Abstract
This chapter describes two types of FRET-based fluorescence assays that can be used to identify and analyze compounds that inhibit the helicase encoded by the hepatitis C virus (HCV). Both assays use a fluorescently labeled DNA or RNA oligonucleotide to monitor helicase-catalyzed strand separation, and they differ from other real-time helicase assays in that they do not require the presence of other nucleic acids to trap the reaction products. The first assay is a molecular beacon-based helicase assay (MBHA) that monitors helicase-catalyzed displacement of a hairpin-forming oligonucleotide with a fluorescent moiety on one end and a quencher on the other. DNA-based MBHAs have been used extensively for high-throughput screening (HTS), but RNA-based MBHAs are typically less useful because of poor signal to background ratios. In the second assay discussed, the fluorophore and quencher are split between two hairpin-forming oligonucleotides annealed in tandem to a third oligonucleotide. This split beacon helicase assay can be used for HTS with either DNA or RNA oligonucleotides. These assays should be useful to the many labs searching for HCV helicase inhibitors in order to develop new HCV therapies that are still desperately needed.
1. Introduction
Specific helicase inhibitors of viral RNA helicases are needed for two reasons. First, they are valuable chemical probes needed to understand the roles that RNA helicases play in biology. Second, inhibitors of viral helicases may be valuable as antiviral agents. Most RNA viruses that replicate outside the cell’s nucleus encode an RNA helicase. If such a virus lacks a functional helicase, neither can it replicate (Kolykhalov et al., 2000) nor can it synthesize its RNA genome (Lam and Frick, 2006). RNA helicases provide medicinal chemists many targets because helicase inhibitors could, at any one of several clearly defined ligand binding sites (or other critical motifs), block ATP binding, ATP hydrolysis, RNA binding, strand separation, or protein translocation. Once a binding site is clearly defined, the many available RNA helicase crystal structures could be used to rationally design more potent derivatives.
Interest in helicases as drug targets peaked about 10 years ago when two classes of compounds targeting a helicase encoded by herpes simplex virus (HSV) were shown to elicit potent antiviral effects in animal models. These novel HSV antiviral drugs target a DNA helicase that coordinates DNA replication and the action of DNA primase (Crute et al., 2002; Katsumata et al., 2011; Kleymann et al., 2002). Inspired by the success of the HSV compounds, several teams have led extensive searches for inhibitors of human helicases (Aggarwal et al., 2011; Yedavalli et al., 2008) and helicases encoded by important human pathogens (Frick, 2006; Kwong et al., 2005; Tuteja, 2007). One of the most frequently targeted RNA helicases is the one encoded by the hepatitis C virus (HCV). The methods discussed in this chapter were specifically designed for use with HCV helicase but they could be used with related helicases with relatively minor changes. Medically relevant RNA helicases related to HCV helicase include enzymes encoded by the flaviviruses (e.g., Dengue virus, Yellow fever virus, and West Nile virus) and the human DEAD-box proteins. The procedures below have been developed and implemented to screen over 290,000 compounds as part of the National Institutes of Health’s Molecular Libraries Probe Production Centers Network (MLPCN). All screening results from this project are posted regularly on PubChem BioAssay (http://www.ncbi.nlm.nih.gov/pcassay).
2. The Need for Additional HCV Drug Targets
HCV infects nearly one in every 50 people alive today causing fibrosis, cirrhosis, and ultimately, liver failure. There are no approved HCV vaccines, but there are effective HCV treatments that all use the broad-acting drugs ribavirin and pegylated recombinant human interferon alpha (INFα). Current HCV drug combinations have an impressive impact on viral proliferation, typically curing more than half of patients, but they are expensive and their considerable side effects make HCV therapy difficult to tolerate (Edlin, 2011). Development of HCV vaccines and less toxic HCV drugs has been slow because it was not possible to study wild-type HCV in the lab until the recent advent of robust cell culture systems and small animal models (Murray and Rice, 2011). For years, HCV drug development focused almost entirely on recombinant HCV proteins that were expressed in model organisms and used to develop assays suitable for high-throughput screening (HTS). “Hits” in these HTS assays were then developed by rationally designing better compounds using high-resolution protein structures of the HCV targets. Eventually, this process led to the discovery of numerous direct acting antivirals (DAAs), which are now being developed to replace INFα and ribavirin in HCV therapy. It is hoped that DAAs will cause fewer side effects because, unlike INFα and ribavirin, DAAs are not designed to modulate the host response to viruses.
The HCV genome contains a single open reading frame encoding an approximately 3000 amino acid long polypeptide. Host and viral proteases cleave the polyprotein into 10 mature HCV proteins. Three HCV proteins are structural, forming the virus particle, and seven are nonstructural (NS) proteins. NS3 is the HCV helicase, but it also has several additional important functions. Upon translation, NS3 combines with NS2 to form an autocatalytic protease that cleaves the NS2/NS3 junction. Processed NS3 then contains another protease active site that is activated after newly translated NS4A binds to the NS3 N-terminal protease domain. This second HCV protease cleaves itself in cis and other HCV and cellular proteins in trans. The most advanced HCV DAAs attack this NS3/NS4A protease. Currently, the most advanced protease inhibitors are the recently approved drugs Telaprevir (Zeuzem et al., 2011) and Boceprevir (Bacon et al., 2011). Triple therapy with INFα, ribavirin, and a protease inhibitor cures up to 88% of patients who have failed prior therapies. There are, however, still several problems with this state-of-the-art HCV therapy. First, the protease inhibitors are only effective when administered with interferon and ribavirin because of a low resistance barrier. In other words, single point mutations confer resistance to Telaprevir and Boceprevir. These mutations evolve rapidly and have only relatively minor effects on HCV fitness or viability (Hiraga et al., 2011). Second, Telaprevir and Boceprevir are only effective against specific HCV strains and genotypes, mainly ones common in North America. Third, additional side effects are associated with the protease inhibitors. Fourth, triple therapy is even more costly, making the new therapy even less accessible to most patients. New drugs are therefore still needed to make HCV treatment more accessible and better tolerated so that it might start to impact the global HCV burden.
It will not likely be possible to decrease HCV therapy cost and toxicity unless drugs are found to replace, rather than supplement, INFα. To this end, other DAAs are being tested alone and in combination with protease inhibitors. None of these drug cocktails contain helicase inhibitors, but, in theory, helicase inhibitors would be particularly attractive additions to DAA cocktails since they would target the same protein as the protease inhibitors already in use. Two drugs targeting the same protein could interact synergistically so that combined they are more effective. The accumulating evidence that the NS3 helicase and protease depend on one another supports the notion that helicase and protease inhibitors might act synergistically (Beran et al.,2007, 2009; Frick et al., 2004).
3. Targeting the NS3 Helicase
HCV helicase was one of the first HCV targets identified with its activity first characterized shortly after HCV was discovered (Choo et al., 1989; Kim et al., 1995; Porter et al., 1998; Preugschat et al., 1996; Suzich et al., 1993). The HCV helicase was also the first RNA helicase crystallized (Yao et al., 1997) and NS3 has been studied extensively both as a model helicase and as a drug target ever since. As discussed extensively in other reviews, tremendous progress has since been made to understand exactly how the helicase unwinds DNA and RNA in an ATP-fueled reaction (Frick, 2007; Pyle, 2008; Raney et al., 2010).
