Abstract
We report rapid-freeze-quench X-ray absorption spectroscopy of a di-zinc metallo-β-lactamase (MβL) reaction intermediate. The Zn(II) ions in the dinuclear active site of the S. maltophilia Class B3 MβL move away from each other, by ~ 0.3 Å after 10 ms of reaction with nitrocefin, from 3.4 to 3.7 Å. Together with our previous characterization of the resting enzyme and its nitrocefin product complex, where the Zn(II) ion separation relaxes to 3.6 Å, these data indicate a scissoring motion of the active site that accompanies the ring-opening step. The average Zn(II) coordination number of 4.5 in the resting enzyme appears to be maintained throughout the reaction with nitrocefin. This is the first direct structural information available on early stage di-zinc metallo-β-lactamase catalysis.
β-Lactam containing compounds remain the most widely used antibiotics, despite the growing number of bacteria that show resistance to this group of compounds. The most common resistance strategy is secretion of β-lactamases, which hydrolyze the antibiotic’s four-membered β-lactam ring, leading to an inactive product. A subset of these enzymes (class B of subclasses A–D) require Zn(II) for activity. These metallo-β–lactamases (MβLs) are further subdivided into three subclasses, based on sequence homology.1, 2 L1, a class B3 MβL from Stenotrophomonas maltophilia requires two Zn(II) ions for maximal activity.3 The crystal structure of L1 (Protein Data Bank entry pdb 1SML) shows two distinct Zn(II) ions: Zn1 is tetrahedrally-coordinated by three histidines and one solvent molecule that bridges to Zn2, which is 5-coordinate, ligated by two histidines, a monodentate aspartate, one terminally-bound water and the bridging solvent molecule.4 The crystallographic Zn1-Zn2 separation of 3.45 Å is similar to that determined by EXAFS (3.42 Å) of the same enzyme.5
The observation of product inhibition6 allows for characterization of product complexes, and a more recent crystal structure of ZnZn-L1, complexed with hydrolyzed moxalactam (pdb 2AIO), showed that the distance between the two Zn(II) ions had increased to 3.68 Å, due to a partial rotation of the bound product, made possible by the ring-opening step.7 A similar increase in metal-metal distance was observed in extended X-ray absorption fine structure (EXAFS) studies of L1 complexed with hydrolyzed nitrocefin (3.62 Å), which rotates to a greater degree with ultimate release of the product’s auxiliary carboxylate in favor of the ring sulfur, as illustrated in Figure 1. The increased M-M distance, without a change in coordination number (as evidenced by a lack of significant changes in the average first shell Zn-L distance), was taken as evidence that the bridging hydroxide was absent in the L1-nitrocefin product complex. A solvent-derived bridge was observed in the moxalactam-bound crystal structure, where the average Zn(II) coordination number had increased from 4.5 to 5.5. 7
Figure 1.

Summary of the time-dependent structure of resting ZnZn-L1 (left), freeze quenched after 10 ms of reaction with nitrocefin (center) and the resulting product complex (right) derived from EXAFS. The structure of nitrocefin, shown above the resting structure, is truncated in the 10 ms and product structures. The Zn-Zn separations are given below each structure.
Rotation of the product, as observed in both the EXAFS and diffraction studies, makes structural characterization of reaction intermediates that precede this rotation critical to understanding their action. Crystal structures of MβLs complexed with substrate or trapped during catalysis have not been reported. To date, the only direct structural information available on MβL catalysis comes from EPR studies of a mixed-metal ZnCo hybrid of L18 and the di-Co(II) substituted form of the related Class B1 enzyme BcII from Bacillus cereus,9 both of which demonstrate a clear interaction of substrate with both metal ions during turnover. However, similar information on the native Zn(II)-containing enzymes is not available. To probe the relative positions of Zn1 and Zn2 during catalysis, we examined the EXAFS of 0.5 mM ZnZn-L1 rapid-freeze-quenched after 10 ms reaction with 2.5 mM nitrocefin at 2° C.10 The 10 ms quench time closely mirrors stopped-flow studies of the same system, which show maximum concentration of the nitrocefin-derived reaction intermediate at 10–20 ms of reaction time.6 The present studies mark the first structural characterization of a true Zn(II)-containing MβL reaction intermediate.
