Abstract
The Qinghai-Tibetan Plateau (QTP) has become one of the hotspots for phylogeographical studies due to its high species diversity. However, most previous studies have focused on the effects of the Quaternary glaciations on phylogeographical structures and the locations of glacial refugia, and little is known about the effects of the aridization of interior Asia on plant population structure and speciation. Here the chloroplast DNA (cpDNA) trnT-trnF and trnS-trnfM sequences were used to investigate the differentiation and phylogeographical history of 14 Ephedra species from the QTP and northern China, based on a sampling of 107 populations. The phylogeographical analysis, together with phylogenetic reconstruction based on combined four cpDNA fragments (rbcL, rpl16, rps4, and trnS-trnfM), supports three main lineages (eastern QTP, southern QTP, and northern China) of these Ephedra species. Divergence of each lineage could be dated to the Middle or Late Miocene, and was very likely linked to the uplift of the QTP and the Asian aridification, given the high drought and/or cold tolerance of Ephedra. Most of the Ephedra species had low intraspecific variation and lacked a strong phylogeographical structure, which could be partially attributed to clonal reproduction and a relatively recent origin. In addition, ten of the detected 25 cpDNA haplotypes are shared among species, suggesting that a wide sampling of species is helpful to investigate the origin of observed haplotypes and make reliable phylogeographical inference. Moreover, the systematic positions of some Ephedra species are discussed.
Introduction
The Late Cenozoic uplift of the Qinghai-Tibetan Plateau (QTP), the highest and largest plateau in the world with a mean elevation of 4500 m and an area of 2.5×106 km2 [1], resulted in Asian aridification [2], [3], and promoted the development of a rich biodiversity in the southern and southeastern QTP, where three world biodiversity hotspots were recognized [4]. For instance, the Himalaya-Hengduan Mountains region harbors over 20,000 species of vascular plants, which represent the richest alpine flora on the earth with a high percentage of endemic species [5], [6]. However, many fewer plants are distributed in the vast QTP platform due to extreme cold and arid environments. The mechanisms underlying species diversification in the QTP have fascinated biologists for a long time (e.g., [7]–[15]). Do the congeneric plant species in the QTP have similar evolutionary and biogeographic history? Did they differ in response to historical climatic changes?
In recent years, the QTP has become one of the hotspots for plant phylogeographical studies, such as first on Pinus densata [16], [17], and then on diverse subalpine and alpine plants (e.g., [18]–[21]). Most of these studies suggested postglacial/interglacial plant colonization/recolonization of the QTP platform and the Himalayas from the adjacent lower-elevation regions, especially from the Hengduan Mountains [18]–[22], or glacial in situ survival in microrefugia on the QTP platform [12], [23], [24]. While most previous studies have focused on the effects of the Quaternary glaciations on phylogeographical structures and the locations of glacial refugia, little is known about the effects of the aridization of interior Asia, partially driven by the QTP uplift and global cooling, on plant population structure and speciation. In addition, the Central Asian plants were very rarely investigated in these studies.
On the other hand, although phylogeographical studies have been conducted in diverse plant groups and in many geographical regions [25]–[29], most of them sampled a single or a couple of closely related species at population level. This makes it difficult to investigate the origin of the haplotypes detected in the studied group. For instance, a haplotype of a species or population, whether rare or common, might be newly evolved or inherited from a common ancestor [10], [20], and could also be obtained by interspecific gene flow. When the evolutionary history of the haplotype and its distribution in other species are unknown, an incorrect phylogeographical inference could be made, especially for studies of groups with high dispersal ability or a complicated evolutionary history.
The genus Ephedra is mainly shrubs and comprises about 50 species [30], most of which are extremely drought and/or cold tolerant. Approximately sixteen Ephedra species occur in the QTP and adjacent regions [31]–[33], and all of them could have originated in the Late Cenozoic by adaptive radiation and are relatively closely related [30], [34], [35], making them ideally suited for the investigation of topographic and climatic effects on plant population dynamics and speciation. For example, Ephedra saxatilis, a species mainly distributed in the Himalayas, has red and fleshy cone bracts adapted to animal dispersal. In contrast, E. przewalskii that is widely distributed in the deserts of Central Asia has winged cone bracts suitable for wind dispersal.
In the present study, we use chloroplast DNA (cpDNA) trnT-trnF and trnS-trnfM sequences to investigate the differentiation and phylogeographical history of the Ephedra species distributed in the QTP and adjacent regions, based on a wide population sampling. The questions to be addressed include: (1) Has the species diversification of Ephedra been driven by the QTP uplift and the Asian aridization? (2) Are the phylogeographical patterns of the Ephedra species similar to those revealed in other QTP plants? (3) How did the Ephedra populations respond to the Quaternary climatic changes? (4) How important is species sampling in phylogeographical studies?
Materials and Methods
Ethics statement
No specific permits were required for the described field studies.
Sampling
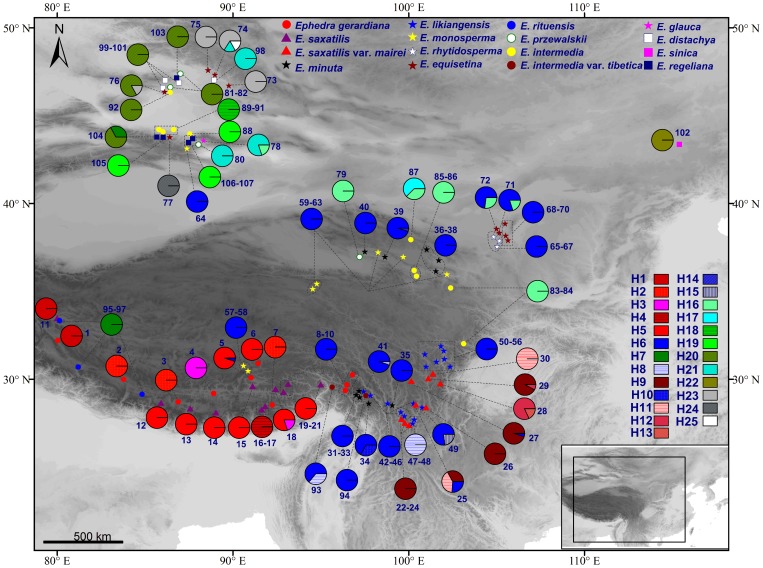
Except E. fedtschenkoae and E. lomatolepis that are difficult to access, all the other 14 Ephedra species distributed in the QTP and adjacent regions were sampled for the phylogeographical study based on cpDNA trnT-trnF and trnS-trnfM sequences (Fig. 1). Young branchlets were collected from 107 populations (totaling 1435 individuals), most of which were represented by 5–29 individuals that were at least 50 m apart from each other. For the species mainly distributed in the QTP, the sample size is larger than 12 for most populations. The population codes, sample sizes, and geographical coordinates are shown in Table S1. Also, one individual of the Mediterranean E. nebrodensis was sampled as outgroup based on the results of previous phylogenetic analyses [30], [35], [36]. To better understand the phylogeographical patterns of the 14 Ephedra species, their evolutionary relationships were also reconstructed from sequence analysis of combined four cpDNA fragments (rbcL, rpl16, rps4, and trnS-trnfM), in which most species were represented by 2–4 individuals and the DNA sequences of other congeneric species available in GenBank were included. In total, 37 species were sampled in the combined cpDNA analysis (Table S2). E. saxatilis var. mairei and E. intermedia var. tibetica were considered as two independent taxa in all analyses due to their unique phylogenetic positions (see discussion). The species status of P. glauca was recognized in a recent revision of the genus Ephedra from China [32], and thus was followed in our study.
Figure 1. Sampling locations and distribution frequencies of the cpDNA haplotypes of 14 Ephedra species.
Population numbers correspond to those in Table 1.
DNA extraction, PCR amplification, and sequencing
Total genomic DNA was isolated from silica gel-dried young branchlets using the modified CTAB method [37]. The cpDNA trnT-trnF and trnS-trnfM regions were amplified with the primer pairs trnT (5′-CATTACAAATGCGAT GCTCT-3′) and trnF (5′-ATTTGAACTGGTGACACGAG-3′) [38], trnS (5′-GAGAGAGAGGGATTCG AACC-3′) and trnfM (5′-CATAACCTTGAGGTCACGGG-3′) [39], respectively. Other primers for the amplification of cpDNA are shown in Table S3. The polymerase chain reaction (PCR) was conducted in a Mastercycler (Eppendorf, Hamburg, Germany) or a Tgradient Thermocycler (Biometra) in a volume of 25 µL, containing 5–50 ng plant DNA, 6.25 pmol of each primer, 200 µmol/L of each dNTP, and 0.75 unit of Taq DNA polymerase (TakaRa Biotech Co., Dalian, China). PCR cycles were as follows: 4 min at 94°C, three cycles of 2 min at 94°C, 30 s at 52°C, and 1–1.5 min at 72°C, followed by 33 cycles of 30 s at 94°C, 30 s at 54°C, and 1–1.5 min at 72°C, with a final extension step of 10 min at 72°C.
