Abstract
Cobalt and zinc binding by the subclass B1 metallo-β-lactamase BcII from Bacillus cereus is examined by X-ray absorption spectroscopy, at various levels of metal loading. The data show that a significant amount of the dinuclear enzyme is formed, even at substoichiometric levels of metal loading, whether the added metal is Zn(II) or Co(II). Increasing metal addition, from 0.5 to 1.0 to 2.0 eq per mole of enzyme, are shown to result in a more ordered active site. While Zn(II) appears to show no preference for the Zn1 (3H) or Zn2 (DCH) sites, the extended X-ray absorption fine structure (EXAFS) suggests that Co(II) shows a slight preference for the DCH site at low levels of added Co(II). The results are discussed in the context of similar metal binding studies of other B1 metallo-β-lactamases.
1. Introduction
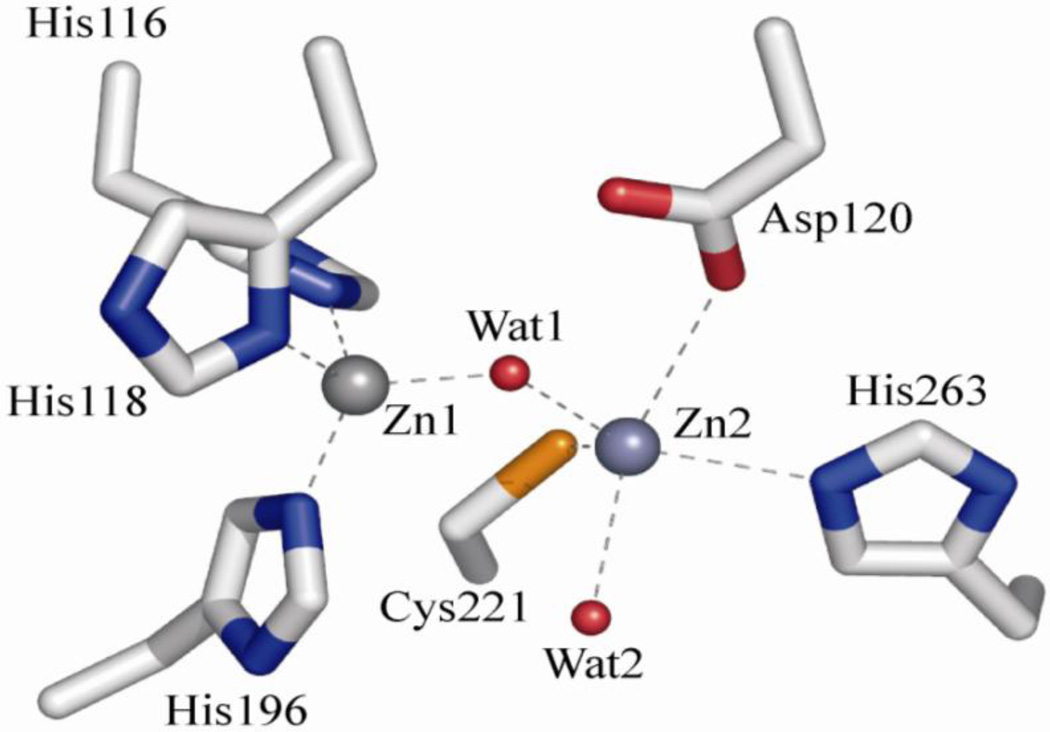
Metallo-β-lactamases (MβLs) pose an ever-increasing threat to human health, as they continue to spread throughout the bacterial world [1]. Unlike the related serine-active β-lactamases (SβLs) [2, 3], which utilize a serine side chain to catalyze antibiotic hydrolysis, no clinically-viable inhibitors for MβLs have been reported. MβLs have been grouped into the three subclasses (B1–B3), based on sequence analysis and enzymatic properties. We will focus here on the B1 enzymes because they are by far the most ubiquitous, as well as the most clinically relevant [4, 5]. The most commonly encountered active site structure in purified B1 enzymes is a solvent-bridged dinuclear cluster, formed from two distinct Zn(II)-binding sites. One site is formed by three histidine side chains (the Zn1 or 3H site), while the other is formed from the side chains of one histidine, one cysteine and one aspartate, and includes a terminal solvent molecule (the Zn2 or DCH site), as shown in Figure 1. Both share an additional solvent molecule that bridges the two metal ions. While it is widely accepted that the metal binding sites in B1 MβLs are capable of binding two equivalents of Zn(II) in a solvent-bridged dinuclear cluster in vitro, the exact nature of the active species in vivo remains the subject of some debate [6–9].
Figure 1.
Metal binding site of di-Zn(II) BcII, a prototypical B1 MβL, from pdb entry 3i13 (González et al, Biochemistry 2010).
