several posttranscriptional blocks (36). Using these GFP reporter viruses, we obtained evidence that Daxx-IN interactions prior to integration, lead to rapid epigenetic repression of ASV-GFP viruses (17). We identified repressive histone deacetylases as participants in this process (17, 37), consistent with the known role of HDACs as functional Daxx-binding partners (29, 38). We also showed that ASV-GFP reporter gene silencing could be reversed by treatment with HDAC inhibitors (18, 37). Other investigators documented a repressive function for Daxx in herpesvirus infection (39), thereby establishing a broad, antiviral role for this cellular protein. The unstructured region of Daxx (40) that engages ASV IN (17) may also underlie the ability of Daxx to interact with other viral proteins (41–46).
In the work described here, we continue to utilize ASV-GFP reporter viruses to investigate retroviral epigenetic silencing in human cells. As the viral replicative genes are intact, these reporter viruses are capable of producing spreading infections in the natural avian host cells, and therefore, they represent biologically relevant agents. Their epigenetic repression in heterologous human cells provides a powerful system to identify viral and host components that account for such differences. In these ASV-GFP viruses, the reporter genes are driven by alternative internal promoters, as well as the native viral long terminal repeat (LTR) promoter (18), with the GFP gene providing a readout for repressive epigenetic effects. The ASV LTR promoter region has been shown previously to be methylated in mammalian cells and functionally linked to ASV epigenetic silencing (22, 47). As we found that viruses using diverse promoters to drive the GFP gene are silenced similarly in human cells (37), the GFP readout appears to serve as a general indicator of the epigenetic state of the provirus.
Using ASV-GFP viruses, we previously observed that ca. one-half of ASV DNA integration events in HeLa cells result in rapid, stable, and complete silencing, with another large fraction of infected cells demonstrating epigenetic muting of viral GFP expression (37). We devised a strategy to isolate HeLa cell populations and clones harboring fully silenced proviruses in order to provide a robust system for identifying repressive host factors. The resulting stable HeLa GFP-silent cell populations were named according to the promoter driving the viral GFP gene: an internal cytomegalovirus (CMV) promoter (TI-C), an internal EF-1α promoter (TI-E), or the native viral LTR promoter (TI-L). In all three cell populations, treatment with HDAC or DNA methyltransferase inhibitors, as well as small interfering RNAs (siRNAs) targeting HDACs and DNMTs, promotes GFP reactivation, indicating cross talk between these two repressive mechanisms (18).
In this report, experiments were undertaken to identify roles for the human antiviral Daxx protein in the initiation and maintenance of retroviral epigenetic transcriptional repression. We found that knockdown of Daxx prior to infection resulted in increased viral GFP reporter expression, confirming a role for Daxx in the initiation of silencing. At early times postinfection, we observed the rapid appearance of repressive DNA methylation and histone modifications, consistent with an active antiviral response. We also found that Daxx is required for both silencing maintenance and full viral DNA methylation and that it is positioned on viral DNA. Last, we confirmed DNMTs as functional binding partners of Daxx in this system. The results provide the first evidence that the antiviral protein Daxx can contribute to DNA methylation-based epigenetic repression of viral DNA.
MATERIALS AND METHODS
Viruses and cells.
The ASV-based viral vectors that utilize the LTR (ASV-EGFP) or an internal human cytomegalovirus (hCMV) immediate-early (IE) promoter (ASV-CMV-EGFP) to drive expression of the enhanced GFP (EGFP) gene have been described previously (36, 37). Cultures of HeLa cells and the chicken embryo fibroblast DF1 cell line were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% heat-inactivated fetal bovine serum. IMR90 cells (human healthy lung fibroblasts) were maintained in Eagle's minimum essential medium with 20% inactivated fetal bovine serum. Cultures of RPE1, a human telomerase reverse transcriptase (hTERT)-immortalized retinal pigment epithelial cell line, were maintained in DMEM/F12 medium, supplemented with fetal bovine serum to a final concentration of 10% and contained 0.01 mg/ml hygromycin B to maintain the hTERT gene. Media for all cell lines contained 100 units/ml penicillin, and 100 μg/ml streptomycin. Preparation and maintenance of HeLa cell populations containing silent GFP genes (TI-C, TI-L, and TI-E) were described previously (37).
Infections, cell sorting, and analysis of GFP expression.
HeLa, RPE1, and IMR90 cells were infected with several dilutions of virus stocks, and GFP expression was quantitated by FACScan as described previously (17, 37). For isolation of a GFP-positive population, the VantageSE flow analyzer was used, and data were analyzed with FlowJo software.
siRNA transfection.
DharmaFECT 1 (T-2001), DharmaFECT 2 (T-2002), and DharmaFECT4 (T-2004) reagents were used according to the manufacturer's protocols for transfection of HeLa, RPE1, and IMR90 cells, respectively, with a final concentration of siRNAs of 50 nM. siRNAs were purchased from Dharmacon (Lafayette, CO). The siRNA SMARTpools used targeted HDAC1 (M-003493-02) and Daxx (M-004420-00); control, nontargeting siRNA#1 (D-001210-01) was used as a negative control for siRNA transfection. The single siRNAs obtained from Qiagen Corporation for these studies included the following: DNMT1 (Hs_DNMT1_4/6/7/8), DNMT2 (Hs_DNMT2_1/2/4/5), DNMT3A (Hs_DNMT3A_3/10/13/15), DNMT3B (Hs_DNMT3B_3/5/6/7), and DNMT3L (Hs_DNMT3L_1/2/5/6).
Immunoblot analysis.
Immunoblotting was performed by standard methods as described previously (17). Anti-Daxx (D7810) and antiactin (A2668) were purchased from Sigma-Aldrich, St. Louis, MO. Antibody against glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (MAB374) was purchased from Chemicon, Temecula, CA. Anti-DNMT1 (H-300), anti-DNMT3A (H-295), and anti-DNMT3B (H-230) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Goat anti-rabbit peroxidase-conjugated secondary antibodies (31462; Pierce, Rockford, IL) and enhanced chemiluminescence reagents were used according to the manufacturer's instructions (Pierce).
ChIP.
Chromatin immunoprecipitation (ChIP) analyses of infected HeLa cell lysates were performed using the EZ-Magna ChIP kit (Millipore, Billerica, MA). Cells were fixed with a final concentration of 1% formaldehyde for 10 min at room temperature, washed with phosphate-buffered saline (PBS), and disrupted with SDS lysis buffer supplemented with protease inhibitor cocktail II. The chromatin was fragmented by sonication to an average length of 1 kb. Cellular debris was removed, and the lysate was cleared with protein G agarose for 1 h. Part of the lysate (10%) was saved as an “input sample.” For immunoprecipitation procedures, anti-RNA polymerase II positive-control antibody and the IgG negative-control antibody were provided with the EZ-Magna ChIP. Anti-H3K9me3 (ab8898) and anti-H4K20me3 (ab9053) were obtained from Abcam, Cambridge, MA, and anti-histone H3 antibody (06-599) was obtained from Upstate. Anti-Daxx (M-112), anti-DNMT1 (H-300), anti-DNMT3A (H-295), and anti-DNMT3B (H-230) antibodies were obtained from Santa Cruz Biotechnology, Santa Cruz, CA. Lysates were incubated first with antibodies overnight and on the next day with 60 μl of protein G agarose for 2 h. Agarose antibody-chromatin complexes were washed according to the EZ-Magna ChIP kit protocol with low-salt, high-salt, LiCl, and Tris-EDTA (TE) buffers, and after elution, cross-linking was reversed by heating, followed by purification using spin columns. DNA detection was performed in triplicate using quantitative real-time PCR, with KAPA SYBR FAST PCR protocol and with primers for the different regions. For the control GAPDH promoter region, the Fw (Fw stands for forward) primer TACTAGCGGTTTTACGGGCG and Rev (Rev stands for reverse) primer TCGAACAGGAGGAGCAGAGAGCGA were used. For the LTR promoter region, the Fw primer CCGATTGGTGGAAGTAAGGTG and the Rev primer AAATACAATATCTCTGCAATGCGG were used. For the CMV promoter region, the Fw primer ATGCCAAGTACGCCCCCTATTGAC and the Rev primer AACCGCTATCCACGCCCATTGATG were used. For the LTR transcription start site region, the Fw primer CCATTTGACCATTCACCACATT and the Rev primer GCTCGTAGTCGTCAGGGAATC were used. For the CMV transcription start site region, the Fw primer GTGGGAGGTCTATATAAGCAGAGC and the Rev primer CAGCTCGACCAGGATGGGCACCAC were used. For the LTR primer binding site, the Fw primer TTGATTCCCTGACGACTACGAGCAC and the Rev primer GCCGACCACTATTCCCTAACTATC were used. Quantifications were performed using the ΔCT method. Biological replicates were performed, and single representative experiments are shown as PCR analyses of triplicate samples with standard deviations.
Coimmunoprecipitation (co-IP).
The Pierce classic IP kit (Thermo Scientific, Rockford, IL) was used according to the manufacturer's protocol. Cells were rinsed with PBS and lysed with cold IP-lysis buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol) supplemented with phosphatase and protease inhibitor cocktails (Thermo Scientific). The soluble fraction was cleared with protein A/G agarose for 1 h, and 5% of whole-cell lysates was saved to measure “total” protein levels. Cell lysates (600 μg) were incubated with anti-DNMT1 (H-300), anti-DNMT3A (H-295), anti-DNMT3B (H-230), or anti-Daxx (M-112) (Santa Cruz Biotechnology, Santa Cruz, CA). Anti-IgG, used as a negative control, was obtained from Millipore. Lysates were incubated with antibodies overnight, followed by 2 h with protein A/G agarose beads. The beads were collected and washed with IP-lysis buffer, Tris-buffered saline (TBS) (pH 7.2), and conditioning buffer (neutral-pH buffer). Immunoprecipitated proteins were extracted with sample buffer (0.3 M Tris HCl, 5% SDS, 50% glycerol, lane marker dye [pH 6.8]) and dithiothreitol (DTT) to a final concentration of 20 mM, subjected to SDS-PAGE, and analyzed by immunoblotting as described in the figure legends.
DNA methylation analysis.
