Summary
Programmmed necrosis is a form of cell death that involves membrane compartment swelling, cell rupture and an immune response. Although long recognized as a normal component of animal development, programmed necrosis remains poorly understood. Recent studies identify MLKL and PGAM5 as factors downstream of the protein kinases RIP1 and RIP3 in programmed necrosis.
Cell death by necrosis, unlike other forms of programmed cell death, is immunogenic and associated with many disease pathologies. Recent work indicates that death cytokines in the TNF family can induce a “programmed” form of necrosis through the kinases RIP1 and RIP3 when caspases are inhibited. Genetic experiments have recently determined that RIP1 and RIP3-driven programmed necrosis is crucial during embryonic development, certain viral infections, tissue injury/inflammation, and clonal expansion of antigen-specific lymphocytes (reviewed in (Oberst and Green, 2011)). Beyond RIP1 and RIP3, however, our understanding of the molecular regulation of programmed necrosis remains limited.
The kinase functions of RIP1 and RIP3 are essential for programmed necrosis. Therefore, identifying their substrates holds the key in understanding the regulation of this poorly defined process. Because pharmacological inhibition of RIP1 abolished RIP3 recruitment and activation of the necrosis inducing complex, RIP1 is likely the upstream activating kinase for RIP3 (Cho et al., 2009; He et al., 2009). By contrast, the substrates of RIP3 have remained elusive. Although mitochondrial enzymes were previously implicated as substrates for RIP3, their roles in programmed necrosis have yet to be independently validated (Zhang et al., 2009). In this issue, Wang and colleagues provide convincing evidence that mixed lineage kinase domain-like (MLKL) and phosphoglycerate mutase 5 (PGAM5) are integral parts of the necrosis signaling machinery downstream of RIP1 and RIP3 activation, and are RIP3 substrates (Sun et al., 2011; Wang et al., 2011).
The authors used a chemical library screening approach to identify the small molecule necrosulfonamide (NSA) that was shown to block programmed necrosis downstream of RIP3. MLKL was then identified as a RIP3 interacting partner using a modified form of NSA and biochemical purification of RIP3 complexes in necrotic cells. Decreased MLKL function by RNAi protected cells against TNF-induced necrosis. MLKL was strongly recruited to RIP3 upon necrosis induction, although a low level of interaction was detected in untreated cells. Phosphorylation of RIP3 at S227 is critical for MLKL binding and subsequent phosphorylation of MLKL at T357 and S358. Alanine substitutions at these phosphorylation sites abrogated the ability of RIP3 and MLKL to signal for necrosis. These results established MLKL as a functional substrate of RIP3. Because MLKL does not possess enzymatic function, it likely serves as an adaptor to bring the RIP1-RIP3 necrosome into proximity with other RIP3 substrates and downstream effectors (Fig. 1).
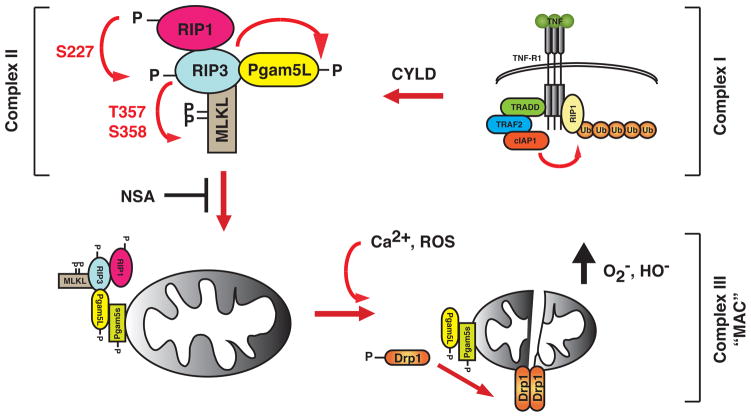
Figure 1.
Sequential recruitment and activation of necrosis signaling complexes. RIP1 is recruited to the activated TNFR-1 undergoes heavy ubiquitination by E3 ligases such as TRAF2 and cIAP-1. This membrane and receptor associated complex, termed Complex I, is responsible for NF-κB activation. As the membrane-associated Complex I become internalized, deubiquitinases such as cylindromatosis (CYLD) removes the polyubiquitin chains on RIP1. The deubiquitination of RIP1 and inhibition of caspase 8 is crucial for the assembly of the secondary signaling complex (Complex II). At this cytoplasmic complex, RIP1 likely phosphorylates RIP3 at S227, which in turn phosphorylates PGAM5L and MLKL at T357 and S358. These phosphorylation events are important for the RIP3 necrosis signaling complex to engage PGAM5s on the mitochondrial membrane, a step that is inhibited by the small molecule inhibitor NSA. Once activated by phosphorylation, the PGAM5L/PGAM5s complex dephosphorylates the mitochondrial fission regulator Drp1 to induce its dimerization and activation. Excessive Drp1 activity could lead to disruption of mitochondrial functions and other organelle and membrane damages that cumulates in programmed necrosis. The PGAM5L-PGAM5s-Drp1 mitochondrial attack complex (MAC) could also be activated by calcium flux and surge of intracellular reactive oxygen species (ROS).