Early HCV helicase studies were performed mainly with truncated NS3 lacking the protease domain (referred to here as NS3h) because such proteins express in Escherichia coli at higher levels than full-length NS3 and they are more stable. In NS3h proteins, NS3 is truncated at a linker connecting the helicase to the NS3/NS4A protease by deleting between 166 and 190 amino acids from the NS3 N-terminus. The protease is then replaced with an affinity tag, or an affinity tag is fused to the C-terminus of NS3h. Most early studies used NS3h as a surrogate for full-length NS3, but more recent studies tend to focus on full-length NS3. Direct comparisons of NS3h to full-length NS3 have revealed that the protease domains and NS4A influence the helicase, and vice versa, suggesting that the NS3 helicase and protease functions do not act independently but instead they are tightly coordinated (Beran et al., 2007, 2009; Frick et al., 2004). The protocols below have been used with a variety of recombinant NS3 and NS3h proteins isolated from a wide array of HCV strains and genotypes (Belon and Frick, 2009b; Belon et al., 2010). We find that results are most consistent with an NS3h lacking the first 166 NS3 amino acids with a His-tag attached to the NS3 C-terminus. The NS3h protein we use most often in screens is the one isolated from the Con1 strain of HCV genotype 1b. The Con1 strain forms the backbone for many common HCV replicons used to study HCV replication in cells (Lohmann et al., 1999).
All NS3 and NS3h proteins unwind both RNA and DNA. This robust DNA helicase activity facilitates in vitro analysis, but it is unusual because HCV has no DNA stage and related proteins act only on RNA. It has been speculated that the activity of NS3 on DNA is somehow related to the fact that HCV infection correlates with high rates of hepatocellular carcinoma. However, only two indirect lines of evidence link NS3 to a role in liver cancer. The first is the observation that, when HCV helicase is overexpressed in human cells, some of the protein has been observed in the nucleus where it might affect host gene expression or transforms cells to a cancerous phenotype (Muramatsu et al., 1997). The second is the biochemical observation that NS3h can catalyze strand exchange reactions, which hints toward a possible role for NS3 in genetic recombination (Rypma et al., 2009).
Regardless of why NS3h unwinds DNA, DNA has already been used in many screens for HCV helicase inhibitors. The major concern with such assays is that compounds inhibiting HCV helicase-catalyzed DNA unwinding might not inhibit the action of NS3 on its natural RNA substrates. The procedures below address this concern by first providing a readout as to whether or not the compound interacts with the DNA substrate and, second, by using a second assay as a counterscreen that uses an RNA-based substrate.
4. HTS for HCV Helicase Inhibitors
Standard helicase assays and some early HTS assays monitor helicase action using radioactive oligonucleotides to observe strand displacement (Kyono et al., 1998). In order to avoid using hazardous radioisotopes, simplify protocols, and provide real-time readouts, newer helicase assays often use fluorescently labeled oligonucleotides and Förster resonance energy transfer (FRET) to monitor strand separation. In most FRET-based helicase assays, one nucleic acid strand is labeled with a donor fluorophore and the complementary oligonucleotide is labeled with an accepter moiety. When a helicase separates the two oligonucleotides upon ATP addition, donor fluorophore fluorescence increases because it is separated from the FRET acceptor (Bjornson et al., 1994; Houston and Kodadek, 1994). FRET-based assays have been used extensively for HCV assays (Boguszewska-Chachulska et al., 2004; Frick et al., 2007; Tani et al., 2009), but we have found they are not ideal for HTS because well-to-well variation in apparent reaction rates and extent makes hit identification difficult. Rypma et al. demonstrated that some of this variability stems from the fact that nucleic acid traps added to the above FRET-based assays to prevent the two labeled strands from reannealing heavily influence observed reaction rates and their extent (Rypma et al., 2009).
To help facilitate HTS for HCV helicase inhibitors, our lab developed a real-time helicase assay that did not require nucleic acid traps to observe helicase action on duplexes. This second-generation FRET-based helicase assay uses a helicase substrate made with a molecular beacon (Tyagi and Kramer, 1996) annealed to a longer DNA oligonucleotide such that a 3′ single-stranded region is available for the helicase to load (Belon and Frick, 2008). Using protocols below (Section 6), this molecular beacon-based helicase assay (MBHA) has been used in mechanistic analyses (Belon and Frick, 2009b), HTS (Belon and Frick, 2010), and for the analysis of known HCV helicase inhibitors (Belon et al., 2010).
One serious limitation of the MBHA is that, when the DNA oligonucleotides are substituted with RNA oligonucleotides, the signal to background (S/B) ratio decreases to a level where the assay is no longer appropriate for HTS (Belon and Frick, 2008). Because the natural substrate for HCV NS3h is most likely RNA, a screen with RNA is needed to identify compounds that act only when DNA is used as a substrate. Compounds that inhibit HCV helicase-catalyzed DNA unwinding but not RNA unwinding might be useful chemical probes to understand if the HCV helicase action on DNA plays any role in HCV biology. However, it is also possible that only compounds that inhibit HCV helicase action on RNA will be effective antivirals. To improve S/B ratios with RNA, we have recently developed a split beacon assay where the fluorophore and quenching moieties are present on separate hairpin- forming oligonucleotides. Unlike this MBHA, this split beacon helicase assay (SBHA) performs similarly when HCV helicase acts on DNA or RNA (Section 7), with Z′ factors in ranges appropriate for HTS (Zhang et al., 1999).
5. Expression and Purification of NS3h
Purified full-length NS3 is typically more active than NS3h, and these differences are most apparent in assays where long stretches of DNA or RNA must be unwound to detect activity. However, full-length NS3 is also notably less stable in solution, losing activity after only a few freeze–thaw cycles and sometimes even during prolonged storage at −80 °C. In contrast, most NS3h proteins we have tested retain activity longer at room temperature or after repeated freeze–thaw cycles. NS3h proteins with fusion tags at the N-terminus, such that the fusion partner replaces the protease domain, generally behave more like full-length NS3 in that they are initially more active (Frick et al., 2004), but they also lose activity more rapidly upon prolonged storage. The assays described here have, therefore, been optimized using an NS3h (isolated from the Con1 strain of HCV genotype 1b; Heck et al., 2008) with a C-terminal His-tag that has been purified using the protocol below. NS3h from other HCV genotypes has also been purified with the below protocol (Lam et al., 2003; Neumann-Haefelin et al., 2008). Our lab’s method to purify full-length NS3 has been published elsewhere (Frick et al., 2010).
- Streak E. coli Rosetta (DE3) cells (EMD Biosciences) harboring the plasmid pET24-Hel-Con1 (Heck et al., 2008) on LB-agar containing kanamycin (50 µg/ml) and tetracycline (50 µg/ml) to isolate single colonies. After overnight incubation at 37 °C, inoculate 5 ml of LB broth containing kanamycin (50 µg/ml) and tetracycline (50 µg/ml) with single colonies. Shake vigorously at 37 °C until slightly turbid. Transfer culture to 1 l of LB containing the same antibiotics. Shake vigorously at 37 °C and periodically monitor OD600. When OD600 is approximately 1.0, add isopropyl β-d-1-thiogalactopyranoside to a final concentration of 1 mM. Incubate 2–3 h at room temperature and harvest cells by centrifugation, wash cells with phosphate buffered saline, and store pellet at −80 °C.Perform all subsequent steps at 4 °C or with all tubes on ice.
Suspend frozen cells in 10 ml 20 mM Tris, pH 8, 0.5 M NaCl, and 5 mM Imidazole (buffer A). Lyse cells using French press or Sonifier Cell disrupter (Branson). Centrifuge at 10,000 g, discard pellet, and filter the supernatant through a 0.8-µm glass fiber filter (Fraction I).
Load Fraction I onto a 5-ml Ni-NTA column (GE Healthcare) equilibrated with buffer A. Wash with buffer A containing 40 mM Imidazole. Elute with buffer A containing an imidazole gradient from 40 to 500 mM Imidazole. NS3h should elute when the imidazole approaches 100 mM. Analyze fractions using 10% SDS-PAGE to identify fractions containing 53 kDa NS3h protein. Combine fractions containing NS3h (Fraction II).
