Abstract
Harpins are produced by Gram-negative phytopathogenic bacteria and typically elicit hypersensitive response (HR) in non-host plants. The characterization of harpins in Xanthomonas species is largely unexplored. Here we demonstrate that Xanthomonas produce a highly conserved single-stranded DNA-binding protein (SSBX) that elicits HR in tobacco as by harpin Hpa1. SSBX, like Hpa1, is an acidic, glycine-rich, heat-stable protein that lacks cysteine residues. SSBX-triggered HR in tobacco, as by Hpa1, is characterized by the oxidative burst, the expression of HR markers (HIN1, HSR203J), pathogenesis-related genes, and callose deposition. Both SSBX- and Hpa1-induced HRs can be inhibited by general metabolism inhibitors actinomycin D, cycloheximide, and lanthanum chloride. Furthermore, those HRs activate the expression of BAK1 and BIK1 genes that are essential for induction of mitogen-activated protein kinase (MAPK) and salicylic acid pathways. Once applied to plants, SSBX induces resistance to the fungal pathogen Alternaria alternata and enhances plant growth. When ssbX was deleted in X. oryzae pv. oryzicola, the causal agent of bacterial leaf streak in rice, the resulting ssbXoc mutant was reduced in virulence and bacterial growth in planta, but retained its ability to trigger HR in tobacco. Interestingly, ssbXoc contains an imperfect PIP-box (plant-inducible promoter) and the expression of ssbXoc is regulated by HrpX, which belongs to the AraC family of transcriptional activators. Immunoblotting evidence showed that SSBx secretion requires a functional type-III secretion system as Hpa1 does. This is the first report demonstrating that Xanthomonas produce a highly-conserved SSBX that functions as a harpin-like protein for plant immunity.
Introduction
Plants employ innate immune systems to overcome microbial pathogen infections [1], [2]. Pathogen-associated molecular patterns (PAMPs) comprise a diverse group of molecules such as flagellin [3], EF-Tu [4], chitin [5] and harpins [6]–[8]. PAMPs are known to elicit plant-triggered immunity (PTI); briefly, PAMPs are recognized by plasma membrane-localized receptor-like kinases (RLKs), which often contain nucleotide-binding domains and leucine-rich repeats [9]–[11]. Examples of RLKs include flagellin-sensitive 2 (FLS2) [12], the EF-Tu receptor EFR [13], and the chitin elicitor receptor kinase 1 (CERK1) [14]. These RLKs take similar roles to proteins encoded by plant resistance (R) genes for pathogen recognition [9], [10], [15], [16].
PTI is a form of basal defense or nonhost-mediated resistance in plants. PTI and effector-triggered immunity (ETI) activate similar signaling pathways and defense responses in plants. However, ETI generally activates a more prolonged, robust resistance than PTI [2]. Signal transduction pathways associated with PTI and ETI include mitogen-activated protein kinase (MAPK) cascades, calcium fluxes, and the activation of reactive oxygen species (ROS). Furthermore, both ETI and PTI are associated with modulations in hormonal signaling pathways including those associated with production of salicylic acid (SA) for systemic acquired resistance (SAR), jasmonic acid (JA) for induced systemic resistance (ISR) and ethylene (Eth) [17]. Unlike ETI, PTI-modulated signaling requires BAK1, which is a BRI1-ASSOCIATED KINASE 1 that regulates plant signaling by functioning as an adaptor for multiple RLKs [2], [17]–[20]. For example, the FLS2/BAK1 complex phosphorylates BIK1 (Botrytis-induced kinase 1) for signal transduction to the MAPK cascade [21]. The latter may then activate the expression of WRKY transcription factors that regulate SA-, JA- or Eth-dependent genes by binding the W-box [22]. However, it is unclear whether the proteins mentioned above are also involved in harpin-triggered immunity.
Recent studies have demonstrated that the α-helical structure of harpins is essential for HR induction, ion-mediated pore formation, development of curvilinear protofibrils or fibrils (amyloidogenesis), membrane-binding activities, ROS production and callose disposition [23]–[27]. Furthermore, multiple genes are activated in harpin-treated tobacco including those involved in hormone signaling [28], [29], HR markers (e.g. HIN1 and HSR203J) [30], [31] and pathogenesis-related (PR1a and PR1b) [31]–[34]. Multiple reports document that harpin application promotes plant growth and induces SAR and ISR both to plant pathogens [28], [34] and insects [29], [35]. However, no reports have shown that harpin-elicited HR has any association with BAK1 in PTI-mediated signaling pathways.
Although the elicitation of HR in resistant host plants is commonly associated with ETI, it also occurs during PTI [36]. Harpins, which are glycine-rich, heat-stable proteins produced by the type-III secretion system (T3SS), are PAMPs that elicit HR and PTI [27], [37]. The first harpin described was HrpN, which is produced by the fire blight pathogen, Erwinia amylovora [7]. Multiple harpins can exist in a single phytopathogenic species; for example, Pseudomonas syringae pv. tomato DC3000 encodes four harpins, which are designated HrpZ1, HrpW1, HopAK1 and HopP1 [25]. In Ralstonia solanacearum, three harpins, PopA1 [38], HrpW [39], and PopW [40] have been identified. The HR elicited by harpins can be suppressed by eukaryotic metabolic inhibitors [6], [41], [42]. In Xanthomonas, the first harpin reported is HpaG in X. axonopodis pv. glycines [43], homologous to Hpa1 of X. oryzae pv. oryzae and X. oryzae pv. oryzicola and to XopA of X. campestris pv. vesicatoria [8], [43], but the latter does not elicit a HR in tobacco [43]. Interestingly, a hpa1 deletion mutant still triggers a HR on nonhost tobacco [8], [44], [45], indicating that uncharacterized HR-elicitors are present in X. oryzae pv. oryzicola.
The genus Xanthomonas contains 307 species or pathovars that infect at least 124 monocotyledonous and 268 dicotyledonous plants and causes enormous agricultural losses [46]. Despite the huge host range of Xanthomonas, few species in this genus are known to cause disease on tobacco, suggesting that tobacco may sense a conserved molecule in Xanthomonas and potentially initiate plant immunity. In this report, we present evidence that a highly-conserved single-stranded DNA-binding protein (SSBX) in Xanthomonas is regulated by HrpX, secreted via the T3SS, required for full virulence in planta, and elicits HR in nonhost plants. These novel results indicate that SSBX functions as a harpin-like protein and modulates plant immunity in tobacco.
Materials and Methods
Bacterial Strains and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Table S1. The wild-type strains X. oryzae pv. oryzicola RS105, X. oryzae pv. oryzae PXO99A, X. campestris pv. vesicatoria 85–10, X. axonopodis pv. citri 306, X. campestris pv. campestris 8004, R. solanacearum ZJ3721 and E. amylovora 0065 (Table S1) were grown on nutrient agar (NA) or in nutrient broth (NB) [44] at 28°C. Pst DC3000 was grown on King’s Medium B [47]; E. coli and A. tumefaciens GV3101 were grown in Luria-Bertani (LB) medium [48] at 37°C and 28°C, respectively. hrp-inducing media included XOM3 for X. oryzae strains [49], XVM2 for X. campestris pv. vesicatoria 85-10 and X. axonopodis pv. citri 306 [50], and MMX for X. campestris pv. campestris 8004 [51]. MS medium was used for germination of plant seeds [52]. Antibiotics were used at the following concentrations (µg/ml): ampicillin (Ap), 100; kanamycin (Km), 50; rifampicin (Rif), 50; and spectinomycin (Sp), 100 µg/ml.
DNA manipulation
DNA isolations, subcloning, transformation, PCR, Northern blot and immunoblotting were conducted using standard procedures [53]. PCR primers are described in Table S2. PCR products were first cloned into pMD18-T (Takara, China) and then verified by sequencing. DNA sequences were analyzed with the VECTOR NTI software (http://www.invitrogen.com).
Determination of SSBX-elicited HR in planta
To investigate whether ssbXoc triggers HR in tobacco, full-length ssbXoc (552 bp) was amplified by PCR with the primer pairs ssbX-F/ssbX-R (Table S2) using the genomic DNA of strain RS105 as template. The amplified product was then cloned into PVX vector pgR107 [54] at ClaI and SalI sites, resulting in pPVXssbX (Table S1). The hpa1 [8] and bax [55] genes were also cloned into pgR107, generating pPVXhpa1 and pPVXbax (Table S1), which were used as controls. These constructs (along with the empty vector) were transferred into A. tumefaciens GV3101, resulting in strains SSBX, Hpa1, Bax and PVX, respectively. Suspensions of A. tumefaciens strains were adjusted to OD600 = 0.5 and infiltrated into N. benthamiana with needleless syringes. HR symptoms were scored 48 hours post inoculation (hpi). Three independent experiments were performed and similar results were yielded. Representative results from one of these experiments are presented here.
SSB Protein Expression and Purification
SSBXoc homologues were amplified from X. oryzae pv. oryzicola RS105, X. oryzae pv. oryzae PXO99A, X. campestris pv. vesicatoria 85-10, X. axonopodis pv. citri 306, X. campestris pv. campestris 8004, R. solanacearum ZJ3721 and E. amylovora 0065, E. coli BL21(DE3), P. syringae pv. tomato DC3000, and P. fluorescens (Table S1). Each ssb homologue was amplified by PCR from corresponding genomic DNAs using the primers listed in Table S2. PCR products were then cloned into pET41a (+) resulting in constructs designated pSSBXoc , pSSBXoo, pSSBXcv, pSSBXac, pSSBXcc, pSSBRs, pSSBEa, pSSBEc, pSSBPst, and pSSBPf respectively (Table S1). These constructs were transformed into E. coli BL21 (DE3) (Table S1) as recommended in the Novagen pET System manual (Novagen, USA). Proteins were expressed as recommended by Novagen. Briefly, a single colony of each recombinant strain was inoculated to 2 ml LB broth containing Km. After incubation at 37°C for 12 h, 2 ml of culture was transferred into 200 ml of fresh LB liquid containing 1.0 mM isopropyl β-D-thiogalactopyranoside (IPTG, Sigma) and incubated for 4 h at 37°C. Cells were harvested by centrifugation, and pellets (1 g) were resuspended in 5 ml PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 2 mM KH2PO4, pH 7.4); the solution also contained 20% glycerol, 5 U/ml DNaseI and 5 µl PMSF (phenylmethanesulfonylfluoride). Bacterial cells were lysed by sonication (20 kHz, 10 min). After centrifugation (15,000×g) for 15 min at 4°C, the supernatants were purified using a GSTrap™ FF column as recommended by the manufacturer (Purification Manual, GE Healthcare, Germany). The purified proteins were digested by thrombin to remove the GST-tag, and the purified proteins were quantified with the Easy Protein Quantitative Kit (TransGen Biotech, China) and a NANODROP 1000 Spectrophotometer (Thermo). Purified proteins were then used for HR induction assays in tobacco. The Hpa1 protein purified and the empty vector preparation (EVP) by the same procedure was used as a positive and negative control, respectively.
HR assays
Purified proteins were tested for their ability to elicit HR on N. benthamiana or N. tabacum cv. Xanthi by infiltration into plant tissues using needleless syringes. Plant responses were observed 48 hpi for the HR. All plants were grown in growth chambers at 25°C with a 12 h photoperiod. Experiments were repeated at least three times.
To measure minimal HR-eliciting concentrations, purified SSBXoc and others were diluted in PBS buffer at the following concentrations: 50, 25, 10, 5.0, 2.5, 1.0, 0.5, 0.1, 0.05 and 0.01 µM, while the purified Hpa1 diluted in PBS at the same concentrations above was used as positive control. All concentrations of the tested SSB proteins and Hpa1 were infiltrated into tobacco leaves and photographed 48 hpi.
To characterize biochemical activity, purified SSBXoc (1 µM) and Hpa1 (1 µM) were heat-treated at 100°C for 10 min and incubated with protease K (0.5 U/ml) at 37°C for 10 min, respectively. To investigate potential susceptibility to eukaryotic metabolic inhibitors, SSBXoc (1 µM) and Hpa1 (1 µM) were mixed with 1 µM LaCl3, 0.71 µM actinomycin D, and 0.1 µM cycloheximide, respectively. Treated and untreated SSBXoc, Hpa1, and EVP were infiltrated into tobacco leaves. Three independent biological experiments were performed and yielded similar results. Representative results from one of these experiments are presented.
To determine whether the HR induced by SSBXoc was dependent on SA accumulation in planta, purified SSBXoc (1.0 and 5.0 µM) was infiltrated into wild-type and NahG tobacco, respectively. Purified Hpa1 (1.0 and 5.0 µM), wild-type strain RS105 (OD600 = 0.5), and EVP were used as controls. Three independent biological experiments were performed and yielded similar results. Representative results from one experiment are presented in this report.
