Abstract
Massively-parallel cDNA sequencing has opened the way to deep and efficient probing of transcriptomes. Current approaches for transcript reconstruction from such data often rely on aligning reads to a reference genome, and are thus unsuitable for samples with a partial or missing reference genome. Here, we present the Trinity methodology for de novo full-length transcriptome reconstruction, and evaluate it on samples from fission yeast, mouse, and whitefly – an insect whose genome has not yet been sequenced. Trinity fully reconstructs a large fraction of the transcripts present in the data, also reporting alternative splice isoforms and transcripts from recently duplicated genes. In all cases, Trinity performs better than other available de novo transcriptome assembly programs, and its sensitivity is comparable to methods relying on genome alignments. Our approach provides a unified and general solution for transcriptome reconstruction in any sample, especially in the complete absence of a reference genome.
Introduction
Recent advances in massively-parallel cDNA sequencing (RNA-Seq) provide a cost-effective way to obtain large amounts of transcriptome data from many organisms and tissue types1, 2. In principle, such data can allow us to identify all expressed transcripts3, as complete and contiguous mRNA sequence from the transcription start site to the transcription end, for multiple alternatively spliced isoforms.
However, reconstruction of all full-length transcripts from short reads with considerable sequencing error rates poses substantial computational challenges4: (1) some transcripts have low coverage, whereas others are highly expressed; (2) read coverage may be uneven across the transcript’s length, due to sequencing biases; (3) reads with sequencing errors derived from a highly expressed transcript may be more abundant than correct reads from a lowly expressed transcript; (4) transcripts encoded by adjacent loci can overlap and thus can be erroneously fused to form a chimeric transcript; (5) data structures need to accommodate multiple transcripts per locus, due to alternative splicing; and (6) sequences that are repeated in different genes introduce ambiguity. A successful method should address each challenge, be applicable to both complex mammalian genomes and gene-dense microbial genomes, and be able to reconstruct transcripts of variable sizes, expression levels and protein-coding capacity.
There are two alternative computational strategies for transcriptome reconstruction4. (1) ‘Mapping first’ approaches5, such as Scripture3 or Cufflinks2, first align all the reads to a reference (unannotated) genome and then merge sequences with overlapping alignment, spanning splice junctions with reads and paired-ends. (2) ‘Assembly first’ (de novo) methods, such as ABySS1, SOAPdenovo6 or Oases (E. Birney, personal communication), use the reads to assemble transcripts directly, which can subsequently be mapped to a reference genome, if available. ‘Mapping-first’ approaches promise, in principle, maximum sensitivity, but depend on correct read-to-reference alignment, a task that is complicated by splicing, sequencing errors, and the lack or incompleteness of many reference genomes. Conversely, ‘assembly first’ approaches do not require any read-reference alignments, important when the genomic sequence is not available, is gapped, highly fragmented, or substantially altered, as in cancer cells.
Successful ‘mapping first’ methods were developed in the past year4, but substantially less progress was made to date in developing effective ‘assembly first’ approaches. As the number of reads grows, it is increasingly difficult to determine which reads should be joined into contiguous sequence contigs. An elegant computational solution is provided by the de Bruijn graph7, 8, the basis for several whole-genome assembly programs9–11. In this graph, a node is defined by a sequence of a fixed length of k nucleotides (“k-mer”, with k considerably shorter than the read length), and nodes are connected by edges, if they perfectly overlap by k-1 nucleotides, and the sequence data supports this connection. This compact representation allows for enumerating all possible solutions by which linear sequences can be reconstructed given overlaps of k-1. For transcriptome assembly, each path in the graph represents a possible transcript. A scoring scheme applied to the graph structure can rely on the original read sequences and mate-pair information to discard nonsensical solutions (transcripts) and compute all plausible ones.
Three critical tasks remain open to date when applying the scheme of de Bruijn graphs to de novo assembly of RNA-Seq data: (1) efficiently constructing this graph from large amounts (billions of base pairs) of raw data; (2) defining a suitable scoring and enumeration algorithm to recover all plausible splice forms and paralogous transcripts; and (3) providing robustness to the noise stemming from sequencing errors and other artifacts in the data. In particular, sequencing errors would introduce a large number of false nodes, resulting in a massive graph with millions of possible (albeit mostly implausible) paths.
Here, we present Trinity, a novel method for the efficient and robust de novo reconstruction of transcriptomes, consisting of three software modules: Inchworm, Chrysalis, and Butterfly, applied sequentially to process large volumes of RNA-Seq reads. We evaluated Trinity on data from two well-annotated species – one microorganism (fission yeast) and one mammal (mouse) – as well as an insect (the whitefly Bemisia tabaci), whose genome has not yet been sequenced. In each case, Trinity recovers most of the reference (annotated) expressed transcripts as full-length sequences, and resolves alternative isoforms and duplicated genes, performing better than other available transcriptome de novo assembly tools, and comparably to methods relying on genome alignments.
RESULTS
Trinity: a method for de novo transcriptome assembly
Trinity is a modular method and software package, combining three components (Fig. 1): Inchworm, Chrysalis and Butterfly. In contrast to de novo assembly of a genome, where few large connected sequence graphs can represent connectivities among reads across entire chromosomes, in assembling transcriptome data we expect to encounter numerous individual disconnected graphs, each representing the transcriptional complexity at non-overlapping loci. Accordingly, Trinity partitions the sequence data into these many individual graphs, and then processes each graph independently to extract full-length isoforms and tease apart transcripts derived from paralogous genes.
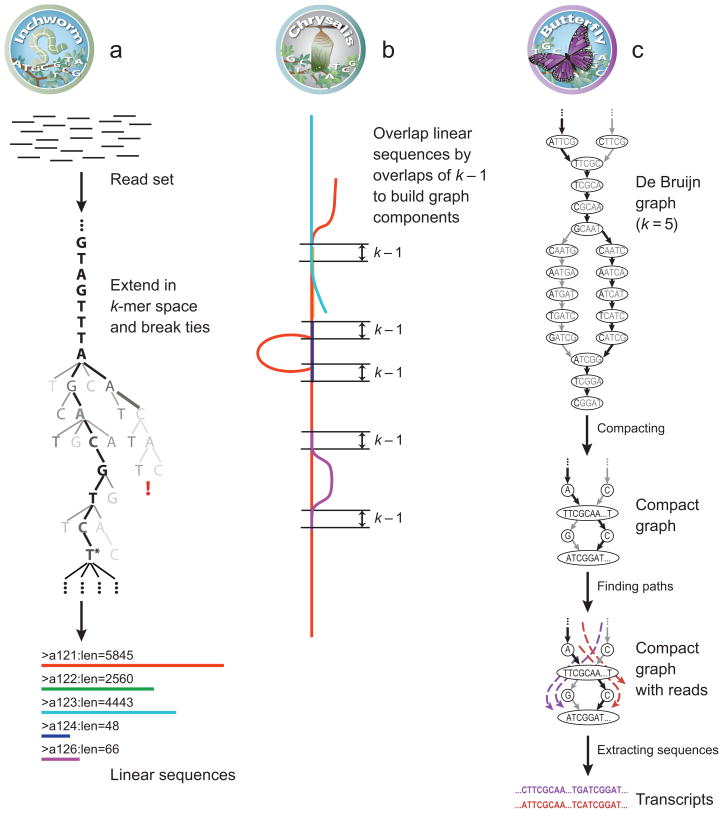
Figure 1. Overview of Trinity.
(a) Inchworm assembles the read data set (short black line, top) by greedily searching for paths in a k-mer graph (middle), resulting in a collection of linear contigs (color lines, bottom), with each k-mer present only once in the contigs. (b) Chrysalis pools contigs if they share at least one k-1-mer and reads span the join, and builds individual de Bruijn graphs from each pool (colored lines). (c) Butterfly takes each de Bruijn graph from Chrysalis (top), and trims spurious edges and compacts linear paths (middle). It then reconciles the graph with reads (dashed colored arrows, bottom) and pairs (not shown), and outputs one linear sequence for each splice form and/or paralogous transcript reflected in the graph (bottom, colored sequences).
