The Rilp-like proteins Rilpl1 and Rilpl2 are novel centrosomal and ciliary proteins. Depletion of Rilp-like proteins leads to the accumulation of signaling proteins in the cilium and disruption of epithelial cell organization, suggesting that Rilpl1 and Rilpl2 regulate ciliary membrane content by promoting protein removal.
Abstract
The primary cilium is a microtubule-based structure found in most cell types in mammals. Disruption of cilium function causes a diverse set of human diseases collectively known as ciliopathies. We report that Rab effector–related proteins Rab-interacting lysosomal protein-like 1 (Rilpl1) and Rilpl2 regulate protein localization in the primary cilium. Rilpl2 was initially identified as up-regulated in ciliating mouse tracheal epithelial cells. Rilpl1 and Rilpl2 both localize to the primary cilium and centrosome, Rilpl1 specifically to the distal end of the mother centriole. Live-cell microscopy reveals that Rilpl2 primary cilium localization is dynamic and that it is associated with tubulovesicular structures at the base of the cilium. Depletion of Rilpl1 and Rilpl2 results in accumulation of signaling proteins in the ciliary membrane and prevents proper epithelial cell organization in three-dimensional culture. These data suggest that Rilp-like proteins function in regulation of ciliary membrane protein concentration by promoting protein removal from the primary cilium.
INTRODUCTION
Most mammalian cell types have a single, sensory primary cilium, whereas some specialized cell types have one or more motile cilia. In all cases, each cilium is nucleated by a centriole, the microtubule structure at the core of the centrosome. Primary cilia are important sensory organelles with functions ranging from mechanosensation and osmosensation to Hedgehog and somatostatin pathway signaling (Pazour and Witman, 2003; Berbari et al., 2009; Oh and Katsanis, 2012). Disruption of cilium function causes dramatic and diverse phenotypes, and human diseases resulting from primary cilium defects are collectively termed ciliopathies. Common features of the ciliopathies include neurological defects, retinal degeneration, obesity, polydactyly, and polycystic kidneys (Hildebrandt et al., 2011).
The importance of cilia in normal development is clear, and many ciliary proteins have been identified, but the mechanisms regulating the localization of signaling proteins to the cilium are not well understood. Many of the signaling events take place at the ciliary membrane, and the content of the ciliary membrane could be regulated at the points of entry, retention, and exit. Entry of membrane proteins into the cilium is regulated by three mechanisms. First, some ciliary membrane proteins such as fibrocystin, somatostatin, and rhodopsin have ciliary targeting sequences that selectively target them to the cilium (Tam et al., 2000; Berbari et al., 2008; Follit et al., 2010). Second, nonciliary membrane proteins are kept out and ciliary membrane proteins in by three diffusion-barrier or gatekeeper complexes: NPHP1, 4, and 8; B9/MKS-JBTS; and septins (Delous et al., 2009; Hu et al., 2010; Otto et al., 2010; Chih et al., 2011; Garcia-Gonzalo et al., 2011; Sang et al., 2011). Third, some proteins are prevented from entering the cilium by linkage to the actin cytoskeleton (Francis et al., 2011).
Once inside the cilium, membrane proteins are guided along the ciliary axoneme by association with the intraflagellar transport complexes IFT-A and IFT-B (Pedersen and Rosenbaum, 2008). This linkage to the microtubule axoneme may also act to selectively retain specific membrane proteins in the cilium (Francis et al., 2011). Removal of membrane proteins from the cilium is likely controlled by endocytosis, as suggested by the association of clathrin with the base of cilia (Molla-Herman et al., 2010; Kaplan et al., 2012). Understanding the contributions of these mechanisms and the proteins involved at each step is critical to understanding regulation of cilium function.
We previously reported on the mouse tracheal epithelial cell (MTEC) culture system as a means to identify and study components of centrosome and cilium formation and function (Vladar and Stearns, 2007; Hoh et al., 2012). Here we report on Rab-interacting lysosomal protein-like 2 (Rilpl2), which is up-regulated during differentiation of multi-ciliated cells, and its related protein Rab-interacting lysosomal protein-like 1 (Rilpl1). Rilpl1 and Rilpl2 are related to a known regulator of membrane traffic, Rab-interacting lysosomal protein (Rilp), a Rab7/Rab34 effector that interacts with dynein–dynactin to direct movement of late endosomes and lysosomes along microtubules (Johansson et al., 2007). Rilpl1 and Rilpl2 do not affect lysosomal trafficking but may also be Rab effectors, as both interact with activated Rab34 and Rab36, and Rilpl1 in addition interacts with Rab12 and Rab40B (Wang et al., 2004; Fukuda et al., 2008; Matsui et al., 2012). Although there is no reported function for Rilpl1, Rilpl2 affects membrane and cytoskeletal dynamics in dendritic spines of hippocampal neurons through an interaction with MyoVa (Lisé et al., 2009).
Here we identify Rilpl1 and Rilpl2 as novel centrosomal and ciliary proteins. Rilpl1 especially localizes to the distal end of the mother centriole of nonciliated cells, and both proteins localize to the primary cilium. Depletion of Rilpl1 and Rilpl2 leads to accumulation of signaling proteins in the cilium and prevents proper epithelial cell organization in three-dimensional culture. These results suggest that the Rilp-like proteins Rilpl1 and Rilpl2 regulate the protein content of the primary cilium.
RESULTS
Rilpl2 is up-regulated during differentiation of multiciliated cells
To identify novel proteins involved in ciliary trafficking, we used a previously described in vitro system recapitulating differentiation of MTECs (You et al., 2002; Vladar and Stearns, 2007). This MTEC culture system was used to profile genes specifically up-regulated in multiciliated cells versus neighboring nonmulticiliated cells (Mahjoub et al., 2010; Hoh et al., 2012). We identified Rilpl2 as up-regulated 9.7-fold (p < 0.05) during the early stage of multiciliated cell differentiation but not significantly up-regulated during the later stage of differentiation. Murine Rilpl2 is 197 amino acids (aa) with two predicted coiled-coil domains (aa 62–95 and 125–149; Marcoil). Rilpl2 is a member of a three-protein family in vertebrates defined by two regions of high sequence similarity, the RH1 (aa 31–66) and RH2 (aa 122–148) domains (Wang et al., 2004; Figure 1A). The related protein Rilpl1 was not significantly up-regulated in the ciliated cell transcriptome at either the early or late time points. The remaining family member, Rilp, was not present on the microarray used, and thus its relative expression is unknown.
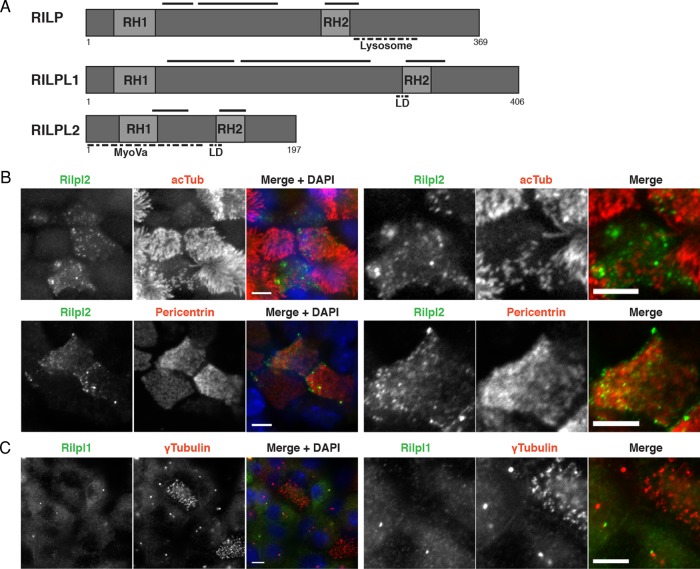
FIGURE 1:
Rilpl2 is up-regulated during ciliogenesis. (A) Schematic diagram comparing Rab-interacting lysosomal protein family members. RH1 and RH2 domains are regions of high sequence similarity. Solid lines represent predicted coiled-coil domains. Dashed lines mark the region specifying the lysosomal function of Rilp, the region of Rilpl2 that interacts with MyoVa, and the region specific to Rilpl1 and Rilpl2 (LD). (B) Immunofluorescence images of the apical surface of MTEC cultures labeled with antibodies for Rilpl2 (green) and the centrosome and cilium markers acTub or pericentrin (red). Images to the right are zoomed in on Rilpl2-expressing cells. Bars, 10 μm. (C) Immunofluorescence image of the apical surface of an MTEC culture (ALI >12 d) labeled with antibodies for Rilpl1 (green) and γ-tubulin (red). Images to the right are zoomed in on Rilpl1-expressing cells. Bar, 10 μm.
To confirm microarray expression data at the protein level, we analyzed Rilp-like protein expression in MTEC cultures by immunofluorescence. We generated a polyclonal rabbit antibody directed against full-length murine Rilpl2 protein. The antibody recognizes Rilpl1 and Rilpl2 by Western blot and Rilpl2 alone by immunofluorescence (Supplemental Figure S1, B and D). This antibody was used to detect endogenous Rilpl2 expression in differentiating MTEC cultures. These cultures contain multiciliated cells at different stages of differentiation, from basal body amplification to maturely ciliated, as well as cells of nonmulticiliated cell lineages (Figure 1, B and C, pericentrin, acTub, and γ-tubulin). Endogenous Rilpl2 expression is observed specifically at the apical surface of a subset of cells in MTEC cultures (Figure 1B). The cells expressing Rilpl2 also have multiple basal bodies (Figure 1B, pericentrin) and multiple cilia (Figure 1B, acTub), suggesting that Rilpl2 is up-regulated in cells of the multiciliated cell lineage. Of interest, Rilpl2 expression is detected in multiciliated cells with clustered basal bodies and fewer, shorter cilia (Figure 1B, right). These features are characteristic of immature multiciliated cells (Vladar and Stearns, 2007). Therefore these data are consistent with the microarray expression data that Rilpl2 is specifically up-regulated in the early stages of multiciliated cell differentiation but not in maturely ciliated cells. In contrast, Rilpl1 is not up-regulated in multiciliated cells but is found at one of the two γ-tubulin foci per cell in neighboring nonmulticiliated cells, as assessed with a rabbit polyclonal antibody directed against a sequence unique to Rilpl1 (Figure 1C and Supplemental Figure S1, A and C). These data are consistent with the microarray expression data and also suggest that Rilpl1 has a centrosomal or ciliary function in nonmulticiliated cells.