The methylation status of selective sites in the integrated proviral DNA was assessed by bisulfite sequencing. Briefly, total genomic DNA was isolated from infected HeLa cells containing integrated viral DNA using the DNeasy kit (Qiagen, Valencia, CA). Bisulfite reactions were performed using the EZ DNA Methylation Gold kit (Zymo Research, Irvine, CA) under conditions that allowed for complete conversations of cytosine to uracil. The bisulfite-modified DNA was amplified by PCR using GoTag Flexi DNA polymerase (Promega, Madison, WI) with primers to the converted sequence of the appropriate region. The following primers and PCR conditions were used for the different regions. For the LTR, the Fw primer GTAATATGTTTTATAAGGAGAGAAAAAG and the Rev primer AAAACCTTCTACTTCATACAAATACTC were used, and 40 cycles of PCR were performed, with 1 cycle consisting of 30 s at 94°C, 40 s at 54°C, and 30 s at 70°C. For CMV, the Fw1 (ATTAGTTTATAGTTTATATATGGAG) and Rev1 (CCATTAATATACTACCAAAACC) primers and the Fw2 (GTTTGGTATTATGTTTAGTATATG) and Rev2 (CCAACTCTACTTATATAAACCTCCC) primers were used, and 40 cycles of PCR were performed, with 1 cycle consisting of 30 s at 94°C, 40 s at 55 or 56°C, and 50 s at 70°C. Amplified products were subcloned in the pGEM-T Easy vector (Promega, Madison, WI), and 10 clones were sequenced per target region. The methylation profile of the region of interest was determined by comparing the sequence of bisulfite-converted DNA with the sequence of unmodified DNA.
RESULTS
Role of the human cellular protein Daxx in initiating a repressed retroviral state.
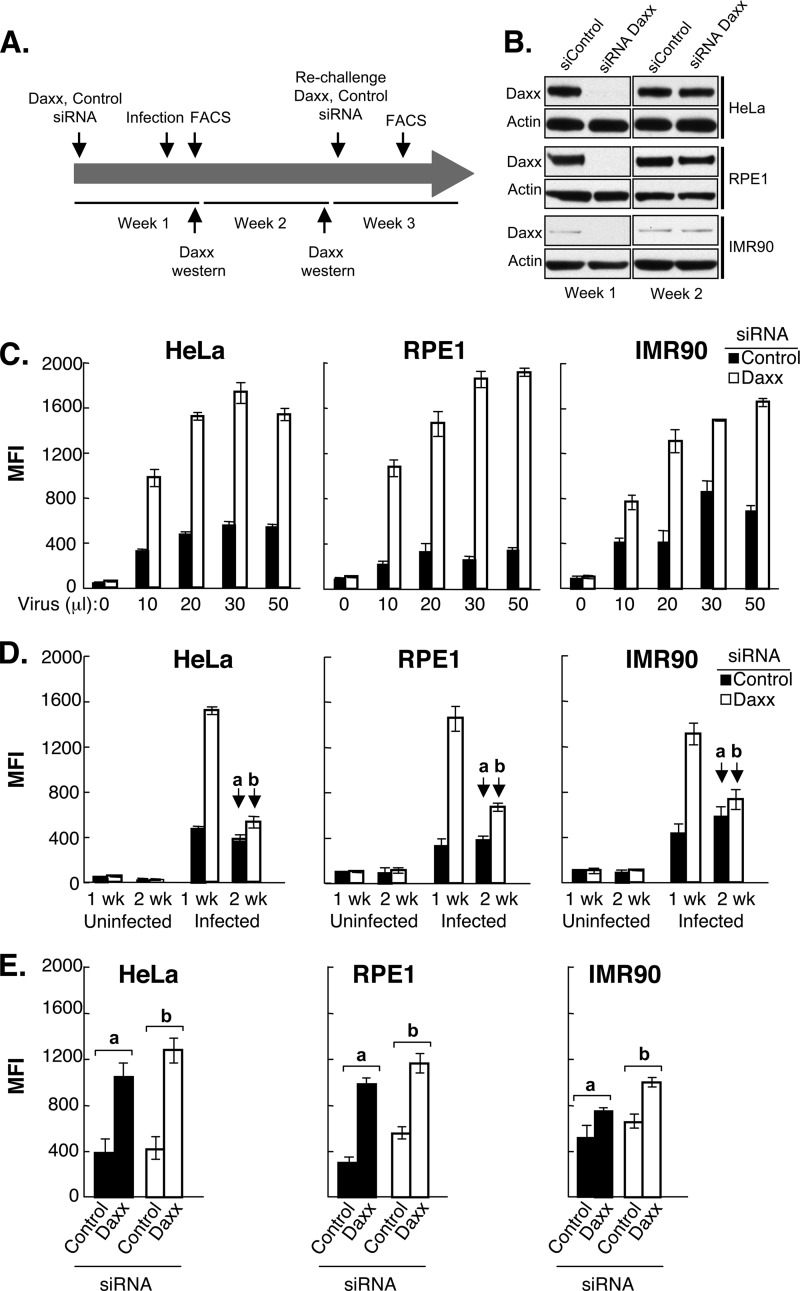
We previously uncovered a functional role for the human Daxx protein in ASV silencing (17). Although Daxx was found to be an ASV IN-interacting protein, we could not identify a role in integration. Rather, we identified a mechanism whereby Daxx-HDAC repressive complexes could be recruited to the integration site to initiate an epigenetically repressed state of ASV proviruses in mammalian cells (17). The experiments described in the legend to Fig. 1 were designed to determine whether Daxx siRNA knockdown in target cells at the time of infection could lead to enhanced and sustained proviral gene expression. We tested this hypothesis using three different human cell lines: the HeLa carcinoma cell line, the RPE1 retinal epithelial line, and IMR90 normal fibroblasts. The outline of the experiments is shown in Fig. 1A. Cells were treated with nonspecific control or Daxx siRNAs, and when Daxx protein was nearly undetected (Fig. 1B, 1 week), cells were infected with the ASV vector encoding a GFP reporter gene under the control of the CMV promoter. GFP expression was measured by fluorescence-activated cell sorting (FACS) at 2 days postinfection. Decreasing dilutions of virus were used both to monitor dose response and reproducibility and to examine the potential for high multiplicities of infection to overcome repression. Substantial increases (2- to 6-fold) in GFP expression were observed with Daxx siRNA-treated cultures versus control siRNA-treated cultures, even at the highest multiplicities of infection (Fig. 1C). Daxx knockdown had no effect on viral integration or histone deposition on viral DNA (data not shown) (see Discussion).
Fig 1.
Daxx siRNA knockdown prior to ASV infection results in an initial increase in viral reporter gene expression (GFP) that is reduced over time after restoration of Daxx expression. (A) Experimental outline. Human cells (HeLa, RPE1, and IMR90) were transfected with Daxx siRNA or control siRNA (Dharmacon) and incubated for 96 h. The cells were then infected with the ASV-GFP vector, and GFP expression was analyzed by FACS at 48 h postinfection (p.i.). At the same time, the cultures were sampled to confirm knockdown efficiency by immunoblotting with Daxx antibody. Selected cultures of each cell type were maintained for an additional week, after which time GFP expression was again measured by FACS, and Daxx protein levels were determined by immunoblotting. (B) Assessment of siRNA knockdown of Daxx and its recovery by immunoblotting. Antiactin antibody was used to monitor loading. (C) Daxx knockdown increases ASV-GFP expression over a range of multiplicity of infections (MOIs). Triplicate samples of cells were treated with Daxx siRNA or control siRNA, incubated for 96 h, and infected as described above for panel A. GFP expression was analyzed by FACS at 48 h p.i. MFI, mean fluorescence intensity. (D) Recovery of Daxx protein levels is correlated with repression of ASV-GFP expression. Triplicate samples from the siRNA-treated cells that were either uninfected or infected with 20 μl of viral stock were maintained for an additional week, after which GFP expression was again analyzed by FACS, as described above for panel A. (E) A second treatment with Daxx siRNA resulted in reactivation of GFP. Cell populations originally treated with either control or Daxx siRNAs prior to infection were maintained for 2 weeks as described above for panel D (see the black arrows labeled a and b) and were treated a second time with either Daxx or control siRNAs. GFP expression was analyzed by FACS after 96 h.
To determine whether the increased GFP expression in Daxx siRNA-treated cells was sustained during continued cell growth, the GFP signal was monitored for one additional week. At this time, GFP expression was found to be reduced to levels near those observed with control siRNA treatment (Fig. 1D, infected IMR90 cells at 2 weeks). This delayed GFP repression corresponded with the restoration of the Daxx protein to normal levels after siRNA treatment (Fig. 1B, 2 weeks). Therefore, either a second mechanism promoted repression over time, or Daxx-mediated repression is not strictly coupled to a time window encompassing the integration step.
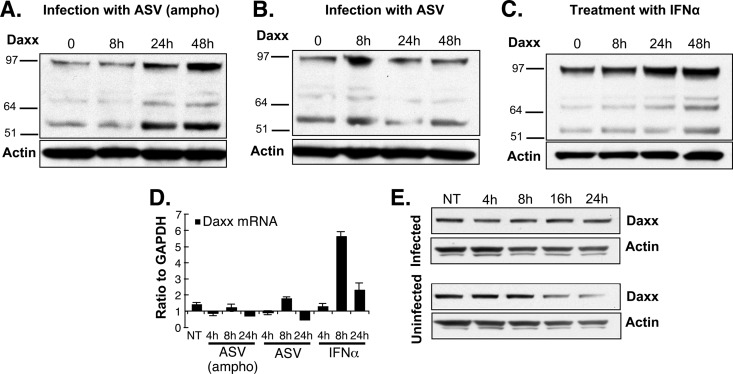
Fig 2.
Daxx protein is stabilized in ASV-infected HeLa cells. (A and B) HeLa cells were incubated with the ASV amphotropic GFP vector capable of infecting mammalian cells (36, 50, 51) (A) or with an analogous ASV subgroup A vector, which is incapable of infecting human cells (B) as a negative control. Lysates were subjected to gel electrophoresis, and the steady-state amounts of Daxx protein were determined by immunoblotting at the indicated times postinfection. Antiactin antibody was used to monitor loading. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gels. (C) Cultures were treated with IFN-α for 4 h (1,000 IU/ml), samples were collected at several intervals, and Daxx protein was detected as described above for panels A and B. (D) RNA was extracted from cultures treated with IFN-α or infected with ASV (ampho) or ASV subgroup A at the indicated time points, and the Daxx mRNA levels were analyzed by quantitative RT-PCR (qRT-PCR). (E) Parallel HeLa cell cultures were infected with the ASV amphotropic GFP vector, and the translation inhibitor emetine was added at a final concentration of 0.5 μg/ml. Infected and uninfected treated cells were harvested at the indicated time points, and the Daxx protein was detected by Western blotting. Prolonged Daxx decay was observed in infected compared to uninfected cells, while actin protein decay was similar under both conditions. NT, not treated.