Early studies of Zn(II) binding by the B1 MβL BcII from Bacillus cereus, the most heavily studied of all MβLs, showed a large disparity in metal affinity, with Kd values of 0.62 nM and 1.50 µM for binding of the first and second equivalents, respectively [10]. Such a large difference would imply sequential loading of the metal site, and indeed the first crystal structure reported for BcII contained a single equivalent of zinc located in the 3H site [11]. However, this structure was obtained at a pH lower than physiological pH, and it was later shown that a decrease in pH gives rise to metal dissociation [12, 13]. Much of what has been learned since that first crystal structure comes from studies of the Co(II)-substituted enzymes. Titration of BcII with substoichiometric cobalt indicates a substantially smaller difference in binding affinity, based on the appearance of d-d band structure associated with Co(II) in the 3H site and a S6Co(II) LMCT band corresponding to cobalt bound at the DCH site, with as few as 0.3 eq of Co(II)/enzyme [6]. Similarly, an EPR titration of apo-BcII with Co(II) showed the initially formed, axial species maximizes at just 0.8 eq of Co(II)/enzyme, while the subsequently formed rhombic species maximizes at 2.0 eq. An observed discrepancy between the quantity of cobalt added and that detected by EPR was attributed to the presence of spin coupled Co(II) present in solvent-bridged binuclear clusters [14]. This contrasts with our preceding studies of Co(II)-binding by the B1 MβLs CcrA and Bla2, which showed that Co(II) loads both sides of the metal site, without formation of a bridged cluster, based on the lack of an ordered metal-metal interaction in the extended X-ray absorption fine structure (EXAFS) [15, 16].
Different kinetic studies of BcII fail to agree on the metal content of the predominant species present under physiological conditions. For example, one study suggests that the enzyme’s physiological state is apo, with substrate binding inducing a conformational change that results in recruitment of metal from the surroundings, activating the enzyme only when necessary [17]. Others suggest, based on kinetic analyses of the zinc and cobalt substituted enzymes, that metal binding is cooperative, with the loss of one metal ion upon turnover [7, 8]. The enzyme would then require the addition of metal from the surroundings to regenerate the active site. Our own stopped-flow kinetic studies of the same enzyme suggest that a mononuclear form, with the metal ion located in the DCH site, and the dinuclear form are both active, while the dizinc form is the physiologically important form of the enzyme [14, 18].
Whereas Co(II) substitution has proven useful for the characterization of the structure and mechanism of MβLs, significantly less information is available regarding the native Zn(II) enzymes. High resolution mass spectrometry of BcII showed that the mass corresponding to dizinc BcII grows linearly from 0 to 2 eq, as the population of apo-BcII steadily decreases [19]. The population of monozinc BcII was shown to grow to a small population (ca. 20% of the total) before disappearing, consistent with the suggestion of metal binding cooperativity. Isothermal calorimetry of Zn(II) binding by apo-BcII showed only one binding event, corresponding to a Kd of 30 nM, which was proposed to indicate equivalence in either the two binding constants or the two binding enthalpies [13]. However, the same study reported a dissociation constant of ca. 20 µM for binding of the second metal ion, based on a kinetic analysis. A difference of three orders of magnitude in binding affinity would necessarily require that metal binding be sequential. A previous EXAFS study of BcII suggested that the binding of Zn(II) is scrambled, loading both sites indiscriminately at 1 and 2 eq of Zn/enzyme [10]. A direct comparison of the native Zn(II) and Co(II)-substituted forms has yet to be reported.
We have previously demonstrated, through multi-faceted spectroscopic studies, significant variability in the metal binding behavior of B1 MβLs. For example, CcrA from Bacillus fragilis binds both Zn(II) and Co(II) sequentially, populating the 3H site completely before loading the DCH site [15, 16]. In contrast, similar studies of Bla2 from Bacillus anthracis showed that metal binding depends on the identity of the metal ion [16]. Zinc clearly binds to Bla2 in the same manner as CcrA, preferentially loading the 3H site first, while cobalt shows little or no preference for one site over the other, populating both sites roughly equally, regardless of stoichiometry. In this study, we report X-ray absorption spectroscopic studies of the BcII active site, as a function of added zinc, or cobalt, from 0.5 to 2.0 eq of metal per enzyme.
