Abstract
The C-terminal portion of hepatitis C virus (HCV) nonstructural protein 3 (NS3) forms a three domain polypeptide that possesses the ability to travel along RNA or single-stranded DNA (ssDNA) in a 3’ to 5’ direction. Fueled by ATP hydrolysis, this movement allows the protein to displace complementary strands of DNA or RNA and proteins bound to the nucleic acid. HCV helicase shares two domains common to other motor proteins, one of which appears to rotate upon ATP binding. Several models have been proposed to explain how this conformational change leads to protein movement and RNA unwinding, but no model presently explains all existing experimental data. Compounds recently reported to inhibit HCV helicase, which include numerous small molecules, RNA aptamers and antibodies, will be useful for elucidating the role of a helicase in positive-sense single-stranded RNA virus replication and might serve as templates for the design of novel antiviral drugs.
Introduction
Hepatitis C (HepC) is a disease that affects about 170 million people worldwide. HepC is frequently called a “silent” killer because it causes few symptoms while the pathogen slowly destroys the liver. After a couple decades of unknown infection, when they might transmit the blood-borne virus to others, many HepC patients develop fibrosis, cirrhosis, or liver cancer. At this late stage, a liver transplant is the only option for survival, and as a result, HCV infection is presently the most common cause for liver transplantation in many countries.
HCV vaccines and treatments have been delayed because the virus is extraordinarily difficult to work with in the laboratory. Although HCV accounts for the vast majority of viral hepatitis not caused by hepatitis A or B viruses, HCV was identified almost two decades after either HAV or HBV, and it was only last year that HCV could be cultivated in cell culture with reliability (Lindenbach et al., 2005; Zhong et al., 2005). HCV is primarily comprised of a single long open reading frame encoding an approximately 3,000 amino acid long protein that is cleaved into 10 mature structural (core, E1, E2) and non-structural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B). The non-structural proteins are responsible for replication and packaging of the viral genome into capsids formed of structural proteins. Since the vast majority of today’s antiviral drugs exert their actions through enzymes involved in viral replication, much attention has focused on studying the non-structural HCV proteins. HCV encodes a polymerase that specifically synthesizes new viral RNA (NS5B), and two proteases that cleave the polyprotein, the NS2/NS3 autocatalytic protease and the NS3-NS4A serine protease. Several compounds that influence the activity of the NS3 protease and the NS5B RNA-dependent RNA polymerase are currently in clinical trials, and will likely become the next generation of anti-HCV drugs.
Even though the NS3-NS4A protease and NS5B have been so far the most successful HCV encoded drug targets, neither was the first HCV enzyme that was purified, characterized and examined at an atomic resolution. The first HCV protein crystallized was the portion of NS3 that acts as a helicase, an enzyme that tracks along a nucleic acid strands displacing annealed strands or RNA-binding proteins. The mature NS3 protein comprises 5 domains: the N-terminal 2 domains form the serine protease along with the NS4A cofactor, and the C-terminal 3 domains form the helicase. The helicase portion of NS3 can be separated form the protease portion by cleaving a linker. Since the protease portion is more hydrophobic, removing it allows the NS3 helicase fragment to be expressed as a more soluble protein at higher levels in E. coli. The fragment of NS3 possessing helicase activity here referred to as “HCV helicase” was shown over a decade ago to hydrolyze ATP (Suzich et al., 1993) to fuel a reaction that unwinds duplex RNA (Kim et al., 1995)or DNA (Tai et al., 1996).
There are numerous reasons why the HCV helicase has not attracted as much attention as the NS5B polymerase and the NS3-NS4A protease. First, until recently the mechanism of action of any helicase was not clear. Second, the HCV helicase resembles helicases and similar motor proteins encoded in all human cells. Third, the role that the HCV helicase plays in viral replication is still not clear. This review focuses on recent progress made in these areas and in identifying inhibitors that target the HCV helicase.
Structure of HCV helicase
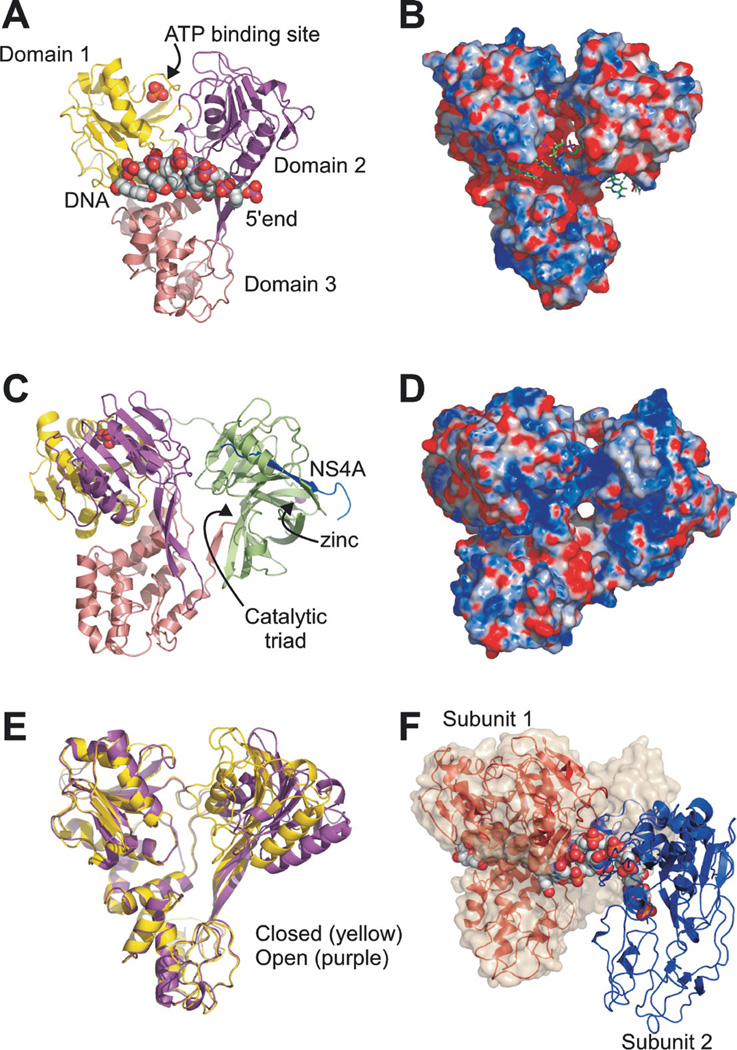
Unlike other systems where mechanistic experiments were carried out long before protein-substrate interactions were viewed at an atomic resolution, the first crystal structures of HCV helicase were solved only a few years after the protein was first purified (Yao et al., 1997; Cho et al., 1998; Kim et al., 1998; Yao et al., 1999; Mackintosh et al., 2006). These structures are shown in Fig. 1. The helicase portion of NS3 forms three domains. When viewed as a Y-shaped molecule, the N-terminal domain (domain 1) and the middle domain (domain 2) are above the C-terminal domain (domain 3). In two structures (Kim et al., 1998; Mackintosh et al., 2006), short DNA oligonucleotides are bound to the helicase in the cleft that separates domain 3 from domains 1 and 2 (Fig. 1A). In several structures, a sulfate molecule is seen bound between domains 1 and 2, in a position where ATP has been seen in high-resolution structures of similar helicases (Soultanas et al., 1999; Velankar et al., 1999; Bernstein et al., 2003) (Fig. 1A, C). When the entire NS3-NS4A complex is viewed with the ATP and DNA binding sites in the front, the protease is in the back, with its active site buried on the back of the helicase domains (Yao et al., 1999) (Fig. 1C). Behind the protease is its NS4Acofactor, which positions the catalytic triad of the protease so it will cleave the NS3-4A junction (Kim et al., 1996). The zinc ion needed for the NS2-3 auto-catalytic protease lies on the same side of the protease as NS4A (Fig. 1C). These crystal structures, along with high resolution NMR structures of HCV helicase domain 2 (Liu et al., 2001; Liu et al., 2003), have greatly influenced proposals explaining how helicases function and have guided experiments designed to test these ideas.
Fig. 1.
HCV Helicase Structures. (A). PDB file 1A1V, showing the HCV helicase with a bound DNA oligonucleotide and sulfate ion (Kim et al., 1998). The N-terminal RecA-like domain (domain 1) is colored yellow, the C-terminal RecA-like domain (domain 2) is purple, and domain 3 is pink. DNA and a sulfate ion (which occupies the ATP binding site) are depicted as spheres. (B) An electrostatic surface of the protein in 1A1V calculated without the DNA using the program APBS (Baker et al., 2001). Note the DNA is held in a negatively-charged pocket. (C) A full-length NS3 complex with the central portion of NS4A covalently tethered to the NS3 N-terminus (Howe et al., 1999), as seen in PDB file 1CU1 (Yao et al., 1999). Helicase domains are colored as in panel A with the protease colored green and NS4A blue. The protein is rotated about 90° relative to panel A. (D) An electrostatic surface of the protein as viewed in panel C. Note that the positively-charged cleft surrounding the protease, which could provide additional RNA binding sites. (E) Comparison of HCV helicase in the closed conformation (PDB file 1HEI (Yao et al., 1997)) and the open conformation (PDB file 8OHM (Cho et al., 1998)). Proteins are superimposed along domains 1 and 3. (F) Two NS3 helicase fragments (red, blue) bound to the same oligonucleotide (Mackintosh et al., 2006). Subunit 1 (red) of the helicase: DNA complex (PDB file 2F55) was aligned with the backbone the helicase portion of 1CU1. The surface of 1CU1, which includes the helicase and protease domains, is shown as transparent amber surface. All structures were rendered using the program Pymol (DeLano Scientific LLC, San Francisco, CA).
Structural variations
Each of the available HCV helicase crystal structures is shown in one of the panels of Fig. 1. Kim et al.’s structure (PDB accession code 1A1V(Kimet al., 1998)) is shown in Fig. 1 panels A and B, and Yao et al.’s structure (PDB 1CU1 (Yao et al., 1999)) is in panels C and D. In panel E, Cho et al.’s structure (PDB 8OHM (Cho et al., 1998)) and Yao et al.’s structure (PDB 1HEI (Yao et al., 1997)) are aligned for comparison. Finally, Mackintosh et al.’s (Mackintosh et al., 2006) structure of a helicase dimer (PDB 2F55) bound to a single oligonucleotide is shown in Fig. 1F. The main difference between the available HCV structures concerns the position of domain 2 relative to domains 1 and 3. Domains 1 and 3 share more of an interface than domain 2 shares with either of the other domains. Domain 2 is connected to domains 1 and 3 via flexible linkers, which allow domain 2 to freely rotate relative to domains 1 and 3. In some structures, domain 2 is rotated away from domain 1 in an “open” conformation, while in other structures domain 2 is closer to domain 1 in a “closed” conformation (Fig. 1E). The pivot point for these rotations is provided by additional contacts between domain 3 and an extended β-hairpin originating from domain 2. An animation showing the rotation of domain 2 is available in the Database of Macromolecular Movements (http://www.molmovdb.org/cgi-bin/morph.cgi?ID=109065-518) (Echols et al., 2003).
How well these structures represent the diverse array of NS3 proteins encoded by all varieties of HCV is not clear because natural variation in the amino acid sequence of NS3 undoubtedly impacts its structure. The known HCV genotypes have remarkably different nucleotide sequences, and the corresponding amino acid substitutions likely would affect protein folding. HCV helicase from only three, very similar, genotypes has been examined at the atomic level. Both Yao et al. (Yao et al., 1997) and Kim et al. (Kim et al., 1998) examined an enzyme isolated from the same genotype 1a H strain, Yao et al. (Yao et al., 1999) examined the helicase from the genotype 1b BK strain, and Cho et al. (Cho et al., 1998) used an enzyme from another genotype 1b strain. Although there are many differences between the structures, no obvious differences appear to be genotype specific; the genotype 1a structures are as different from each other as they are from the genotype 1b structures.
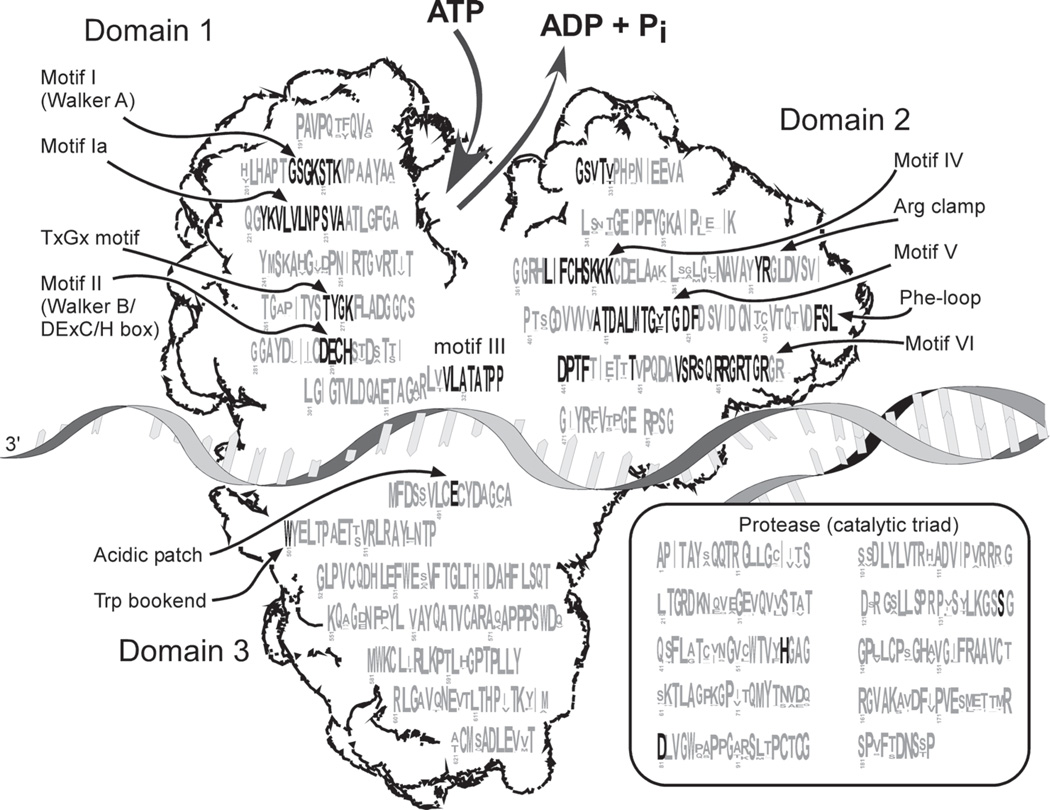
Nevertheless, variation in HCV helicase residues clearly influences its activity, as evidenced by the fact that adaptive mutations in HCV replicons frequently arise in the helicase region (Blight et al., 2000; Krieger et al., 2001; Grobler et al., 2003). To concisely depict helicase sequence variability among various HCV genotypes, a consensus sequence of the NS3 peptide is superimposed on a cartoon of the helicase structure in Fig. 2. Figure 2 was generated by rendering an alignment of all the NS3 sequences deposited in the hepatitis C virus (HCV) database project (http://hcv.lanl.gov/) using a program called Weblogo (http://weblogo.berkeley.edu/) (Crooks et al., 2004). The original alignment can be downloaded from the HCV database by choosing “NS3” under the subheading “alignments.” Weblogo depicts an alignment as a sequence logo (Schneider and Stephens, 1990), in which each NS3 residue is represented as a stack of one letter amino acid codes. The height of each stack corresponds to the amino acid conservation at that position. When the residue is invariant, only one letter is shown, and the most common substitutions are noted when the residue is variable.
