Abstract
Embryogenic suspensions that involve extensive cell division are risky in respect to genome and epigenome instability. Elevated frequencies of somaclonal variation in embryogenic suspension-derived plants were reported in many species, including coffee. This problem could be overcome by using culture conditions that allow moderate cell proliferation. In view of true-to-type large-scale propagation of C. arabica hybrids, suspension protocols based on low 2,4-D concentrations and short proliferation periods were developed. As mechanisms leading to somaclonal variation are often complex, the phenotypic, genetic and epigenetic changes were jointly assessed so as to accurately evaluate the conformity of suspension-derived plants. The effects of embryogenic suspensions and secondary embryogenesis, used as proliferation systems, on the genetic conformity of somatic embryogenesis-derived plants (emblings) were assessed in two hybrids. When applied over a 6 month period, both systems ensured very low somaclonal variation rates, as observed through massive phenotypic observations in field plots (0.74% from 200 000 plant). Molecular AFLP and MSAP analyses performed on 145 three year-old emblings showed that polymorphism between mother plants and emblings was extremely low, i.e. ranges of 0–0.003% and 0.07–0.18% respectively, with no significant difference between the proliferation systems for the two hybrids. No embling was found to cumulate more than three methylation polymorphisms. No relation was established between the variant phenotype (27 variants studied) and a particular MSAP pattern. Chromosome counting showed that 7 of the 11 variant emblings analyzed were characterized by the loss of 1–3 chromosomes. This work showed that both embryogenic suspensions and secondary embryogenesis are reliable for true-to-type propagation of elite material. Molecular analyses revealed that genetic and epigenetic alterations are particularly limited during coffee somatic embryogenesis. The main change in most of the rare phenotypic variants was aneuploidy, indicating that mitotic aberrations play a major role in somaclonal variation in coffee.
Introduction
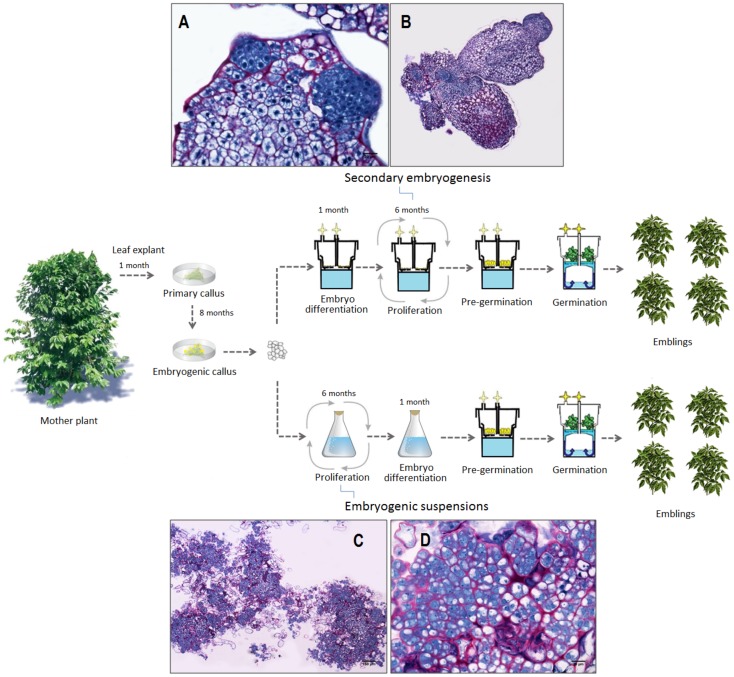
Among micropropagation methods, somatic embryogenesis has the best potential for rapid and large-scale multiplication of selected varieties in a wide range of economically important species. Schematically, the initial step of dedifferentiation leading to the acquisition of embryogenic competence is common to most plant species, whereas for the following step of proliferation of embryogenic material, efficient procedures can be classified under two main strategies. The first is the proliferation through secondary embryogenesis (SCE) which involves first differentiating the somatic embryos before enhancing their proliferation by adventitious budding (Figure 1). The second consists of establishing embryogenic suspensions (ESP) to favor large-scale embryogenic cell proliferation before the subsequent embryo differentiation step. In order to come up with an industrial procedure, the development of ESP represents the best option to ensure synchronous and massive somatic embryo production [1]. In addition, ESP allows the production of large numbers of embryogenic-competent cells and this process can be easily scaled up. Nevertheless, tissue culture systems such as somatic embryogenesis that involve the acquisition of competence for pluripotentiality and extensive cell division are more risky with respect to genome and epigenome instability [2]. The use of ESP has frequently been associated with an increased risk of genetic instability and somaclonal variation in the regenerated plants [3]–[5]. Although ESP has been developed for some important crops, it has therefore not been widely applied for commercial purposes. Somaclonal variation in ESP-derived plants is probably related to the presence of 2,4-dichlorophenoxyacetic acid (2,4-D), which is often essential for maintaining proliferating cells in an embryogenic, undifferentiated state [6], [7]. This auxin could enhance somaclonal variation through the stimulation of rapid disorganized growth that can influence the mitotic process, resulting in chromosomal aberrations [8], [9].
Figure 1. Schematic representation of two somatic embryogenesis processes applied at the industrial level.
The first somatic embryogenesis process (upper section of the flow diagram) involved a proliferation step based on secondary embryogenesis in RITA® temporary immersion bioreactors (photos 1A, 1B). The second process (lower section of flow diagram) included a proliferation step based on embryogenic suspensions (photos 1C, 1D). 1A, initial developmental stages of secondary embryos at the root pole of primary somatic embryos; 1B, clusters of primary and secondary embryos; 1C, clusters of embryogenic cells in suspension; 1D, embryogenic cells in suspension.
The term ‘somaclonal variation’ (SV) describes the tissue culture-induced stable genetic, epigenetic or phenotypic variation in clonally propagated plant populations [10]. Somaclonal variation is considered to be one of the main bottlenecks in the development of micropropagation procedures, especially in view of large-scale commercial operations, for which the strict maintenance of genetic and agronomic traits from selected individuals is required. An analysis of the progeny of phenotypic variants showed that some of the variations produced by somatic embryogenesis can occur in the form of stable and heritable mutations [8], [11]. In maize, Kaeppler and Phillips [12] also reported stable segregation of somaclonal variant phenotypic qualities in several seed generations. It has also been well documented that somaclonal variants commonly present cytological aberrations such as chromosomal rearrangements (deletions, duplications, inversions and translocations), and sometimes more severe alterations like aneuploidy or polyploidy [12]–[16].
Although most mutants segregate in a Mendelian fashion upon selfing and outcrossing [12], SV is sometimes present in the form of transient mutations, suggesting the involvement of epigenetic events [11]. Epigenetic traits are heritable changes associated with chemical modification of DNA without alteration of the primary DNA sequence [17]. Cytosine methylation has been proposed as a possible cause of SV [11], [12]. Epigenetic modifications (methylation) can mediate the transmission of an active or silent gene in the short-term (mitotic cell division) or long-term (meiotic divisions leading to transmission across generations) [18]. DNA methylation in plants commonly occurs at cytosine (5-methylcytosine, m5C) bases in all sequence contexts: the symmetric CG and CHG (in which H could be A, T or C) and the asymmetric CHH contexts [17], [18]. Molecular marker approaches like methylation-sensitive amplified polymorphism (MSAP) and Met-AFLP have proved efficient in the analysis of methylation patterns [19], [20]. The existence of zones susceptible to methylation variations was recently shown in somatic embryogenesis-derived plants (emblings) in grapevine [21] and barley [20]. SV was also associated with the activity of mobile DNA elements or retroelements [22], [23]. Novel mechanisms such as RNAi directed demethylation have recently been proposed to explain retrotransposon activation [2], [24].
Coffea arabica is an allotetraploid tree species (2n = 4X = 44) characterized by low molecular polymorphism [25]. Somatic embryogenesis is currently applied industrially for large-scale and rapid dissemination of selected F1 hybrids that provide a highly significant increase in the yield of high quality coffee [26], [27]. Regarding industrial-scale micropropagation, upgrading production to several million vitroplants per production unit would undoubtedly boost economic profitability. This would require switching from an SCE- to an ESP-based protocol. However, former field observations revealed that SV occurs at relatively high rates in ESP-derived C. arabica plants [28], [29]. Apart from different phenotypic variants easily identifiable through morphological characteristics, we did not discover in trees showing a normal phenotype any variations involving agronomically important quantitative and physiological characteristics [30]. In view of true-to-type propagation of selected C. arabica hybrid varieties, we previously developed improved ESP protocols based on the use of low exogenous 2,4-D concentrations and short proliferation periods, allowing reliable somatic embryo mass regeneration [27]. For potential commercial applications, here using two C. arabica hybrids we verified the conformity of suspension-derived plants with that of plants obtained by secondary embryogenesis, i.e. the industrial process currently in use. The objectives were to assess large-scale phenotype conformity in commercial field plots, to quantify genetic and epigenetic modifications in the regenerated plants through AFLP (Amplified fragment length polymorphism) and MSAP molecular markers, and to cytologically characterize the karyotype of different phenotypic variants detected in the study.