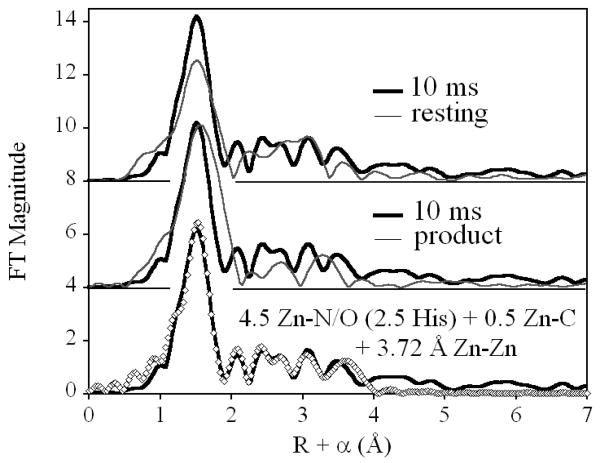
Comparing the Fourier transformed EXAFS of the 10 ms freeze-quenched sample with the EXAFS of the resting enzyme (Figure 2, top) shows (i) an increase in the amplitude associated with the first shell, without a change in peak position or FWHM, (ii) the loss of a feature at R + α. 3.0 Å, and (iii) increased magnitude at R + α. 2.2 and 3.4 Å. This qualitative description is consistent with curve fitting results (Figure S1 and Table S1), which indicate that the average coordination number of Zn is retained (i), and that the Zn-Zn distance increases by nearly 0.3 Å (ii and iii). The larger feature at 2.2 Å in the freeze-quenched FT is attributed to a more ordered Zn-CCO2− interaction, compared to the resting enzyme.
Figure 2.
Comparison of the Fourier transformed EXAFS of ZnZn-L1, freeze quenched after 10 ms of reaction with nitrocefin (bold line), with resting ZnZn-L1 (top) and the ZnZn-L1 product complex with nitrocefin (center), and the best fit to the data (bottom). The data for ZnZnL1 and the ZnZnL1-nitrocefin product complex are reproduced, for comparison, from Costello, et al.5
The lack of any apparent change in coordination number has significant mechanistic implications. Without changing the coordination number of either Zn ion, the presence of a bridging reaction intermediate, as implied by a relatively well-ordered Zn-Zn vector, requires the loss of two Zn ligands from the resting structure. The first most likely comes from the loss of one bond to the bridging solvent, which is incorporated into the product as part of the newly formed carboxylate and remains coordinated in the product complex. 7
In several mechanistic models,11 the bond between Zn2 and its terminal solvent ligand is lost during the catalytic cycle, though the bond is re-formed on product release. Loss of this solvent ligand is required in order to maintain the Zn coordination number, and still form a bond to the anionic nitrogen that results from ring opening. We show these two bonds as dashed in Figure 1, only in that we have no direct evidence for one over the other. However, given the well-established relationship between the protonation state of the ring nitrogen and the blue color of the intermediate12 (the same color displayed by the present set of samples), we must at present favor loss of the solvent ligand.
Comparison with the product complex shows the intermediate has similar first shell amplitude, but the main peak in the product complex FT is shifted to higher R, due to incorporation of the product sulfur atom into the Zn2 coordination sphere. With no evidence of a large atom scatterer in the first shell of the intermediate, we must conclude that product rotation, leading to a measurable Zn-S interaction, has not yet occurred at 10 ms, and that the apparent increase in first shell amplitude for the intermediate is therefore due to lower disorder in the first shell of the intermediate. The outer shell scattering of the product complex is of lower amplitude than in the intermediate, also consistent with a more ordered site in the intermediate. This may be reflective of a less planar orientation of the product, relative to substrate and/or intermediate.
Meanwhile, the dramatic increase in Zn-Zn separation to 3.72 Å suggests that even at 10 ms, the Zn-(OH)-Zn bridge has been lost (this nearly linear arrangement is expected to lead to more dramatic metal-oxygen-metal scattering than is observed). We note that a second, much shallower minimum was observed, with a Zn-Zn separation of 3.37 Å. This distance is more in line with that of the resting enzyme, although it is closer to that observed in similar systems with a bridging bidentate carboxylate,13 as opposed to the terminal monodentate carboxylate expected in the MβL active site. In either case, inclusion of two Zn-Zn populations, suggests that such a species represents no more than ~ 11 % of the total enzyme present.
Motion of the Zn ions has potentially large mechanistic implications. The two metals are expected to be anchored to the auxiliary carboxylate of the fused ring (Zn2) and the lactam carbonyl (Zn1). The 0.3 Å movement of the metal ions, away from each other, suggests that binding of substrate, and possibly release of the bridging hydroxyl, is accompanied by a scissoring motion that exerts added pressure on the N-C(=O) bond, helping facilitate the ring opening reaction. This proposal is consistent with prior observations that suggest all mono-Zn MβLs are active,14, 15 and that inclusion of the second metal ion increases the enzyme’s catalytic efficiency, without affecting the overall rates of reaction.5 Further experiments are planned for the mixed-metal hybrid, which will allow us to assess structural changes at each metal site, independently.
