Abstract
This protocol describes an EDTA-based passaging procedure to be used with chemically defined E8 medium that serves as a tool for basic and translational research into human pluripotent stem cells (iPSCs). In this protocol, passaging one six-well or 10 cm plate of cells takes about 6–7 min. This enzyme-free protocol achieves maximum cell survival without enzyme neutralization, centrifugation, or drug treatment. It also allows for higher throughput, requires minimal material and limits contamination. Here we describe how to produce a consistent E8 medium for routine maintenance and reprogramming and how to incorporate the EDTA-based passaging procedure into human induced PSC (iPSC) derivation, colony expansion, cryopreservation and teratoma formation. This protocol has been successful in routine cell expansion, and efficient for expanding large-volume cultures or a large number of cells with preferential dissociation of PSCs. Effective for all culture stages, this procedure provides a consistent and universal approach to passaging human pluripotent stem cells in E8 medium.
Keywords: Cell culture, human embryonic stem cells, induce pluripotent stem cells Cell survival, cell dissociation
Introduction
The study of human embryonic stem cells (ESCs), the first established human PSC lines 1,2, led to the derivation of iPSCs from human somatic cells3–5. Human ESCs and iPSCs are both capable of generating cells from all three germ lines and have thus gathered tremendous interest for their practical and scientific values. Maintained in cell culture, both are affected by different culture conditions and cell handling techniques.
Cell culture conditions for human ESCs and iPSCs have evolved from feeder-dependent and feeder-free medium to defined medium on defined extracellular matrix (ECM) 1,2,6–9. Improved understanding of self-renewal and differentiation means that growth media and ECM are becoming increasingly defined and simplified. At the same time, multiple cell handling techniques have evolved to facilitate PSC research. However, no single technique can be used at every stage of culture, from expansion through differentiation and cryopreservation.
Dissociation of ESCs
For regular cell culture practices, cells are often individualized during passaging to achieve even distribution and uniform treatments. However, human ESCs survive poorly after individualization (i.e., being made single cell), because these cells are more sensitive to treatments and are prone to cell death, a fact that has made the development of a universal dissociation method particularly challenging. In routine experiments, dissociation methods are chosen on the basis of either cell survival or sensitivity. In regular expansion, cell survival is the priority. ESCs and iPSCs are usually passaged as aggregates with enzymatic dissociation, with collagenase used for culture on feeder cells 1,2 and Dispase used for culture on feeder-free cells8. Mechanical approaches, such as cell scrapers and other passaging tools, have also been developed to dissociate cells as aggregates. Furthermore, in a differentiation or transfection experiment, TryPLE and Accutase can be used to individualize ESCs, but poor survival often leads to abnormal karyotypes 10,11,12; small chemicals, such as Rho-associated protein kinase (ROCK) inhibitors, must be used to boost cell survival in this process13. All these methods require specialized tools or reagents that are costly for long-term or large-scale experiments. At the same time, the consistency of enzymatic methods is usually affected by the quality of enzymes from batch to batch. Given the variability of these methods, it is highly desirable to find a universal approach for all purposes.
The quality of culture conditions is also crucial to the maintenance and expansion of the PSCs. The medium components related to feeder cells or animal products often greatly affect the consistency of the cell culture, which could be even more problematic when cells have potential applications in translational research. To improve the consistency of cell culture, we systematically analyzed the functions of individual medium components, and finally formulated a xeno-free, chemically defined stem cell medium E8, which contains eight essential factors for human ESCs14. With minimal modification of E8, human iPSCs can be directly derived from skin biopsies in chemically defined conditions. This culture system could have great applications in future PSC research. In companion with the development of cell culture medium, a consistent xeno-free dissociation method is important to fulfilling the potential of the defined culture conditions.
We previously studied cell death mechanisms after individualization and found that myosin-actin dependent contraction leads to cell death, and that cell-cell adhesions promote survival by inhibiting the contraction15. Cells survived by reforming small aggregates in the first few hours after dissociation. This observation provided the theoretical foundation for a method that generates aggregates strong enough to survive, but small enough to be accessed by growth factors or transfection reagents. In combination with the development of chemically defined cell culture, we set out to devise a universal dissociation technique for the expansion, cryopreservation, and experimentation of human PSCs in defined conditions. We found that, after a specific EDTA treatment, cells can be partially dissociated to generate small aggregates that survive as well as Dispase-treated cells, and which are more accessible to various treatments. When used with the chemically defined E8 medium, this dissociation method can be used to handle human PSCs during derivation, expansion, and preservation. In this protocol we describe how to derive iPSCs from fibroblasts as an example of how to use our EDTA dissociation method and defined chemical culture conditions in each stage of the iPSC derivation process.
Development of the protocol
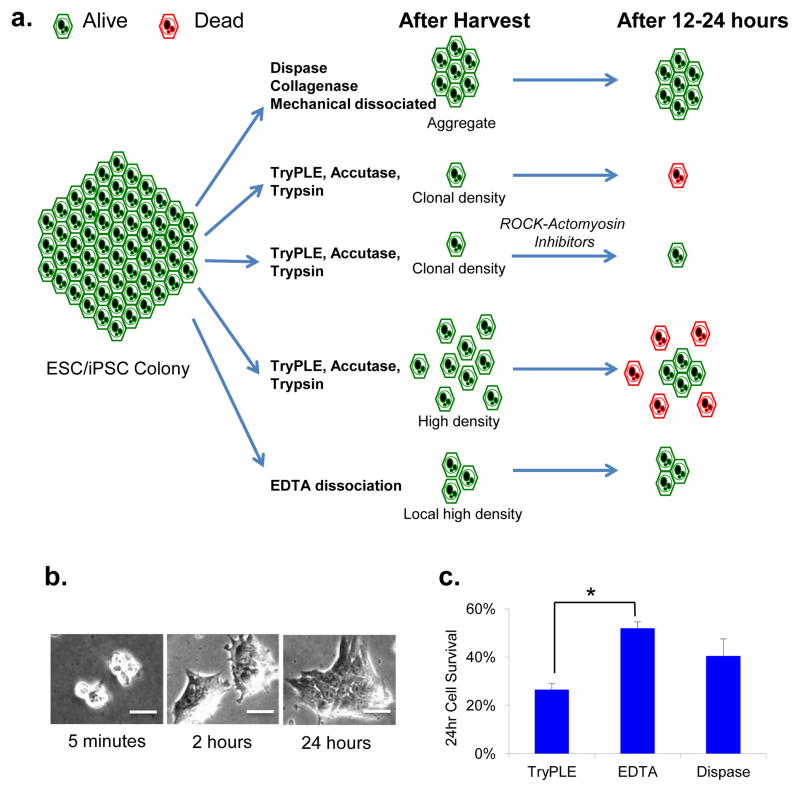
Conventionally, human PSCs are passaged as aggregates with enzymes, with individualized cells typically dying from actomyosin contraction. However, individualized cells can survive through the inhibition of ROCK-actin-myosin pathway components. Alternatively, cells can survive naturally through reaggregation of adjacent cells, indicating that higher local density could lead to reassociation efficiency independent of drug treatment (Fig. 1a). We found that EDTA treatment could partially dissociate human ESCs, and that ESCs were easily washed off with medium (Supplementary Fig. 1a). Loose adhesions between the cells generated very high local density beneficial to cell survival. Most cells were in small aggregates, which attached to a Matrigel-coated plate within minutes (5 min), spread in 2 h and survived as colonies (24 h; Fig. 1b). EDTA-dissociated ESCs survive significantly better than individualized cells by TryPLE, and the survival efficiency is similar to passaging cells as large aggregates using a Dispase protocol (Fig. 1c and Supplementary Methods). Cell survival when using this EDTA method is comparable to that observed in cells treated by ROCK inhibitor (Supplementary Fig. 1b), and it is E-cadherin dependent (Supplementary Fig. 1c). This indicates that cells harvested by EDTA survive without drug treatment, but require direct cell-cell adhesion.
