Abstract
Friedreich ataxia (FRDA) is an autosomal recessive disorder characterized by neurodegeneration and cardiomyopathy. The presence of a GAA trinucleotide repeat expansion in the first intron of the FXN gene results in the inhibition of gene expression and an insufficiency of the mitochondrial protein frataxin. There is a correlation between expansion length, the amount of residual frataxin and the severity of disease. As the coding sequence is unaltered, pharmacological up-regulation of FXN expression may restore frataxin to therapeutic levels. To facilitate screening of compounds that modulate FXN expression in a physiologically relevant manner, we established a cellular genomic reporter assay consisting of a stable human cell line containing an FXN-EGFP fusion construct, in which the EGFP gene is fused in-frame with the entire normal human FXN gene present on a BAC clone. The cell line was used to establish a fluorometric cellular assay for use in high throughput screening (HTS) procedures. A small chemical library containing FDA-approved compounds and natural extracts was screened and analyzed. Compound hits identified by HTS were further evaluated by flow cytometry in the cellular genomic reporter assay. The effects on FXN mRNA and frataxin protein levels were measured in lymphoblast and fibroblast cell lines derived from individuals with FRDA and in a humanized GAA repeat expansion mouse model of FRDA. Compounds that were established to increase FXN gene expression and frataxin levels included several anti-cancer agents, the iron-chelator deferiprone and the phytoalexin resveratrol.
Introduction
Friedreich ataxia (FRDA) is an autosomal recessive disorder characterized by neurodegeneration and cardiomyopathy. It is the most common form of hereditary ataxia with an incidence of approximately 1 in 50,000 in Caucasian populations [1]. About 98% of individuals with FRDA are homozygous for an expansion of a GAA trinucleotide repeat sequence within the first intron of the FXN gene. The remaining individuals are compound heterozygotes for a GAA expansion and a point mutation, deletion and/or insertion. Pathogenic GAA expansion alleles are in the size range of 60 to more than 1300 repeats. The presence of a GAA repeat expansion results in the inhibition of FXN gene expression, reduced levels of full length FXN transcript and an insufficiency of the mitochondrial protein frataxin [2]–[5].
Frataxin deficiency results in mitochondrial dysfunction including the loss of iron-sulfur cluster (ISC)-containing enzymes, increased oxidative damage and mitochondrial iron accumulation [6]–[8]. Frataxin has been implicated to function as a multimeric iron storage protein that also possesses ferroxidase activity [9], as an iron chaperone which modulates mitochondrial aconitase activity [10]–[12], as a mediator of iron delivery to ferrochelatase [13] and in the early stages of ISC biogenesis by direct interaction with ISU-type proteins [14]–[17]. The main sites of pathology include the large sensory neurons of the dorsal root ganglia and the dentate nucleus of the cerebellum [18], [19].
The mechanism by which the GAA expansion results in reduced FXN gene expression is not clear. There is evidence that suggests that the GAA repeat expansion may form an unusual and stable triple helical non-B DNA structure or DNA/RNA hybrid that impedes transcription elongation [4], [5], [20]. It is now apparent that the GAA repeat expansion generates a heterochromatin-mediated gene silencing effect [21], [22]. Changes in both DNA methylation and histone modification have been described [22]–[28].
Overall, an inverse correlation has been found between the size of the smaller GAA repeat expansion and transcript levels, the amount of residual frataxin produced and the age of onset of disease symptoms [29]–[33]. Heterozygous carriers of a GAA repeat expansion produce about half the normal level of frataxin and are asymptomatic. As the GAA repeat expansion mutation does not alter the coding sequence of the gene, it is hypothesized that any increase in frataxin levels should prove beneficial, while a several-fold increase could be sufficient to halt disease progression.
We were the first group to propose the pharmacological up-regulation of FXN gene expression as a novel therapeutic approach for the treatment of FRDA [34]. Unlike pharmacological treatments focused on secondary disease effects by using antioxidant and iron chelation therapy [35]–[37] our approach is aimed at directly addressing the primary issue of frataxin deficiency.
We previously reported the identification of a 188 kb BAC clone (RP11-265B8) containing exons 1–5b of the human FXN locus and extensive flanking regions both upstream and downstream of the gene [34]. The clone contains a (GAA)6 sequence in the region that undergoes expansion within the first intron of the FXN gene. We demonstrated that the genomic insert is able to successfully complement the embryonic lethal phenotype of homozygous Fxn knockout mice, indicating that key regulatory elements required for normal expression of the FXN gene are present within this clone [38]. The BAC clone was used for the generation of an FXN-EGFP genomic reporter construct in which the EGFP gene was fused in-frame immediately following the final codon of exon 5a of the human FXN gene [34]. The FXN-EGFP genomic reporter preserves the normal location and spacing of many regulatory elements that may be positioned over large distances in the surrounding chromosomal region, and facilitates the recapitulation of normal gene expression patterns [39]. The construct was shown to drive the expression of EGFP from the regulatory elements of the FXN locus in cell lines and transgenic mice, with the frataxin-EGFP fusion protein targeted to the mitochondria [34], [40]. A stable BHK-21 cell line containing the genomic reporter fusion was established and used to demonstrate the induction of FXN gene expression by hemin and butyric acid [34].
In this study we report the development of a stable human cell line containing the FXN-EGFP genomic reporter fusion. The sensitive cellular assay for FXN gene expression was used to evaluate pharmacological agents for their ability to modulate FXN gene expression and adapted for the high throughput screening (HTS) of a small compound library. The evaluation of hit compounds in cells from individuals with FRDA and in a humanized GAA repeat expansion mouse model of FRDA identified a number of agents that were able to increase FXN gene expression and frataxin protein, including several anti-cancer compounds, the iron-chelator deferiprone and the phytoalexin resveratrol.
Materials and Methods
Ethics Statement
Lymphoblasts and fibroblasts from individuals with FRDA were obtained from Coriell Cell Repositories (Camden, New Jersey, USA) or collected following written informed consent with approval from the Royal Children’s Hospital Human Research Ethics Committee (project 22009). All procedures involving the treatment of mice were carried out in accordance with the UK Home Office ‘Animals (Scientific Procedures) Act 1986’ under project license PPL70/6716. All efforts were made to minimize suffering.
Tissue Culture and Transfection
HeLa and HeLa (FXN-EGFP) cell lines were maintained in adherent culture in DMEM supplemented with 10% fetal calf serum. Lymphoblasts were maintained in suspension culture in RPMI 1640 supplemented with 20% fetal calf serum. Fibroblasts were maintained in adherent culture in BME supplemented with 10% fetal calf serum. Media contained antibiotics (100 U/ml penicillin and 0.1 mg/ml streptomycin) for routine cell maintenance but were omitted during pharmacological compound assays. Cells were cultured at 37°C in 5% CO2 atmosphere.
For transfection studies, HeLa cells were plated in 6-well culture plates at a density of 2×105 cells per well and allowed to reach 50–80% confluency. Cells were transfected with 10 µg of purified BAC DNA (linearized by digestion with AscI and BsiWI) using LipofectAMINE (Life Technologies, Grand Island, NY, USA), at a 4∶1 lipid to DNA ratio, according to the manufacturer’s protocol. For the selection of stable clones, cells were transferred to 96-well flat-bottom plates at two days post-transfection and grown in medium containing 300 µg/ml G418 (Life Technologies). G418-resistant clones were isolated via limiting cell dilution at 100–500 cells per ml. Cells were examined for EGFP expression by fluorescence microscopy.
Fluorescence In Situ Hybridization (FISH) Analysis
Cell cultures were harvested after exposure to colcemid for 4 hours. Chromosome preparations were obtained using standard techniques. Cells were spread onto slides and subsequently denatured by immersion in 70% formamide in 2×SSC at 73°C for 3 minutes. The probe was prepared using purified DNA from RP11-265B8, which was labeled by nick translation with digoxigenin, according to the manufacturer’s instructions (Roche, Mannheim, Germany). The labeled DNA was ethanol precipitated together with human COT1 DNA and resuspended in 50% formamide, 10% dextran sulfate, and 2×SSC to a concentration of 40 ng/µl. The probe was denatured by heating at 75°C for 8 minutes, followed by preannealing at 37°C for 20 minutes. Hybridization was at 37°C for 16 hours followed by washing in 1×SSC at 70°C for 5 minutes. The probe was detected with mouse anti-digoxigenin antibody (Roche) followed by rhodamine conjugated anti-mouse antibody (Rockland, Gilbertsville, PA, USA). The slides were mounted in Vectashield (Vector Laboratories, Burlingame, CA, USA) containing DAPI counterstain.
Pharmacological Compound Analysis In Vitro
HeLa (FXN-EGFP) cells were plated at a density of 1×104 cells per ml, lymphoblasts were plated at a density of 1×105 cells per ml and fibroblasts were plated at a density of 5×104 cells per ml in appropriate media in 6-well culture plates. Cultures were incubated overnight prior to removal of growth medium and replacement with fresh medium containing various concentrations of test compounds ( Table 1 ). Cultures were incubated for a further 72 hrs. Adherent cells were collected by trypsinization. All cells were washed twice in phosphate-buffered saline (PBS). Assays were performed on triplicate cultures in at least three independent experiments.