Results
Frequency of phenotypic variants
Embling batches of hybrids HI and H3 obtained from both SCE and ESP were checked for phenotype variation at both nursery and field levels. The frequency of phenotypic variants assessed among more than 600 000 plants in the nursery was very low (approx. 0.1%) and not significantly affected by the proliferation system nor the hybrid variety (Table 1). Observation of around 200 000 emblings in the field two years after planting revealed roughly an additional 0.74% of abnormal phenotypes, still without any significant difference between the two proliferation systems and hybrids. Apart from these phenotypic variants, all the other studied trees flowered, grew and produced normally. In conclusion, the overall phenotypic variation rate obtained by pooling the data obtained both in the nursery and in the field was less than 1% and no significant differences were noted between the proliferation systems or between the hybrids used.
Table 1. Frequency of coffee phenotypic variants detected in the nursery and field on three year-old plants from two C. arabica hybrids depending on the type of proliferation system used in the industrial somatic embryogenesis process.
| Proliferation system in the somatic embryogenesis process | Hybrid | Observations after 10 months in nursery | Observations after 36 months in field | ||||||
| No. of observed emblings | No. of variants | Somaclonal variation frequency (%) | 3σ confidence interval* | No. of emblings | No. of variants | Somaclonal variation frequency (%) | 3σ confidence interval* | ||
| Secondary embryogenesis | H1 | 117.115 | 148 | 0.13 | [0.09–0.16] | 51.131 | 373 | 0.73 | [0.62–0.84] |
| H3 | 121.894 | 159 | 0.13 | [0.10–0.16] | 49.126 | 390 | 0.79 | [0.67–0.91] | |
| Total | 239.009 | 307 | 0.13 | [0.10–0.15] | 100.257 | 763 | 0.76 | [0.67–0.84] | |
| Embryogenic suspension | H1 | 204.871 | 206 | 0.10 | [0.08–0.12] | 54.218 | 402 | 0.74 | [0.63–0.85] |
| H3 | 197.705 | 183 | 0.09 | [0.07–0.11] | 54.566 | 394 | 0.72 | [0.61–0.83] | |
| Total | 402.576 | 389 | 0.09 | [0.08–0.11] | 108.784 | 796 | 0.73 | [0.65–0.80] | |
The variable analyzed was the proportion (p) of variant (p = X/n), where X was the number of variant and n the number of plants observed. A 3σ confidence limit for binomial distribution was calculated using the formula 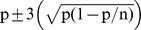 with a level of confidence of 99%.
with a level of confidence of 99%.
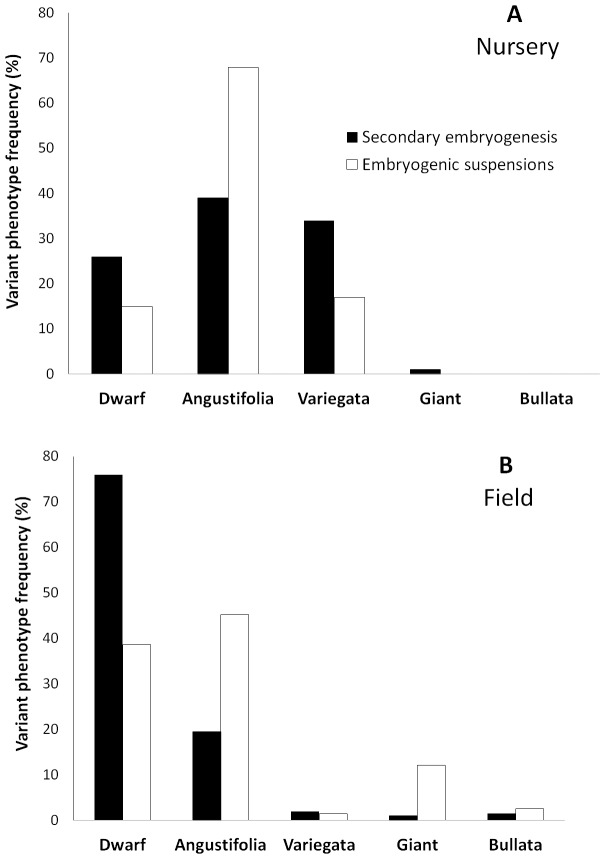
Both proliferation systems generated the same kind of phenotypic variants (Figures 2A, B), with the Dwarf and Angustifolia types (Figures 3E, B) being the most frequent. Note that the secondary embryogenesis proliferation system specifically enhanced the occurrence of Dwarf variants, whereas the embryogenic suspensions favored the occurrence of the Angustifolia type. This latter phenotype can easily be detected and eliminated at the nursery level along with the Variegata variant (Figure 3C). A comparison of Figures 2A and 2B clearly shows that elimination in the nursery is not efficient for the Dwarf type. This somaclonal variation is more easily observable 2–3 years after planting in the field thanks to the characteristic grouped canopy morphology and low yield. Similarly, the Giant and Bullata (Figure 3F) phenotypic variants could only be detected in the field on well-developed trees.
Figure 2. Proportions (%) of the different types of phenotypic variants in comparison to the total number of variants.
Variants representing less than 1% of somatic embryogenesis-derived plants were observed in C. arabica embling batches at nursery (A) and field (B) levels, depending on the proliferation system used, i.e. secondary embryogenesis (SCE) or embryogenic suspension (ESP). In the nursery, the data were obtained from 239 009 emblings derived from SCE and 402 576 emblings from ESP. In the field, the data were obtained through the observation of 100 257 emblings derived from secondary embryogenesis and 108 784 from embryogenic suspensions.
Figure 3. Examples of different C. arabica phenotypic variants in plants mass propagated through somatic embryogenesis.
A, plant exhibiting a normal phenotype in the nursery; B, Angustifolia variant in the nursery; C, Variegata variant in the field; D, plant showing a normal phenotype in the field; E, Dwarf variant in the field; F, Bullata variant in nursery.
Locus specific polymorphisms revealed by AFLP
In order to verify the induction of molecular polymorphism by the somatic embryogenesis process, AFLP analysis (four primer combinations, Table 2) was carried out on mother plants and their derived emblings. From a total of 204 bands obtained, only one polymorphic fragment of 173 bp in size (Eco-ACT/Mse-AGT) shared by two emblings of hybrid H1 and exhibiting a normal phenotype, was found (Table 3). From a total of 198 bands obtained, no polymorphism was found in emblings of hybrid H3. All variants had the same AFLP pattern as the mother plants. For both hybrids, no significant quantitative effect on AFLP was detected when comparing SCE and ESP.
Table 2. Primer combinations used for AFLP and MSAP analyses.
| AFLP primer combinations (Eco+3 labeled/Mse+3) | MSAP primer combinations (Eco−3/Hpa+2 labeled) |
| Eco-ACT/Mse-AGT | C1 Eco-AAC/HPA-AA |
| Eco-AGG/Mse-AGT | C2 Eco-AAC/HPA-AT |
| Eco-CGC/Mse-CCA | C3 Eco-AGG/HPA-AA |
| Eco-CAC/Mse-CCA | C4 Eco-AGG/HPA-AT |
| C5 Eco-ACT/HPA-CA | |
| C6 Eco-ACT/HPA-CT | |
| C7 Eco-AGA/HPA-CA | |
| C8 Eco-AGA/HPA-CT |
Fluorescent dyes for marked primers correspond to 5'-FAM TM and 5'-HEX TM
Table 3. Summary of AFLP data and observed polymorphisms among mother plants and emblings derived from secondary embryogenesis or embryogenic suspensions.
| Proliferation system in the somatic embryogenesis process | Hybrid | No. of analyzed emblings | No. of fragments | Polymorphic fragments | Emblings showing polymorphisms | Total polymorphism ** | ||
| No. | (%) | No. | (%) | (%) | ||||
| Secondary embryogenesis | H1 | 33 | 204 | 1 * | 0.5 | 2 | 6 | 0.03 |
| H3 | 45 | 198 | 0 | 0 | 0 | 0 | 0 | |
| Embryogenic suspensions | H1 | 26 | 204 | 0 | 0 | 0 | 0 | 0 |
| H3 | 41 | 198 | 0 | 0 | 0 | 0 | 0 | |
Data were obtained for two C. arabica hybrids (H1 and H3) and compared with the patterns of the mother plants.