The PCR products were purified using a Gel Band Purification Kit (Amersham Biosciences, Buckinghamshire, UK) or PEG 8000, and then were directly sequenced using ABIPrism BigDye Terminator Cycle Sequencing Ready Reaction Kit or DYEnamic Energy Transfer (ET) Terminator Reagent Premix Kit. After precipitation with 95% EtOH and 3M NaAc (pH 5.3), the sequencing products were separated on an ABI PRISM 3730xl analyzer (Applied Biosystems) or a MegaBACE 1000 automatic sequencer (Amersham Biosciences). The sequences reported in this study are deposited in GenBank under accession numbers KC222975–KC223020 (rbcL), KC223021–KC223066 (rpL16), KC223067–KC223112 (rps4), KC223113–KC223158 and KC407804–KC407829 (trnS-trnfM), and KC407778–KC407803 (trnT-trnF).
Data analyses
The DNA sequences were aligned using the program Clustal X [40] and manually adjusted in BioEdit v. 7.0.9 [41]. Arlequin 3.11 [42] was used to estimate the molecular diversity indices, including the number of segregating sites (S), number of haplotypes (Nh), haplotype diversity (H d) and nucleotide diversity (π), for each population and species. The haplotype richness (A) was calculated by dividing the number of haplotypes by the number of sampled individuals (N). The program PERMUT [43] was used to calculate average gene diversity within populations (HS), total gene diversity (HT), and two measures of population differentiation, i.e., GST [44] and NST (equivalent coefficient taking into account sequence similarities between haplotypes). An analysis of molecular variance (AMOVA) [42] and a Mantel test [45] were performed in Arlequin 3.11 to partition variation within and among populations and to assess the correlation between genetic and geographic distances, respectively. Also, the DNA divergence between populations (FST, [42]) was measured, and the significance was tested using 10,000 permutations. A network of the cpDNA haplotypes (chlorotypes) was constructed using TCS 1.21 [46], with a default parsimony connection limit of 95% and each insertion/deletion (indel) treated as a single mutation event.
The evolutionary relationships of the chlorotypes were also reconstructed with maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI), using PAUP*4.0b10 [47], PhyML 3.0 [48], and MrBayes 3.1.2 [49], respectively. All phylogenetically informative gaps (indels) were coded as single mutation events in the final alignment. In the MP analysis, all characters were treated as unordered and equally weighted, and a heuristic search was implemented with 1000 random addition sequence replicates, tree-bisection-reconnection (TBR) branch swapping and MULTREES on. To examine the robustness of clades in the most parsimonious trees, a bootstrap analysis was conducted with 1000 replicates using the same heuristic search settings as described above. In the ML analysis, we chose the K81uf+I model, which was determined to be the best-fit model for the cpDNA dataset by the Akaike Information Criterion (AIC) implemented in Modeltest 3.06 [50]. The Bayesian analysis used the best-fit model HKY+I determined by AIC in MrModeltest 2.3 [51] and random starting trees. One cold and three incrementally heated Markov chain Monte Carlo (MCMC) chains were run for 1,000,000 generations each, sampling one tree per 100 generations with the first 300 samples discarded as burn-in. The trees sampled after generation 30,000 were used for phylogenetic inference. In the phylogenetic reconstruction of the Ephedra species based on the combined four chloroplast genes, the ML and BI analyses were performed with the best-fit models TVM+I+G and GTR+R+G, respectively.
To detect whether historical population expansion events occurred in the Ephedra species, mismatch distributions were calculated using the program Arlequin 3.11 [42]. Based on 1000 parametric bootstrap replicates, an expected distribution was generated under a model of sudden demographic expansion [52]. The sum-of-squared deviations (SSD) between observed and expected mismatch distributions were calculated, with the P-values representing the proportion of simulations producing a larger SSD than the observed SSD.
Divergence times of the chlorotypes were estimated by molecular clock analysis. The rate constancy among lineages was evaluated by a likelihood ratio test (LRT), which compared log likelihood ratios of the chosen model with and without an enforced molecular clock [53]. Significance was assessed by comparing two times the difference in log likelihood to a chi-square distribution, with the degree of freedom equal to the number of taxa minus two. Since the clock assumption was rejected (δ = 41.5215, df = 24, P<0.05), divergence times were estimated with BEAST v1.7.2 [54], using the HKY+I+G substitution model, an uncorrelated lognormal relaxed clock model, the Yule model of speciation and a user-specified starting tree (ML tree). The root age was set to a median value of 29.56 Ma based on the time of the most recent common ancestor of ‘Core Ephedra’ [30] and in consideration of the fact that the New World Ephedra species were nested within the Asian clade in some previously published phylogenies [30], [55], [56] and the combined cpDNA phylogeny constructed in the present study, and minimally 23.03 Ma according to the fossil record (pollen) of Ephedra in the Late Oligocene sediments of the QTP and neighboring regions [57]–[59]. The MCMC analysis was run for 10,000,000 generations to estimate the mean posterior divergence times with standard deviations based on the variance-covariance matrix, a sampling frequency of every 1000 generations and a burn in of 1000. The program TRACER v.1.5 [60] was used to check convergence of chains to the stationary distribution. The MCMC output was analyzed with TreeAnnotator v1.5.4, and the chronological tree was visualized by FigTree v1.3.1.
Results
Distributions and evolutionary relationships of cpDNA haplotypes
In the phylogeographical study, we obtained trnT-trnF and trnS-trnfM sequences from all of the 1435 individuals surveyed. The alignment of the combined two cpDNA fragments was 1177 bp in length, including 20 nucleotide substitutions and 8 indels (1–27 bp in size) that were used to designate 25 haplotypes (H1–H25) (Tables 1, 2). Ephedra equisetina had the most haplotypes (7), although the total sample size (N = 64) for this species is not very large. The number of detected haplotypes was five in E. gerardiana (N = 196), E. saxatilis var. mairei (N = 183) and E. intermedia (N = 96), four in E. saxatilis (N = 197), three in E. minuta (N = 158), E. likiangensis (N = 183), E. przewalskii (N = 22) and E. regeliana (N = 27), and two in E. intermedia var. tibetica (N = 19), E. glauca (N = 5) and E. distachya (N = 15), respectively. No intraspecific variation was found in E. rhytidosperma (N = 17), E. monosperma (N = 96), E. rituensis (N = 66), and E. sinica (N = 13) (Fig. 1; Table 1). The haplotype H6 was the most widely distributed, and was shared by eight species (E. equisetina, E. gerardiana, E. intermedia var. tibetica, E. likiangensis, E. minuta, E. monosperma, E. rhytidosperma and E. saxatilis var. mairei). Also, each of the nine haplotypes (H1, H3, H5, H7, H8, H16, H19–H21) was shared by two or four species. The rest haplotypes were species-specific (Fig. 1; Table S4).
Table 1. The cpDNA trnT-trnF+trnS-trnfM haplotypes detected in the sampled populations of 14 Ephedra species.