Table 1. Properties of selected compounds.
| Compound | CAS number | Molecular formula | Molecular weight | Suppliera |
| δ-Aminolevulinic acid | 106-60-5 | C5H9NO3 | 131.13 | Sigma Aldrich |
| Ascorbic acid | 50-81-7 | C6H8O6 | 176.12 | Sigma Aldrich |
| Benzalkonium chloride | 8001-54-5 | C22H40ClN | 354.02 | Sigma Aldrich |
| Betamethasone | 378-44-9 | C22H29FO5 | 392.46 | Sigma Aldrich |
| Bupropion hydrochloride | 34841-39-9 | C13H18ClNO | 239.74 | Sigma Aldrich |
| Camptothecin | 7689-03-4 | C20H16N2O4 | 348.35 | Sigma Aldrich |
| Cefdinir | 91832-40-5 | C14H13N5O5S2 | 395.42 | Sigma Aldrich |
| Chlorpropham | 101-21-3 | C10H12ClNO2 | 213.67 | Sigma Aldrich |
| Chrysanthemic acid | 4638-92-0 | C10H16O2 | 168.23 | Sigma Aldrich |
| Cisplatin | 15663-27-1 | H6Cl2N2Pt | 301.1 | Mayne Pharma |
| Clozapine | 5786-21-0 | C18H19ClN4 | 326.82 | Sigma Aldrich |
| Convallatoxin | 508-75-8 | C29H42O10 | 550.65 | Sigma Aldrich |
| Deferiprone | 30652-11-0 | C7H9NO2 | 139.15 | Lipomed |
| Desferrioxamine | 70-51-9 | C25H48N6O8 | 560.68 | Novartis |
| Dimethyl sulfoxide | 67-68-5 | C2H6OS | 78.13 | Sigma Aldrich |
| Ethanol | 64-17-5 | C2H6O | 46.07 | Merck |
| Ferric ammonium sulfate | 10138-04-2 | FeNH4(SO4)2 | 266.07 | Sigma Aldrich |
| Ferrous ammonium sulfate | 10045-89-3 | (NH4)2Fe(SO4)2 | 284.05 | Sigma Aldrich |
| Hemin | 16009-13-5 | C34H32ClFeN4O4 | 651.94 | Sigma Aldrich |
| Molsidomine | 25717-80-0 | C9H14N4O4 | 242.24 | Sigma Aldrich |
| Nimustine | 42471-28-3 | C9H13ClN6O2 | 272.70 | Sigma Aldrich |
| Phenothrin | 26002-80-2 | C23H26O3 | 350.46 | Sigma Aldrich |
| Resveratrol | 501-36-0 | C14H12O3 | 228.24 | Sigma Aldrich |
| Rifampin | 13292-46-1 | C43H58N4O12 | 822.96 | Sigma Aldrich |
| Sulfameter | 651-06-9 | C11H12N4O3S | 280.30 | Sigma Aldrich |
Lipomed (Arlesheim, Switzerland), Mayne Pharma (Indianapolis, IN, USA), Merck (Darmstadt, Germany), Novartis (East Hanover, NJ, USA), Sigma Aldrich (St. Louis, MO, USA).
Flow Cytometry
The proportion of EGFP-positive HeLa (FXN-EGFP) cells and the relative levels of EGFP expression were measured by flow cytometry. Cells were resuspended in PBS with 2% fetal calf serum. Flow cytometry was performed using a LSR II flow cytometer (Becton-Dickinson, Franklin Lakes, NJ, USA) and analyzed using the FACSDiva Software Package.
Compound Library
The library screened was the Spectrum Collection (MicroSource Discovery Inc., Gaylordsville, CT, USA). The library contains 2,000 biologically active and structurally diverse compounds that are primarily Food and Drug Administration (FDA)-approved compounds, bioactive compounds or natural products. The compounds are supplied as 10 mM solutions in dimethyl sulfoxide (DMSO). More details of the library can be found at http://www.msdiscovery.com/spectrum.html.
High Throughput Screening Procedures
High throughput screening procedures were conducted at the Walter and Eliza Hall Institute High-Throughput Chemical Screening Facility (Bundoora, Victoria, Australia).
On Day 1 HeLa (FXN-EGFP) cells were seeded into 96-well optical-grade NUNC Cytowell Special Optics microtiter plates (Thermo Fisher Scientific, Rochester, NY, USA) at a density of 104 cells per well in a total volume of 200 µl of culture media (DMEM supplemented with 10% fetal calf serum) using a Multidrop 384 high-speed reagent dispenser (Thermo Electron Corporation, Waltham, MA, USA). Cells were allowed to adhere overnight at 37°C in 5% CO2 atmosphere.
On Day 2 each test compound was dispensed robotically from the library into individual wells at a volume of 0.2 µl to achieve a final concentration of 10 µM using a Zymark Mini Staccato system (SOTAX Corporation, Westborough, MA, USA). Cultures were incubated for a further 72 hours. Control rows and columns were set up containing: media only, cells without any test compounds, and cells exposed to the positive control drug cisplatin (3.3 µM) which is known to increase FXN gene expression. In addition, parental HeLa cells not containing the FXN-EGFP genomic reporter were set up as a measure of background cell fluorescence.
On Day 5 the cells were washed twice with 200 µl of PBS via a MiniTrak IX automated liquid handling system (PerkinElmer, Waltham, MA, USA) and overlaid with 200 µl of PBS prior to the fluorometric measurement of EGFP levels in an Envision 2100 Multilabel plate reader (PerkinElmer) configured for EGFP bottom-plate measurements at multiple locations per well.
Cell Viability Assay
To correct for variations in cell number from well to well due to possible toxicity of different test compounds a measurement of cell viability was performed. This was done following measurement of EGFP levels using the CellTiter-Blue cell viability assay (Promega, Alexandria, NSW, Australia). Culture medium containing the CellTiter-Blue reagent (resazurin) was added to each well and plates incubated at 37°C for 90 minutes. The signal produced by conversion of resazurin to resorufin is directly proportional to viable cell number and was detected fluorometrically in an Envision 2100 plate reader set up for top-plate measurements. A standard curve of cell number versus viability was established for each run.
Post High Throughput Screening Data Analysis
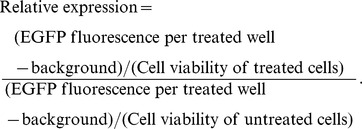
All fluorescence readings were exported into a set of Microsoft Excel spread sheets programmed to perform the necessary data analysis. The viability readings were converted to cell numbers for each well using the standard curve described above. The average background fluorescence of parental HeLa cells was subtracted from the EGFP readings of HeLa (FXN-EGFP) cell exposed to the chemical library. A correction factor for the number of viable cells was applied to the EGFP levels in each corresponding well. This was compared to the average value obtained from cells not exposed to any test compound to determine the level of change in FXN gene expression (represented as fold change in expression). This is represented as:
 |
Pharmacological Compound Analysis In Vivo
The YG8R FRDA mice [41] were housed in conventional open cages with Litaspen Premium 8/20 bedding, paper wool nesting and standard fun tunnel environmental enrichment, with 13 hours light, 11 hours dark, 20–23°C and 45–60% humidity. The mice were given a diet of SDS RM3 Expanded food pellets and standard drinking water. YG8R mice at an age of 9–12 months were given subcutaneous injections for three consecutive days of 25–300 mg/kg resveratrol in a vehicle solution of 6.25% DMSO, 20% glycerol, 20% PEG400, 20% propylene glycol, 5 mM sodium acetate (pH 5.2). Each dose of resveratrol or vehicle was evaluated in four mice. After either four hours or 24 hours mice were culled and tissues were collected and snap frozen in liquid nitrogen.
Quantitative Real-time Reverse Transcription PCR
Total cellular mRNA was isolated from cultured cell lines using the Dynabeads mRNA DIRECT Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Reverse transcription was performed using the SuperScript II First Strand Synthesis System (Invitrogen) with oligo (dT)12–15 primers. Real-time PCR was performed on an ABI7300 Sequence Detection System (Applied Biosystems, Carlsbad, CA, USA) using TaqMan Gene Expression Assays (Applied Biosystems) with primers and probes specific for the human FXN gene (Hs00175940_m1) and human hypoxanthine phosphoribosyltransferase 1 (HPRT1) (Hs99999909_m1) as the reference gene. FXN gene expression was determined using the comparative Ct method (ΔΔCt) relative to the endogenous control HPRT1. Assays were performed in triplicate in at least three independent experiments.
Total RNA was isolated from snap frozen YG8R mouse brain tissues by homogenization with Trizol (Invitrogen) and cDNA was prepared by using AMV reverse transcriptase (Invitrogen) with oligo-dT primers. Levels of human transgenic FXN expression were assessed by quantitative RT-PCR using an ABI7900 sequencer and SYBR Green (Applied Biosystems) with the primers FRT-I (5′-TTGAAGACCTTGCAGACAAG-3′) and RRT-II (5′-AGCCAGATTTGCTTGTTTGG-3′). Mouse Gapdh RT-PCR primers used for normalization were GapdhmF (5′-ACCCAGAAGACTGTGGATGG-3′) and GapdhmR (5′-GGATGCAGGGATGATGTTCT-3′).
Frataxin Protein Measurements
The levels of frataxin protein were measured by lateral flow immunoassay with the Frataxin Protein Quantity Dipstick Assay Kit (MitoSciences, Eugene, Oregon, USA) according to the manufacturer’s instructions [42]. Signal intensity was measured with a Hamamatsu ICA-1000 Immunochromatographic Reader (MitoSciences).
