Abstract
Study Objectives:
To determine the association between common genetic variation in the clock gene pathway and objectively measured acti-graphic sleep and activity rhythm traits.
Design:
Genetic association study in two population-based cohorts of elderly participants: the Study of Osteoporotic Fractures (SOF) and the Osteoporotic Fractures in Men (MrOS) study.
Setting:
Population-based.
Participants:
SOF participants (n = 1,407, 100% female, mean age 84 years) and MrOS participants (n = 2,527, 100% male, mean age 77 years) with actigraphy and genotype data.
Interventions:
N/A.
Measurements and Results:
Common genetic variation in 30 candidate genes was captured using 529 single nucleotide polymorphisms (SNPs). Sleep and activity rhythm traits were objectively measured using wrist actigraphy. In a region of high linkage disequilibrium on chromosome 12p13 containing the candidate gene GNB3, the rs1047776 A allele and the rs2238114 C allele were significantly associated with higher wake after sleep onset (meta-analysis: rs1047776 PADD = 2 × 10-5, rs2238114 PADD = 5 × 10-5) and lower LRRC23 gene expression (rs1047776: ρ = -0.22, P = 0.02; rs2238114: ρ = -0.50, P = 5 × 10-8). In MrOS participants, SNPs in ARNTL and NPAS2, genes coding for binding partners, were associated with later sleep and wake onset time (sleep onset time: ARNTL rs3816358 P2DF = 1 × 10-4, NPAS2 rs3768984 P2DF = 5 × 10-5; wake onset time: rs3816358 P2DF = 3 × 10-3, rs3768984 P2DF = 2 × 10-4) and the SNP interaction was significant (sleep onset time PINT = 0.003, wake onset time PINT = 0.001). A SNP association in the CLOCK gene replicated in the MrOS cohort, and rs3768984 was associated with sleep duration in a previously reported study. Cluster analysis identified four clusters of genetic associations.
Conclusions:
These findings support a role for common genetic variation in clock genes in the regulation of inter-related sleep traits in the elderly.
Citation:
Evans DS; Parimi N; Nievergelt CM; Blackwell T; Redline S; Ancoli-Israel S; Orwoll ES; Cummings SR; Stone KL; Tranah GJ. Common genetic variants in ARNTL and NPAS2 and at chromosome 12p13 are associated with objectively measured sleep traits in the elderly. SLEEP 2013;36(3):431-446.
Keywords: Genetic, aging, circadian, actigraphy, SNP
INTRODUCTION
Circadian clocks influence many aspects of physiology and behavior such as body temperature, release of hormones, and sleep-wake cycles. Sleep consolidation and the timing of sleep-wake cycles are regulated by the complex interplay between a circadian process and a homeostatic process.1 The central circa-dian pacemaker located in the suprachiasmatic nucleus (SCN) of the hypothalamus entrains peripheral clocks in extra-SCN brain regions to regulate sleep-wake cycles.2
Aging is accompanied by multiple changes in sleep quality and sleep timing.3 In more than 9,000 participants 65 yr and older, 43% reported difficulty initiating or maintaining sleep.4 Cross-sectional analysis has demonstrated that older age is associated with lower sleep efficiency (SE) and lower percentage of slow wave sleep.5–9 Longitudinal analysis also points to an age-related decline of SE and slow wave sleep. Over a 3-yr follow-up, SE and percentage of slow wave sleep declined more significantly in subjects age 75 to 87 yr than in subjects age 61 to 74 yr, and this decline was not correlated with changes in chronic medical burden.10 Comparisons of sleep timing across age groups indicate that older age is also associated with sleep occurring at earlier clock times and increased morning preference; however, this relationship has not been examined longitudinally.11–14
Studies in humans measuring the output of the circadian pacemaker, e.g., core body temperature or melatonin, have provided strong evidence for an age-related advancement of the phase of circadian rhythms; however, the evidence for an age-related reduction of circadian rhythm amplitude is mixed.12,13,15–18 Circadian period, on the other hand, does not appear to differ by age.19 Multiple lines of evidence indicate that age-related changes in sleep are related to an interaction between age-related changes in the circadian system and the sleep homeostatic process.12,13,20–22 Contributing to an early habitual wake time in older people is a shorter interval between the core body temperature minimum (CBTmin) and wake time compared with young adults.13 In addition, age-related changes in the circadian regulation of sleep-wake propensity results in decreased sleep consolidation at CBTmin among older people, which could result in early awakening and exposure to light at an earlier circadian phase, which in turn could shift circadian phase to an earlier hour.13,21–24 Accompanying these age-related changes in sleep are alterations in the neural organization of the SCN.25 There is also evidence from a transgenic rat study that age-related deterioration of rhythmic clock gene expression is more prominent in some peripheral tissues than the SCN, suggesting that the SCN loses the ability to entrain some peripheral clocks with age.26 Results from a human sleep laboratory study using a 90-min sleep-wake cycle are consistent with the notion of an age-related decline in the SCN's ability to entrain peripheral clocks.24 In summary, age-related changes in the circadian and sleep homeostatic processes and in central and peripheral clocks accompany age-related changes in sleep.
A highly conserved set of clock genes regulates the timing of the central pacemaker in the SCN through a transcription-translation feedback loop.27 CLOCK and ARNTL heterodimerize and activate transcription of the period (PER1, PER2, and PER3) and cryptochrome (CRY1 and CRY2) genes.27 NPAS2 plays a similar role to that of CLOCK in the forebrain and possibly in the SCN.28,29 PER and CRY protein levels accumulate over the course of the day, leading to PER:CRY heterodimers that trans-locate back into the nucleus where they repress CLOCK and ARNTL activity, thus repressing their own transcription. Degradation of the PER:CRY heterodimers during the night allows CLOCK:ARNTL to activate a new cycle of PER and CRY gene transcription. Posttranslational modification of the components of the molecular clock regulates clock function. Subcellular localization and ubiquitin-dependent degradation of PER and CRY is regulated by CSNK1D/CSNK1E/GSK3β-mediated phosphorylation. FBXL3 is involved in the ubiquitination of phosphorylated CRY proteins, and βTrCP1 and βTrCP2 are involved in the ubiquitination of phosphorylated PER proteins. An auxiliary clock feedback loop regulates ARNTL levels and makes the cyclic pattern of clock gene expression more robust. This auxiliary loop is composed of REV-ERBα/NR1D1 and REV-ERBβ/NR1D2, which repress ARNTL expression, and RORα, RORβ, and RORγ, which activate ARNTL expression. PGC1α enhances RORα's activation of ARNTL. A second auxiliary feedback loop consists of DBP, TEF, HLF, and E4BP4/ NFIL3. Several additional molecular factors are involved in the regulation of the mammalian molecular clock, as reviewed by Lowrey and Takahashi.27 In addition to exhibiting a cyclic pattern of gene expression within an SCN neuron, clock gene expression is also synchronized among the SCN neurons. Specifically, VIP and VIPR2 are required to couple SCN neurons to produce synchronized rhythms of clock gene expression.27
Clock genes have been shown to influence sleep traits in model organisms, and there is growing evidence that genetic variants in clock genes are associated with sleep traits in humans. A missense mutation in PER2 disrupting the phosphorylation site of CSNK1E has been linked to familial advanced sleep phase syndrome, a rare mendelian sleep disorder.30 A variant in CSNK1D, a paralog of CSNK1E, has also been found to be associated with familial advanced sleep phase syndrome.30 Genetic variants in PER2, PER3, CLOCK, CSNK1E, DBP, and DEC2 have been found to be associated with various sleep traits in humans, including diurnal preference and delayed sleep phase.30–36 Variants in NPAS2 and ARNTL have been associated with seasonality and seasonal affective disorder, phenotypes that could reflect circadian rhythm disruption.33,37,38 However, some clock gene SNP associations with sleep traits have failed to replicate.39 Furthermore, many of the previously reported genetic association studies of sleep traits have examined a small number of candidate polymorphisms in clock genes rather than systematically assaying common genetic variation within the entire gene region; thus, allelic heterogeneity has not been explored in many studies. To date, three studies have attempted to systematically capture common genetic variation in the known clock genes to examine genetic associations with sleep traits, but subjective measures of sleep-related traits were analyzed in all three studies.32,40,41 SNPs in TIMELESS were significantly associated with case-control status where cases were defined using trait combinations of depression, early morning awakening, and fatigue.40 However, the combination of traits used in the case definition presents a challenge in selecting appropriate replication cohorts. The other studies that systematically captured common genetic variation in clock genes examined self-reported sleep duration using the Munich ChronoType Questionnaire (MCTQ).32,41 Objectively measured sleep traits have not been examined in genetic association studies with SNPs designed to systematically capture common genetic variation in clock genes. Moreover, study populations composed entirely of elderly individuals have not been used in genetic association studies examining variation in clock genes and sleep traits measured objectively or subjectively.
In this study, we aim to examine the association between common genetic variation within clock gene regions and objectively measured sleep and activity rhythm traits in the elderly, a population group in which sleep continuity is frequently disrupted and the circadian and homeostatic sleep processes have undergone multiple age-related changes. We investigated the association between these sleep and activity rhythm traits that were objectively measured using actigraphy and 529 tagSNPs capturing common genetic variation in 30 clock genes in two large population-based cohorts of elderly participants 65 yr and older. The 30 clock genes that were examined included the canonical circadian pathway genes and genes previously shown to be associated with sleep traits in humans. Hierarchical cluster analysis was performed to further elucidate the relationship between genetic associations of correlated sleep traits.
METHODS
Study Participants
Data from two studies, the Osteoporotic Fractures in Men (MrOS) study and the Study of Osteoporotic Fractures (SOF), contributed to this analysis. The design of the MrOS study was based on the SOF study to allow for comparable analyses in both men and women. Both are large prospective studies recruited from communities in the United States, with the primary aim of studying osteoporotic fractures. Both had the limited exclusion criteria at enrollment of age younger than 65 yr, history of a bilateral hip replacement, or inability to walk without assistance. In the MrOS study, 5,994 men 65 yr and older were recruited from 2000 to 2002.42,43 The MrOS Sleep Study, an ancillary study of the parent MrOS cohort, was conducted between December 2003 and March 2005 and recruited 3,135 MrOS participants for a comprehensive sleep assessment. Among the participants of the MrOS Sleep Study, 2,527 self-identified white MrOS participants had DNA extracted, SNPs genotyped, and technically adequate sleep actigraphy data collected. In the SOF study, 9,704 self-identified white women 65 yr and older were recruited and baseline examinations were conducted from 1986 to 1988.44 From January 2002 to February 2004, 4,727 women attended the eighth SOF clinic visit. Among these 4,727 women, 3,219 were provided with an actigraph, as described in Tranah et al.,45 and technically adequate sleep actigraphy data were collected from 2,799 women. Among these 2,799 participants, DNA was extracted and SNPs were genotyped for 1,407 participants. All data were collected with written informed consent as approved by the review boards of the participating institutions.
Actigraphic Sleep Phenotypes
Sleep-wake patterns were assessed using actigraphy in both cohorts (Sleep-Watch-O, Ambulatory Monitoring, Inc., Ardsley, NY), as was described previously.46,47 Briefly, the actigraph was similar in size to a wristwatch and was worn on the non-dominant wrist. Activity was detected by a piezoelectric bimorph-ceramic cantilever beam that generated a voltage each time the actigraph was moved. Data were collected in three modes but are reported here in proportional integration mode given our work showing that this mode correlates best with the gold standard of polysomnography for measurement of sleep traits.47,48 While the actigraph was worn, participants completed sleep diaries that included time into and out of bed and times the actigraph was removed. This information was used when editing the actigraphy data files to set intervals for when the participant was in bed trying to sleep (after “lights off”), and to delete time when the actigraph was removed. A sleep scoring algorithm was applied to the data to determine sleep-wake boundaries. Actigraphy data were available in MrOS and SOF participants for 5.2 ± 0.8 nights (mean ± standard deviation [SD]) and 4.1 ± 0.7 nights, respectively.