DNA laddering assays
Genomic DNA of cv. Xanthi leaves infiltrated with purified SSBXoc (1 µM), Hpa1 (0.5 µM) and PBS buffer was isolated at 3, 6, 12, 24, 36 and 48 hpi, respectively. DNase-free RNase A was then used to digest existing RNA. Genomic DNA (10 µg) from each sample was subjected to electrophoresis in 2% agarose gels for at least 10 h under low voltage. Three independent experiments were performed and similar results were yielded. Representative results from one experiment are shown here.
H2O2 assays
Tobacco leaves (N. benthamiana) were infiltrated with purified SSBXoc (1 µM), Hpa1 (0.5 µM), and EVP, respectively, using needleless syringes. Eight hours later, treated leaves were collected and incubated in diaminobenzidine (DAB) for 8 h at 25°C, and then boiled in 95% ethanol for 10 min to remove the dye [56]. After 4 h further incubation in ethanol, leaves were fully bleached and brown precipitates were observed, indicating H2O2 accumulation and the production of ROS. Epidermal peels were performed at the injection sites 0 and 8 hpi with purified SSBXoc (1 µM), Hpa1 (0.5 µM), and EVP; these were then stained with DAB for 8 h at 25°C, and visualized with an OLYMPUS IX71 microscope. Three independent experiments were performed and similar results were yielded. Representative results from one experiment are presented here.
Callose deposition assays
To observe callose deposition, tobacco leaves (N. benthamiana) were infiltrated with purified SSBXoc (1 µM), Hpa1 (0.5 µM), or EVP with a needleless syringe. After infiltration (0 and 8 h), leaf epidermal peels of the infiltrated area were removed and incubated with aniline blue (0.1% in 0.15% K2HPO4, pH 8.2) for 0.5 h. Fluorescence (400 nm excitation and 510 nm emission) and bright-field images were obtained with an OLYMPUS IX71 microscope. Three independent experiments were performed and similar results were yielded. Representative results from one experiment are displayed here.
Northern blot assays
Total RNA of N. benthamiana leaves infiltrated with SSBXoc (1 µM), Hpa1 (0.5 µM) and EVP, was extracted 8 hpi. Prior to electrophoresis, RNA samples were treated with RNase-free DNaseI (TaKaRa, China) to remove potential traces of genomic DNA. RNA samples (30 µg) were then separated by electrophoresis in 1% agarose gels. Biotin-labeled DNA probes were prepared with the BrightStar Psoralen-Biotin Labeling kit (Ambion, USA) as recommended by the manufacturer. The primers for DNA probes are listed in Table S2. RNA was transferred to Hybond N+ membranes (Amersham Pharmacia Biotech, USA), hybridized with specific probes (Table S2) at 42°C using Northern Max (Ambion, USA), and detected using BrightStar BioDetect (Ambion) according to the manufacturer’s instruction.
Assays for Plant Growth Promotion and Disease Prevention by SSBXoc
To detect potential plant growth promoting activity of SSBXoc, seeds of tobacco cv. Xanthi and Arabidopsis. thaliana (Col-0) were treated with SSBXoc (1 µM), Hpa1 (0.5 µM), EVP, and sterile water (DDW) for 8 h at 4°C. Treated seeds were placed on MS agar medium and measured for root length and fresh weight two weeks after treatment.
The potential effect of SSBXoc in enhancing plant disease resistance was investigated on tobacco inoculated with A. alternata strain TBA28A (Table S1), the causal agent of brown spot disease. Ten plants of two-month-old tobacco were spray-inoculated with SSBXoc (1 µM in 0.5% Tween 20 solution) or Hpa1 (0.5 µM); plants were sprayed twice in three-day intervals. EVP was used as a negative control. Three days after the second spray, plants were inoculated with A. alternata TBA28A fresh disc. Infection was measured as the diameter of necrotic brown spots by statistical analysis.
Construction of ssbXoc deletion mutants
Experiments were designed to generate nonpolar deletion mutants of ssbX in X. oryzae pv. oryzicola RS105 and RΔhpa1, a hpa1 deletion mutant [45]. The DNA sequences flanking ssbX were amplified from RS105 genomic DNA using primer pairs ssbI-F/ssbI-R and ssbII-F/ssbII-R (Table S2), cloned into pMD18-T (Takara, China), and verified by sequencing. After digestion with BamHI/XhoI and XhoI/PstI, the two fragments were cloned into the suicide vector pKMS1 [57] at BamHIand PstI sites, resulting in pKΔssbX (Table S1). This construct was introduced into the wild-type RS105 and the hpa1 deletion mutant RΔhpa1, and the isolation of ssbX deletion mutants was performed as described previously [57]. The ssbXoc deletion mutant RΔssbX and the double mutant RΔhpa1ΔssbX were verified by PCR using primers ssbI-F/ssbII-R (Table S2) and by Southern blot analysis using ssbX as a probe.
Bacterial pathogenicity and HR assays
Pathogenicity assays were performed as described previously [8]. X. oryzae pv. oryzicola derivatives were examined for their ability to cause disease symptoms in rice IR24 (Oryza sativa ssp. indica) or to trigger a HR in tobacco cv. Xanthi. Rice adult plants (two-months-old) were inoculated by leaf-needling and fully-expanded tobacco leaves were infiltrated by needleless syringes with bacterial suspensions (∼3×108 cfu/ml). Lesion lengths in rice were scored 14 days post-inoculation (dpi) and the HR in tobacco 2 dpi. All plants were maintained in growth chambers at 25°C with a 12 h photoperiod. Experiments were repeated at least three times.
Measurement of bacterial growth in rice
Bacterial cell suspensions (3×108 cfu/ml; OD600 = 0.3) were infiltrated into recently expanded leaves of two-week-old rice IR24 with needleless syringes at three spots per leaf. Three 0.8 cm diameter leaf discs were harvested with a cork borer from each infiltrated area. The leaf discs were surface sterilized with 70% ethanol first and then with 30% hypochlorite, macerated with a sterile mortar and pestle in 1 ml of distilled water, serial dilutions were plated in triplicate on NA with appropriate antibiotics. Plates were incubated at 28°C for 3–4 days until single colonies could be counted. Bacterial numbers (cfu/cm2) were calculated, and standard deviations were determined using colony counts from three triplicate spots in each of three samples per time point per inoculum. Experiments were repeated at least three times.
Promoter activity assays and quantitative real-time PCR
To construct a transcriptional fusion between the ssbX promoter and glucuronidase (GUS), the promoter region (−1 to −350 bp) upstream of ssbX was amplified from the genomic DNA of X. oryzae pv. oryzicola RS105 with the primer pair pssb-F/pssb-R (Table S2). This PCR product was then fused with the promoter-less gusA gene, which was obtained with primers gusA-F/gusA-R (Table S2). The ssbX-gusA fusion was then cloned into pUFR034 [58] at the EcoRI site, resulting in pPIPAGUS (Table S1). In another experiment, a mutation was introduced into the PIP-box of the ssbX promoter using primers mpssb-F/pssb-R (Table S2) and fused with gusA, resulting in pPIPBGUS (Table S1).
For GUS activity assays, X. oryzae pv. oryzicola RS105 strain and hrp mutants were cultured in XOM3 to OD600 = 0.5. Bacterial cells were diluted and disrupted in sonication buffer (20 mM Tris-HCl, pH 7.0, 10 mM 2-mercaptoethanol, 5 mM EDTA, and 1% Triton X-100). GUS activities were determined every 30 min over a 3-h time period by measuring absorbance (A 415) with p-nitrophenyl-D-glucuronide as the substrate [59]. One unit (U) was defined as 1 nmol of 4-methyl-umbelliferone produced per min per bacterium.
For quantitative real-time PCR analysis (qRT-PCR), the bacteria were cultured as described for the GUS activity assay in this report or cultured in rice suspension cells as described by Li and her colleagues [44]. Total RNA was extracted using Trizol reagent according to the manufacturer’s instructions (Invitrogen, USA). Total RNA was quantified by measuring the OD260/OD280, and the quality was examined by gel electrophoresis. Before synthesis of the first stranded, total RNA was treated with RNase-free DNaseI (TaKaRa, China) to remove genomic DNA. Removal of contaminating DNA was confirmed by using extracted RNA as a template to amplify selected target genes using the primers listed in Table S2. cDNA synthesis and PCR were conducted with AMV (TaKaRa) and Ex-Taq DNA polymerase (TaKaRa, China) using the primers listed in Table S2. Semi-quantitative RT-PCR was performed using the following program: step 1, 95°C for 3 min; step 2, 95°C for 20 s; step 3, 55°C for 30 s; step 4, 72°C for 40 s; 35 cycles of steps 2–4; and step 5, 72°C for 10 min. Quantitative real-time PCR was performed on an Applied Biosystems 7500 Real-Time thermocycler using SYBR Premix Ex Taq™ (Takara, China). Conditions for quantitative RT-PCR were as follows: denaturation at 95°C for 30 s and 41 cycles for 95°C, 5 s; 60°C, 34 s. The results were analyzed using Applied Biosystems 7300 System SDS software and the RQ study application. Expression of the 16S rRNA gene was used as an internal standard to verify the absence of significant variation in cDNA levels. The comparative-threshold method by log2 value was used to calculate the relative mRNA level with respect to the corresponding transcript of ssbXoc in the wild-type RS105 and the hrpG and hrpX mutants (Table S1) cultured in NA medium and rice suspension cells, respectively. All the real-time quantitative RT-PCRs were performed in triplicate.
SSBXoc secretion assays
To determine whether the secretion of SSBXoc was dependent on the T3SS, X. oryzae pv. oryzicola strains, containing SsbXoc-c-myc or hpa1-c-myc fusion (as a positive control) (Table S1), were pre-incubated in NB medium to logarithmic phase. Bacterial cells were harvested and adjusted to OD600 = 2.0 with sterilize water and washed twice. Then, 40 µl of bacterial suspension was poured into 1 ml of the hrp-inducing medium XOM3 [49] and incubated at 28°C for 8 h. Cell and supernatant fractions were separated by centrifugation, and the protein in the supernatant fraction was precipitated with 12.5% trichloroacetic acid. Proteins were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to membranes for immunoblotting using anti-c-Myc primary antibody (Genescript, China). Primary antibodies were recognized by anti-rabbit secondary antibodies (Genescript, China) and visualized on autoradiographs with the Western-Light Chemiluminescence System (Transgene, Beijing, China). Three independent experiments were performed and yielded similar results. Representative results from one of these experiments are presented here.
Results
ssbXoc Encodes a Single-stranded DNA-binding Protein Eliciting HR in Tobacco
Mutagenesis of hrpG or hrpX in X. oryzae pv. oryzicola abolishes the elicitation of HR in tobacco and pathogenicity in rice [8]. Thus, we assumed that the expression of HR-eliciting genes, including hpa1, are also regulated by HrpG and HrpX [60]. Using cDNA microarrays of X. oryzae pv. oryzicola strain RS105 and the hrpG & hrpX mutants (unpublished), we discovered that the expression of XOC_1514, which encodes a single-stranded DNA-binding protein (AEQ95695.1) [61], was positively regulated by HrpG and HrpX in pathogen-infected rice cells (Fig. 1). This protein, which was designated SSBXoc, is rich in glycine (20% of the total amino acids) but lacks cysteine residues (Table S3, Fig. S1); these are characteristics typical of the harpin protein family. To confirm this, we expressed ssbXoc in PVX vector pgR107, which is typically used to screen HR elicitors in tobacco [54]. SSBXoc triggered HR in N. benthamiana that was similar to Hpa1 [8] and Bax [55] (Fig. 2A), suggesting that SSBXoc functions as a harpin in X. oryzae pv. oryzicola.
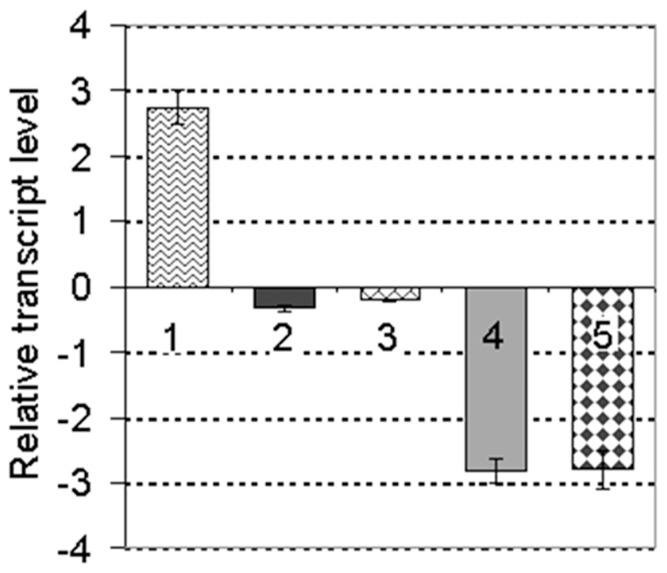
Figure 1. Expression of ssbXoc is induced in rice suspension cells.
