Abstract
The spindle assembly checkpoint (SAC) is a ‘wait-anaphase' mechanism that has evolved in eukaryotic cells in response to the stochastic nature of chromosome–spindle attachments. In the recent past, different aspects of the SAC regulation have been described. However, the role of microRNAs in the SAC is vaguely understood. We report here that Mad1, a core SAC protein, is repressed by human miR-125b. Mad1 serves as an adaptor protein for Mad2 – which functions to inhibit anaphase entry till the chromosomal defects in metaphase are corrected. We show that exogenous expression of miR-125b, through downregulation of Mad1, delays cells at metaphase. As a result of this delay, cells proceed towards apoptotic death, which follows from elevated chromosomal abnormalities upon ectopic expression of miR-125b. Moreover, expressions of Mad1 and miR-125b are inversely correlated in a variety of cancer cell lines, as well as in primary head and neck tumour tissues. We conclude that increased expression of miR-125b inhibits cell proliferation by suppressing Mad1 and activating the SAC transiently. We hypothesize an optimum Mad1 level and thus, a properly scheduled SAC is maintained partly by miR-125b.
Keywords: miR-125b, spindle assembly checkpoint, MAD1, apoptosis, mitotic delay, chromosomal abnormalities
MicroRNAs (miRNAs) are small (∼22 nucleotides) non-coding RNAs that modulate gene expression through translational inhibition or messenger RNA (mRNA) degradation.1 A single miRNA can target hundreds of genes and hence, it is predicted that miRNAs regulate expression of most of the protein-coding genes in humans.2 Currently, about 2000 miRNAs have been identified in humans3 (http://www.mirbase.org). Accumulating evidence suggest that miRNAs have a critical role in cell proliferation and survival,4, 5 defects in which form one of the hallmarks of cancer. The emerging view is that deregulated miRNA expression occurs frequently in diverse cancer types. In fact, miRNA expression profiles are distinct biomarkers for classifying human cancers and identifying tumour-tissue origin.6, 7 These studies highlight the need for elucidating miRNA function in tumorigenic pathways.4 Presently, however, experimentally validated targets of miRNAs affecting cell survival are few.
Chromosomal instability (CIN) has a key role in aneuploidy and tumorigenesis,8 where as aneuploidy is known to frequently result from a defective mitotic checkpoint.9 In this context, spindle assembly checkpoint (SAC) is crucial to ensure fidelity of chromosome segregation during mitosis. Any defect in kinetochore–spindle attachment between sister chromatids fails to satisfy SAC as a result of which metaphase remains halted until the defect is corrected.10 Once all chromatids are bi-oriented at metaphase, E3 ubiquitin-ligase anaphase-promoting complex/cyclosome (APC/C) catalyses the proteasomal degradation of securin, an inhibitory chaperone of separase. Activated separase then cleaves the cohesin complex required for the physical linkage of sister chromatids. Owing to loss of cohesion, separated chromatids move to opposite spindle poles and cells enter anaphase.11, 12 Furthermore, APC/C activity depends on an adaptor-protein Cdc20 (cell division cycle 20).13 When SAC is ‘on', Cdc20 remains sequestered by Mad2 and BubR1/Bub3 in the form of mitotic checkpoint complex (MCC), owing to which, APC/C remains inactive. Premature anaphase progression is thus prevented by the ‘wait-anaphase' signal generated from the diffusible MCC.10 In this context, one of the first events in SAC activation is the recruitment of an adaptor protein Mad1 to the kinetochore. Mad1 is crucial in Mad2-transportation from cytosol to nucleus and also in its kinetochore localization.14 According to ‘template' model, Mad2 exists in two states: open/free (O-Mad2), and closed (bound to either Mad1 or Cdc20 (C-Mad2)). Mad1 traps O-Mad2 and this Mad1–C-Mad2 complex gets transported to unoccupied kinetochores. Bound Mad2 functions as template for conversion of other O-Mad2 molecules to C-Mad2 that can then bind Cdc20. Moreover, Mad1–C-Mad2 complex facilitates further interaction of Cdc20 and Mad2 away from kinetochores, thus providing scope for checkpoint signal-amplification.12 The importance of Mad1 in regulating SAC is accentuated by the observation that excess Mad1 reduces free-Mad2 level in Xenopus extracts,15 and weakens SAC by disrupting mitotic-timing.16 Conversely, excess Mad2 can arrest cells in metaphase, in spite of all chromosomes being bi-oriented successfully.15 This underlines the need for a proper Mad1/Mad2 ratio to maintain the integrity of SAC.15
Head and neck/oral cancer (HNOC) is the sixth most common cancer worldwide. In the Indian subcontinent, it comprises of about 50% of all cancers.17 CIN is a consistent property of primary head and neck tumours,18 which makes it pertinent to describe SAC defects in HNOC. Meanwhile, 14 miRNAs are reported to be downregulated, while 29 are upregulated in HNOC.19 To the best of our knowledge, the role of miRNAs in SAC regulation has not been elucidated yet.
In the present study, we have identified MAD1 (mitotic-arrest deficient) as a novel target of human miR-125b, a downregulated miRNA in HNOC. Importantly, we show in an oral cancer cell line model that this regulation of Mad1 delays mitotic exit by transient activation of SAC. This delay results in accumulation of CIN, which culminates in apoptotic cell death. We have also verified the expression status of miR-125b and Mad1 in HNOC patients to obtain the in vivo relevance of the cell line observations.
Results
The 3′ untranslated region (UTR) of MAD1 is a putative target of hsa-miR-125b
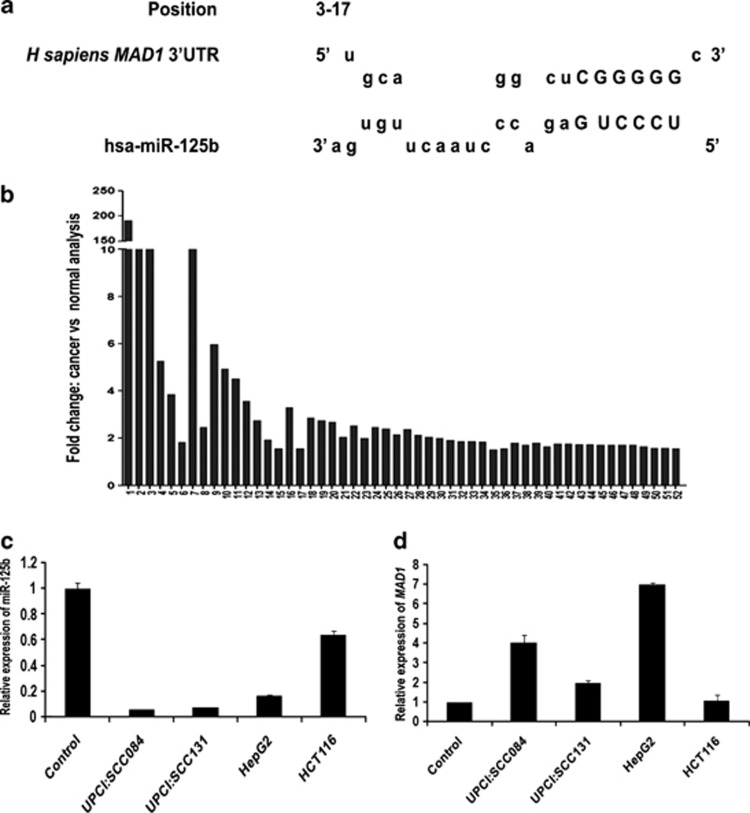
The in silico strategy to identify miRNAs that exhibit altered expression in HNOC and their putative mitotic targets has been illustrated (Supplementary Figure S1). Online target prediction of 43 miRNAs deregulated in HNOC (29 upregulated and 14 downregulated)19 by miRBase gave us an initial data set of a large number of putative targets (Supplementary Table S1). Neumann et al.20 have reported a number of genes, whose knockdown lead to one or more mitotic defects. We used these data to identify mitotic genes from the predicted targets listed in Supplementary Table S1 (Supplementary Table S2). Next, differential expression of this set of genes was analysed in HNOC using the Oncomine data set for nine studies (Supplementary Table S3). These genes were further filtered on the basis of, (1) experimentally validated mitotic roles and (2) inverse relation with the corresponding miRNA with respect to their expressions in HNOC (Supplementary Table S4). This data set revealed that the essential SAC gene, MAD1, is a potential target of Homo sapiens miRNA (hsa-miR)-125b. Indeed, RNAhybrid revealed that MAD1 has a miR-125b recognition site at 3′UTR position 3–17 (Figure 1a and Supplementary Figure S2). Simultaneously, it was found that Mad1 levels are elevated in various cancers including HNOC (Figure 1b and Supplementary Table S5). This prompted us to select MAD1 as gene of interest.
Figure 1.