Found in 2 emblings with normal phenotype (N°210 and 232) showing a new AFLP band Eco-ACT/Mse-AGT 173 bp
Total polymorphism = [No. of polymorphic fragments/(No. of fragments × No. emblings)]×100
Methylation changes revealed by MSAP
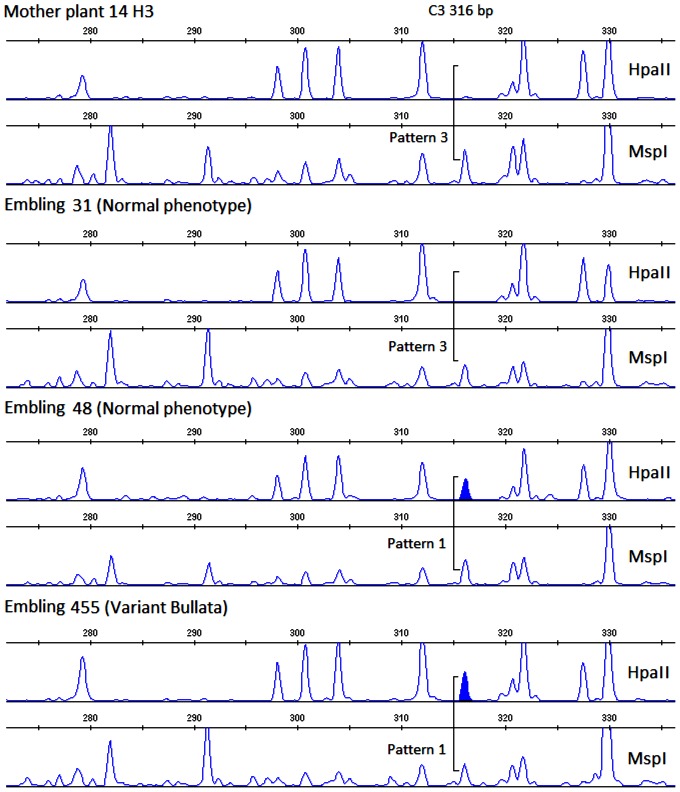
In order to evaluate the occurrence of possible epigenetic modifications in the micropropagated plants, a study on the alteration of methylation patterns was performed by MSAP analysis using eight primer combinations (Table 2). Only clear and reproducible bands were selected for the analysis. More than 395 fragments were considered. First, MSAP patterns were obtained from DNA digested by the two isoschizomers (HpaII and MspI), as illustrated in Figure 4. They were further compared with those of mother plants to classify the amplified fragments according to the methylation pattern, as shown in Table 4. The percentages of monomorphic fragments (pattern 1) were elevated and similar for both hybrids at nearly 91%. The remaining 9% of fragments (8.5% for H1 and 9.1% for H3) almost exclusively corresponded to pattern 3.
Figure 4. Representation of MSAP electropherograms observed for coffee mother plants and embling progeny using the isoschizomers HpaII and MspI.
Illustration of the pattern variation obtained for normal and variant phenotypes within the embling progeny.
Table 4. MSAP patterns corresponding to different methylation states of the symmetric sequence CCGG, as revealed by the specificity of the restriction enzymes Hpa II and Msp I.
| Restriction enzymes | MSAP patterns after enzymatic digestion | |||
| Pattern 1 | Pattern 2 | Pattern 3 | Pattern 4 | |
| Hpa II | 1 | 1 | 0 | 0 |
| Msp I | 1 | 0 | 1 | 0 |
| Methylation state | Unmethylated | Hemi-methylated | Fully-methylated | Fully-methylated |
| Methylation position | None | External cytosine | Internal cytosine | External cytosine |
| Schematic representation | CCGGGGCC | CH3CCGGGGCC | CH3CCGGGGCCCH3 | CH3CCGGGGCCCH3 |
A comparison of amplification patterns in mother plants and their respective progeny are reported in Table 5. All differences between mother and derived plants were switches between patterns 1 and 3 and vice versa, i.e. likely modifications in the restriction ability of HpaII. Among the polymorphic bands, eight bands corresponded to a change from pattern 3 in mother plants towards the unmethylated pattern 1 in emblings, suggesting demethylation. Changes associated with certain polymorphic MSAP bands were more frequent than others (Table 5). Seventy percent of the changes were linked to only five polymorphic bands. The detected polymorphism was very low and similar for both hybrids (Table 6) but slightly higher (0.07–0.18%) when compared to AFLP molecular markers. Similar to the AFLP results, the total MSAP polymorphism was not significantly different between the two proliferation systems nor between the two hybrid varieties.
Table 5. MSAP methylation patterns in mother plants and modified patterns in emblings.
| Polymorphic MSAP fragments (size in bp) | MSAP methylation patterns | Proliferation system affected by the methylation change | Presence of the methylation change depending on the plant phenotype | No. of methylation changes for each fragment | ||||
| Mother plants | Emblings | Hybrid H1 | Hybrid H3 | Normal | Variant | No. | (%) | |
| C3- 107 bp | Pattern 3 | Pattern 1 | SCE, ESP | SCE, ESP | +** | − | 8 | 9.8 |
| C2 -127 bp | Pattern 3 | Pattern 1 | 0 | ESP | + | − | 1 | 1.2 |
| C4 -134 bp | Pattern 3 | Pattern 1 | 0 | SCE | + | − | 4 | 4.8 |
| C1- 251 bp | Pattern1* | Pattern 3 | SCE, ESP | 0 | + | − | 12 | 14.6 |
| C3- 253 bp | Pattern 3 | Pattern 1 | 0 | SCE | + | − | 2 | 2.4 |
| C2- 302 bp | Pattern 3 | Pattern 1 | 0 | SCE, ESP | + | + | 16 | 19.5 |
| C3- 316 bp | Pattern 3 | Pattern 1 | 0 | SCE | + | + | 4 | 4.8 |
| C6 -370 bp | Pattern 3 | Pattern 1 | SCE, ESP | ESP | + | + | 10 | 12.2 |
| C8- 370 bp | Pattern 3 | Pattern 1 | SCE, ESP | SCE, ESP | + | + | 12 | 14.6 |
| C5- 387 bp | Pattern 1 | Pattern 3 | SCE | SCE | + | − | 13 | 15.8 |
| No. changes | 82 | |||||||
Relation with the type of C. arabica hybrid, type of proliferation system [secondary embryogenesis (SCE) and embryogenic suspension (ESP)] and regenerant phenotype.
Pattern 1: Fragment present in both HpaII and MspI restriction digests (1∶1); Pattern 3: Fragment absent in HpaII digests and present in MspI digests (0∶1). ** Relationship with a particular phenotype is indicated with (+) for presence and (−) for absence.
Table 6. Overall MSAP data and methylation polymorphism among mother plants and emblings of C. arabica hybrids derived from secondary embryogenesis or embryogenic suspensions.
| Proliferation in the somatic embryogenesis process | Hybrid | No. of emblings analyzed | No. of fragments | Methylation polymorphic fragments | Total polymorphism*(%) | 3σ confidence intervals** | |
| No. | Percentage (%) | ||||||
| Secondary embryogenesis | H1 | 33 | 399 | 5 | 1.2 | 0.18 | [0.071–0.294] |
| H3 | 45 | 396 | 7 | 1.7 | 0.16 | [0.068–0.246] | |
| 78 | 12 | 1.5 | 0.17 | [0.098–0.238] | |||
| Embryogenic suspensions | H1 | 26 | 399 | 4 | 1.0 | 0.18 | [0.057–0.309] |
| H3 | 41 | 396 | 5 | 1.0 | 0.07 | [0.006–0.129] | |
| 67 | 9 | 1.1 | 0.11 | [0.051–0.174] | |||
Total polymorphism = [No. of methylation polymorphic fragments/(No. of fragments × No. emblings)]×100.
The variable analyzed was the proportion (p) of methylation polymorphisms (p = X/n), where X was the number of methylation polymorphisms and n the total number of fragments. A 3σ confidence limit for binomial distribution was calculated using the formula 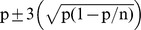 with a level of confidence of 99%.
with a level of confidence of 99%.
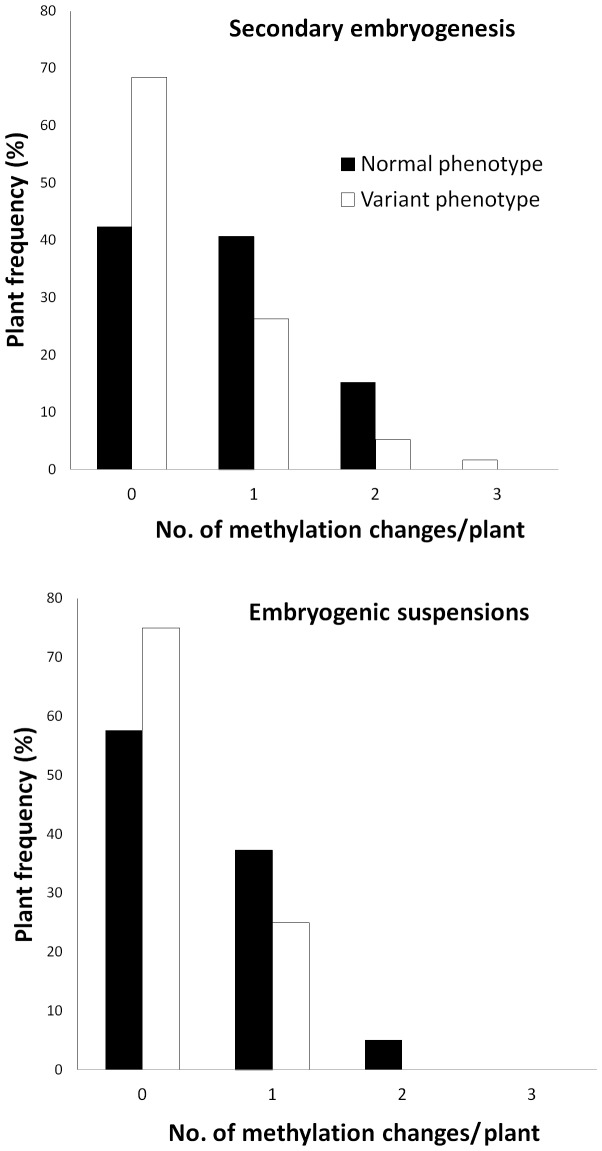
Most emblings showing changes in MSAP pattern had only one or two methylation polymorphisms (Figure 5). We did not find any emblings with more than three methylation polymorphisms. The same polymorphic bands were shared by plants from both proliferation systems and/or both hybrids in approximately half of the cases (Table 5). No relation was established between the variant phenotype and a particular MSAP pattern (Table 6).