Fig. 2.
HCV NS3 sequence conservation. A sequence logo (Schneider and Stephens, 1990) of an NS3 sequence alignment is overlaid on a cartoon of the HCV helicase. Each residue of NS3 is depicted as a stack of letters, the height of which correlates with how well it is conserved in 138 NS3 sequences deposited in the HCV database (http://hcv.lanl.gov/). The height of the letters in each stack correlates with how frequently that amino acid occurs at that position. Conserved superfamily 2 helicase motifs and other key residues are noted and highlighted with bold type.
Lam et al. (Lam et al., 2003b) have explored the impact of genotypic variation on the various activities of HCV helicase by examining recombinant proteins that were isolated from infectious clones of HCV genotype 1a (Yanagi et al., 1997), 1b (Yanagi et al., 1998), and 2a (Yanagi et al., 1999). Although there are some differences between the genotypes, the proteins are surprisingly similar. The main difference between genotype 1 and 2 strains can be attributed to variation at residue 450, which is normally a Thr (Fig. 2), but in the genotype 2a infectious clone it is an Ile (Yanagi et al., 1999). When Thr450 alone is changed to Ile, the protein binds ssDNA differently and unwinds DNA faster, suggesting that the interaction of Thr450 with DNA observed in the crystal structure somehow modulates the rate of helicase movement. Upon close examination of sequences that were later deposited in the HCV database, it appears now that only the particular genotype 2a strain used in our study (Lam et al., 2003b) contains the Ile substitution. Since this was an infectious clone isolated from a chimpanzee (Yanagi et al., 1999), we now believe that T450I is an adaptive mutation that permits the virus to efficiently replicate in chimpanzees, but it is not normally seen in HCV infecting humans.
Conserved motifs
The sequence logo in Fig. 2 also reveals that there are numerous stretches of amino acids that do not vary in known isolates. The numbered sequence motifs are shared with related helicases (Gorbalenya and Koonin, 1993; Hall and Matson, 1999). Some of these motifs, such as the DExC/H-box portion of motif II and motif IV are characteristic only of helicases closely related to HCV helicase, while others, such as the Walker A motif (Motif I), are conserved among all helicases and in a wide variety of other proteins that hydrolyze ATP. Other motifs are found only in HCV and closely related viruses, including the Arg-clamp, the Phe loop (Lam et al., 2003a), and all motifs in domain 3.
It was not possible to precisely depict all the conserved residues in Fig. 2 relative to their position within each domain of HCV helicase, but most are close. Diagrams noting exact motif positions on actual crystal structures have been published elsewhere (Hall and Matson, 1999; Kwong et al., 2000; Lam et al., 2003a; Frick, 2004). Motifs I, Ia, II, III, IV, V, and VI, which are conserved in similar helicases encoded by both viruses and cellular organisms, line the ATP binding cleft, and some of these motifs project residues into the nucleic acid binding site. These seven helicase motifs essentially form the motor which converts the chemical energy derived from ATP hydrolysis into a mechanical force that drives helicase movements leading to the disruption of DNA or RNA base pairs.
The roles of most of the key conserved residues in motifs I through VI have been investigated using site-directed mutagenesis. The various studies are tabulated in Table 1 along with a phenotype of each mutant. The phenotype listed in Table 1 simply depicts whether helicase activity (i.e. DNA or RNA unwinding), ATPase, or nucleic acid binding properties of each mutant is unchanged (normal), diminished, or enhanced. Mutations in motifs I–VI normally impact the ability of the protein to both unwind DNA and hydrolyze ATP, showing that the two activities are coupled. However, sometimes mutations result in decreases of ATP hydrolysis rate, but not a similar corresponding decrease in DNA unwinding. The results have been interpreted by some authors as evidence that ATP hydrolysis is not absolutely required for unwinding, but they could instead be due to the different sensitivities of ATPase and helicase assays under the particular conditions utilized.
Table 1.
HCV helicase mutants.
An, Normal ATPase; A+, Enhanced ATPase; A−, Poor ATPase; Hn, Normal duplex unwinding; H+, Enhanced duplex unwinding; H−, Poor duplex unwinding; Bn, Normal nucleic acid binding; B+, Enhanced nucleic acid binding; B−, Poor nucleic acid binding
Outside motifs I to VI, domains 1 and 2 have regions that are both variable and conserved. With a hope of discovering regions that might provide binding sites for novel anti-HCV therapeutics, our lab examined the role of two motifs in domain 2 that are conserved in all HCV isolates but not related proteins. The rationale was that compounds that bind such sites would be relatively non-toxic because similar sites are not present on related cellular helicases. The first motif identified centered on Arg393, a residue that contacts the nucleic acid backbone. When Arg393 is changed to Ala, the protein still catalyzes RNA-stimulated ATP hydrolysis but does not unwind DNA or RNA. The R393A protein also binds DNA weaker both in the presence and absence of a non-hydrolyzable ATP analog, suggesting that this Arg-clamp motif functions to tether the protein to the nucleic acid strand on which it is translocating (Lam et al., 2003a).
The second motif characteristic of only helicases from HCV strains forms a loop connecting two β-sheets that extend from domain 2. The β-loop structure is composed of residues Thr430 to Ala452, and a pair of residues, Phe438 and Phe444, and is located in a highly conserved region at the loop’s tip. The turn of the loop is composed of NS3 amino acids 438 to 444. At the time, the function of this “Phe-loop” was a curiosity. Kim et al. (Kim et al., 1998) had proposed that this loop functions like a DNA binding loop found in ssDNA binding proteins. Alternately, Yao et al. (Yao et al., 1997) proposed that Phe438 and Phe444 could pack into a hydrophobic pocket together with Phe531, Phe536, and Trp532, allowing the loop to take on a more structural role. Both Phe438 and Phe444 were altered to Ala to assess these two very different possibilities. Mutagenesis of the Phe’s that flank the Phe-loop demonstrates that the loop is not involved in nucleic acid binding. Rather, Phe438 and Phe444 are important both for proper protein folding and for modulating conformational changes leading to the release of DNA upon ATP binding (Lam et al., 2003a).
All helicases crystallized to date contain domains that resemble domains 1 and 2, but none share a domain that resembles domain 3. In some helicases, such as PcrA (Subramanya et al., 1996) and Rep (Korolev et al., 1997), two domains replace domain 3, one which extends from domain 1 (called domain 1B) and one that extends from domain 2 (called domain 2B). In several helicase structures that share domains similar to domains 1 and 2 of HCV helicase, such as the RecQ protein (Bernstein et al., 2003), DnaG (Singleton et al., 2001), and eukaryotic translation initiation factor 4A (Caruthers et al., 2000) domain 3 is missing entirely, suggesting that domain 3 might not be required for HCV helicase movements. This is not the case, however, and although its role in unwinding is only beginning to be understood, domain 3 is clearly essential. Deletion of 97 amino acids from the C-terminus of NS3 results in an inactive helicase (Jin and Peterson, 1995; Kim et al., 1997a). Two key residues in domain 3 are Trp501, which stacks against a nucleic acid base to act like a bookend (Lin and Kim, 1999; Preugschat et al., 2000; Kim et al., 2003), and Glu493 which helps repel nucleic acids from the binding cleft upon ATP binding (Frick et al., 2004a).
Mechanism of action
There is presently no consensus on exactly how the HCV helicase unwinds RNA. Debate about the HCV helicase mechanism continues largely because in some experiments, HCV helicase appears to function as a monomer, but in others it appears to be a dimer or a higher order oligomer. Below, attempts will be made to reconcile this and some other controversies, but first, to understand the intricacies of these molecular models, it will be necessary to review a few fundamental characteristics of all helicases.
Basic properties of all helicases
All helicases can be divided into two basic groups. Some form rings that encircle DNA (or RNA) while others, like HCV helicase, do not form rings. Both ring and non-ring helicases, are primarily associated with one strand of a double helix and can be classified based on the polarity of that strand. The protein either shifts from the 3’-end to the 5’-end or from the 5’-end to the 3’-end on the strand to which it is mainly bound. The most common method to diagnose the direction of movement is to determine whether the helicase requires a 5’-ssDNA tail or a 3’-ssDNA tail to initiate unwinding. 5’-3’ helicases need a 5’-ssDNA tail, and 3’-5’ helicases require a 3’-ssDNA tail. HCV helicase is a 3’-5’ helicase (Tai et al., 1996; Morris et al., 2002). As a consequence, if the oligonucleotide bound to HCV helicase in PDB file 1A1V (Fig. 1A) (Kim et al., 1998) represents the strand on which HCV helicase translocates, then the duplex portion of the helix would likely be positioned to the right of the protein in Fig. 1A (see cartoon in Fig. 2). Helicases are thirdly classified based on their evolutionary relationships. Gorbalenya and Koonin have used protein sequence comparisons to classify most helicase families into one of three large superfamilies (Gorbalenya and Koonin, 1993). Non-ring helicases are generally members of helicase superfamily 1 (SF1) or superfamily 2 (SF2), while ring helicases are in superfamily 3 (SF3) or in other families not in the three main superfamilies. HCV helicase is a member of SF2, and like all helicases in SF2 shares conserved motifs I through VI described above.
The ring formed by ring helicases usually is composed of six identical subunits assembled in a head-to-tail manner. The rings surround the strand on which the helicase is translocating and the complementary strand passes outside the ring (Egelman et al., 1995). ATP binds between the subunits, to the head of one subunit and the tail of an adjacent protomer. There are consequently six ATP binding sites per hexameric ring (Singleton et al., 2000). Each subunit of a ring helicase contains a single domain that resembles a domain first seen in the structure of a protein called RecA, which plays a key role in E. coli DNA recombination (Story and Steitz, 1992). In ring helicases, ATP hydrolysis leads to rotation of the RecA-like domains which in turn leads to movements of positively-charged loops that protrude into the center of the ring. The positively charged loops bind DNA (Notarnicola et al., 1995; Washington et al., 1996), and the sequential interaction of the DNA-binding loops with DNA is thought to lead to ring helicase movement (Singleton et al., 2000).
In non-ring helicases, like HCV, there are two RecA-like domains in a single protein subunit, and ATP binds between these subunits. In HCV helicase, domains 1 and 2, fold into similar structures although they share no apparent sequence homology. The core of both domains is composed of a series of beta sheets sandwiched between sets of alpha helices. Both domains 1 and 2 are similar to RecA, form the ATP binding site, and contact DNA. The main structural difference between domains 1 and 2 is that domain 2 contains two long beta sheets that project towards domain 3 (the Phe-loop discussed above), which are not present in domain 1.
Mechanism of ATP hydrolysis
Although the position of ATP bound to HCV helicase has not yet been visualized, the mechanism of its hydrolysis most likely resembles that seen in other helicases. The approximate configuration of ATP in the binding site can be seen by comparing a HCV helicase structure with one of a similar helicase that has been crystallized in the presence of a non-hydrolyzable ATP analog. Figure 3A shows the results of a structural alignment of HCV helicase (PDB file 1A1V) with the SF2 helicase RecQ bound to ATPγS (PDB file 1OYY) (Bernstein et al., 2003). Shown only are the ATPγS (from 1OYY), the HCV helicase, and its bound oligonucleotide (both from 1A1V). Residues that likely play key roles in ATP hydrolysis are highlighted as sticks.