MAD1 is a putative target of miR-125b. (a) The nucleotide position 3–17 of MAD1-3′UTR is the recognition site for miR-125b as predicted by RNAhybrid. Letters in upper case denote the seed region. (b) Mad1 expression is upregulated in different cancers. Expression data of Mad1 in 52 different cancers from Oncomine database were analysed. Cancer versus normal data sets of Mad1 overexpression with fold change ≥1.5 and P-value≤0.05 were selected. Bars represent fold overexpression of Mad1 in these cancers. (c) Endogenous miR-125b expression in different cancer cell lines as compared with control oral epithelial cells. Total RNA was isolated from oral swab (control), UPCI:SCC084, UPCI:SCC131, HepG2 and HCT116 cells and reverse transcribed using miR-125b and miR-17-5p specific stem-loop primers. cDNAs were subjected to RT-PCR. Relative expression values were normalized to those of miR-17-5p. (d) Corresponding endogenous Mad1 levels in the above cell lines as compared with control oral epithelial cells. Total RNA was reverse transcribed and RT-PCR was done using MAD1-specific primers. Relative expression values were normalized to those of GAPDH. For both (c) and (d), data represent three independent experiments (N=3) and are shown as average±S.E. N=number of times the experiment was performed
miR-125b expression is inversely correlated with Mad1 expression in human oral cancer cell lines
It is reported that miR-125b is downregulated in squamous cell carcinoma (SCC) cell lines.21 Initially, we tested and reconfirmed that miR-125b expression is much lower in two SCC cell lines, UPCI:SCC084 and UPCI:SCC131, than in normal oral epithelial cells (Figure 1c). Conversely, we observed that MAD1 transcript levels were higher in these cell lines (Figure 1d). Moreover, this inverse expression pattern between MAD1 and miR-125b was also observed in other cancer cell lines (Figures 1c and d). Hence, UPCI:SCC084 cell line was chosen to study the possible role of miR-125b in MAD1 regulation.
miR-125b negatively regulates MAD1 expression by binding to its 3′UTR
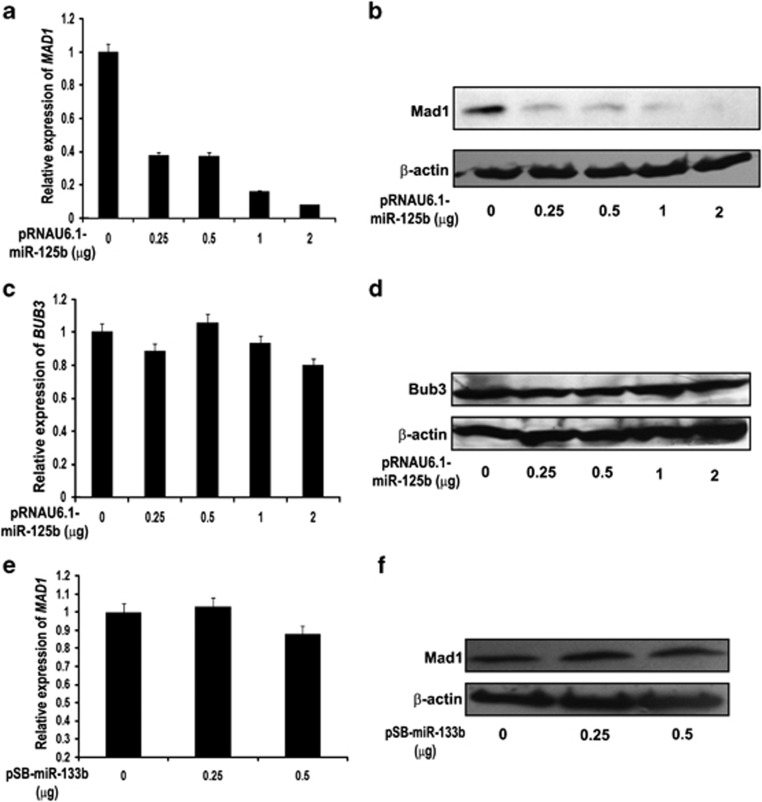
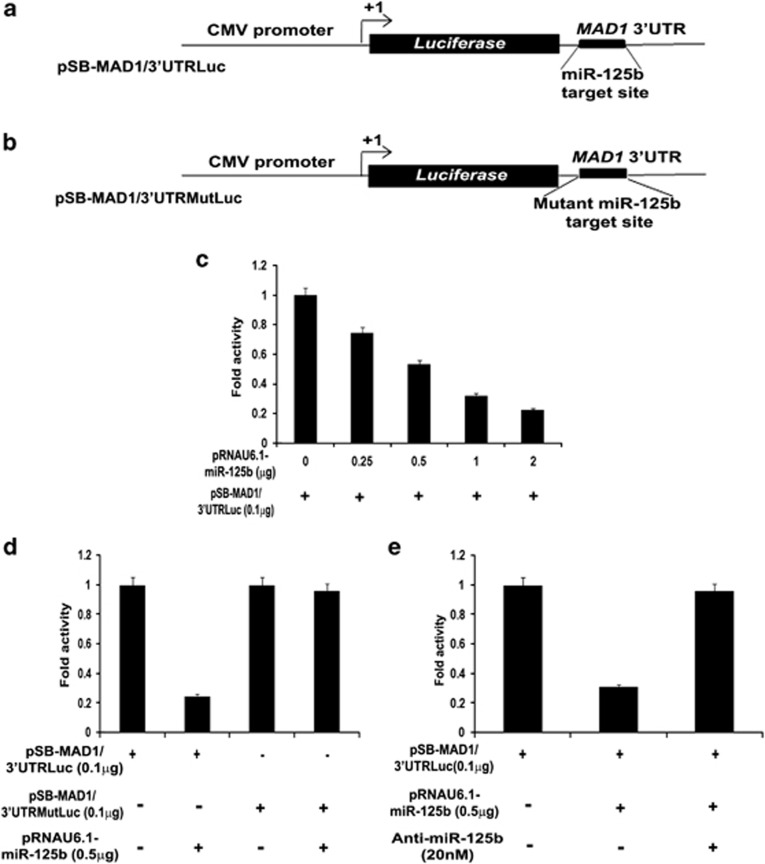
To validate whether MAD1 is a bona fide target of miR-125b, we transfected UPCI:SCC084 cells with increasing doses of miR-125b expression plasmid (Supplementary Figure S3A) and measured the mRNA and protein levels of MAD1. We found that MAD1 transcript levels decreased in a dose-dependent manner indicating that miR-125b regulates MAD1 post-transcriptionally (Figure 2a). Concordantly, Mad1 protein levels also declined upon ectopic miR-125b expression (Figure 2b and Supplementary Figure S3B). That ectopic miR-125b suppressed MAD1 transcript levels was checked in other cell lines (HepG2 and HCT116) and similar observations were made (Supplementary Figures S3C and D). Additionally, specificity of this interaction was supported by the observation that levels of another SAC gene BUB3 (budding uninhibited by benzimidazole; that does not have a recognition site for miR-125b) remained unaffected by miR-125b (Figures 2c and d and Supplementary Figure S4A). On the other hand, Mad1 levels also remained unaltered in presence of another unrelated miRNA miR-133b, which does not have a binding site on MAD1 (Figures 2e and f and Supplementary Figure S4B). Next, we co-transfected UPCI:SCC084 and HepG2 cells with either pSB-MAD1/3′UTRLuc (Figure 3a) or pSB-MAD1/3′UTRMutLuc (Figure 3b) with increasing doses of miR-125b and measured the luciferase (Luc) activity. As expected, a concordant dose-dependent decrease in Luc activity was observed for the 3′UTR containing wild-type miR-125b recognition site (Figure 3c and Supplementary Figure S5). In contrast, there was no significant change in Luc activity for mutant (Mut) MAD1-3′UTR (Figure 3d). To complement these observations, we co-transfected UPCI:SCC084 cells with antisense inhibitor against miR-125b along-with the miRNA and pSB-MAD1/3′UTRLuc. Suitably, miR-125b could not bind to its target site and consequently, was unable to suppress Luc activity (Figure 3e). Overall, these results prove that miR-125b binds to MAD1-3′UTR and specifically represses its expression.