Precipitate NS3h from fraction II by slowly adding solid (NH4)2SO4 to 60% saturation (0.361 g/ml). Centrifuge at 12,000 g for 20 min. Discard supernatant. Dissolve pellet in 2 ml storage buffer (20 mMTris, pH8, 50 mM NaCl, 1 mM EDTA, 0.1 mM DTT, 25% glycerol) (Fraction III).
Load Fraction III onto a 100-ml gel filtration column (Sephacryl S-300 HR, GE Healthcare) that has been previously equilibrated with 20 mM Tris, pH 8, 50 mM NaCl, 1 mM EDTA, and 0.1 mM DTT (GF buffer). Elute protein with GF buffer by collecting 2 ml fractions at 0.1 ml/min. Analyze fractions using a 10% SDS-PAGE. Combine fractions containing NS3h (fraction IV).
Load fraction IV on a 1-ml DEAE Sepharose FF column (GE Healthcare) that has been equilibrated with GF buffer. After washing with GF buffer, elute with a GF buffer containing a gradient of NaCl from 0 to 500 mM. NS3h should elute around 150 nM NaCl. Analyze the fractions with a 10% SDS-PAGE, and combine fractions containing NS3h (Fraction V).
Dialyze protein with GF buffer (1 l). Protein may be concentrated at this point by sprinkling dialysis tubing with polyethylene glycol (average molecular weight > 20,000) and allowing liquid to absorb at 4 °C. Let desired buffer absorb and return bag to GF buffer. After two changes of GF buffer, dialyze with storage buffer (prepared in step 4).
Determine protein concentration from absorbance at 280 nm using an extinction coefficient calculated from the protein sequence (51,890 M−1 cm−1 for NS3h_1b(Con1)). Store aliquots at −80 °C.
6. The Molecular Beacon-Based Helicase Assay (MBHA)
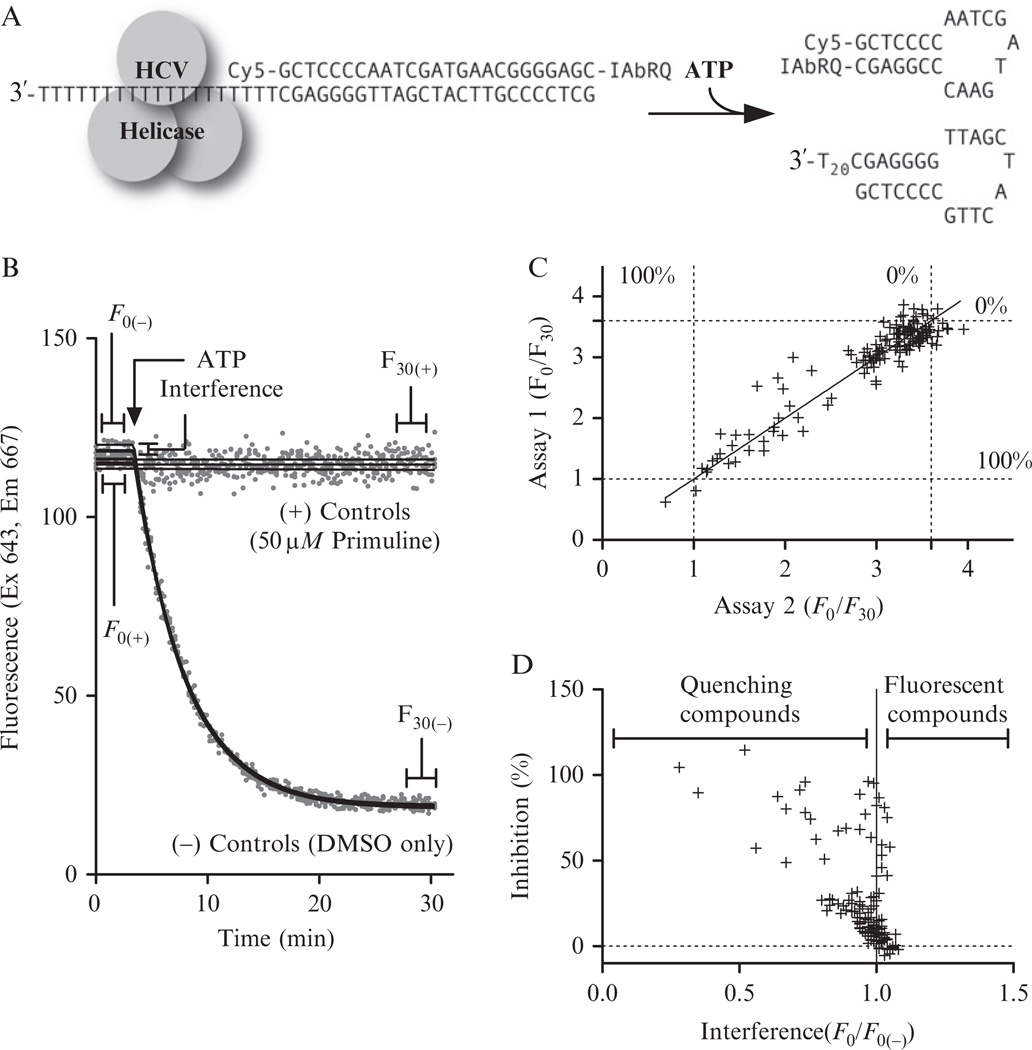
The MBHA most commonly used in our laboratory employs a substrate designed to mimic a hairpin-forming region at the 3′ end of the HCV polyprotein reading frame (Fig. 21.1A). The shorter (top) oligonucleotide in this substrate is modified by attaching a cyanine 5 (Cy5) at the 5′ end and attaching an Iowa black RQ (IAbRQ) to its 3′ end. The longer DNA oligonucleotide is complementary to the shorter strand and also has a 20-nucleotide long 3′ tail. The assay works by monitoring the fluorescence of the DNA probe before and after the addition of ATP, which is needed to fuel helicase movement. As the reaction proceeds, the helicase rearranges the nucleic acids such that the complementary nucleotides near the ends of the hybridization probe bind, forming a hairpin loop structure (Fig. 21.1A). The hairpin loop allows the fluorophore and quencher molecule to come into contact, with the result being a reduction in Cy5 fluorescence. This drop in fluorescence is plotted versus time to determine both the rate and extent of the reaction (Fig. 21.1B). For screening, each MBHA needs to be read only twice, before ATP addition (F0) and at the completion of the reaction (F30). The F0/F30 ratio reflects the extent of the reaction. An inhibitor will cause a decrease in the F0/F30 ratio to a limit of one, where, by definition, no reaction takes place.
Figure 21.1.
The molecular beacon-based helicase assay and its use in HTS. (A) A Cy5-labeled MBHA substrate based on the sequence of the HCV genome. (B) Four MBHAs performed in the absence of test compounds and four performed with a saturating concentration (50 µM) of a known HCV helicase inhibitor (primuline). (C) Reaction extents (F30/F0) for duplicate reactions performed with the same concentration (20 µM) of 125 different test compounds. (D) Average compound interference and inhibition obtained from the duplicate reactions shown in panel C. All reactions contained in 25 mM MOPS, pH 6.5, 1.25 mM MgCl2, 5 nM MBHA substrate, 2% (v/v) DMSO, 12.5 nM NS3h_1b(con1), and 1 mM ATP.