Figure 5. Methylation polymorphism accumulation in coffee emblings showing a normal vs. variant phenotype depending on the somatic embryogenesis process used.
For the secondary embryogenesis process, data were derived from the analysis of 59 phenotypically normal and 19 variant emblings. For the embryogenic suspension process, 59 phenotypically normal and 8 variant emblings were studied.
Chromosome counting of somaclonal variants
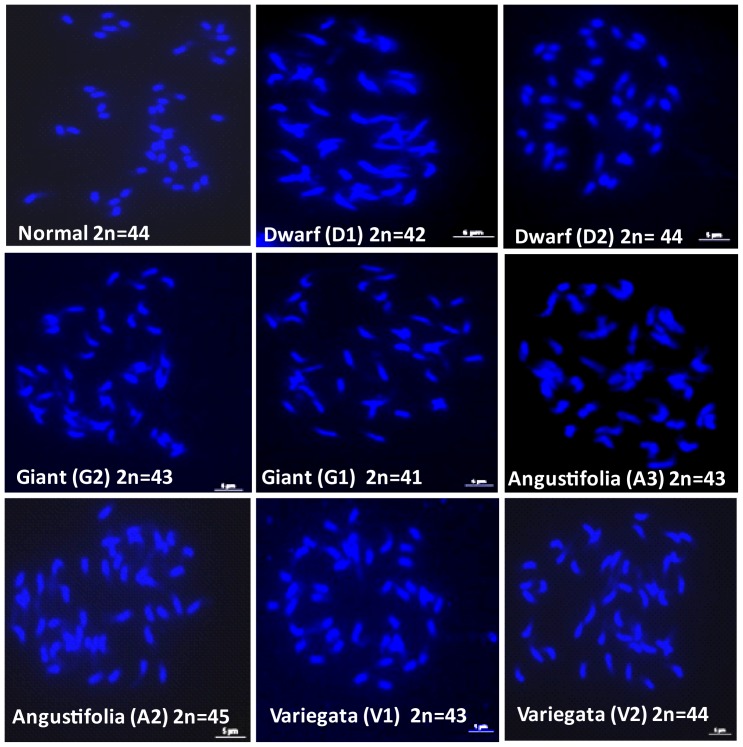
Chromosome numbers were assessed in 2 phenotypically normal emblings and 11 somaclonal variants (Fig. 6). Table 7 shows that the two normal regenerants exhibited the expected chromosome number for the allotetraploid C. arabica species (2n = 4x = 44) whereas 7 of the 11 variant emblings showed a different chromosome number (aneuploids). In almost all cases, the aneuploid karyotypes were characterized by the loss of 1–3 chromosomes. One Angustifolia variant had an extra chromosome. Moreover, the results showed that different chromosome numbers - including the normal number - could be observed for the same variant phenotype and that abnormal chromosome numbers were obtained for most of the variant phenotypes.
Figure 6. Mitotic cells at metaphase or prometaphase stages and observed ploidy levels of some normal and variant emblings from the allotetraploid C. arabica species (2n = 44).
Karyotype analyses were performed by counting chromosomes on four to eight clear metaphase spreads obtained from root tips of three year-old plants.
Table 7. Summary of chromosome counting in some normal versus variant C. arabica hybrids derived from somatic embryogenesis.
| Embling phenotype (normal or variant) | Somatic embryogenesis process | Code | No. of metaphases analyzed | No. of chromosomes |
| Normal | ESP | N1 | 8 | 44 |
| Normal | SCE | N2 | 7 | 44 |
| Angustifolia | ESP | A1 | 4 | 44 |
| Angustifolia | SCE | A2 | 8 | 45 |
| Angustifolia | ESP | A3 | 8 | 43 |
| Bullata | ESP | B1 | 4 | 44 |
| Dwarf | SCE | D1 | 5 | 42 |
| Dwarf | ESP | D2 | 4 | 44 |
| Giant | ESP | G1 | 6 | 41 |
| Giant | ESP | G2 | 6 | 43 |
| Giant | SCE | G3 | 6 | 43 |
| Variegata | ESP | V1 | 6 | 43 |
| Variegata | SCE | V2 | 4 | 44 |
The chromosome numbers obtained from root tips are indicated for 3 year-old emblings derived from embryogenic suspensions (ESP) or secondary embryogenesis (SCE) showing normal or abnormal phenotypes.
Discussion
Until now, relatively high somaclonal variation rates have been reported in C. arabica emblings, particularly with embryogenic suspensions [28], [29], [30]. The variant frequency was found to strongly increase in embryogenic suspensions after 6 months proliferation (25% after 12 months) in the presence of 4.52 μM 2,4-D [29]. The presence of disorganized rapid growth phases in tissue culture, such as callus and cell suspension cultures, has often been considered as one of the factors that cause SV [8], [31]. In view of true-to-type propagation, we further established processes with limited disorganized rapid growth phases. The first was based on SCE proliferation in temporary immersion bioreactors enhanced by the addition of cytokinin, similar to methods described in other woody plants such as rubber [32], oak [33] and tea [34] and is currently used on a commercial scale for coffee. The second involves the proliferation of ESP in the presence of both auxin and cytokinin albeit at a very low level of 2,4-D (1.36 µM) with a short proliferation time (6 months). These conditions allow sufficient amplification of embryogenic material to ensure the cost-effectiveness of the industrial process. A large-scale phenotypic evaluation for each process: 230 000–400 000 emblings in the nursery and 100 000 in the field was performed to obtain valuable and accurate information on genetic stability. The present study showed for the first time that, for both somatic embryogenesis processes, the variant phenotype frequencies were extremely low (less than 1%) and not statistically different. Moreover no statistical difference could be noted between the two studied hybrids. This clearly demonstrates that it is possible to control SV by optimizing the ESP culture conditions. In another cultivated Coffea species, i.e. the diploid C. canephora, Ducos et al. [35] found 2–4% of a low-vigor phenotype off-type among 5 067 emblings derived from 5–7-month-old ESP. In contrast to the C. arabica protocol, all somatic embryogenesis steps for C. canephora were performed with cytokinins as sole source of growth regulator. Hence, in coffee, both auxin and cytokinin could probably be involved in the generation of SV. The Angustifolia, Variegata and Dwarf variants were the most frequent types, irrespective of the proliferation strategy employed, confirming previous studies conducted on a lesser scale [29], [30]. Interestingly, except for the Dwarf variant which seems to be tissue culture specific, all the phenotypic variants observed in the somatic embryogenesis progenies were also observed among C. arabica seed-derived progeny. Indeed, phenotypic mutations are frequent in coffee nurseries, and Krug and Carvalho [36] previously characterized the numerous different morphologies in detail.
In several cases, AFLP markers have proved useful in the detection of genetic variation in tissue culture derived plants [20], [37]–[40]. In coffee, the genetic stability of coffee emblings has been poorly evaluated by molecular markers and limited to plants derived from experimental protocols. The present study revealed no or extremely limited mutations at the DNA level in large-scale somatic embryogenesis-propagated plants. From a total sample of 145 plants analyzed belonging to two hybrid varieties, only 1 out of 204 bands was polymorphic in only two SCE-produced plants. ESP-derived plants showed no AFLP polymorphism when compared with mother plants. Our results significantly differed from those previously reported in C. arabica. In a first approach on only 27 plants, Rani et al. [41] used different DNA markers (RAPD, random amplified polymorphic DNA and SSR, simple sequence repeat) to assess the genetic integrity of C. arabica emblings obtained from embryogenic calli, and they found a higher polymorphism level (4%) in the nuclear genome. By performing RAPD analyses on Norway spruce emblings, Heinze and Schmidt [42] concluded that gross somaclonal variation was absent in their plant regeneration system. In contrast, RAPD and SSR markers allowed the detection of high genetic variation in cotton emblings regenerated in the presence of 2,4-D [39]. AFLP analysis of 24 rye emblings led to the scoring of 887 AFLP markers, among which 8.8% identified the same polymorphism in plants obtained independently, revealing putative mutational hot spots [38].
DNA cytosine methylation plays an important role in plant regulation and development [43], [44]. Since the pioneer studies on maize emblings [12], substantial evidence has been obtained which indicates that demethylation can occur at a high frequency during somatic embryogenesis and can be an important cause of tissue culture induced variation [45]. DNA methylation has also been implicated in gene silencing and transposable element reactivation [24], [46]. To our knowledge, epigenetic deregulation during coffee micropropagation has not yet been studied. The very low total methylation polymorphism values obtained for both somatic embryogenesis processes and both hybrids (0.07–0.18% range) indicated that the tissue culture procedures employed in coffee weakly affected DNA methylation of the regenerated plants. This finding is in accordance with the 0.87% total polymorphism recently found in Freesia hybrid emblings by Gao et al. [40]. Moreover, the low number of methylation polymorphisms per embling (range 1–3) confirmed that neither SCE nor ESP induced additional stress at the methylation level during embryogenic material proliferation. In contrast, a significantly higher accumulation of methylation changes in some emblings has been regularly observed in other species [20], [21], [47], [48]. For example, in grapevine, most emblings showed between zero and three changes, similar to our findings, but a few accumulated up to 18 [21]. It has also been well demonstrated that auxin levels strongly alter DNA methylation of embryogenic cell cultures [49]. However, similar to our results, examples of stable MSAP patterns have already been reported using bamboo tissues at different developmental stages of somatic embryogenesis [50]. The timing of plant regeneration from proliferating callus cultures could be crucial for the appearance of variation. In callus-derived hop plants, an increase in the variation was detected by MSAP in prolonged callus cultures [20]. Our results demonstrated that very few changes are possible by limiting both the auxin level and culture duration.