Statistical Analyses
For the evaluation of pharmacological compounds in cell lines, the experimental data is reported as the mean ± the standard error of the mean (SEM) of triplicate assays in at least three independent experiments. Comparisons were made between groups of equal size by Student’s t-test for paired data. For the evaluation of pharmacological compounds in mice, reactions were carried out in triplicate for each biological sample (n = 4) and data from the vehicle and drug-treated groups were compared using the Student’s t-test. Data were considered significantly different at p<0.05.
Results
Generation and Characterization of HeLa (FXN-EGFP) Stable Cell Lines
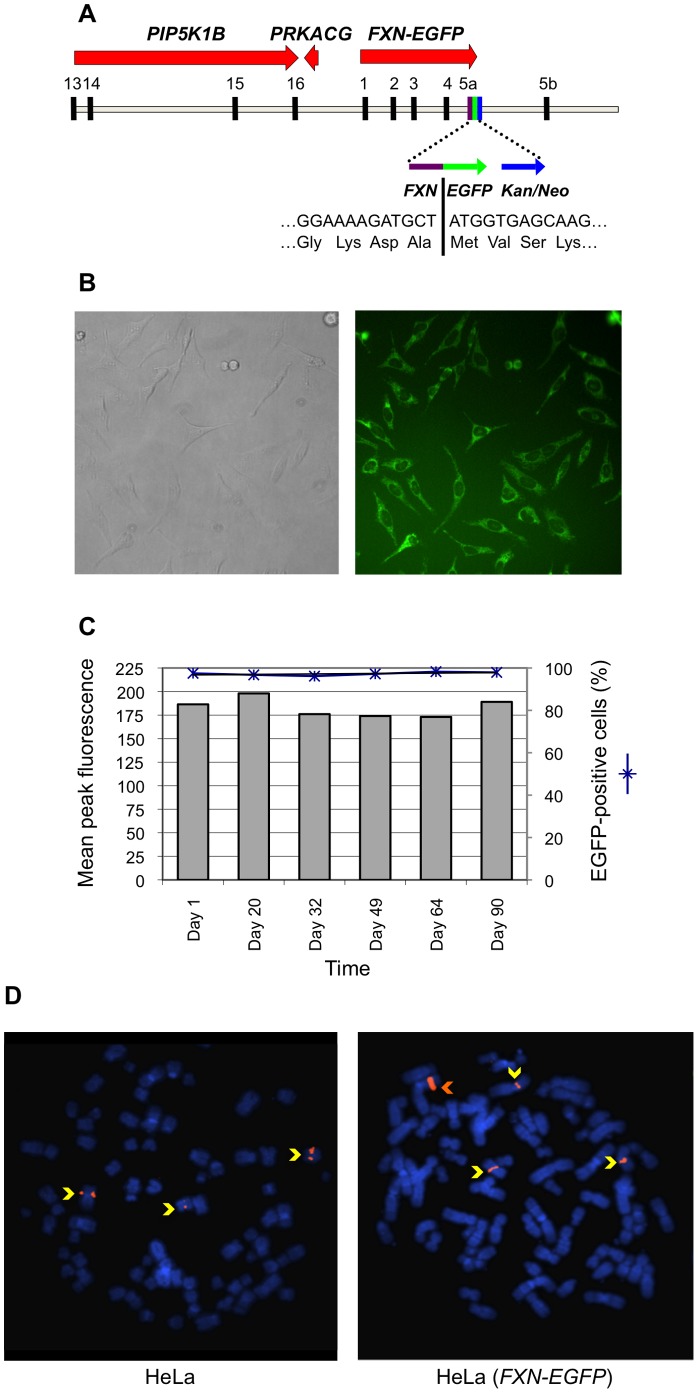
We previously generated an FXN-EGFP genomic reporter fusion by using homologous recombination [43] to insert an EGFP-Kan/Neo cassette in-frame immediately following the final codon of exon 5a of the normal human FXN gene present on the 188 kb BAC clone RP11-265B8 [34]. The modified genomic insert from the BAC clone ( Figure 1A ) was isolated from most of the vector sequence by digestion at unique sites with AscI and BsiWI and transfected into HeLa cells. A number of stable cell lines were produced and selected for the Kan/Neo resistance determinant using G418, followed by limiting dilution to establish single cell clones ( Figure 1B ). Examination of a number of independent stable clones by flow cytometry indicated that the proportion of EGFP-positive cells in each clone was greater than 96%. These clones exhibited a tight fluorescence peak, indicating high homogeneity of the cell population, and varying values of median peak fluorescence (data not shown). The proportion of EGFP-positive cells and the relative levels of EGFP expression were found to be stable following growth in continuous culture for over 90 days (25 passages) ( Figure 1C ). Fluorescent in situ hybridization (FISH) using the entire RP11-265B8 sequence as a probe was performed on metaphase spreads of a selected HeLa (FXN-EGFP) stable cell line. HeLa cells have altered ploidy and three hybridization signals were observed corresponding to the endogenous FXN gene. The stable clone displayed an additional and brighter hybridization signal establishing the presence of a single integration site containing multiple copies of the FXN-EGFP transgene ( Figure 1D ).
Figure 1. Characterization of HeLa (FXN-EGFP) stable cell lines.
(A) Diagrammatic representation of the BAC genomic DNA fragment containing the FXN-EGFP genomic reporter construct. The sequence includes exons 13–16 of the PIP5K1B gene and the PRKACG gene upstream of the FXN locus, and about 23 kb of additional sequence downstream of exon 5b. The exon 5a–EGFP–Kan/Neo region is shown in greater detail. (B) Microscopic imaging. Transmitted light (left) and fluorescence (right) images of a HeLa (FXN-EGFP) stable cell line. EGFP expression produced by the FXN-EGFP genomic reporter is evident in all cells. (C) Flow cytometric analysis. The levels of EGFP expression (left Y-axis) and the proportion of EGFP-positive cells (right Y-axis) were stable following growth in continuous culture. (D) Determination of transgenic fragment integration site by FISH. Rhodamine-labeled RP11-265B8 was hybridized onto metaphase chromosomes (DAPI stained) of HeLa (left) and HeLa (FXN-EGFP) (right) cells. Three hybridization signals (yellow arrows) corresponded to the endogenous FXN gene. The presence of one additional brighter signal (orange arrow) establishes the presence of a single integration site containing multiple copies of the FXN-EGFP transgene.
Primary Compound Screening Procedures
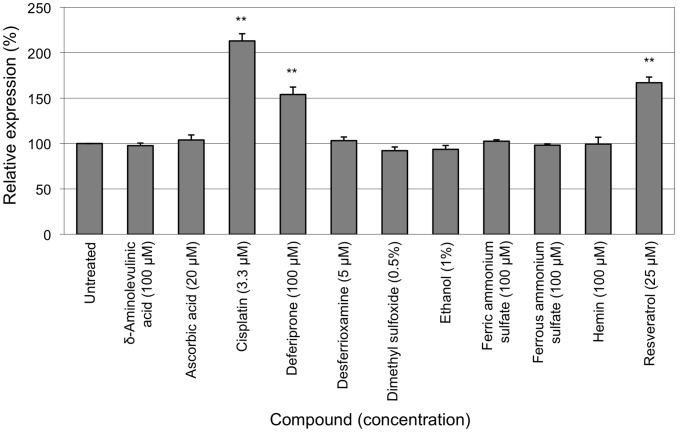
A small-scale non-automated primary screen was conducted in which selected compounds were evaluated over a 10,000-fold concentration range on HeLa (FXN-EGFP) cells and EGFP fluorescence was measured by flow cytometry. Compounds were selected based on the known pathophysiology of FRDA including those involved in iron metabolism and antioxidants. Compounds were dissolved in water, ethanol or dimethyl sulfoxide (DMSO), as appropriate. None of the solvents had a significant effect on cell viability or the expression of the FXN-EGFP reporter at the concentrations used in the cellular assay ( Figure 2 ). The anti-cancer compound cisplatin (3.3 µM) elicited a 2.2-fold increase in FXN-EGFP expression. The iron-chelator deferiprone (100 µM) was found to increase expression by 1.6-fold and the phytoalexin resveratrol (25 µM) increased expression by 1.7-fold. The combination of any two of these three compounds did not result in an additive effect on FXN-EGFP expression (data not shown). The other compounds tested had no notable effect on the level of FXN-EGFP expression.
Figure 2. Primary compound screen.
The HeLa (FXN-EGFP) stable cell line was exposed to various concentrations of selected test compounds for 72 hours. The levels of EGFP expression were measured by flow cytometry. For each compound the lowest concentration that produced the greatest change in FXN-EGFP expression is shown. Assays were performed in triplicate on at least three independent occasions. Error bars represent standard error of the mean. **p<0.01 in comparison to the untreated control.
High Throughput Screening Procedures
The HeLa (FXN-EGFP) cellular genomic reporter assay was adapted for use in high throughput screening procedures. Cells were cultured in 96-well optical-quality microtiter plates and measurement of EGFP levels was via a fluorometer. To correct for well to well variations in cell number a measurement of cell viability was incorporated. HTS procedures were undertaken at the Walter and Eliza Hall Institute High-Throughput Chemical Screening Facility. The Spectrum Collection was screened which is a library containing 2,000 biologically active and structurally diverse compounds that are primarily FDA-approved compounds and some natural products. Each test compound was evaluated at a final concentration of 10 µM. Fluorometric measurement of EGFP fluorescence was performed using a bottom-plate reader followed by cell viability assays. A standard curve of cell number versus viability was also established for each run. The average background fluorescence of parental HeLa cells was subtracted from the EGFP readings of HeLa (FXN-EGFP) cells exposed to the chemical library. A correction factor for the number of viable cells was applied to the EGFP levels in each corresponding well. This was compared to the average value obtained from cells not exposed to any test compound to determine the level of change in FXN gene expression.