Figure 2.
miR-125b downregulates Mad1 in a specific manner. (a) MAD1 mRNA level is reduced in presence of ectopic miR-125b. UPCI:SCC084 cells were transiently transfected with 0, 0.25, 0.5, 1 and 2 μg of miR-125b expression plasmid. Total RNA isolated was reverse transcribed and the cDNA subjected to RT-PCR using MAD1-specific primers. Relative expression values were normalized to those of GAPDH. (b) Representative picture showing that Mad1 protein level decreases upon ectopic expression of miR-125b. UPCI:SCC084 cells were transiently transfected with 0, 0.25, 0.5, 1 and 2 μg of miR-125b expression plasmid. Cell lysates were prepared followed by western blot with antibodies against Mad1 and β-actin. (c) BUB3 mRNA level is unaffected by ectopic miR-125b. UPCI:SCC084 cells were transiently transfected with 0, 0.25, 0.5, 1 and 2 μg of miR-125b expression plasmid. Total RNA isolated was reverse transcribed and the cDNA subjected to RT-PCR using BUB3-specific primers. Relative expression values were normalized to those of GAPDH. (d) Representative picture showing that Bub3 protein level is unaffected upon ectopic expression of miR-125b. UPCI:SCC084 cells were transiently transfected with 0, 0.25, 0.5, 1 and 2 μg of miR-125b. Cell lysates were prepared followed by western blot with antibodies against Bub3 and β-actin. (e) MAD1 mRNA level remains unaltered in presence of an unrelated miRNA, miR-133b. UPCI:SCC084 cells were transiently transfected with 0, 0.25 and 0.5 μg of pSB-miR-133b. Total RNA isolated was reverse transcribed and the cDNA subjected to RT-PCR using MAD1-specific primers. Relative expression values were normalized to those of GAPDH. (f) Representative picture showing that Mad1 protein level is unaffected upon ectopic expression of pSB-miR-133b. UPCI:SCC084 cells were transiently transfected with 0, 0.25 and 0.5 μg of pSB-miR-133b. Cell lysates were prepared followed by western blot with antibodies against Mad1 and β-actin. For a, c and e, data represent three independent experiments (N=3) and are shown as average±S.E. For b, d and f, images are representative of three independent experiments
Figure 3.
miR-125b and Mad1 interaction is specific. (a and b) Cartoons showing the two MAD1-3′UTR clones used for Luc assay. (a) represents wild type MAD1-3′UTR downstream of Luc gene while (b) shows MAD1-3′UTR in which the target site for miR-125b is mutated. (c) miR-125b decreases Luc activity in presence of MAD1-3′UTR. UPCI:SCC084 cells were transiently co-transfected with 0.1 μg pSB-MAD1/3′UTRLuc and 0, 0.25, 0.5, 1 or 2 μg of miR-125b expression plasmid. Protein lysates were prepared for Luc assay. (d) Mutant MAD1-3′UTR prevents downregulation by miR-125b. UPCI:SCC084 cells were co-transfected with either 0.1 μg pSB-MAD1/3′UTRLuc, or pSB-MAD1/3′UTRMutLuc along-with 0 or 0.5 μg miR-125b expression plasmid. Protein lysates were prepared for Luc assay. (e) Blocking miR-125b prevents MAD1 repression. UPCI:SCC084 cells were transiently transfected with either only pSB-MAD1/3′UTRLuc (0.1 μg), or with pSB-MAD1/3′UTRLuc (0.1 μg) and miR-125b expression plasmid (0.5 μg), or with pSB-MAD1/3′UTRLuc (0.1 μg), miR-125b expression plasmid (0.5 μg) and anti-miR-125b (20 nM). Protein lysates were prepared for Luc assay. For (c–e), data represent three independent experiments (N=3) and are shown as average±S.E.
Ectopic miR-125b delays mitotic exit through MAD1 repression
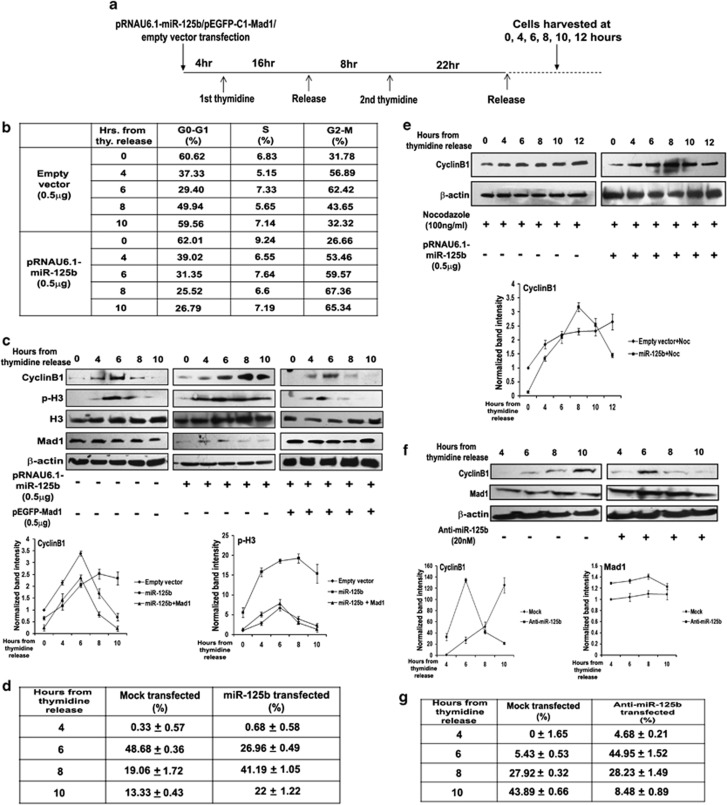
We next investigated the functional consequences of MAD1 repression by miR-125b. As Mad1 is a core factor in SAC regulation, initially we examined the pattern of mitotic progression of UPCI:SCC084 cells with ectopic miR-125b. This was done in three different ways in cells synchronized by thymidine treatment and released from G1 block (Figure 4a). First, FACS analysis of vector-transfected cells revealed that their peak accumulation at G2/M phase (62.42%) occurred at 6 h post-release. By 10 h from release, percentage of cells in G2/M reduced to 32.32% (Figure 4b and Supplementary Figure S6, upper panels). In comparison, 67.36% of cells overproducing miR-125b were in G2/M at 8 h from release, and remained marginally changed (65.34%) even at 10 h post-release (Figure 4b and Supplementary Figure S6, lower panels). Second, we followed the mitotic progression by monitoring the levels of two marker proteins, cyclinB1 and phosphorylated-histone H3 (p-H3). For vector-transfected cells, cyclinB1 and p-H3 levels peaked at 6 h post-release, after which they began to decline, reflecting mitotic exit (Figure 4c, left panel and graphs). In contrast, levels of both proteins were maximal at 8 h post-release upon ectopic miR-125b expression and little degraded even at the 10 h time-point implying that cells were still in mitosis (Figure 4c, middle panel and graphs). Furthermore, specificity of miR-125b-mediated mitotic regulation was proved by the observation that rapid degradation of cyclinB1 and p-H3 levels were rescued by Mad1 expression even in presence of miR-125b (Figure 4c, right panel and graphs). Besides, Cdc20 overexpression along-with miR-125b was also able to retrieve cyclinB1 degradation and p-H3 level by overriding the SAC arrest (Supplementary Figure S7). It is noteworthy that the Mad1 expression plasmid does not carry the 3′UTR recognition site for miR-125b. So, Mad1 can restore the original phenotype even in presence of miR-125b. Thirdly, we evaluated the mitotic indices (MI) of miR-125b-transfected cells. The data showed that MI for miR-125b-overproducing cells was highest at the 8-h time-point (Figure 4d, third column) in comparison with vector-transfected cells, which showed highest MI at 6 h post-release (Figure 4d, middle column). Even at 10 h post-release, MI was significantly higher in presence of miR-125b than for vector-transfected cells (Figure 4d). Moreover, many miR-125b-transfected cells were still in metaphase at the 10-h time-point, while most of the vector-transfected cells had already proceeded to anaphase (Supplementary Figures S8A and B). Together, these results prove that Mad1 downregulation by miR-125b delays mitotic exit. However, previous studies have reported that partial downregulation of Mad1 leads to functional inactivation of SAC.22 To analyse this dichotomy, we monitored the effect of miR-125b-mediated Mad1 downregulation on mitotic progression of synchronized cells in presence of the spindle poison nocodazole. Here, we followed cyclinB1 degradation pattern up to 12 h post-release. As expected, cyclinB1 levels were stabilised for vector-treated cells as a result of nocodazole-induced mitotic arrest (Figure 4e, left panel and graph). On the contrary, in presence of exogenous miR-125b, forceful degradation of cyclinB1 was observed at the 12-h time-point (Figure 4e, right panel and graph). This implies that upon Mad1 downregulation, there is transient activation of SAC, which delays cells in mitosis. Subsequently, delayed cells undergo forced mitotic exit, overriding even nocodazole-induced SAC arrest.