MSAP markers have already been successfully used to demonstrate epigenetic instabilities (methylation alteration) induced by somatic embryogenesis in a great variety of plants, such as the ornamental flower Freesia, banana, barley, grapevine and maize [20], [21], [40], [51]–[53], also indicating that demethylation events were generally the most frequent. Although occurring at low frequency, our results also indicated demethylation events and mainly the loss of methylation in the internal cytosine of the 5'-CCGG-3′ sequence to produce a new HpaII band not detected in mother plants but present in the amplification pattern of the isoschizomer MspI. The detection of the same MSAP polymorphic fragments in independent plant samples from different hybrids and somatic embryogenesis processes suggests the existence of hotspots of DNA methylation changes in the genome. The existence of non-randomly behaving methylation polymorphic fragments in micropropagated plants has already been described using Met-AFLP [20], [48] and MSAP [21], [53]–[55].
Gross changes, such as variation in ploidy level, number of chromosomes and structural changes, are mitotic aberrations that represent major genomic alterations of in vitro plants often generated during in vitro proliferation and differentiation [12], [56]–[58]. Variations in chromosome number and structure have been described among emblings for several species [59]–[62]. We demonstrated that gross changes occur during coffee somatic embryogenesis and are related to SV, whereas genetic and epigenetic (methylation) alterations are very weak. Until now, by using flow cytometry analysis, normal ploidy levels were reported in coffee emblings [29], [63] but chromosome counting was not performed in these studies. The sensitivity of flow cytometry analysis was probably not sufficient to identify aneuploid plants. Nevertheless, the presence of mitotic aberrations, including double prophase, lagging chromosomes, aneuploids and polyploid cells, has previously been described in leaves [64], [65] and embryogenic calli [64] of C. arabica but not in the later steps of somatic embryogenesis, and without establishing any relation with SV. The presence of aneuploidy has also been well documented in embryogenic calli of Hordeum vulgare [66] and sweet orange [60].
The mechanisms underlying SV remain largely theoretical and unclear [16]. Thus it is often difficult to correlate a well-described genetic or epigenetic mechanism to a variant phenotype. For example, although DNA methylation has often been suggested as a possible cause of SV, a number of studies have reported high levels of methylation variation with no effect on the plant phenotype [20], [21], [52]. Another example is given by oil palm emblings, approximately 5% of which exhibit the ‘mantled’ phenotype affecting the formation of floral organs in both male and female flowers. Although a decrease in DNA methylation was observed, it was not possible to determine the nature of the epigenetic deregulation [67]. In the present study, it was possible to reveal a large proportion of aneuploid karyotypes in different variant phenotypes, hence showing that chromosomal rearrangements could be directly involved in the occurrence of phenotypic variation. The addition or subtraction of a single chromosome has a greater impact on phenotype than whole genome duplication, i.e. polyploidy [68]. It was clearly demonstrated in Arabidopsis thaliana that certain phenotypic traits are strongly associated with the dosage of specific chromosomes and that chromosomal effects can be additive [69]. Similarly, in maize seedlings, trisomic plants showed characteristic features such as reduced stature, tassel morphology changes and the presence of knots on the leaves, suggesting a phenotypic effect caused by the altered copy of specific chromosome related genes [70]. A similar mechanism could explain most of the variant phenotypes observed in C. arabica. The observation of a variant phenotype in plants with the expected chromosome number could be explained by the coexistence of monosomic and trisomic chromosomes in the same genome or by other chromosomal-like structural changes associated with undetected deletions, duplications, inversions or translocations of specific chromosomal segments [10], [71]. The karyotype analyses performed in the present study were limited to chromosome counting and did not enable observation of such chromosomal alterations.
Conclusions
This report shows that somatic embryogenesis is reliable for true-to-type and large-scale propagation of elite varieties in the C. arabica species. Both ESP and SCE ensured high proliferation rates along with very low SV rates, as observed through massive phenotypic observations in a commercial nursery and field plots. Molecular analysis (AFLP and MSAP) performed on 145 emblings derived from two proliferation processes and two different F1 hybrids showed that polymorphism between mother plants and emblings was extremely low. Consequently, it can be concluded that genetic and epigenetic alterations were also particularly limited during somatic embryogenesis in our controlled culture conditions. C. arabica is a young allopolyploid still having the most of its genes in duplicated copies [72]. It could be hypothesized that the impact of genetic or epigenetic variations on phenotype was restricted because of the buffer effect due to polyploidy. The main change in most of the rare phenotypic variants was aneuploidy. Although further studies are necessary for an accurate understanding of the chromosome anomalies involved in the acquisition of a particular phenotype, it is now obvious that mitotic aberrations play a major role in SV in coffee. The identification and use of molecular markers at the heterozygous state in mother plants (i.e. polymorphic between the two parental lines) would be required to further investigate this type of chromosome abnormality. Current studies based on the use of long-term embryogenic cultures [73] are aimed at establishing the full range of cytological, genetic and epigenetic (with a special focus on transposable elements) mechanisms underlying SV.
Materials and Methods
Plant material and somatic embryogenesis
Selected F1 hybrids of C. arabica [26] obtained by crossing traditional dwarf American varieties (Caturra, Sarchimor T5296) and wild accessions originating from Ethiopia and Sudan are disseminated in Central America through somatic embryogenesis. In the present study, emblings derived from the two hybrids Sarchimor T5296 x Rume Sudan and Caturra x ET531, named respectively H1 and H3, were analyzed to assess the SV level. Large-scale phenotypic observations were performed in Nicaragua both at the nursery (more than 600 000 young emblings) and field level (more than 200 000 three year-old emblings) on 11 coffee plots belonging to the ‘La Cumplida’ coffee research experimental sites in the Matagalpa region (Nicaragua). Nursery and field phenotypic observations were done on balanced amounts of plants from hybrids H1 and H3 (Table 1). Field observations were performed for all trees by visual evaluation of growth and morphology, flowering and fruit yield during the first and second production years. No specific permits were required for the described field studies that were performed with the authorization of the Coffee Research Department of ‘La Cumplida’, owner of the experimental sites. These sites are not protected and the studies did not involve endangered or protected species. Molecular marker analysis was applied on F1 hybrid mother plants propagated by rooted horticultural cuttings (four for each hybrid) and used as source material for in vitro propagation, as well as on adult emblings (3 years after planting) for which plants exhibiting an abnormal phenotype (phenotypic variant) were distinguished from plants with a normal phenotype and productivity (Table 8). Molecular marker patterns obtained for emblings were systematically compared with those obtained with mother plants (four plants per hybrid propagated by horticultural cutting) used as reference.
Table 8. Plant material used in molecular marker analyses.
| Somatic embryogenesis proliferation step | Type of plant material | No. of plants per C. arabica hybrid | Total no. of plants | |
| H1 | H3 | |||
| Secondary embryogenesis | Emblings normal phenotype | 28 | 31 | 59 |
| Emblings variant phenotype | 5 | 14 | 19 | |
| Angustifolia (A) | 0 | 4 | ||
| Bullata (B) | 2 | 4 | ||
| Dwarf (D) | 0 | 2 | ||
| Variegata (V) | 3 | 4 | ||
| Total | 33 | 45 | 78 | |
| Embryogenic suspension | Emblings normal phenotype | 25 | 34 | 59 |
| Emblings variant phenotype | 1 | 7 | 8 | |
| Angustifolia (A) | 1 | 2 | ||
| Dwarf (D) | 0 | 1 | ||
| Variegata (V) | 0 | 4 | ||
| Total | 26 | 41 | 67 | |
Total number of plants analyzed from two F1 Coffea arabica hybrid lines (H1 and H3) corresponding to somatic-embryo derived 3 year-old plants (emblings) with normal or variant phenotypes along with their respective mother plants as reference. The numbers of emblings for each variant phenotype are also given.
The somatic embryogenesis process (Figure 1) involved the following stages:
1) Production of embryogenic callus: pieces of young leaves were surface-sterilized and used as explants. The explants were cultured for 1 month on a 1/2 strength MS [74] “C” callogenesis medium [75] containing 2.3 µM 2,4-D, 4.9 µM indole-3-butyric acid (IBA) and 9.8 μM iso-pentenyladenine (iP), and then transferred for 6 months to MS/2 “ECP” embryogenic callus production medium [75] containing 4.5 µM 2,4-D and 17.7 µM benzylaminopurine (6-BA). All media were solidified using 2.4 g/l Phytagel (Sigma, Steinheim, Germany). These steps were carried out at 26–27°C in the dark.
2) 6-month multiplication step and embryo regeneration. Fully developed somatic embryos were mass regenerated via two distinct multiplication processes, i.e. either secondary (or repetitive) embryogenesis or embryogenic suspensions (Figure 1).