In the first step, Inchworm assembles reads into the unique sequences of transcripts. Inchworm (Fig. 1a) uses a greedy k-mer based approach for fast and efficient transcript assembly, recovering only a single (best) representative for a set of alternative variants that share k-mers (due to alternative splicing, gene duplication, or allelic variation). Next, Chrysalis (Fig. 1b) clusters related contigs that correspond to portions of alternatively-spliced transcripts or otherwise unique portions of paralogous genes. Chrysalis then constructs a de Bruijn graph for each cluster of related contigs, each reflecting the complexity of overlaps between variants. Finally, Butterfly (Fig. 1c) analyzes the paths taken by reads and read pairings in the context of the corresponding de Bruijn graph and reports all plausible transcript sequences, resolving alternatively spliced isoforms and transcripts derived from paralogous genes. Below, we describe each of Trinity’s modules.
Inchworm assembles contigs greedily and efficiently
Inchworm efficiently reconstructs linear transcript contigs in six steps (Methods, Fig. 1a). Inchworm (1) constructs a k-mer dictionary from all sequence reads (in practice, k=25); (2) removes likely error-containing k-mers from the k-mer dictionary; (3) selects the most frequent k-mer in the dictionary to seed a contig assembly, excluding both low-complexity and singleton k-mers; (4) extends the seed in each direction by finding the highest occurring k-mer with a k-1 overlap with the current contig terminus and concatenating its terminal base to the growing contig sequence (once a k-mer has been used for extension, it is removed from the dictionary); (5) extends the sequence in either direction until it cannot be extended further, then reporting the linear contig; (6) repeats steps 3–5 starting with the next most highly abundant k-mer, until the entire k-mer dictionary has been exhausted.
Inchworm does not capture the full complexity of the transcriptome; for example, only one alternatively spliced variant can be reported at full length per locus, with partial sequences reported for unique regions of any alternatively spliced transcripts. However, its contigs do maintain the information required by subsequent Trinity components to reconstruct and search the entire graph containing all possible sequences. Indeed, these contigs provide a complete representation of the sequence-overlap-based de Bruijn graph, with each k-mer being unique in the set, and the k-1 sub-sequences implicitly defining the edges in the graph. This approach is much more efficient than computing a full graph from all reads at once, and it quickly provides a meaningful intermediate output of the strong-supported contigs.
Chrysalis builds de Bruijn transcript graphs
Chrysalis clusters minimally-overlapping Inchworm contigs into sets of connected components, and constructs complete de Bruijn graphs for each component (Fig. 1b, Methods). Each component defines a collection of inchworm contigs that are likely to be derived from alternative splice forms or closely-related paralogs. Chrysalis works in three phases (Methods). (1) It recursively groups inchworm contigs into connected components. Contigs are grouped if there is a perfect overlap of k-1 bases between them and if there is a minimal number of reads that span the junction across both contigs with a (k-1)/2 bases match on each side of the (k-1)-mer junction. (2) It builds a de Bruijn graph for each component using a word size of k-1 to represent nodes, and k to define the edges connecting the nodes. It weights each edge of the de Bruijn graph with the number of (k-1)-mers in the original read set that support it. (3) It assigns each read to the component with which it shares the largest number of k-mers, and determines the regions within each read that contribute k-mers to the component.
Butterfly resolves alternatively spliced and paralogous transcripts
Butterfly reconstructs plausible full-length, linear transcripts by reconciling the individual de Bruijn graphs generated by Chrysalis with the original reads and paired-ends. It reconstructs distinct transcripts for splice isoforms and paralogous genes, and resolves ambiguities stemming from errors or from sequences of length greater than k bases that are shared between transcripts.
Butterfly consists of two parts (Fig. 1c, Methods). During graph simplification, Butterfly iterates between (1) merging consecutive nodes in linear paths in the de Bruijn graph to form nodes that represent longer sequences; and (2) pruning edges that represent minor deviations (supported by comparatively few reads), which likely correspond to sequencing errors. Diploid polymorphisms are expected to be more frequent than sequencing errors and will likely be maintained. In plausible path scoring, Butterfly identifies those paths that are supported by actual reads and read pairs, using a dynamic programming procedure that traverses potential paths in the graph while maintaining the reads (and pairs) that support them (Methods). Since reads and sequence fragments (paired reads) are typically much longer than k, they can resolve ambiguities and reduce the combinatorial number of paths to a much smaller number of actual transcripts, enumerated as linear sequences.
RNA-Seq of Schizosaccharomyces pombe
We first generated RNA-Seq data from the fission yeast Schizosaccharomyces pombe. The S. pombe transcriptome12 has relatively substantial splicing for a eukaryotic microorganism, with short introns (mean intron length = 80.6 bp) and dense transcripts (mean intergenic region = 938 bp based on coding genes only). To maximize transcript coverage, we pooled ~154 million pairs of strand-specific13, 14 76 base Illumina read sequences from four biological conditions: mid-log growth, growth as all glucose has been consumed, late stationary phase, and heat shock (Methods, 15).
Sensitivity limit for full-length reconstruction
We next estimated the upper limit for which annotated transcripts can possibly be perfectly reconstructed given a particular dataset of sequences. Any k-mer-based assembly approach is limited to those sequences that are represented by the exact k-mer composition of the RNA-Seq read set. To determine this empirical upper sensitivity limit (“Oracle Set”), we built a k-mer dictionary from all the reads and identified all known reference protein-coding sequences (CDS) that are “reconstructable to full length (FL) given the read set”, as those sequences that can be populated by adjacent and overlapping k-mers across their entire length. Since this Oracle set also contains transcript sequences that are covered by k-mers, but not entire reads, some transcripts will appear reconstructable but are not. Conversely, the Oracle set reflects only annotated known genes and known isoforms, which are likely an underestimate, especially in mammals16. Nevertheless, the Oracle set provides a useful sensitivity benchmark.
In the S. pombe data set, nearly all (91%, 4600/5064) reference CDS sequences exist in the Oracle Set (25-mer dictionary, 154M paired-reads), since almost all encoded transcripts (98%) are expressed in the measured conditions (>= 0.5 Fragments Per transcript Kilobase per Million fragments mapped (FPKM)), consistent with previous studies in yeasts5, 17, 18. When reducing the coverage by random sub-sampling, the size of the Oracle Set is saturated at 50M paired reads (4494/5064, Supplementary Fig. 1 online), which we chose as our subsequent benchmarking set.
Trinity recovered most S. pombe transcripts
From the 50M pairs of reads, Trinity fully reconstructed 86% of annotated transcripts (4338/5064, Supplementary Table 1 online) at full length, including 94% of the stringently-defined oracle transcripts (4218/4494). Of the 276 oracle transcripts not fully reconstructed, 90 (33%) are reconstructed over at least 90% of their length, and 177 (64%) are reconstructed over at least 50% of their length.
Overall, Trinity generated 27,841 linear contigs longer than 100 bases, grouped into 23,232 components (Supplementary Note online). Only 2,454 of the 27,841 Trinity contigs did not align to the genome using GMAP19. Of those, 30% match a Uniref9020 protein (BLASTX E<=1e-10), almost invariably (90%) a Schizosaccharomyces protein, and likely reflect assemblies with error-rich reads.