The variables estimated by actigraphy were time in bed (the time spent in bed trying to sleep at night, from “lights off” to the time of getting out of bed), total sleep time (TST, the hours per night spent sleeping during the time spent in bed), sleep onset time (the start of the first 20 minute continuous block scored as sleeping after “lights off”), wake onset time (the last minute during the in-bed interval scored as sleeping), number of long wake episodes (NWAK, number of awakenings 5 minutes or longer in duration while in bed), sleep latency (SL, minutes from “lights off” to sleep onset), wake after sleep onset (WASO, minutes spent awake during the in-bed interval between sleep and wake onset times), sleep efficiency (SE, the percentage of time in bed spent sleeping between sleep and wake onset times), and nap minutes (minutes scored as sleep for continuous blocks of at least 5 minutes while not in bed at night trying to sleep). These actigraphy variables reflect data averaged over all nights or days, as appropriate, that participants wore the actigraph in order to obtain a more representative characterization of usual sleep patterns.
The activity data gathered by actigraphy was used to compute measures of activity rhythms using an extension to the traditional cosine curve to allow the activity data to fit a more squared wave rather than a cosine curve.49 Nonlinear least squares regression was used to estimate the activity parameters. The following activity rhythm traits were computed: acrophase (the time of day of peak activity, measured in portions of hours), amplitude (the difference between the minimum and maximum of the function, measured in arbitrary units of activity as counts/ minute), mesor (the minimum of the function plus half of the amplitude, which represents the middle of the fitted activity peak with units of counts/minute), and the pseudo-F statistic (a measure of overall fit of the activity data to the extended cosine curve, with higher values indicating stronger activity rhythms).
Measurement of Other Nongenetic Characteristics
All participants completed questionnaire data, which included questions about alcohol use, caffeine intake, and place of residence. Prescription and nonprescription medications used within the preceding 30 days were identified, recorded by the clinics, and stored in an electronic medications inventory database (San Francisco Coordinating Center, San Francisco, CA); each medication was matched to its ingredient(s) based on the Iowa Drug Information Service Drug Vocabulary (College of Pharmacy, University of Iowa, Iowa City, IA).50 A comprehensive examination included measurements of body weight and height; body mass index was calculated as weight in kilograms divided by the square of height in meters.
Clock Gene Selection, Genotyping and Quality Control of TagSNPs, and Analysis of Population Stratification
The California Pacific Medical Center investigators and the University of California, San Diego group collaborated to develop a custom Illumina Golden Gate assay (Illumina, San Diego, CA) to genotype polymorphisms in circadian and sleep related genes. Candidate genes (Table S1) were selected after review of experimental studies involving circadian rhythms in common model organisms (e.g., Drosophila melanogaster and Mus musculus) and association studies performed in human participants. The role of the canonical circadian pathway genes listed in Table S1 has been previously described.27 In addition, FMR1 and FXR2 regulate the circadian rhythm in mice, but their role in the regulation of sleep in humans has not been examined.51 Evidence for the association between the C825T GNB3 allele and seasonality is mixed.52,53 It has been reported that the C825T GNB3 allele interacts with CLOCK variants in an association with diurnal preference.54 PGC1B has been found to be associated with bipolar disorder,55 and its paralog, PGC1A, regulates ARNTL.56 Evidence for the role of TIMELESS in the mammalian molecular clock is mixed; nevertheless, TIMELESS was included as a candidate gene.57
TagSNPs were selected using Tagger58 (r2 ≥ 0.8, minor allele frequency (MAF) ≥ 0.01) with HapMap CEU Phase II (release 22) genotype data in the candidate gene regions including 10 kb upstream and downstream of transcript boundaries. Using these Tagger settings, 314 tagSNPs would have been selected from the 798 SNPs found within the 761 kb of genomic DNA that includes the RORA gene and 20 kb of flanking DNA. To reduce our RORA genotyping burden, the linkage disequilibrium (LD) threshold in Tagger was lowered to 0.6 and a maximum of 100 tagSNPs were selected for genotyping, resulting in 81% of the 798 RORA SNPs being captured at r2 ≥ 0.6. In total, 658 SNPs within the candidate gene regions were selected for genotyping. Genotypes were called using Beadstudio software (Illumina, San Diego, CA). Genotype concordance rate was > 0.99 (8% of MrOS samples and three SOF samples were plated in duplicate). Samples with < 90% SNP call rate were excluded. SNPs with missing call rate frequency > 0.05 and MAF < 0.01 were excluded. Autosomal SNPs with a Hardy-Weinberg equilibrium (HWE) exact P value < 8 × 10-5 (Bonferroni-corrected P value for 658 SNPs) were excluded.59 Among the all-male MrOS samples, X-linked SNPs were excluded if they were found to have heterozygous genotypes. Among the 658 geno-typed SNPs, 529 in MrOS and 508 in SOF passed quality control (QC) filters (Table S1).
To correct for residual population stratification in these self-identified European American cohorts, 195 independent, autosomal SNPs were used in multidimensional scaling analysis (MDS) as implemented in PLINK for the MrOS and SOF participants, and the resulting first two MDS components were included as covariates in all regression models.60
Statistical Analysis
All actigraphic sleep traits, except for NWAK, were continuous variables and linear regression analysis was performed. Effect size was reported as the β parameter estimate. Interaction analysis was performed by inclusion of a multiplicative term in linear regression models. Poisson regression models were explored for the count variable NWAK, but there was evidence of overdispersion (P < 0.0001 for the likelihood ratio test of the dispersion parameter in negative binomial regression models); thus, negative binomial regression was used to model NWAK. To obtain rate ratios for NWAK, time in bed (h) was included as an offset variable in models. Results presented here were based on regression models adjusted for age, body mass index, clinic site as indicator variables, and the first two MDS components. Secondary analysis was performed to assess the sensitivity of our results to various factors. In the secondary analysis, each of the following steps was performed individually: exclusion of participants who reported taking nonbenzodiazepine nonbarbiturate sedative hypnotic prescription sleep medication (Chloral Hydrate, Dexmedetomidine, Eszopiclone, Zopiclone, Zolpidem, Zaleplon, or Ramelteon) (MrOS n = 51, SOF n = 18), adjustment for current alcohol consumption, adjustment for current caffeine consumption, adjustment for residence type, and adjustment for season of the year when actigraphy was performed. SNP associations whose significance exceeded the multiple testing threshold in primary analysis did not change in secondary analysis (data not shown).
To correct for multiple hypothesis testing in the presence of LD, the effective number of independent SNPs using our QC-filtered genotype data was estimated and a multiple testing significance threshold of 1.7 × 10-4 was adopted.61,62 Results for all three modes of inheritance (additive, dominant, and recessive) were shown for SNPs whose association significance exceeded the multiple testing threshold under the additive inheritance mode or the model-free two degrees of freedom (2DF) test. Regression analysis was performed in each cohort separately, and then combined by fixed-effect meta-analysis using inverse variance weighting of effect estimates. P values from the 2DF test were combined by a Z-statistic based approach weighted by the square root of the sample size. Heterogeneity between studies was assessed using the I2 statistic and the P value from the Q-test. Association analysis was performed using R (www.r-project.org), meta-analysis was performed using METAL,63 and power calculations were performed using QUANTO.64
Hierarchical agglomerative cluster analysis (pvclust R package) was performed using the complete linkage clustering method on the distance matrix of the absolute value of the sample correlation of the additive SNP test statistics from the meta-analysis for all examined actigraphic traits. Cluster significance was assessed using multiscale bootstrap resampling with 10,000 replications. Principal component analysis was performed on the correlation matrices of the actigraphic traits and the additive SNP test statistics from the meta-analysis.
BioGPS was used to determine the distribution of LRRC23 gene expression across 79 tissues in a human microarray panel.65,66 Genevar was used to access HapMap CEU expression quantitative trait loci (eQTL) data, and the association between SNP genotype and gene expression was estimated using the Spearman rank correlation coefficient.67,68
RESULTS
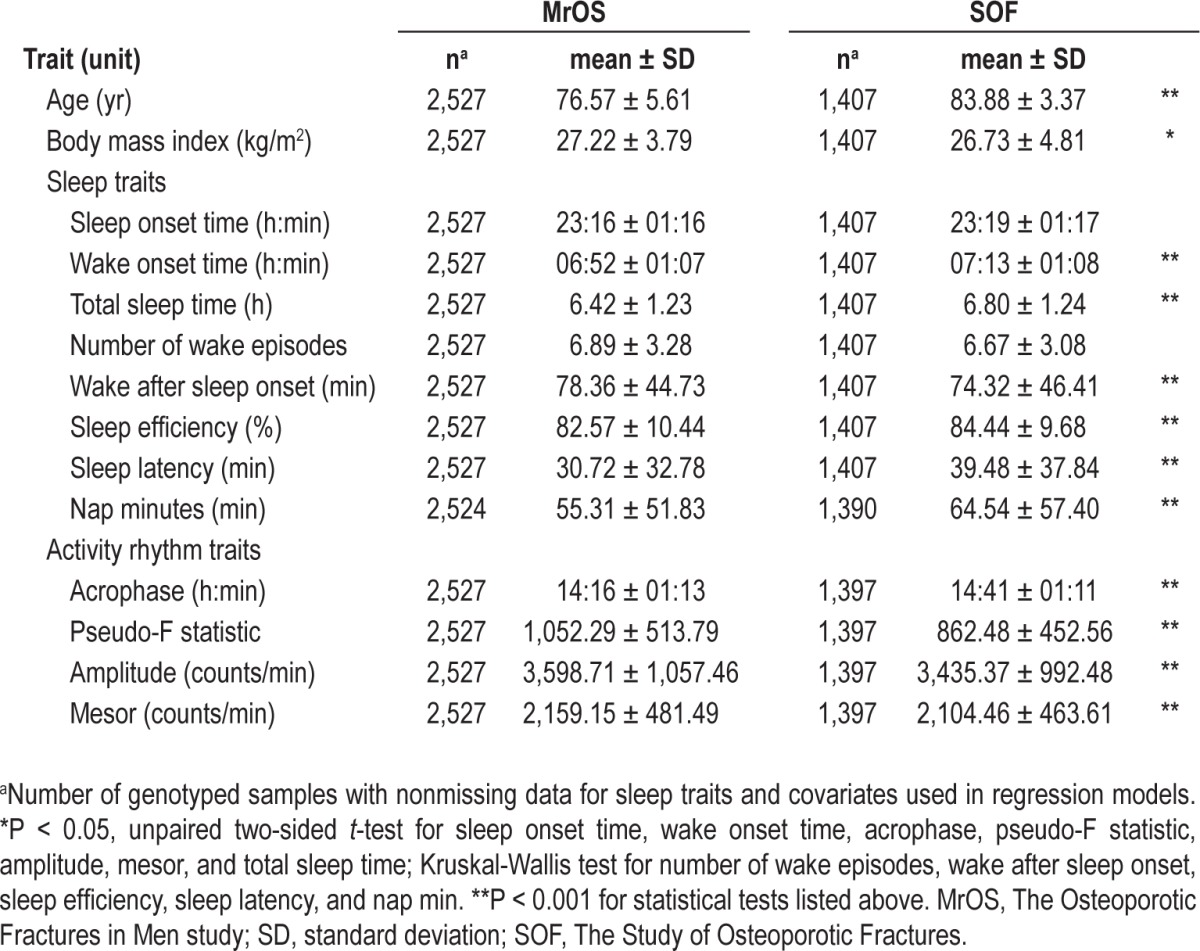
In this genetic association study of common genetic variation in clock genes and objectively measured actigraphic sleep traits, two population-based cohort studies (MrOS and SOF) of elderly participants were examined. Age was significantly lower and body mass index was significantly higher in the all-male MrOS cohort compared with the all-female SOF cohort (Table 1). The age range of the MrOS and SOF participants was 67 to 96 yr and 79 to 98 yr, respectively. Except for sleep onset time and NWAK, all other sleep and activity rhythm traits were significantly different between the two cohorts (Table 1). Actigraphy-based TST and SE were reported in a population cohort (Rotterdam Study) with a similar participant age distribution as the MrOS and SOF cohorts, and the TST and SE estimates from the three cohorts were remarkably similar (Rotterdam mean TST in h: 6.4 in men, 6.7 in women; mean SE %: 77.8 in men, 79.0 in women).69
Table 1.