Figure 4.
miR-125b delays cells at metaphase by downregulating Mad1. (a) Schematic diagram outlining the cell synchronization protocol. Time-points for transfection, thymidine treatment and incubation are shown. (b) miR-125b blocks UPCI:SCC084 cells at G2/M phase. Cells were transiently transfected with 0.5 μg miR-125b expression plasmid or empty vector, synchronized and harvested at 0, 4, 6, 8 and 10 h from second thymidine release. DNA content was analysed by flow cytometry. (c) Representative picture showing that miR-125b delays cyclinB1 degradation and dephosphorylation of H3 while Mad1 rescues the normal pattern even in presence of miR-125b. UPCI:SCC084 cells were transiently transfected with either empty vector (0.5 μg), miR-125b expression plasmid (0.5 μg), or miR-125b expression plasmid (0.5 μg) and pEGFP-C1-Mad1 (0.5 μg), and synchronized. Cell lysates were prepared at 0, 4, 6, 8 and 10 h from second thymidine release followed by western blot with antibodies against cyclinB1, p-H3, H3, Mad1 and β-actin. Densitometric values of bands by ImageJ are plotted: cyclinB1 was normalized to β-actin levels while p-H3 was normalized to H3 levels. Both proteins were further normalized to the respective protein expressions at 0 h in empty vector-treated cells. (d) miR-125b-mediated Mad1 repression influences mitotic progression. UPCI:SCC084 cells were transiently transfected with 0.5 μg miR-125b expression plasmid or empty vector, synchronized and at 4, 6, 8 and 10 h from second thymidine release, they were fixed, stained with DAPI and visualized under a fluorescence microscope. Mitotic indices represent the frequencies of mitotic cells. (e) Representative picture showing that miR-125b-treated cells forcefully exit mitosis in presence of nocodazole. UPCI:SCC084 cells were transiently transfected with 0.5 μg miR-125b expression plasmid or empty vector and synchronized. They were treated with nocodazole (100 ng/ml) 16 h prior to second thymidine release. Cell lysates were prepared at 0, 4, 6, 8, 10 and 12 h from second thymidine release followed by western blot with antibodies against cyclinB1 and β-actin. Densitometric values of bands by ImageJ are plotted: cyclinB1 was normalized to β-actin levels and further normalized to protein expression at 0 h in empty vector-treated cells. (f) Representative picture showing that inhibition of endogenous miR-125b accelerates cyclinB1 degradation. HCT116 cells were transiently transfected with or without 20 nℳ anti-miR-125b and synchronized. Cell lysates were prepared at 4, 6, 8 and 10 h from second thymidine release followed by western blot with antibodies against cyclinB1, Mad1 and β-actin. Densitometric values of bands by ImageJ are plotted: cyclinB1 and Mad1 were normalized to β-actin levels. Both proteins were further normalized to the respective protein expressions at 4 h in control cells. (g) Mitotic progression is accelerated upon inhibition of endogenous miR-125b. HCT116 cells were transiently transfected with or without 20 nM anti-miR-125b, synchronized and at 4, 6, 8 and 10 h from second thymidine release, they were fixed, stained with DAPI and visualized under a fluorescence microscope. For b–g, data represent three independent experiments (N=3) and shown as average±S.D.
Inhibition of endogenous miR-125b accelerates mitotic exit in HCT116 cells
To find out whether the above results are reversed upon blocking endogenous miR-125b, we followed cyclinB1 degradation pattern in synchronized HCT116 cells in presence of anti-miR-125b. The HCT116 cell line was selected because, compared with other tumour cell lines, relative expression of miR-125b in this cell line was higher and MAD1 expression was low (see Figures 1c and d). This time, we found that cyclinB1 level was maximal at 6 h post-release for anti-miR-125b-transfected cells (Figure 4f, right panel and graph). Moreover, while its level was stable at 10 h post-release for control cells, cyclinB1 had undergone near-complete degradation at the same time-point in presence of anti-miR-125b (Figure 4f, left and right panels and graph). This implies that cells exit mitosis faster when miR-125b is downregulated and consequently Mad1 levels are elevated. Similar observations were made upon evaluation of MI of HCT116 cells treated with anti-miR-125b. For control cells, MI steadily increased from 4–10 h of release. However, in presence of the inhibitor, MI was maximal at the 6 h time-point after which it was found to decline (Figure 4g and Supplementary Figures S9A and B). This implies that when endogenous level of Mad1 is increased by downregulating miR-125b, cells progress through mitosis at a faster rate. Here we also checked that the inhibitor has no effect on the levels of another SAC gene Mad2 (Supplementary Figure S10, right panel). Incidentally, Mad2 also remained unaltered upon ectopic miR-125b expression in UPCI:SCC084 cells (Supplementary Figure S10, left panel). These data confirmed our earlier results that mitotic exit is delayed by ectopic miR-125b, while inhibiting miR-125b switches SAC ‘off' and accelerates mitotic exit.
miR-125b-induced mitotic delay initiates chromosomal abnormalities and consequent cell death
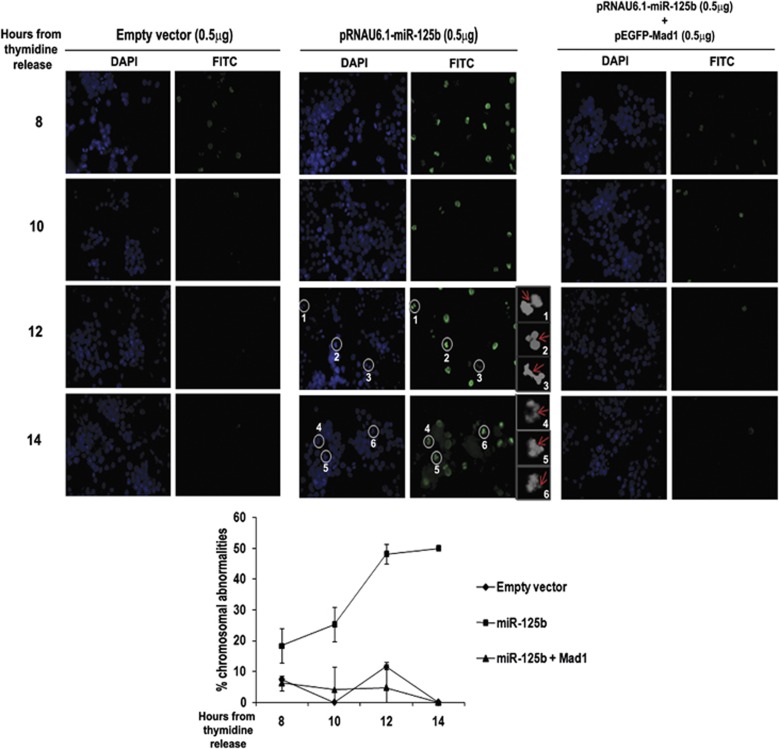
We hypothesized that miR-125b might cause chromosomal abnormalities as a consequence of the transiently activated SAC and delayed mitotic exit. Addressing this in synchronized UPCI:SCC084 cells revealed that chromosomal abnormalities were indeed pronounced in miR-125b-overproducing cells at later time-points (12 and 14 h from release) as compared with vector-transfected cells (Figure 5, left and middle panels and graph). Immunostaining of p-H3 in these abnormal chromosomes strengthened our conjecture that miR-125b-mediated delay in mitotic exit induces chromosomal aberrations (shown in greyscale images 1-6 of Figure 5, middle panel, third and fourth rows). A number of them were found to be lagging chromosomes (Figure 5, middle panel, greyscale images 1, 2 and 3). Lagging chromosomes are seen as connecting threads between separating chromatids of a defective anaphase. As these chromosomes are misaligned at metaphase and forcefully enter anaphase, they retain the H3-phosphorylation, detectable by immunostaining. Besides this, ectopic Mad1 expression along-with miR-125b was also able to reverse enhanced chromosomal aberrations that were observed in presence of miR-125b, thus establishing the specificity of our results (Figure 5, right panel and graph).
Figure 5.
miR-125b induces CIN. Chromosomal abnormalities are enhanced upon ectopic expression of miR-125b. UPCI:SCC084 cells were transiently transfected with either empty vector (0.5 μg), miR-125b expression plasmid (0.5 μg), or miR-125b expression plasmid (0.5 μg) and pEGFP-C1-Mad1 (0.5 μg) and synchronized. Immunostaining was done with antibody against p-H3 and nuclei were stained with DAPI at 8, 10, 12 and 14 h from second thymidine release. Cells were visualized under a fluorescence microscope. Representative images are shown and the defects are indicated by white circles in the full images and by red arrows in the cropped greyscale images (1–6). Lagging chromosomes can be visualized clearly in the cropped greyscale images 1, 2 and 3 (middle panel). Scale bar represents 500 μm. Images represent × 40 magnification. Percentages of mitotic abnormalities were calculated and plotted as shown. Data represents three independent experiments (N=3) and shown as average±S.D.