Rilp-like proteins localize to the centrosome and primary cilium
To investigate centrosomal and ciliary functions of Rilpl1 and Rilpl2, we examined their localization in NIH 3T3 and IMCD3 cells, two mouse cell lines that form a primary cilium under serum-starvation conditions. Both Rilpl1 and Rilpl2 localized to primary cilia, as marked by polyglutamylated tubulin labeling of the axoneme (Figure 2, A and B). In nonciliated cells, Rilpl2 also localized to the centrosome in a small fraction of cells (<5%). Rilpl2 localization was not specific for a particular subdomain of the centrosome (Figure 2A). In agreement with localization in MTEC cultures, Rilpl1 localized to one of two γ-tubulin foci in NIH 3T3 cells (Figure 2B). We also generated IMCD3 cell lines stably expressing Rilpl1 or Rilpl2 with a localization and affinity purification (LAP) tag. Both proteins localized to the centrosome and cilium at similar frequencies to endogenous protein, with additional cytoplasmic localization (Figure 2C). Further, endogenous Rilpl1 colocalized with Rilpl2-LAP to primary cilia (Figure 2D). In contrast to the localization of Rilpl1 and Rilpl2, green fluorescent protein (GFP)–tagged Rilp did not localize to cilia or centrosomes but to perinuclear lysosomal structures, as previously reported (Cantalupo et al., 2001; Supplemental Figures S1, C and D, and 3F). These data suggest that Rilpl1 and Rilpl2 function, at least in part, at the centrosome and cilium.
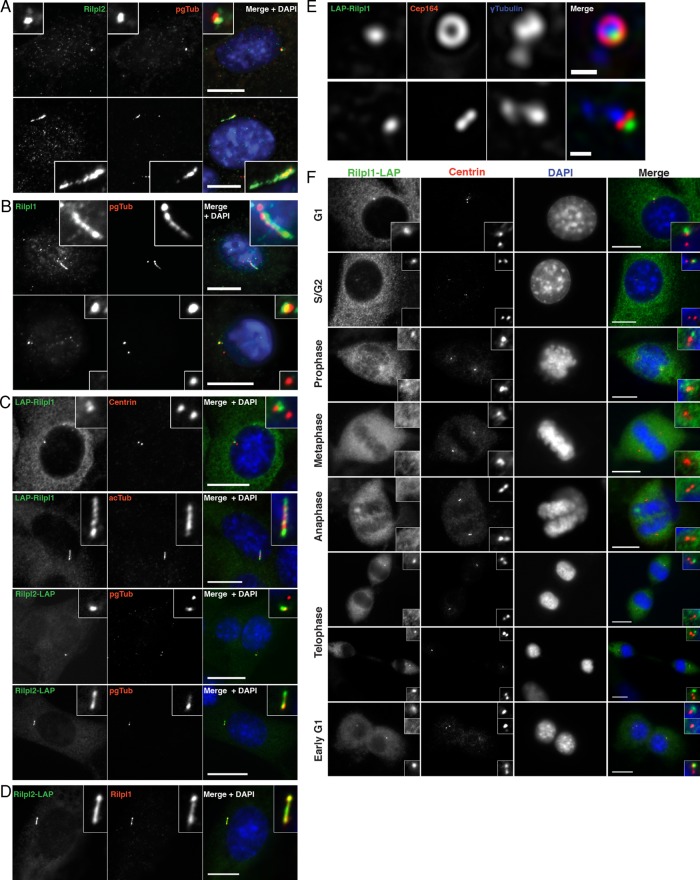
FIGURE 2:
Rilp-like proteins localize to the centrosome and primary cilium. (A) IMCD3 cells were fixed and stained for endogenous Rilpl2 (green), polyglutamylated tubulin (pgTub, red), and DNA (DAPI, blue). Bottom, cells were serum starved. Bars, 10 μm. (B) NIH 3T3 cells were fixed and stained for endogenous Rilpl1 (green), pgTub (red), and DNA (DAPI, blue). Top, cells were serum starved. Bars, 10 μm. Note that Rilpl1 localizes to one of the two γ-tubulin spots. (C) Serum-starved IMCD3 cells stably expressing LAP-Rilpl1 or Rilpl2-LAP were fixed and stained for GFP (Rilpl1/Rilpl2, green), centrioles or cilia (centrin, acTub, pgTub; red), and DNA (DAPI, blue). Bars, 10 μm. (D) IMCD3 FlpIN cells stably expressing Rilpl2-LAP were serum starved, fixed, and stained for GFP (Rilpl2, green), endogenous Rilpl1 (red), and DNA (DAPI, blue). Bar, 10 μm. (E) N2A cells were transfected with LAP-Rilpl1, fixed, and stained for LAP-Rilpl1 (GFP, green), γ-tubulin (blue), and the distal appendage protein Cep164 (red). Images are maximum projections of image stacks obtained by deconvolution microscopy. LAP-Rilpl1 localizes to the distal end of the mother centriole. Bars, 0.5 μm. (F) An asynchronous population of NIH 3T3 cells expressing Rilpl1-LAP was fixed and stained for Rilpl1-LAP (GFP, green), centrin (red), and DNA (DAPI, blue). Rilpl1 is lost at the mother centriole in early mitosis, but asymmetric distribution returns at telophase. Bars, 10 μm. Insets are enlarged images of centrosome/cilium regions.
Rilpl1 specifically localizes to the mother centriole
The localization of Rilpl1 to only one of two γ-tubulin foci led us to investigate the nature of this asymmetric localization. In ciliating cells, the mother centriole becomes the basal body that nucleates the primary cilium. Appendage proteins located at the distal end of the mother centriole are important for cilium formation and function. To determine the suborganelle localization of Rilpl1 at the centrosome, we expressed LAP-Rilpl1 in mouse N2A cells, which were costained for γ-tubulin and Cep164, a distal appendage protein. Deconvolution microscopy showed that Rilpl1 localized to the mother centriole, marked by Cep164 (Figure 2E). Further, it was distal to Cep164 along the centriole, based on the relationship to γ-tubulin. These data demonstrate that Rilpl1 is a distal, mother centriole–specific protein.
The primary cilium forms in G1 of the cell cycle, is disassembled before mitosis, and re-forms in G1 (Ishikawa and Marshall, 2011). The timing of Rilpl1 localization to the mother centriole was examined during cell cycle progression of Rilpl1-LAP NIH 3T3 cells (Figure 2F). During G1, Rilpl1 localizes to a focus at one centriole in ∼70% of cells. After centriole duplication, S/G2, Rilpl1 localizes to only one of four centrioles. At prophase, Rilpl1 remains localized to only one centriole. By metaphase and through anaphase, Rilpl1 no longer localizes to either centrosome at the spindle poles. At telophase/cytokinesis, Rilpl1 is in one focus at the centrosomes of each new daughter cell. This demonstrates that Rilpl1 localization to the mother centriole is cell cycle dependent, and the timing of its localization is consistent with a role in cilium formation or function.
The related C-terminal domains of Rilpl1 and Rilpl2 are necessary and sufficient for centrosomal localization
To identify the region of the Rilp-like proteins responsible for localization to the centrosome, we generated constructs in which either the C-terminus (ΔC) or N-terminus (ΔN) of each protein was deleted. The Rilpl2 constructs were identical to those used in a previous study (ΔC, aa 1–113; ΔN, aa 114–197; Lisé et al., 2009). Sequence alignment was used to divide Rilpl1 at positions equivalent to those in Rilpl2 (ΔC, aa 1–288; ΔN, aa 289–406). The ΔC construct of both proteins contains the RH1 domain and in Rilpl2 the MyoVa interaction domain (Lisé et al., 2009). The ΔN construct of both proteins contains the RH2 domain and the Rab36 interaction domain (Figure 1A; Matsui et al., 2012). GFP-tagged deletion constructs were expressed in NIH 3T3 and IMCD3 cells and assessed for localization. ΔC-Rilpl1 and ΔC-Rilpl2 were both diffusely cytoplasmic, and ΔC-Rilpl2 also localized to the nucleus. In contrast, both ΔN-Rilpl1 and ΔN-Rilpl2 localized to the centrosome (Figure 3, A and B). Very few cells expressing ΔN-Rilpl1 or ΔN-Rilpl2 had cilia (see later discussion), but both proteins were seen in the primary cilium (Figure 3, A and B). Thus the C-termini of Rilpl1 and Rilpl2 are necessary and sufficient for centrosomal and ciliary localization.