Characteristics of the study populations
This candidate gene study examined 529 SNPs in 30 clock genes (Table S1). SNP associations that passed multiple testing under the additive inheritance mode or the model-free 2DF test in either cohort alone or in the meta-analysis are described in detail. Briefly, two SNPs (rs1047776 and rs2238114) at chromosome 12p13 were significantly associated with sleep continuity in the meta-analysis of results from MrOS and SOF. In MrOS, rs3816358 in ARNTL and rs3768984 in NPAS2 were significantly associated with later sleep timing, and the SNP interaction was significant. In SOF, the NPAS2 SNP rs895520 was significantly associated with the pseudo-F statistic, a measure of activity rhythm robustness, and rs1047776 at chromosome 12p13 was significantly associated with sleep latency.
SNP Associations With Sleep Continuity at Chromosome 12p13
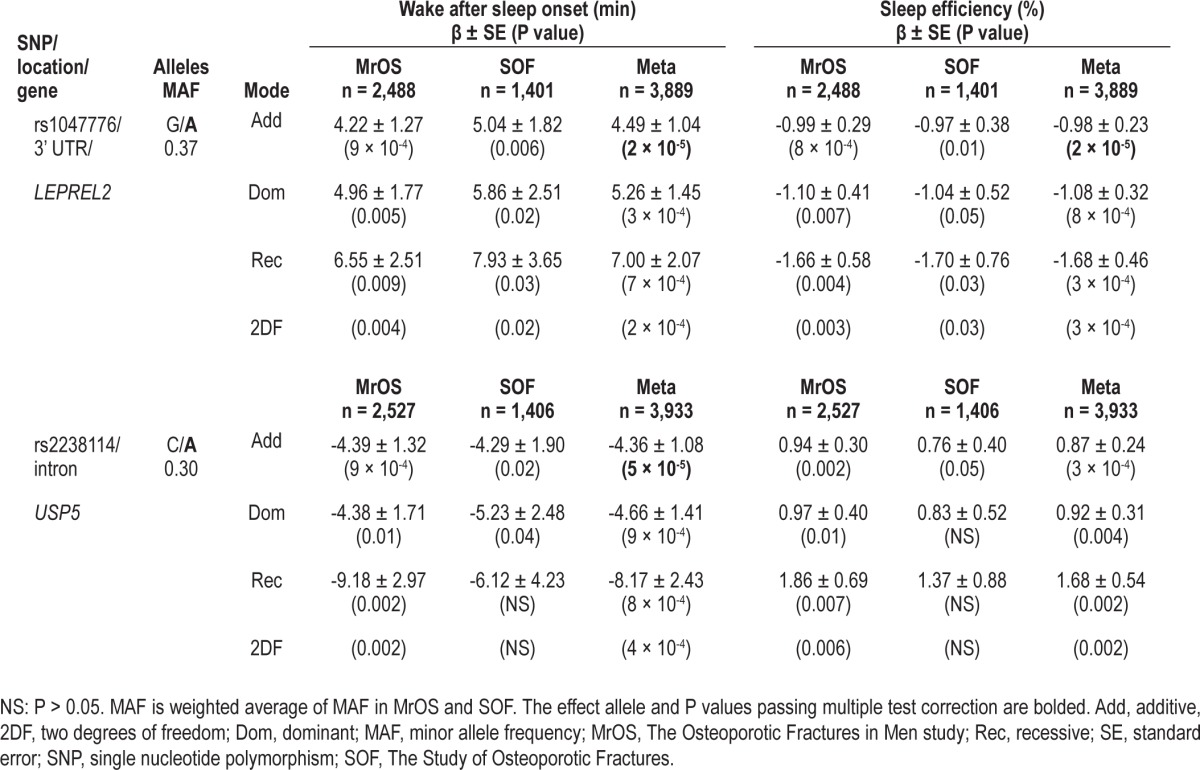
In an attempt to tag common genetic variation in and around the GNB3 gene at chromosome 12p13, tagSNPs were geno-typed within GNB3 and two surrounding genes, LEPREL2 and USP5. The LEPREL2 SNP rs1047776 and the USP5 SNP rs2238114 (Figure 1) were both significantly associated with multiple sleep continuity traits after multiple test correction (Table 2, Table S2). Under an additive mode of inheritance, rs1047776 in the 3' untranslated region (UTR) of LEPREL2 was associated with higher WASO in MrOS (βADD ± SE, PADD: 4.22 ± 1.27, 9x10-4) and SOF (βADD ± SE, PADD: 5.04 ± 1.82, 0.006), and the meta-analysis P value passed multiple test correction (βMETA ± SE, PMETA: 4.49 ± 1.04, 2 × 10-5) (Table 2). Consistent with an increase in WASO, rs1047776 was also associated with a decrease in SE under an additive mode of inheritance in MrOS (βADD ± SE, PADD: -0.99 ± 0.29, 8 × 10-4) and SOF (βADD ± SE, PADD: -0.97 ± 0.38, 0.01), and the meta-analysis P value passed multiple test correction (βMETA ± SE, PMETA: -0.98 ± 0.23, 2 × 10-5) (Table 2). Consistent with an increase in WASO and a decrease in SE, meta-analysis indicated that rs1047776 was also significantly associated with a higher rate of long (> 5 min) wake episodes while in bed (Table S2). Similarly, the intronic SNP rs2238114 within the USP5 gene was also significantly associated with WASO and NWAK, but the effect of the minor allele was in the opposite direction to that of rs1047776 (Table 2, Table S2). The association significance between rs2238114 and SE nearly exceeded the multiple testing threshold (Table 2). There was no evidence for interaction between rs1047776 and rs2238114 for WASO or SE (Pint > 0.05).
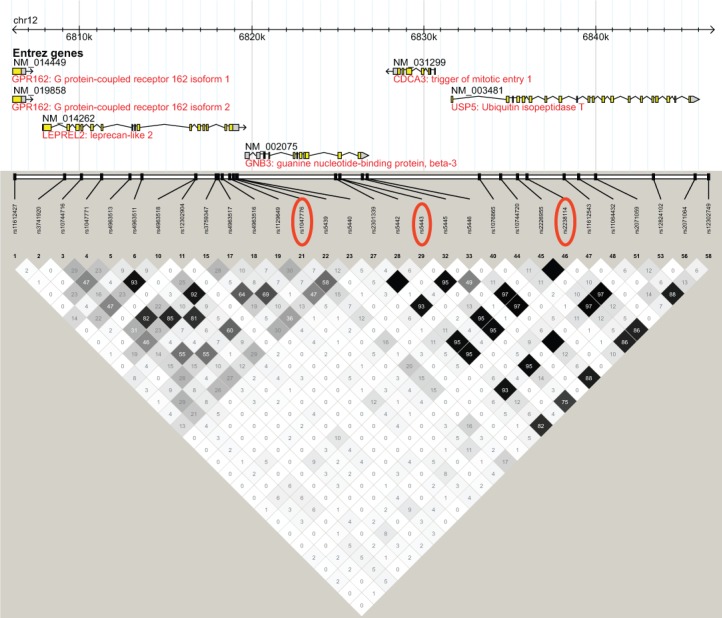
Figure 1.
Linkage disequilibrium (LD) plot of the GNB3 locus. LD plot generated using Haploview with data from CEU HapMap phase 2 and 3, release 28, Aug10, genome build 36, dbSNP b126 from chromosome 12, position 6,806,200 – 6,846,900. LD measured in r2 units. Single nucleotide polymorphisms described in text are circled in red.
Table 2.
Genetic association with sleep continuity traits at chromosome 12p13
Although rs2238114 was in high LD (HapMap CEU r2 = 0.95, Figure 1) with the GNB3 SNP rs5443 that was previously reported to interact with a CLOCK SNP in the association with diurnal preference,54 rs1047776 in LEPREL2 was selected to capture common genetic variation upstream of GNB3 and was not in high LD with any of the HapMap SNPs within GNB3 (maximum HapMap CEU r2 = 0.16), including rs5443 (HapMap CEU r2 = 0.12) (Figure 1). The SNPs rs1047776 and rs2238114 were not in high LD with each other in MrOS participants (r2 = 0.12) or in HapMap CEU individuals (r2 = 0.15, Figure 1). Despite the low LD between rs1047776 and rs2238114, the effect sizes and P values for the WASO and SE SNP associations were shifted toward the null in conditional analysis that included both SNPs in the same regression model (Table S3).
In addition to being in high LD with SNPs in GNB3, the USP5 SNP rs2238114 was also in high LD (HapMap CEU r2 ≥ 0.8) with SNPs in many neighboring genes. In fact, rs2238114 and the previously reported rs5443 reside within a 101kb haplotype block (HapMap CEU) that spans the following genes: GNB3, CDCA3, USP5, TPI1, SPSB2, LRRC23, ENO2, ATN1, and PTPN6 (Figure S1).
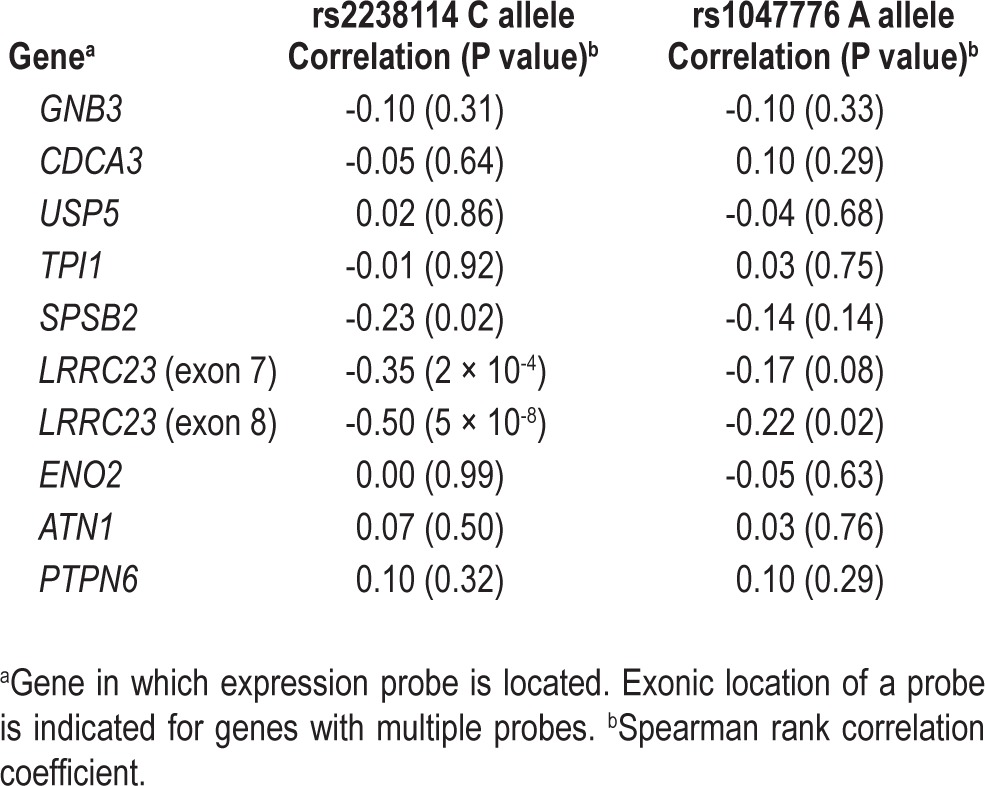
Publicly available eQTL data from HapMap CEU lymphoblastoid cell lines were used to determine whether the sleep continuity-associated SNPs that we identified (rs2238114 and rs1047776) were associated with expression of any of the genes in the 101 kb haplotype block. The rs2238114 and rs1047776 alleles associated with higher WASO were significantly associated with lower LRRC23 gene expression in HapMap CEU lymphoblastoid cell lines (Table 3). Significant association was observed between rs2238114 and both LRRC23 gene expression probes (P = 5 × 10-8 and 2 × 10-4) (Table 3, Figure S2A). Although the significance of the correlation between LRRC23 gene expression and rs1047776 was not as significant as that for rs2238114, rs1047776 was more correlated with LRRC23 gene expression than with the expression of any other gene in the LD block (Table 3). LRRC23 gene expression is found in a broad range of human tissues, including multiple brain regions (Figure S2B).