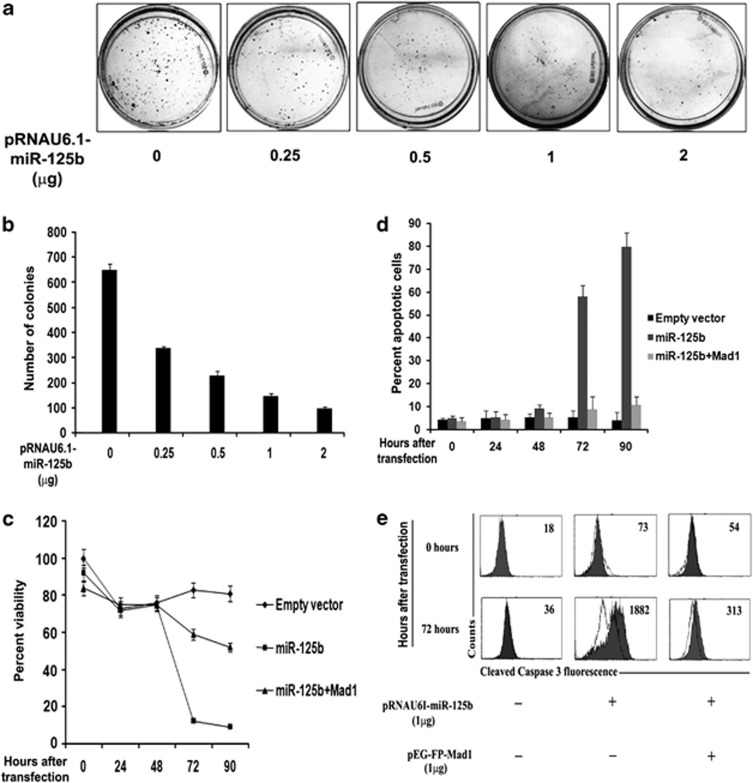
We then evaluated the phenotypic relevance of miR-125b-mediated Mad1 downregulation by examining its effect on clonogenicity and viability of UPCI:SCC084 cells. Surprisingly, the number of colonies formed after a week by miR-125b-overproducing cells was significantly less than those formed by vector-transfected cells (Figures 6a and b and Supplementary Figure S11A). Conversely, we found no significant difference in colony numbers formed by anti-miR-125b-treated HCT116 cells (Supplementary Figures S11B and C). Similarly, MTT assay also confirmed that miR-125b reduced the viability of UPCI:SCC084 cells 72 h post-transfection (Figure 6c). To elucidate whether apoptosis was the underlying cause for this cell death, we adopted two different approaches with miR-125b-treated UPCI:SCC084 cells. First, time-course FACS analysis after annexin V-FITC/propidium iodide (PI) staining revealed that cells underwent considerable apoptotic death 72 h after miR-125b treatment (58.3%) as compared with vector-transfected cells (5.4%) (Figure 6d and Supplementary Figure S11D, left and middle panels). Secondly, we evaluated caspase-3 cleavage by flow cytometry using cleaved caspase-3-specific antibody. Our data indicate that caspase-3 activation increased 72 h post-transfection in presence of ectopic miR-125b (Figure 6e). Camptothecin-treated cells (100μg/ml for 48 h) were used as positive control (Supplementary Figure S11E). Interestingly, Mad1 co-expression was able to rescue cell viability and prevent apoptosis by miR-125b (Figures 6c, d and e and Supplementary Figure S11D, right panel). Collectively, these data confirm that miR-125b induces chromosomal abnormalities, which in turn render the cells non-viable through apoptosis in a Mad1-specific manner.
Figure 6.
Excess miR-125b leads to cell death. (a) Clonogenicity of cells is affected by ectopic miR-125b. UPCI:SCC084 cells were seeded at a density of 103 and transiently transfected with 0, 0.25, 0.5, 1 and 2 μg miR-125b expression plasmid or empty vector. Colonies were stained with methylene blue after a week. (b) Colonies were counted from (a) and plotted as shown. (c) Ectopic miR-125b reduces cell viability. 105 UPCI:SCC084 cells were transiently transfected with either empty vector (1 μg), miR-125b expression plasmid (1 μg), or miR-125b expression plasmid (1 μg) and pEGFP-C1-Mad1 (1 μg) (N=3). MTT reagent was added at a final concentration of 50 μg/ml at 0, 24, 48, 72 and 90 h post-transfection Dye was dissolved in DMSO and read in a microplate reader. (d) Excess miR-125b induces apoptosis. 105 UPCI:SCC084 cells were transiently transfected with either empty vector (1 μg), miR-125b expression plasmid (1 μg), or miR-125b expression plasmid (1 μg) and pEGFP-C1-Mad1 (1 μg). At 0, 24, 48, 72, 90 h post-transfection, cells were subjected to annexin V-FITC/PI staining followed by flow cytometry analysis. (e) Ectopic miR-125b increases cleaved caspase-3 levels. 105 UPCI:SCC084 cells were transiently transfected with either empty vector (1 μg), miR-125b expression plasmid (1 μg), or miR-125b expression plasmid (1 μg) and pEGFP-C1-Mad1 (1 μg). Cells were fixed, permeabilized and stained with control antibodies or cleaved caspase-3 antibody at 0 and 72 h post-transfection followed by flow cytometric analysis. Open and filled histograms represent staining with normal rabbit serum and anti-cleaved caspase-3 antibody, respectively. Values within histograms represent specific mean fluorescence intensity upon subtracting respective control values. For (b–d), data represent three independent experiments (N=3) and shown as average±S.D. For (a) and (e), images are representative of three independent experiments
Mad1 expression is inversely related to miR-125b expression in oral SCC tissues
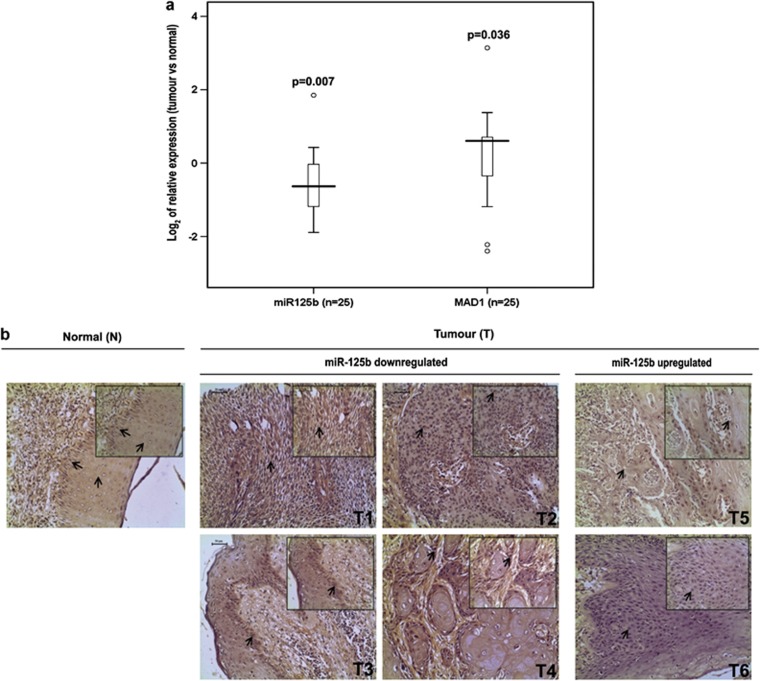
Finally, we examined MAD1 and miR-125b expression profiles in 25 tumour and 20 adjacent normal tissues obtained from HNOC patients. Pathological information for these samples are provided in Supplementary Table S7. The pooled data demonstrated that MAD1 expression was inversely related to miR-125b expression in these samples (Figure 7a). In particular, MAD1 expression was high in 13 of 19 tumour tissues in which miR-125b was downregulated (Supplementary Table S6). A median fivefold decrease of miR-125b and fourfold increase of MAD1 expression was observed from the relative-expression data (Supplementary Table S6). In addition, quantitative immunohistochemical study in 16 primary HNOC tissues confirmed that number of Mad1-expressing cells and staining-intensity were indeed greater in samples in which miR-125b levels were low and vice versa (Figure 7b (shows six representative samples) and Supplementary Table S8). Moreover, Mad1 and miR-125b expressions in these 16 tissues were found to be significantly associated (χ2 P-value=0.017). Altogether, these results confer that the in vitro observations are reflected, at least in part, in vivo as well.
Figure 7.