Another important parameter obtained with an MBHA concerns compound interference. Three classes of compounds tend to interfere with the MBHA. The first interfering class contains fluorescent compounds that absorb and emit light at wavelengths similar to Cy5. The second quenches Cy5 fluorescence by absorbing light at the Cy5 excitation or emission wavelength, and the third class alters substrate fluorescence by binding the MBHA substrate and changing the orientation of Cy5 relative to IAbRQ. Simply scanning compound absorbance and fluorescence in the absence of the MBHA substrate can identify compounds in the first two classes. Examining the interaction of the compounds and DNA using other assays can identify compounds in the third class. All such compounds would alter both F0 and F30 in such a way that they might be misidentified as helicase inhibitors because they lower the F0/F30 ratio. To identify interfering compounds, the F0 of each particular compound is compared to the F0 of DMSO-only controls (F0(−)). A compound with an F0/F0(−) near 1 generally does not interfere, quenching compounds and DNA-binders have F0/F0(−) < 1, and fluorescent compounds have F0/F0(−) > 1.
6.1. MBHA-based high-throughput screens
The following protocol is routinely used in our lab to monitor helicase activity in either low-volume 96-well microplates or standard 384-well microplates. The protocol is optimized for NS3h_con(1b), but it can be used with other NS3 proteins by simply changing the amount of NS3h in each assay, using less of more active helicases and more of less active helicases. We typically use the yellow dye primuline (MP Biochemicals) as a positive control in these assays. Belon and Frick previously reported that a similar dye called thioflavine S is a HCV helicase inhibitor (Belon and Frick, 2010). Thioflavine S is a more heterogeneous mixture of compounds than is primuline, which we have since found to give more consistent results in MBHAs. The protocol below is for 60 µl reactions, but the reaction can be scaled down to conserve reagents. Precision at low volumes is limited only by sensitivity of the micro plate reader used and the precision of available liquid handlers. A protocol for 5 µl reactions suitable for ultra-high-throughput screening (uHTS) performed in 1536-well plates is available on PubChem BioAssay (AID #1800).
Anneal substrate by combining oligonucleotide as shown in Fig. 21.1A (Integrated DNA Technologies, Coralville, IA) at 50 µM each in 10 mM Tris–HCl, pH 8.0. Heat to 95 °C, and cool slowly to room temperature. Store concentrated substrate in the dark at −20 °C.
Prepare 0.5 M3-(N-morpholino)propanesulfonic acid (MOPS) adjusting pH to 6.5 with NaOH. Also prepare 25 mM MOPS, pH 6.5, 25 mM MgCl2, 10mM ATP, 1 mM Primuline (MP Biochemicals) in dimethyl sulfoxide (DMSO), and NS3h dilution buffer (25 mM MOPS, pH 6.5, 1 mM DTT, 0.2% Tween20, 0.1 mg/mL BSA).
Dilute the Cy5-MBHA substrate to 100 nM in 25 mM MOPS.
Dilute NS3h to 250 nM in NS3h dilution buffer.
Assemble enough reaction mixture for the desired number of assays. For each assay, combine 3 µl of 0.5 M MOPS, 3 µl of 25 mM MgCl2, 3 µl of 100 nMCy5-MBHA substrate, 3 µl of 250 nM NS3h, and 39 µl of nuclease free water.
Dispense 51 µl of the reaction mix to each well of a polystyrene white low-volume 96-well microplate (Corning) at 23 °C. Add 3 µl of DMSO or compounds dissolved in DMSO. (Note: The assay tolerates up to 35% DMSO, so more or less DMSO can be added depending on compound solubility.)
Read Cy5 fluorescence (Ex 643 Em 667) of each well.
Add 6 µl of 10 mM ATP, and read fluorescence until values in negative control reactions remain constant (typically 20–30 min) (Fig. 21.1B). Record endpoint (F30). Note: After ATP addition each reaction will contain 25 mM MOPS, pH 6.5, 1.25 mM MgCl2, 0.05 mM DTT, 5% DMSO, 0.01% Tween20, 5 µg/mL BSA, 5 nM MBHA substrate, 12.5 nM NS3h, and 1 mM ATP.
Calculate F0/F30 ratios for each reaction. Figure 21.1C shows results of duplicate MBHAs that were performed with a library of 150 known or suspected HCV helicase inhibitors and DNA binding compounds that we use to evaluate HCV helicase assays in our lab. The duplicate assays are plotted against each other to evaluate reproducibility. Dotted lines mark average F0/F30 ratios for negative controls (DMSO only) and positive control reactions (50 µM primuline).
- Calculate percent inhibition by normalizing F0/F30 ratios for each reaction to the ratio obtained with positive (+) and negative (−) controls (Eq. 21.1):
(21.1) Calculate compound interference by calculating the ratio of F0 for each compound and F0 for the negative controls (F0/F0(−)). Figure 21.1D shows the average percent inhibition and interference ratio values of the duplicate assays performed with our helicase inhibitor library, where all library compounds were tested at 10 µM.
6.2. Examining effects of inhibitors on the kinetics of DNA strand separation
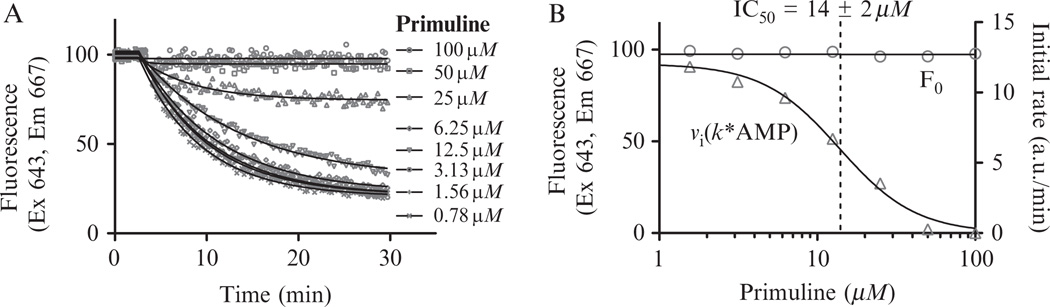
Once a compound is identified to influence the MBHA, the next step we take is to examine the apparent affinity of the compound for the MBHA substrate and the apparent affinity of the compound for the unwinding complex. Affinity of the compound for the substrate is estimated by first examining the effect of compound concentration on substrate fluorescence before ATP is added (if there is an effect, direct DNA binding is measured with another assay). The apparent affinity of the compound for the unwinding complex is estimated from effect of the compound on the initial rates of unwinding reactions. Both parameters can be estimated from time courses obtained in the presence of various compound concentrations, by first calculating the average fluorescence of the substrate during the time before and after ATP is added, and then fitting data obtained after ATP addition to a rate equation. There are many commercially available software packages, including some supplied with more sophisticated plate readers, which will fit kinetic readouts directly to first order rate equations. However, the protocol below uses the program Graphpad Prism (La Jolla, CA) software because it can be used with data exported from any reader. As demonstrated with primuline (Fig. 21.2), the protocol below fits time course data to calculate F0 and initial velocities (Fig. 21.2A), export the values, and fit them directly to a dose response equation (Fig. 21.2B).
Add 2 mM solutions of each inhibitor to the first tube in an eight-tube strip and serially dilute the compound 1:2 into DMSO in remaining tubes. This creates an eight-step 1:2 dilution series beginning at 2 mM. To a second eight-tube strip, add DMSO to four tubes and a positive control inhibitor (e.g., 1 mM primuline in DMSO) to the other four tubes.
Assemble MBHA reactions as described above, except add diluted compounds and positive/negative control series instead of the chemical library.
Perform the assay using a microplate reader equipped with a reagent dispenser capable of precisely adding the 6 µl of ATP to each well needed to start the reaction. Collect fluorescence data as rapidly as the plate reader allows for ~2 min before ATP injection, and then until fluorescence no longer changes in the negative control reactions.