Table 3.
Single nucleotide polymorphism associations with gene expression in HapMap CEU lymphoblastoid cell lines at chromosome 12p13
SNP Associations with Sleep Timing in the ARNTL and NPAS2 Genes
The SNPs rs3816358 in ARNTL and rs3768984 in NPAS2 were associated with later sleep timing in the MrOS cohort but not the SOF cohort. The intronic SNP rs3816358 within ARNTL was significantly associated with later sleep onset time in MrOS participants (βADD ± SE, PADD: 0.19 ± 0.06, 7 × 10-4) and the P value for the 2DF test (P2DF = 1.2 × 10-4) exceeded the multiple testing threshold (Table 4). The SNP rs3816358 was also nominally associated with a later wake onset time in MrOS participants (βADD ± SE, PADD: 0.16 ± 0.05, 0.001; P2DF = 0.003) (Table 4).
Table 4.
Genetic association with sleep timing-related traits
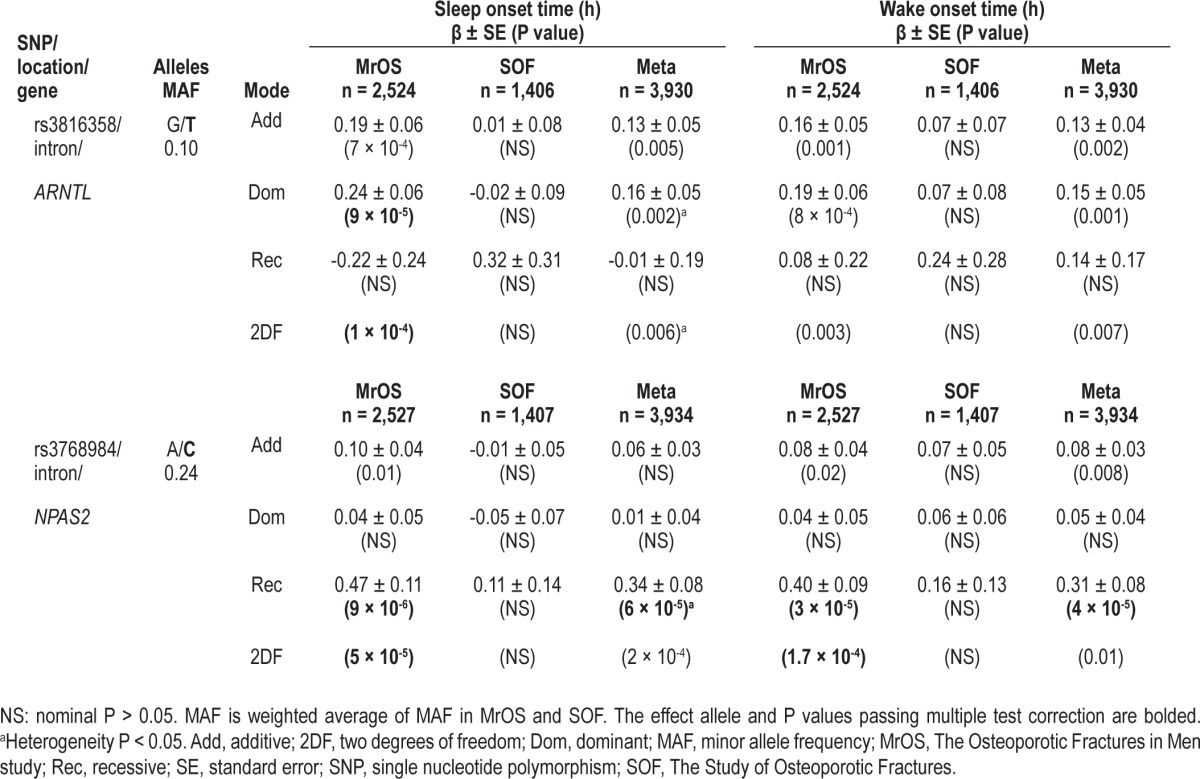
Consistent with a nearly equal association effect size with sleep onset time and wake onset time, rs3816358 was not associated (P > 0.05) with TST in the MrOS cohort (data not shown). In addition, rs3816358 was not associated with WASO, SE, or NWAK in the MrOS cohort (P > 0.05, additive mode of inheritance, data not shown). The SNP rs3816358 was nominally associated with sleep latency in the MrOS cohort (βADD ± SE in units of h, PADD: 0.05 ± 0.03, 0.04), raising the possibility that the association between rs3816358 and a later sleep onset time could be mediated by an increase in sleep latency. Although the association in the MrOS cohort between rs3816358 and sleep onset time was slightly attenuated after adjustment for sleep latency (βADD ± SE, PADD: 0.15 ± 0.05, 0.004; P2DF = 0.001), the SNP association with sleep onset time remained nominally significant. The association between rs3816358 and wake onset time was essentially unchanged after adjustment for sleep latency (βADD ± SE, PADD: 0.16 ± 0.05, 0.002; P2DF = 0.006).
An intronic SNP rs3768984 within NPAS2 was also associated with later sleep onset time and wake onset time in the MrOS cohort, and the P value for the 2DF test exceeded the multiple testing threshold (Figure 2, Table 4). Plots of adjusted mean sleep onset time and mean wake onset time by genotype indicated that a recessive genetic model is most appropriate for this SNP (Figure S3). Under a recessive mode of inheritance, rs3768984 was significantly associated with a later sleep onset time in MrOS participants (βREC ± SE, PREC: 0.47 ± 0.11, 9 × 10-6) (Table 4). The direction of effect was the same in SOF participants, but the association failed to reach a nominal significance level (Table 4). This SNP also passed multiple test correction after meta-analysis under the recessive mode of inheritance (βMETA ± SE, PMETA: 0.34 ± 0.08, 6 × 10-5) (Table 4). Similarly, rs3768984 was significantly associated with a later wake onset time in MrOS participants (βREC ± SE, PREC: 0.40 ± 0.09, 3 × 10-5; P2DF = 1.7 × 10-4) but not in SOF participants, and the meta-analysis results were also significant (βMETA ± SE, PMETA: 0.31 ± 0.08, 4 × 10-5) (Table 4). Power was 0.56 to detect the meta-analysis effect size for sleep onset time (0.34 h) among 3,930 participants (two-sided α = 1.7x10-4, recessive mode of inheritance). Both rs3816358 in ARNTL and rs3768984 in NPAS2 were nominally associated with a later acrophase (peak activity time later in the day) in the MrOS cohort and in the meta-analysis, but the association P value did not pass multiple test correction (Table S4). The NPAS2 SNP rs3768984 was not significantly associated with TST, WASO, SE, sleep latency, or NWAK in the MrOS cohort (P > 0.05, recessive mode of inheritance, data not shown).
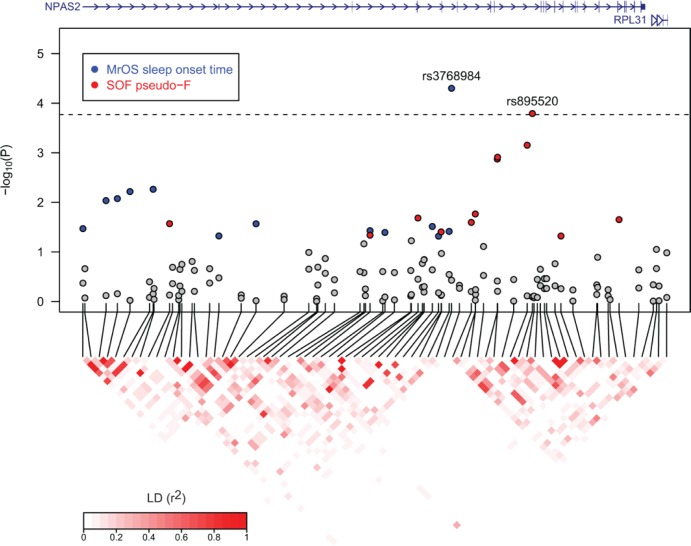
Figure 2.
Regional association plot of the NPAS2 gene region. Gene region from UCSC RefSeq gene track, chromosome 2, 100,794,551 – 100,989,160, genome build 36. Association P values are from the model-free two degrees of freedom test. Gray circles mark single nucleotide polymorphisms (SNP) association P > 0.05, blue circles mark The Osteoporotic Fractures in Men (MrOS) sleep onset time SNP association P ≤ 0.05, and red circles mark The Study of Osteoporotic Fractures (SOF) pseudo-F statistic SNP association P ≤ 0.05. Linkage disequilibrium (LD) was calculated using phased chromosomes and measured in r2 units. Dashed line indicates significance threshold using multiple testing procedures as described in methods.
As rs3816358 and rs3768984 were significantly associated with later sleep timing and are located in genes coding for protein binding partners, we investigated the statistical interaction between these two SNPs. With an additive mode of inheritance for rs3816358 and a recessive mode of inheritance for rs3768984, there was a significant interaction between these SNPs for sleep onset time and wake onset time (βINT ± SE, PINT = 0.72 ± 0.24, 0.003 and 0.71 ± 0.22, 0.001, respectively) in MrOS participants (Figure 3). The effect size of the association between the ARNTL SNP rs3816358 and sleep onset time was much larger among MrOS participants with an NPAS2 SNP rs3768984 C/C genotype (βADD ± SE, PADD = 0.78 ± 0.34, 0.02) than those with A/A or A/C rs3768984 genotypes (βADD ± SE, PADD = 0.15 ± 0.06, 0.007). Consistently, the effect size of the association between rs3768984 and sleep onset time increased with the number of minor alleles of rs3816358 (rs3816358 G/G: βREC ± SE, PREC = 0.32 ± 0.11, 0.006; rs3816358 G/T: βREC ± SE, PREC = 1.03 ± 0.27, 0.00014; rs3816358 T/T: βREC ± SE, PREC = 2.36 ± 1.86, NS). These two SNPs also interacted in their association with acrophase in the MrOS cohort (βINT ± SE, PINT = 0.96 ± 0.24, 5 × 10-5).
Figure 3.
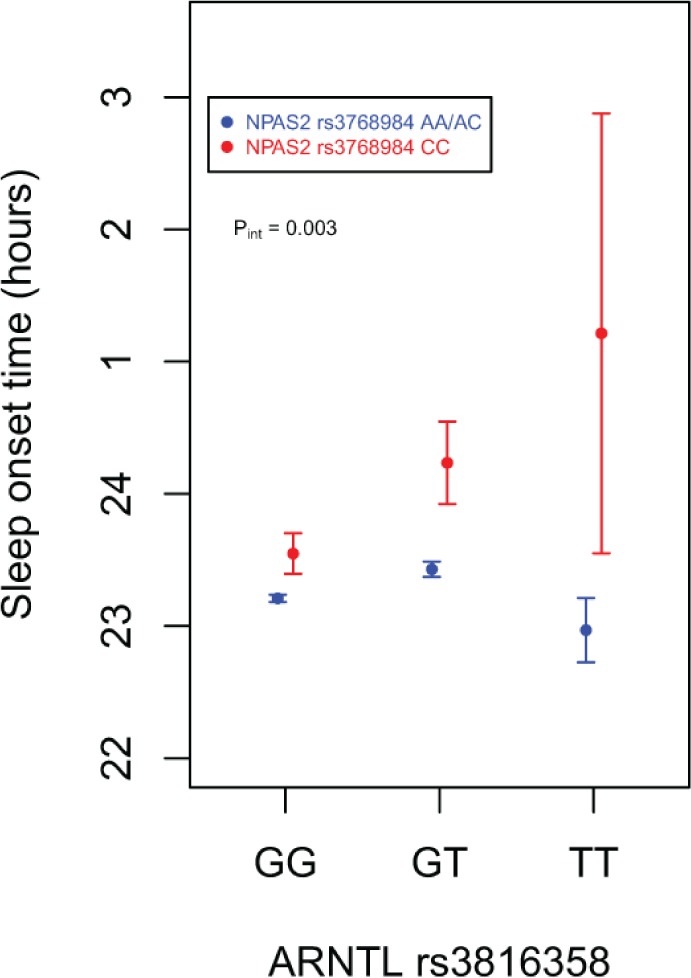
Interaction between rs3768984 (NPAS2) and rs3816358 (ARNTL) on sleep onset time. Adjusted means (filled circles) with standard errors of the predicted means are shown using results from The Osteoporotic Fractures in Men study participants. Sleep onset time adjusted for the same covariates as regression models, as described in the Methods section.