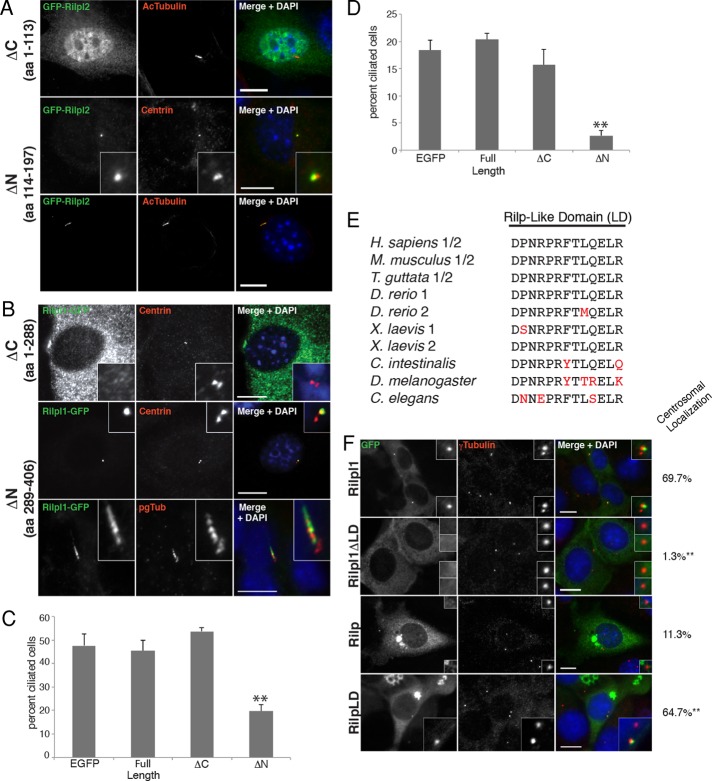
FIGURE 3:
The C-terminal regions of Rilpl1 and Rilpl2 are necessary and sufficient for centrosomal localization. (A) NIH 3T3 cells were transfected with GFP-ΔC-Rilpl2 or GFP-ΔN-Rilpl2, fixed, and stained for GFP (green), acTub or centrin (red), and DNA (DAPI, blue). Top and bottom, cells were serum starved. ΔN-Rilpl2 is sufficient for centrosomal and ciliary localization. Bars, 10 μm. (B) NIH 3T3 (top and middle) or IMCD3 (bottom) cells were transfected with ΔC-Rilpl1-GFP or ΔN-Rilpl1-GFP, serum starved, fixed, and stained for GFP (green), centrin/pgTub (red), and DNA (DAPI, blue). ΔN-Rilpl1 is sufficient centrosomal and ciliary localization. Bars, 10 μm. (C) Cilium formation in IMCD3 cells transfected with GFP, Rilpl2-GFP (full-length), GFP-ΔC-Rilpl2, or GFP-ΔN-Rilpl2. Results shown are the mean of three independent experiments ± SEM (100 cells/experiment; **p < 0.01). (D) Cilium formation in IMCD3 cells transfected with GFP, Rilpl1-GFP (full-length), ΔC-Rilpl1-GFP, or ΔN-Rilpl1-GFP. Results shown are the mean of three independent experiments ± SEM (100 cells/experiment; **p < 0.01). (E) Amino acid sequence alignment of the 13–amino acid Rilp-like domain (LD) of Rilpl1 and Rilpl2 from several vertebrates and the Rilp homologue of invertebrates. Amino acid changes are noted in red. (F) IMCD3 cells were transfected with GFP-tagged Rilpl1, Rilpl1-deltaLD, Rilp, or Rilp-LD, fixed, and stained for GFP (green), γ-tubulin (red), and DNA (DAPI, blue). Bars, 10 μm. Insets are enlarged images of the centrosomal/ciliary regions. The average percentage of transfected cells with centrosomal localization of GFP-tagged protein from three independent experiments is noted to the right (100 cells/experiment; **p < 0.01, t test).
Cells expressing the Rilpl1- and Rilpl2-deletion constructs were also assessed for phenotypes associated with overexpression. IMCD3 cells expressing ΔN-Rilpl2 had a lower frequency of ciliation compared with controls (Figure 3, A and C): 19.7 ± 4.7% of ΔN-Rilpl2–expressing cells were ciliated compared with 45.3 ± 8.0% of cells expressing full length Rilpl2 or 47.7 ± 8.4% of cells expressing GFP (Figure 3C). IMCD3 cells expressing ΔN-Rilpl1 also had a lower frequency of ciliation compared with controls: 2.7 ± 1.5% of ΔN-Rilpl1–expressing cells were ciliated compared with 20.3 ± 2.1% of cells expressing full-length Rilpl1 or 18.3 ± 3.2% of cells expressing GFP (Figure 3D). These data suggest that the Rilp-like proteins have a ciliary function and that ΔN-Rilpl1 and ΔN-Rilpl2 may dominantly interfere with a function critical to cilium formation.
In addition to the RH1 and RH2 domains shared among the Rilp-family proteins, Rilpl1 and Rilpl2 contain a third region of high sequence homology that is absent from Rilp. This 13–amino acid sequence is directly upstream of the RH2 domain (Rilpl1, aa 291–303; Rilpl2, aa 116–128; Figure 1A). The amino acid sequence is highly conserved between Rilpl1 and Rilpl2 in vertebrates (Figure 3E). To determine whether this conserved 13–amino acid Rilp-like domain (LD) is important for directing centrosomal localization, we removed the LD from Rilpl1 and inserted it into an equivalent position in Rilp. These GFP-tagged domain-swap constructs were expressed in IMCD3 cells (Figure 3F). Centrosome localization was scored as positive if GFP-tagged protein was within one γ-tubulin radius of the centrosome. As previously observed, Rilpl1-GFP localized to the centrosome in 69.7 ± 10.5% of transfected cells. In contrast, Rilpl1-ΔLD localized with the centrosome in only 1.3 ± 0.6% of transfected cells, indicating that the LD sequence is necessary for centrosome localization (p < 0.01, t test). In the case of Rilp-LD, overexpression resulted in the aggregation of lysosomes in the perinuclear region, as previously reported for Rilp (Cantalupo et al., 2001) and consistent with retention of the 62–amino acid domain responsible for the lysosomal function of Rilp in the Rilp-LD construct (Wang et al., 2004). To assess centrosomal localization independent of lysosome aggregation, we only evaluated localization in Rilp-GFP and Rilp-LD-GFP cells in which the large lysosome cluster was well-separated from the centrosome. Rilp-GFP localized with the centrosome in 11.3 ± 3.5% of transfected cells. In contrast, the Rilp-LD protein localized with the centrosome in 64.7 ± 7.5% of cells (Figure 3F). These data suggest that the LD sequence is sufficient for centrosome localization in the context of the Rilp protein backbone (p < 0.01, t test) but is not dominant over the lysosomal function of Rilp.
Rilpl2 ciliary localization is dynamic
Localization of Rilpl1 and Rilpl2 to only a subset of primary cilia suggested that they are not structural components of cilia but may be transiently localized as a part of their function. To determine the dynamics of ciliary localization of Rilp-like proteins, we assessed the localization of Rilpl2-LAP in live IMCD3 cells also expressing tdTomato-Inversin, a marker of the proximal end of the cilium. Cells were subconfluent for optimal imaging and serum starved to enhance ciliogenesis. At each time point, images in several focal planes were acquired, and z-slices with the cilium in focus were assembled into a time-lapse movie. Figure 4A (Supplemental Movie S1) shows a single primary cilium imaged over 90 min. Rilpl2-LAP is initially absent from the cilium, and then enters, exits, and reenters at 44 min and remains until the end of imaging.
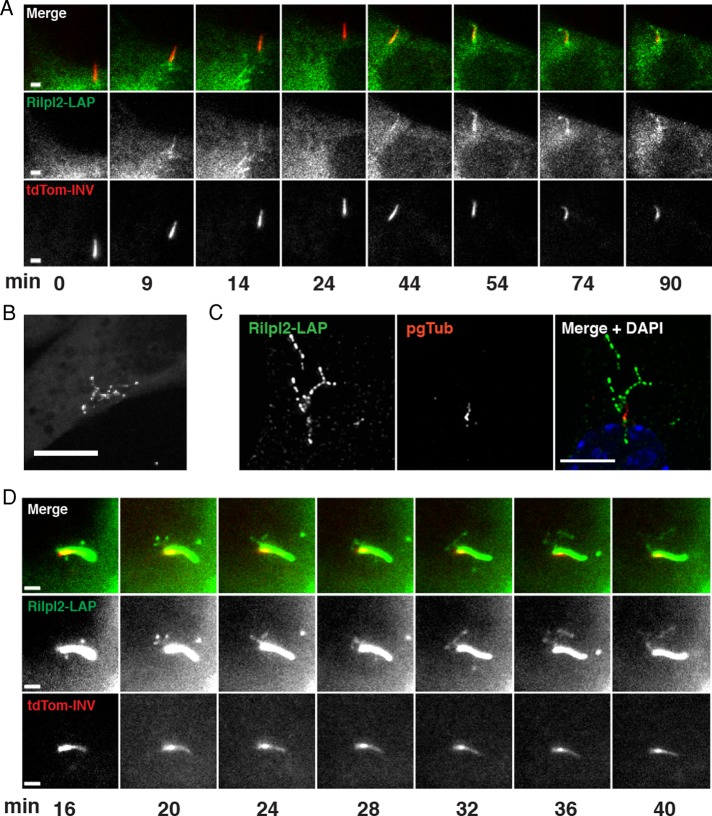
FIGURE 4:
Rilpl2 ciliary localization is dynamic. (A) IMCD3 cells expressing Rilpl2-LAP (green) and tdTom-Inv (proximal cilium, red) were imaged for 90 min under serum starvation conditions. Images shown were taken at the marked time points after the start of imaging, keeping the primary cilium in focus. Note the absence of Rilpl2-LAP from the cilium at t = 0, its appearance at t = 9 min, and its presence from t = 44 min until the end of imaging (t = 90 min). Still frames from Supplemental Movie S1. Bar, 2 μm (B) Rilpl2-LAP tubulovesicular structure in a live IMCD3 cell. Still frame from Supplemental Movie S2. Bar, 10 μm (C) IMCD3 cells expressing Rilpl2-LAP were serum starved, fixed, and stained for Rilpl2-LAP (GFP, green), pgTub (red), and DNA (DAPI, blue). Image is a maximum projection of a z-stack obtained by deconvolution microscopy. Bar, 5 μm (D) Serum-starved IMCD3 cell expressing Rilpl2-LAP (green) and tdTom-Inv (proximal cilium, red) was imaged for 90 min. Images shown are from the marked time points. Note the tubulovesicular structure at the base of the cilium. Still frames from Supplemental Movie S3. Bar, 1 μm.