Supplementary Material
Table 1.
EXAFS-derived structural models for ZnZn-L1 in the resting state,a freeze-quenched after 10 ms of reaction with nitrocefinb and its product complexa with hydrolyzed nitrocefin.
| sample | Zn-Lc | Zn-His | Zn-Zn (Å) |
|---|---|---|---|
| restinga | 4 N/O @ 2.02 Å | 2.5 | 3.42 |
| 10 msb | 4.5 N/O @ 2.03 Å | 2.5 | 3.72 |
| producta | 4 N/O @ 2.04 Å | 2.5 | 3.62 |
| 0.5 S @ 2.29 Å |
From Costello, et al.5
This work. See Supporting Information for details.
Zn-L distances ± 0.01 Å; Zn-Zn distances ± 0.02 Å.
Acknowledgments
This work was supported by the National Institutes of Health (NCRR P20RR-16480 to DLT and AI056231 to BB), by the Volwiler Professorship at Miami University (to MWC) and by the National Science Foundation (CHE0809985 to DLT).
Footnotes
Supporting Information Available: Experimental procedures, including sample preparation and EXAFS data collection and analysis, along with one Figure (S1) and one Table (S1), showing detailed EXAFS curve fitting results, are included as Supporting Information. This information is available free of charge at http://pubs.acs.org/.
References
- 1.Galleni M, Lamotte-Brasseur J, Rossolini GM, Spencer J, Dideberg O, Frere JM. Antimicrob Agents Chemo. 2001;45:660–663. doi: 10.1128/AAC.45.3.660-663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bebrone C. Biochem Pharmacol. 2007;74:1686–1701. doi: 10.1016/j.bcp.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 3.Periyannan G, Shaw PJ, Sigdel T, Crowder MW. Prot Sci. 2004;13:2236–2243. doi: 10.1110/ps.04742704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ullah JH, Walsh TR, Taylor IA, Emery DC, Verma CS, Gamblin SJ, Spenser J. J Mol Biol. 1998;287:125–136. doi: 10.1006/jmbi.1998.2148. [DOI] [PubMed] [Google Scholar]
- 5.Costello AL, Periyannan G, Yang KW, Crowder MW, Tierney DLJ. Biol Inorg Chem. 2006;11:351–358. doi: 10.1007/s00775-006-0083-z. [DOI] [PubMed] [Google Scholar]
- 6.McManus-Munoz S, Crowder MW. Biochemistry. 1999;38:1547–1553. doi: 10.1021/bi9826512. [DOI] [PubMed] [Google Scholar]
- 7.Spencer J, Read J, Sessions RB, Howell S, Blackburn GM, Gamblin SJ. J Am Chem Soc. 2005;127:14439–14444. doi: 10.1021/ja0536062. [DOI] [PubMed] [Google Scholar]
- 8.Hu Z, Periyannan G, Bennett B, Crowder MW. J Am Chem Soc. 2008;130:14207–14216. doi: 10.1021/ja8035916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tioni MF, Llarrull LI, Poeylaut-Palena AA, Marti MA, Saggu M, Periyannan GR, Mata EG, Bennett B, Murgida DH, Vila AJ. J Am Chem Soc. 2008;130:15852–15863. doi: 10.1021/ja801169j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrity JD, Bennett B, Crowder MW. Biochemistry. 2005;44:1078–1087. doi: 10.1021/bi048385b. [DOI] [PubMed] [Google Scholar]
- 11.Xu D, Guo H, Cui Q. J Am Chem Soc. 2007;129:10814–10822. doi: 10.1021/ja072532m. [DOI] [PubMed] [Google Scholar]
- 12.Kaminskaia NV, Spingler B, Lippard SJ. J Am Chem Soc. 2001;123:6555–6563. doi: 10.1021/ja002699e. [DOI] [PubMed] [Google Scholar]
- 13.Thomas PW, Stone EM, Costello AL, Tierney DL, Fast W. Biochemistry. 2005;44:7559–7565. doi: 10.1021/bi050050m. [DOI] [PubMed] [Google Scholar]
- 14.Cricco JA, Orellano EG, Rasia RM, Ceccarelli EA, Vila AJ. Coord Chem Rev. 1999;190–192:519–535. [Google Scholar]
- 15.Llarrull LI, Tioni MF, Vila AJ. J Am Chem Soc. 2008;130:15842–15851. doi: 10.1021/ja801168r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.