Abstract
Kisspeptin is essential for reproductive functions in humans. As a model for the human we have used the female guinea pig, which has a long ovulatory cycle similar to that of primates. Initially, we cloned a guinea pig kisspeptin cDNA sequence and subsequently explored the distribution and 17β-estradiol (E2) regulation of kisspeptin mRNA (Kiss1) and protein (kisspeptin) by using in situ hybridization, real-time PCR and immunocytochemistry. In ovariectomized females, Kiss1 neurons were scattered throughout the preoptic periventricular areas (PV), but the vast majority of Kiss1 neurons were localized in the arcuate nucleus (Arc). An E2 treatment that first inhibits (negative feedback) and then augments (positive feedback) serum luteinizing hormone (LH) increased Kiss1 mRNA density and number of cells expressing Kiss1 in the PV at both time points. Within the Arc, Kiss1 mRNA density was reduced at both time points. Quantitative real-time PCR confirmed the in situ hybridization results during positive feedback. E2 reduced the number of immunoreactive kisspeptin cells in the PV at both time points, perhaps an indication of increased release. Within the Arc, the kisspeptin immunoreactivity was decreased during negative feedback but increased during positive feedback. Therefore, it appears that in guinea pig both the PV and the Arc kisspeptin neurons act cooperatively to excite gonadotropin-releasing hormone (GnRH) neurons during positive feedback. We conclude that E2 regulation of negative and positive feedback may reflect a complex interaction of the kisspeptin circuitry, and both the PV and the Arc respond to hormone signals to encode excitation of GnRH neurons during the ovulatory cycle.
INDEXING TERMS: In situ hybridization, quantitative real-time PCR, immunocytochemistry
The guinea pig, similarly to primates, has a long ovulatory cycle (17–21 days) with an estrogen-dominated follicular phase and a progesterone-dominated luteal phase (Czaja and Goy, 1975). Guinea pigs are also diurnal animals, with the active period at dusk and dawn (Roepke et al., 2010). As in other species, gonadotropin-releasing hormone (GnRH) neurons in guinea pig are distributed through specific regions of the preoptic area (POA). In addition, GnRH neurons are located in the arcuate nucleus (Arc) and adjacent basal hypothalamic (BH) regions in guinea pig, as in primates (Kelly et al., 1985; Rønnekleiv and Resko, 1990; Pau et al., 1990; Grove-Strawser et al., 2002; Kim et al., 2009). GnRH neurons are potently excited by kisspeptin through activation of nonselective cationic channels and inhibition of potassium channels (Liu et al., 2008; Pielecka-Fortuna et al., 2008; Zhang et al., 2008).
Kisspeptins, encoded by the Kiss1 gene, are currently recognized as key factors in the regulation of reproductive development and functions (Tena-Sempere, 2006; Kuohung and Kaiser, 2006; Gottsch et al., 2006; Plant, 2006). The Kiss1 gene encodes a 145-amino-acid protein, which is proteolytically processed to produce a 54-aminoacid peptide called kisspeptin-54 and several other smaller peptide fragments (Kotani et al., 2001). Kisspeptin-54 is the endogenous ligand of a G-protein-coupled receptor, GPR54, also called the kisspeptin receptor (Kotani et al., 2001; Stafford et al., 2002; Oakley et al., 2009). Mutations in GPR54 cause autosomal recessive idiopathic hypogonadotropic hypogonadism in humans (Seminara et al., 2003; De Roux et al., 2003), whereas deletion of GPR54 in mice causes defective sexual development and reproductive failures (Seminara et al., 2003). In addition, targeted deletion of the Kiss1 gene in mice causes a phenotype similar to that of GPR54 gene mutation (d’Anglemont de Tassigny et al., 2007). Collectively, these findings suggest that kisspeptins and their GPR54 receptor are essential for normal reproductive physiology.
The distribution and estrogen regulation of kisspeptin mRNA (Kiss1) expression have been extensively described for the mouse and rat brain (Smith et al., 2005, 2006; Oakley et al., 2009). For these rodent species, it is known that Kiss1 mRNA is expressed primarily in the anteroventral periventricular nucleus (AVPV) and adjacent periventricular (PV) areas, as well as in the Arc of the hypothalamus (Smith, 2008; Clarkson et al., 2009). Importantly, 17β-estradiol (E2) increases the mRNA expression of Kiss1 in the AVPV but decreases the expression in the Arc (Smith et al., 2005, 2006). These findings are consistent with data showing that the AVPV is necessary for E2 positive feedback on GnRH and luteinizing hormone (LH) secretion in these species (Wiegand et al., 1980; Petersen and Barraclough, 1989; Ma et al., 1990). In other species such as the guinea pig, sheep, and rhesus monkey the POA appears not to be the main region responsible for E2 positive feedback (Terasawa and Wiegand, 1978; Plant et al., 1978; Weick, 1981; King et al., 1998; Caraty et al., 1998). Thus, it appears that the basal hypothalamus may be sufficient for maintaining positive feedback regulation of GnRH and LH secretion in these species.
The distribution of kisspeptin has not been ascertained in the guinea pig, but in rhesus monkey and also in sheep most kisspeptin neurons are located in the Arc, although kisspeptin neurons (or mRNA) are also observed in the POA (Rometo et al., 2007; Ramaswamy et al., 2008; Kim et al., 2009; Smith et al., 2010; Hoffman et al., 2011). In rhesus monkey and human females, a consistent finding is that Arc (infundibular) kisspeptin neurons or mRNA expression is highly increased in menopausal compared with young individuals (Rometo et al., 2007; Kim et al., 2009; Eghlidi et al., 2010), suggesting that gonadal steroids exert an inhibitory effect on Arc Kiss1 expression in human and primates, as in other species. For the ewe (a seasonal breeder), several publications have addressed the hypothesis that Arc kisspeptin neurons are involved in positive feedback regulation of GnRH/LH secretion (Smith, 2008; Hoffman et al., 2011). Measurements of Kiss1 mRNA and kisspeptin have provided evidence for increased kisspeptin expression in the caudal Arc during positive feedback but also increased expression in the POA, leading to the hypothesis that both neuronal populations are involved in positive feedback regulation of GnRH in the ewe (Smith et al., 2009; but see Hoffman et al., 2011).
To address this issue further, we investigated the distribution and estrogen regulation of Kiss1 mRNA and immunoreactive kisspeptin during negative as well as positive feedback regulation of LH secretion in guinea pig. The studies revealed that most Kiss1 neurons are located in the Arc of guinea pig, although a few scattered cells can be detected in the POA. The mRNA expression of the POA Kiss1 population is positively regulated by E2, and the Arc mRNA expression is negatively regulated by E2 irrespective of feedback status. In contrast, kisspeptin protein (immunoreactive kisspeptin) is reduced in the Arc during negative feedback and increased during positive feedback, suggesting a role for both nuclear groups in positive feedback regulation of LH by E2.
MATERIALS AND METHODS
Animals
All procedures performed with animals were in accordance with the NIH guidelines for the care and use of laboratory animals and were approved by our local committee on animal care and use. Adult female multicolor guinea pigs (Topeka from our own colony or from Elm Hill, Chelmsford, MA; 470–660 g) were given free access to food and water and were maintained under a 14-hour light, 10-hour dark lighting schedule, with lights on at 0630 hours and lights off at 2030 hours. The animals were ovariectomized (OVX) under ketamine/xylazine anesthesia (50 mg and 5 mg/kg, respectively, s.c.). The analgesic meloxicam was administered orally for 3 days postoperatively (0.2 mg/kg) during the 1-week recovery period.
Experimental design
Experiment 1: induction of LH surge in guinea pig
OVX females (n = 7) were injected s.c. with 17β-estradiol benzoate (here referred to as E2; 20–25 μg in 100 μl sesame oil) at 2030 hours (at the time of lights out) using a procedure modified from that described previously (Terasawa et al., 1979). Thirty-three hours later, serial blood samples were collected from an indwelling catheter that had been implanted in the jugular vein as described previously (Condon et al., 1988). The sampling periods ranged from 12 to 14 hours. Serum LH levels were determined in duplicate with a heterologous radioimmunoassay using an anti-ovine LH antiserum (GDN-15; kindly provided by Dr. Gordon Niswender) as reported previously (Condon et al., 1988).
Experiment 2: distribution and E2 regulation of Kiss1 mRNA by in situ hybridization
Before experimentation, the OVX animals were injected with E2; 20–25 μg in 100 μl sesame oil or the sesame oil vehicle (100 μl) and killed at approximately 24 hours or 42 hours while sedated with ketamine. Trunk blood was collected for measurements of E2 by radioimmunoassay (RIA) as described previously (Jamali et al., 2003), and the E2 injection produced late-follicular-phase levels of E2. We used in situ hybridization to analyze the distribution and estrogen regulation of kisspeptin neuronal mRNA expression in the guinea pig brain during negative and positive feedback regulation of LH.
Coronal hypothalamic blocks (2 mm each) obtained following 24-hour vehicle, 24-hour E2 (negative feedback), or 42-hour vehicle and 42-hour E2 treatment (positive feedback) were dissected and fixed by immersion in 4% paraformaldehyde in Sorensen buffer (0.03 M; pH 7.4; Sigma, St Louis, MO) for 5–7 hours at 4°C, rinsed overnight in 20% buffered-sucrose solution (pH 7.4) using RNAse free reagents, embedded in O.C.T. (Sakura Finetek, Torrence, CA), and frozen in isopentane at −55°C. Coronal sections (20 μm) were cut on a cryostat and thaw-mounted onto Superfrost Plus glass slides (Fisher Scientific, Pittsburgh, PA). The sections were stored at −80°C until used for in situ hybridization (or immunocytochemistry; see below).
Experiment 3: distribution and E2 regulation of Kiss1 mRNA by real-time PCR
In additional experiments, we used real-time PCR analysis of RNA extracted from microdissected hypothalamic tissues for a more quantitative assessment of distribution and estrogen regulation of Kiss1 mRNA expression. In this study, we were particularly interested in exploring changes in mRNA expression during positive feedback, i.e., 42 hours E2 (25 μg) or vehicle treatment.
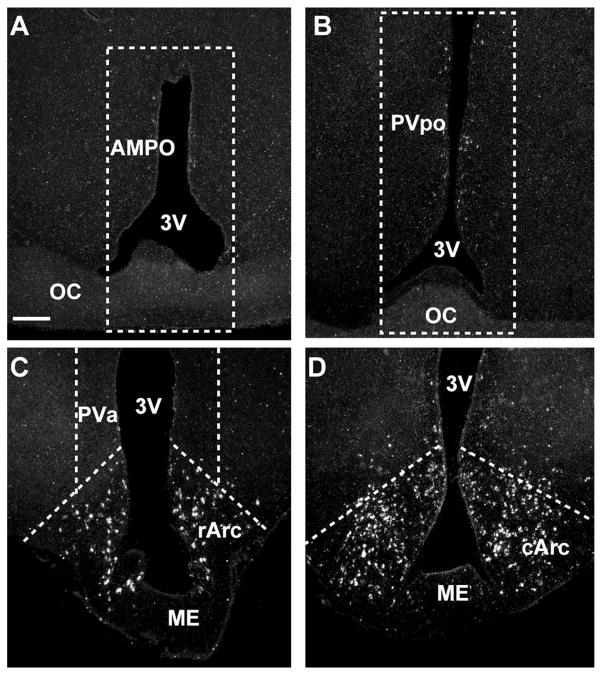
For quantitative real-time PCR analysis, a brain slicer (EM Corporation, Chestnut Hill, MA) was utilized to produce 1-mm frontal slices. Each brain was sliced quickly under chilled conditions, and the slices were placed in an RNA-stabilizing solution, RNAlater (Ambion, Austin, TX), which prevents RNA degradation. The correct rostral and caudal border of each slice was confirmed using a dissecting microscope and a guinea pig brain atlas (Bleier, 1983). On completion of the slicing, hypothalamic nuclei were microdissected under the guidance of a dissecting microscope as follows (Fig. 1): anterior medial preoptic (AMPO) nucleus (Fig. 1A), preoptic periventricular (PVpo) area (Fig. 1B), anterior PV (PVa) area (Fig. 1C), rostral Arc (rArc; Fig. 1C), and middle to caudal remainder of the Arc (cArc; Fig. 1D). Microdissected tissues were rapidly frozen and stored at −80°C.