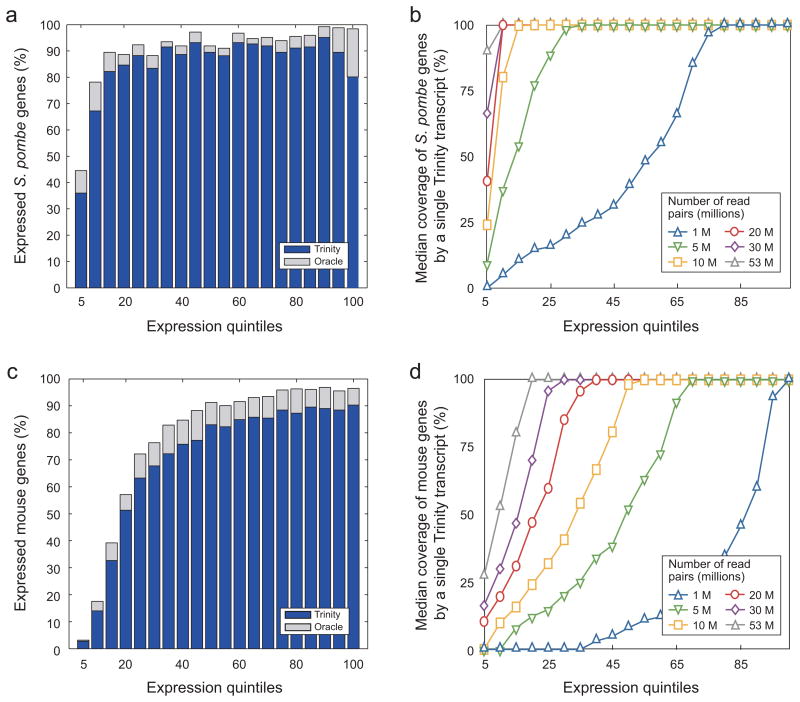
Trinity reconstructs full-length transcripts across a broad range of expression levels and sequencing depths (Fig. 2a). For example, it accurately captured the full-length transcript of 71% of genes from the second quintile (5–10%), and 81–95% full-length coverage of annotated transcripts in the remaining quintiles. Considering both full-length and partial reconstructions, Trinity reconstructed a large fraction of the bases in each transcript (Fig. 2b).
Figure 2. Trinity correctly reconstructs the majority of full-length transcripts in fission yeast and mouse.
(a,c) Shown is the fraction of Oracle genes fully reconstructed in different expression quintiles (5% increments) in fission yeast (50M pairs assembly) (a) and the fraction of Oracle genes with at least one transcript fully reconstructed in different expression quintiles in mouse (53M pairs assembly) (c). Each bar represents a 5% quintile of read coverage for genes expressed. Bar height is the fraction of annotated genes in that quintile and among the Oracle set (grey) or the subset of the Oracle set that are fully reconstructed by Trinity (blue). For example, ~36% of the S. pombe transcripts at the bottom 5% of expression levels are fully reconstructed by Trinity; ~45% of the transcripts in this quintile are in the Oracle set. (b, d) Shown are the median values for coverage by length of reference transcripts by the longest corresponding Trinity-assembled transcript, according to expression quintiles in yeast (b) and mouse (d), depending on the number of read pairs that went into each assembly.
In many cases, Trinity accurately resolved the sequences of closely-related paralogous transcripts. Out of 77 gene families containing 185 paralogs21, Trinity recovered at full length all members of 33 families (68 genes), at least one member from an additional 33 families (46 genes found, 45 genes missing), and missed all 26 genes in the remaining 11 families, often involving lowly expressed genes. Some of the most highly expressed transcripts in S. pombe are derived from paralogous genes with very similar sequences (e.g., those encoding ribosomal proteins21), yet were resolved by Trinity.
Extended UTRs and long anti-sense in S. pombe
Compared to the existing annotation, Trinity extended the 5′UTR of 312 transcripts (median extension = 80bp; average = 176bp), and the 3′UTR of 543 transcripts (median = 72bp; average = 172bp) (Supplementary Fig. 2a,b online). It also found 3,726 previously unannotated 5′ UTRs (median length 183bp, average length = 288bp), and 3,416 3′ UTRs (median length = 272bp, average length = 397bp).
Trinity identified 2319 transcripts at 1235 intergenic loci as novel transcribed sequences (Fig. 3a, Methods) and 612 long antisense transcripts that covered more than 75% of the length of the corresponding sense transcript (Fig. 3b), and were not likely to be derived from extended transcription of a neighboring gene. 113 of the intergenic transcripts and 612 long antisense transcripts were multiexonic. Although both were more lowly expressed on average than annotated protein-coding genes (Supplementary Fig. 3 online), 49 long antisense transcripts (at 35 loci) were at least 5-fold more highly expressed than the corresponding sense coding transcript (e.g., an antisense transcript to the meiotic gene Mug27/Slk1 (SPCC417.06c) was >100-fold more highly expressed, Supplementary Fig. 4 online). This supports a role for antisense transcriptional regulation in meiosis for S. pombe15, 22–24, and is consistent with previous findings in S. cerevisiae25.
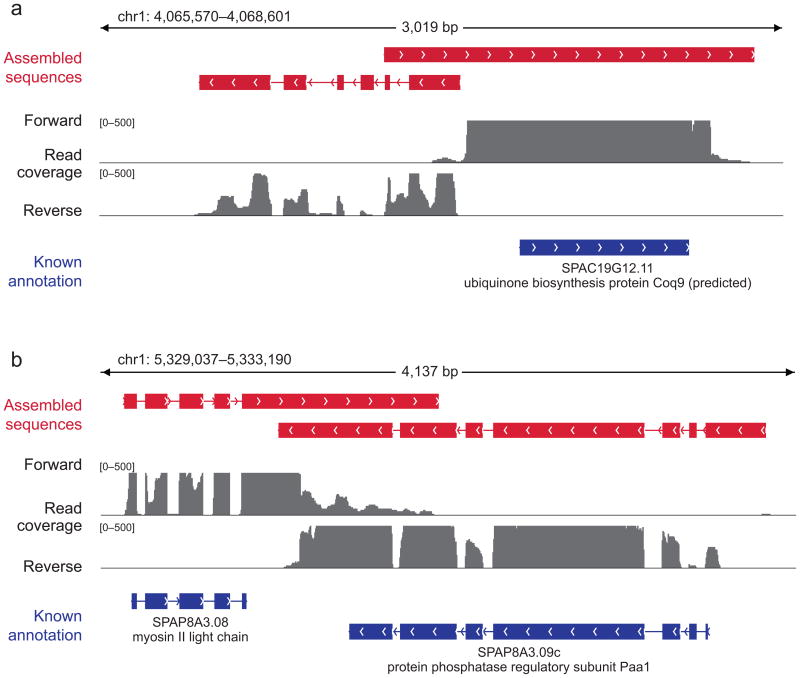
Figure 3. Trinity improves the yeast annotation.
Shown are examples of Trinity assemblies (red) along with the corresponding annotated transcripts (blue) and underlying reads (grey) all aligned to the S. pombe genome (read alignment shown for graphical clarity; no alignments were used to generate the assemblies). (a) Trinity identifies a new multi-exonic transcript (left) and extends the 5′ and 3′ UTRs of the Coq9 gene (right). (b) Trinity extends the UTRs of two convergently transcribed and overlapping genes.
Trinity recovered most expressed annotated mouse transcripts
Compared to yeasts, mammalian transcriptomes exhibit substantially more complex patterns of alternative splicing26. To test Trinity’s ability to identify different isoforms, we sequenced ~52.6 million 76b read pairs from C567BL/6 mouse primary immune dendritic cells (DCs). Unlike in S. pombe, only 54% of known mouse genes (10,724) were identified as expressed (>= 0.5 FPKM), and of those, the Oracle set determined 8,358 to be full-length reconstructable (727 loci have two or more isoforms variable in the CDS, totaling 9,258 transcripts).