| Species | Population No. | N | Haplotypes (Individuals) | Species | Population No. | N | Haplotypes (Individuals) |
| Ephedra gerardiana | 1 | 24 | H1 (24) | 55 | 10 | H6 (10) | |
| 2 | 27 | H2 (27) | 56 | 22 | H6 (22) | ||
| 3 | 16 | H2 (16) | E. monosperma | 57 | 14 | H6 (14) | |
| 4 | 24 | H3 (24) | 58 | 8 | H6 (8) | ||
| 5 | 23 | H5 (22), H6 (1) | 59 | 16 | H6 (16) | ||
| 6 | 25 | H5 (25) | 60 | 23 | H6 (23) | ||
| 7 | 3 | H2 (3) | 61 | 17 | H6 (17) | ||
| 8 | 25 | H6 (25) | 62 | 6 | H6 (6) | ||
| 9 | 12 | H6 (12) | 63 | 6 | H6 (6) | ||
| 10 | 17 | H6 (17) | 64 | 6 | H6 (6) | ||
| E. saxatilis | 11 | 14 | H1 (14) | E. rhytidosperma | 65 | 5 | H6 (5) |
| 12 | 24 | H5 (24) | 66 | 6 | H6 (6) | ||
| 13 | 6 | H5 (6) | 67 | 6 | H6 (6) | ||
| 14 | 25 | H5 (25) | E. equisetina | 68 | 6 | H6 (6) | |
| 15 | 29 | H5 (29) | 69 | 6 | H6 (6) | ||
| 16 | 6 | H4 (6) | 70 | 6 | H6 (6) | ||
| 17 | 3 | H4 (3) | 71 | 6 | H6 (5), H16 (1) | ||
| 18 | 25 | H3 (5), H5 (20) | 72 | 5 | H6 (4), H16 (1) | ||
| 19 | 29 | H5 (29) | 73 | 10 | H23 (10) | ||
| 20 | 16 | H5 (16) | 74 | 6 | H21 (1), H23 (4), H25 (1) | ||
| 21 | 20 | H5 (20) | 75 | 7 | H23 (7) | ||
| E. saxatilis var. mairei | 22 | 24 | H9 (24) | 76 | 6 | H20 (5), H23 (1) | |
| 23 | 29 | H9 (29) | 77 | 6 | H24 (6) | ||
| 24 | 12 | H9 (12) | E. glauca | 78 | 5 | H16 (1), H21 (4) | |
| 25 | 12 | H6 (3), H9 (4), H11 (5) | E. przewalskii | 79 | 5 | H16 (5) | |
| 26 | 17 | H9 (17) | 80 | 5 | H21 (5) | ||
| 27 | 23 | H6 (1), H9 (22) | 81 | 6 | H20 (6) | ||
| 28 | 28 | H12 (23), H13 (5) | 82 | 6 | H20 (6) | ||
| 29 | 26 | H9 (24), H12 (2) | E. intermedia | 83 | 5 | H16 (5) | |
| 30 | 12 | H11 (12) | 84 | 5 | H16 (5) | ||
| E. minuta | 31 | 25 | H6 (25) | 85 | 27 | H16 (27) | |
| 32 | 17 | H6 (17) | 86 | 22 | H16 (22) | ||
| 33 | 22 | H6 (22) | 87 | 16 | H16 (6), H17 (10) | ||
| 34 | 3 | H6 (2), H10 (1) | 88 | 2 | H19 (2) | ||
| 35 | 6 | H6 (6) | 89 | 5 | H18 (5) | ||
| 36 | 20 | H6 (20) | 90 | 6 | H18 (6) | ||
| 37 | 3 | H6 (3) | 91 | 6 | H18 (6) | ||
| 38 | 26 | H6 (26) | 92 | 2 | H20 (2) | ||
| 39 | 20 | H6 (19), H14 (1) | E. intermedia var. tibetica | 93 | 14 | H6 (9), H8 (5) | |
| 40 | 16 | H6 (16) | 94 | 5 | H6 (5) | ||
| E. likiangensis | 41 | 22 | H6 (21), H8 (1) | E. rituensis | 95 | 12 | H7 (12) |
| 42 | 5 | H6 (5) | 96 | 30 | H7 (30) | ||
| 43 | 24 | H6 (24) | 97 | 24 | H7 (24) | ||
| 44 | 13 | H6 (13) | E. distachya | 98 | 3 | H21 (3) | |
| 45 | 25 | H6 (25) | 99 | 5 | H20 (5) | ||
| 46 | 22 | H6 (22) | 100 | 4 | H20 (4) | ||
| 47 | 25 | H8 (25) | 101 | 3 | H20 (3) | ||
| 48 | 4 | H8 (4) | E. sinica | 102 | 13 | H22 (13) | |
| 49 | 13 | H6 (10), H15 (3) | E. regeliana | 103 | 6 | H20 (6) | |
| 50 | 21 | H6 (21) | 104 | 6 | H7 (2), H20 (4) | ||
| 51 | 11 | H6 (11) | 105 | 5 | H19 (5) | ||
| 52 | 5 | H6 (5) | 106 | 5 | H19 (5) | ||
| 53 | 23 | H6 (23) | 107 | 5 | H19 (5) | ||
| 54 | 16 | H6 (16) | All species | Total | 1435 |
N, number of sampled individuals.
Table 2. The cpDNA haplotypes detected in Ephedra species.
| Hap. | trnT-trnF (668 bp) | trnS-trnfM (509 bp) | ||||||||||||||||||||||||||||||||
| 34 | 66 | 77 | 100 | 106 | 111 | 180 | 224 | 234 | 298 | 314 | 324 | 333 | 398 | 437 | 489 | 524 | 543 | 615 | 616 | 627 | 666 | 672 | 698 | 944–960 | 973 | 991 | 994–1008 | 1011 | 1089–1096 | 1103 | 1113–1139 | 1140–1158 | 1161 | |
| H1 | C | A | A | - | G | C | G | A | A | A | C | G | A | T | T | T | C | A | T | A | - | - | G | G | - | T | T | § | A | □ | T | - | - | A |
| H2 | C | A | A | - | G | C | G | A | A | A | C | G | A | T | T | T | C | A | T | A | - | - | G | G | - | T | G | § | A | □ | T | - | - | A |
| H3 | C | A | A | - | G | C | G | A | A | C | C | G | A | T | T | T | C | A | T | A | - | - | G | G | - | T | T | ◊ | A | □ | T | - | - | A |
| H4 | C | A | A | - | G | C | G | A | A | A | C | G | A | T | T | T | C | A | T | A | G | - | G | G | - | T | T | ◊ | A | □ | T | - | - | A |
| H5 | C | A | A | - | G | C | G | A | A | A | C | G | A | T | T | T | C | A | T | A | - | - | G | G | - | T | T | ◊ | A | □ | T | - | - | A |
| H6 | C | A | A | - | G | G | G | A | A | A | A | G | A | T | T | T | C | G | T | A | - | - | G | T | - | T | T | * | G | - | T | - | - | T |
| H7 | C | A | A | A | G | G | G | A | A | A | C | G | A | T | T | T | C | G | T | A | - | - | G | T | # | T | T | ◊ | G | - | T | - | - | A |
| H8 | C | A | A | - | G | G | G | A | A | A | A | G | A | T | T | T | C | G | T | A | - | - | G | T | - | T | T | Δ | A | - | T | - | - | T |
| H9 | C | A | A | - | G | C | G | A | A | A | C | G | A | T | G | T | C | A | T | A | - | - | G | G | - | T | T | ◊ | A | T | - | - | A | |
| H10 | C | A | A | - | G | G | G | A | A | A | A | G | A | C | T | T | C | G | T | A | - | - | G | T | - | T | T | * | G | - | T | - | - | T |
| H11 | C | A | A | - | G | C | G | A | A | A | C | G | A | T | G | T | C | A | T | A | - | C | G | G | - | T | T | ◊ | A | □ | T | - | - | A |
| H12 | C | A | A | - | G | C | G | A | A | A | C | G | A | T | G | C | C | A | T | A | - | - | G | G | - | T | T | ◊ | A | □ | T | - | - | A |
| H13 | A | A | A | - | G | C | G | A | A | A | C | G | A | T | G | C | C | A | T | A | - | - | G | G | - | T | T | ◊ | A | □ | T | - | - | A |
| H14 | C | A | A | - | G | G | G | A | A | A | A | G | A | T | T | T | C | G | T | A | - | - | G | T | - | T | T | * | G | - | T | † | - | T |
| H15 | C | A | A | - | G | G | G | A | A | A | A | G | C | T | T | T | C | G | T | A | - | - | G | T | - | T | T | * | G | - | T | - | - | T |
| H16 | C | A | A | A | G | G | A | A | A | A | C | G | A | T | T | T | C | G | T | A | - | - | G | T | - | A | T | ◊ | G | - | T | - | - | A |
| H17 | C | A | A | A | G | G | G | A | A | A | C | G | A | T | T | T | C | G | T | A | - | - | G | T | - | T | T | ◊ | G | - | T | - | - | A |
| H18 | C | A | A | A | G | G | A | A | A | A | C | G | A | T | T | T | C | G | T | A | - | - | G | T | # | T | T | ◊ | G | - | T | - | - | A |
| H19 | C | G | A | A | G | G | G | A | A | A | C | G | A | T | T | T | C | G | T | A | - | - | G | T | # | T | T | ◊ | G | - | T | - | - | A |
| H20 | C | A | T | A | G | G | G | A | A | A | C | G | A | T | T | T | C | G | T | C | - | - | G | T | - | T | T | ◊ | G | - | T | - | - | A |
| H21 | C | A | T | A | G | G | G | A | A | A | C | G | A | T | T | T | C | G | T | A | - | - | G | T | - | T | T | ◊ | G | - | T | - | - | A |
| H22 | C | A | A | A | G | G | G | A | A | A | C | G | A | T | T | T | C | G | T | A | - | - | A | T | - | T | T | ◊ | G | - | T | - | - | A |
| H23 | C | A | A | - | G | G | G | A | A | A | C | G | A | T | T | T | C | G | T | A | - | - | G | T | - | T | T | ◊ | G | - | C | - | - | A |
| H24 | C | A | A | - | G | G | G | A | A | A | C | G | A | T | T | T | C | G | T | A | - | - | G | T | - | T | G | ◊ | G | - | C | - | - | A |
| H25 | C | A | A | - | G | G | G | A | A | A | C | G | A | T | T | T | C | G | T | A | - | - | G | T | - | T | T | ◊ | G | - | C | - | ‡ | A |
| H26 | C | A | A | - | A | G | G | G | T | A | C | - | A | T | T | T | T | A | G | A | - | - | G | T | - | T | G | ◊ | G | □ | T | - | - | A |
#, TCACAAACTTAATAAGT; §, TTGAACGGATTAAAA; ◊, TTGAACGGA------; *, TTAAACGGA------; Δ, ---------------; □, TAAGAATA; †, TATTAAGACAATAAGAATAAGACATAT; ‡, TAATAAGACATAATATGTC. Numbers above lanes indicate positions of the nucleotides in the alignment.