To determine whether Daxx-dependent or Daxx-independent mechanisms played a role in delayed repression, cultures that were treated with control siRNA or Daxx siRNA for 2 weeks and analyzed in Fig. 1D (infected IMR90 cells at 2 weeks) were rechallenged with both siRNAs, and GFP expression was monitored at 96 h posttreatment (Fig. 1E). The results showed that rechallenge with Daxx siRNA led to increased GFP expression in cultures initially treated with Daxx siRNA, as well as those initially treated with the control siRNA. These findings indicate that after an initial enhancement of GFP expression by Daxx knockdown, muting of the GFP signal was resumed over time and that this process was dependent on Daxx. From the results of these experiments, we conclude that the depletion of Daxx at the time of viral DNA integration results in increased viral GFP gene expression but that Daxx can also promote repression long after the provirus is established. Having previously shown that Daxx can promote repression through interaction with the viral IN protein (17), these data indicate that a second, IN-independent but Daxx-dependent mechanism is operative weeks after infection, at which time the incoming viral IN protein has dissipated.
Daxx protein levels are increased after ASV infection.
Retroviral infections are countered by a variety of sometimes inducible host cell proteins that participate in intrinsic host defense mechanisms (20, 48). We carried out experiments to investigate further the finding that Daxx protein levels increase in response to retroviral infection (17). We measured Daxx steady-state protein levels after exposure to either the ASV-GFP virus encoding the MLV amphotropic envelope gene that allows entry into mammalian cells or a natural subgroup A strain of ASV that cannot infect mammalian cells (Fig. 2). The Daxx protein is 740 amino acids in length, with a predicted molecular mass of 81 kDa. The prominent form of Daxx identified in mammalian cells migrates as a ca. 110-kDa species, with additional forms migrating as 70-kDa and 97-kDa species. Previous studies showed that the 110-kDa species is phosphorylated (49), but differences in migration may also reflect the highly unstructured nature of the protein (40). In Fig. 1 (and Fig. 5 and 6 below), we highlight the 110-kDa form, as the antibody used in these experiments primarily recognizes this form. The antibody used in the Western blots in Fig. 2 recognizes all forms.
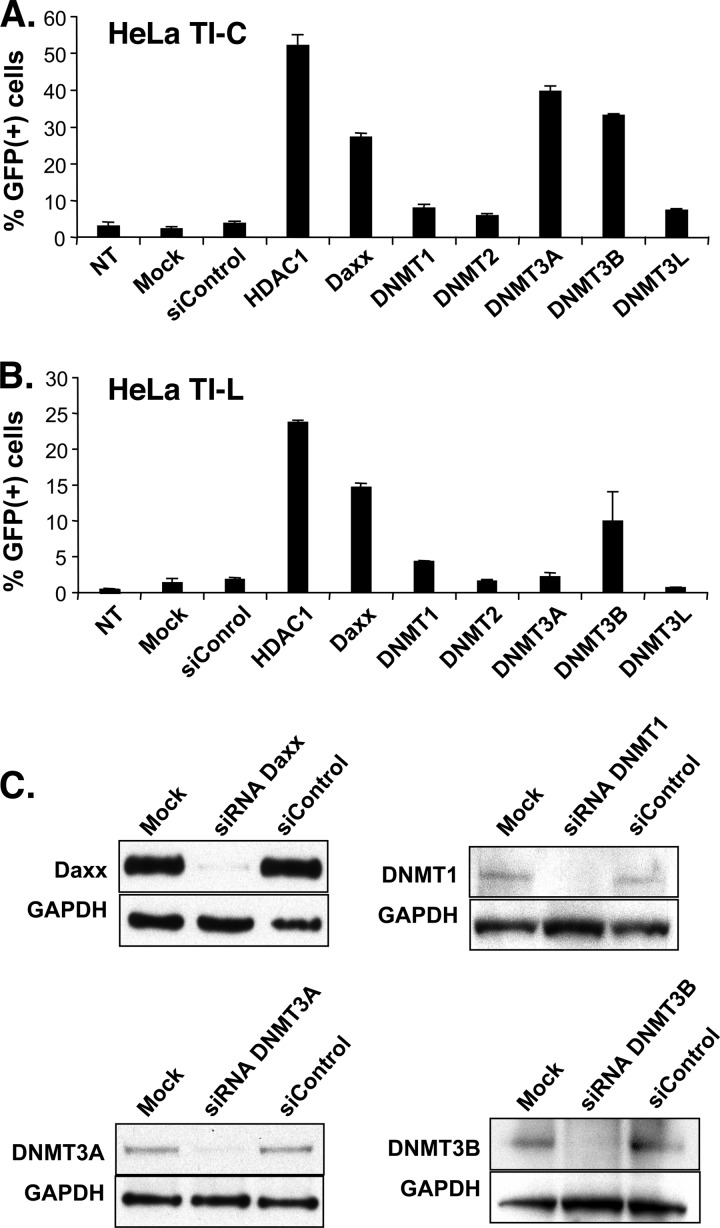
Fig 5.
siRNA knockdown of Daxx, HDAC1, and DNMT3A and -3B leads to reactivation of reporter gene expression in long-term ASV-GFP-silent populations. (A) Results of siRNA treatment of a population of HeLa cells that contain a silent ASV-GFP provirus with the GFP gene under the control of an internal CMV promoter (HeLa TI-C). (B) Results of siRNA treatment of a population of HeLa cells that contain a silent ASV-GFP provirus with GFP reporter gene expression regulated by the proviral LTR (HeLa TI-L). In panels A and B, GFP expression was determined by FACS 96 h after siRNA addition. NT, not transfected; Mock, mock treated (treated with transfection reagent without siRNA). (C) Assessment of knockdown efficiencies by immunoblotting. Samples were analyzed 96 h after siRNA treatment. GAPDH was used as a loading control.
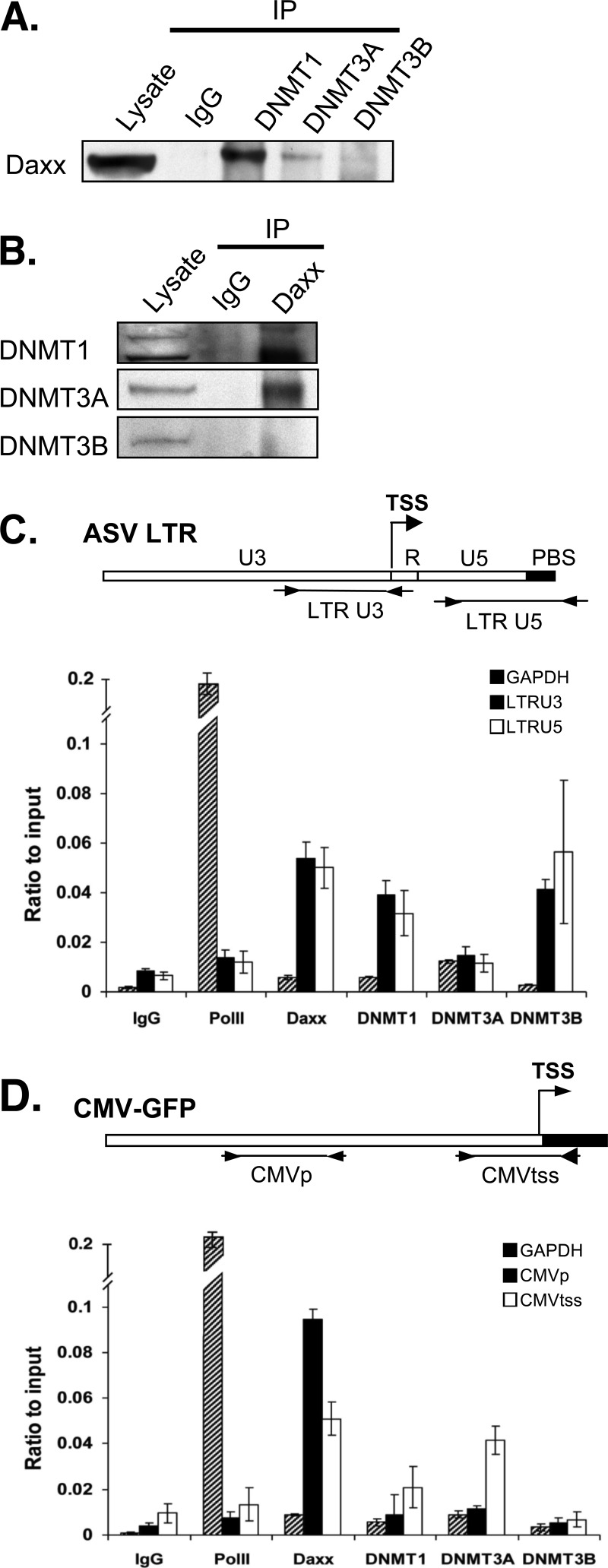
Fig 6.
Evidence for physical interaction between Daxx and DNMTs and their association with the proviral DNA. (A) DNMT1 and DNMT3A interact with Daxx. HeLa TI-C cell lysates were treated with anti-DNMT1, anti-DNMT3A, anti-DNMT3B antibody, or control IgG. The resulting immune complexes were analyzed by SDS-PAGE, followed by immunoblotting with Daxx antibodies. (B) Daxx interacts with DNMT1 and DNMT3A. HeLa TI-C cell lysates were treated with anti-Daxx or control IgG antibody. The resulting immune complexes were analyzed as described above for panel A and probed with the anti-DNMT antibodies. Lysates were loaded directly onto gels, as controls for total protein content in both panels A and B. (C) DNMT1, DNMT3B, and Daxx are associated with the proviral LTR. HeLa TI-C cells were subjected to ChIP with antibodies specific for Daxx, DNMT1, DNMT3A, and DNMT3B. Anti-RNA polymerase II (anti-PolII) antibody was used as a control to measure expected high RNA PolII occupancy on the GAPDH promoter and low occupancy on the LTR and the internal CMV promoter in HeLa TI-C cells. Normal IgG served as a background control. Primer sets that target the viral LTR U3 and U5 regions flanking the transcriptional start site (TSS) are illustrated in the map above the graph. Primers that flank the active cellular GAPDH promoter region were used as a control to monitor the expected low occupancy of silencing factors. (D) DNMT1, DNMT3A, and Daxx are associated with the internal CMV transcription start site for the GFP reporter in the provirus. ChIP was performed with HeLa TI cells as described above for panel C. The locations of the primer sets that target the CMV promoter region and transcription start site (TSS) are illustrated in the map above the graph. Error bars show the standard deviation among triplicates. Results shown are representative of biological duplicates.