MAD1 expression negatively correlates with that of miR-125b in HNOC. (a) Reciprocal relation of miR-125b and MAD1 in HNOC tumours. Total RNA was isolated from primary HNOC tissues (n=25) and adjacent normal tissues (n=20). cDNA was prepared by stem-loop primers specific for miR-125b and miR-17-5p and subjected to RT-PCR. Values were normalized to those of miR-17-5p. For MAD1, the above RNA was reverse transcribed and cDNA was subjected to RT-PCR using MAD1-specific primers. GAPDH was used as endogenous control. ΔΔCt values were calculated and data plotted in terms of log2 of relative expressions. P-values are indicated. ‘n' represents the number of tumour samples and circles represent outliers. (b) Representative images showing inverse relation between Mad1 and miR-125b in primary HNOC tissues. Paraffin-embedded tissues samples (in which miR-125b expression was found to be low and Mad1 expression was high and vice versa; n=16) were processed for IHC with antibody against Mad1, stained with 3,3′diaminobenzidine and counterstained with hematoxylin. Nuclei, which stained mild or deep brown represent moderate or high expression of Mad1, respectively. Blue/bluish-purple staining of nuclei represents low Mad1 expression. Images represent × 20 magnification, while inset images represent × 40 magnification. Scale bars represent 50 μm. T1, T2, T3, T4, T5 and T6 are representative tumour samples taken from six individual patients
Discussion
Numerous reports suggest the role of miRNAs in cell cycle control.23 However, recent studies have brought miRNA-mediated mitotic regulation under the spotlight.24, 25 Most recently, cyclinB1 was shown to be upregulated by miR-744 and miR-1186, prolonged overexpression of which causes CIN and inhibits tumour growth.26 Central to mitosis is SAC, which monitors accurate chromosome segregation. Although Mad1, Mad2, BubR1/Mad3 and Bub3 are major SAC components, they have been joined by players like Cdc20, Bub1, Mps1 and AuroraB.10 In this study, we have presented evidence that miRNAs, like bona fide SAC proteins, are capable of regulating this checkpoint. MiR-125b is a highly conserved miRNA that functions both as tumour suppressor and as oncogene depending upon cellular contexts. For instance, in hepatic carcinogenesis, miR-125b is tumour suppressive by downregulating oncogene Lin28B27 whereas in megakaryoblastic leukaemia, it is oncogenic by repressing tumour suppressors such as ST18.28 Interestingly, we observed that not only does miR-125b have a recognition site on MAD1-3′UTR, its expression is also reciprocally related with that of Mad1 in HNOC tumours and in several cancer cell lines. Subsequently, we demonstrated that miR-125b targets MAD1 specifically. Given that Mad1 is a core SAC protein, these results bring forward a new perspective of SAC regulation – that by miRNAs.
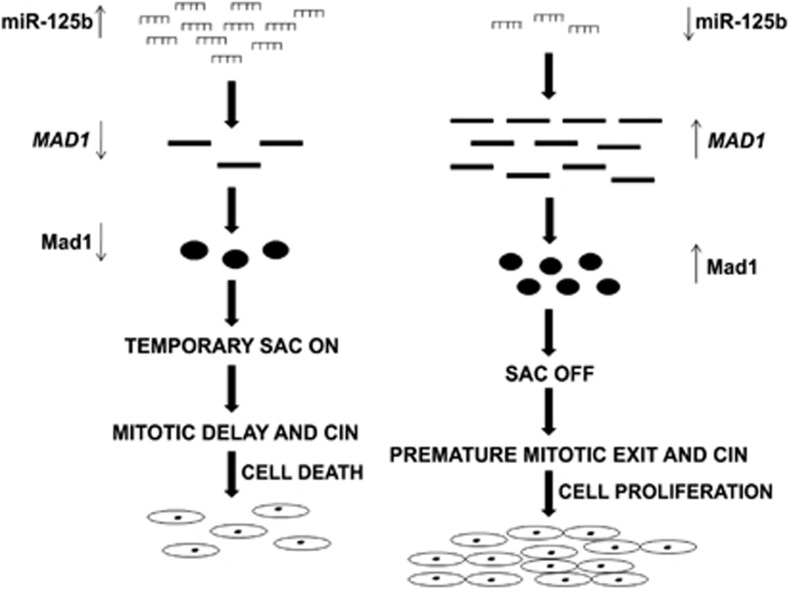
We then showed how miR-125b-mediated Mad1 downregulation affects mitotic progression of UPCI:SCC084 cells, which were earlier shown to be chromosomally stable with modal chromosome number of 49.29 A consistently delayed anaphase entry of cells overexpressing miR-125b implied that in absence of Mad1, SAC is activated transiently, delaying metaphase-to-anaphase transition. Notably, because MAD1 lacks its 3′UTR in the expression-vector, co-expression with miR-125b did not alter Mad1's ability to change the cellular phenotype. This also indicates that other known targets of miR-125b (like TP53)30 might not have any role in the Mad1-mediated phenotype observed here. Our observations were underscored when Cdc20 overexpression along-with miR-125b gave the same rescue pattern as Mad1. This was significant because Cdc20 is a direct target of SAC and hence, it confirmed the involvement of SAC. It is noteworthy that Cdc20 levels impact both mitotic slippage rate and cell fate.29, 31, 32 Meanwhile, inhibiting endogenous miR-125b resulted in faster mitotic exit, possibly by inactivating SAC through increased Mad1 levels. This is reminiscent of a recent report that Mad1 upregulation might promote tumour formation and manifest resistance to therapies.16 It is well-known that accelerated mitotic progression can induce CIN.29, 16 Our data also reveal a higher incidence of chromosomal defects in presence of ectopic miR-125b, mostly in metaphase-to-anaphase transition. Cell viability too was considerably reduced by increased apoptotic death. Hence, it is conceivable that miR-125b up- or downregulation could both be harmful to cells as both give rise to CIN and lead to either accelerated proliferation or cell death, respectively (Figure 8). Therefore, a priori, in normal circumstances miR-125b exerts it tumour suppressive role whenever Mad1 levels are elevated in response to some stimuli. This would be important to maintain the extremely delicate Mad1–Mad2 balance. In fact, this could be extended to all SAC genes—most of which are both up or downregulated in different tumours.33
Figure 8.
Proposed model for the regulation of Mad1 and cell fate by miR-125b. On the left, it is shown that high expression of miR-125b downregulates Mad1 as a result of which SAC is transiently activated. This leads to mitotic delay and accumulation of chromosomal abnormalities. The cells fail to maintain themselves and finally undergo apoptotic cell death. On the right, the reverse situation is illustrated. Mad1 levels increase when miR-125b levels are low. As a result, SAC is overridden and premature mitotic exit occurs. This leads to elevated proliferation of cells
Although aneuploidy is recognized to promote tumorigenesis, it is also proposed to be tumour suppressive in some contexts.34 We speculate that segregation defects arising from miR-125b-induced Mad1 repression and transient SAC activation lead to accumulation of aneuploid cells and finally reduce cell growth and viability. Indeed, it was earlier reported that Mad1 expression is heightened in cancer cells and that this correlates with cellular proliferation.35 At the molecular level, Mad1-bound and free-Mad2 are both essential to maintain SAC. Moreover, excess Mad1 can outdo Cdc20 in binding free-Mad2 owing to which Cdc20 can bind and activate APC/C. This leads to rapid mitotic exit and SAC abolishment.15, 16 Through our study, we hypothesize that reduced Mad1 levels, due to excess miR-125b, increases free-Mad2 pool, which can bind more Cdc20 molecules, as a result of which APC/C remains inactive and cells are halted at metaphase.
In summary, (1) the SAC gene MAD1 is a novel target of miR-125b, (2) Mad1 downregulation brings about transient SAC activation and mitotic delay and (3) this delay gives rise to chromosomal abnormalities, presumable accumulation of which leads to apoptosis. All these cellular effects of miR-125b appear to be Mad1-specific. These findings might help to understand the molecular mechanisms that contribute towards high degree of CIN associated with HNOC. The fact that HNOC frequently exhibits alterations in mitotic checkpoint genes mirrors the criticality of SAC integrity in this disease.18 As alteration of Mad1 levels by miR-125b can cause distinct cellular events such as apoptosis, it can be comprehended that suppressing high Mad1 levels might prove to be therapeutic in HNOC. However, it is crucial to reckon that miR-125b has other known and unknown targets across the cell cycle whereas Mad1 regulation could also be controlled by hitherto unknown miRNAs. Hence, a systemic study of miRNAs involved in regulation of Mad1 and other SAC genes is warranted to fully understand the impact of miRNAs on SAC function, as well as on development and progression of HNOC.
Materials and Methods
Cell culture, synchronization, drug treatment and transfection
Human oral cancer cell lines UPCI:SCC084 and UPCI:SCC131 were kind gifts from Dr. Susanne M. Gollin (University of Pittsburgh, USA). HCT116 cell line was purchased from American Type Culture Collection (Manassas, VA, USA) and HepG2 cell line was gifted by Dr. Samit Adhya (Indian Institute of Chemical Biology, India). All cell lines were cultured in Dulbecco's Modified Eagle's Medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal calf serum and antibiotics (1% Pen Strep Glutamine and 0.006% Gentamicin, Invitrogen) at 37 °C in a 5% C02 incubator. For synchronization, cells were treated with thymidine (2.5 mℳ; USB, Cleveland, OH, USA) for 16 h followed by an 8-h release. A second thymidine treatment was given for another 22 h. Following this double thymidine block, cells were released in thymidine free complete medium and harvested at various time-points. Transient transfections were done with various plasmids and miRNA inhibitors in different cell lines using Lipofectamine 2000 reagent (Invitrogen) according to manufacturer's protocol. Except in case of synchronization experiments, all transient transfections were performed for 48 h. Transfection of synchronized cells was done 2 h before first thymidine addition. To induce mitotic arrest, cells were treated with the spindle disrupting drug, nocodazole (100 ng/ml, Sigma, St. Louis, MO, USA) for 16 h before second thymidine release. For induction of apoptosis by camptothecin, cells were treated with the drug (100 μg/ml, Sigma) for 48 h.