Trinity reported 48,497 contigs longer than 350bp, capturing 8,185 transcripts to full-length (Supplementary Table 2, Supplementary Note online), corresponding to 7,749 loci (including 7947 (86%) transcripts at 7573 (91%) loci in the mouse Oracle set). The percentage of transcripts recovered to full-length and the fraction of length captured were high across a broad range of expression levels (Fig. 2c,d).
Trinity resolved splice isoforms and gene paralogs in a manner consistent with the mouse Oracle set. Trinity found 872 full length, alternatively spliced, isoforms from 385 loci (53% of the loci with alternatively spliced variants in the Oracle set), and matched the full-length transcripts for 463 (61.6%) of 752 paralogous transcripts in the Oracle set (> 70% identity between paralogs, Methods, Fig. 4).
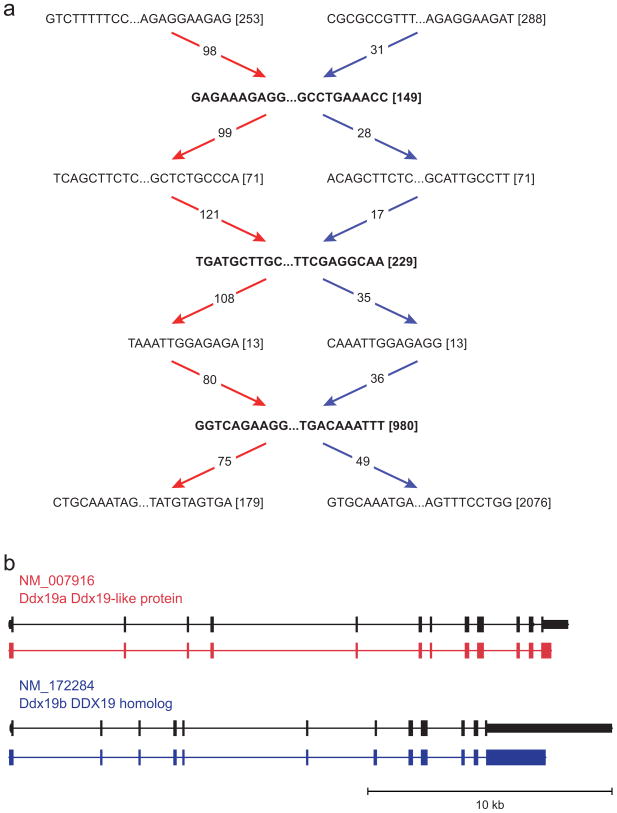
Figure 4. Trinity resolves closely paralogous genes.
(a) Shown is the compacted component graph for two paralogous mouse genes, Ddx19a and Ddx19b (93% identity), highlighting the two paths (red and blue) chosen by Trinity out of the 64 possible paths in this portion alone. (b) Shown are the alignments between the transcripts represented by the red and blue paths in (a) and the paralogous genes Ddx19a and Ddx19b relative to the mouse reference genome (genome alignment shown for graphical clarity only; no alignments were used to generate the assemblies).
Trinity extended the annotated 5′UTR for 5,265 transcripts (5,036 loci, median length = 43, average = 91, Supplementary Fig. 2c online), and included one or more additional 5′UTR exons in 305 cases (Supplementary Fig. 5 online). It extended the 3′UTR in 2918 transcripts (2819 loci, median length = 20, average = 248, Supplementary Fig. 2d online), adding 3′UTR exons in 62 cases (Supplementary Fig. 2b online). UTR length differences were often due to alternative splicing events restricted to the UTR.
High sequence fidelity of reconstructed transcripts
We measured the assembled transcript base error rate by aligning the full-length transcripts to the corresponding reference genome (using BLAT), and capturing mismatches, insertions, and deletions from the highest scoring alignment (Supplementary Table 3 online). In fission yeast, rates of mismatches, insertions, and deletions are each less than 1 in 10,000. In mouse, rates were approximately twice as high, reflecting the lesser transcript fold-coverage. Since the raw read error rate is ~1%, Trinity thus resolved ~99% of sequencing errors.
Comparing Trinity’s performance to other methods
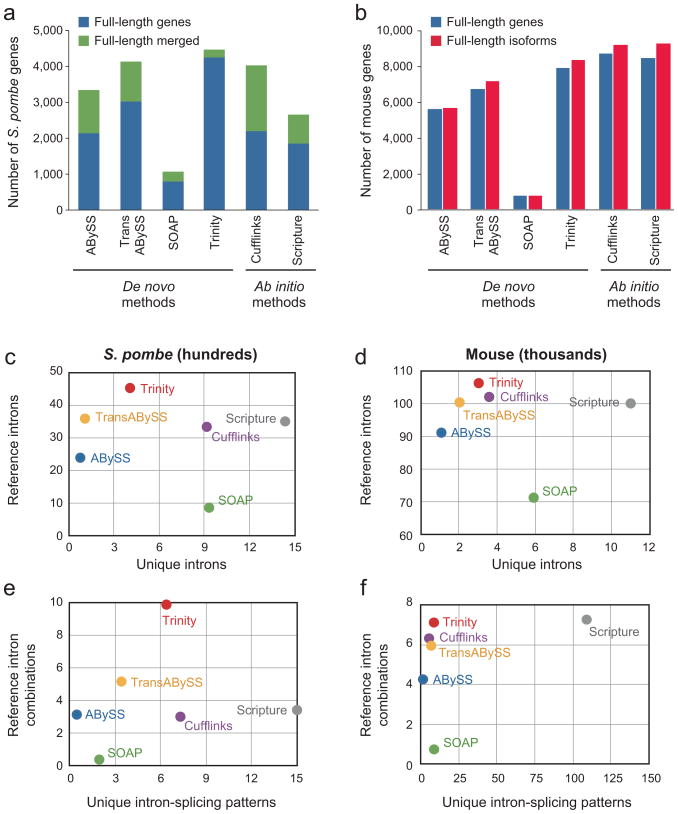
We compared Trinity’s performance to that of other assemblers by several measures. First, we examined the number of reference transcripts reconstructed to full-length by each method (‘sensitivity’). In S. pombe, Trinity out-performed the de novo sequence assemblers, ABySS1, Trans-ABySS27 and SOAPdenovo6, as well as the ‘mapping first’ programs Scripture3 and Cufflinks2 (Fig. 5a). Trinity performed well across a range of 10M to the full 150M input sequence reads, while the alternative methods tended to peak at ~50M pairs or smaller inputs (Supplementary Fig. 6a online). In mouse (RefSeq annotation set, Fig. 5b), Trinity (8185 transcripts, 7749 genes) outperformed the other de novo assembly methods ABySS (5561, 5500), Trans-ABySS (7025, 6598) and SOAPdenovo (761, 760), with the mapping-first programs Cufflinks (9010, 8536) and Scripture (9086, 8293) exhibiting better sensitivity. Furthermore, Trinity and Cufflinks appear best-tuned in their sensitivity across the broadest range of expression levels (Supplementary Fig. 7 online). Unlike Trinity, several of the de novo methods did not perform well in fully reconstructing transcripts within the highest expression quintiles (Supplementary Fig. 7 online).
Figure 5. Comparison of Trinity to other mapping-first and assembly-first methods.
(a,b) Evaluation based on number of full-length annotated transcripts reconstructed by each method in in S. pombe (50M read pair assemblies) (a) and mouse (53M read pair assemblies) (b). Shown is the number of genes reconstructed in full length (blue) or as fusions of two full-length genes (green, yeast only) and the number of full length reconstructed transcript isoforms (red, mouse only) in each of four ‘assembly first’ (de novo) and two ‘mapping first’ approaches. (c,d) Evaluation based on the number of introns defined by the transcripts from each method for S. pombe (c) and mouse (d). Shown is the number of distinct introns consistent with the reference annotation (y axis) versus the number of uniquely predicted introns (x axis), based on mapping to the genome of the transcripts reconstructed by each of Trinity (red), Trans-ABySS (yellow), ABySS (blue), SOAPdenovo (green), Scripture (purple) and Cufflinks (grey). (e,f) Evaluation based on the number of splicing patterns (complete sets of introns in multi-intronic transcripts) defined by the transcripts from each method for S. pombe (e) and mouse (f). Shown are the numbers of distinct splicing patterns (y axis) consistent with the reference annotation versus the number of unique splicing patterns (x axis), for each method (methods are colored as above).