While imaging Rilpl2-LAP in live cells, we observed that Rilpl2 occasionally forms dynamic tubule structures. To examine these Rilpl2-LAP tubules more closely, we used spinning-disk confocal microscopy to collect z-stacks of live cells every ∼15 s. Supplemental Movie S2 (Figure 4B) follows a single Rilpl2-LAP–expressing cell over ∼5 min, showing a dynamic tubulovesicular structure. Similar cytoplasmic tubules have been observed to be formed by Rab8, a known component of ciliary membrane trafficking machinery (Hattula et al., 2006). These Rab8 tubules are dependent on microtubule and actin dynamics. To determine whether Rilpl2 tubules are also dependent on cytoskeletal dynamics, we treated Rilpl2-LAP–expressing IMCD3 cells with cytochalasin D (1 μM) or vehicle (dimethyl sulfoxide [DMSO]) for 30 min to disrupt the actin cytoskeleton. The frequency of cells containing Rilpl2-LAP tubules was significantly higher in cytochalasin D–treated cells (6.4 ± 0.5% vs. 2.2 ± 0.6%, p < 0.01, t test) (Supplemental Figure S2). In contrast, when the same cells were treated with nocodazole to disrupt the microtubule network before cytochalasin D treatment there was a significant decrease in tubule formation (0.7 ± 0.3%, p < 0.01, t test). These data suggest that formation of the Rilp-like tubules is dependent on microtubule and actin dynamics. In fixed cells, some of the Rilpl2-positive tubule structures were associated with the primary cilium (Figure 4C), and time-lapse microscopy was used to assess this association in more detail. Figure 4D (Supplemental Movie S3) shows a 40-min sequence focusing on the base of a primary cilium, acquired using wide-field microscopy as described for Figure 4A. Rilpl2 is present in the cilium at the start of imaging and then appears to form a dynamic tubulovesicular structure from the base of the cilium. These data are consistent with Rilpl2 involvement in ciliary membrane dynamics.
Rilp-like proteins are required for epithelial cell organization
To test whether Rilpl1 and Rilpl2 are required for cilium formation or function, we generated short hairpin RNA (shRNA) constructs directed against unique regions of each protein. These shRNA constructs were introduced into IMCD3 cells by lentiviral infection. Western blot analysis of lysate from these cells shows that the shRNAs successfully deplete Rilpl1 and Rilpl2 (Figure 5A). We created IMCD3 stable cell lines depleted of Rilpl1 or Rilpl2 individually or together. Cells depleted of either or both of the Rilp-like proteins were indistinguishable from control cells for centriole duplication and cilium formation (Supplemental Figure S3).
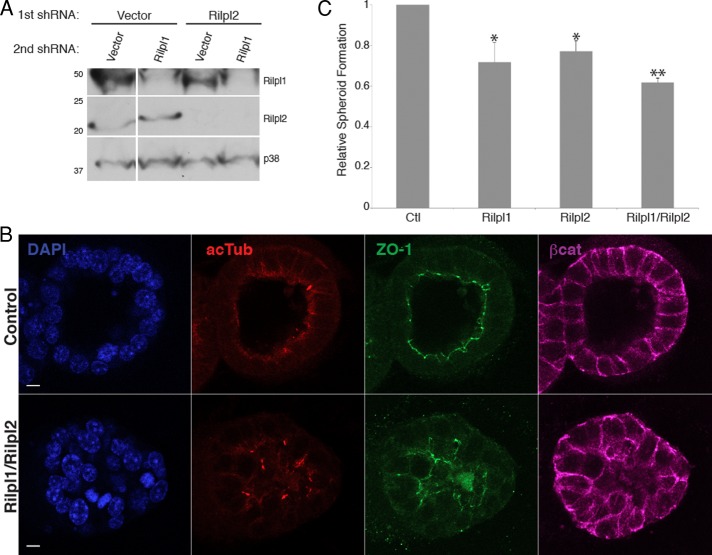
FIGURE 5:
Loss of Rilp-like proteins prevents spheroid formation. (A) Rilpl1 and Rilpl2 depleted individually or together from IMCD3 cells by lentiviral expression of shRNAs. Lysates were probed for Rilpl1, Rilpl2, and p38 as a loading control. Numbers to the left are molecular weights in kilodaltons. (B) Representative images of cell clusters from control and Rilpl1/Rilpl2–depleted IMCD3 cells grown in Matrigel, fixed, and stained for nuclei (DAPI, blue), apical junctions (ZO-1, green), cilia (acTub, red), and basolateral surfaces (β-catenin, purple). Bars, 10 μm. (C) Quantification of the frequency of spheroid formation of IMCD3 cells depleted of the indicated Rilp-like protein(s) normalized to the frequency of spheroid formation of control cells. Results shown are the mean of three independent experiments ± SEM (200 cells/experiment, *p < 0.05, **p < 0.01, t test).
We examined cells depleted of Rilp-like proteins for evidence of cilium dysfunction. The formation of epithelial spheroids is sensitive to cilium function and has been used as an assay to reveal ciliary defects (Delous et al., 2009; Otto et al., 2010; Sang et al., 2011). IMCD3 cells form spheroids when grown in three-dimensional culture, progressing from unorganized clusters of cells to organized spheroids with basal surfaces facing outward and apical domains facing inward. Primary cilia project from the apical surfaces into the spheroid lumen. Defects in spheroid formation are seen with loss of some ciliary proteins that are required for cilia function but not ciliogenesis (Delous et al., 2009; Otto et al., 2010; Sang et al., 2011). To determine whether Rilpl1 and Rilpl2 are required for spheroid formation, IMCD3 cells depleted of these proteins were grown in Matrigel and stained for cilia and markers of polarization. To control for the effect of lentiviral infection on spheroid formation, control cells were mock infected with lentivirus containing empty shRNA vectors. Clusters of cells were examined for proper organization of apical and basal surfaces (ZO-1 and β-catenin), presence of a lumen, and orientation of cilia (acetylated tubulin). Clusters that lacked a hollow lumen, had multiple lumens, or had ZO-1 staining outside of a central ring (the apical surface of a proper spheroid) were scored as defective. Depletion of Rilpl1 and/or Rilpl2 resulted in the decreased frequency of proper spheroid formation relative to control cells, with a slightly stronger defect observed in the double-depleted cells (Figure 5, B and C). The defects observed were the lack of a hollow lumen, ZO-1 staining outside of a central ring, and misoriented cilia (Figure 5B). We did not observe clusters with multiple lumens. The same cell lines depleted of Rilpl1 and/or Rilpl2 when grown on Transwell filters did not display defects in polarization (Supplemental Figure S3). These data suggest that Rilpl1 and Rilpl2 are important for epithelial cell organization in three-dimensional culture, consistent with a function at the primary cilium.
Rilp-like proteins regulate ciliary protein content
To determine whether Rilpl1 and Rilpl2 regulate ciliary membrane protein transport, we examined the effect of their depletion on the localization of signaling proteins to the primary cilium membrane. First, we infected IMCD3 cells stably expressing GFP fused to the membrane-anchoring and ciliary-targeting sequence domain of fibrocystin (PKHDCTS-GFP) with lentivirus containing empty vectors or shRNA against Rilpl1 and/or Rilpl2 and then serum starved them to induce ciliation (Figure 6A). The percentage of PKHDCTS-GFP–positive cilia in cells depleted of Rilpl1 (67.7 ± 7.5%), Rilpl2 (71 ± 5.2%), or both (74.7 ± 6.33%) was significantly higher than in control cells (60.0 ± 13.2%; p < 0.01, χ2 on pooled data; Figure 6, A and B). We confirmed this result with another marker of the cilium membrane, GFP fused to the full-length serotonin receptor 5-HT6 (5-HT6-GFP). In this case, the fraction of 5-HT6-GFP–positive cilia was similar in control and Rilp-like–depleted cells; however, the intensity of ciliary 5-HT6-GFP signal in Rilpl1-, Rilpl2-, or Rilpl1/Rilpl2–depleted cells was significantly higher than that in control cells (26.76 ± 15.83, 27.57 ± 19.77, and 27.71 ± 16.80 vs. 20.94 ± 13.86, respectively; p < 0.01, t test; Figure 6, C and D). Thus, for both 5-HT6-GFP and PKHDCTS-GFP, depletion of Rilpl1 and Rilpl2 alone or together led to a significant increase of signaling protein in the cilium, suggesting that the Rilp-like proteins are involved in removal of membrane proteins from the cilium.
FIGURE 6:
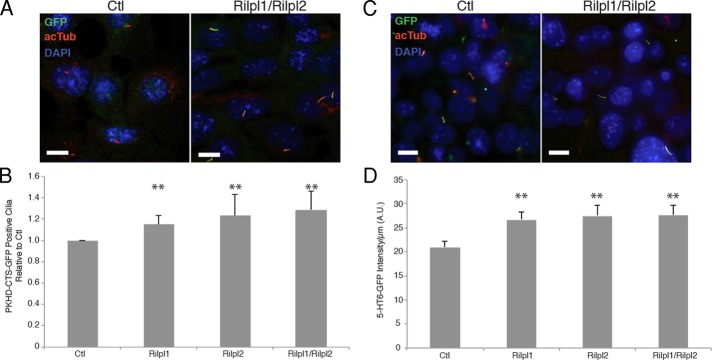
Loss of Rilp-like proteins increases signaling proteins in the primary cilium. (A) Representative images of control and Rilpl1/Rilpl2–depleted IMCD3 cells serum starved, fixed, and stained for PKHDCTS-GFP (green), acTub (red), and DNA (DAPI, blue). Bars, 10 μm. (B) Quantification of frequency of PKHDCTS-GFP positive cilia in IMCD3 cells depleted of the indicated protein(s) normalized to control frequency. Results shown are the mean of three independent experiments ± SEM (200 cells/experiment, **p < 0.01, χ2 on pooled data). (C) Representative images of control and Rilpl1/Rilpl2 depleted IMCD3 cells serum starved, fixed, and stained for 5-HT6-GFP (green), acTub (red), and DNA (DAPI, blue). Bars, 10 μm. (D) Quantification of 5-HT6-GFP in cilia (fluorescence intensity/micrometer). Results shown are the mean of cilia from 20 random fields of cells for each of three independent experiments ± SEM (n = 124, 89, 67, 106, respectively; **p < 0.01, t test).