SNP Associations With Sleep Continuity and Activity Rhythm Traits in the NPAS2 Gene and at Chromosome 12p13
In the SOF cohort, P values from the recessive inheritance mode and the 2DF test exceeded multiple testing thresholds for the association between the NPAS2 SNP rs895520 and the pseudo-F statistic, a measure of activity rhythm robustness (Table 5, Figure 2). Results from the recessive model were based on 252 TT SOF participants (18% homozygous TT genotype frequency). The two NPAS2 SNPs that exceeded multiple testing thresholds in this study (rs895520 and rs3768984) are not in LD (r2 = 0.02 based on SNP genotypes from MrOS and SOF participants).
Table 5.
Genetic association with sleep latency and activity rhythm
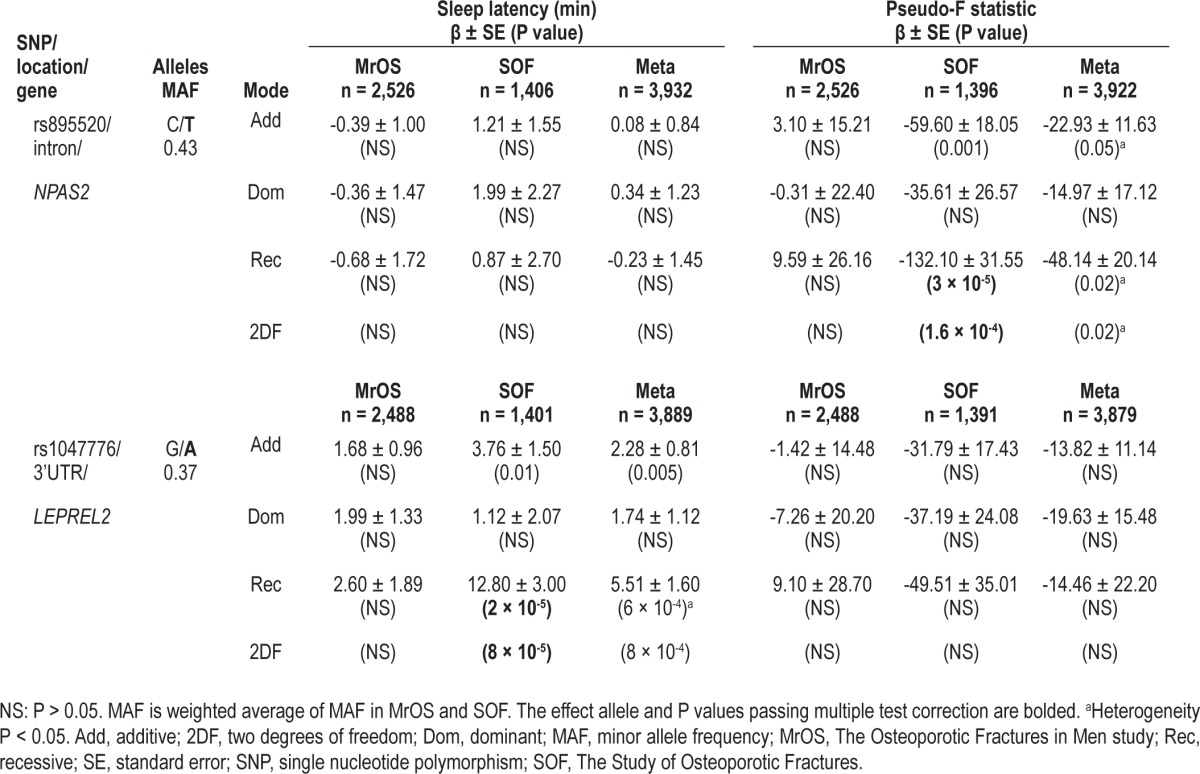
Also in the SOF cohort, P values from the recessive inheritance mode and the 2DF test exceeded multiple testing thresholds for the association between rs1047776 at chromo-some 12p13 and sleep latency (Table 5). The homozygous AA genotype frequency was 13% (183 AA genotypes) in the SOF cohort. Given the rs1047776 SNP association with WASO and SE (Table 2), conditional analysis was performed. Adjustment for WASO or SE did not greatly change the association between rs1047776 and sleep latency in the SOF cohort (WASO adjusted P2DF = 3 × 10-4, SE adjusted P2DF = 4 × 10-4).
Rare RORC SNP Associated With Sleep Continuity
The SNP rs4284267 within the RORC 5' flanking region was significantly associated with WASO and SE in the MrOS cohort after multiple test correction, but the MAF was 5%, resulting in low power and potentially unstable, biased effect estimates (Table S5). Given the low MAF of rs4284267 combined with the recessive genetic model, this result should be considered preliminary.
Phenotypic and Genotypic Correlations
It could be expected that rs3768984 would be associated with sleep onset time, wake onset time, and acrophase, as these three traits are highly correlated. To explore the relationship between genetic associations of multiple correlated traits, the correlation matrices of phenotypes and genetic associations were compared (Figure 4). Genetic associations are represented by SNP association test statistics so that significance level and effect direction are considered, and the genetic correlation is the correlation of these test statistics for different actigraphic traits. Traits with strong phenotypic correlation also showed strong genetic correlation, as depicted by nearly complete overlap between black and blue ellipses (Figure 4, upper triangle) and nearly identical correlation values (Figure 4, lower triangle).
Figure 4.
Phenotypic and genetic correlation of actigraphic sleep and activity rhythm traits. The upper triangle depicts correlation graphically, and the lower triangle shows correlation values, with phenotypic correlations in black and genetic correlations in blue. Phenotypic correlations (Pearson's r) are calculated using The Osteoporotic Fractures in Men study data; similar results are seen using The Study of Osteoporotic Fractures phenotypic data. Genetic correlations (Pearson's r) are based on the meta-analysis single nucleotide polymorphisms test-statistics using the additive genetic model. A correlation of 0 is depicted with a circle, and correlations approaching 1 or -1 are depicted with more narrow ellipses to the right or left, respectively. Gray triangles mark the clusters formed by hierarchical cluster analysis of genetic associations. NWAK, number of awakenings; TST, total sleep time; WASO, wake after sleep onset.
Hierarchical cluster analysis was performed to identify clusters such that SNP associations within clusters were more similar to each other than to those in other clusters. Genetic associations for the actigraphic traits can be clustered into four groups: wake onset time and acrophase; sleep onset time and TST; pseudo-F statistic, amplitude, mesor, and nap min; and NWAK, WASO, SE, and sleep latency (Figure 4, gray triangles, and Figure S4). The existence of four clusters identified by hierarchical cluster analysis was supported by principal component analysis of the correlation matrices. The first four principal components explained 97% and 96% of the phenotypic and genotypic variance, respectively.
Replication Analysis
A previously reported candidate gene study of 194 SNPs (Bonferroni corrected P value = 2.6 × 10-4) in clock genes and self-reported sleep duration using the MCTQ was conducted in a discovery sample of approximately 360 participants, and the top two SNP associations from the discovery stage (rs12649507 and rs11932595, both located in the CLOCK gene) were tested for replication in 1,011 participants.32 In a subsequent meta-analysis of genome-wide association studies (GWAS) of MCTQ-based self-reported sleep duration, these two CLOCK SNPs were not imputed in many of the cohorts, and as such, association statistics for the replication of these two CLOCK SNPs were not reported.41 In the original candidate gene study, the association between sleep duration and rs12649507 was significant in the discovery stage (P = 0.0051), the replication stage (P = 0.045), and the meta-analysis (P = 0.0087).32 In the same report, the association between sleep duration and rs11932595 was signifi-cant in the discovery stage (P = 0.0080) and the replication stage (P = 0.047), but apparently the direction of effect was not the same, as the association in the meta-analysis was not significant (P = 0.24).32 Unfortunately, the effect size and effect direction were not reported, thus preventing the determination of the power to replicate or whether the effect direction replicates.
Of the top two SNPs, rs11932595 was genotyped in our study but rs12649507 was not. The SNP rs9312661 was in high LD with rs12649507 based on HapMap CEU release 22 geno-types (r2 = 0.80) and was selected as a proxy SNP. There was no evidence that rs11932595 was associated with TST in the MrOS or SOF cohorts, but the A allele of the proxy SNP for rs12649507 was associated with less TST at a nominal level of significance (P ≤ 0.05) in the MrOS cohort and the meta-analysis but not in the SOF cohort (Table S6). Associations between the two CLOCK SNPs and all sleep traits and activity rhythm traits listed in Table 1 were also examined. In addition to TST, the A allele of the proxy SNP for rs12649507 was associated (P ≤ 0.05) with more napping minutes and lower mesor of the activity rhythm in the MrOS cohort and in the meta-analysis (Table S6). The decrease in TST (-4.06 min) associated with rs12649507 was similar to the increase in nap minutes (3.59 min) (Table S6). The rs11932595 G allele was associated (P ≤ 0.05) with higher activity rhythm amplitude and higher pseudo-F statistic (indicative of a more robust activity rhythm) in the SOF cohort (Table S6).
The previously published GWAS of MCTQ-based self-reported sleep duration reported SNPs near and within clock genes associated at a nominal significance level (P ≤ 0.05). Although we did not identify significant SNP associations with TST, we investigated whether the SNPs associated with traits other than TST in our current study might have been previously reported to be associated with MCTQ-based sleep duration.41 The C allele of the NPAS2 SNP rs3768984, which was associated with later sleep timing in the MrOS cohort, was significantly associated with lower MCTQ-based sleep duration (β ± SE, P = -0.07 ± 0.03, 0.008). None of the other SNPs we identified were reported to be associated at a nominal significance level with MCTQ-based sleep duration.
DISCUSSION
We present evidence from the analysis of two large population-based cohorts of elderly participants that common genetic variation in ARNTL and NPAS2 and at chromosome 12p13 is significantly associated with objectively measured actigraphic sleep and activity rhythm traits. Two SNPs (rs1047776 and rs2238114) at chromosome 12p13 were associated with sleep continuity traits in both cohorts, and the meta-analysis P value exceeded the multiple testing significance threshold. SNP rs1047776 was also significantly associated with sleep latency in the SOF cohort. SNP rs2238114 resided in a region of high LD that included nine genes, and the rs1047776 and rs2238114 alleles that were associated with higher WASO were also significantly associated with lower LRRC23 gene expression in HapMap CEU lymphoblastoid cell lines. SNP associations that were significant in a single cohort were also identified. The ARNTL SNP rs3816358 and the NPAS2 SNP rs3768984 were significantly associated with later sleep and wake onset time in the MrOS cohort. ARNTL and NPAS2 biochemically interact to form a heterodimeric transcription factor that acts at the core of the circadian rhythm pathway, and the ARNTL SNP rs3816358 and the NPAS2 SNP rs3768984 statistically interact as well (sleep onset time PINT = 0.003, wake onset time PINT = 0.001). In addition, a second NPAS2 SNP (rs895520) was significantly associated with the pseudo-F statistic, a measure of activity rhythm robustness, in the SOF cohort. Our study achieved adequate power (power > 0.8) to detect an effect size of 0.15 SD units at MAF values even lower than 0.20 and approached adequate power to detect an effect size of 0.10 SD units at an MAF of 0.40 (Table S7); thus, undetected SNP associations are likely to have small effect sizes. Cluster analysis revealed that the genetic associations for all examined actigraphic traits could be grouped into four clusters. A previously identified SNP association in the CLOCK gene with self-reported sleep duration replicated in the MrOS cohort, and the NPAS2 SNP rs3768984 we discovered to be associated with later sleep timing was also associated with sleep duration from a previous GWAS meta-analysis.32,41
The GNB3 locus was included in our investigation based on the previous report that the 825C/T allele (rs5443) of GNB3 interacted with the 3111C/T allele of CLOCK in an association with diurnal preference among healthy Korean college students.54 We attempted to directly genotype rs5443, but the Illumina designability score (range 0 - 1.1, designability cutoff = 0.6) was only 0.65 for rs5443, and not surprisingly, this SNP failed our genotyping assay. The HapMap project also had difficulty genotyping this SNP (50% missing rate in CEU samples). Although the GNB3 SNP rs5443 was not successfully genotyped in our samples, a proxy SNP, rs2238114, was successfully genotyped. The rs2238114 SNP was in high LD with rs5443 (HapMap CEU r2 = 0.95), but rather than being within the GNB3 gene, it was located downstream of GNB3 in a nearby gene, USP5 (Figure 1). The SNP rs2238114 was not associated with sleep timing in the MrOS or SOF cohorts, but it was significantly associated with sleep continuity traits in the meta-analysis of MrOS and SOF results. The SNP rs1047776 located in LEPREL2, a gene upstream of GNB3, was also observed to be associated with sleep continuity traits in both cohorts. The low LD between the LEPREL2 SNP rs1047776 and the USP5 SNP rs2238114 raised the possibility that genetic variation in LEPREL2 might be independently associated with sleep traits, but conditional analysis proved otherwise. Thus, the LEPREL2 and USP5 SNP associations appear to represent the same genetic signal.