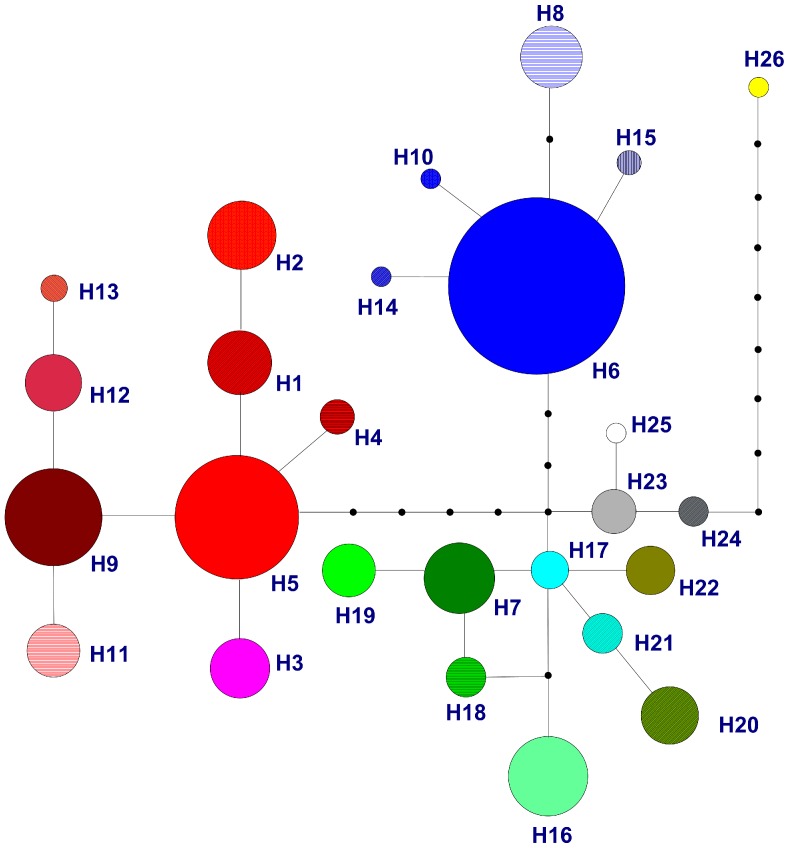
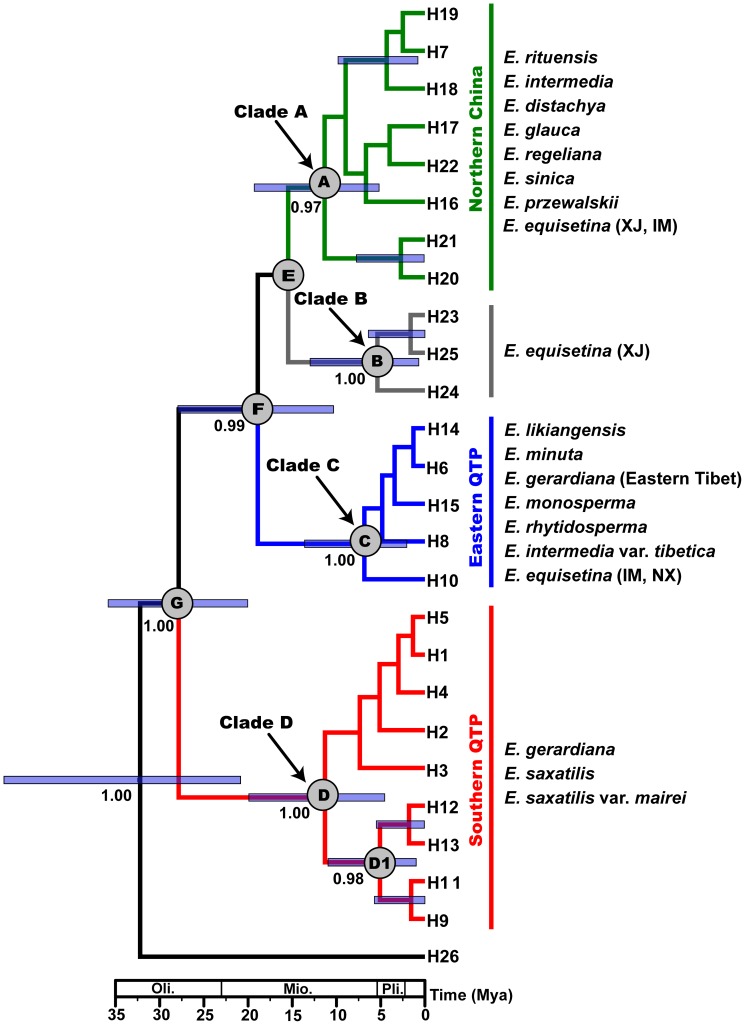
Four main lineages were resolved in the network of the cpDNA haplotypes when the sequence of the Mediterranean E. nebrodensis (H26) was used as outgroup (Fig. 2). One comprised three haplotypes (H23–25) from E. equisetina, a species widely distributed in northern China and most closely related to the outgroup. The other three lineages were mainly distributed in eastern QTP (H6, H8, H10, H14–15), southern QTP (H1–5, H9, H11–13), and northern China (H7, H16–22), respectively. The four lineages were also supported by all of the three phylogenetic analyses (MP, ML, BI) of the cpDNA haplotypes (see clades A–D in Figs. 3 and S1). It is interesting that E. rituensis, a species distributed in western Himalayas, had pure haplotype H7 of clade A, the northern China lineage. Moreover, all but four individuals of E. saxatilis var. mairei, which is sympatrically distributed with E. likiangensis in the Hengduan Mountains (SE QTP), harbored haplotypes (H9, H11–13) of clade D, the southern QTP lineage (Figs. 1, 3; Table 1).
Figure 2. A network of the cpDNA haplotypes constructed by using TCS 1.21.
The sizes of the circles in the network are proportional to the observed frequencies of the haplotypes.
Figure 3. Phylogenetic chronogram of the cpDNA (trnT-trnF+trnS-trnfM) haplotypes generated from BEAST.
Numbers below the branches indicate the Bayesian posterior probabilities. Median ages of nodes are shown, with horizontal bars indicating the 95% highest posterior density intervals (for details, see Table 3).
Divergence time estimation
According to the estimates by BEAST (Fig. 3; Table 3), the most recent common ancestor (MRCA) of the cpDNA haplotypes (H1–H25) detected from the 14 Ephedra species distributed in the QTP and adjacent regions could be dated to 27.85 Ma with the 95% highest posterior density (HPD) interval of 35.82-20.04 Ma (Fig. 3, node G). The MRCAs of the three main clades, i.e., northern China (node A), eastern QTP (node C) and southern QTP (node D), were dated to 11.35 Ma (95% HPD: 19.30-5.18 Ma), 6.84 Ma (95% HPD: 13.60-2.05 Ma) and 11.31 Ma (95% HPD: 19.92-4.55 Ma), respectively. It is interesting that the lineage D1 of clade D distributed in the east (Hengduan Mountains) also diverged early from the western lineage (Himalayas), with its MRCA dated to 5.08 Ma (95% HPD: 10.95-0.99 Ma). In general, the main clades diverged from each other in the Middle to Late Miocene.
Table 3. Estimates of divergence times for main lineages of cpDNA haplotypes.
| Node | Median age (million years) | 95% Highest posterior density intervals |
| A | 11.35 | 5.18–19.30 |
| B | 5.37 | 0.69–12.98 |
| C | 6.84 | 2.05–13.60 |
| D | 11.31 | 4.55–19.92 |
| D1 | 5.08 | 0.99–10.95 |
| E | 15.46 | — |
| F | 18.90 | 10.34–27.94 |
| G | 27.85 | 20.04–35.82 |
Genetic differentiation within and among populations of some Ephedra species
The amount of cpDNA (trnT-trnF+trnS-trnfM) variation within and among populations was calculated using AMOVA for nine species and two varieties with a sampling size larger than two populations, separately. The three species E. monosperma, E. rhytidosperma and E. rituensis were excluded from the analysis due to the lack of intraspecific variation. The results showed that, for most species, genetic variation mainly occurred among populations with high FST values (Table 4), such as E. gerardiana (F ST = 0.98285), E. saxatilis (F ST = 0.85572), E. likiangensis (F ST = 0.92549), E. equisetina (F ST = 0.77895), E. intermedia (F ST = 0.86907), and E. regeliana (F ST = 0.84163). Particularly, the populations of E. przewalskii and E. distachya fixed different haplotypes with F ST = 1, although this could be partially attributed to a small sample size. In contrast, a low F ST value (0.15317) was found in E. minuta, since all but two individuals of the species shared the haplotype H6. In the PERMUT analysis, no significant phylogeographic structure was detected in the species analyzed, although NST>GST (not significant) was observed in some species (Table 5). In addition, the Mantel test did not detect a significant correlation between genetic and geographical distances in most species (Table 4).