Secondary embryogenesis (SCE). Two hundred mg of embryogenic aggregates were placed in a 1 liter-RITA® temporary immersion bioreactor (CIRAD, Montpellier, France; [76]) along with 200 ml of “R” MS/2 regeneration medium [75] containing 17.76 µM 6-BA, in darkness for 6 weeks. An immersion frequency of 1 min every 12 h was applied. Proliferating embryo masses were then placed in 1/2 strength MS [74] regeneration medium containing 5.6 µM 6-BA and subcultured once every 6 weeks for three proliferation cycles. Secondary embryos were produced with an immersion frequency of 1 min every 12 h and a high culture density (approx. 10 000 embryos). The cultures were kept at 27°C, with a 12 h/12 h photoperiod and 50 µmol m−2 s−1 photosynthetic photon flux density.
Embryogenic cell suspensions (ESP). Embryogenic calli were transferred to 100 ml Erlenmeyer flasks at a density of 1 g/L in 1/4 MS strength Yasuda liquid proliferation medium [77] with 1.36 µM 2,4-D and 4.4 µM 6-BA. Suspension cultures were maintained by the monthly transfer of 1 g/L of embryogenic aggregates into fresh medium. Six month-old suspensions were used for somatic embryo regeneration. Embryo differentiation was initiated by transferring embryogenic aggregates at a density of 1 g/L in 250 ml Erlenmeyer flasks in a full strength MS medium containing 1.35 µM 6-BA. Fully developed torpedo-shaped embryos were obtained after two 4 week subcultures in such conditions. All suspension cultures were shaken at 110 rpm at 27 °C under 50 µmol m−2 s−1 photosynthetic photon flux density.
3) Pre-germination in a bioreactor. Germination was triggered by applying a low culture density of around 800–900 embryos per 1 l-RITA® bioreactor. An "EG" embryo germination medium [75] containing 1.33 μM BA was used for 2 months and finally, for 2 weeks, the “EG” culture medium was supplemented with 234 mM sucrose. By the end of the in vitro culture stage, each bioreactor contained around 700 pre-germinated cotyledonary somatic embryos with an elongated embryonic axis and a pair of open, chlorophyllous cotyledons. Pre-germination was conducted in the light (12/12 h, 50 µmol m−2 s−1).
4) Plantlet conversion was obtained after direct sowing of mature somatic embryos in the nursery. Mature embryos were sown vertically on top of the substrate (two parts soil, one part sand, one part coffee pulp) sterilized by chemical treatment (Dazomet (DMTT), Union Carbide). The somatic embryo culture density in the plastic boxes (l.w.h = 30/21/10 cm) was approximately 3600 m−2. The cultures were placed under a transparent roof that provided 50% shade, and were watered for 2 min twice daily. Conversion of somatic embryos into plants was generally observed 12 weeks after sowing, and characterized by the emergence of a stem bearing at least two pairs of true leaves.
5) Growth and hardening in the nursery (21 weeks). Plantlets grown from somatic embryos were transferred to 0.3 L plugs on a substrate comprising peat-based growing medium (Pro-mix, Premier Tech Ltd, Canada) and coffee pulp (3/1, v/v) under conventional nursery conditions until they reached the required size for planting in the field (approx. 30 cm). During this stage, the shade (50% light interception) and relative humidity (80%) were gradually reduced over 4 weeks to 0% light interception, with natural RH ranging from 65 to 90%.
Molecular analysis
DNA extraction
Young fully expanded leaves were selected on three year-old plants for molecular analysis. Genomic DNA was isolated from 100 mg of lyophilized leaves using Dellaporta buffer [78] containing sodium dodecyl sulfate (SDS) detergent and sodium bisulfite 1% w/v to avoid leaf oxidation. DNA was purified in spin-column plates as described in the DNeasy plant kit protocol from QIAGEN.
AFLP markers
AFLP analysis was carried out as described by Vos et al. [79], with a total of four primer combinations (Table 2), using 5-FAM or 5-HEX fluorescently labeled EcoR1 (+3) and unlabeled MseI (+3) primers. A touchdown PCR program for selective amplification was performed in an Eppendorf thermocycler under the following conditions: 3 min at 94°C, 12 cycles of 45 s at 94°C, 12 cycles of 45 s at 65°C and 1 min at 72°C; the annealing temperature was decreased by 0.7°C per cycle from a starting point of 65°C during this stage, with a final round of 25 cycles of 94°C for 45 s, 56°C for 45 s, 72°C for 1 min and a final elongation step of 72°C for 1 min. The same PCR conditions were found to be appropriate for MSAP in preliminary tests.
MSAP markers
MSAP analysis was carried out as described by Reyna-López et al. [19] with minor adaptations for capillary electrophoresis. The MSAP protocol is an adaptation of the AFLP method for the evaluation of different states of methylation in the symmetric sequence CCGG. In the MSAP protocol, the frequent cutting endonuclease (MseI) was replaced by the two isoschizomeric restriction enzymes HpaII and MspI with different sensitivity to the methylation state of the symmetric sequence CCGG (Table 4). Specifically, HpaII is able to recognize and cut only when the CCGG sequence is unmethylated or hemi-methylated on the external cytosine. MspI is able to cut when CCGG sequence is unmethylated or if the internal cytosine is fully or hemi-methylated. Both HpaII and MspI are unable to cut if the external cytosine presents full methylation. DNA methylation in plants commonly occurs at cytosine bases in all sequence contexts: the symmetric CG and CHG, in which (H = A, T or C) and the asymmetric CHH contexts [17]. Selective amplification included a total of eight primer combinations per isoschizomer (Table 2). HpaII (+2) primers were fluorescently labeled with 5-FAM or 5-HEX while EcoR1 (+3) remained unlabeled. In order to reduce the possibility of technical artifacts, two repetitions using different DNA extractions were performed for each primer combination.
Capillary electrophoresis and data analysis
PCR products were separated by capillary electrophoresis with Pop 7™ polymer in a 16 capillary 3130 XL Genetic Analyzer from Applied Biosystems using an internally manufactured 524 ROX fluorophore as sizing standard. The fragments used for fingerprinting were visualized as electropherograms in applied Biosystems software GeneMapper® version 3.7. Informative fragments were mostly found in the 100–450 bp range. All amplified fragments were classified based on the primer combination used and their size. The sample fingerprint data was converted to binary code, with “1” denoting the presence of the fragment and “0” the absence. Different binary matrices were constructed for comparative analysis depending on the kind of molecular marker.
As shown in Table 4, the MSAP patterns were classified as follows: Pattern 1 when a comigrating amplified fragment was obtained from the DNA template digested by both restriction enzymes HpaII and MspI; Pattern 2 and 3 when an amplification fragment was obtained only from the DNA template digested by HpaII or MspI, respectively.
Before analysis of the embling versus mother plant population, we successfully verified, on a set of plants from the Caturra variety, that the same MSAP patterns were systematically generated whatever the plant age and the leaf developmental stage (data not shown). Hence, a possible developmental variability in the studied plant material does not seem to introduce any additional source of variation in the methylation state. Nevertheless, in all experiments, only leaves from the same developmental stage were chosen.
Slide preparation and karyotyping
Root tips were harvested from individual adult emblings and placed in an aqueous solution of 8-hydroxyquinoline (2.5 mM) used as pre-treatment, for 4 h in darkness (2 h at 4°C plus 2 h at room temperature). A solution of Carnoy (absolute ethanol and glacial acetic acid, 3∶1 v/v) was used to fix the tissues for at least 24 h at −20°C. Fixed material was then stored in 70% ethanol at 4°C until use for slide preparation. The stored root tips were used for slide preparations by employing the technique for cell dissociation of enzymatically macerated roots, as described previously by Herrera et al. [80]. Preparations were frozen in liquid nitrogen in order to remove the coverslips, stained with 4',6-diamidino-2-phenylindole, DAPI (1µg/mL), and mounted in Vectashield (Vector Laboratories, Peterborough, UK).
In order to determine the occurrence of chromosome modifications, individual plants of C. arabica regenerated by somatic embryogenesis showing a normal (2 plants) or variant (11 plants) phenotype were submitted to karyotype analysis. The Angustifolia, Bullata, Dwarf, Giant and Variegata phenotypic variants were analyzed. During slide examination, mitotic cells at metaphase or prometaphase stages were used for chromosome counting. Between 4 and 8 mitotic cells from each individual were analyzed to determine the chromosome number. The best examples were photomicrographed at metaphase to document the chromosome number and morphology. A Nikon Eclipse 90i epi-fluorescence microscope equipped with a digital, cooled B/W CCD camera (VDS 1300B Vosskühler ®) was used with the appropriate filter (UV-2E/C excitation wavelength 340–380).
Acknowledgments
The authors thank Clément Poncon, owner of the Coffee Research Center ‘La Cumplida’, and José Martin Hidalgo for their interest and invaluable assistance during this work.