Small molecule inhibitors have significantly advanced our understanding of programmed necrosis. The RIP1 kinase inhibitor necrostatin-1 (Nec-1) (Degterev et al., 2008) and NSA inhibit distinct steps of programmed necrosis and provide critical insight into the hierarchical relationships among the factors controlling this process. Unlike Nec-1, NSA inhibits necrosis by covalently modifying MLKL. In the presence of NSA, RIP3 puncta failed to enlarge, yet MLKL and PGAM5L binding to RIP3 was enhanced. This finding suggests that RIP1-RIP3-MLKL-PGAM5L form a dynamic and transient complex that dissolves over time as the tertiary complex of PGAM5s-Drp-1 (Complex III) becomes activated (Fig. 1). The covalent modification of MLKL by NSA likely stabilizes the RIP3 complex and prevents it from engaging PGAM5s. Interestingly, NSA inhibits necrosis in human cells, but not in mouse cells. This is due to the absence of a critical cysteine in the mouse MLKL sequence. The covalent modification of MLKL by NSA can potentially be useful for positive identification of necrotic cells, since biotinylated NSA also inhibits necrosis.
PGAM5, a mitochondrial phosphoglycerate mutase that can also function as a serine/threonine phosphatase, was identified as another substrate of RIP3 and a component of the RIP3-containing complex. Genetic evidence in Drosophila suggests that PGAM5 regulates the function of the Parkinson’s disease gene PINK1, suggesting a possible link between necrosis and neurodegeneration (Imai et al., 2010). PGAM5 exists in two isoforms, and each one appears to have a distinct function in necrosis. PGAM5L, but not PGAM5s, was detected in mild detergent-soluble RIP3-MLKL complex. NSA increased rather than inhibited the recruitment of PGAM5L to RIP3. Moreover, it inhibited PGAM5s, but not PGAM5L phosphorylation. Thus, PGAM5L is the first PGAM5 isoform to be recruited to the RIP3 complex. In contrast, PGAM5s was only found in SDS-soluble heavy membrane fraction. NSA inhibited the redistribution of PGAM5L to the SDS-soluble membrane fraction. Thus, PGAM5L tethers the RIP1-RIP3-MLKL necrosome (Complex II) to PGAM5s on the mitochondrial membrane (Fig. 1). This event appears to be critical for activation of Drp1, a GTPase that is essential for mitochondrial fission. PGAM5L dephosphorylates Drp-1 at S637 and activates its GTPase activity. Strikingly, either NSA or knock-down of MLKL by RNAi prevented mitochondria fragmentation upon necrosis induction. RNAi-mediated silencing of either PGAM5 or Drp-1, or pharmacological inhibition of Drp-1 prevented mitochondrial fragmentation and necrosis. These results suggest a tantalizing mechanism in which the RIP1-RIP3 necrosome complex signals for necrosis by engaging the mitochondrial fission machinery. Significantly, the “mitochondria attack complex” consisting of PGAM5 and Drp-1, but neither RIP1, RIP3 nor MLKL, are also required for necrosis induced by signals such as calcium flux and oxidative stress. Thus, these results provide strong evidence that extrinsic (e.g. TNF-like death cytokines) and intrinsic signals (e.g. oxidative stress) converge upon the PGAM5-Drp-1 axis to induce necrosis.
These studies provide a significant advance in our understanding of programmed necrosis by identifying 2 downstream effectors of this process. Necrotic cell death has long been neglected as a process lacking physiological relevance even though early studies observed the morphological hallmarks of this process in developing mammalian embryos. A blueprint for programmed necrosis is now emerging, but much remains to be learned about how different forms of cell death are regulated, how they may influence each other, and whether we can augment certain types of cell death as a therapeutic approach to compensate for mutations in cell death pathways. Bla bla
References
- Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A, Hitomi J, Germscheid M, Ch’en IL, Korkina O, Teng X, Abbott D, Cuny GD, Yuan C, Wagner G, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He SD, Wang L, Miao L, Wang T, Du FH, Zhao LP, Wang XD. Receptor Interacting Protein Kinase-3 Determines Cellular Necrotic Response to TNF-alpha. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Imai Y, Kanao T, Sawada T, Kobayashi Y, Moriwaki Y, Ishida Y, Takeda K, Ichijo H, Lu BW, Takahashi R. The Loss of PGAM5 Suppresses the Mitochondrial Degeneration Caused by Inactivation of PINK1 in Drosophila. Plos Genet. 2010;6 doi: 10.1371/journal.pgen.1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst A, Green DR. It cuts both ways: reconciling the dual roles of caspase 8 in cell death and survival. Nat Rev Mol Cell Bio. 2011;12:757–763. doi: 10.1038/nrm3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wang H, Wang Z, He SD, Chen S, Liao D, Wang L, Yan J, Liu W, et al. Mixed Lineage Kinase Domain-Like Protein Mediates Necrosis Signaling Downstream of Receptor-Interacting Serine-Threonine Kinase 3. Cell. 2011 doi: 10.1016/j.cell.2011.11.031. This issue. [DOI] [PubMed] [Google Scholar]
- Wang ZG, Jiang H, Chen S, Du FH, Wang XD. The MItochondrial Phosphatase PGAM5 Functions at the Convergent Point of Multiple Necrotic Death Pathways. Cell. 2011 doi: 10.1016/j.cell.2011.11.030. This issue. [DOI] [PubMed] [Google Scholar]
- Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC, Dong MQ, Han JH. RIP3, an Energy Metabolism Regulator That Switches TNF-Induced Cell Death from Apoptosis to Necrosis. Science. 2009;325:332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]