To further examine the GNB3 genetic association region, we characterized the LD structure of the GNB3 region from the HapMap CEU population of European descent, which revealed that the GNB3 SNP rs5443 and the USP5 SNP rs2238114 reside on a 101 kb haplotype at chromosome 12p13 that spans multiple genes (GNB3, CDCA3, USP5, TPI1, SPSB2, LRRC23, ENO2, ATN1, and PTPN6) (Figure S1). The fact that SNPs in the GNB3 region reside on a large haplotype aids in the interpretation of our results and previously published associations between rs5443 and multiple diverse traits. The initial discovery that the coding synonymous GNB3 SNP rs5443 alters GNB3 transcript splicing, resulting in an in-frame deletion of 41 amino acids, has led to the hypothesis that this SNP represents the causal allele for this locus.70 Subsequent genetic analysis of the GNB3 locus has focused on this single SNP, and a wide variety of phenotypes, including hypertension, circadian-related traits, depression, response to antidepressant medication, obesity, and insulin resistance have been reported to be associated with rs5443.54,70–73 Certainly, these traits could have a common molecular etiology that involves GNB3. Considering the large LD block that includes rs5443, genetic associations with rs5443 could also reflect the influence of SNPs in the other genes located in the large haplotype block in this genomic region. In fact, rs5443 is in high LD (HapMap CEU r2 = 0.96) with rs2226955, a coding synonymous SNP within the fourth exon of USP5, a protein involved in ubiquitin-mediated proteolysis.74 The GNB3 SNP rs5443 is also in high LD (HapMap CEU r2 = 0.73) with rs710415, an E276G missense SNP (glutamine to glycine) in the LRRC23 gene. In an attempt to narrow down the list of genes that might be regulated by the chromosome 12p13 SNPs associated with sleep continuity in the MrOS and SOF cohorts, publicly available eQTL data were examined. The rs1047776 and rs2238114 alleles associated with higher WASO were significantly associated with lower LRRC23 gene expression in HapMap CEU lymphoblastoid cell lines, suggesting a link between lower levels of LRRC23 gene expression and higher levels of sleep fragmentation. Little is known about LRRC23 (leucine-rich repeat containing 23) gene function. LRRC23 is expressed in a wide range of tissues (Figure S2B). The leucine-rich repeat (LRR) protein domain is involved in protein-protein interactions, and a recent bioinformatic categorization of 375 human LRR-containing proteins grouped LRRC23 into a nuclear and centrosome cluster.75 Publicly available eQTL data along with our SNP association results point toward LRRC23 as an interesting candidate for further study, however, human lymphoblastoid cell lines might not faithfully reproduce gene expression regulation in human tissue relevant to the regulation of sleep-wake patterns. Follow-up eQTL studies in neural tissues relevant to sleep-wake regulation will be necessary to strengthen the evidence for LRRC23's role in sleep regulation. Furthermore, future studies conducted in populations of African descent where the LD in this region is greatly reduced can help to disentangle the associations between multiple traits and SNPs in this region. Our examination of the chromosome 12p13 region surrounding the SNPs we genotyped near our candidate gene GNB3 contributes to a more complete understanding of genetic associations in this region.
To our knowledge, our study is the first to identify signifi-cant associations between NPAS2 SNPs and sleep traits and activity rhythm traits in humans. SNPs in NPAS2 and ARNTL were significantly associated with a shift toward later sleep timing in the MrOS cohort, a shift that was particularly striking given the well-established age-related shift toward earlier sleep timing.3,21 Replication analysis in well-powered studies of younger participants will inform whether these SNP associations are specific to particular age groups. Although NPAS2 has been shown to regulate sleep in the mouse,29,76,77 its role in the regulation of sleep in humans has been less clear. For instance, seasonal affective disorder, a condition that is related to circadian rhythm disruption but is not a direct measure of sleep, has been associated with NPAS2 genetic variants.33,37 It is thought that ARNTL:CLOCK heterodimers control clock gene expression in the SCN, but in the forebrain, ARNTL partners with NPAS2.28,30 NPAS2 might also partner with ARNTL in the SCN, but NPAS2 likely plays a less prominent role than CLOCK in the SCN.29 Our observation of a statistical interaction between rs3768984 in NPAS2 and rs3816358 in ARNTL is consistent with the biochemical interaction between the two proteins and strengthens the case that these SNP associations reflect a functional alteration in the circadian clock. Germline polymorphisms such as SNPs cannot provide insight into the tissue-specific role of NPAS2; thus, further studies will be needed to determine whether NPAS2:ARNTL heterodimers in central clocks (SCN) and/or peripheral clocks (forebrain) regulate sleep timing.
In addition to examining TST in the replication analysis of the CLOCK SNPs rs12649507 and rs11932595,32 we also examined whether these two SNPs were associated with any of the actigraphic sleep and activity rhythm traits that were analyzed in our genetic association study. In the MrOS cohort but not the SOF cohort, the CLOCK SNP rs12649507 was associated with less TST, more nap minutes, and lower mesor (mean activity level from the fitted activity rhythm curve). Although the association between TST and the other CLOCK SNP, rs11932595, did not replicate, the association between this SNP and two activity rhythm traits (higher amplitude and higher pseudo-F statistic) in the SOF cohort was indicative of a more robust activity rhythm. Examination of these SNP associations in cohorts of younger participants may help to determine whether the associations are age-dependent.
We also assessed whether the SNPs we discovered in the MrOS and SOF cohorts were reported in a GWAS of MCTQ-based average weekly sleep duration.41 The weighted mean age of the female and male participants in the discovery cohorts of the GWAS meta-analysis was 45.8 yr and 46.9 yr, respectively, and the participants in the replication cohorts were even younger.41 Sleep duration was the only trait examined in the previous GWAS, and although we did not discover SNPs that were significantly associated with total sleep time in our study, we did identify SNPs associated with related traits. The NPAS2 SNP rs3768984, which was significantly associated with later sleep timing in the MrOS cohort, was reported to be associated with less MCTQ-based self-reported sleep duration. Later sleep timing, i.e., later chronotype, is related to sleep duration when work and free days are examined separately, although the relationship is nearly abolished when sleep duration is averaged across all days.14 A later chronotype is associated with less sleep on work days, which is normally compensated for by more sleep on free days.14 In the cohorts contributing to the previously reported GWAS meta-analysis of sleep duration, the NPAS2 SNP rs3768984 might be associated with a later chronotype and shorter sleep duration on work days that is not fully compensated for on free days, resulting in shorter average weekly sleep duration. Under this model, rs3768984 would not be associated with sleep duration in the elderly who do not typically have work days, and indeed, rs3768984 was not associated with TST in the MrOS or SOF cohorts. Alternatively, the NPAS2 SNP might regulate different sleep traits (sleep timing and sleep quantity) in different age groups, or the association might differ based on subjective and objective sleep measurements. Follow-up of the NPAS2 SNP with chronotype measures in young and old individuals should help to resolve these questions.
Replication will be critical to establish the validity and generalizability of the SNP associations discovered in the MrOS and SOF participants. However, the SNP associations discovered in the MrOS and SOF cohorts of elderly participants might not replicate in cohorts of younger participants if the SNP associations with sleep traits differ by age, i.e., interaction between SNPs and age. Age-dependent relationships between sleep traits and circadian rhythm markers and age-related changes in the circadian and sleep homeostatic processes make SNP interactions with age plausible. For instance, the strong relationship found in young adults between circadian period and morningness-eveningness and the related trait, self-selected wake times, was found to be much weaker in older adults.20 Thus, genetic variants affecting circadian period in the same manner across all age ranges could still be differentially associated with morningness-eveningness across age ranges, simply due to the nature of the age-dependent relationship between circadian period and morningness-eveningness. Other sleep traits that are related to circadian parameters in an age-dependent manner could also exhibit age-dependent associations with genetic variants. In addition, age-related changes in the circadian and sleep homeostatic processes are well documented. For instance, the amplitude of circadian gene expression and other circadian outputs decreases with age.25 Age-related decline of circadian rhythms potentially places the circadian clock on a threshold where subtle genetic variants that had little to no effect during the early years of life could have an effect during the later years of life. If age-specific genetic associations with sleep traits are identified, and the corresponding causal variants are discovered, the characterization of the effect of causal variants on the function of the molecular clock and the resultant circadian parameters could help to elucidate the fundamental clock properties that regulate sleep in different age groups. Our study represents a first step toward this goal, and further studies in cohorts composed of elderly and nonelderly participants will be needed to establish the existence of age-specific genetic associations with sleep traits.
The examination of SNP associations with multiple correlated actigraphic traits provided a more complete view of SNP associations compared with the conventional analysis of a single sleep trait. For instance, the consistency of SNP associations among correlated traits could be examined. Not surprisingly, SNP associations for correlated traits were similar, and this applied to all significance levels (nearly complete overlap of black and blue ellipses in Figure 4). In addition, we explored whether SNP associations were mediated by related sleep traits, e.g., whether ARNTL and NPAS2 SNP associations with later sleep timing were mediated by sleep latency. We also performed cluster analysis and found that genetic associations fell into four clusters that largely mirrored phenotypic correlations. The existence of four clusters of genetic associations supports the notion that rather than examining 12 independent traits, our study examined four groups of traits. Even if multiple test adjustment was based on the number of independent SNPs (310) and clusters of traits (four), the significance threshold would be 4 × 10-5, and many of the SNP associations reported here would remain significant.
One strength of this study is the use of objectively measured sleep and activity rhythm traits. A second strength is the examination of these traits in two cohorts composed of elderly individuals, an age group that experiences high rates of sleep disturbance. Sleep and activity rhythm traits have been associated with multiple adverse outcomes in the elderly, and thus are relevant to health in the elderly.45,78,79 The disadvantages of the candidate gene approach have been discussed previously.80 Briefly, candidate gene studies that were commonly performed prior to GWAS were plagued by the following problems: risk factors (candidate genes) chosen based on diverse considerations, studies were underpowered (especially considering what is now known about the effect size of common genetic variants), nominal significance thresholds of 0.05 were adopted, and confounding by population stratification was unaccounted for.80 Our study avoids many of the pitfalls that plagued previously performed candidate gene studies. The clock gene pathway is well established from work in multiple model organisms, our choice of clock genes was clearly documented, and we systematically surveyed common genetic variation in these genes rather than choosing individual variants to test within these genes. Our sample size was sufficient to achieve adequate power to observe small effect sizes (Table S7). Achieving adequate power also means that our reported effect sizes are less likely to be inflated.81 Multiple test correction was applied using modern techniques that take LD into account. We accounted for population stratification by restricting our analysis to self-identified Caucasian participants and by adjusting for genetic ancestry using components from multidimensional scaling analyses in our regression models. Candidate gene studies can also miss genetic associations because common genetic variation in the entire genome is not surveyed. Indeed, genetic associations outside of the candidate gene regions may have been missed in this study. Despite this limitation, we made the serendipitous discovery that SNPs near the candidate gene GNB3 reside on a large LD block on chromosome 12 that includes genes not previously known to be associated with sleep traits, which could eventually lead to the potential identification of novel sleep regulatory genes.