Figure 1.
Outline of microdissection of hypothalamic nuclei. Darkfield photomicrograph of coronal sections through the anterior medial preoptic nucleus (AMPO; A), preoptic periventricular (PVpo) area (B), rostral arcuate (rArc) nucleus (C), and caudal arcuate (cArc) nucleus (D), illustrating the location of the microdis-sected tissues and the location of kisspeptin mRNA (white grains) in the different hypothalamic nuclei. The dissection along the dashed lines in A included the AMPO nucleus. The dissection along the dashed lines in B included the PVpo. The rArc dissection along the ventral dashed lines in C included the rostral part of the Arc but also extended into the retrochiasmatic area. The periventricular dissection along the dashed lines in C included the anterior periventricular nucleus (PVa). The remainder of the Arc was dissected along the dashed lines in D and extended to the caudal parts of the Arc. Scale bar = 250 μm.
Experiment 4: distribution and E2 regulation of immunoreactive kisspeptin in the diencephalon
We used immunocytochemistry to analyze the distribution and E2 regulation of kisspeptin expression in the guinea pig brain during E2 negative (24 hours) and positive (42 hours) feedback regulation of LH. This analysis was performed on alternate sections compared with those used for the in situ hybridization analysis (see above). Initially, we used immunocytochemistry combined with in situ hybridization to document that the kisspeptin polyclonal antibody against mouse kisspeptin (see below) was specific for guinea pig kisspeptin neurons.
Cloning of the guinea pig Kiss1 gene
Degenerate primers were designed for guinea pig Kiss1 by aligning known sequences for Kiss1 using the Clone Manager software (version 8; Scientific and Educational Software, Cary NC). Areas of high homology were found among cattle (Bos tarus; Genbank accession No. XM_867473), mouse (NM_178260), rat (NM_181692), human (NM_002256), and chimpanzee (Pan trogloydyte; XM_514123). Degenerate primers were designed based on the human sequence that also incorporated all the base combinations of the aligned species. Primer sequences were as follows: forward primer 5′-YTTCYTGG-CAGCTRMTGCTTYTCC-3′ (corresponding to human sequence 169–191 nt), reverse primer 5′-AGGCCRAAG-GAGTTCCAGTTGTAG-3′ (corresponding to human sequence 505–485 nt; degenerate bases were Y = C and T, R = A and G, M = A and C).
Two hundred nanograms of total RNA from guinea pig basal hypothalamus was reverse transcribed into cDNA in a 20-μl reaction containing 4 μl 5 × GoTaq Flexi buffer (Promega, Madison, WI), 5 mM MgCl2, 0.625 mM dNTPs, 100 ng random primers, 15 U RNasin, 10 mM dithiothreitol (DTT), and 50 U MuLV reverse transcriptase (Applied Biosystems, Foster City, CA). The reaction parameters were 42°C for 60 minutes, 99°C for 5 minutes, and 4°C for 5 minutes. PCR was performed in a 30-μl reaction containing 6 μl 5 × Go Taq flexi buffer, 2 mM MgCl2, 0.33 mM dNTPs, 0.5 μM forward and reverse primers, 0.22 μg TaqStart Antibody (Clontech, Mountain View, CA), 2 U Go Taq DNA polymerase (Promega), and 2 μl cDNA. Taq polymerase and TaqStart antibody were combined in a tube at room temperature for 5 minutes, and then the remaining reagents were added to the tube. Step-down PCR was performed for an initial denaturation at 94°C for 2 minutes; 22 cycles of amplification at 94°C for 30 seconds, 65–55°C for 1 minute (for 2 cycles at each degree), and 72°C for 2 minutes; followed by 36 cycles of amplification at 94°C for 30 seconds, 50°C for 1 minute, and 72°C for 2 minutes; with a final 72°C extension for 5 minutes. A single band at the predicted size (334 bp) was visualized with ethidium bromide on a 1.5% agarose gel. The remainder of the PCR product was run on a low-melting-point gel and extracted from the gel using the Qiaex II gel extraction kit (Qiagen, Valencia, CA). The product was subcloned into the pGEM-T Easy vector (Promega) and sequenced, revealing a 334-bp guinea pig-specific sequence (accession No. HM030726).
Quantitative real-time PCR
This analysis was performed as described in detail previously (Malyala et al., 2008). Briefly, total RNA was extracted from microdissected tissue samples using the RNaqueous Micro kit (Ambion) and quantified on a spectrophotometer (Nanodrop Technologies, Wilmington, DE). Total RNA was DNase I treated (DNAfree; Ambion) at 37°C for 30 minutes, and the DNase I was heat inactivated at 65°C for 10 minutes. Two hundred nanograms of total RNA was reverse transcribed into cDNA using 100 ng random hexamers (Promega) and Superscript III reverse transcriptase (100 U; Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. As a negative control, 200 ng total RNA was reacted without reverse transcriptase (−RT) and used in the real-time PCR assay. Quantitative real-time PCR (qPCR) for guinea pig Kiss1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; used as control) was accomplished with primers designed in Clone Manager 8 software and the use of SYBR green chemistry (Applied Biosystems, Foster City, CA) on the ABI 7500 fast real-time PCR system (Applied Biosystems). The primer sequences were as follows: guinea pig Kiss1 (accession No. HM030726; 67 bp product) forward primer (5′-GTGTCACCTCCTTGGGA-GAAC-3′, 22–42 nt) and reverse primer (5′-TGGCCTGTGGGTCTAGGAT-3′, 70–88 nt); guinea pig GAPDH (accession No. NM_0011172951; 212 bp product) forward primer (5′-CATCCACTGGTGCTGCCAAG-3′, 676–695 nt) and reverse primer (5′-GTCCTCGGTGTAG CCCAAGA-3′, 868–887 nt). Forward and reverse primers were used at a final concentration of 0.3 μM in a reaction that also contained 10 μl 2 × mastermix, 3 μl cDNA, and nuclease-free water to a final volume of 20 μl. Samples for the target gene were run in triplicate and GAPDH was run in duplicate on the ABI 7500 under the following thermal cycling conditions: 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute, followed by a dissociation step for melting curve analysis. To exclude the possibility of contamination, nuclease-free water and a sample from the −RT control were run in parallel to every qPCR run. Amplification efficiency for the primer pairs was assessed using serial dilutions (from 1:50 to 1:12,800) of basal hypothalamic cDNA. A standard curve was constructed and the efficiency was calculated according to the following formula: E = 10(−1/m) − 1, where m = slope (Livak and Schmittgen, 2001; Pfaffl, 2001). The PCR amplification efficiencies were 96% for Kiss1 and 98% for GAPDH, which allowed us to use the 2−ΔΔCt method to determine differences in our treatment groups (Livak and Schmittgen, 2001). GAPDH was used as an internal reference for normalization. (We have previously determined that GAPDH does not change with E2 treatment in the brain regions analyzed (Roepke et al., 2007). The expression value of each gene was quantified on the basis of the interpolated cycle number in which the fluorescence reached a threshold value (Ct value), using background fluorescence in cycles 1–15 as baseline. The Kiss1 transcripts were normalized to the amount of GAPDH in that sample to calculate a relative amount of transcript present for each gene. The normalized expression values for all control and E2-treated samples were averaged, and an average fold change was calculated using 2−ΔΔCt (Livak and Schmittgen, 2001). The relative abundance of Kiss1 was compared with the oil-treated rostral Arc (rArc; calibrator) expression level. A two-tailed Student’s t-test was conducted between the normalized relative expression values for each oil- and E2-treated sample to determine statistical significance (P < 0.05).
Kiss1 probe preparation
Radioactive antisense cRNA probe incorporating the full-length 334-bp guinea pig-specific Kiss1 sequence was transcribed in vitro with T7 RNA polymerase from the BSTX1 linearized construct in the presence of 35S-uridine 5′ (-thio)triphosphate (35S-UTP). The sense probe was prepared using SP6 polymerase from the Nco1 linearized guinea pig Kiss1 construct.
Residual DNA was digested with 10 U DNase I (Roche Diagnostics Corp., Indianapolis, IN). The sense and anti-sense RNA probes were separated from free nucleotides by using a G-50 Sephadex column (Amersham Biosciences, Piscataway, NJ).
In situ hybridization
Slides from oil- and E2-treated females were simultaneously reacted. These slides were postfixed in fresh 4% paraformaldehyde in Sorensen’s buffer for 15 minutes, rinsed with Sorenson’s phosphate buffer (0.03 M, pH 7.4), and treated with proteinase K (1.0 μg/ml) for 4 minutes at 37°C. Sections were then treated with 0.1 M triethanolamine (3–5 minutes), followed by 0.25% acetic anhydride in 0.1 M triethanolamine (10 min; Sigma, St. Louis, MO) and rinsed briefly in 2 × SSC. Sections were prehybridized for 1 hour at 57°C with hybridization buffer (50% formamide,1 × Denhardt’s solution, 10% dextran sulfate, 100 μM DTT, 200 mM sodium chloride, 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, pH 8.0, 125 μg/ml tRNA; Sigma) and then quickly rinsed in 2 × SSC buffer. The 35S-labeled antisense, as well as sense, riboprobes were heat-denatured, diluted with hybridization buffer, and used at a final concentration of 1.6 × 104 dpm/μl. Subsequently, the sections were covered with glass cover-slips, sealed, and hybridized in a moist chamber for at least 18 hours at 58°C. After hybridization, the slides with the coverslips removed were rinsed in 2× SSC buffer, reacted with RNAse (20 μg/ml) for 30 minutes at 37°C, and washed in decreasing concentrations of SSC (2×, 1×, 0.5×, 0.1×) at 55°C, with a final wash at 63°C for 1 hour in 0.1 × SSC containing DTT. The slides from oil- and E2-treated females were dehydrated in ethanol, placed side by side on a flat surface together with autoradiographic 14C microscales, and exposed to Hyperfilm-βmax (Amersham Biosciences, Piscataway, NJ) for 5–7 days at 4°C. The slides were then dipped in Kodak NTB-2 nuclear track emulsion and exposed for up to 21 days at 4°C. Thereafter, slides were developed in D19 developer, fixed in Kodak fixer, counterstained with Gills 3 × hematoxylin, dehydrated, cleared in xylene, and coverslipped with polymount.
Data analysis of film images
Quantification of film images was performed with a Macintosh G4 computer equipped with NIH Image 1.61. Film images of an average of five sections from each brain region in oil control and E2-treated females were scanned (CanoScan 5600F; Cannon) and analyzed, and an average density value was obtained for each subregion and each treatment, which was used for further analysis. The 14C microscale (present on each film) was used as a standard to calculate the density of mRNA signals, and the optical density was converted to nanocuries (nCi) per gram. Comparison between the vehicle- and the E2-treated groups were analyzed by either two-tailed Student’s t-test or two-way analysis of variance (ANOVA; main factors treatment and hypothalamic nuclear subregions), followed by Newman Keuls post hoc test. Differences were considered statistically significant if the probability of error was less than 5%.