2. Experimental Procedures
BcII containing 0.5, 1.0 and 2.0 eq of Zn (II) and Co (II) were prepared and characterized according to published procedures [6]. Samples for EXAFS (0.5 – 2 mM) were prepared containing ca. 30% (v/v) glycerol as glassing agent, loaded into Lucite cuvettes with 6 µm polypropylene windows, and flash frozen in liquid nitrogen. X-ray absorption spectra were measured at the National Synchrotron Light Source (NSLS), beamline X3B, using a Si(111) double crystal monochromator; harmonic rejection was accomplished using a Ni focusing mirror. Fluorescence excitation spectra were measured with a 13-element solid-state Ge array detector. Samples were held at ~ 15 K in a Displex cryostat during XAS measurements. X-ray energies were calibrated by reference to the absorption spectrum of the appropriate metal foil, measured concurrently with the protein spectra. All of the data shown represent the average of ~ 12 total scans, from two independently prepared samples for each stoichiometry. Data collection and reduction were performed according to published procedures [15, 20], with E0 set to 9675 eV for Zn and 7730 eV for Co. The Fourier-filtered EXAFS were fit to Equation 1 using the nonlinear least-squares engine of IFEFFIT that is distributed with SixPack [21, 22].
| (1) |
In Eq. 1, Nas is the number of scattering atoms within a given radius (Ras, ± σas), As(k) is the backscattering amplitude of the absorber-scatterer (as) pair, Sc is a scale factor, ϕas(k) is the phase shift experienced by the photoelectron, λ is the photoelectron mean free-path, and the sum is taken over all shells of scattering atoms included in the fit. Theoretical amplitude and phase functions, As(k)exp(−2Ras/λ) and ϕas(k), were calculated using FEFF v. 8.00 [23]. The scale factor (Sc) and ΔE0 for Zn-N (Sc = 0.78, ΔE0 = −16 eV), Zn-S (0.85, −16 eV), Co-N (0.74, −21 eV) and Co-S (0.85, −21 eV) scattering were determined previously and held fixed throughout this analysis [15, 20]. Fits to the current data were obtained for all reasonable integer or half-integer coordination numbers, refining only Ras and σas2 for a given shell. Multiple scattering (ms) contributions from histidine ligands were approximated according to published procedures [15, 20], fixing the number of imidazole ligands per metal ion at half-integral values while varying Ras and σas2 for each of the four combined ms pathways (see Table 1 and Tables S1–S6). Metal-metal (Co-Co and Zn-Zn) scattering was modeled by fitting calculated amplitude and phase functions to the experimental EXAFS of Co2(salpn)2 and Zn2(salpn)2.
Table 1.
EXAFS curve fitting results for Zn- and Co-BcII.a
| Fit | 4 M-N/O 0.5 M-S 2 | 2 M-Hisb | M-M | % ISc | % IMd | Refined M-M CN |
|---|---|---|---|---|---|---|
| Zn 0.5 1.0 2.0 |
2.02 (5.0) 2.28 (8.0) 2.03 (6.6) 2.28 (6.0) 2.03 (6.3) 2.27 (2.6) |
2.95 (1.6) 3.21 (5.0) 4.12 (9.0) 4.41 (21) 2.93 (0.6) 3.18 (2.0) 4.08 (13) 4.44 (14) 2.90 (3.1) 3.18 (5.8) 4.08 (11) 4.43 (15) |
3.41 (13) 3.41 (10) 3.42 (8.3) |
37 36 41 |
24 30 47 |
0.12 0.30 0.91 |
| Co 0.5 1.0 2.0 |
2.03 (6.9) 2.30 (1.7) 2.06 (6.1) 2.30 (3.7) 2.09 (1.3) 2.31 (5.4) |
2.94 (4.2) 3.24 (15) 4.03 (12) 4.28 (18) 2.94 (2.2) 3.25 (14) 4.02 (10) 4.28 (16) 2.93 (1.8) 3.19 (14) 4.08 (14) 4.32 (12) |
3.56 (8.8) 3.55 (6.0) 3.55 (5.4) |
79 61 23 |
39 43 27 |
0.15 0.33 0.96 |
Distances (Å) and disorder parameters (in parentheses, σ2 (10−3 Å2)) shown derive from integer or half-integer coordination number fits to filtered EXAFS data [k = 1.5–12.5 Å−1; R = 0.2–4.2 Å].
Imidazole multiple scattering paths represent combined paths, as described previously (see Materials and Methods).
Percentage reduction of the fit residual for a first shell fit that includes 0.5 S scatterers over one that utilizes only N/O scatterers (% IS). See Supporting Information for details.
Percentage reduction of the fit residual for a fit that includes 4 N/O and 0.5 S scatterers in the first shell, the outer shell scattering from two coordinated imidazoles and a metal-metal vector, compared to one that lacks the metal-metal vector (% IM). See Supporting Information for details.