Figure 1. EDTA dissociation is an effective method for pluripotent stem cell passaging.
a)Summary of cell survival efficiency by different methods. After dissociation, enzyme/mechanic-generated large aggregates survive efficiently; individualized cells die at clonal density, but survive in the presence of inhibitors or high loading density; EDTA-generated small aggregates survive efficiently by high local density without the help of drug treatment. b)Human ESC colony morphology after plating. H1 cells were partially dissociated with EDTA, and they attached to the plate in a few minutes, spread in 2 h, and survived as colonies (24 h). Scale bars, 50 μm. c)Comparison of cell survival of different passaging methods. Cells were dissociated by TrypLE, EDTA or Dispase and added onto Matrigel-coated plates; next, live cells were counted after 24 h. Asterisk (*) signifies that the survival by EDTA differed significantly (P < 0.05, n=3) from TrypLE at 24 h.
Because of the survival efficiency of EDTA dissociation, we have used it extensively in long-term culture expansion of human PSCs, on more than 50 different human ESCs and iPSCs in different feeder-free media, including TeSR and E8. In each of these cases, normal karyotypes were effectively maintained in long-term culture. The longest iPSC culture conducted in our research lasted more than 51 passages and longer than 6 months, and the cells maintained normal karyotypes (Supplementary Table 1 lists some of these ESC and iPSC lines). Cells dissociated by EDTA efficiently survived not only on Matrigel and vitronectin surfaces, but also on synthetic surface independent of ROCK inhibitors (Supplementary Fig. 1d). Because of its simplicity and consistency, this protocol has been used for our routine cell culture practices14–17, and other groups have successfully used EDTA dissociation to maintain human PSCs18.
EDTA dissociation is also useful in transfection and differentiation experiments (Supplementary Fig. 2 and Supplementary Methods). In transfection experiments, EDTA-dissociated cells were transfected efficiently while maintaining high survival rate (Supplementary Fig. 2a–c). At the same time, these cells were sensitive to growth factor-induced differentiation (Supplementary Fig. 2d). We hypothesize that EDTA dissociation enables small aggregates to survive better than individualized cells, and that the use of this method allows easier access to reagents used in differentiation experiments when compared with conventional aggregates collected with enzymes such as Dispase. The EDTA method can also be used to harvest PSCs for effective teratoma formation in immune-deficient mice. EDTA dissociation has been successfully used for all culture stages of human PSCs.
EDTA dissociation also improves the handling of PSCs mixed with differentiated cells. In reprogramming experiments iPSC colonies are mechanically isolated for colony expansion, and in the process they are often contaminated with unreprogrammed somatic cells. Conventionally, colony picking is used to enrich stem cells, but this approach is prone to bacterial and fungal contamination, and is not practical when too many lines are involved. We were able to avoid contamination by combining the advantages of E8 medium and the EDTA dissociation method. First, defined E8 medium allows stem cells to grow faster than fibroblasts, needing additional factors such as hydrocortisone. Even without colony isolation, PSCs could potentially outgrow somatic cells in a few passages with high passaging efficiency. Second, we found that PSCs respond to EDTA differently from somatic cells, such as fibroblasts. EDTA treatment preferentially harvests ESC colonies, leaving most fibroblast cells on the original plate (Supplementary Fig. 3a,b). In comparison, TrypLE ubiquitously dissociates both ESCs and fibroblasts.
These observations led us to use EDTA dissociation for iPSC derivation and colony expansion, which requires both human PSCs and somatic cells. In a conventional iPSC derivation procedure, cells are often passaged as individual cells, with ROCK inhibitors boosting iPSC survival before individual colonies are picked. However, we have used EDTA differential dissociation and harvested iPSCs with high survival rates without the help of a ROCK inhibitor (Supplementary Fig. 3c); we found that most EDTA-dissociated cells expressed the stem cell marker SSEA-4 (Supplementary Fig. 3d). EDTA dissociation thus enables the enrichment of potential iPSCs in an overcrowded reprogramming culture in which a secondary passaging is needed. We have also used EDTA dissociation to expand isolated iPSC colonies. We have often left differentiated cells in the original plate and found that stem cells reached a high purity in one or two passages. In addition, in EDTA-based cell expansion, we routinely maintained established ESC and iPSC lines with high purity (i.e., >95% of the population is positive for the stem cell marker Oct4) without enrichment by manual picking. It is possible that spontaneous differentiation is suppressed by the easy access to growth factors by small aggregates created by EDTA passaging, and that differential dissociation allows potential enrichment of stem cells after each passage while leaving some differentiated cells behind.
In addition to preferential dissociation, EDTA passaging has another advantage: it is a quick procedure with minimal opportunity for contamination. The procedure can be performed in a biosafety cabinet, and it does not require enzymatic neutralization and centrifugation. We are thus able to passage 24 individual lines in 15 minutes with minimal risk of contamination. In addition, as EDTA treatment does not damage the ECM, a portion of the iPSCs can be maintained in the original wells after dissociation, providing a backup or duplicate plate for maintenance or further characterizations.