Second, we assessed the accuracy of splice pattern detection by examining individual introns and the combinations of introns (‘splicing patterns’) defined by mapping all the reconstructed transcripts (annotated or not) back to the reference genome (Fig. 5c–f, Methods). We compared the number of annotated reference introns (or splicing patterns) captured by each method, and the number of previously unannotated introns (or extended splicing patterns). Unannotated introns or splice patterns captured by more than one method are less likely to be false positives. In S. pombe, Trinity identified the largest number of reference introns (4,543) (Fig. 5c) and 1,582 unannotated introns, most of which are in putative, unannotated UTRs. 1,174 of these (74%) are also identified by at least one other method and thus are more likely genuine. Trinity also identifies the largest number of annotated splicing patterns in S. pombe (Fig. 5e). The large numbers of falsely-fused S. pombe transcripts reported by the alternative methods contributes to their lack of sensitivity.
In mouse, most methods had comparably high sensitivity for detecting individual annotated introns (Fig. 5d), but varied in detecting complete splicing patterns (Fig. 5f). Scripture identifies the most annotated splicing patterns (7,274), closely followed by Trinity (7,127). However, Scripture reports over 110,000 unique splicing patterns, ~10-fold more than Trinity and all other methods (each less than 10,000 unique patterns), suggesting many false positives in Scripture, and excellent precision in Trinity. Overall, relatively few of the non-annotated splicing patterns predicted by each method are supported by at least one other method (18–25%, except Scripture (2%) and ABySS (66%)).
Finally, we examined the number of distinct contigs that mapped to each reference genomic locus, as well as the coverage (tiers) of reconstructed transcripts per locus. This accounts for multiple reported transcripts that represent the same region of a locus, for example due to alternative splicing, captured allelic variation, or enumerating transcripts with otherwise undetected sequencing errors. In S. pombe, Trinity reports 7,057 transcripts that map to 4,874 genes with an average coverage of 1.37 tiers per gene, similar to the alternative methods except Scripture (4.37 tiers per gene) and trans-ABySS (5.08 tiers per gene). Similarly, in mouse, Trinity performs comparably well to all other methods (31,706 contigs map to 11,334 genes, 2.05 tiers per gene on average), except trans-ABySS (111,000 contigs, 10,685 genes, 5.93 tiers). The large numbers of trans-ABySS transcripts covering similar regions of loci is not reflected in the number of distinct splicing patterns, indicating that multiple similar transcript sequences are being generated at individual loci, rather than many different splice isoforms. ABySS alone, although lacking the higher sensitivity of trans-ABySS, reports a smaller number of contigs (~1 transcript tiers per locus).
De novo assembly of the whitefly transcriptome
In the absence of a sequenced genome, de novo assembly of RNA-Seq is the only viable option to study the transcriptomes of most organisms to date. For example, although the highly diverse class Insecta contains several key model organisms, it is not densely covered by high quality draft genome sequences. In addition, insect transcriptomes exhibit complex alternative splicing patterns28. The whitefly B. tabaci is one such example, for which the genome was not sequenced, and where RNA-Seq samples are genetically polymorphic, since they are derived from a mixture of individuals from an outbred population28.
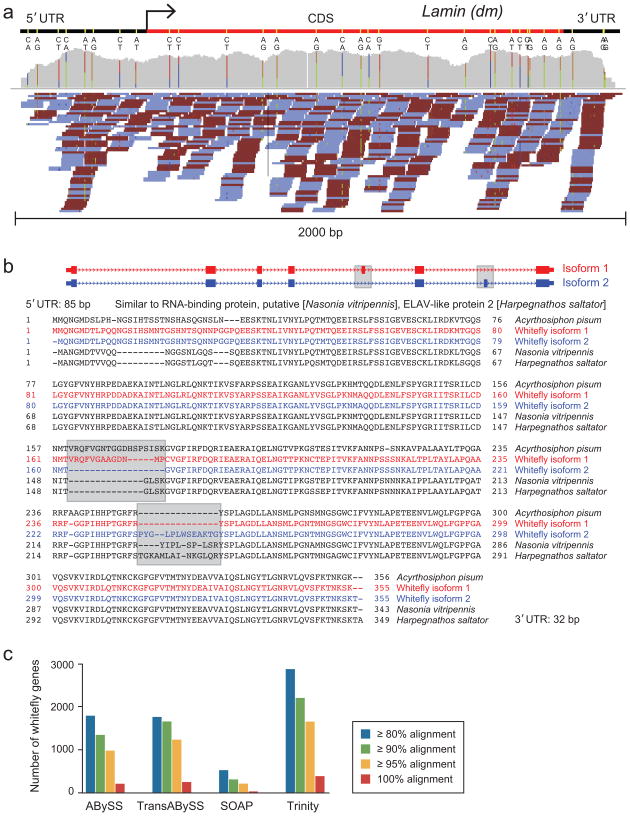
We applied Trinity to a published RNA-Seq dataset from whitefly, consisting of ~21.9 million pairs of 76 base Illumina reads, sequenced using conventional non-strand-specific methods29. Trinity produced 196,000 transcripts, 14,522 longer than 1,000 base pairs, capturing allelic variants (e.g., Fig. 6a). Of those, 4,323 had top blastx matches (E <= 1−10) to 2,880 unique Uniref9020 protein sequences, along at least 80% of the corresponding homologous protein sequence. This number of approximately full-length Trinity-assembled transcripts is substantially higher than achieved by other de novo assemblers (Fig. 6c).
Figure 6. Trinity reconstructs polymorphic transcripts in whitefly.
(a) Allelic variation evident from mapping RNA-Seq reads to a Trinity-reconstructed full-length whitefly transcript. Top: Shown is a single transcript (top, red bar), orthologous to the D. melanogaster Lamin gene, determined by grouping of allelic variant transcripts generated by Trinity. SNPs: yellow bars, Middle: Cummulative read coverage along the transcripts; colored bars: SNPs; bar height: relative proportions of SNP variants. Blue: C, red: T, orange: G, green: A. Bottom: Individual read coverage. (b) Example of two alternatively spliced transcripts resolved even in the absence of a reference genome. Top: Shown are two isoforms of an ELAV-like gene (top) reconstructed by Trinity (grey boxes, alternative exons). Exon structure is determined for visualization by the D. melanogaster ortholog. Bottom: shown is the protein sequence alignment of the two whitefly isoforms to orthologous proteins from other insects, confirming the splice variants (grey boxes). (c) Comparison of performance in de novo assembly of the whitefly transcriptome. For each of the methods, shown is the number of unique top-matching (blastx) uniref9020 protein sequences aligned across the corresponding minimum percent protein length value at >= 80% (blue), >= 90% (green), >= 95% (orange) and 100% (red).
To assess the extent to which alternative splice forms are captured by the Trinity assembly, we self-aligned all contigs within individual graph components, requiring at least one alternative internal exon of minimum length 21 bp and a multiple of 3. By this definition, 325 components contain at least two different isoforms. One such example (Fig. 6b), is a highly conserved ortholog to an ELAV-like protein in the ant Harpegnathos saltator, which is present as two different isoforms involving inclusion of two different alternatively-spliced exons.
DISCUSSION
We presented Trinity, a method for de novo reconstruction of the majority of full-length transcripts in a sample from RNA-Seq reads directly, across a broad range of expression levels. Trinity resolved ~99% of the initial sequencing errors, determined splice isoforms, distinguished transcripts from recently duplicated, and identified allelic variants.