Data analysis of emulsion-coated slides
Kiss1 mRNA images from emulsion-coated slides were evaluated under darkfield and brightfield illumination with a Nikon eclipse 800 microscope equipped with a digital camera. In addition to the densitometry measurements from film images (see above), the total number of cells in the POA that contained autoradiographic grains (at least 4 × above background levels) was counted with a 20 × objective and an eyepiece square grid reticle. This was done in part to compare densitometry measures with cell counts. (Cells were not counted in the Arc, because it was difficult to distinguish the labeling of individual cells accurately). Four to eight sections from each animal, spaced an average of 90 μm apart, were counted from rostral to caudal periventricular POA, and the number of cells per section and per animal (n = 5 animals in each group) was determined and used for further analysis. Comparison between the two groups (oil and 42 hours E2) was performed via two-tailed Student’s t-test. Differences were considered statistically significant if the probability of error was less than 5%. For visualization purposes, Kiss1 images were photographed under darkfield or brightfield illumination with a Nikon Eclipse 800 microscope. Contrast and brightness were adjusted in digitized images in Adobe Photoshop (Adobe, Mountain View, CA) to match the images observed in the microscope.
Immunocytochemistry alone or in combination with in situ hybridization
Immunocytochemistry for kisspeptin was performed as described previously (Qiu et al., 2011) using the Caraty kisspeptin 10 antibody (No. 564; Franceschini et al., 2006). This antibody against amino acids 43–52 of mouse kisspeptin was produced in rabbit and found to be highly specific for mouse kisspeptin based on peptide preabsorption experiments and inhibition curves generated in a radioimmunoassay for mouse kisspeptin (Franceschini et al., 2006). We had previously determined that this antibody worked well in guinea pig, and that immunoreactive kisspeptin within the guinea pig basal hypothalamic (BH) area was completely eliminated when the antiserum was preabsorbed with mouse kisspeptin 10 (Qiu et al., 2011). In additional experiments, we used serial diluted kisspeptin antiserum (1:2,500–1:7,500) on Arc tissue sections from 24-hour oil- and E2-treated animals. Based on this preliminary analysis, the kisspeptin antiserum was used at dilutions of 1:2,500–1:5,000, which consistently gave robust kisspeptin immunoreactivity in the oil-treated animals but more faint immunoreactivity in the 24-hour E2-treated group. In all instances, slides from oil- and E2-treated animals were always reacted together. To ascertain that the kisspeptin immunoreactivity was specific for guinea pig Kiss1 neurons, we performed coexpression analysis of Kiss1 mRNA and immunoreactive kisspeptin on mediobasal hypothalamic sections from oil-treated females, because arcuate Kiss1 mRNA expression is elevated in these animals. The combined in situ hybridization and immunocytochemistry was performed essentially as described previously (Zheng et al., 2005). Briefly, sections reacted with the antisense probe (see above) were incubated at 4°C overnight with kisspeptin primary antiserum diluted 1:2,500 in 0.01 M phosphate-buffered saline (PBS) containing 2% bovine serum albumin (BSA). A 30-minute postprimary wash in PBS was followed by a 2-hour incubation at 37°C with biotinylated IgG (1:300; Vector Laboratories, Burlingame, CA), another 30-minute wash, and a 2-hour incubation at 37°C with streptavidin-Cy3 (1:1,000; Jackson Immunoresearch, West Grove, PA), followed by a final wash for 1 hr. Then, sections were briefly dipped in 70% and 95% ethanol and air dried. The slides were exposed to hyperfilm-βmax (Amersham Biosciences) for 5–7 days at 4°C and were then dipped in Kodak NTB-2 nuclear track emulsion and exposed for up to 28 days at 4°C. Slides were developed in D19 developer, fixed in Kodak fixer, and coverslipped with a glycerin solution as described previously (Zheng et al., 2005).
Image analysis
Analysis of Kiss1 mRNA and immunoreactive kisspeptin colocalization
Immunoreactive cells and Kiss1 mRNA images were evaluated and photographed under fluorescent and dark-field illumination using a Nikon Eclipse 800 microscope. Darkfield together with fluorescent microscopy allowed direct visualization of autoradiographic grains and immuno-reactive cells and, therefore, the determination of immuno-reactive cells that colocalized clusters of autoradiographic grains. Three to five sections from each animal were evaluated from the rostral to the caudal BH. Darkfield, fluorescent, and combined views were photographed. Images were illustrated in Adobe Photoshop and Macromedia FreeHand (Macromedia, San Francisco, CA).
Immunocytochemistry
Sections throughout the POA-BH region from oil- and E2-treated animals were evaluated and photographed with a Nikon Eclipse 800 microscope equipped with a digital Nikon DS-L1 color cooled camera. Within the POA, scattered PV-area kisspeptin cells were counted in four to six sections from each animals, and the mean number of cells per section was determined for each animal and used for further analysis. In addition, the immunoreactive kisspeptin integrated density in the same areas was calculated in NIH Image J (W.S. Rasband, Image J, U.S. National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/, 1997–2008). The threshold was set the same for all images, and the free-hand selection tool was used to outline the area to be measured. Within the Arc, individual cells were often difficult to discriminate because of the dense population of cells and intense fiber stain. With this caveat in mind, the number of cells was counted blindly in two to four sections from each Arc subregion (bilateral) in oil-and E2-treated animals at the 24-hour and 42-hour time points. In addition, immunoreactive kisspeptin was analyzed by densitometry as follows: Arc sections (four to six sections/animal) reacted together for immunoreactive kisspeptin were evaluated using a Zeiss fluorescent microscope equipped with the Slidebook 4 (3I, Denver, CO) program. This semiquantitative analysis was done blindly on randomized sections from oil-and E2-treated animals. Images were acquired with a Marianas Stereological imaging workstation (3I, Denver, CO) consisting of a Zeiss Axiovert 200M, Lambda DG-4 175-W xenon lamp, excitation S565/28×, emission S617/73m, 10 × NA 0.5 fluar objective, ASI motorized stage, and Coolsnap EZ camera, all controlled by Slidebook 4.2. Exposure time was 1 second for all images. Montages of several (typically 2 × 2) fields of view, covering the entire Arc in each section, were thresholded at the same intensity level (300) chosen to separate the bright immunofluorescence signal from the dark background. Slidebook 4.2 was used for image acquisition and for measuring the overall fluorescence intensity (sum intensity) of the kisspeptin immunostaining in arbitrary units.
RESULTS
Negative and positive feedback regulation of LH in guinea pig
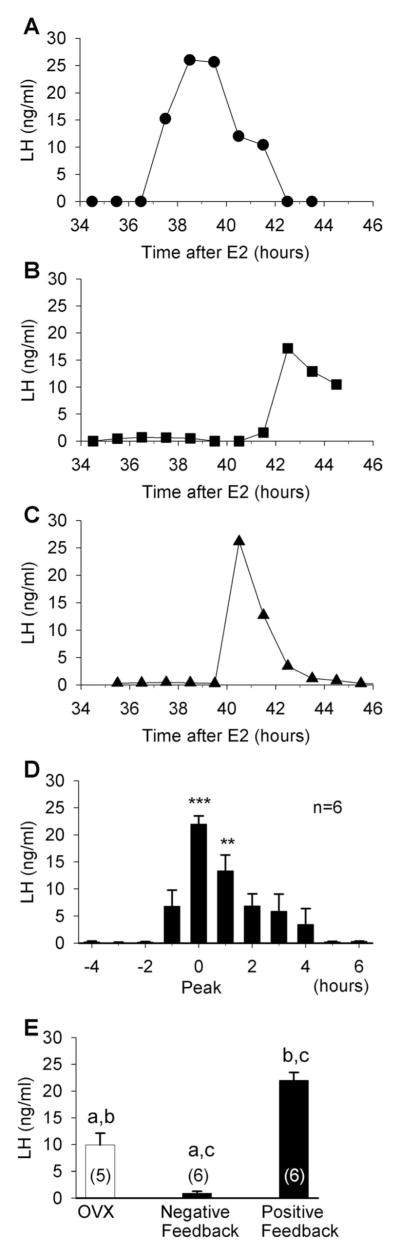
We used a guinea pig model developed previously (Terasawa et al., 1979) and modified by us for induction of positive and negative feedback (Condon et al., 1988; Wagner et al., 2001). An LH surge was induced in six of seven females as early as 37 hours and as late as 44 hours following the E2 treatment (Fig. 2A–D). The mean levels of serum LH during the surge, normalized to the individual peak level, were elevated above basal levels for at least 1 hour (Fig. 2D). As expected, systemic E2 administration to OVX females resulted in a long-lasting inhibition of LH, as measured at 24 hours following the injection, below levels in OVX controls and peak LH surge levels (Fig. 2E). For subsequent experiments, we used approximately 24 hours and 42 hours after E2 administration (the mean surge time) as the times of euthanasia for studies of negative and positive feedback, respectively.
Figure 2.
LH levels in guinea pig during positive and negative feedback. Serum LH levels in representative individual ovariectomized (OVX) guinea pigs that were injected with E2 (25 μg) to induce an LH surge (A–C). Mean LH levels in six animals during positive feedback normalized to the peak LH level in each animal (D). One-way ANOVA, **P < 0.01, ***P < 0.001. E: Mean LH levels in OVX animals (left bar) and E2-treated animals at 24 hours (negative feedback; middle bar) and peak levels during positive feedback (right bar). One-way ANOVA, aa, bb, cc, P < 0.001.
Guinea pig kisspeptin clone
To study the mRNA expression of Kiss1 in the female guinea pig brain, we used PCR to clone a guinea pig-specific Kiss1 product that was 334 nucleotides in length. The PCR clone corresponded to nucleotides 169–505 in the human sequence (NM_002256). The nucleotide sequence homology with corresponding Kiss1 sequences in different species was 76% homologous to the corresponding sequences in human, 77% homologous to cattle (Bos taurus), 75% homologous to chimpanzee (Pan troglodytes), 72% homologous to mouse, and 71% homologous to rat. The amino acid alignment of the cloned guinea pig kisspeptin in comparison with that of other species is illustrated (Fig. 3) and reveals a high degree of amino acid similarity in the C-terminal region but less amino acid similarity across species in other parts of the molecule. Of importance is that the guinea pig kisspeptin C-terminal sequence has complete homology with the mouse kisspeptin sequence that was used by Caraty and coworkers to generate their kisspeptin antiserum (Franceschini et al., 2006), which was used in this study (see below) for measurement of immunoreactive kisspeptin in guinea pig (Fig. 3).
Figure 3.

Comparison of kisspeptin amino acid sequences among different species. Alignment of amino acid sequences of guinea pig, human, chimpanzee, cow, rat, and mouse kisspeptin. Numbers indicate amino acid position, and identical amino acid residues are shaded. The underlined sequence in mouse is the sequence used to produce the kisspeptin-10 polyclonal antibody (Franceschini et al., 2006) used in this study and was found previously to be specific for kisspeptin neurons in guinea pig (Qiu et al., 2011). This 10-amino-acid sequence is highly conserved across all species shown. The three underlined amino acids (LRY) in guinea pig are the predicted sequence from the UCSC genome browser (scaffold_12.640).