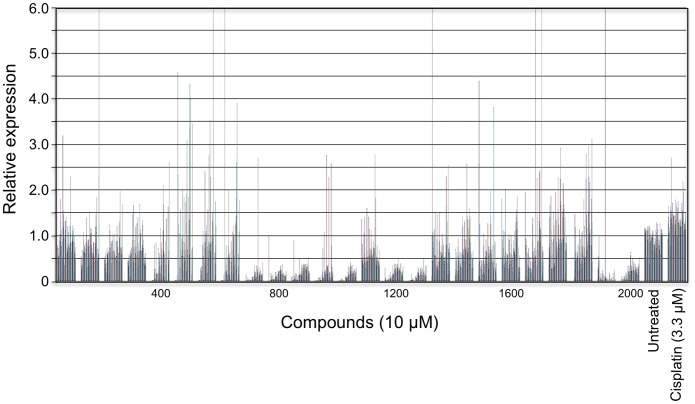
Following the complete screening of the 2,000 compound library, 18 hit compounds were identified that elicited a greater than three-fold increase in FXN-EGFP expression at a concentration of 10 µM in the HeLa (FXN-EGFP) genomic reporter assay ( Figure 3 ). A total of 74 compounds were identified that elicited a greater than 1.5-fold increase in EGFP expression (30 compounds increased expression by 1.5–2-fold; 15 by 2–2.5-fold; and 11 by 2.5–3-fold).
Figure 3. High throughput screening.
The HeLa (FXN-EGFP) stable cell line was used in high throughput screening procedures to screen the Spectrum Collection compound library. Each test compound was evaluated at a final concentration of 10 µM. Controls included untreated cells and cells exposed to 3.3 µM cisplatin. Cultures were incubated for 72 hours. The fluorometric measurement of EGFP fluorescence was performed followed by the fluorometric measurement of cell viability. The average background fluorescence of parental HeLa cells was subtracted from the EGFP readings of HeLa (FXN-EGFP) cells exposed to the chemical library. A correction factor for the number of viable cells was applied to the EGFP levels in each corresponding well. This was compared to the average value obtained from cells not exposed to any test compound to determine the level of change in FXN gene expression.
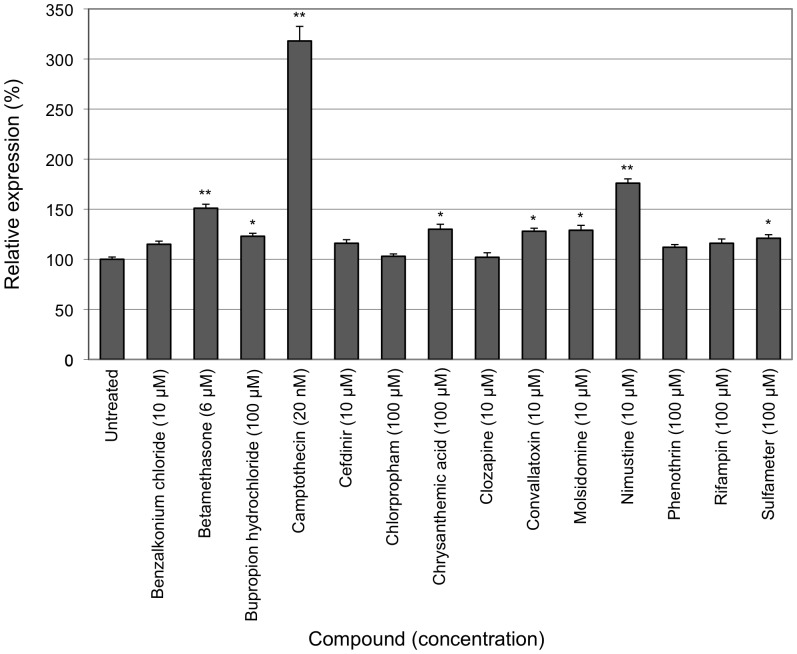
Most of the primary hit compounds that increased expression by greater than 1.5-fold that were available for commercial purchase or closely related analogs that were available were subjected to dose-response experiments over a large concentration range on HeLa (FXN-EGFP) cells and EGFP fluorescence was measured by flow cytometry. Three compounds were validated to reproducibly increase EGFP levels by greater than 1.5-fold in the flow cytometric analysis ( Figure 4 ). The greatest level of induction was observed with the anti-cancer compound camptothecin, which elicited a 3.3-fold increase in FXN-EGFP expression at a concentration of 20 nM. This is the greatest level of induction that we have observed with the HeLa (FXN-EGFP) genomic reporter assay. The glucocorticoid betamethasone (6 µM) and the alkylating agent nimustine (10 µM) increased FXN-EGFP expression by 1.5-fold and 1.7-fold, respectively. Bupropion hydrochloride (100 µM), chrysanthemic acid (100 µM), convallatoxin (10 µM), molsidomine (10 µM) and sulfameter (100 µM) elicited modest (less than 1.5-fold) but significant levels of induction of FXN-EGFP expression.
Figure 4. High throughput screening compound validation.
The HeLa (FXN-EGFP) stable cell line was exposed to various concentrations of compounds identified by high throughput screening. Cultures were incubated for 72 hours. The levels of EGFP expression were measured by flow cytometry. For each compound the lowest concentration that produced the greatest change in FXN-EGFP expression is shown. Assays were performed in triplicate on at least three independent occasions. Error bars represent standard error of the mean. *p<0.05, **p<0.01 in comparison to the untreated control.
Compound Evaluation in Friedreich Ataxia Cells
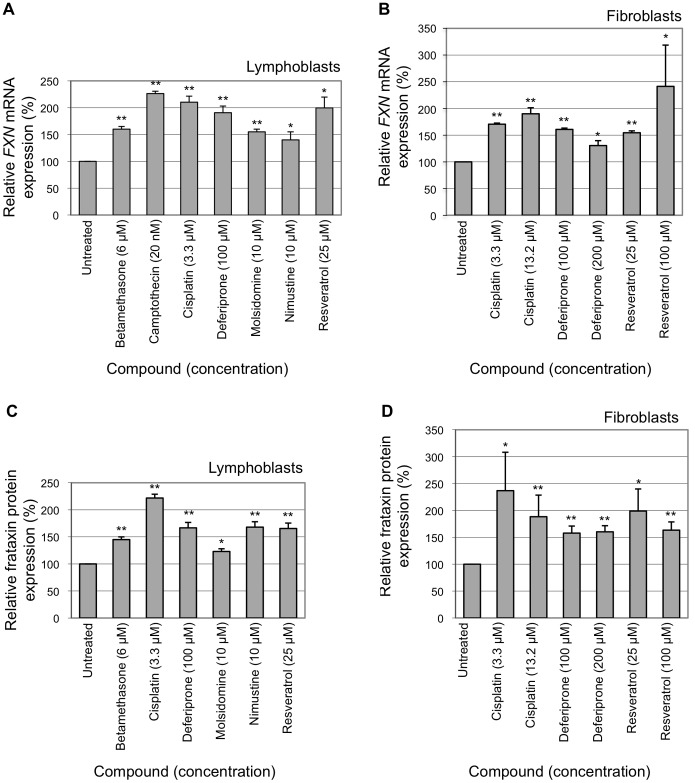
The FXN-EGFP genomic reporter contains a normal copy of the FXN gene without a GAA trinucleotide repeat expansion. Selected compounds were therefore subsequently evaluated on cells derived from individuals with FRDA. Transformed lymphoblasts and primary fibroblasts containing a GAA trinucleotide expansion on both FXN alleles were used. The levels of FXN mRNA were measured by quantitative real time RT-PCR and the levels of frataxin protein were measured by lateral flow immunoassay (dipstick assay).
The anti-cancer compounds cisplatin (3.3 µM) and camptothecin (20 nM) were found to increase the level of FXN mRNA by over 2-fold in FRDA lymphoblasts. Resveratrol (25 µM) increased mRNA levels by 2-fold in FRDA lymphoblasts. Betamethasone (6 µM), deferiprone (100 µM) and molsidomine (10 µM) elicited increases in the level of FXN mRNA by over 1.5-fold in FRDA lymphoblasts ( Figure 5A ). Cisplatin (13.2 µM) and deferiprone (100 µM) were found to increase the level of FXN mRNA by over 1.5-fold in FRDA fibroblasts. Resveratrol (100 µM) elicited a 2.4-fold increase in FXN mRNA levels in FRDA fibroblasts ( Figure 5B ).
Figure 5. Compound evaluation in Friedreich ataxia cells.
The modulation of FXN gene expression by selected test compounds was evaluated in lymphoblast and fibroblast cell lines derived from individuals with FRDA. All cell lines were homozygous for a GAA repeat expansion in the first intron of the FXN gene. Cultures were incubated for 72 hours. FXN mRNA levels were measured by real-time RT-PCR. Frataxin protein levels were measured by lateral flow immunoassay. (A) FXN mRNA levels measured in FRDA lymphoblast cell lines. (B) FXN mRNA levels measured in FRDA fibroblast cell lines. (C) Frataxin protein levels measured in FRDA lymphoblast cell lines. (D) Frataxin protein levels measured in FRDA fibroblast cell lines. Assays were performed in triplicate on at least three independent occasions. Error bars represent standard error of the mean. *p<0.05, **p<0.01 in comparison to the untreated control.