The identification of significant associations between objectively measured sleep and activity rhythm traits and SNPs in the highly conserved circadian rhythm gene pathway supports the notion that common genetic variation in these genes plays a role in sleep regulation in humans. SNPs at chromosome 12p13 were significantly associated with sleep continuity traits in both cohorts. Although associations between SNPs in the ARNTL/ NPAS2 genes and delayed sleep timing were limited to a single cohort, the statistical interaction between SNPs mirrored the known biochemical interaction between these proteins, suggesting that these SNPs or genetic variants in LD might be functionally relevant. The significance of NPAS2 and ARNTL SNP associations in the MrOS cohort but not the SOF cohort could indicate that these SNP associations are sex-specific. Alternatively, the lack of association significance in the SOF cohort could indicate a lack of replication in an independent cohort or it could be due to the smaller sample size in SOF. Replication studies using sex-stratified analysis can help to resolve this question. Our results highlight the relevance of common genetic variation in clock genes in the regulation of sleep traits in the elderly and could motivate the further exploration of genetic associations with objectively measured sleep traits in multiple human populations.
ACKNOWLEDGMENTS
The Osteoporotic Fractures in Men (MrOS) Study is supported by National Institutes of Health (NIH) funding. The following institutes provide support: the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), the National Institute on Aging (NIA), the National Center for Research Resources (NCRR), and NIH Roadmap for Medical Research under the following grant numbers: U01 AR45580, U01 AR45614, U01 AR45632, U01 AR45647, U01 AR45654, U01 AR45583, U01 AG18197, U01-AG027810, and UL1 RR024140. The National Heart, Lung, and Blood Institute
(NHLBI) provides funding for the MrOS Sleep ancillary study “Outcomes of Sleep Disorders in Older Men” under the following grant numbers: R01 HL071194, R01 HL070848, R01 HL070847, R01 HL070842, R01 HL070841, R01 HL070837, R01 HL070838, and R01 HL070839. NIAMS provides funding for the MrOS ancillary study “Replication of candidate gene associations and bone strength phenotype in MrOS” under the grant number R01-AR051124, which funded the creation and maintenance of MrOS DNA collections.
The Study of Osteoporotic Fractures (SOF) is supported by NIH funding. The NIA provides support under the following grant numbers: AG05407, AR35582, AG05394, AR35584, AR35583, R01 AG005407, R01 AG027576-22, 2 R01 AG005394-22A1, and 2 R01 AG027574-22A1 and AG026720.
Gregory J. Tranah, PhD was supported by NIA grant R01AG030474. We thank Dr. Dan Kripke for his guidance in the selection of circadian genes and variants for this study.
Footnotes
A commentary on this article appears in this issue on page 309.
DISCLOSURE STATEMENT
This was not an industry supported study. Dr. Redline has received grant support from ResMed Foundation to supplement data collection in a NIH study, CPAP units from Philips Respironics and ResMed Inc. for use in NIH funded studies. She has also received an honorarium from Johnson and Johnson to present at a scientific meeting. Dr. Ancoli Israel has served as a consultant or a member of the Scientific Advisory Board for the following: Astra-Zeneca, Merck, NeuroVigil, Inc., Pfizer, Philips, Purdue Pharma LP, Sanofi Aventis, and Somaxon. The other authors have indicated no financial conflicts of interest.
SUPPLEMENTAL MATERIAL
Number of genotyped SNPs passing quality control filters per gene
Genetic association with number of wake episodes at chromosome 12p13
Conditional analysis of rs1047776 and rs2238114
Genetic association with acrophase
RORC SNP with MAF ≤ 0.05 associated with sleep continuity traits
Replication analysis of CLOCK SNPs
Power to detect SNP associations in the present study
Linkage disequilibrium (LD) plot of single nucleotide polymorphisms (SNPs) at chromosome 12p13. LD plot generated using Haploview with data from CEU HapMap phase 2 and 3, release 28, Aug10, genome build 36, dbSNP b126 from chromosome 12, position 6,806,200 – 6,950,000. LD measured in r2 units. Haplotype blocks estimated using the confidence interval method.1
LRRC23 gene expression association and tissue distribution. (A) SNP rs2238114 association with LRRC23 gene expression in HapMap CEU lymphoblastoid cells. (B) Tissue distribution of LRRC23 gene expression.
Adjusted mean sleep timing related traits by NPAS2 rs3768984 genotype. Adjusted means (filled circles) with standard errors of the predicted means are shown. Sleep traits adjusted for the same covariates as regression models, as described in methods. (A) Sleep onset time. (B) Wake onset time. MrOS, Osteoporotic Fractures in Men; SOF, Study of Osteoporotic Fractures.
Hierarchical cluster dendrogram of genetic association results. Multiscale bootstrap resampling-based P values shown in red above edges. For clusters with P > 95, the hypothesis that the cluster does not exist is rejected with significance level 0.05. Red boxes mark clusters with multiscale bootstrap resampling- based P > 95. Height on the y-axis indicates the complete clustering method values. NWAK, number of awakenings; TST, total sleep time; WASO, wake after sleep onset.
SUPPLEMENTAL REFERENCE
- 1.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
REFERENCES
- 1.Dijk DJ, von Schantz M. Timing and consolidation of human sleep, wakefulness, and performance by a symphony of oscillators. J Biol Rhythms. 2005;20:279–90. doi: 10.1177/0748730405278292. [DOI] [PubMed] [Google Scholar]
- 2.Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005;437:1257–63. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- 3.Bliwise DL. Sleep in normal aging and dementia. Sleep. 1993;16:40–81. doi: 10.1093/sleep/16.1.40. [DOI] [PubMed] [Google Scholar]
- 4.Foley DJ, Monjan AA, Brown SL, Simonsick EM, Wallace RB, Blazer DG. Sleep complaints among elderly persons: an epidemiologic study of three communities. Sleep. 1995;18:425–32. doi: 10.1093/sleep/18.6.425. [DOI] [PubMed] [Google Scholar]
- 5.Walsleben JA, Kapur VK, Newman AB, et al. Sleep and reported daytime sleepiness in normal subjects: the Sleep Heart Health Study. Sleep. 2004;27:293–8. doi: 10.1093/sleep/27.2.293. [DOI] [PubMed] [Google Scholar]
- 6.Redline S, Kirchner HL, Quan SF, Gottlieb DJ, Kapur V, Newman A. The effects of age, sex, ethnicity, and sleep-disordered breathing on sleep architecture. Arch Intern Med. 2004;164:406–18. doi: 10.1001/archinte.164.4.406. [DOI] [PubMed] [Google Scholar]
- 7.Unruh ML, Redline S, An MW, et al. Subjective and objective sleep quality and aging in the sleep heart health study. J Am Geriatr Soc. 2008;56:1218–27. doi: 10.1111/j.1532-5415.2008.01755.x. [DOI] [PubMed] [Google Scholar]
- 8.Ohayon MM, Carskadon MA, Guilleminault C, Vitiello MV. Meta-analysis of quantitative sleep parameters from childhood to old age in healthy individuals: developing normative sleep values across the human lifespan. Sleep. 2004;27:1255–73. doi: 10.1093/sleep/27.7.1255. [DOI] [PubMed] [Google Scholar]
- 9.Van Cauter E, Leproult R, Plat L. Age-related changes in slow wave sleep and REM sleep and relationship with growth hormone and cortisol levels in healthy men. JAMA. 2000;284:861–8. doi: 10.1001/jama.284.7.861. [DOI] [PubMed] [Google Scholar]
- 10.Hoch CC, Dew MA, Reynolds CF, 3rd, et al. Longitudinal changes in diary- and laboratory-based sleep measures in healthy “old old” and “young old” subjects: a three-year follow-up. Sleep. 1997;20:192–202. doi: 10.1093/sleep/20.3.192. [DOI] [PubMed] [Google Scholar]
- 11.Carrier J, Monk TH, Buysse DJ, Kupfer DJ. Sleep and morningness-eveningness in the ‘middle’ years of life (20-59 y) J Sleep Res. 1997;6:230–7. doi: 10.1111/j.1365-2869.1997.00230.x. [DOI] [PubMed] [Google Scholar]
- 12.Czeisler CA, Dumont M, Duffy JF, et al. Association of sleep-wake habits in older people with changes in output of circadian pacemaker. Lancet. 1992;340:933–6. doi: 10.1016/0140-6736(92)92817-y. [DOI] [PubMed] [Google Scholar]
- 13.Duffy JF, Dijk DJ, Klerman EB, Czeisler CA. Later endogenous circadian temperature nadir relative to an earlier wake time in older people. Am J Physiol. 1998;275:R1478–87. doi: 10.1152/ajpregu.1998.275.5.r1478. [DOI] [PubMed] [Google Scholar]
- 14.Roenneberg T, Kuehnle T, Juda M, et al. Epidemiology of the human circadian clock. Sleep Med Rev. 2007;11:429–38. doi: 10.1016/j.smrv.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Monk TH. Aging human circadian rhythms: conventional wisdom may not always be right. J Biol Rhythms. 2005;20:366–74. doi: 10.1177/0748730405277378. [DOI] [PubMed] [Google Scholar]
- 16.Vitiello MV, Smallwood RG, Avery DH, Pascualy RA, Martin DC, Prinz PN. Circadian temperature rhythms in young adult and aged men. Neurobiol Aging. 1986;7:97–100. doi: 10.1016/0197-4580(86)90146-6. [DOI] [PubMed] [Google Scholar]
- 17.Weitzman ED, Moline ML, Czeisler CA, Zimmerman JC. Chronobiology of aging: temperature, sleep-wake rhythms and entrainment. Neurobiol Aging. 1982;3:299–309. doi: 10.1016/0197-4580(82)90018-5. [DOI] [PubMed] [Google Scholar]
- 18.Monk TH, Buysse DJ, Reynolds CF, 3rd, Kupfer DJ, Houck PR. Circadian temperature rhythms of older people. Exp Gerontol. 1995;30:455–74. doi: 10.1016/0531-5565(95)00007-4. [DOI] [PubMed] [Google Scholar]
- 19.Czeisler CA, Duffy JF, Shanahan TL, et al. Stability, precision, and near-24-hour period of the human circadian pacemaker. Science. 1999;284:2177–81. doi: 10.1126/science.284.5423.2177. [DOI] [PubMed] [Google Scholar]
- 20.Duffy JF, Czeisler CA. Age-related change in the relationship between circadian period, circadian phase, and diurnal preference in humans. Neurosci Lett. 2002;318:117–20. doi: 10.1016/s0304-3940(01)02427-2. [DOI] [PubMed] [Google Scholar]
- 21.Dijk DJ, Duffy JF, Czeisler CA. Contribution of circadian physiology and sleep homeostasis to age-related changes in human sleep. Chronobiol Int. 2000;17:285–311. doi: 10.1081/cbi-100101049. [DOI] [PubMed] [Google Scholar]
- 22.Dijk DJ, Duffy JF, Riel E, Shanahan TL, Czeisler CA. Ageing and the circadian and homeostatic regulation of human sleep during forced desynchrony of rest, melatonin and temperature rhythms. J Physiol. 1999;516:611–27. doi: 10.1111/j.1469-7793.1999.0611v.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haimov I, Lavie P. Circadian characteristics of sleep propensity function in healthy elderly: a comparison with young adults. Sleep. 1997;20:294–300. doi: 10.1093/sleep/20.4.294. [DOI] [PubMed] [Google Scholar]
- 24.Buysse DJ, Monk TH, Carrier J, Begley A. Circadian patterns of sleep, sleepiness, and performance in older and younger adults. Sleep. 2005;28:1365–76. doi: 10.1093/sleep/28.11.1365. [DOI] [PubMed] [Google Scholar]
- 25.Hofman MA, Swaab DF. Living by the clock: the circadian pacemaker in older people. Ageing Res Rev. 2006;5:33–51. doi: 10.1016/j.arr.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 26.Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Effects of aging on central and peripheral mammalian clocks. Proc Natl Acad Sci U S A. 2002;99:10801–6. doi: 10.1073/pnas.152318499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowrey PL, Takahashi JS. Genetics of circadian rhythms in mammalian model organisms. Adv Genet. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: an analog of clock operative in the mammalian forebrain. Science. 2001;293:506–9. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- 29.DeBruyne JP, Weaver DR, Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007;10:543–5. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–75. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi J, Wittke-Thompson JK, Badner JA, et al. Clock genes may influence bipolar disorder susceptibility and dysfunctional circadian rhythm. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1047–55. doi: 10.1002/ajmg.b.30714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allebrandt KV, Teder-Laving M, Akyol M, et al. CLOCK gene variants associate with sleep duration in two independent populations. Biol Psychiatry. 2010;67:1040–7. doi: 10.1016/j.biopsych.2009.12.026. [DOI] [PubMed] [Google Scholar]