Expression of Kiss1 mRNA by in situ hybridization in the guinea pig hypothalamus
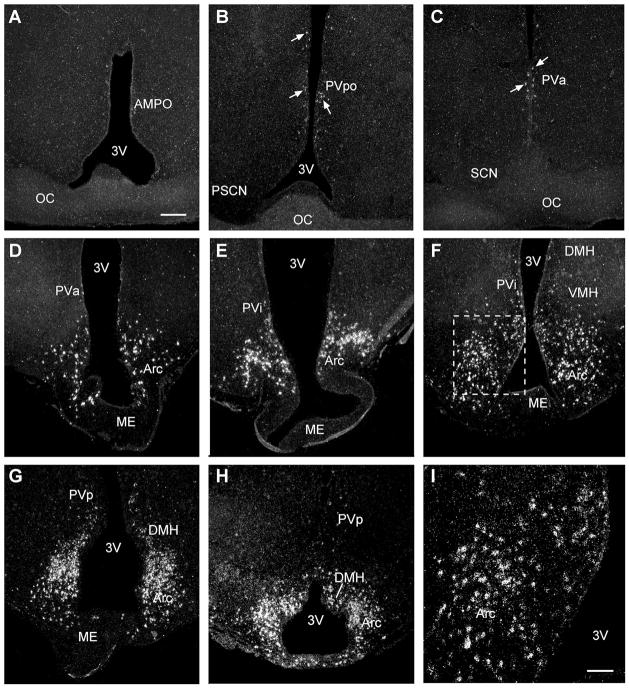
The expression of Kiss1 mRNA was documented in the POA and BH in OVX oil- and E2-treated guinea pigs. There was little Kiss1 mRNA within the POA. Specifically, there were cells scattered throughout the preoptic periventricular (PVpo) area and anterior PV (PVa) area, whereas essentially no cells were found in the anterior medial pre-optic nucleus (AMPO), believed to be equivalent to the anteroventral periventrivular (AVPV) nucleus in mouse and rat (Fig. 4A–C). Within the BH, Kiss1 mRNA was expressed in the rostral to caudal Arc, with the majority of labeled cells in the middle and caudal parts of the Arc (Fig. 4D–I). In addition, labeled cells were detected in the intermediate PV area (PVi), the posterior PV area (PVp), and the caudal part of the dorsomedial nucleus of the hypothalamus (DMH; Fig. 4E–H). Sections reacted with the sense RNA probe exhibited no signal above background in any of the brain regions analyzed (data not shown).
Figure 4.
Expression and distribution of kisspeptin mRNA. Emulsion autoradiograms of kisspeptin (Kiss1) mRNA by in situ hybridization. Darkfield photomicrographs illustrating kisspeptin mRNA signal (white grains) in coronal sections through the preoptic area (POA) from rostrally to caudally (A–C) and Arc from rostrally to caudally (D–H). Boxed area from the Arc (F) is shown at higher magnification (I). For abbreviations see list. Scale bars = 250 μm A (applies to A–H); 100 μm in I.
17β-Estradiol regulation of Kiss1 mRNA by in situ hybridization
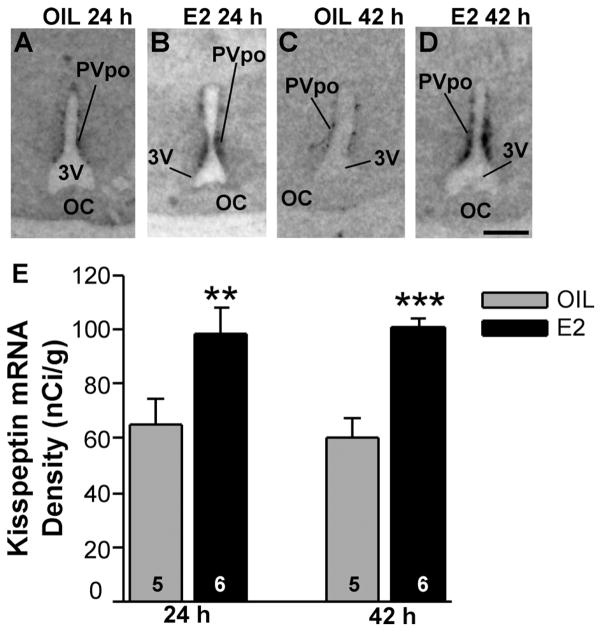
OVX female guinea pigs, which had been treated with E2 for 24 hours or 42 hours to induce negative and positive feedback regulation of LH (GnRH), respectively, were used to explore E2 regulation of Kiss1 mRNA expression in the POA via in situ hybridization. Densitometry analysis of film images revealed that Kiss1 mRNA density was significantly increased in the POA already at 24 hours (negative feedback; P < 0.01, n = 5–6, Student’s t-test) and remained elevated in animals analyzed at 42 hours after E2 treatment (positive feedback; P < 0.001, n = 5–6, Student’s t-test; Fig. 5). Kiss1 analysis of emulsion-coated slides revealed that only a few Kiss1-positive cells were detected in the AMPO area of either group of animals. More caudally in the PVpo and PVa areas, darkfield and brightfield images revealed that the number of cells expressing Kiss1 mRNA increased in sections from E2-treated females. Quantification of the number of cells in the rostral to caudal PV areas in comparing oil and E2-treated animals (42-hour time point; emulsion slides from the 24-hour group were not available) revealed a significant increase in the number of Kiss1 neurons in the E2-treated group (Fig. 6A–C; P < 0.01, n = 5), supporting the film densitometry analysis.
Figure 5.
Kiss1 mRNA expression in POA from oil- and E2-treated females. Matched film images of Kiss1 mRNA expression in the PVpo (periventricular POA) in 24-hour and 42-hour ovariectomized oil (A,C)- and E2 (B,D)-treated animals. The darkness of the image represents the density of mRNA expression. For abbreviations see list. E: Quantitative analysis of Kiss1 mRNA in the PVpo from oil-and E2-treated females at 24 hours (negative feedback) and 42 hours (positive feedback); n = 5–6 animals (two-tailed Student’s t-test, **P < 0.01, ***P < 0.001). Scale bar = 1 mm.
Figure 6.
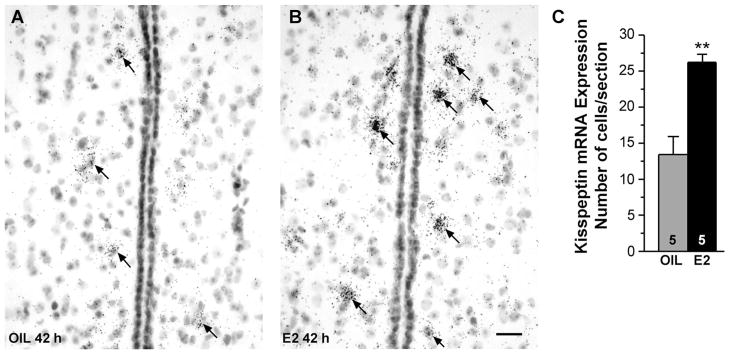
Brightfield images of Kiss1 mRNA in the POA. Brightfield photomicrographs of representative coronal sections through the medial POA in oil-treated (A) and E2-treated (B) females illustrating more clearly the periventricular distribution of Kiss1 mRNA (dark grains). C: Cells expressing autoradiographic grains at least four times above background were counted in sections throughout the POA, and the mean number of cells/section was quantified. This analysis revealed that the number of cells/section expressing Kiss1 mRNA was increased in the OVX E2-treated group (two-tailed Student’s t-test, **P < 0.01, n = 5). Scale bar = 20 μm.
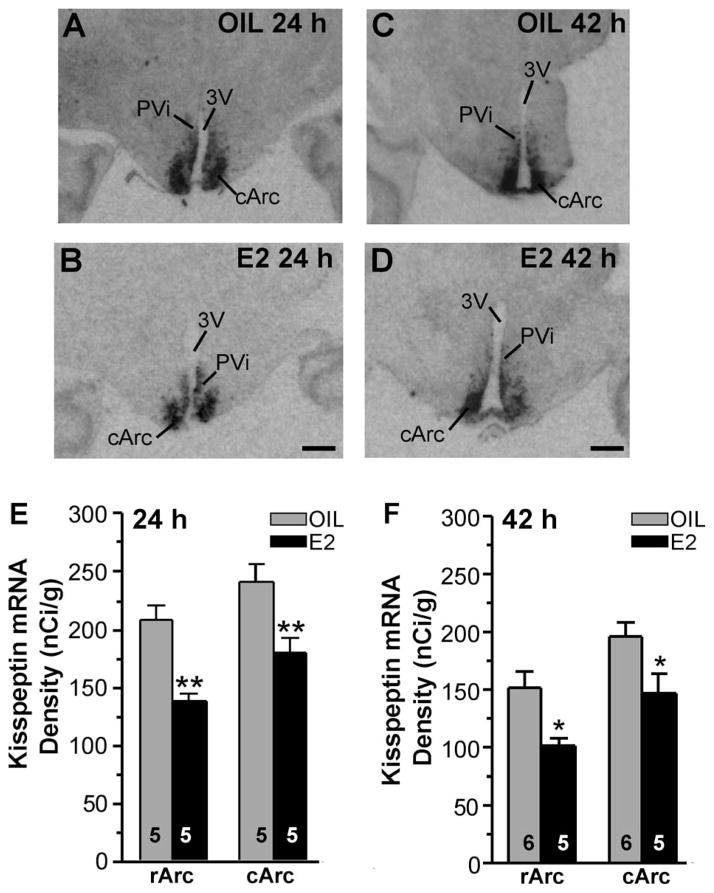
In the guinea pig, positive feedback regulation of LH secretion can be mediated by the BH (Terasawa and Wiegand, 1978; King et al., 1998), which argues for the involvement of Arc kisspeptin neurons in positive feedback regulation of LH (GnRH) by E2. Therefore, we examined the effects of E2 on Kiss1 mRNA expression by in situ hybridization in our guinea pig model of negative and positive feedback at 24 hours and 42 hours, respectively. Analysis of images from emulsion-coated slides of the distribution of Kiss1 mRNA in the rostral (rArc) and middle to caudal (cArc) areas of the Arc in the four groups of animals revealed an equivalent distribution pattern irrespective of treatment. For quantification, film images from OVX oil-and E2-treated groups were matched and analyzed via densitometry (Fig. 7A–D). Quantitative analysis of film images, revealed that E2 treatment in the 24-hour group reduced Kiss1 mRNA density in both rArc and cArc areas (two-way ANOVA, F1,16 = 27.93 for treatment, F1,16 = 9.73 for area; P < 0.01, n = 5; Fig. 7E). Kiss1 mRNA density was also decreased in the Arc in the 42-hour E2-treated group (n = 5) vs. the oil-treated group (n = 6). Quantitative analysis of film images revealed that E2 treatment reduced Kiss1 mRNA density in rArc and cArc areas (two-way ANOVA, F1,18 = 14.85 for treatment, F1,18 = 11.99 for area; P < 0.05, n = 5–6; Fig. 7F). We also measured the Kiss1 density in the PVi area and found no difference between the oil- and the E2-treated groups (oil, 76.1% ± 7.2%, n = 6; E2, 67.1% ± 9.9%, n = 5; Student’s t-test).
Figure 7.
Kiss1 mRNA expression in oil- and E2-treated females during negative feedback and positive feedback. Film images of Kiss1 mRNA expression in 24-hour (A,B) and 42-hour (C,D) ovariectomized oil (A,C)- and E2 (B,D)-treated animals. Images of film autoradiograms illustrate the distribution of Kiss1 mRNA in representative matched coronal sections from caudal Arc (cArc) in 24-hour and 42-hour oil- and E2-treated animals. The darkness of the image represents the density of mRNA expression. For abbreviations see list. Distribution and quantitative analysis of Kiss1 mRNA in the rostral (rArc) and caudal Arc (cArc) from 24 hours (E) and 42 hours (F) oil- and E2-treated females revealed decreased Kiss1 mRNA expression in rArc and cArc in the E2-treated groups (two-way ANOVA, **P < 0.01, n = 5; *P < 0.05, n = 5–6). Scale bar = 1 mm.