Real-time quantitative PCR analysis of ssbXoc transcript levels in X. oryzae pv. oryzicola wild-type RS105 and mutants RΔhrpG and RΔhrpX. Strains were grown in NB or rice suspension cells, and designated as (−) and (+), respectively. The ratios (shown in units of log2) reflect ssbXoc transcript levels between different strains in two different growth conditions. 1. +RS105/−RS105; 2. −RΔhrpG/−RS105; 3. −RΔhrpX/−RS105; 4. +RΔhrpG/+RS105; 5. +RΔhrpX/+RS105. Data represent the means ± standard deviations (SD) from three replicates.
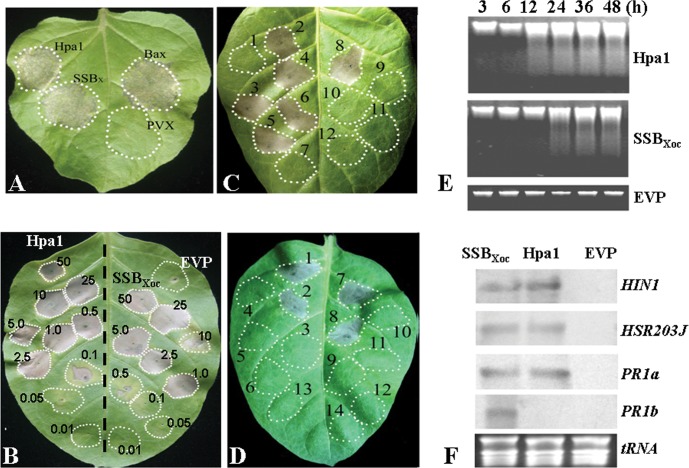
Figure 2. A highly conserved single-stranded DNA-binding protein (SSB) triggers a HR in tobacco.
(A) HR induction by the SSBXoc protein of X. oryzae pv. oryzicola. A. tumefaciens GV3101 (OD600 = 0.5) containing hpa1, ssbX and bax genes in the PVX vector pgR107 was inoculated into N. benthamiana tobacco leaves with a needleless syringe. Hpa1 and Bax were used as positive controls, and A. tumefaciens containing the empty vector PVX was used as a negative control. (B) Concentration of SSBXoc required for HR induction in tobacco cv. Xanthi. Purified SSBXoc was diluted in PBS buffer and inoculated into tobacco with needleless syringes. Hpa1, which functions as a harpin of X. oryzae pv. oryzicola, was used as a positive control and EVP as a negative control. (C) HR assays in tobacco inoculated with SSBX homologues obtained from various bacterial species. SSB proteins were overproduced in E. coli, purified (see Methods), and diluted in PBS buffer to different concentrations from 0.01 to 50 µM. A typical image of HRs on tobacco leaves caused by the proteins at 1 µM was taken in this report. Numbers represent sections of leaves inoculated with the following: 1, EVP; 2, SSBXoc from X. oryzae pv. oryzicola RS105; 3, SSBXoo from X. oryzae pv. oryzae PXO99A; 4, SSBXac from X. axonopodis pv. citri 306; 5, SSBXcv from X. campestris pv. vesicatoria 85-10; 6, SSBXcc from X. campestris pv. campestris 8004; 7, SSBPf from P. fluorescens Pf-5; 8, Hpa1Xoc, from X. oryzae pv. oryzicola RS105; 9, SSBEa from E. amylovora 0065; 10, SSBEc from E. coli BL21 (DES); 11, SSBRs from R. solanacearum ZJ2731; and 12, SSBPst from P. syringae pv. tomato DC3000. (D) Assays for SSBx and Hpa1-induced HR in response to various metabolic inhibitors. Tobacco plants were infiltrated with SSBX (1 µM) or Hpa1 (0.5 µM), which was heat-treated or incubated (see methods) with one of the following: 1 µM LaCl3, 0.71 µM actinomycin D, 0.1 µM cycloheximide or protease K (0.5 U/ml). Leaf panels: 1, Hpa1; 2, heat-treated Hpa1; 3, protease K-treated Hpa1; 4, Hpa1 plus 1 µM LaCl3; 5, Hpa1 plus 0.71 µM actinomycin D; 6, Hpa1 plus 0.1 µM cycloheximide; 7, SSBX; 8, heat-treated SSBXoc; 9, protease K-treated SSBXoc; 10, SSBXoc plus LaCl3; 11, SSBXoc plus 0.1 µM actinomycin D; 12, SSBXoc plus cycloheximide; 13, distilled water; and 14, EVP. Leaves in panels A to D were photographed 24–48 h after infiltration. (E) Analysis of DNA laddering in SSBXoc-treated tobacco leaves. Total genomic DNA was isolated from tobacco leaves 3, 6, 12, 24, 36 and 48 hpi with Hpa1 (0.5 µM), SSBXoc (1 µM) and EVP. DNA laddering was evaluated in 2% agarose gels. (F) Northern blot analysis in tobacco inoculated with SSBX, Hpa1, or EVP. The marker genes, HIN1, HSR203J, PR1a and PR1b, were chosen as the targets. Total RNAs were extracted from tobacco leaves infiltrated with SSBXoc (1 µM), Hpa1 (0.5 µM), or PBS buffer. Aliquots (10 µg each) of the extracted RNAs were separated in 1% agarose gels, transferred onto membranes, and analyzed by northern blotting. Blots were hybridized with digoxigenin-labeled HIN1, HSR203J, PR1a and PR1b cDNA. The experiment was conducted twice with similar results.
Previously, we reported that the minimum concentration of Hpa1 for HR induction is 0.1 µM [8]. To determine the concentration of SSBXoc required for HR induction, we over-expressed the protein in E. coli BL21 (DE3) (Table S1). Purified SSBXoc was infiltrated into tobacco at concentrations ranging from 0.01 to 50 µM. The minimum concentration of SSBXoc needed for HR induction in tobacco cv. Xanthi was 1.0 µM, approximately 10-fold higher than the minimum effective concentration of Hpa1 (Fig. 2B).
Nucleotide and protein searches using the NBCI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) indicate that SSBXoc homologues exist in other bacteria. Protein sequence alignment of SSBXoc with homologues from other Gram-negative bacteria indicated that the differences of SSB proteins between Xanthomonas and other prokaryotic bacteria mainly existed in the glycine-rich regions (see rectangle marked with dashes, Fig. S1). A phylogenetic analysis showed that SSB proteins could be classified into one of three groups (Fig. S2). Group I contained SSB proteins from closely related Xanthomonas species, group II SSB homologues from Xyllela fastidiosa, R. solanacearum, Thermus aquaticus, P. aeruginosa, and P. syringae pv. tomato, and group III from Candidatus Liberibacter asiaticus, P. fluorescens, E. amylovora, Dickeya dadantii, Escherichia coli and Shigellia dysenteriae (Fig. S2). Percentages of glycine-rich amino acids of SSBX in X. oryzae pv. oryzicola RS105 strain and other bacteria are also shown in Table S3.
The bioinformatics analysis described above prompted us to investigate whether the SSB proteins from various bacterial species could elicit HR in tobacco. PCR was used to amplify ssb genes from X. oryzae pv. oryzae PXO99A, X. campestris pv. campestris 8004, X. axonopodis pv. citri (Xac) 306, X. campestris pv. vesicatoria 85-10, R. solanacearum ZJ3271, P. syringae pv. tomato DC3000, P. fluorecens Pf-5, E. amylovora 0065, and E. coli BL21 (DE3) (Table S1). ssb genes were amplified using the primers listed in Table S2, and then cloned into pET30a, generating pSSB constructs (Table S1) harbored by E. coli BL21 (DE3). The overproduced and purified SSB proteins at concentrations from 0.01 to 50 µM were infiltrated into tobacco cv. Xanthi with needleless syringes. Only did the SSB proteins from Xanthomonas elicited HR in tobacco, whereas those from other bacterial species did not ((Fig. 2C), suggesting that only SSBX homologues, which are closely related and highly conserved in Xanthomonas (Fig. S1; Table S3), function as harpin.
Electrophoretic mobility shift assays (EMSA) demonstrated that SSBXoc from X. oryzae pv. oryzicola, as the representative of these proteins in Xanthomonas bound randomly synthesized single-stranded DNAs (DNA1 and DNA2, Fig. S3). This is consistent with that the single-stranded DNA-binding protein is for ssDNA protection from nucleolytic digestion in bacterial cell viability [62], implying that SSBX of Xanthomonas rather than other plant pathogenic bacteria is coevolved to be recognized as a potential HR-elicitor by plants.
Previous reports indicate that harpin proteins are heat-stable and protease-sensitive [6], [7], [34], [41], [42]. To investigate these characteristic for SSBX., purified protein (1 µM) was incubated in a water bath at 100°C for 10 min and with protease K (0.5 U/ml) at 37°C for 10 min, while Hpa1 was used as positive control. Heat- or protease-treated SSBXoc was then inoculated into tobacco cv. Xanthi. At 48 hpi, heat-treated SSBXoc still triggered HR in tobacco (Fig. 2D panel 8), but protease-treated SSBXoc did not (Fig. 2D, panel 9).
SSBXoc-elicited HR is a form of Programmed Cell Death
The next experiments were designed to determine whether SSBXoc, like Hpa1, is toxic to plant cells or not and SSBXoc leads to a metabolic response that triggers HR. Three metabolic inhibitors were used: actinomycin D (inhibits eukaryotic RNA polymerase II), cycloheximide (targets 80S ribosomes), and LaCl3 (a calcium channel blocker). These inhibitors were incubated with purified SSBXoc (see Methods) and then assayed for HR induction in tobacco. All three inhibitors prevented the SSBXoc-elicited HR in tobacco when co-infiltrated with the purified SSBXoc (Fig. 2D, panels 12–14). These results indicated that the SSBXoc-elicited HR is an active process and requires de novo gene expression, protein synthesis and calcium flux across membranes. Thus, SSBXoc acts as an elicitor, like Hpa1, of HR but is not directly toxic to plant cells.
It is well-documented that harpin-elicited HR is a form of programmed cell death (PCD), which is accompanied by DNA laddering [63]. To determine whether the SSBXoc-elicited HR is a form of PCD, DNA laddering experiments were performed. Total genomic DNA from SSBXoc-infiltrated tobacco leaves were extracted at different time points after infiltration and analyzed on 2% agarose gels. As shown in Fig. 2E, DNA ladders were clearly observed in SSBXoc-inoculated leaves at 24 hpi, 12 h later than that in Hpa1-inoculated leaves. Thus, SSBXoc, like Hpa1 (Fig. 2E), elicits PCD that is characterized by DNA laddering.
We then investigated whether SSBXoc-elicited HR occurs with the activation of known HR marker genes including HIN1 (harpin-induced 1) [64], HSR203J [65], and the SA-dependent marker, PR1a [66]; the JA-dependent gene, PR1b [67], was also conducted. The expression of these genes was evaluated in tobacco leaves infiltrated with SSBXoc, Hpa1, and EVP 6 hpi. All four genes were induced in response to SSBXoc; however, Hpa1 did not induce the expression of PR1b (Fig. 2F). Transcripts started to accumulate 6 hpi with SSBXoc and Hpa1 and were expressed at high levels up to 24 h (data not shown). These findings indicate that SSBXoc-elicited HR was accompanied by the expression of HR markers and plant defense genes.
SSBXoc-elicited HR is Dependent on SA Accumulation
It has been reported that HR induction by harpins requires SA accumulation [28], [68]. To investigate whether this is valid for SSBXoc-elicited HR, we utilized transgenic tobacco expressing NahG; this line produces salicylate hydroxylase which degrades SA and blocks its accumulation [69]. Purified SSBX and Hpa1 produced a HR in wild-type tobacco (Fig. 3B), but not in the NahG line (Fig. 3A). Thus, SSBXoc-induced HR relies on SA accumulation in planta, which is the case for other harpins. It is noteworthy that the wild-type RS105 of X. oryzae pv. oryzicola still elicited HR in SA-deficient tobacco (Fig. 3A). Thus, in addition to SSBXoc and Hpa1, other unidentified HR-elicitor(s) exist(s) in X. oryzae pv. oryzicola to trigger HR on tobacco in SA-independence.