The results in Fig. 2 show an increase in steady-state levels of all Daxx protein species at 24 and 48 h after infection with the ASV (ampho) virus, but no significant change after exposure to the subgroup A virus that is unable to enter human cells. These observations are consistent with those of our earlier studies, showing an increase in Daxx steady-state levels after infection of human cells with ASV (17). To determine whether the increase in Daxx protein after ASV infection is due to transcriptional or posttranscriptional mechanisms, Daxx mRNA levels were measured. For a positive control, Daxx mRNA and protein levels were examined after treatment with alpha interferon (IFN-α). As expected (50, 51), increases in Daxx protein (Fig. 2C) and Daxx mRNA (Fig. 2D) were detected after IFN-α treatment. These findings are consistent with participation of Daxx in the mobilization of intrinsic host cell defenses. However, analysis by quantitative real-time reverse transcription-PCR (RT-PCR) showed that the amount of Daxx mRNA in the cells infected with ASV (ampho) did not increase significantly during the first 24 h (Fig. 2D), indicating that the increase in Daxx protein following ASV infection is likely due to posttranscriptional stabilization mechanisms. Treatment with the protein translation inhibitors emetine (Fig. 2E) and cycloheximide (not shown) resulted in higher levels of Daxx protein over time in infected cells compared to uninfected cells. These data indicate that the increase in steady-state levels of Daxx protein in infected cells observed in Fig. 2A is due, at least in part, to increased protein stability.
Rapid accumulation of repressive epigenetic marks on LTRs.
Our published findings and those described above suggest that retroviral protein-DNA complexes are recognized in infected cells and that an epigenetically repressed state is established rapidly as part of an antiviral response. As ASV-GFP epigenetic repression appears to be highly pervasive in HeLa cells (37), we expected that the appearance of repressive epigenetic marks could be tracked even in populations that were not sorted for complete GFP silencing.
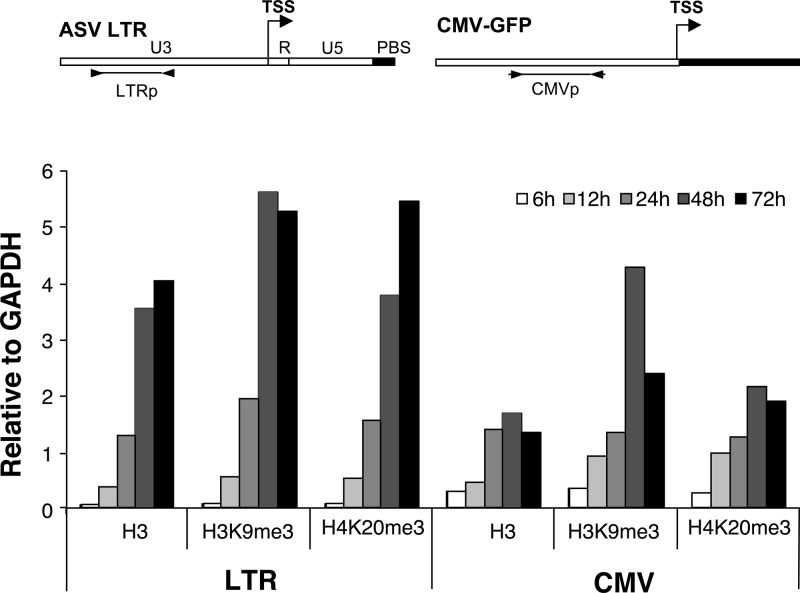
To monitor the rate of appearance of histones (i.e., histone H3) and repressive H3K9me3 and H4K20me3 histone marks (52) on newly integrated DNA in unsorted cultures, we used a chromatin immunoprecipitation (ChIP) assay. The ASV-GFP vector in which the GFP gene is under the control of the internal CMV promoter was used to infect HeLa cells, and both the viral LTR and the CMV promoter region were monitored for the assembly of nucleosomal histones and histone repressive marks. All of the measurements were normalized against the values for the GAPDH gene. As shown in Fig. 3, we observed a rapid and continuous increase in the association of histone H3 on the viral LTR, with occupancy detected in as few as 6 h, and a maximum occupancy at about 48 h postinfection. An association of histone H3 with the internal CMV promoter was also observed. We conclude that viral DNA is chromatinized rapidly postinfection, and the timing is consistent with an integration event (53). Furthermore, the data in Fig. 3 show that the repressive histone modifications H3K9me3 and H4K20me3 are detected on the LTR and the internal CMV promoter by 12 h, nearly concomitantly with chromatization.
Fig 3.
Deposition of core histones on viral DNA and accumulation of repressive histone marks. HeLa cells were infected with the ASV-GFP vector, and chromatin samples were collected for ChIP analysis at 6, 12, 24, 48, and 72 h postinfection. The indicated antibodies were used to detect the association of integrated proviral DNA with histone H3, and nucleosomes bearing the major repressive histone marks: histone H3 Lys 9 trimethyl (H3K9me3) and histone H4 Lys 20 trimethyl (H4K20me3). DNA was quantitated by real-time PCR using primers corresponding to the viral LTR region (LTRp) or the viral CMV promoter (CMVp). Primers for the promoter region of the GAPDH promoter region were used as a control. Values for H3 and histone marks associated with viral sequences were normalized to those obtained with the GAPDH promoter, with the ratios plotted on the y axis.
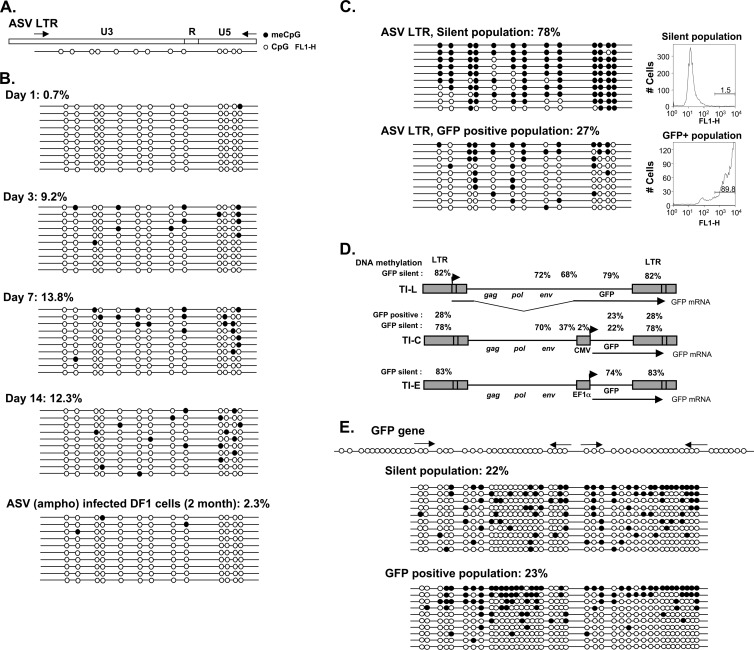
The appearance of 5MeCpG repressive marks on the ASV LTR (Fig. 4A) was also monitored over time after infection of unsorted HeLa cells, using the standard bisulfite conversion method. As shown in Fig. 4B, significant methylation of the LTR was detected by day 3, and the percentage continued to increase over time to day 7, with a plateau at about 13%. For a technical control, as well as a relevant biological comparison, LTR methylation analysis was performed after long-term infection of avian DF1 cells with the same ASV-GFP virus. In this case, LTR methylation levels were very low (ca. 2% 5MeCpG) (Fig. 4B) consistent with the lack of detectable ASV silencing in these natural avian host cells (54, 55).
Fig 4.
DNA methylation of the ASV proviral LTR. (A) Diagram of the ASV LTR showing the distribution of CpG dinucleotides within the U3, R, and U5 regions and the locations of primers for PCR. (B) Methylation patterns of the LTR in newly infected HeLa cells and long-term-infected DF1 cells as determined by bisulfite conversion and cloning of converted PCR product. Analysis of 10 clones is shown as a linear array, with empty circles representing nonmethylated CpG residues and filled circles representing methylated CpG residues. (C) Methylation pattern of the LTR in long-term populations of infected sorted GFP-silent (HeLa TI-C) and GFP-expressing cells. FACS profiles and the percentage of GFP-positive cells (GFP+) in each population are shown at the right. (D) Summary of proviral methylation patterns of proviruses in sorted GFP-silent HeLa TI-L, TI-C, and TI-E cells and GFP-positive HeLa cells. (E) Methylation pattern of the GFP reporter gene in long-term populations of infected, sorted GFP-silent and GFP-positive cells. The locations of primers are shown above.
Viral DNA methylation in long-term-silent cells.
The experiments described above demonstrate that Daxx-mediated epigenetic repression and ASV LTR DNA methylation occur during early stages of ASV infection in human cells. To begin to assess the functional relationship between these findings, we examined sorted long-term-passaged cells in which the ASV-GFP provirus is stably silent. For these experiments, we analyzed proviral DNA methylation in three HeLa cell populations harboring silent proviruses in which the silent GFP gene is controlled by either the LTR promoter (TI-L), the CMV internal promoter (TI-C), or an elongation factor 1 alpha subunit (EF1α) internal promoter (TI-E) (37). An example of results with the LTR region, in the TI-C population, is provided in the top panel of Fig. 4C, revealing high levels of DNA methylation (78%). Figure 4D shows a summary of the complete analyses in which several regions of the viral genomes in TI-C, TI-L, and TI-E cells were analyzed, including the LTRs, two areas of the env gene, the internal CMV promoter, and the GFP-coding region. In TI-L cells, all four regions show ca. 70 to 80% methylation. High LTR methylation levels were also observed in TI-E cells (83%), in which an internal cell-derived promoter controls the GFP gene. In these analyses, primer sets for methylation analyses did not distinguish between the upstream and downstream LTRs. In a separate experiment with TI-C cells, in which specific primer sets were used to distinguish upstream and downstream LTRs, the levels were 83% and 76%, respectively. We conclude from Fig. 4D that heavy LTR methylation is prominent in silent proviruses.
Differences in the DNA methylation patterns of the three proviruses in the TI-L, TI-C, and TI-E GFP-silent HeLa populations were highly informative (Fig. 4D). As the TI-C and TI-E cells were selected for epigenetic silencing of the GFP genes, we expected that the internal promoters driving GFP would display DNA methylation marks. Conversely, as the LTR promoter has no direct role in GFP transcription in these two constructs, there should be no selection for LTR promoter-based silencing. The results summarized in Fig. 4D show, however, that the TI-C and TI-E proviral LTRs were indeed, heavily methylated. These results together with the data in Fig. 4B are consistent with a mechanism in which LTRs are targeted for methylation independently of selection for LTR promoter activity. Furthermore, analysis of the methylation patterns in TI-C proviruses showed, unexpectedly, that the internal CMV promoter was essentially devoid of DNA methylation (Fig. 4D). The regions flanking the CMV promoter in the env and GFP genes also showed dramatically reduced methylation compared to TI-L and TI-E proviruses (Fig. 4D). These results suggest that the strong CMV promoter and environs are protected from DNA methylation. Figure 3 shows that the CMV promoter region is subject to repressive histone marks, which may be sufficient to initiate and maintain silencing of the GFP gene. It is also possible that LTR DNA methylation can lead to repression of the internal CMV promoter (see Discussion). In either case, the results in Fig. 3 and 4 clearly indicate that repressive histone marks and DNA methylation are established on the ASV genome early after infection of human cells and that heavy LTR DNA methylation correlates with GFP silencing, despite the fact that the GFP gene is under the control of an internal promoter.