3. Results
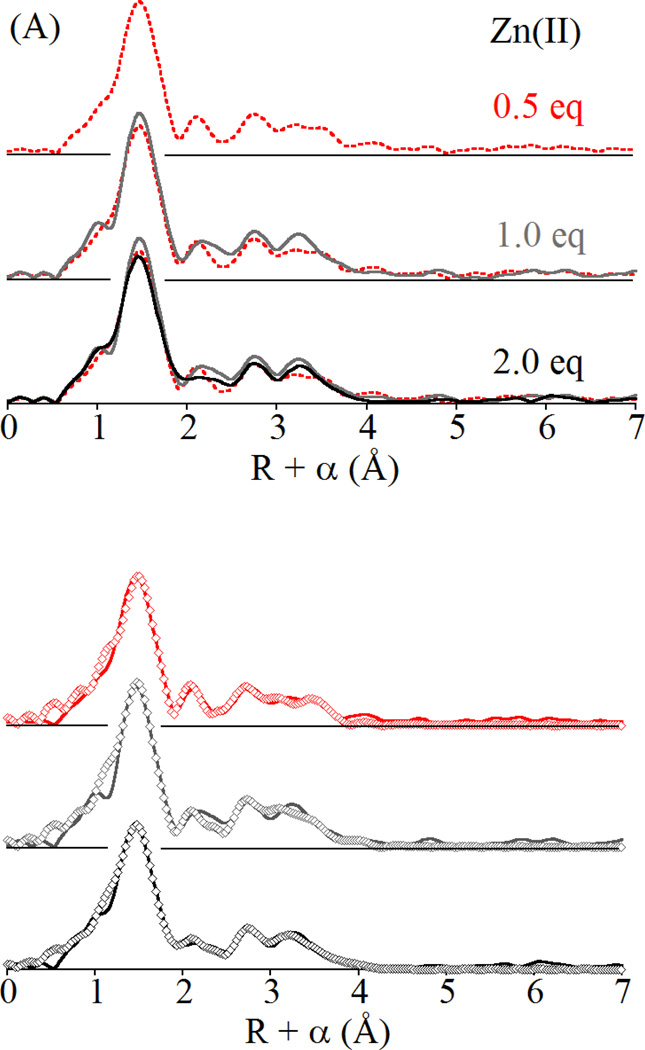
Previous spectroscopic studies of Co(II) binding by BcII indicate little preference for one site over the other, with both the 3H and DCH sites populated with as few as 0.3 eq of added Co(II) [6, 24]. The preceding XAS study of Zn(II) binding by BcII suggested similar behavior, with no appreciable difference in the EXAFS of mono- and di-Zn(II) BcII [10]. As can be seen in Figure 2, the EXAFS of BcII changes little from 0.5 to 1.0, to 2.0 eq of added Zn(II). All three spectra are nearly superimposable, aside from a notable increase in the outermost feature in the Fourier transforms, which we attribute to ordering of the metal-metal interaction as the site is loaded. The curve fits, summarized in Table 1, and presented in detail in Figures S1 – S3 and Tables S1 – S3, are consistent with this interpretation. At 0.5 eq of Zn(II), the data are best fit with an average of 4 N/O and 0.5 S in the primary coordination sphere. Inclusion of the relatively small sulfur contribution leads to a 37% improvement in the fit residual, warranting its inclusion in modeling the data (see Figure S1 and Table S1). Multiple-scattering fits indicate an average of two coordinated imidazoles per zinc, also consistent with roughly equal population of the 3H and DCH sites. The multiple scattering fit residual decreases 24% with addition of a Zn-Zn scattering interaction at 3.41 Å, suggesting that even at 0.5 eq of Zn, a significant amount of di-Zn BcII is present.
Figure 2.
(A) Comparison of Fourier transformed EXAFS data for BcII loaded with 0.5 (red), 1.0 (gray) and 2.0 (black) equivalents of Zn(II) and (B) corresponding best fits (open symbols) including a Zn-Zn vector. For complete fitting details, see Supporting Information, Figures S1 – S3 and Tables S1 – S3.
Increasing the amount of added Zn(II) gives very similar results. At one eq of added Zn(II) (Figure 2, center), the best fit remains a mixture of the 3H and DCH sites, with inclusion of a Zn-S path improving the first shell fit by 36% and the addition of a Zn-Zn interaction improving the full multiple scattering fit by 30%. At two eq of Zn(II), the best fit employs the same structural model, where the addition of 0.5 S scatterers improves the first shell fit by 41% over the corresponding N/O only fit, and the Zn-Zn path improves the full fit by 47% (Figure 2, bottom). As illustrated in Table 1, increased zinc loading appears to monotonically decrease the amount of static disorder in the site, based on a steady decline in σ2 for the Zn-S shell, and a steady increase in fit improvement from inclusion of the Zn-Zn vector.