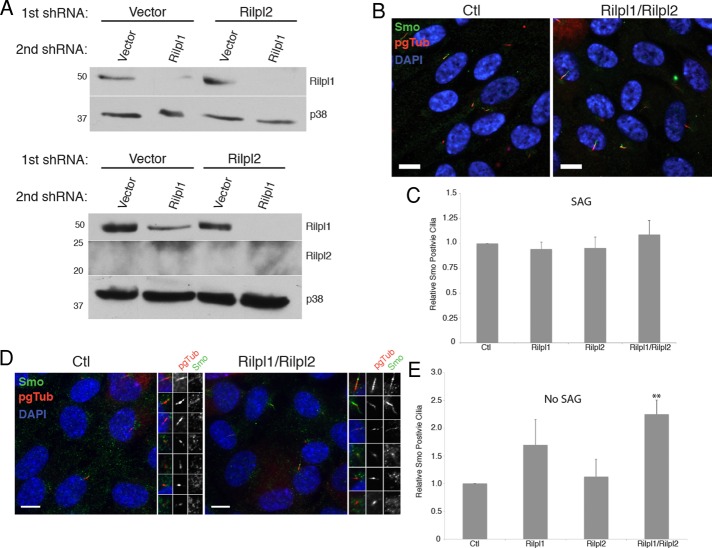
The 5-HT6-GFP and PKHDCTS-GFP proteins just described are overexpressed and constitutively localize to the primary cilium, and it is possible that any trafficking defect is dampened by a saturation effect (Mahjoub and Stearns, 2012). We circumvented these limitations by examining Smoothened (Smo), a Hedgehog-pathway protein that accumulates in primary cilia in a regulated manner. When the pathway is off, Smo does not accumulate in the primary cilium, although evidence suggests that Smo does traffic through the cilium in this state (Kim et al., 2009; Ocbina et al., 2011). In the presence of the natural ligand Shh or the Smo agonist SAG, the pathway is activated and Smo accumulates in the cilium. To determine whether Rilpl1 and Rilpl2 also affect regulated ciliary accumulation of endogenous Smo, we used shRNA to deplete both proteins from Hedgehog-responsive mouse embryonic fibroblasts (MEFs; Figure 7A). In ciliated MEFs treated with SAG (Hedgehog pathway on), the percentage of Smo-positive cilia in control MEFs was not significantly different from that in MEFs depleted of Rilpl1 and Rilpl2 (Figure 7, B and C). However, in ciliated MEFs not treated with SAG (Hedgehog pathway off) the percentage of Smo-positive cilia in Rilpl1/Rilpl2–depleted cells was 2.25 ± 0.44–fold greater than that of control cells (Figure 7, D and E). Cells depleted of Rilpl1 alone also had more Smo-positive cilia (1.7 ± 0.79–fold increase) but did not reach statistical significance. The increase in the basal amount of Smo in cilia strongly suggests that Rilp-like proteins control removal of ciliary membrane proteins.
FIGURE 7:
Loss of Rilp-like proteins increases basal level of Smo in cilia. (A) Rilpl1 and Rilpl2 were depleted individually or together from MEFs by lentiviral expression of shRNAs. Lysates were probed for Rilpl1, Rilpl2, and p38 as a loading control. Numbers to the left are molecular weights in kilodaltons. Protein loaded: 150 μg (top) or 500 μg (bottom). Note that Rilpl2 expression is below detectable levels in all samples. (B) Representative immunofluorescence images of control and Rilpl1/Rilpl2–depleted MEFs serum starved, treated with 100 nM SAG for 24 h, fixed, and stained for Smo (green), pgTub (red), and DNA (DAPI, blue). Bars, 10 μm. (C) Quantification of the frequency of Smo-positive cilia in SAG-treated MEFs depleted of the indicated proteins relative to control cells. Results shown are the mean of three independent experiments ± SEM (200 cells/experiment). (D) Representative immunofluorescence images of control and Rilpl1/Rilpl2 MEFs serum starved, treated with vehicle, fixed, and stained for Smo (green), pgTub (red), and DNA (DAPI, blue). Individual cilia from each image are highlighted to the right. Bars, 10 μm. (E) Quantification of the frequency of Smo-positive cilia in MEFs depleted of the indicated proteins relative to control cells. Results shown are the mean of three independent experiments ± SEM (200 cells/experiment, **p < 0.01, t test).
DISCUSSION
The Rilp-like proteins Rilpl1 and Rilpl2 share two regions of high sequence homology with Rilp, but unlike Rilp, they do not function in lysosomal trafficking (Wang et al., 2004). We showed that Rilpl1 and Rilpl2 localize to the primary cilium and centrosome and that the localization to the cilium is characterized by dynamic entry and exit and by formation of a tubulovesicular structure at the base of the cilium. Depletion of Rilpl1 and Rilpl2 increases the concentration of signaling proteins in the ciliary membrane and prevents proper epithelial cell organization. These results suggest that the Rilp-like proteins Rilpl1 and Rilpl2 regulate the protein content of the primary cilium.
We found that depletion of Rilpl1 and Rilpl2 increased the concentration of Smo in the ciliary membrane in unstimulated cells. This suggests that in the absence of these proteins, the balance of trafficking of Smo is shifted toward retention in the cilium and, more generally, that the Rilp-like proteins are involved in the removal of proteins from the ciliary membrane (Figure 8). This phenotype is similar to that reported in cells with disrupted retrograde ciliary transport (Kim et al., 2009; Ocbina et al., 2011). This putative function is further supported by the Rilp-like protein tubulovesicular structures observed forming at the base of the primary cilium in live and fixed cells (Figure 4). These structures might represent membrane invaginations resulting from endocytic events. Endocytosis is known to occur at the base of the primary cilium in mammalian cells and trypanosomatids (Brown et al., 1965; Duszenko et al., 1988; Bates et al., 1989; Field and Carrington, 2009; Molla-Herman et al., 2010; Rattner et al., 2010; Rich and Clark, 2012). The ciliary pocket has been the focus of much of this work, although clathrin-mediated endocytosis components have also been observed at the base of cilia and flagella lacking morphologically identifiable pockets (Molla-Herman et al., 2010; Kaplan et al., 2012). The Rilp protein has been shown to promote the formation of membrane tubules from phagosomes for fusion with late endosomes/lysosomes (Harrison et al., 2003). This tubulation represents a connection between the phagosome membrane and the microtubule network provided by the interaction of Rilp with Rab proteins and the dynein/dynactin motor complex. Recently Rilpl1 and Rilpl2 were shown to directly interact with active Rab36, and, although the function of Rab36 is unclear, it is the most highly up-regulated Rab protein in our ciliated cell transcriptome (Hoh et al., 2012), suggesting that it might have a ciliary function (Mori et al., 1999; Chen et al., 2010; Kanno et al., 2010; Matsui et al., 2012; Nottingham et al., 2012). Thus Rilp-like proteins may function in removing membrane proteins from the cilium and directing their distribution to endosomal compartments by bridging the ciliary membrane and cytoskeletal motors.
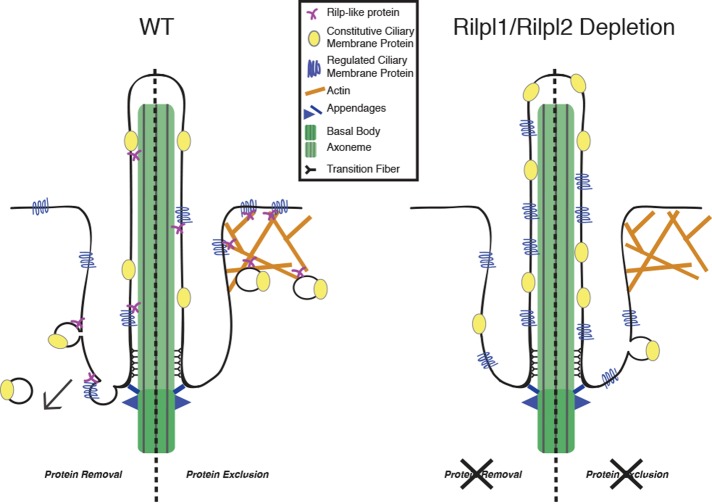
FIGURE 8:
Model of Rilp-like protein function. The data presented in this study fit two models for Rilp-like protein control of ciliary membrane content. 1) Protein removal: Rilp-like proteins may promote the removal of membrane and membrane proteins from the primary cilium. 2) Protein exclusion: Rilp-like proteins may prevent entry of membrane proteins into the cilium. Loss of the Rilp-like proteins increases the ciliary membrane protein concentration by either decreasing protein removal or increasing protein entry.
Although we favor the proposed model in which the Rilp-like proteins mediate endocytic removal of proteins from the ciliary membrane, we cannot rule out the possibility that they work instead by excluding proteins from the cilium (Figure 8). For example, Rilp-like proteins could function similarly to NHERF, which has been shown to link podocalyxin to the actin cytoskeleton, thereby holding it outside of the periciliary membrane domain and the cilium, consistent with Rilpl2’s known interaction with MyoVa (Francis et al., 2011). It is unlikely, however, that Rilp-like proteins are gatekeepers or part of a diffusion barrier at the cilium like the septin, NPHP, or MKS-JBTS diffusion barriers (Hu et al., 2010; Chih et al., 2011; Garcia-Gonzalo et al., 2011; Sang et al., 2011). In the absence of these components, membrane proteins normally localized to the cilium are present at lower levels, the opposite of what we observe with depletion of Rilp-like proteins.
Although we initially identified Rilpl2 by its transcriptional up-regulation during ciliogenesis, we found that in many respects Rilpl1 and Rilpl2 are similar to each other and distinct from the defining family member Rilp. Rilpl1 and Rilpl2 are more closely related to each other than to Rilp, and the proteins share a 13–amino acid sequence just upstream of the RH2 domain that is absent from Rilp. We showed that this short sequence is necessary and sufficient for centrosomal localization of the Rilp family proteins. Next, both Rilpl1 and Rilpl2 interact with active Rab34 and Rab36 but not Rab7, whereas Rilp interacts with all three (Matsui et al., 2012). Finally, Rilpl1 and Rilpl2 are both cytoplasmic and membrane associated, whereas Rilp is strictly associated with the membrane (Wang et al., 2004).