Our previous results (37), as well as those reported here, indicate that in human cells, ASV proviruses are subject to a continuum of epigenetic effects, which range from complete silencing to muting of GFP reporter gene expression. To address whether there exists a gradient of underlying epigenetic marks, we analyzed resident proviruses in GFP-positive HeLa cells (Fig. 4C). Long-term-passaged GFP-positive cells, harboring the same provirus that is present in TI-C silent cells, were sorted from the brightest GFP decade. A significant level of LTR methylation (∼27%) was detected in this HeLa GFP-positive cell population, but this value was much lower than that observed in TI-C silent cells (78%) (Fig. 4C). As discussed above, LTR DNA methylation is very low in chronically infected, permissive avian cells (Fig. 4B, bottom). The latter finding establishes a biologically relevant baseline with which the HeLa GFP-positive LTR DNA methylation levels can be compared. In contrast to the differences in LTR methylation between TI-C and GFP-positive cell populations, methylation levels of the GFP gene body were the same (22 to 23%) in both populations (Fig. 4E). Together, these findings indicate that the extent of LTR methylation correlates with the epigenetically repressed state and suggests that LTR methylation can exert long-range effects on the internal CMV promoter (see Discussion).
Silent proviruses are reactivated after knockdown of Daxx or DNMTs.
We used siRNA knockdown to identify host factors that participate in the observed DNA LTR methylation and repression in TI-C and TI-L long-term-silent cells, focusing on the role of Daxx and DNMTs as potential functional partners. The results (Fig. 5A and B) show that siRNAs that target Daxx or HDAC1 promote GFP reactivation in both TI-C and TI-L populations, as expected from previous studies (17, 18). In HeLa TI-C cells, knockdown of either DNMT3A or DNMT3B also promoted GFP reactivation, whereas knockdown of other DNMTs had little effect (Fig. 5A). In TI-L cells, only knockdown of DNMT3B resulted in a detectable increase in GFP expression (Fig. 5B). Efficient knockdown of DNMTs was confirmed by Western blotting (Fig. 5C). From these results, we conclude that Daxx and the de novo DNMTs (DNMT3A and DNMT3B) are required to maintain long-term ASV silencing in human cells and that Daxx-DNMT interactions may play a role in this process. Furthermore, as the internal CMV promoter driving GFP is unmethylated in TI-C cells, we speculated that DNMTs may be modulating GFP expression through long-range effects of LTR methylation. It is also possible that DNMTs play a nonenzymatic scaffolding role at the internal CMV promoter. Last, in TI-L cells, the LTR drives the GFP gene, and therefore, the effects of knockdown on GFP expression are predicted to be mediated by direct action of Daxx and DNMTs on the LTR.
Evidence for physical interactions between Daxx and DNMTs and their association with proviral DNA.
To determine whether Daxx represses proviral gene expression via effects on LTR methylation, we first asked whether Daxx associates with DNMTs in HeLa cells, as detected by coimmunoprecipitation. Cell lysates were treated with antibodies against DNMT1, DNMT3A, or DNMT3B, and the immunoprecipitates were subjected to Western blotting with anti-Daxx antibodies. Daxx was found to be associated with both DNMT1 and DNMT3A, but no interaction with DNMT3B was detected (Fig. 6A). The interactions were confirmed by a reciprocal strategy, using anti-Daxx antibody for immunoprecipitation, and DNMT antibodies for the Western blot analyses (Fig. 6B). These results are consistent with previous reports that DNMT1 and DNMT3A interact directly or indirectly with Daxx (32). An interaction of Daxx with DNMT3B could not be excluded in these experiments, as the level of DNMT3B in these cells was low.
To investigate potential Daxx-DNMT interactions on the viral LTR, we performed ChIP experiments followed by quantitative real-time PCR using two primer sets that flank the LTR transcription start site (TSS). The GAPDH cellular promoter served as a control. For these experiments, HeLa TI-C cells harboring silent ASV-GFP proviruses were used. As shown in Fig. 6C, high RNA polymerase II (RNA PolII) occupancy and low Daxx and DNMT occupancy were observed on the active GAPDH promoter, as expected. In contrast, low RNA PolII occupancy and high Daxx and DNMT occupancy were detectable on the viral LTR. Daxx, DNMT1, and DNMT3B were detected on LTR U3 and U5 sites (Fig. 6C). Analysis of the CMV-GFP region in the provirus (Fig. 6D) revealed low occupancy by RNA PolII consistent with GFP silencing. Daxx and, to a lesser extent, DNMT3A were associated with the CMV TSS. As the CMV promoter is unmethylated in TI-C cells, the role of the DNMT3A at the CMV promoter is unclear. However, the presence of Daxx and DNMTs at the LTR could signify roles in maintaining LTR DNA methylation, and this possibility was tested as described below.
Daxx siRNA knockdown results in loss of full LTR DNA methylation in long-term-silent cells.
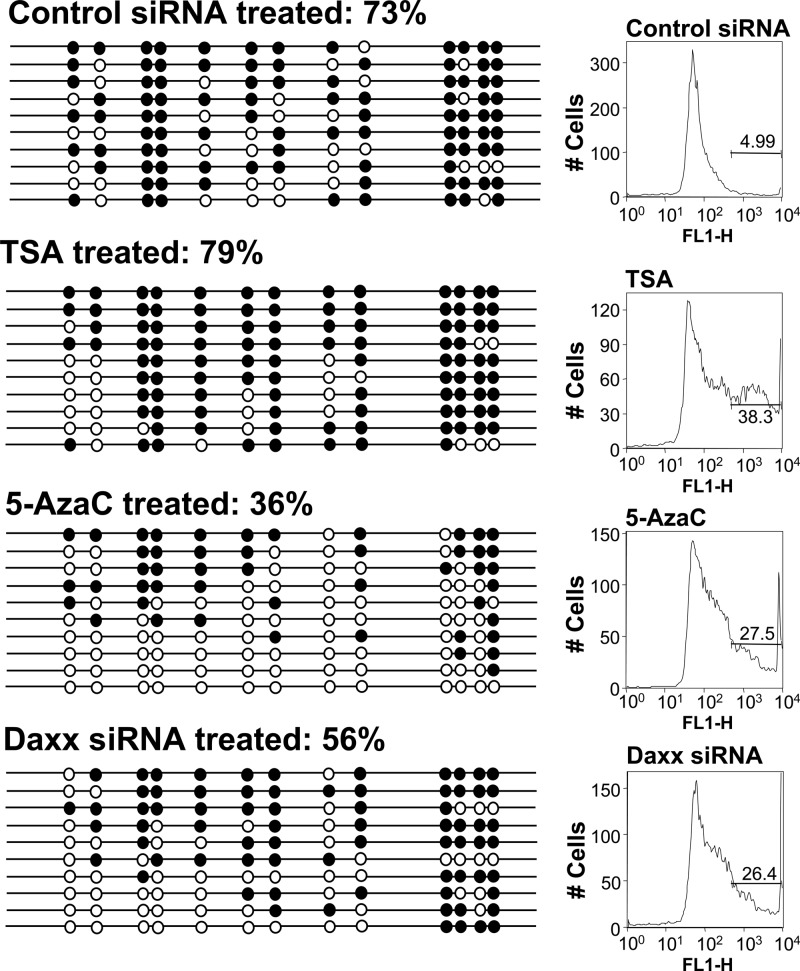
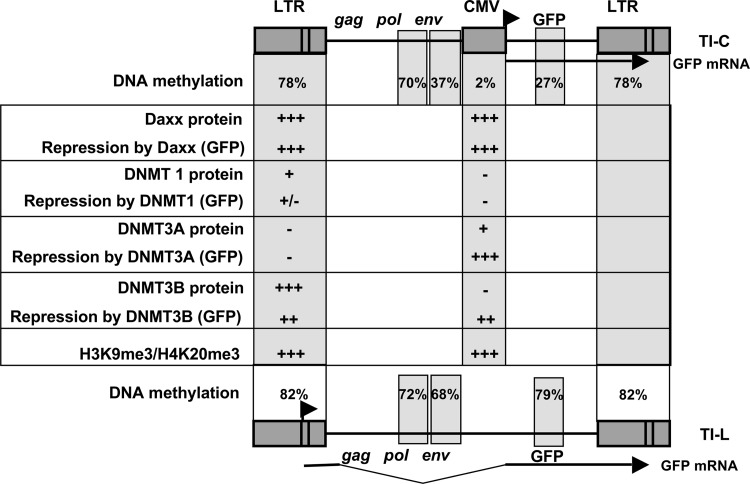
As Daxx occupancy was detected on the LTR (Fig. 6), and this protein participates in complexes that contain DNMTs (32, 38), we asked whether Daxx knockdown in long-term-silent TI-C cells could affect the DNA methylation profile of the ASV LTR (Fig. 7). For these experiments, an inhibitor of DNMTs, 5-azacytidine (5-azaC), was used as a positive control. Inhibitor treatment resulted in a reduction of LTR CpG methylation from 73% to 36%. Daxx knockdown caused a decrease in LTR methylation from 73% to 56%. For negative controls, a nonspecific siRNA and the HDAC inhibitor trichostatin A (TSA) were used. TSA reactivates GFP expression in TI-C cells but is not expected to affect DNA methylation. GFP expression following each of these treatments was monitored, and the FACS profiles confirmed the expected reversal of silencing (Fig. 7). These results are consistent with a model in which Daxx and DNMT(s) collaborate to control long-term retroviral silencing. Figure 8 summarizes findings relevant to GFP reactivation in response to siRNA knockdown, DNA methylation levels, Daxx binding, and repressive histone marks.
Fig 7.
Daxx siRNA knockdown reverses LTR DNA methylation in long-term-passaged GFP-silent populations. Methylation patterns of the LTR in HeLa TI-C cells was determined 96 h following treatment with control or Daxx siRNAs or 72 h following treatment with 0.5 μM TSA (negative control) and 5 μM 5-azaC (positive control). Analysis and data representation are as described in the legend to Fig. 4. FACS profiles and GFP expression levels are shown on the right.
Fig 8.
Summary of protein factor binding and mapping of repressive DNA methylation and histone marks on silent proviruses. Functional roles in repression were indicated by reactivation of GFP in response to factor knockdown with siRNA. For silent proviruses in TI-L cells (bottom), only the DNA methylation analysis was performed. Arrows indicate transcriptional start sites for GFP mRNA in TI-C cells and TI-L cells, as controlled by the internal CMV or LTR promoter, respectively.