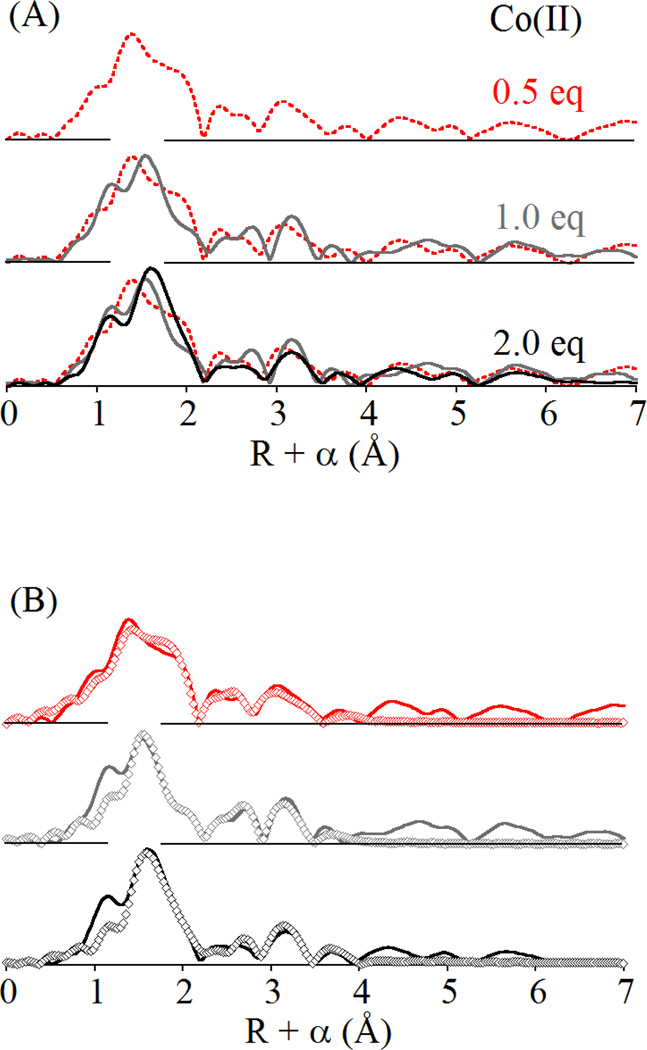
Comparison with the EXAFS of the Co(II)-substituted enzyme tells a similar story, while perhaps pointing out subtle differences between Zn(II) and Co(II) binding. The Co(II)-BcII fits are summarized in Figures S4 – S6 and Tables S4 – S6. At 0.5 equivalents of Co (II), the best fit to the EXAFS data is a mixture of the 3H and DCH sites, including an average of four low Z (N/O) donors and one half of a sulfur donor per cobalt (Figure 3, top). The 0.5 S path reduces the first shell fit residual by 79% (see Tables 1 and S4). Multiple scattering fits indicate an average of two histidine ligands per cobalt, while the ms fit residual improves by 39% with the inclusion of a Co-Co interaction at 3.56 Å. This distance is 0.14 Å longer than the 3.42 Å reported for the dizinc enzyme, consistent with the increase of 0.06 Å in covalent radius for Co(II) relative to Zn(II) [25].
Figure 3.
(A) Comparison of Fourier transformed EXAFS data for BcII loaded with 0.5 (red), 1.0 (gray) and 2.0 (black) equivalents of Co(II) and (B) corresponding best fits (open symbols) including a Co-Co vector. For complete fitting details, see Supporting Information, Figures S4 – S6 and Tables S4 – S6.
At one equivalent of added cobalt, the BcII EXAFS show some subtle differences from 0.5 eq, chiefly in the shape of the main peak in the FT, corresponding to the cobalt ion’s primary coordination sphere. The prominent shoulder on the high-R side of this peak is substantially reduced in the 1.0 eq FT. In addition, there is a notable shift in the main component of this peak to longer distance. However, the data are best fit with nearly the same structural model as the 0.5 eq EXAFS (Figure 3, center). The N/O bond distance increases slightly, and the Co-S disorder increases slightly, suggesting that the average coordination number of the protein-bound Co(II) may be increasing as more Co(II) is added. In this case, the fit residual improves by 61% when the Co-S contribution is included in the first shell, and 43% upon addition of a Co-Co vector at 3.55 Å. EXAFS of the di-cobalt enzyme shows a still longer N/O distance that encompasses the shoulder attributed to Co-S scattering at 0.5 and 1.0 eq of added Co(II) (Figure 3, bottom). Addition of a Co-Co distance of 3.55 Å improves the fit by 27%, significantly less than observed in fits to the 0.5 and 1.0 eq data. Overall, despite some cosmetic differences, the Co(II)-substituted BcII EXAFS data are best fit with a structural model representing the average of the DCH and 3H sites, regardless of the level of added Co(II). Similar to titration of the enzyme with Zn(II) (above), increasing addition of Co(II) appears to lead to an overall ordering of the site, while the decreasing importance of the Co-S path to the first shell fits seems to indicate that indeed Co shows some slight preference for the DCH site at substoichiometric concentrations.