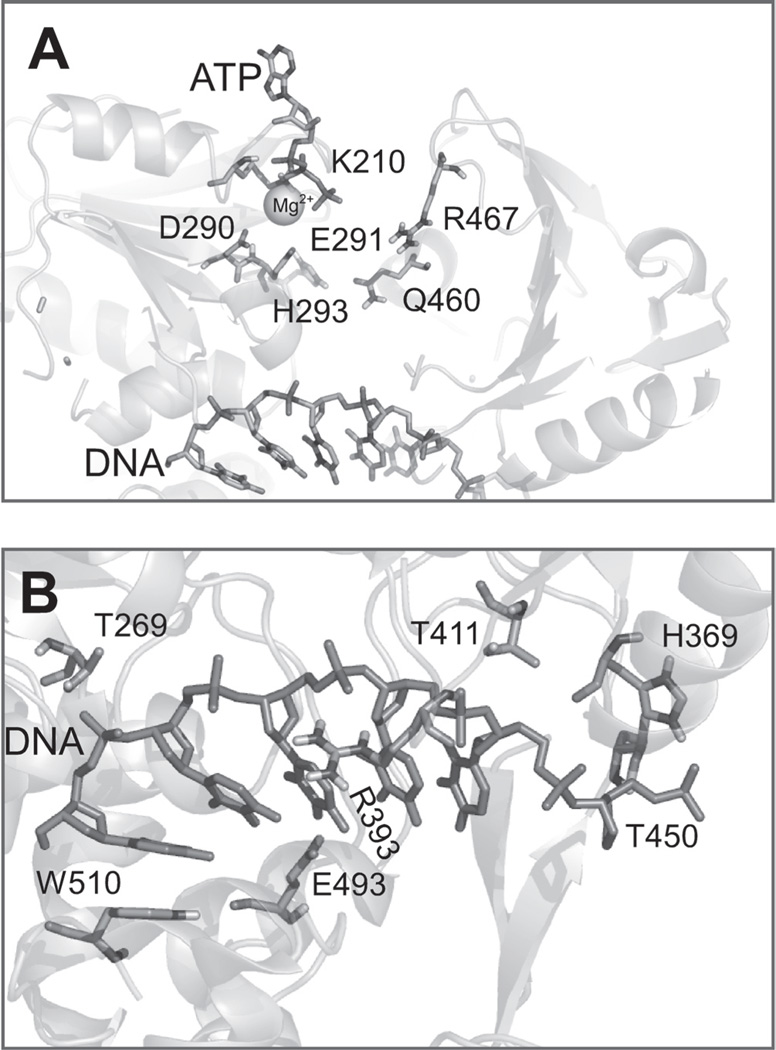
Fig. 3.
Key residues in HCV helicase. (A) HCV helicase residues likely involved in modulating ATP binding and hydrolysis. The approximate position of ATP bound to HCV helicase is revealed by a structural alignment of HCV helicase (PDB file 1A1V) (Kim et al., 1998) with the SF2 helicase RecQ bound to ATPγS (PDB file 1OYY) (Bernstein et al., 2003). (B) Key residues contacting the oligonucleotide bound to HCV helicase in PDB file 1A1V (Kim et al., 1998).
The configuration of residues at the ATP-binding site depicted in Fig. 3A is reminiscent of that seen in all other helicases that have been studied bound to NTPs (Sawaya et al., 1999; Soultanas et al., 1999; Velankar et al., 1999; Singleton et al., 2000; Bernstein et al., 2003; Gai et al., 2004; James et al., 2004). ATP and a required metal ion cofactor (depicted as Mg2+ in Fig. 3A) normally bind to a helicase in the cleft that separates two adjacent RecA-like domains. The most critical residues for ATP binding arise from the Walker A and B motifs (Walker et al., 1982). The Walker A motif of HCV helicase forms a phosphate binding loop (P-loop) with the conserved Lys210 likely contacting the γ phosphate of ATP. The Walker B motif contains acidic residues that coordinate the positively charged divalent metal cation, which in turn contacts the phosphates of ATP. In the alignment in Fig. 3A, Asp290 seems to be ideally suited to coordinate the catalytic metal. In or near the Walker B motif of helicases and related proteins, there is normally a residue which acts as a catalytic base by accepting a proton from the water molecule that attacks the γ phosphate of ATP. Normally, the catalytic base in this class of enzymes is a glutamate (Goetzinger and Rao, 2003; Orelle et al., 2003), and Glu291 seems to be properly positioned to perform this function.
A more detailed analysis of HCV structures suggests that the roles of particular residues might be somewhat more complicated than assumed above. For example, to function as a catalytic base, the pKa of Glu291 would need to be much higher than that of a typical Glu in a protein. However, electrostatic analysis of all HCV helicase structures reveals that neither Glu291, nor any nearby Glu, has an abnormally high pKa. In contrast, Asp290 has a pKa as high as 10 in some structures and as low as 3 in others. Interestingly, in structures in the open conformation (such as 8OHM), the pKa of Asp290 is low, and in the closed conformation (ex. 1A1V), the pKa of Asp290 is higher than 7, suggesting that Asp290 picks up a proton (like a catalytic base) when the protein changes from the open to the closed conformation. Thus, Asp290 could serve as a catalytic base instead of, or in addition to, coordinating the magnesium-ATP complex (Frick et al., 2004a).
Once the water molecule is activated, it acts as a nucleophile, most likely attacking the terminal phosphate of ATP. The pentavalent transition state, where the γ phosphate is bound to 5 oxygen atoms, then breaks down into ADP and inorganic phosphate. In similar ATP hydrolyzing enzymes, the transition state is stabilized by one or more positively charged residues, normally arginines, that function in concert with the lysine of the Walker A motif. In many enzymes, including helicases, these “arginine fingers” are frequently part of adjacent protein subunits. For example, in small GTPases like Ras, the Arg-finger that activates GTP hydrolysis is part of the GTPase-Activating Protein (GAP) (Ahmadian et al., 1997). In F1ATPase, an Arg-finger on the alpha subunit stabilizes the transition state (Nadanaciva et al., 1999). Recently, it was shown that in ring helicases, the Arg-finger and P-loop are part of different polypeptide chains in the hexamer (Crampton et al., 2004), demonstrating why ring helicases need to oligomerize to cleave ATP. In HCV helicase, several arginines are present in domain 2, and they line the ATP binding cleft. These residues are part of conserved motif IV and include Arg461, Arg462, Arg464, and Arg467. In the model shown in Fig. 3, Arg467 and Arg464 are nearest the phosphates of ATP. Either could rotate even closer to ATP if the cleft between domains 1 and 2 closes. Of the two, only Arg467 is shown because it is of the most interest. Arg467 is methylated by cellular protein arginine-methyltransferase I (Rho et al., 2001). Although it is still unclear how methylation influences helicase activity, such a modification should eliminate all activity if Arg467 acts as an Arg-finger. Site-directed mutagenesis supports this contention. When Arg467 is changed to Lys (Kim et al., 1997b; Wardell et al., 1999) or Ala (Kwong et al., 2000), the proteins do not unwind RNA, and ATPase activity is decreased over 10-fold. An R464A mutant has a similar effect (Kim et al., 1997b; Min et al., 1999; Kwong et al., 2000).
Many of the other conserved residues help to properly position the above groups by forming networks of hydrogen bonds, ionic bonds, and hydrophobic interactions. In addition, some conserved residues help coordinate the rotation of domain 2. Two such amino acids are noted on Fig. 3A. His293 in motif II (domain 1) and Gin460 in motif VI (domain 2) are near each other in many structures and could interact. Kim et al, called these residues “gatekeepers” and propose that they might provide a switch modulating the opening and closure of the cleft between domains 1 and 2 upon ATP binding (Kim et al., 1998). Mutation of either residue has a profound and interesting effect on ATPase. Mutation of Gin460 abolishes detectable ATPase, but an H293A mutation results in a protein with a significantly higher level of ATPase in the absence of RNA, and the protein still unwinds RNA. In the presence of RNA, the H293A mutant hydrolyzes ATP slower than wildtype, to such an extent that RNA appears to inhibit ATP hydrolysis (Kim et al., 1997b). These data further support the idea that rotation of domain 2 is related to ATP hydrolysis, and that domain closure leads to a completion of the active site and hydrolysis of ATP. The question of how ATP hydrolysis is translated into helicase movement on nucleic acid still remains unanswered, however. The first two models that were applied to HCV helicase to explain its movements suggest that either the protein operates as a monomer like an inchworm (Kim et al., 1998) or as a dimer, which rolls along nucleic acid (Cho et al., 1998).
The inchworm and rolling models
If ATP binding and hydrolysis leads to movement of domain 2 relative to domain 1, then this conformational change would in turn impact interactions between residues in these two domains with RNA. A close-up of the DNA binding site of structure 1A1V is shown in Fig. 3B, with key amino acids highlighted. Unlike SF1 helicases, which have many interactions with nucleic acid bases (Velankar et al., 1999), most of the contacts occur with protein side chains and the sugar-phosphate backbone of DNA (Kim et al., 1998). One key hydrogen bond is donated from domain 1 residue Thr269, which is part of the conserved TxGx motif, and an analogous interaction arises from motif V residue Thr411 in domain 2. Mutagenesis of either residue affects both RNA binding affinity and unwinding rates (Lin and Kim, 1999). Also noted on Fig. 3B are residues Thr450, Arg393, Trp501, and Glu493, whose roles were alluded to above.
Unlike ring helicases that need to oligomerize to cleave ATP because the Arg-finger is located on the opposite side of a protein monomer relative to the Walker A motif, all the residues necessary for ATP hydrolysis are present in a single polypeptide chain in HCV helicase. Monomeric models for HCV helicase action state that upon rotation, ATP binding leads to a closure of the cleft between domains 1 and 2 by a rotation of domain 2 relative to the rest of the protein, a movement first observed in the structures of Yao et al. (Yao et al., 1997). Such conformational changes conceivably could cause the protein to act like an inchworm to move along RNA. Most monomeric models are variations on the “ratcheting inchworm” model first proposed for HCV helicase by Kim et al. (Kim et al., 1998). Based on the observation that the oligonucleotide appears to be locked into the binding cleft because a residue in domain 3, Trp501, is stacked against the 3’-terminal base, Kim et al. proposed that ATP binding, and the subsequent closure of the cleft between domains 1 and 2, will lead to a ratcheting of Trp501 past 1 or 2 nucleotides. Consequently, the protein would move towards the 5’-end of the bound nucleic acid. After ATP is hydrolyzed and Trp501 is again locked into place acting as a bookend, the cleft opens and RNA slides through the other side of the protein. Kim et al. proposed that the residue that acts as a 5’-bookend, analogous to the 3’-bookend Trp501, might be Val432 in domain 2 (Kim et al., 1998).
The dimeric models for HCV helicase action are essentially variations on Wong and Lohman’s rolling dimer hypothesis that was used to explain the actions of a dimer formed by the E. coli Rep helicase upon DNA binding (Wong and Lohman, 1992). In the rolling dimer, each subunit alternates between a form that prefers to bind ssDNA and a form that preferentially binds a double helix. Switching between the states is modulated by ATP binding and hydrolysis. In theory, both forms are bound to a DNA fork, with one subunit bound to the ssDNA tail, and the other bound to the duplex region. When the trailing subunit changes conformation so that it prefers to bind duplex DNA, it will roll toward the double helix causing the subunit bound to the duplex to wrench one strand away from its complement so that it can then bind the resulting ssDNA (for review see (Lohman and Bjornson, 1996)). A modified rolling model was applied to the HCV helicase by Cho et al. (Cho et al., 1998), who observed that two HCV helicase monomers could pack together. Cho et al. called their model a “descending molecular see-saw” and proposed that RNA could thread through a long cleft formed between domains 1 and 2 of adjacent subunits (Cho et al., 1998). However, the later structure by Kim et al. (Kim et al., 1998) showing DNA bound in another cleft (Fig. 1A), coupled with the fact that ATP likely binds in the cleft between domains 1 and 2 (Fig. 3), makes such an orientation seem unlikely.
More recently Mackintosh et al. have crystallized two HCV helicase monomers bound to the same oligonucleotide revealing an interface between the two subunits. When this interface is perturbed using site-directed mutagenesis, HCV sub-genomic replicons fail to replicate in cells but there are only small effects observed in unwinding assays, suggesting that the interface is more important for inter-protein interactions than for unwinding (Mackintosh et al., 2006). When this structure is aligned with the structure of the full-length NS3 protein (Fig. 1F), it is apparent that much of second subunit occupies the same space as the protease domains of NS3 (the NS3 fragment used in the Mackintosh et al. study lacks the protease). Thus, in order for the helicase to oligomerize in such a manner, the protease domain would need to rotate away from the helicase, as has been proposed by Yao et al. (Yao et al., 1999).
Evidence for a functional monomer (the inchworm model)
Ever since the HCV helicase portion of NS3 was first purified, it was apparent that it behaved as a monomer and did not need to oligomerize to cleave ATP. Initial studies found HCV helicase to act as a monomer in solution based on gel filtration (Preugschat et al., 1996) and analytical ultracentrifugation (Porter et al., 1998). As discussed above, the monomeric enzyme has all the residues necessary to catalyze ATP hydrolysis, and as a result, no decrease in turnover number (kcat) is observed when HCV helicase is diluted (Levin and Patel, 2002). In contrast, diluting a ring helicase leads to a loss of the ability to hydrolyze ATP at low protein concentrations (Guo et al., 1999).
Soon after the ratcheting inchworm model was introduced for HCV helicase (Kim et al., 1998), it was tested by several groups using site-directed mutagenesis (Table 1). Most initial interest focused on the residues that act as bookends, or the teeth of the ratchet. Several groups have confirmed the importance of Trp501 in both nucleic acid binding and unwinding (Lin and Kim, 1999; Paolini et al., 2000; Preugschat et al., 2000; Kim et al., 2003). Without a bulky aromatic amino acid at position 501, HCV helicase is unable to unwind RNA (Lin and Kim, 1999; Tai et al., 2001; Kim et al., 2003) but retains some ability to unwind DNA (Kim et al., 2003), albeit more slowly than wild type (Preugschat et al., 2000). The data is less clear regarding the residue that might bookend the 5’-end of the RNA. Kim et al. propose that this residue is Val432 in Domain 2 (Kim et al., 1998), but Paolini et al. have suggested that Tyr392 could play a similar role (Paolini et al., 2000). Some of these reports have suggested that mutation of these residues leads to decreases in helicase activity (Paolini et al., 2000; Preugschat et al., 2000; Tai et al., 2001; Kim et al., 2003).