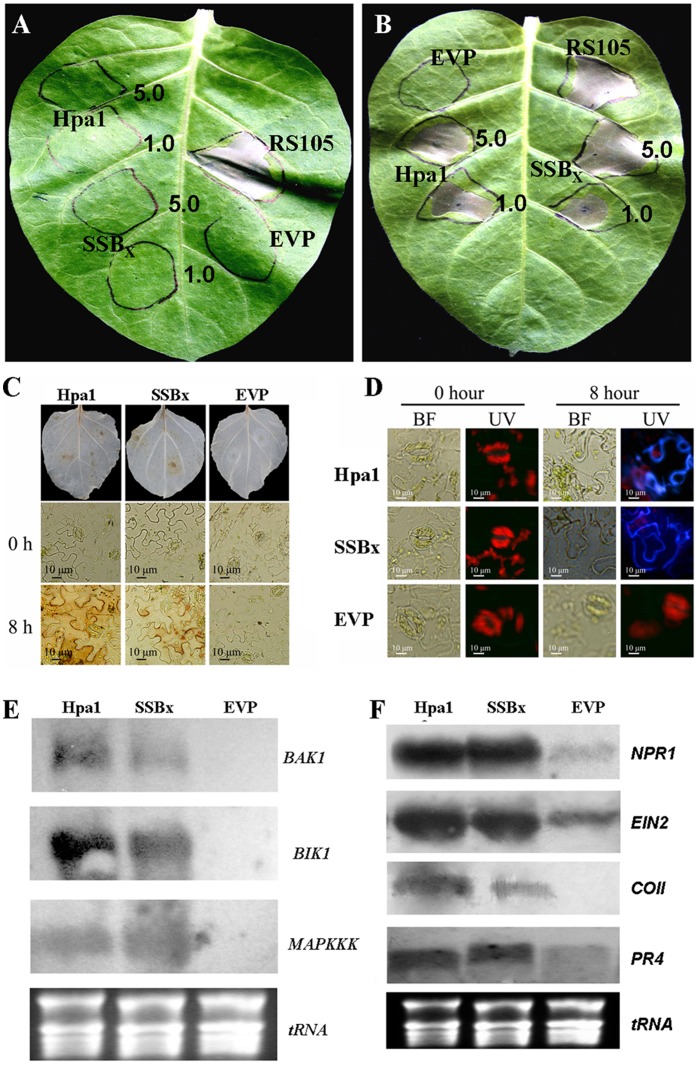
Figure 3. SSBXoc from X. oryzae pv. oryzicola may function as a PAMP and activates PTI in tobacco.
SSBXoc-triggered HR depends on the accumulation of salicylic acid (SA). X. oryzae pv. oryzicola strain RS105 (OD600 = 0.5), SSBXoc (1.0 and 5.0 µM), Hpa1 (1.0 and 5.0 µM), and EVP were inoculated into a NahG tobacco leaves (A) or wild-type tobacco cv. Xanthi (B). Photographs were taken 48 hpi. (C) SSBXoc-triggered HR is accompanied by the oxidative burst. The production of H2O2 was evaluated in tobacco leaves by staining with 3,3′-diaminobenzidine tetrahydrochloride (DAB). The reaction mixture contained 200 ml of 0.5 mM DAB in 50 mM Tris acetate buffer (pH 6.0) with purified SSBXoc (1 µM) or Hpa1 (0.5 µM). Fully-expanded tobacco leaves were infiltrated with needleless syringe, incubated at room temperature for 0 and 8 h, and decolorized in 80% (v/v) ethanol for 10 min at 70°C. Leaves were examined with an OLYMPUS IX71 microscope. PBS buffer was included as a negative control. (D) SSBXoc elicits callose deposition in tobacco cell walls. Callose deposition in tobacco leaves was observed using fluorescence microscopy (OLYMPUS IX71) and staining with aniline blue at 0 and 8 hpi. Purified SSBXoc (1 µM) or Hpa1 (0.5 µM) was infiltrated into tobacco leaves (N. benthamiana) using needleless syringes, and EVP was used as a negative control. Inoculated epidermal peels were incubated with aniline blue for 0.5 h. Fluorescence images were captured using a 400 nm exposure for 510 nm absorbed light (UV), and bright-field (BF) images were captured using general bright light. Panels (E) and (F) show Northern blot analysis of PTI signaling pathways in tobacco treated with SSBXoc, Hpa1, and PBS buffer (control). (E) The expression of BAK1, BIK1 and MAP3K; (F) The expression of NPR1, EIN2, COl1, and PR4. Purified SSBXoc (1 µM) or Hpa1 (0.5 µM) was infiltrated into tobacco leaves using needleless syringe, and PBS buffer was used as a negative control. RNA was extracted 8 hpi and 10 µg aliquots were separated on 1% agarose gels and transferred to nylon membranes. Blots were hybridized with digoxigenin-labeled cDNA probes of the indicated genes.
SSBXoc Activates Plant Basal Defense
The oxidative burst, which involves the generation of ROS in response to microbial elicitors, occurs quite quickly in resistant plant cells [70], [71]. Thus we investigated whether the oxidative burst is generated in SSBXoc-treated tobacco cells. At 8 hpi, DAB staining resulted in necrotic brown spots indicative of H2O2 production in both SSBXoc- and Hpa1-infiltrated leaves (Fig. 3C).
Along with the oxidative burst, plants often mobilize multiple forms of basal defense to inhibit pathogen ingress, including callose deposition in cell walls [72]. To determine whether SSBXoc elicits callose deposition in tobacco, epidermal peels from SSBXoc-infiltrated tissue were stained with aniline blue and examined by fluorescence microscopy. Both SSBXoc- and Hpa1-infiltrated leaves showed evidence of callose deposition beginning at 8 hpi (Fig. 3D). Thus, SSBXoc, like Hpa1, functions as an elicitor of basal defense responses and stimulates callose deposition.
The oxidative burst and callose deposition in tobacco infiltrated with SSBXoc prompted us to speculate that SSBXoc may function as a PAMP and activate the expression of genes involved in PTI signaling pathways. Our results indicate that BAK1, BIK1 and MAP3K (Fig. 3E), and NPR1, EIN2, COl1 and PR4 (Fig. 3F) genes are activated in response to SSBXoc or Hpa1. The data show that SSBXoc triggers a cascade of events similar to those triggered by Flg22 [12], [71], [73], [74], which leads to the oxidative burst and callose deposition and activates the expression of PR genes. These results support our presumption that SSBXoc acts as a PAMP like Hpa1.
SSBXoc Induces Plant Disease Resistance and Promotes Plant Growth
Tobacco infiltrated with SSBXoc shows elevated expression of SA- and JA-dependent genes, along with the oxidative burst and callose deposition (Fig. 2F; Fig. 3C, D). Therefore, we hypothesized that SSBXoc may stimulate induced resistance to pathogen infection. For this, we inoculated a fungal pathogen, A. alternata TBA28A (Table S1), causal agent of tobacco brown spot disease, to fully-expanded tobacco leaves that were previously spray-inoculated twice with SSBXoc (1 µM) in three-day intervals. The necrotic areas in tobacco leaves treated with SSBXoc were significantly smaller (P = 0.01, t test) than those observed on leaves inoculated with EVP ( Fig. 4 ). Like SSBXoc, we found that Hpa1 (0.5 µM) also induced similar resistance to A. alternata. The data suggest that both SSBX and Hpa1 induce SAR against pathogen infection.
Figure 4. SSBXoc induces resistance to tobacco brown spot disease caused by A. alternata.
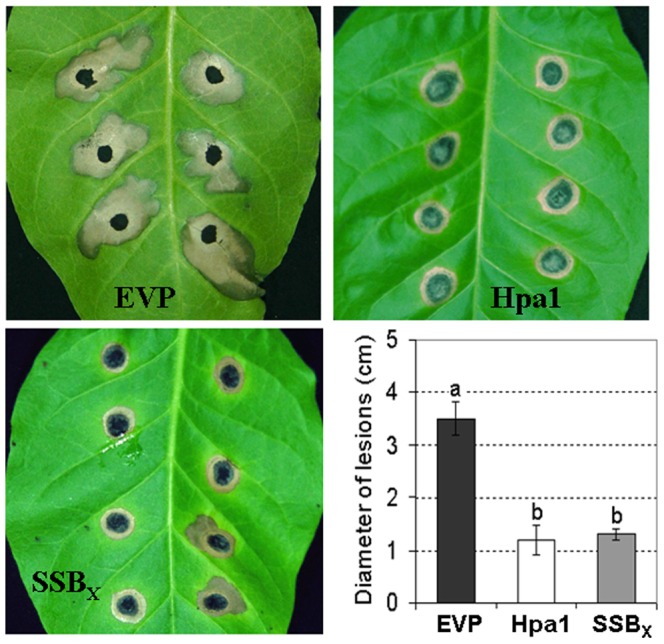
Fully-expanded tobacco leaves (cv. Xanthi) were sprayed twice in three-day intervals with purified SSBXoc (1 µM), Hpa1 (0.5 µM) and EVP (negative control). Three days after the second application of SSBXoc, leaves were inoculated with A. alternata strain TBA28A. Diameters of brown spot lesions were measured and photographed 14 dpi. Lesion size (diameter) are shown ± SD of triplicate measurements. Different letters above columns indicate significant differences at P = 0.01 using the Student’s t test.
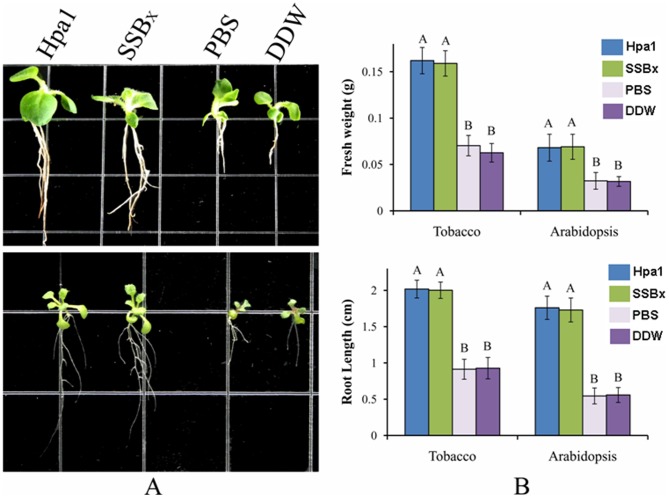
The application of a harpin, HrpN from E. amylovora, enhances plant growth, particularly because the Eth-dependent genes are activated [30], [75]. The activation of Eth-dependent genes, e.g. EIN2 and PR4 (Fig. 3F), led us to determine whether SSBXoc promotes plant growth or not. For this, germinating seeds of Arabidopsis thaliana Col-0 and tobacco cv. Xanthi were soaked in a solution containing 1 µM SSBXoc for 8 h and then transferred to MS medium for 14 days. The results showed that root lengths of SSBXoc-treated Arabidopsis and tobacco plants were nearly 2-fold longer than plants treated with EVP or water. There was no significant difference in root lengths between SSBXoc and Hpa1 treatments (P = 0.01, t test; Fig. 5). In addition, fresh weight of SSBXoc–treated plants, like those treated with Hpa1, was nearly three times more than those treated with EVP or water (Fig. 5B). Thus, SSBXoc promotes plant growth, possibly through the activation of the Eth signaling pathway (Fig. 3F).
Figure 5. SSBXoc enhances plant growth.