Distribution of Kiss1 mRNAs by real-time PCR in the guinea pig hypothalamus
To substantiate further the quantitative changes in expression of Kiss1 mRNA within the hypothalamus, we measured Kiss1 mRNA expression in microdissected hypothalamic nuclei from OVX guinea pigs by using quantitative real-time PCR (qPCR; Fig. 8A–C). We determined from cDNA serial dilutions that the amplification efficiencies for Kiss1 and GAPDH were 96% and 98% respectively, which allowed us to use the 2−ΔΔCt method for quantification (Livak and Schmittgen, 2001). We also documented the amplification of a specific, single product for each of the two transcripts by single-product melting curves, which further validated the analyses (Fig. 8B). In agreement with the in situ hybridization measurement, Kiss1 mRNA levels were low in the AMPO, PVpo, and PVa areas compared with the Arc (Fig. 8A,D). The rostral Arc (rArc) had moderate levels of Kiss1, whereas the middle to caudal Arc (cArc) in comparison expressed high levels of Kiss1 mRNA (Fig. 8D). Kiss1 mRNA expression was significantly increased in the PVpo and PVa but not in the AMPO area in E2-treated (42 hours) vs. oil-treated females (Fig. 8D; P < 0.05, n = 6). Within the microdissected Arc, Kiss1 mRNA was significantly reduced in cArc but not in rArc of E2-treated vs. oil-treated females (Fig. 8D; P < 0.01, n = 6).
Figure 8.
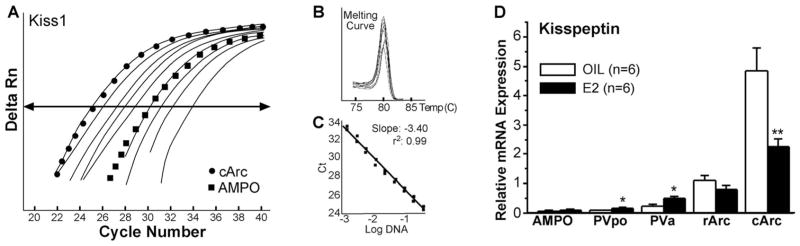
Kiss1 mRNA distribution by real-time PCR. Quantitative real-time PCR measurements of Kiss1 mRNA in microdissected hypothalamic nuclei in oil- and E2-treated female guinea pigs. A standard curve was prepared with basal hypothalamic (BH) cDNA serial dilutions from 1:50 to 1:12,800 (A). Cycle number was plotted against the normalized fluorescence intensity (Delta RN) to visualize the PCR amplification. The cycle threshold (Ct; arrow) is the point in the amplification at which the sample values were calculated. The BH cDNA serial dilutions and two representative samples (cArc and AMPO) are illustrated in A. The superimposed dissociation (melting) curves depict a single Kiss1 product (B). The standard curve regression line and slope from serial dilution data are illustrated (C). The amplification efficiency calculated from the slope was 96% for Kiss1. GAPDH was used as a control, and its amplification efficiency was 98% (data not shown). The expression values were calculated using the ΔΔCt method where the calibrator was the ΔCt of rArc oil-treated samples (D). Number of animals (n) = 6 in each group; *P < 0.05, **P < 0.01, two-tailed Student’s t-test.
Distribution of immunoreactive kisspeptin in the guinea pig diencephalon
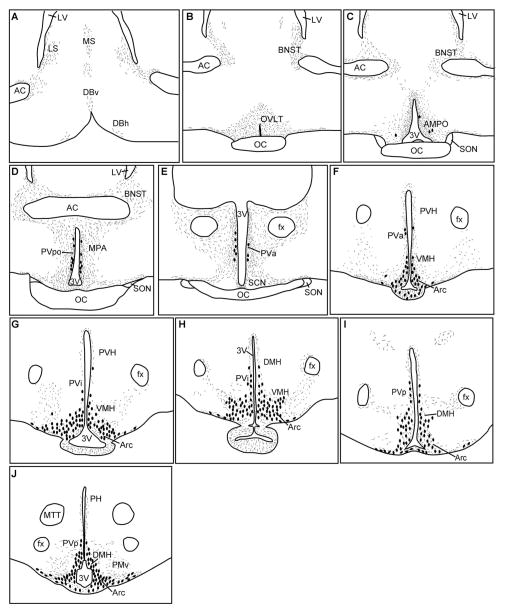
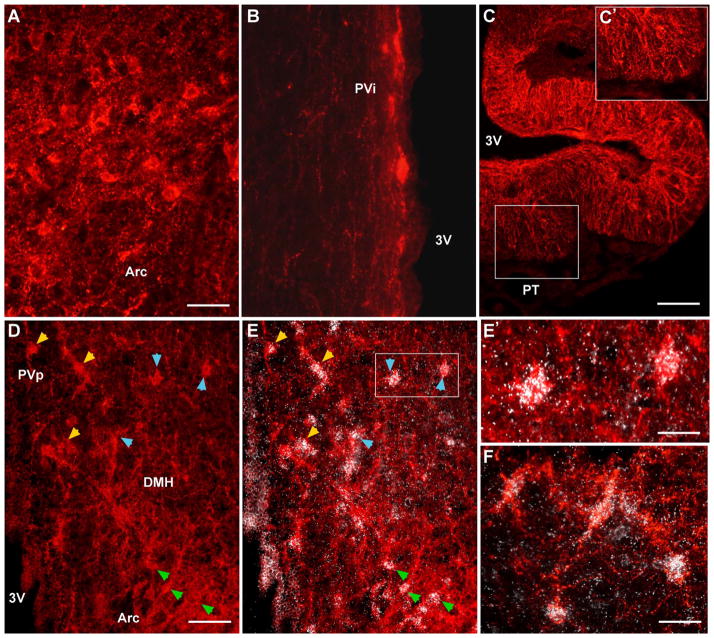
We used the kisspeptin antibody generated against a 10-amino-acid sequence that is highly conserved among different species, including guinea pig, to analyze the kisspeptin distribution (Franceschini et al., 2006; Fig. 3). Kisspeptin-immunoreactive fibers were observed in the organum vasculosum of the lamina terminalis (OVLT) and medial preoptic areas from rostrally to caudally (Fig. 9). Kisspeptin fibers were also present in the medial and lateral septum and in the bed nucleus of the stria terminalis (BNST; Fig. 9). In contrast to the fiber stain, kisspeptin cell bodies were rarely detected in the AMPO area, but scattered cells were present in the PVpo and PVa areas (Fig. 9). The highest concentration of kisspeptin immunoreactivity, cells together with dense fiber staining, was observed in the rostral to caudal Arc (Figs. 9F–J, 10A). Cells and fibers were also present in the intermediate periventricular area (PVi; Figs. 9G,H, 10B). Moreover, kisspeptin fibers were dense in both the internal and the external zones of the median eminence (ME; Fig. 10C), which is similar to kisspeptin fiber distribution in the sheep and rhesus monkey but differs from that in rat and mouse (Franceschini et al., 2006; Ramaswamy et al., 2008; Clarkson et al., 2009; True et al., 2010). Kisspeptin fibers, but not cells, were also present within the ventral premammillary nucleus (Fig. 9J). Finally, a population of kisspeptin neurons was located in the caudal part of the DMH (Figs. 9I,J, 10D). Thus, overall, the distribution pattern of cells positive for immunoreactive kisspeptin and Kiss1 mRNA was similar within the guinea pig hypothalamus (Fig. 9 vs. Fig. 4). In support of these observations, Kiss1 mRNA and immunoreactive kisspeptin were colocalized in all areas tested, including the Arc and the caudal part of the DMH (Fig. 10D–F), suggesting that the rabbit-generated kisspeptin antibody used is specific for guinea pig kisspeptin. Moreover, we have shown previously that the kisspeptin staining in guinea pig BH is completely eliminated when the antibody is preabsorbed with the peptide used to generate the kisspeptin antibody (Qiu et al., 2011).
Figure 9.
Distribution of kisspeptin in guinea pig POA-hypothalamus. Schematic drawings of the distribution of kisspeptin immunoreactive fibers (stippled areas) and cell bodies (large dots) from rostral to caudal POA (A–E) and from rostral to caudal BH region (F–J) in the ovariectomized guinea pig. For abbreviations see list.
Figure 10.
Immunoreactive kisspeptin and coexpression with Kiss1 mRNA. Photomicrographs of coronal sections illustrating immunoreactive kisspeptin cells and fibers in the Arc (A), intermediate periventricular area (PVi; B), and caudal dorsomedial nucleus (D). C: Low-power view of immunoreactive kisspeptin fibers in the internal and external zones of the median eminence (ME). Boxed area in C is magnified in C′ to illustrate the kisspeptin fibers in the external zone of the ME. D,E: Representative caudal BH section reacted for kisspeptin (immunocytochemistry) and Kiss1 mRNA (in situ hybridization). D: Kisspeptin-immunoreactive cells are localized in PVp, caudal DMH, and Arc (Arc). E: Cells within all of these regions also express Kiss1 mRNA (white grains) as demarcated by yellow arrowheads (PVp), blue arrowheads (DMH), and green arrowheads (Arc). The boxed area in E is magnified in E′ to illustrate clearly autoradiographic grains (Kiss1 mRNA) coexpressed in immunoreactive kisspeptin cells in the caudal DMH. F: High-power view of Arc immunoreactive kisspeptin cells that also express Kiss1 mRNA. Scale bars = 45 μm in A (applies to A,B); 165 μm in C; 85 μm in D (applies to D,E); 30 μm in E′; 40 μm in F.
17β-Estradiol regulation of immunoreactive kisspeptin
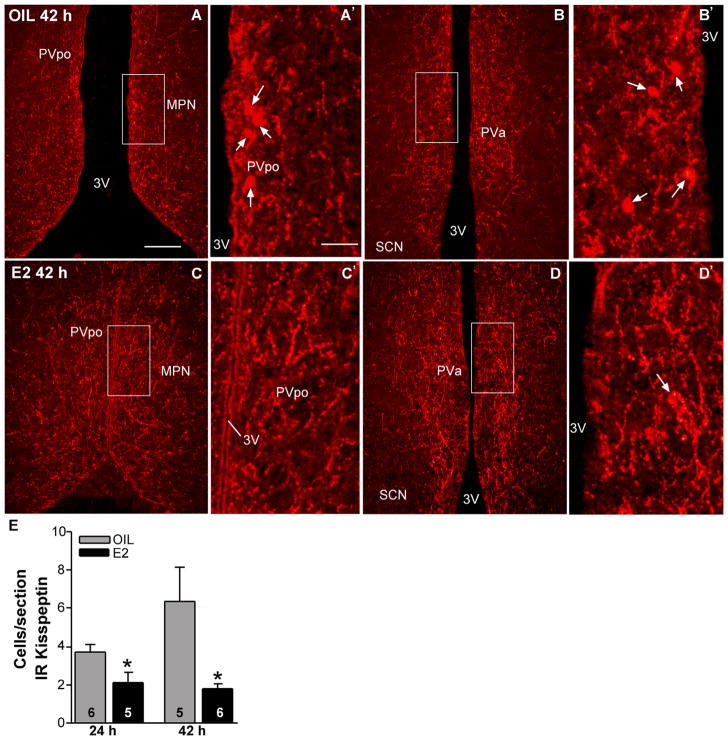
OVX female guinea pigs, which had been treated with E2 for 24 hours or 42 hours to induce negative and positive feedback regulation of LH (GnRH), respectively, were used to explore E2 regulation of kisspeptin in the POA via immunocytochemistry. Again, only a few kisspeptin-positive cells were detected in the AMPO area in either group of animals. Farther caudally in the PVpo and PVa areas, scattered cells varied in number from one to four in each section in E2-treated animals and from two to 19 in each section in oil-treated animals in both the 24-hour and the 42-hour treatment groups. Low-power images from the 42-hour oil- and E2-treated females illustrate the extensive kisspeptin fiber distribution in the medial preoptic area from rostrally to caudally (Fig. 11A–D). High-power images illustrate kisspeptin cells in the PVpo and PVa areas in a representative oil-treated female and very few cells in similar regions in an E2-treated female (Fig. 11A′–D′). By using densitometry analysis, we found no difference in the overall immunoreactive kisspeptin density in the preoptic periventricular area between 42-hour oil-and 42-hour E2-treated animals (oil, 101.2 ± 23.1 arbitrary units [AU]; E2 102.5 ± 8.4 AU; n = 4–5). However, quantification of kisspeptin cells during E2-induced negative and positive feedback revealed that the number of immunoreactive cells in the POA was significantly reduced in the E2- compared with oil-treated females, both at 24 hours and at 42 hours following treatment (Fig. 11E; P < 0.05, n = 5–6).