More recently, other predictions made by the inchworm model have been tested. One basic prediction is that binding of ssDNA to the cleft separating domain 3 from domains 1 and 2 activates ATP hydrolysis. Supporting this theory, several mutations have been made in the DNA binding cleft that decrease the affinity of the protein for both DNA and RNA and affect rates of ATP hydrolysis (see Table 1) (Kim et al., 1997b; Lin and Kim, 1999; Tai et al., 2001). However, there exists an alternate explanation for such results. The mutant proteins might not fold properly or may be less stable than the wildtype, explaining the decreased binding and unwinding activity. In contrast to these negative results, our lab has recently found that substitution of one of two residues in the DNA binding cleft, His369 or Glu493, enhances binding and lowers the amount of nucleic acid needed to stimulate ATP hydrolysis. For example, an E493K mutant enhances binding to RNA in the presence of ATP by several orders of magnitude. This positive result provides the clearest evidence that DNA/RNA binding to this region activates ATP hydrolysis (Frick et al., 2004a).
Another prediction made by the inchworm model is that the ssDNA bound between domain 3 and domains 1 and 2 is the strand on which the helicase is translocating in a 3’ to 5’ direction. We have tested this idea by analyzing a helicase in which Arg393 is changed to Ala. Without this Arg-clamp in the DNA binding cleft (Fig. 3B), the protein cannot unwind DNA or displace proteins bound to ssDNA. The R393A protein retains full RNA-stimulated ATPase activity, and still binds ssDNA with the same stoichiometry as wildtype, albeit more weakly. These data provide strong evidence that the protein moves along the strand seen in the crystal structure in a 3’ to 5’ direction and the duplex region would lie as diagramed in Fig. 2 (Lam et al., 2003a).
The inchworm model lastly predicts that ATP binding should modulate the affinity of the protein for RNA (or ssDNA). Early kinetic studies of nucleic acid stimulation of ATP hydrolysis suggest that, indeed, ATP binding weakens the affinity of the protein for RNA (Preugschat et al., 1996). However, it was difficult to confirm this observation by directly measuring dissociation constants because HCV helicase only weakly interacts with most common non-hydrolyzable ATP analogs. A breakthrough came when Levin et al. found that the presence of BeF3 tightly locks the reaction product, ADP, on the enzyme (Levin et al., 2003). In other systems, BeF3 coordinates like the γ phosphate of ATP, so that ADP(BeF3) is essentially a non-hydrolyzable ATP analog (Xu et al., 1997). Using ADP(BeF3), Levin et al. showed that when ATP binds HCV helicase, affinity for DNA falls by almost two orders of magnitude. More recently, Lam et al. have shown that nucleic acid binding to HCV helicase is pH dependent in the presence of ADP(BeF3) but not in the absence of the analog, demonstrating that a conformational change occurs upon ATP binding (Lam et al., 2004), as is also predicted by the inchworm model.
Evidence for a functional oligomer (the rolling model)
While the evidence that HCV helicase acts as a monomer is convincing, there is also evidence that multiple subunits interact with each other to efficiently unwind RNA. Yeast two-hybrid assays provide the most persuasive evidence that NS3 interacts with itself (Flajolet et al., 2000; Khu et al., 2001). In such experiments, the minimum peptide required for an NS3-NS3 interaction contains only domain 1 residues 162–335. This peptide surrounds conserved motifs I, II, and III (Khu et al., 2001), suggesting domain 1 would interact with domain 1 of another monomer. Three residues, which were identified using a reverse two hybrid screen, are critical for dimer formation, Thr266, Tyr267 and Met288 (Khu et al., 2001). Met288 is not conserved and is normally an Ile in all but a few HCV isolates (Fig. 2). Mutations of these residues not only influence dimer formation that can be assayed using gel filtration, but also the ability of the protein to unwind DNA (Khu et al., 2001).
Oligomerization of NS3 seems to be dependent on nucleic acids. Before the yeast two-hybrid data were reported, Levin and Patel demonstrated that DNA aids the ability to chemically crosslink HCV helicase into high molecular weight species (Levin and Patel, 1999). Dimerization of NS3 has also been visualized using analytical gel filtration, but only in the presence of an oligonucleotide (Khu et al., 2001). Nucleic acid binding data can sometimes be fit to models that do not take into account subunit interactions (Porter, 1998b; Porter, 1998a; Porter et al., 1998; Levin and Patel, 2002), but under certain conditions, cooperative models fit the data better (Locatelli et al., 2002; Frick et al., 2004b). Taken together, these data suggest that two or more HCV helicase protomers cooperatively assemble onto ssDNA (or RNA) in a controlled manner.
Other evidence for oligomerization has emerged from measurements of rates of HCV helicase catalyzed DNA and RNA unwinding. Notably, unwinding rates are not linearly dependent on the amount of protein present in the reaction, but rather, accelerate greatly once a critical protein concentration is reached (Lam et al., 2003a; Frick et al., 2004b). By measuring unwinding under single-turnover conditions, several groups have presented kinetic models explaining this cooperativity (Levin et al., 2004; Serebrov and Pyle, 2004; Tackett et al., 2005). As reviewed elsewhere (Bianco, 2004), these models all take into account the interaction of multiple protomers aligned on a ssDNA (or RNA) strand and attempt to calculate the number of base pairs unwound in a single turnover event (called “step size”). The theory holds that an oligomer would unwind many base pairs (10 or more) in a single event while a monomer would only unwind a few base pairs at a time. Levin et al. (Levin et al., 2004) have calculated a step size of 9 base pairs using DNA, and using a long RNA substrate, Serebrov and Pyle have determined that 18 base pairs are unwound by HCV helicase in a single step (Serebrov and Pyle, 2004). These values support a rolling dimer model that undergoes a periodic cycle of pausing and unwinding and stand in stark contrast with an older calculation by Porter et al. that only a few base pairs of fluorescently-labeled DNA are unwound by HCV helicase in a single turnover event (Porter et al., 1998). This periodic cycle of pausing and unwinding was recently confirmed in a collaborative study between Pyle’s group and Carlos Bustamante’s group using optical tweezers to examine the action of single molecules of NS3 on RNA(Dumont et al., 2006). Notably, however the single molecule study did not confirm that NS3 need to oligomerize to unwind RNA.
Other mechanisms explaining helicase movement
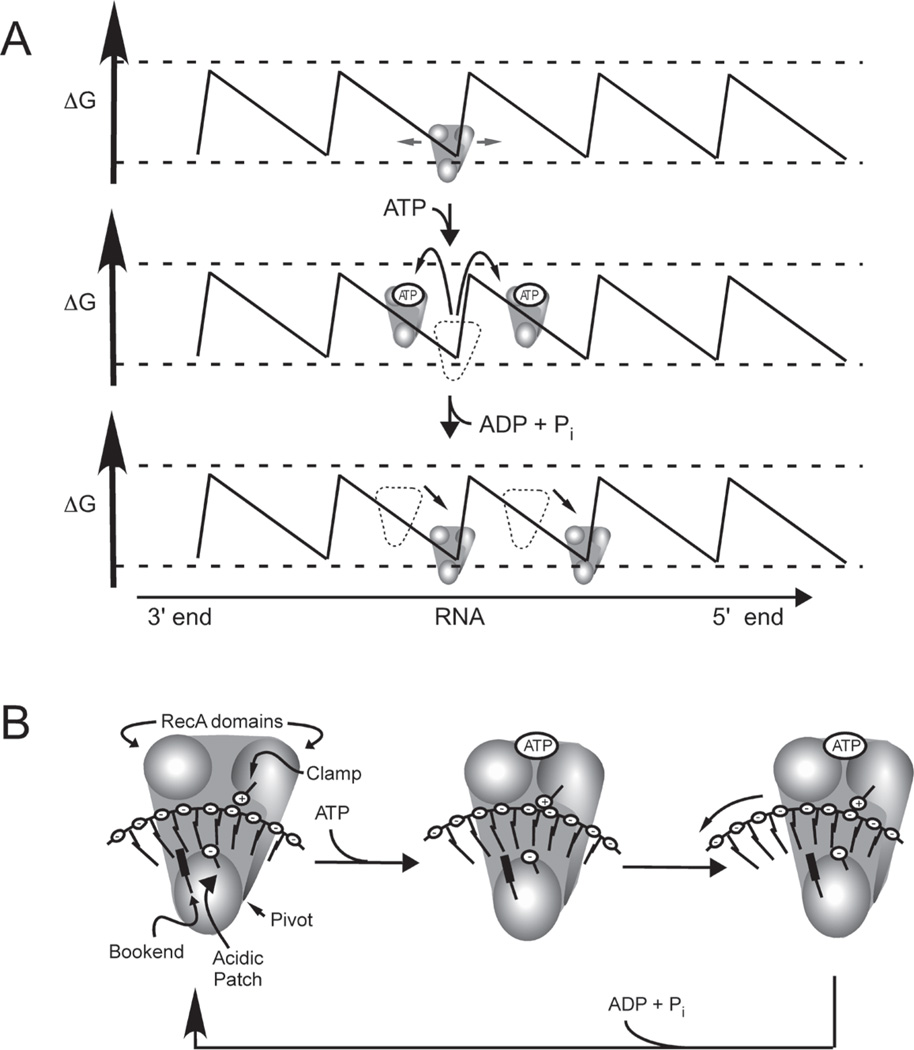
Because neither the inchworm nor the rolling model fully explains all elements of helicase action, additional models have been recently proposed. One such model states that HCV helicase acts like a Brownian motor (Astumian, 1997, Levin et al., 2005). A Brownian motor exploits random movements that constantly occur on the molecular level (Brownian motion) and an asymmetrical path to shift an object in a single direction. As diagrammed in Fig. 4A, collisions that occur between HCV helicase, water, and other small molecules constantly transfer small amounts of momentum to the protein so that it wobbles slightly relative to the RNA to which it is bound. In the absence of ATP, the helicase is constrained in a certain location due to molecular barriers. However, when HCV helicase binds ATP, it releases its grip on RNA by changing conformation so that it is free to move along RNA. Random collisions will then be more likely to transfer enough momentum that the protein clears the barrier constraining it to its original position. The key to the Brownian motor model is the asymmetry of the path on which the motor is traveling. Because the path is asymmetrical in the Brownian motor model, the protein will be more likely to move in one direction than the other, and the net result of many movements will be movement in a single direction. If the path were symmetrical, then the protein would be equally likely to return to the original position as it would be to move to either adjacent position, and the net result would be no movement.
Fig. 4.
Two possible mechanisms for HCV helicase translocation on RNA. (A) The Brownian motor model (Levin et al., 2005). In the absence of ATP, HCV helicase is confined in a single location on an asymmetrical path of RNA. When ATP binds, binding releases the protein from RNA, allowing random movement (Brownian motion) to transport the helicase either in a 5’ or 3’ direction. Because the path is asymmetrical, molecules moving in the 3’ direction will return to their original position, whereas molecules moving in the 5’ direction will change positions. Net movement will be in a 5’ direction. (B) The propulsion-by-repulsion model (Frick et al., 2004a; Lam et al., 2004). ATP binding rotates domain 2 so that a positively charged Arg-clamp (Lam et al., 2003a) moves the RNA so that it clears Trp501, which is holding the RNA in a negatively charged cleft. When ATP is bound, the protein repels RNA past Trp501 so that the protein moves in a 5’ direction until ATP is hydroiyzed and the protein returns to its original conformation.
In Fig. 4, this irregularity is depicted as an asymmetrical free energy diagram. If the helicase moves in a 5’ direction, then it will likely clear a free energy peak and descend to the base of another valley towards the 5’ end of the RNA. On the other hand, if the protein moves toward the 3’ end of the RNA by the same amount, it will not clear the peak and will return to the position at which it began the cycle. Thus, even though movements occur randomly in either direction and many molecules will remain in the same location, the net result will be that most molecules will move in the same 3’ to 5’ direction. This model has been applied to HCV helicase by Levin et al. (Levin et al., 2003), who recently observed that HCV helicase has a higher affinity for a partially duplex DNA substrate with 3’-ssDNA tails, than it does for either ssDNA alone, or DNA with a 5’-ssDNA tail. They propose that interaction with the fork of the DNA leads to asymmetry of the free energy diagram (Levin et al., 2005). While movements towards the fork likely play some role in helicase movement, the fact remains that two groups have independently observed translocation of HCV helicase in a 3’ to 5’ direction in systems containing no duplex portion on the ssDNA substrate (Morris et al., 2002; Lam et al., 2003a). Thus, if the Brownian model holds true for HCV helicase, then all asymmetry in the free energy diagram should be intrinsic to the helicase and the nucleic acid strand on which it is traveling.
Our lab has proposed another model to explain HCV helicase movement that suggests that HCV helicase utilizes electrostatic forces to move along DNA and RNA (Frick et al., 2004a; Lam et al., 2004). This “propulsion-by-repulsion” model (Fig. 4B) is based on two observations. First, DNA is tightly bound in a pocket of the enzyme that is highly negatively charged (see Fig. 1B). Second, release of DNA from the enzyme is pH dependent; the enzyme binds weaker to DNA in the presence of ATP at a higher pH. The first observation hints that there is a potential energy buildup when the protein is locked onto DNA in the absence of ATP. The second observation suggests that ionizable residues come in contact with DNA upon ATP binding. We have shown using mutagenesis that one of these key residues is Glu493 in the ssDNA binding cleft (Frick et al., 2004a). In our model, ATP binding leads to a conformational change such that the nucleic acid bases can clear the Trp501 bookend (Lam et al., 2004). In the absence of ATP, RNA cannot exit the enzyme because it is blocked by Trp501 and clamped in the cleft by the Arg-clamp on domain 2 (Lam et al., 2003a). When ATP binds, domain 2 rotates bringing with it the positively-charged Arg-clamp. The Arg-clamp attracts the negatively-charged phosphodiester backbone so that RNA moves free from the bookend. The negatively-charged RNA is then repelled by the negatively charged binding cleft, so it moves through the protein until ATP is hydrolyzed, and the protein clamps it tightly again. In such a model, the step size of the helicase would depend on the nature of the nucleic acid on which the protein is translocating explaining, in part, why different step sizes have been calculated using different substrates (Porter et al., 1998; Levin et al., 2004; Serebrov and Pyle, 2004; Dumont et al., 2006).