Cisplatin (3.3 µM) was found to increase the levels of frataxin protein by 2.2-fold in FRDA lymphoblasts. Deferiprone (100 µM), nimustine (10 µM) and resveratrol (25 µM) each increased the levels of frataxin protein by1.7-fold in FRDA lymphoblasts. Betamethasone (6 µM) elicited a 1.4-fold increase in frataxin levels in FRDA lymphoblasts ( Figure 5C ). Cisplatin (3.3 µM), deferiprone (100 µM) and resveratrol (25 µM) increased frataxin protein levels by 2.4-fold, 1.6-fold and 2-fold, respectively, in FRDA fibroblasts ( Figure 5D ).
Compound Evaluation in a Friedreich Ataxia Mouse Model
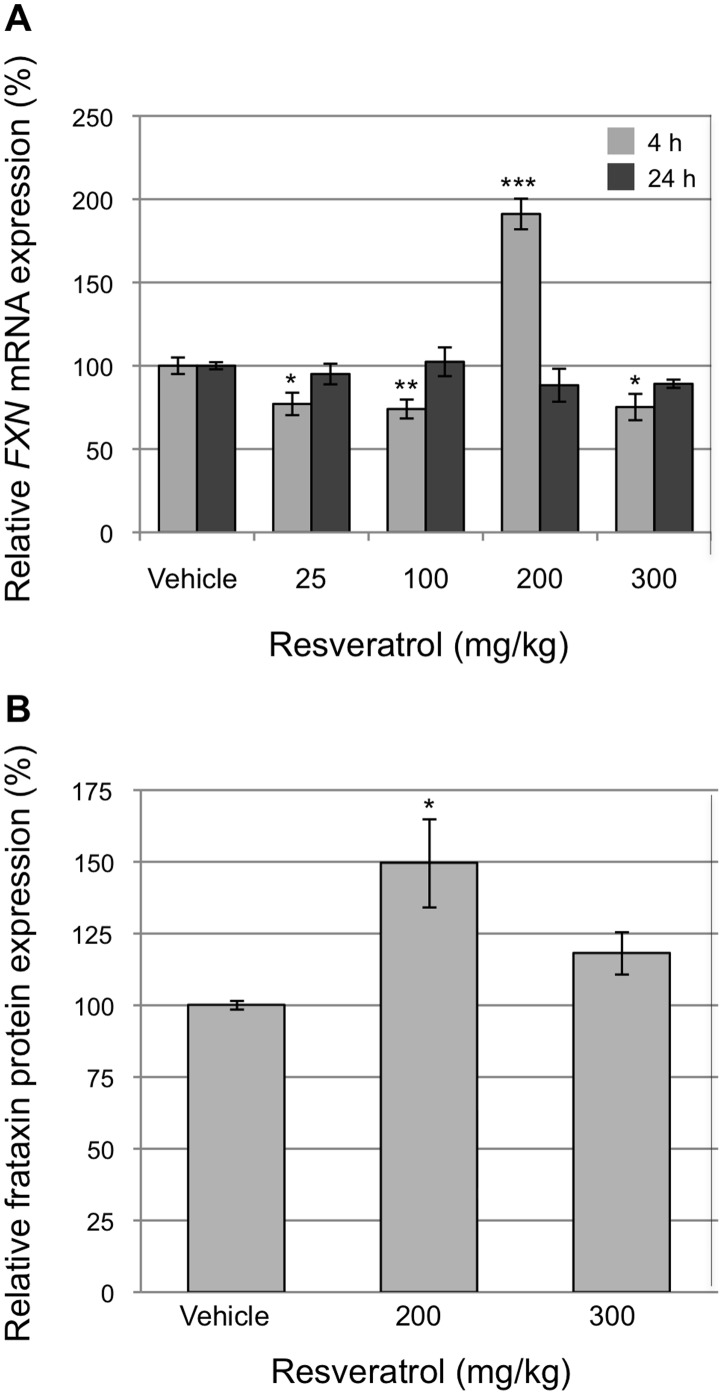
As resveratrol is a safe drug and has a number of proposed modes of action that may be beneficial in FRDA including neuroprotection, cardioprotection and antioxidant properties, an in vivo evaluation of this compound was conducted in a YAC-based GAA expansion humanized mouse model of FRDA [44]. The YG8R transgenic mice contain the entire human genomic FXN gene containing a GAA repeat expansion in the first intron of the gene, and also contain a homozygous knockout of the murine Fxn gene. Only transgenic human frataxin protein is expressed in the mice. The mice provide the same underlying molecular cause of the disease as found in individuals with FRDA and exhibit decreased frataxin levels, progressive neuronal and cardiac pathology that recapitulates that seen in individuals with FRDA and measurable neurobehavioral deficits consistent with that observed in FRDA [41].
Resveratrol was administered daily for three days by subcutaneous injection at doses ranging from 25–300 mg/kg. Samples were collected at four and 24 hours after the final dose. The levels of human FXN mRNA were measured by real-time RT-PCR. An increase in human FXN gene expression of almost 2-fold was observed in mouse brain at a dose of 200 mg/kg ( Figure 6A ). The levels of human frataxin protein were measured by lateral flow immunoassay. Human frataxin protein was found to increase by 1.5-fold in transgenic mouse brain at the same dose ( Figure 6B ), indicating the ability of resveratrol to cross the blood-brain barrier and to exert a frataxin-inducing effect. Interestingly, no other dose of resveratrol resulted in an increase in FXN mRNA or frataxin protein levels in mice and some doses resulted in a reduction in mRNA.
Figure 6. Evaluation of resveratrol in a Friedreich ataxia mouse model.
The modulation of FXN gene expression by resveratrol was evaluated in the YG8R humanized FRDA mouse model. Resveratrol was administered daily for three days by subcutaneous injection. (A) The levels of human FXN mRNA were measured by real-time RT-PCR. Brain samples were collected at four and 24 hours after the final dose. (B) The levels of human frataxin protein were measured by lateral flow immunoassay. Brain samples were collected four hours after the final dose. Reactions were carried out in triplicate for each biological sample (n = 4). Error bars represent standard error of the mean. *p<0.05, **p<0.01, ***p<0.001 in comparison to the untreated control (vehicle).
Discussion
The clinical manifestations of FRDA are due to the presence of a GAA trinucleotide repeat expansion within the first intron of the FXN gene that results in decreased gene expression and an insufficiency of frataxin protein. An overall inverse correlation exists between the length of the smaller GAA repeat expansion and the level of FXN transcript, the amount of frataxin protein and the age of onset of disease symptoms [29]–[33]. Individuals who are heterozygous for a GAA repeat expansion produce about half the normal level of frataxin and are asymptomatic. The GAA repeat expansion mutation does not alter the coding sequence of the gene and the low levels of frataxin protein produced retain normal function. In theory, any increase in frataxin levels should prove beneficial, while a several-fold increase could be sufficient to halt disease progression. Initiation of treatment at the onset of symptoms or prior to onset has the potential to delay or prevent the manifestation of disease pathology. The pharmacological up-regulation of FXN gene expression is a therapeutic approach for the treatment of FRDA that directly addresses the primary issue of frataxin deficiency rather than secondary disease effects.
In order to rapidly and accurately monitor human FXN gene expression and evaluate compounds for their ability to modulate the expression of the gene, we developed an FXN-EGFP genomic reporter that consists of the fusion of the EGFP gene in-frame immediately following the final codon of exon 5a of the normal human FXN gene present on a BAC clone [34]. Genomic reporters preserve the normal location and spacing of many regulatory elements that may be positioned over large distances in the surrounding chromosomal region and more accurately recapitulate expression patterns of tagged genes than can be obtained with conventional small vector transgenic constructs. We have now developed a cellular genomic reporter assay for FXN gene expression by the generation of a stable HeLa cell line containing the FXN-EGFP genomic reporter that permits the precise measurement of human FXN gene expression by flow cytometry and fluorometry.
The FXN-EGFP reporter construct could be stably maintained and expressed in HeLa cells. We did not observe reduced cell viability, reduction in fluorescence or variegation of expression in the stable cell line after repeated passages of cell cultures. The HeLa cell line was selected because a robust and rapidly growing human cell type with undemanding growth conditions was required for HTS procedures. As the FXN gene appears to be expressed in all cell types analyzed we did not believe that it was necessary to introduce the FXN-EGFP genomic reporter into cell lines derived from tissues affected in the disorder, namely neuronal and cardiac cells. In addition, the more fastidious growth requirements of such cell types would negatively impact on the feasibility of HTS procedures.
EGFP expression in the cellular assay is under the control of the natural FXN promoter and regulatory elements present on the BAC clone. As the FXN gene is not highly expressed the levels of EGFP expressed by the FXN-EGFP genomic reporter were relatively low. EGFP expression was sufficiently robust for accurate and reproducible measurement by flow cytometry but required careful optimization of the fluorometric detection system used in HTS procedures. The specific HeLa (FXN-EGFP) stable cell line that was selected for compound screening procedures produced the highest levels of mean peak fluorescence among the clones isolated and was shown by FISH analysis to contain multiple copies of the FXN-EGFP transgene at a single integration site. This clone was used in order to maximize the signal-to-noise ratio.