Table 4. Results of analyses of molecular variance (AMOVA) and Mantel tests for different Ephedra species.
| Species | Source of variation | df | SS | VC | Variation (%) | Fixation index | Mantel test |
| E. gerardiana | Among populations | 9 | 410.144 | 2.35820 | 98.29 | FST = 0.98285** | r = 0.462, p = 0.001** |
| Within populations | 186 | 7.652 | 0.04114 | 1.71 | |||
| Total | 195 | 417.796 | 2.39934 | ||||
| E. saxatilis | Among populations | 10 | 22.467 | 0.12755 | 85.57 | FST = 0.85572** | r = 0.326, p = 0.179 |
| Within populations | 186 | 4.000 | 0.02151 | 14.43 | |||
| Total | 196 | 26.467 | 0.14906 | ||||
| E. saxatilis var. mairei | Among populations | 8 | 42.851 | 0.25626 | 54.17 | FST = 0.54167** | r = 0.305, p = 0.025 |
| Within populations | 174 | 37.729 | 0.21683 | 45.83 | |||
| Total | 182 | 80.579 | 0.47309 | ||||
| E. minuta | Among populations | 9 | 0.371 | 0.00198 | 15.32 | FST = 0.15317 | r = −0.094, p = 0.849 |
| Within populations | 148 | 1.617 | 0.01092 | 84.68 | |||
| Total | 157 | 1.987 | 0.01290 | ||||
| E. likiangensis | Among populations | 15 | 51.852 | 0.21377 | 92.55 | FST = 0.92549** | r = 0.062, p = 0.274 |
| Within populations | 245 | 4.217 | 0.01721 | 7.45 | |||
| Total | 260 | 56.069 | 0.23099 | ||||
| E. equisetina | Among populations | 9 | 77.265 | 1.28994 | 77.90 | FST = 0.77895** | r = 0.758, p = 0.000** |
| Within populations | 54 | 19.767 | 0.36605 | 22.10 | |||
| Total | 63 | 97.031 | 1.65599 | ||||
| E. intermedia var. tibetica | Among populations | 1 | 0.940 | 0.07623 | 16.78 | FST = 0.16777 | — |
| Within populations | 17 | 6.429 | 0.37815 | 83.22 | |||
| Total | 18 | 7.368 | 0.45438 | ||||
| E. intermedia | Among populations | 9 | 46.562 | 0.57885 | 86.91 | FST = 0.86907** | r = 0.596, p = 0.005* |
| Within populations | 86 | 7.500 | 0.08721 | 13.09 | |||
| Total | 95 | 54.062 | 0.66606 | ||||
| E. przewalskii | Among populations | 3 | 20.909 | 1.27072 | 100 | FST = 1** | r = 0.570, p = 0.180 |
| Within populations | 18 | 0 | 0 | 0 | |||
| Total | 21 | 20.909 | 1.27072 | ||||
| E. distachya | Among populations | 3 | 2.400 | 0.21687 | 0 | FST = 1* | r = 0.898, p = 0.239 |
| Within populations | 11 | 0 | 0 | 0 | |||
| Total | 14 | 2.400 | 0.21687 | ||||
| E. regeliana | Among populations | 4 | 21.556 | 0.96626 | 84.16 | FST = 0.84163** | r = 0.141, p = 0.284 |
| Within populations | 22 | 4.000 | 0.18182 | 15.84 | |||
| Total | 26 | 25.556 | 1.14808 |
df, degrees of freedom; SS, sum of squares; VC, variance components; ** P≤0.001; * P≤0.01; —, not calculated.
Table 5. Estimates of genetic diversity and population differentiation (± SE in parentheses) for Ephedra species.
| Species | HS | HT | GST | NST |
| E. gerardiana | 0.009 (0.0087) | 0.843 (0.0541) | 0.990 (0.0102) | 0.985 (0.0162)ns |
| E. saxatilis | 0.030 (0.0303) | 0.498 (0.1548) | 0.939 (0.0652) | 0.944 (0.0599)ns |
| E. saxatilis var. mairei | 0.139 (0.0795) | 0.532 (0.1697) | 0.739 (0.1270) | 0.455 (0.1819)ns |
| E. minuta | 0.077 (0.0663) | 0.076 (0.0616) | −0.005 (NC) | −0.000 (NC) |
| E. likiangensis | 0.030 (0.0243) | 0.262 (0.1243) | 0.887 (0.1042) | 0.930 (0.0634)ns |
| E. equisetina | 0.288 (0.1012) | 0.777 (0.0726) | 0.629 (0.1239) | 0.630 (0.1242)ns |
| E. intermedia var. tibetica | NC | NC | NC | NC |
| E. intermedia | 0.063 (0.0625) | 0.625 (0.0814) | 0.900 (0.0920) | 0.900 (0.0920)ns |
| E. przewalskii | 0 | 0.833 (0.1443) | 1 (NC) | 1 (NC) |
| E. distachya | 0 | 0.500 (0.2500) | 1 (NC) | 1 (NC) |
| E. regeliana | 0.107 (0.1067) | 0.633 (0.1550) | 0.832 (0.1346) | 0.855 (0.1359)ns |
HS, average genetic diversity within populations; HT, total gene diversity; GST, interpopulation haplotype differentiation; NST, interpopulation haplotype differentiation taking into account sequence difference; ns, NST not significantly different from GST (P>0.05); NC, not computed due to small sample size or low variation among populations.
Mismatch distributions
Among the 14 Ephedra species used in the phylogeographical study, only four of them (E. gerardiana, E. likiangensis, E. minuta, and E. saxatilis) were used in the mismatch distribution analysis (Fig. S2). The other species were excluded from the analysis due to small sample size, lack of intraspecific variation, or putative historical interspecific hybridization (such as E. saxatilis var. mairei, see discussion). The results showed that the hypothesis of demographic expansion was only rejected for E. likiangensis (P SSD = 0.030) (Fig. S2a). The mismatch distributions for E. saxatilis (P SSD = 0.310) and E. minuta (P SSD = 0.120) were unimodal (Fig. S2b, c), suggesting that the two species could have experienced recent population expansion. The two peaks in the mismatch distribution of E. gerardiana could be attributed to the occurrence of haplotypes from two main lineages in the species (Fig. S2d).
Chloroplast DNA phylogeny of Ephedra
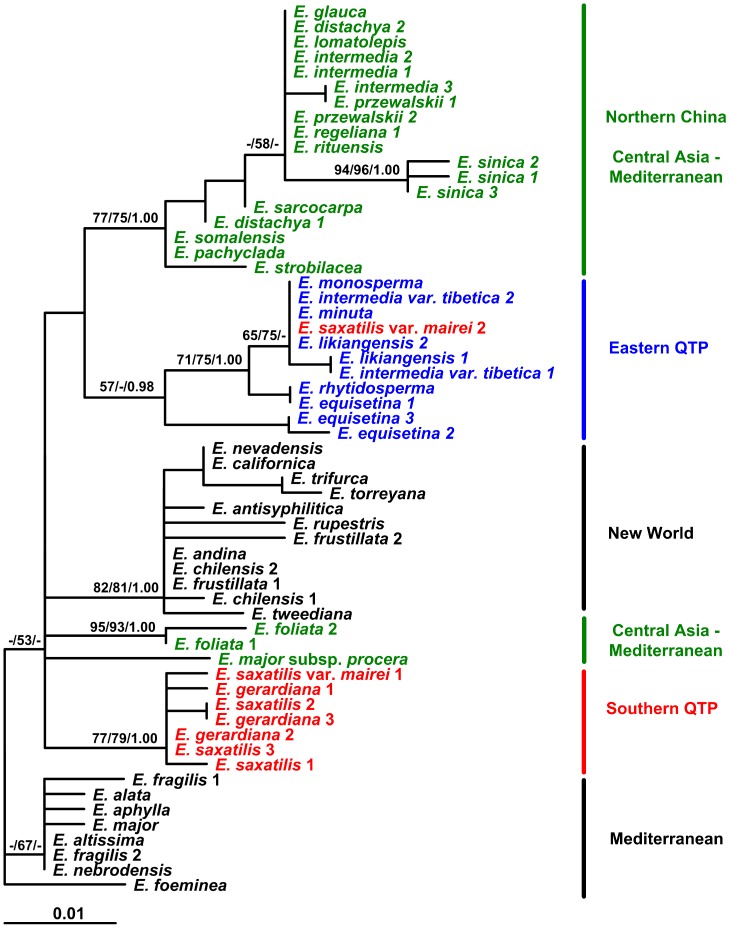
Length of the four cpDNA fragments (rbcL, rps4, rpL16, trnS-trnfM) was 758 bp, 443 bp, 496–516 bp and 387–458 bp, respectively. The combined alignment was 2266 bp, including 60 nucleotide substitutions and one phylogenetically informative indel (8 bp). When the Mediterranean species E. foeminea was used as outgroup, following Rydin et al. [36], the ML and BI trees generated from the combined four cpDNA fragments were generally consistent in topology, supporting five main clades that correspond well to their geographical distributions. That is, species distributed in the New World, southern QTP and eastern QTP formed monophyletic clades, respectively; Most species from the Mediterranean region clustered together; The species from northern China and Central Asia formed a clade together with a couple of species from West Asia and the Mediterranean region (E. pachyclada, E. somalensis). Like in the phylogeny of the cpDNA haplotypes (Fig. 3), E. rituensis from the western Himalayas was nested in the northern China clade, E. intermedia var. tibetica was located in the eastern QTP clade, and different individuals of E. saxatilis var. mairei were placed in two clades (eastern QTP and southern QTP), respectively (Fig. 4).