The genes for Rilpl1 and Rilpl2 are adjacent to each other in both mammalian and Xenopus genomes, separated by one gene, suggesting that a gene duplication event occurred early in vertebrate evolution. The maintenance of both genes after the duplication is consistent with a divergence in function or with a divergence of regulation. Consistent with this latter possibility, the genes are differentially expressed in different tissues and cell types (Wang et al., 2004). We found that Rilpl1, unlike Rilpl2, is not transcriptionally up-regulated in multiciliated cells but is localized to the centrosome in neighboring nonmulticiliated cells (see Figure 1C), and we also found differences in relative expression in different cell lines (see Figures 5A and 7A). There are, however, some indications that Rilpl1 and Rilpl2 differ in more than their expression. Rilpl1 contains a large 175-aa protein sequence between the RH1 and RH2 domains that is absent from Rilpl2. Also, although both proteins localized to a subset of cilia and centrosomes, Rilpl1 specifically localized to the distal end of the mother centriole, whereas Rilpl2 had no specific suborganelle localization (Figure 2, A–C). Despite these differences, analysis of cells depleted of Rilpl1 and/or Rilpl2 suggests they may have redundant function. In both spheroid formation and smoothened translocation assays, depletion of both proteins gave a more significant phenotype than depletion of either protein individually. Our results suggest that the main differences between these proteins relate to their expression, but more work will be needed to precisely define the relationship between Rilpl1 and Rilpl2.
Rilpl2 was previously identified in a screen for proteins interacting with MyoVa. Rilpl2 controls cell shape and dendritic spine formation in neurons in a Rac1-dependent mechanism. The interaction of Rilpl2 with MyoVa occurs via the N-terminus of Rilpl2 (Lisé et al., 2009). Our domain analysis demonstrated that this interaction is not required for Rilpl2’s localization to the centrosome or cilium. However, expression of ΔN-Rilpl2 had a negative affect on ciliogenesis, as it did on dendritic spine formation, suggesting that the interaction with MyoVa might be important for the cilium function as well. Dendritic spines, like the primary cilium, are protrusions of the plasma membrane, and there might be mechanistic links between the two. Several of the proteins involved in dendritic spine formation also have connections to ciliary function; MyoVa itself is associated with the recycling endosome and interacts with Rab11, both of which are involved with the ciliary membrane (Correia et al., 2008). As with almost all membrane-associated process, the actin cytoskeleton is an important factor in ciliogenesis and in dendritic spine formation and might be the link between the functions of Rilp-like proteins in these two processes (Tada and Sheng, 2006; Bershteyn et al., 2010; Galletta et al., 2010; Kim et al., 2010; Molla-Herman et al., 2010).
MATERIALS AND METHODS
Plasmids
Full-length cDNA clones of mouse Rilp, Rilpl1, and Rilpl2 were obtained from Open Biosystems (Huntsville, AL; clones 40126069, 6403995, 3156139, respectively). The open reading frame (ORF) of each protein was amplified by PCR and cloned into pDONR221 (pTS1950) using Gateway technology (Invitrogen, Carlsbad, CA), creating pTS2124, pTS2125, and pTS2034 without stop codons for C-terminal tags and pTS3043, pTS2601, and pTS2050 with stop codons for N-terminal tags. C-terminal GFP-tagged constructs were created by Gateway cloning into pDEST 47 (pTS1953; Invitrogen), making Rilp-GFP (pTS2128), Rilpl1-GFP (pTS2129), and Rilpl2-GFP (pTS2041). For creation of Rilpl2-LAP (-S-Prescission-eGFP; pTS2049) pTS2034 was Gateway cloned into pTS1937 provided by M. Nachury (Stanford University, Stanford, CA). To create LAP, (eGFP-Prescission-Strep-HA)-Rilpl1 (pTS2607) pTS2601 was Gateway cloned into pTS2497. To create a lentiviral transfer vector with Rilpl1-LAP (pTS2921), pTS2125 was cloned into pTS1954, pLenti6.2 DEST eGFP-LAP provided by M. Nachury, using Gateway technology.
For Rilpl1- and Rilpl2-deletion constructs, the codons for the indicated amino acid sequences were amplified from full-length constructs and cloned into pDONR221 using Gateway technology. ΔC-Rilpl1 (aa 1–288, pTS2533 and pTS2534), ΔN-Rilpl1 (aa 289–406, pTS2535 and pTS2536), ΔC-Rilpl2 (aa 1–113, pTS2227 [stop]), ΔN-Rilpl2 (aa 114–197, pTS2229 [stop]). C-terminal GFP-tagged deletion constructs of Rilpl1 (pTS2550 and pTS2552) were made by Gateway cloning into pTS1953 (pDest 47, Invitrogen). N-terminal GFP-tagged deletion constructs of Rilpl2 (pTS2228 and pTS2230) were made by Gateway cloning into pTS1951 (pDest 53; Invitrogen). The Rilp-LD construct was generated by swapping the coding sequence for the LD (DPNRPRFTLQELR) from human Rilpl2 into the equivalent position upstream of the RH2 domain in mouse Rilp by overlap PCR (pTS3042). The Rilpl1-ΔLD construct was generated by removing the coding sequence for the LD from mouse Rilpl1 by overlap PCR (pTS3040). Gateway cloning into pTS1953 generated C-terminally GFP-tagged constructs (pTS3048 and pTS3049).
For bacterial expression of Rilpl1 and Rilpl2, glutathione S-transferase (GST)–Rilpl2 and GST-Rilpl1 were created by Gateway cloning of pTS2050 and pTS2601 into pDest15 (pTS1952; Invitrogen) to create pTS3052 and pTS2290. Histidine (His)-tagged Rilpl1 and Rilpl2 were created by PCR amplifying the ORF of Rilpl1 or Rilpl2 and cloning by restriction enzyme digest and ligation into pET28aFA (pTS1668) provided by the G. Fang (Stanford University, Stanford, CA).
For shRNA lentivirus constructs, 19-mer shRNAs targeting mouse Rilpl1 and Rilpl2 were chosen: Rilpl1_1 (gacgaggctaatgaagatc) and Rilpl2_4 (cgccttgactgatatttga). shRNA oligos were designed using pSicoOligomaker 1.5. Oligos were annealed and cloned into pSicoR-puro (Ventura et al., 2004; pTS1613) for Rilpl2 (pTS2427) or pLentiRFP3.7 (pLentiLox3.7 with GFP replaced by mRFP, pTS1770) for Rilpl1 (pTS2501).
Cell culture and transfection
NIH 3T3, MEF, and HEK 293T cells were grown in DMEM (Cellgro, Manassas, VA) with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA). IMCD3 cells were grown in DMEM/F12 (Cellgro) with 10% FBS. N2A cells were grown in DMEM/OptiMEM 1:1 with 5% FBS. MTECs were cultured as previously described (Vladar and Stearns, 2007). Cells were maintained at 37°C with 5% CO2. To induce ciliation in IMCD3, 3T3, and MEF cells, the medium was switched to serum starvation medium containing 0.5% FBS for 16–24 h. IMCD3 cell lines stably expressing PKHDCTS-GFP or 5-HT6-GFP were obtained from M. Nachury (Stanford University).
Transfection of all cells, except for virus generation and antibody testing, was performed using the recommended protocol of either Lipofectamine2000 or LipofectamineLTX (Invitrogen). Briefly cells were grown to ∼80% confluence, Lipofectamine–DNA mixture was added for 4–8 h, and expression of proteins was assayed 24–48 h after transfection. HEK 293T cells were transfected by calcium phosphate precipitation.
IMCD3 cell lines expressing Rilpl2-LAP or LAP-Rilpl1 were generated as follows. IMCD3 FlpIN cells (M. Nachury) were transfected with the FlpIN-compatible vector containing Rilpl2-LAP or LAP-Rilpl1 along with a vector encoding Flp recombinase (pOG44, pTS2064) at a ratio of 1:5 using Lipofectamine2000. Cells were transfected for 4–8 h, washed, and allowed to recover for 24 h before being selected with 150 μg/ml hygromycin B (Invitrogen). Pools of selected cells were seeded at a low density to allow for single-colony isolation. Several colonies were picked and screened for expression of the LAP-tagged protein by immunofluorescence and Western blot before proceeding. IMCD3 Rilpl2-LAP/tdTom-Inversin cells were generated by Lipofectamine transfection of pTS1641 into previously derived IMCD3 Rilpl2-LAP cells and selected for resistance to 450 μg/ml Geneticin (Invitrogen). NIH 3T3 cells stably expressing Rilpl1-LAP were generated by infecting cells with lentivirus containing an Rilpl1-LAP transfer vector, followed by fluorescence-activated cell sorting for enrichment.
For spheroid formation assays, IMCD3 cells were plated at 5000 cells/well in 2% FBS and 2% Matrigel (BD Biosciences, San Diego, CA) in eight-well chamber slides coated with Matrigel. Cells were grown for 6–8 d until spheroid formation was evident, replacing the medium every 4 d. For polarization of IMCD3 cells, cells were plated on Transwell-Clear permeable filters (Corning, Tewksbury, MA) and grown 7 d postconfluence, changing medium every 2 d.
Drug treatments
For disruption of the microtubule and actin cytoskeletons, IMCD3 cells stably expressing Rilpl2-LAP were grown to 80% confluence and serum starved for 24 h. Cells were then treated with 5 μg/ml nocodazole (US Biological, Swampscott, MA) or DMSO for 1 h. The medium was then changed to 5 μg/ml nocodazole plus 1 μM cytochalasin D (Sigma-Aldrich, St. Louis, MO), 5 μg/ml nocodazole plus DMSO, DMSO + 1 μM cytochalasin D, or DMSO alone for 30 min. Cells were then fixed and processed for imaging.
For smoothened translocation experiments, MEFs were grown to near confluence (80–100%) and switched to serum starvation medium for 24 h. The medium was changed again to serum starvation medium plus 100 nM SAG (Smoothened Agonist; EMD/Calbiochem, La Jolla, CA). At 24 h later cells were fixed and processed for imaging.