Plasmids and miRNA inhibitors
The cloning of human precursor miR-125b is described by Ghose et al.36 The same strategy was followed to clone the human precursor miR-133b (Chromosome 6: 52011721-52015839, + strand). This clone will now be referred to as pSB-miR-133b. The MAD1-3′UTR (NM_003550.2, +2422 to +2727) was amplified from human genomic DNA with the primers listed in Supplementary Table S9. This region was cloned into the linearized pMIR-REPORT vector (Applied Biosystems, Foster City, CA, USA) downstream of the Luc gene using the MluI and HindIII restriction sites (New England Biolabs, Beverly, MA, USA). This clone will henceforth be referred to as pSB-MAD1/3′UTRLuc. The mutant version of this clone was constructed using the primers listed in Supplementary Table S9 and Quikchange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) according to the manufacturer's protocol. This clone will be referred to as pSB-MAD1/3′UTRMutLuc. Full length FLAG-tagged Cdc20 expression plasmid pCDNA5/FRT/TO FLAG-CDC20 was a kind gift from Dr. Jonathon Pines (Gurdon Institute, Cambridge, UK). Full length GFP-tagged Mad1 expression plasmid pEGFP-C1-Mad1 was also gifted by Dr. Kuan-Teh Jeang (National Institute of Allergy and Infectious Diseases, Bethesda, MD, USA). Anti-miR-125b (Ambion, Austin, TX, USA) was used at a final concentration of 20 nℳ or as indicated.
Quantitative real-time PCR
Total RNA from different cell lines, patient tissues and oral swab was isolated using TRIZOL (Invitrogen) according to manufacturer's protocol. 5 μg isolated RNA was treated with DNase (Promega, Madison, WI, USA) and 1 μg of the DNase-treated RNA was used for cDNA preparation using random hexamer (Invitrogen) and MMLV-RT (Promega). For miRNAs, 200 ng isolated RNA was used for cDNA preparation in a master mix containing stem-loop primers specific for the desired miRNAs (Sigma), dNTPs (Invitrogen) and MMLV-RT (Promega). Real-time PCR was performed in the 7500 Fast Real-Time PCR System (Applied Biosystems) using power SYBR Green PCR Master Mix (Applied Biosystems). The comparative threshold cycle method (ΔΔCt) was used to quantify relative amounts of product transcripts with GAPDH (for mRNAs) and hsa-miR-17-5p (for miRNAs) as endogenous reference controls. Primer sets for MAD1, BUB3 and GAPDH are listed in Supplementary Table S10. Fold activation values were calculated as mean of three independent experiments.
Western blotting and antibodies
Whole cell lysates having equal protein concentrations were resolved by SDS/PAGE (8–12% gel) and transferred onto a PVDF membrane (Millipore, Billerica, MA, USA). Various primary antibodies used are mouse monoclonal Mad1 (Millipore), mouse monoclonal cyclinB1 (Cell Signaling Technology, Beverly, MA, USA), rabbit polyclonal p-H3 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), rabbit polyclonal H3 (Santa Cruz Biotechnology), mouse monoclonal Cdc20 (E-7, Santa Cruz Biotechnology), mouse monoclonal β-actin antibody (Sigma). Bands were detected using Super Signal West Pico chemiluminescent substrate (Thermo Scientific, Rockford, IL, USA) after treating with HRP-conjugated secondary antibody (Sigma).
Luciferase assay
Cells were lysed with luciferase cell culture lysis reagent supplied with the luciferase assay kit (Promega). After a short vortex, whole cell lysates were centrifuged at 4 °C at 13 000 r.p.m. for 2 min and 15–30 μl of supernatants was mixed with 30–60 μl of luciferase assay substrate. Luminescence was measured as relative luciferase unit in GLOMAX luminometer (Promega). Total protein concentration in each lysate was measured by Bio-Rad protein assay reagent (Bio-Rad Laboratories, Hercules, CA, USA) and then used to normalize the Luc activity of each lysate. Fold activation values were calculated as mean of three independent experiments.
Determination of MI
Synchronized UPCI:SCC084 or HCT116 cells were harvested at different time-points (as mentioned in respective cases) after thymidine release. They were then fixed with ice-cold acetomethanol (1 : 1) and stained with DAPI (4,6-diamidino-2-phenylindole) (Invitrogen). Frequencies of mitotic cells were counted under fluorescence microscope (Leica DM 3000, Leica Microsystems, Heerbrugg, Switzerland) among 100–200 cells each time. MI was calculated as mean of three independent experiments.
Cell cycle, cell survival and apoptotic assays
UPCI:SCC084 cells were synchronized and after release, approximately 106 cells were harvested at respective time-points and resuspended in 0.25 ml of cold phosphate-buffered saline (PBS). Cells were fixed by adding cold 70% ethanol dropwise into the samples, while vortexing gently and then incubated at 4 °C for minimum 24 h. After fixation, cells were resuspended in 1 ml PBS containing 100 μg/ml PI (Sigma) and 20 μg/ml RNase A (Invitrogen). Fixed cells were kept at room temperature for 40 min and then analysed by FACS (BD FACSARIA III, Becton-Dickinson, San Jose, CA, USA).
For annexin V-PI binding assay, UPCI:SCC084 cells were seeded at a density of 105 cells and transfected as mentioned. Apoptosis was measured at the indicated time-points using FITC AnnexinV Apoptosis Detection Kit I (BD Pharmingen, Franklin Lakes, NJ, USA) according to the manufacturer's protocol. Analysis was done by BD FACSARIA (Becton-Dickinson).
To measure caspase-3 cleavage, 105 UPCI:SCC084 cells were seeded and transiently transfected as mentioned. At indicated time-points, cells were fixed with 4% paraformaldehyde, permeabilized by fluorescence-activated cell sorter permeabilizing solution (BD Biosciences, San Jose, CA, USA). Before staining, permeabilized cells were treated with heat-inactivated 2% normal goat serum to block non-specific staining. Cells were then stained with normal rabbit sera or rabbit anti-human-cleaved caspase-3 antibody (Cell Signalling Technology). After washing, cells were incubated with multiple adsorbed FITC-conjugated secondary antibody (goat anti-rabbit immunoglobulin; BD Biosciences), washed and analysed by BD FACSARIA (Becton-Dickinson).
For colony formation assay, UPCI:SCC084 cells were counted and seeded at different cell densities (as mentioned) after which they were transiently transfected with hsa-miR-125b expression plasmid. For dose-dependent assay, 103 UPCI:SCC084 and HCT116 cells were seeded and transiently transfected with indicated concentrations of plasmid or inhibitor, respectively. After 7 days, cells were stained with 0.2% Methylene Blue (Sisco Research laboratories (SRL), Mumbai, India) and washed with distilled water. The colonies were then counted and the number of colonies was calculated as mean of three independent experiments.
For MTT assay, 105 UPCI:SCC084 cells were seeded and transfected as mentioned. At the indicated time-points, MTT solution was added to each dish at a final concentration of 50 μg/ml. After 4–5 h incubation at 37 °C, MTT solution was removed and 500 μl DMSO was added to dissolve dye. 100 μl of each sample was taken in a 96-well plate and absorbance at 595 nm was measured using a microplate reader (355 Thermo Multiskan Ex, Thermo Scientific). Percent viability was calculated as a mean of three independent experiments.
Immunofluorescence
Synchronized UPCI:SCC084 cells were harvested at different time-points (as mentioned in respective cases) after thymidine release. They were then fixed with ice-cold acetomethanol (1 : 1) and permeabilized with 0.03% saponin (Calbiochem, Darmstadt, Germany). After blocking in 3% BSA, rabbit polyclonal antibody against p-H3 (Santa Cruz Biotechnology) was added. Secondary antibody conjugated with FITC (Sigma) was added and the nuclei were stained with DAPI (Invitrogen). Slides were observed under a fluorescence microscope (Leica DM 3000). Abnormal chromosomes were counted among 100–200 cells each time from three independent experiments and plotted as percent abnormality. Lagging chromosomes are observed as thread-like structures between the separating sets of chromatids at anaphase, which stain positive for p-H3-FITC.
Immunohistochemistry (IHC)
About 3–5 μm paraffin-embedded sections of normal oral and primary HNOC tissues were deparaffinized and rehydrated in gradient alcohol. Antigen retrieval was followed by blocking with 1% BSA, addition of mouse monoclonal antibody against Mad1 (Santa Cruz Biotechnology) and incubation overnight at 4 °C. Slides were developed using 3,3′diaminobenzidine as the chromogen after treating with HRP-conjugated secondary antibody (Sigma) and then counterstained with hematoxylin. Negative controls lacked primary antibody. Data were scored as described by Perrone et al.37
Tumour samples
Matched oral tumour (n=25) and normal (n=20) tissues were obtained from the archive samples of the hospital section, Chittaranjan National Cancer Institute (CNCI), Kolkata, India. For five tumour samples, adjacent-normal tissues were unavailable. Out of the 25 tumour samples, 16 were available as paraffin blocks for IHC study. Prior to sample collection, written informed consent was taken from each individual and approved by the Research Ethics Committee of CNCI. The pathological history of the tumours is provided in Supplementary Table S7.