4. Discussion
Metal binding by the metallo-β-lactamases has been the subject of some debate since they were first discovered. The EXAFS analysis presented here marks the first direct structural information regarding BcII metal binding at substoichiometric concentrations, and the first direct comparison of Zn(II) and Co(II) binding by this, the most well-studied MβL. Competition and spectroscopic studies have shown that two eq of Co(II) bind to BcII with similar affinity, giving rise to a mixed population of mononuclear (both 3H and DCH) and dinuclear species, even at substoichiometric concentrations of metal [6]. BcII has been shown bind Zn(II) cooperatively, based on activity assays of the Zn protein [19]. The data presented here are not inconsistent with this picture, although there is better agreement with the model of simultaneous binding reported for Co(II)-BcII. With substoichiometric amounts of metal present (Zn or Co), the XAS clearly provides evidence that both sites are populated, and that a significant fraction is part of a bridged dinuclear cluster. The improvement of the fits upon inclusion of a metal-metal interaction at 0.5 eq of metal per protein (24% for Zn, 39% for Co), is significant in the face of only a 14% increase in the number of variable parameters. Additional added zinc serves to order the site, as evidenced by the increasing fit improvement from inclusion of the metal-metal interaction (from 24 to 30 to 37%), and the general decrease in σ2 for the Zn-S and Zn-Zn scattering paths. The values of σ2 reported in Tables 1 and S1–S6 indicate that the uncertainty in the Zn-Zn distance is as much as 0.1 Å. Given that the B1 MβL active site contains a single hydroxo bridge, it is not unreasonable to expect a relatively large distribution in distance, and we note here that we have previously reported substantial motion of the metal ions during substrate turnover [26]. However, it is also important to bear in mind that the value of σ2 for the metal-metal interaction is refined along with σ2 for 6 other scattering paths, and that they are not completely uncorrelated. For this reason, we prefer to discuss the Zn-Zn σ2 in terms of the general disorder in the metal site. In each case, allowing the coordination number of the metal-metal scattering path to float follows a similar trend, refining to 0.12, 0.30 and 0.91, respectively, for 0.5, 1.0 and 2.0 eq of added Zn(II). However, this general trend also argues against a completely cooperative binding model, as fully cooperative binding would lead to the expectation of no discernable differences from 0.5 to 2.0 eq of added Zn(II) and refined coordination numbers of 0.25, 0.5 and 1.0 (Table 1).
These trends are more pronounced when the added metal is cobalt. While the first shell fits improve dramatically when the Co-S scattering pathway is included, and the full ms fits do the same when the Co-Co interaction is added, the effect is not linear as it is with Zn(II) addition. In contrast, the importance of the metal-sulfur path appears to decrease with increasing added Co(II), from 79% improvement in the fit residual at 0.5 eq to just 23% when the site is fully loaded, suggesting that the Co(II) ion initially populates the DCH site at very low metal concentrations. This is consistent with our earlier optical studies [6]. The greatest improvement on addition of the Co-Co path is seen at 1.0 eq of added Co(II), rather than at 2.0 eq, as seen for Zn(II), which may reflect the tendency of Co(II) to adopt a higher coordination number, as we have noted previously for the MβLs CcrA (B1) and Bla2 (B1) [15, 16]. In support of this suggestion, we note that the first shell N/O bond length increases by 0.06 Å from 0.5 to 2.0 eq of added Co(II). When the Co-Co coordination number is refined, the trend is similar to that seen with Zn(II), giving 0.15 at 0.5 eq of added Co(II), 0.33 at 1.0 eq and 0.96 at 2.0 eq (Table 1). The similar features of the dinuclear adducts observed with Zn(II) and Co(II) are in excellent agreement with the recently reported crystal structure of Co(II)-substituted BcII [27] and the previously reported di-Zn(II) structure [28].
The XAS metal-binding titration of BcII presented here, in the context of our earlier studies, shows three distinct patterns of metal binding behavior among three closely related examples from subclass B1. While metal binding by BcII displays results in the formation of the di-Zn(II) or di-Co(II) proteins even at substoichiometric metal content, CcrA binds both Zn(II) and Co(II) sequentially, loading the 3H site fully before filling the DCH site [15, 16]. Further, Co(II) binds sequentially, but the EXAFS gives no indication of a bridged structure, and both Co(II) ions adopt a higher coordination number than Zn(II) in the same site. Bla2 is the most unusual, binding Zn(II) like CcrA (sequentially), while binding Co(II) like BcII [16]. Despite this diversity in metal binding, it is important to note that all three examples display identical structures when the metal site is fully loaded.