Figure 11.
Immunoreactive kisspeptin in the POA from oil- and E2-treated females. Representative photomicrographs of immunoreactive kisspeptin in coronal sections through the POA from rostrally (A,C) to caudally (B,D) in oil-treated (A,B) and E2-treated (C,D) animals (42 hours after treatment). The boxed areas in A–D are amplified in A′–D′, respectively, to illustrate immunoreactive cells (arrows). E: Quantitative analysis of the effects of E2 on the number of kisspeptin-immunoreactive cells in POA during negative (24 hours) and positive (42 hours) feedback. *P < 0.05; n = 5–6, two-tailed Student’s t-test. Scale bars = 165 μm in A (applies to A–D); 40 μm in A′ (applies to A′–D′). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
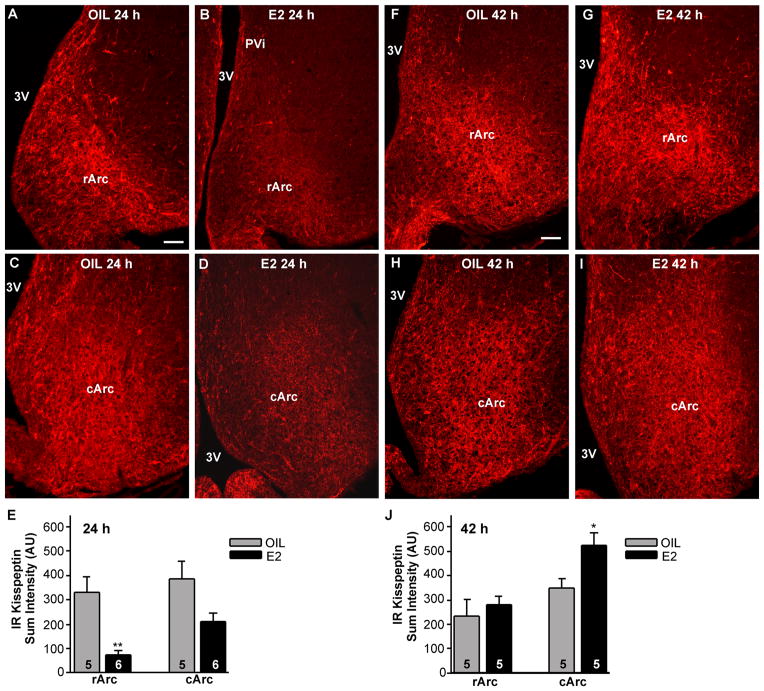
Within the Arc, the overall intensity of immunoreactive kisspeptin was decreased in the E2-treated females at 24 hours following treatment (negative feedback; Fig. 12A–D) and increased at 42 hours following treatment (positive feedback; Fig. 12F–I). In contrast, there was no change in the oil-treated animals (Fig. 12A,C,F,H). A semi-quantitative analysis of the immunoreactive kisspeptin sum intensity in the Arc revealed an E2-induced decrease in the rostral part of the nucleus (rArc) at 24 hours (Fig. 12E; two-way ANOVA, F1,18 = 19.94 for treatment, F1,18 = 3.99 for area; P < 0.01) and an E2-induced increase in the middle to caudal part (cArc) at 42 hours (Fig. 12J; two-way ANOVA, F1,16 = 5.01 for treatment, F1,16 = 12.61 for area; P < 0.05) in comparison with the respective analysis in the oil-treated groups. As an additional measure of changes in immunoreactive kisspeptin, we also counted kisspeptin cell bodies in rostral and caudal Arc tissue sections from oil- and E2-treated animals at both time points. This analysis revealed that the number of immunoreactive kisspeptin cells in the 24-hour E2-treated compared with the oil-treated group was reduced in both rArc (oil, 146 ± 19/section; E2, 73 ± 3/section; P < 0.01, two-way ANOVA) and cArc (oil, 258 ± 15 cells/section; E2, 145 ± 5 cells/section; P < 0.01, two-way ANOVA). In contrast, the number of immunoreactive kisspeptin cells in the 42-hour E2-treated compared with the oil-treated group was not altered in either the rArc (oil, 117 ± 18 cells/section; E2, 143 ± 18 cells/section; P > 0.05) or the cArc (oil, 220 ± 11 cells/section; E2, 241 ± 25 cells/section; P > 0.05).
Figure 12.
Immunoreactive kisspeptin in the Arc during negative feedback and positive feedback. Photomicrographs of representative coronal sections through the rostral Arc (rArc; A,B,F,G) and caudal Arc (cArc; C,D,H,I) in 24-hour oil (A,C)-, 42-hour oil (F,H)-, 24-hour E2 (B,D)-, and 42-hour E2- (G,I)-treated females. E,J: Quantitative analysis of immunoreactive kisspeptin sum intensity in arbitrary units (AU). *P < 0.05, n = 5; **P < 0.01, n = 5–6, two-way ANOVA. Scale bars = 100 μm. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
DISCUSSION
E2 treatment augmented Kiss1 mRNA expression in the rostral PV area and decreased Kiss1 mRNA levels in the Arc at both 24-hour (negative feedback) and 42-hour (positive feedback) in OVX female guinea pigs. This was associated with reduced number of POA kisspeptin neurons at both time points but no change in overall immuno-reactive kisspeptin in this brain region. Within the Arc, immunoreactive kisspeptin was reduced during negative feedback and increased during positive feedback. Thus, in the guinea pig, a small population of rostral PV kisspeptin neurons is activated by E2, leading to increased mRNA expression irrespective of gonadal steroid feedback status. Within the Arc, reduced Kiss1 mRNA levels were associated with reduced immunoreactive kisspeptin during negative feedback but increased immunoreactivity during E2-induced positive feedback. Therefore, the steroid regulation of hypothalamic kisspeptin neurons might reflect not simply one group (rostral PV area) involved in positive feedback and the other (Arc) involved in negative feedback but a complex interaction of the kisspeptin circuitry and the steroid hormone signals to ascertain both inhibition and activation of GnRH neurons during the ovulatory cycle.
We have documented here that an E2 injection in ovariectomized guinea pigs results in an initial inhibition of serum LH levels as measured at 24 hours following E2 (negative feedback), with subsequent increased serum LH levels 37–45 hours later (positive feedback), which largely confirms previous findings in this species (Terasawa et al., 1979). It should be noted that, in guinea pig, similarly to animal models such as the sheep and monkey, the LH (GnRH) surge is not as tightly coupled to a circadian signal but rather is dependent on the time after E2 treatment (Terasawa et al., 1979, 1984; Caraty et al., 1989). Also, in these species, in contrast to the rat and mouse, lesion or E2 brain-implant studies have documented that naturally occurring or E2-induced LH and GnRH surges are dependent on neural substrates within the BH and not just the POA (Terasawa and Wiegand, 1978; Wiegand et al., 1980; Weick, 1981; Petersen and Barraclough, 1989; Ma et al., 1990; King et al., 1998; Caraty et al., 1998). Therefore, the guinea pig is an alternative model for the human in addition to the ewe (a seasonal breeder) and the rhesus monkey (a more costly animal model). With the discovery of the kisspeptin neuronal system and its importance for reproductive function in humans, we hypothesized that these neurons would be localized in the Arc in guinea pig and that different subgroups of Arc neurons would be responsible for negative and positive feedback regulation of GnRH neurons.
Indeed, the present studies have revealed that, in the adult female guinea pig, Kiss1-expressing neurons are located primarily in the Arc, with a minor population in the periventricular POA. Similarly, in humans, only a few scattered Kiss1 mRNA neurons are localized in the mPOA, with the majority of cells in the infundibular (Arc) nucleus (Rometo et al., 2007). Although in other species, including the sheep, pig, and mouse, there appears to be a more substantial POA population of Kiss1 neurons, most Kiss1 mRNA-expressing cells are located in the Arc of most species (Smith et al., 2005, 2006, 2007; Tomikawa et al., 2010). Analyses of kisspeptin distribution using immunocytochemistry have produced mixed results because of difficulties with the specificity of antibodies (Brailoiu et al., 2005; Mikkelsen and Simonneaux, 2009; Iijima et al., 2011). However, the current study in guinea pig showed, for the most part, a similar distribution pattern of Kiss1 mRNA-expressing cells and immunoreactive kisspeptin cells. Both mRNA and peptide were localized in the periventricular POA and more caudal periventricular areas, the Arc, and the caudal part of the dorsomedial nucleus of the hypothalamus (DMH). Moreover, we have documented, within the regions examined (arcuate, DMH and PVi), Kiss1 mRNA and immunoreactive kisspeptin colocalization, suggesting that the kisspeptin antibody used for immunostaining (Franceschini et al., 2006) was specific for guinea pig kisspeptin. A similar distribution of immunoreactive kisspeptin neuronal cell bodies has been documented in the mouse brain, with the exception that most cells are located in the AVPV and periventricular POA, with fewer cells in the Arc and a low-density group of cells within the DMH (Clarkson et al., 2009). All of the immunoreactivity was lost in kisspeptin knockout mice, supporting the specificity of the kisspeptin immunoreactivity (Clarkson et al., 2009). With the rat it has been difficult to identify immunoreactive kisspeptin cells in the POA without the use of colchicine (Adachi et al., 2007), suggesting that kisspeptin is not being stored in the POA kisspeptin cell bodies in this species. However, after colchicine pretreatment and antibody preabsorption with cross-reacting neuropeptides, the kisspeptin-specific immunoreactivity exhibits a distribution similar to that of Kiss1 mRNA also in the rat; i.e., cell bodies in the periventricular POA and Arc but not in the DMH (Iijima et al., 2011). Thus, there appear to be species differences in the distribution of kisspeptin neurons (Clarkson et al., 2009; Tomikawa et al., 2010; Hoffman et al., 2011; Iijima et al., 2011; current Fig. 9). There also appear to be species differences in the kisspeptin fiber distribution to the median eminence (ME) region. In guinea pig, there are kisspeptin fibers in both the internal and the external zones of the ME (Fig. 10C,C′), which is consistent with previous findings in the sheep and rhesus monkey (Franceschini et al., 2006; Ramaswamy et al., 2008; Smith et al., 2011) but not in the rat or mouse (Clarkson et al., 2009; True et al., 2010). Although it has been hypothesized that kisspeptin fibers in the ME provide functional input to GnRH fibers in rhesus monkey and ewe (Ramaswamy et al., 2008; Smith et al., 2011), the mechanism for such an interaction is currently unknown. Interestingly, recent studies in mouse, based on anterograde and retrograde tracing techniques, showed little evidence for kisspeptin–GnRH interaction in the ME (Yeo and Herbison, 2011). Rather, kisspeptin neurons in the rostal part of the Arc, similarly to POA kisspeptin neurons, may send projections to GnRH neuronal cell bodies to control GnRH neurosecretion. In general, the kisspeptin neuronal systems appears to be more complex than previously realized and may be involved in a number of homeostatic functions, including transmitting metabolic signals to GnRH neurons (Qiu et al., 2011; Yeo and Herbison, 2011).
It is well documented that increased plasma E2 levels, during the ovulatory cycle or following E2 treatment, augment the mRNA expression of Kiss1 in the POA (Smith et al., 2005, 2006; Adachi et al., 2007). Similarly, with guinea pig, we have found that Kiss1 mRNA was increased in the preoptic PV areas as measured by two different methods. Interestingly, the same E2 treatments resulted in decreased cellular protein levels (immunoreactive kisspeptin) within POA kisspeptin cell bodies, whereas the overall kisspeptin immunoreactivity did not change. This may indicate an E2-induced increased synthesis and transport of kisspeptin within the POA kisspeptin neuronal population in guinea pig, which may also account in part for the detection of fewer immunoreactive kisspeptin cells than Kiss1 mRNA-expressing cells in the POA.