Experimental Design
E8 medium production
This protocol describes how to produce chemically defined stem cell medium E8 with the formulas listed in Tables 1 and 2. To maintain consistency in the cell culture, we recommend making large quantities of medium each time, storing it in the freezer and performing a batch test before applying the medium in real experiments. A base medium, which can be used for all stages of stem cell culture, is first prepared according to Table 1 (see also Supplementary Table 2). Full E8 medium can be prepared with additional self-renewal essential factors (Table 2). The medium can also be prepared on a small scale with the reagents listed in Tables 1 and 2.
Table 1.
E8 Base Medium for 5% CO2.
| Reagent | Stock | Amount required for 1 liter of medium | Amount required for 50 liters of medium | Final concentration | Source |
|---|---|---|---|---|---|
| DMEM-F12 | 1 liter | 50 liters | Gibco, cat. no. 11330-032 | ||
| L-ascorbic acid | 64 mg | 3200 mg | 64 mg per liter | Sigma, cat. no. A8960 | |
| Sodium Selenite | 0.7 mg per ml | 19.4 μl | 970 μl | 13.6 μg per liter | Sigma, cat. no. S5261 |
| HCl or NaOH | Adjust pH to 7.4 | ||||
| Sodium Chloride | 1 g | 50 g | Sigma, cat. no. S5886 |
Freeze at −20°C for storage
Formula suitable for 10% CO2 is listed in Supplementary Table 2.
Table 2.
E8-related media for reprogramming and maintenance
| Reagent | E8(TGF-β1) | Reprogramming 1 | Reprogramming 2 | Final Concentration |
|---|---|---|---|---|
| E8 base medium | 500 ml | 500 ml | 500 ml | |
| Holo-transferrin | 500 μl | 500 μl | 500 μl | 10 μg per ml |
| bFGF | 500 μl | 500 μl | 500 μl | 100 ng per ml |
| TGF-β1 | 500 μl | --- | --- | 1.74 ng per ml |
| Insulin | 1 ml | 1 ml | 1 ml | 20 μg per ml |
| Hydrocortisone | --- | 50 μl | --- | 1 μM |
| Sodium butyrate | --- | 500 μl | 100 μM |
Routine cell culture expansion with EDTA dissociation
When the E8 medium is ready, we test it with H1 ESCs and one control fibroblast iPSC line. EDTA dissociation is used to passage the cells for at least five passages, and cells are then analyzed with flow cytometry and karyotyping. After the E8 medium passes the test, the frozen stock can be used for routine cell culture expansion and other experiments. We find that OCT4-positive cells consistently compose >95% of the whole population when we use the EDTA passaging method with E8 medium.
Derivation of human iPSCs from fibroblast cells
After E8 components pass the quality test, a set of E8-based media are used to reprogram human skin fibroblast cells. After transduction, cells are first cultured in E8-based reprogramming medium 1, and then in reprogramming medium 2 (Table 2). We usually perform the reprogramming experiment in a six-well plate so that multiple treatments can be applied to the cells in the same experiment (Fig. 2). Sodium butyrate treatment is usually used to improve reprogramming efficiency.
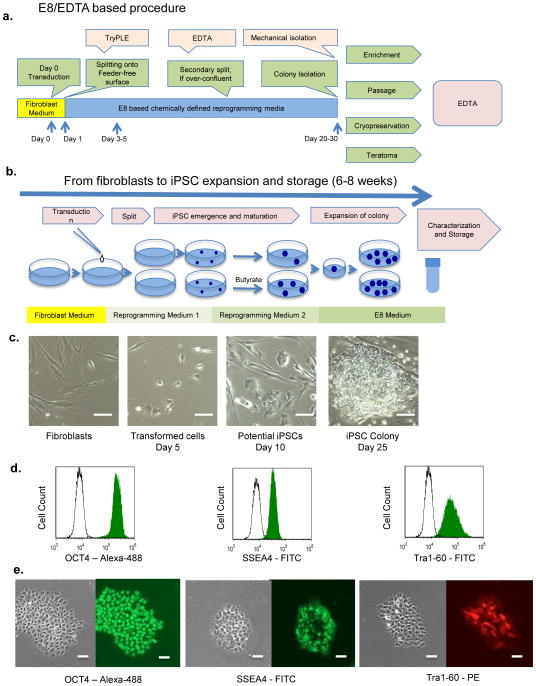
Figure 2. iPSC derivation from human fibroblast cells in chemically defined medium.
a)In the EDTA/E8-based iPSC derivation procedure, EDTA is the major handling method necessary for most cell culture stages. In comparison, the traditional procedure uses multiple dissociation methods (Supplementary Fig. 4a). b)A brief summary of the steps described in this protocol. c) Cell morphology changes in iPSC derivation process. Scale bars, 40 μm. d) FACS analysis of pluripotency markers on iPSCs derived from fibroblasts. Cells were derived in defined medium, and then expanded by the EDTA method. IgG control: unfilled peak; FITC-conjugated antibody: green peak. Antibodies: OCT4-Alexa-488 (Millipore, cat. no. FCMAB113A4); SSEA4-FITC (Biolegend, cat. no. 330410); Tra1-60-FITC (Millipore, cat. no. FCMAB115F); msIGg-FITC and nonimmune control (Millipore, cat. no. MABC006F). e)Immunostaining of pluripotency markers on iPSCs derived from fibroblasts. Left, phase-contrast images; right, fluorescence images. Scale bars, 20μm. Antibodies: OCT4-Alexa-488 (Millipore, cat. no. FCMAB113A4); SSEA4-FITC (Biolegend, cat. no. 330410); Tra1-60-PE (Biolegend, cat. no. 330610).
Reprogramming cells usually show up about 3–5 d after transduction, and the colonies mature 20–30 d after transduction. During this period, cells usually need to be passaged to avoid overconfluence, which can inhibit the emergence of true iPSC colonies. The EDTA method is applied to dissociate cells after the emergence of reprogramming cells, which preferentially enrich potential iPSCs.
The procedure has been successfully used in lentiviral-, sendai virus- and episomal DNA-based reprogramming. For simplicity, we used commercially available STEMCCA polycistron lentivirus in this protocol. This procedure could also be modified to reprogram human umbilical vein endothelial cells (HUVECs) and adipocyte cells.