Figure 4. The ML tree of Ephedra constructed from the combined cpDNA fragments (rbcL, rps4, rpL16 and trnS-trnfM).
Numbers above the branches are bootstrap values ≥50% for MP (left) and ML (middle) analyses, and Bayesian posterior probabilities ≥0.90 (right).
Discussion
Diversification of Ephedra in the QTP and adjacent regions was associated with uplift of the plateau and Asian aridification in the Miocene
The mechanisms underlying the development of high species diversity in the QTP are still largely unknown due to the complicated geological and climatic history of the plateau [3], [61]–[64]. Phylogeography provides a good link between population genetics and phylogenetics, and is very helpful to study speciation [65]. However, most previous phylogeographical studies of the QTP plants sampled populations of a single species and focused on the response of plant populations to the Quaternary glacial-interglacial cycles, phylogeographical structure and location of glacial refugia [18], [19], [29], [66]. In the present study, we sampled all but two of the Ephedra species distributed in the QTP and adjacent regions (totaling 14 species, 107 populations). Both phylogeographical analysis based on the cpDNA (trnT-trnF, trnS-trnfM) haplotypes and phylogenetic reconstruction from the combined four cpDNA fragments (rbcL, rps4, rpL16, trnS-trnfM) support three main lineages, i.e., eastern QTP, southern QTP, and northern China lineages of Ephedra (Figs. 3, 4). It is particularly interesting that the MRCA of each lineage can be dated back to the Middle or Late Miocene (Fig. 3, Table 3), a period during which the fast uplift of the QTP occurred [61], [63], [64]. Correspondingly, in the Miocene, the climate of Asia transformed from a zonal pattern to a monsoon-dominated pattern, and the aridification and desertification intensified in the Asian interior, including the QTP and northern China [2], [3], [67]. Given the high drought and/or cold tolerance of Ephedra, it could be inferred that the divergence of its three lineages was triggered by the Asian aridification driven by the QTP uplift. This inference is corroborated by a rich record of Ephedra or Ephedra-like fossil pollen from the late Oligocene and Miocene sediments of the QTP and neighboring areas (e.g., [58], [59], [67]). Loera et al. [68] also reported that the expansion of arid lands due to orogenetic and climatic changes played a role in the diversification of the North American Ephedra in the Late Miocene and Pliocene.
The study of Ickert-Bond et al. [30] indicated that Ephedra possibly originated in the Mediterranean region, and then dispersed into Asia, and further into the New World. If this biogeographic scenario is true, the Ephedra species in the QTP and neighboring regions could have evolved from a Central Asian ancestor, which possibly diverged during the QTP uplift and dispersed eastward along two routes. One was along the Himalayas, giving rise to the southern QTP lineage (Fig. 3, clade D), while the other was from Central Asia to North China, giving rise to the northern China and eastern QTP lineages (Fig. 3, clades A and C). One may argue that the eastern QTP and southern QTP lineages were once closely related, even with a sympatric distribution in the QTP, but separated afterwards due to the Quaternary climatic changes. However, this explanation is not supported by the ancient divergence between the two lineages and the sister relationship between the northern China lineage and the eastern QTP lineage (Fig. 3). Although the three haplotypes H23–H25 of E. equisetina are most closely related to the outgroup according to the haplotype network (Fig. 2), they show a close relationship with northern China and eastern QTP lineages in the ML trees (Figs. 3, S1). This discrepancy could be due to low resolution of the DNA markers and different algorithms of the methods. As to the distribution of E. saxatilis var. mairei (a taxon of the southern QTP lineage) in eastern QTP (Figs. 1, 3), it could be resulted from interspecific hybridization (see discussion later). It is worthy to mention that great genetic differentiation was found to have occurred between eastern QTP and southern QTP populations of single plant species (e.g., [21], [69]), but rarely between groups of congeneric species as reported in the present study.
Low intraspecific variation and lack of strong phylogeographical structure: Implications for phylogeographical studies
Although 25 cpDNA (trnT-trnF and trnS-trnfM) haplotypes were detected in the 14 Ephedra species sampled from the QTP and adjacent regions, only the widely distributed E. equisetina has relatively rich haplotypes (7). The other species harbor 2–5 haplotypes, respectively, or lack intraspecific variation such as in E. rhytidosperma, E. monosperma and E. rituensis (Fig. 1; Table 1). In addition, 15 haplotypes (60%) are species-specific (Fig. 1; Table S4). Among the studied 107 populations, 89 (83%) are pure in cpDNA haplotype (Fig. 1; Table 1). Obviously, most of the Ephedra species have low genetic diversity (HT = 0.076–0.633; HS = 0.030–0.139) (Table 5), compared to other plants from the same region [29], such as Pinus densata (HT = 0.929; HS = 0.812) [22] and Hippophae tibetana (HT = 0.956; HS = 0.372) [69]. Although the three species E. equisetina, E. gerardiana and E. przewalskii have relatively high HT values (0.777–0.843), their HS values are also very low (0–0.288) (Table 5). Moreover, unlike in most previous studies that found a strong phylogeographic structure in the QTP plants [29], no significant phylogeographic structure was detected in the Ephedra species analyzed, although NST>GST (not significant) and a high FST value were observed in some species (Tables 4, 5). This is consistent with the result of the Mantel test that genetic and geographical distances are not significantly correlated in most species (Table 4). The low intraspecific variation and lack of strong phylogeographical structure could be attributed to the prevalence of clonal reproduction, recent population expansion and a relatively recent origin of most extant species (Fig. 3), although the genus Ephedra could have an ancient origin [56], [70], [71]. Actually, low interspecific variation of Ephedra has been consistently reported in previous studies (e.g., [30], [34], [56], [68], [72]).
It seems that the level of genetic (haplotype) diversity is not obviously correlated to the wideness of distribution or ploidy level of a species. For example, among the species with a wide distribution and a good population sampling, the three species E. equisetina (2n = 2x = 14), E. gerardiana (2n = 14, 28, 56) and E. intermedia (2n = 4x = 28) harbor a relatively high genetic diversity, but E. monosperma (2n = 14, 28) lacks genetic variation (Fig. 1; Tables 1, S1). The current distribution of the QTP plants has been greatly shaped by the climatic oscillations in the late Cenozoic, especially by the Quaternary glacial-interglacial cycles, such as in Tsuga dumosa [20] and Pedicularis longiflora [19]. Despite that the mismatch distributions were only estimated for four Ephedra species, the results indicate different demographic histories of the congeneric species (Fig. S2). That is, unlike E. likiangensis from the southeastern edge of the QTP (Fig. S2a), the two species E. minuta and E. saxatilis from the eastern and southern QTP, respectively, could have experienced recent population expansion (Fig. S2b, c). This difference could be caused by different responses to the Quaternary climatic changes, although the time of population expansion needs to be further studied.
Among the 25 cpDNA haplotypes, nine (H1, H3, H5, H7, H8, H16, H19–H21) are shared by two or four species, respectively, and the haplotype H6 is even shared by eight species, suggesting that a wide sampling of species is helpful to investigate the origin of observed haplotypes and make reliable phylogeographical inference. For instance, the species E. gerardiana harbors five haplotypes, including H1–H3, H5, and H6 (Fig. 1; Table S4). If we only sample this species in the study, the haplotype H6 may be incorrectly considered as a newly evolved type due to its limited distribution in the species. Actually, this haplotype widely occurs in other species and is the most frequent haplotype in the eastern QTP lineage. For another example, the haplotype H7 is shared between E. rituensis from southern QTP and E. regeliana from northern China (Fig. 1; Table S4). If we only study one of the two species, it will be difficult for us to reveal the evolutionary history of this haplotype.
Systematic positions of some Ephedra species
In the phylogenies of cpDNA haplotypes and combined four chloroplast genes, the western Himalayan E. rituensis is nested in the northern China lineage rather than in the southern QTP lineage, and E. intermedia var. tibetica is closely related to species of the eastern QTP lineage rather that to E. intermedia of the northern China lineage. Morphologically, according to our field investigation, E. intermedia var. tibetica sometimes has both straight and spirally twisted integument tubes in the same individual, which is different from E. intermedia. To reveal systematic positions of E. rituensis and E. intermedia var. tibetica, the biparentally inherited nuclear genes should be used in future studies.