-
Export time course data to Prism, and fit fluorescence time courses to the following Prism nonlinear regression equation series that calculates F0 and uses the calculated F0 as a starting fluorescence that decays according to a first order rate constant (Fig. 21.2A).
In the above equations, X is time, Y is observed fluorescence, Y1 is the data before ATP injection, Y2 is data collected after ATP injection, START is the time that ATP was added, AMP is reaction amplitude, and K is a first order rate constant describing the loss of fluorescence. Constrain “START” to the time ATP was added, and AMP, F0, and K so they remain greater than zero. Set initial estimates for F0, AMP, and K to Ymax, Ymin, and the 1/value of X at Ymid, respectively. When entering this equation, set Prism to calculate initial velocities for each reaction by defining a “V” under “transforms to report” as K × AMP. While setting parameters for nonlinear regression, set Prism to output F0, V, and AMP to a summary table and plot each on an XY graph. Notes: if a program other than Prism is used to fit reaction time courses, calculate initial rates for each reaction by multiplying observed first order rate constant by the reaction amplitudes. If Prism or another nonlinear regression software is not available, estimates of initial velocities can be obtained from the slopes of the initial linear phase of each reaction time course.
- Fit plots of initial velocities versus compound concentration to a dose response equation such as the Prism equation below:
where X is compound concentration, Y is velocity in the presence of inhibitor, V0 is velocity in the absence of inhibitor, h is the Hill coefficient, and IC50 is the concentration of compound that inhibits the unwinding reaction by 50%. Note: several other dose response equations are available in Prism, but data must first be transformed to the LOG of inhibitor concentration. Examine plots of F0’s versus compound concentration. A negative slope to the F0 graph indicates that the compounds quench the MBHA substrate fluorescence. Data obtained with compounds that interact with DNA, or quench by inner filter effects, will sometimes also fit the equation in step 5 and yield IC50 values that mirror those obtained if velocities are plotted. Such compounds likely exert their effect by binding DNA rather than the helicase itself. Compounds that do not affect F0 in a dose response manner are assumed not to significantly interfere with the assay (Fig. 21.2B); however, this does not mean that they do not interact with the DNA substrate. To identify all potential DNA binding compounds, we subject all hits to a counterscreen that monitors DNA binding potential. One such assay monitors a compound’s ability to displace a fluorescent DNA intercalator like ethidium bromide (Boger and Tse, 2001; Boger et al., 2001).
Figure 21.2.
Effect of various primuline concentrations on a HCV helicase-catalyzed MBHA. (A) MBHAs were performed with the indicated concentration of the known helicase inhibitor primuline. Solid lines show nonlinear regression fits described in text. (B) Secondary plot of initial velocities (triangles) and F0s (circles) fit to a dose response equation as described in text. Dotted line indicates the IC50 value determined by nonlinear regression.
6.3. Evaluating alternate MBHAs for HTS
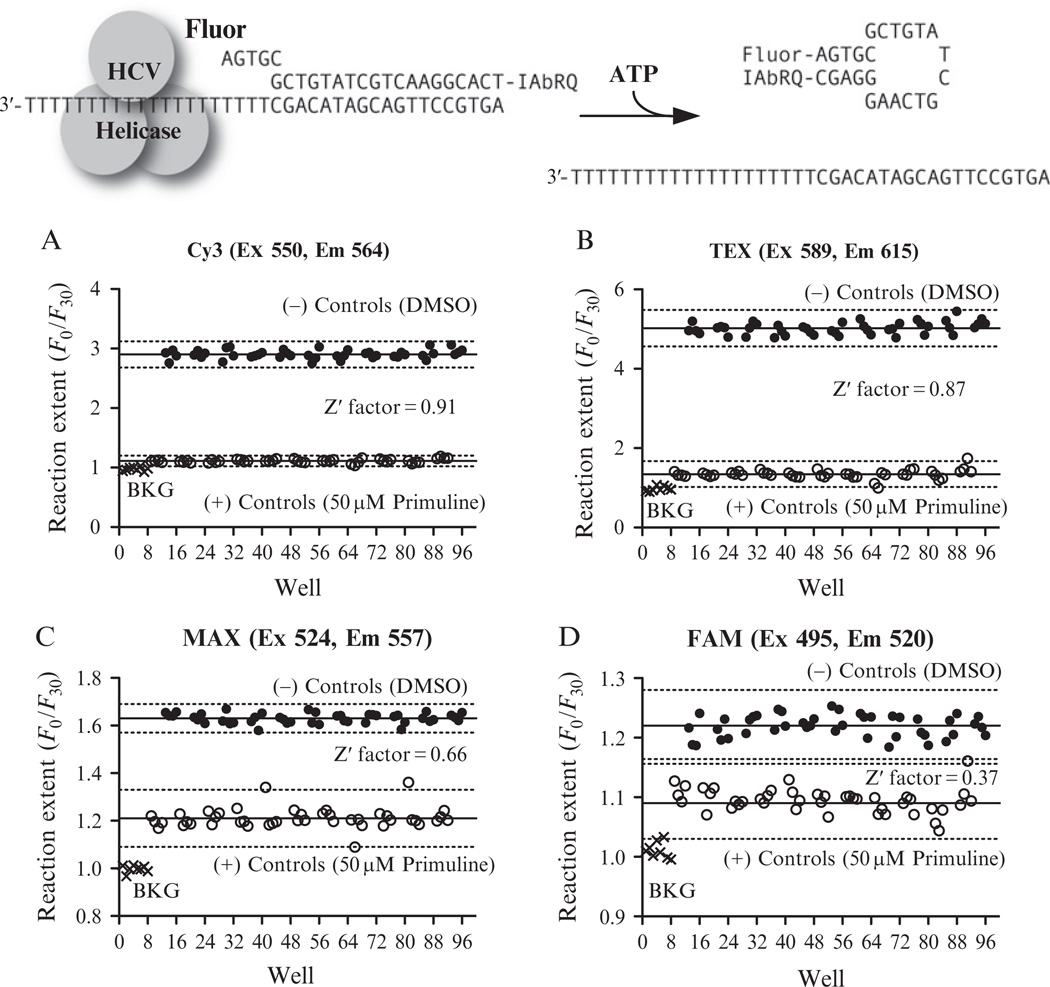
The brief protocol below is designed to evaluate the quality of an MBHA or related assay. When analyzing hits from a helicase HTS, we typically examine a compound’s behavior in helicase assays that use a variety of DNA sequences and fluorophores because it is possible that hits might act only by interacting with certain DNA sequences or the fluorophore used to monitor the reaction (Belon et al., 2010). One alternate MBHA substrate we use is shown in Fig. 21.3A, which differs from the one in Fig. 21.1 in that the bottom strand does not form a hairpin when separated from the top strand. Figure 21.3A–D show results obtained when the sequence is labeled with fluorophores with different chemistry. Although primuline inhibits the observed reactions with all four beacons, S/B varies among the assays. All but one assay has a Z′ factor in the range appropriate for HTS.
Assemble an eight-tube strip containing four tubes of DMSO and four tubes of a known helicase inhibitor (e.g., 1 mM primuline).
Assemble reaction mixtures as described in Section 6.1 and dispense 51 µl into all wells of a low-volume white 96-well plate. Add 3 µl of the controls from the eight-tube strip to each column.
Monitor DNA fluorescence at appropriate wavelength (F0); dispense 6 µl of 10 mM ATP to each well to start the reactions. Continue to monitor fluorescence of each well until negative control reaction fluorescence no longer changes. Record F30 for each well.