An advantage of using EGFP as a readout of gene expression is that EGFP reporter expression can be visualized in living cells without the need for cell fixation, cell disruption or addition of enzymatic substrates. For flow cytometry purposes, cells are collected and readings are made on a per cell basis. For HTS procedures the detection of EGFP fluorescence can be made in situ, but HeLa cells and derivatives grow as a monolayer attached to the bottom of culture vessels and the EGFP signal is not uniformly distributed in solution throughout each well. It was therefore necessary to adapt the optics of the plate reader to make measurements from beneath the growth chamber focused on a plane corresponding to the cell monolayer. Optical-grade microtiter plates were necessary in order to minimize quenching of the signal read through the base of the plates. In order to account for the possible non-uniform growth of cells exposed to certain compounds, readings were taken at multiple positions for each well. To increase the signal-to-noise ratio in future screens the assay system could be modified to express an alternative reporter with greater signal intensity or by using a two-step transcriptional amplification (TSTA) system [45] to amplify the signal.
Changes in frataxin-EGFP content per cell in response to exposure to pharmacological compounds may reflect not only changes at the transcriptional level, but also changes in the processing and export of mRNA and in mRNA and protein turnover. This is a significant advantage of the FXN-EGFP assay system, since changes in any one of these processes could affect frataxin levels in a therapeutic manner. It is possible that the induction of frataxin expression quantified by the FXN-EGFP cellular genomic reporter assay may be underestimated due to turnover of the fusion protein during the induction period. EGFP has been reported to have a half-life of about 26 hours [46] but the half-life of the frataxin-EGFP fusion protein has not been determined. Cells are typically incubated with test compounds for a period of 72 hours in the assay system. The effects of compounds with a short half-life may be diminished at the time of readout. Some of these issues could be alleviated by replenishing cultures with fresh test compound periodically throughout the assay incubation period or taking more frequent readings. However, these modifications would increase the complexity and costs of HTS procedures.
The level of increase in EGFP expression induced by the positive control drug cisplatin quantified via fluorometric measurement in the HTS assay (1.4-fold) was lower than that measured via flow cytometry (2.2-fold). This likely represents inherent differences in the quantification of fluorescence by the two detection techniques and could contribute to false negative readouts. Other factors leading to potential false negative results include compounds causing complete cell death at the tested concentration (10 µM) or those not sufficiently potent at this concentration. False positive readouts could be attributed to compounds that were intrinsically fluorescent or to stochastic variability inherent in the HTS assay that was performed a single time. Post-HTS verification evaluation identified false positive compounds and excluded them from further consideration.
The entire 2,000 compound Spectrum Collection library was subjected to HTS procedures. The relatively small size of the chemical library made it cost-effective and suitable for use as an initial proof-of-principle screen. In addition, as the majority of the compounds in this library are FDA-approved, it was envisaged that promising hit compounds could be more rapidly moved into a clinical setting. The screening of such a library also lends itself to the Selective Optimization of Side Activities (SOSA) approach in which known pharmacological agents are structurally modified to give increased affinity for a new target [47].
Of therapeutic relevance, it is important to underscore that although the FXN-EGFP genomic reporter contains a normal copy of the FXN gene without a GAA trinucleotide repeat expansion, its use has allowed the identification of a number of agents that are capable of increasing FXN gene expression and frataxin protein in FRDA cells. However, classes of compounds that are specifically able to overcome the effects of the GAA repeat expansion on FXN gene expression would not be detected using this assay. This limitation could be overcome by the development of an FXN genomic reporter construct containing a long GAA repeat expansion.
Two anticancer agents, cisplatin and camptothecin, were found to elicit the highest levels of induction in the HeLa (FXN-EGFP) genomic reporter and in FRDA lymphoblasts and fibroblasts. Cisplatin is a platinum-containing compound that binds to and cross-links DNA most commonly acting at adjacent guanine residues [48]. It was previously shown to increase FXN gene expression in a cisplatin-resistant ovarian carcinoma cell line [49]. Camptothecin is a cytotoxic quinoline alkaloid that inhibits DNA topoisomerase I [50]. The mechanisms of action that modulate FXN gene expression are not known but likely due to non-specific effects. Both drugs have toxic side-effects that limit their use for the long-term treatment of FRDA.
Deferiprone is a membrane-permeable iron-chelator that has the ability to shuttle iron between cellular organelles and across membranes [51]. Deferiprone binds to ferric ions to form neutral 3∶1 (deferiprone:iron) complexes. It is in clinical use for the treatment of thalassemia. In an open-label clinical trial examining individuals with FRDA deferiprone appeared to reduce accumulated iron in the dentate nucleus and reduced neuropathy and ataxic gait [35]. Our findings indicate that deferiprone is also able to induce FXN gene expression and increase the levels of frataxin suggesting that intracellular iron levels can modulate FXN gene regulation. Besides the direct effects of deferiprone on iron mobilization in FRDA it may confer the additional benefit of increased frataxin levels.
A number of compounds were found to elicit modest but measurably reproducible increases in FXN-EGFP expression. These include: bupropion, a norepinephrine-dopamine reuptake inhibitor used as an antidepressant; chrysanthemic acid, a compound related to pyrethroids; onvallatoxin, a glycoside used in the treatment of cardiac conditions; and sulfameter, a long-acting sulfonamide antibacterial. Additional compounds that were identified to result in small increases in FXN gene expression and frataxin protein in FRDA cells include: betamethasone, a glucocorticoid with anti-inflammatory and immunosuppressive properties; nimustine, a nitrosourea alkylating agent with antineoplastic activity; and molsidomine, an orally active and long acting vasodilating drug.
Resveratrol was identified in the HeLa (FXN-EGFP) cellular genomic reporter to increase FXN gene expression by 1.7-fold. It was subsequently shown to increase FXN mRNA levels by 2-fold in FRDA lymphoblasts and by 2.4-fold in FRDA fibroblasts. It elicited an increase in frataxin protein by 1.7-fold in FRDA lymphoblasts and by 2-fold in FRDA fibroblasts. Resveratrol (3,4′,5-trihydroxy-trans-stilbene) is a polyphenolic compound that is synthesized naturally by several plant species. It is commonly found in red grape skin, berries and nuts [52]. Resveratrol is a phytoalexin; a toxic compound produced by higher plants in response to infection or other stresses. Resveratrol appears to have a wide spectrum of targets as a pharmacological agent. It has been proposed as a treatment for cancers, cardiac disease, neurodegenerative conditions and metabolic disorders including diabetes [53], [54]. It has also been reported to possess antioxidant, anti-inflammatory, anti-angiogenic and antiviral properties. There is evidence that resveratrol contributes to enhanced longevity in S. cerevisiae [55], C. elegans [56], D. melanogaster [56] and mice consuming a high caloric diet [57]. The effects on life extension appear to mimic several of the biochemical effects of calorie restriction.
There is divergent evidence regarding the ability of resveratrol to interact with sirtuin proteins. Sirtuins are NAD+-dependent deacetylases that have been implicated to influence aging, transcription regulation, apoptosis and stress resistance. The deacetylase activity of sirtuins is directed to histone proteins and transcription factors including p53, FOXO proteins and the peroxisome proliferator activated receptor gamma (PPAR-γ) coactivator 1 alpha (PGC-1α) [58]. There are seven sirtuin genes in humans (SIRT1-7). There is some debate regarding the ability of resveratrol to directly activate Sirtuin 1 or increase expression of the SIRT1 gene [53], [55], [59], [60].
PGC-1α plays a role in the regulation of mitochondrial function and fatty acid oxidation [61]. It is activated by deacetylation by Sirtuin 1 which has also been shown to repress PPAR-γ [62]. There is evidence of cross regulation mechanisms that act between frataxin and PGC-1α. Reduced expression of PGC-1α has been shown in FRDA cells, in several tissues of an FRDA mouse model and upon inhibition of FXN gene expression by shRNA [63]. The downregulation of PGC-1α by siRNA results in decreased FXN transcript and frataxin protein in human control and FRDA fibroblasts [64]. Conversely, the overexpression of PGC-1α in skeletal muscle cells has been shown to increase frataxin levels [65], and the PPAR-γ agonist Azelaoyl PAF is able to increase FXN gene expression and frataxin protein levels in FRDA cells [66]. These findings suggest a mechanism that accounts for the observed increased levels of FXN gene expression and frataxin protein levels mediated by resveratrol. It can be hypothesized that the activation of Sirtuin 1 by resveratrol enables it to modulate the activity of the PPAR-γ and PGC-1α pathway which in turn influences the regulation of FXN gene expression.
Resveratrol has a good safety profile and has not been reported to have any major adverse side-effects in humans [67]. Of the pharmacological compounds that we identified to increase FXN gene expression, resveratrol had the most favorable safety criteria for human use and is expected to be well tolerated by individuals with FRDA. It is also able to cross the blood-brain barrier [68], [69]. Pharmacokinetic studies indicate that the metabolization of resveratrol into conjugated forms may influence its bioavailability in higher organisms [70], [71]. We therefore further evaluated resveratrol pre-clinically in the YG8R humanized mouse model of FRDA that contains the entire human FXN gene incorporating a GAA trinucleotide repeat expansion. At a dose of 200 mg/kg resveratrol was shown to increase human FXN gene transcription by almost 2-fold and frataxin protein was found to increase by 1.5-fold in transgenic mouse brain. It is curious that this was the only dose at which FXN mRNA and frataxin protein were increased. On the basis of this data, an open-label, proof of principle study of resveratrol in individuals with FRDA is currently underway.