Role of the protease domain and NS4A
Reviewing the HCV helicase literature is perplexing because frequently the data reported in one study differs from that reported in other studies. Rates of ATP hydrolysis, DNA/RNA unwinding, and dissociation frequently differ by more than 10-fold. The most likely explanation for such differences (when the same protocol is used) is that most labs utilize different recombinant versions of HCV helicase. Many of these proteins are quite different because they either (1) include different portions of NS3, or (2) have been isolated from different HCV strains. Because full-length NS3 is difficult to express in E. coli, a truncated protein containing only NS3 residues 166–631 is frequently used. However, some studies have used helicase constructs with more or fewer N-terminal NS3 residues. Furthermore, some studies use a helicase lacking fusion proteins, whereas other studies utilize a helicase with a N-terminal or C-terminal His-tags, a T7-tag, a GST-tag, or combinations of multiple tags. Frequently, the tags are not removed before analysis.
We have compared numerous NS3 constructs in our laboratory to try to understand why different studies have reported such different results. Initially, we thought that such variation might be due to intrinsic differences between the HCV genotypes. Our comparison of three helicases isolated using the same procedure from three different genotypes noted some differences, but these tended to be small (less than 2-fold) and did not explain the widely divergent data in the literature (Lam et al., 2003b). We then set out to compare the effect of fusion proteins, which are attached to the helicase to aid expression and purification, and found that these modifications led to major changes in activity (Frick et al., 2004b). While modifications to the C-terminus did not effect most assays, modification to the N-terminus did, suggesting that the protease domain and the conformation of the region linking it to the helicase could have a major role in aiding the cooperative assembly of the protein on RNA and in unwinding (Frick et al., 2004b).
In our hands (Frick et al., 2004b), full-length NS3 (with NS4A) unwinds RNA better than versions lacking the protease, but hydrolyzes ATP slower, suggesting that it is a more efficient molecular motor. Some of the effects of the protease could be substituted for by GST or His-tag fusion proteins, but several could not, suggesting that RNA makes specific contacts with the protease region. As discussed above, it is not clear where the complementary strand or the duplex region of RNA interacts with HCV helicase. Nevertheless, an electrostatic analysis of the full-length protein (Fig. 1D) reveals that a positively-charged cleft is formed between the protease and domain 2 of the helicase. Residues in this cleft could tether the protein to the negatively-charged phosphate backbone of RNA. It is possible that a similar cleft could be formed when the protease is replaced with a fusion protein, explaining why such proteins have a higher apparent processivity than the helicase domain alone (Frick et al., 2004b).
Not all other studies have noted as clear differences when the protease domain is removed from NS3. For example, even though Kuang et al. found that a NS3-NS4A complex unwinds RNA better than an isolated helicase domain, they also noted NS3 lacking NS4A is a poor helicase relative to an isolated helicase domain, suggesting that the protease without its NS4A cofactor might actually inhibit helicase movements (Kuang et al., 2004). Similarly perplexing data showing relatively poor helicase activity for full-length NS3 have been reported by others (Heilek and Peterson, 1997; Gallinari et al., 1998). The poor helicase activity of some full-length NS3 constructs could be explained by the conformational flexibility of the protein. In order to cleave the rest of the polyprotein, Yao et al. proposed that the protease domain swings away from the helicase via the flexible linker that connects the two regions (Yao et al., 1999). If this occurs, then the putative RNA binding cleft proposed above would be disrupted and the helicase would more rapidly dissociate from RNA substrates.
A model in which RNA binds a cleft between domain 2 of the helicase and the protease would also provide a plausible role for the NS4A peptide in facilitating helicase action. NS4A could hold the protein in a conformation so that the RNA binding cleft between the protease and helicase remains intact, explaining why some investigators find that a NS3-NS4A complex unwinds RNA better than NS3 alone. For example, Pang et al. (Pang et al., 2002) compared the activities of a NS3-NS4A complex expressed in insect cells (Sali et al., 1998) with a His-tagged, full-length NS3 protein expressed and purified from E. coli, and found that the NS3-NS4A complex requires less time to form a functional complex on RNA. Based on the structure of the NS3-NS4A complex (Fig. 1C), it is difficult to envision a direct interaction between NS4A and RNA, as has been proposed by others (Silverman et al., 2003). Thus, we prefer a model where NS4A stabilizes the formation of an RNA binding cleft on NS3 (Frick et al., 2004b).
In addition to being a better RNA helicase, we also find that the full-length protein oligomerizes more readily than the truncated protein lacking a protease domain, indicating that the protease domain properly configures the protein for oligomerization. As evidence, we find twice as many protomers of full-length NS3 bound to a single oligonucleotide as recombinant proteins containing the helicase domain only (Frick et al., 2004b). This key observation explains why early studies using isolated helicase domains lacking the protease failed to detect cooperative assembly (Preugschat et al., 1996; Levin and Patel, 2002; Lam et al., 2003a), while later studies using the full-length NS3 often detect oligomers (Khu et al., 2001; Locatelli et al., 2002; Frick et al., 2004b).
HCV helicase inhibitors
Because compounds that inhibit a helicase encoded by herpes simplex virus (HSV) have been recently shown to moderate disease symptoms (Crute et al., 2002; Kleymann et al., 2002), there has been great interest in finding inhibitors of HCV helicase. Many compounds that inhibit HCV helicase have been reported, and they can be broadly classified as small molecules, nucleic acids, or antibodies. There are, however, many obstacles that must be overcome before developing helicase inhibitors into viable antiviral agents. The main problem will likely be toxicity because the motor domains of HCV helicase are conserved in a vast array of cellular proteins. Consequently, there is more focus on finding inhibitors that bind sites that are not conserved with cellular enzymes, such as the RNA binding site(s) described above and possible allosteric regulatory sites. Even if these inhibitors are never developed into drugs, they should still be useful for elucidating the role of HCV helicase in the viral lifecycle.
Small molecules
Many of the small molecules that were initially examined as HCV helicase inhibitors were nucleoside analogs. Although nucleoside analogs might also inhibit cellular proteins by interacting with conserved Walker sequences, there is some potential for these compounds because there is a possibility that nucleotides could bind to a second site on the helicase in addition to the conserved Walker site. Such a site could be formed, for example, if the protein oligomerizes and ATP binds to an interface between the RecA-like domains of adjacent subunits. Porter et al. first detected a possible second nucleotide binding site when they studied product inhibition in the presence of NaF. In their studies, about two moles of ADP bound per protein monomer (Porter, 1998a). In contrast, as discussed above, when beryllium fluoride is added to the reaction, only one mole of ADP is bound per protomer (Lam et al., 2003a; Levin et al., 2003). One model explaining these data assumes that the helicase functions as a dimer with ATP bound tightly to the interface between domains 1 and 2 and ADP bound more weakly to a second interface. ADP fluoride complexes likely do not resemble the substrate, ATP, as closely as ADP(BeF3), so they might bind both active and allosteric sites. Also in support of a second NTP-binding site, Locatelli et al. have demonstrated that nucleotides bind HCV helicase cooperatively (Locatelli et al., 2002).
There is also some evidence that the second potential nucleotide binding site on HCV helicase is more specific than the nucleotide binding site between domains 1 and 2. The alignment in Fig. 3 reveals few contacts are made between HCV helicase and the sugar or base of an NTP, explaining the observed non-specificity of this site. HCV helicase hydrolyzes all eight canonical nucleoside triphosphates (Preugschat et al., 1996; Wardell et al., 1999; Lam et al., 2003b). The seven other (d)NTPs are competitive inhibitors of ATP hydrolysis (Lam et al., 2003b) and most studies find that they all support unwinding. However, one study found that only some NTPs fuel unwinding with efficiency comparable to that seen with ATP (Locatelli et al., 2001). Other (d)NTPs, particularly dATP, were found to be poor substrates and potent inhibitors of unwinding (Locatelli et al., 2001). Such results can be explained if NTP binding to a regulatory site is more specific than NTP binding to the catalytic site. For example, the regulatory site might only bind dATP but not the NTPs that fail to inhibit unwinding.
Regardless of whether nucleoside analogs will ever be developed into anti-HCV therapeutics, the effects of such compounds on HCV helicase have been extensively studied. Examples include ribavirin triphosphate (Borowski et al., 2001), 5’-O-(4-fluorosulphonylbenzoyl)-esters of ribavirin (FSBR), adenosine (FSBA), guanosine (FSBG) and inosine (FSBI) (Bretner et al., 2004), and ring-expanded (“fat”) nucleosides and nucleotides (Zhang et al., 2003). Much of the data has been previously reviewed (Borowski et al., 2002a; Borowski et al., 2002b). Generally, such compounds inhibit only at very low ATP concentrations, and are competitive with ATP, so that under physiological conditions little or no inhibition is observed.
Compounds resembling nucleoside bases have also been reported to be HCV helicase inhibitors. For example, tetrachlorobenzotriazole (TCBT) and tetrabromobenzotriazole (TBBT) were recently analyzed as helicase inhibitors. Both compounds inhibit unwinding catalyzed by helicases from related viruses (such as West Nile virus) with IC50’s in the low micromolar range, but only TBBT inhibits RNA unwinding by HCV helicase (IC50 ~60 µM). Neither compound inhibits helicase-catalyzed ATP hydrolysis (Borowski et al., 2003), but it is still not clear whether these compounds bind a true allosteric site or if they inhibit unwinding by non-specific interactions with the nucleic acid substrate. Also a compound that resembles nucleotides (called QU663) was recently reported to be a potent HCV helicase inhibitor that competes with the nucleic acid substrate but not ATP with a Ki of 0.75 µM (Maga et al.,,2005)
Many groups have reported non-nucleoside based inhibitors of HCV helicase, primarily in the patent literature. These compounds include a piperidine derivative, heterocyclic carboxamide, antracycline antibiotics (Borowski et al., 2002b), paclitaxel, trifluoperazine (Borowski et al., 2002a), and aminophenylbenzimidazole derivatives (Phoon et al., 2001). Many of these compounds intercalate in nucleic acids and likely act via that non-specific mechanism. Whether or not any of these compounds or other small molecules decrease HCV replication measured using replicons or animal models is still yet to be reported.
Nucleic acid based inhibitors
One of the unique properties of HCV helicase is that, unlike other helicases, the protein binds RNA and DNA in a sequence specific manner. Even the first studies of the protein noted that HCV helicase has a distinctive nucleic acid stimulation profile (Suzich et al., 1993). This means that ATP hydrolysis is stimulated by some nucleic acid polymers much better than it is stimulated by others. The range is quite dramatic. Poly(G) RNA does not stimulate at any measurable level, and poly(U) RNA (or DNA) stimulates best (up to 50 fold). Interestingly, differential stimulation is not entirely due to differences in binding affinity. Direct binding assays confirm that poly(U) binds HCV helicase tighter than polymers composed of the other bases (Gwack et al., 1996), but at saturating nucleic acid concentrations, not all sequences support the same maximum rate of ATP hydrolysis, suggesting that the protein assumes different conformations when bound to different sequences (Lam et al., 2003b). RNA specificity has also been proposed to play a role in directing the helicase, and possibly the entire HCV replication complex, to certain regions of the viral genome. For example, HCV helicase specifically binds both to the 3’-UTR and the 3’-end of the negative strand viral transcript (the complement of the 5’-UTR). This might be necessary during the viral lifecycle to allow the NS5B polymeraseto synthesize RNA in these regions that contain stable secondary structures (Banerjee and Dasgupta, 2001).
Nucleic acid interactions with HCV helicase depend not only on the base composition but also on the composition of the nucleic acid backbone. It is widely recognized that HCV helicase unwinds a DNA duplex more efficiently than an RNA duplex. The biological reason for this, if there is one, is still a mystery because there is no DNA stage in the viral lifecycle, and replication likely occurs on the endoplasmic reticulum (Wolk et al., 2000). However, some reports have detected NS3 in the nucleus (Muramatsu et al., 1997; Errington et al., 1999), where the helicase could modify host gene expression (Sakamuro et al., 1995). Whereas loss of the 2’-OH group from RNA permits the helicase to unwind substrates faster (DNA is unwound faster than RNA), adding a methyl group to this position (2’-O-methyl RNA) weakens helicase interaction with RNA and prevents unwinding (Hesson et al., 2000). The effects are strand specific in that the helicase only appears to sense the chemical composition of the strand with the 3’-overhang (the longer strand in the helicase substrate). Composition of the shorter strand does not affect unwinding rates as drastically, suggesting that interactions are made primarily with the nucleic acid sugars in the single-stranded region not in the double-stranded region of the helicase substrate. When the longer strand (with the 3’ overhang) is DNA, the shorter strand can be composed of RNA, 2’-O-methyl RNA, morpholino-DNA, or phosphorothioate-DNA without affecting unwinding (Hesson et al., 2000; Tackett et al., 2001; Pang et al., 2002). However, if the shorter strand is composed of peptide nucleic acid (where a N-(2-aminoethyl)glycine backbone replaces the deoxyribose phosphates), then unwinding is slower than with natural substrates (Tackett et al., 2001). Whether peptide nucleic acids are poor substrates because of the lack of specific interactions with HCV helicase, or simply because they form more stable duplexes, is still unclear (Tackett et al., 2001).
Two groups have tried to exploit the nucleic acid specificity of HCV helicase with a goal of developing RNA-based inhibitors. Both groups have used SELEX (systematic evolution of ligands by exponential amplification) to find RNA aptamers that tightly bind HCV helicase. In the SELEX procedure, an RNA library is screened for sequences that bind a macromolecule. Only those sequences that bind tightly are amplified to create a new library, and the selection process is repeated with the new library. Although directly using RNA as an antiviral drug will be challenging because of its cellular instability, the information derived from aptamer studies could be used to make more stable derivatives or by delivering RNA directly to infected cells using gene therapy.