Funding Statement
Financial support for this study was provided by the Mexican Government through a grant to Roberto Bobadilla Landey by the Consejo Nacional de Ciencia y Tecnología (CONACyT) (CVU:1623391) program (http://www.conacyt.mx/), by another grant from the PCP France-Mexico (http://www.pcp-mexique.com/) and by the CIRAD funds for doctoral support (http://www.cirad.fr/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Etienne H, Dechamp E, Barry Etienne D, Bertrand B (2006) Bioreactors in coffee micropropagation (Review). Braz J Plant Physiol 18: 45–54. [Google Scholar]
- 2. Miguel C, Marum L (2011) An epigenetic view of plant cells cultured in vitro: somaclonal variation and beyond. J Exp Bot 62: 3713–3725. [DOI] [PubMed] [Google Scholar]
- 3. Jähne A, Lazzeri PA, Jäger Gussen M, Lörz H (1991) Plant regeneration from embryogenic cell suspensions derived from anther cultures of barley (Hordeum vulgare L.). Theor Appl Genet 82: 74–80. [DOI] [PubMed] [Google Scholar]
- 4. Rival A, Beulé T, Barre P, Hamon S, Duval Y, et al. (1997) Comparative flow cytometric estimation of nuclear DNA content in oil palm (Elaeis guineensis Jacq.) tissue cultures and seed-derived plants. Plant Cell Rep 16: 884–887. [DOI] [PubMed] [Google Scholar]
- 5. Lu S, Wang Z, Peng X, Guo Z, Zhang G, et al. (2006) An efficient callus suspension culture system for triploid bermudagrass (Cynodon transvaalensis x C. dactylon) and somaclonal variations. Plant Cell Tiss Org Cult 87: 77–84. [Google Scholar]
- 6. Lambé P, Mutambel HSN, Fouche JG, Deltour R, Foidart JM, et al. (1997) DNA methylation as a key process in regulation of organogenic totipotency and plant neoplastic progression. In Vitro Cell Dev Biol Plant 33: 155–162. [Google Scholar]
- 7. Von Aderkas P, Bonga J (2000) Influencing micropropagation and somatic embryogenesis in mature trees by manipulation of phase change, stress and culture environment. Tree Physiol 20: 921–928. [DOI] [PubMed] [Google Scholar]
- 8.Karp A (1994) Origins, causes and uses of variation in plant tissue cultures. In: Vasil IK, Thorpe TA, editors. Plant cell and tissue culture. Dordrecht: Kluwer Academic Publishers. pp. 139–152. [Google Scholar]
- 9. Bukowska B (2006) Toxicity of 2,4-Dichlorophenoxyacetic acid – Molecular mechanisms. Polish J of Environ Stud 15: 365–374. [Google Scholar]
- 10. Larkin PJ, Scowcroft WR (1981) Somaclonal variation: a novel source of variability from cell cultures for plant improvement. Theor Appl Genet 60: 197–214. [DOI] [PubMed] [Google Scholar]
- 11. Kaeppler SM, Kaeppler HF, Rhee Y (2000) Epigenetic aspects of somaclonal variation in plants. Plant Mol Biol 43: 179–188. [DOI] [PubMed] [Google Scholar]
- 12. Kaeppler SM, Phillips RL (1993) DNA methylation and tissue culture-induced DNA methylation variation in plants. In Vitro Cell Dev Bio Plant 29: 125–130. [Google Scholar]
- 13. Peschke VM, Phillips RL (1992) Genetic implications of somaclonal variation in plants. Adv Genet 30: 41–75. [Google Scholar]
- 14. Duncan RR (1997) Tissue culture-induced variation and crop improvement. Adv Agron 58: 201–240. [Google Scholar]
- 15. Sahijram L, Soneji J, Bollamma K (2003) Analyzing somaclonal variation in micropropagated bananas (Musa spp.). In Vitro Cell Dev Bio Plant 39: 551–556. [Google Scholar]
- 16. Bairu MW, Aremu OA, Van Staden J (2011) Somaclonal variation in plants: causes and detection methods. Plant Growth Regul 63: 147–173. [Google Scholar]
- 17. Law J, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saze H (2008) Epigenetic memory transmission through mitosis and meiosis in plants. Sem Cell Dev Biol 19: 527–536. [DOI] [PubMed] [Google Scholar]
- 19. Reyna Lopez GE, Simpson J, Ruiz Herrera J (1997) Differences in DNA methylation pattern are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Mol Gen Genet 253: 703–710. [DOI] [PubMed] [Google Scholar]
- 20. Bednarek PT, Orłowska R, Koebner RMD, Zimny J (2007) Quantification of tissue-culture induced variation in barley (Hordeum vulgare L.). BMC Plant Biol 7: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schellenbaum P, Mohler V, Wenzel G, Walter B (2008) Variation in DNA methylation patterns of grapevine somaclones (Vitis vinifera L.). BMC Plant Biol 8: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peschke VM, Phillips RL, Gegenbach BG (1987) Discovery of transposable element activity among progeny of tissue culture-derived maize plants. Science 238: 804–807. [DOI] [PubMed] [Google Scholar]
- 23. Mckenzie NL, Wen LY, Dale J (2002) Tissue-culture enhanced transposition of the maize transposable element Dissociation in Brassica oleracea var. ‘Italica’. Theor Appl Genet 105: 23–33. [DOI] [PubMed] [Google Scholar]
- 24. Slotkin RK, Martienssen R (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285. [DOI] [PubMed] [Google Scholar]
- 25. Anthony F, Bertrand B, Quiros O, Lashermes P, Berthaud J, et al. (2001) Genetic diversity of wild coffee (Coffea arabica L.) using molecular markers. Euphytica 118: 53–65. [Google Scholar]
- 26. Bertrand B, Alpizar E, Llara L, Santacreo R, Hidalgo M, et al. (2011) Performance of Arabica F1 hybrids in agroforestry and full-sun cropping systems in comparison with pure lines varieties. Euphytica 181: 147–158. [Google Scholar]
- 27. Etienne H, Bertrand B, Montagnon C, Bobadilla Landey R, Dechamp E, et al. (2012) Un exemple de transfert technologique réussi en micropropagation: la multiplication de Coffea arabica par embryogenèse somatique. Cah Agric 21: 115–24. [Google Scholar]
- 28.Söndahl MR, Lauritis JA (1992) Coffee. In: Hammerschlag FA, Litz RE, editors. Biotechnology of Perennial Fruit Crops. Wallingford: CAB International. pp. 401–420. [Google Scholar]
- 29. Etienne H, Bertrand B (2003) Somaclonal variation in Coffea arabica: effects of genotype and embryogenic cell suspension age on frequency and phenotype of variants. Tree Physiol 23: 419–426. [DOI] [PubMed] [Google Scholar]
- 30. Etienne H, Bertrand B (2001) The effect of the embryogenic cell suspension micropropagation technique on the trueness to type, field performance, bean biochemical content and cup quality of Coffea arabica trees. Tree Physiol 21: 1031–1038. [DOI] [PubMed] [Google Scholar]
- 31. Rani V, Raina S (2000) Genetic fidelity of organized meristem-derived micropropagated plants: a critical reappraisal. In Vitro Cell Dev Bio Plant 36: 319–330. [Google Scholar]
- 32. Hua HW, Huang TD, Huang HS (2010) Micropropagation of self-rooting juvenile clones by secondary somatic embryogenesis in Hevea brasiliensis . Plant Breeding 129: 202–207. [Google Scholar]
- 33. Mallón R, Covelo P, Vieitez AM (2012) Improving secondary embryogenesis in Quercus robur: Application of temporary immersion for mass propagation. Trees Structure and Function 26: 731–741. [Google Scholar]
- 34. Akula A, Becker D, Bateson M (2000) High-yielding repetitive somatic embryogenesis and plant recovery in a selected tea clone, ‘TRI-2025’, by temporary immersion. Plant Cell Rep 19: 1140–1145. [DOI] [PubMed] [Google Scholar]
- 35. Ducos JP, Alenton R, Reano JF, Kanchanomai C, Deshayes A, et al. (2003) Agronomic performance of Coffea canephora P. trees derived from large-scale somatic embryo production in liquid medium. Euphytica 131: 215–223. [Google Scholar]
- 36. Krug CA, Carvalho A (1951) The genetics of Coffea . Advances in Genetics 4: 127–158. [DOI] [PubMed] [Google Scholar]
- 37. Hornero J, Martínez I, Celestino C, Gallego FJ, Torres V, et al. (2001) Early checking of genetic stability of cork oak somatic embryos by AFLP analysis. Int J Plant Sci 162: 827–833. [Google Scholar]
- 38. De la Puente R, Gonzalez AI, Ruiz ML, Polanco C (2008) Somaclonal variation in rye (Secale cereale L.) analyzed using polymorphic and sequenced AFLP markers. In Vitro Cell Dev Biol Plant 44: 419–426. [Google Scholar]
- 39. Jin S, Mushke R, Zhu H, Tu L, Lin Z, et al. (2008) Detection of somaclonal variation of cotton (Gossypium hirsutum) using cytogenetics, flow cytometry and molecular markers. Plant Cell Rep 27: 1303–1316. [DOI] [PubMed] [Google Scholar]
- 40. Gao X, Yang D, Cao D, Ao M, Sui X, et al. (2010) In vitro micropropagation of Freesia hybrida and the assessment of genetic and epigenetic stability in regenerated plantlets. J Plant Growth Regul 29: 257–267. [Google Scholar]
- 41. Rani V, Singh KP, Shiran B, Nandy S, Goel S, et al. (2000) Evidence for new nuclear and mitochondrial genome organizations among high-frequency somatic embryogenesis-derived plants of allotetraploid Coffea arabica L. (Rubiaceae). Plant Cell Rep 19: 1013–1020. [DOI] [PubMed] [Google Scholar]