(A) Phenotype of tobacco (cv. Xanthi) and Arabidopsis thaliana (Col-0) grown on MS medium, 14 days after seed treatment with Hpa1 (0.5 µM), SSBXoc (1 µM), EVP, or double distilled water (DDW). Upper panel, tobacco; lower panel, Arabidopsis. (B) Fresh weight and root length of treated plants. Upper panel, fresh weight; lower panel, root length. Data are means ± SD of 50 randomly selected plants. Different letters above columns represent significant differences between treatments (P = 0.01 by t test).
ssbXoc is Required for Full Virulence and Bacterial Growth in Rice
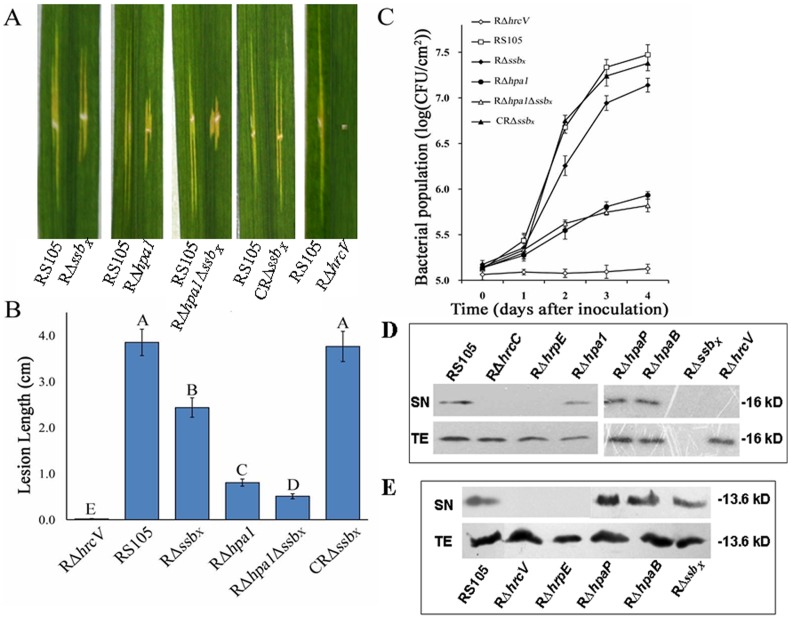
To investigate the potential contribution of SSBX to virulence, ssbXoc was deleted both in X. oryzae pv. oryzicola RS105 and the hpa1 deletion mutant, RΔhpa1 (Table S1). The RΔssbXΔhpa1 double mutant (Table S1) was constructed using a two-step integration procedure [57]. Inoculation studies were conducted by inoculating one half of a rice leaf with wild-type RS105 and the remaining half with one of the following: ssbXoc deletion mutant RΔssbX, hpa1 mutant RΔhpa1 [44], the double mutant RΔssbXΔhpa1 (Table S1), the complemented strain CRΔssbX and the T3SS mutant RΔhrcV [76]. Symptoms in RΔssbX-inoculated leaves were reduced relative to the wild-type strain, but were not as attenuated as RΔhpa1-mediated symptoms (Fig. 6A). Lesion lengths in RΔssbX-inoculated leaves were significantly smaller than those induced by the wild-type RS105 (P = 0.01, t test) but larger than those induced by RΔhpa1 (Fig. 6B). The double mutant RΔssbXΔhpa1 did not lose pathogenicity in rice (Fig. 6A), but lesions were significantly smaller than those induced by the wild-type and single mutants (RΔssbX and RΔhpa1) (Fig. 6B). As expected, the T3SS mutant, RΔhrcV, produced no obvious disease symptoms in rice (Fig. 6A, B). Disease lesion lengths for the complemented strain CRΔssbX were equivalent to those induced by the wild-type RS105 (Fig. 6A, B), indicating that the mutant could be complemented for symptoms with the ssbXoc gene in trans.
Figure 6. Secretion of SSBXoc depends on the T3SS and is required for full virulence and bacterial growth in rice.
(A) Symptoms and (B) lesions lengths were used to assess the virulence of X. oryzae pv. oryzicola RS105 and selected mutants. One half of a rice leaf (IR24, two-months old) was inoculated with wild-type RS105, and the remaining half was inoculated with one of the following deletion mutants: ssbXoc deletion mutant RΔssbX, hpa1 mutant RΔhpa1, the double mutant RΔssbXΔhpa1, the complemented mutant CRΔssbX, and the T3SS mutant RΔhrcV. Ten leaves were inoculated with each strain (OD600 = 0.3; approximately 3×108 cfu/ml) by leaf-needling, and the assay was conducted in triplicate. Bacterial leaf streak symptoms were photographed 14 dpi, and representative symptoms are shown (A). The average lesion lengths formed by the wild-type and mutants were measured 14 dpi (B), and data represent means ± SD from three replicates. Different letters in each data column indicate significant differences at P = 0.01 (t test). (C) Bacterial growth assays in planta. Strains (OD600 = 0.3) were infiltrated into leaves of rice seedlings (IR24, two-weeks old) with blunt-end plastic syringes, and the cfu/cm2 of tissue was evaluated as described in Methods. Data represent means ± SD from three replications. (D) and (E) demonstrated the secretion of SSBXoc (D) and Hpa1 (E) are dependent on a functional T3SS of X. oryzae pv. oryzicola. This experiment utilized X. oryzae pv. oryzicola RS105 and strains containing mutations in the following genes: hrcV (RΔhrcV), hrcC (RΔhrcC), hrpE (RΔhrpE), hpaB (RΔhpaB), hpaP (RΔhpaP), hpa1 (RΔhpa1) and ssbXoc (RΔssbX) to express ssbXoc-c-myc or hpa1-c-myc fusion (as a positive control). After incubation (8 h) in hrp-inducing medium XOM3, total cell extracts (TEs) and culture supernatants (SNs) were analyzed by SDS-PAGE and immunoblotted with an anti-c-Myc antibody. The immunoblotting assay was conducted twice, and similar results were obtained each time. For the detection of SSBXoc, the strain RΔssbX with the empty vector pUFR034 was used as a negative control (D).
To determine whether ssbxoc contributes to growth of X. oryzae pv. oryzicola in rice, we compared the population dynamics of the wild-type RS105, RΔssbX, CRΔssbX, RΔhpa1, RΔssbXΔhpa1, and RΔhrcV. The populations of RΔssbX were significantly lower than the wild-type RS105 beginning 2 dpi, but higher than the population of RΔhpa1 and RΔssbXΔhpa1 (Fig. 6C). Growth of RΔssbX was restored to wild-type levels when ssbXoc was present in trans (Fig. 6C). These results indicated that ssbXoc, like hpa1, contributes to bacterial growth in planta, although the effect was not as pronounced as seen with the T3SS mutant (RΔhrcV) or RΔhpa1. Furthermore, mutations in ssbXoc and hpa1 did not abolish the ability of the pathogen to elicit HR in tobacco (data not shown), implying that other HR-elicitor(s) exist in X. oryzae pv. oryzicola.
SSBX Secretion is Dependent on a Functional TTSS
The T3SS deficient mutant RΔhrcV did not trigger HR in tobacco implies that HR elicitors, including SSBXoc and Hpa1, may be secreted via the T3SS. Bioinformatics analysis of SSBXoc did not show obvious T3SS secretion signals that are commonly found in T3SS effector proteins [77], [78]; thus it was not clear whether SSBXoc secretion required a functional T3SS. We used immunoblotting and SSBXoc tagged with a c-Myc epitope to explore whether SSBxoc secretion was T3SS-dependent. The construct for expressing c-Myc-tagged SSBXoc was transferred into the wild-type and mutants defective in hrcV (encodes an inner membrane component of the T3SS), hrcC (encodes an outer membrane component), hrpE (encodes protein subunits of the Hrp pilus), hpaB and hpaP (encode exit control proteins for T3SE secretion) [8]. These mutants were designated RΔhrcV, RΔhrcC, RΔhrpE, RΔhpaB and RΔhpaP (Table S1), respectively. RΔssbX with the empty vector pUFR034 was used as a negative control. When the wild-type RS105 and mutants RΔhrcC, RΔhrcV, RΔhrpE, were incubated in a hrp-inducing medium XOM3 [50] and examined by immunoblotting, the SSBXoc-c-Myc protein, like Hpa1-c-Myc protein (as a positive control), was found in the supernatants (SN) of the wild-type and RΔhrpE, RΔhpaB, and RΔhpaP but absent from the SN fraction of RΔhrcV, RΔhrcC and RΔhrpE (Fig. 6D, E). These results indicate that a functional T3SS is needed for secretion of SSBXoc and Hpa1. The SSBXoc-c-Myc protein was detected in the SNs and total extracts (TE) of RΔhpaB and RΔhpaP, indicating that HpaB and HpaP are not required for secretion of SSBXoc and Hpa1 (Fig. 6D, E). Moreover, the secretion of Hpa1 was not impaired by the mutagenesis in ssbXoc, and vice versa (Fig. 6D, E).
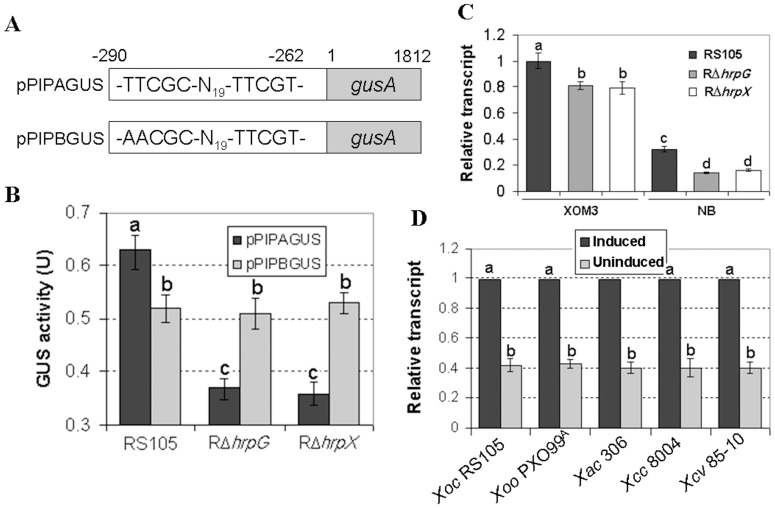
ssbX is positively regulated by HrpX
The down-regulated expression of ssbXoc in the RΔhrpX and RΔhrpG mutants indicates that ssbXoc is positively regulated by HrpX and HrpG (Fig. 1). To investigate this further, we used promoter prediction software (HUhttp://www.fruitfly.org/seq_tools/promoter.html) to analyze the ssbXoc promoter region in the X. oryzae pv. oryzicola BLS256 genome [61]. This analysis revealed an imperfect PIP-box (TTCGC-N19-TTCGT) upstream of the ssbXoc start codon (Fig. 7A), suggesting that ssbXoc may be regulated by HrpX [79]. To determine whether ssbXoc expression depends on the putative PIP-box and HrpX, we constructed a recombinant plasmid pPIPAGUS, which contains the ssbXoc promoter region fused to a promoter-less gusA in pUFR034, resulting in pPIPAGUS (Fig. 7A, Table S1). A mutated ssbXoc promoter (first two TT nucleotides replaced with AA, see Fig. 7A) was also fused to gusA in pUFR034, generating pPIPBGUS (Fig. 7A, Table S1). Plasmids pPIPAGUS and pPIPBGUS were transformed into the wild-type RS105 and mutants RΔhrpX and RΔhrpG, incubated in the hrp-inducing medium XOM3, and GUS activities were measured. GUS activity of pPIPAGUS was significantly lower in the hrpG and hrpX mutants than in the wild-type strain (P = 0.01, t test). GUS activity of the mutated ssbXoc transcriptional fusion (pPIPBGUS) was similar in the wild-type, RΔhrpG, and RΔhrpX strains (P = 0.01, t test) (Fig. 7B). We also used real-time PCR to evaluate expression levels of ssbXoc in strains RS105, RΔhrpG, and RΔhrpX. The expression of ssbXoc was higher in the hrp-inducing medium XOM3, but significantly reduced in the nutrient-rich NB, regardless of the genetic background (Fig. 7C). ssbXoc expression levels were consistently lower in RΔhrpG and RΔhrpX than the wild-type (Fig. 7C). Collectively, results indicate that ssbX transcription is positively regulated by HrpG and HrpX and suggest that ssbXoc contains a PIP-box that is likely regulated by HrpX.
Figure 7. ssbX is expressed in hrp-inducing conditions and regulated by HrpG and HrpX.
(A) Schematic map of a transcriptional fusion where the ssbXoc promoter of X. oryzae pv. oryzicola RS105 is fused to the gusA reporter gene. Upper panel shows pPIPAGUS containing the ssbXoc promoter and an imperfect PIP-box (TTCGC-N19-TTCGT) fused with a promoter-less gusA gene. Lower panel shows pPIPBGUS with a mutated ssbXoc promoter (the first TT nucleotides replaced with AA) fused with gusA. (B) â-glucuronidase (GUS) activity in the hrp-inducing medium XOM3. Plasmids pPIPAGUS and pPIPBGUS were transferred into the wild-type RS105 and mutants RΔhrpG and RΔhrpX. The recombinant strains were then grown in hrp-inducing medium XOM3 for 16 h. GUS activity was determined by measuring the OD at 415 nm using ρ-nitrophenyl-â-D-glucuronide as a substrate. Data represent the mean ± SD of triplicate measurements. The different letters above each horizontal column indicate significant differences at P = 0.01 (t test). (C) Expression of ssbXoc in hrp-inducing and nutrient-rich media. Real-time quantitative RT-PCR was used to compare relative expression of ssbXoc in X. oryzae pv. oryzicola strains RS105, RΔhrpG, and RΔhrpX. RNA was isolated from strains grown in a nutrient-rich medium (NB) and the hrp-inducing medium (XOM3) for 16 h. The relative mRNA levels of ssbXoc in the hrpG and hrpX mutants were calculated with respect to the wild-type strain. Values given are the means ± SD of triplicate measurements from a representative experiment, and similar results were obtained in two other experiments. Different letters above horizontal columns represent significant differences at P = 0.01 using the Student’s t test. (D) Real-time RT-PCR evaluation of ssbX expression in Xanthomonas species. Strains were grown at 28°C for 16 h in NB or one of the following hrp-inducing media: XOM3 for X. oryzae pv. oryzicola RS105 and X. oryzae pv. oryzae PXO99A (Xiao et al. 2007), XVM2 for X. axonopodis pv. citri 306 & X. campestris pv. vesicatoria 85-10, and MMX for X. campestris pv. campestris 8004 (see methods). Relative mRNA quantitative of ssbX was calculated with respect to the levels observed for wild-type strains grown in NB. Genes encoding 16S rRNA were used as internal controls. Data represent means ± SD of triplicate measurements (P = 0.01, t test).