Colony expansion and preservation of human iPSCs
When iPSCs are mature, individual colonies are mechanically isolated into a 24-well plate for colony expansion. Cells are subsequently expanded with the EDTA method, and can be cryopreserved in 3–5 d. The cells can be expanded and should be characterized by alkaline phosphatase staining (APS) and other appropriate characterizations. After further confirmation by karyotyping and immunostaining for pluripotency markers, validated iPSC lines can be used for teratoma formation with the EDTA method.
MATERIALS
REAGENTS
Cells
Human ESCs: H1 cells, WA01 (US National Institutes of Health (NIH), human ESC registry no. 0043)
Human iPSCs from fibroblasts: ND1 (NIH control iPSC line)
Human foreskin fibroblasts: CCD-1079Sk (ATCC cat. no. CRL-2097)
-
SCID-Beige mice (optional for teratoma only, step 28C;Taconic cat. no. CBSCBG-F)
Critical: Multiple mice are usually needed for each injection, and female mice are often used.
Caution: Experiments involving live rodents must conform to Local and National regulations.
Human STEMCCA Cre-Excisable Constitutive Polycistronic (OKSM) Lentivirus reprogramming kit (Millipore cat. no. SCR545)
Sodium Chloride (Sigma S5886 or equivalent)
Sodium Chloride (NaCl, Sigma, cat. no. S5886 or equivalent)
DMSO (Sigma, cat. no. D4540 or equivalent)
-
Matrigel, growth factor reduced (BD Biosciences, cat. no. 354230)
Critical: Always store in a non-antifreeze freezer.
Rock Inhibitor Y-27623 (Tocris, cat. no. 1254)
Holo-transferrin (Sigma, cat. no. T0665 or equivalent)
Recombinant human fibroblast growth factor (FGF, Peprotech, cat. no.100-18B or equivalent)
Recombinant human transforming growth factor (TGF-β1, R&D Systems, cat. no. 240-B/CF)
Insulin, human, 10 mg/mL solution (Sigma, cat. no. I9278-5ML)
Hydrocortisone (Sigma, cat. no. H-0396)
Sodium butyrate (Sigma, cat. no. 303410)
Dulbecco’s PBS, calcium and magnesium Free (Invitrogen, cat. no. 14190-250 or equivalent)
Polybrene (Sigma, cat. no. H9268)
TrypLE (Invitrogen, cat. no. 12563)
BCIP/NBT alkaline phosphatase substrate kit IV (Vector Laboratories, cat. no. SK-5400)
Tris-HCl, 100 mM, pH 9.5
EDTA, 0.5M, pH8.0 stock solution (K&D Medical cat. no. RGF-3130 or equivalent)
Human Serum Albumin(HSA, 0.05% (wt/vol); Sigma, cat. no. A6608; see Reagent Setup)
HSA in HCl in PBS (0.05% (wt/vol); Sigma, cat. no. A6608; see Reagent Setup)
Liquid Nitrogen
Ethanol
Dulbecco’s modified Eagle medium (DMEM)-F1
Sodium Selinite
L-ascorbic acid
Sodium Hydroxide
Table 3.
Troubleshooting table
| Step | Problem | Possible Causes | Solutions |
|---|---|---|---|
| 5 | Colonies not coming off | Pipetting too gently | Use a strong motion to rinse most cells off in one shot. |
| Insufficient treatment | Extend treatment time | ||
| Too many colonies come off during EDTA treatment | Treating for too long | Shorten treatment time or skip one washing step. | |
| Plate is disturbed too much during buffer change | Don’t shake or swirl the plate during the treatment. | ||
| Cells can be neutralized and collected with E8 medium. Either spin down and plate, or directly plate the suspension and change medium again after 1 h | |||
| 8 | Poor cell survival | Cells have not been passaged frequently enough | Cells need to be passaged around every 4 d. If the culture is beyond 5 d, add ROCK inhibitor |
| Cells are overconfluent | Passage the cells when the cells reach to 80% confluence. ROCK inhibitors could be added to help the survival. | ||
| Too much pipetting | Don’t repetitively pipet the cells. After sufficient treatment, cells are easily washed off the plate. | ||
| Overtreatment leads to too many single cells | Shorten the treatment time or skip one washing step. | ||
| EDTA concentration is too high | Decrease the EDTA concentration from 5 to 0.5–2 mM. | ||
| 9 | Poor pluripotency marker expression | Reagent quality | Batch test for reagents listed in Table 2. |
| 17 | Putative iPSCs do not come off the plate | Confluent plates need more EDTA to remove calcium | Increase EDTA treatment time or add more washing steps. |
| If iPSC colonies are rare, aim at potential iPSC colonies to specifically remove iPSCs. |
|||
| Poor cell survival | Cells are overconfluent | Add ROCK inhibitor. | |
| 26 | No attached colony | Colony is too big | Cut the colony to small pieces before harvest. |
| Non-iPSC morphology | Colonies picked may not have been iPSC colonies, improve iPSC identification | ||
| 27 | Poor survival | Cells are overconfluent | Add ROCK inhibitor |
| Move the culture to hypoxia condition (5% O2) |
Table 4.
Comparison of human PSC dissociation protocols.
| Method | Dispase | TrypLE/Accutase | EDTA |
|---|---|---|---|
| Cell Survival | Good | Poor | Good |
| ROCK inhibitor treatment | No | Essential | Not necessary |
| Rinsing or neutralization of the enzymes | Yes | Yes | No |
| Centrifugation | No | Yes | No |
| Time needed for one six-well plate | 7–10 min | 10–15 min | 5–6 min |
| Damage to extracellular matrix or membrane proteins | Yes | Yes | No |
| Dependency on different plate coating materialss14 | Not Applicable | High | Low |
| Accessible to transfection | Less Efficient | Efficient | Efficient |
| Differentiation | Less sensitive | Sensitive | Sensitive |
| Cryopreservation | Good19 | Good with ROCK inhibitors20 | Good, ROCK inhibitor optional |
| Stem cell enrichment | No | No | Yes |
| Batch Consistency | Poor | Good | Good |
| Teratoma formation | Good | Poor | Good |
| Source | Commercial | Commercial | Homemade |
| Cost | High | High | Low |
EQUIPMENT
Laminar Flow Hood with Vacuum
Centrifuge (Beckman Coulter Allegra X15-R with SX4750 swinging bucket rotor or equivalent)
Cell Culture Incubator (37°C, 95% humidity, 5% CO2 and 5%O2 atmosphere, Heracell, Thermo Scientific)
Inverted phase contrast microscope (x4 and x10 objectives, Zeiss or equivalent)
Freezer −80°C (Thermal Scientific)
Waterbath, 37°C Isopropanol Cell-Freezing Container
Cryovials, 1.2 ml (USA Scientific, cat. no. 1412-9100 or equivalent)
Six-well Nunclon Delta Surface tissue culture dishes (Thermo Scientific cat. no. 140675 or equivalent)
P1000 pipetman and sterile tips with filter
Pipet-aid and sterile 5- and 10-ml plastic disposable pipettes
Sterile filter units, 500 ml (Millipore, Stericup, cat. no. SCGPU05RE)
Conical tubes, 15 and 50 ml (Falcon, cat. nos. 352097 and 352098 or equivalent)
Eppendorf tube
Gauge needle (20.5)
Reagent Setup
EDTA dissociation solution
Add 500 μl 0.5 M EDTA (pH 8.0) stock into 500 ml calcium/magnesium-free PBS. Add 0.9g of NaCl and adjust the osmolarity to 340 mOsm. Sterilize the solution by filtration, and store it at 4°C for up to 6 months.