Given identical metal site structures, these differences in metal binding mode must arise due to secondary interactions. The sequence homology between BcII and CcrA is only modest (32%), but both retain the HXHXD motif present in all metallo-β-lactamases and a number of other conserved sequences near the active site, including thirteen conserved glycines [11]. Bla2 provides a much more compelling comparison, as its sequence is nearly identical to that of BcII (92% homology; 89% identity) with only three changes in the active site pocket [29, 30], all in the secondary shell of the active site. These include replacement of Ile39 with valine, shortening the aliphatic side chain by one methylene. While, this change should do little to alter overall protein structure, this side chain is only 4.1 Å from His210 in BcII, and may alter the orientation of the imidazole sufficiently to affect metal binding. In addition, Gly151 in BcII is replaced with glutamate in Bla2, introducing a large charged side chain in place of a hydrogen atom, on the surface of the protein. However, this residue is only two amino acids away from a metal binding histidine of the 3H site. The associated loss of flexibility in the protein backbone could affect metal binding. Finally, Thr182 in BcII is replaced with alanine in Bla2. This is perhaps the most likely source of the differences in metal binding behavior, introducing a hydrogen bonding residue on the edge of the active site pocket, which can drastically alter the hydrogen-bonding network that penetrates the active site.
We have shown previously that second sphere mutations arising from directed evolution experiments give rise to significant changes in the activity of MβLs [31, 32]. The present data show a natural example of the effect of outer sphere residues on active site structure and reactivity. With only three alterations responsible for such different metal-binding behavior, BcII and Bla2 offer a unique opportunity to probe the effect of outer sphere residues on active site structure and reactivity which can be probed through site-directed mutagenesis.
Highlights.
Cobalt and zinc binding by the subclass B1 metallo-β-lactamase BcII from Bacillus cereus is examined by X-ray absorption spectroscopy, at various levels of metal loading. The data show that a significant amount of the dinuclear enzyme is formed, even at substoichiometric levels of metal loading, whether the added metal is Zn(II) or Co(II). Increasing metal addition, from 0.5 to 1.0 to 2.0 eq per mole of enzyme, are shown to result in a more ordered active site. While Zn(II) appears to show no preference for the 3H or DCH sites, the EXAFS suggests that Co(II) shows a slight preference for the DCH site at low levels of added Co(II). The results are discussed in the context of similar metal binding studies of other B1 metallo-β-lactamases.
Supplementary Material
Acknowledgements
This work was supported by the U. S. National Institutes of Health (1R15GM093987 from NIGMS to DLT and P30-EB-009998 to the Center for Synchrotron Biosciences from the NIBIB, which supports beamline X3B at the NSLS). Work in Rosario was supported by grants to AJV from ANPCyT and HHMI. AJV, MFT and LIL are staff scientists at CONICET.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information Available: Six Figures and six Tables providing detailed fits to EXAFS data for 0.5, 1.0 and 2.0 eq of added Zn(II) or Co(II) are provided, including both R- and k-space presentations.
References
- 1.Perez F, Endimiani A, Hujer KM, Bonomo RA. Curr. Opin. Pharmacol. 2007;7:459–469. doi: 10.1016/j.coph.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush K, Jacoby GA, Medeiros AA. Antimicrob. Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush K. Clin. Infect. Dis. 1998;27:S48–S53. doi: 10.1086/514922. [DOI] [PubMed] [Google Scholar]
- 4.Cricco JA, Orellano EG, Rasia RM, Ceccarelli EA, Vila AJ. Coord. Chem. Rev. 1999;190–192:519–535. [Google Scholar]
- 5.Heinz U, Adolph HW. Cell. Mol. Life Sci. 2004;61:2827–2839. doi: 10.1007/s00018-004-4214-9. [DOI] [PubMed] [Google Scholar]