In conclusion, we have demonstrated the utility of using an FXN-EGFP cellular genomic reporter assay in HTS procedures for the identification of pharmacological compounds able to increase FXN gene expression and frataxin protein. Any compound that specifically increases frataxin levels by several-fold in individuals with FRDA could serve as a potential pharmacological therapy for the disorder. Candidate agents were processed along the drug development pipeline from drug screening to lead identification and resveratrol was selected for evaluation in pre-clinical animal testing and is currently being evaluated in a human therapeutic trial. Ultimately, a cocktail of drugs may be required to provide an effective therapy for FRDA. This may include pharmacological agents that address the primary defect of frataxin deficiency by increasing FXN gene expression and/or frataxin protein such as those identified in this study, HDAC inhibitors [72]–[74], erythropoietin [75]–[77], PPAR-γ agonists [66] or IFNγ [78], and those directed at secondary effects, such as antioxidants and iron-chelators.
Acknowledgments
We are grateful to Ian Street and John Parisot for advice and assistance in conducting high throughput screening procedures. We thank members of the Cell and Gene Therapy Research Group and the Bruce Lefroy Centre for Genetic Health Research for informative discussions. Panos Ioannou passed away in 2005.
Funding Statement
This work was supported by the Muscular Dystrophy Association (USA) (grants 3657, 3725, 69033), the National Health and Medical Research Council (Australia) (grant 491234), the Friedreich’s Ataxia Research Alliance (USA), the Brockhoff Foundation (Australia), the Friedreich Ataxia Research Association (Australasia), Seek A Miracle (USA) and the Victorian Government’s Operational Infrastructure Support Program. MBD is a National Health and Medical Research Council Practitioner Fellow. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pandolfo M (2006) Friedreich ataxia: Detection of GAA repeat expansions and frataxin point mutations. Methods Mol Med 126: 197–216. [DOI] [PubMed] [Google Scholar]
- 2. Bidichandani SI, Ashizawa T, Patel PI (1998) The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. Am J Hum Genet 62: 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ohshima K, Montermini L, Wells RD, Pandolfo M (1998) Inhibitory effects of expanded GAA.TTC triplet repeats from intron I of the Friedreich ataxia gene on transcription and replication in vivo. J Biol Chem 273: 14588–14595. [DOI] [PubMed] [Google Scholar]
- 4. Grabczyk E, Usdin K (2000) The GAA*TTC triplet repeat expanded in Friedreich’s ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res 28: 2815–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sakamoto N, Ohshima K, Montermini L, Pandolfo M, Wells RD (2001) Sticky DNA, a self-associated complex formed at long GAA.TTC repeats in intron 1 of the frataxin gene, inhibits transcription. J Biol Chem 276: 27171–27177. [DOI] [PubMed] [Google Scholar]
- 6. Delatycki MB, Camakaris J, Brooks H, Evans-Whipp T, Thorburn DR, et al. (1999) Direct evidence that mitochondrial iron accumulation occurs in Friedreich ataxia. Ann Neurol 45: 673–675. [PubMed] [Google Scholar]
- 7. Cavadini P, Gellera C, Patel PI, Isaya G (2000) Human frataxin maintains mitochondrial iron homeostasis in Saccharomyces cerevisiae. Hum Mol Genet 9: 2523–2530. [DOI] [PubMed] [Google Scholar]
- 8. Puccio H, Simon D, Cossee M, Criqui-Filipe P, Tiziano F, et al. (2001) Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat Genet 27: 181–186. [DOI] [PubMed] [Google Scholar]
- 9. Park S, Gakh O, Mooney SM, Isaya G (2002) The ferroxidase activity of yeast frataxin. J Biol Chem 277: 38589–38595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cavadini P, O’Neill HA, Benada O, Isaya G (2002) Assembly and iron-binding properties of human frataxin, the protein deficient in Friedreich ataxia. Hum Mol Genet 11: 217–227. [DOI] [PubMed] [Google Scholar]
- 11. Lesuisse E, Santos R, Matzanke BF, Knight SA, Camadro JM, et al. (2003) Iron use for haeme synthesis is under control of the yeast frataxin homologue (Yfh1). Hum Mol Genet 12: 879–889. [DOI] [PubMed] [Google Scholar]
- 12. Bulteau AL, O’Neill HA, Kennedy MC, Ikeda-Saito M, Isaya G, et al. (2004) Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science 305: 242–245. [DOI] [PubMed] [Google Scholar]
- 13. Yoon T, Cowan JA (2004) Frataxin-mediated iron delivery to ferrochelatase in the final step of heme biosynthesis. J Biol Chem 279: 25943–25946. [DOI] [PubMed] [Google Scholar]
- 14. Ramazzotti A, Vanmansart V, Foury F (2004) Mitochondrial functional interactions between frataxin and Isu1p, the iron-sulfur cluster scaffold protein, in Saccharomyces cerevisiae. FEBS Lett 557: 215–220. [DOI] [PubMed] [Google Scholar]
- 15. Yoon T, Cowan JA (2003) Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J Am Chem Soc 125: 6078–6084. [DOI] [PubMed] [Google Scholar]
- 16. Foury F, Pastore A, Trincal M (2007) Acidic residues of yeast frataxin have an essential role in Fe-S cluster assembly. EMBO Rep 8: 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang T, Craig EA (2008) Binding of yeast frataxin to the scaffold for Fe-S cluster biogenesis, Isu. J Biol Chem 283: 12674–12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koeppen AH, Michael SC, Knutson MD, Haile DJ, Qian J, et al. (2007) The dentate nucleus in Friedreich’s ataxia: the role of iron-responsive proteins. Acta Neuropathol 114: 163–173. [DOI] [PubMed] [Google Scholar]
- 19. Koeppen AH, Morral JA, Davis AN, Qian J, Petrocine SV, et al. (2009) The dorsal root ganglion in Friedreich’s ataxia. Acta Neuropathol 118: 763–776. [DOI] [PubMed] [Google Scholar]
- 20. Grabczyk E, Mancuso M, Sammarco MC (2007) A persistent RNA.DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res 35: 5351–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saveliev A, Everett C, Sharpe T, Webster Z, Festenstein R (2003) DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature 422: 909–913. [DOI] [PubMed] [Google Scholar]
- 22. Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, et al. (2006) Histone deacetylase inhibitors reverse gene silencing in Friedreich’s ataxia. Nat Chem Biol 2: 551–558. [DOI] [PubMed] [Google Scholar]
- 23. Greene E, Mahishi L, Entezam A, Kumari D, Usdin K (2007) Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res 35: 3383–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Al-Mahdawi S, Pinto RM, Ismail O, Varshney D, Lymperi S, et al. (2008) The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum Mol Genet 17: 735–746. [DOI] [PubMed] [Google Scholar]
- 25. Castaldo I, Pinelli M, Monticelli A, Acquaviva F, Giacchetti M, et al. (2008) DNA methylation in intron 1 of the frataxin gene is related to GAA repeat length and age of onset in Friedreich’s ataxia patients. J Med Genet 45: 808–812. [DOI] [PubMed] [Google Scholar]
- 26. Rai M, Soragni E, Jenssen K, Burnett R, Herman D, et al. (2008) HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLoS ONE 3: e1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu C, Soragni E, Chou CJ, Herman D, Plasterer HL, et al. (2009) Chemical probes identify a role for histone deacetylase 3 in Friedreich’s ataxia gene silencing. Chem Biol 16: 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Evans-Galea MV, Carrodus N, Rowley SM, Corben LA, Tai G, et al. (2012) FXN methylation predicts expression and clinical outcome in Friedreich ataxia. Ann Neurol 71: 487–497. [DOI] [PubMed] [Google Scholar]
- 29. Filla A, De Michele G, Cavalcanti F, Pianese L, Monticelli A, et al. (1996) The relationship between trinucleotide (GAA) repeat length and clinical features in Friedreich ataxia. Am J Hum Genet 59: 554–560. [PMC free article] [PubMed] [Google Scholar]
- 30. Monros E, Molto MD, Martinez F, Canizares J, Blanca J, et al. (1997) Phenotype correlation and intergenerational dynamics of the Friedreich ataxia GAA trinucleotide repeat. Am J Hum Genet 61: 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. De Michele G, Filla A, Criscuolo C, Scarano V, Cavalcanti F, et al. (1998) Determinants of onset age in Friedreich’s ataxia. J Neurol 245: 166–168. [DOI] [PubMed] [Google Scholar]
- 32. Delatycki MB, Paris DB, Gardner RJ, Nicholson GA, Nassif N, et al. (1999) Clinical and genetic study of Friedreich ataxia in an Australian population. Am J Med Genet 87: 168–174. [DOI] [PubMed] [Google Scholar]
- 33. Santoro L, Perretti A, Lanzillo B, Coppola G, De Joanna G, et al. (2000) Influence of GAA expansion size and disease duration on central nervous system impairment in Friedreich’s ataxia: contribution to the understanding of the pathophysiology of the disease. Clin Neurophysiol 111: 1023–1030. [DOI] [PubMed] [Google Scholar]
- 34. Sarsero JP, Li L, Wardan H, Sitte K, Williamson R, et al. (2003) Upregulation of expression from the FRDA genomic locus for the therapy of Friedreich ataxia. J Gene Med 5: 72–81. [DOI] [PubMed] [Google Scholar]