- 33.Johansson C, Willeit M, Smedh C, et al. Circadian clock-related polymorphisms in seasonal affective disorder and their relevance to diurnal preference. Neuropsychopharmacology. 2003;28:734–9. doi: 10.1038/sj.npp.1300121. [DOI] [PubMed] [Google Scholar]
- 34.He Y, Jones CR, Fujiki N, et al. The transcriptional repressor DEC2 regulates sleep length in mammals. Science. 2009;325:866–70. doi: 10.1126/science.1174443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Archer SN, Robilliard DL, Skene DJ, et al. A length polymorphism in the circadian clock gene Per3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep. 2003;26:413–5. doi: 10.1093/sleep/26.4.413. [DOI] [PubMed] [Google Scholar]
- 36.Katzenberg D, Young T, Finn L, et al. A CLOCK polymorphism associated with human diurnal preference. Sleep. 1998;21:569–76. doi: 10.1093/sleep/21.6.569. [DOI] [PubMed] [Google Scholar]
- 37.Partonen T, Treutlein J, Alpman A, et al. Three circadian clock genes Per2, Arntl, and Npas2 contribute to winter depression. Ann Med. 2007;39:229–38. doi: 10.1080/07853890701278795. [DOI] [PubMed] [Google Scholar]
- 38.Kovanen L, Saarikoski ST, Aromaa A, Lonnqvist J, Partonen T. ARNTL (BMAL1) and NPAS2 gene variants contribute to fertility and seasonality. PLoS One. 2010;5:e10007. doi: 10.1371/journal.pone.0010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kripke DF, Shadan FF, Dawson A, et al. Genotyping sleep disorders patients. Psychiatry Investig. 2010;7:36–42. doi: 10.4306/pi.2010.7.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Utge SJ, Soronen P, Loukola A, et al. Systematic analysis of circadian genes in a population-based sample reveals association of TIMELESS with depression and sleep disturbance. PLoS One. 2010;5:e9259. doi: 10.1371/journal.pone.0009259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allebrandt KV, Amin N, Muller-Myhsok B, et al. A K(ATP) channel gene effect on sleep duration: from genome-wide association studies to function in Drosophila. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.142. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 42.Blank JB, Cawthon PM, Carrion-Petersen ML, et al. Overview of recruitment for the osteoporotic fractures in men study (MrOS) Contemp Clin Trials. 2005;26:557–68. doi: 10.1016/j.cct.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 43.Orwoll E, Blank JB, Barrett-Connor E, et al. Design and baseline characteristics of the osteoporotic fractures in men (MrOS) study--a large observational study of the determinants of fracture in older men. Contemp Clin Trials. 2005;26:569–85. doi: 10.1016/j.cct.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 44.Cummings SR, Nevitt MC, Browner WS, et al. Risk factors for hip fracture in white women. Study of Osteoporotic Fractures Research Group. N Engl J Med. 1995;332:767–73. doi: 10.1056/NEJM199503233321202. [DOI] [PubMed] [Google Scholar]
- 45.Tranah GJ, Blackwell T, Ancoli-Israel S, et al. Circadian activity rhythms and mortality: the study of osteoporotic fractures. J Am Geriatr Soc. 2010;58:282–91. doi: 10.1111/j.1532-5415.2009.02674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehra R, Stone KL, Blackwell T, et al. Prevalence and correlates of sleep-disordered breathing in older men: osteoporotic fractures in men sleep study. J Am Geriatr Soc. 2007;55:1356–64. doi: 10.1111/j.1532-5415.2007.01290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blackwell T, Redline S, Ancoli-Israel S, et al. Comparison of sleep parameters from actigraphy and polysomnography in older women: the SOF study. Sleep. 2008;31:283–91. doi: 10.1093/sleep/31.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blackwell T, Ancoli-Israel S, Redline S, Stone KL. Factors that may influence the classification of sleep-wake by wrist actigraphy: the MrOS Sleep Study. J Clin Sleep Med. 2011;7:357–67. doi: 10.5664/JCSM.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marler MR, Gehrman P, Martin JL, Ancoli-Israel S. The sigmoidally transformed cosine curve: a mathematical model for circadian rhythms with symmetric non-sinusoidal shapes. Stat Med. 2006;25:3893–904. doi: 10.1002/sim.2466. [DOI] [PubMed] [Google Scholar]
- 50.Pahor M, Chrischilles EA, Guralnik JM, Brown SL, Wallace RB, Carbonin P. Drug data coding and analysis in epidemiologic studies. Eur J Epidemiol. 1994;10:405–11. doi: 10.1007/BF01719664. [DOI] [PubMed] [Google Scholar]
- 51.Zhang J, Fang Z, Jud C, et al. Fragile X-related proteins regulate mammalian circadian behavioral rhythms. Am J Hum Genet. 2008;83:43–52. doi: 10.1016/j.ajhg.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee HJ, Sung SM, Han CS, et al. G-protein beta3 subunit C825T polymorphism tends to be associated with seasonal variation in young male college students. Neuropsychobiology. 2005;52:135–9. doi: 10.1159/000087559. [DOI] [PubMed] [Google Scholar]
- 53.Johansson C, Willeit M, Aron L, et al. Seasonal affective disorder and the G-protein beta-3-subunit C825T polymorphism. Biol Psychiatry. 2004;55:317–9. doi: 10.1016/s0006-3223(03)00640-1. [DOI] [PubMed] [Google Scholar]
- 54.Lee HJ, Paik JW, Kang SG, Lim SW, Kim L. Allelic variants interaction of CLOCK gene and G-protein beta3 subunit gene with diurnal preference. Chronobiol Int. 2007;24:589–97. doi: 10.1080/07420520701534632. [DOI] [PubMed] [Google Scholar]
- 55.Kripke DF, Nievergelt CM, Joo E, Shekhtman T, Kelsoe JR. Circadian polymorphisms associated with affective disorders. J Circadian Rhythms. 2009;7:2. doi: 10.1186/1740-3391-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–81. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 57.Barnes JW, Tischkau SA, Barnes JA, et al. Requirement of mammalian Timeless for circadian rhythmicity. Science. 2003;302:439–42. doi: 10.1126/science.1086593. [DOI] [PubMed] [Google Scholar]
- 58.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 59.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–93. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–9. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–7. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 63.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gauderman W, Morrison J. QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. [Accessed October 8, 2012]. http://hydra.usc.edu/gxe.
- 65.Su AI, Wiltshire T, Batalov S, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu C, Orozco C, Boyer J, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang TP, Beazley C, Montgomery SB, et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. 2010;26:2474–6. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stranger BE, Montgomery SB, Dimas AS, et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van den Berg JF, Miedema HM, Tulen JH, Hofman A, Neven AK, Tie-meier H. Sex differences in subjective and actigraphic sleep measures: a population-based study of elderly persons. Sleep. 2009;32:1367–75. doi: 10.1093/sleep/32.10.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siffert W, Rosskopf D, Siffert G, et al. Association of a human G-protein beta3 subunit variant with hypertension. Nat Genet. 1998;18:45–8. doi: 10.1038/ng0198-45. [DOI] [PubMed] [Google Scholar]
- 71.Brand E, Wang JG, Herrmann SM, Staessen JA. An epidemiological study of blood pressure and metabolic phenotypes in relation to the Gbeta3 C825T polymorphism. J Hypertens. 2003;21:729–37. doi: 10.1097/00004872-200304000-00016. [DOI] [PubMed] [Google Scholar]
- 72.Keers R, Bonvicini C, Scassellati C, et al. Variation in GNB3 predicts response and adverse reactions to antidepressants. J Psychopharmacol. 2011;25:867–74. doi: 10.1177/0269881110376683. [DOI] [PubMed] [Google Scholar]
- 73.Klenke S, Kussmann M, Siffert W. The GNB3 C825T polymorphism as a pharmacogenetic marker in the treatment of hypertension, obesity, and depression. Pharmacogenet Genomics. 2011;21:594–606. doi: 10.1097/FPC.0b013e3283491153. [DOI] [PubMed] [Google Scholar]
- 74.Dayal S, Sparks A, Jacob J, Allende-Vega N, Lane DP, Saville MK. Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J Biol Chem. 2009;284:5030–41. doi: 10.1074/jbc.M805871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ng AC, Eisenberg JM, Heath RJ, et al. Human leucine-rich repeat proteins: a genome-wide bioinformatic categorization and functional analysis in innate immunity. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4631–8. doi: 10.1073/pnas.1000093107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dudley CA, Erbel-Sieler C, Estill SJ, et al. Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science. 2003;301:379–83. doi: 10.1126/science.1082795. [DOI] [PubMed] [Google Scholar]
- 77.Franken P, Dudley CA, Estill SJ, et al. NPAS2 as a transcriptional regulator of non-rapid eye movement sleep: genotype and sex interactions. Proc Natl Acad Sci U S A. 2006;103:7118–23. doi: 10.1073/pnas.0602006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Paudel ML, Taylor BC, Ancoli-Israel S, et al. Rest/activity rhythms and mortality rates in older men: MrOS Sleep Study. Chronobiol Int. 2010;27:363–77. doi: 10.3109/07420520903419157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paudel ML, Taylor BC, Ancoli-Israel S, et al. Rest/activity rhythms and cardiovascular disease in older men. Chronobiol Int. 2011;28:258–66. doi: 10.3109/07420528.2011.553016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ioannidis JP, Tarone R, McLaughlin JK. The false-positive to false-negative ratio in epidemiologic studies. Epidemiology. 2011;22:450–6. doi: 10.1097/EDE.0b013e31821b506e. [DOI] [PubMed] [Google Scholar]
- 81.Ioannidis JP. Why most discovered true associations are inflated. Epidemiology. 2008;19:640–8. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Number of genotyped SNPs passing quality control filters per gene
Genetic association with number of wake episodes at chromosome 12p13
Conditional analysis of rs1047776 and rs2238114
Genetic association with acrophase
RORC SNP with MAF ≤ 0.05 associated with sleep continuity traits
Replication analysis of CLOCK SNPs
Power to detect SNP associations in the present study
Linkage disequilibrium (LD) plot of single nucleotide polymorphisms (SNPs) at chromosome 12p13. LD plot generated using Haploview with data from CEU HapMap phase 2 and 3, release 28, Aug10, genome build 36, dbSNP b126 from chromosome 12, position 6,806,200 – 6,950,000. LD measured in r2 units. Haplotype blocks estimated using the confidence interval method.1
LRRC23 gene expression association and tissue distribution. (A) SNP rs2238114 association with LRRC23 gene expression in HapMap CEU lymphoblastoid cells. (B) Tissue distribution of LRRC23 gene expression.
Adjusted mean sleep timing related traits by NPAS2 rs3768984 genotype. Adjusted means (filled circles) with standard errors of the predicted means are shown. Sleep traits adjusted for the same covariates as regression models, as described in methods. (A) Sleep onset time. (B) Wake onset time. MrOS, Osteoporotic Fractures in Men; SOF, Study of Osteoporotic Fractures.
Hierarchical cluster dendrogram of genetic association results. Multiscale bootstrap resampling-based P values shown in red above edges. For clusters with P > 95, the hypothesis that the cluster does not exist is rejected with significance level 0.05. Red boxes mark clusters with multiscale bootstrap resampling- based P > 95. Height on the y-axis indicates the complete clustering method values. NWAK, number of awakenings; TST, total sleep time; WASO, wake after sleep onset.