Our results suggest that the HR-eliciting SSBX protein is highly conserved in Xanthomonas species (Fig. S1, Fig. 2C), leading us to investigate whether PIP-box promoters drive ssbX expression in other xanthomonads or not. Interestingly, PIP-box promoters were identified upstream of the ssb coding sequences in X. oryzae pv. oryzae PXO99A, X. campestris pv. vesicatoria 85-10, X. axonopodis pv. citri 306, and X. campestris pv. campestris 8004 (data not shown), implying that ssb expression in these bacteria is also induced in planta. Thus, we performed real-time PCR to investigate the expression levels of ssbX in various Xanthomonas species grown in the hrp-inducing media (see Methods). ssbX expression was significantly higher in hrp-inducing media than in the nutrient-rich media, which suggests that these genes have a functional PIP-box and are regulated by HrpX.
Discussion
In this report, we demonstrate that single-stranded DNA-binding proteins from Xanthomonas elicit HR in tobacco. This activity was not demonstrated with SSB proteins obtained from other prokayrotes, so it may be a unique feature of Xanthomonas. Like Hpa1, SSBXoc contributes both to bacterial growth and virulence in rice (Fig. 6) and also triggers programmed cell death (Fig. 2, 3). This is the first report that Xanthomonas produce a highly-conserved SSBX protein that functions as a harpin-like protein. Furthermore, we showed that SSBX binds nonspecifically to single-stranded DNAs (Fig. S3), perhaps a potential role in protecting ssDNA from nucleases [62]. Unlike some PAMPs with a very narrow distribution, such as Ax21 in X. oryzae pv. oryzae [80], SSBx may be widely distributed in Xanthomonas species. It has been proposed that PAMPs are conserved throughout classes or genera of microbes and contribute to general microbial fitness [81].
Harpins are generally highly constrained structures that are difficult for plant pathogenic bacteria to alter because they have evolved to help bacteria avoid recognition in plants. The first identified harpin, HrpN from E. amylovora [7], has been identified in related pathogens including Pantoea stewartii subsp. stewartii [82] and D. dadantii [83]. Another harpin, HrpZ, first identified in P. syringae pathovars [6], [84], but was later shown to be present in nonpathogenic pseudomonads including P. putida and P. fluorescens [85]. The harpin HrpW, which contains harpin and pectate-lyase domains, is widely conserved across genera and has been identified in E. amylovora [41], P. syringae [42], D. dadantii [86], R. solanacearum [39], and X. campestris pv. campestris [46]. Interestingly, there is no HrpW homologue in the genomes of X. oryzae pv. oryzae, X. oryzae pv. oryzicola, X. campestris pv. vesicatoria, or X axonopodis pv. citri [46], [61], [87]. Thus, some harpins may have a more narrow distribution. For example, R. solanacearum contains an SSB protein but this does not elicit HR in tobacco (Fig. 2D), possibly because SSBRs lacks the conserved glycine-rich region of SSBX (Fig. S1) and the pathogen causes bacterial wilt in tobacco. It is also interesting to recall that flg15, a truncated version of flagellin-derived flg22, does not act as an elicitor in Arabidopsis or N. benthamiana, while it is fully active in tomato [88]. In Xanthomonas, only Hpa1 [8], [43] and SSBX (Fig. 2; this study) have been identified as harpins that elicit HRs in tobacco. However, the hpa1-ssbXoc double mutant RΔhpa1ΔssbX still elicited HR in the wild-type and SA-deficient (NahG) tobacco plants (data not all shown), implying that Xanthomonas produce other elicitors that trigger a SA-independent HR. This double mutant is a valuable resource for identifying other harpin(s) that exist in Xanthomonas.
Mutations in the T3SS of Gram-negative phytopathogenic bacteria often disrupt the ability of the pathogen to elicit HR in tobacco, presumably because harpin proteins are secreted via the T3SS [6]–[8], [41]–[43]. In Hrp group II, the cis-acting PIP-box promoter is followed by a −10 box, and collectively these elements indicate regulation via HrpX [76], [79], [89]. However, the PIP-box in ssbX genes is not followed by the −10 box, possibly because proteins in addition to HrpX are involved in regulation [44], [60]. At the amino acid level, harpin proteins possess T3SS signals that are characterized by at least 20% Pro and Ser residues in the first 50 amino acids at the N-terminus; this protein signature is required for secretion through the T3SS [78], [90]. Intriguingly, SSBX protein does not have the T3SS signal (Fig. S1), but is secreted, as Hpa1, through the T3SS independently of HpaB and HpaP (Fig. 6D, 6E). Thus, the compelling topics for future studies include understanding of how ssbX is regulated by HrpX and the mechanism of SSBx secretion.
Hpa1 and SSBX function for HR induction in tobacco (Fig. 2) and are required for full virulence in rice (Fig. 6); these dual functions are difficult to reconcile. Harpins may activate defense by entering into plant membranes and modulating ion channels or may be recognized by unidentified receptor(s) [24]. Recent reports show that the key α-helical domain in harpins, including HpaG of X. campestris pv. glycines that is orthologous to Hpa1, is required for amyloidogenesis [23], [27]. However, the Hpa1 α-helix does not show any similarity to SSBX proteins (data not shown). We speculate that the conserved domain of SSBX proteins (Fig. S1) is required for HR induction, and experiments to investigate this hypothesis are underway in our lab. The fact that harpins do not elicit HR in host plants suggests that the recognition of harpins differs in host and non-host plants. It is also important to mention that harpins function in the translocation of T3SEs, and this function is required for a full level of virulence in host plants [91]. Whether SSBX plays a role in T3SE translocation or not, remains unknown; however, the mutation in ssbXoc did not impaired the secretion of Hpa1 (Fig. 6E) and the ssbXoc-hpa1 double mutant still caused disease in rice (Fig. 6A, B, C), so at least some effectors were delivered to rice cells inoculated with the double mutant (Fig. 6). These findings are reminiscent of those reported for P. syringae pv. tabaci flagellin mutants, which were abrogated in elicitor activity and displayed reduced virulence due to impaired motility [92], [93].
Plant immune responses triggered by harpins are often associated with HR and SAR [6], [7], [24], [28]. In the present study, we show that SSBx induces PCD (Fig. 2E), the oxidative burst (Fig. 3C), the expression of HR and SAR marker genes (Fig. 2F and 3F), and callose deposition (Fig. 3D), which stimulate plant defense. SSBx-induced HR, like Hpa1-induced, could be blocked by eukaryotic metabolic inhibitors ( Fig. 2D ). It will be interesting to determine whether SSB proteins from diverse genera can elicit HR, and such studies will help us understand how pathogens recruit molecules that are instrumental for bacterial fitness and re-deploy them as agents for plant defense.
Although plant-associated microbes can potentially produce many molecules with conserved signatures, only a few PAMPs have been identified, and most of these trigger a similar set of responses. In the current study, we evaluated the expression of PTI signature genes, e.g. BAK1, BIK1 and MAP3K [19], [73], [94]. The activation of these genes by Hpa1 and SSBXoc (Fig. 3) further supports the contention that PTI is a variant of ETI [18], [81]. Recently, Ax21 of X. oryzae pv. oryzae, which is perceived by Xa21 in rice [80], was shown to be recognized by FLS2 in Arabidopsis [95]. Thus it remains possible that Hpa1 and SSBXoc, like Ax21, may recruit FLS2 in a receptor complex together with other receptors and adaptors that modulate PTI. The identity of receptors for Hpa1 and SSBXoc and whether these PAMPs interact with FLS2 remains unclear.
There is abundant evidence in the literature showing that harpins display pleiotropic effects both on HR & SAR and also impact plant growth [28], [29], [96]. Our results also showed that Hpa1 and SSBXoc enhanced growth of Arabidopsis and tobacco (Fig. 5); this was correlated with increased expression of EIN2 and PR4 genes (Fig. 3F) that are essential for Eth-signaling [75]. This is consistent with the contention that Eth-signaling regulates the accumulation of the FLS2 receptor and is required for the oxidative burst leading to PTI [97]; thus, Hpa1- and SSBXoc-mediated plant immunity may also require Eth-signaling. Eth- and SA- signaling may be regulated by WRKY transcription factors that are phosphorylated by the MAPK cascade [98], [99]. Nevertheless, SSBx may possibly have pleiotropic effects in plants.
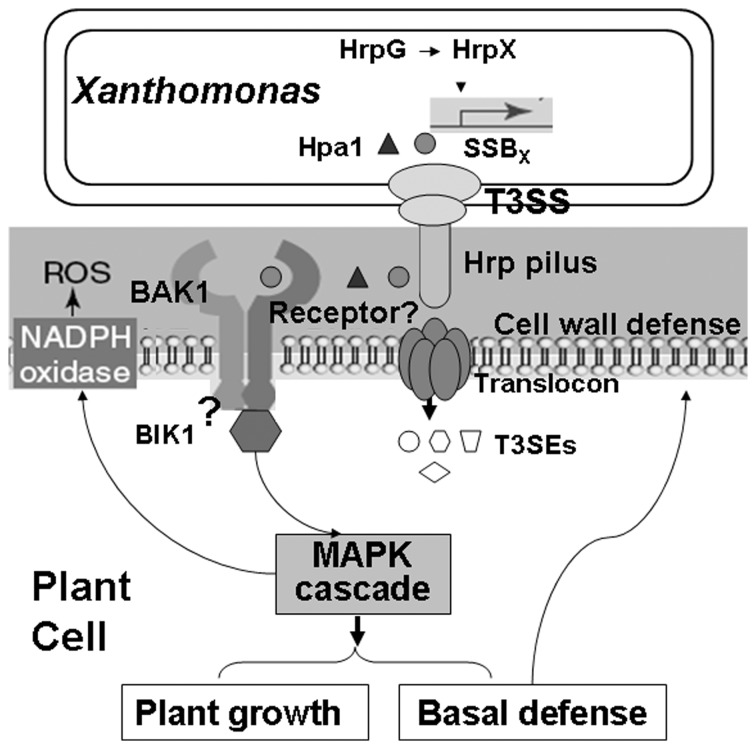
Based on the results of this study, we propose a working model for SSBX function that is also applicable to Hpa1 (Fig. 8). The ssbx gene in Xanthomonas is regulated by HrpG and HrpX; the latter protein potentially binds to the PIP-box promoter and activates transcription. Although the mechanistic basis of secretion is not totally understood, SSBx secretion depends on the functional T3SS, but does not depend on the presence of HpaB and HpaP. We hypothesize that, besides protecting ssDNAs from nucleases in bacterial cells, SSBx, possibly like Hpa1, is secreted through the T3SS, but not translocated into plant cells, and perceived in plant apoplast where it is recognized by an unknown receptor, possibly a plasma membrane-localized PAMP receptor-like kinase (RLKs) that recruits other proteins, like BAK1, and activates downstream signal transduction cascades for HR induction (Fig. 8). We speculate that signaling leads to expression of Eth-dependent genes for plant growth and SA- or JA-dependent genes for plant defense. These hypotheses are the subject of ongoing experiments in our laboratory by undertaking the investigation of an unknown SSBX-interacting protein in plants.
Figure 8. Working model of SSBX function.
SSBXoc, like Hpa1, which are regulated at the transcriptional level by HrpG and HrpX, are potentially secreted into the plant apoplast via the T3SS. The grey circle and black triangle indicate SSBX and Hpa1 proteins that may be secreted through the Hrp pilus (encoded by hrpE), but not translocated through the translocon as other T3SEs (different white shapes), and then are possibly recognized by an unidentified receptor (question mark) which associates with BAK1. This interaction may result in phosphorylation of BIK1 and subsequent phosphotransfer to the MAPK cascade to activate the expression of genes involving in SA-, JA- and Eth-signaling pathways that lead to induced resistance (SAR and/or ISR) accompanied by callose deposition on cell walls and enhanced plant growth. MAPK signaling regulates NADPH oxidase-dependent oxidative burst in the early stages of plant defense.