One set of aptamers specific to HCV helicase was generated by modifying aptamers that bind tightly and inhibit the NS3-NS4A serine protease. Such aptamers were found to bind truncated NS3 lacking the helicase domains using SELEX. They all share the conserved sequence GA(A/U)UGGGAC (Fukuda et al., 2000), bind NS3 protease over one thousand times tighter than random RNA sequences and are effective at inhibiting the HCV NS3 protease (Fukuda et al., 2000; Nishikawa et al., 2003). When positions Arg130, Arg161, and Lys165 are substituted with Ala, the aptamers no longer bind NS3, suggesting that they interact near these NS3 residues, which are located in the region that links the protease to the helicase (Hwang et al., 2000). To create an aptamer that inhibits protease and helicase activity of NS3, a 14-mer uridine tail was added to one of the most effective HCV protease-binding RNA aptamers. The new, longer aptamer interacts with both the protease and helicase domains of the full-length NS3 protein (Fukuda et al., 2004), binds to the helicase portion of NS3 with high affinity (Kd ~4 nM) (Fukuda et al., 2004), and inhibits the NS3 helicase activity with an EC50 of ~500 nM. The same group has recently reported a new “advanced dual functional” aptamer in which another aptamer, selected for helicase binding (Nishikawa et al., 2004), is tethered to a protease-binding aptamer using a poly(U) linker. This new aptamer is about five times more effective than either aptamer when they are not covalently linked (Umehara et al., 2005).
A second group has also selected for aptamers using the helicase portion of NS3 as the bait in the SELEX procedure (Hwang et al., 2004). This aptamer (called SE RNA) folds to form four stem loops with GC pairs that are similar to the stem loop located at the 3’-terminal of the negative strand HCV RNA. This observation suggests that the SE aptamer might bind the helicase in a similar manner as the stem loop located at the 3’-terminal of the negative strand HCV RNA (Banerjee and Dasgupta, 2001). SE RNA binds the HCV helicase tightly (Kd ~990 pM), efficiently competes with poly(U), stimulates ATP hydrolysis, and potently inhibits RNA unwinding (IC50 ~12.5 nM). When delivered to human liver cells (Huh 7) infected with HCV replicons, the SE aptamer slows HCV RNA synthesis, and interestingly, labeled SE aptamers can also be used as a diagnostic tool to detect the NS3 protein in cells from HCV patients (Nishikawa et al., 2004).
Antibodies
The third, and possibly most ambitious, method that is currently being explored to inhibit HCV helicase is to generate antibody-like molecules that, when expressed intracellularly, will bind and inhibit HCV helicase activities. Almost all HCV patients produce antibodies directed against the NS3 protein, and the vast majority of these bind to the helicase portion of the protein (Chen et al., 1998). Several groups are working toward the goal of introducing recombinant antibodies into cells for “cellular immunization,” a procedure which has been used experimentally with HIV (Goncalves et al., 2002). In this approach, HCV infected cells are transfected with a gene expressing a portion of an antibody selected for reactivity with NS3. One method is to use single chain fragment (ScFv) antibodies. A ScFv is composed of the immunoglobulin heavy chain variable domain connected to the variable region of the light chain by a polypeptide linker. Such a molecule can be constructed using PCR. The other principal method uses an antibody fragment (Fab), which contains the complete light chain and the variable and first constant domains of the heavy chain. A Fab is larger and usually more stable than a ScFv.
To construct a ScFv, immunoglobulin specific PCR is first used to construct a library of human antibody fragments using plasma cells from HCV patients as the PCR template. To identify which antibodies react with HCV helicase, the fragments are fused to a bacteriophage coat protein for phage display, and phages with a high affinity for HCV helicase are purified. Tessman et al. have used this technique to isolate a series of high affinity ScFv’s that specifically interact with HCV helicase (Tessmann et al., 2002). ScFv’s that bind HCV helicase have also been constructed by splicing together the variable domains of monoclonal antibodies (Zhang et al., 2000; Sullivan et al., 2002), and after expression and purification, several of these recombinant proteins inhibit HCV helicase-catalyzed DNA unwinding (Sullivan et al., 2002; Artsaenko et al., 2003). One particular ScFv consists of the variable regions of the human monoclonal antibody CM3.B6, which recognizes an epitope that spans conserved SF2 helicase motifs IV and V (Mondelli et al., 1994). The CM3. B6 ScFv has been expressed in HCV infected hepatocytes (HepG2 cells), immunoblots of which reveal an intra-cellular interaction between the antibody and NS3. HCV RNA synthesis within primary hepatocytes infected with HCV is also reduced by 10-fold when the cells contain a vector carrying the CM3.B6 ScFv gene (Sullivan et al., 2002).
Phage display has also been used to isolate an anti-HCV helicase Fab from a patient infected with HCV genotype 1b. Prabhu et al. have isolated this human Fab, called HFab-aNS3, and demonstrated that it has HCV antiviral activity (Prabhu et al., 2004). HFab-aNS3 recognizes an epitope that spans motifs I to V of the protein, and when purified and pre-incubated with HCV helicase, HFab-aNS3 abolishes detectable DNA unwinding. Intracellular expression of HFab-aNS3 within replicon-transfected Huh 7 cells suppresses NS3 protein expression and significantly inhibits viral RNA synthesis of both subgenomic and full-length HCV replicons (Prabhu et al., 2004).
Conclusions and future directions
HCV helicase has attracted the attention not only of researchers interested in developing novel antiviral drugs, but also those studying how proteins interact with nucleic acids. As one of only three helicases that have been crystallized bound to an oligonucleotide, HCV helicase has become one of the best model proteins to study how helicases unwind duplexes and move on DNA and RNA. Different theories on how chemical energy stored in ATP is transformed into the mechanical force necessary to move a protein from one position to another are currently hotly debated, and many have been tested using the HCV NS3 protein. Obviously, work will continue until HCV helicase’s precise mechanism of action is defined.
While it is still uncertain if HCV helicase functions using an inchworm, rolling, Brownian, or electrostatic mechanism, extensive work has uncovered its basic properties and the roles of several key residues. ATP binds HCV helicase between two RecA-like domains, causing a conformational change that leads to a decrease in the affinity of the protein for nucleic acids. Key residues contacting ATP include Lys210, which likely coordinates the phosphates, Asp290, which could coordinate a divalent metal ion, Glu291, which might act as a catalytic base, and one or more arginines on the adjacent domain. One strand of RNA binds in a second cleft formed perpendicular to the ATP-binding cleft and its binding leads to stimulation of ATP hydrolysis. RNA and/or ATP binding likely causes rotation of domain 2 of the enzyme relative to domains 1 and 3, and somehow this conformational change allows the protein to move like a motor. Key residues involved in RNA binding include Trp501, which locks the protein in position in the absence of ATP, and Glu493, which repels RNA when ATP binds.
Clearly, the biological role of HCV helicase needs to be investigated in more detail. It has long been a mystery why RNA viruses that replicate outside the nucleus encode a helicase. A protein that resolves duplex RNA and DNA structures and displaces proteins bound to nucleic acids, like HCV helicase, could be valuable to [1] provide single stranded templates to the viral polymerase, [2] resolve secondary structure blocking translation or transcriptions, [3] strip proteins from viral RNA, or [4] regulate cellular gene expression. Based on three lines of information, the HCV helicase most likely assists RNA dependent RNA replication by tracking along RNA and resolving double stranded intermediates that form either as secondary structures in a single strand or between (+) sense and (−) sense RNA molecules. First, one group has shown in vitro that NS3 stimulates the ability of NS5B to synthesize long RNAs (Piccininni et al., 2002). Second, the motor action of NS3 on RNA is clearly established. Third, recent studies with subgenomic replicons show that HCV genomes with mutations in the helicase domains of NS3 replicate HCV RNA poorly in cells (Lam and Frick, 2006; Mackintosh et al., 2006). More specifically, our study with subgenomic replicons determined that replicons lacking a functional helicase translated and processed the polyprotein normally and showed reduction in both (−) strand and (+) strand RNA synthesis (Lam and Frick, 2006). Similarly, when an antibody against HCV helicase is co-expressed in cells expressing HCV replicons, there is not only diminished positive strand synthesis but also less synthesis of HCV negative strand RNA and HCV proteins, suggesting that the helicase plays numerous complex and important roles in the viral lifecycle (Prabhu et al., 2004).
While presently it appears that HCV protease and HCV polymerase inhibitors will be developed as the next generation of anti-HCV drugs, compounds inhibiting HCV helicase might also someday prove therapeutically useful. Both NS5B and NS3 protease have clearer roles in HCV replication, and unlike helicases, the mechanisms of serine proteases and RNA polymerases have been understood for decades. Consequently, it is not surprising that HCV helicase inhibitor development lags behind that for the other HCV enzymes. As long as its mechanism and role in replication are not clearly understood, development of antiviral drugs targeting HCV helicase will remain difficult. Nevertheless, rapid progress is being made in the helicase field, and it will not be surprising if HCV helicase inhibitors someday enter clinical trials.
Acknowledgments
This work was supported by National Institutes of Health grant AI052395. I am grateful to Angela M. I. Lam and Ryan S. Rypma for reviewing the manuscript and Christopher M. Frenz for help preparing the figures.
References
- Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Struct. Biol. 1997;4:686–689. doi: 10.1038/nsb0997-686. [DOI] [PubMed] [Google Scholar]
- Artsaenko O, Tessmann K, Sack M, Haussinger D, Heintges T. Abrogation of hepatitis C virus NS3 helicase enzymatic activity by recombinant human antibodies. J. Gen. Virol. 2003;84:2323–2332. doi: 10.1099/vir.0.19299-0. [DOI] [PubMed] [Google Scholar]
- Astumian RD. Thermodynamics and kinetics of a Brownian motor. Science. 1997;276:917–922. doi: 10.1126/science.276.5314.917. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R, Dasgupta A. Specific Interaction of Hepatitis C Virus Protease/Helicase NS3 with the 3’-Terminal Sequences of Viral Positive- and Negative-Strand RNA. J. Virol. 2001;75:1708–1721. doi: 10.1128/JVI.75.4.1708-1721.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein DA, Zittel MC, Keck JL. High-resolution structure of the E.coli RecQ helicase catalytic core. EMBO J. 2003;22:4910–4921. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco PR. Hepatitis C NS3 helicase unwinds RNA in leaps and bounds. Lancet. 2004;364:1385–1387. doi: 10.1016/S0140-6736(04)17238-3. [DOI] [PubMed] [Google Scholar]
- Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1975. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- Borowski P, Deinert J, Schalinski S, Bretner M, Ginalski K, Kulikowski T, Shugar D. Halogenated benzimidazoles and benzotriazoles as inhibitors of the NTPase/helicase activities of hepatitis C and related viruses. Eur. J. Biochem. 2003;270:1645–1653. doi: 10.1046/j.1432-1033.2003.03540.x. [DOI] [PubMed] [Google Scholar]
- Borowski P, Lang M, Niebuhr A, Haag A, Schmitz H, zur Wiesch JS, Choe J, Siwecka MA, Kulikowski T. Inhibition of the helicase activity of HCV NTPase/helicase by 1-beta-D-ribofuranosyl-1,2,4-triazole-3-carboxamide-5 ‘-triphosphate (ribavirin-TP) Acta Biochim. Pol. 2001;48:739–744. [PubMed] [Google Scholar]
- Borowski P, Niebuhr A, Schmitz H, Hosmane RS, Bretner M, Siwecka MA, Kulikowski T. NTPase/helicase of Flaviviridae: inhibitors and inhibition of the enzyme. Acta Biochim. Pol. 2002a;49:597–614. [PubMed] [Google Scholar]
- Borowski P, Schalinski S, Schmitz H. Nucleotide triphosphatase/helicase of hepatitis C virus as a target for antiviral therapy. Antiviral. Res. 2002b;55:397–412. doi: 10.1016/s0166-3542(02)00096-7. [DOI] [PubMed] [Google Scholar]
- Bretner M, Schalinski S, Haag A, Lang M, Schmitz H, Baier A, Behrens SE, Kulikowski T, Borowski P. Synthesis and evaluation of ATP-binding site directed potential inhibitors of nucleoside triphosphatases/helicases and polymerases of hepatitis C and other selected Flaviviridae viruses. Antivir. Chem. Chemother. 2004;15:35–42. doi: 10.1177/095632020401500104. [DOI] [PubMed] [Google Scholar]