Another interesting observation is that the cellular expression of kisspeptin in the guinea pig POA might not be tightly coupled to E2 induction of the LH surge, insofar as we found Kiss1 mRNA in the POA increased during both negative and positive feedback and the number of immunoreactive kisspeptin cells reduced at both time points. A careful circadian analysis of Kiss1 mRNA expression in E2-treated mice found evidence for increased Kiss1 mRNA levels at the time of the LH surge (Robertson et al., 2009). However in most animal models, including mouse, rat, and sheep, E2-induced expression of cFos in POA kisspeptin neurons appears to be a more reliable index of increased kisspeptin activity coupled with the timing of the GnRH and LH surges compared with Kiss1 mRNA levels (Smith et al., 2006; Adachi et al., 2007; Robertson et al., 2009; Hoffman et al., 2011). Although these and other studies emphasize the importance of the POA population of kisspeptin cells for the E2 induction of the GnRH and LH surges, the augmented expression of Kiss1 mRNA might not be a reliable index of positive feedback in guinea pig. Additionally, we found in guinea pig that Kiss1 mRNA was reduced in the Arc during both negative and positive feedback, but immunoreactive kisspeptin was reduced during negative feedback in the rostral part of the Arc and increased during positive feedback in the caudal part of the Arc. This may indicate that the caudal arcuate population of kisspeptin cells in guinea pig is involved in positive feedback regulation of GnRH neurons. Similarly in sheep and rhesus monkey, the caudal part of the Arc appears to be involved in positive feedback regulation of the female reproductive cycle (Smith et al., 2009, 2010; Clarke et al., 2009). Therefore, in certain species, including guinea pig, sheep, and perhaps primates, kisspeptin neurons in the Arc may transmit positive feedback of the steroid signal to GnRH neurons by an as yet unknown mechanism.
In summary, we have shown that the kisspeptin neuronal population in the guinea pig extends from the PV area rostrally to the caudal parts of the BH, with most kisspeptin expression within the Arc. During E2-induced negative and positive feedback, Kiss1 mRNA levels were increased in the rostral PV area and reduced in the Arc. Immunoreactive kisspeptin was reduced in the Arc during negative feedback and increased during positive feedback. Therefore, the guinea pig has a small population of rostral PV kisspeptin neurons that is positively regulated by E2 and, as in most other species, is most likely involved in positive feedback regulation of GnRH neurons. We also provide evidence that the Arc population of kisspeptin neurons may be involved in the E2 induction of the LH surge. Indeed, recently we have documented that kisspeptin neurons in the middle to caudal Arc of late-follicular (high-E2)-phase guinea pigs, express pacemaker currents that allow them to burst fire rhythmically (Qiu et al., 2011). However, future studies aimed at more detailed investigation of the signaling properties of these cells are necessary for a more complete understanding of the differences in subpopulations of kisspeptin neurons and their role in reproductive and other functions.
Acknowledgments
The authors thank Taiping Jia, PhD, for his assistance with the in situ hybridization procedure and Martin J. Kelly, PhD, for his helpful comments on drafts of the manuscript. We thank Anda Cornea, PhD, at the Neuro-science Imaging Center, West Campus Confocal Core, for her expert assistance with the immunoreactive kisspeptin quantification analysis. We are also grateful to Dr. Caraty, UMR 6175 INRA/CNRS/University of Tours, Nouzilly, France, for providing the kisspeptin antiserum.
Grant sponsor: National Institutes of Health; Grant number: NS43330; Grant number: DK68098; Grant number: P30-NS061800.
Abbreviations
- AC
anterior commisure
- Arc
arcuate nucleus
- rArc
rostral arcuate nucleus
- cArc
caudal arcuate nucleus
- AMPO
anterior medial preoptic nucleus
- BNST
bed nuclei stria terminalis
- DBh
diagonal band of Broca horizontal portion
- DBv
diagonal band of Broca vertical portion
- DMH
dorsomedial nucleus hypothalamus
- fx
fornix
- LS
lateral septum
- LV
lateral ventricle
- ME
median eminence
- MPA
medial preoptic area
- MPN
medial preoptic nucleus
- MS
medial septum
- MTT
mammillothalamic tract
- OC
optic chiasm
- OVLT
organum vasculosum lamina terminalis
- PH
posterior hypothalamic nucleus
- PMv
ventral premammillary nucleus
- POA
preoptic area
- PSCH
suprachiasmatic preoptic nucleus
- PV
periventricular
- PVpo
preoptic periventricular nucleus
- PVa
anterior periventricular nucleus hypothalamus
- PVi
intermediate periventricular nucleus hypothalamus
- PVp
posterior periventricular nucleus hypothalamus
- PVH
paraventricular nucleus hypothalamus
- SCN
suprachiasmatic nucleus
- VMH
ventromedial nucleus hypothalamus
- 3V
third ventricle
LITERATURE CITED
- Adachi S, Yamada S, Takatsu Y, Matsui H, Kinoshita M, Takase K, Sugiura H, Ohtaki T, Matsumoto H, Uenoyama Y, Tsukamura H, Inoue K, Maeda K-I. Involvement of anteroventral periventricular metastin/kisspeptin neurons in estrogen positive feedback action on luteinizing hormone release in female rats. J Reprod Dev. 2007;53:367–378. doi: 10.1262/jrd.18146. [DOI] [PubMed] [Google Scholar]
- Bleier R. The hypothalamus of the guinea pig: a cytoarchitectonic atlas. Madison: University of Wisconsin Press; 1983. [Google Scholar]
- Brailoiu GC, Dun SL, Ohsawa M, Yin D, Yang G, Chang JK, Brailoiu E, Dun NJ. Kiss1 expression and metastin-like immunoreactivity in the rat brain. J Comp Neurol. 2005;481:314–329. doi: 10.1002/cne.20350. [DOI] [PubMed] [Google Scholar]
- Caraty A, Locatelli A, Martin GB. Biphasic response in the secretion of gonadotrophin-releasing hormone in ovariectomized ewes injected with oestradiol. J Endocrinol. 1989;123:375–382. doi: 10.1677/joe.0.1230375. [DOI] [PubMed] [Google Scholar]
- Caraty A, Fabre-Nys C, Delaleu B, Locatelli A, Bruneau G, Karsch FJ, Herbison A. Evidence that the mediobasal hypothalamus is the primary site of action of estradiol in inducing the preovulatory gonadotropin releasing hormone surge in the ewe. Endocrinlogy. 1998;139:1752–1760. doi: 10.1210/endo.139.4.5904. [DOI] [PubMed] [Google Scholar]
- Clarke IJ, Smith JT, Caraty A, Goodman RL, Lehman MN. Kisspeptin and seasonality in sheep. Peptides. 2009;30:154–163. doi: 10.1016/j.peptides.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson J, d’Anglemont de Tassigny X, Colledge WH, Caraty A, Herbison AE. Distribution of kisspeptin neurones in the adult female mouse brain. J Endocrinol. 2009;21:673–682. doi: 10.1111/j.1365-2826.2009.01892.x. [DOI] [PubMed] [Google Scholar]
- Condon TP, Dykshoorn-Bosch MA, Kelly MJ. Episodic LH release in the ovariectomized guinea pig: Rapid inhibition by estrogen. Biol Reprod. 1988;38:121–126. doi: 10.1095/biolreprod38.1.121. [DOI] [PubMed] [Google Scholar]
- Czaja JA, Goy RW. Ovarian hormones and food intake in female guinea pigs and rhesus monkeys. Horm Behav. 1975;6:329–349. doi: 10.1016/0018-506x(75)90003-3. [DOI] [PubMed] [Google Scholar]
- d’Anglemont de Tassigny X, Fagg LA, Dixon JPC, Day K, Leitch HG, Hendrick AG, Zahn D, Franceschini I, Caraty A, Carlton MBL, Aparicio SAJR, Colledge WH. Hypogonadotropic hypogonadism in mice lacking a functional Kiss1 gene. Proc Natl Acad Sci U S A. 2007;104:10714–10719. doi: 10.1073/pnas.0704114104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roux N, Genin E, Carel J-C, Matsuda F, Chaussain J-L, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the Kiss1-derived peptide receptor GPR54. Proc Natl Acad Sci U S A. 2003;100:10972–10976. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eghlidi DH, Haley GE, Noriega NC, Kohama SG, Urbanski HF. Influence of age and 17β-estradiol on kisspeptin, neurokinin B, and prodynorphin gene expression in the arcuate-median eminence of female rhesus macaques. Endocrinology. 2010;151:3783–3794. doi: 10.1210/en.2010-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini I, Lomet D, Cateau M, Delsol G, Tillet Y, Caraty A. Kisspeptin immunoreactive cells of the ovine pre-optic area and Arc co-express estrogen receptor alpha. Neurosci Lett. 2006;401:225–230. doi: 10.1016/j.neulet.2006.03.039. [DOI] [PubMed] [Google Scholar]
- Gottsch ML, Clifton DK, Steiner RA. Kisspeptin-GPR54 signaling in the neuroendocrine reproductive axis. Mol Cell Endocrinol. 2006;254–255:91–96. doi: 10.1016/j.mce.2006.04.030. [DOI] [PubMed] [Google Scholar]
- Grove-Strawser D, Sower SA, Ronsheim PM, Connolly JB, Bourn CG, Rubin BS. Guinea pig GnRH: localization and physiological activity reveal that it, not mammalian GnRH, is the major neuroendocrine form in guinea pigs. Endocrinology. 2002;143:1602–1612. doi: 10.1210/endo.143.5.8803. [DOI] [PubMed] [Google Scholar]
- Hoffman GE, Le WW, Franceschini I, Caraty A, Advis JP. Expression of Fos and in Vivo median eminence release of LHRH identifies an active role for preoptic area kisspeptin neurons in synchronized surges of LH and LHRH in the ewe. Neuroendo. 2011;152:214–222. doi: 10.1210/en.2010-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima N, Takumi K, Sawai N, Ozawa H. An immunohistochemical study on the expressional dynamics of kisspeptin neurons relevant to GnRH neurons using a newly developed anti-kisspeptin antibody. J Mol Neurosci. 2011;43:146–154. doi: 10.1007/s12031-010-9433-y. [DOI] [PubMed] [Google Scholar]
- Jamali K, Naylor BR, Kelly MJ, Rønnekleiv OK. Effect of 17β-Estradiol on mRNA expression of large-conductance, voltage dependent, and calcium-sensitive potassium channel α and β subunits in the guinea pig. Endocrine. 2003;20:227–237. doi: 10.1385/ENDO:20:3:227. [DOI] [PubMed] [Google Scholar]
- Kelly MJ, Condon TP, Levine JE, Rønnekleiv OK. Combined electrophysiological, immunocytochemical and peptide release measurements in the hypothalamic slice. Brain Res. 1985;345:264–270. doi: 10.1016/0006-8993(85)91002-9. [DOI] [PubMed] [Google Scholar]
- Kim W, Jessen HM, Auger AP, Terasawa E. Postmenopausal increase in Kiss1, GPR54, and luteinizing hormone releasing hormone (LHRH-1) mRNA in the basal hypothalamus of female rhesus monkeys. Peptides. 2009;30:103–110. doi: 10.1016/j.peptides.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JC, Ronsheim PM, Liu E, Powers L, Slonimski M, Rubin BS. Fos expression in luteinizing hormone-releasing hormone neurons of guinea pigs, with knife cuts separating the preoptic area and the hypothalamus, demonstrating luteinizing hormone surges. Biol Repro. 1998;58:323–329. doi: 10.1095/biolreprod58.2.323. [DOI] [PubMed] [Google Scholar]