Trinity’s transcripts substantially enhance our annotation of the mouse and fission yeast transcriptomes. In yeast, we identified a large number of UTR extensions, antisense transcripts, and novel intergenic transcripts. In mouse, we identified many novel transcripts and novel exons for reference transcripts. Trinity reconstructed many full-length transcripts from the whitefly transcriptome in the presence of substantial polymorphisms, as well as alternatively spliced variants.
Paired-reads are important to increase the distance at which Trinity can resolve ambiguities. For example, a component representing two paralogous genes (e.g., Fig. 4) or alternative isoforms can have an enormous number of possible paths, but often only very few of them represent real transcripts. Read pairs, representing longer fragments allow us to resolve differences (e.g., two pairs of SNPs, or two different exons) that occur at that distance or below. At longer distances, there is no physical unit to support alternative paths, although similarity in expression levels could be used in the future, as well as longer reads and fragments from improved high-throughput sequencing technologies.
Evaluating the performance of transcript assemblers introduces several challenges, mostly since many transcripts, especially alternative isoforms, are not thoroughly defined as part of genome annotations. To address these challenges we used several complementary benchmarks. Our Oracle Set allowed us to assess sensitivity, by defining a ‘gold standard’ of expressed annotated transcripts present at full length. To assess our ability to reconstruct other reference transcripts, we considered the number of reference loci to which reconstructed transcripts map, and the coverage (tiers) of reconstructed transcripts per locus. Finally, we assessed precision by considering all the reconstructed transcripts and the number of ‘correct’ intron boundaries and splice patterns. Each measure represents a useful benchmark, and showed that Trinity performs better than other de novo methods and comparably well to mapping-first methods depending on the organism.
Trinity is important for both genome annotation and the study of non-model organisms. For example, all but two vertebrate genomes are only available as unfinished “drafts”, containing sequence gaps, scaffolds that cannot be anchored to chromosomes, and assembly errors30, each negatively impacting genome annotation and read mapping. We expect that new genomes, assembled from next-generation high-throughput sequencing data, will be even more fragmented. Thus, high-quality de novo transcriptome reconstruction, as implemented in Trinity, featuring low base error rates and the ability to capture multiple isoforms, will prove crucial to maintain acceptable levels of accuracy when characterizing genes. Finally, genomic sequences are available for only a tiny fraction of the enormous variety of organisms. Trinity provides an effective starting point to examine the transcriptomes of such species.
Trinity and its open source code are publicly available at http://TrinityRNASeq.sourceforge.net
Materials and Methods
Inchworm
Inchworm decomposes each sequence read into overlapping k-mers (default k = 25). Each k-mer is stored in a hash table as a key/value pair, where the key is the k-mer sequence and the value is the abundance of that k-mer in the input data set. The k-mer key is stored as a 64-bit unsigned integer with 2-bit nucleotide encoding. Likely sequencing error-containing k-mers are identified by examining k-mers that have identical k-1 prefixes, differing only at their terminal nucleotide, and removing those k-mers that are less than 5% abundant as compared to the most highly abundant k-mer of the group. After processing the entire read set into a set of k-mers and pruning the likely error k-mers, the most frequently occurring k-mer is identified as a seed k-mer for reconstruction of draft transcript contigs. The information content of the seed k-mer is computed as Shannon’s Entropy31, and only k-mers having entropy H >= 1.5, occurring at least twice in the complete set of input reads, and not palindromic, are allowed as seed k-mers. The seed k-mer is extended at both ends in a coverage-guided manner, first from 5′ to 3′, followed by extension from 3′ to 5′. Seed selection by Inchworm was largely inspired by similar methods implemented in the RepeatScout algorithm32. Extension from the seed is performed greedily based on the frequencies of candidate overlapping k-mers, with the single most abundant k-mer with (k-1) overlap chosen to provide a single base extension. In the case of tied extensions, paths are recursively explored to identify the extension yielding the cumulatively maximal coverage. Extension continues until no k-mer exists in the data set to provide an extension. The sequence yielded from the bidirectional seed k-mer extension is reported as a draft transcript contig, and the set of overlapping k-mers comprising the contig are removed from the hash table. The entire cycle of seed selection and bidirectional k-mer extension continues until all k-mers in the hash table have been exhausted.
In strand-specific mode (default), k-mers are derived from only the sense strand of the RNA-Seq read. Double-stranded mode, used with non-strand-specific RNA-Seq data involves several modifications: both the sense and the reverse-complemented read sequence are parsed into overlapping k-mers; during Inchworm contig extension, a k-mer chosen to extend a given path has the reverse-complemented k-mer sequence disabled for further k-mer extensions; and when an Inchworm contig is reported at the end of one iteration of contig assembly, both the sense and reverse-complemented k-mers are removed from the k-mer dictionary.
Only Inchworm contigs with an average k-mer coverage of 2 and length at least 48 (2*(k-1), k=25), the minimal contig length required to capture variation anchored by (k-1) at each terminus, are used by Chrysalis, as described below.
Chrysalis
To convert the linear contigs into a proper de Bruijn graph, Chrysalis first builds a k-1-mer lookup table and recursively pools contigs that share sequences (excluding low-complexity sequence, as above in Inchworm) into components, given that there are reads that span across a potential junction (the “welds”) and extend perfect matches by (k-1)/2 bases on each side. The number of welds must exceed 0.04 times the average k-1-mer coverage of each contig, as computed by inchworm. In addition, the k-1-mer coverage of one contig cannot exceed the coverage of the other by a factor of 100. Next, Chrysalis processes each component individually and computes a de Bruijn graph from the linear inchworm contigs. The reads are then mapped to components by selecting the component that shares the most k-1-mers with the read, with a single k-1 mer being sufficient for assignment. Chrysalis also counts all k-mers and stores them as “edge weight” to indicate their support in the read set. Components with fewer than 276 nodes are discarded by default.
Butterfly
The input to butterfly is a de Bruijn graph component as built by Chrysalis. First, Butterfly trims edges in the de Bruijn graph. It uses two criteria. (1) We reasoned that if there is a node with several outgoing edges, such that one of them has a much smaller read support than the total outgoing reads (less than 5%), then it probably represents a sequencing error or a very lowly expressed variant (Supplementary Fig. 8a online). (2) If the outgoing edge has less than 2% support from the total incoming reads, then it is more likely a spurious transcript extension (Supplementary Fig. 8b online). Outgoing or incoming edges that fail according to one of these criteria are removed (both these numbers are parameters to the program, and can be changed for specific requirements).
Second, Butterfly transforms the modified graph into a weighted sequence graph, where each node is a sequence, rather than an individual k-mer providing a single base path extension as in the de Bruijn graph. In this step, Butterfly generates a compact graph – the set of paths in the compacted graph is identical to that of the original de Bruijn graph. As a result, linear paths will be compacted into a single node, and polymorphisms will be minimized. The weight on each edge of the modified graph corresponds to the number of reads supporting the edge in the original de Bruijn graph. For each compound node, we compute the average coverage, which corresponds to the weights of the original edges that made up the sequence divided by the length of the node.
We then repeat the trimming step, except that when examining compound nodes of length > 1, we also use the node coverage as a measure of opposite flow in the second criterion. These two steps (trimming and graph compaction) are reiterated until convergence. The resulting graph represents possible transcripts as paths through the graph.
Finally, Butterfly uses read sequences, read-pairings and Chrysalis’ read mappings to the graph to select the paths that are best supported by read sequences. The goal is to look for paths with physical evidence for contiguity, by either reads or read pairings. To do so, we first represent all the reads that contributed to the de Bruijn graph by the list of the nodes that they traverse. We then use a dynamic programming algorithm for finding supported path prefixes. The procedure is initialized with source nodes in the graphs (one without incoming edges), and at each step one path prefix is extended by an additional node.