- Caruthers JM, Johnson ER, McKay DB. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl. Acad. Sci. USA. 2000;97:13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, Cheng JC, Kou YH, Kao CH, Chiu CH, Wu HY, Chang MF. Roles of the AX(4)GKS and arginine-rich motifs of hepatitis C virus RNA helicase in ATP- and viral RNA-binding activity. J. Virol. 2000;74:9732–9737. doi: 10.1128/jvi.74.20.9732-9737.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Sallberg M, Sonnerborg A, Jin L, Birkett A, Peterson D, Weiland O, Milich DR. Human and murine antibody recognition is focused on the ATPase/helicase, but not the protease domain of the hepatitis C virus nonstructural 3 protein. Hepatology. 1998;28:219–224. doi: 10.1002/hep.510280128. [DOI] [PubMed] [Google Scholar]
- Cho HS, Ha NC, Kang LW, Chung KM, Back SH, Jang SK, Oh BH. Crystal structure of RNA helicase from genotype 1b hepatitis C virus. A feasible mechanism of unwinding duplex RNA. J. Biol. Chem. 1998;273:15045–15052. doi: 10.1074/jbc.273.24.15045. [DOI] [PubMed] [Google Scholar]
- Crampton DJ, Guo S, Johnson DE, Richardson CC. The arginine finger of bacteriophage T7 gene 4 helicase: role in energy coupling. Proc. Natl. Acad. Sci. USA. 2004;101:4373–4378. doi: 10.1073/pnas.0400968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crute JJ, Grygon CA, Hargrave KD, Simoneau B, Faucher AM, Bolger G, Kibler P, Liuzzi M, Cordingley MG. Herpes simplex virus helicase-primase inhibitors are active in animal models of human disease. Nat. Med. 2002;8:386–391. doi: 10.1038/nm0402-386. [DOI] [PubMed] [Google Scholar]
- Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Jr, Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols N, Milburn D, Gerstein M. MolMovDB: analysis and visualization of conformational change and structural flexibility. Nucleic Acids Res. 2003;31:478–482. doi: 10.1093/nar/gkg104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH, Yu X, Wild R, Hingorani MM, Patel SS. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc. Natl. Acad. Sci. USA. 1995;92:3869–3873. doi: 10.1073/pnas.92.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington W, Wardell AD, McDonald S, Goldin RD, McGarvey MJ. Subcellular localisation of NS3 in HCV-infected hepatocytes. J. Med. Virol. 1999;59:456–462. doi: 10.1002/(sici)1096-9071(199912)59:4<456::aid-jmv6>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Flajolet M, Rotondo G, Daviet L, Bergametti F, Inchauspe G, Tiollais P, Transy C, Legrain P. A genomic approach of the hepatitis C virus generates a protein interaction map. Gene. 2000;242:369–379. doi: 10.1016/s0378-1119(99)00511-9. [DOI] [PubMed] [Google Scholar]
- Frick DN. The Hepatitis C Virus Replicase: Insights into RNA-dependent RNA Replication and Prospects for Rational Drug Design. Curr. Org. Chem. 2004;8:223–241. [Google Scholar]
- Frick DN, Rypma RS, Lam AM, Frenz CM. Electrostatic analysis of the hepatitis C virus NS3 helicase reveals both active and allosteric site locations. Nucleic Acids Res. 2004a;32:5519–5528. doi: 10.1093/nar/gkh891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick DN, Rypma RS, Lam AM, Gu B. The nonstructural protein 3 protease/helicase requires an intact protease domain to efficiently unwind duplex RNA. J. Biol. Chem. 2004b;279:1269–1280. doi: 10.1074/jbc.M310630200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K, Umehara T, Sekiya S, Kunio K, Hasegawa T, Nishikawa S. An RNA ligand inhibits hepatitis C virus NS3 protease and helicase activities. Biochem. Biophys. Res. Commun. 2004;325:670–675. doi: 10.1016/j.bbrc.2004.10.089. [DOI] [PubMed] [Google Scholar]
- Fukuda K, Vishnuvardhan D, Sekiya S, Hwang J, Kakiuchi N, Taira K, Shimotohno K, Kumar PK, Nishikawa S. Isolation and characterization of RNA aptamers specific for the hepatitis C virus nonstructural protein 3 protease. Eur. J. Biochem. 2000;267:3685–3694. doi: 10.1046/j.1432-1327.2000.01400.x. [DOI] [PubMed] [Google Scholar]
- Gai D, Zhao R, Li D, Finkielstein CV, Chen XS. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell. 2004;119:47–60. doi: 10.1016/j.cell.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Gallinari P, Brennan D, Nardi C, Brunetti M, Tomei L, Steinkuhler C, De Francesco R. Multiple enzymatic activities associated with recombinant NS3 protein of hepatitis C virus. J. Virol. 1998;72:6758–6769. doi: 10.1128/jvi.72.8.6758-6769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetzinger KR, Rao VB. Defining the ATPase center of bacteriophage T4 DNA packaging machine: requirement for a catalytic glutamate residue in the large terminase protein gp17. J. Mol. Biol. 2003;331:139–154. doi: 10.1016/s0022-2836(03)00636-3. [DOI] [PubMed] [Google Scholar]
- Goncalves J, Silva F, Freitas-Vieira A, Santa-Marta M, Malho R, Yang X, Gabuzda D, Barbas C., 3rd Functional neutralization of HIV-1 Vif protein by intracellular immunization inhibits reverse transcription and viral replication. J. Biol. Chem. 2002;277:32036–32045. doi: 10.1074/jbc.M201906200. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- Grobler JA, Markel EJ, Fay JF, Graham DJ, Simcoe AL, Ludmerer SW, Murray EM, Migliaccio G, Flores OA. Identification of a key determinant of hepatitis C virus cell culture adaptation in domain II of NS3 helicase. J. Biol. Chem. 2003;278:16741–16746. doi: 10.1074/jbc.M212602200. [DOI] [PubMed] [Google Scholar]
- Guo S, Tabor S, Richardson CC. The linker region between the helicase and primase domains of the bacteriophage T7 gene 4 protein is critical for hexamer formation. J. Biol. Chem. 1999;274:30303–30309. doi: 10.1074/jbc.274.42.30303. [DOI] [PubMed] [Google Scholar]
- Gwack Y, Kim DW, Han JH, Choe J. Characterization of RNA binding activity and RNA helicase activity of the hepatitis C virus NS3 protein. Biochem. Biophys. Res. Commun. 1996;225:654–659. doi: 10.1006/bbrc.1996.1225. [DOI] [PubMed] [Google Scholar]
- Hall MC, Matson SW. Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol. 1999;34:867–877. doi: 10.1046/j.1365-2958.1999.01659.x. [DOI] [PubMed] [Google Scholar]
- Heilek GM, Peterson MG. A point mutation abolishes the helicase but not the nucleoside triphosphatase activity of hepatitis C virus NS3 protein. J. Virol. 1997;71:6264–6266. doi: 10.1128/jvi.71.8.6264-6266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesson T, Mannarino A, Cable M. Probing the relationship between RNA-stimulated ATPase and helicase activities of HCV NS3 using 2’-O-methyl RNA substrates. Biochemistry. 2000;39:2619–2625. doi: 10.1021/bi992127a. [DOI] [PubMed] [Google Scholar]
- Howe AY, Chase R, Taremi SS, Risano C, Beyer B, Malcolm B, Lau JY. A novel recombinant single-chain hepatitis C virus NS3-NS4A protein with improved helicase activity. Protein Sci. 1999;8:1332–1341. doi: 10.1110/ps.8.6.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B, Cho JS, Yeo HJ, Kim JH, Chung KM, Han K, Jang SK, Lee SW. Isolation of specific and high-affinity RNA aptamers against NS3 helicase domain of hepatitis C virus. RNA. 2004;10:1277–1290. doi: 10.1261/rna.7100904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Fauzi H, Fukuda K, Sekiya S, Kakiuchi N, Shimotohno K, Taira K, Kusakabe I, Nishikawa S. The RNA aptamer-binding site of hepatitis C virus NS3 protease. Biochem. Biophys. Res. Commun. 2000;279:557–562. doi: 10.1006/bbrc.2000.4007. [DOI] [PubMed] [Google Scholar]
- James JA, Aggarwal AK, Linden RM, Escalante CR. Structure of adeno-associated virus type 2 Rep40-ADP complex: insight into nucleotide recognition and catalysis by superfamily 3 helicases. Proc. Natl. Acad Sci USA. 2004;101:12455–12460. doi: 10.1073/pnas.0403454101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Peterson DL. Expression, isolation, and characterization of the hepatitis C virus ATPase/RNA helicase. Arch. Biochem. Biophys. 1995;323:47–53. doi: 10.1006/abbi.1995.0008. [DOI] [PubMed] [Google Scholar]
- Khu YL, Koh E, Lim SP, Tan YH, Brenner S, Lim SG, Hong WJ, Goh PY. Mutations that affect dimer formation and helicase activity of the hepatitis C virus helicase. J. Virol. 2001;75:205–214. doi: 10.1128/JVI.75.1.205-214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Gwack Y, Han JH, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem. Biophys. Res. Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- Kim DW, Gwack Y, Han JH, Choe J. Towards defining a minimal functional domain for NTPase and RNA helicase activities of the hepatitis C virus NS3 protein. Virus Res. 1997a;49:17–25. doi: 10.1016/s0168-1702(97)01452-4. [DOI] [PubMed] [Google Scholar]
- Kim DW, Kim J, Gwack Y, Han JH, Choe J. Mutational analysis of the hepatitis C virus RNA helicase. J. Virol. 1997b;71:9400–9409. doi: 10.1128/jvi.71.12.9400-9409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- Kim JL, Morgenstern KA, Lin C, Fox T, Dwyer MD, Landro JA, Chambers SP, Markland W, Lepre CA, O’Malley ET, et al. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [published erratum appears in Cell 1997 Apr 4;89(1):159] [DOI] [PubMed] [Google Scholar]
- Kim JW, Seo MY, Shelat A, Kim CS, Kwon TW, Lu HH, Moustakas DT, Sun J, Han JH. Structurally conserved amino acid W501 is required for RNA helicase activity but is not essential for DNA helicase activity of hepatitis C virus NS3 protein. J. Virol. 2003;77:571–582. doi: 10.1128/JVI.77.1.571-582.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleymann G, Fischer R, Betz UA, Hendrix M, Bender W, Schneider U, Handke G, Eckenberg P, Hewlett G, Pevzner V, et al. New helicase-primase inhibitors as drug candidates for the treatment of herpes simplex disease. Nat. Med. 2002;8:392–398. doi: 10.1038/nm0402-392. [DOI] [PubMed] [Google Scholar]
- Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- Krieger N, Lohmann V, Bartenschlager R. Enhancement of hepatitis C virus RNA replication by cell culture- adaptive mutations. J. Virol. 2001;75:4614–4624. doi: 10.1128/JVI.75.10.4614-4624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang WF, Lin YC, Jean F, Huang YW, Tai CL, Chen DS, Chen PJ, Hwang LH. Hepatitis C virus NS3 RNA helicase activity is modulated by the two domains of NS3 and NS4A. Biochem. Biophys. Res. Commun. 2004;317:211–217. doi: 10.1016/j.bbrc.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Kwong AD, Kim JL, Lin C. Structure and function of hepatitis C virus NS3 helicase. Curr. Top. Microbiol. Immunol. 2000;242:171–196. doi: 10.1007/978-3-642-59605-6_9. [DOI] [PubMed] [Google Scholar]
- Lam AM, Frick DN. Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase. J. Virol. 2006;80:404–411. doi: 10.1128/JVI.80.1.404-411.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam AM, Keeney D, Frick DN. Two novel conserved motifs in the hepatitis C virus NS3 protein critical for helicase action. J. Biol. Chem. 2003a;278:44514–44524. doi: 10.1074/jbc.M306444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam AMI, Keeney D, Eckert PQ, Frick DN. Hepatitis C virus NS3 ATPases/helicases from different genotypes exhibit variations in enzymatic properties. J. Virol. 2003b;77:3950–3961. doi: 10.1128/JVI.77.7.3950-3961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam AMI, Rypma RS, Frick DN. Enhanced nucleic acid binding to ATP-bound hepatitis C virus NS3 helicase at low pH activates RNA unwinding. Nucl. Acids Res. 2004;32:4060–4070. doi: 10.1093/nar/gkh743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin MK, Gurjar M, Patel SS. A Brownian motor mechanism of translocation and strand separation by hepatitis C virus helicase. Nat. Struct. Mol. Biol. 2005 doi: 10.1038/nsmb920. [DOI] [PubMed] [Google Scholar]
- Levin MK, Gurjar MM, Patel SS. ATP binding modulates the nucleic acid affinity of hepatitis C virus helicase. J. Biol. Chem. 2003;278:23311–23316. doi: 10.1074/jbc.M301283200. [DOI] [PubMed] [Google Scholar]
- Levin MK, Patel SS. The helicase from hepatitis C virus is active as an oligomer. J. Biol. Chem. 1999;274:31839–31846. doi: 10.1074/jbc.274.45.31839. [DOI] [PubMed] [Google Scholar]
- Levin MK, Patel SS. Helicase from hepatitis C virus, energetics of DNA binding. J. Biol. Chem. 2002;277:29377–29385. doi: 10.1074/jbc.M112315200. [DOI] [PubMed] [Google Scholar]
- Levin MK, Wang YH, Patel SS. The functional interaction of the hepatitis C virus helicase molecules is responsible for unwinding processivity. J. Biol. Chem. 2004;279:26005–26012. doi: 10.1074/jbc.M403257200. [DOI] [PubMed] [Google Scholar]
- Lin C, Kim JL. Structure-based mutagenesis study of hepatitis C virus NS3 helicase. J. Virol. 1999;73:8798–8807. doi: 10.1128/jvi.73.10.8798-8807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Liu D, Wang YS, Gesell JJ, Wyss DF. Solution structure and backbone dynamics of an engineered arginine-rich subdomain 2 of the hepatitis C virus NS3 RNA helicase. J. Mol. Biol. 2001;314:543–561. doi: 10.1006/jmbi.2001.5146. [DOI] [PubMed] [Google Scholar]
- Liu D, Windsor WT, Wyss DF. Double-stranded DNA-induced localized unfolding of HCV NS3 helicase subdomain 2. Protein Sci. 2003;12:2757–2767. doi: 10.1110/ps.03280803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli GA, Gosselin G, Spadari S, Maga G. Hepatitis C virus NS3 NTPase/helicase: different stereoselectivity in nucleoside triphosphate utilisation suggests that NTPase and helicase activities are coupled by a nucleotide-dependent rate limiting step. J. Mol. Biol. 2001;313:683–694. doi: 10.1006/jmbi.2001.5088. [DOI] [PubMed] [Google Scholar]