Calculate F0/F30 ratios. Plot data versus well number and inspect. A slope to the negative control data will result if all the reactions have not gone to completion.
Average the ratios for the positive control reactions (M+) and negative control reaction (M−), and the standard deviation for positive controls (SD+) and negative controls (SD−).
-
Calculate a Z′ factor (Zhang et al., 1999) from the means and standard deviations (Eq. 21.2).
(21.2) A Z′ value of 1.0 indicates an ideal assay, excellent assays yield values between 0.5 and 1.0, a value between 0.5 and 0 indicates a marginal assay needing improvement, and assays with Z′ factors below 0 are generally not useful for HTS.
Figure 21.3.
Evaluation of alternate fluorophores for use in MBHA-based screens. Each substrate was labeled with Iowa Black RQ (IAbRQ) and one of the four indicated fluorophores available from Integrated DNA technologies (Coralville, IA). Each panel shows results from a single 96-well low-volume plate containing 8 reactions without DNA (BKG), 44 reactions with 2% DMSO, and 44 reactions containing the HCV helicase inhibitor primuline at a final concentration of 50 µM in 2% DMSO. Solid lines show means of positive and negative controls and the dotted lines show three times the standard deviations.
7. An RNA-Based Split Beacon Helicase Assay (SBHA)
We have performed many of the above assays using substrates where RNA forms one or both strands of the MBHA substrate. While such assays provide valuable results when performed carefully under analytical conditions, their usefulness in HTS is limited. Specifically, the F30/F0 ratios obtained from negative control reactions (no inhibitors) with RNA substrates is usually two to four times lower than it is for the same negative control reactions performed with DNA substrates. As a result, difference is smaller between the positive and negative controls in RNA assays and Z′ factors are usually only in the marginal range at best.
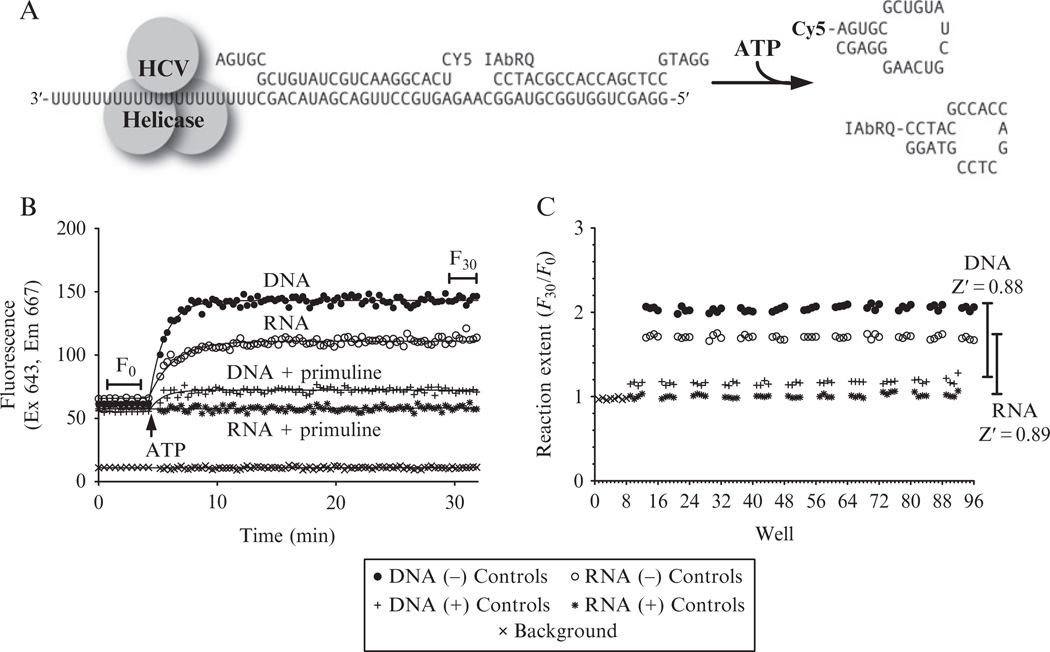
To monitor HCV helicase action on RNA in HTS, we instead use an RNA-based assay where the fluorophore and quencher are split between two different hairpin-forming oligonucleotides that both anneal to a third strand at adjacent positions. In this SBHA, the two oligonucleotides that the helicase must separate for a signal change are made of RNA, while the third oligonucleotide containing the quenching moiety is made of DNA (Fig. 21.4A). In SBHAs, F0 and F30 signals are similar when either DNA or RNA is used as a substrate (Fig. 21.4B), but more enzyme is needed with RNA because the same amount of NS3h unwinds the RNA substrate more slowly. Similar rates are obtained if five times more NS3h (i.e., 62.5 nM) is added to the reactions with RNA SBHA substrate than is added to DNA SBHA substrates (i.e., 5 nM NS3h). Unlike the MBHA, fluorescence increases when the helicase unwinds an SBHA substrate, and reaction extent is therefore calculated as an F30/F0 ratio (rather than an F0/F30 ratio). Reaction extents and Z′ factors are similar for RNA- and DNA-based SBHA (Fig. 21.4C). The following protocol can be used to perform RNA-based SBHAs.
Anneal substrate by combining three oligonucleotides shown in Fig. 21.1A (Integrated DNA Technologies, Coralville, IA) at 50 µM each in RNAse-free 10 mM Tris–HCl, pH 8.5. Heat to 95 °C. Cool slowly to room temperature. Store concentrated substrate in the dark at −20 °C.
Assemble reaction mixtures, controls, and inhibitors as described above for the MBHA (Section 6.1), except that NS3h_1b(Con1) should be included at 60 nM in each reaction. Add 51 µl of reaction mix and 3 µl of either control compounds or dilutions of compounds to be tested.
Monitor Cy5 fluorescence before ATP addition. Add 6 µl of ATP. Record final fluorescence (F30) after fluorescence in control reactions no longer changes. Unlike the MBHA, fluorescence will increase upon ATP addition. Calculate F30/F0 ratios and normalize data with ratios obtained with positive and negative control reactions to determine percent inhibition.
-
For kinetic analyses, exported data can be analyzed as discussed in Section 6.2, except that the data obtained after ATP addition should be fit a rate equation describing substrate appearance rather than decay. The following set of equations can be used to fit SBHA data using Prism.
In the above equations, X is time, Y is observed fluorescence, Y1 is the data before ATP injection, Y2 is data collected after ATP injection, START is the time that ATP was added, AMP is reaction amplitude, and K is a first order rate constant describing the gain of fluorescence. Constrain “START” to the time ATP was added, and AMP, F0, and K so they remain greater than zero. Set initial estimates for F0, AMP, and K to Ymin, Ymax, and the 1/the value of X at Ymid, respectively.
Figure 21.4.
An RNA-based HCV helicase assay suitable for HTS. (A) Design of the split beacon helicase assay (SBHA). In the RNA substrate, the bottom strand and the Cy5-labeled strand are RNA, while the IAbRQ-labeled strand is composed of DNA. In the DNA-labeled substrate, all three strands are composed of DNA, with Ts replacing Us in the bottom strand and the Cy5-labeled strand. (B) Time courses for sample negative control reactions using the DNA (closed circles) and the RNA (open circles) substrate, positive control reactions containing DNA plus 50 µM primuline (+), and the RNA substrate with 50 µM primuline (*). Lines show fit to the equations described in the text. The fluorescence observed in a well without substrate (×) is shown for comparison. (C) Reaction extent for two 96-well plates, one of which contained an RNA substrate and one a DNA substrate.