DISCUSSION
Inserted retroviral DNA is initially epigenetically naive, and very little is known about the mechanisms by which the epigenetic state of retroviral DNA is established. Non-mutually exclusive models include the passive influence of the epigenetic environment at the chromosomal integration site (13–16) or the active recruitment of repressive complexes as part of an antiviral response (2, 17, 18, 20, 21).
Daxx as an antiviral factor.
We previously identified Daxx as an antiviral host factor that participates in ASV epigenetic repression in mammalian cells (17), possibly in response to interspecies infection. The engagement of Daxx with the ASV IN protein appears to represent an antiviral mechanism through which repressive epigenetic factors are recruited to viral DNA. A simple model, consistent with its known functions, is that Daxx acts as a scaffold to target such factors to viral IN-DNA complexes. The model implies that Daxx complexes are present at the site of viral DNA integration, and thereby could mediate rapid repression of viral transcription (Fig. 9).
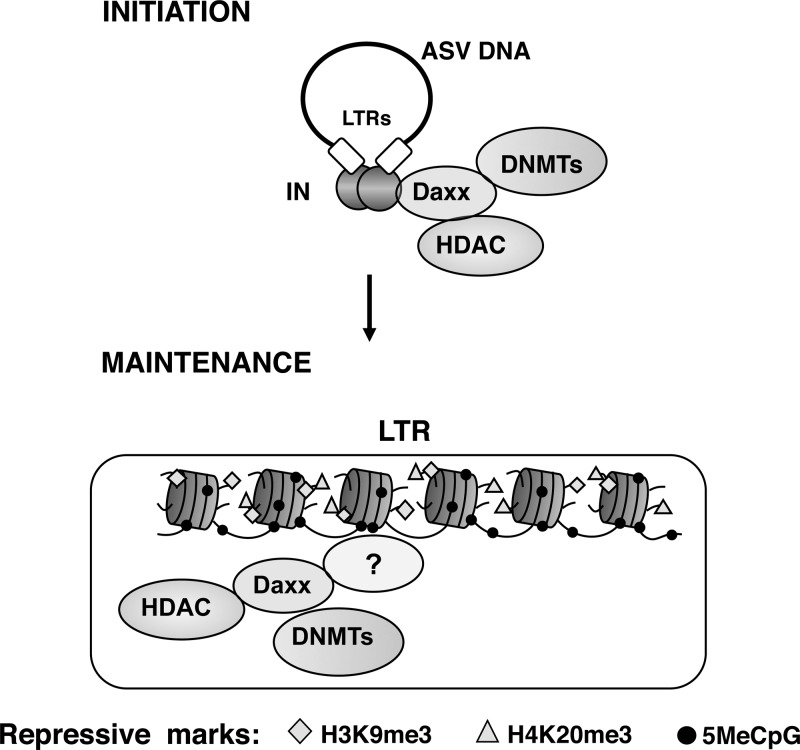
Fig 9.
Model for the roles of Daxx in the initiation and maintenance of retroviral silencing. (Top) The role of Daxx as a scaffolding protein to recruit DNMTs and HDACs to viral DNA is depicted. (Bottom) Postintegration repressive epigenetic marks are depicted, along with Daxx, HDAC, and DNMTs. The mechanism by which Daxx is positioned on viral DNA during silencing maintenance is unknown, as indicated by the question mark.
In addition to its antiviral function, Daxx has now been found to have multiple cellular roles (56, 57). However, the fact that Daxx interacts with a large number of diverse viral proteins (33, 39, 41–46, 58) indicates that functions relevant to host-virus interactions are prominent. This large collective of interactions between Daxx and viral proteins seem to highlight opposing strategies for the host and virus. One strategy, as exemplified by the Daxx-IN interaction, signifies the recognition of a virus-encoded protein, IN, by the host antiviral protein Daxx. In a second strategy, the antiviral Daxx protein is recognized and disabled by virus-encoded proteins; several of these Daxx countermeasure proteins have been identified (39, 41, 44, 46, 58, 59). Recently, it has been determined that Daxx is a highly unstructured protein (40). This disorder may provide structural plasticity, allowing initial engagement with these diverse viral proteins with subsequent formation of organized, stable interfaces.
Unlike large DNA viruses, simple retroviruses such as ASV do not encode accessory proteins that function as countermeasures to disable antiviral host factors such as Daxx. Despite this apparent lack of ASV countermeasures, Daxx activity is not sufficient to completely repress ASV DNA expression in human cells, and it is unclear how some ASV proviruses escape full Daxx-mediated epigenetic repression. It is likely that Daxx functions as a part of an ensemble of intrinsic antiviral factors that target various steps in retroviral replication (2). It is also possible that the chromosomal integration site can modulate the role of Daxx as an antiviral factor. We note, however, that the epigenetic repression and silencing that we have observed do not preclude the effective use of ASV vectors for gene transfer into mammalian cells (36, 60).
DNA methylation and histone modification cross talk.
DNA methylation is a well-recognized silencing mechanism for retroviruses and other retrotransposons (22, 61), frequently targeting the LTR promoter. As long-term ASV silencing in mammalian cells was known to be mediated by DNA methylation (22, 47, 62), an unexpected finding from our initial studies was that treatment with HDAC inhibitors was sufficient to reactivate epigenetically silent ASV proviruses in freshly infected or long-term-silent human cells (37). Furthermore, Daxx-HDAC complexes were found to have a prominent role in rapid silencing that we observed (17). In the work described here, we reinvestigated the role of DNA methylation in retroviral silencing, in this case focusing on the participation of Daxx in this process, as well as monitoring the appearance of DNA methylation on viral DNA postinfection (Fig. 4B).
With respect to overall epigenetic control of cellular genes, cross talk between histone modifications and DNA methylation is well recognized, but highly complex and poorly understood (63). Several studies have shown that DNA methylation and histone modifications coordinately contribute to retroviral silencing (6, 64–68). In the case of HIV latency, HDAC-mediated epigenetic repression is prominent, while CpG methylation may contribute only to a more stable silent state of the provirus (64). Results of another study indicate that sequence-specific transcription factors and DNA methylation cooperate to maintain HIV latency (69).
Rapid appearance of repressive histone modifications and DNA methylation.
The results in Fig. 3 show that repressive histone marks (H3K9me3 and H4K20me3) could be detected as early as 12 h postinfection on both the viral LTR and the internal CMV promoter controlling the GFP gene. Although this approach cannot be used to determine the fraction of viral histones harboring repressive marks, an apparent plateauing of repressive mark accumulation was observed in several cases, suggesting that maximum modification levels are achieved within 12 to 24 h of infection. The finding that H3K9me3 repressive marks accumulate on viral chromatin early after infection is consistent with the previously detected role for HDACs; the HDACs facilitate removal of the activating H3K9 acetylation marks, thus allowing formation of the H3K9me3 marks.
The question of which epigenetic marks (DNA methylation and histone modifications) are the founders, or “drivers,” of the epigenetically repressed proviral state has been elusive, due to inherent differences in detection methods, time scales, and sensitivities. However, we can conclude that repressive histone marks are formed on viral chromatin within 24 h postinfection (Fig. 3).
Our experiments also revealed fundamental features of the assembly of retroviral DNA into chromatin. An association of histone H3 with the proviral LTR could be detected as early as 12 h postinfection, and its relative occupancy appeared to increase until it reached a maximum in about 48 h (Fig. 3). These kinetics are similar to those expected for retroviral DNA integration (53, 70, 71) and suggest that the provirus is rapidly chromatinized upon insertion into host DNA. The accumulation of repressive histone H3 and H4 marks on LTR-associated chromatin follows similar kinetics, indicating that formation of repressive chromatin is coincident with viral DNA integration. It is currently unknown whether histone deposition on newly integrated retroviral DNA must await passage of a replication fork during S phase. The kinetics for histone assembly on viral DNA shown in Fig. 3 indicate that near maximal histone occupancy occurs between 24 and 48 h, consistent with a DNA replication-dependent H3 deposition mechanism. The histone variant H3.1 is deposited in nucleosomes during S phase, while H3.3 is deposited in a DNA replication-independent manner (72). Interestingly, Daxx was recently identified as a histone chaperone for deposition of histone H3.3 (56, 57). It is therefore possible that in addition to recruiting epigenetic repressors, this large scaffold protein can participate in histone deposition on viral DNA. As the antibodies used in these experiments do not distinguish between H3.1 and H3.3, additional studies will be required to address the mechanism of histone deposition and the potential role of Daxx in this process.
Daxx facilitates silencing of proviruses in both newly infected and long-term-passaged cells.
Our earlier findings (17) suggested a “hit-and-run” repression mechanism whereby IN-Daxx-HDAC complexes directed the initiation of an epigenetically repressed retroviral state that was subsequently maintained by the cellular epigenetic machinery in a Daxx-independent manner. However, further studies identified a role for Daxx in long-term ASV provirus silencing (18). To temporally uncouple roles for Daxx in the initiation and maintenance of a repressive epigenetic viral state, here we asked whether Daxx depletion at the time of infection was sufficient for durable viral GFP reporter gene expression. The results in Fig. 1 show that Daxx knockdown at the time of infection with ASV-GFP viruses resulted in a significant increase in GFP intensity, as monitored by both mean fluorescence intensity (MFI) (Fig. 1C) and percentage of GFP-positive cells (not shown). We also show that ASV infection of human cells results in an increase in Daxx protein levels and that posttranscriptional mechanisms, including protein stabilization, contribute to this response (Fig. 2). These results suggest a surveillance role for Daxx, with rapid stabilization and recruitment of Daxx repressive complexes to the chromosomal integration sites (Fig. 9).
We note that the knockdown of Daxx at the time of infection (Fig. 1) is expected to interrupt both HDAC- and DNMT-mediated silencing. We have found that treatment with either the DNMT inhibitor 5-azaC or the HDAC inhibitor TSA just prior to infection did not enhance viral GFP expression. However, combined treatment did result in an increased GFP signal (not shown). These findings suggest that cross talk between DNMTs and HDACs is required for initiating epigenetic repression. We believe that knockdown of Daxx interrupts both pathways, as its scaffolding role recruits both HDACs and DNMTs. As such, we have not attempted to discern a role for individual DNMTs in Daxx-dependent initiation of silencing using siRNA knockdown. From the literature and our current experiments (Fig. 6), Daxx is capable of interacting with all three DNMTs. In terms of roles for individual DNMTs in silencing maintenance, the picture is complex, as summarized in Fig. 8. However, DNMT3B and Daxx are detected on the LTR during long-term silencing, and both are functionally required for silencing maintenance.