Supporting Information
Comparison of single-stranded DNA-binding proteins in Xanthomonas species and other prokaryotes by multiple sequence alignment. The sequences within the black dashed-line rectangle represent conserved region in Xanthomonas but variable in other prokaryotes. Protein accession numbers are indicated. The abbreviations are as follows: Xoc, X. oryzae pv. oryzicola; Xoo, X. oryzae pv. oryzae; Xcv, X. campestris pv. vesicatoria; Xcc, X. campestris pv. campestris; Rs, Ralstonia solanacearum; Ya, Yersinia aldovae; Ea, Erwinia amylovora; Pst, Pseudomonas syringae pv. tomato; Ec, Eschericha coli, and Xf, Xylella fastidiosa.
(TIFF)
Phylogenetic analysis of SSB proteins in various bacterial species. A neighbor-joining bootstrap tree was derived from the amino acid sequences of SSB proteins using the Vector NTI Align program (http://www.invitrogen.com). Protein accession numbers are indicated after the bacterial species or strain designation. Based on phylogenetic analysis, SSB proteins were classified into one of three groups (I, II and III) for HR induction in nohost tobacco.
(TIFF)
SSBXoc binds to single-stranded DNAs in electrophoretic mobility shift assays (EMSA). Randomly synthesized DNA1 and DNA2 (Table S2) were labeled with the Biotin 3′ End DNA Labeling Kit (Thermo, USA). EMSA was performed using protocols supplied with the LightShift Chemoluminescent EMSA Kit (Thermo, USA). Five µg of purified SSBXoc protein was mixed with 20 µl of the binding buffer and 20 fmol of biotin-labeled DNA1 (left panel) or DNA2 (right panel); in competition assays (lanes marked with*), labeled DNA was mixed with a 200-fold molar excess of unlabeled DNA1 or DNA2. The mixtures were incubated at room temperature for 20 min. Samples were then loaded on 5% polyacrylamide gels in 0.5X TBE buffer (pH 8.3). Gels were transferred to Hybond N+ membranes (Amersham, Pharmacia), and signals were detected by chemoluminescence according to the manufacturer’s instructions. The experiment was repeated twice and similar results were obtained. Lanes that are labeled (−) do not contain SSBx; lanes labeled (+) contain SSBXoc and DNA. The middle lane in each panel clearly shows the retardation of DNA mobility due to SSBXoc binding.
(TIFF)
Strains and plasmids used in this study.
(DOC)
Primers used in this study.
(DOC)
Amino acid identity between SSBX in X. oryzae pv. oryzicola RS105 and homologues in other bacteria.
(DOC)
Acknowledgments
We thank A. Collmer at Cornell University for his critical suggestions and helpful discussions on the manuscript when GC worked in his lab as a Tang Scholar. We are grateful to Dr. Carol Bender, Oklahoma State University, USA, for her critical reading and editing the manuscript prior to submission.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (31230059 and 31071656 to GC and 31171832 to HZ), the State Key Basic Research and Development Project of China (2012CB114003), and the Key Basic Research Project of the Shanghai Committee of Science and Technology (11JC1406300). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- 2. Tsuda K, Katagiri F (2010) Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr Opin Plant Biol 13: 459–465. [DOI] [PubMed] [Google Scholar]
- 3. Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276. [DOI] [PubMed] [Google Scholar]
- 4. Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, et al. (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Felix G, Regenass M, Boller T (1993) Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J 4: 307–316. [Google Scholar]
- 6. He SY, Huang HC, Collmer A (1993) Pseudomonas syringae pv. syringae harpinPss: a protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell 73: 1255–1266. [DOI] [PubMed] [Google Scholar]
- 7. Wei ZM, Laby RJ, Zumoff CH, Bauer DW, He SY, et al. (1992) Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora . Science 257: 85–88. [DOI] [PubMed] [Google Scholar]
- 8. Zou LF, Wang XP, Xiang Y, Zhang B, Li YR, et al. (2006) Elucidation of the hrp clusters of Xanthomonas oryzae pv. oryzicola that control the hypersensitive response in nonhost tobacco and pathogenicity in susceptible host rice. Appl Environ Microbiol 72: 6212–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Altenbach D, Robatzek S (2007) Pattern recognition receptors: from the cell surface to intracellular dynamics. Mol Plant Microbe Interact 20: 1031–1039. [DOI] [PubMed] [Google Scholar]
- 10. Boller T, He SY (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwessinger B, Ronald PC (2012) Plant innate immunity: perception of conserved microbial signatures. Annu Rev Plant Biol 63: 451–482. [DOI] [PubMed] [Google Scholar]
- 12. Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, et al. (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci U S A 104: 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, et al. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760. [DOI] [PubMed] [Google Scholar]
- 14. Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, et al. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis . Proc Natl Acad Sci U S A 104: 19613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Afzal AJ, Wood AJ, Lightfoot DA (2008) Plant receptor-like serine threonine kinases: roles in signaling and plant defense. Mol Plant Microbe Interact 21: 507–517. [DOI] [PubMed] [Google Scholar]
- 16. Postel S, Kemmerling B (2009) Plant systems for recognition of pathogen-associated molecular patterns. Semin Cell Dev Biol 20: 1025–1031. [DOI] [PubMed] [Google Scholar]
- 17. Gimenez-Ibanez S, Rathjen JP (2010) The case for the defense: plants versus Pseudomonas syringae . Microbes Infect 12: 428–437. [DOI] [PubMed] [Google Scholar]
- 18. Block A, Alfano JR (2011) Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr Opin Microbiol 14: 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chinchilla D, Shan L, He P, de Vries S, Kemmerling B (2009) One for all: the receptor-associated kinase BAK1. Trends in plant science 14: 535–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li J, Wen J, Lease KA, Doke JT, Tax FE, et al. (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222. [DOI] [PubMed] [Google Scholar]
- 21. Lu D, Wu S, Gao X, Zhang Y, Shan L, et al. (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci U S A 107: 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oh J, Kim JG, Jeon E, Yoo CH, Moon JS, et al. (2007) Amyloidogenesis of type III-dependent harpins from plant pathogenic bacteria. J Biol Chem 282: 13601–13609. [DOI] [PubMed] [Google Scholar]
- 24. Reboutier D, Bouteau F (2008) Harpins and ion channels modulations: Many ways to die. Plant Signal Behav 3: 314–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kvitko BH, Ramos AR, Morello JE, Oh HS, Collmer A (2007) Identification of harpins in Pseudomonas syringae pv. tomato DC3000, which are functionally similar to HrpK1 in promoting translocation of type III secretion system effectors. J Bacteriol 189: 8059–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Engelhardt S, Lee J, Gabler Y, Kemmerling B, Haapalainen ML, et al. (2009) Separable roles of the Pseudomonas syringae pv. phaseolicola accessory protein HrpZ1 in ion-conducting pore formation and activation of plant immunity. Plant J 57: 706–717. [DOI] [PubMed] [Google Scholar]
- 27. Haapalainen M, Engelhardt S, Kufner I, Li CM, Nurnberger T, et al. (2011) Functional mapping of harpin HrpZ of Pseudomonas syringae reveals the sites responsible for protein oligomerization, lipid interactions and plant defence induction. Mol Plant Pathol 12: 151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dong H, Delaney TP, Bauer DW, Beer SV (1999) Harpin induces disease resistance in Arabidopsis through the systemic acquired resistance pathway mediated by salicylic acid and the NIM1 gene. Plant J 20: 207–215. [DOI] [PubMed] [Google Scholar]
- 29. Dong HP, Peng J, Bao Z, Meng X, Bonasera JM, et al. (2004) Downstream divergence of the ethylene signaling pathway for harpin-stimulated Arabidopsis growth and insect defense. Plant Physiol 136: 3628–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee J, Klessig DF, Nurnberger T (2001) A harpin binding site in tobacco plasma membranes mediates activation of the pathogenesis-related gene HIN1 independent of extracellular calcium but dependent on mitogen-activated protein kinase activity. Plant Cell 13: 1079–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takahashi Y, Uehara Y, Berberich T, Ito A, Saitoh H, et al. (2004) A subset of hypersensitive response marker genes, including HSR203J, is the downstream target of a spermine signal transduction pathway in tobacco. Plant J 40: 586–595. [DOI] [PubMed] [Google Scholar]
- 32. Mei C, Qi M, Sheng G, Yang Y (2006) Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol Plant Microbe Interact 19: 1127–1137. [DOI] [PubMed] [Google Scholar]
- 33. Kim CY, Zhang S (2004) Activation of a mitogen-activated protein kinase cascade induces WRKY family of transcription factors and defense genes in tobacco. Plant J 38: 142–151. [DOI] [PubMed] [Google Scholar]
- 34.Che YZ, Li YR, Zou HS, Zou LF, Zhang B, et al. (2011) A novel antimicrobial protein for plant protection consisting of a Xanthomonas oryzae harpin and active domains of cecropin A and melittin. Microbe Biotechnol. [DOI] [PMC free article] [PubMed]
- 35. Reboutier D, Frankart C, Briand J, Biligui B, Laroche S, et al. (2007) The HrpN(ea) harpin from Erwinia amylovora triggers differential responses on the nonhost Arabidopsis thaliana cells and on the host apple cells. Mol Plant Microbe Interact 20: 94–100. [DOI] [PubMed] [Google Scholar]
- 36. Taguchi F, Shimizu R, Inagaki Y, Toyoda K, Shiraishi T, et al. (2003) Post-translational modification of flagellin determines the specificity of HR induction. Plant Cell Physiol 44: 342–349. [DOI] [PubMed] [Google Scholar]
- 37. Nurnberger T, Brunner F, Kemmerling B, Piater L (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev 198: 249–266. [DOI] [PubMed] [Google Scholar]
- 38. Arlat M, Van Gijsegem F, Huet JC, Pernollet JC, Boucher CA (1994) PopA1, a protein which induces a hypersensitivity-like response on specific Petunia genotypes, is secreted via the Hrp pathway of Pseudomonas solanacearum . EMBO J 13: 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Salanoubat M, Genin S, Artiguenave F, Gouzy J, Mangenot S, et al. (2002) Genome sequence of the plant pathogen Ralstonia solanacearum . Nature 415: 497–502. [DOI] [PubMed] [Google Scholar]
- 40. Li JG, Liu HX, Cao J, Chen LF, Gu C, et al. (2010) PopW of Ralstonia solanacearum, a new two-domain harpin targeting the plant cell wall. Mol Plant Pathol 11: 371–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim JF, Beer SV (1998) HrpW of Erwinia amylovora, a new harpin that contains a domain homologous to pectate lyases of a distinct class. J Bacteriol 180: 5203–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Charkowski AO, Alfano JR, Preston G, Yuan J, He SY, et al. (1998) The Pseudomonas syringae pv. tomato HrpW protein has domains similar to harpins and pectate lyases and can elicit the plant hypersensitive response and bind to pectate. J Bacteriol 180: 5211–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim JG, Park BK, Yoo CH, Jeon E, Oh J, et al. (2003) Characterization of the Xanthomonas axonopodis pv. glycines Hrp pathogenicity island. J Bacteriol 185: 3155–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li YR, Zou HS, Che YZ, Cui YP, Guo W, et al. (2011) A novel regulatory role of HrpD6 in regulating hrp-hrc-hpa genes in Xanthomonas oryzae pv. oryzicola . Mol Plant Microbe Interact 24: 1086–1101. [DOI] [PubMed] [Google Scholar]
- 45. Li YR, Che YZ, Zou HS, Cui YP, Guo W, et al. (2011) Hpa2 is required by HrpF to translocate Xanthomonas oryzae TAL effectors into rice for pathogenicity. Appl Environ Microbiol 77: 3809–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. da Silva AC, Ferro JA, Reinach FC, Farah CS, Furlan LR, et al. (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417: 459–463. [DOI] [PubMed] [Google Scholar]
- 47. Johnsen K, Nielsen P (1999) Diversity of Pseudomonas strains isolated with King’s B and Gould’s S1 agar determined by repetitive extragenic palindromic-polymerase chain reaction, 16S rDNA sequencing and fourier transform infrared spectroscopy characterisation. FEMS Microbiol Lett 173: 155–162. [DOI] [PubMed] [Google Scholar]
- 48.Miller JH (1972) Experiments in molecular genetics: Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 49. Xiao YL, Li YR, Liu ZY, Xiang Y, Chen GY (2007) Establishment of the hrp-inducing systems for the expression of the hrp genes of Xanthomonas oryzae pv. oryzicola . Wei Sheng Wu Xue Bao 47: 396–401. [PubMed] [Google Scholar]
- 50. Tondo ML, Petrocelli S, Ottado J, Orellano EG (2010) The monofunctional catalase KatE of Xanthomonas axonopodis pv. citri is required for full virulence in citrus plants. PLoS One 5: e10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dow JM, Clarke BR, Milligan DE, Tang JL, Daniels MJ (1990) Extracellular proteases from Xanthomonas campestris pv. campestris, the black rot pathogen. Appl Environ Microbiol 56: 2994–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, et al. (1996) Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol 37: 49–59. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual: Cold spring harbor laboratory press.