- 6.Llarrull LI, Tioni MF, Kowalski J, Bennett B, Vila AJ. Biol. Chem. 2007;282:30586–30595. doi: 10.1074/jbc.M704613200. [DOI] [PubMed] [Google Scholar]
- 7.Badarau A, Damblon C, Page ML. Biochem. J. 2007;401:197–203. doi: 10.1042/BJ20061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badarau A, Page MI. Biochemistry. 2006;45:10654–10666. doi: 10.1021/bi060934l. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez JM, Medrano FJ, Costello AL, Tierney DL, Vila AJ. J. Mol. Biol. 2007;373:1141–1156. doi: 10.1016/j.jmb.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 10.deSeny D, Heinz U, Wommer S, Kiefer M, Meyer-Klaucke W, Galleni M, Frere JM, Bauer R, Adolph HW. J. Biol. Chem. 2001;276:45065–45078. doi: 10.1074/jbc.M106447200. [DOI] [PubMed] [Google Scholar]
- 11.Carfi A, Pares S, Duee E, Galleni M, Duez C, Frere J, Dideberg O. EMBO J. 1995;14:4914–4921. doi: 10.1002/j.1460-2075.1995.tb00174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rasia RM, Vila A. Biochemistry. 2002;41:1853–1860. doi: 10.1021/bi010933n. [DOI] [PubMed] [Google Scholar]
- 13.Badarau A, Page MI. J. Biol. Inorg. Chem. 2008;13:919–928. doi: 10.1007/s00775-008-0379-2. [DOI] [PubMed] [Google Scholar]
- 14.Tioni MF, Llarrull LI, Poeylaut-Palena AA, Marti MA, Saggu M, Periyannan GR, Mata EG, Bennett B, Murgida DH, Vila AJ. J. Am. Chem. Soc. 2008;130:15852–15863. doi: 10.1021/ja801169j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Periyannan G, Costello AL, Tierney DL, Yang K-W, Bennett B, Crowder MW. Biochemistry. 2006;45:1313–1320. doi: 10.1021/bi051105n. [DOI] [PubMed] [Google Scholar]
- 16.Hawk MJ, Breece RM, Hajdin CE, Bender KM, Hu Z, Costello AL, Bennett B, Tierney DL, Crowder MW. Am. Chem. Soc. 2009;131:10753–10762. doi: 10.1021/ja900296u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wommer S, Rival S, Heinz U, Galleni M, Frere JM, Franceschini N, Amicosante G, Rasmussen B, Bauer R, Adolph HW. J. Biol. Chem. 2002;277:24142–24147. doi: 10.1074/jbc.M202467200. [DOI] [PubMed] [Google Scholar]
- 18.Llarrull LI, Tioni MF, Vila AJ. J. Am. Chem. Soc. 2008;130:15842–15851. doi: 10.1021/ja801168r. [DOI] [PubMed] [Google Scholar]
- 19.Jacquin O, Balbeur D, Damblon C, Marchot P, De Pauw E, Roberts GCK, Frere J, Matagne A. J. Mol. Biol. 2009;392:1278–1291. doi: 10.1016/j.jmb.2009.07.092. [DOI] [PubMed] [Google Scholar]
- 20.Thomas PW, Stone EM, Costello AL, Tierney DL, Fast W. Biochemistry. 2005;44:7559–7565. doi: 10.1021/bi050050m. [DOI] [PubMed] [Google Scholar]
- 21.Newville M. J. Synchrotron Rad. 2001;8:322–324. doi: 10.1107/s0909049500016964. [DOI] [PubMed] [Google Scholar]
- 22. Sixpack is available free of charge from http://www-sssrl.slac.stanford.edu/~swebb/index.htm.
- 23.Ankudinov AL, Ravel B, Rehr JJ, Conradson SD. Phys. Rev. B. 1998;58:7565–7576. [Google Scholar]
- 24.Orellano EG, Girardini JE, Cricco JA, Ceccarelli EA, Vila AJ. Biochemistry. 1998;37:10173–10180. doi: 10.1021/bi980309j. [DOI] [PubMed] [Google Scholar]
- 25.Huheey JE. Inorganic Chemistry, Harper and Row. New York, NY: 1983. [Google Scholar]
- 26.Breece RM, Hu Z, Bennett B, Crowder MW, Tierney DL. J. Am. Chem. Soc. 2009;131:11642–11643. doi: 10.1021/ja902534b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez JM, Buschiazzo A, Vila AJ. Biochemistry. 2010;49:7930–7938. doi: 10.1021/bi100894r. [DOI] [PubMed] [Google Scholar]
- 28.Fabiane SM, Sohi MK, Wan T, Payne DJ, Bateson JH, Mitchell T, Sutton BJ. Biochemistry. 1998;37:12404–12411. doi: 10.1021/bi980506i. [DOI] [PubMed] [Google Scholar]
- 29.Materon IC, Queenan AM, Koehler TM, Bush K, Palzkill T. Antimicrob. Agents Chemother. 2003;47:2040–2042. doi: 10.1128/AAC.47.6.2040-2042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Tenover FF, Koehler TM. Antimicrob. Agents Chemother. 2004;48:4873–4877. doi: 10.1128/AAC.48.12.4873-4877.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomatis PE, Fabiane SM, Simona F, Carloni P, Sutton BJ, Vila AJ. Proc. Nat. Acad. Sci. 2008;105:20605–20610. doi: 10.1073/pnas.0807989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomatis PE, Rasia RM, Segovia L, Vila AJ. Proc. Natl. Acad. Sci. 2005;102:13761–13766. doi: 10.1073/pnas.0503495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.