When extending a path prefix that ends at node n, we consider all outgoing edges from n, and evaluate the support for the extension. By construction, each edge in the graph is supported by reads. We however, further require that the last L nucleotides of the path be supported by reads. We define a path as L-supported at coverage c if at each extension of this path, we have at least c reads supporting the L nucleotide suffix of this path (Supplementary Fig. 8c online). A read supports a path fragment either if it contains that fragment as a subsequence, or in the case of paired-reads, if the fragment lies on all paths from nodes that correspond to the first sequence mate to the second sequence mate. In addition, to avoid combinatorial explosion due to small variations (most likely caused by sequence errors), once we extend a path prefix, we examine other paths ending at the same node, and merge the new path with previous path prefix ending at the same node if they are more than 95% identical.
In the results here we used L = 250 and c = 2. The requirement for 250-supported paths emerges from the expected insert size of our library, since we do not expect to have support for a longer suffix if our read pairs (derived from a single fragment) do not span that far. We note that the resolution of ambiguities, which includes alternative splicing and allelic variation, is limited to the insert size of the read pairs, or the read lengths for unpaired data. Although this program can be in theory exponential in size, in practice, its cost is defined by the number of supported paths.
Yeast and mouse cells growth conditions
We used the S. pombe strain SPY73 975h+ and dendritic cells isolated from C57BL/6J mice. Details of cell isolation and growth conditions are in the Supplementary Methods online.
RNA isolation for yeast samples
Total yeast RNA was isolated using Qiagen RNeasy kit following manufacturers’ protocol for mechanical lysis using 0.5 mm zirconia/silica beads (Biospec). PolyA+ RNA was isolated from total RNA using Poly(A) purist kit (Ambion) or Dynabeads mRNA purification kit (Invitrogen). Total RNA and polyA+ RNA were treated with Turbo DNA-free (Ambion) as described. The integrity of the RNA was confirmed using the Agilent 2100 Bioanalyzer and quantified using RNA Quant-It assay for the Qubit® Fluorometer (Invitrogen).
RNA preparation for mouse RNA
Dendritic cells where lysed using QIAzol reagent and total RNA was extracted the miRNeasy kit’s procedure (Qiagen), sample quality was controlled on a 2100 Bioanalyzer (Agilent).
RNA-Seq library preparation
For the mouse dendritic cell sample, we created a dUTP second strand library starting from 200 ng of Turbo DNase treated and poly(A)+ RNA using a previously described method14 except that we fragmented RNA in 1x fragmentation buffer (Affymetrix) at 80°C for 4 min, purified and concentrated it to 6 μl after ethanol precipitation. For the S. pombe samples, we prepared dUTP second strand libraries similarly, with the following additional modifications. We added an index (8 base barcode) to each library to enable pooling of these libraries (S. Fisher et al. manuscript in preparation). In addition, the adaptor ligation step was done with 1.2 μl of index adaptor mix and 4000 cohesive end units of T4 DNA Ligase (New England Biolabs) overnight at 16°C in a final volume of 20 μl. Finally, we generated libraries with an insert size ranging from 225 to 425 bp.
RNA-Seq library sequencing
We sequenced all the cDNA libraries with an Illumina Genome Analyzer IIx. We pooled the four S. pombe libraries together with four other indexed libraries and sequenced them using eight lanes of 76 nucleotide paired reads. We sequenced the mouse library using two lanes of 76 nucleotide paired reads.
Defining empirical limits of full-length transcript reconstruction
Inchworm was used to construct a k-mer dictionary based on the input reads as described above. Reference CDS sequences were examined by searching for each overlapping k-mer sequence in the dictionary. Reference CDS sequences lacking at least one k-mer in the Inchworm Kmer graph were classified as inaccessible for full-length reconstruction via the k-mer graph method. Those reference sequences fully represented within the k-mer dictionary were included in the oracle set.
Finding paralogous genes in mouse
To determine paralogous transcripts, we aligned all isoforms of all genes present in the Oracle set against each other, using the alignment program Satsuma33 We required alignments to be longer than half of the shorter of both sequences and at sequence identity of 70% and up. If at least one pair of transcripts from two genes met the criteria, we called both genes paralogous.
Short read spliced alignments and transcript reconstructions using Cufflinks and Scripture
The S. pombe genome was obtained from the Sanger Institute (http://www.sanger.ac.uk/Projects/S_pombe/download.shtml). The mouse genome version 9 was obtained from the UCSC mouse genome browser gateway (http://genome.ucsc.edu/cgi-bin/hgGateway?db=mm9). Left and right fragment reads were separately aligned to the genomes using TopHat (version 1.1.4)34 with mouse RNA-Seq reads, and BLAT with S. pombe RNA-Seq reads; the BLAT short read alignment pipeline is provided at http://inchworm.sourceforge.net/blat_short_read_alignment.html. We found BLAT to provide more accurate short read alignment with S. pombe, with TopHat lacking sensitive detection of the very short introns in S. pombe. In addition, both Scripture and Cufflinks demonstrated better performance using the BLAT alignments for S. pombe as compared to the TopHat alignments (Supplementary Fig. 9a online). Conversely, performance of Scripture and Cufflinks using TopHat alignments in mouse exceeded that using BLAT alignments (Supplementary Fig. 9b online). Hence, for evaluation purposes, we leveraged BLAT short read spliced alignments in S. pombe and TopHat alignments in mouse.
BLAT alignments of short reads to the S. pombe genome were performed using the pipeline described above with the following settings: maximum intron length set to 500 bases, maximum distance between read pairs of 500, and only the single best alignment was reported per read. TopHat alignments to the mouse genome were performed using the following parameters: minimum intron length of 50 bases, maximum intron of 100kb, and mate inner distance set to 300 bases. Transcribed strand information was assigned to the individual reads based on knowledge of the fragment type (left or right) and the aligned strand of the genome. Both Cufflinks (version 0.9.3)2 and Scripture3 (version VPaperR3, obtained from Scripture author Manuel Garber) were executed on these alignments.
Evaluation of published de novo methods
Illumina reads were de novo assembled using ABySS1 (version 1.2.1), SOAPdenovo6 (version 1.04) or Trans-ABySS27. Command-line parameters used with ABySS were “abyss-pe k=25 E=0 n=10 in= ‘left.fa right.fa’ ”, employing a K-mer length of 25. Likewise, a 25-mer length was employed with SOAPdenovo along with other default parameters. Trans-ABySS27 was run on Mouse and S. pombe using a set of k-mers including 26, 31, 36, 41, and 46 followed by merging the results by running the 1st stage of the trans-ABySS analysis pipeline. In the case of Whitefly, all k-mers from 26 through 46 were used so as to maximize sensitivity given the smaller input number of reads.
Comparisons to reference transcripts
Current gene annotations for S. pombe were downloaded as file ‘pombe_290110.gff’ from GeneDB (http://old.genedb.org/genedb/pombe/). RefSeq transcript gene annotations were downloaded for mouse at the UCSC mouse genome browser gateway (http://genome.ucsc.edu/cgi-bin/hgGateway?db=mm9) in BED format. Protein coding nucleotide sequences were extracted from the genome sequences based on the gene annotations using custom PERL scripts. The mouse reference coding sequences were further distilled to remove entirely identical sequences corresponding to isoforms encoding identical proteins and paralogous sequences: the original 19,947 genes encoding 23,881 transcripts were reduced to 19,857 genes encoding 22,717 on-identical coding transcripts.
Reconstructed transcript sequences (via de novo assembly, Scripture, or Cufflinks) were mapped to the reference coding sequences using BLAT35. Full-length reference annotation mappings were defined as having at least 95% sequence identity covering the entire reference coding sequence and containing at most 5% insertions or deletions (cumulative gap content). In evaluating methods that leverage the strand-specific data (Trinity and Cufflinks), proper sense-strand mapping of sequences was required. Transcripts reconstructed by the alternative methods (Scripture, ABySS, and SOAPdenovo) were allowed to map to either strand. Fusion transcripts were identified as individual reconstructed transcripts that mapped as full-length to multiple reference coding sequences and lacked overlap among the matching regions within the reconstructed transcript. One-to-one mappings were required between reconstructed transcripts and reference transcripts, including alternatively spliced isoforms, with the exception of fusion transcripts.