Critical: To achieve the least disturbance of cells during dissociation, the osmolarity of EDTA solution is designed to be the same as that of E8 medium.
Matrigel
See Box 1 for Matrigel Preparation
ROCK inhibitor
Y-27632 is the most commonly used ROCK inhibitor, and it can be dissolved in H2O or DMSO. Dissolve the Y-27632 in sterile H2O or sterile DMSO with a final concentration of 10 mM (1000x), then aliquot and store it at −80°C. The solution is stable for at least 1 year.
Critical: With the EDTA protocol, ROCK inhibitor is not as essential as it is in enzymatic protocols. In routine maintenance of iPSC lines, ROCK inhibitor treatment is optional. However, if the stem cell cultures are too confluent, or have not been passaged for 4 d or more, ROCK inhibitor can greatly increase cell survival. If most of the cells are individualized, ROCK inhibitor treatment is recommended.
Holo-transferrin (1000x)
Dissolve 500 mg of Holo-transferrin in 50 mL of Dulbecco’s PBS and filter to sterilize. This makes a 1000x (10 mg/ml) stock that can be divided into 500-μl aliquots and frozen at −80°C, at which temperature it is stable for at least 1 year.
FGF (1000x)
One milligram of FGF should be dissolved in 10 mL of sterile 0.05% HSA in PBS. This 1000x FGF (0.1 mg/ml) should be divided into 500-μl aliquots and frozen at −80°C, at which temperature it is stable for at least 1 year.
TGF-β1 (1000x)
TGFβ1 should be resuspended to a concentration of 1.74μg/ml (1000x) in sterile 0.05% HSA in 4 mM HCl in PBS. This is then aliquotted into 500-μl aliquots and frozen at −80°C; it is stable at this temperature for at least 1 year.
Hydrocortisone (10,000x)
A measure of 3.625 mg of hydrocortisone is dissolved in 1 ml of sterile water (10mM stock). This solution is then divided into 50-μl aliquots and frozen at −80°C; it is stable at this temperature for at least 1 year.
Sodium butyrate (1,000x)
A measure of 110 mg of sodium butyrate is dissolved in 10 ml of sterile water and filtered (100 mM stock). This is then divided into 500-μl aliquots and frozen at −80°C; it is stable at this temperature for at least 1 year.
Cryopreservation media (2x)
Add 2 ml of sterile DMSO and 20 μl 10 mM ROCK inhibitor stock into 8 ml E8 medium to make 2x cryopreservation medium for iPSCs in colony expansion. Medium can be stored at 4°C for up to 1 week.
Polybrene Stock (1000x)
Dissolve the polybrene in PBS to 1000x (6 μg/ml) and filter to sterilize. Freeze the solution in small aliquots at −80°C and store them for up to 1 year before use. The final concentration on cells during infection will be 6 ng/ml.
E8 base medium
Pour 50 liters of DMEM-F12 medium into a suitable container, and stir it on a stir plate. Save the empty bottles. Add 3,200 mg L-ascorbic acid and 970μl sodium selenite (0.7 mg/ml stock) into medium and mix well. Adjust the pH to 7.4 with 10 N NaOH. Add NaCl to adjust the osmolarity to 340 mOsm. Aliquot the medium back into the original bottles. The final composition of the medium should contain 64 mg/l L-ascorbic acid and 13.6μg/l sodium selenite. The details of how to make this medium are also given in Table 1. This base E8 medium should be frozen and stored at −20°C, it is stable at this temperature for at least 1 year.
Full E8 (TGFβ1) medium
Thaw one bottle (500 ml) of E8 base medium. Add 500μl of 1000x holo-transferrin, 500μl of 1000x FGF, 500μl of 1000x TGFβ1 and 1 ml 10 mg/ml insulin. Filter the E8 (TGFβ1) medium through a 500-ml Millipore Stericup filter. Store the medium at 4°C before use. The medium is stable for 2–3 weeks at 4°C. Do not warm this medium in a 37°C water bath before use, as it will shorten the life of the growth factors. Note that full E8 medium can now be purchased as Essential 8 media from Life Technologies (cat. no. A14666SA) or from Stem Cell Technologies (cat. no. 05840) as TeSR-E8. The details of how to make this medium are also given in Table 2.
Caution: These purchased media are only for stem cell maintenance. They contain TGF-β1, which can interfere with reprogramming.
Reprogramming media 1 and 2
Prepare 500 ml of reprogramming medium with the same formula as Full E8 medium, but leave out the TGF-β1. In place of the TGF-β1, either add 50μl 10,000s hydrocortisone (reprogramming Medium 1) or 500μl of 1000x sodium butyrate (reprogramming media 2) before filtering.
The details of how to make these media are also given in Table 2. Media can be stored at 4°C for up to 2 weeks.
HSA in PBS
Add 25 mg HAS to 50 ml of PBS and filter to sterilize. Store it for 1–2 months at 4°C or make aliquots and store them at −20°C for 1 year.
HAS in HCl-PBS
Dissolve 25 mg of HAS in 50 ml of 4 mM HCl in PBS and filter to sterilize. Store it for 1–2 months at 4°C or make aliquots and store them at −20°C for 1 year.
Procedure
Steps 1–9:Human PSC expansion with EDTA in full E8 Medium (TGF-β1 medium)
Timing, 6–7 minutes per plate
-
1|
Place a new six-well plate that has been coated with Matrigel (Box 1) and warmed to room temperature (RT, 20–25°C) in the tissue culture hood. In addition, keep the EDTA solution ready, which has been warmed to RT.