- 35. Boddaert N, Le Quan Sang KH, Rotig A, Leroy-Willig A, Gallet S, et al. (2007) Selective iron chelation in Friedreich ataxia: biologic and clinical implications. Blood 110: 401–408. [DOI] [PubMed] [Google Scholar]
- 36. Pandolfo M (2008) Drug Insight: antioxidant therapy in inherited ataxias. Nat Clin Pract Neurol 4: 86–96. [DOI] [PubMed] [Google Scholar]
- 37. Schulz JB, Boesch S, Burk K, Durr A, Giunti P, et al. (2009) Diagnosis and treatment of Friedreich ataxia: a European perspective. Nat Rev Neurol 5: 222–234. [DOI] [PubMed] [Google Scholar]
- 38. Sarsero JP, Li L, Holloway TP, Voullaire L, Gazeas S, et al. (2004) Human BAC-mediated rescue of the Friedreich ataxia knockout mutation in transgenic mice. Mamm Genome 15: 370–382. [DOI] [PubMed] [Google Scholar]
- 39. Puspasari N, Rowley SM, Gordon L, Lockhart PJ, Ioannou PA, et al. (2011) Long range regulation of human FXN gene expression. PLoS One 6: e22001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sarsero JP, Holloway TP, Li L, McLenachan S, Fowler KJ, et al. (2005) Evaluation of an FRDA-EGFP genomic reporter assay in transgenic mice. Mamm Genome 16: 228–241. [DOI] [PubMed] [Google Scholar]
- 41. Al-Mahdawi S, Pinto RM, Varshney D, Lawrence L, Lowrie MB, et al. (2006) GAA repeat expansion mutation mouse models of Friedreich ataxia exhibit oxidative stress leading to progressive neuronal and cardiac pathology. Genomics 88: 580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Willis JH, Isaya G, Gakh O, Capaldi RA, Marusich MF (2008) Lateral-flow immunoassay for the frataxin protein in Friedreich’s ataxia patients and carriers. Mol Genet Metab 94: 491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Narayanan K, Williamson R, Zhang Y, Stewart AF, Ioannou PA (1999) Efficient and precise engineering of a 200 kb beta-globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system. Gene Ther 6: 442–447. [DOI] [PubMed] [Google Scholar]
- 44. Al-Mahdawi S, Pinto RM, Ruddle P, Carroll C, Webster Z, et al. (2004) GAA repeat instability in Friedreich ataxia YAC transgenic mice. Genomics 84: 301–310. [DOI] [PubMed] [Google Scholar]
- 45. Iyer M, Wu L, Carey M, Wang Y, Smallwood A, et al. (2001) Two-step transcriptional amplification as a method for imaging reporter gene expression using weak promoters. Proc Natl Acad Sci U S A 98: 14595–14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Corish P, Tyler-Smith C (1999) Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng 12: 1035–1040. [DOI] [PubMed] [Google Scholar]
- 47. Wermuth CG (2006) Selective optimization of side activities: the SOSA approach. Drug Discov Today 11: 160–164. [DOI] [PubMed] [Google Scholar]
- 48. Boulikas T, Vougiouka M (2003) Cisplatin and platinum drugs at the molecular level. (Review). Oncol Rep 10: 1663–1682. [PubMed] [Google Scholar]
- 49. Ghazizadeh M (2003) Cisplatin may induce frataxin expression. J Nippon Med Sch 70: 367–371. [DOI] [PubMed] [Google Scholar]
- 50. Ulukan H, Swaan PW (2002) Camptothecins: a review of their chemotherapeutic potential. Drugs 62: 2039–2057. [DOI] [PubMed] [Google Scholar]
- 51. Sohn YS, Breuer W, Munnich A, Cabantchik ZI (2008) Redistribution of accumulated cell iron: a modality of chelation with therapeutic implications. Blood 111: 1690–1699. [DOI] [PubMed] [Google Scholar]
- 52. Pirola L, Frojdo S (2008) Resveratrol: one molecule, many targets. IUBMB Life 60: 323–332. [DOI] [PubMed] [Google Scholar]
- 53. Baur JA, Sinclair DA (2006) Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov 5: 493–506. [DOI] [PubMed] [Google Scholar]
- 54. Cucciolla V, Borriello A, Oliva A, Galletti P, Zappia V, et al. (2007) Resveratrol: from basic science to the clinic. Cell Cycle 6: 2495–2510. [DOI] [PubMed] [Google Scholar]
- 55. Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, et al. (2003) Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425: 191–196. [DOI] [PubMed] [Google Scholar]
- 56. Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, et al. (2004) Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 430: 686–689. [DOI] [PubMed] [Google Scholar]
- 57. Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, et al. (2006) Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444: 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang T, Fu M, Pestell R, Sauve AA (2006) SIRT1 and endocrine signaling. Trends Endocrinol Metab 17: 186–191. [DOI] [PubMed] [Google Scholar]
- 59. Beher D, Wu J, Cumine S, Kim KW, Lu SC, et al. (2009) Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem Biol Drug Des 74: 619–624. [DOI] [PubMed] [Google Scholar]
- 60. Dai H, Kustigian L, Carney D, Case A, Considine T, et al. (2010) SIRT1 activation by small molecules: kinetic and biophysical evidence for direct interaction of enzyme and activator. J Biol Chem 285: 32695–32703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, et al. (2005) Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434: 113–118. [DOI] [PubMed] [Google Scholar]
- 62. Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, et al. (2004) Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 429: 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Coppola G, Marmolino D, Lu D, Wang Q, Cnop M, et al. (2009) Functional genomic analysis of frataxin deficiency reveals tissue-specific alterations and identifies the PPARgamma pathway as a therapeutic target in Friedreich’s ataxia. Hum Mol Genet 18: 2452–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Marmolino D, Manto M, Acquaviva F, Vergara P, Ravella A, et al. (2010) PGC-1alpha down-regulation affects the antioxidant response in Friedreich’s ataxia. PLoS One 5: e10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. O’Hagan KA, Cocchiglia S, Zhdanov AV, Tambuwala MM, Cummins EP, et al. (2009) PGC-1alpha is coupled to HIF-1alpha-dependent gene expression by increasing mitochondrial oxygen consumption in skeletal muscle cells. Proc Natl Acad Sci U S A 106: 2188–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Marmolino D, Acquaviva F, Pinelli M, Monticelli A, Castaldo I, et al. (2009) PPAR-gamma agonist Azelaoyl PAF increases frataxin protein and mRNA expression: new implications for the Friedreich’s ataxia therapy. Cerebellum 8: 98–103. [DOI] [PubMed] [Google Scholar]
- 67. Almeida L, Vaz-da-Silva M, Falcao A, Soares E, Costa R, et al. (2009) Pharmacokinetic and safety profile of trans-resveratrol in a rising multiple-dose study in healthy volunteers. Mol Nutr Food Res 53 Suppl 1S7–15. [DOI] [PubMed] [Google Scholar]
- 68. Wang Q, Xu J, Rottinghaus GE, Simonyi A, Lubahn D, et al. (2002) Resveratrol protects against global cerebral ischemic injury in gerbils. Brain Res 958: 439–447. [DOI] [PubMed] [Google Scholar]
- 69. Mokni M, Elkahoui S, Limam F, Amri M, Aouani E (2007) Effect of resveratrol on antioxidant enzyme activities in the brain of healthy rat. Neurochem Res 32: 981–987. [DOI] [PubMed] [Google Scholar]
- 70. Walle T, Hsieh F, DeLegge MH, Oatis JE Jr, Walle UK (2004) High absorption but very low bioavailability of oral resveratrol in humans. Drug Metab Dispos 32: 1377–1382. [DOI] [PubMed] [Google Scholar]
- 71. la Porte C, Voduc N, Zhang G, Seguin I, Tardiff D, et al. (2010) Steady-State pharmacokinetics and tolerability of trans-resveratrol 2000 mg twice daily with food, quercetin and alcohol (ethanol) in healthy human subjects. Clin Pharmacokinet 49: 449–454. [DOI] [PubMed] [Google Scholar]
- 72. Rai M, Soragni E, Chou CJ, Barnes G, Jones S, et al. (2010) Two new pimelic diphenylamide HDAC inhibitors induce sustained frataxin upregulation in cells from Friedreich’s ataxia patients and in a mouse model. PLoS One 5: e8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sandi C, Pinto RM, Al-Mahdawi S, Ezzatizadeh V, Barnes G, et al. (2011) Prolonged treatment with pimelic o-aminobenzamide HDAC inhibitors ameliorates the disease phenotype of a Friedreich ataxia mouse model. Neurobiol Dis 42: 496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Soragni E, Xu C, Cooper A, Plasterer HL, Rusche JR, et al. (2011) Evaluation of histone deacetylase inhibitors as therapeutics for neurodegenerative diseases. Methods Mol Biol 793: 495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Acquaviva F, Castaldo I, Filla A, Giacchetti M, Marmolino D, et al.. (2008) Recombinant Human Erythropoietin Increases Frataxin Protein Expression Without Increasing mRNA Expression. Cerebellum. [DOI] [PubMed]
- 76. Boesch S, Sturm B, Hering S, Scheiber-Mojdehkar B, Steinkellner H, et al. (2008) Neurological effects of recombinant human erythropoietin in Friedreich’s ataxia: a clinical pilot trial. Mov Disord 23: 1940–1944. [DOI] [PubMed] [Google Scholar]
- 77. Sacca F, Piro R, De Michele G, Acquaviva F, Antenora A, et al. (2011) Epoetin alfa increases frataxin production in Friedreich’s ataxia without affecting hematocrit. Mov Disord 26: 739–742. [DOI] [PubMed] [Google Scholar]
- 78. Tomassini B, Arcuri G, Fortuni S, Sandi C, Ezzatizadeh V, et al. (2012) Interferon gamma upregulates frataxin and corrects the functional deficits in a Friedreich ataxia model. Hum Mol Genet 21: 2855–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]