Analysis of alignment-inferred introns and splicing patterns from reconstructed transcripts
Reconstructed transcripts were mapped to genome sequences using GMAP, reporting only the single top scoring alignment per sequence. Individual introns and complete splicing patterns were extracted from each of the alignments and compared to reference annotations using custom PERL scripts. Unique introns (missing from the reference annotations) were required to contain consensus dinucleotide splice sites (GT or GC donors and AG acceptors).
Locus coverage (tiering) by reconstructed transcripts
The BLAT alignments between reference coding sequences (loci) and reconstructed transcripts described above were organized into locus-level coverage tiers as follows. Given a set of different reconstructed transcripts that have a best match to a reference sequence, the first match is selected and applied to that reference contig at the first coverage tier. The remaining matches are then examined for placement in the first tier. If a subsequent reference-matching region in common between two matches exceeds 30% of the shorter match length, then this subsequent match is propagated to the next highest tier lacking such restrictive match overlap. Tier placement continues until all matches are placed. The maximal tier level defines the locus-level coverage for that reference sequence and can be at most equal to the number of reconstructed transcripts mapped to that locus. Strand-specific transcript reconstructions were tiered in a strand-specific manner (as in the case of Trinity and Cufflinks). In the case of a highly fragmented transcriptome assembly, it is possible for many reconstructed transcripts to populate the first tier yielding a coverage of 1. In the case of alternatively spliced isoforms or redundant transcript generation at a given locus, the coverage value will exceed 1.
Running Trinity on data sets varying read depth
We randomly sub-sampled pairs in the mouse data set to generate such subsets. Inchworm and Chrysalis were run on a server with 256GB of RAM, Butterfly on a server (LSF) farm in parallel. Wall-clock runtimes are: ~17 hours (10M pairs set), ~36 hours (30M pairs set), and ~60 hours (full 50 M pairs set). All experiments were performed with Trinity using parameters: minimum contig length of 100 bases and average fragment length of 300 bases.
Computing gene expression values from aligned RNA-Seq reads
The aligned reads (by TopHat in the case of mouse leveraging the full 52.6M read pairs, and by BLAT in the case of S. pombe leveraging the 50M read pairs) were used for computing gene (and other feature) expression values. The number of fragments mapped to segments (exons) of a genome-mapped feature were tallied based on overlap of the segment’s coordinates by either read from a sequenced fragment, counting fragments as opposed to counting individual reads. Expression was computed as the normalized value of Fragments per Kilobase of feature sequence Per Million fragments mapped or FPKM2. Calculations were performed using custom PERL scripts. Genes were defined as ‘expressed’ if observed to have expression values of at least 0.5 FPKM, and these genes were divided into expression quintiles at 5% intervals for purposes of analysis.
Supplementary Material
Acknowledgments
We thank Leslie Gaffney for help with figure preparation, Jim Bochicchio for project management, the Broad Sequencing Platform for all sequencing work, Alexie Papanicolaou and Michael Ott for Inchworm software testing and code enhancements, and Filipe Ribeiro for helpful discussions regarding error-pruning. The work was supported in part by a grant from the National Human Genome Research Institute (NIH 1 U54 HG03067, Lander), HHMI, an NIH PIONEER award, a BWF CASI (AR), the US-Israel Binational Science Foundation (NF and AR), and funds from the National Institute of Allergy and Infectious Diseases under Contract No. HHSN27220090018C. MY was supported by a Clore Fellowship. KLT is an EYRYI award recipient. AR is a researcher of the Merkin Foundation for Stem Cell Research at the Broad Institute.
Footnotes
Author Contributions. MGG, MY, BJH, KLT, NF and AR conceived and designed the study. BJH, MGG and MY developed the Inchworm, Chrysalis, and Butterfly components, respectively. NR, FDP, BB, CN, KLT contributed to the study’s conception and execution. JZL, DAT, XA, LF, RR, IA, NH, AR and AG designed and performed all experiments. QZ, ZC and EM contributed computational analyses. MGG, BJH and MY designed, implemented, and evaluated all methods. AR, NF, MGG, BJH and MY wrote the manuscript, with input from all authors.
The authors declare no competing financial interest.
Data Availability. Data is available in the gene expression omnibus or SRA (Fission yeast data is SRP005611 15, mouse is GSEXXX).
References
- 1.Birol I, et al. De novo transcriptome assembly with ABySS. Bioinformatics. 2009;25:2872–2877. doi: 10.1093/bioinformatics/btp367. [DOI] [PubMed] [Google Scholar]
- 2.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haas BJ, Zody MC. Advancing RNA-Seq analysis. Nat Biotechnol. 2010;28:421–423. doi: 10.1038/nbt0510-421. [DOI] [PubMed] [Google Scholar]
- 5.Yassour M, et al. Ab initio construction of a eukaryotic transcriptome by massively parallel mRNA sequencing. Proc Natl Acad Sci USA. 2009;106:3264–3269. doi: 10.1073/pnas.0812841106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li R, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 7.De Bruijn NG. A Combinatorical Problem. Koninklijke Nederlandse Akademie v Wetenschappen. 1946;46:758–764. [Google Scholar]
- 8.Good IJ. Normal recurring decimals. Journal of the London Mathematical Society. 1946;21:167–169. [Google Scholar]
- 9.Pevzner PA, Tang H, Waterman MS. An Eulerian path approach to DNA fragment assembly. Proc Natl Acad Sci USA. 2001;98:9748–9753. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler J, et al. ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res. 2008;18:810–820. doi: 10.1101/gr.7337908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hertz-Fowler C, et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levin JZ, et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkhomchuk D, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhind N, et al. Comparative Functional Genomics of the Fission Yeasts. Science. 2011 doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 18.Xu Z, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 20.Wu CH, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- 22.Molnar M, et al. Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics. 2001;157:519–532. doi: 10.1093/genetics/157.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura T, Kishida M, Shimoda C. The Schizosaccharomyces pombe spo6+ gene encoding a nuclear protein with sequence similarity to budding yeast Dbf4 is required for meiotic second division and sporulation. Genes Cells. 2000;5:463–479. doi: 10.1046/j.1365-2443.2000.00343.x. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe T, et al. Comprehensive isolation of meiosis-specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res. 2001;29:2327–2337. doi: 10.1093/nar/29.11.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yassour M, et al. Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol. 2010;11:R87. doi: 10.1186/gb-2010-11-8-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matlin AJ, Clark F, Smith CWJ. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 27.Robertson G, et al. De novo assembly and analysis of RNA-seq data. Nat Methods. 2010;7:909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- 28.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 29.Wang XW, et al. De novo characterization of a whitefly transcriptome and analysis of its gene expression during development. BMC Genomics. 2010;11:400. doi: 10.1186/1471-2164-11-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salzberg SL, Yorke JA. Beware of mis-assembled genomes. Bioinformatics. 2005;21:4320–4321. doi: 10.1093/bioinformatics/bti769. [DOI] [PubMed] [Google Scholar]
- 31.Shannon CE. Prediction and entropy of printed English. The Bell System Technical Journal. 1951;30:50–64. [Google Scholar]
- 32.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21 (Suppl 1):i351–358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 33.Grabherr MG, et al. Genome-wide synteny through highly sensitive sequence alignment: Satsuma. Bioinformatics. 2010;26:1145–1151. doi: 10.1093/bioinformatics/btq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kent WJ. BLAT--the BLAST-like alignment tool. Genome research. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.