-
2|
Label the new Matrigel-coated plate and aspirate the Matrigel from the well. Replace with 1.5–2 ml full E8 medium (or E8 medium + ROCK Inhibitor if desired) per well. Critical Step: This step can also be performed during the EDTA incubation, but if a large number of cells are being passed, it is better to prepare plates ahead, as time is limited.
-
3|
Aspirate media from the ESCs or iPSCs that are about to split. Rinse the wells with 1 ml per well of EDTA solution to remove the magnesium and calcium from the medium. Critical Step: When rinsing, add the EDTA solution to the wells and then immediately aspirate the EDTA off again without incubating. Critical Step: When washing with EDTA, add the EDTA to the wall of the well slowly to avoid washing the colonies off the plate too early.
-
4|
Repeat the EDTA wash (as described in Step 3) a second time to ensure that all magnesium and calcium is removed. Add another 1 ml per well of the EDTA solution and let it sit for 2–5 min at RT within the hood. Critical Step: The longer the cells sit in the EDTA, the smaller the colonies that will result. In addition, it is important to keep the movement of the plate to a minimum to avoid lifting the colonies completely off the plate during this step.
-
5|
After 2–5 min, carefully aspirate the EDTA solution from the cells. Use 1–4 ml of E8 medium (Or E8 medium + ROCK inhibitor) and add it rapidly to wash the colonies off the plate and disperse the colonies as quickly as possible. The calcium and magnesium in the medium will neutralize any remaining EDTA. Critical Step: Do not break apart the colonies too much by excess pipetting. Critical Step: This and the next step should be performed quickly, as the colonies start to reattach to the plate very rapidly after the addition of calcium and magnesium.
-
6|
Ensure that the cell suspension is well mixed and then plate the desired cell amount per well in the newly prepared plate.
-
7|
Shake the plate back and forth and side to side to distribute the cells. Place the plate in an incubator. Let it sit overnight for maximum attachment.
-
8|
On the next day, take E8 medium out from a 4°C refrigerator, let it sit at RT to warm, if desired. Remove the medium from cells, and add 2 ml of E8 medium to each well in the six-well plate.
-
9|
On each following day, repeat Step 8, monitor cells daily, and when confluence reaches ~80% repeat Steps 1–7 (usually between 3 and 5 d after seeding). Passage the cells for the desired number of passages. After five passages we recommend collecting some cells for FACS to measure pluripotency markers and for karyotyping. If the FACS and karyotyping results are satisfactory, the media components are suitable for iPSC maintenance and reprogramming. This enzyme-free passaging protocol can be used during the derivation of iPS cells from other cell types. When you are ready to begin reprogramming, continue to the next section.
Steps 10–11:Transduction of human fibroblasts
Timing: 3 d
-
10|
Take a low-passage-number fibroblast culture and plate it in one well of a six-well dish so that cells will be about 80% confluent the next day (100,000 or 150,000 cells per well) in fibroblast medium (usually 10% (vol/vol) FBS and Penicillin-streptomycin, L-glutamine and non-essential amino acids in DMEM; however, if your fibroblasts are growing in another medium, use that medium). Incubate the cells overnight.
-
11|
Late on the next day, mix together 1.5 ml of fibroblast medium with polybrene (to obtain 6 ng/ml final concentration). Add Millipore STEM-CCA virus at the desired titer, add to mixture to the cells, and leave on the cells overnight.. Our current protocol utilizes a multiplicity of infection (MOI) of ~5–10. On the next day, if cells are nearly confluent, change medium to fresh fibroblast medium to remove polybrene. Coat two six-well plates with Matrigel, as described in Box 1.
Steps 12–18: Culture of transduced fibroblasts on Matrigel
Timing:3–4 weeks
-
12|
Still on the same day, passage the well of reprogramming cells using TrypLE. TrypLE dissociation preserves cells better than regular trypsin-EDTA, and it does not require serum-based neutralization. To do this, remove the medium from the cells and add 0.5 ml of TrypLE to the wells. Incubate for 5 min in an incubator, and then wash the cells off the plate with fibroblast medium into a centrifuge tube and dilute them fivefold with fibroblast medium to inactivate TrypLE. Critical Step: Some cell lines do not come off the plate well with TrypLE. With these lines, you may want to wash the plate twice with EDTA before adding TrypLE.
-
13|
Spin cells down for 5 min at RT at 200g in a swinging bucket rotor and resuspend them in 24 ml of fibroblast medium. Remove the Matrigel/DMEM-F12 from the two six-well plates and add 2 ml of cells per well. This gives a final passage of one well of infected cells into two six-well plates coated with Matrigel in fibroblast medium or directly into reprogramming medium 1.
-
14|
Maintain the cells in reprogramming medium 1, replacing medium every other day for 3–5d.
-
15|
Remove the medium and replace it with reprogramming medium 2. To improve reprogramming efficiency, add 100 μM sodium butyrate into medium 2 on one plate. Critical Step: The time at which to replace reprogramming media 1 with 2 is based on cell confluence. Medium 1 acts to promote fibroblast growth, and should be switched to medium 2 when the confluence reaches ~20–30%. If cells are still very sparse you may want to keep them in reprogramming medium 1 for a few more days.
-
16|
Continue replacing the medium every other day, using reprogramming medium 2 with or without sodium butyrate.
-
17|
Monitor the cells daily, if cells become too confluent while you are waiting for iPSC colonies to mature, cells may need to be passaged with EDTA at some point during the 2 weeks. Choose three or four wells to passage with EDTA onto two new Matrigel-coated plates (1:3 ratio). Use the EDTA passaging method as described in Steps 1–7 above, but using reprogramming medium 2 instead of full E8 (TGF-β1) medium.
-
18|
Twenty to twenty-five days after transduction, colonies should be ready for picking. At this point, proceed to Step 19. Around this time, begin feeding the original reprogramming plates full E8 (TGF-β1) medium daily.
Steps 19–27: Mechanical isolation of human iPSc colonies
Timing: 2–3 d
-
19|
For picking, prepare a 24-well plate by coating with Matrigel (as described in Box 1, using 250 ul per well of the Matrigel stock for a total of 1 mg of Matrigel per 24-well plate).
-
20|
After the 30-min Matrigel incubation, replace Matrigel/DMEM-F12 with full E8 (TGF-β1) medium containing 1x Rock inhibitor. Critical Step: Although Rock inhibitor is described as optional in the EDTA passaging method, ROCK inhibitor will greatly increase the survival chances of newly picked colonies.