8. Discussion
The above protocols are being used to analyze HCV helicase inhibitors reported by others (Belon et al., 2010), to screen compound libraries for HCV helicase inhibitors, and to analyze hits (Belon and Frick, 2010). These assays, however, are only the first step in the process of finding a compound that could be used as a molecular probe or as a lead compound for drug discovery. After identifying compounds that inhibit HCV helicase activity on both DNA and RNA, we next examine whether or not they can inhibit HCV replication in cells. The assay most useful in this regard employs an HCV replicon in which a reporter gene is linked to a marker that can be used to select cells in which HCV RNA replicates (Hao et al., 2007). Although we have studied helicase inhibitors that are not active against the HCV replicon (Belon et al., 2010), our lab now primarily focuses on inhibitors that display some antiviral potency in HCV replicon-based assay. These compounds are now being studied using a variety of techniques to elucidate how they inhibit the unwinding reaction on a molecular level and how they effect HCV replication in cells. It should be appreciated that many other assays are available to identify inhibitors of HCV helicase, which have been the subject of other recent reviews (Belon and Frick, 2009a, 2011). There are also other resources available that summarize compounds that have been discovered to inhibit HCV helicase (Belon and Frick, 2009a, 2011; Borowski et al., 2000; Briguglio et al., 2011; Lemon et al., 2010) and compounds that target other helicases (Frick, 2003; Frick and Lam, 2006; Kwong et al., 2005).
ACKNOWLEDGMENTS
This work was supported by NIH grant RO1 AI088001 and a Research Growth Initiative Award (101X219) from the UWM Research Foundation. We would also like to thank all our collaborators at the MLPCN for their help with this project, particularly Dmitriy Minond of the Scripps Research Institute for helpful advice with assay design and Kelin Li, Kevin J. Frankowski, and Ben Neuenswander of the University of Kansas for providing the compounds used in our helicase inhibitor library.
REFERENCES
- Aggarwal M, Sommers JA, Shoemaker RH, Brosh RMJ. Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proc. Natl. Acad. Sci. USA. 2011;108:1525–1530. doi: 10.1073/pnas.1006423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon BR, Gordon SC, Lawitz E, Marcellin P, Vierling JM, Zeuzem S, Poordad F, Goodman ZD, Sings HL, Boparai N, Burroughs M, Brass CA, et al. Boceprevir for previously treated chronic HCV genotype 1 infection. N. Engl. J. Med. 2011;364:1207–1217. doi: 10.1056/NEJMoa1009482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belon CA, Frick DN. Monitoring helicase activity with molecular beacons. Biotechniques. 2008;45(433–40):442. doi: 10.2144/000112834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belon CA, Frick DN. Helicase inhibitors as specifically targeted antiviral therapy for hepatitis C. Future Virol. 2009a;4:277–293. doi: 10.2217/fvl.09.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belon CA, Frick DN. Fuel specificity of the hepatitis C virus NS3 helicase. J. Mol. Biol. 2009b;388:851–864. doi: 10.1016/j.jmb.2009.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belon C, Frick DN. Thioflavin S inhibits hepatitis C virus RNA replication and the viral helicase with a novel mechanism. FASEB J. 2010;24:lb202. [Google Scholar]
- Belon CA, Frick DN. NS3 helicase inhibitors. In: He Y, Tan SL, editors. Hepatitis C: Antiviral Drug Discovery and Development. Norfolk, UK: Caister Academic Press; 2011. pp. 327–356. [Google Scholar]
- Belon CA, High YD, Lin TI, Pauwels F, Frick DN. Mechanism and specificity of a symmetrical benzimidazolephenylcarboxamide helicase inhibitor. Biochemistry. 2010;49:1822–1832. doi: 10.1021/bi901974a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beran RK, Serebrov V, Pyle AM. The serine protease domain of hepatitis C viral NS3 activates RNA helicase activity by promoting the binding of RNA substrate. J. Biol. Chem. 2007;282:34913–34920. doi: 10.1074/jbc.M707165200. [DOI] [PubMed] [Google Scholar]
- Beran RK, Lindenbach BD, Pyle AM. The NS4A protein of hepatitis C virus promotes RNA-coupled ATP hydrolysis by the NS3 helicase. J. Virol. 2009;83:3268–3275. doi: 10.1128/JVI.01849-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson KP, Amaratunga M, Moore KJ, Lohman TM. Single-turnover kinetics of helicase-catalyzed DNA unwinding monitored continuously by fluorescence energy transfer. Biochemistry. 1994;33:14306–14316. doi: 10.1021/bi00251a044. [DOI] [PubMed] [Google Scholar]
- Boger DL, Tse WC. Thiazole orange as the fluorescent intercalator in a high resolution fid assay for determining DNA binding affinity and sequence selectivity of small molecules. Bioorg. Med. Chem. 2001;9:2511–2518. doi: 10.1016/s0968-0896(01)00243-7. [DOI] [PubMed] [Google Scholar]
- Boger DL, Fink BE, Brunette SR, Tse WC, Hedrick MP. A simple, high-resolution method for establishing DNA binding affinity and sequence selectivity. J. Am. Chem. Soc. 2001;123:5878–5891. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]
- Boguszewska-Chachulska AM, Krawczyk M, Stankiewicz A, Gozdek A, Haenni AL, Strokovskaya L. Direct fluorometric measurement of hepatitis C virus helicase activity. FEBS Lett. 2004;567:253–258. doi: 10.1016/j.febslet.2004.04.072. [DOI] [PubMed] [Google Scholar]
- Borowski P, Mueller O, Niebuhr A, Kalitzky M, Hwang LH, Schmitz H, Siwecka MA, Kulikowsk T. ATP-binding domain of NTPase/helicase as a target for hepatitis C antiviral therapy. Acta Biochim. Pol. 2000;47:173–180. [PubMed] [Google Scholar]
- Briguglio I, Piras S, Corona P, Carta A. Inhibition of RNA Helicases of ssRNA+ Virus Belonging to Flaviviridae, Coronaviridae and Picornaviridae Families. Int. J. Med. Chem. 2011;2011 doi: 10.1155/2011/213135. Article ID 213135, 22 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Crute JJ, Grygon CA, Hargrave KD, Simoneau B, Faucher AM, Bolger G, Kibler P, Liuzzi M, Cordingley MG. Herpes simplex virus helicase-primase inhibitors are active in animal models of human disease. Nat. Med. 2002;8:386–391. doi: 10.1038/nm0402-386. [DOI] [PubMed] [Google Scholar]
- Edlin BR. Perspective: Test and treat this silent killer. Nature. 2011;474:S18–S19. doi: 10.1038/474S18a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick DN. Helicases as antiviral drug targets. Drug News Perspect. 2003;16:355–362. doi: 10.1358/dnp.2003.16.6.829307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick DN. HCV helicase: Structure, function, and inhibition. In: Tan SL, editor. Hepatitis C Viruses: Genomes and Molecular Biology. Chapter 7. Norfolk (UK): Horizon Bioscience; 2006. [PubMed] [Google Scholar]
- Frick DN. The hepatitis C virus NS3 protein: A model RNA helicase and potential drug target. Curr. Issues Mol. Biol. 2007;9:1–20. [PMC free article] [PubMed] [Google Scholar]