In addition, there was debate about whether E. saxatilis var. mairei should be placed in E. likiangensis or E. saxatilis [31]. According to the present cpDNA analysis, E. saxatilis var. mairei is closely related to E. saxatilis-E. gerardiana rather than to E. likiangensis. However, our preliminary nuclear gene analysis (unpublished) seems to suggest a close relationship of E. saxatilis var. mairei to both E. likiangensis and E. saxatilis. That is, E. saxatilis var. mairei could have originated from hybridization between E. likiangensis and E. saxatilis. Actually, interspecific hybridization has been reported from the New World species [73], [74], such as the formation of ×E. arenicola (E. torreyana×E. coryi var. viscida [E. cutleri]) and ×E. intermixta (E. torreyana×E. trifurca). Moreover, E. equisetina harbors very different cpDNA haplotypes that are located in different clades (Fig. 3). This species has a very wide distribution in Central Asia and northern China. It would be interesting to investigate whether this species has evolved into several cryptic species given the greatly reduced morphological characters of Ephedra.
Supporting Information
The ML tree showing evolutionary relationships of cpDNA haplotypes with H26 (Ephedra nebrodensis) as outgroup. Numbers above branches are bootstrap values ≥50% for MP (left) and ML (middle) analyses, and the Bayesian posterior probabilities ≥0.90 (right), respectively. Arrows denote Clades A, B, C and D.
(TIF)
Mismatch distributions for Ephedra species.
(TIF)
Population sampling information and cpDNA (trnT-trnF+trnS-trnfM) variation of the studied Ephedra species.
(DOC)
Sources of materials for phylogenetic reconstruction of Ephedra based on combined cpDNA.
(DOC)
Primers used in the present study.
(DOC)
The distribution of cpDNA haplotypes in Ephedra species.
(DOC)
Acknowledgments
The authors thank Drs. Dun-Yan Tan and Shi-Long Chen for their help in sample collection. We also thank Ms. Wan-Qing Jin for her assistance in DNA sequencing.
Funding Statement
This work was supported by by the National Natural Science Foundation of China (grants 31170197, 30730010) and the Chinese Academy of Sciences (the 100-Talent Project). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zhang YL, Li BY, Zheng D (2002) A discussion on the boundary and area of the Tibetan Plateau in China. Geograph Res 21: 1–8. [Google Scholar]
- 2. An Z, Kutzbach JE, Prell WL, Porter SC (2001) Evolution of Asian monsoons and phased uplift of the Himalaya-Tibetan plateau since Late Miocene times. Nature 411: 62–66. [DOI] [PubMed] [Google Scholar]
- 3. Guo ZT, Ruddiman WF, Hao QZ, Wu HB, Qiao YS, et al. (2002) Onset of Asian desertification by 22 Myr ago inferred from loess deposits in China. Nature 416: 159–163. [DOI] [PubMed] [Google Scholar]
- 4.Mittermeier RA, Gil PR, Hoffman M, Pilgrim J, Brooks T, et al.. (2005) Hotspots revisited: Earth's biologically richest and most endangered terrestrial ecoregions. Monterrey, Mexico: Conservation International and Agrupación Sierra Madre.
- 5. Wu ZY (1988) Hengduan Mountain flora and her significance. J Jap Bot 63: 297–311. [Google Scholar]
- 6. Li XW, Li J (1993) A preliminary floristic study on the seed plants from the region of Hengduan Mountains. Acta Bot Yunnan 15: 217–231. [Google Scholar]
- 7. Wang XR, Szmidt AE, Savolainen O (2001) Genetic composition and diploid hybrid speciation of a high mountain pine, Pinus densata, native to the Tibetan plateau. Genetics 159: 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wei XX, Wang XQ, Hong DY (2003) Marked intragenomic heterogeneity and geographical differentiation of nrDNA ITS in Larix potaninii (Pinaceae). J Mol Evol 57: 623–635. [DOI] [PubMed] [Google Scholar]
- 9. Wei XX, Wang XQ (2004) Recolonization and radiation in Larix (Pinaceae): evidence from nuclear ribosomal DNA paralogues. Mol Ecol 13: 3115–3123. [DOI] [PubMed] [Google Scholar]
- 10. Ran JH, Wei XX, Wang XQ (2006) Molecular phylogeny and biogeography of Picea (Pinaceae): Implications for phylogeographical studies using cytoplasmic haplotypes. Mol Phylogenet Evol 41: 405–419. [DOI] [PubMed] [Google Scholar]
- 11. Li Y, Stocks M, Hemmilä S, Källman T, Zhu H, et al. (2010) Demographic histories of four spruce (Picea) species of the Qinghai-Tibetan Plateau and neighboring areas inferred from multiple nuclear loci. Mol Biol Evol 27: 1001–1014. [DOI] [PubMed] [Google Scholar]
- 12. Xu TT, Abbott RJ, Milne RI, Mao KS, Du FK, et al. (2010) Phylogeography and allopatric divergence of cypress species (Cupressus L.) in the Qinghai-Tibetan Plateau and adjacent regions. BMC Evol Biol 10: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gao J, Wang BS, Mao JF, Ingvarsson P, Zeng QY, et al. (2012) Demography and speciation history of the homoploid hybrid pine Pinus densata on the Tibetan Plateau. Mol Ecol 21: 4811–4827. [DOI] [PubMed] [Google Scholar]
- 14. Sun Y, Wang A, Wan D, Wang Q, Liu J (2012) Rapid radiation of Rheum (Polygonaceae) and parallel evolution of morphological traits. Mol Phylogenet Evol 63: 150–158. [DOI] [PubMed] [Google Scholar]
- 15. Yang FS, Qin AL, Li YF, Wang XQ (2012) Great genetic differentiation among populations of Meconopsis integrifolia and its implication for plant speciation in the Qinghai-Tibetan Plateau. PLoS ONE 7: e37196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Song BH, Wang XQ, Wang XR, Ding KY, Hong DY (2003) Cytoplasmic composition in Pinus densata and population establishment of the diploid hybrid pine. Mol Ecol 12: 2995–3001. [DOI] [PubMed] [Google Scholar]
- 17. Song BH, Wang XQ, Wang XR, Sun LJ, Hong DY, et al. (2002) Maternal lineages of Pinus densata, a diploid hybrid. Mol Ecol 11: 1057–1063. [DOI] [PubMed] [Google Scholar]
- 18. Meng LH, Yang R, Abbott RJ, Miehe G, Hu TH, et al. (2007) Mitochondrial and chloroplast phylogeography of Picea crassifolia Kom. (Pinaceae) in the Qinghai-Tibetan Plateau and adjacent highlands. Mol Ecol 16: 4128–4137. [DOI] [PubMed] [Google Scholar]
- 19. Yang FS, Li YF, Ding X, Wang XQ (2008) Extensive population expansion of Pedicularis longiflora (Orobanchaceae) on the Qinghai-Tibetan Plateau and its correlation with the Quaternary climate change. Mol Ecol 17: 5135–5145. [DOI] [PubMed] [Google Scholar]
- 20. Cun YZ, Wang XQ (2010) Plant recolonization in the Himalaya from the southeastern Qinghai-Tibetan Plateau: Geographical isolation contributed to high population differentiation. Mol Phylogenet Evol 56: 972–982. [DOI] [PubMed] [Google Scholar]
- 21. Li Y, Zhai SN, Qiu YX, Guo YP, Ge XJ, et al. (2011) Glacial survival east and west of the ‘Mekong-Salween Divide’ in the Himalaya-Hengduan Mountains region as revealed by AFLPs and cpDNA sequence variation in Sinopodophyllum hexandrum (Berberidaceae). Mol Phylogenet Evol 59: 412–424. [DOI] [PubMed] [Google Scholar]
- 22. Wang BS, Mao JF, Gao J, Zhao W, Wang XR (2011) Colonization of the Tibetan Plateau by the homoploid hybrid pine Pinus densata . Mol Ecol 20: 3796–3811. [DOI] [PubMed] [Google Scholar]
- 23. Opgenoorth L, Vendramin GG, Mao KS, Miehe G, Miehe S, et al. (2010) Tree endurance on the Tibetan Plateau marks the world's highest known tree line of the Last Glacial Maximum. New Phytol 185: 332–342. [DOI] [PubMed] [Google Scholar]
- 24. Shimono A, Ueno S, Gu S, Zhao X, Tsumura Y, et al. (2010) Range shifts of Potentilla fruticosa on the Qinghai-Tibetan Plateau during glacial and interglacial periods revealed by chloroplast DNA sequence variation. Heredity 104: 534–542. [DOI] [PubMed] [Google Scholar]
- 25. Schönswetter P, Stehlik I, Holderegger R, Tribsch A (2005) Molecular evidence for glacial refugia of mountain plants in the European Alps. Mol Ecol 14: 3547–3555. [DOI] [PubMed] [Google Scholar]
- 26. Soltis DE, Morris AB, McLachlan JS, Manos PS, Soltis PS (2006) Comparative phylogeography of unglaciated eastern North America. Mol Ecol 15: 4261–4293. [DOI] [PubMed] [Google Scholar]
- 27. Hickerson MJ, Carstens BC, Cavender-Bares J, Crandall KA, Graham CH, et al. (2010) Phylogeography's past, present, and future: 10 years after. Mol Phylogenet Evol 54: 291–301. [DOI] [PubMed] [Google Scholar]
- 28. Shafer ABA, Cullingham CI, Côté SD, Coltman DW (2010) Of glaciers and refugia: a decade of study sheds new light on the phylogeography of northwestern North America. Mol Ecol 19: 4589–4621. [DOI] [PubMed] [Google Scholar]
- 29. Qiu YX, Fu CX, Comes HP (2011) Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of Quaternary climate and environmental change in the world's most diverse temperate flora. Mol Phylogenet Evol 59: 225–244. [DOI] [PubMed] [Google Scholar]
- 30. Ickert-Bond SM, Rydin C, Renner SS (2009) A fossil-calibrated relaxed clock for Ephedra indicates an Oligocene age for the divergence of Asian and New World clades and Miocene dispersal into South America. J Syst Evol 47: 444–456. [Google Scholar]
- 31.Fu L, Yu Y, Riedl H (1999) Ephedraceae. In: Wu CY, Raven P, editors. Flora of China: Science Press, Beijing, and Missouri Botanical Garden Press, St. Louis. pp. 97–101.