-
21|
Spray 70% (vol/vol) ethanol on a microscope and on the surrounding area, as well as on a pipet and box of tips. Critical Step: We pick the colonies on a benchtop in the laboratory using a normal inverted light microscope. If space allows, a microscope can be placed inside a laminar flow hood or a bench top PCR clean hood to allow for a sterile field while picking colonies. If preferred, a dissection microscope can be used to find and manually passage the colonies.
-
22|
While wearing a facemask, find iPSC colonies under the microscope with the x4 objective. By using a P20 pipette with a tip, circle around the colony until it is loosened from surrounding cells.
-
23|
While still using a pipette tip, cross-hatch the colony so that it will come off the plate in smaller pieces.
-
24|
Next, use the pipette to push the colony off the plate and suck it into a pipette tip. Transfer the colony pieces into one well of the 24-well plate.
-
25|
Repeat Steps 22–24 with other colonies, placing one colony in each well of the 24-well plate. Incubate the cells overnight in the tissue culture incubator.
-
26|
On the day after picking, remove the medium and replace it with full E8 (TGF-β1) medium without rock inhibitor.
-
27|
Replace the E8 medium daily until the attached colony is big enough to passage. The colony is usually ready to passage after 2–3 d.
Step 28: Ongoing passage and analysis of colonies
Timing, 5–7 days
-
28|
When the colony is ready for passaging, use EDTA to passage as described in Steps 1–7. However, leave some of the cells in the original well of the 24-well plate while transferring most or them to a new Matrigel-coated well on a 12 - or 6-well plate (again in 1x Rock inhibitor). Continue to incubate the cells, replacing the medium daily and passing as described in Steps 1–7 when cells become confluent. Once you have one well in a six-well plate ready to passage, freeze two vials of cells (option A) and leave some cells (~10–20%) in the well to continue growing. Cells can now be further expanded for further cryopreservation (option A), APS staining (option B), analysis of teratoma formation (option C) and other characterizations such as FACS staining. Critical Step: When expanding each colony, keep track of passage number (manually picking and transferring into the 24-well plate, as described in Steps 19–24, is passage 1).
A) Cryopreservation and recovery of human iPSC colonies
Timing, 6–10 min per line
Split cells with 1:4 ratio 2–3 d before cryopreservation as described in Steps 1–7.
When cells reach to ~70–80% confluence, dissociate the cells with the EDTA method (as described in Steps 3 and 4).
After 2–5 min, carefully aspirate the EDTA solution from the cells and quickly add 1 ml of full E8 medium per well of a six-well plate.
In a dropwise manner, add an equal volume (1 ml) of 2x cryopreservation medium (20% (vol/vol) DMSO in full E8 medium) into the EDTA-harvested cell suspension to yield a final concentration of 10% (vol/vol) DMSO.
Aliquot 250–500 ul of cells into each labeled cryotube. For one well from a six-well plate with 70–80% confluence, it is possible to freeze four to eight vials. During early reprogramming, if iPSCs are more sparse, it is more common to freeze two vials from one well.
Freeze cells at −80°C in an isopropanol freezing container for at least 2 h.
-
Transfer the cells to liquid nitrogen for long-term storage.
Pause Point: Cells can be stored in liquid nitrogen for more than 5 years.
To thaw cells, take the cryotube out of the liquid nitrogen tank and place it directly in a 37°C water bath.
Gently stir the water with the tube, and check closely for the disappearance of ice.
When only a small ice particle is floating, spray the vial thoroughly with 70% (vol/vol) ethanol and transfer the tube into a biosafety cabinet.
Transfer the cells into a 15-ml conical tube
Add 10 ml of E8 medium dropwise while continuously mixing the solution in the tube.
Centrifuge cells at 200g for 5 minutes at 4°C
Carefully aspirate the supernatant and use E8 medium to resuspend the cells. Plate cells into one to three wells of a Matrigel coated six-well plate. ROCK inhibitors can be added into E8 medium when plating, if desired.
B) APS staining
Timing 1–2 h
When passaging colonies by the EDTA method (Steps 1–7), take a small portion of the cells and seed a 12-well Matrigel-coated plate.
When cells have grown and are ready to passage, they are ready for APS staining. By using the BCIP/NBT alkaline phospatase substrate kit IV, stain the cells for APS. First mix enough reagents for the wells you are staining (0.5 ml per well).
To 5 ml of 100 mM Tris-HCl, pH 9.5, add two drops of Reagent A and mix well. Add two drops of Reagent B and mix, then add two drops of reagent C and mix (note that Reagents A, B and C are part of the kit).
Remove the medium from the cells and add 0.5 ml per well of the APS staining reagent mix. Colonies are usually visible after 20 min. When the colonies are visible, remove the liquid; otherwise, wait until the colonies are stained with distinctive signal in contrast to surrounding cells.
C) Teratoma formation assay with the EDTA method
Timing 6–9 weeks
After iPSCs are confirmed by different characterization, select a cell line for teratoma formation assay. Split selected cells in a 1:4 ratio (as described in Steps 1–7). This should be done 2–3 d before you plan to inject.
When cells reach ~70–80% confluence, dissociate the cells with EDTA (as described in Steps 3–4). Meanwhile prethaw 200 μl Matrigel on ice and dilute it with 400 μl ice-cold E8 medium.
Collect the cells after the EDTA incubation with 1 ml of E8 medium per well (six-well plate).
Spin cells in a 1.5-ml Eppendorf tube at 100g for 5 minutes at RT.
Remove the supernatant and resuspend the pellet with 600 μl of the E8 medium/Matrigel mixture.
Take up the cell suspension into a 1000-μl syringe, and keep it on ice before injection.
By using a 20.5 gauge needle, inject 100–300 μl of suspension per mouse as soon as possible. For intramuscular injection, the cell suspension should be injected into the muscle center in the hind leg quadriceps along its long axis.
House mice normally and assess them regularly for the formation of teratomas. These should be seen at ~4 weeks after injection.
Around 2 weeks after teratomas are first detected, remove the teratoma for histological analysis. Caution: Ensure that the method used for removal follows all relevant animal handling and experimentation guidelines.
Troubleshooting
Troubleshooting advice can be found in Table 3.