Bioinformatic and statistical analysis
Target prediction of miRNAs was done by miRBase (version 17). Complementarity between MAD1-3′UTR and the seed sequence of miR-125b was obtained from RNAhybrid (version 2.1)38 (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/). Oncomine 4.4 research edition database39 (http://www.oncomine.org/resource/login.html) was used for the data set of Mad1 overexpression in primary tumours. Cancer versus normal data sets of Mad1 overexpression with fold change ≥1.5 and P-value≤0.05 were selected. Densitometric scanning for western blots was done by ImageJ software (http://rsb.info.nih.gov/ij/index.html). SPSS 16.0 (SPSS Inc., Chicago, IL, USA; http://www.spss.com) was used to perform Mann–Whitney t-test in order to determine significant differences between individual groups (normal and tumour) with respect to miR-125b and Mad1 expressions. Fisher's exact χ2-test was performed to determine the significant association between Mad1 level (as quantified from IHC data) and miR-125b expression in tumour-tissue samples by GraphPad QuickCalcs online software (http://www.graphpad.com/quickcalcs/contingency1.cfm). Statistically significant differences were defined by two-sided P<0.05.
Acknowledgments
We thank Dr. Chitra Mandal and Dr. Keya Chaudhuri (Indian Institute of Chemical Biology, India) for FACS and microscopic analyses, respectively. We are also grateful to Saikat Mukhopadhyay and Eashita Das (Saha Institute of Nuclear Physics, India) for their invaluable help in the bioinformatic-based miRNA studies and FACS analyses, respectively. This work was supported in part by Department of Biotechnology Grants BT/PR/5524/Med/14/649/2004, BT/01/COE/05/04 and Council of Scientific and Industrial Research Grant IAP 001 awarded to Dr. Susanta Roychoudhury. SB is supported by predoctoral fellowship from the University Grants Commission (New Delhi, India). SN, GPM and NB are supported by predoctoral fellowships from the Council of Scientific and Industrial Research (New Delhi, India), while JG is supported by predoctoral fellowship from the Department of Atomic Energy (Mumbai, India).
Glossary
- miRNA
microRNA
- hsa-miR
Homo sapiens miRNA
- mRNA
messenger RNA
- SAC
spindle assembly checkpoint
- MAD
mitotic-arrest deficient
- CIN
chromosomal instability
- APC/C
anaphase-promoting complex/cyclosome
- Cdc20
cell division cycle 20
- BUB
budding uninhibited by benzimidazole
- MCC
mitotic checkpoint complex
- C-Mad2
closed Mad2
- O-Mad2
open Mad2
- HNOC
head and neck/oral cancer
- UTR
untranslated region
- SCC
squamous cell carcinoma
- Mut
mutant
- Luc
luciferase
- p-H3
phosphorylated histone H3
- MI
mitotic index
- PI
propidium iodide
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
Supplementary Material
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Brit J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene. 2006;25:6170–6175. doi: 10.1038/sj.onc.1209911. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Yu H. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol Cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Sironi L, Melixetian M, Faretta M, Prosperini E, Helin K, Musacchio A. Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J. 2001;20:6371–6382. doi: 10.1093/emboj/20.22.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E, Chen RH. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol Biol Cell. 2002;13:1501–1511. doi: 10.1091/mbc.02-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan SD, Britigan EM, Zasadil LM, Witte K, Audhya A, Roopra A, et al. Up-regulation of the mitotic checkpoint component Mad1 causes chromosomal instability and resistance to microtubule poisons. Proc Natl Acad Sci USA. 2012;109:E2205–E2214. doi: 10.1073/pnas.1201911109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barwad A, Sood S, Gupta N, Rajwanshi A, Panda N, Srinivasan R. Human papilloma virus associated head and neck cancer: a PCR based study. Diagn Cytopathol. 2011;40:893–897. doi: 10.1002/dc.21667. [DOI] [PubMed] [Google Scholar]
- Minhas KM, Singh B, Jiang WW, Sidransky D, Califano JA. Spindle assembly checkpoint defects and chromosomal instability in head and neck squamous cell carcinoma. Int J Cancer. 2003;107:46–52. doi: 10.1002/ijc.11341. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Kowarsch A, Schmidl D, Buggenthin F, Brauner B, Dunger I, et al. PhenomiR: a knowledgebase for microRNA expression in diseases and biological processes. Genome Biol. 2010;11:R6. doi: 10.1186/gb-2010-11-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson BJ, Bhattacharjee S, O'Dee DM, Feingold E, Gollin SM. Decreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancy. Genes Chromosomes Cancer. 2009;48:569–582. doi: 10.1002/gcc.20666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienitz A, Vogel C, Morales I, Muller R, Bastians H. Partial downregulation of MAD1 causes spindle checkpoint inactivation and aneuploidy, but does not confer resistance towards taxol. Oncogene. 2005;24:4301–4310. doi: 10.1038/sj.onc.1208589. [DOI] [PubMed] [Google Scholar]
- Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys acta. 2011;1812:592–601. doi: 10.1016/j.bbadis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Shi W, Alajez NM, Bastianutto C, Hui AB, Mocanu JD, Ito E, et al. Significance of Plk1 regulation by miR-100 in human nasopharyngeal cancer. Int J Cancer. 2010;126:2036–2048. doi: 10.1002/ijc.24880. [DOI] [PubMed] [Google Scholar]
- Nakada C, Tsukamoto Y, Matsuura K, Nguyen TL, Hijiya N, Uchida T, et al. Overexpression of miR-210, a downstream target of HIF1alpha, causes centrosome amplification in renal carcinoma cells. J Pathol. 2011;224:280–288. doi: 10.1002/path.2860. [DOI] [PubMed] [Google Scholar]
- Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, et al. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. 2011;40:1695–1707. doi: 10.1093/nar/gkr934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Wong CM, Ying Q, Fan DN, Huang S, Ding J, et al. MicroRNA-125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology. 2010;52:1731–1740. doi: 10.1002/hep.23904. [DOI] [PubMed] [Google Scholar]
- Klusmann JH, Li Z, Bohmer K, Maroz A, Koch ML, Emmrich S, et al. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev. 2010;24:478–490. doi: 10.1101/gad.1856210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal G, Sengupta S, Panda CK, Gollin SM, Saunders WS, Roychoudhury S. Overexpression of Cdc20 leads to impairment of the spindle assembly checkpoint and aneuploidization in oral cancer. Carcinogenesis. 2007;28:81–92. doi: 10.1093/carcin/bgl100. [DOI] [PubMed] [Google Scholar]
- Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath S, Banerjee T, Sen D, Das T, Roychoudhury S. Spindle assembly checkpoint protein Cdc20 transcriptionally activates expression of ubiquitin carrier protein UbcH10. J Biol Chem. 2011;286:15666–15677. doi: 10.1074/jbc.M110.160671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J. Cdc20 control of cell fate during prolonged mitotic arrest: do Cdc20 protein levels affect cell fate in response to antimitotic compounds. Bioessays. 2011;33:903–909. doi: 10.1002/bies.201100094. [DOI] [PubMed] [Google Scholar]
- Perez de Castro I, de Carcer G, Malumbres M. A census of mitotic cancer genes: new insights into tumor cell biology and cancer therapy. Carcinogenesis. 2007;28:899–912. doi: 10.1093/carcin/bgm019. [DOI] [PubMed] [Google Scholar]
- Weaver BA, Cleveland DW. Aneuploidy: instigator and inhibitor of tumorigenesis. Cancer Res. 2007;67:10103–10105. doi: 10.1158/0008-5472.CAN-07-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanaga Y, Jeang KT. Expression of mitotic spindle checkpoint protein hsMAD1 correlates with cellular proliferation and is activated by a gain-of-function p53 mutant. Cancer Res. 2002;62:2618–2624. [PubMed] [Google Scholar]
- Ghose J, Sinha M, Das E, Jana NR, Bhattacharyya NP. Regulation of miR-146a by RelA/NFkB and p53 in STHdh(Q111)/Hdh(Q111) cells, a cell model of Huntington's disease. PloS One. 2011;6:e23837. doi: 10.1371/journal.pone.0023837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone F, Suardi S, Pastore E, Casieri P, Orsenigo M, Caramuta S, et al. Molecular and cytogenetic subgroups of oropharyngeal squamous cell carcinoma. Clin Cancer Res. 2006;12:6643–6651. doi: 10.1158/1078-0432.CCR-06-1759. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.