Lentivirus production and infection
Recombinant lentivirus was made by cotransfection of HEK 293T cells with packaging and envelope vectors (pCMVDR8.74 and pMD2.VSVG; Dull et al., 1998) and the appropriate transfer vector by calcium phosphate transfection. Medium was changed 6–8 h after transfection. Viral supernatant was harvested after another 48 h and titered on IMCD3 or MEF cells. For infection, 4 × 104 cells were plated in 24-well tissue culture plates. The next day the medium was replaced with medium containing the appropriate lentivirus(es) each at a multiplicity of infection of 1. After 16–24 h of infection, cells were washed with fresh medium and infected for a second round. For long-term depletion in IMCD3 cells, cells infected with shRNA plasmids with puromycin resistance were selected with 1 μg/ml puromycin (InvivoGen, San Diego, CA). For depletion, MEF cells were assayed 7–8 d after initial infection, and IMCD3 cells were assayed 6 d after initial infection.
Antibodies
Rabbit anti–mouse Rilpl2 antibody was made by immunizing rabbits with bacterially expressed GST-Rilpl2 (Cocalico Biologicals, Reamstown, PA). Rilp-like dual-affinity Rilpl1/Rilpl2 antibody was purified against His-Rilpl2 immobilized on nitrocellulose membrane (Bio-Rad, Hercules, CA). An Rilpl2 higher-specificity antibody was first incubated with His-Rilpl1 immobilized on nitrocellulose, and then spent serum was purified against His-Rilpl2 immobilized on nitrocellulose membrane. Specificity was determined by antibody blocking experiments. Rilp-like antibody recognized both Rilpl1 and Rilpl2 with high affinity by Western blot, whereas Rilpl2-specific antibody had higher affinity for Rilpl2 than Rilpl1 by Western blot. Both anti–Rilp-like and anti–Rilpl2 antibodies were specific for Rilpl2 by immunofluorescence. Rabbit anti–Rilp-like was used at 1.1 μg/ml for Western blot and 0.275–1.1 μg/ml for immunofluorescence. Rabbit anti-Rilpl2 was used at 0.08 μg/ml for Western blot and 0.04–0.08 μg/ml for immunofluorescence. For antibody preabsorption, rabbit anti–Rilp-like antibody was diluted to working concentration, 10× the molar amount of His-Rilpl1 was added, and the solution was mixed for 1 h at room temperature.
Affinity-purified rabbit anti–human Rilpl1 antibody was purchased from Sigma-Aldrich (HPA041314, peptide antigen, residues 82–169). Anti-Rilpl1 was diluted 1:500 to 1:1000 for immunofluorescence and 1:250 for Western blot. Rabbit anti-GFP antibody was generated and used as previously described (Hatch et al., 2010). Additional antibodies used for immunofluorescence were mouse anti–α-tubulin (DM1α, Sigma-Aldrich) at 1:4000, mouse anti–polyglutamylated tubulin (GT335; provided by C. Janke, Center de Recherches de Biochimie Macromoléculaire, Montpellier, France) at 1:5000, mouse anti–acetylated α-tubulin (6-11B-1; Abcam, Cambridge, MA) at 1:15,000, mouse anti-GFP (3e6; Invitrogen) at 1:750, mouse anti–centrin-2 (20H5; provided by J. Salisbury, Mayo Clinic, Rochester, MN) at 1:1000, mouse anti–centrin-3 (Cetn3; Abnova, Taipei City, Taiwan) at 1:4000, mouse anti-myc (9e10) at 1:500, mouse anti–γ-tubulin (GTU-88; Sigma-Aldrich) at 1:5000, rabbit anti-Smoothened (38686, Abcam) at 1:1000, rat anti–ZO-1 at 1:1000 (American Research Products, Waltham, MA), rabbit anti–β-catenin at 1:1000 (Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti-pericentrin at 1:200 (BD Biosciences), and rabbit anti-Cep164 at 1:2000 (E. Nigg, Biozentrum, University of Basel, Basel, Switzerland). For immunofluorescence secondary antibodies against rabbit immunoglobulin G (IgG), rat IgG, or specific mouse IgG isotypes and conjugated to Alexa Fluor 488, 594, 649, or 680 were obtained from Invitrogen and used at 1:500 to 1:1000 for 488 and 594 or at 1:100 to 1:200 for 649 and 680. For Western blot, rabbit anti-p38 (C-20; Santa Cruz Biotechnology) was used at 1:5000 and goat anti-GFP (Rockland Immunochemicals, Gilbertsville, PA) was used at 1:5000.
Western blots
To obtain cell lysate for protein depletion analysis, cells were trypsinized off the plate, washed in phosphate-buffered saline (PBS), and lysed in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (50 mM HEPES, pH 7.5, 1 mM MgCl2, 1 mM EDTA, and 150 mM NaCl) with protease inhibitors and 0.5% Triton X-100 for 1 h at 4°C. Lysate was cleared by centrifugation at 16,000 × g at 4°C for 15 min. The protein concentration was determined using Bradford Reagent (Bio-Rad). Equivalent amounts of total protein for each sample were mixed with sample buffer, boiled for 5 min, and loaded on polyacrylamide gels for SDS–PAGE. For antibody-specificity testing, cells were lysed on the plate in sample buffer and harvested. Samples were boiled for 5 min, and equivalent volumes of lysate were loaded on polyacrylamide gels. After SDS–PAGE, proteins were transferred to nitrocellulose membranes, blocked in TBS-T (Tris-buffered saline with 0.1% Tween-20) with 5% milk, and then incubated with primary antibodies overnight (see Antibodies). Membranes were washed with TBS-T and incubated with a 1:10,000 dilution of horseradish peroxidase–conjugated goat anti-rabbit or 1:5000 donkey anti-goat secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA, and Santa Cruz Biotechnology) for 1 h at room temperature. Membranes were washed, developed using SuperSignal West Pico Chemiluminescent Substrate (ThermoFisher, Waltham, MA), and exposed to film.
Immunofluorescence and live-cell microscopy
Cells were grown on polylysine-coated coverslips and fixed using either ice-cold methanol or 4% paraformaldehyde, followed by quenching with 1 mg/ml NaBH4. Cells were blocked in 3% BSA (Sigma-Aldrich) in PBS plus 0.1% Triton X-100. Coverslips were incubated in primary antibodies diluted in blocking solution at the previously indicated concentrations (see Antibodies) for 30 min overnight, washed in blocking buffer, and incubated in secondary antibodies diluted as described (see Antibodies) for 30 min to 1 h. Nuclei were stained by brief incubation with 4′,6-diamidino-2-phenylindole (1 μg/ml). Coverslips were mounted on slides using MOWIOL mounting medium containing 1,4,-diazobicycli-[2.2.2]-octane (Sigma-Aldrich) and p-phenylenediamine (Sigma-Aldrich). MTECs grown on filters were fixed, stained, and prepared for imaging as previously described (Vladar and Stearns, 2007). Images were acquired using OpenLab software (PerkinElmer, Waltham, MA) on a microscope (Axiovert 200M; Carl Zeiss, Jena, Germany) with PlanNeofluar 100×/1.3 numerical aperture (NA) and PlanApoChromat 63×/1.40 NA objectives and a cooled, charge-coupled device camera (Orca ER; Hamamatsu Photonics, Hamamatsu, Japan). Images were processed using Photoshop (Adobe, San Jose, CA) and ImageJ (National Institutes of Health, Bethesda, MD). Spheroids were imaged on a confocal laser scanner (LSM510; Carl Zeiss) using a 40× oil objective (NA 1.3). For suborganelle localization of Rilpl1, Rilpl2-LAP tubule imaging, and polarization of IMCD3s on filters, a z-stack of digital optical sections at 0.2-μm intervals was collected using a 100×/1.35 NA or 60×1.40 NA objective (Olympus, Tokyo, Japan) on an epifluorescence inverted microscope (IX70; Olympus) in an imaging station (DeltaVision; Applied Precision, Issaquah, WA) with a charge-coupled device camera (CH350; Photometrics, Tucson, AZ) in the Cell Sciences Imaging Facility (Stanford, CA). Deconvolution of images was performed using the constrained iterative algorithm and point-spread functions provided with the DeltaVision system. Images in the stacks were merged into a single maximum-intensity projection image. Brightness and contrast were adjusted in Photoshop, except when intensity values were collected (see below).
For wide-field, live-cell microscopy, IMCD3 cells were seeded on 32 mm or 42 mm poly-l-lysine–coated coverslips. Coverslips were mounted in a sealed chamber with DMEM medium without phenol red (Invitrogen) and 0–0.5% FBS on the Axiovert microscope described earlier, at 37°C. Stacks of images were taken with a PlanApoChromat 63×/1.40 NA objective at the stated interval. To create time-lapse movies, the in-focus planes from each time point were combined in a time-lapse series. Brightness and contrast were adjusted in Photoshop and ImageJ. For spinning-disk confocal microscopy, IMCD3 cells were seeded on chamber slides and imaged in DMEM medium without phenol red (Invitrogen) and 0.5% FBS on a spinning disk confocal microscope (PerkinElmer UltraVIEW VoX with a Zeiss AxioObserver) using a 63×/1.40 NA objective and a Hamamatsu C100600-108 camera (ORCA-R2). Images were assembled into a time-lapse movie of z-stack maximum projections using Volocity (PerkinElmer).
For quantification of fluorescence intensity of 5-HT6-GFP in the primary cilium, images of 20 random fields of cells were taken at identical exposures for each sample. ImageJ was used to analyze images. The total intensity of the GFP signal in the cilium was measured in addition to the background fluorescence intensity and length of the cilium based on the GFP signal. The intensity per unit length is reported in arbitrary units.
Supplementary Material
Acknowledgments
We gratefully acknowledge members of the Stearns lab for helpful advice and Joanna Lee and Yin Loon Lee (Stanford University) for critical reading of the manuscript. This work was supported by a National Science Foundation Graduate Research Fellowship (J.R.S.); PHS Grant Number CA09302, awarded by the National Cancer Institute, Department of Health and Human Services (J.R.S.); and National Institutes of Health Grant GM52022 (T.S.).
Abbreviation used:
- LD
Rilp-like domain
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-08-0598) on December 21, 2012.