- 32.Yang Y (2002) Systematics and evolution of Ephedra L. (Ephedraceae) from China. Beijing: Chinese Academy of Sciences.
- 33. Yang Y, Fu D, Zhu G (2003) A new species of Ephedra (Ephedraceae) from China. Novon 13: 153–155. [Google Scholar]
- 34. Huang J, Price RA (2003) Estimation of the age of extant Ephedra using chloroplast rbcL sequence data. Mol Biol Evol 20: 435–440. [DOI] [PubMed] [Google Scholar]
- 35. Rydin C, Korall P (2009) Evolutionary relationships in Ephedra (Gnetales), with implications for seed plant phylogeny. Int J Plant Sci 170: 1031–1043. [Google Scholar]
- 36. Rydin C, Khodabandeh A, Endress PK (2010) The female reproductive unit of Ephedra (Gnetales): comparative morphology and evolutionary perspectives. Bot J Linn Soc 163: 387–430. [DOI] [PubMed] [Google Scholar]
- 37. Rogers SO, Bendich AJ (1988) Extraction of DNA from plant tissues. Plant Mol Biol (Manual) A6: 1–10. [DOI] [PubMed] [Google Scholar]
- 38. Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17: 1105–1109. [DOI] [PubMed] [Google Scholar]
- 39. Demesure B, Sodzi N, Petit RJ (1995) A set of universal primers for amplification of polymorphic noncoding regions of mitochondrial and chloroplast DNA in plants. Mol Ecol 4: 129–131. [DOI] [PubMed] [Google Scholar]
- 40. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98. [Google Scholar]
- 42. Excoffier L, Laval G, Schneider S (2005) Arlequin version 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- 43. Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144: 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nei M (1975) Molecular population genetics and evolution. Amsterdam, Oxford: North-Holland Publishing Company.
- 45. Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27: 209–220. [PubMed] [Google Scholar]
- 46. Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9: 1657–1659. [DOI] [PubMed] [Google Scholar]
- 47.Swofford DL (2002) PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods) 4.0 Beta. Sunderland, MA: Sinauer Associates.
- 48. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- 49. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 50. Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- 51.Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden.
- 52. Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9: 552–569. [DOI] [PubMed] [Google Scholar]
- 53. Felsenstein J (1988) Phylogenies from molecular sequences: Inference and reliability. Ann Rev Genet 22: 521–565. [DOI] [PubMed] [Google Scholar]
- 54. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ickert-Bond SM, Wojciechowski MF (2004) Phylogenetic relationships in Ephedra (Gnetales): evidence from nuclear and chloroplast DNA sequence data. Syst Bot 29: 834–849. [Google Scholar]
- 56. Rydin C, Pedersen KR, Friis EM (2004) On the evolutionary history of Ephedra: Cretaceous fossils and extant molecules. Proc Natl Acad Sci USA 101: 16571–16576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang KF, Yang JW, Z L, Li ZR (1975) On the Tertiary sporo-pollen assemblages from Lunpola Basin of Xizang, China and their palaeogeographic significance. Sci Geol Sin 1975-04: 366–374. [Google Scholar]
- 58. Song ZC, Wang WM, Mao FY (2008) Palynological implications for relationship between aridification and monsoon climate in the Tertiary of NW China. Acta Palaeontol Sin 47: 265–272. [Google Scholar]
- 59. Sun J, Zhang Z (2008) Palynological evidence for the mid-Miocene climatic optimum recorded in Cenozoic sediments of the Tian Shan Range, northwestern China. Glob Planet Change 64: 53–68. [Google Scholar]
- 60.Rambaut A, Drummond AJ (2007) Tracer v1.4. Molecular Evolution, Phylogenetics and Epidemiology. Available: http://beast.bio.ed.ac.uk/Tracer. Accessed 2012.
- 61. Spicer RA, Harris NBW, Widdowson M, Herman AB, Guo SX, et al. (2003) Constant elevation of southern Tibet over the past 15 million years. Nature 421: 622–624. [DOI] [PubMed] [Google Scholar]
- 62. Dupont-Nivet G, Krijgsman W, Langereis CG, Abels HA, Dai S, et al. (2007) Tibetan plateau aridification linked to global cooling at the Eocene-Oligocene transition. Nature 445: 635–638. [DOI] [PubMed] [Google Scholar]
- 63. Royden LH, Burchfiel BC, van der Hilst RD (2008) The geological evolution of the Tibetan Plateau. Science 321: 1054–1058. [DOI] [PubMed] [Google Scholar]
- 64. Wang C, Zhao X, Liu Z, Lippert PC, Graham SA, et al. (2008) Constraints on the early uplift history of the Tibetan Plateau. Proc Natl Acad Sci USA 105: 4987–4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Avise JC (2009) Phylogeography: retrospect and prospect. J Biogeogr 36: 3–15. [Google Scholar]
- 66. Liu JQ, Sun YS, Ge XJ, Gao LM, Qiu YX (2012) Phylogeographic studies of plants in China: Advances in the past and directions in the future. J Syst Evol 50: 267–275. [Google Scholar]
- 67. Jiang H, Ding Z (2008) A 20 Ma pollen record of East-Asian summer monsoon evolution from Guyuan, Ningxia, China. Palaeogeogr Palaeoclimatol Palaeoecol 265: 30–38. [Google Scholar]
- 68. Loera I, Sosa V, Ickert-Bond SM (2012) Diversification in North America arid lands: Niche conservatism, divergence and expansion of habitat explain speciation in the genus Ephedra . Mol Phylogenet Evol 65: 437–450. [DOI] [PubMed] [Google Scholar]
- 69. Wang H, Qiong L, Sun K, Lu F, Wang YG, et al. (2010) Phylogeographic structure of Hippophae tibetana (Elaeagnaceae) highlights the highest microrefugia and the rapid uplift of the Qinghai-Tibetan Plateau. Mol Ecol 19: 2964–2979. [DOI] [PubMed] [Google Scholar]
- 70. Wang X, Zheng S (2010) Whole fossil plants of Ephedra and their implications on the morphology, ecology and evolution of Ephedraceae (Gnetales). Chin Sci Bull 55: 1511–1519. [Google Scholar]
- 71. Ickert-Bond SM, Rydin C (2011) Micromorphology of the seed envelope of Ephedra L. (Gnetales) and its relevance for the timing of evolutionary events. Int J Plant Sci 172: 36–48. [Google Scholar]
- 72. Huang J, Giannasi DE, Price RA (2005) Phylogenetic relationships in Ephedra (Ephedraceae) inferred from chloroplast and nuclear DNA sequences. Mol Phylogenet Evol 35: 48–59. [DOI] [PubMed] [Google Scholar]
- 73. Cutler HC (1939) Monograph of the North American species of the genus Ephedra . Ann Mo Bot Gard 26: 373–428. [Google Scholar]
- 74. Wendt T (1993) A new variety of Ephedra torreyana (Ephedraceae) from west Texas and Chihuahua, with notes on hybridization in the E. torreyana complex. Phytologia 74: 141–150. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The ML tree showing evolutionary relationships of cpDNA haplotypes with H26 (Ephedra nebrodensis) as outgroup. Numbers above branches are bootstrap values ≥50% for MP (left) and ML (middle) analyses, and the Bayesian posterior probabilities ≥0.90 (right), respectively. Arrows denote Clades A, B, C and D.
(TIF)
Mismatch distributions for Ephedra species.
(TIF)
Population sampling information and cpDNA (trnT-trnF+trnS-trnfM) variation of the studied Ephedra species.
(DOC)
Sources of materials for phylogenetic reconstruction of Ephedra based on combined cpDNA.
(DOC)
Primers used in the present study.
(DOC)
The distribution of cpDNA haplotypes in Ephedra species.
(DOC)