- 54. Sharma PC, Ito A, Shimizu T, Terauchi R, Kamoun S, et al. (2003) Virus-induced silencing of WIPK and SIPK genes reduces resistance to a bacterial pathogen, but has no effect on the INF1-induced hypersensitive response (HR) in Nicotiana benthamiana . Mol Genet Genomics 269: 583–591. [DOI] [PubMed] [Google Scholar]
- 55. Upton JP, Valentijn AJ, Zhang L, Gilmore AP (2007) The N-terminal conformation of Bax regulates cell commitment to apoptosis. Cell Death Differ 14: 932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Samuel MA, Hall H, Krzymowska M, Drzewiecka K, Hennig J, et al. (2005) SIPK signaling controls multiple components of harpin-induced cell death in tobacco. Plant J 42: 406–416. [DOI] [PubMed] [Google Scholar]
- 57. Zou LF, Li YR, Chen GY (2011) Novel phenotypes revealed by multiple mutagenesis in hrp genes of a rice pathogen Xanthomonas oryzae pv. oryzicola with a new suicide vector pKMS1. J Integ Agric 10: 1139–1150. [Google Scholar]
- 58. DeFeyter R, Kado CI, Gabriel DW (1990) Small, stable shuttle vectors for use in Xanthomonas . Gene 88: 65–72. [DOI] [PubMed] [Google Scholar]
- 59. Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li YR, Xiao YL, Zou LF, Zou HS, Chen GY (2012) Identification of HrpX regulon genes in Xanthomonas oryzae pv. oryzicola using a GFP visualization technique. Arch Microbiol 194: 281–291. [DOI] [PubMed] [Google Scholar]
- 61. Bogdanove AJ, Koebnik R, Lu H, Furutani A, Angiuoli SV, et al. (2011) Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp. J Bacteriol 193: 5450–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fedorov R, Witte G, Urbanke C, Manstein DJ, Curth U (2006) 3D structure of Thermus aquaticus single-stranded DNA-binding protein gives insight into the functioning of SSB proteins. Nucleic Acids Res 34: 6708–6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Danon A, Delorme V, Mailhac N, Gallois P (2000) Plant programmed cell death: a common way to die. Plant Physiol Biochem 38: 647–655. [Google Scholar]
- 64. Gopalan S, Wei W, He SY (1996) hrp gene-dependent induction of hin1: a plant gene activated rapidly by both harpins and the avrPto gene-mediated signal. Plant J 10: 591–600. [DOI] [PubMed] [Google Scholar]
- 65. Pontier D, Tronchet M, Rogowsky P, Lam E, Roby D (1998) Activation of hsr203, a plant gene expressed during incompatible plant-pathogen interactions, is correlated with programmed cell death. Mol Plant Microbe Interact 11: 544–554. [DOI] [PubMed] [Google Scholar]
- 66. Durner J, Shah J, Klessig DF (1997) Salicylic acid and disease resistance in plants. Trend Plant Sci 2: 266–274. [Google Scholar]
- 67. Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5: 325–331. [DOI] [PubMed] [Google Scholar]
- 68. Abreu ME, Munne-Bosch S (2009) Salicylic acid deficiency in NahG transgenic lines and sid2 mutants increases seed yield in the annual plant Arabidopsis thaliana. J Exp Bot 60: 1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. van Wees SC, Glazebrook J (2003) Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J 33: 733–742. [DOI] [PubMed] [Google Scholar]
- 70. Bolwell GP, Bindschedler LV, Blee KA, Butt VS, Davies DR, et al. (2002) The apoplastic oxidative burst in response to biotic stress in plants: a three-component system. J Exp Bot 53: 1367–1376. [PubMed] [Google Scholar]
- 71. Asai S, Ohta K, Yoshioka H (2008) MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana . Plant Cell 20: 1390–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Luna E, Pastor V, Robert J, Flors V, Mauch-Mani B, et al. (2011) Callose deposition: a multifaceted plant defense response. Mol Plant-Microbe Interact 24: 183–193. [DOI] [PubMed] [Google Scholar]
- 73. Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, et al. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500. [DOI] [PubMed] [Google Scholar]
- 74. Veronese P, Nakagami H, Bluhm B, Abuqamar S, Chen X, et al. (2006) The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18: 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang KL, Li H, Ecker JR (2002) Ethylene biosynthesis and signaling networks. Plant Cell 14 Suppl: S131–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang YP, Zou LF, Zhou D, Chen GY (2009) Key roles of hrpE gene of Xanthomonas oryzae pv. oryzicola in formation of Hrp pilus and pathogenicity in rice. Act Phytopathol Sin 39: 392–398. [Google Scholar]
- 77. Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S (2005) Prediction of twin-arginine signal peptides. BMC Bioinform 6: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Furutani A, Takaoka M, Sanada H, Noguchi Y, Oku T, et al. (2009) Identification of novel type III secretion effectors in Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact 22: 96–106. [DOI] [PubMed] [Google Scholar]
- 79. Furutani A, Nakayama T, Ochiai H, Kaku H, Kubo Y, et al. (2006) Identification of novel HrpXo regulons preceded by two cis-acting elements, a plant-inducible promoter box and a -10 box-like sequence, from the genome database of Xanthomonas oryzae pv. oryzae . FEMS Microbiol Lett 259: 133–141. [DOI] [PubMed] [Google Scholar]
- 80. Lee SW, Han SW, Sririyanum M, Park CJ, Seo YS, et al. (2009) A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 326: 850–853. [DOI] [PubMed] [Google Scholar]
- 81. Thomma BP, Nurnberger T, Joosten MH (2011) Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23: 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ahmad M, Majerczak DR, Pike S, Hoyos ME, Novacky A, et al. (2001) Biological activity of harpin produced by Pantoea stewartii subsp. stewartii . Mol Plant Microbe Interact 14: 1223–1234. [DOI] [PubMed] [Google Scholar]
- 83. Yap MN, Rojas CM, Yang CH, Charkowski AO (2006) Harpin mediates cell aggregation in Erwinia chrysanthemi 3937. J Bacteriol 188: 2280–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Preston G, Huang HC, He SY, Collmer A (1995) The HrpZ proteins of Pseudomonas syringae pvs. syringae, glycinea, and tomato are encoded by an operon containing Yersinia ysc homologs and elicit the hypersensitive response in tomato but not soybean. Mol Plant Microbe Interact 8: 717–732. [DOI] [PubMed] [Google Scholar]
- 85. Studholme DJ, Ibanez SG, MacLean D, Dangl JL, Chang JH, et al. (2009) A draft genome sequence and functional screen reveals the repertoire of type III secreted proteins of Pseudomonas syringae pathovar tabaci 11528. BMC Genomics 10: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yang S, Peng Q, Zhang Q, Zou L, Li Y, et al. (2010) Genome-wide identification of HrpL-regulated genes in the necrotrophic phytopathogen Dickeya dadantii 3937. PLoS One 5: e13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Salzberg SL, Sommer DD, Schatz MC, Phillippy AM, Rabinowicz PD, et al. (2008) Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A . BMC Genomics 9: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Robatzek S, Bittel P, Chinchilla D, Köchner P, Felix G, et al. (2007) Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Mol Biol 64: 539–547. [DOI] [PubMed] [Google Scholar]
- 89. Tsuge S, Terashima S, Furutani A, Ochiai H, Oku T, et al. (2005) Effects on promoter activity of base substitutions in the cis-acting regulatory element of HrpXo regulons in Xanthomonas oryzae pv. oryzae . J Bacteriol 187: 2308–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Collmer A, Lindeberg M, Petnicki-Ocwieja T, Schneider DJ, Alfano JR (2002) Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trend Microbiol 10: 462–469. [DOI] [PubMed] [Google Scholar]
- 91. Alfano JR, Collmer A (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu Rev Phytopathol 42: 385–414. [DOI] [PubMed] [Google Scholar]
- 92. Taguchi F, Takeuchi K, Katoh E, Murata K, Suzuki T, et al. (2006) Identification of glycosylation genes and glycosylated amino acids of flagellin in Pseudomonas syringae pv. tabaci . Cell Microbiol 8: 923–938. [DOI] [PubMed] [Google Scholar]
- 93. Taguchi F, Yamamoto M, Ohnishi-Kameyama M, Iwaki M, Yoshida M, et al. (2010) Defects in flagellin glycosylation affect the virulence of Pseudomonas syringae pv. tabaci 6605. Microbiol 156: 72–80. [DOI] [PubMed] [Google Scholar]
- 94. Yeam I, Nguyen HP, Martin GB (2010) Phosphorylation of the Pseudomonas syringae effector AvrPto is required for FLS2/BAK1-independent virulence activity and recognition by tobacco. Plant J 61: 16–24. [DOI] [PubMed] [Google Scholar]
- 95. Danna CH, Millet YA, Koller T, Han SW, Bent AF, et al. (2011) The Arabidopsis flagellin receptor FLS2 mediates the perception of Xanthomonas Ax21 secreted peptides. Proc Natl Acad Sci U S A 108: 9286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Oh CS, Beer SV (2007) AtHIPM, an ortholog of the apple HrpN-interacting protein, is a negative regulator of plant growth and mediates the growth-enhancing effect of HrpN in Arabidopsis. Plant Physiol 145: 426–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Mersmann S, Bourdais G, Rietz S, Robatzek S (2010) Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol 154: 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Menke FL, Kang HG, Chen Z, Park JM, Kumar D, et al. (2005) Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR-like cell death in tobacco. Mol Plant Microbe Interact 18: 1027–1034. [DOI] [PubMed] [Google Scholar]
- 99. Pitzschke A, Schikora A, Hirt H (2009) MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol 12: 421–426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of single-stranded DNA-binding proteins in Xanthomonas species and other prokaryotes by multiple sequence alignment. The sequences within the black dashed-line rectangle represent conserved region in Xanthomonas but variable in other prokaryotes. Protein accession numbers are indicated. The abbreviations are as follows: Xoc, X. oryzae pv. oryzicola; Xoo, X. oryzae pv. oryzae; Xcv, X. campestris pv. vesicatoria; Xcc, X. campestris pv. campestris; Rs, Ralstonia solanacearum; Ya, Yersinia aldovae; Ea, Erwinia amylovora; Pst, Pseudomonas syringae pv. tomato; Ec, Eschericha coli, and Xf, Xylella fastidiosa.
(TIFF)
Phylogenetic analysis of SSB proteins in various bacterial species. A neighbor-joining bootstrap tree was derived from the amino acid sequences of SSB proteins using the Vector NTI Align program (http://www.invitrogen.com). Protein accession numbers are indicated after the bacterial species or strain designation. Based on phylogenetic analysis, SSB proteins were classified into one of three groups (I, II and III) for HR induction in nohost tobacco.
(TIFF)
SSBXoc binds to single-stranded DNAs in electrophoretic mobility shift assays (EMSA). Randomly synthesized DNA1 and DNA2 (Table S2) were labeled with the Biotin 3′ End DNA Labeling Kit (Thermo, USA). EMSA was performed using protocols supplied with the LightShift Chemoluminescent EMSA Kit (Thermo, USA). Five µg of purified SSBXoc protein was mixed with 20 µl of the binding buffer and 20 fmol of biotin-labeled DNA1 (left panel) or DNA2 (right panel); in competition assays (lanes marked with*), labeled DNA was mixed with a 200-fold molar excess of unlabeled DNA1 or DNA2. The mixtures were incubated at room temperature for 20 min. Samples were then loaded on 5% polyacrylamide gels in 0.5X TBE buffer (pH 8.3). Gels were transferred to Hybond N+ membranes (Amersham, Pharmacia), and signals were detected by chemoluminescence according to the manufacturer’s instructions. The experiment was repeated twice and similar results were obtained. Lanes that are labeled (−) do not contain SSBx; lanes labeled (+) contain SSBXoc and DNA. The middle lane in each panel clearly shows the retardation of DNA mobility due to SSBXoc binding.
(TIFF)
Strains and plasmids used in this study.
(DOC)
Primers used in this study.
(DOC)
Amino acid identity between SSBX in X. oryzae pv. oryzicola RS105 and homologues in other bacteria.
(DOC)