- Kotani M, Detheux M, Vandenbogaerde A, Communi D, Vanderwinden J-M, Le Poul E, Brezillon S, Tyldesley R, Suarez-Huerta N, Vandeput F, Blanpain C, Schiffmann SN, Vassart G, Parmentier M. The metastasis suppressor gene Kiss1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J Biol Chem. 2001;276:34631–34636. doi: 10.1074/jbc.M104847200. [DOI] [PubMed] [Google Scholar]
- Kuohung W, Kaiser UB. GPR54 and Kiss1: role in the regulation of puberty and reproduction. Rev Endocr Metab Disord. 2006;7:257–263. doi: 10.1007/s11154-006-9020-2. [DOI] [PubMed] [Google Scholar]
- Liu X, Lee K, Herbison AE. Kisspeptin excites gonadotropin-releasing hormone (GnRH) neurons through a phospholipase C/calcium-dependent pathway regulating multiple ion channels. Endocrinology. 2008;149:4605–4614. doi: 10.1210/en.2008-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma YJ, Kelly MJ, Rønnekleiv OK. Pro-gonadotropin-releasing hormone (ProGnRH) and GnRH content in the preoptic area and the basal hypothalamus of anterior medial preoptic nucleus/suprachiasmatic nucleus-lesioned persistent estrous rats. Endocrinology. 1990;127:2654–2664. doi: 10.1210/endo-127-6-2654. [DOI] [PubMed] [Google Scholar]
- Malyala A, Zhang C, Bryant D, Kelly MJ, Rønnekleiv OK. PI3K signaling effects in hypothalamic neurons mediated by estrogen. J Comp Neurol. 2008;506:895–911. doi: 10.1002/cne.21584. [DOI] [PubMed] [Google Scholar]
- Mikkelsen JD, Simonneaux V. The neuroanatomy of the kisspeptin system in the mammalian brain. Peptides. 2009;30:26–33. doi: 10.1016/j.peptides.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Oakley AE, Clifton DK, Steiner RA. Kisspeptin signaling in the brain. Endocr Rev. 2009;30:713–743. doi: 10.1210/er.2009-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pau K-YF, Gliessman PM, Hess DL, Rønnekleiv OK, Levine JE, Spies HG. Acute administration of estrogen suppresses LH secretion without altering GnRH release in ovariectomized rhesus macaques. Brain Res. 1990;517:229–235. doi: 10.1016/0006-8993(90)91031-b. [DOI] [PubMed] [Google Scholar]
- Petersen SL, Barraclough CA. Suppression of spontaneous LH surges in estrogen-treated ovariectomized rats by microimplants of antiestrogens into the preoptic brain. Brain Res. 1989;484:279–289. doi: 10.1016/0006-8993(89)90371-5. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acid Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielecka-Fortuna J, Chu Z, Moenter SM. Kisspeptin acts directly and indirectly to increase GnRH neuron activity and its effects are modulated by estradiol. Endocrinology. 2008;149:1979–1986. doi: 10.1210/en.2007-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant TM. The role of Kiss1 in the regulation of puberty in higher primates. Eur J Endocrinol. 2006;155:S11–S16. doi: 10.1530/eje.1.02232. [DOI] [PubMed] [Google Scholar]
- Plant TM, Krey LC, Moosy J, McCormack JT, Hess DL, Knobil E. The arcuate nucleus and the control of gonadotropin and prolactin secretion in the female rhesus monkey (Macaca mulatta) Endocrinology. 1978;102:52–62. doi: 10.1210/endo-102-1-52. [DOI] [PubMed] [Google Scholar]
- Qiu J, Fang Y, Bosch MA, Rønnekleiv OK, Kelly MJ. Guinea pig kisspeptin neurons are depolarized by leptin via activation of TRPC channels. Endocrinology. 2011;152:1503–1514. doi: 10.1210/en.2010-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Guerriero KA, Gibbs RB, Plant TM. Structural interactions between kisspeptin and GnRH neurons in the mediobasal hypothalamus of the male rhesus monkey (Macaca mulatta) as revealed by double immunofluorescence and confocal microscopy. Endocrinology. 2008;149:4387–4395. doi: 10.1210/en.2008-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JL, Clifton DK, de la Iglesia HO, Steiner RA, Kauffman AS. Circadian regulation of Kiss1 neurons: implications for timing the preovulatory gonadotropin-releasing hormone/luteninizing hormone surge. Neuroendocrinology. 2009;150:3664–3671. doi: 10.1210/en.2009-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepke TA, Malyala A, Bosch MA, Kelly MJ, Rønnekleiv OK. Estrogen regulation of genes important for K+ channel signaling in the arcuate nucleus. Endocrinology. 2007;148:4937–4951. doi: 10.1210/en.2007-0605. [DOI] [PubMed] [Google Scholar]
- Roepke TA, Bosch MA, Rick EA, Lee B, Wagner EJ, Seidlová-Wuttke D, Wuttke W, Scanlan TS, Rønnekleiv OK, Kelly MJ. Contribution of a membrane estrogen receptor to the estrogenic regulation of body temperature and energy homeostasis. Endocrinology. 2010;151:4926–4937. doi: 10.1210/en.2010-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rometo AM, Krajewski SJ, Voytko ML, Rance NE. Hypertrophy and increased kisspeptin gene expression in the hypothalamic infundibular nucleus of postmenopausal women and ovariectomized monkeys. J Clin Endorinol Metab. 2007;92:2744–2750. doi: 10.1210/jc.2007-0553. [DOI] [PubMed] [Google Scholar]
- Rønnekleiv OK, Resko JA. Ontogeny of gonadotropin-releasing hormone-containing neurons in early fetal development of rhesus macaques. Endocrinology. 1990;126:498–511. doi: 10.1210/endo-126-1-498. [DOI] [PubMed] [Google Scholar]
- Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O’Rahilly S, Carlton MBL, Crowley WF, Aparicio SAJR, Colledge WH. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- Smith JT. Kisspeptin signalling in the brain: steroid regulation in the rodent and ewe. Brain Res Rev. 2008;57:288–298. doi: 10.1016/j.brainresrev.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Smith JT, Cunningham MJ, Rissman EF, Clifton DK, Steiner RA. Regulation of Kiss1 gene expression in the brain of the female mouse. Endocrinology. 2005;146:3686–3692. doi: 10.1210/en.2005-0488. [DOI] [PubMed] [Google Scholar]
- Smith JT, Popa SM, Clifton DK, Hoffman GE, Steiner RA. Kiss1 neurons in the forebrain as central processors for generating the preovulatory luteinizing hormone surge. J Neurosci. 2006;26:6687–6694. doi: 10.1523/JNEUROSCI.1618-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JT, Clay CM, Caraty A, Clarke IJ. Kiss1 messenger ribonucleic acid expression in the hypothalamus of the ewe is regulated by sex steroids and season. Endocrinology. 2007;148:1150–1157. doi: 10.1210/en.2006-1435. [DOI] [PubMed] [Google Scholar]
- Smith JT, Pereira LA, Clarke IJ. Kisspeptin neurons in the ovine arcuate nucleas and preoptic area are involved in the preovulatory luteinizing hormone surge. Neuroendocrinology. 2009;150:5530–5538. doi: 10.1210/en.2009-0712. [DOI] [PubMed] [Google Scholar]
- Smith JT, Shahab M, Pereira A, Pau K-YF, Clarke IJ. Hypothalamic expression of Kiss1 and gonadotropin inhibitory hormone genes during the menstrual cycle of a non-human primate. Biol Repro. 2010;83:568–577. doi: 10.1095/biolreprod.110.085407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JT, Li Q, Yap KS, Shabab M, Roseweir AK, Millar RP, Clarke IJ. Kisspeptin is essential for the full preovulatory LH surge and stimulates GnRH release from the isolated ovine median eminence. Endocrinology. 2011;152:0000. doi: 10.1210/en.2010-1225. [DOI] [PubMed] [Google Scholar]
- Stafford LJ, Xia C, Ma W, Cai Y, Liu M. Identification and characterization of mouse metastasis-suppressor Kiss1 and its G-protein-coupled receptor. Cancer Res. 2002;62:5399–5404. [PubMed] [Google Scholar]
- Tena-Sempere M. Kiss-1 and reproduction: focus on its role in the metabolic regulation of fertility. Neuroendocrinology. 2006;83:275–281. doi: 10.1159/000095549. [DOI] [PubMed] [Google Scholar]
- Terasawa E, Wiegand SJ. Effects of hypothalamic deaf-ferentation on ovulation and estrous cyclicity in the female guinea pig. Neuroendocrinology. 1978;26:229–248. doi: 10.1159/000122830. [DOI] [PubMed] [Google Scholar]
- Terasawa E, Rodriguez JS, Bridson WE, Wiegand SJ. Factors influencing the positive feedback action of estrogen upon luteinizing hormone surge in the ovariectomized guinea pig. Endocrinology. 1979;104:680–686. doi: 10.1210/endo-104-3-680. [DOI] [PubMed] [Google Scholar]
- Terasawa E, Yeoman RR, Schultz NJ. Factors influencing the progesterone-induced luteinizing hormone surge in rhesus monkeys: diurnal influence and time interval after estrogen. Biol Reprod. 1984;31:732–741. doi: 10.1095/biolreprod31.4.732. [DOI] [PubMed] [Google Scholar]
- Tomikawa J, Homma T, Tajima S, Shibata T, Inamoto Y, Takase K, Naoko I, Ohkura S, Uenoyama Y, Maeda K, Tsukamura H. Molecular characterization and estrogen regulation of hypothalamic KISS1 gene in the pig. Biol Reprod. 2010;82:313–319. doi: 10.1095/biolreprod.109.079863. [DOI] [PubMed] [Google Scholar]
- True C, Kirigiti M, Ciofi P, Grove KL, Smith MS. Characterisation of arcuate nucleus kisspeptin/neurokinin B neuronal projections and regulation during lactation in the rat. J Neuroendocrinol. 2010;22:1–13. doi: 10.1111/j.1365-2826.2010.02076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Rønnekleiv OK, Bosch MA, Kelly MJ. Estrogen biphasically modifies hypothalamic GABAergic function concomitantly with negative and positive control of luteinizing hormone release. J Neurosci. 2001;21:2085–2093. doi: 10.1523/JNEUROSCI.21-06-02085.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weick RF. Induction of the luteinizing hormone surge by intrahypothalamic application of estrogen in the rhesus monkey. Biol Reprod. 1981;24:415–422. doi: 10.1095/biolreprod24.2.415. [DOI] [PubMed] [Google Scholar]
- Wiegand SJ, Terasawa E, Bridson WE, Goy RW. Effects of discrete lesions of preoptic and suprachiasmatic structures in the female rat: alterations in the feedback regulation of gonadotropin secretion. Neuroendocrinology. 1980;31:147–157. doi: 10.1159/000123066. [DOI] [PubMed] [Google Scholar]
- Yeo S-H, Herbison AE. Projections of arcuate nucleus and rostral periventricular kisspeptin neurons in the adult female mouse brain. Endocrinology. 2011;152:2387–2399. doi: 10.1210/en.2011-0164. [DOI] [PubMed] [Google Scholar]
- Zhang C, Roepke TA, Kelly MJ, Rønnekleiv OK. Kisspeptin depolarizes gonadotropin-releasing hormone neurons through activation of TRPC-like cationic channels. J Neurosci. 2008;28:4423–4434. doi: 10.1523/JNEUROSCI.5352-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SX, Bosch MA, Rønnekleiv OK. Mu-opioid receptor mRNA expression in identified hypothalamic neurons. J Comp Neurol. 2005;487:332–344. doi: 10.1002/cne.20557. [DOI] [PubMed] [Google Scholar]