Timing
Reagent Preparation, Large-scale medium preparation: 2 d
Steps 1–7, medium certification with EDTA passaging:6–7 min per plate
Steps 8 and 9, feeding and iPSC maintenance passaging every 3–4 d: (optional) continue for at least 20 d to certify a batch of medium
Steps 10 and 11, fibroblast culture and viral transduction: 3 d
Steps 12–18, reprogramming in chemically defined medium: 3–4 weeks
Steps 19–28, mechanical isolation and colony expansion:1–2 weeks
Step 28A, cryopreservation, 6–10 min per line
Step 28B, APS staining: 1–2 h
Step 28C, teratoma formation: 6–9 weeks.
ANTICIPATED RESULTS
Chemically defined E8 medium provides ideal cell culture conditions for human PSC research. By modifying a few growth factors, we are able to create cell culture conditions for human iPSC derivation and expansion (Tables 1 and 2). EDTA dissociation also provides a simple and efficient way to handle stem cells in different circumstances (Table 4). This combination of medium and handling method enables effective culture expansion and other treatments. All these experiments can be performed without drug treatment, but ROCK inhibitor is included in this protocol for those who prefer its use.
When human fibroblasts are reprogrammed in the E8-based feeder-free system, transformed cells are visible 5–6 d after transduction. It takes 20–30 d before true iPSC colonies mature enough for colony expansion (Fig. 2c). Overcrowded fibroblast cells often suppress the emergence of iPSC colonies, and thus EDTA dissociation could be used to differentially enrich iPSCs.
After mechanical isolation of iPSC colonies, EDTA dissociation enables effective expansion and cryopreservation in 1–2 weeks, markedly decreasing the time needed to preserve the cells. Owing to its simplicity, we are able to passage 24 individual lines from a 24-well plate in 15 minutes.
Expanded iPSCs should be first confirmed by APS staining and by pluripotency marker staining using flow cytometry or immunostaining (Fig. 2d,e and Supplementary Fig. 4b). Teratoma formation assays should be performed on selected iPSC lines by injecting cells into SCID mice. Teratomas usually emerge after 4 weeks, and are ready to be analyzed 6–7 weeks after injection.
Supplementary Material
Acknowledgments
This work was supported by NHLBI, NIH Common Fund through the Center for Regenerative Medicine (to G.C. and J.B.), the Charlotte Geyer Foundation, the Morgridge Institute for Research, NIH Grant UO1ES017166 (to J.A.T.), NIH contract RR-05-19 (to J.A.T.), NIH contract No. HHSN309200582085C (to J.J.) and private funds from the Wisconsin Alumni Research Foundation (to J.J.). We thank M. Boehm, T. Finkel and M. Rao for their suggestions. We thank K. Eastman for editorial assistance.
Footnotes
Contributions
G.C. and J.A.T. conceived the experiments and supervised the project; G.C. developed dissociation protocol; J.B. and G.C. performed the reprogramming experimental procedure in the paper; G.C. and D.R.G. demonstrated preferential dissociation of EDTA; N.G, J.J., D.R.G. and G.C. performed long-term culture; G.C., D.R.G. and J.B. performed teratoma formation assay; L.I.S. performed immunostaining and EDTA sequential dissociation imaging; J.B. and G.C. wrote the paper.
Competing Financial interests
J.A.T. is a founder, stockowner, consultant, and board member of Cellular Dynamics International. He also serves as scientific advisor to and has financial interests in Tactics II Stem Cell Ventures.
Contributor Information
Jeanette Beers, Email: beersj@nhlbi.nih.gov.
Daniel R. Gulbranson, Email: daniel.gulbranson@colorado.edu.
Nicole George, Email: ngeorge@wicell.org.
Lauren I. Siniscalchi, Email: lsiniscalchi.22@gmail.com.
Jeffrey Jones, Email: jjones@wicell.org.
James A. Thomson, Email: jthomson@morgridgeinstitute.org.
References
- 1.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 3.Yu JY, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu JY, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Xu RH, et al. Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat Methods. 2005;2:185–190. doi: 10.1038/nmeth744. [DOI] [PubMed] [Google Scholar]
- 7.Levenstein ME, et al. Basic fibroblast growth factor support of human embryonic stem cell self-renewal. Stem Cells. 2006;24:568–574. doi: 10.1634/stemcells.2005-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ludwig TE, et al. Feeder-independent culture of human embryonic stem cells. Nat Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- 9.Ludwig TE, et al. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol. 2006;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- 10.Ellerstrom C, Strehl R, Noaksson K, Hyllner J, Semb H. Facilitated expansion of human embryonic stem cells by single-cell enzymatic dissociation. Stem Cells. 2007;25:1690–1696. doi: 10.1634/stemcells.2006-0607. [DOI] [PubMed] [Google Scholar]
- 11.Bajpai R, Lesperance J, Kim M, Terskikh AV. Efficient propagation of single cells Accutase-dissociated human embryonic stem cells. Mol Reprod Dev. 2008;75:818–827. doi: 10.1002/mrd.20809. [DOI] [PubMed] [Google Scholar]
- 12.Thomson A, et al. Human embryonic stem cells passaged using enzymatic methods retain a normal karyotype and express CD30. Cloning Stem Cells. 2008;10:89–106. doi: 10.1089/clo.2007.0072. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe K, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 14.Chen G, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen G, Hou Z, Gulbranson DR, Thomson JA. Actin-myosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell. 2010;7:240–248. doi: 10.1016/j.stem.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howden SE, et al. Genetic correction and analysis of induced pluripotent stem cells from a patient with gyrate atrophy. Proc Natl Acad Sci USA. 2011;108:6537–6542. doi: 10.1073/pnas.1103388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen G, Gulbranson DR, Yu P, Hou Z, Thomson JA. Thermal stability of fibroblast growth factor protein is a determinant factor in regulating self-renewal, differentiation, and reprogramming in human pluripotent stem cells. Stem Cells. 2012;30:623–630. doi: 10.1002/stem.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng X, et al. Cnot1, Cnot2, and Cnot3 maintain mouse and human ESC identity and inhibit extraembryonic differentiation. Stem Cells. 2012;30:910–922. doi: 10.1002/stem.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ware CB, Nelson AM, Blau CA. Controlled-rate freezing of human ES cells. Biotechniques. 2005;38:879–880. 882–883. doi: 10.2144/05386ST01. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Krawetz R, Liu S, Meng G, Rancourt DE. ROCK inhibitor improves survival of cryopreserved serum/feeder-free single human embryonic stem cells. Hum Reprod. 2009;24:580–589. doi: 10.1093/humrep/den404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.