- Frick DN, Lam AM. Understanding helicases as a means of virus control. Curr. Pharm. Des. 2006;12:1315–1338. doi: 10.2174/138161206776361147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick DN, Rypma RS, Lam AM, Gu B. The nonstructural protein 3 protease/helicase requires an intact protease domain to unwind duplex RNA efficiently. J. Biol. Chem. 2004;279:1269–1280. doi: 10.1074/jbc.M310630200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick DN, Banik S, Rypma RS. Role of divalent metal cations in ATP hydrolysis catalyzed by the hepatitis C virus NS3 helicase: Magnesium provides a bridge for ATP to fuel unwinding. J. Mol. Biol. 2007;365:1017–1032. doi: 10.1016/j.jmb.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick DN, Ginzburg O, Lam AM. A method to simultaneously monitor hepatitis C virus NS3 helicase and protease activities. Methods Mol. Biol. 2010;587:223–233. doi: 10.1007/978-1-60327-355-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W, Herlihy KJ, Zhang NJ, Fuhrman SA, Doan C, Patick AK, Duggal R. Development of a novel dicistronic reporter-selectable hepatitis C virus replicon suitable for high-throughput inhibitor screening. Antimicrob. Agents Chemother. 2007;51:95–102. doi: 10.1128/AAC.01008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck JA, Lam AM, Narayanan N, Frick DN. Effects of mutagenic and chain-terminating nucleotide analogs on enzymes isolated from hepatitis C virus strains of various genotypes. Antimicrob. Agents Chemother. 2008;52:1901–1911. doi: 10.1128/AAC.01496-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga N, Imamura M, Abe H, Nelson Hayes C, Kono T, Onishi M, Tsuge M, Takahashi S, Ochi H, Iwao E, Kamiya N, Yamada I, et al. Rapid emergence of telaprevir resistant hepatitis C virus strain from wild type clone in vivo. Hepatology. 2011;54:781–788. doi: 10.1002/hep.24460. [DOI] [PubMed] [Google Scholar]
- Houston P, Kodadek T. Spectrophotometric assay for enzyme-mediated unwinding of double-stranded DNA. Proc. Natl. Acad. Sci. USA. 1994;91:5471–5474. doi: 10.1073/pnas.91.12.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumata K, Chono K, Sudo K, Shimizu Y, Kontani T, Suzuki H. Effect of ASP2151, a herpesvirus helicase-primase inhibitor, in a guinea pig model of genital herpes. Molecules. 2011;16:7210–7223. doi: 10.3390/molecules16097210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Gwack Y, Han JH, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem. Biophys. Res. Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- Kleymann G, Fischer R, Betz UA, Hendrix M, Bender W, Schneider U, Handke G, Eckenberg P, Hewlett G, Pevzner V, Baumeister J, Weber O, et al. New helicase-primase inhibitors as drug candidates for the treatment of herpes simplex disease. Nat. Med. 2002;8:392–398. doi: 10.1038/nm0402-392. [DOI] [PubMed] [Google Scholar]
- Kolykhalov AA, Mihalik K, Feinstone SM, Rice CM. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3’ nontranslated region are essential for virus replication in vivo. J. Virol. 2000;74:2046–2051. doi: 10.1128/jvi.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong AD, Rao BG, Jeang KT. Viral and cellular RNA helicases as antiviral targets. Nat. Rev. Drug Discov. 2005;4:845–853. doi: 10.1038/nrd1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyono K, Miyashiro M, Taguchi I. Detection of hepatitis C virus helicase activity using the scintillation proximity assay system. Anal. Biochem. 1998;257:120–126. doi: 10.1006/abio.1998.2560. [DOI] [PubMed] [Google Scholar]
- Lam AM, Frick DN. Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase. J. Virol. 2006;80:404–411. doi: 10.1128/JVI.80.1.404-411.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam AM, Keeney D, Eckert PQ, Frick DN. Hepatitis C virus NS3 ATPases/helicases from different genotypes exhibit variations in enzymatic properties. J. Virol. 2003;77:3950–3961. doi: 10.1128/JVI.77.7.3950-3961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon SM, McKeating JA, Pietschmann T, Frick DN, Glenn JS, Tellinghuisen TL, Symons J, Furman PA. Development of novel therapies for hepatitis C. Antiviral Res. 2010;86:79–92. doi: 10.1016/j.antiviral.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- Muramatsu S, Ishido S, Fujita T, Itoh M, Hotta H. Nuclear localization of the NS3 protein of hepatitis C virus and factors affecting the localization. J. Virol. 1997;71:4954–4961. doi: 10.1128/jvi.71.7.4954-4961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CL, Rice CM. Turning hepatitis C virus into a real virus. Annu. Rev. Microbiol. 2011;65:307–327. doi: 10.1146/annurev-micro-090110-102954. [DOI] [PubMed] [Google Scholar]
- Neumann-Haefelin C, Frick DN, Wang JJ, Pybus OG, Salloum S, Narula GS, Eckart A, Biezynski A, Eiermann T, Klenerman P, Viazov S, Roggendorf M, et al. Analysis of the evolutionary forces in an immunodominant CD8 epitope in hepatitis C virus at a population level. J. Virol. 2008;82:3438–3451. doi: 10.1128/JVI.01700-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter DJ, Short SA, Hanlon MH, Preugschat F, Wilson JE, Willard DHJ, Consler TG. Product release is the major contributor to kcat for the hepatitis C virus helicase-catalyzed strand separation of short duplex DNA. J. Biol. Chem. 1998;273:18906–18914. doi: 10.1074/jbc.273.30.18906. [DOI] [PubMed] [Google Scholar]
- Preugschat F, Averett DR, Clarke BE, Porter DJ. A steady-state and pre-steady-state kinetic analysis of the NTPase activity associated with the hepatitis C virus NS3 helicase domain. J. Biol. Chem. 1996;271:24449–24457. doi: 10.1074/jbc.271.40.24449. [DOI] [PubMed] [Google Scholar]
- Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- Raney KD, Sharma SD, Moustafa IM, Cameron CE. Hepatitis C virus non-structural protein 3 (HCV NS3): A multifunctional antiviral target. J. Biol. Chem. 2010;285:22725–22731. doi: 10.1074/jbc.R110.125294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rypma RS, Lam AM, Frick DN. Effect of substrate traps on hepatitis C virus NS3 helicase catalyzed DNA unwinding: Evidence for enzyme catalyzed strand exchange. In: Knudsen WD, Bruns SS, editors. Bacterial DNA, DNA Polymerase and DNA Helicases. New York: Nova Science Publishers, Inc.; 2009. pp. 389–407. [Google Scholar]
- Suzich JA, Tamura JK, Palmer-Hill F, Warrener P, Grakoui A, Rice CM, Feinstone SM, Collett MS. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J. Virol. 1993;67:6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Akimitsu N, Fujita O, Matsuda Y, Miyata R, Tsuneda S, Igarashi M, Sekiguchi Y, Noda N. High-throughput screening assay of hepatitis C virus helicase inhibitors using fluorescence-quenching phenomenon. Biochem. Biophys. Res. Commun. 2009;379:1054–1059. doi: 10.1016/j.bbrc.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Tuteja R. Helicases—Feasible antimalarial drug target for Plasmodium falciparum. FEBS J. 2007;274:4699–4704. doi: 10.1111/j.1742-4658.2007.06000.x. [DOI] [PubMed] [Google Scholar]
- Tyagi S, Kramer FR. Molecular beacons: Probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- Yao N, Hesson T, Cable M, Hong Z, Kwong AD, Le HV, Weber PC. Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- Yedavalli VS, Zhang N, Cai H, Zhang P, Starost MF, Hosmane RS, Jeang KT. Ring expanded nucleoside analogues inhibit RNA helicase and intracellular human immunodeficiency virus type 1 replication. J. Med. Chem. 2008;51:5043–5051. doi: 10.1021/jm800332m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeuzem S, Andreone P, Pol S, Lawitz E, Diago M, Roberts S, Focaccia R, Younossi Z, Foster GR, Horban A, Ferenci P, Nevens F, et al. Telaprevir for retreatment of HCV infection. N. Engl. J. Med. 2011;364:2417–2428. doi: 10.1056/NEJMoa1013086. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]