Knockdown of Daxx at the time of infection resulted in only transient viral derepression (Fig. 1D). Rechallenge with Daxx siRNA confirmed that this delayed repression was Daxx dependent (Fig. 1E). There is no known mechanism or biological requirement for retention of IN molecules at the integration site. Thus, our findings suggest that Daxx-mediated viral silencing and repression can also occur independently of IN-Daxx interactions (Fig. 9). These observations are consistent with results in Fig. 5, as well as the previous studies (18), which demonstrate a role for Daxx in long-term silencing.
We show here that Daxx is bound to both the LTR and the internal viral CMV promoter during long-term silencing (Fig. 6C and D and Fig. 8). Similar roles for Daxx as part of an antiviral repressor complex have been documented in other viral systems (33, 46, 73). Furthermore, the observation that large DNA viruses encode proteins that disable Daxx (e.g., herpes simplex virus [HSV] pp71) (39, 46, 58) further supports our original proposal that Daxx complexes act as antiviral transcriptional repressors (17, 18).
Methylation of viral LTR DNA sequences is correlated with silencing.
The ASV LTR is highly methylated (ca. 80% of CpGs) in long-term-passaged GFP-silent HeLa cells (Fig. 4C), consistent with numerous studies that show methylation of ASV DNA and silencing in mammalian cells (22). We detected extensive methylation of viral LTRs regardless of whether the silent GFP gene was under transcriptional control of the LTR or the internal EF1α or CMV promoters (Fig. 4 and 8). The use of internal promoters to drive GFP provides a powerful means to uncouple the effects of LTR methylation on the LTR promoter activity versus potential long-range effects within the provirus. In contrast to the heavy methylation on the LTRs, methylation was largely undetectable in the CMV promoter region of proviruses in TI-C cells, which were selected for GFP silencing (Fig. 4D). As GFP expression is controlled by the CMV promoter, and not by the LTR in this provirus, there was no selection for LTR promoter silencing in these long-term-passaged cells. These findings suggest two non-mutually exclusive models. (i) The LTR is targeted for methylation, and the internal CMV promoter is repressed only by histone marks. (ii) LTR methylation exerts a long-range effect on the internal CMV promoter. The finding that the CMV promoter is devoid of methylation yet is responsive to the DNMT inhibitor 5-azaC (Fig. 4D and 7) supports the idea that long-range effects of LTR methylation modulate epigenetic effects at the internal promoter.
LTR methylation was also observed in stable infected GFP-positive HeLa cells, but the levels were significantly lower than those observed in long-term-silent TI-C cells (27% versus 78%). These results were not unexpected, as ASV-GFP infection of HeLa cells is characterized by a broad range of moderate-to-weak GFP intensities (37). The level of LTR methylation in GFP-positive cells is apparently not sufficient to promote full epigenetic silencing, or alternatively, chromosomal positioning of proviruses in this GFP-positive population may contribute to overcoming repressive methylation. In either case, the fact that the extent of LTR methylation in TI-C and GFP-positive HeLa cells correlates with GFP repression (Fig. 4C) again points to a functional relationship between LTR methylation and activity of the internal CMV promoter. The mechanism of this apparent long-range effect is unknown.
In contrast to long-term-passaged GFP-silent and GFP-positive human HeLa cells, only a very low level of ASV LTR CpG methylation (2%) was detected in chronically infected avian cells (Fig. 4). Because ASV viral DNA integration site selection is similar in human and avian cells (23–25), chromosomal position is unlikely to account for such repression. Rather, our findings are consistent with the idea that DNA methylation of the LTR and concomitant GFP repression in human cells represents an antiviral response to interspecies infection of human cells. Our earlier work showed that a CMV promoter-matched HIV-1-based vector was not silenced in HeLa cells (37), also supporting the idea that heterologous ASV components specifically trigger proviral epigenetic repression in human cells.
Daxx mediates proviral DNA methylation.
We previously showed that the antiviral protein Daxx is recruited to viral DNA soon after infection through IN binding (17) (Fig. 9). As DNMTs are known Daxx-binding partners (32, 74), we hypothesized that the rapid initiation of proviral DNA methylation and silencing could be mediated by Daxx-DNMT complexes (Fig. 9). We show that ASV LTR DNA methylation can be detected within several days postinfection of HeLa cells (Fig. 4), consistent with the presence of Daxx at the integration site and acting as a facilitator of DNA methylation. This idea is difficult to test due to the initial low levels of LTR DNA methylation early after infection. However, a role for Daxx in DNA methylation was investigated with the TI-C long-term-silent cells. Our results (Fig. 7) show that Daxx is required to maintain full LTR DNA methylation in these cells. Furthermore, we confirmed that Daxx and DNMTs interact (32) in HeLa cells and showed that both Daxx and DNMTs are bound to the LTR (Fig. 6 and 8). Daxx-DNMT complexes are known to mediate repression of cellular genes (32), and our results demonstrate that Daxx can also promote both LTR DNA methylation and long-term retroviral silencing. These findings provide the first evidence that the antiviral protein Daxx can contribute to DNA methylation-based epigenetic repression of viral DNA.
The mechanism by which Daxx is targeted to the viral genome in long-term-silent cells is unknown (Fig. 9). We detected occupancy of Daxx on both the LTR and internal CMV promoters (Fig. 6C and D and Fig. 8), suggesting that the targeting mechanism is not highly localized. Whether positioning of Daxx is limited to promoter regions on viral DNA remains to be explored. As Daxx knockdown triggers reactivation of GFP in TI-L cells in which GFP gene expression is controlled by the LTR promoter, we hypothesize that positioning of Daxx on the LTR likely reflects its functional role in methylation and transcriptional repression.
Epigenetic repression as a broad antiretroviral mechanism.
Several studies, including our own (18), have identified host epigenetic factors that maintain retroviral silencing (2, 7, 20, 21, 66). In the case of MLV, a host zinc finger protein expressed in embryonic mouse cells has been shown to recognize a viral DNA sequence, the primer binding site, and thereby recruit the assembly of repressive epigenetic complexes. In other studies, lentivirus and MLV-based vector proviruses have been shown to undergo rapid DNA methylation in undifferentiated mouse cells (66, 67). Thus far, there has been a focus on such silencing as a protective antiviral mechanism in embryonic cells. Both rapid appearance of repressive marks and DNA methylation-histone modification cross talk appear to be common features in retroviral silencing in undifferentiated mouse cells and in human cells infected with a heterologous retrovirus (ASV). The repressive epigenetic effects on ASV in human cells that we have observed appear to indicate an activated antiviral response to interspecies infection. Such activities may represent a novel and underappreciated facet of the intrinsic antiretroviral response that may surveil human cells.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI40385, CA71515, and CA006927.
The contents of the manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute or any other sponsoring organization.
We thank Sid Balachandran and Bill Mason for critical comments. We are also grateful to Marie Estes and Karen Trush for assistance in preparing the manuscript. The Fox Chase Cancer Center DNA Sequencing and Cell Sorting Facilities provided support for this work. We also thank Jim Oesterling for help with cell sorting.
Footnotes
Published ahead of print 5 December 2012
REFERENCES
- 1. Ellis J, Hotta A, Rastegar M. 2007. Retrovirus silencing by an epigenetic TRIM. Cell 131:13–14 [DOI] [PubMed] [Google Scholar]
- 2. Wolf D, Goff SP. 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458:1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ellis J. 2005. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum. Gene Ther. 16:1241–1246 [DOI] [PubMed] [Google Scholar]
- 4. Hakre S, Chavez L, Shirakawa K, Verdin E. 2011. Epigenetic regulation of HIV latency. Curr. Opin. HIV AIDS 6:19–24 [DOI] [PubMed] [Google Scholar]
- 5. Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, Lorincz MC. 2011. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8:676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leung DC, Lorincz MC. 2012. Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem. Sci. 37:127–133 [DOI] [PubMed] [Google Scholar]
- 7. Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464:927–931 [DOI] [PubMed] [Google Scholar]
- 8. Reichmann J, Crichton JH, Madej MJ, Taggart M, Gautier P, Garcia-Perez JL, Meehan RR, Adams IR. 2012. Microarray analysis of LTR retrotransposon silencing identifies Hdac1 as a regulator of retrotransposon expression in mouse embryonic stem cells. PLoS Comput. Biol. 8:e1002486 doi:10.1371/journal.pcbi.1002486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weiss RA. 2006. The discovery of endogenous retroviruses. Retrovirology 3:67 doi:10.1186/1742-4690-3-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boyer LA, Mathur D, Jaenisch R. 2006. Molecular control of pluripotency. Curr. Opin. Genet. Dev. 16:455–462 [DOI] [PubMed] [Google Scholar]
- 11. Jones PA, Liang G. 2009. Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 10:805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klose RJ, Bird AP. 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31:89–97 [DOI] [PubMed] [Google Scholar]
- 13. Lewinski MK, Bisgrove D, Shinn P, Chen H, Hoffmann C, Hannenhalli S, Verdin E, Berry CC, Ecker JR, Bushman FD. 2005. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 79:6610–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Plachy J, Kotab J, Divina P, Reinisova M, Senigl F, Hejnar J. 2010. Proviruses selected for high and stable expression of transduced genes accumulate in broadly transcribed genome areas. J. Virol. 84:4204–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Senigl F, Auxt M, Hejnar J. 2012. Transcriptional provirus silencing as a crosstalk of de novo DNA methylation and epigenomic features at the integration site. Nucleic Acids Res. 40:5298–5312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shan L, Yang HC, Rabi SA, Bravo HC, Shroff NS, Irizarry RA, Zhang H, Margolick JB, Siliciano JD, Siliciano RF. 2011. Influence of host gene transcription level and orientation on HIV-1 latency in a primary-cell model. J. Virol. 85:5384–5393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Greger JG, Katz RA, Ishov AM, Maul GG, Skalka AM. 2005. The cellular protein daxx interacts with avian sarcoma virus integrase and viral DNA to repress viral transcription. J. Virol. 79:4610–4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poleshko A, Palagin I, Zhang R, Boimel P, Castagna C, Adams PD, Skalka AM, Katz RA. 2008. Identification of cellular proteins that maintain retroviral epigenetic silencing: evidence for an antiviral response. J. Virol. 82:2313–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wolf D, Cammas F, Losson R, Goff SP. 2008. Primer binding site-dependent restriction of murine leukemia virus requires HP1 binding by TRIM28. J. Virol. 82:4675–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wolf D, Goff SP. 2008. Host restriction factors blocking retroviral replication. Annu. Rev. Genet. 42:143–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wolf D, Goff SP. 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131:46–57 [DOI] [PubMed] [Google Scholar]