REFERENCES
- Bates PA, Hermes I, Dwyer DM. Leishmania donovani: immunochemical localization and secretory mechanism of soluble acid phosphatase. Exp Parasitol. 1989;68:335–346. doi: 10.1016/0014-4894(89)90115-x. [DOI] [PubMed] [Google Scholar]
- Berbari NF, Johnson AD, Lewis JS, Askwith CC, Mykytyn K. Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol Biol Cell. 2008;19:1540–1547. doi: 10.1091/mbc.E07-09-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari NF, O'Connor AK, Haycraft CJ, Bradley K Yoder. The primary cilium as a complex signaling center. Curr Biol. 2009;19:R526–R535. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershteyn M, Atwood SX, Woo W-M, Li M, Oro AE. MIM and cortactin antagonism regulates ciliogenesis and hedgehog signaling. Dev Cell. 2010;19:270–283. doi: 10.1016/j.devcel.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KN, Armstrong JA, Valentine RC. The ingestion of protein molecules by blood forms of Trypanosoma rhodesiense. Exp Cell Res. 1965;39:129–135. doi: 10.1016/0014-4827(65)90015-7. [DOI] [PubMed] [Google Scholar]
- Cantalupo G, Alifano P, Roberti V, Bruni CB, Bucci C. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 2001;20:683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Hu J, Yun Y, Wang T. Rab36 regulates the spatial distribution of late endosomes and lysosomes through a similar mechanism to Rab34. Mol Membr Biol. 2010;27:24–31. doi: 10.3109/09687680903417470. [DOI] [PubMed] [Google Scholar]
- Chih B, Liu P, Chinn Y, Chalouni C, Komuves LG, Hass PE, Sandoval W, Peterson AS. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol. 2011;14:61–72. doi: 10.1038/ncb2410. [DOI] [PubMed] [Google Scholar]
- Correia SS, Bassani S, Brown TC, Lisé M-F, Backos DS, El-Husseini A, Passafaro M, Esteban JA. Motor protein–dependent transport of AMPA receptors into spines during long-term potentiation. Nat Neurosci. 2008;11:457–466. doi: 10.1038/nn2063. [DOI] [PubMed] [Google Scholar]
- Delous M, Hellman NE, Gaudé H-M, Silbermann F, Le Bivic A, Salomon R, Antignac C, Saunier S. Nephrocystin-1 and nephrocystin-4 are required for epithelial morphogenesis and associate with PALS1/PATJ and Par6. Hum Mol Genet. 2009;18:4711–4723. doi: 10.1093/hmg/ddp434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duszenko M, Ivanov IE, Ferguson MA, Plesken H, Cross GA. Intracellular transport of a variant surface glycoprotein in Trypanosoma brucei. J Cell Biol. 1988;106:77–86. doi: 10.1083/jcb.106.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MC, Carrington M. The trypanosome flagellar pocket. Nat Rev Microbiol. 2009;7:775–786. doi: 10.1038/nrmicro2221. [DOI] [PubMed] [Google Scholar]
- Follit JA, Li L, Vucica Y, Pazour GJ. The cytoplasmic tail of fibrocystin contains a ciliary targeting sequence. J Cell Biol. 2010;188:21–28. doi: 10.1083/jcb.200910096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis SS, Sfakianos J, Lo B, Mellman I. A hierarchy of signals regulates entry of membrane proteins into the ciliary membrane domain in epithelial cells. J Cell Biol. 2011;193:219–233. doi: 10.1083/jcb.201009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Kanno E, Ishibashi K, Itoh T. Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Mol Cell Proteom. 2008;7:1031–1042. doi: 10.1074/mcp.M700569-MCP200. [DOI] [PubMed] [Google Scholar]
- Galletta BJ, Mooren OL, Cooper JA. Actin dynamics and endocytosis in yeast and mammals. Curr Opin Biotechnol. 2010;21:604. doi: 10.1016/j.copbio.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43:776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RE, Bucci C, Vieira OV, Schroer TA, Grinstein S. Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: role of Rab7 and RILP. Mol Cell Biol. 2003;23:6494–6506. doi: 10.1128/MCB.23.18.6494-6506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol. 2010;191:721–729. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattula K, Furuhjelm J, Tikkanen J, Tanhuanpää K, Laakkonen P, Peränen J. Characterization of the Rab8-specific membrane traffic route linked to protrusion formation. J Cell Sci. 2006;119:4866–4877. doi: 10.1242/jcs.03275. [DOI] [PubMed] [Google Scholar]
- Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N. Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh RA, Stowe TR, Turk E, Stearns T. Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PLoS One. 2012;7:e52166. doi: 10.1371/journal.pone.0052166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Milenkovic L, Jin H, Scott MP, Nachury MV, Spiliotis ET, Nelson WJ. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science. 2010;329:436–439. doi: 10.1126/science.1191054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. Ciliogenesis: building the cell's antenna. Nat Rev Mol Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- Johansson M, Rocha N, Zwart W, Jordens I, Janssen L, Kuijl C, Olkkonen VM, Neefjes J. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno E, Ishibashi K, Kobayashi H, Matsui T, Ohbayashi N, Fukuda M. Comprehensive screening for novel rab-binding proteins by GST pull-down assay using 60 different mammalian Rabs. Traffic. 2010;11:491–507. doi: 10.1111/j.1600-0854.2010.01038.x. [DOI] [PubMed] [Google Scholar]
- Kaplan OI, Doroquez DB, Cevik S, Bowie RV, Clarke L, Sanders AAWM, Kida K, Rappoport JZ, Sengupta P, Blacque OE. Endocytosis genes facilitate protein and membrane transport in C. elegans sensory cilia. Curr Biol. 2012;22:451–460. doi: 10.1016/j.cub.2012.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kato M, Beachy PA. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus; Proc Natl Acad Sci USA; 2009. pp. 21666–21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee JE, Heynen-Genel S, Suyama E, Ono K, Lee K, Ideker T, Aza-Blac P, Gleeson JG. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464:1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisé M-F, Srivastava DP, Arstikaitis P, Lett RL, Sheta R, Viswanathan V, Penzes P, O'Connor TP, El-Husseini A. Myosin-Va-interacting protein, RILPL2, controls cell shape and neuronal morphogenesis via Rac signaling. J Cell Sci. 2009;122:3810–3821. doi: 10.1242/jcs.050344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub MR, Stearns T. Supernumerary centrosomes nucleate extra cilia and compromise primary cilium signaling. Curr Biol. 2012;22:1628–1634. doi: 10.1016/j.cub.2012.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub MR, Xie Z, Tim Stearns. Cep120 is asymmetrically localized to the daughter centriole and is essential for centriole assembly. J Cell Biol. 2010;191:331. doi: 10.1083/jcb.201003009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Ohbayashi N, Fukuda M. The Rab interacting lysosomal protein (RILP) homology domain functions as a novel effector domain for small GTPase Rab36. J Biol Chem. 2012;287:28619–28631. doi: 10.1074/jbc.M112.370544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla-Herman A, et al. The ciliary pocket: an endocytic membrane domain at the base of primary and motile cilia. J Cell Sci. 2010;123:1785–1795. doi: 10.1242/jcs.059519. [DOI] [PubMed] [Google Scholar]
- Mori T, Fukuda Y, Kuroda H, Matsumura T, Ota S, Sugimoto T, Nakamura Y, Inazawa J. Cloning and characterization of a novel Rab-family gene, Rab36, within the region at 22q11.2 that is homozygously deleted in malignant rhabdoid tumors. Biochem Biophys Res Commun. 1999;254:594–600. doi: 10.1006/bbrc.1998.9968. [DOI] [PubMed] [Google Scholar]
- Nottingham RM, Pusapati GV, Ganley IG, Barr FA, Lambright DG, Pfeffer SR. RUTBC2 protein, a Rab9A effector and GTPase-activating protein for Rab36. J Biol Chem. 2012;287:22740–22748. doi: 10.1074/jbc.M112.362558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocbina PJR, Eggenschwiler JT, Moskowitz I, Anderson KV. Complex interactions between genes controlling trafficking in primary cilia. Nat Genet. 2011;43:547–553. doi: 10.1038/ng.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh EC, Katsanis N. Cilia in vertebrate development and disease. Development. 2012;139:443–448. doi: 10.1242/dev.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto EA, et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet. 2010;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Witman GB. The vertebrate primary cilium is a sensory organelle. Curr Opin Cell Biol. 2003;15:105–110. doi: 10.1016/s0955-0674(02)00012-1. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT): role in ciliary assembly, resorption and signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- Rattner JB, Sciore P, Ou Y, van der Hoorn FA, Lo IKY. Primary cilia in fibroblast-like type B synoviocytes lie within a cilium pit: a site of endocytosis. Histol Histopathol. 2010;25:865–875. doi: 10.14670/HH-25.865. [DOI] [PubMed] [Google Scholar]
- Rich DR, Clark AL. Chondrocyte primary cilia shorten in response to osmotic challenge and are sites for endocytosis. Osteoarthr Cartil. 2012;20:923–930. doi: 10.1016/j.joca.2012.04.017. [DOI] [PubMed] [Google Scholar]
- Sang L, et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol. 2006;16:95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Tam BM, Moritz OL, Hurd LB, Papermaster DS. Identification of an outer segment targeting signal in the COOH terminus of rhodopsin using transgenic Xenopus laevis. J Cell Biol. 2000;151:1369–1380. doi: 10.1083/jcb.151.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci USA. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar EK, Stearns T. Molecular characterization of centriole assembly in ciliated epithelial cells. J Cell Biol. 2007;178:31–42. doi: 10.1083/jcb.200703064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wong KK, Hong W. A unique region of RILP distinguishes it from its related proteins in its regulation of lysosomal morphology and interaction with Rab7 and Rab34. Mol Biol Cell. 2004;15:815–826. doi: 10.1091/mbc.E03-06-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y, Richer EJ, Huang T, Brody SL. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am J Physiol Lung Cell Mol Physiol. 2002;283:L1315–L1321. doi: 10.1152/ajplung.00169.2002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.