- Locatelli GA, Spadari S, Maga G. Hepatitis C virus NS3 ATPase/helicase: an ATP switch regulates the cooperativity among the different substrate binding sites. Biochemistry. 2002;41:10332–10342. doi: 10.1021/bi026082g. [DOI] [PubMed] [Google Scholar]
- Lohman TM, Bjornson KP. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- Mackintosh SG, Lu JZ, Jordan JB, Harrison MK, Sikora B, Sharma SD, Cameron CE, Raney KD, Sakon J. Structural and biological identification of residues on the surface of NS3 helicase required for optimal replication of the hepatitis C virus. J. Biol. Chem. 2006;281:3528–3535. doi: 10.1074/jbc.M512100200. [DOI] [PubMed] [Google Scholar]
- Maga G, Gemma S, Fattorusso C, Locatelli GA, Butini S, Persico M, Kukreja G, Romano MP, Chiasserini L, Savini L, et al. Specific targeting of hepatitis C virus NS3 RNA helicase. Discovery of the potent and selective competitive nucleotide-mimicking inhibitor QU663. Biochemistry. 2005;44:9637–9644. doi: 10.1021/bi047437u. [DOI] [PubMed] [Google Scholar]
- Min KH, Sung YC, Choi SY, Ahn BY. Functional interactions between conserved motifs of the hepatitis C virus RNA helicase protein NS3. Virus Genes. 1999;19:33–43. doi: 10.1023/a:1008184522153. [DOI] [PubMed] [Google Scholar]
- Mondelli MU, Cerino A, Boender P, Oudshoorn P, Middeldorp J, Fipaldini C, La Monica N, Habets W. Significance of the immune response to a major, conformational B-cell epitope on the hepatitis C virus NS3 region defined by a human monoclonal antibody. J. Virol. 1994;68:4829–4836. doi: 10.1128/jvi.68.8.4829-4836.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris PD, Byrd AK, Tackett AJ, Cameron CE, Tanega P, Ott R, Fanning E, Raney KD. Hepatitis C virus NS3 and simian virus 40 T antigen helicases displace streptavidin from 5’-biotinylated oligonucleotides but not from 3’-biotinylated oligonucleotides: evidence for directional bias in translocation on single-stranded DNA. Biochemistry. 2002;41:2372–2378. doi: 10.1021/bi012058b. [DOI] [PubMed] [Google Scholar]
- Muramatsu S, Ishido S, Fujita T, Itoh M, Hotta H. Nuclear localization of the NS3 protein of hepatitis C virus and factors affecting the localization. J. Virol. 1997;71:4954–4961. doi: 10.1128/jvi.71.7.4954-4961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadanaciva S, Weber J, Wilke-Mounts S, Senior AE. Importance of F1-ATPase residue alpha-Arg-376 for catalytic transition state stabilization. Biochemistry. 1999;38:15493–15499. doi: 10.1021/bi9917683. [DOI] [PubMed] [Google Scholar]
- Nishikawa F, Funaji K, Fukuda K, Nishikawa S. In vitro selection of RNA aptamers against the HCV NS3 helicase domain. Oligonucleotides. 2004;14:114–129. doi: 10.1089/1545457041526335. [DOI] [PubMed] [Google Scholar]
- Nishikawa F, Kakiuchi N, Funaji K, Fukuda K, Sekiya S, Nishikawa S. Inhibition of HCV NS3 protease by RNA aptamers in cells. Nucleic Acids Res. 2003;31:1935–1943. doi: 10.1093/nar/gkg291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notarnicola SM, Park K, Griffith JD, Richardson CC. A domain of the gene 4 helicase/primase of bacteriophage T7 required for the formation of an active hexamer. J. Biol. Chem. 1995;270:20215–20224. doi: 10.1074/jbc.270.34.20215. [DOI] [PubMed] [Google Scholar]
- Orelle C, Dalmas O, Gros P, Di Pietro A, Jault JM. The conserved glutamate residue adjacent to the Walker-B motif is the catalytic base for ATP hydrolysis in the ATP-binding cassette transporter BmrA. J. Biol. Chem. 2003;278:47002–47008. doi: 10.1074/jbc.M308268200. [DOI] [PubMed] [Google Scholar]
- Pang PS, Jankowsky E, Planet PJ, Pyle AM. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 2002;21:1168–1176. doi: 10.1093/emboj/21.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolini C, Lahm A, De Francesco R, Gallinari P. Mutational analysis of hepatitis C virus NS3-associated helicase. J Gen Virol. 2000;81(Pt 7):1649–1658. doi: 10.1099/0022-1317-81-7-1649. [DOI] [PubMed] [Google Scholar]
- Phoon CW, Ng PY, Ting AE, Yeo SL, Sim MM. Biological evaluation of hepatitis C virus helicase inhibitors. Bioorg. Med. Chem. Lett. 2001;11:1647–1650. doi: 10.1016/s0960-894x(01)00263-3. [DOI] [PubMed] [Google Scholar]
- Piccininni S, Varaklioti A, Nardelli M, Dave B, Raney KD, McCarthy JE. Modulation of the hepatitis C virus RNA-dependent RNA polymerase activity by the non-structural (NS) 3 helicase and the NS4B membrane protein. J. Biol. Chem. 2002;277:45670–45679. doi: 10.1074/jbc.M204124200. [DOI] [PubMed] [Google Scholar]
- Porter DJ. Inhibition of the hepatitis C virus helicase-associated ATPase activity by the combination of ADP, NaF, MgCl2, and poly(rU). Two ADP binding sites on the enzyme-nucleic acid complex. J. Biol. Chem. 1998a;273:7390–7396. doi: 10.1074/jbc.273.13.7390. [DOI] [PubMed] [Google Scholar]
- Porter DJ. A kinetic analysis of the oligonucleotide-modulated ATPase activity of the helicase domain of the NS3 protein from hepatitis C virus. The first cycle of interaction of ATP with the enzyme is unique. J. Biol. Chem. 1998b;273:14247–14253. doi: 10.1074/jbc.273.23.14247. [DOI] [PubMed] [Google Scholar]
- Porter DJ, Short SA, Hanlon MH, Preugschat F, Wilson JE, Willard DH, Jr, Consler TG. Product release is the major contributor to kcat for the hepatitis C virus helicase-catalyzed strand separation of short duplex DNA. J. Biol. Chem. 1998;273:18906–18914. doi: 10.1074/jbc.273.30.18906. [DOI] [PubMed] [Google Scholar]
- Prabhu R, Khalap N, Burioni R, Clementi M, Garry RF, Dash S. Inhibition of hepatitis C virus nonstructural protein, helicase activity, and viral replication by a recombinant human antibody clone. Am. J. Pathol. 2004;165:1163–1173. doi: 10.1016/S0002-9440(10)63377-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preugschat F, Averett DR, Clarke BE, Porter DJT. A steady-state and pre-steady-state kinetic analysis of the NTPase activity associated with the hepatitis C virus NS3 helicase domain. J. Biol. Chem. 1996;271:24449–24457. doi: 10.1074/jbc.271.40.24449. [DOI] [PubMed] [Google Scholar]
- Preugschat F, Danger DP, Carter LH, 3rd, Davis RG, Porter DJ. Kinetic analysis of the effects of mutagenesis of W501 and V432 of the hepatitis C virus NS3 helicase domain on ATPase and strand-separating activity. Biochemistry. 2000;39:5174–5183. doi: 10.1021/bi9923860. [DOI] [PubMed] [Google Scholar]
- Rho J, Choi S, Seong YR, Choi J, Im DS. The arginine-1493 residue in QRRGRTGR1493G motif IV of the hepatitis C virus NS3 helicase domain is essential for NS3 protein methylation by the protein arginine methyltransferase 1. J. Virol. 2001;75:8031–8044. doi: 10.1128/JVI.75.17.8031-8044.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamuro D, Furukawa T, Takegami T. Hepatitis C virus nonstructural protein NS3 transforms NIH 3T3 cells. J. Virol. 1995;69:3893–3896. doi: 10.1128/jvi.69.6.3893-3896.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali DL, Ingram R, Wendel M, Gupta D, McNemar C, Tsarbopoulos A, Chen JW, Hong Z, Chase R, Risano C, et al. Serine protease of hepatitis C virus expressed in insect cells as the NS3/4A complex. Biochemistry. 1998;37:3392–3401. doi: 10.1021/bi972010r. [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Guo S, Tabor S, Richardson CC, Ellenberger T. Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell. 1999;99:167–177. doi: 10.1016/s0092-8674(00)81648-7. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebrov V, Pyle AM. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature. 2004;430:476–480. doi: 10.1038/nature02704. [DOI] [PubMed] [Google Scholar]
- Silverman E, Edwalds-Gilbert G, Lin RJ. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Scaife S, Wigley DB. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107:79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- Soultanas P, Dillingham MS, Velankar SS, Wigley DB. DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase. J. Mol. Biol. 1999;290:137–148. doi: 10.1006/jmbi.1999.2873. [DOI] [PubMed] [Google Scholar]
- Story RM, Steitz TA. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- Subramanya HS, Bird LE, Brannigan JA, Wigley DB. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- Sullivan DE, Mondelli MU, Curiel DT, Krasnykh V, Mikheeva G, Gaglio P, Morris CB, Dash S, Gerber MA. Construction and characterization of an intracellular single-chain human antibody to hepatitis C virus non-structural 3 protein. J. Hepatol. 2002;37:660–668. doi: 10.1016/s0168-8278(02)00270-2. [DOI] [PubMed] [Google Scholar]
- Suzich JA, Tamura JK, Palmer-Hill F, Warrener P, Grakoui A, Rice CM, Feinstone SM, Collett MS. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J. Virol. 1993;67:6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tackett AJ, Chen Y, Cameron CE, Raney KD. Multiple full-length NS3 molecules are required for optimal unwinding of oligonucleotide DNA in vitro. J. Biol. Chem. 2005 doi: 10.1074/jbc.M407971200. [DOI] [PubMed] [Google Scholar]
- Tackett AJ, Wei L, Cameron CE, Raney KD. Unwinding of nucleic acids by HCV NS3 helicase is sensitive to the structure of the duplex. Nucleic Acids Res. 2001;29:565–572. doi: 10.1093/nar/29.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai CL, Chi WK, Chen DS, Hwang LH. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3) J. Virol. 1996;70:8477–8484. doi: 10.1128/jvi.70.12.8477-8484.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai CL, Pan WC, Liaw SH, Yang UC, Hwang LH, Chen DS. Structure-based mutational analysis of the hepatitis C virus NS3 helicase. J. Virol. 2001;75:8289–8297. doi: 10.1128/JVI.75.17.8289-8297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessmann K, Erhardt A, Haussinger D, Heintges T. Cloning and molecular characterization of human high affinity antibody fragments against hepatitis C virus NS3 helicase. J. Virol. Methods. 2002;103:75–88. doi: 10.1016/s0166-0934(02)00017-4. [DOI] [PubMed] [Google Scholar]
- Umehara T, Fukuda K, Nishikawa R, Kohara M, Hasegawa T, Nishikawa S. Rational design of dual-functional aptamers that inhibit the protease and helicase activities of HCV NS3. J. Biochem. (Tokyo) 2005;137:339–347. doi: 10.1093/jb/mvi042. [DOI] [PubMed] [Google Scholar]
- Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardell AD, Errington W, Ciaramella G, Merson J, McGarvey MJ. Characterization and mutational analysis of the helicase and NTPase activities of hepatitis C virus full-length NS3 protein. J. Gen. Virol. 1999;80:701–709. doi: 10.1099/0022-1317-80-3-701. [DOI] [PubMed] [Google Scholar]
- Washington MT, Rosenberg AH, Griffin K, Studier FW, Patel SS. Biochemical analysis of mutant T7 primase/helicase proteins defective in DNA binding, nucleotide hydrolysis, and the coupling of hydrolysis with DNA unwinding. J. Biol. Chem. 1996;271:26825–26834. doi: 10.1074/jbc.271.43.26825. [DOI] [PubMed] [Google Scholar]
- Wolk B, Sansonno D, Krausslich HG, Dammacco F, Rice CM, Blum HE, Moradpour D. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 2000;74:2293–2304. doi: 10.1128/jvi.74.5.2293-2304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong I, Lohman TM. Allosteric effects of nucleotide cofactors on Escherichia coli Rep helicase-DNA binding. Science. 1992;256:350–355. doi: 10.1126/science.256.5055.350. [DOI] [PubMed] [Google Scholar]
- Xu YW, Morera S, Janin J, Cherfils J. AIF3 mimics the transition state of protein phosphorylation in the crystal structure of nucleoside diphosphate kinase and MgADP. Proc. Natl. Acad. Sci. USA. 1997;94:3579–3583. doi: 10.1073/pnas.94.8.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi M, Purcell RH, Emerson SU, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi M, Purcell RH, Emerson SU, Bukh J. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology. 1999;262:250–263. doi: 10.1006/viro.1999.9889. [DOI] [PubMed] [Google Scholar]
- Yanagi M, St Claire M, Shapiro M, Emerson SU, Purcell RH, Bukh J. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology. 1998;244:161–172. doi: 10.1006/viro.1998.9092. [DOI] [PubMed] [Google Scholar]
- Yao N, Hesson T, Cable M, Hong Z, Kwong AD, Le HV, Weber PC. Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- Yao N, Reichert P, Taremi SS, Prosise WW, Weber PC. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure Fold. Des. 1999;7:1353–1363. doi: 10.1016/s0969-2126(00)80025-8. [DOI] [PubMed] [Google Scholar]
- Zhang N, Chen HM, Koch V, Schmitz H, Liao CL, Bretner M, Bhadti VS, Fattom AI, Naso RB, Hosmane RS, Borowski P. Ring-expanded (“fat”) nucleoside and nucleotide analogues exhibit potent in vitro activity against flaviviridae NTPases/helicases, including those of the West Nile virus, hepatitis C virus, and Japanese encephalitis virus. J. Med. Chem. 2003;46:4149–4164. doi: 10.1021/jm030842j. [DOI] [PubMed] [Google Scholar]
- Zhang ZX, Lazdina U, Chen M, Peterson DL, Sallberg M. Characterization of a monoclonal antibody and its single-chain antibody fragment recognizing the nucleoside Triphosphatase/Helicase domain of the hepatitis C virus nonstructural 3 protein. Clin. Diagn. Lab Immunol. 2000;7:58–63. doi: 10.1128/cdli.7.1.58-63.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]