- 22. Svoboda J, Hejnar J, Geryk J, Elleder D, Vernerova Z. 2000. Retroviruses in foreign species and the problem of provirus silencing. Gene 261:181–188 [DOI] [PubMed] [Google Scholar]
- 23. Barr SD, Leipzig J, Shinn P, Ecker JR, Bushman FD. 2005. Integration targeting by avian sarcoma-leukosis virus and human immunodeficiency virus in the chicken genome. J. Virol. 79:12035–12044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2:e234 doi:10.1371/journal.pbio.0020234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Narezkina A, Taganov KD, Litwin S, Stoyanova R, Hayashi J, Seeger C, Skalka AM, Katz RA. 2004. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 78:11656–11663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang X, Khosravi-Far R, Chang HY, Baltimore D. 1997. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell 89:1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF, III, Maul GG. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Torii S, Egan DA, Evans RA, Reed JC. 1999. Human Daxx regulates Fas-induced apoptosis from nuclear PML oncogenic domains (PODs). EMBO J. 18:6037–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. 2002. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J. Cell Sci. 115:3319–3330 [DOI] [PubMed] [Google Scholar]
- 30. Li R, Pei H, Watson DK, Papas TS. 2000. EAP1/Daxx interacts with ETS1 and represses transcriptional activation of ETS1 target genes. Oncogene 19:745–753 [DOI] [PubMed] [Google Scholar]
- 31. Morozov VM, Massoll NA, Vladimirova OV, Maul GG, Ishov AM. 2008. Regulation of c-met expression by transcription repressor Daxx. Oncogene 27:2177–2186 [DOI] [PubMed] [Google Scholar]
- 32. Puto LA, Reed JC. 2008. Daxx represses RelB target promoters via DNA methyltransferase recruitment and DNA hypermethylation. Genes Dev. 22:998–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang L, Xu GL, Zhang JQ, Tian L, Xue JL, Chen JZ, Jia W. 2008. Daxx interacts with HIV-1 integrase and inhibits lentiviral gene expression. Biochem. Biophys. Res. Commun. 373:241–245 [DOI] [PubMed] [Google Scholar]
- 34. Saffert RT, Kalejta RF. 2007. Human cytomegalovirus gene expression is silenced by Daxx-mediated intrinsic immune defense in model latent infections established in vitro. J. Virol. 81:9109–9120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salomoni P, Michod D. 2009. End of the Daxx diatribe? Cell Cycle 8:1467–1468 [PubMed] [Google Scholar]
- 36. Barsov EV, Hughes SH. 1996. Gene transfer into mammalian cells by a Rous sarcoma virus-based retroviral vector with the host range of the amphotropic murine leukemia virus. J. Virol. 70:3922–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Katz RA, Jack-Scott E, Narezkina A, Palagin I, Boimel P, Kulkosky J, Nicolas E, Greger JG, Skalka AM. 2007. High-frequency epigenetic repression and silencing of retroviruses can be antagonized by histone deacetylase inhibitors and transcriptional activators, but uniform reactivation in cell clones is restricted by additional mechanisms. J. Virol. 81:2592–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li H, Leo C, Zhu J, Wu X, O'Neil J, Park EJ, Chen JD. 2000. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol. Cell. Biol. 20:1784–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saffert RT, Kalejta RF. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 80:3863–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Escobar-Cabrera E, Lau DK, Giovinazzi S, Ishov AM, McIntosh LP. 2010. Structural characterization of the DAXX N-terminal helical bundle domain and its complex with Rassf1C. Structure 18:1642–1653 [DOI] [PubMed] [Google Scholar]
- 41. Hwang J, Kalejta RF. 2007. Proteasome-dependent, ubiquitin-independent degradation of Daxx by the viral pp71 protein in human cytomegalovirus-infected cells. Virology 367:334–338 [DOI] [PubMed] [Google Scholar]
- 42. Li XD, Makela TP, Guo D, Soliymani R, Koistinen V, Vapalahti O, Vaheri A, Lankinen H. 2002. Hantavirus nucleocapsid protein interacts with the Fas-mediated apoptosis enhancer Daxx. J. Gen. Virol. 83:759–766 [DOI] [PubMed] [Google Scholar]
- 43. Lukashchuk V, Everett RD. 2010. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J. Virol. 84:4026–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schreiner S, Wimmer P, Sirma H, Everett RD, Blanchette P, Groitl P, Dobner T. 2010. Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells. J. Virol. 84:7029–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tang Q, Maul GG. 2003. Mouse cytomegalovirus immediate-early protein 1 binds with host cell repressors to relieve suppressive effects on viral transcription and replication during lytic infection. J. Virol. 77:1357–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tsai K, Thikmyanova N, Wojcechowskyj JA, Delecluse HJ, Lieberman PM. 2011. EBV tegument protein BNRF1 disrupts DAXX-ATRX to activate viral early gene transcription. PLoS Pathog. 7:e1002376 doi:10.1371/journal.ppat.1002376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Senigl F, Plachy J, Hejnar J. 2008. The core element of a CpG island protects avian sarcoma and leukosis virus-derived vectors from transcriptional silencing. J. Virol. 82:7818–7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hatziioannou T, Bieniasz PD. 2011. Antiretroviral restriction factors. Curr. Opin. Virol. 1:526–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hollenbach AD, Sublett JE, McPherson CJ, Grosveld G. 1999. The Pax3-FKHR oncoprotein is unresponsive to the Pax3-associated repressor hDaxx. EMBO J. 18:3702–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Scherbik SV, Stockman BM, Brinton MA. 2007. Differential expression of interferon (IFN) regulatory factors and IFN-stimulated genes at early times after West Nile virus infection of mouse embryo fibroblasts. J. Virol. 81:12005–12018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shimoda K, Kamesaki K, Numata A, Aoki K, Matsuda T, Oritani K, Tamiya S, Kato K, Takase K, Imamura R, Yamamoto T, Miyamoto T, Nagafuji K, Gondo H, Nagafuchi S, Nakayama K, Harada M. 2002. tyk2 is required for the induction and nuclear translocation of Daxx which regulates IFN-alpha-induced suppression of B lymphocyte formation. J. Immunol. 169:4707–4711 [DOI] [PubMed] [Google Scholar]
- 52. Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837 [DOI] [PubMed] [Google Scholar]
- 53. Daniel R, Ramcharan J, Rogakou E, Taganov KD, Greger JG, Bonner W, Nussenzweig A, Katz RA, Skalka AM. 2004. Histone H2AX is phosphorylated at sites of retroviral DNA integration but is dispensable for postintegration repair. J. Biol. Chem. 279:45810–45814 [DOI] [PubMed] [Google Scholar]
- 54. Hejnar J, Svoboda J, Geryk J, Fincham VJ, Hak R. 1994. High rate of morphological reversion in tumor cell line H-19 associated with permanent transcriptional suppression of the LTR, v-src, LTR provirus. Cell Growth Differ. 5:277–285 [PubMed] [Google Scholar]
- 55. Searle S, Gillespie DA, Chiswell DJ, Wyke JA. 1984. Analysis of the variations in proviral cytosine methylation that accompany transformation and morphological reversion in a line of Rous sarcoma virus-infected Rat-1 cells. Nucleic Acids Res. 12:5193–5210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. 2010. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24:1253–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. 2010. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. U. S. A. 107:14075–14080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ullman AJ, Hearing P. 2008. Cellular proteins PML and Daxx mediate an innate antiviral defense antagonized by the adenovirus E4 ORF3 protein. J. Virol. 82:7325–7335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schreiner S, Martinez R, Groitl P, Rayne F, Vaillant R, Wimmer P, Bossis G, Sternsdorf T, Marcinowski L, Ruzsics Z, Dobner T, Wodrich H. 2012. Transcriptional activation of the adenoviral genome is mediated by capsid protein VI. PLoS Pathog. 8:e1002549 doi:10.1371/journal.ppat.1002549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Du YC, Lewis BC, Hanahan D, Varmus H. 2007. Assessing tumor progression factors by somatic gene transfer into a mouse model: Bcl-xL promotes islet tumor cell invasion. PLoS Biol. 5:e276 doi:10.1371/journal.pbio.0050276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yoder JA, Walsh CP, Bestor TH. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335–340 [DOI] [PubMed] [Google Scholar]
- 62. Hejnar J, Hajkova P, Plachy J, Elleder D, Stepanets V, Svoboda J. 2001. CpG island protects Rous sarcoma virus-derived vectors integrated into nonpermissive cells from DNA methylation and transcriptional suppression. Proc. Natl. Acad. Sci. U. S. A. 98:565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cedar H, Bergman Y. 2009. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 10:295–304 [DOI] [PubMed] [Google Scholar]
- 64. Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A, Verdin E, Olive D, Van Lint C, Hejnar J, Hirsch I. 2009. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 5:e1000554 doi:10.1371/journal.ppat.1000554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cherry SR, Biniszkiewicz D, van Parijs L, Baltimore D, Jaenisch R. 2000. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol. Cell. Biol. 20:7419–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Golding MC, Zhang L, Mann MR. 2010. Multiple epigenetic modifiers induce aggressive viral extinction in extraembryonic endoderm stem cells. Cell Stem Cell 6:457–467 [DOI] [PubMed] [Google Scholar]
- 67. He J, Yang Q, Chang LJ. 2005. Dynamic DNA methylation and histone modifications contribute to lentiviral transgene silencing in murine embryonic carcinoma cells. J. Virol. 79:13497–13508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pannell D, Osborne CS, Yao S, Sukonnik T, Pasceri P, Karaiskakis A, Okano M, Li E, Lipshitz HD, Ellis J. 2000. Retrovirus vector silencing is de novo methylase independent and marked by a repressive histone code. EMBO J. 19:5884–5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. 2009. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5:e1000495 doi:10.1371/journal.ppat.1000495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Roe T, Chow SA, Brown PO. 1997. 3′-end processing and kinetics of 5′-end joining during retroviral integration in vivo. J. Virol. 71:1334–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vandegraaff N, Kumar R, Burrell CJ, Li P. 2001. Kinetics of human immunodeficiency virus type 1 (HIV) DNA integration in acutely infected cells as determined using a novel assay for detection of integrated HIV DNA. J. Virol. 75:11253–11260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51–61 [DOI] [PubMed] [Google Scholar]
- 73. Woodhall DL, Groves IJ, Reeves MB, Wilkinson G, Sinclair JH. 2006. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J. Biol. Chem. 281:37652–37660 [DOI] [PubMed] [Google Scholar]
- 74. Muromoto R, Sugiyama K, Takachi A, Imoto S, Sato N, Yamamoto T, Oritani K, Shimoda K, Matsuda T. 2004. Physical and functional interactions between Daxx and DNA methyltransferase 1-associated protein, DMAP1. J. Immunol. 172:2985–2993 [DOI] [PubMed] [Google Scholar]