- 42. Heinze B, Schmidt J (1995) Monitoring genetic fidelity vs somaclonal variation in Norway spruce (Picea abies) somatic embryogenesis by RAPD analysis. Euphytica 85: 341–345. [Google Scholar]
- 43. Zhang M, Kimatu JN, Xu K, Liu B (2010) DNA cytosine methylation in plant development. Review. J Genet Genomics 37: 1–12. [DOI] [PubMed] [Google Scholar]
- 44. Vanyushin BF, Ashapkin VV (2011) DNA methylation in higher plants: Past, present and future. Biochim Biophys Acta 1809: 360–368. [DOI] [PubMed] [Google Scholar]
- 45. Smulders MJM, de Klerk GJ (2011) Epigenetics in plant tissue culture. Plant Growth Reg 63: 137–146. [Google Scholar]
- 46. Feschotte C, Jiang N, Wessler SR (2002) Plant transposable elements: when genetics meets genomics. Nat Rev Genet 3: 329–341. [DOI] [PubMed] [Google Scholar]
- 47. Li X, Yu X, Wang N, Feng Q, Dong Z, et al. (2007) Genetic and epigenetic instabilities induced by tissue culture in wild barley (Hordeum brevisubulatum (Trin.) Link). Plant Cell Tiss Org Cult 90: 153–168. [Google Scholar]
- 48. Fiuk A, Bednarek PT, Rybczyński JJ (2010) Flow cytometry, HPLC-RP and met AFLP analyses to assess genetic variability in somatic embryo-derived plantlets of Gentiana pannonica scop. Plant Mol Biol Rep 28: 413–420. [Google Scholar]
- 49. LoSchiavo F, Pitto L, Giuliano G, Torti G, Nuti Ronchi V, et al. (1989) DNA methylation of embryogenic carrot cell cultures and its variations as caused by mutation, differentiation, hormones, and hypomethylating drugs. Theor Appl Genet 77: 325–331. [DOI] [PubMed] [Google Scholar]
- 50. Gillis K, Gielis J, Peters H, Dhooghe E, Orpins J (2007) Somatic embryogenesis from mature Bambusa balcooa Roxburgh as basis for mass production of elite forestry bamboos. Plant Cell Tiss Org Cult 91: 115–123. [Google Scholar]
- 51. Peraza Echeverria S, Herrera Valencia VA, Kay AJ (2001) Detection of DNA methylation changes in micropropagated banana plants using methylation-sensitive amplification polymorphism (MSAP). Plant Sci 161: 359–367. [DOI] [PubMed] [Google Scholar]
- 52. Yu X, Li X, Zhao X, Jiang L, Miao G, et al. (2011) Tissue culture-induced genomic alteration in maize (Zea mays) inbred lines and F1 hybrids. Ann Appl Biol 158: 237–247. [Google Scholar]
- 53. Baránek M, Křižan B, Ondružíková E, Pidra M (2010) DNA-methylation changes in grapevine somaclones following in vitro culture and thermotherapy. Plant Cell Tiss Org Cult 101: 11–22. [Google Scholar]
- 54. Guo WL, Wu R, Zhang YF, Liu XM, Wang HY, et al. (2007) Tissue culture-induced locus-specific alteration in DNA methylation and its correlation with genetic variation in Codonopsis lanceolata Benth et Hook. Plant Cell Rep 26: 1297–1307. [DOI] [PubMed] [Google Scholar]
- 55. Díaz Martínez M, Nava Cedillo A, Guzmán López JA, Escobar Guzmán R, Simpson J (2012) Polymorphism and methylation patterns in Agave tequilana Weber var. ‘Azul’ plants propagated asexually by three different methods. Plant Sci 185–186: 321–330. [DOI] [PubMed] [Google Scholar]
- 56.Karp A (1991) On the current understanding of somaclonal variation. In: Miflin BJ, editor. Oxford surveys of plant molecular and cell biology. Vol 17 .Oxford : Oxford University Press. pp. 1–58. [Google Scholar]
- 57. Larkin PJ (1987) Somaclonal variation: history, method and meaning. Iowa State 1 Res 61: 393–43. [Google Scholar]
- 58. Neelakandan AK, Wang K (2012) Recent progress in the understanding of tissue culture-induced genome level changes in plants and potential applications. Plant Cell Rep 31: 597–620. [DOI] [PubMed] [Google Scholar]
- 59. Al Zahim MA, Ford Loyd BV, Newbury HJ (1999) Detection of somaclonal variation in garlic (Allium sativum L.) using RAPD and cytological analysis. Plant Cell Rep 18: 473–477. [Google Scholar]
- 60. Hao YI, Deng XX (2002) Occurrence of chromosomal variations and plant regeneration from long-term-cultured citrus callus. In Vitro Cell Dev Biol Plant 38: 472–476. [Google Scholar]
- 61. Mujib A, Banerjee S, Dev Ghosh P (2007) Callus induction, somatic embryogenesis and chromosomal instability in tissue culture-raised hippeastrum (Hippeastrum hybridum cv. United Nations). Propag Ornam Plants 7: 169–174. [Google Scholar]
- 62. Leal F, Loureiro J, Rodriguez E, Pais MS, Santos C, et al. (2006) Nuclear DNA content of Vitis vinifera cultivars and ploidy level analyses of somatic embryo-derived plants obtained from anther culture. Plant Cell Rep 25: 978–985. [DOI] [PubMed] [Google Scholar]
- 63. Sanchez Teyer LF, Quiroz Figueroa F, Loyola Vargas V, Infante D (2003) Culture-induced variation in plants of Coffea arabica cv. Caturra rojo, regenerated by direct and indirect somatic embryogenesis. Mol Biotechnol 23: 107–115. [DOI] [PubMed] [Google Scholar]
- 64. Ménendez Yuffá A, Da Silva R, Rios L, Xena de Enrech N (2000) Mitotic aberrations in coffee (Coffea arabica cv. 'Catimor') leaf explants and their derived embryogenic calli. Electron J Biotechnol 3: 0–5 ISSN 0717–3458 [Google Scholar]
- 65. Zoriniants SE, Nosov AV, Monforte Gonzalez M, Mendes Zeel M, Loyolas Vargas VM (2003) Variation of nuclear DNA content during somatic embryogenesis and plant regeneration of Coffea arabica L. using cytophotometry. Plant Sci 164: 141–146. [Google Scholar]
- 66. Gözükirmizi N, Ari S, Oraler G, Okatan Y, Ünsal N (1990) Callus induction, plant regeneration and chromosomal variations in barley. Acta Botanica Neerlandica 39: 379–387. [Google Scholar]
- 67. Jaligot E, Beulé T, Baurens FC, Billote N, Rival A (2004) Search for methylation-sensitive amplification polymorphism associated with the ‘mantled’ variant phenotype in oil palm (Elaeis guineensis Jacq.). Genome 47: 224–228. [DOI] [PubMed] [Google Scholar]
- 68. Birchler JA, Veitia RA (2007) The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19: 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Henry IM, Dilkes BP, Miller ES, Burkart Waco D, Comai L (2010) Phenotypic consequences of aneuploidy in Arabidopsis thaliana . Genetics 186: 1231–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Makarevitch I, Harris C (2010) Aneuploidy causes tissue specific qualitative changes in global gene expression patterns in maize. Plant Physiol 152: 927–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fukuoka H, Kawata M, Tkaiwa M (1994) Molecular changes of organelle DNA sequences in rice through dedifferentiation, long-term culture or the morphogenesis process. Plant Mol Biol 26: 899–907. [DOI] [PubMed] [Google Scholar]
- 72. Cenci A, Combes MC, Lashermes P (2012) Genome evolution in diploid and tetraploid Coffea species as revealed by comparative analysis of orthologous genome segments. Plant Mol Biol 78: 135–145. [DOI] [PubMed] [Google Scholar]
- 73. Ribas A, Dechamp E, Bertrand B, Champion A, Verdeil JL, et al. (2011) Agrobacterium tumefaciens-mediated genetic transformation of Coffea arabica (L.) is highly enhanced by using long-term maintained embryogenic callus. BMC Plant Biol 11: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Murashige T, Skoog FA (1962) Revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497. [Google Scholar]
- 75.Etienne H (2005) Protocol of somatic embryogenesis: Coffee (Coffea arabica L. and C. canephora P.). In: Jain SM, Gupta PK, editors. Protocols for somatic embryogenesis in woody plants. Series: Forestry Sciences. Vol 77 . The Netherlands: Springer edn. pp. 167–179. [Google Scholar]
- 76.Teisson C, Alvard D (1995) A new concept of plant in vitro cultivation liquid medium: temporary immersion. In: Terzi M et al.., editors. Current Issues in Plant Molecular and Cellular Biology. Dordrecht: Kluwer Academic Publishers. pp. 105–110. [Google Scholar]
- 77. Yasuda T, Fujii Y, Yamaguchi T (1985) Embryogenic callus induction from Coffea arabica leaf explants by benzyladenine. Plant Cell Physiol 26: 595–597. [Google Scholar]
- 78. Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA miniprepration: version II. Plant Mol Biol Rep 1: 19–21. [Google Scholar]
- 79. Vos P, Hogers R, Bleeker M, Rijans M, Van de Lee T, et al. (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Herrera JC, D'hont A, Lashermes P (2007) Use of fluorescence in situ hybridization as a tool for introgression analysis and chromosome identification in coffee (Coffea arabica L.). Genome 50: 619–626. [DOI] [PubMed] [Google Scholar]